Abstract
The surface of the mature dengue virus (DENV) particle is covered with 180 envelope (E) proteins arranged as homodimers that lie relatively flat on the virion surface. Each monomer consists of three domains (ED1, ED2, and ED3), of which ED3 contains the critical neutralization determinant(s). In this study, a large panel of DENV-2 recombinant ED3 mutant proteins was used to physically and biologically map the epitopes of five DENV complex-specific monoclonal antibodies (MAbs). All five MAbs recognized a single antigenic site that includes residues K310, I312, P332, L389, and W391. The DENV complex antigenic site was located on an upper lateral surface of ED3 that was distinct but overlapped with a previously described DENV-2 type-specific antigenic site on ED3. The DENV complex-specific MAbs required significantly higher occupancy levels of available ED3 binding sites on the virion, compared to DENV-2 type-specific MAbs, in order to neutralize virus infectivity. Additionally, there was a great deal of variability in the neutralization efficacy of the DENV complex-specific MAbs with representative strains of the four DENVs. Overall, the differences in physical binding and potency of neutralization observed between DENV complex- and type-specific MAbs in this study demonstrate the critical role of the DENV type-specific antibodies in the neutralization of virus infectivity.
The dengue (DEN) viruses (DENVs) are members of the genus Flavivirus, family Flaviviridae, and cause DEN, a mosquito-borne disease found throughout the tropics. DENV infection can result in clinical manifestations ranging from an asymptomatic state to DEN fever and the more severe forms of disease, which are DEN hemorrhagic fever and DEN shock syndrome. DEN is the most significant arboviral disease affecting humans, with 50 to 100 million infections estimated to occur annually, including several hundred thousand cases of DEN hemorrhagic fever and DEN shock syndrome (7, 20). The DENVs consist of four antigenically related viruses (DENV-1, DENV-2, DENV-3, and DENV-4), and infection with one DENV provides life-long immunity to that particular DENV but only temporary cross-protective immunity against the other DENVs (25).
The DENVs have a positive-sense, single-stranded RNA genome of approximately 11 kb which is packaged in a relatively small enveloped virion that is 50 nm in diameter and consists of a capsid protein (C), a membrane protein (M), and the major envelope glycoprotein (E) (14). The surface of the mature virion is covered with 180 E proteins arranged as homodimers (14, 35). Each monomeric unit is approximately 500 amino acids in length, of which the N-terminal 400 amino acids form the ectodomain. The E protein ectodomain consists of three structural domains referred to as envelope protein domain I (ED1), ED2, and ED3, respectively (19). ED1 is the central domain within the folded monomer, while ED2 contains the fusion loop that is responsible for low-pH-triggered fusion with the endosomal membrane (reviewed in reference 29). ED3 is the putative receptor binding domain based on several factors: ED3 has an immunoglobulin-like fold characteristic of many cell receptors, ED3 has loops that project further from the virion surface than either ED1 and ED2, and various soluble forms of ED3 have been shown to block infection of cells by West Nile virus (WNV) and DENV (1, 2, 11).
Among the flaviviruses, ED3 contains the critical, virus-specific, neutralization site(s) (23). DENV neutralizing epitopes have been mapped to specific amino acids within ED3 by generating monoclonal antibody (MAb)-resistant viruses (10, 16, 18, 27). Yeast surface display and random mutagenesis of DENV-2 ED3 have been used to map the epitopes of a panel of DENV-2 type-specific and DENV subcomplex-specific MAbs (30). Site-directed mutagenesis of recombinant ED3s has been used to identify amino acid residues involved in antigenic site recognized by a panel of DENV-2 type-specific neutralizing MAbs (6), as well as an epitope recognized by a DENV complex-specific MAb (17).
Antibody-mediated neutralization of animal viruses, in general, is thought to occur by a “multihit” mechanism of neutralization (reviewed in reference 13), which means that multiple antibodies need to engage individual virus particles in order for neutralization to occur. Evidence for a multihit mechanism of neutralization has been demonstrated with ED3-specific MAbs against two different flaviviruses: WNV (22) and DENV-2 (6). Antibodies have the potential to directly neutralize flaviviruses at several different stages early in infection, including blocking attachment and/or receptor interactions, blocking endocytosis, or directly or indirectly blocking fusion with the endosomal membrane. Blocking of attachment has been shown for a number of DENV neutralizing antibodies (3, 8). DENV and WNV neutralizing antibodies have also been observed to potentially block postattachment steps of infection (5, 12, 21). In addition, for tick-borne encephalitis virus, the fusion-blocking activity of several neutralizing antibodies specific for all three domains of the E protein has been demonstrated (28).
In this study, we expanded on work by others who have identified one DENV ED3 complex-specific neutralizing epitope (17) and two DENV ED3 subcomplex-specific neutralizing epitopes (30) by mapping, in detail, the epitopes of five unique DENV ED3 complex-specific MAbs by using a large panel of DENV-2 rED3 mutant proteins. The antigenic site recognized by these MAbs is in a location similar to that of previously mapped epitopes but, significantly, differs in the critical residues required for binding. Furthermore, the antigenic site is distinct but overlaps with a DENV-2 type-specific antigenic site previously mapped with a similar panel of rED3 mutant proteins (6). Interestingly, this study shows that the DENV complex-specific MAbs bound rED3 of DENV-2 with greater affinity than the type-specific MAbs but neutralized the virus considerably less efficiently, requiring much higher occupancy levels in order to neutralize the same level of virus infectivity. These data further our understanding of the physical and biological differences between DENV-2 ED3 complex- and type-specific neutralizing epitopes.
MATERIALS AND METHODS
Cells and viruses.
Monkey kidney Vero cells were maintained at 37°C in a 5% CO2 incubator in Dulbecco's modified essential medium containing 8% fetal bovine serum (FBS). Mosquito C6/36 cells were maintained at 28°C in Dulbecco's modified essential medium containing 10% FBS and supplemented with tryptose phosphate buffer. The DENV strains used in this study were DENV-1 OBS7690, DENV-2 New Guinea C (NGC), DENV-3 H87, and DENV-4 703-4. Primary virus stocks were passaged twice in C6/36 mosquito cells to obtain sufficient titers.
Purification of virus.
C6/36 mosquito cells were grown in an 850-cm2 roller bottle and infected with DENV-2 NGC at a multiplicity of infection of approximately 0.1. The infected-cell supernatant was harvested on day 7 postinfection. Cell debris was removed by centrifugation at 5,000 rpm for 30 min at 4°C. The supernatant was layered on top of a 30% sucrose solution containing 10 mM Tris, 100 mM NaCl, and 1 mM EDTA. The virus was pelleted by ultracentrifugation in a swinging-bucket rotor at 26,000 rpm for 4 h at 4°C to remove low-molecular-weight contaminants such as soluble proteins. The supernatant was poured off, and the tubes were briefly left upside down on chromatography paper in order to remove excess liquid from the side of the tubes. The virus pellet was resuspended in phosphate-buffered saline (PBS). The infectious titer was determined by a standard plaque assay on Vero cells. The purity of the virus preparations was verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown).
MAbs.
Five commercially available MAbs were used in this study: GTX29202 (GeneTex), GTX77557 (GeneTex), MDVP-55A (Immunology Consultants Laboratory, Inc.), MA1-27093 (Affinity BioReagents), and 20-783-74014 (GenWay). These five MAbs were affinity-purified immunoglobulin G2A (IgG2A) mouse MAbs and were all reported to be DENV complex specific in the product literature. MAb 3H5 (Chemicon) (4) is an affinity-purified, DENV-2 type-specific immunoglobulin G1 MAb that was used as a control for the 50% plaque reduction neutralization (PRNT50) test and enzyme-linked immunosorbent assay (ELISA) experiments to ensure that the results obtained were consistent with previously collected data for the DENV-2 type-specific MAbs (6).
Expression and purification of recombinant DENV-2 ED3.
The region corresponding to ED3 of DENV-2 strain NGC was reverse transcription-PCR amplified for cloning and expression as maltose binding protein (MBP) fusions with the pMal-c2x system (New England BioLabs [NEB], Beverly, MA). RNA extraction of virus-infected cell culture supernatant was done with a Qiagen viral RNA extraction kit (Qiagen). Reverse transcription-PCR was done with the Titan kit (Roche). The primers used (forward, 5′-CGAGGGAAGGATTTCAAAAGGAATGTCATACTCTATG-3′; reverse, 5′-GCCAAGCTTTCATCCTTTCTTAAACCAGTTGAGC-3′) were designed for cloning into the pMal-c2x vector and contained XmnI and HindIII restriction sites, respectively. Appropriate primers were also designed for constructing rED3s for DENV-1 strain OBS7690, DENV-3 strain H87, and DENV-4 strain 703-4, and these gene fragments were also cloned into pMal-c2x. The rED3 protein, comprising amino acids 295 to 395 of the E protein tagged to MBP, was expressed in Escherichia coli DH5α by a method similar to that described previously (6, 15). Briefly, 20-ml cultures of bacteria were grown in LB medium containing 50 μg/ml ampicillin to an optical density at 600 nm of approximately 0.6 and induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 37°C for 3 h. Bacterial cells were pelleted and stored at −20°C overnight. The following day, cells were lysed in 1 ml of MBP column buffer (20 mM Tris-HCl, 200 mM NaCl, 1 mM EDTA) by freezing in liquid nitrogen and thawing in a 37°C water bath. Lysates were centrifuged at 12,000 rpm at 4°C for 30 min, and the supernatant was mixed with 500 μl amylose resin (NEB) in a 1.5-ml Eppendorf tube and incubated at 4°C on a rocker for 1 h. Tubes were centrifuged at 3,000 rpm for 1 min, and the supernatant was removed. The resin was washed three times with 1 ml MBP column buffer, and bound protein was eluted twice with 500 μl of MBP column buffer containing 10 mM maltose. Concentrations of proteins were determined by spectrophotometric analysis.
Mutagenesis of recombinant DENV-2 ED3.
Site-directed mutagenesis of the DENV-2 NGC ED3 gene fragment in the pMal-c2x vector was done with the QuikChange kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions.
PRNT50.
MAbs were diluted to 320 nM for DENV-1 OBS7690 and DENV-2 NGC or to 2.0 μM for DENV-3 H87 and DENV-4 and then serially diluted twofold in minimal essential medium (MEM) containing 2% FBS (160 nM and 1.0 μM, respectively, once diluted with virus). Virus was diluted to approximately 1 PFU/μl in MEM containing 2% FBS. A 400-μl volume of virus (∼ 400 PFU) was mixed with an equal volume of MAb dilution or 400 μl of MEM containing 2% FBS (control) and then incubated at room temperature (25°C) for 1 h. Following this incubation, 200 μl of each virus-MAb mixture, or controls, was added in triplicate to wells of a six-well plate containing ∼80% confluent monkey kidney Vero cells. Infection was allowed to take place for 1 h at room temperature, at which point the cells were washed twice with PBS, overlaid with MEM containing 2% FBS and 1% agar, and incubated at 37°C. Plaques were visualized on days 6 to 8 by staining with neutral red. PRNT50 data were converted to percent neutralization relative to controls in the absence of MAb, and PRNT50 concentrations were calculated by doing a nonlinear regression analysis with Sigmaplot (version 9.01; Systat Software, Inc., CA). The data are fitted by a standard four-parameter logistic curve (i.e., dose-response curve) by the equation y = minimum + (maximum − minimum)/[1 + (x/50% effective concentration)slope factor]. The results are an average of three experiments.
Affinity measurements by indirect ELISA with rED3.
The wells of 96-well microtiter plates (Corning Inc., Corning, NY) were coated with 200 ng of rED3 diluted in borate saline (pH 9.0) at 37°C for 2 h. This amount of rED3 was determined to saturate the wells by undertaking a checkerboard serial dilution of anti-MBP antisera (NEB) and rED3, which yielded peak absorbance values that were below the maximum range of the plate reader (data not shown). Wells were washed twice with distilled and deionized water (ddH2O), blocked for 30 min at room temperature (25°C) with blocking buffer (PBS, 0.1% [vol/vol] Tween 20, 0.25% [wt/vol] bovine serum albumin [BSA]), and subsequently washed twice with ddH2O. The DENV complex-specific MAbs were diluted to 10 nM for the DENV-1 and -2 rED3 proteins, 200 nM for the DENV-3 rED3 protein, and 3 μM for the DENV-4 rED3 protein and added to duplicate wells, followed by 11 twofold serial dilutions in blocking buffer. This range of concentrations was determined to cover the full range of binding to the respective rED3 proteins, from undetectable binding to saturation. Antibodies were allowed to bind overnight at room temperature (25°C) for 15 to 18 h. Subsequently, the microtiter plates were washed twice with ddH2O and twice with PBS-T (PBS containing 0.1% Tween 20), followed by the addition of a 1/1,000 dilution of horseradish peroxidase-conjugated goat anti-mouse immunoglobulins (Sigma) and incubated for 1 h at room temperature. Following this incubation, microtiter plates were washed twice with ddH2O and twice with PBS-T. Antibody binding was visualized by addition of 3,3′,5,5′-tetramethylbenzidene substrate (Sigma). After 30 min of incubation at room temperature, the absorbance was read at 655 nm on a model 3550 UV plate reader (Bio-Rad, Hercules, CA). Binding curves and KD values were determined by doing a nonlinear regression analysis with Sigmaplot (version 9.01; Systat Software, Inc., CA). The results are an average of two experiments.
Affinity measurements and estimates of occupancy by antibody sandwich ELISA with purified virus.
The wells of a 96-well microtiter plate (Corning Inc., Corning, NY) were coated with 50 μl of a 1/5 dilution of rabbit anti-DENV ED3 polyclonal sera (capture antibody) for 2 h at 37°C. The plates were washed twice with PBS-T and twice with ddH2O. Purified DENV-2 NGC (∼5 × 107 PFU/ml) was diluted 1/50 in blocking buffer, and 50 μl was added to each well and incubated at 37°C for 2 h. The remainder of the assay was undertaken as described above with the rED3 protein, except that the primary MAbs were only incubated for 1 h at room temperature, in accordance with the length of incubation used for the virus neutralization assay. The entire experiment was carried out in a biological safety cabinet, as the virus was presumed to be infectious throughout the experiment. Binding curves and KD values were determined by doing a nonlinear regression analysis with Sigmaplot (version 9.01; Systat Software, Inc., CA). The results are an average of two experiments. The generation of a colored substrate, and thus the absorbance reading in the indirect ELISA, is proportional to the amount of antibody bound to antigen in the well. At saturation (i.e., maximum absorbance), it is predicted that all possible ED3 binding sites on the virion are occupied by MAb. Therefore, absorbance readings below saturation can be calculated as fractional occupancies of the total available ED3 proteins by MAb.
Hemagglutination (HA) inhibition (HAI) assay.
The DENV complex- and type-specific MAbs were diluted to 40 and 320 nM concentrations (four times that used for the affinity measurements with rED3/virus), respectively, in borate saline (pH 9.0) containing 0.4% (wt/vol) bovine serum albumin, and 11 twofold serial dilutions were performed across the plate in 50 μl borate saline (pH 9.0) containing 0.4% (wt/vol) bovine serum albumin. Control wells contained no MAb. DENV-2 NGC virus-infected C6/36 cell culture supernatant was diluted to 8 HA units/50 μl (i.e., if the HA titer was 1/128, then a 1/16 dilution of virus was used), and 50 μl was added to each of the MAb wells and no-MAb control wells and incubated for 1 h at room temperature (25°C). Following the incubation, 100 μl of a 0.125% suspension of goose red blood cells (RBC) diluted in HA buffer (150 mM NaCl, 31 mM Na2HPO4, 169 mM NaH2PO4), pH 6.2, was added to each well. The plate was covered and incubated at 37°C for 1 h. The MAb concentration of the last well having a button (i.e., complete HAI) was recorded as the HAI titer.
Statistical analysis.
The KD values and PRNT50 concentrations are presented as the mean ± the standard error of the mean (SEM) and were analyzed with SigmaStat (version 3.1; Systat Software, Inc., CA). Statistical analyses were done by one-way analysis of variance, followed by a Bonferroni post hoc test with P < 0.05 required for statistical significance.
RESULTS
Characterization of MAbs.
Five DENV complex-specific MAbs (i.e., GTX29202, GTX77557, MDVP-55A, MA1-27093, and 20-783-74014, recognizing DENV-1, DENV-2, DENV-3, and DENV-4 only) were tested for their affinity for the DENV-2 NGC rED3 protein. The KD values obtained with the DENV-2 rED3 protein ranged from 0.13 ± 0.02 to 0.43 ± 0.09 nM (Table 1). The DENV complex-specific MAbs bound to the DENV-2 rED3 protein approximately 2- to 33-fold more tightly than DENV-2 type-specific MAbs characterized in a previous study (6) (Table 1). The five DENV complex-specific MAbs were also tested for their affinity for virion-associated ED3 as measured by an antibody sandwich ELISA with purified virus. The KDs ranged from 0.30 ± 0.02 to 0.67 ± 0.09 nM (Table 1). There was no statistically significant difference between the KDs obtained for virion-associated ED3 compared with the rED3 protein. Therefore, presentation of ED3 epitopes is similar for both the rED3 protein and virion-associated ED3 for this particular set of MAbs. The same result was seen with a panel of DENV-2 type-specific MAbs in a previous study (6) (Table 1).
TABLE 1.
Properties of DENV ED3-specific MAbs
| MAb clone | Isotype | DENV rED3 specificity | Mean KD with DENV-2 rED3 (nM) | Mean KD with DENV-2 NGC (nM) | Mean [PRNT50] concn with DENV-2 NGC (nM) | HAI titer (nM) |
|---|---|---|---|---|---|---|
| GTX29202 | IgG2A | 1, 2, 3 | 0.20 ± 0.02 | 0.39 ± 0.02 | 5.9 ± 0.3 | 0.94 |
| GTX77557 | IgG2A | 1, 2, 3 | 0.19 ± 0.05 | 0.30 ± 0.02 | 4.0 ± 0.4 | 0.63 |
| MDVP-55A | IgG2A | 1, 2, 3 | 0.13 ± 0.02 | 0.30 ± 0.03 | 3.0 ± 0.2 | 0.63 |
| MA1-27093 | IgG2A | 1, 2, 3 | 0.22 ± 0.05 | 0.45 ± 0.04 | 7.5 ± 0.5 | 1.25 |
| 20-783-74014 | IgG2A | 1, 2, 3 | 0.43 ± 0.09 | 0.67 ± 0.09 | 11.8 ± 1.3 | 1.25 |
| 3H5a | IgG1 | 2 | 4.3 ± 0.7 | 2.8 ± 0.1 | 4.6 ± 0.8 | 1.3 |
| M8051122a | IgG1 | 2 | 2.5 ± 0.3 | 4.1 ± 1.1 | 3.6 ± 0.4 | 5.0 |
| 9F16a | IgG1 | 2 | 3.2 ± 0.3 | 4.1 ± 0.7 | 5.2 ± 0.8 | 5.0 |
| 2Q1899a | IgG1 | 2 | 4.0 ± 0.5 | 7.2 ± 2.1 | 5.4 ± 0.5 | 10.0 |
| IP-05-143a | IgG1 | 2 | 1.8 ± 0.2 | 3.0 ± 0.5 | 5.0 ± 0.6 | 2.5 |
| GTX77558a | IgG1 | 2 | 0.9 ± 0.1 | 0.9 ± 0.4 | 1.5 ± 0.2 | 1.3 |
| 5C36a | IgG1 | 2 | 2.2 ± 0.2 | 3.9 ± 0.3 | 11.6 ± 1.2 | 5.0 |
The KD values with DENV-2 rED3 and NGC and the PRNT50 concentrations were previously reported (6).
The ability of each MAb to neutralize DENV infectivity was quantified with a PRNT50 test. The PRNT50 concentration was determined for each of the five DENV complex-specific MAbs with DENV-2 strain NGC and found to range from 3.0 ± 0.2 to 11.8 ± 1.3 nM (Table 1). These PRNT50 concentrations were comparable to the PRNT50 concentrations previously determined for a panel of DENV-2 type-specific MAbs (6) (Table 1). However, these values were approximately 10- to 18-fold higher than the KD for virion-associated ED3, with statistically significant differences between the PRNT50 concentration and the KD for virion-associated ED3 for all five DENV complex-specific MAbs. This differs from the results obtained with DENV-2 type-specific MAbs (6) (Table 1).
The HAI titers were determined for the five DENV complex-specific and seven DENV-2 type-specific MAbs with DENV-2 strain NGC (Table 1). There was no statistically significant difference between the HAI titer and the KD for virion-associated ED3 for all 12 MAbs. Thus, 50% relative occupancy of ED3 on the virus particle by a MAb seems to be the minimum requirement for HAI for this panel of DENV ED3-specific MAbs. Interestingly, the HAI titer was only predictive of the neutralization efficiency of 5 of the 12 MAbs (3H5, M8051122, 9F16, 2Q1899, and GTX77558), all of which were DENV-2 type-specific MAbs.
Binding affinity of MAbs with rED3 mutant proteins.
A total of 52 single amino acid substitutions were introduced into the rED3 protein at 36 different surface-accessible residues. This includes 41 previously described substitutions (6), as well as 11 additional single amino acid substitutions (Table 2). This makes a total of 18 naturally occurring amino acid substitutions and 34 amino acid substitutions of conserved residues (Fig. 1 and Table 2). Conserved residues were mutated to glycine and/or alanine to essentially remove the amino acid side chain as described previously (6).
TABLE 2.
Relative KDs of mutant rED3s compared to the wild-type NGC rED3 with all MAb-rED3 mutant combinations
| rED3 |
KD for DENV complex-specific MAb:
|
||||
|---|---|---|---|---|---|
| GTX29202 | GTX77557 | MDVP-55A | MA1-27093 | 20-783-74014 | |
| Wild type | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| K295G | 2.5 | 1.5 | 2.3 | 0.7 | 1.0 |
| S298G | 1.8 | 1.1 | 1.4 | 1.0 | 1.3 |
| Y299I | 1.6 | 0.9 | 1.4 | 1.0 | 1.1 |
| S300L | 2.0 | 1.7 | 2.0 | 1.0 | 0.7 |
| M301G | ≥50.0 | ≥53.0 | ≥79.0 | ≥46.0 | ≥23.0 |
| M301A | 1.3 | 1.1 | 1.1 | 1.2 | 1.4 |
| T303G | 1.8 | 1.3 | 1.4 | 1.3 | 1.6 |
| K305G | 3.9 | 2.4 | 3.3 | 0.9 | 1.3 |
| K305A | 1.2 | 0.8 | 1.0 | 1.1 | 2.0 |
| K307G | 5.1 | 7.0 | 14.0 | 3.0 | 5.4 |
| K307A | 3.5 | 3.7 | 4.4 | 2.8 | 2.8 |
| V309G | 2.8 | 2.1 | 2.0 | 1.8 | 2.4 |
| K310E | ≥50.0 | ≥53.0 | ≥79.0 | ≥46.0 | ≥23.0 |
| K310G | ≥50.0 | ≥53.0 | ≥79.0 | ≥46.0 | ≥23.0 |
| K310A | ≥50.0 | ≥53.0 | ≥79.0 | ≥46.0 | ≥23.0 |
| E311G | 2.2 | 1.1 | 2.2 | 1.6 | 2.1 |
| E311A | 3.1 | 2.0 | 2.0 | 1.7 | 2.1 |
| I312F | 15.0 | 8.3 | 5.9 | 5.7 | ≥23.0 |
| Q325A | 1.4 | 1.0 | 1.0 | 1.1 | 1.4 |
| E327G | 2.1 | 0.5 | 3.3 | 1.4 | 1.9 |
| D329E | 1.5 | 1.4 | 1.5 | 1.2 | 2.1 |
| D329G | 1.2 | 1.9 | 1.5 | 0.8 | 1.1 |
| G330D | 1.3 | 0.9 | 1.0 | 1.0 | 2.3 |
| S331A | 2.4 | 1.2 | 2.1 | 1.2 | 0.8 |
| S331P | 1.5 | 1.5 | 1.5 | 1.5 | 1.3 |
| P332G | 11.0 | 13.0 | 13.0 | 7.8 | 16.0 |
| P332A | 18.0 | 10.0 | 15.0 | 12.0 | 0.9 |
| K334Q | 2.3 | 1.4 | 2.2 | 1.0 | 1.2 |
| E338G | 1.8 | 1.2 | 1.5 | 1.3 | 2.1 |
| K344N | 1.9 | 1.0 | 1.4 | 1.3 | 2.3 |
| R345K | 1.6 | 1.3 | 1.5 | 0.9 | 0.7 |
| R345G | 1.8 | 1.1 | 1.4 | 1.6 | 2.8 |
| T359I | 1.3 | 0.8 | 1.2 | 0.9 | 0.8 |
| T359G | 2.0 | 1.1 | 1.6 | 1.5 | 2.7 |
| E360G | 1.0 | 0.7 | 1.2 | 0.5 | 0.6 |
| K361G | 1.7 | 2.3 | 2.1 | 1.3 | 1.4 |
| D362G | 0.9 | 1.4 | 1.4 | 0.7 | 1.4 |
| S363R | 1.6 | 0.9 | 2.0 | 0.8 | 0.9 |
| P364A | 1.8 | 1.4 | 1.3 | 1.6 | 1.9 |
| E383G | ≥50.0 | ≥53.0 | ≥79.0 | ≥46.0 | ≥23.0 |
| E383A | 1.7 | 0.8 | 1.2 | 1.1 | 1.9 |
| P384G | 3.9 | 4.4 | 6.6 | 3.5 | 6.9 |
| P384A | 5.2 | 3.7 | 4.8 | 2.4 | 3.9 |
| G385E | 0.8 | 0.4 | 2.2 | 0.6 | 0.5 |
| L387A | 7.2 | 3.0 | 3.1 | 2.8 | 5.7 |
| K388G | 0.7 | 2.7 | 1.6 | 0.5 | 0.6 |
| K388A | 1.6 | 1.2 | 1.3 | 1.3 | 1.5 |
| L389A | 11.0 | 6.6 | 6.3 | 5.0 | 3.9 |
| N390D | 3.0 | 0.5 | 3.6 | 1.1 | 1.1 |
| N390H | 0.5 | 0.3 | 0.7 | 0.5 | 0.4 |
| N390S | 1.8 | 1.0 | 1.1 | 1.3 | 1.4 |
| W391A | 23.0 | 25.0 | 21.0 | 1.6 | 0.8 |
FIG. 1.
ED3 alignment of 11 strains of DENV-2, representing the majority of surface-accessible amino acid sequence diversity, compared to DENV-2 strain NGC. Naturally occurring amino acid substitutions and conserved residues that were mutated to glycine or alanine are in bold and underlined. All of the mutated residues are surface accessible on the DENV-2 virion. The DENV-2 NGC and DAKHD10674 sequences were reported previously (6). The rest of the sequences were acquired from GenBank, i.e., 16681 (U87411), Malaysia86 (X15214), Malaysia87a (X15433), Malaysia87b (X15434), PR159 (L10046), P81407 (AF231717), SriLanka85b (L10040), Taiwan87 (L10052), Thailand58 (D10514), and Trinidad53 (L10053).
The KD for each of the five DENV complex-specific MAbs was determined by titration in an indirect ELISA with all 52 rED3 mutant proteins. The majority of the mutations tested resulted in no change or only a small change (less than fourfold) in the KD compared to the KD for the wild-type rED3 protein (Table 2). Mutant proteins resulting in changes in the KD of between 4- and 10-fold were considered “weak” changes in affinity. These mutations were K307G for MAbs GTX29202, GTX77557, and 20-783-74014; K307A for MAb MDVP-55A; I312F for MAbs GTX77557, MDVP-55A, and MA1-27093; P332G for MAb MA1-27093; P384G for MAbs GTX77557, MDVP-55A, and 20-783-74014; P384A for MAbs GTX29202 and MDVP-55A; L387A for MAbs GTX29202 and 20-783-74014; and L389A for MAbs GTX77557, MDVP-55A, and MA1-27093. Mutations that resulted in a greater-than-10-fold change in the KD were considered “strong” changes in binding affinity. These mutations were M301G for all five MAbs; K307G for MAb MDVP-55A; K310E/G/A for all five MAbs; I312F for MAbs GTX29202 and 20-783-74014; P332G for MAbs GTX29202, GTX77557, MDVP-55A, and 20-783-74014; P332A for MAbs GTX29202, GTX77557, MDVP-55A, and MA1-27093; E383G for all five MAbs; L389A for MAb GTX29202; and W391A for MAbs GTX29202, GTX77557, and MDVP-55A.
Epitope analysis.
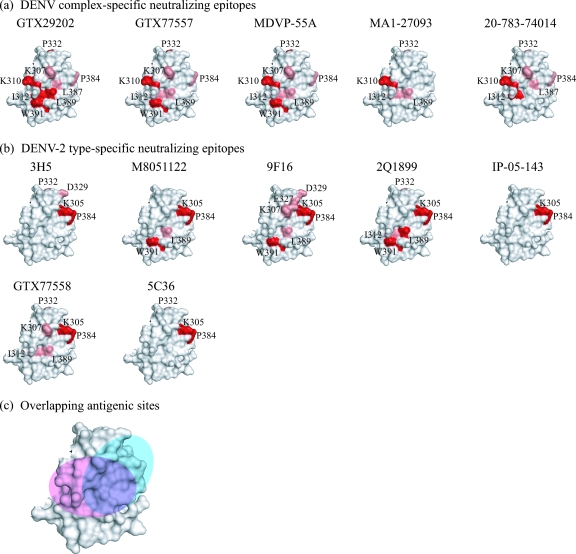
Those mutations that had a strong effect (i.e., a greater-than-10-fold change in the KD) on MAb binding affinity were considered to be critical epitope residues. The mutations listed above with discrepancies between strong and weak effects, depending on the substitution, were considered to be important but not necessarily critical. The critical epitope residues identified for each MAb (Fig. 2a, in red) are as follows: K310 for all five MAbs; I312 for MAbs GTX29202 and 20-783-74014; P332 for MAbs GTX29202, GTX77557, and MDVP-55A; L389 for MAb GTX29202; and W391 for MAbs GTX29202, GTX77557, and MDVP-55A.
FIG. 2.
Predicted epitopes for five DENV complex-specific and seven DENV-2 type-specific MAbs. For panels a and b, the critical epitope residues are in red and residues predicted to be located on the periphery of the epitope are in pink. The DENV complex-specific epitopes, shown in panel a, overlap with one another and center on residue K310. In contrast, the DENV-2 type-specific epitopes, shown in panel b, overlap with one another and center on residues K305 and P384. The approximate location of the DENV complex-specific and DENV-2 type-specific antigenic sites (pink and cyan, respectively) are shown in panel c. The estimated amount of overlap between these antigenic sites is shown in purple. The diagrams are based on the crystal structure of the DENV-2 E protein (PDB-ID, 10KE). The epitopes for the DENV-2 type-specific MAbs have been reported previously (6). However, residues M301 and E383 were omitted as peripheral epitope residues (see Discussion). Additionally, residues I312, L389, and W391 were identified as epitope residues for subsets of the DENV-2 type-specific MAbs in this study (data not shown).
The mutations that had weak effects on MAb binding affinity, as well as those having minor discrepancies between strong and weak effects on affinity, depending on the substitution (listed above), were considered to be located on the periphery of the epitope (Fig. 2a, in pink). These residues include K307 for MAbs GTX29202, GTX77557, MDVP-55A, and 20-783-74014; I312 for MAbs GTX77557, MDVP-55A, and MA1-27093; P332 for MAbs MA1-27093 and 20-783-74014; P384 for MAbs GTX29202, GTX77557, MDVP-55A, and 20-783-74014; L387 for MAbs GTX29202 and 20-783-74014; and L389 for MAbs GTX77557, MDVP-55A, and MA1-27093. Residues M301 and E383 were not considered epitope residues due to the difficulty in interpreting the results of the glycine substitution at each position (see Discussion).
Conservation of DENV complex-specific epitopes in ED3.
The critical DENV complex-specific epitope residues identified are not completely conserved among representative strains of the four DENVs in the linear sequence of ED3 (Fig. 3). Residue K310 was critical for all five MAbs and is conserved among representative strains of all four DENVs. Residue I312 was critical for MAbs GTX29202 and 20-783-74014 but is not conserved among any of the DENVs. Residue P332 is conserved among the four DENVs and was critical for MAbs GTX29202, GTX77557, and MDVP-55A. Residue L389 was critical for MAb GTX29202 and is conserved in DENV-1, -2, and -4. Residue W391 is conserved among the four DENVs and was critical for MAbs GTX29202, GTX77557, and MDVP-55A. The conservation of critical epitope residues among representative DENVs based on the DENV-2 epitope mapping in this study is summarized in Table 3.
FIG. 3.
ED3 alignment of representative strains of each of the four DENVs. The locations of critical DENV complex-specific residues K310, I312, P332, L389, and W391 are in bold and underlined. Residue K310 was critical for all of the DENV complex-specific MAbs and is conserved in the linear sequence of ED3 among the four DENVs. Residues P332 and W391 were critical for subsets of MAbs and are also conserved. However, residues I312 and L389 are not conserved among the four DENVs but were critical for subsets of the DENV complex-specific MAbs. Numbering is based on the DENV-2 ED3 sequence. All four sequences were done by us.
TABLE 3.
Conservation of critical epitope residues in the linear sequence of ED3 among the four DENVs
| Critical residue based on DENV-2 epitope mapping | Epitope use by MAb:
|
DENV conservation | ||||
|---|---|---|---|---|---|---|
| GTX 29202 | GTX 77557 | MDVP- 55A | MA1- 27093 | 20-783- 74014 | ||
| K310 | X | X | X | X | X | 1, 3, 4 |
| I312 | X | X | None | |||
| P332 | X | X | X | 1, 3, 4 | ||
| L389 | X | 1, 4 | ||||
| W391 | X | X | X | 1, 3, 4 | ||
MAb binding affinity for rED3 from four representative DENV strains.
The five DENV complex-specific MAbs were tested by titration in an indirect ELISA for their affinity to rED3 proteins from representative strains of each of the four DENVs (DENV-1 OBS7690, DENV-2 NGC, DENV-3 H87, and DENV-4 703-4). The MAbs bound with similar affinities to the DENV-1 rED3 protein, with KDs ranging from 0.06 ± 0.00 to 0.21 ± 0.05 nM (Table 4). There was no statistically significant difference between the KDs of any of these MAbs for the DENV-1 rED3 protein and the DENV-2 rED3 protein. All five MAbs bound with statistically significantly weaker affinities to the DENV-3 rED3 protein (4- to 100-fold weaker binding) compared to the DENV-1 and -2 rED3 proteins, with KDs ranging from 0.94 ± 0.1 to 8.0 ± 1.1 nM (Table 4). For all five MAbs, there was no detectable binding to the DENV-4 rED3 protein in the range of MAb concentrations tested, suggesting that the binding of these MAbs to DENV-4 is very weak.
TABLE 4.
Dissociation constants (KD) with representative DENV-1, -2, -3, and -4 rED3s
| MAb | Mean KD (nM) ± SEMb
|
|||
|---|---|---|---|---|
| DENV-1 | DENV-2 | DENV-3 | DENV-4 | |
| GTX29202a | 0.08 ± 0.00 | 0.20 ± 0.02 | 8.0 ± 1.1 | >3,000 |
| GTX77557 | 0.21 ± 0.05 | 0.13 ± 0.02 | 0.94 ± 0.1 | >3,000 |
| MDVP-55Aa | 0.06 ± 0.00 | 0.19 ± 0.05 | 6.1 ± 1.1 | >3,000 |
| MA1-27093 | 0.06 ± 0.00 | 0.22 ± 0.05 | 2.5 ± 0.38 | >3,000 |
| 20-783-74014 | 0.08 ± 0.00 | 0.43 ± 0.09 | 3.3 ± 0.52 | >3,000 |
These KD values were reported previously (34).
Strains: DENV-1, OBS7690; DENV-2, NGC; DENV-3, H87; DENV-4, 703-4.
PRNT50 concentrations with four representative DENV strains.
The PRNT50 concentrations for each of the DENV complex-specific MAbs were determined with representatives of the four DENVs (DENV-1 OBS7690, DENV-2 NGC, DENV-3 H87, and DENV-4 703-4). These DENV strains are identical to those used to make the rED3 proteins for DENV-1 to -4. The PRNT50 concentrations with DENV-1 OBS7690 ranged from 8.0 ± 1.4 to 14.4 ± 1.5 nM (Table 5). There was no statistically significant difference between the PRNT50 concentrations obtained with DENV-1 OBS7690 and DENV-2 NGC for any of the DENV complex-specific MAbs. The five DENV complex-specific MAbs had statistically significantly higher PRNT50 concentrations (14- to 44-fold higher, depending on the MAb) with DENV-3 H87 compared with either DENV-1 or DENV-2 (Table 5). There was no detectable neutralization with DENV-4 703-4 up to a 1.0 μM concentration for any of the DENV complex-specific MAbs.
TABLE 5.
PRNT50 concentrations with representative DENV-1, -2, -3, and -4 strains
| MAb | Mean PRNT50 concn (nM) ± SEMa
|
|||
|---|---|---|---|---|
| DENV-1 | DENV-2 | DENV-3 | DENV-4 | |
| GTX29202 | 14.4 ± 1.5 | 5.9 ± 0.3 | 196 ± 18 | >1,000 |
| GTX77557 | 9.1 ± 1.9 | 4.0 ± 0.4 | 139 ± 7 | >1,000 |
| MDVP-55A | 8.0 ± 1.4 | 3.0 ± 0.2 | 133 ± 15 | >1,000 |
| MA1-27093 | 11.8 ± 1.7 | 7.5 ± 0.5 | 194 ± 20 | >1,000 |
| 20-783-74014 | 10.0 ± 1.6 | 11.8 ± 1.3 | 278 ± 69 | >1,000 |
Strains: DENV-1, OBS7690; DENV-2, NGC; DENV-3, H87; DENV-4, 703-4.
Occupancy of ED3 on the virion and neutralization of infectivity.
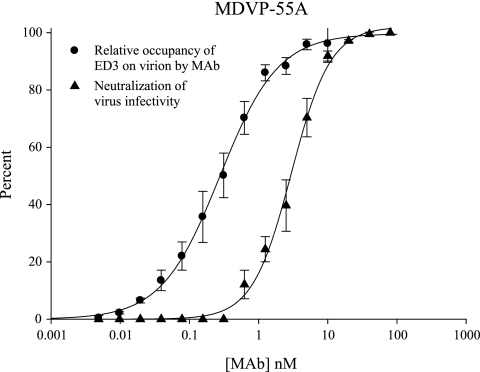
The ELISA binding data with purified virus as the antigen was translated to percent relative occupancy as described previously (6) and was compared to the curve of percent neutralization of virus infectivity as a function of the MAb concentration. This comparison is shown for MAb MDVP-55A (Fig. 4). The occupancy data best fit a standard ligand binding curve, and the neutralization data best fit a dose-response curve having a variable slope factor. For MAb MDVP-55A, the curve for relative occupancy of ED3 on the virion as a function of the MAb concentration had a slope factor of −1.0 ± 0.0 and a KD value of 0.30 ± 0.03 nM (Fig. 4). The curve for percent neutralization of virus infectivity as a function of the MAb concentration had a slope factor of −1.5 ± 0.1 and a PRNT50 concentration of 3.0 ± 0.2 nM (Fig. 4). The slope factor for the occupancy curves was −1.0 ± 0.0 for all of the MAbs, and the KD values for these curves are listed in Table 1 (i.e., the KD with DENV-2 NGC). The slope factors for the neutralization curves were −1.7 ± 0.1 for GTX29202, −1.6 ± 0.2 for GTX77557, −1.2 ± 0.1 for MA1-27093, and −1.1 ± 0.1 for 20-783-74014. The PRNT50 concentrations for all of these curves are reported in Table 1.
FIG. 4.
Percent relative occupancy of available ED3 binding sites on the DENV-2 virion by MAb and percent neutralization of virus infectivity as a function of the concentration of MAb. This comparison is shown for DENV complex-specific MAb MDVP-55A as an example. The relative occupancy curve has a slope factor of −1.0 ± 0.0, while the neutralization curve has a much steeper slope factor of −1.5 ± 0.1. Additionally, the neutralization curve is shifted to the right and away from the occupancy curve, revealing that the majority of virus infectivity is neutralized at very high occupancy levels. Error bars represent the SEM.
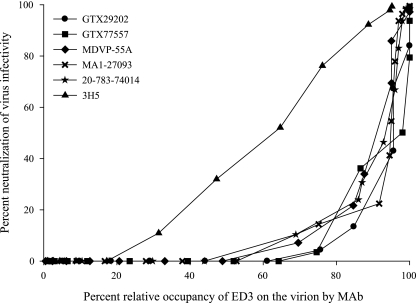
The data for all five MAbs were also plotted with percent neutralization of virus infectivity as a function of percent relative occupancy of ED3 on the virion. For all of the MAbs tested, the majority of virus infectivity was neutralized at very high relative occupancy levels compared to a similar curve for DENV-2 type-specific MAb 3H5 (6) (shown in Fig. 5 for comparison). The threshold for detectable neutralization had the widest range of occupancy values, from 44 to 64%, depending on the MAb, with an average of 54% ± 4% occupancy. Relative occupancies to achieve 10% neutralization had a wide range, from 68 to 81%, with an average of 73% ± 2%. The relative occupancy to achieve 50% neutralization ranged from 91 to 98%, with an average of 95% ± 1%. Finally, the relative occupancy to achieve 90% neutralization had the narrowest range, from 97 to 99%, with an average of 98% ± 0.4%.
FIG. 5.
Percent virus infectivity neutralized as a function of percent relative occupancy of available ED3 binding sites on the virion by MAb. The five DENV complex-specific MAbs (GTX29202, GTX77557, MDVP-55A, MA1-27093, and 20-783-74014) neutralize the majority of virus infectivity at very high relative occupancy levels. Similar data for DENV-2 type-specific MAb 3H5 (triangles), reported previously (6), are shown for comparison. MAb 3H5 neutralizes virus infectivity over a broad range of occupancies, and there is a linear relationship between occupancy and neutralization. In comparison, the DENV complex-specific MAbs neutralize virus considerably less efficiently.
Overall, the efficiency of neutralization (i.e., fractions of virus infectivity neutralized at particular relative occupancy levels) for the DENV complex-specific MAbs in this study was considerably less compared to previous data for a panel of DENV-2 ED3 type-specific MAbs (6). The average relative occupancies for the DENV-2 type-specific MAbs reported in reference 6 were 25% ± 5% for the threshold of neutralization, 32% ± 5% for 10% neutralization, 59% ± 4% for 50% neutralization, and 86% ± 3% for 90% neutralization of infectivity. For the DENV complex-specific MAbs in this study, on average, 29% more occupancy of ED3 at the threshold, 41% more occupancy at 10% neutralization, 36% more occupancy at 50% neutralization, and 12% more occupancy at 90% neutralization were required. There were statistically significant differences between the averages of DENV complex-specific versus DENV-2 type-specific occupancy levels required at both the threshold for neutralization and 50% neutralization of infectivity. The absorbance values at saturation with purified virus were compared for the DENV complex- and type-specific MAbs and were 1.39 ± 0.07 and 1.47 ± 0.03, respectively, with no statistically significant difference between these values. This observation suggests that both the complex- and type-specific MAbs are capable of engaging identical numbers of ED3 epitopes on the DENV-2 virion, which makes the relative occupancy differences, stated above, equal to differences in actual occupancy.
DISCUSSION
The overall objective of this study was to characterize the epitopes recognized by DENV ED3 complex-specific MAbs and examine their potency of neutralization. The epitope mapping data indicate that all five DENV complex-specific MAbs recognize a set of overlapping epitopes that form an antigenic site on a lateral surface of ED3. Amino acid residue K310 was critical for all five MAbs, with residues I312, P332, L389, and W391 being critical for various subsets of the MAbs. Specifically, residue P332 is located on the upper surface of ED3 and does not cluster with the other critical residues. Therefore, the mutations at residue P332 may have long-range effects on the epitope of some DENV complex-specific MAbs, but this requires further investigation. Additional residues that appeared to be involved in the epitope of different sets of MAbs were K307, P384, and L387. However, mutation of these residues had only a weak effect on MAb binding affinity and therefore these residues were considered to be peripheral to the epitope.
The epitope mapping data are consistent with the general location of epitopes mapped for other DENV ED3 complex- and subcomplex-specific neutralizing MAbs. DENV complex-specific MAb 4E11 has an epitope on the DENV-1 ED3 protein that includes residues K307, L308, E309, K310, E311, V312, L387, L389, and W391 (17, 31, 32). Similarly DENV subcomplex-specific MAbs 9D12 and 1A1D-2 were reported to involve residues G304, K305, K307, K310, and P384 (for 9D12 only) in the DENV-2 ED3 epitopes they recognize (30). In all three studies, including this one, residue K310 was a common critical residue for DENV complex-specific MAbs. This residue is not conserved among all DENV-2 strains, as a K310E mutation has been previously identified in a human isolate from Malaysia (26). We incorporated this substitution into our DENV-2 rED3 protein, and it abolished the binding of all five DENV complex-specific MAbs and may similarly affect the binding of other DENV complex- and subcomplex-specific MAbs that recognize this residue. Interestingly, in this study, residue K307 was not a critical epitope residue for any of the MAbs while residue P332 was critical for three of the five MAbs (GTX29202, GTX77557, and MDVP-55A). These differences distinguish the DENV complex-specific antigenic site mapped here from epitopes that have been mapped previously.
There were notable discrepancies in the relative change in KD between glycine and alanine mutations in the rED3 protein at residues 301 and 383, which was observed previously (6). Mutations M301G and E383G result in essentially no detectable binding over the range of concentrations tested for all of the MAbs, whereas the alanine substitution at these residues had no effect. Therefore, these residues do not appear to be critical for the binding of these MAbs to ED3. It is, however, interesting that substituting a glycine at these positions completely ablates the binding of 12 different MAbs, irrespective of the particular epitope they recognize. This seems to imply that there are “global” effects on the rED3 protein when mutations M301G and E383G are incorporated. One possibility is that these mutant rED3 proteins are not properly folded. Minor discrepancies in KD occurred for MAb MDVP-55A with mutations K307G (14-fold change) and K307A (4.4-fold change) and for MAbs MA1-27093 and 20-783-74014 with mutations P332G (7.8-fold and 16-fold changes, respectively) and P332A (12-fold and 0.9-fold changes, respectively). In most cases, the glycine substitution results in a greater change in affinity, with the exception of residue P332, where for MAb MA1-27093 the alanine substitution resulted in a greater relative change in affinity than did the glycine substitution.
When comparing the linear sequence of ED3 for representatives of each of the four DENVs (Fig. 3), three of the residues are completely conserved (K310, P332, and W391), one (L389) is conserved in DENV-1, -2, and -4, and one (I312) is not conserved among the representatives of the four DENVs. There were three MAbs (GTX77557, MDVP-55A, and MA1-27093) that each had completely conserved critical residues in the epitope that they recognized. However, when comparing the binding affinity of these MAbs to rED3 proteins from DENV-1 to -4, all three MAbs bound with very different affinities or not at all in the range of concentrations tested, as was the case with DENV-4 (Table 4). As expected, similar results were seen for the MAbs that did not have completely conserved critical residues among representative strains of the four DENVs. Therefore, identifying similarities in the linear sequence of ED3 among the four DENVs seems to have very little ability to predict the affinities of these DENV complex-specific MAbs for different DENV ED3 proteins and is consistent with the hypothesis that the DENV complex-specific epitopes are structurally conserved and/or that interactions with different “critical” amino acid residues may occur, depending on the DENV. This agrees with the structural data of Volk et al. (34), who found that the electrostatic charges on the surfaces of the ED3 proteins of DENV-2, -3, and -4 were different.
There was an excellent correlation between the binding affinity for the rED3 and neutralization of virus infectivity for representative strains of the four DENVs. All five DENV complex-specific MAbs bound with the highest affinity to the rED3 proteins of DENV-1 OBS7690 and DENV-2 NGC, and these two viruses were also neutralized the best. The binding affinity for the DENV-3 H87 rED3 protein, however, was much weaker, and so was the ability to neutralize this virus. No detectable binding or neutralization of DENV-4 703-4 occurred in the range of concentrations tested, suggesting that these antibodies may bind and potentially neutralize DENV-4 only at very high concentrations. This correlation between affinity for the rED3 protein and neutralization would be predicted considering that the dissociation constant (KD) determines the relative occupancy of ED3 epitopes on the virion at any given MAb concentration and consequently neutralization of virus infectivity.
The DENV complex-specific antigenic site mapped by the panel of MAbs in this study is adjacent to a dominant DENV-2 type-specific antigenic site previously mapped to ED3 (6). This DENV-2 type-specific antigenic site included residues K305 and P384 as critical components of the site and residues K307, E327, D329, and P332 as peripheral residues for different subsets of MAbs. Further mapping of the DENV-2 type-specific MAbs in this study revealed that W391 and L389 were critical for some of these MAbs (Fig. 2b, in red), while residues I312 and L389 were found to be on the periphery of the epitope for a few MAbs (Fig. 2b, in pink). Overall, both DENV complex- and type-specific MAbs recognize distinct antigenic sites that center on critical residues located on the lateral surface of ED3. However, there is some overlap between critical and/or peripheral epitope residues of both complex-specific and type-specific MAbs, including K307, I312, P332, P384, L389, and W391, with some MAbs having more overlap than others, depending on the epitopes they recognize. The approximate location and amount of overlap between the DENV complex-specific and DENV-2 type-specific antigenic sites are shown in Fig. 2c.
This study has revealed that differences in neutralization efficiency can be attributed to the particular epitope that a MAb recognizes on ED3. The neutralization efficiency of the DENV complex-specific MAbs tested in this study was much lower than the efficiency of neutralization of DENV type-specific MAbs previously studied (6). On average, much higher occupancy levels were needed by the DENV complex-specific MAbs, in comparison to the DENV-2 type-specific MAbs, in order to neutralize similar levels of virus infectivity. This is an intriguing result considering how close the DENV complex- and type-specific antigenic sites are on the surface of ED3. Previous studies have shown that the particular epitope recognized by MAbs on ED3 can affect their mechanism of neutralization. WNV ED3-specific MAbs were tested for the ability to inhibit virus attachment or a postattachment step of infection, and results varied, depending on the MAb (21). Interestingly, WNV MAb E16, which recognizes a region of ED3 analogous to that of a panel of DENV-2 type-specific MAbs described by Gromowski and Barrett (6), was shown to predominantly neutralize WNV at a postattachment step (21). Stiasny et al. (28) examined the fusion blocking/inhibition of four ED3-specific tick-borne encephalitis virus MAbs, and two of these MAbs had no effect on fusion, while the other two affected both the rate and extent of fusion. It was proposed by the authors of that study (28) that fusion inhibition by these ED3 MAbs could occur by interfering with the relocation of ED3 that takes place during fusion, a mechanism that has been proposed previously for WNV MAb E16 (21). All of these observations demonstrate that subtle variations in the ED3 epitope recognized by different MAbs can result in major differences in the mechanism and overall efficiency of virus neutralization.
Previous studies have shown that there was little correlation between the ability of an antibody to neutralize virus and HAI activity for the flaviviruses, including the DENVs (33). Our data agree with these previous observations in that HAI activity was only predictive of the neutralization efficiency of 5 of 12 MAbs (Table 1). There was, however, a strong statistical correlation between the HAI titer and the KD for virion-associated ED3. Since 50% relative occupancy of available binding sites on the virion occurs at the KD, it follows that this is also the minimum occupancy requirement for complete inhibition of HA of RBC by these DENV ED3-specific MAbs. Antibodies with HAI activity are not limited to ED3, however, as many DENV E protein-specific MAbs and antisera have been shown to have HAI activity (4, 9, 24, 33). Therefore, it is difficult to attribute the HA activity of DENV to a single epitope on the E protein; it more likely reflects the ability of MAbs to sufficiently coat virus particles so that necessary interactions with RBC are blocked or inhibited. This probably explains why MAbs with little or no neutralizing activity can readily inhibit HA activity by virus. On the other hand, a lack of HAI activity may reflect the inability of a MAb to adequately coat the virus particle, which may correlate well with an inability to neutralize virus.
In this study, the epitopes of five DENV complex-specific MAbs were determined; they recognize what is likely to be a dominant antigenic site on ED3. Higher levels of occupancy of the DENV complex-specific antigenic site by MAbs were required for neutralization than have been previously described for an adjacent type-specific antigenic site on the DENV-2 ED3. This could indicate that DENV-2 ED3 type-specific MAbs neutralize virus by a more potent mechanism than DENV complex-specific MAbs. Additionally, the DENV complex-specific MAbs neutralized the four DENVs with variable efficacy. Taken together, these results for DENV-2 suggest that targeting antibodies to the type-specific antigenic site on ED3 rather than the DENV complex-specific antigenic site described in this study would be advantageous for DEN vaccine development.
Acknowledgments
This work was supported by the Pediatric Dengue Vaccine Initiative. G.D.G. is supported by a NIAID T32 predoctoral fellowship (AI060549). N.D.B. was supported by the UTMB Summer Undergraduate Research Program.
We thank John Roehrig and Jim Lee for helpful discussions.
Footnotes
Published ahead of print on 18 June 2008.
REFERENCES
- 1.Chin, J. F., J. J. Chu, and M. L. Ng. 2007. The envelope glycoprotein domain III of dengue virus serotypes 1 and 2 inhibit virus entry. Microbes. Infect. 91-6. [DOI] [PubMed] [Google Scholar]
- 2.Chu, J. J., R. Rajamanonmani, J. Li, R. Bhuvanakantham, J. Lescar, and M. L. Ng. 2005. Inhibition of West Nile virus entry by using a recombinant domain III from the envelope glycoprotein. J. Gen. Virol. 86405-412. [DOI] [PubMed] [Google Scholar]
- 3.Crill, W. D., and J. T. Roehrig. 2001. Monoclonal antibodies that bind to domain III of dengue virus E glycoprotein are the most efficient blockers of virus adsorption to Vero cells. J. Virol. 757769-7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gentry, M. K., E. A. Henchal, J. M. McCown, W. E. Brandt, and J. M. Dalrymple. 1982. Identification of distinct antigenic determinants on dengue-2 virus using monoclonal antibodies. Am. J. Trop. Med. Hyg. 31548-555. [DOI] [PubMed] [Google Scholar]
- 5.Gollins, S. W., and J. S. Porterfield. 1986. A new mechanism for the neutralization of enveloped viruses by antiviral antibody. Nature 321244-246. [DOI] [PubMed] [Google Scholar]
- 6.Gromowski, G. D., and A. D. Barrett. 2007. Characterization of an antigenic site that contains a dominant, type-specific neutralization determinant on the envelope protein domain III (ED3) of dengue 2 virus. Virology 366349-360. [DOI] [PubMed] [Google Scholar]
- 7.Halstead, S. B. 1988. Pathogenesis of dengue: challenges to molecular biology. Science 239476-481. [DOI] [PubMed] [Google Scholar]
- 8.He, R. T., B. L. Innis, A. Nisalak, W. Usawattanakul, S. Wang, S. Kalayanarooj, and R. Anderson. 1995. Antibodies that block virus attachment to Vero cells are a major component of the human neutralizing antibody response against dengue virus type 2. J. Med. Virol. 45451-461. [DOI] [PubMed] [Google Scholar]
- 9.Henchal, E. A., J. M. McCown, D. S. Burke, M. C. Seguin, and W. E. Brandt. 1985. Epitopic analysis of antigenic determinants on the surface of dengue-2 virions using monoclonal antibodies. Am. J. Trop. Med. Hyg. 34162-169. [DOI] [PubMed] [Google Scholar]
- 10.Hiramatsu, K., M. Tadano, R. Men, and C. J. Lai. 1996. Mutational analysis of a neutralization epitope on the dengue type 2 virus (DEN2) envelope protein: monoclonal antibody resistant DEN2/DEN4 chimeras exhibit reduced mouse neurovirulence. Virology 224437-445. [DOI] [PubMed] [Google Scholar]
- 11.Hung, J. J., M. T. Hsieh, M. J. Young, C. L. Kao, C. C. King, and W. Chang. 2004. An external loop region of domain III of dengue virus type 2 envelope protein is involved in serotype-specific binding to mosquito but not mammalian cells. J. Virol. 78378-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hung, S. L., P. L. Lee, H. W. Chen, L. K. Chen, C. L. Kao, and C. C. King. 1999. Analysis of the steps involved in Dengue virus entry into host cells. Virology 257156-167. [DOI] [PubMed] [Google Scholar]
- 13.Klasse, P. J., and Q. J. Sattentau. 2002. Occupancy and mechanism in antibody-mediated neutralization of animal viruses. J. Gen. Virol. 832091-2108. [DOI] [PubMed] [Google Scholar]
- 14.Kuhn, R. J., W. Zhang, M. G. Rossmann, S. V. Pletnev, J. Corver, E. Lenches, C. T. Jones, S. Mukhopadhyay, P. R. Chipman, E. G. Strauss, T. S. Baker, and J. H. Strauss. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, L., A. D. Barrett, and D. W. Beasley. 2005. Differential expression of domain III neutralizing epitopes on the envelope proteins of West Nile virus strains. Virology 33599-105. [DOI] [PubMed] [Google Scholar]
- 16.Lin, B., C. R. Parrish, J. M. Murray, and P. J. Wright. 1994. Localization of a neutralizing epitope on the envelope protein of dengue virus type 2. Virology 202885-890. [DOI] [PubMed] [Google Scholar]
- 17.Lisova, O., F. Hardy, V. Petit, and H. Bedouelle. 2007. Mapping to completeness and transplantation of a group-specific, discontinuous, neutralizing epitope in the envelope protein of dengue virus. J. Gen. Virol. 882387-2397. [DOI] [PubMed] [Google Scholar]
- 18.Lok, S. M., M. L. Ng, and J. Aaskov. 2001. Amino acid and phenotypic changes in dengue 2 virus associated with escape from neutralisation by IgM antibody. J. Med. Virol. 65315-323. [DOI] [PubMed] [Google Scholar]
- 19.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2003. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. USA 1006986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monath, T. P. 1994. Dengue: the risk to developed and developing countries. Proc. Natl. Acad. Sci. USA 912395-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nybakken, G. E., T. Oliphant, S. Johnson, S. Burke, M. S. Diamond, and D. H. Fremont. 2005. Structural basis of West Nile virus neutralization by a therapeutic antibody. Nature 437764-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pierson, T. C., Q. Xu, S. Nelson, T. Oliphant, G. E. Nybakken, D. H. Fremont, and M. S. Diamond. 2007. The stoichiometry of antibody-mediated neutralization and enhancement of West Nile virus infection. Cell Host Microbe 1135-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roehrig, J. T. 2003. Antigenic structure of flavivirus proteins. Adv. Virus Res. 59141-175. [DOI] [PubMed] [Google Scholar]
- 24.Roehrig, J. T., R. A. Bolin, and R. G. Kelly. 1998. Monoclonal antibody mapping of the envelope glycoprotein of the dengue 2 virus, Jamaica. Virology 246317-328. [DOI] [PubMed] [Google Scholar]
- 25.Sabin, A. B. 1952. Research on dengue during World War II. Am. J. Trop. Med. Hyg. 130-50. [DOI] [PubMed] [Google Scholar]
- 26.Samuel, S., C. L. Koh, J. Blok, T. Pang, and S. K. Lam. 1989. Nucleotide sequence of the envelope protein gene of a Malaysian dengue-2 virus isolated from a patient with dengue shock syndrome. Nucleic Acids Res. 178888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Serafin, I. L., and J. G. Aaskov. 2001. Identification of epitopes on the envelope (E) protein of dengue 2 and dengue 3 viruses using monoclonal antibodies. Arch. Virol. 1462469-2479. [DOI] [PubMed] [Google Scholar]
- 28.Stiasny, K., S. Brandler, C. Kossl, and F. X. Heinz. 2007. Probing the flavivirus membrane fusion mechanism by using monoclonal antibodies. J. Virol. 8111526-11531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stiasny, K., and F. X. Heinz. 2006. Flavivirus membrane fusion. J. Gen. Virol. 872755-2766. [DOI] [PubMed] [Google Scholar]
- 30.Sukupolvi-Petty, S., S. K. Austin, W. E. Purtha, T. Oliphant, G. E. Nybakken, J. J. Schlesinger, J. T. Roehrig, G. D. Gromowski, A. D. Barrett, D. H. Fremont, and M. S. Diamond. 2007. Type- and subcomplex-specific neutralizing antibodies against domain III of dengue virus type 2 envelope protein recognize adjacent epitopes. J. Virol. 8112816-12826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thullier, P., C. Demangel, H. Bedouelle, F. Megret, A. Jouan, V. Deubel, J. C. Mazie, and P. Lafaye. 2001. Mapping of a dengue virus neutralizing epitope critical for the infectivity of all serotypes: insight into the neutralization mechanism. J. Gen. Virol. 821885-1892. [DOI] [PubMed] [Google Scholar]
- 32.Thullier, P., P. Lafaye, F. Megret, V. Deubel, A. Jouan, and J. C. Mazie. 1999. A recombinant Fab neutralizes dengue virus in vitro. J. Biotechnol. 69183-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trent, D. W., C. L. Harvey, A. Qureshi, and D. LeStourgeon. 1976. Solid-phase radioimmunoassay for antibodies to flavivirus structural and nonstructural proteins. Infect. Immun. 131325-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Volk, D. E., Y. C. Lee, X. Li, V. Thiviyanathan, G. D. Gromowski, L. Li, A. R. Lamb, D. W. Beasley, A. D. Barrett, and D. G. Gorenstein. 2007. Solution structure of the envelope protein domain III of dengue-4 virus. Virology 364147-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, W., P. R. Chipman, J. Corver, P. R. Johnson, Y. Zhang, S. Mukhopadhyay, T. S. Baker, J. H. Strauss, M. G. Rossmann, and R. J. Kuhn. 2003. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat. Struct. Biol. 10907-912. [DOI] [PMC free article] [PubMed] [Google Scholar]