Abstract
Membrane fusion promoted by human metapneumovirus (HMPV) fusion (F) protein was suggested to require low pH (R. M. Schowalter, S. E. Smith, and R. E. Dutch, J. Virol. 80:10931-10941, 2006). Using prototype F proteins representing the four HMPV genetic lineages, we detected low-pH-dependent fusion only with some lineage A proteins and not with lineage B proteins. A glycine at position 294 was found responsible for the low-pH requirement in lineage A proteins. Only 6% of all HMPV lineage A F sequences have 294G, and none of the lineage B sequences have 294G. Thus, acidic pH is not a general trigger of HMPV F proteins for activity.
The human metapneumovirus (HMPV) was first described in 2001 when it was classified as the first mammalian member of the Paramyxoviridae family, subfamily Pneumovirinae, genus Metapneumovirus (16). Sequence analyses of the surface glycoprotein genes of several HMPV isolates revealed the existence of two main genetic HMPV lineages (A and B), each divided into two sublineages (A1, A2, B1, and B2) (17).
One of the major HMPV glycoproteins is the fusion (F) protein, which is relatively conserved among HMPV strains (17) and shares structural features with other paramyxovirus F proteins. These proteins are class I viral fusion proteins (18), which are synthesized as inactive precursors, F0, that must be cleaved to yield fusion-competent disulfide-linked F2-F1 chains (reviewed in references 7 and 8). They mediate fusion of the viral and cell membranes to facilitate virus entry into the cell and commonly promote cell-cell fusion leading to syncytium formation (SF).
The fusion proteins of viruses from virus families that enter the cell via acidic endosomes require low pH for triggering membrane fusion (13, 18). In contrast, membrane fusion promoted by paramyxovirus F proteins generally occurs at the cell surface and at neutral pH (6, 11). It is believed that in viruses of the Paramyxovirinae subfamily, the F protein is maintained in the virion in a metastable prefusion conformation by specific interactions with the attachment protein. Following attachment to the target cell receptor, conformational changes in the attachment protein are transduced to the F protein to activate it for fusion “at the right time and in the right place” (6, 10). However, it is unlikely that this applies to the Pneumovirinae, including human respiratory syncytial virus (HRSV) and HMPV, since spontaneous mutants of HRSV (5) or genetically engineered recombinants of HRSV (14, 15) and HMPV (1) expressing F as the only surface glycoprotein are infectious. In addition, whereas Paramyxovirinae F proteins require cooperation of the attachment protein for SF, expression of HRSV (3, 20) or HMPV (12) F proteins alone in transfected cells is sufficient to induce SF. Thus, the activating mechanism of the F protein of viruses belonging to the Pneumovirinae subfamily remains unclear.
Schowalter et al. (12) reported that SF promoted by the F protein of HMPV, strain CAN97-83, was detected only when cells transfected with an F-expressing plasmid were treated with trypsin (required to cleave the F0 precursor) and exposed to low pH. This low-pH dependency suggested a unique way of triggering fusion among paramyxovirus F proteins. To test the generality of this statement, HMPV F-mediated membrane fusion was assayed with proteins derived from prototype strains isolated in The Netherlands and representative of the four HMPV genetic lineages, as shown in Fig. 1 (17). The F genes were cloned in plasmid pCAGGS (9) and used in an SF assay, essentially as described by Schowalter et al. (12). Briefly, Vero cells growing in eight-well plates were transfected with 1 μg of pCAGGS-F plasmids, using Fugene. The next day, cells were incubated for 2 hours in medium with 1 μg/ml trypsin and subsequently exposed to phosphate-buffered saline at pH 5 or pH 7.4 for 4 minutes. This pH pulse was repeated three times, 2 hours apart. Twenty-four hours after the last pH pulse, the cells were fixed, and the syncytia were visualized with HMPV F-specific antibodies.
FIG. 1.
HMPV strains used in this study. A phylogenetic tree of prototype strains of HMPV isolated in The Netherlands (prefix NL) and representing the four genetic lineages (indicated in parentheses) was constructed on the basis of sequences reported by van den Hoogen et al. (17). The CAN97-83 strain used by Schowalter et al. (12) was also included in the analysis for comparison (underlined and in italics). Also indicated is the amino acid at residue 294 of the F protein in each of the viruses. Finally, the low-pH dependency of each F protein for membrane fusion is indicated (+, increased membrane fusion at low pH; −, membrane fusion not increased at low pH), as inferred from the results of Fig. 2 and the results reported by Schowalter et al. (12) for strain CAN97-83. The bar denotes 0.01 nucleotide substitution per site.
Fusion was also tested in a cell content mixing (CM) assay in which two wells of a six-well plate containing Vero cells were each transfected with 2 μg of pCAGGS-F and 0.4 μg of pTS27 plasmid, a constitutive β-galactosidase (β-Gal) expression vector, using Lipofectamine 2000. The cells in one well were cotransfected with 2 μg of pLTR-CAT (containing the chloramphenicol acetyltransferase [CAT] gene under the control of the human immunodeficiency virus type 1 [HIV-1] long terminal repeat [LTR]), and the cells in the other well were cotransfected with 2 μg of pTat (expressing the HIV-1 transactivator of transcription Tat) (4). One day after transfection, both cell populations were harvested using trypsin-EDTA, mixed, and plated in three wells of a six-well plate. The next morning, cells were exposed to three pH pulses as described above. Cell lysates were harvested 24 hours after the last pH pulse, and CAT and β-Gal expression was quantified by an enzyme-linked immunosorbent assay. In this assay, CM, as a result of F-mediated fusion, resulted in Tat-mediated transactivation of the HIV-1 LTR, and hence induction of CAT expression.
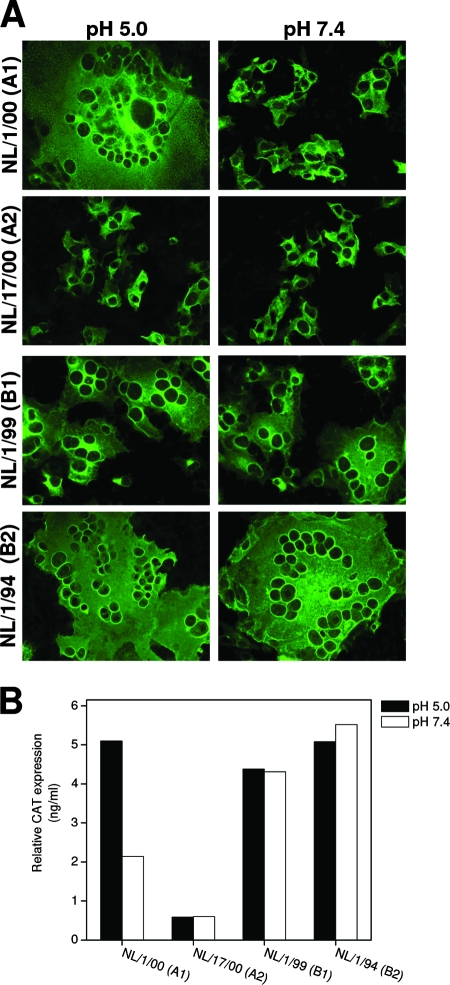
Figure 2 shows SF and CM assays performed with the four prototype HMPV proteins. Transfection of empty pCAGGS vector or exclusion of trypsin resulted in no measurable membrane fusion in both assays (not shown). A clear low-pH dependency could be observed for FNL/1/00(A1) (F protein from isolate NL/1/00 of HMPV sublineage A1), since large syncytia were observed upon exposure to pH 5, whereas only small syncytia were visible after exposure to pH 7.4 (Fig. 2A). In the CM assay, low-pH treatment also resulted in significant enhanced CAT expression (Fig. 2B). Very small syncytia and low levels of CAT expression were observed for the FNL/17/00(A2) protein at low and neutral pH. In contrast, transfection of FNL/1/99(B1) and particularly FNL/1/94(B2) resulted in large syncytia at low and neutral pH (Fig. 2A), indicating that fusion promoted by HMPV lineage B F proteins was highly efficient and not enhanced at low pH, a finding that was confirmed in the CM assay (Fig. 2B). These initial results, summarized in Fig. 1, demonstrated that low-pH dependency is not a general phenomenon for the F proteins of HMPV. Similar results were obtained with LLC-MK2, ruling out the possibility that they were cell type specific (not shown).
FIG. 2.
Evaluation of cell-cell fusion mediated by F proteins representing the genetic lineages A1, A2, B1 and B2 of HMPV. (A) Syncytium formation in Vero cells transfected with pCAGGS expression vectors carrying the F genes of the HMPV viruses shown at the left margin (genetic lineage indicated in parentheses). The cells were exposed to low pH (pH 5.0) or neutral pH (pH 7.4), before being stained with a rabbit polyclonal anti-F antibody. Expression levels were measured by flow cytometry and found to be essentially the same under all conditions and for all F proteins (not shown). (B) Cell content mixing assays were performed in Vero cells transfected with plasmids carrying F, β-Gal, and LTR-CAT genes or F, β-Gal, and tat genes. The next day, the two cell populations were mixed, followed by low-pH (pH 5.0) or neutral-pH (pH 7.4) treatments. Twenty-four hours later, the CAT values were measured and normalized to β-Gal expression to correct for differences in transfection efficiency and sample processing. The results presented in panels A and B are representative of five independent experiments.
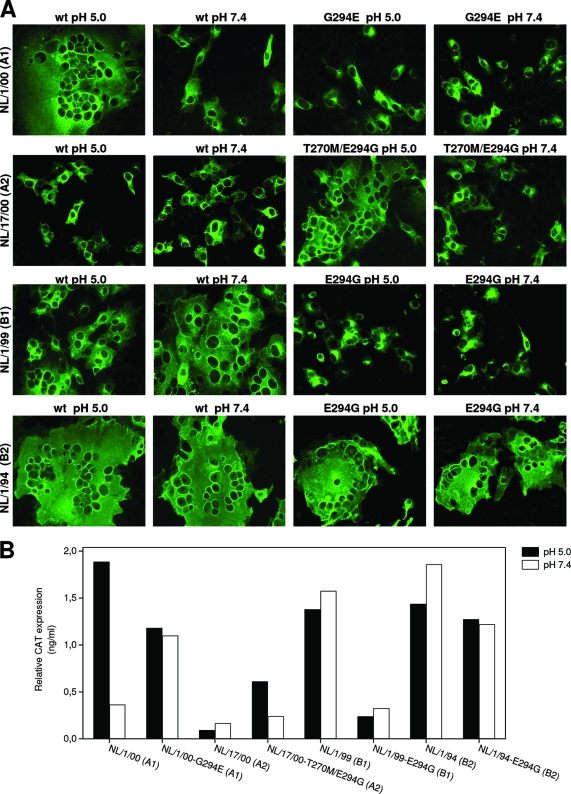
The results obtained with FNL/17/00(A2) were unexpected, since Schowalter et al. (12) had made use of an almost identical F protein, FCAN97-83(A2), which differs only in two amino acids, T270M and E294G, from FNL/17/00(A2) (Fig. 1). Site-directed mutagenesis was used to change T270M in FNL/17/00(A2) because all HMPV F sequences in public databases, except this strain, have Met at that position. This change had no effect on F-protein activity (not shown). Therefore, the difference (E294G) between FNL/17/00(A2) and FCAN97-83 (A2) was apparently solely responsible for the different pH requirements of the two proteins. To test this hypothesis, E294G was further substituted in FNL/17/00(A2). When assayed for membrane fusion, the double mutant T270M E294G showed increased activity in both SF and CM assays, but only at low pH (Fig. 3), mimicking the results reported for FCAN97-83(A2) (12).
FIG. 3.
Evaluation of cell-cell fusion with F-protein mutants. Syncytium formation assays (A) and cell content mixing assays (B) were done with the wild-type (wt) and mutant F proteins shown in each panel, as described in the legend to Fig. 2. The results of this figure are representative of four independent experiments.
It was interesting to note that the other F protein that displayed enhanced fusion upon low-pH treatment in Fig. 2 [FNL/1/00(A1)] also had Gly at residue position 294 (Fig. 1). Although FNL/1/00(A1) and FNL/17/00(A2) sequences differ at 10 positions, including position 294 (17), we decided to test whether the G294E change was sufficient to revert the low-pH-dependent phenotype of FNL/1/00(A1). Indeed, as shown in Fig. 3, the G294E change inhibited the fusogenic activity of FNL/1/00(A1) in both SF and CM assays at pH 5.0 and at pH 7.4, making it very similar to the protein derived from FNL/17/00(A2). Therefore, the amino acid residue at position 294 is a key determinant of membrane fusion activity in a low-pH-dependent manner, for HMPV lineage A F proteins.
The E294G change was also tested in the F proteins of prototype HMPV lineage B strains. In contrast to the results obtained with lineage A proteins, the mutation E294G had contrasting effects in FNL/1/99(B1) and FNL/1/94(B2). In the B1 strain, it inhibited F-protein-induced fusion at low and neutral pH, and in the B2 strain, it only marginally reduced F-mediated fusion at either pH. The different responses of lineage A and B proteins to the E294G change are likely determined by other amino acid differences between the two lineages that may also have a dominant effect on the fusogenic properties of the respective F proteins.
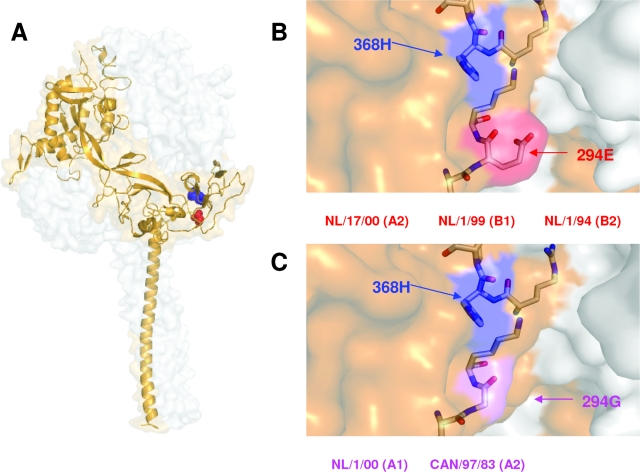
There is no clear explanation for the dominant effect of residue 294 in the fusogenic properties of HMPV lineage A F proteins. Since a three-dimensional structure of HMPV F protein is still lacking, a model of its preactive conformation was built, based on the structure determined for the parainfluenza virus 5 F protein (19). In this model, residue 294 is located at the base of the globular head of the F trimer (Fig. 4). Given that residue 294 is near 368H in the three-dimensional model, it is possible that the presence of either Glu or Gly at position 294 could influence protonation of 368H. Protonation of histidines is important for activation of certain fusion proteins that require low pH for fusion (2). Therefore, 294E could facilitate protonation of 368H in HMPV F proteins even at neutral pH, whereas protonation of 368H may require low pH when it is 294G. This protonation of 368H could be necessary for the conformational changes associated to the activation of HMPV F protein for membrane fusion, even if this is triggered by a pH-independent mechanism. This explanation for the low-pH requirement for certain lineage A F proteins cannot be applied to lineage B proteins, as inferred from the results of Fig. 2 and 3. It is likely that other amino acid differences between HMPV lineages A and B may override the dominant effect of residue 294 for membrane fusion in lineage B proteins. Further mutagenesis based on sequence differences between lineage A and B strains may shed light on this aspect of HMPV F-protein activity.
FIG. 4.
Locations of critical residues in a model of HMPV F protein. (A) The model represents the prefusion conformation of the HMPV F trimer, built with the atomic coordinates of the preactive structure of the parainfluenza virus 5 F protein, reported by Yin et al. (19) and using the SWISS-MODEL server facilities (http://swissmodel.expasy.org/). Details of the model-building process will be published elsewhere. One of the monomers is shown as a golden ribbon in which residues 368 (blue) and 294 (red) are shown as balls. (B and C) Partial blowups of the structure showing the lateral side chains of 368H and 294E or 294G. The strains mentioned in the text, which have either 294E or 294G, are indicated below panels B and C, respectively.
Since HMPV strains with 294G in the F protein represent only a small proportion of all HMPV lineage A sequences in public databases (27 of 433 lineage A sequences), low-pH-dependent fusion is probably a rare characteristic of HMPV. In conclusion, the data reported by Schowalter et al. (12) and the data presented here indicate that some HMPV F proteins require low pH for efficient fusion, but it is unlikely that exposure to low pH is a general trigger of HMPV F protein for membrane fusion.
Acknowledgments
We thank Eefje Schrauwen and Miranda de Graaf for technical support, M. Malim for providing plasmid pTS27, L. Martínez-Sobrido for providing plasmid pCAGGS, and R. Gruters for providing plasmids pLTR-CAT and pTat.
This work was sponsored in part by the FP5 grant “Hammocs” from the European Union. V.M. was funded by the VIRUSHOST consortium (Comunidad de Madrid), and L.S.V. was the recipient of a predoctoral fellowship from Ministerio Educación y Ciencia (Spain).
Footnotes
Published ahead of print on 2 July 2008.
REFERENCES
- 1.Biacchesi, S., M. H. Skiadopoulos, L. Yang, E. W. Lamirande, K. C. Tran, B. R. Murphy, P. L. Collins, and U. J. Buchholz. 2004. Recombinant human metapneumovirus lacking the small hydrophobic SH and/or attachment G glycoprotein: deletion of G yields a promising vaccine candidate. J. Virol. 7812877-12887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carneiro, F. A., F. Stauffer, C. S. Lima, M. A. Juliano, L. Juliano, and A. T. Da Poian. 2003. Membrane fusion induced by the vesicular stomatitis virus depends on histidine protonation. J. Biol. Chem. 27813789-13794. [DOI] [PubMed] [Google Scholar]
- 3.Gonzalez-Reyes, L., M. B. Ruiz-Arguello, B. Garcia-Barreno, L. Calder, J. A. Lopez, J. P. Albar, J. J. Skehel, D. C. Wiley, and J. A. Melero. 2001. Cleavage of the human respiratory syncytial virus fusion protein at two distinct sites is required for activation of membrane fusion. Proc. Natl. Acad. Sci. USA 989859-9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gruters, R. A., S. A. Otto, B. J. Al, A. J. Verhoeven, C. L. Verweij, R. A. Van Lier, and F. Miedema. 1991. Non-mitogenic T cell activation signals are sufficient for induction of human immunodeficiency virus transcription. Eur. J. Immunol. 21167-172. [DOI] [PubMed] [Google Scholar]
- 5.Karron, R. A., D. A. Buonagurio, A. F. Georgiu, S. S. Whitehead, J. E. Adamus, M. L. Clements-Mann, D. O. Harris, V. B. Randolph, S. A. Udem, B. R. Murphy, and M. S. Sidhu. 1997. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc. Natl. Acad. Sci. USA 9413961-13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamb, R. A. 1993. Paramyxovirus fusion: a hypothesis for changes. Virology 1971-11. [DOI] [PubMed] [Google Scholar]
- 7.Lamb, R. A., and T. S. Jardetzky. 2007. Structural basis of viral invasion: lessons from paramyxovirus F. Curr. Opin. Struct. Biol. 17427-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamb, R. A., R. G. Paterson, and T. S. Jardetzky. 2006. Paramyxovirus membrane fusion: lessons from the F and HN atomic structures. Virology 34430-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108193-199. [DOI] [PubMed] [Google Scholar]
- 10.Russell, C. J., T. S. Jardetzky, and R. A. Lamb. 2001. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J. 204024-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Russell, C. J., and L. E. Luque. 2006. The structural basis of paramyxovirus invasion. Trends Microbiol. 14243-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schowalter, R. M., S. E. Smith, and R. E. Dutch. 2006. Characterization of human metapneumovirus F protein-promoted membrane fusion: critical roles for proteolytic processing and low pH. J. Virol. 8010931-10941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69531-569. [DOI] [PubMed] [Google Scholar]
- 14.Techaarpornkul, S., N. Barretto, and M. E. Peeples. 2001. Functional analysis of recombinant respiratory syncytial virus deletion mutants lacking the small hydrophobic and/or attachment glycoprotein gene. J. Virol. 756825-6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Techaarpornkul, S., P. L. Collins, and M. E. Peeples. 2002. Respiratory syncytial virus with the fusion protein as its only viral glycoprotein is less dependent on cellular glycosaminoglycans for attachment than complete virus. Virology 294296-304. [DOI] [PubMed] [Google Scholar]
- 16.van den Hoogen, B. G., J. C. de Jong, J. Groen, T. Kuiken, R. de Groot, R. A. Fouchier, and A. D. Osterhaus. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 7719-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van den Hoogen, B. G., S. Herfst, L. Sprong, P. A. Cane, E. Forleo, R. L. de Swart, A. D. M. E. Osterhaus, and R. A. M. Fouchier. 2004. Antigenic and genetic variability of human metapneumoviruses. Emerg. Infect. Dis. 10658-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weissenhorn, W., A. Hinz, and Y. Gaudin. 2007. Virus membrane fusion. FEBS Lett. 5812150-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin, H.-S., X. Wen, R. G. Paterson, R. A. Lamb, and T. S. Jardetzky. 2006. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature 43938-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zimmer, G., L. Budz, and G. Herrler. 2001. Proteolytic activation of respiratory syncytial virus fusion protein. J. Biol. Chem. 27631642-31650. [DOI] [PubMed] [Google Scholar]