Abstract
Hepatitis C virus (HCV) core protein has shown to be localized in the detergent-resistant membrane (DRM), which is distinct from the classical raft fraction including caveolin, although the biological significance of the DRM localization of the core protein has not been determined. The HCV core protein is cleaved off from a precursor polyprotein at the lumen side of Ala191 by signal peptidase and is then further processed by signal peptide peptidase (SPP) within the transmembrane region. In this study, we examined the role of SPP in the localization of the HCV core protein in the DRM and in viral propagation. The C terminus of the HCV core protein cleaved by SPP in 293T cells was identified as Phe177 by mass spectrometry. Mutations introduced into two residues (Ile176 and Phe177) upstream of the cleavage site of the core protein abrogated processing by SPP and localization in the DRM fraction. Expression of a dominant-negative SPP or treatment with an SPP inhibitor, L685,458, resulted in reductions in the levels of processed core protein localized in the DRM fraction. The production of HCV RNA in cells persistently infected with strain JFH-1 was impaired by treatment with the SPP inhibitor. Furthermore, mutant JFH-1 viruses bearing SPP-resistant mutations in the core protein failed to propagate in a permissive cell line. These results suggest that intramembrane processing of HCV core protein by SPP is required for the localization of the HCV core protein in the DRM and for viral propagation.
The hepatitis C virus (HCV), which has infected an estimated 170 million people worldwide, leads to chronic hepatitis, which in turn causes severe liver diseases, including steatosis, cirrhosis, and eventually hepatocellular carcinoma (47). HCV possesses a positive-sense single-stranded RNA with a nucleotide length of 9.6 kb, which encodes a single large precursor polyprotein composed of about 3,000 amino acids. The viral polyprotein is processed by cellular and viral proteases into structural and nonstructural proteins (24). The development of efficient therapies for hepatitis C had been hampered by the lack of a reliable cell culture system, as well as by the absence of a small-animal model. Lohmann et al. established an HCV replicon, which consisted of an antibiotic selection marker and a genotype 1b HCV RNA, and showed that it replicated autonomously in the intracellular compartments of a human hepatoma cell line, Huh7 (16). The replicon system has been used as an important tool in the investigation of HCV replication, and it has served as a cell-based assay system for the evaluation of antiviral compounds. Recently, cell culture systems for in vitro replication and infectious-virus production were established based on the full-length HCV genome of a genotype 2a isolate, which was recovered from a fulminant hepatitis C patient (15, 45, 50). However, the molecular mechanism of the HCV life cycle in host cells has not been well characterized.
Several viruses have been reported to utilize a lipid raft composed of cholesterol and sphingolipids upon entry (34). The lipid raft is characterized by resistance to nonionic detergents at 4°C and includes caveolin, glycolipids, and other substances (40). Several nonenveloped viruses enter cells through a caveola/raft-mediated endosome, designated the caveosome, and then translocate to the endoplasmic reticulum (ER), endosome, or nucleus (34, 35), although enveloped viruses generally enter host cells through a clathrin-dependent pathway (18). HCV is enclosed by a host cell-derived membrane and belongs to the family Flaviviridae. Several reports suggest that HCV enters host cells through general endocytosis, such as by a clathrin-mediated pathway (5, 6, 22). However, HCV has been suggested to replicate on a detergent-resistant membrane (DRM), including some characteristic membrane structures such as lipid rafts and membranous webs (8, 9, 38). In a previous report, an HCV replication complex prepared from a cell fraction treated with a nonionic detergent was shown to be enzymatically active (2). HCV nonstructural proteins remodel the intracellular membrane to form a replication complex that includes several host proteins (8, 46). The HCV core protein has a C-terminal transmembrane region that is anchored on intracellular compartments such as the ER and mitochondria and on the surfaces of lipid droplets (10, 30, 42). Recent studies have indicated that assembly of HCV particles occurs around lipid droplets that are surrounded by the remodeled membranes (23). Although the HCV core protein functions as a capsid protein, it is found in the DRM fraction, which is distinct from the classical lipid rafts (20). However, the biological function of the HCV core protein localized in the DRM has not been clarified.
The HCV core protein is cleaved from a precursor polyprotein by a signal peptidase (SP) to liberate it from the envelope protein E1 and is then further processed by a signal peptide peptidase (SPP) (21). However, the biological significance of the intramembrane processing of the HCV core protein by SPP remains largely unknown. Furthermore, the C-terminal end of the mature HCV core protein expressed in insect cells has been reported to be Phe177 or Leu179 (12, 29), while that in mammalian cells has not been determined. Expression of SPP enhanced the accumulation of nonenveloped nucleocapsid and reduced that of enveloped nucleocapsid in yeast cells, suggesting that maturation of core protein is carried out after the formation of enveloped particles (17). However, the effect of SPP cleavage on viral assembly in mammalian cells has not been well characterized. Randall et al. have reported that introduction of a small interfering RNA targeted to SPP reduced the production of infectious HCV particles (36), suggesting that SPP is required for the production of HCV particles. In this study, we determined the cleavage site of the mature HCV core protein expressed in human cells and examined the biological significance of the intramembrane processing of the core protein by SPP for the localization of the core protein in the DRM and the production of infectious particles.
MATERIALS AND METHODS
Cell lines and HCV infection.
HCV subgenomic RNA was removed from the replicon cell line 9-13 (16) by treatment with alpha interferon. A cell line that was highly permissive for JFH-1 infection was cloned from the resulting crude populations by the limited-dilution method and designated Huh7OK1 (32). The Huh7OK1 cell line retained the ability to produce type I interferons through the RIG-I-dependent signaling pathway upon infection with RNA viruses and exhibited a cell surface expression level of human CD81 comparable to that of the parental cell line. The detailed characteristics of this cell line will be described in a future communication. The HuhOK1 and Huh7.5.1 cell lines (the latter was kindly provided by F. Chisari) and the human embryonic kidney cell line 293T were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and nonessential amino acids (Sigma, St. Louis, MO). Huh7OK1 or Huh7.5.1 cells were infected with HCV strain JFH-1 as described by Wakita et al. (45). The plasmid carrying strain JFH-1 cDNA under the control of the polI promoter (19) was transfected into Huh7OK1 or Huh7.5.1 cells, and propagation of the JFH-1 virus was determined by the production of HCV core protein (as described below) and by the titration of infectious particles (39). The persistently infected Huh7OK1 cells were maintained under normal conditions after 8 passages before use. The 9-13 cell line, which possesses an HCV subgenomic replicon (16), was cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and 1 mg/ml G418.
Plasmids.
Genes encoding the N-terminally FLAG-tagged and/or C-terminally hemagglutinin (HA)-tagged core proteins derived from the HCV genotype 1b strain J1 or its mutants were introduced into plasmid vector pcDNA3.1 (Invitrogen, Carlsbad, CA) as described previously (30). Each insert gene was transferred into a pCAGGS vector (28) at the PmeI site. The resulting plasmids encoded the HCV core protein (amino acid residues 1 to 191) with or without FLAG and HA tags at the N and C termini, respectively. All of the core proteins with these tags (FLAG-core-HA proteins) had a mutation of Ala191 to Arg in order to prevent cleavage by the SP (7). Plasmid pHH21/JFH-1, carrying a full genomic cDNA of strain JFH-1 under the control of the polI promoter, was used to produce the infectious JFH-1 virus (19). An adaptive mutation of Leu to Val at amino acid position 758 in the p7 region was introduced during a long-term passage of the JFH-1 virus into Huh7.5.1 cells (data not shown). To improve the replication efficiency of the JFH-1 virus, a mutation of Leu to Val was introduced into pHH21/JFH-1 by site-directed mutagenesis, and the resulting plasmid was designated pHH21/JFH-1/L758V. To generate plasmids encoding the mutant JFH-1 viruses, the following substitutions were introduced into pHH21/JFH-1/L758V: Val139, Val140, and Leu144 were replaced with Ala (JFH-1/VVL/3A); Ile176and Phe177 were replaced with Ala and Leu, respectively (JFH-1/IF/AL); Ala180, Ser183, and Cys184 were replaced with Val, Leu, and Val, respectively (JFH-1/ASC/VLV); and Asp2736 was replaced with Asn (JFH-1/GND).
Antibodies and reagents.
Antisera against HCV genotype 1 or 2a core proteins were raised in rabbits by immunization with peptides corresponding to the region spanning residues 103 to 115, conserved among genotypes 1a and 1b, or to the region from residue 101 to 119 of genotype 2a (strain JFH-1). These peptides were synthesized and conjugated with keyhole limpet hemocyanin (Scrum Inc., Tokyo, Japan). Antisera were purified with an affinity column conjugated with the antigenic peptides. A monoclonal antibody to HCV NS5A (5A27) was prepared from BALB/c mice (CLEA Japan, Tokyo, Japan) immunized with the recombinant domain I of NS5A by a method described previously (31). Antibodies to caveolin-1, calreticulin, and the FLAG tag (M2) were purchased from Sigma. Antibodies to the HA tag and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were purchased from Babco (Richmond, CA) and Santa Cruz Biotechnology (Santa Cruz, CA), respectively. The aspartic protease inhibitors (Z-LL)2 ketone and L685,458 were purchased from the Peptide Institute (Osaka, Japan). These inhibitors were dissolved in dimethyl sulfoxide and stored at −20°C until use.
Transfection, SDS-PAGE, and Western blotting.
Huh7.5.1 and 293T cells were transfected with plasmids by lipofection with Trans IT LT-1 (Mirus, Madison, WI) and Lipofectamine 2000 (Invitrogen), respectively, according to the manufacturers' protocols. Cells were lysed on ice in Triton lysis buffer (20 mM Tris-HCl [pH 7.4], 135 mM NaCl, 1% Triton-X 100, 10% glycerol) supplemented with a protease inhibitor mix (Nacalai Tesque, Kyoto, Japan) at 24 or 48 h after transfection and were then subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using Tris-glycine buffer and Western blotting using appropriate antibodies as previously described (30). The stained protein bands were visualized using the SuperSignal West Femto enhanced-chemiluminescence substrate (Pierce, Rockford, IL) and an LAS3000 imaging system (Fuji Photo Film, Tokyo, Japan).
Determination of the expression of the C terminus of the mature HCV core protein in mammalian cells.
Two million 293T cells cultured in a collagen-coated dish (diameter, 10 cm) were transfected with pCAGGS-FLAG-core (26) by lipofection, harvested at 20 h posttransfection with a rubber policeman after two washes with ice-cold phosphate-buffered saline (PBS), and collected by centrifugation at 1,000 × g for 5 min. The cells were lysed with 0.1 ml of triple-detergent lysis buffer (45 mM Tris-HCl [pH 7.4] containing 0.5% sodium deoxycholate, 0.1% SDS, 1% Triton X-100, 135 mM NaCl, and a protease inhibitor mix [Nacalai Tesque]) (24). The lysate was stored at −80°C until use. The lysate was thawed on ice and then centrifuged at 20,000 × g for 10 min at 4°C. The supernatant was mixed with 20 μl of 50% (vol/vol) anti-FLAG M2 affinity gel (Sigma) and then rotated at 4°C for 90 min. The gel beads were washed with the triple-detergent lysis buffer and then suspended in 30 μl of the loading buffer. The suspended gel beads were boiled for 5 min and then centrifuged at 20,000 × g for 5 min at room temperature. The resulting supernatant was subjected to SDS-PAGE, and the gel was stained with Sypro Ruby dye (Invitrogen). The portion of the gel including proteins with an expected molecular size of 20 kDa was excised from the stained gel, washed twice with 200 μl of 50 mM NH4HCO3 dissolved in 50% acetonitrile (vol/vol), and then immersed in 100 μl of 100% acetonitrile for dehydration. The dehydrated gel was incubated in 10 mM dithiothreitol and 100 mM NH4HCO3 at 56°C for 1 h. To prevent the digestion of Cys residues at the C termini by endoproteinase Asp-N, alkylation of the gels was carried out in 55 mM iodoacetamide and 100 mM NH4HCO3 at 25°C for 45 min in the dark. Finally, gel pieces were washed twice with 100 mM ammonium carbonate dissolved in acetonitrile and were dried completely before digestion. An immersed volume of endoproteinase Asp-N solution (10 μg/ml Asp-N and 50 mM NH4HCO3) was added to the dried gel and incubated at 37°C overnight, and the supernatant (the digested solution) after centrifugation was transferred to a new centrifuge tube. The precipitated gels were washed first with 20 μl of 20 mM NH4HCO3 and then with 20 μl of 50% (vol/vol) acetonitrile in 5% (vol/vol) formic acid, and the washed solutions were mixed with the digested solution and dried completely under a vacuum. The digested mixtures were applied to a ZipTip C18 column (Millipore, Tokyo, Japan). After a wash with 0.1% (vol/vol) trifluoroacetic acid, the peptides were eluted with 1 μl of 0.1% (vol/vol) trifluoroacetic acid dissolved in 75% (vol/vol) acetonitrile. Samples with 10 mg of 2,5-dihydroxybenzoic acid per ml of 33% acetonitrile matrix were analyzed by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS) using a MALDI-quadrupole-TOF tandem MS (MS-MS) QStar Pulsar i system (Applied Biosystems, Foster City, CA) in the linear positive-ion mode following the method of Hitachi Science Systems (Ibaraki, Japan).
Flotation assay.
The flotation assay was carried out according to the method of Lecat et al. (14). Briefly, 10 million transfected or infected cells were washed with ice-cold PBS and then harvested with a rubber policeman. Collected cells were suspended in 0.6 ml of TNE buffer (25 mM Tris-HCl [pH 7.4] containing 150 mM NaCl, a protease inhibitor mix [Nacalai Tesque], and 5 mM EDTA) and then homogenated with a Dounce homogenizer or suspended with a 24-gauge needle. Each homogenate was incubated for 30 min on ice with or without 1% Triton X-100. The lysates were mixed with 0.4 ml of Optiprep (Sigma) to a final concentration of 40%. This mixture was overlaid with 1.2 ml of 30%, 1.2 ml of 25%, and 0.8 ml of 5% Optiprep and was then centrifuged at 42,000 rpm and 4°C for 5 h in an SW50 rotor (Beckman Coulter, Fullerton, CA). Each fraction was collected as 0.4 ml from the top of the centrifuging tube and was then precipitated with 4 volumes of cold acetone. The pellets were resolved in the loading buffer, boiled, and then subjected to SDS-PAGE and Western blotting. The fractions containing calreticulin, which is resident in the ER, in the absence and presence of the detergent were defined as the membrane and detergent-soluble fractions, respectively. In the presence of the detergent, the fractions with caveolin-1 were defined as the detergent-resistant fractions.
Quantitative real-time PCR.
Total RNA was prepared from Huh7OK1 cells persistently infected with the JFH-1 virus or 9-13 cells by using an RNeasy minikit (Qiagen, Tokyo, Japan). The HCV genomic RNA was reverse transcribed and amplified by using a TaqMan EZ RT-PCR reagent kit (Applied Biosystems) with sense (5′-GAG TGT CGT GCA GCC TCC A-3′) and antisense (5′-CAC TCG CAA GCA CCC TAT CA-3′) primers corresponding to nucleotides 98 to 116 and 294 to 313, respectively. The kinetics of cDNA amplification were monitored by an ABI Prism 7000 sequence detection system (Applied Biosystems) using a reporter probe corresponding to nucleotides 238 to 267 of the 5′-conserved region for the HCV genotypes (5′-GCC CGC AAG ACT GCT AGC CGA GTA GTG TTG G-3′) conjugated with 6-carboxyfluorescein and 6-carboxytetramethylrhodamine at the 5′ and 3′ termini, respectively. A serial dilution of the partial HCV RNA synthesized by in vitro transcription from plasmids encoding the 5′-terminal region of HCV cDNA under the control of a T7 promoter was used as the standard for HCV genomic RNA. Intracellular GAPDH mRNA was also amplified using the TaqMan Pre-Developed Assay Reagent human GAPDH (Applied Biosystems). The values for HCV genomic RNA were normalized to those for GAPDH mRNA.
Quantitative detection of HCV core protein by ELISA.
HCV core protein was quantified by using an Ortho HCV antigen enzyme-linked immunosorbent assay (ELISA) (Ortho Clinical Diagnostics, Tokyo, Japan) according to the manufacturer's instructions. Huh7.5.1 cells were transfected with pHH21/JFH-1/L758V or its mutants by lipofection. Cells and culture supernatants were harvested at 2, 4, 6, or 8 days after transfection. To determine the amounts of the intracellular core protein, cells were lysed with Triton lysis buffer on ice and subjected to the ELISA after 100- to 10,000-fold dilutions with PBS. Total protein levels were determined with a Micro BCA protein assay reagent kit (Pierce). Amounts of intracellular and extracellular core protein were normalized to total-protein amounts.
Immunofluorescent assay.
Transfected Huh7.5.1 cells were fixed with a cold acetone-and-methanol mixture (50:50, vol/vol). After being blocked with 1% normal goat serum, cells were incubated with a mouse monoclonal antibody to NS5A at 4°C for 16 h, washed three times with PBS containing 0.5% Tween 20, and then incubated with an Alexa Fluor 594-conjugated antibody to mouse immunoglobulin G (Invitrogen). Cell nuclei were stained with Hoechst dye. The stained cells were washed three times with PBS containing 0.5% Tween 20 and then observed with a FluoView FV1000 laser scanning confocal microscope (Olympus, Tokyo, Japan).
RESULTS
Mutation in the HCV core protein confers resistance to SPP cleavage.
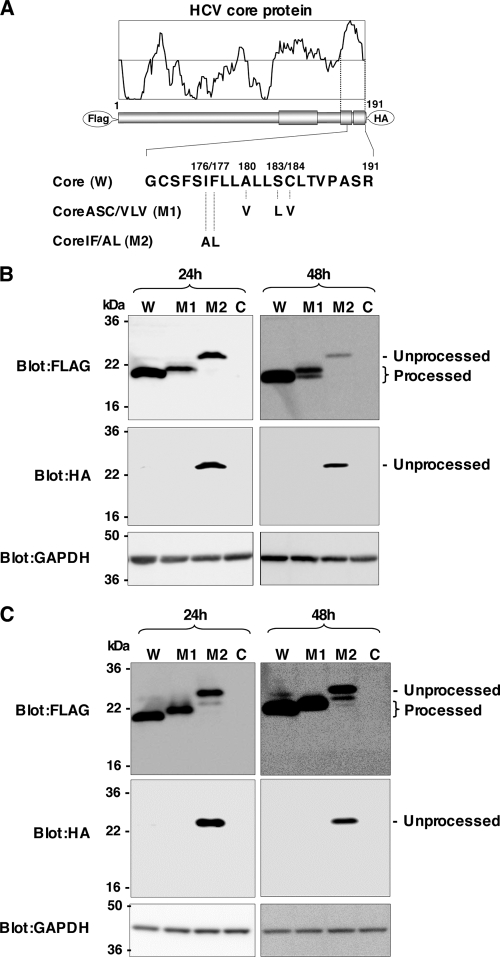
Amino acid residues Ala180, Ser183, and Cys184 of the HCV core protein have been shown by others to be essential for intramembrane processing by SPP (10, 21), although our data suggested that Ile176 and Phe177, but not Ala180, Ser183, and Cys184, were required for the processing of the HCV core protein by SPP (30). To clarify this discrepancy, we constructed an N-terminally FLAG-tagged and C-terminally HA-tagged wild-type HCV core protein and similarly tagged mutant core proteins in which Ala180, Ser183, and Cys184 were replaced with Val, Leu, and Val, respectively (referred to below as Core ASC/VLV, or M1) (21), or Ile176 and Phe177 were replaced with Ala and Leu, respectively (referred to below as Core IF/AL, or M2) (30). We then expressed these core proteins in 293T cells (Fig. 1). Ala191 was replaced with Arg in these FLAG-core-HA constructs to prevent cleavage by SP (7), and only the SPP-resistant core protein was detected by an anti-HA antibody in this experimental setting. Core IF/AL was detected in cells by both anti-FLAG and anti-HA antibodies at 24 h and 48 h posttransfection, whereas the wild-type core and Core ASC/VLV were detected by an anti-FLAG antibody but not by an anti-HA antibody (Fig. 1B). These results indicate that Core IF/AL is resistant to SPP cleavage, in contrast to the complete processing of the wild-type core and Core ASC/VLV. Although Core ASC/VLV exhibited a single band that was slightly larger than the wild-type core protein at 24 h posttransfection, an extra band with the same mobility as the wild-type core protein appeared at 48 h posttransfection (Fig. 1B), suggesting that the introduction of mutations in Ala180, Ser183, and Cys184 induces multiple processing in the signal sequence of the mutant core protein. To exclude the possibility that unprocessed Core ASC/VLV is degraded by a proteasome due to misfolding, each of the core constructs or the empty vector was transfected into 293T cells and treated with a proteasome inhibitor for 5 h. The unprocessed band of Core IF/AL, but not that of Core ASC/VLV, was detected by the anti-HA antibody (Fig. 1C). These results further support the notion that Core ASC/VLV is sensitive to SPP-dependent processing. Bands observed between unprocessed and processed proteins in cells expressing wild-type core or Core IF/AL in the presence of a proteasome inhibitor were not detected by the anti-HA antibody, suggesting that these products are generated by C-terminal truncation and are sensitive to proteasome degradation.
FIG. 1.
Effects of mutations in the HCV core protein on cleavage by SPP. (A) cDNA constructs encoding the N-terminally FLAG- and C-terminally HA-tagged wild-type HCV core protein (W), Core ASC/VLV (M1), and Core IF/AL (M2). The Ala at amino acid residue 191 of all constructs was mutated to Arg in order to prevent the processing of an HA tag by SP. (B) Each of the core constructs or an empty vector (lane C) was transfected into 293T cells. Cell lysates harvested at 24 or 48 h posttransfection were subjected to Western blotting using antibodies against the indicated proteins. (C) Cells transfected with each of the core constructs or an empty vector were treated with 15 μM MG132 for 5 h and examined as described for panel B.
Identification of the C-terminal residue of the mature HCV core protein.
Previous reports have suggested that the C terminus of the mature HCV core protein expressed in insect cells by using a baculovirus expression system is Phe177 (29) or Leu179 (12). To clarify the C-terminal amino acid residue of the mature HCV core protein expressed in human cells, a purified fragment of the HCV core protein was analyzed by MALDI-TOF MS. The FLAG-tagged HCV core protein was expressed under the control of a CAG promoter in 293T cells, purified by immunoprecipitation with beads conjugated with the anti-FLAG antibody, and then released from the beads by the addition of free FLAG peptide. The purified FLAG-tagged core protein was digested with Asp-N protease, and the final sample was subjected to MALDI-TOF MS for determination of the C-terminal residue. The N-terminal amino acid of the peptide fragment including the C terminus of the mature HCV core protein was expected to be Asp160 (Fig. 2A). The peptide fragment with an m/z of 1,918.0452, which is close to the calculated value (m/z 1,918.8) of the sequence DGVNYATGNLPGCSFSIF (Fig. 2A), was detected, and no larger peak was evident (Fig. 2B). MS-MS analysis showed that the fragment has the amino acid sequence DGVNYATGNLPGCSFSIF (Fig. 2C). These results indicate that the C terminus of the mature HCV core protein expressed in human cells is Phe177. This is consistent with our previous observation (30) and with the data shown in Fig. 1, which indicate that the M2 mutation completely abrogated the processing of core protein by SPP. Both Ile176 and Phe177 may play crucial roles in recognition by SPP for intramembrane cleaving activity.
FIG. 2.
Determination of the C termini of the mature HCV core protein. (A) Schematic representation of the junction between the core and E1 proteins. The cleavage sites for the exogenous Asp-N protease and the host SP were the N-terminal residue Asp160 and the C-terminal residue Ala191, respectively. The cleavage site of the host SPP was determined to be the C-terminal residue Phe177 in this study. The expected m/z of the peptide fragment (spanning residues 160 to 177) processed by the Asp-N protease and SPP is indicated. (B) The FLAG-core protein was purified with an anti-FLAG antibody, digested with Asp-N, and analyzed on a 2,5-dihydroxybenzoic acid matrix by MALDI-TOF MS in the linear positive-ion mode. The peak at m/z 1,918.0452 corresponded to the expected fragment (m/z 1,918.8) derived from the Asp-N- and SPP-digested core protein, DGVNYATGNLPGCSFSIF. (C) The peak at m/z 1,918.0452 was subjected to MS-MS analysis with a MALDI-Qq-TOF MS-MS QStar Pulsar i system. The resulting spectrum was applied to MASCOT to determine the amino acid sequence. The analyzed peak at m/z 1,918.0452 corresponded to the sequence DGVNYATGNLPGCSFSIF.
SPP processing is required for the localization of HCV core protein in the DRM.
Based on confocal microscopy observations, Matto et al. reported that the HCV core protein associates with a DRM that is distinct from the classical raft fraction, as evidenced by the lack of colocalization of typical raft markers, including caveolin-1 and the B subunit of the cholera toxin, which binds to glycosphingolipid GM1 in the plasma membrane (20). We have previously suggested that intramembrane processing by SPP affects the intracellular localization of the HCV core protein, and the replacement of Leu139, Val140, and Leu144 with Ala in the HCV core protein (Core LVL/3A [M3]) (Fig. 3A) abrogated SPP-mediated processing and ER retention (30). In this study, we examined the effect of SPP cleavage on the DRM localization of the HCV core protein. The wild-type or mutant HCV core protein was expressed in 293T cells, solubilized at 4°C in the presence or absence of 1% Triton X-100, and subjected to sucrose gradient centrifugation. Fractions were collected after ultracentrifugation and analyzed by immunoblotting. The wild-type core protein was partially detected in fraction 3, which corresponded to the DRM fraction, and was mainly detected in the detergent-soluble fraction (Fig. 3B). However, the mutant core proteins Core LVL/3A (M3) and Core IF/AL (M2) were localized in the membrane fraction but not in the DRM fraction (Fig. 3B). Although the M2 mutant exhibits clear resistance to SPP-dependent cleavage, as shown in Fig. 1B, processed core proteins of M2 and M3 mutants were detected by flotation analyses (Fig. 3B), suggesting that the M2 and M3 mutants are cleaved by unknown mechanisms during the concentration step. These results suggest that processing by SPP is required for the DRM localization of the HCV core protein.
FIG. 3.
HCV core protein partially migrates to the DRM after SPP processing. (A) cDNAs encoding authentic wild-type (W) and Core LVL/3A (M3) HCV core proteins. (B) Each plasmid was transfected into 293T cells, lysed with or without 1% Triton X-100, and then subjected to a flotation assay. Proteins in each fraction were concentrated with cold acetone and then subjected to Western blotting using antibodies against core protein, caveolin-1, and calreticulin. Membrane (left panels, lanes 1 to 9), DRM (right panels, lanes 1 to 7), and detergent-soluble (right panels, lanes 8 to 11) fractions were identified based on the localization of the marker proteins.
A dominant-negative SPP mutant inhibits the intramembrane processing and DRM localization of the HCV core protein.
SPP belongs to the family of aspartic proteases, which share two Asp residues for the active sites of protease activity. Asp219 and Asp264 have been identified as active sites for the protease activity of SPP (48). Overexpression of the SPP mutant in which Asp219 was replaced with Ala (SPPD219A) resulted in a dominant-negative activity that prevented the intramembrane processing of the HCV core protein (30). To examine the relationship between intramembrane processing by SPP and the localization of the HCV core protein in the DRM fraction, a C-terminally HA-tagged wild-type (SPP-HA) or D219A mutant (SPPD219A-HA) SPP was coexpressed with FLAG-core-HA in 293T cells (Fig. 4A). Overexpression of SPP-HA showed no effect on the localization of the HCV core protein, and the processed HCV core protein was partially localized in the DRM fraction (Fig. 4B, left). In contrast, overexpression of SPPD219A-HA inhibited the processing of the HCV core protein by endogenous SPP, and the level of unprocessed core protein, which was detected in the detergent-soluble fraction but not in the DRM fraction, was increased, whereas part of the processed core protein was localized in the DRM fraction (Fig. 4B, right). These results suggest that SPP cleavage is a prerequisite for the localization of HCV core protein in the DRM fraction. We have previously shown that the HCV core protein is degraded through proteasome pathways (26, 39, 43). To rule out the possibility of proteasome-dependent degradation of the unprocessed HCV core protein in the DRM fraction, we examined the effect of the proteasome inhibitor MG132 on the localization of HCV core protein. The processed HCV core protein, but not the unprocessed core protein, was partially localized in the DRM fraction, irrespective of treatment with MG132 (Fig. 4C). These results indicate that the failure of the unprocessed HCV core protein to localize in the DRM fraction was not due to selective degradation of the unprocessed core protein by proteasomes.
FIG. 4.
The dominant-negative mutant of SPP prevents the cleavage of HCV core protein by SPP and its localization in the DRM. (A) Schematic representation of the processing of FLAG-core-HA by a wild-type SPP (SPP-HA) (top) and the dominant-negative effect of SPPD219A-HA (bottom). (B) FLAG-core-HA was coexpressed with SPP-HA or SPPD219A-HA in 293T cells, lysed in the presence or absence of detergent, and subjected to a flotation assay. (C) Effect of a proteasome inhibitor, MG132, on the DRM localization of the HCV core protein. Proteins in each fraction were concentrated with acetone and analyzed by immunoblotting using antibodies against the FLAG epitope tag, caveolin-1, calreticulin, and the HA epitope tag. The membrane (lanes 1 to 9 in the absence of Triton X-100), DRM (lanes 1 to 7 in the presence of Triton X-100), and detergent-soluble (lanes 8 to 11 in the presence of Triton X-100) fractions were identified based on the localization of the marker proteins.
An SPP inhibitor prevents the processing of HCV core protein and its localization in the DRM.
To further assess the role of intramembrane processing by SPP on the localization of HCV core protein in the DRM, we examined the effect of the SPP inhibitors (Z-LL)2 ketone and L685,458 on the processing of the HCV core protein. Although (Z-LL)2 ketone was insoluble at a concentration of 10 μM and was highly toxic to 293T, Huh7, and Huh7-derived cell lines (data not shown), L685,458 was capable of penetrating the plasma membrane (49) and showed no visible cytotoxicity to the cell lines examined. Treatment with L685,458 inhibited the cleavage of the HCV core protein by SPP in a dose-dependent manner (Fig. 5A). As determined by flotation analyses of 293T cells expressing HCV core protein, the processed core protein was no longer localized in the DRM fraction following treatment with 25 or 50 μM L685,458 (Fig. 5B). These results further support the notion that intramembrane processing by SPP is required for the localization of HCV core protein in the DRM.
FIG. 5.
Effect of an SPP inhibitor on the cleavage of HCV core protein by SPP. The HCV core protein was expressed in 293T cells, and L685,458 was added to the culture supernatant, at the indicated concentrations, at 5 h posttransfection. Cells harvested at 29 h posttransfection were lysed with 1% Triton X-100 and subjected to Western blotting (A) or a flotation assay (B). DMSO, dimethyl sulfoxide. In the flotation assay, proteins in each fraction were concentrated with acetone and analyzed by immunoblotting using antibodies against core protein, caveolin-1, and calreticulin. The membrane (left panels, lanes 1 to 9), DRM (right panels, lanes 1 to 7), and detergent-soluble (right panels, lanes 8 to 11) fractions were identified based on the localization of the marker proteins.
Processing of the HCV core protein by SPP participates in viral propagation.
To examine the effect of the processing of the HCV core protein by SPP on the propagation of strain JFH-1, Huh7OK1 cells persistently infected with the JFH-1 virus were treated with 25 μM L685,458, and the cells were examined for processing of the HCV core protein and replication of viral RNA. The processed core protein of strain JFH-1 was clearly detected in the DRM fraction in untreated control cells, whereas processing of the core protein was impaired by treatment with L685,458, corresponding to the decrease in the level of processed core protein in the DRM (Fig. 6A). In Huh7OK1 cells infected with strain JFH-1, intracellular viral RNA levels were reduced 30% by treatment with L685,458 at 2 days posttreatment but showed no reduction at 1 day (Fig. 6B, left), and viral RNA levels in the culture supernatant were reduced 60% to 70% by treatment with the compound at 1 and 2 days posttreatment (Fig. 6B, center). To exclude the possibility of deleterious effects of L685,458 on cellular proteins involved in viral replication, we determined the effect of L685,458 on viral RNA replication by using HCV subgenomic-replicon cells. The replication of the RNA lacking the region coding for structural proteins showed a slight enhancement rather than suppression at 1 and 2 days after treatment with L685,485 (Fig. 6B, right), suggesting that the SPP inhibitor treatment used in this study is not toxic to the cellular proteins involved in HCV RNA replication. The slight decrease in the level of intracellular HCV RNA in infected cells after treatment with L685,458 (Fig. 6B, left), but not in replicon cells, may be attributable to the ER stress induced by the accumulation of unprocessed core proteins in infected cells. Although no effect of the inhibitor treatment on the expression of the intracellular core was observed, the secretion of core protein was slightly reduced (Fig. 6C). Furthermore, the production of infectious viral particles in the culture supernatants was clearly impaired by treatment with the SPP inhibitor (Fig. 6D).
FIG. 6.
Effect of the processing of HCV core protein by SPP on the propagation of JFH-1 virus. (A) L685,458 was added, at a concentration of 25 μM, to the culture supernatant of Huh7OK1 cells persistently infected with HCV strain JFH-1. Cells harvested at 24 h after treatment were lysed with 1% Triton X-100 and subjected to a flotation assay. DRM (lanes 1 to 7) and detergent-soluble (lanes 8 to 11) fractions were identified based on the localization of the marker proteins (data not shown). Asterisks indicate processed core proteins. DMSO, dimethyl sulfoxide. (B to D) Cells persistently infected with HCV strain JFH-1 were harvested at 1 or 2 days after treatment with the inhibitor. The data shown in each panel are representative of three independent experiments. (B) Total RNA was prepared from the cells (left) and the culture supernatant (center). Levels of HCV viral RNA and GAPDH mRNA were determined by real-time quantitative PCR. Values for the levels of viral DNA were normalized to that for GAPDH mRNA as described in Materials and Methods. The subgenomic-replicon cell line 9-13 was treated with the inhibitor, and total RNA was prepared from the cells (right). The amount of RNA is represented as a percentage of the amount in the untreated sample at 24 h after treatment (taken as 100%). (C) The amounts of intracellular (left) and extracellular (right) core protein were quantified by a quantitative ELISA. (D) Virus production in the culture supernatants was determined by a focus-forming assay. FFU, focus-forming units. (E) Plasmids coding for the full-length of the wild-type (WT) JFH-1 virus or a mutant (VVL/3A, IF/AL, ASC/VLV, or GND) were transfected into Huh7.5.1 cells. (Left) The amounts of intracellular and extracellular core protein were quantified by a quantitative ELISA at 2, 4, 6, and 8 days posttransfection. (Right) Virus production in the culture supernatants of Huh7.5.1 cells at 8 days after transfection with each plasmid was determined by a focus-forming assay. The data in each panel are representative of three independent experiments. (F) Detection of HCV RNA replication by NS5A immunofluorescence. At 6 days after transfection, NS5A and nuclei were stained red and blue, respectively.
The amino acid residues Val140, Leu144, Ile176, Phe177, Ala180, Ser183, and Cys184 were conserved within the core proteins of the genotype 1b strain J1 and the genotype 2a strain JFH-1, while the hydrophobic amino acid residues Leu and Val were found at position 139 in the core proteins of strains J1 and JFH-1, respectively. In order to examine the role of SPP-mediated cleavage of the HCV core protein on the growth of HCV strain JFH-1, mutations of Val139, Val140, and Leu144 to Ala (JFH-1/VVL/3A), of Ile176 and Phe177 to Ala and Leu (JFH-1/IF/AL), or of Ala180, Ser183, and Cys184 to Val, Leu, and Val, respectively (JFH-1/ASC/VLV), in the core protein, or mutation of the Gly-Asp-Asp motif to Gly-Asn-Asp in NS5B (JFH-1/GND) as a negative control, were introduced into cDNAs encoding strain JFH-1. The plasmid carrying each cDNA under the control of the polI promoter (19) was transfected into Huh7.5.1 cells, and the propagation of the JFH-1 viruses was determined. The expression of the core protein both in the culture medium and in cells transfected with the wild-type strain JFH-1 was increased during incubation, whereas it was severely impaired in the culture medium and cells transfected with JFH-1/VVL/3A, JFH-1/IF/AL, or the replication-deficient mutant JFH-1/GND. In contrast to JFH-1/VVL/3A and JFH-1/IF/AL, JFH-1/ASC/VLV was still capable of producing the core protein at a lower level than the wild-type strain JFH-1 (Fig. 6E). Furthermore, production of infectious particles was completely abrogated in the culture supernatants of cells transfected with JFH-1/VVL/3A, JFH-1/IF/AL, or the replication-deficient mutant JFH-1/GND, whereas JFH-1/ASC/VLV was still capable of producing infectious particles at a lower level than the wild-type strain JFH-1 (Fig. 6E, right). Expression of NS5A proteins was detected by immunofluorescent analyses in cells transfected with wild-type JFH-1, JFH-1/VVL/3A, JFH-1/IF/AL, or JFH-1/ASC/VLV but not in those transfected with JFH-1/GND, suggesting that JFH-1/VVL/3A and JFH-1/IF/AL are capable of replicating in cells but incapable of generating infectious particles (Fig. 6F). The propagation of JFH-1/ASC/VLV, bearing mutations in Ala180, Ser183, and Cys184, residues that are suggested to be essential for the processing of the HCV core protein by SPP (10, 30), further supports our notion that mutation of these residues is unable to completely abrogate the intramembrane cleavage of the core protein (30). Collectively, these results suggest that the processing of the HCV core protein by SPP plays crucial roles in viral propagation.
DISCUSSION
A previous report has suggested that the amino acid residues Ala180, Ser183, and Cys184 in the signal sequence are essential for the intramembrane proteolysis by SPP of the core protein of the HCV genotype 1a strain Glasgow expressed in the BHK and Huh7 cell lines by using the Semliki Forest virus expression system (21). However, we have shown that Leu139, Val140, and Leu144 in the hydrophobic region and Ile176 and Phe177 in the region upstream of the cleavage site, but not Ala180, Ser183, or Cys184, are required for the ER retention and SPP cleavage of the core proteins derived from the genotype 1b strain J1 and the genotype 1a strain H77 expressed in 293T cells by transfection of expression plasmids (30). Subsequently, Hope et al. suggested that these discrepancies were attributable to differences in the SDS-PAGE systems used to separate the processed and unprocessed core proteins, not to any difference in the HCV strains or expression systems, indicating that the core protein cleaved by SPP could be separated by a Tris/Bicine-buffered system but not by a Tris/glycine system (10). In this study, we added an HA tag at the C terminus of each core protein in order to easily distinguish between the cleaved and uncleaved HCV core proteins, and we then examined the processing of the wild-type and mutant core proteins by SDS-PAGE using Tris/glycine buffer. The resistance of Core IF/AL to SPP cleavage was consistent with the finding that Ile176 and Phe177 are located just upstream of the SPP cleavage site identified in this study. In contrast, Core ASC/VLV was not detected by the anti-HA antibody, indicating that Ala180, Ser183, and Cys184 in the signal sequence of the HCV core protein are not required for processing by SPP. A similar result was also obtained by immunoblotting using a Tris/Bicine-buffered system (data not shown). Furthermore, treatment with the SPP inhibitor L685,458 suppressed the cleavage of the core protein and abrogated both the localization of the mature core protein in the DRM and the propagation of strain JFH-1, suggesting that the intramembrane cleavage of the HCV core protein by SPP plays crucial roles in the DRM localization of the HCV core protein and the propagation of HCV. To further confirm the biological significance of the cleavage of the HCV core protein with respect to infectivity, we generated mutant viruses carrying mutations identical to each mutation of core protein described above. A JFH-1 mutant virus carrying the same mutation as Core ASC/VLV, but not other mutants, was still sufficiently viable to propagate in Huh7.5.1 cells. These findings clearly indicate that mutation of Ala180, Ser183, and Cys184 to Val, Leu, and Val, respectively, in the signal sequence of the HCV core protein is not able to completely abrogate the cleavage of the core protein by SPP.
Interestingly, the Core ASC/VLV mutant exhibited an extra band that was identical in size to the band of the wild-type core protein, in addition to a slow-migrating band, on the SDS-PAGE gel at 48 h posttransfection (Fig. 1B). Vauloup-Fellous et al. also reported that the Core ASC/VLV mutant expressed by a recombinant Semliki Forest virus in mammalian cells or by a baculovirus in insect cells exhibited bands between the mature (21 kDa) and the immature (23 kDa) core protein (44). If Core ASC/VLV was cleaved at the same site as the wild-type core protein, the processed core protein should have the same molecular size as the processed wild-type core protein, because the mutations in Core ASC/VLV were introduced into the region downstream of the cleavage site. These results suggest that Core ASC/VLV is first processed downstream of the authentic SPP cleavage site and is then further processed at the residue close to Phe177. Presenilins, which are involved in the cleavage of amyloid β protein precursor (APP), belong to the same aspartic protease family as SPP, which contains two Asp residues in the enzymatic active site (48). SPP might be able to cleave a substrate at multiple sites, as observed in the processing of APP by presenilins (33, 37). The Core ASC/VLV mutant may exhibit a preference for cleaving at the site between Asp178 and Ala191 rather than at that between Phe177 and Leu178. However, we still do not know whether SPP can cleave multiple sites within the C-terminal transmembrane region of the wild-type HCV core protein, because our mass spectrometry data show that there was no peptide larger than m/z 1,918.0452, the size corresponding to the amino acid residues from position 160 to 177 (Fig. 2).
Although the wild-type HCV core protein is known to be partially localized in the DRM fraction (20), Core LVL/3A and Core IF/AL, which are resistant to cleavage by SPP, were detected in the detergent-soluble fraction. Furthermore, overexpression of a dominant-negative SPP mutant or treatment with an SPP inhibitor increased the amount of unprocessed core protein in the detergent-soluble fraction irrespective of the presence of the proteasome inhibitor. These results suggest that processing of the HCV core protein by SPP is a prerequisite for stable localization of the mature core protein in the DRM. Indeed, the biological significance of the DRM localization of the mature HCV core protein is still unclear. In addition, we still do not know how HCV core protein migrates into the DRM fraction, and we could not exclude the possibility of involvement of other cellular and viral proteins in the DRM localization of HCV core protein. The DRM fraction is suggested to consist of various membrane microdomains that include lipid rafts, which are enriched in cholesterol and sphingolipids. The immunofluorescent analyses by Matto et al. showed that the DRM fraction containing the HCV core protein in replicon cells harboring a full genomic HCV RNA was different from the classical lipid raft, as evidenced by the lack of colocalization of the typical lipid raft markers, including caveolin-1 and the cholera toxin B subunit (20). However, Aizaki et al. suggested that the HCV replication complex was localized in a lipid-raft-like DRM fraction that included sphingolipids (2). Previous studies have indicated that the HCV core protein is localized in lipid droplets (1, 10, 20, 21, 23) and that processing by SPP is essential for the localization of the HCV core protein in lipid droplets (21). Furthermore, it was shown that the HCV core protein of strain JFH-1 recruits the replication complex to the lipid-droplet-associated membranes, and HCV particles were detected in close proximity to the lipid droplets, suggesting that the lipid droplets and the lipid-droplet-associated membranes induced by the core protein participate in the assembly of HCV particles (23). In addition, lipid droplets including the core protein surrounded by nonstructural proteins were also detected in cells expressing the nonstructural proteins of strain JFH-1 (23). Based on these observations, it might be feasible to speculate that the HCV core protein is matured through processing by the SP and SPP and is then translocated to the DRM and to the lipid droplets for viral assembly. A recent report by Aizaki et al. shows that HCV particles are enriched with cholesterol and sphingolipids (3), suggesting that the DRM is involved in viral assembly. On the other hand, some fraction of the core protein has been shown to migrate into the nucleus, where it is degraded by nuclear proteasomes (26, 41).
An alanine-scanning mutagenesis study of the HCV core protein has suggested that numerous residues within the carboxy-terminal two-thirds of the core protein are dispensable for RNA replication but essential for efficient infectious-virus production and that alanine substitution of the residues between positions 137 and 144 or 177 and 180 abrogated the extracellular release and intracellular stability of the mutant core proteins of chimeric JFH-1 viruses (27). This is consistent with the severe impairment of virus production by the JFH-1/VVL/3A mutant, in which Val139, Val140, and Leu144 are all replaced with Ala, and by the JFH-1/IF/AL mutant, in which Ile176 and Phe177 are replaced with Ala and Leu, respectively, in spite of the substantial RNA replication in the cells (Fig. 6E and F). The impairment of viral assembly by the introduction of SPP-resistant mutations in the core protein and the reduction of viral production by treatment with an SPP inhibitor, without any effect on subgenomic-RNA replication, also support the notion that SPP-dependent cleavage of the HCV core protein is required for viral assembly rather than for viral replication. Furthermore, the lack of significant effects on viral production and on the stability of the core protein in cells infected with JFH-1 mutants in which residues from 181 to 190 were replaced with Ala (27) is also consistent with the incomplete inhibition of the replication of the JFH-1/ASC/VLV mutant, in which Ala180, Ser183, and Cys184 are replaced with Val, Leu, and Val, respectively.
Increases in the levels of saturated and monounsaturated fatty acids enhance HCV RNA replication, in contrast to its suppression by polyunsaturated fatty acids (13), suggesting that enzymes associated with lipid biosynthesis are also involved in HCV replication. SREBP-1c regulates the transcription of acetyl coenzyme A carboxylase, fatty acid synthase, and stearoyl coenzyme A desaturase, leading to the production of saturated and monounsaturated fatty acids and triglycerides (11). Expression of the HCV core protein induces the production of lipid droplets composed mainly of triglycerides (4). Our recent study suggests that SREBP-1c was upregulated in the livers of transgenic mice expressing the HCV core protein through the LXRα/RXRα-dependent pathway, which leads to the development of fatty liver (25). The upregulation of SREBP-1c in the transgenic mice was required for the expression of PA28γ, an HCV core-binding host protein involved in the activation of nuclear proteasome activity (26). The HCV core protein cleaved by SPP may play a role in the formation of lipid droplets associated with the core protein, leading to an enhancement of viral assembly.
In summary, we determined the C-terminal end of the mature HCV core protein expressed in human cells and demonstrated that SPP processing is essential for the DRM localization and stability of the mature core protein. Furthermore, both mutation in the core protein resistant to cleavage by SPP and treatment with an SPP inhibitor abrogated the propagation of strain JFH-1 in the permissive cell line. These results suggest that SPP is a promising target for the development of novel antiviral drugs for the treatment of chronic hepatitis C.
Acknowledgments
We thank H. Murase for secretarial work. We also thank R. Bartenschlager and T. Wakita for providing cell lines and plasmids.
This work was supported in part by grants-in-aid from the Ministry of Health, Labor, and Welfare; the Ministry of Education, Culture, Sports, Science, and Technology; the 21st Century Center of Excellence Program; and the Foundation for Biomedical Research and Innovation.
Footnotes
Published ahead of print on 18 June 2008.
REFERENCES
- 1.Ait-Goughoulte, M., C. Hourioux, R. Patient, S. Trassard, D. Brand, and P. Roingeard. 2006. Core protein cleavage by signal peptide peptidase is required for hepatitis C virus-like particle assembly. J. Gen. Virol. 87855-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aizaki, H., K. J. Lee, V. M. Sung, H. Ishiko, and M. M. Lai. 2004. Characterization of the hepatitis C virus RNA replication complex associated with lipid rafts. Virology 324450-461. [DOI] [PubMed] [Google Scholar]
- 3.Aizaki, H., K. Morikawa, M. Fukasawa, H. Hara, Y. Inoue, H. Tani, K. Saito, M. Nishijima, K. Hanada, Y. Matsuura, M. Lai, T. Miyamura, T. Wakita, and T. Suzuki. 2008. A critical role of virion-associated cholesterol and sphingolipid in hepatitis C virus infection. J. Virol. 825715-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barba, G., F. Harper, T. Harada, M. Kohara, S. Goulinet, Y. Matsuura, G. Eder, Z. Schaff, M. J. Chapman, T. Miyamura, and C. Brechot. 1997. Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proc. Natl. Acad. Sci. USA 941200-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanchard, E., S. Belouzard, L. Goueslain, T. Wakita, J. Dubuisson, C. Wychowski, and Y. Rouille. 2006. Hepatitis C virus entry depends on clathrin-mediated endocytosis. J. Virol. 806964-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Codran, A., C. Royer, D. Jaeck, M. Bastien-Valle, T. F. Baumert, M. P. Kieny, C. A. Pereira, and J. P. Martin. 2006. Entry of hepatitis C virus pseudotypes into primary human hepatocytes by clathrin-dependent endocytosis. J. Gen. Virol. 872583-2593. [DOI] [PubMed] [Google Scholar]
- 7.Dubuisson, J., S. Duvet, J. C. Meunier, A. Op De Beeck, R. Cacan, C. Wychowski, and L. Cocquerel. 2000. Glycosylation of the hepatitis C virus envelope protein E1 is dependent on the presence of a downstream sequence on the viral polyprotein. J. Biol. Chem. 27530605-30609. [DOI] [PubMed] [Google Scholar]
- 8.Gao, L., H. Aizaki, J. W. He, and M. M. Lai. 2004. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J. Virol. 783480-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gosert, R., D. Egger, V. Lohmann, R. Bartenschlager, H. E. Blum, K. Bienz, and D. Moradpour. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 775487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hope, R. G., M. J. McElwee, and J. McLauchlan. 2006. Efficient cleavage by signal peptide peptidase requires residues within the signal peptide between the core and E1 proteins of hepatitis C virus strain J1. J. Gen. Virol. 87623-627. [DOI] [PubMed] [Google Scholar]
- 11.Horton, J. D., J. L. Goldstein, and M. S. Brown. 2002. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Investig. 1091125-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hussy, P., H. Langen, J. Mous, and H. Jacobsen. 1996. Hepatitis C virus core protein: carboxy-terminal boundaries of two processed species suggest cleavage by a signal peptide peptidase. Virology 22493-104. [DOI] [PubMed] [Google Scholar]
- 13.Kapadia, S. B., and F. V. Chisari. 2005. Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids. Proc. Natl. Acad. Sci. USA 1022561-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lecat, S., P. Verkade, C. Thiele, K. Fiedler, K. Simons, and F. Lafont. 2000. Different properties of two isoforms of annexin XIII in MDCK cells. J. Cell Sci. 1132607-2618. [DOI] [PubMed] [Google Scholar]
- 15.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309623-626. [DOI] [PubMed] [Google Scholar]
- 16.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285110-113. [DOI] [PubMed] [Google Scholar]
- 17.Majeau, N., V. Gagne, M. Bolduc, and D. Leclerc. 2005. Signal peptide peptidase promotes the formation of hepatitis C virus non-enveloped particles and is captured on the viral membrane during assembly. J. Gen. Virol. 863055-3064. [DOI] [PubMed] [Google Scholar]
- 18.Marsh, M., and A. Helenius. 2006. Virus entry: open sesame. Cell 124729-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masaki, T., R. Suzuki, M. Matsuda, T. Miyamura, T. Wakita, and T. Suzuki. 2006. Production of infectious hepatitis C virus by using RNA polymerase I-mediated transcription, abstr. 209, p. 59. In Abstracts of the 13th International Meeting on Hepatitis C Virus and Related Viruses. Nola Miles-Clark, Cairns, Australia.
- 20.Matto, M., C. M. Rice, B. Aroeti, and J. S. Glenn. 2004. Hepatitis C virus core protein associates with detergent-resistant membranes distinct from classical plasma membrane rafts. J. Virol. 7812047-12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McLauchlan, J., M. K. Lemberg, G. Hope, and B. Martoglio. 2002. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 213980-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meertens, L., C. Bertaux, and T. Dragic. 2006. Hepatitis C virus entry requires a critical post-internalization step and delivery to early endosomes via clathrin coated vesicles. J. Virol. 8011571-11578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyanari, Y., K. Atsuzawa, N. Usuda, K. Watashi, T. Hishiki, M. Zayas, R. Bartenschlager, T. Wakita, M. Hijikata, and K. Shimotohno. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 91089-1097. [DOI] [PubMed] [Google Scholar]
- 24.Moriishi, K., and Y. Matsuura. 2003. Mechanisms of hepatitis C virus infection. Antivir. Chem. Chemother. 14285-297. [DOI] [PubMed] [Google Scholar]
- 25.Moriishi, K., R. Mochizuki, K. Moriya, H. Miyamoto, Y. Mori, T. Abe, S. Murata, K. Tanaka, T. Miyamura, T. Suzuki, K. Koike, and Y. Matsuura. 2007. Critical role of PA28γ in hepatitis C virus-associated steatogenesis and hepatocarcinogenesis. Proc. Natl. Acad. Sci. USA 1041661-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moriishi, K., T. Okabayashi, K. Nakai, K. Moriya, K. Koike, S. Murata, T. Chiba, K. Tanaka, R. Suzuki, T. Suzuki, T. Miyamura, and Y. Matsuura. 2003. Proteasome activator PA28γ-dependent nuclear retention and degradation of hepatitis C virus core protein. J. Virol. 7710237-10249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murray, C. L., C. T. Jones, J. Tassello, and C. M. Rice. 2007. Alanine scanning of the hepatitis C virus core protein reveals numerous residues essential for production of infectious virus. J. Virol. 8110220-10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108193-199. [DOI] [PubMed] [Google Scholar]
- 29.Ogino, T., H. Fukuda, S. Imajoh-Ohmi, M. Kohara, and A. Nomoto. 2004. Membrane binding properties and terminal residues of the mature hepatitis C virus capsid protein in insect cells. J. Virol. 7811766-11777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okamoto, K., K. Moriishi, T. Miyamura, and Y. Matsuura. 2004. Intramembrane proteolysis and endoplasmic reticulum retention of hepatitis C virus core protein. J. Virol. 786370-6380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okamoto, T., Y. Nishimura, T. Ichimura, K. Suzuki, T. Miyamura, T. Suzuki, K. Moriishi, and Y. Matsuura. 2006. Hepatitis C virus RNA replication is regulated by FKBP8 and Hsp90. EMBO J. 255015-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okamoto, T., H. Omori, Y. Kaname, T. Abe, Y. Nishimura, T. Suzuki, T. Miyamura, T. Yoshimori, K. Moriishi, and Y. Matsuura. 2008. A single amino acid mutation in hepatitis C virus NS5A disrupting FKBP8 interaction impairs viral replication. J. Virol. 823480-3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okochi, M., S. Eimer, A. Bottcher, R. Baumeister, H. Romig, J. Walter, A. Capell, H. Steiner, and C. Haass. 2000. A loss of function mutant of the presenilin homologue SEL-12 undergoes aberrant endoproteolysis in Caenorhabditis elegans and increases Aβ42 generation in human cells. J. Biol. Chem. 27540925-40932. [DOI] [PubMed] [Google Scholar]
- 34.Pelkmans, L. 2005. Secrets of caveolae- and lipid raft-mediated endocytosis revealed by mammalian viruses. Biochim. Biophys. Acta 1746295-304. [DOI] [PubMed] [Google Scholar]
- 35.Pelkmans, L., D. Puntener, and A. Helenius. 2002. Local actin polymerization and dynamin recruitment in SV40-induced internalization of caveolae. Science 296535-539. [DOI] [PubMed] [Google Scholar]
- 36.Randall, G., M. Panis, J. Cooper, T. Tellinghuisen, K. Sukhodolets, S. Pfeffer, M. Landthaler, P. Landgraf, S. Kan, B. Lindenbach, M. Chien, D. Weir, J. Russo, J. Ju, M. Brownstein, R. Sheridan, C. Sander, M. Zavolan, T. Tuschl, and C. Rice. 2007. Cellular cofactors affecting hepatitis C virus infection and replication. Proc. Natl. Acad. Sci. USA 10412884-12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sastre, M., H. Steiner, K. Fuchs, A. Capell, G. Multhaup, M. M. Condron, D. B. Teplow, and C. Haass. 2001. Presenilin-dependent γ-secretase processing of β-amyloid precursor protein at a site corresponding to the S3 cleavage of Notch. EMBO Rep. 2835-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi, S. T., K. J. Lee, H. Aizaki, S. B. Hwang, and M. M. Lai. 2003. Hepatitis C virus RNA replication occurs on a detergent-resistant membrane that cofractionates with caveolin-2. J. Virol. 774160-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shirakura, M., K. Murakami, T. Ichimura, R. Suzuki, T. Shimoji, K. Fukuda, K. Abe, S. Sato, M. Fukasawa, Y. Yamakawa, M. Nishijima, K. Moriishi, Y. Matsuura, T. Wakita, T. Suzuki, P. M. Howley, T. Miyamura, and I. Shoji. 2007. E6AP ubiquitin ligase mediates ubiquitylation and degradation of hepatitis C virus core protein. J. Virol. 811174-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simons, K., and E. Ikonen. 1997. Functional rafts in cell membranes. Nature 387569-572. [DOI] [PubMed] [Google Scholar]
- 41.Suzuki, R., Y. Matsuura, T. Suzuki, A. Ando, J. Chiba, S. Harada, I. Saito, and T. Miyamura. 1995. Nuclear localization of the truncated hepatitis C virus core protein with its hydrophobic C terminus deleted. J. Gen. Virol. 7653-61. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki, R., S. Sakamoto, T. Tsutsumi, A. Rikimaru, K. Tanaka, T. Shimoike, K. Moriishi, T. Iwasaki, K. Mizumoto, Y. Matsuura, T. Miyamura, and T. Suzuki. 2005. Molecular determinants for subcellular localization of hepatitis C virus core protein. J. Virol. 791271-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki, R., K. Tamura, J. Li, K. Ishii, Y. Matsuura, T. Miyamura, and T. Suzuki. 2001. Ubiquitin-mediated degradation of hepatitis C virus core protein is regulated by processing at its carboxyl terminus. Virology 280301-309. [DOI] [PubMed] [Google Scholar]
- 44.Vauloup-Fellous, C., V. Pene, J. Garaud-Aunis, F. Harper, S. Bardin, Y. Suire, E. Pichard, A. Schmitt, P. Sogni, G. Pierron, P. Briand, and A. R. Rosenberg. 2006. Signal peptide peptidase-catalyzed cleavage of hepatitis C virus core protein is dispensable for virus budding, but destabilizes the viral capsid. J. Biol. Chem. 28127679-27692. [DOI] [PubMed] [Google Scholar]
- 45.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, C., M. Gale, Jr., B. C. Keller, H. Huang, M. S. Brown, J. L. Goldstein, and J. Ye. 2005. Identification of FBL2 as a geranylgeranylated cellular protein required for hepatitis C virus RNA replication. Mol. Cell 18425-434. [DOI] [PubMed] [Google Scholar]
- 47.Wasley, A., and M. J. Alter. 2000. Epidemiology of hepatitis C: geographic differences and temporal trends. Semin. Liver Dis. 201-16. [DOI] [PubMed] [Google Scholar]
- 48.Weihofen, A., K. Binns, M. K. Lemberg, K. Ashman, and B. Martoglio. 2002. Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science 2962215-2218. [DOI] [PubMed] [Google Scholar]
- 49.Weihofen, A., M. K. Lemberg, E. Friedmann, H. Rueeger, A. Schmitz, P. Paganetti, G. Rovelli, and B. Martoglio. 2003. Targeting presenilin-type aspartic protease signal peptide peptidase with γ-secretase inhibitors. J. Biol. Chem. 27816528-16533. [DOI] [PubMed] [Google Scholar]
- 50.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 1029294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]