Abstract
Changes in the envelope proteins of retroviruses can alter the ability of these viruses to infect the central nervous system (CNS) and induce neurological disease. In the present study, nine envelope residues were found to influence neurovirulence of the Friend murine polytropic retrovirus Fr98. When projected on a three-dimensional model, these residues were clustered in two spatially separated groups, one in variable region B of the receptor binding site and the other on the opposite side of the envelope. Further studies indicated a role for these residues in virus replication in the CNS, although the residues did not affect viral entry.
Amino acid sequence variations in retroviral envelope proteins play a critical role in altering viral pathogenesis and the host response. Human immunodeficiency virus envelope variants vary in chemokine induction in vitro and may influence neurovirulence in vivo (4, 18, 19). Similarly, simian immunodeficiency virus, feline immunodeficiency virus, and murine retrovirus envelopes play a role in regulating virus infection, host response, and neurological disease (1, 3, 8, 11, 15). Determining which amino acid residues are required for neurovirulence may provide important information on how retroviruses induce damage in the central nervous system (CNS).
The neurovirulence of the polytropic murine retrovirus Fr98 is encoded within the SphI-ClaI restriction sites of the viral genome, which contain the 3′ end of the polymerase and most of the viral envelope gene (16). The polytropic Fr54, which differs from Fr98 by multiple nucleotide substitutions in the SphI-ClaI region, does not induce neurovirulence, despite neuroinvasion and infection of similar brain cell types (16). Two separate areas of the SphI-ClaI region influence neurovirulence, one region within the SphI-EcoRI (SE) restriction sites and one within the EcoRI-ClaI (EC) restriction sites (6). These regions mediate pathogenesis by separate mechanisms, as viruses encoding only the SE or EC region of the Fr98 genome induce disease more slowly than Fr98 does (6).
Previous studies mapped the residues in the EC region responsible for neurovirulence to two residues at positions 165 and 168 (17) in the receptor binding domain (RBD) (5). However, the Fr98 residues in the SE region which are associated with neurovirulence have not been identified. In the present study, we analyzed which amino acids encoded by the SE fragment of the Fr98 envelope gene were necessary or sufficient for the induction of neurological disease.
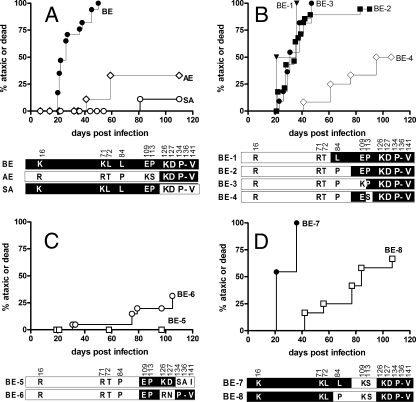
A common restriction site, BbsI, found in the 5′ end of the envelope gene for both Fr54 and SE was used to generate a chimeric virus, BE, that coded for Fr98 residues in the envelope region, but not in the polymerase gene. Newborn inbred Rocky Mountain White (IRW) mice injected with BE by intraperitoneal inoculation developed clinical signs of ataxia and/or seizures at 20 to 50 days postinoculation (Fig. 1A), similar to that of mice injected with SE (17). Thus, the neurovirulent determinants of SE were encoded within the BE region of the envelope gene.
FIG. 1.
Survival curve analyses of mice inoculated with viral clones. (A to D) Mice were inoculated with viral clones BE (n = 24), AE (n = 12), and SA (n = 8) (A), BE-1 (n = 4), BE-2 (n = 31), BE-3 (n = 12), and BE-4 (n = 11) (B), BE-5 (n = 10) and BE-6 (n = 24) (C), and BE-7 (n = 11) and BE-8 (n = 55) (D). IRW mice were infected with 104 focus-forming units of virus within 24 h of birth by intraperitoneal inoculation. Mice were monitored for clinical signs of severe ataxia and seizures. Variant residues encoded between BbsI and EcoRI are shown for all viruses. The amino acid residues encoded by BE (white letters on a black background) and the amino acid residues encoded by Fr54 (black letters on a white background) are indicated. The numbers indicate the amino acid residue positions in the gp70 SU protein.
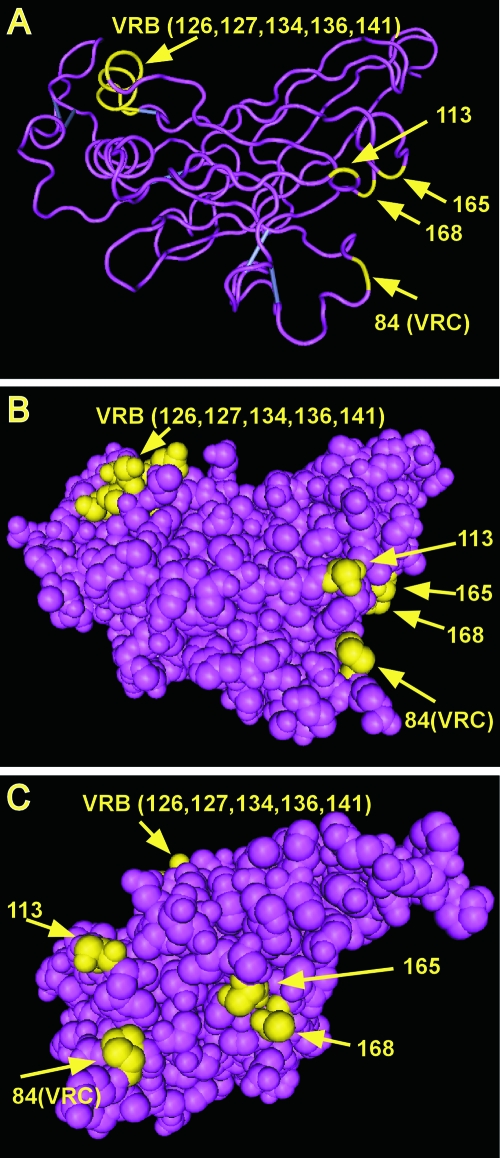
The envelope region encoded by BE contains most of the receptor binding domain. The RBDs of type C retroviruses share regions of homology interspersed with three variable regions (variable regions A, B, and C [VRA, VRB, and VRC, respectively]), which are believed to influence receptor specificity (2). Because of the strong sequence homology between Friend murine leukemia virus (MLV) and Fr98 outside the variable regions, we were able to predict the locations of the VRA, VRB, and VRC regions in the Fr98 amino acid sequence (Fig. 2) as well as the putative three-dimensional structure of the Fr98 RBD based on the crystal structure of Friend MLV (Fig. 3). Between the BbsI-EcoRI restriction sites, Fr98 and Fr54 encode 11 different residues: 2 in VRA, 1 in VRC, 5 in VRB, and 3 outside these variable regions (Fig. 2).
FIG. 2.
Alignment of amino acid sequence of SU (gp70) envelope proteins of Fr98 and Fr54 for the receptor binding domain. Residues 1 to 200 are shown, and the locations of BbsI, AflII, and EcoRI restriction sites are indicated. Residue 1 is the first residue in the mature gp70 SU protein after cleavage of the signal peptide. The positions of variable regions VRA, VRB, and VRC are shown (2). Residues encoded between BbsI and EcoRI that differed in Fr98 and Fr54 are in bold type. Boxed residues are those shown to influence neurovirulence in this paper. Boxed residues 165 and 168 in white type on a black background were identified as important for neurovirulence in a previous paper (17).
FIG. 3.
Three-dimensional representation of the crystal structure of the RBD of Friend ecotropic murine leukemia virus (Fr-MLV) (5). The crystal structure of the RBD of Fr-MLV (5) was obtained from GenBank (Structure: accession number 1AOL) and visualized with the Cn3D 4.1 program. The RBDs of Fr98 and Fr-MLV are highly homologous except at the variable regions (VRA, VRB, and VRC), and this allowed modeling of the locations of variant Fr98 residues important for neurovirulence in this three-dimensional structure. Virulence-associated residues in VRB are on the upper left part of the molecule, whereas residues 84, 113, 165, and 168 are opposite on the lower right external surface. Because VRA and VRB sequences vary extensively between Fr-MLV and Fr98, it is likely that the details of this portion of the RBD model, such as the two helices and loop structures, are not accurate for Fr98.
To begin to dissect the roles of these regions, using restriction mapping, we made two complementary viruses that carried genes encoding VRA/VRC or VRB regions of the Fr98. Mice inoculated either with SA, which encoded the VRA/VRC regions of Fr98, or with AE, which encoded the VRB region of Fr98, had a low incidence of neurological disease compared to mice infected with the BE virus (Fig. 1A). Thus, residues from both regions were necessary for neurovirulence.
To delineate which combination of envelope amino acids encoded by BE contributed to disease, we created chimeric viruses with sequential changes in this region using site-directed mutagenesis of the SphI-to-ClaI fragment of the BE or AE viruses (Fig. 1B to D). Confirmation of the correct sequence was completed using sequencing prior to insertion of the mutated SphI-ClaI region into the parent virus (16, 21). The chimeric viruses BE-1 and BE-2 induced clinical disease at a similar incidence as BE virus did, indicating that Fr98 residues at positions 16, 71, 72, and 84 were not necessary for neurovirulence (Fig. 1B). Further analysis of the E and P residues at positions 109 and 113 indicated that the P residue at position 113 was important for neurovirulence, but the E residue at position 109 was not (Fig. 1B).
The five variant residues in the VRB region were also tested for their influence on disease. Two mutant viruses, BE-5 and BE-6, with genes that encoded opposing Fr98 residues in the VRB region induced only minimal disease (Fig. 1C). Thus, both of these groups of residues in the VRB region contained at least one amino acid involved in neurovirulence. These VRB residues probably have receptor-specific interactions and are likely to influence neurovirulence in this process.
Although the results of the above studies indicated that residue 113 was critical for neurovirulence, the results of testing two additional mutant viruses suggested that this effect could be modulated in the presence of additional Fr98 residues at upstream positions. For example, BE-7 was highly virulent despite having the Fr54 residues at positions 109 and 113 (Fig. 1D). However, this virulence was reduced by substitution of the Fr54 residue at position 84 in mutant clone BE-8. Thus, the presence of the Fr98 residue at either position 84 or 113 appeared to be sufficient for neurovirulence in the context of these clones.
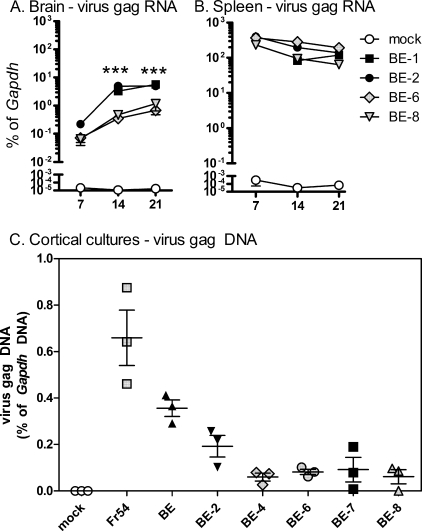
Previous studies indicated that virulence of the SE virus correlated with high virus levels in the CNS and infection of microglial cells (20). Therefore, we analyzed the kinetics of virus infection of two high-neurovirulence (BE-1 and BE-2) and two low-neurovirulence (BE-6 and BE-8) mutant viruses in the brain using real-time reverse transcriptase PCR as previously described (9). Viral RNA was readily detected in the brain in all virus-infected mice by 7 days postinfection (dpi), with expression increasing approximately 10-fold by 14 dpi and remaining high at 28 dpi (Fig. 4A). Interestingly, viral RNA levels were higher in brain tissue from BE-1- and BE-2-infected mice than in brain tissue from BE-6- or BE-8-infected mice at all three time points. Thus, the BE neurovirulent amino acid residues were associated with increased virus levels in the CNS. In contrast, splenic levels of viral RNA were similar for all four viruses (Fig. 4B).
FIG. 4.
Influence of residues on virus replication. (A and B) Brain (A) and spleen (B) tissue from mock- or virus-infected mice were removed at the indicated time points (7, 14, and 21 days postinfection) and processed for gag RNA analysis. Real-time reverse transcriptase PCR was used to analyze the expression of individual genes. Data are calculated as gene expression relative to glyceraldehyde-3-phosphate dehydrogenase (Gapdh) expression per sample (as a percentage of Gapdh expression). Data are the means ± standard errors (error bars) for three mice per group per time point. Statistical analysis was completed by two-way analysis of variance. Values that were significantly different between virulent and nonvirulent viruses are indicated as follows: ***, P < 0.001. (C) Viruses with different degrees of neurovirulence were analyzed for their ability to enter cells. Primary mixed cortical cells were cultured with ∼105 focus-forming units of each virus for 16 h. DNA was then purified from each sample and analyzed for viral DNA by real-time PCR analysis. Virus gag DNA levels were calculated as a percentage of cellular Gapdh DNA levels to control for the input of DNA. Heat-inactivated samples were used as a negative control. Samples were analyzed in triplicate wells. Data are shown as individual samples with error bars for each virus. Black symbols represent highly neurovirulent viruses, and gray symbols represent low-virulence viruses.
The difference in virus levels between neurovirulent and nonneurovirulent mutant viruses may be due to the ability of the virus to infect cells in the CNS. Therefore, we tested viral entry of several mutant viral clones using a biochemical entry assay (24) using primary cells prepared from cortical tissue from 1- or 2-day-old IRW mice. There was no consistent correlation between virus entry and neurovirulence (Fig. 3C), indicating that the residues did not influence neurovirulence by affecting viral entry in primary cortical cells, which were mostly astroglia, microglia, and fibroblasts.
Previous studies identified two residues at positions 165 and 168 in the EcoRI-ClaI region of the Fr98 envelope that contributed to neurovirulence (17). In the present study, we identified seven virulent residues in the BbsI-EcoRI region of the Fr98 envelope (Fig. 1). Of the nine Fr98 residues that influence neurovirulence, one residue at position 84 is in VRC, five residues are in VRB, and three residues are not located in any variable region. In the predicted crystal structure model of Fr98 (Fig. 3), these residues group into two different clusters, one in VRB and the other encompassing VRC residue 84 plus the nonvariable region residues at positions 113, 165, and 168. Together this latter cluster of residues appears to form a pocket on the surface of the RBD some distance away from the VRB cluster (Fig. 3A, B, and C). BE neurovirulence requires residues in both of these clusters on both sides of the envelope, whereas EC neurovirulence requires residues only in the second cluster region and does not require specific Fr98 residues in VRB (17). This correlates with distinct mechanisms of pathogenesis for BE and EC (14).
One of the primary associations with neurovirulence of the BE envelope was virus load in the CNS. Interestingly, viruses with altered residues either in VRB (BE-6) or in the second pocket (BE-8) had reduced replication in the CNS compared to viruses with the correct residues in both regions (Fig. 4A). The residues in VRB may interact directly with the polytropic receptor (xenotropic/polytropic receptor 1), perhaps affecting receptor binding. The location of these residues on the other side of the envelope protein may influence virus replication by other mechanisms, perhaps influencing trimerization of the envelope protein or interactions with other cellular proteins.
The influence of envelope amino acid residues on neuropathogenesis is a common finding for MuLVs. Point mutations in the virus envelope protein influence the neurovirulence of the ecotropic MuLV Ts1, which induces spongiform degeneration, and the ecotropic MuLV TR1.3, which induces intracerebral hemorrhage and stroke (7, 10, 12, 13, 22, 23). In contrast to Ts1 and TR1.3 infection, Fr98 or BE infection of the CNS induces only minimal pathological changes, despite the severe clinical symptoms making it difficult to determine the downstream effects of these mutations. The amino acid residues associated with BE neurovirulence appear to alter virus spread in the brain, although they do not alter the ability of virus to enter cells or infect other tissues, such as the spleen (Fig. 4). The early differences in viral RNA levels at 7 and 14 dpi suggest that the influence of these residues on virus load in the CNS may be related to variation in transport from the spleen to the brain as well as in neuroinvasion and infection of endothelia and microglia.
Acknowledgments
We thank Scott Hughes for technical assistance, Anita Mora and Gary Hettrick for graphics assistance, and Kim Hasenkrug and John Portis for helpful suggestions with the manuscript.
This research was supported in part by the Intramural Research Program of the NIAID, NIH, and in part by the National Center for Research Resources (grant IP20RR020159).
Footnotes
Published ahead of print on 25 June 2008.
REFERENCES
- 1.Babas, T., D. Munoz, J. L. Mankowski, P. M. Tarwater, J. E. Clements, and M. C. Zink. 2003. Role of microglial cells in selective replication of simian immunodeficiency virus genotypes in the brain. J. Virol. 77208-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Battini, J. L., J. E. Rasko, and A. D. Miller. 1999. A human cell-surface receptor for xenotropic and polytropic murine leukemia viruses: possible role in G protein-coupled signal transduction. Proc. Natl. Acad. Sci. USA 961385-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dimcheff, D. E., S. Askovic, A. H. Baker, C. Johnson-Fowler, and J. L. Portis. 2003. Endoplasmic reticulum stress is a determinant of retrovirus-induced spongiform neurodegeneration. J. Virol. 7712617-12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunfee, R. L., E. R. Thomas, P. R. Gorry, J. Wang, J. Taylor, K. Kunstman, S. M. Wolinsky, and D. Gabuzda. 2006. The HIV Env variant N283 enhances macrophage tropism and is associated with brain infection and dementia. Proc. Natl. Acad. Sci. USA 10315160-15165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fass, D., R. A. Davey, C. A. Hamson, P. S. Kim, J. M. Cunningham, and J. M. Berger. 1997. Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 angstrom resolution. Science 2771662-1666. [DOI] [PubMed] [Google Scholar]
- 6.Hasenkrug, K. J., S. J. Robertson, J. Portis, F. McAtee, J. Nishio, and B. Chesebro. 1996. Two separate envelope regions influence induction of brain disease by a polytropic murine retrovirus (FMCF98). J. Virol. 704825-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang, Y., V. L. Scofield, M. Yan, W. Qiang, N. Liu, A. J. Reid, W. S. Lynn, and P. K. Wong. 2006. Retrovirus-induced oxidative stress with neuroimmunodegeneration is suppressed by antioxidant treatment with a refined monosodium α-luminol (Galavit). J. Virol. 804557-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnston, J. B., C. Silva, and C. Power. 2002. Envelope gene-mediated neurovirulence in feline immunodeficiency virus infection: induction of matrix metalloproteinases and neuronal injury. J. Virol. 762622-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khaleduzzaman, M., J. Francis, M. E. Corbin, E. McIlwain, M. Boudreaux, M. Du, T. W. Morgan, and K. E. Peterson. 2007. Infection of cardiomyocytes and induction of left ventricle dysfunction by neurovirulent polytropic murine retrovirus. J. Virol. 8112307-12315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim, H. T., K. Waters, G. Stoica, W. Qiang, N. Liu, V. L. Scofield, and P. K. Wong. 2004. Activation of endoplasmic reticulum stress signaling pathway is associated with neuronal degeneration in MoMuLV-ts1-induced spongiform encephalomyelopathy. Lab. Investig. 84816-827. [DOI] [PubMed] [Google Scholar]
- 11.Lynch, W. P., and A. H. Sharpe. 2000. Differential glycosylation of the Cas-Br-E Env protein is associated with retrovirus-induced spongiform neurodegeneration. J. Virol. 741558-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park, B. H., E. Lavi, K. J. Blank, and G. N. Gaulton. 1993. Intracerebral hemorrhages and syncytium formation induced by endothelial cell infection with a murine leukemia virus. J. Virol. 676015-6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park, B. H., B. Matuschke, E. Lavi, and G. N. Gaulton. 1994. A point mutation in the env gene of a murine leukemia virus induces syncytium formation and neurologic disease. J. Virol. 687516-7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peterson, K. E., S. Hughes, D. E. Dimcheff, K. Wehrly, and B. Chesebro. 2004. Separate sequences in a murine retroviral envelope protein mediate neuropathogenesis by complementary mechanisms with differing requirements for tumor necrosis factor alpha. J. Virol. 7813104-13112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peterson, K. E., S. J. Robertson, J. L. Portis, and B. Chesebro. 2001. Differences in cytokine and chemokine responses during neurological disease induced by polytropic murine retroviruses map to separate regions of the viral envelope gene. J. Virol. 752848-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Portis, J. L., S. Czub, S. Robertson, F. McAtee, and B. Chesebro. 1995. Characterization of a neurologic disease induced by a polytropic murine retrovirus: evidence for differential targeting of ecotropic and polytropic viruses in the brain. J. Virol. 698070-8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poulsen, D. J., S. J. Robertson, C. A. Favara, J. L. Portis, and B. W. Chesebro. 1998. Mapping of a neurovirulence determinant within the envelope protein of a polytropic murine retrovirus: induction of central nervous system disease by low levels of virus. Virology 248199-207. [DOI] [PubMed] [Google Scholar]
- 18.Power, C., J. C. McArthur, R. T. Johnson, D. E. Griffin, J. D. Glass, R. Dewey, and B. Chesebro. 1995. Distinct HIV-1 env sequences are associated with neurotropism and neurovirulence. Curr. Top. Microbiol. Immunol. 20289-104. [DOI] [PubMed] [Google Scholar]
- 19.Power, C., J. C. McArthur, R. T. Johnson, D. E. Griffin, J. D. Glass, S. Perryman, and B. Chesebro. 1994. Demented and nondemented patients with AIDS differ in brain-derived human immunodeficiency virus type 1 envelope sequences. J. Virol. 684643-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robertson, S. J., K. J. Hasenkrug, B. Chesebro, and J. L. Portis. 1997. Neurologic disease induced by polytropic murine retroviruses: neurovirulence determined by efficiency of spread to microglial cells. J. Virol. 715287-5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sitbon, M., B. Sola, L. Evans, J. Nishio, S. F. Hayes, K. Nathanson, C. F. Garon, and B. Chesebro. 1986. Hemolytic anemia and erythroleukemia, two distinct pathogenic effects of Friend MuLV: mapping of the effects to different regions of the viral genome. Cell 47851-859. [DOI] [PubMed] [Google Scholar]
- 22.Szurek, P. F., E. Floyd, P. H. Yuen, and P. K. Wong. 1990. Site-directed mutagenesis of the codon for Ile-25 in gPr80env alters the neurovirulence of ts1, a mutant of Moloney murine leukemia virus TB. J. Virol. 645241-5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szurek, P. F., P. H. Yuen, R. Jerzy, and P. K. Wong. 1988. Identification of point mutations in the envelope gene of Moloney murine leukemia virus TB temperature-sensitive paralytogenic mutant ts1: molecular determinants for neurovirulence. J. Virol. 62357-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walter, B. L., K. Wehrly, R. Swanstrom, E. Platt, D. Kabat, and B. Chesebro. 2005. Role of low CD4 levels in the influence of human immunodeficiency virus type 1 envelope V1 and V2 regions on entry and spread in macrophages. J. Virol. 794828-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]