Abstract
We previously reported that a mutant Rous sarcoma virus (RSV) with an alternate polypurine tract (PPT), DuckHepBFlipPPT, had unexpectedly high titers and that the PPT was miscleaved primarily at one position following a GA dinucleotide by the RNase H of reverse transcriptase (RT). This miscleavage resulted in a portion of the 3′ end of the PPT (5′-ATGTA) being added to the end of U3 of the linear viral DNA. To better understand the RNase H cleavage by RSV RT, we made a number of mutations within the DuckHepBFlipPPT and in the sequences adjacent to the PPT. Deleting the entire ATGTA sequence from the DuckHepBFlipPPT increased the relative titer to wild-type levels, while point mutations within the ATGTA sequence reduced the relative titer but had minimal effects on the cleavage specificity. However, mutating a sequence 5′ of ATGTA affected the relative titer of the virus and caused the RNase H of RSV RT to lose the ability to cleave the PPT specifically. In addition, although mutations in the conserved stretch of thymidine residues upstream of the PPT did not affect the relative titer or cleavage specificity, the mutation of some of the nucleotides immediately upstream of the PPT did affect the titer and cleavage specificity. Taken together, our studies show that the structure of the PPT in the context of the cognate RT, rather than a specific sequence, is important for the proper cleavage by RSV RT.
During retroviral infection, the virally encoded enzyme reverse transcriptase (RT) converts the single-stranded RNA genome of the virus into a linear double-stranded DNA that can be integrated into the host genome (31, 32). RT has two enzymatic activities: a DNA polymerase that can copy either an RNA or DNA template and an RNase H that cleaves RNA if, and only if, it is part of an RNA-DNA hybrid. Both activities are required for the synthesis of the linear viral DNA. Like many other DNA polymerases, RT cannot initiate DNA synthesis de novo. A host tRNA, hybridized to the viral RNA genome near the 5′ end of the genome, is used to prime the synthesis of minus-strand DNA. As the polymerase activity of RT synthesizes minus-strand DNA, it generates an RNA-DNA hybrid that is a substrate for RNase H. After RT has copied the short segment of the retroviral RNA genome between the tRNA primer and the 5′ end of the viral genome and RNase H has degraded the RNA strand, minus-strand DNA synthesis is translocated to the 3′ end of the viral genome via the repeat or R sequence found at both the 5′ and 3′ ends of the viral RNA genome. The synthesis of minus-strand DNA is then able to proceed toward the 5′ end of the RNA genome. The viral RNA genome contains a sequence called the polypurine tract (PPT), which is relatively resistant to degradation by RNase H. Because the PPT is relatively stable, it is used to prime plus-strand DNA synthesis. In general, the cleavages made by RNase H are thought to be relatively nonspecific; however, there are several steps during reverse transcription in which the cleavages are known to be specific. One specific cleavage removes the tRNA primer, which defines the right end of the viral genome. Most retroviruses remove the entire tRNA primer, but human immunodeficiency virus type 1 (HIV-1) is the exception; RNase H cleavage leaves the last nucleotide from the 3′ end of the tRNA on the viral DNA (4, 12, 20, 30, 33). The second step in which the cleavages are specific involves the generation and subsequent removal of the PPT primer, which defines the left end of the viral genome. The cleavages that generate and remove the PPT are made with considerable precision, despite the fact that RT does not contain specific motifs that recognize defined sequences in nucleic acids.
It is possible to use model substrates to measure the specificity of RNase H cleavage in vitro; however, it is also useful to determine the specificity of RNase H cleavage in vivo. It is technically difficult to analyze the ends of the linear viral DNA accurately. However, a portion of the viral DNA that is not integrated is ligated by host enzymes to produce two-long-terminal-repeat (2-LTR) circular DNA (14). The sequence of the 2-LTR circle junctions can be used to infer information about the ends of the linear viral DNA from which the circles derive; the inferred sequence of the linear viral DNA can, in turn, be used to understand the effects of mutations on the specificity of RNase H cleavage. Consensus 2-LTR circle junctions are formed by ligation of the ends of a linear viral DNA in which both ends are normal.
Considerable effort has been made to understand how the PPT is able to resist degradation and to determine how RT makes the specific cleavages that generate and remove the PPT. Mutational analysis has been used to determine what is an acceptable PPT sequence. Experiments with murine leukemia virus (MLV) containing a large pool of random PPT sequences showed that positions −2, −5, −10, and −11 from the PPT-U3 junction were strongly selected; the other positions in the PPT were less important (25). We measured the effects of mutations in the 5′ and 3′ ends of the HIV-1 PPT on titer and cleavage specificity in vivo (9, 15). The sequence of the HIV-1 PPT is AAAAGAAAAGGGGGG. In the analysis of mutations in the 5′ end of the PPT, substituting one or two C residues for the A residues at the 5′ end of the PPT had modest effects on the titer of an HIV-1 vector (two- to fivefold) and, using the 2-LTR circle junction assay, on the specificity of RNase H cleavage. Miles et al. reported that a complex multiple mutation in the 5′ end of the HIV-1 PPT had a relatively modest effect on the titer (about 40% of the wild type [WT]) (16). This region has also been analyzed using in vitro assays; the results imply that weak base pairing plays a role in determining cleavage specificity (3, 13, 35).
Based on the 2-LTR circle junction assay, mutations in the 5′ portion of the PPT caused an increase in the retention of portions of the (mutant) PPT sequences. The in vivo analysis could not determine whether the miscleavages that gave rise to the retained PPT sequences happened when the PPT primer was generated, when it was removed, or both. We generated and tested a mutant (Multi A box) that had seven substitutions in the nine positions at the 5′ end of the PPT. Given the large number of changes, the effect on the titer was moderate (20-fold). The Multi A box mutation did not appear to have a significantly greater effect on the specificity of cleavage than some of the simpler mutations in the 5′ end of the PPT, suggesting that the larger effect on the titer might be due to an effect on the efficiency or extent of PPT cleavage rather than an effect on the specificity of PPT cleavage. We also made single and double transition and transversion mutations in the G tract in the 3′ end of the PPT. The single mutants, and most of the double mutants, had a small effect on the titer (twofold or less); the exception was T2-5, in which the second and fifth positions of the G tract (numbered from the 5′ end) were substituted by T residues; the titer of this mutant was 10% of that of the WT. We did two sets of experiments in which we asked whether mutations in the PPT caused similar defects in RNase H cleavages in vitro and in vivo; we were able to show that there is good agreement in the in vivo and in vitro results (8, 23). Miles et al. reported that substituting Cs either for the last four or all six Gs in the G tract had almost as great an effect on the titer as replacing the entire PPT with a non-PPT sequence (16). We sequenced the 2-LTR circle junctions that resulted from infections with vectors carrying these PPT mutations; these assays showed that the A3, A5, T2, T5, A2-5, C2-5, and T2-5 mutations affected cleavage specificity (the number represents the position or positions in the G tract that were mutated, and the letter designates the nucleotide substitution). This suggested that the second and fifth positions of the G tract are particularly important for specific cleavage of the HIV-1 PPT; these are two of the five positions that were strongly selected in a random pool of MLV PPTs in vivo (25). Rausch and Le Grice also did in vitro assays with mutant PPTs with A substitutions in the G tract of the HIV-1 PPT and showed that the A1, A2, A3, and A5 mutations affected the specificity or extent of cleavage (22). Schultz et al. reported, based on in vitro cleavage of non-PPT sequences, that the RNase H of HIV-1 RT cleaves poorly if the substrate has the equivalent of an A3, T5, or A5 substitution (29).
HIV-1 PPT, in a complex with its cognate RT, has an unusual structure (26). This brings up the question of whether the PPTs of different retroviruses, which have different sequences, have some common shared properties or structure that allows them to be cleaved accurately by the RNase Hs of their respective RTs. If these features (i.e., structural properties) are shared, then the RNase H of a particular RT might be able to cleave noncognate PPTs with some specificity. We previously showed that the RSV RT is able to use alternative PPTs derived from other retroviruses sufficiently well to support a reduced level of viral replication (2). In addition to showing a reduction in the viral titer, the viruses that replicated using these noncognate PPTs showed significant increases in the miscleavages by the RNase H of RSV RT that resulted in an increase in the proportion of the viral DNAs that retained a portion of the PPT. Experiments with mutant viruses that had either a mutant RSV PPT or a PPT derived from MLV also showed that the RNase H of RSV RT preferentially cleaves after a GA dinucleotide. This preference for cleavage after a GA dinucleotide is also reflected by the fact that the last two nucleotides in the RSV PPT are GA.
Testing the RSV-based mutant viruses with PPTs derived from other viruses, we showed that a virus with a sequence derived from a polypyrimidine sequence from duck hepatitis virus B, when inserted in the reverse orientation (DuckHepBFlipPPT), had an unexpectedly high titer. Because the sequence in the duck hepatitis B genome is a polypyrimidine tract, there was no reason to expect that it could function as a PPT, even if the opposite strand was used (i.e., the purine-rich strand). Because the sequence was not derived from a functional PPT, the decision of what to include in the “PPT” was somewhat arbitrary.
We examined the cleavage of the DuckHepBFlipPPT by the RNase H of RSV RT, using the 2-LTR circle junction assay. The predominant cleavage caused the addition of 5 nucleotides (5′-ATGTA) from the 3′ end of the PPT onto the U3 end of the viral DNA. This added sequence was an exact duplication of the +2 to +6 nucleotides found in the U3 of WT RSV. The remainder of the DuckHepBFlipPPT is similar to that of the WT RSV PPT in that both are 11 nucleotides in length and that there is a stretch of G residues at the 3′ end of the PPT. This means that the primary RNase H cleavage site in the DuckHepBFlipPPT creates a new PPT-U3 junction in the PPT, and the 5′-ATGTA portion of the DuckHepBFlipPPT becomes part of U3. In most WT linear retroviral DNAs, there is a CA 2 nucleotides from each of the 3′ ends of the linear DNA. Integrase (IN) joins this CA dinucleotide to the host genomic DNA to create the integrated provirus. The 2 nucleotides that are lost from the 3′ ends of the viral DNA are removed by IN before it joins the viral and host sequences. The ATGTA is in the 5′ strand of the U3 end of the linear viral DNA, and the complementary (3′) strand does contain a CA dinucleotide; however, this CA is 1 nucleotide from the 3′ end of the linear viral DNA (the CA is the reverse complement of the underlined TG in the 5′-ATGTA sequence). Despite the fact that the CA is 1 nucleotide from the U3 end of the viral DNA, RSV IN is able to remove 1 nucleotide and then join the CA to host DNA (17).
Although it is clear that the sequence of the PPT has a major role in determining where RNase H will cleave, which in turn determines the ends of the linear viral DNA, there are reports that sequences outside the LTR also play a role in controlling the cleavage of the PPT. In the genomes of many retroviruses, there is a run of Us near the 5′ end of the PPT. Ilyinskii and Desrosiers showed, for simian immunodeficiency virus, that these sequences were essential for the replication of that virus and, based on the phenotype of the mutant (which had defects in viral DNA synthesis) and the nature of compensatory mutations (some of which expanded the length of the PPT), speculated about the role these sequences had in viral replication (6). We were interested in asking whether similar sequences present in the genomes of avian sarcoma/leukosis viruses (ASLVs) are equally important and, if they are, seeing if we could specifically determine whether these sequences have a role in the generation or removal of the PPT primer. We were also interested in determining if the sequences at the 5′ end of U3 sequences, near the PPT, have any significant role in the generation or removal of the PPT primer.
The structure of HIV-1 RT in a complex with an RNA-DNA duplex that includes the PPT suggests that there are few important contacts between RT and the RNA 3′ of the cleavage site (26), and a similar result would be anticipated for the U5 sequences adjacent to the tRNA primer. As would be expected from the structure, mutating the U5 sequences adjacent to the primer binding site in ASLV did not affect the removal of the tRNA primer in vivo (17); however, making similar mutations in the genome of an HIV-1 vector did affect the cleavage that removes the tRNA primer (19). This suggests that it might be possible for mutations in the 5′ end of U3 to affect the cleavage at the PPT-U3 junction; in particular, Kvaratskhelia et al. (13) suggested the A-T base pair immediately adjacent to the HIV-1 PPT is weak and that this weak base pair might affect cleavage.
In the experiments reported here, we made mutations in the DuckHepBFlipPPT and adjacent sequences to determine what factors contribute to the ability of the RSV RT to recognize and properly cleave this foreign PPT. We showed that the U3 sequences do not make a significant contribution to the specific cleavage at the PPT-U3 junction by RSV RT, and we found that the U tract 5′ of the PPT, which has been reported to play a role in the efficient replication of primate lentiviruses (6), plays no measurable role in the replication of ASLVs or in the RNase H cleavages that determine the ends of the linear viral DNA. We did find that sequences that lie between the PPT and the U tract do make a modest contribution to cleavage specificity in vivo.
MATERIALS AND METHODS
Cell culture.
DF-1, a cell line derived from EV-O line chicken embryonic fibroblasts (5, 27), was grown in Dulbecco's modified Eagle's medium (Gibco) supplemented with 5% newborn calf serum, 5% fetal bovine serum, 100 U of penicillin per ml, and 100 μg of streptomycin per ml (Gibco) at 39°C. The cells were passaged at a 1:5 dilution for routine propagation.
Construction of RSVP(A)Z1-LTRgfp-based viruses.
The RSVP(A)Z1-LTRgfp-based viruses were generated as previously described (2) using the SapI cassette cloning strategy. In brief, complementary oligonucleotides that specified the various mutations in the PPT, in sequences upstream of the PPT, or in U3 were prepared that had SapI restriction enzyme cleavage site overhangs appropriate for the vector (Fig. 1A). These oligonucleotides were phosphorylated with polynucleotide kinase. A total of 5 M NaCl was added to the complementary oligonucleotides, and the primers were heated in a water bath for 2 min at 95°C and allowed to anneal at room temperature. The annealed oligonucleotides were ligated into the SapI transfer cassette using two matching SapI cleavage sites in the cassette. The different PPTs in the SapI cassette were cloned into RSVP(A)Z1-LTRgfp using the MluI and PvuI sites.
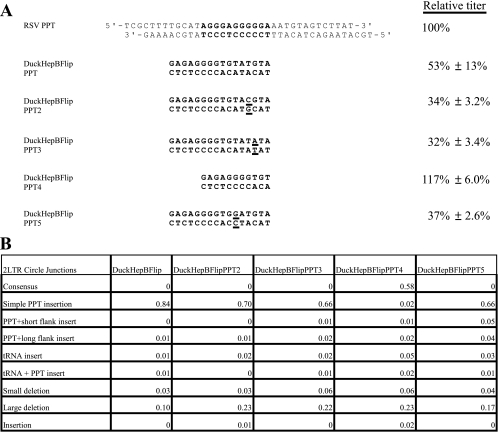
FIG. 1.
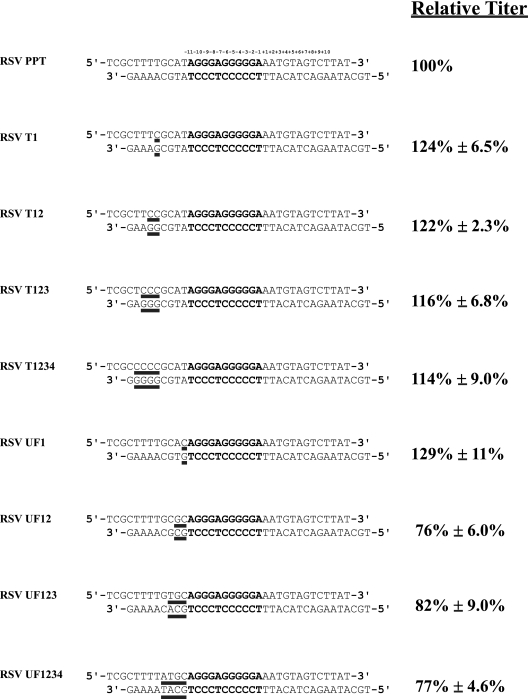
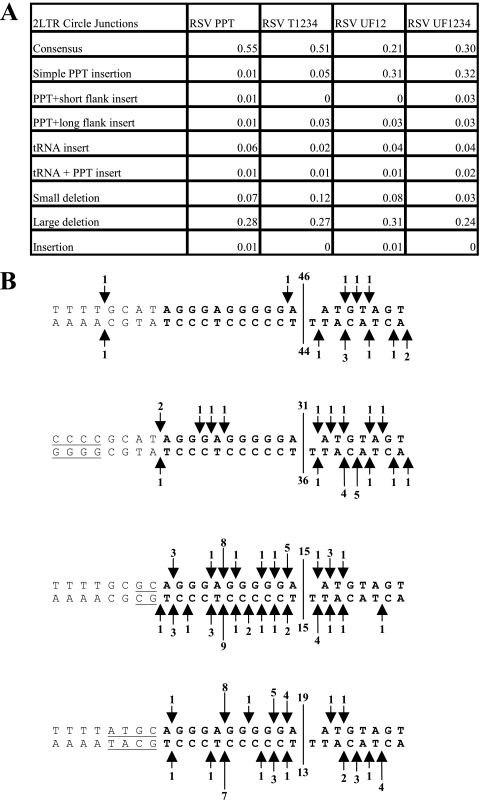
Sequences of the DuckHepBFlipPPT mutants, their relative titers and 2-LTR circle junction analysis. (A) The PPT (bold) and flanking sequences of the WT RSV (RSV PPT) and the various DuckHepBFlipPPT mutants are shown. The specific nucleotide changes are underlined in the sequences of the DuckHepBFlipPPT mutants. The relative titers of the vectors encoding the mutations are shown on the right. The relative titers were determined by infecting 293-tva cells and normalizing the titer based on the amount of p27 antigen (capsid) present in the viral stock used for the infection. The results were normalized to the WT RSV. Shown are the averages of three independent infections ± the standard deviation. (B) 2-LTR circle junction data for the DuckHepBFlipPPT mutants. Also shown are the percentages of the different classes of sequences detected in the 2-LTR circle junction assay. Consensus sequences represent a joining of the correct ends of the linear viral DNA. Simple PPT insert mutations ranged from 1 nucleotide to the insertion of the entire PPT. The PPT sequences could be inserted either into a consensus sequence junction or into a junction in which a portion of the U5 sequence was deleted. PPT plus short flank inserts were insertions of the PPT with no more than 10 nucleotides from the segment immediately upstream of the PPT. The PPT plus long flank inserts were identical to the PPT plus short flank inserts, except that the insertions contained more than 10 nucleotides from the flanking region immediately 5′ of the PPT. The tRNA insertions consisted of 1 or more nucleotides from the tryptophan tRNA primer inserted at the circle junction. The tRNA plus PPT inserts were the same as the tRNA inserts, except that they also contained either a portion of PPT or the entire PPT. Small-deletion mutants had deletions of no more than 10 nucleotides in U5 and/or U3. Large-deletion mutants had deletions larger than 10 nucleotides in the U5 and/or U3. Insertion mutants had nucleotide insertions at the circle junction of sequences that were not derived from either the PPT or tRNA primer. In the cases in which there were both insertions of nucleotides not derived from the PPT or the tRNA and deletions of either U5 and/or U3, the mutants were classified based on whether the deletion or the insertion involved more nucleotides. The data represent the average of two independent experiments.
Virus production.
Viral stocks were generated by calcium phosphate-mediated transfection of DF-1 cells seeded at 2 × 106 cells per 100-mm dish (Falcon). A total of 10 μg of the plasmids encoding the retroviral vectors was transfected into cells on 100-mm dishes for 4 h at 39°C. The transfected cells were subjected to glycerol shock for 3 min using culture medium supplemented with 15% sterile glycerol. Following the glycerol shock, the cells were washed twice with 1× phosphate-buffered saline and the normal culture media was replaced. Two days posttransfection, supernatants were collected from the cells and clarified by low-speed centrifugation at 3,000 rpm for 15 min. The clarified supernatants were used to infect fresh DF-1 cells. The relative titer of the viral stocks was measured on a 293-tva cell line (a kind gift from John A. T. Young). The number of green fluorescent protein-positive cells was determined by flow cytometry and normalized to the amount of p27 (capsid) antigen present in the infecting viral stock. The amount of p27 antigen was measured by p27 antigen-capture enzyme-linked immunosorbent assay (ELISA) (1). The results were then normalized to the titers from the WT vector, RSVP(A)Z1-LTRgfp.
2-LTR circle junction analysis.
DF-1 cells were seeded at 5 × 105 cells per 60-mm dish (Falcon) and infected overnight at 39°C with viral stocks, using the same amount of p27 for each infection. Two days postinfection, DNA was isolated from the cells by using the EZ1 DNA tissue protocol and the Qiagen BioRobot. The 2-LTR circle junctions were amplified by PCR using the primers 2LTRFOR2 (5′-CGAACCACTGAATTCCGCATTGCAG-3′) and 2LTRREV2 (5′-ACCATTCTTCTAGACAATCCATGTCAGACCCGTCTGTTGC-3′), and the resulting PCR products were subcloned into pBluescript II KS+ through the EcoRI and XbaI restriction sites introduced by the primers. DNA was isolated from the clones, and approximately 90 clones were sequenced for each mutant.
Passage of DuckHepBFlipPPT virus.
Falcon tissue culture dishes (100 mm) that had been seeded on the previous day with 1.5 × 106 DF-1 cells per dish were infected overnight at 39°C with DuckHepBFlipPPT virus produced by transfection. The next day, the inoculum was replaced with fresh culture medium, and the supernatant was collected 2 days later. The supernatant was clarified of cellular debris by centrifugation at 3,000 rpm for 15 min and used to infect fresh DF-1 cells. An aliquot of the supernatant was used to measure the amount of virus by p27 antigen-capture ELISA. The infected DF-1 cells were washed once with 1× phosphate-buffered saline and trypsinized. The DNA was extracted from the trypsinized cells by using the EZ1 DNA tissue protocol and Qiagen BioRobot. The virus was passaged by repeating this procedure 12 times. The primers RSVPZEOFOR3 (5′-CGGGACGCGTATCTGAGGG-3′) and ASEREV3 (5′-ACTAGTTCTAGAGGTATTAATGAAGCCTTCTGCT-3′) were used to amplify a portion of the viral DNA containing the PPT and U3 by PCR. The resulting PCR products were subcloned into pBluescript II KS+ through the MluI and XbaI restriction enzyme cleavage sites for DNA sequencing.
Statistical methods.
The data were analyzed by log-linear categorical analysis, contingency table analysis, and related methods. I-by-J circle junction-by-mutant tables were decomposed through the use of likelihood ratio chi-square statistics into independent partitions to show associations between circle junction and/or mutant groupings and categories. Subsets of pertinent groupings were followed up with traditional two-by-two chi-square analyses and Fisher's exact tests.
RESULTS
Mutations of the DuckHepBFlipPPT affect the relative viral titer.
To further investigate how RSV RT is able to process the DuckHepBFlipPPT, specific mutations were introduced into the DuckHepBFlipPPT (see Materials and Methods). The data we previously obtained suggest that RSV RNase H cleaves the DuckHepBFlipPPT predominantly at one position within the PPT (Fig. 2), adding a 5′-ATGTA sequence onto the U3 end of the linear viral DNA (2). Coincidentally, the sequence of this added sequence matches positions +2 to +6 in U3. The complementary (3′) strand of the 5′-ATGTA does contain a CA dinucleotide (the CA is the reverse complement of the underlined TG in the 5′-ATGTA sequence), but it will be 1 nucleotide from the U3 end of the linear viral DNA.
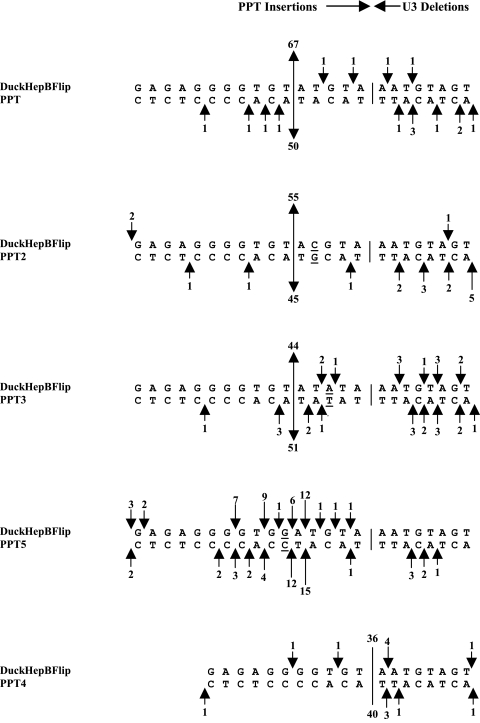
FIG. 2.
Cleavages of the DuckHepBFlipPPT mutants by the RNase H of RSV RT. The arrows depict the positions of the PPT miscleavages, which result in insertions of part or all of the PPT (simple PPT inserts) or small deletions of the U3. The positions of the cleavages are inferred from the analysis of the 2-LTR circle junctions. The arrows at the top represent the results from one experiment, and the arrows at the bottom represent the results from a second, independent experiment. The sizes of the arrows are related to the number of events (denoted by a number above and/or below the arrows). The line delineates the original border between the PPT and the U3 and, in the case of the DuckHepBFlipPPT4 mutant, where there were cleavages at this junction. The numbers above and below the line are the numbers of consensus sequences found in two experiments. The underlined nucleotides were mutated.
In order to determine whether altering the CA dinucleotide in the 3′ strand of the U3 end had any effects on the titer of the DuckHepBFlipPPT virus, the cleavage of the PPT-U3 junction, or whether these changes affected integration, each of the 2 nucleotides in the CA was individually mutated to convert the A to a G (DuckHepBFlipPPT2) or the C to a T (DuckHepBFlipPPT3) (Fig. 1A). Mutating either the A or the C reduced the relative titer from 53% ± 13% (DuckHepBFlipPPT) to 34% ± 3.2% (DuckHepBFlipPPT2) and 32% ± 3.4% (DuckHepBFlipPPT3), respectively (Fig. 1A). As described in Materials and Methods, the titer was measured by infecting 293-tva cells. Using a modified mammalian cell in which the vector cannot replicate avoids the problem of viral spread; to get the relative titer, the titer was corrected for the amount of capsid protein. Previous work from our laboratory showed that the RNase H of RSV RT preferentially cleaves after the GA dinucleotide at the 3′ end of the WT RSV PPT and that it preferentially cleaves after a GA in mutant vectors that contain PPTs from other retroviruses (2). At the sequence in the PPT of DuckHepBFlipPPT that is recognized as the PPT-U3 junction, there is a GT dinucleotide instead of a GA. In order to determine if the T immediately 5′ of the RNase H cleavage site contributes to the ability of RSV RT to specifically cleave at that particular position, the T was mutated to a G (DuckHepBFlipPPT5). This change also reduced the relative titer from 53% ± 13% to 37% ± 2.6% compared to the DuckHepBFlipPPT virus (Fig. 1A). Finally, because a 5′-ATGTA sequence is left when the RSV RT cleaves the DuckHepBFlipPPT, we removed the 5′-ATGTA sequence from the PPT, which, if the RSV RT still cleaves preferentially after the GT in the PPT, would generate a normal (consensus) U3 end in the linear viral DNA. Removing the ATGTA sequence from the DuckHepBFlipPPT (DuckHepBFlipPPT4) increased the titer to that of the WT RSV (Fig. 1A).
Effects of mutations in the DuckHepBFlipPPT on the 2-LTR circle junctions and RNase H cleavage.
Aberrant 2-LTR circle junctions can have either insertions or deletions or both insertions and deletions. Aberrant U3 ends can be generated by miscleavages by RNase H, when it either generates or removes the PPT primer. In the HIV-1 system, we showed that errors in the RNase H cleavage at the U3/PPT in the generation of the PPT primer led to the majority of the aberrant U3 ends (8). We have not repeated this type of in vitro assay with RSV RT; however, it is likely that the majority of the aberrant U3 ends are caused by miscleavages at the PPT-U3 junction.
Mutating either the C (DuckHepBFlipPPT3) or the A (DuckHepBFlipPPT2) of the CA dinucleotide (the underlined TG in the 5′-ATGTA sequence) or the T (DuckHepBFlipPPT5) immediately 5′ of the cleavage site in the PPT of the DuckHepBFlipPPT did not significantly affect the percentage of consensus 2-LTR circle junctions when the results were compared to the results obtained with 2-LTR circle junctions from DuckHepBFlipPPT-infected samples (Fig. 1B). No consensus U3 ends were found in the 2-LTR circles from cells infected with the DuckHepBFlipPPT2, DuckHepBFlipPPT3, or DuckHepBFlipPPT5 viruses. However, there were decreases in the fraction of simple PPT insertions in the 2-LTR circle junctions derived from infections with the DuckHepBFlipPPT2 mutant (70%; P = 0.0048), DuckHepBFlipPPT3 (66%; P = 0.0003), and DuckHepBFlipPPT5 (66%; P = 0.0004) samples compared to DuckHepBFlipPPT (84%). Furthermore, significant increases in the fraction of sequences with large deletions were detected for DuckHepBFlipPPT2 (23%; P = 0.0023) and DuckHepBFlipPPT3 (22%; P = 0.0052) compared to those for DuckHepBFlipPPT (10%); the percentage of aberrant 2-LTR circle junctions can be affected if integration removes some of the linear viral DNAs from the pool that gives rise to the 2-LTR circles. However, we showed previously that ASLV linear viral DNAs that have one normal and one aberrant end can still integrate with surprising efficiency (approximately 30% of the efficiency of a fully normal linear viral DNA). This means that both normal and some aberrant DNAs can be removed from the pool that gives rise to 2-LTR circle junctions by integration (17).
Based on the results obtained by analyzing 2-LTR circle junctions, mutating the CA dinucleotide in the 5′-ATGTA sequence did not affect the position of the RNase H cleavage of the DuckHepBFlipPPT. The position of RNase H cleavage was the same as that seen with the DuckHepBFlipPPT when either the C or the A was individually mutated (Fig. 2). However, mutating the T just 5′ of the cleavage site to a G (DuckHepBFlipPPT5) altered the cleavage pattern such that the RSV RT no longer cleaved predominantly at one position within the DuckHepBFlipPPT; this mutation appears to reduce the cleavage specificity of the RNase H of RSV RT (Fig. 2). However, this mutation generated a GA dinucleotide in the viral RNA, and there was a modest preferential cleavage following the newly created GA. This is not a surprising result; as has already been discussed, we previously reported that the RNase H domain of RSV RT prefers to cleave after a GA. This result and the fact that RNase H predominantly cleaves the DuckHepBFlipPPT after a GT, whereas the RNase H loses the cleavage specificity if the T of the GT is mutated, suggest that the T can function in a similar manner to the A in the GA dinucleotide at the 3′ end of the WT RSV PPT and also suggest that it may be important to have a weak base pair at this position.
Support for this interpretation comes from experiments that showed that removing the entire 5′-ATGTA sequence (DuckHepBFlipPPT4) significantly increased the fraction of consensus sequences (to 58%) in cells infected with DuckHepBFlipPPT4 (P < 0.0001) (Fig. 1B). In addition, there was a significant decrease in the percentage of 2-LTR circle junction sequences that contained part or all of the PPT (using a simple PPT insertion; P < 0.001), from 84% to 2% compared to the DuckHepBFlipPPT virus, and a significant increase (using Fisher's exact test; P = 0.0052) in junction sequences with deletions of either U3 and/or U5 larger than 10 nucleotides (large deletions) with DuckHepBFlipPPT4-infected samples (23%) compared to DuckHepBFlipPPT-infected samples (10%) (Fig. 1B). The mechanisms by which these deletions arise are not known; however, deletions in U3 could have been caused by the generation of a plus-strand primer in U3, rather than from the mutant PPT. There is no expectation that mutations in the PPT would affect the initiation of minus-strand DNA synthesis; however, it is possible that a mutation in or near the PPT could affect the ability of RT to complete the synthesis of plus-stranded DNA, which might lead to a deleted U5 end. In this mutant, the RNase H of RSV RT cleaves at the PPT-U3 junction immediately after the (mutant) PPT (Fig. 2) with good efficiency and specificity, despite the fact that the 2 nucleotides at the 3′ end of the mutant PPT are GT, not GA.
Passage of the DuckHepBFlipPPT virus.
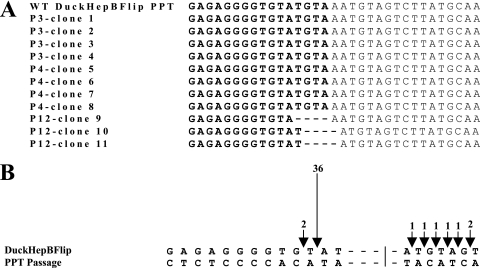
The DuckHepBFlipPPT virus was serially passaged in DF-1 cells to see if passaging the virus would select for mutations in the PPT that would enhance the level of viral replication. The virus was passaged 12 times, and after three to four passages, the levels of P27 antigen began to increase (data not shown). PCR was used to amplify the region that contains the PPT, the resulting PCR products were subcloned into pBluescript II KS+, and three or four of the clones isolated from passages 3, 4, and 12 were sequenced. The clones sequenced at passages 3 (clones 1 to 4) and 4 (clones 5 to 8) did not have changes in DuckHepBFlipPPT. However, there were changes in the PPT and adjacent U3 sequences in clones derived from passage 12 (clones 9 to 11) (Fig. 3A). Clone 9 had a deletion of the last 4 nucleotides at the 3′ end of the PPT (5′-TGTA), while clones 10 and 11 had a deletion of the last 3 nucleotides of the DuckHepBFlipPPT (5′-GTA), and the terminal nucleotide of U3 (A) was deleted. The mutations in clones 10 and 11 were recreated in the DuckHepBFlipPPT virus, and the titer was measured in 293-tva cells. The titer of the virus was relative to that of the WT RSV (93% ± 15%) (data not shown). When the RNase H cleavage of this PPT was examined by 2-LTR circle junction analysis, we found that the PPT was cleaved in the same position as in the DuckHepBFlipPPT, immediately after a T (Fig. 3B). This result provides additional support for the idea that the nucleotide (T) 5′ of the cleavage site in this foreign PPT sequence might be involved in a weak base pair, and this may be important for cleavage specificity by the RNase H of RSV RT.
FIG. 3.
Sequence of PPT-U3 junctions from passaged DuckHepBFlipPPT viruses and their cleavage by RNase H. (A) The sequences from viruses at passage 3 (P3), passage 4 (P4), and passage 12 (P12) are shown. The PPTs from the passaged viruses are shown in bold, and the dashed lines represent the deleted nucleotides. (B) Cleavage of the passaged DuckHepBFlipPPT by the RNase H of RSV RT. The arrows represent the position of the (mis)cleavages of the PPT by RT. The preferential cleavage creates an insertion, and a smaller number of miscleavages causes deletions of U3. The number of events is denoted above the arrows and by the sizes of the arrows. The line represents the position of the PPT-U3 junction in WT RSV.
The U3 sequences are not required for specific cleavage of the PPT-U3 junction by the RNaseH of RSV RT.
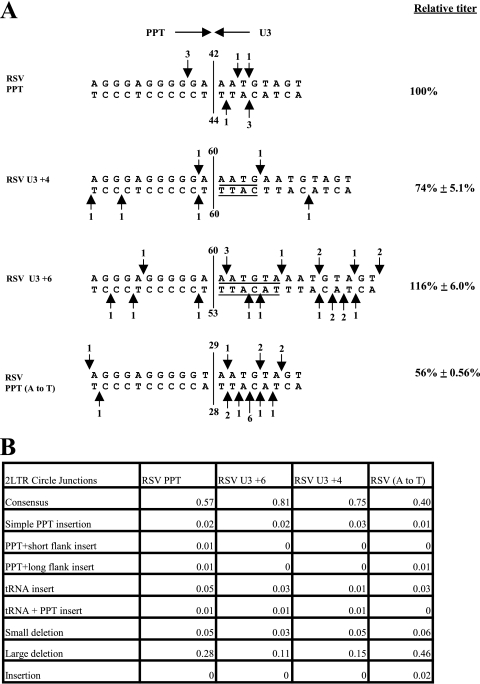
The data from the vectors with mutations in the DuckHepBFlipPPT suggested that the sequences 3′ of the cleavage site in the PPT are dispensable for the specific cleavage at this position by RSV RT. However, these observations were made in the context of the DuckHepBFlipPPT, a foreign PPT. In order to determine the effects of similar mutations in the WT RSV PPT-U3 sequence, additional mutants were generated. Duplications of the 4 (RSV U3 +4) and 6 (RSV U3 +6) terminal nucleotides of U3 were added to the end of U3 in a vector that contains the RSV PPT (Fig. 4A). Furthermore, to determine whether T could substitute for the A that is normally present at the 3′ end of the WT RSV PPT, a mutant with that substitution was generated [RSV PPT (A to T)]. The RSV U3 +4 mutant had a titer of 74% ± 5.1% relative to that of the WT RSV, while the RSV U3 +6 mutant had a titer comparable to that of the WT (Fig. 4A). 2-LTR circle junction data of RSV U3 +4 mutants (75%) showed that the percentage of sequences with consensus ends was significantly higher than that of the WT RSV (57%; P < 0.0001), and a similar result was seen with the DNA with the U3 +6 insert (81%; P = 0.0008). There was a significant decrease in the percentage of large deletions for RSV U3 +4 (15%; P = 0.0004) and RSV U3 +6 (11%; P = 0.0053) compared to that for the WT (28%) (Fig. 4B). In both mutants, regardless of whether there was a 4 or 6 nucleotide duplication, the RSV RT was still cleaved immediately after the PPT (Fig. 4A). These data suggest that the reduction in titer relative to that seen with the RSV U3 +4 mutant could result from a reduction in the efficiency with which IN was able to insert viral DNA that contained the duplication seen in RSV U3 +4. This idea is supported by the observation that there is an increase in the consensus 2-LTR circle junctions for the mutants. However, it is also possible that there is an effect on the initiation of plus-strand DNA synthesis. Because the effects on the relative titer are small (less than twofold), real-time PCR is not accurate enough to determine whether there is an effect on plus-strand DNA synthesis. We previously mutated the nucleotide at the 5′ end of U3 and showed that altering this nucleotide did not affect the specificity of cleavage at the PPT-U3 junction (18). Taken together, our data suggest that the U3 sequences near the PPT-U3 junction do not make significant contributions to the specific cleavage at the PPT-U3 junction by the RNase H of RSV RT.
FIG. 4.
(Mis)cleavages of the RSV U3 and RSV PPT (A to T) mutants by the RNase H of RSV RT and the relative titer. (A) The arrows depict the positions of the PPT (mis)cleavages, shown the same way as in Fig. 2. (B) 2-LTR circle junction analysis of RSV U3 and RSV (A to T) mutants. The categories of the 2-LTR circle junctions are the same as those in Fig. 1B.
However, the RSV PPT (A to T) mutant had a titer of 56% ± 0.56% relative to that of the WT. There was a statistically significant decrease in the percentage of consensus ends (40%; P = 0.0034) compared to that of the WT (57%), and there was a statistically significant increase in the percentage of 2-LTR circle junctions with large deletions (46%; P = 0.0011) compared to that of the WT virus (28%) (Fig. 4B). Despite the reduction in titer, the percentage of simple PPT insertions was not significantly increased by this mutation. Examination of the 2-LTR circle junctions derived from infections with this mutant showed that the RNase H of RSV RT (Fig. 4A) made relatively few miscleavages near the PPT-U3 junction. There was a significant increase in large deletions in U3 and/or U5; this could contribute to the decrease in titer.
Effects of mutations immediately 5′ of the PPT.
To determine whether sequences immediately 5′ of the PPT influence the specificity of the cleavages of the PPT-U3 junction, we sequentially mutated the conserved stretch of four Ts [sometimes called the poly(U) tract], that is, 4 nucleotides 5′ of the PPT in the RSV genome to Cs (Fig. 5). Mutating the poly(U) tract had no measurable effect on the relative titer (Fig. 5). The RSV T1, RSV T12, RSV T123, and RSV T1234 mutants all had titers comparable to those of the parental WT vector (RSV PPT). Nor was the relative titer affected by mutating the first of the 4 nucleotides that lie between the poly(U) tract and the PPT (the RSV UF1 mutation) (Fig. 5). However, mutating two or more of the nucleotides immediately upstream of the PPT caused a modest reduction in the relative titer. Mutating two nucleotides (RSV UF12) reduced the titer to 76% ± 6.0% of the WT; however, mutating three or all four of the nucleotides immediately upstream of the PPT (RSV UF123 and RSV UF1234) did not cause any further reduction in the relative titer (Fig. 5).
FIG. 5.
Relative titers of RSV upstream PPT and poly(U) tract mutants. The relative titers obtained from infection of 293-tva cells and normalized to the amount of p27 (capsid) measured by p27 antigen-capture ELISA are shown. The results were normalized to the WT RSV. The values are the averages of three independent experiments ± standard deviation.
The specificities of the RNase H cleavages in the RSV T1234, RSV UF12, and RSV UF1234 mutants were analyzed by isolating and sequencing 2-LTR circle junctions. As expected from the titer, the percentages of the different classes of 2-LTR circle junction sequences found for the RSV T1234 mutant were quite similar to what was seen with a vector with the WT RSV PPT (Fig. 6A). However, there were significant decreases of consensus sequences seen with both the RSV UF12 (21%; P < 0.001) and RSV UF1234 (30%; P < 0.0001) mutants compared to those with the WT (55%) and significant increases of simple PPT insertions seen with the RSV UF12 (31%; P < 0.0001) and RSV UF1234 (32%; P < 0.0001) mutants compared to those with the WT (1%). The percentages of the consensus sequences and various classes of aberrant 2-LTR circles found with the RSV UF 12 and RSV UF1234 mutants were not significantly different from each other (Fig. 6A). However, an examination of the number and location of the miscleavages of the PPT-U3 junction from RSV UF12- and RSV UF1234-derived 2-LTR circle junctions suggests that the RSV UF12 mutation appears to have a greater effect on miscleavages of the PPT (Fig. 6B). In both the RSV UF12 and UF1234 PPTs, the RNase H of RSV RT appeared to preferentially miscleave near the middle of the PPT (Fig. 6B). This preferential miscleavage occurs after a GA dinucleotide sequence, consistent with our previous finding that the RNase H of RSV RT preferentially cleaves after a GA dinucleotide (2).
FIG. 6.
2-LTR circle junction analysis of mutants 5′ of the PPT. (A) 2-LTR circle junction data. The data are the average of two independent experiments. The categories of the 2-LTR circle junctions are the same as those in Fig. 1B. (B) (Mis)cleavages of the PPT-U3 junctions derived from the mutants 5′ of the PPT. Shown are the PPT (bold), sequences 5′ of the PPT, and the 5′ end of U3. The line delineates the PPT-U3 junction, and the underlined nucleotides indicate the changes made to the WT sequence. The arrows depict the positions of the PPT (mis)cleavages, shown the same way as in Fig. 2.
DISCUSSION
The cleavages that generate and remove the PPT primer define the left end of the linear viral DNA that is inserted into the host genome to form the provirus. Mutation and/or substitution of the WT PPT with exogenous PPTs can lead to miscleavages by RNase H, including miscleavages near the PPT-U3 junction. In turn, these miscleavages can lead to the production of linear viral DNAs with aberrant U3 ends that are integrated somewhat less efficiently than the normal linear viral DNA; the aberrant ends can cause aberrant integrations (17, 18).
Our previous studies showed that replacing the endogenous PPT from RSV with the DuckHepBFlipPPT gave relatively high titers even though the sequence of the DuckHepBFlipPPT differs considerably from the sequence of the endogenous RSV PPT (2). The DuckHepBFlipPPT is longer than the RSV PPT, and the RNaseH preferentially cleaves the DuckHepBFlipPPT at one position within the PPT, which causes the 3′ end of the PPT (5′-ATGTA) to be added to the U3 end of the linear viral DNA (Fig. 2). The addition of the 5′-ATGTA would introduce a CA 1 nucleotide from the U3 3′ end of the linear viral DNA. We previously showed, using mutations in the vectors that have a WT PPT, that RSV IN can remove a single nucleotide from the end of a linear DNA that has the CA 1 nucleotide from the end of the linear viral DNA, and then properly insert the viral DNA into the host genome (17, 18). However, we also found in mutant viruses in which IN can process and insert only one of the two ends of the linear viral DNA into the host genome that the titer can be as high as 30% of WT. In the absence of recovered proviruses, it was unclear whether IN processed and integrated some or all of the U3 ends of these mutants. Single-nucleotide mutations in the CA dinucleotide in the 3′ strand complementary to the 5′-ATGTA sequence decreased the relative titer to 32% of WT compared to 53% of WT with the DuckHepBFlipPPT virus. When the cleavage of the PPT from these mutants was examined, the position of the predominant cleavage site was not altered from what was observed for the DuckHepBFlipPPT, suggesting that these two mutations do not affect the specificity of RNase H cleavage. Because the titers were decreased when the CA was mutated, it is likely that the CA is used for IN-mediated integration; however, we have not ruled out the possibility that one or both of these mutations could have affected the initiation of plus-strand DNA synthesis.
The results from the DuckHepBFlipPPT and CA dinucleotide mutants suggested that RSV can replicate reasonably efficiently with a duplication of the sequences at the 5′ end of U3. We confirmed the fact that a modest duplication of the sequences at the 5′ end of U3 has little effect on the specificity of cleavage by introducing U3 duplications into the genome of the WT vector; these mutant viruses all have relatively high titers. The RNase H of RSV RT ignored the duplications and cleaved correctly at the PPT-U3 junction.
We previously reported that the RNase H of RSV RT preferentially cleaves after the GA dinucleotide at the 3′ end of the RSV PPT (2). Both structural analysis of the HIV-1 PPT in a complex with HIV-1 RT (26) and biochemical studies (13) have suggested that the HIV-1 PPT has several weak A-T base pairs. In particular, Kvaratskhelia et al. (13) suggested that the A-T base pair immediately adjacent to the PPT in the U3 was weak, indicating a possible distortion in the RNA-DNA hybrid at this position. We suggested that the A-T base pair in the GA dinucleotide at the 3′ end of the PPT of RSV may also be weak, and the resulting distortion could help RNase H cleave immediately after the GA. In the DuckHepBFlipPPT, there is a GT instead of GA immediately upstream of the preferred cleavage site. If the critical element is that the last base pair before the cleavage site is easy to distort, a T-A base pair might also work. To test this idea, a mutant was generated that had a T-to-G substitution (DuckHepBFlipPPT5), which should create a stronger G-C base pair. This mutation decreased the titer relative to that of the DuckHepBFlipPPT virus. Instead of making one predominant cleavage in the PPT of the DuckHepBFlipPPT5, RNase H made multiple cleavages within the PPT, although there did appear to be a preferred cleavage site following a GA dinucleotide. This suggests that, at least in the context of the DuckHepBFlipPPT, a T-A base pair can take the place of the A-T base pair just 5′ of the cleavage site. To test the idea that a critical element determining RNase H cleavage at the PPT-U3 junction is a weak base pair just before the cleavage site, we changed the terminal A in the RSV PPT to a T [RSV PPT (A to T)]; however, this reduced the relative titer to 56% of that of the WT. This shows that a T-A base pair cannot fully substitute for an A-T base pair at the PPT-U3 junction of the WT RSV PPT and suggests that the specificity of cleavage is determined by more than the strength of the base pair adjacent to the cleavage site. The reduction in titer for the RSV PPT (A to T) mutant was greater than that seen with RSV PPT2, a previously generated mutant where the A at the 3′ end of the RSV PPT was replaced with a G (73% of WT) (2). A possible explanation for this difference is that the first nucleotide in U3 is an A. Converting the 3′ terminal A of the PPT to a G (RSV PPT2) generated a GA dinucleotide 1 nucleotide from the original PPT-U3 junction. The 2-LTR circle junction analysis shows that this mutation shifts the PPT-U3 junction by 1 nucleotide, following the newly created GA dinucleotide, which appears to explain the relatively high titer of this mutant.
When the RSV RNase H cleaves the DuckHepBFlipPPT, the resulting linear DNA has a small portion of the PPT (5′-ATGTA) appended to U3. Removing the 5′-ATGTA sequence produced a virus that replicated like the WT and cleaved the PPT-U3 junction appropriately. To determine whether passage of the DuckHepBFlipPPT virus would select for viruses that lost the 5′-ATGTA portion of the PPT, we serially passaged the virus in DF-1 cells. Contrary to our expectations, the viruses isolated after 12 passages did not lose the entire 5′-ATGTA from the PPT. Instead, some of the viruses had the last 3 nucleotides of the 3′ end of the PPT (GTA) and the terminal adenine residue in U3 adjacent to the PPT deleted (Fig. 3A). This mutation was reconstructed in the DuckHepBFlipPPT virus and the titer of this mutant was comparable to that of the WT RSV vector. 2-LTR circle junction analysis of the RNase H cleavage of the PPT showed that cleavage predominantly occurred in the same position as that of the DuckHepBFlipPPT vector (Fig. 3B). This results in a U3 end that has an additional nucleotide relative to the U3 end of the linear DNA from the WT RSV vector. Because the titer for this mutant was comparable to that for the WT RSV vector, it would appear that the RSV IN is able to correctly and efficiently remove 3 nucleotides from the U3 end of linear viral DNA during integration. This result differs markedly from the results seen with an RSV mutant that had an additional nucleotide on the U5 end of the linear viral DNA; the titer of this mutant was 3.5% relative to that of the WT RSV (17). At the present time, we cannot easily explain this difference; however, the sequences of the U3 and U5 ends of the viral DNA are different, and we have found that linear viral DNAs with similar changes in the U3 and U5 ends are not treated the same way by IN (17, 18).
Other groups have shown that mutations in sequences just 5′ of the PPT can affect the replication of some retroviruses. Ilyinskii et al. reported that mutations of the U box, a conserved stretch of T residues (Us in the retroviral RNA) found 5′ of the PPT in many retroviruses, affected the replication of simian immunodeficiency virus (6). A mutant of MLV that had a substitution of the entire T-rich region upstream of the PPT had low viral titers (24). Four adjacent T (U) residues are found 5′ of the RSV PPT. Mutating all four Ts (RSV T1234) did not affect either the titer or the specificity of the RNase H cleavage. However, mutating two, three, or four nucleotides that lie between the four T residues and the PPT did cause a reduction in the titer relative to that of the WT RSV. Based on the 2-LTR circle junction analysis, it appears that these mutations affect the cleavage specificity at the PPT-U3 junction. There was a significant increase in the percentage of 2-LTR circle junctions with a simple PPT insertion compared to that of the WT RSV (from 0.01% to 0.3%). In addition, there was a modest preference for cleavage after a GA sequence, which provides additional support for the idea that a GA 5′ of the cleavage site is important for the specific cleavage of RNase H of RSV RT. However, it is also possible that the mutations, which are near the site where the 5′ end of the PPT primer is cleaved from the viral RNA genome, may also affect the cleavage at the 5′ end of the PPT.
X-ray crystallography of an RNA-DNA hybrid of the HIV-1 PPT in a complex with HIV-1 RT showed that there are several bases at the 5′ end of the PPT that are weakly paired, mispaired, or unpaired (26). The study also showed that there are interactions between RT and the DNA primer strand in the complex (26), and subsequent biochemical and virological analyses showed that these interactions were important (10, 21). The structural analysis also led to the proposal the unpaired/mispaired nucleotides in the PPT alter the trajectory of the template primer and affect the positioning at the RNase H active site. KMnO4 chemical footprinting studies by Kvaratskhelia et al. showed that some of the As in the template strand in the HIV PPT do not have standard Watson-Crick base pairing, even in the absence of the HIV-1 RT (13). Substitution mutations in the HIV-1 PPT have shown that these regions of the PPT are important for the specificity of the in vitro cleavage of the PPT-U3 junction in reactions done with purified HIV-RT (21, 35). Furthermore, a crystal structure of an RNA-DNA hybrid of the first 10 base pairs of the HIV PPT (in the absence of HIV-1 RT) showed that the second A residue adjacent to the 5′ end of the PPT had an unusual sugar conformation (11). The authors proposed that this unusual structure may cause the RT to pause at the 5′ end of the HIV PPT, helping to direct the RNase H cleavage at the PPT-U3 junction. Taking these data together, it is possible that the A residue (and/or the A-T base pair) 2 nucleotides 5′ of the RSV PPT may have a similar effect on the structure of this PPT in a complex with the RSV RT. Mutating the A could potentially alter the contacts between the RNase H and the nucleic acid, affecting cleavage specificity.
As has already been discussed, a truncated DuckHepBFlipPPT, DuckHepBFlipPPT4, was cleaved normally, which restored the normal U3 end of the linear viral DNA, and the virus replicated as well as the WT. However, when we selected for viruses that could replicate faster than the original DuckHepBFlipPPT mutant, we selected viruses that deleted only a portion of the “extra” sequences. Clone 9 from passage 12 also had a 4-nucleotide deletion at the PPT-U3 junction, but the deletion was shifted 1 nucleotide in comparison to the clone 10 and 11 sequences (Fig. 3A). This mutant could have been selected to conform to the distance between the RSV polymerase and RNase H active sites. This distance has not been accurately defined for RSV RT, but is 17 to 18 nucleotides in HIV-1 (7, 26, 28, 34).
It is clear from our studies that there are a number of sequences for the PPT-U3 region that allow RSV isolates to replicate reasonably efficiently. This strengthens the idea that what is required is not a specific sequence but a structure, which, in the context of the cognate RT, allows RNase H to make the correct cleavages that create and remove the PPT primer, thus producing a linear DNA that is a good substrate for IN.
Acknowledgments
We thank Teresa Burdette for help with the preparation of the manuscript and Tammy L. Schroyer from the Scientific Publications, Graphics and Media, SAIC-Frederick office for help with the preparation of the figures.
Research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Published ahead of print on 18 June 2008.
REFERENCES
- 1.Barsov, E. V., W. S. Payne, and S. H. Hughes. 2001. Adaptation of chimeric retroviruses in vitro and in vivo: isolation of avian retroviral vectors with extended host range. J. Virol. 754973-4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang, K. W., J. G. Julias, W. G. Alvord, J. Oh, and S. H. Hughes. 2005. Alternate polypurine tracts (PPTs) affect the Rous sarcoma virus RNase H cleavage specificity and reveal a preferential cleavage following a GA dinucleotide sequence at the PPT-U3 junction. J. Virol. 7913694-13704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dash, C., J. W. Rausch, and S. F. Le Grice. 2004. Using pyrrolo-deoxycytosine to probe RNA/DNA hybrids containing the human immunodeficiency virus type-1 3′ polypurine tract. Nucleic Acids Res. 321539-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Furfine, E. S., and J. E. Reardon. 1991. Human immunodeficiency virus reverse transcriptase ribonuclease H: specificity of tRNA(Lys3)-primer excision. Biochemistry 307041-7046. [DOI] [PubMed] [Google Scholar]
- 5.Himly, M., D. N. Foster, I. Bottoli, J. S. Iacovoni, and P. K. Vogt. 1998. The DF-1 chicken fibroblast cell line: transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses. Virology 248295-304. [DOI] [PubMed] [Google Scholar]
- 6.Ilyinskii, P. O., and R. C. Desrosiers. 1998. Identification of a sequence element immediately upstream of the polypurine tract that is essential for replication of simian immunodeficiency virus. EMBO J. 173766-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacobo-Molina, A., J. Ding, R. G. Nanni, A. D. Clark, Jr., X. Lu, C. Tantillo, R. L. Williams, G. Kamer, A. L. Ferris, P. Clark, et al. 1993. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc. Natl. Acad. Sci. USA 906320-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones, F. D., and S. H. Hughes. 2007. In vitro analysis of the effects of mutations in the G-tract of the human immunodeficiency virus type 1 polypurine tract on RNase H cleavage specificity. Virology 360341-349. [DOI] [PubMed] [Google Scholar]
- 9.Julias, J. G., M. J. McWilliams, S. G. Sarafianos, W. G. Alvord, E. Arnold, and S. H. Hughes. 2004. Effects of mutations in the G tract of the human immunodeficiency virus type 1 polypurine tract on virus replication and RNase H cleavage. J. Virol. 7813315-13324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Julias, J. G., M. J. McWilliams, S. G. Sarafianos, E. Arnold, and S. H. Hughes. 2002. Mutations in the RNase H domain of HIV-1 reverse transcriptase affect the initiation of DNA synthesis and the specificity of RNase H cleavage in vivo. Proc. Natl. Acad. Sci. USA 999515-9520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kopka, M. L., L. Lavelle, G. W. Han, H. L. Ng, and R. E. Dickerson. 2003. An unusual sugar conformation in the structure of an RNA/DNA decamer of the polypurine tract may affect recognition by RNase H. J. Mol. Biol. 334653-665. [DOI] [PubMed] [Google Scholar]
- 12.Kulkosky, J., R. A. Katz, and A. M. Skalka. 1990. Terminal nucleotides of the preintegrative linear form of HIV-1 DNA deduced from the sequence of circular DNA junctions. J. Acquir. Immune Defic. Syndr. 3852-858. [PubMed] [Google Scholar]
- 13.Kvaratskhelia, M., S. R. Budihas, and S. F. Le Grice. 2002. Pre-existing distortions in nucleic acid structure aid polypurine tract selection by HIV-1 reverse transcriptase. J. Biol. Chem. 27716689-16696. [DOI] [PubMed] [Google Scholar]
- 14.Li, L., J. M. Olvera, K. E. Yoder, R. S. Mitchell, S. L. Butler, M. Lieber, S. L. Martin, and F. D. Bushman. 2001. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 203272-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McWilliams, M. J., J. G. Julias, S. G. Sarafianos, W. G. Alvord, E. Arnold, and S. H. Hughes. 2003. Mutations in the 5′ end of the human immunodeficiency virus type 1 polypurine tract affect RNase H cleavage specificity and virus titer. J. Virol. 7711150-11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miles, L. R., B. E. Agresta, M. B. Khan, S. Tang, J. G. Levin, and M. D. Powell. 2005. Effect of polypurine tract (PPT) mutations on human immunodeficiency virus type 1 replication: a virus with a completely randomized PPT retains low infectivity. J. Virol. 796859-6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oh, J., K. W. Chang, and S. H. Hughes. 2006. Mutations in the U5 sequences adjacent to the primer binding site do not affect tRNA cleavage by Rous sarcoma virus RNase H but do cause aberrant integrations in vivo. J. Virol. 80451-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oh, J., K. W. Chang, R. Wierzchoslawski, W. G. Alvord, and S. H. Hughes. 2008. Rous sarcoma virus (RSV) integration in vivo: a CA dinucleotide is not required in U3, and RSV linear DNA does not autointegrate. J. Virol. 82503-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oh, J., M. J. McWilliams, J. G. Julias, and S. H. Hughes. 2008. Mutations in the U5 region adjacent to the primer binding site affect tRNA cleavage by human immunodeficiency virus type 1 reverse transcriptase in vivo. J. Virol. 82719-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pullen, K. A., L. K. Ishimoto, and J. J. Champoux. 1992. Incomplete removal of the RNA primer for minus-strand DNA synthesis by human immunodeficiency virus type 1 reverse transcriptase. J. Virol. 66367-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rausch, J. W., and S. F. Le Grice. 2004. ‘Binding, bending and bonding’: polypurine tract-primed initiation of plus-strand DNA synthesis in human immunodeficiency virus. Int. J. Biochem. Cell Biol. 361752-1766. [DOI] [PubMed] [Google Scholar]
- 22.Rausch, J. W., and S. F. Le Grice. 2007. Purine analog substitution of the HIV-1 polypurine tract primer defines regions controlling initiation of plus-strand DNA synthesis. Nucleic Acids Res. 35256-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rausch, J. W., D. Lener, J. T. Miller, J. G. Julias, S. H. Hughes, and S. F. Le Grice. 2002. Altering the RNase H primer grip of human immunodeficiency virus reverse transcriptase modifies cleavage specificity. Biochemistry 414856-4865. [DOI] [PubMed] [Google Scholar]
- 24.Robson, N. D., and A. Telesnitsky. 1999. Effects of 3′ untranslated region mutations on plus-strand priming during Moloney murine leukemia virus replication. J. Virol. 73948-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robson, N. D., and A. Telesnitsky. 2000. Selection of optimal polypurine tract region sequences during Moloney murine leukemia virus replication. J. Virol. 7410293-10303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarafianos, S. G., K. Das, C. Tantillo, A. D. Clark, Jr., J. Ding, J. M. Whitcomb, P. L. Boyer, S. H. Hughes, and E. Arnold. 2001. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 201449-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schaefer-Klein, J., I. Givol, E. V. Barsov, J. M. Whitcomb, M. VanBrocklin, D. N. Foster, M. J. Federspiel, and S. H. Hughes. 1998. The EV-O-derived cell line DF-1 supports the efficient replication of avian leukosis-sarcoma viruses and vectors. Virology 248305-311. [DOI] [PubMed] [Google Scholar]
- 28.Schatz, O., J. Mous, and S. F. Le Grice. 1990. HIV-1 RT-associated ribonuclease H displays both endonuclease and 3′—-5′ exonuclease activity. EMBO J. 91171-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schultz, S. J., M. Zhang, and J. J. Champoux. 2006. Sequence, distance, and accessibility are determinants of 5′-end-directed cleavages by retroviral RNases H. J. Biol. Chem. 2811943-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith, J. S., and M. J. Roth. 1992. Specificity of human immunodeficiency virus-1 reverse transcriptase-associated ribonuclease H in removal of the minus-strand primer, tRNA(Lys3). J. Biol. Chem. 26715071-15079. [PubMed] [Google Scholar]
- 31.Telesnitsky, A., and S. P. Goff. 1997. Reverse transcriptase and the generation of retroviral DNA, p. 121-160. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- 32.Whitcomb, J. M., and S. H. Hughes. 1992. Retroviral reverse transcription and integration: progress and problems. Annu. Rev. Cell Biol. 8275-306. [DOI] [PubMed] [Google Scholar]
- 33.Whitcomb, J. M., R. Kumar, and S. H. Hughes. 1990. Sequence of the circle junction of human immunodeficiency virus type 1: implications for reverse transcription and integration. J. Virol. 644903-4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wöhrl, B. M., and K. Moelling. 1990. Interaction of HIV-1 ribonuclease H with polypurine tract containing RNA-DNA hybrids. Biochemistry 2910141-10147. [DOI] [PubMed] [Google Scholar]
- 35.Yi-Brunozzi, H. Y., and S. F. Le Grice. 2005. Investigating HIV-1 polypurine tract geometry via targeted insertion of abasic lesions in the (−)-DNA template and (+)-RNA primer. J. Biol. Chem. 28020154-20162. [DOI] [PubMed] [Google Scholar]