Abstract
Human immunodeficiency virus type 2 (HIV-2) infection results in slower CD4+ T-cell decline, lower plasma viral load levels, and hence slower progression of the disease than does HIV-1 infection. Although the reasons for this are not clear, it is possible that HIV-2 replication is more effectively controlled by host responses. We used aligned pools of overlapping HIV-1 and HIV-2 Gag peptides in an enhanced gamma interferon enzyme-linked immunospot assay to compare the levels of homologous and cross-reactive Gag-specific T-cell responses between HIV-1- and HIV-2-infected patients. HIV-2-infected patients showed broader and stronger homologous Gag-specific T-cell responses than HIV-1-infected patients. In contrast, the cross-reactive T-cell responses in HIV-2-infected patients were both narrower and weaker than those in HIV-1-infected patients, in line with overall weaker correlations between homologous and heterologous T-cell responses among HIV-2-infected patients than among HIV-1-infected patients. Cross-reactive responses in HIV-2-infected patients tended to correlate directly with HIV-1/HIV-2 Gag sequence similarities; this was not found in HIV-1-infected patients. The CD4+ T-cell counts of HIV-2-infected patients correlated directly with homologous responses and inversely with cross-reactive responses; this was not found in HIV-1-infected patients. Our data support a model whereby high-level HIV-2-specific T-cell responses control the replication of HIV-2, thus limiting viral diversification and priming of HIV-1 cross-reactive T-cell responses over time. However, we cannot exclude the possibility that HIV-2 replication is controlled by other host factors and that HIV-2-specific T-cell responses are better maintained in the context of slow viral divergence and a less damaged immune system. Understanding the nature of immune control of HIV-2 infection could be crucial for HIV vaccine design.
Human immunodeficiency virus type 1 (HIV-1) and HIV-2 are closely related lentiviruses with different biological and epidemiological characteristics. Like HIV-1 infection, HIV-2 infection leads to immune suppression and AIDS, but with slower CD4+ T-cell decline, lower plasma viral load levels, and hence slower progression of the disease (15, 24, 27). In addition, HIV-2 shows lower transmission rates than HIV-1 (1, 18). While HIV-1 has spread worldwide, HIV-2 has remained mainly confined to West Africa, with most countries now reporting a decrease in HIV-2 prevalence (32).
Several observations suggest that HIV-2 is not simply an attenuated virus. The large differences in plasma viral load levels between HIV-1- and HIV-2-infected patients are less pronounced for the proviral load levels in peripheral blood mononuclear cells (PBMC) (3, 13, 26). HIV-1- and HIV-2-infected patients matched for plasma viral load levels showed equal rates of CD4+ T-cell decline (11). At the time of AIDS diagnosis, the mortality rate was found to be more influenced by the CD4+ T-cell count than by the HIV type (25). HIV-1 and HIV-2 also show comparable levels of cytopathicity in vitro (29). Together, these findings suggest that HIV-2 replication is more effectively controlled by host responses than HIV-1.
To date, the nature of these host defenses remains uncertain. The presence of more vigorous or effective HIV-specific T-cell responses in HIV-2-infected patients than in HIV-1-infected patients has been hypothesized, but the findings thus far have been controversial (7, 9, 16, 33). One study found that HIV-2-infected patients display a more diverse T-cell receptor repertoire, resulting in an enhanced potential to cross-recognize mutant variants, including HIV-1 variants, of HIV-2 epitopes (22). However, that study only tested a limited number of epitopes in expanded T-cell cultures, and whether this reflects the in vivo situation in HIV-2-infected patients is unclear. Therefore, in the present study, we compared levels of homologous and cross-reactive T-cell responses between HIV-1- and HIV-2-infected patients by using aligned pools of overlapping peptides spanning the entire HIV-1 and HIV-2 Gag proteins in ex vivo gamma interferon (IFN-γ) enzyme-linked immunospot (ELISPOT) assays. ELISPOT assays were enhanced by addition of the cytokines interleukin-7 (IL-7) and IL-15, which are shown to reverse HIV-1-specific T-cell anergy (12, 17). Using this methodology, we show significantly higher homologous Gag-specific T-cell responses in HIV-2-infected patients than in HIV-1-infected patients. In addition, and surprisingly, we found lower cross-reactive T-cell responses in HIV-2-infected patients than in HIV-1-infected patients.
MATERIALS AND METHODS
Patient samples.
Seventeen HIV-1- and seventeen HIV-2-infected patients were included in the study. All HIV-1-infected patients were enrolled at the Ambulatory Treatment Centre of the Centre Hospitalier Universitaire de Fann in Dakar, Senegal. Ten HIV-2-infected patients were enrolled at the Institut d'Hygiène Sociale in Dakar, Senegal, and seven were enrolled at the Institute of Tropical Medicine in Antwerp, Belgium. In addition, samples from 14 healthy HIV-seronegative blood donors were collected, 7 samples were from the National Blood Transfusion Centre in Dakar, Senegal, and 7 samples were from the blood transfusion center in Antwerp, Belgium. The study was approved by the ethical committees of the Institute of Tropical Medicine in Antwerp, Belgium, and the Ministry of Health in Dakar, Senegal. All subjects gave informed consent prior to enrolment.
Laboratory methods.
Blood samples were drawn in EDTA tubes, and plasma was separated from whole blood by centrifugation. The HIV status of all subjects was determined in plasma by testing algorithms based on enzyme-linked immunosorbent assays (ELISAs) and Western blot analyses. In Senegal, samples testing positive for an HIV screening ELISA or rapid test were confirmed for HIV-1 or HIV-2 infection by Western blotting (New Lav Blot I/II; Bio-Rad). Indeterminate samples were retested by a membrane immunoassay (Immunocomb II; Orgenics). In Belgium, plasma samples testing positive for an HIV screening ELISA were confirmed for HIV-1 or HIV-2 infection by using a line immunoassay (InnoLIA; Innogenetics). Indeterminate or dually reactive samples were retested by Western blotting (New Lav Blot I/II). Patients with a confirmed HIV-1/HIV-2 dual infection were excluded from the study. HIV-1 viral load levels were assessed by the Amplicor HIV-1 monitor test (version 1.5; Roche), HIV-2 viral load levels by an in house semiquantitative reverse transcription-PCR test. CD4+ and CD8+ T-cell counts were measured in fresh whole blood by using a FACSCount flow cytometer (Becton Dickinson). PBMC were isolated from whole blood by density gradient centrifugation using Ficoll-Hypaque (Pharmacia). Cells were washed twice, resuspended in fetal bovine serum (Biochrom AG, Berlin, Germany) containing 10% dimethyl sulfoxide (Axis-Shield PoC AS), and kept frozen in liquid nitrogen.
Peptides and peptide pools.
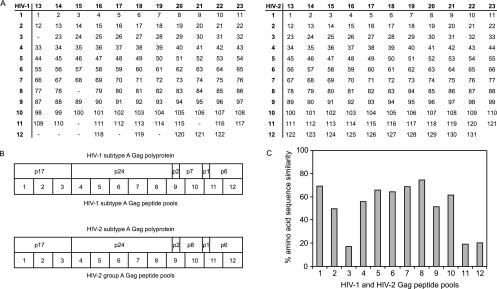
A total of 122 15-mer peptides overlapping by 11 amino acids spanning HIV-1 consensus subtype A Gag were obtained from the AIDS Research and Reference Reagent Program (Division of AIDS, National Institute of Allergy and Infectious Disease, National Institutes of Health). A total of 131 15-mer peptides overlapping by 11 amino acids spanning HIV-2 group A Gag were obtained from Pepscan Systems (The Netherlands). The peptides were dissolved in water, dimethyl sulfoxide, or acetonitrile according to the instructions of the suppliers. HIV-1 and HIV-2 Gag sequences were aligned by using CLUSTAL W (http://www.ebi.ac.uk/clustalw/) and manual editing. HIV-1 and HIV-2 peptides were mixed together in 12 consecutive and 11 perpendicular peptide pools (Fig. 1A). Empty spaces in the HIV-1 peptide matrix represent insertions in HIV-2 Gag relative to HIV-1 Gag in agreement with the alignment. Peptide pools 1 to 12 thus represent comparable consecutive parts of the Gag polyprotein of the two viruses (Fig. 1B). The percentages of amino acid sequence similarities between pairs of aligned HIV-1 and HIV-2 peptide pools were calculated by using CLUSTAL W (Fig. 1C).
FIG. 1.
HIV-1 and HIV-2 Gag peptide pools. (A) Twelve consecutive and eleven perpendicular peptide pools containing 122 HIV-1 subtype A Gag peptides (left) aligned to 131 HIV-2 group A Gag peptides (right). Peptides are 15-mers overlapping by 11 amino acids. Empty spaces in the HIV-1 peptide matrix represent insertions in HIV-2 Gag relative to HIV-1 Gag in agreement with the alignment. (B) Positions of the 12 consecutive peptide pools within the Gag polyprotein of HIV-1 (top) and HIV-2 (bottom). (C) Percentages amino acid sequence similarity between corresponding and aligned HIV-1 and HIV-2 peptide pools.
Enhanced IFN-γ ELISPOT assay.
We used an enhanced IFN-γ ELISPOT assay with addition of the cytokines IL-7 and IL-15 as described elsewhere (17). Briefly, 96-well polyvinylidene fluoride-bottom plates (Millipore) were coated with 5 μg/ml of anti-human IFN-γ (1-D1K; Mabtech) overnight at 4°C. Cryopreserved PBMC were thawed, washed twice, and incubated overnight at 37°C and 5% CO2 at a concentration of 2 × 106 cells/ml in RPMI containing 10% fetal bovine serum, 100 U/ml of penicillin, and 100 μg/ml of streptomycin (both from Roche) (referred to as medium). The next day, plates were washed four times with phosphate-buffered saline (PBS) and blocked with medium for at least 1 h. Peptides and peptide pools at final concentrations of 2 μg/ml were added to single wells in the presence of 1 ng of both cytokines IL-7 and IL-15 (R&D Systems)/ml. Medium containing the cytokines alone was used as a negative control, and staphylococcal enterotoxin B (Sigma) at a final concentration of 5 μg/ml was used as a positive control. Between 50,000 and 200,000 cells were added per well. ELISPOT plates were then incubated overnight at 37°C in 5% CO2. Plates were washed, incubated with 1 μg/ml of biotinylated anti-human IFN-γ (7-B6-1; Mabtech), washed again, and incubated with streptavidin-conjugated alkaline phosphatase (Mabtech). Plates were washed again, and developed with BCIP (5-bromo-4-chloro-3-indolylphosphate) and nitroblue tetrazolium substrates (Bio-Rad). Spots were counted by using an AID ELISPOT reader (Autoimmun Diagnostika GmbH) and normalized to spot-forming cells (SFC) per million PBMC. Negative-control spots were subtracted from stimulated spots, and negative values were scored as zero. Peptide and peptide pool responses were considered to be positive when (i) there was IFN-γ production in staphylococcal enterotoxin B-stimulated wells, (ii) the numbers of spots in stimulated wells was at least twice that in negative-control wells, and (iii) higher than the negative cutoff value calculated as the average peptide pool response plus two times the standard deviation among 14 HIV-seronegative blood donors—175 SFC/106 PBMC for HIV-1 peptide pools and 125 SFC/106 PBMC for HIV-2 peptide pools.
Homologous responses were defined as responses to peptides derived from the homologous virus, i.e., to HIV-1 peptide pools in HIV-1-infected patients and to HIV-2 peptide pools in HIV-2-infected patients. Heterologous responses were defined as responses to peptides derived from the heterologous virus, i.e., to HIV-2 peptide pools in HIV-1-infected patients and to HIV-1 peptide pools in HIV-2-infected patients. Cross-reactive responses were defined as responses to heterologous peptide pools that confirm responses to homologous peptide pools.
Intracellular staining for IFN-γ and IL-2.
PBMC were stimulated with 2 μg/ml of HIV-1 or HIV-2 peptides in 96-well U-bottom plates, followed by incubation for 6 h at 37°C and 5% CO2. Stimulation with 0.02 μg/ml of phorbol myristate acetate and 1 μg/ml of ionomycin (both from Sigma) during 6 h served as a positive control. After 1 h of incubation, 10 μg/ml of brefeldin A (Sigma) was added to the cell cultures. Intracellular staining was performed as described before with minor modifications (31). In brief, PBMC were washed with PBS by centrifugation at 500 × g for 10 min and decanting the supernatant. Next, PBMC were incubated for 10 min at 37°C and 5% CO2 with 0.02% EDTA in PBS and washed again. The T lymphocytes were stained by adding anti-CD3 allophycocyanin and anti-CD8 phycoerythrin monoclonal antibodies (Becton Dickinson) to the cell suspension for 15 min at 4°C. The cells were washed with PBS containing 1% bovine serum albumin and 0.05% azide, fixed using Leucoperm reagent A (Serotec) for 15 min at room temperature, and washed again. The cells were then permeabilized by using Leucoperm reagent B (Serotec) and stained with fluorescein isothiocyanate-conjugated anti-IL-2 and phycoerythrin-conjugated anti-IFN-γ monoclonal antibodies (Becton Dickinson) for 30 min at 4°C. The cells were washed twice, resuspended in PBS containing 1% paraformaldehyde, and stored at 4°C until acquisition with a FACScalibur flow cytometer and CellQuest software (Becton Dickinson).
Statistical analysis.
Differences in continuous and categorical variables between HIV-1- and HIV-2-infected patients were analyzed with nonparametric Mann-Whitney U tests and Fisher exact tests, respectively. Differences between HIV-1- and HIV-2-infected patients for homologous, heterologous, and cross-reactive responses to aligned pairs of HIV-1 and HIV-2 peptide pools were analyzed with Wilcoxon signed rank tests. Linear regression analysis was used to calculate the cross-reactive response ratios. Correlation analyses were performed with Spearman rank correlation tests. The level of significance for all statistical tests was set at P < 0.05. Statistical analyses were performed with SPSS version 15.0 and R (http://www.R-project.org).
RESULTS
Study population.
A total of 17 HIV-1- and 17 HIV-2-infected patients were enrolled (Table 1, all patients). Nine HIV-1-infected patients and five HIV-2-infected patients were receiving antiretroviral therapy at the time of sample collection. HIV-2-infected patients had significantly higher CD4+ T-cell counts (∼2.5-fold) and significantly lower plasma viral load levels (∼50-fold) than did HIV-1-infected patients. Seven HIV-1-infected patients were matched to seven HIV-2-infected patients based on CD4 counts (Table 1, CD4-matched patients). Five of the seven patient pairs also matched for antiretroviral therapy status. CD4 count-matched HIV-1- and HIV-2-infected patients showed comparable viral load levels.
TABLE 1.
Clinical and demographic characteristics of HIV-1- and HIV-2-infected patients
| Characteristic | All patients
|
CD4-matched patients
|
||||
|---|---|---|---|---|---|---|
| HIV-1 infected (n = 17) | HIV-2 infected (n = 17) | Pe | HIV-1 infected (n = 7) | HIV-2 infected (n = 7) | Pe | |
| No. (%) of patients in Senegalese cohorta | 17 (100) | 10 (59) | 7 (100) | 2 (29) | ||
| No. (%) of patients in Belgian cohort | 0 (0) | 7 (41)b | 0 (0) | 5 (71)c | ||
| Median age in yr (IQR) | 35 (30-39) | 43 (41-47) | <0.001 | 32 (35-40) | 41 (41-43) | 0.047 |
| No. (%) of male subjects | 6 (35) | 5 (29) | 0.921 | 3 (43) | 4 (57) | 1.000 |
| No. (%) of patients with use of ARV | 9 (53) | 5 (29) | 0.125 | 5 (71) | 5 (71) | 1.000 |
| Median CD4+ T cells/μl (IQR) | 302 (109-486) | 781 (440-1,088) | 0.002 | 304 (426-571) | 309 (456-616) | 0.848 |
| No. of patients with CD4+ T cells/μl: | ||||||
| Missing data | 1 | 3 | 0 | 0 | ||
| <200 | 6 | 1 | 1 | 1 | ||
| 200-500 | 6 | 3 | 3 | 3 | ||
| >500 | 4 | 10 | 3 | 3 | ||
| Median CD8+ T cells/μl (IQR) | 1,093 (765-1,398) | 1,108 (948-1,378) | 0.708 | 790 (1,184-1,247) | 601 (1,095-1,516) | 0.949 |
| Median log10 RNA copies/ml (IQR)d | 4.03 (1.70-5.10) | 2.36 (2.36-2.36) | 0.018 | 1.70 (1.70-1.70) | 2.36 (2.36-3.11) | 0.528 |
| No. of patients with log10 RNA copies/ml: | ||||||
| Missing data | 4 | 2 | 1 | 1 | ||
| Undetectable | 6 | 13 | 5 | 4 | ||
| Undetectable, −10,000 | 0 | 1 | 0 | 1 | ||
| 10,000-100,000 | 4 | 0 | 1 | 0 | ||
| 100,000-1,000,000 | 2 | 1 | 0 | 1 | ||
| >1,000,000 | 1 | 0 | 0 | 0 | ||
All patients are of West African origin.
Three patients are West African, three are European, and one is Central American.
Two patients are West African, two are European, and one is Central American.
The viral load detection limits were 1.7 log10 RNA copies/ml for HIV-1 and 2.36 log10 RNA copies/ml for HIV-2.
P, level of significance for Mann-Whitney U tests or Fisher exact tests.
HIV-2-infected patients showed broader and stronger homologous Gag responses than HIV-1-infected patients.
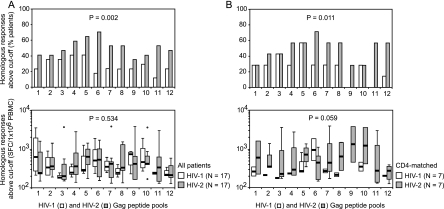
An overview of homologous peptide pool responses is shown in Fig. 2, top panels. Fifteen (88%) of seventeen HIV-1-infected patients and fourteen (82%) of seventeen HIV-2-infected patients showed ELISPOT responses above the negative cutoff value for at least one homologous peptide pool (P = 1.000). For the 12 aligned HIV-1 and HIV-2 Gag peptide pools, the percentages of HIV-2-infected patients responding to HIV-2 peptide pools were consistently and significantly higher than the percentages of HIV-1-infected patients responding to HIV-1 peptide pools (Fig. 3A, top panel). Among the patients with detectable responses, median patient responses to the 12 aligned peptide pools were similar for HIV-1- and HIV-2-infected patients (Fig. 3A, bottom panel). After matching for CD4 counts, the percentages of HIV-2-infected patients responding to homologous peptide pools remained significantly higher (Fig. 3B, top panel). In addition, HIV-2-infected patients now showed higher median homologous responses than HIV-1-infected patients (Fig. 3B, bottom panel).
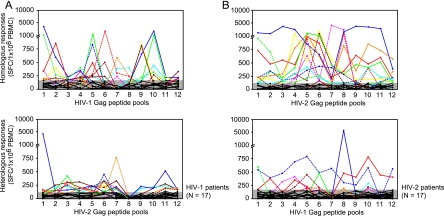
FIG. 2.
Overview of ELISPOT responses to the 12 consecutive HIV-1 and HIV-2 Gag peptide pools. (A) HIV-1-infected patients. Top panel, homologous responses to HIV-1 Gag peptide pools; bottom panel, heterologous responses to HIV-2 Gag peptide pools. (B) HIV-2-infected patients. Top panel, homologous responses to HIV-2 Gag peptide pools; bottom panel, heterologous responses to HIV-1 Gag peptide pools. Shaded areas represent negative cutoff values for the HIV-1 and HIV-2 peptide pools, respectively, calculated as the average responses plus two times the standard deviation among 14 HIV-seronegative blood donors. Different line patterns and colors represent responses above the negative cutoff value for individual HIV-1-infected patients (A) and HIV-2-infected patients (B).
FIG. 3.
Homologous peptide pool responses in HIV-1 and HIV-2-infected patients. (A) All HIV-1- and HIV-2-infected patients. (B) HIV-1 and HIV-2-infected patients matched for CD4 counts. Top graphs show percentages of patients with responses above the cutoff value to the 12 consecutive HIV-1 and HIV-2 peptide pools. Bottom graphs show patient response levels above the cutoff value to the 12 consecutive HIV-1 and HIV-2 peptide pools. Box plots represent the lowest value, the 25th percentile, the 50th percentile (median), the 75th percentile, and the highest value, respectively. Outliers, defined as values smaller than the 25th percentile minus 1.5 times the IQR or larger than the 75th percentile plus 1.5 times the IQR, are represented by open circles. Differences in response frequencies (top graphs) and median response levels (bottom graphs) between HIV-1 and HIV-2-infected patients were analyzed for the 12 aligned peptide pool pairs with Wilcoxon signed-rank tests.
HIV-1-infected patients showed broader but weaker heterologous Gag responses than HIV-2-infected patients.
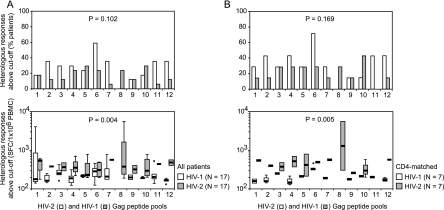
An overview of heterologous peptide pool responses is shown in Fig. 2 (bottom panels). Thirteen (76%) of seventeen HIV-1-infected patients and ten (59%) of seventeen HIV-2-infected patients showed ELISPOT responses above the negative cutoff value for at least one heterologous peptide pool (P = 0.465). For the 12 aligned HIV-1 and HIV-2 Gag peptide pools tested, the percentages of HIV-1-infected patients responding to HIV-2 peptide pools tended to be higher than the percentages of HIV-2-infected patients responding to HIV-1 peptide pools (Fig. 4A, top panel). Among the patients with detectable responses, median responses to the 12 aligned peptide pools were significantly higher for HIV-2-infected patients than for HIV-1-infected patients (Fig. 4A, bottom panel). These findings remained valid after matching for CD4 counts (Fig. 4B).
FIG. 4.
Heterologous peptide pool responses in HIV-1 and HIV-2-infected patients. (A) All HIV-1- and HIV-2-infected patients. (B) HIV-1- and HIV-2-infected patients matched for CD4 counts. Top graphs show percentages of patients with responses above the cutoff value to the 12 consecutive HIV-2 and HIV-1 peptide pools. Bottom graphs show patient response levels above the cutoff value to the 12 consecutive HIV-2 and HIV-1 peptide pools. Box plots represent the lowest value, the 25th percentile, the 50th percentile (median), the 75th percentile, and the highest value, respectively. Outliers, defined as values smaller than the 25th percentile minus 1.5 times the IQR or larger than the 75th percentile plus 1.5 times the IQR, are represented by open circles. Differences in response frequencies (top graphs) and median response levels (bottom graphs) between HIV-1- and HIV-2-infected patients were analyzed for the 12 aligned peptide pool pairs with Wilcoxon signed-rank tests.
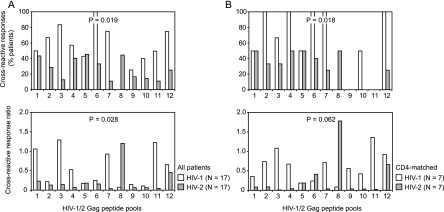
HIV-1-infected patients showed broader and stronger cross-reactive Gag responses than HIV-2-infected patients.
As a measure for cross-reactivity, we calculated the percentages of patients with heterologous peptide pool responses above the cutoff value, confirming homologous peptide pool responses above the cutoff value. However, for the 12 corresponding HIV-1 and HIV-2 Gag peptide pools tested, HIV-1-infected patients showed significantly higher percentages of cross-reactivity than HIV-2-infected patients (Fig. 5A, top panel). This remained valid after matching for CD4 counts (Fig. 5B, top panel). Next, for the 12 corresponding peptide pool pairs and for both groups of HIV-1- and HIV-2-infected patients, we performed linear regression analyses using the homologous peptide pool responses as the independent variables and the heterologous peptide pool responses as the dependent variables. This resulted in statistically significant regression lines for all 12 peptide pool pairs among HIV-1-infected patients but only for 5 of 12 peptide pool pairs among HIV-2-infected patients (P < 0.001, Table 2). The slope of the linear regression equation was used as a measure for the cross-reactive response ratio. A slope of <1 for a given peptide pool pair would indicate on average weaker heterologous responses than homologous responses, which was the case for most peptide pool pairs for both HIV-1- and HIV-2-infected patient groups. However, for the 12 corresponding peptide pool pairs, HIV-1-infected patients showed significantly higher cross-reactive response ratios than HIV-2-infected patients (Table 2 and Fig. 5A, bottom panel). These findings remained valid after matching for CD4 counts (Table 2 and Fig. 5B, bottom panel). In three cases, linear regression slopes higher than 1 were found (Table 2). The value of 1.06 obtained for peptide pool pair 1 in HIV-1-infected patients resulted from one patient with high responses (>3,000 SFC/106 PBMC) to both HIV-1 and HIV-2 peptide pool 1. The value of 1.23 obtained for peptide pool pair 11 in HIV-1-infected patients was supported by the majority of patients. The value of 1.20 obtained for peptide pool pair 8 among HIV-2-infected patients resulted from one patient with a response to the HIV-1 peptide pool that was nearly fourfold higher than the response to the HIV-2 peptide pool. The reason for this last outlier is unknown.
FIG. 5.
Cross-reactive peptide pool responses in HIV-1 and HIV-2-infected patients. (A) All HIV-1- and HIV-2-infected patients. (B) HIV-1- and HIV-2-infected patients matched for CD4 counts. Top graphs show percentages of patients with cross-reactive responses, i.e., heterologous responses above the cutoff value confirming homologous responses above the cutoff value, for the 12 consecutive peptide pool pairs. Bottom graphs show the cross-reactive response ratios, i.e., the linear regression slopes using the homologous responses as the independent variables and the heterologous responses as the dependent variables, for the 12 consecutive peptide pool pairs. Differences between HIV-1- and HIV-2-infected patients were analyzed for the 12 aligned peptide pool pairs with Wilcoxon signed-rank tests.
TABLE 2.
Linear regression analysis of heterologous versus homologous Gag peptide pool responses in HIV-1 and HIV-2 patientsa
| Gag peptide pool pair | Linear regression
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All patients
|
CD4-matched patients
|
|||||||||||
| HIV-1 infected (n = 17)
|
HIV-2 infected (n = 17)
|
HIV-1 infected (n = 7)
|
HIV-2 infected (n = 7)
|
|||||||||
| B | R2 | P | B | R2 | P | B | R2 | P | B | R2 | P | |
| 1 | 1.06 | 0.91 | <0.001 | 0.23 | 0.30 | 0.018 | 0.36 | 0.39 | 0.100 | 0.08 | 0.05 | 0.580 |
| 2 | 0.21 | 0.28 | 0.024 | 0.13 | 0.15 | 0.107 | 0.74 | 0.58 | 0.028 | 0.08 | 0.07 | 0.519 |
| 3 | 1.29 | 0.81 | <0.001 | 0.15 | 0.08 | 0.717 | 1.08 | 0.85 | 0.001 | 0.01 | 0.00 | 0.918 |
| 4 | 0.52 | 0.62 | <0.001 | 0.07 | 0.06 | 0.315 | 0.67 | 0.85 | 0.001 | 0.04 | 0.02 | 0.757 |
| 5 | 0.17 | 0.30 | 0.018 | 0.18 | 0.24 | 0.039 | 0.18 | 0.26 | 0.198 | 0.18 | 0.13 | 0.346 |
| 6 | 0.25 | 0.30 | 0.019 | 0.16 | 0.34 | 0.011 | 0.23 | 0.33 | 0.140 | 0.42 | 0.71 | 0.009 |
| 7 | 0.93 | 0.62 | <0.001 | 0.04 | 0.06 | 0.321 | 0.71 | 0.83 | 0.002 | 0.03 | 0.06 | 0.568 |
| 8 | 0.07 | 0.27 | 0.028 | 1.20 | 0.33 | 0.012 | 0.08 | 0.52 | 0.043 | 1.78 | 0.47 | 0.059 |
| 9 | 0.15 | 0.42 | 0.004 | 0.06 | 0.19 | 0.067 | 0.56 | 0.42 | 0.085 | 0.04 | 0.27 | 0.185 |
| 10 | 0.12 | 0.46 | 0.002 | 0.06 | 0.05 | 0.363 | 0.42 | 0.83 | 0.002 | 0.03 | 0.03 | 0.669 |
| 11 | 1.23 | 0.73 | <0.001 | 0.05 | 0.10 | 0.213 | 1.36 | 0.93 | <0.001 | 0.02 | 0.15 | 0.349 |
| 12 | 0.65 | 0.84 | <0.001 | 0.46 | 0.30 | 0.019 | 0.92 | 0.87 | 0.001 | 0.66 | 0.40 | 0.095 |
Data are calculated with linear regression analysis through the origin. B, slope of the regression line; R2, proportion of the variability in the dependent variable about the origin explained by regression; P, level of significance. Regressions with P < 0.05 are in boldface.
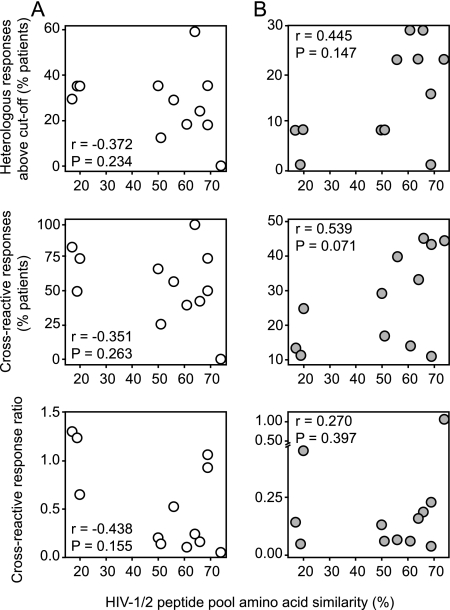
HIV-1/HIV-2 Gag sequence similarities correlated directly with levels of cross-reactivity among HIV-2-infected patients but not among HIV-1-infected patients.
Amino acid sequence similarities of the 12 corresponding HIV-1 and HIV-2 peptide pools ranged from 17 to 69% (Fig. 1C). The highest levels of cross-reactivity are expected for peptide pools with the highest percentages of sequence similarity. Among HIV-2-infected patients, heterologous and cross-reactive responses, but not the cross-reactive response ratios, tended to correlate directly with the HIV-1/HIV-2 Gag sequence similarities. These correlations were not present or tended to be inversed among HIV-1-infected patients (Fig. 6).
FIG. 6.
Cross-reactivity of peptide pool responses and HIV-1/HIV-2 Gag sequence similarity. (A) HIV-1-infected patients. (B) HIV-2-infected patients. Graphs show the correlations of percentages amino acid sequence similarities of the 12 consecutive peptide pool pairs with percentages of patients with heterologous responses (top graphs), percentages of patients with cross-reactive responses (middle graphs), and cross-reactive response ratios (bottom graphs). Correlation analyses were performed with Spearman's rank correlation test.
CD4+ T-cell counts among HIV-2-infected patients correlated directly with homologous responses and inversely with heterologous responses.
Among HIV-2-infected patients, the amplitude of the homologous peptide pool responses tended to correlate directly with the CD4+ T-cell counts (r = 0.591, P = 0.056), while the median heterologous peptide pool responses tended to correlate inversely with CD4+ T-cell counts (r = −0.667, P = 0.071). Two HIV-2-infected patients with detectable viral load levels showed a trend toward higher total homologous and heterologous responses compared to HIV-2-infected patients with undetectable viral load levels (P = 0.143 and P = 0.143, respectively). No correlations of homologous and heterologous peptide pool responses with CD4+ T-cell counts or plasma viral load levels were observed among HIV-1-infected patients (data not shown).
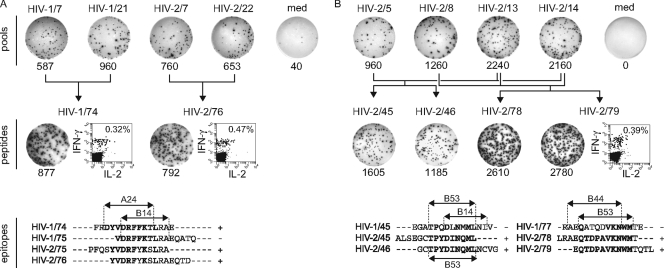
Gag peptide pool responses in one HIV-1-infected patient and one HIV-2-infected patient consisted of IFN-γ-positive, IL-2-negative CD8+ T cells targeting both known and novel epitopes.
For one HIV-1-infected patient and one HIV-2-infected patient, ELISPOT responses to the 12 consecutive and 11 perpendicular HIV-1 and HIV-2 peptide pools were analyzed (Fig. 1A). Potentially reactive HIV-1 and HIV-2 peptides were identified in the peptide matrix and retested by ELISPOT assay and intracellular cytokine staining. HIV-1-infected patient HCR1/05 showed responses to HIV-1 peptide 74 and HIV-2 peptide 76, both consisting of IFN-γ+, IL-2− CD8+ T cells (Fig. 7A). HIV-2 peptide 75 was also recognized but with lower magnitude, while HIV-1 peptide 75 was not recognized; hence, the most likely epitope was the previously described HLA A24-restricted HIV-1 Gag DYVDRFFKTL sequence (5), with SYVDRFYKSL being a cross-recognized HIV-2 Gag variant. HIV-2-infected patient HCR2/09 responded to HIV-2 peptides 45, 46, 78, and 79 (Fig. 7B). Responses to peptide 79 were confirmed as IFN-γ+, IL-2− CD8+ T cells. Peptides 45 and 46 share the previously described HLA B53-restricted HIV-2 Gag TPYDINQML epitope (10). Its B53-restricted HIV-1 Gag TPQDLNMML variant was not recognized, as previously reported (10). Peptides 78 and 79 share a novel HIV-2 Gag QTDPAVKNWM epitope which is probably also HLA B53 restricted, in agreement with the previously described HIV-1 Gag QATQDVKNWM variant (6). This HIV-1 variant was not recognized. The apparent predominance of CD8+ T-cell responses upon Gag peptide stimulation was supported by direct correlations between CD8+ T-cell counts and peptide pool ELISPOT responses among HIV-1-infected patients (heterologous: r = 0.500, P = 0.082) and HIV-2-infected patients (homologous: r = 0.709, P = 0.015; heterologous: r = 0.762, P = 0.028).
FIG. 7.
Specificity of Gag peptide pool responses. (A) HIV-1-infected patient HCR1/05 with responses to HIV-1 and HIV-2 peptides. (B) HIV-2-infected patient HCR2/09 with responses to HIV-2 peptides. Peptide and peptide pool codes are indicated above the ELISPOT images, numbers of IFN-γ SFC/106 PBMC are shown below the ELISPOT images. Flow cytometry dot plots represent gated CD3+ CD8+ lymphocytes, percentages of IFN-γ+ cells within CD8+ T cells are shown. Dot plots with gated CD4+ T cells (as CD3+ CD8− lymphocytes) are not shown but were negative for IFN-γ and IL-2 for all peptides tested. HLA class I alleles shown on peptide alignments are taken from the literature (see Results for details).
DISCUSSION
It remains unknown why infection with HIV-2 is associated with slower CD4+ T-cell decline, lower plasma viral load levels, and hence slower disease progression than infection with HIV-1. In the present study, by using aligned pools of overlapping HIV-1 and HIV-2 Gag peptides in an enhanced IFN-γ ELISPOT assay, we compared the levels of homologous and cross-reactive Gag-specific T-cell responses in HIV-1- and HIV-2-infected patients. We found that HIV-2-infected patients showed homologous Gag-specific T-cell responses that were broader and stronger than those among HIV-1-infected patients. In contrast to this, cross-reactive Gag-specific T-cell responses among HIV-2-infected patients were narrower and weaker than those among HIV-1-infected patients.
Our data are the first to show significantly higher levels of homologous HIV-specific T-cell responses in HIV-2-infected patients than in HIV-1-infected patients. Although previous studies also detected significant homologous T-cell responses in HIV-2-infected patients, the levels were either comparable to (16, 33) or lower than (9) those in HIV-1-infected patients. Our findings could suggest that HIV-2-specific T-cell responses are directly responsible for the better control of HIV-2 infection. This is supported by the observed direct correlation between homologous HIV-2-specific T-cell responses and CD4+ T-cell counts among HIV-2-infected patients. However, we cannot exclude the possibility that HIV-2 replication is controlled by another host factor(s) and that HIV-2-specific T-cell responses are simply better maintained in the context of a less-damaged immune system. In addition, in concert with lower viral load levels, HIV-2 displays significantly lower viral divergence and diversification rates than does HIV-1 (21, 23), which could support the development of strong HIV-2-specific T-cell responses over time by virtue of limited immune evasion. Nevertheless, our data comparing nonviremic HIV-2-infected patients with viremic HIV-1-infected patients are in agreement with two recent studies describing stronger Gag-specific T-cell responses in HIV-2-infected patients with undetectable viral loads compared to those with detectable viral loads (2, 20). Surprisingly, the two HIV-2-infected patients with detectable viral loads in our study did not fit this model since they actually showed relatively high T-cell responses; the reason for this is unclear. Thus, HIV-2-specific T-cell responses among HIV-2-infected patients are higher than HIV-1-specific T-cell responses among HIV-1-infected patients, but it remains to be proven whether these responses are the true cause of the attenuated pathogenesis of HIV-2 infection.
Interestingly, we found significantly lower levels of cross-reactive T-cell responses in HIV-2-infected patients than in HIV-1-infected patients, confirming the results of a previous study (33). Our data do not confirm the findings by Lopes et al., who proposed higher levels of HIV-1 cross-recognition by HIV-2-specific CD8+ T cells as a mechanism of immune control of HIV-2 (22). This discrepancy may not be unexpected in the light of the different methods that were used. In the present study, we analyzed circulating cross-reactive effector memory T cells in short-term stimulated ex vivo ELISPOT assays. The study by Lopes et al. rather analyzed the capacity of naive and central memory T cells to mount cross-reactive responses in 10-day-stimulated lymphocyte cultures. Thus, despite a higher potential of T cells in HIV-2-infected patients to cross-react with HIV-1 epitopes, possibly as a result of greater T-cell receptor heterogeneity (22), these cells appear not to be present as such in HIV-2-infected patients in vivo. Instead, low levels of circulating cross-reactive effector memory T cells in HIV-2-infected patients also appear to be consistent with the lower divergence and diversification rates of HIV-2 (21, 23). Indeed, as a result of lower diversification, HIV-2-infected patients probably sustain a relatively stable pool of HIV-2-specific T-cell clones that are mainly directed to well-conserved HIV-2 epitopes. We found that cross-reactive responses correlated directly with HIV-1/HIV-2 sequence similarities and thus require a certain level of epitope identity. Therefore, they probably consist of HIV-2-specific T-cell clones that recognize HIV-1 variants of HIV-2 epitopes within the common HLA peptide-binding motif. In contrast, higher rates of HIV-1 diversification are expected to lead to priming of larger numbers of HIV-1-specific T-cell clones recognizing a broader range of variant and probably also HIV-2 cross-reactive epitopes. Cross-reactive responses in HIV-1-infected patients did not correlate with HIV-1/HIV-2 sequence similarities; therefore, they probably consist of de novo-primed T-cell clones recognizing HIV-2 or near-HIV-2 sequences evolved within the diverse HIV-1 quasispecies.
Cross-reactive T-cell responses among HIV-2-infected patients inversely correlated with CD4+ T-cell counts, suggesting that they do not contribute to HIV-2 control but instead may be a sign of disease progression. This is in agreement with the notion of higher HIV-2 diversification rates among HIV-2-infected patients with decreasing CD4 counts in the study by MacNeil et al. (23). Neither do HIV-2 cross-reactive T-cell responses appear to play a role in the control of HIV-1, as witnessed by the lack of correlations with CD4 counts and viral load, which is in contrast with the conclusion of a previous study (33). The broad HIV-2-cross-reactive T-cell responses in HIV-1-infected patients in the present study are in agreement though with the broad cross-clade T-cell responses observed in HIV-1-infected patients infected with B and non-B subtypes (4, 8) and with the notion that broad HIV-specific CD8+ T-cell responses often represent footprints left by viral escape rather than effective immune control (19).
The present study has some limitations related to the composition of the patient groups which could have led to a number potential biases. First of all, the HIV-2-infected patient group consisted of patients enrolled in Senegal and Belgium, while all included HIV-1-infected patients were from Senegal. This could have resulted in differences in the HLA repertoire, and thus HLA-restricted T-cell responses, between the two patient groups. It is unlikely, however, that this has influenced our conclusions since HIV-2-infected patients from the Senegalese and Belgian cohorts showed comparable levels of homologous, heterologous, and cross-reactive responses (data not shown). On the other hand, the heterogeneous composition of the HIV-2-infected patient group helped us to control for differences in stage of disease progression between HIV-1- and HIV-2-infected patients. Indeed, because HIV-2-infected patients from the Belgian cohort showed lower CD4 counts, higher viral load levels, and more frequent use of antiretroviral therapy than the Senegalese HIV-2-infected patients, they were primarily used for the mutual CD4-matching with the Senegalese HIV-1-infected patients (Table 1). Second, a number of HIV-1- and HIV-2-infected patients were receiving antiretroviral therapy, which could have influenced the detection of HIV-specific T-cell responses due to its effect on viral antigen levels. However, this is not always evident from the literature (30), and it was also not obvious in our study since we did not observe differences in homologous, heterologous, and cross-reactive responses between treated and untreated HIV-1- or HIV-2-infected patients (data not shown). The fact that viral antigen is not a prerequisite for the detection of antigen-specific T-cell responses can also be deduced from the HIV-2-infected patients in this and previous studies, who, despite undetectable viral load levels, often show high-level HIV-2-specific T-cell responses. Thus, although the heterogeneous study group composition in terms of patient origin and use of antiretroviral therapy may not have been ideal, it is unlikely that this has influenced our conclusions. Furthermore, the observed differences in immune responses between HIV-1- and HIV-2-infected patients were consistently confirmed in patient subgroups that were matched for CD4 counts, viral load levels, and use of therapy.
Otherwise, no other obvious technical biases occurred in the comparison of the HIV-1- and HIV-2-infected patients. PBMC sample viabilities after overnight incubation were comparable for HIV-1- and HIV-2-infected patients (data not shown), as were the numbers of input PBMC per ELISPOT well (data not shown). Both HIV-1 consensus subtype A and HIV-2 group A Gag peptide sets were representative for the studied HIV-1- and HIV-2-infected patients. HIV-1 CRF02_AG and subtype A predominate in Senegal (14), and the used HIV-1 consensus subtype A Gag peptides corresponded well to Gag sequences from 16 Senegalese HIV-1 CRF02_AG strains and 7 Senegalese HIV-1 subtype A strains from the Los Alamos HIV database (median similarity, 94%, interquartile range [IQR], 93 to 95%). HIV-2 group A is the predominant HIV-2 type in Senegal (28), and the used HIV-2 group A Gag peptides corresponded well to Gag sequences from 2 Senegalese and 4 Gambian HIV-2 group A strains from the Los Alamos HIV database (median similarity, 92%; IQR, 90 to 93%). The higher homologous responses among HIV-2-infected patients are thus not a simple consequence of higher sequence similarities between the used peptide pools and the circulating virus strains; in fact, these similarities were higher for the HIV-1 peptides.
In the present study we used an IFN-γ ELISPOT assay enhanced by the addition of the cytokines IL-7 and IL-15 and 15-mer peptides as antigens, which are expected to stimulate both CD4+ and CD8+ T-cell responses. Because the IFN-γ ELISPOT assay does not identify the responding T-cell subset, we retested three peptides that gave detectable ELISPOT responses in one HIV-1-infected and one HIV-2-infected patient with intracellular cytokine flow cytometry. All three peptides gave clear CD8+ T-cell responses in the absence of CD4+ T-cell responses (Fig. 7). In addition, we observed direct correlations between peptide pool ELISPOT responses and CD8+ T-cell counts among HIV-1- and HIV-2-infected patients. In agreement with a previous study (20), these data suggest that the majority of the observed ELISPOT responses consisted of CD8+ T cells.
In conclusion, we have demonstrated significantly higher homologous and significantly lower cross-reactive Gag-specific T-cell responses in HIV-2-infected patients than in HIV-1-infected patients. These data could support a model whereby high-level HIV-2-specific T-cell responses control the replication of HIV-2, thus limiting viral diversification and the induction of HIV-1 cross-reactive T-cell responses over time. However, we cannot exclude the possibility that HIV-2 replication is controlled by other host factors and that HIV-2-specific T-cell responses are simply better maintained in the context of slow viral divergence and a less-damaged immune system. Understanding the nature of immune control of HIV-2 infection may be crucial for the design of HIV-protective vaccines.
Acknowledgments
This study was supported by Belgian Fund for Scientific Research (FWO-Vlaanderen) grant G.0660.06.
We thank Ndèye Fatou Ngom and Ann De Roo for blood samples from HIV-1- and HIV-2-infected patients and Katrien Fransen for viral load testing. The HIV-1 consensus subtype A Gag peptides were obtained through the AIDS Research and Reference Reagent Program (Division of AIDS, NIAID, NIH, Rockville, MD).
Footnotes
Published ahead of print on 18 June 2008.
REFERENCES
- 1.Adjorlolo-Johnson, G., K. M. De Cock, E. Ekpini, K. M. Vetter, T. Sibailly, K. Brattegaard, D. Yavo, R. Doorly, J. P. Whitaker, L. Kestens, C. Y. Ou, J. R. George, and H. D. Gayle. 1994. Prospective comparison of mother-to-child transmission of HIV-1 and HIV-2 in Abidjan, Ivory Coast. JAMA 272462-466. [PubMed] [Google Scholar]
- 2.Alatrakchi, N., F. Damond, S. Matheron, S. Beretta-Tempelhoff, P. Campa, G. Carcelain, F. Brun-Vezinet, and B. Autran. 2006. Proliferative, IFNγ and IL-2-producing T-cell responses to HIV-2 in untreated HIV-2 infection. AIDS 2029-34. [DOI] [PubMed] [Google Scholar]
- 3.Ariyoshi, K., N. Berry, A. Wilkins, D. Ricard, P. Aaby, A. Naucler, P. T. Ngom, O. Jobe, S. Jaffar, F. Dias, R. S. Tedder, and H. Whittle. 1996. A community-based study of human immunodeficiency virus type 2 provirus load in rural village in West Africa. J. Infect. Dis. 173245-248. [DOI] [PubMed] [Google Scholar]
- 4.Currier, J. R., W. E. Dowling, K. M. Wasunna, U. Alam, C. J. Mason, M. L. Robb, J. K. Carr, F. E. McCutchan, D. L. Birx, and J. H. Cox. 2003. Detection of high frequencies of HIV-1 cross-subtype reactive CD8 T lymphocytes in the peripheral blood of HIV-1-infected Kenyans. AIDS 172149-2157. [DOI] [PubMed] [Google Scholar]
- 5.Dorrell, L., T. Dong, G. S. Ogg, S. Lister, S. McAdam, T. Rostron, C. Conlon, A. J. McMichael, and S. L. Rowland-Jones. 1999. Distinct recognition of non-clade B human immunodeficiency virus type 1 epitopes by cytotoxic T lymphocytes generated from donors infected in Africa. J. Virol. 731708-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dorrell, L., B. E. Willcox, E. Y. Jones, G. Gillespie, H. Njai, S. Sabally, A. Jaye, K. DeGleria, T. Rostron, E. Lepin, A. McMichael, H. Whittle, and S. Rowland-Jones. 2001. Cytotoxic T lymphocytes recognize structurally diverse, clade-specific and cross-reactive peptides in human immunodeficiency virus type-1 gag through HLA-B53. Eur. J. Immunol. 311747-1756. [DOI] [PubMed] [Google Scholar]
- 7.Duvall, M. G., A. Jaye, T. Dong, J. M. Brenchley, A. S. Alabi, D. J. Jeffries, M. van der Sande, T. O. Togun, S. J. McConkey, D. C. Douek, A. J. McMichael, H. C. Whittle, R. A. Koup, and S. L. Rowland-Jones. 2006. Maintenance of HIV-specific CD4+ T-cell help distinguishes HIV-2 from HIV-1 infection. J. Immunol. 1766973-6981. [DOI] [PubMed] [Google Scholar]
- 8.Geels, M. J., S. A. Dubey, K. Anderson, E. Baan, M. Bakker, G. Pollakis, W. A. Paxton, J. W. Shiver, and J. Goudsmit. 2005. Broad cross-clade T-cell responses to gag in individuals infected with human immunodeficiency virus type 1 non-B clades (A to G): importance of HLA anchor residue conservation. J. Virol. 7911247-11258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gillespie, G. M., S. Pinheiro, M. Sayeid-Al-Jamee, A. Alabi, S. Kaye, S. Sabally, R. Sarge-Njie, H. Njai, K. Joof, A. Jaye, H. Whittle, S. Rowland-Jones, and L. Dorrell. 2005. CD8+ T-cell responses to human immunodeficiency viruses type 2 (HIV-2) and type 1 (HIV-1) gag proteins are distinguishable by magnitude and breadth but not cellular phenotype. Eur. J. Immunol. 351445-1453. [DOI] [PubMed] [Google Scholar]
- 10.Gotch, F., S. N. McAdam, C. E. Allsopp, A. Gallimore, J. Elvin, M. P. Kieny, A. V. Hill, A. J. McMichael, and H. C. Whittle. 1993. Cytotoxic T cells in HIV2 seropositive Gambians: identification of a virus-specific MHC-restricted peptide epitope. J. Immunol. 1513361-3369. [PubMed] [Google Scholar]
- 11.Gottlieb, G. S., P. S. Sow, S. E. Hawes, I. Ndoye, M. Redman, A. M. Coll-Seck, M. A. Faye-Niang, A. Diop, J. M. Kuypers, C. W. Critchlow, R. Respess, J. I. Mullins, and N. B. Kiviat. 2002. Equal plasma viral loads predict a similar rate of CD4+ T-cell decline in human immunodeficiency virus (HIV) type 1- and HIV-2-infected individuals from Senegal, West Africa. J. Infect. Dis. 185905-914. [DOI] [PubMed] [Google Scholar]
- 12.Gu, X. X., F. Y. Yue, C. M. Kovacs, and M. A. Ostrowski. 2007. The role of cytokines which signal through the common gamma chain cytokine receptor in the reversal of HIV specific CD4+ and CD8+ T-cell anergy. PLoS ONE 2e300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gueudin, M., F. Damond, J. Braun, A. Taieb, V. Lemee, J. C. Plantier, G. Chene, S. Matheron, F. Brun-Vezinet, and F. Simon. 2008. Differences in proviral DNA load between HIV-1- and HIV-2-infected patients. AIDS 22211-215. [DOI] [PubMed] [Google Scholar]
- 14.Hamel, D. J., J. L. Sankale, G. Eisen, S. T. Meloni, C. Mullins, A. Gueye-Ndiaye, S. Mboup, and P. J. Kanki. 2007. Twenty years of prospective molecular epidemiology in Senegal: changes in HIV diversity. AIDS Res. Hum. Retrovir. 231189-1196. [DOI] [PubMed] [Google Scholar]
- 15.Jaffar, S., A. Wilkins, P. T. Ngom, S. Sabally, T. Corrah, J. E. Bangali, M. Rolfe, and H. C. Whittle. 1997. Rate of decline of percentage CD4+ cells is faster in HIV-1 than in HIV-2 infection. J. Acquir. Immune. Defic. Syndr. Hum. Retrovirol. 16327-332. [DOI] [PubMed] [Google Scholar]
- 16.Jaye, A., R. Sarge-Njie, M. F. Schim van der Loeff, J. Todd, A. Alabi, S. Sabally, T. Corrah, and H. Whittle. 2004. No differences in cellular immune responses between asymptomatic HIV type 1- and type 2-infected Gambian patients. J. Infect. Dis. 189498-505. [DOI] [PubMed] [Google Scholar]
- 17.Jennes, W., L. Kestens, D. F. Nixon, and B. L. Shacklett. 2002. Enhanced ELISPOT detection of antigen-specific T-cell responses from cryopreserved specimens with addition of both IL-7 and IL-15: the Amplispot assay. J. Immunol. Methods 27099-108. [DOI] [PubMed] [Google Scholar]
- 18.Kanki, P. J., K. U. Travers, S. Mboup, C. C. Hsieh, R. G. Marlink, A. Gueye-Ndiaye, T. Siby, I. Thior, M. Hernandez-Avila, J. L. Sankale, I. Ndoye, and M. E. Essex. 1994. Slower heterosexual spread of HIV-2 than HIV-1. Lancet 343943-946. [DOI] [PubMed] [Google Scholar]
- 19.Karlsson, A. C., A. K. Iversen, J. M. Chapman, T. de Oliviera, G. Spotts, A. J. McMichael, M. P. Davenport, F. M. Hecht, and D. F. Nixon. 2007. Sequential broadening of CTL responses in early HIV-1 infection is associated with viral escape. PLoS ONE 2e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leligdowicz, A., L. M. Yindom, C. Onyango, R. Sarge-Njie, A. Alabi, M. Cotten, T. Vincent, C. da Costa, P. Aaby, A. Jaye, T. Dong, A. McMichael, H. Whittle, and S. Rowland-Jones. 2007. Robust Gag-specific T-cell responses characterize viremia control in HIV-2 infection. J. Clin. Investig. 1173067-3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemey, P., S. L. Kosakovsky Pond, A. J. Drummond, O. G. Pybus, B. Shapiro, H. Barroso, N. Taveira, and A. Rambaut. 2007. Synonymous substitution rates predict HIV disease progression as a result of underlying replication dynamics. PLoS Comput. Biol. 3e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopes, A. R., A. Jaye, L. Dorrell, S. Sabally, A. Alabi, N. A. Jones, D. R. Flower, A. De Groot, P. Newton, R. M. Lascar, I. Williams, H. Whittle, A. Bertoletti, P. Borrow, and M. K. Maini. 2003. Greater CD8+ TCR heterogeneity and functional flexibility in HIV-2 compared to HIV-1 infection. J. Immunol. 171307-316. [DOI] [PubMed] [Google Scholar]
- 23.MacNeil, A., J. L. Sankale, S. T. Meloni, A. D. Sarr, S. Mboup, and P. Kanki. 2007. Long-term intrapatient viral evolution during HIV-2 infection. J. Infect. Dis. 195726-733. [DOI] [PubMed] [Google Scholar]
- 24.Marlink, R., P. Kanki, I. Thior, K. Travers, G. Eisen, T. Siby, I. Traore, C. C. Hsieh, M. C. Dia, E. H. Gueye, J. Hellinger, A. Gueyendiaye, J. L. Sankale, I. Ndoye, S. Mboup, and M. Essex. 1994. Reduced rate of disease development after HIV-2 infection as compared to HIV-1. Science 2651587-1590. [DOI] [PubMed] [Google Scholar]
- 25.Martinez-Steele, E., A. A. Awasana, T. Corrah, S. Sabally, M. van der Sande, A. Jaye, T. Togun, R. Sarge-Njie, S. J. McConkey, H. Whittle, and M. F. Schim van der Loeff. 2007. Is HIV-2-induced AIDS different from HIV-1-associated AIDS? Data from a West African clinic. AIDS 21317-324. [DOI] [PubMed] [Google Scholar]
- 26.Popper, S. J., A. D. Sarr, A. Gueye-Ndiaye, S. Mboup, M. E. Essex, and P. J. Kanki. 2000. Low plasma human immunodeficiency virus type 2 viral load is independent of proviral load: low virus production in vivo. J. Virol. 741554-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Popper, S. J., A. D. Sarr, K. U. Travers, A. Gueye-Ndiaye, S. Mboup, M. E. Essex, and P. J. Kanki. 1999. Lower human immunodeficiency virus (HIV) type 2 viral load reflects the difference in pathogenicity of HIV-1 and HIV-2. J. Infect. Dis. 1801116-1121. [DOI] [PubMed] [Google Scholar]
- 28.Sarr, A. D., J. L. Sankale, A. Gueye-Ndiaye, M. Essex, S. Mboup, and P. J. Kanki. 2000. Genetic analysis of HIV type 2 in monotypic and dual HIV infections. AIDS Res. Hum. Retrovir. 16295-298. [DOI] [PubMed] [Google Scholar]
- 29.Schramm, B., M. L. Penn, E. H. Palacios, R. M. Grant, F. Kirchhoff, and M. A. Goldsmith. 2000. Cytopathicity of human immunodeficiency virus type 2 (HIV-2) in human lymphoid tissue is coreceptor dependent and comparable to that of HIV-1. J. Virol. 749594-9600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Streeck, H., H. Jessen, G. Alter, N. Teigen, M. T. Waring, A. Jessen, I. Stahmer, J. van Lunzen, M. Lichterfeld, X. Gao, T. M. Allen, M. Carrington, B. D. Walker, J. K. Rockstroh, and M. Altfeld. 2006. Immunological and virological impact of highly active antiretroviral therapy initiated during acute HIV-1 infection. J. Infect. Dis. 194734-739. [DOI] [PubMed] [Google Scholar]
- 31.Tavernier, A., W. Jennes, K. Fransen, A. De Roo, and L. Kestens. 2006. Dominant ex vivo cross-stimulation of CD8+ T cells with whole soluble gag protein in HIV-infected subjects. J. Acquir. Immune. Defic. Syndr. 41548-556. [DOI] [PubMed] [Google Scholar]
- 32.van der Loeff, M. F., A. A. Awasana, R. Sarge-Njie, M. van der Sande, A. Jaye, S. Sabally, T. Corrah, S. J. McConkey, and H. C. Whittle. 2006. Sixteen years of HIV surveillance in a West African research clinic reveals divergent epidemic trends of HIV-1 and HIV-2. Int. J. Epidemiol. 351322-1328. [DOI] [PubMed] [Google Scholar]
- 33.Zheng, N. N., N. B. Kiviat, P. S. Sow, S. E. Hawes, A. Wilson, H. Diallo-Agne, C. W. Critchlow, G. S. Gottlieb, L. Musey, and M. J. McElrath. 2004. Comparison of human immunodeficiency virus (HIV)-specific T-cell responses in HIV-1- and HIV-2-infected individuals in Senegal. J. Virol. 7813934-13942. [DOI] [PMC free article] [PubMed] [Google Scholar]