Abstract
A generally accepted view of norovirus replication is that capsid expression requires production of a subgenomic transcript, the presence of capsid often being used as a surrogate marker to indicate the occurrence of viral replication. Using a polymerase II-based baculovirus delivery system, we observed capsid expression following introduction of a full-length genogroup 3 norovirus genome into HepG2 cells. However, capsid expression occurred as a result of a novel translation termination/reinitiation event between the nonstructural-protein and capsid open reading frames, a feature that may be unique to genogroup 3 noroviruses.
Noroviruses are nonenveloped, positive-strand RNA viruses belonging to the family Caliciviridae and are the single most frequent cause of human nonbacterial gastroenteritis worldwide (6). Their highly infectious nature means that outbreaks are difficult to contain. No vaccine exists for noroviruses, and although there is a cell culture system for mouse norovirus (MNV) (17), routine propagation of enteron-specific noroviruses has yet to be achieved. Indeed, information regarding norovirus replication has mostly been obtained by drawing parallels with the more distantly related but cultivatable pathogen feline calicivirus (1).
Caliciviruses possess certain features that distinguish them from other positive-strand RNA viruses. The 5′ end of their genome is covalently linked to a viral protein (VPg), which both serves as a primer for genome replication (13) and directs translation of viral proteins (2, 3, 5). The 3′ end contains a short untranslated region as well as a poly(A) tail. Several open reading frames (ORFs) exist within a calicivirus genome. In noroviruses, there are three verified ORFs, with the largest of these (ORF1) being translated from the viral genomic RNA and encoding nonstructural proteins (Fig. 1a). Translation of the major structural protein, encoded by ORF2, is currently believed to be dependent on production of a subgenomic RNA, which also allows for ORF3 expression due to translation termination reinitiation (TTR) that occurs between ORF2 and ORF3 (4, 11, 16).
FIG. 1.
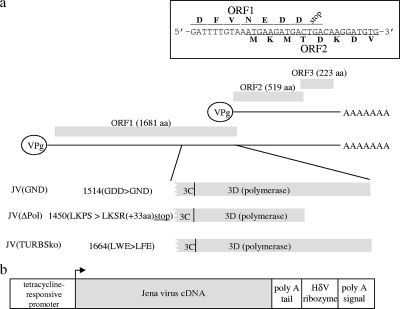
Diagram detailing the organization of the JV genome which highlights mutations introduced into the viral transcripts used in this study and has an insert displaying the region of the genome comprising the end of ORF1 and the beginning of ORF2 (a). In addition, an overview of the baculovirus constructs used to deliver these transcripts into cells is provided (b).
We recently developed a baculovirus-based delivery system for recovery of infectious MNV (15). A parallel attempt to develop a reverse genetics system for bovine genogroup 3 noroviruses, which are more closely related to human noroviruses (7), is in progress. As a result of this work, we now report a novel observation, that bovine norovirus can express capsid protein as a result of TTR between ORF1 and ORF2.
Characterization of cells transduced with genogroup 3-containing baculovirus.
A cDNA of a genotype 3 norovirus genome, derived from the Jena virus (JV) (7), was cloned into the polymerase II-based baculovirus delivery system (9, 10, 15) (Fig. 1b). Recovered baculovirus, referred to as JV(wt), was used to transduce HepG2 cells in combination with BACtTA, a construct expressing the tetracycline repressor/VP16 transactivator. At 20 hours posttransduction, fully processed and proteolytic precursors of VPg and 3C were detected by Western blot analysis (Fig. 2a, lane 2), confirming ORF1 expression in these cells. To assess whether viral replication might be occurring, Western blotting was used to detect capsid expression from ORF2. As work with the related MNV reverse genetics system had failed to demonstrate capsid expression, despite recovery of infectious virus (15), it was surprising that JV capsid expression was detectable (Fig. 2a, lane 2). While the capsid monoclonal antibody used cross-reacted with other cellular antigens of various molecular weights, detection of capsid was nonetheless specific, as it was also observed using a rabbit anti-capsid antiserum and was absent when cells were maintained in the presence of tetracycline (data not shown).
FIG. 2.
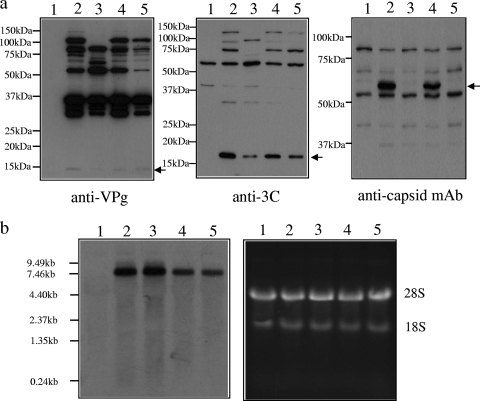
Production of viral protein and RNA transcripts in transduced cells. HepG2 cells were either transduced with BACtTA alone (lane 1) or cotransduced with BACtTA and either JV(wt), JV(ΔPol), JV(GND), or JV(TURBSko) (lane 2, 3, 4, or 5, respectively). Western blot analysis (a) was used to confirm expression of VPg, 3C, and capsid. An arrow indicates the location of capsid as well as fully proteolytically processed VPg and 3C. Additional bands on the blots that are not seen in the control lanes (lane 1) represent expected proteolytic precursors. Concomitant Northern blot analysis (b) was performed using a double-stranded DNA probe directed at a JV ORF2 encoding region (nucleotides 5587 to 7197; left), the integrity of the RNA being assessed by ethidium bromide staining of total cellular RNA on a separate gel (right).
Capsid expression indicated that viral replication might be occurring; therefore, cellular RNA was analyzed by Northern blotting for the presence of a subgenomic transcript. While a major single transcript of the expected size for the full-length genome was present, no subgenomic RNA production was observed (Fig. 2b, lane 2).
Generation of replication-defective constructs where capsid expression is maintained.
To establish further whether low-level viral replication and subgenomic RNA production were the cause of capsid expression, two different replication-defective JV constructs were generated (Fig. 1a). The first construct, JV(ΔPol), had a frameshift mutant at the 3′ end of ORF1 so that premature translational termination occurred within the 3D viral polymerase-encoding region. The second construct, JV(GND), was mutated at the GDD active site within 3D to inactivate polymerase activity (14). Both constructs expressed ORF1 products, based on Western blot analysis (Fig. 2a, lanes 3 and 4), and showed proteolytic processing of ORF1 similar to that shown by the parental JV construct, albeit with the expected generation of some shorter precursor products in JV(ΔPol) transduced cells. However, while capsid was not expressed from JV(ΔPol), it was readily detectable in cells transduced with JV(GND), demonstrating that the presence of capsid was not due to viral replication. Furthermore, Northern blot analysis (Fig. 2b, lanes 3 and 4) showed that both constructs resulted in the expression of a single major transcript that was identical in size to that produced from JV(wt), indicating that the changes introduced into the JV genome had not facilitated unanticipated RNA splicing that might account for differential capsid expression.
Identification of a functional TURBS1 motif at the 3′ end of ORF1.
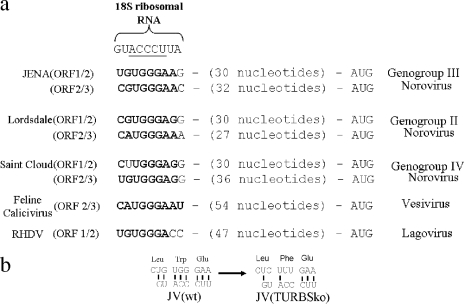
One explanation that would link capsid expression with a requirement for translation of an intact ORF1 is that JV employs a TTR mechanism at the ORF1/2 boundary: a mechanism that was thought to be restricted to the ORF2/3 boundary in noroviruses. Two RNA elements at the end of ORF2 that allow TTR to occur have recently been described (8, 12). They have been named translational upstream ribosome binding sites (TURBS), and the presence of both is essential for TTR. While the sequence of the second TURBS is highly variable, preventing identification based on sequence analysis, the sequence of the first TURBS, which is typically found approximately 30 to 60 nucleotides upstream of ORF3, has a conserved motif that is speculated to base pair with the 18S ribosomal subunit. Interestingly, examination of the nucleotide sequence at the end of ORF1 of JV identified a putative TURBS motif that was appropriately positioned with respect to the start of ORF2 (Fig. 3a). Therefore, a further Jena construct, JV(TURBSko), was generated; in this construct, the TURBS motif was disrupted through introduction of a conservative, nonsynonymous mutation within ORF1 (Fig. 1a and 3b). Transduction of cells with this construct resulted in expression of an intact ORF1 locus, based on Western blot analysis (Fig. 2a, lane 5). However, the mutation introduced into the TURBS1 motif resulted in a loss of capsid expression (Fig. 2a, lane 5), indicating that capsid expression was due to translational termination/reinitiation.
FIG. 3.
The upper panel (a) shows the sequence of the 18S ribosome site proposed to interact with the calicivirus TURBS1 motif, with the five core nucleotides thought to be critical for this interaction underlined. Underneath are experimentally determined functional TURBS1 motifs from feline calicivirus and rabbit hemorrhagic disease virus (RHDV) as well as TURBS1-like sequences found at the boundaries between ORF1/2 and ORF2/3 for three different genogroups of norovirus. Nucleotides highlighted in bold indicate the ability to hydrogen bond with 18S rRNA. The distances between these motifs and the downstream ORF start codon are also provided. The lower panel (b) details codon usage at the possible TURBS1 site within JV and the changes introduced at this location in order to disrupt it.
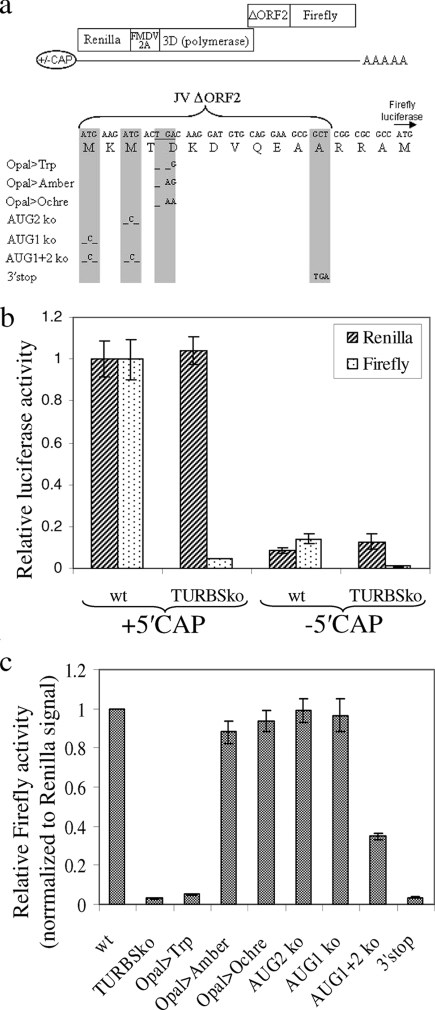
To further substantiate this hypothesis, additional T7-based constructs were generated. These contained both Renilla and firefly luciferase sequences separated by a JV ORF1/2 encoding region such that the two luciferases were expressed in-frame with ORF1 and ORF2, respectively, and served as a marker for ORF translation (Fig. 4a). When transfected into cells as capped RNA transcripts, both Renilla and firefly luciferase activities could be detected (Fig. 4b). However, when the same mutation used to disrupt the TURBS1 motif in JV(TURBko) was present, firefly activity relative to Renilla activity was reduced by ∼95%, reconfirming the importance of this motif for ORF2 expression. More importantly, when cells were transfected with noncapped versions of the same RNAs, the activities of both Renilla and firefly luciferases fell by equivalent amounts, demonstrating that ORF2 expression is directly dependent on cap-dependent translation of ORF1.
FIG. 4.
(a) Diagram summarizing the structure of RNA transcripts used to examine ORF1-dependent ORF2 expression from JV-derived sequences where the putative TURBS1 motif was either maintained or disrupted. The foot-and-mouth disease virus (FMDV) 2A sequence was included to allow separation of Renilla luciferase from norovirus 3D(polymerase). Details of the ΔORF2/firefly luciferase fusion are also provided, as are the additional nucleotide changes introduced into the TURBS1-containing construct to examine the importance of the stop and start codons at this boundary. (b) Luciferase results from transfecting 293T cells with 5 μg of the above-described RNA transcripts, synthesized either with or without a 5′ cap. Renilla and firefly luciferase activities have been plotted as relative values compared to the activity seen in cells transfected with the 5′ cap JV(wt) transcript. wt, wild type. (c) Relative firefly activities from the constructs containing mutations at the stop and start codons were determined after transfection of 293T cells with 2 μg of RNA transcripts, the results of which have been normalized to the Renilla signal obtained from the wt construct. Both graphs show the means ± standard errors of the means from three separate experiments.
Characterization of TTR between ORF1 and ORF2.
The characteristic features of previously characterized TTR events in caliciviruses are that they require a correctly positioned stop codon in the upstream ORF but that the AUG start codon in the downstream ORF is at least partially dispensable (8, 11). To investigate whether the TTR event between JV ORF1 and ORF2 behaved in a comparable fashion, a series of mutations were introduced into the above-described luciferase-based construct (Fig. 4a). Disruption of the ORF1 stop codon by a single nucleotide change to generate a Trp codon (lane Opal>Trp), which extended ORF1 by a further 38 codons, reduced firefly expression by 95% (Fig. 4c). However, this was restored by an additional nucleotide change that reintroduced the stop codon at the end of ORF1 (lane Opal>Amber) or by an exchange of the original stop codon for an Ochre stop codon (lane Opal>Ochre), demonstrating that the location of this stop codon is indeed important for ORF2 expression. In contrast, the two potential AUG start codons in ORF2 were less influential in determining whether firefly expression occurred. Disruption of either of them individually did not affect firefly expression, and when both were disrupted, a significant albeit reduced level of ORF2 translation was still observed, confirming that AUG-independent translation initiation indeed contributes to ORF2 protein production. To exclude the possibility that this residual expression was due to AUG-dependent initiation elsewhere, a stop codon was introduced at the end of the ORF2 sequence (lane 3′stop). Firefly expression from this construct was effectively reduced to levels comparable to that of the TURBSko construct, demonstrating that AUG-independent translation initiation was occurring. Therefore, the features of the TTR event at the JV ORF1/2 boundary are essentially identical to those of other caliciviruses.
In summary, the experimental results demonstrate that TTR is an additional novel mechanism that genogroup 3 noroviruses use to express capsid. While sequence analysis suggests that a putative TURBS1 motif might also exist at the end of ORF1 for other genogroups (Fig. 3a), experimental data suggest the opposite, at least for genogroup 2 viruses (see Fig. S1 in the supplemental material). Possible advantages that might be gained from the use of TTR by genogroup 3 noroviruses are unclear, as translation of ORF2 from the viral genome is unlikely to make a significant contribution to overall levels of capsid expression in an infected cell since expression of the same protein from subgenomic RNA transcripts is likely to be more effective. However, other genera of the Caliciviridae family, comprising sapoviruses and lagoviruses, also achieve capsid expression independently of subgenomic RNA production by having the nonstructural-protein- and capsid-encoding regions contained within the same ORF. Therefore, it is tempting to speculate that in some caliciviruses, capsid plays a role beyond that of a structural protein, which is critical for earlier steps in the virus life cycle occurring before subgenomic RNA production, and the existence of TTR between ORF1 and ORF2 in genogroup 3 noroviruses reflects this.
Supplementary Material
Acknowledgments
This work was supported by a pump-priming fund provided by Southampton School of Medicine to C.J.M. and by Welcome Trust Grant 069233.
Footnotes
Published ahead of print on 25 June 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Clarke, I. N., and P. R. Lambden. 1997. The molecular biology of caliciviruses. J. Gen. Virol. 78291-301. [DOI] [PubMed] [Google Scholar]
- 2.Daughenbaugh, K. F., C. S. Fraser, J. W. Hershey, and M. E. Hardy. 2003. The genome-linked protein VPg of the Norwalk virus binds eIF3, suggesting its role in translation initiation complex recruitment. EMBO J. 222852-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goodfellow, I., Y. Chaudhry, I. Gioldasi, A. Gerondopoulos, A. Natoni, L. Labrie, J. F. Laliberte, and L. Roberts. 2005. Calicivirus translation initiation requires an interaction between VPg and eIF 4 E. EMBO Rep. 6968-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herbert, T. P., I. Brierley, and T. D. Brown. 1996. Detection of the ORF3 polypeptide of feline calicivirus in infected cells and evidence for its expression from a single, functionally bicistronic, subgenomic mRNA. J. Gen. Virol. 77123-127. [DOI] [PubMed] [Google Scholar]
- 5.Herbert, T. P., I. Brierley, and T. D. Brown. 1997. Identification of a protein linked to the genomic and subgenomic mRNAs of feline calicivirus and its role in translation. J. Gen. Virol. 781033-1040. [DOI] [PubMed] [Google Scholar]
- 6.Hutson, A. M., R. L. Atmar, and M. K. Estes. 2004. Norovirus disease: changing epidemiology and host susceptibility factors. Trends Microbiol. 12279-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu, B. L., P. R. Lambden, H. Gunther, P. Otto, M. Elschner, and I. N. Clarke. 1999. Molecular characterization of a bovine enteric calicivirus: relationship to the Norwalk-like viruses. J. Virol. 73819-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luttermann, C., and G. Meyers. 2007. A bipartite sequence motif induces translation reinitiation in feline calicivirus RNA. J. Biol. Chem. 2827056-7065. [DOI] [PubMed] [Google Scholar]
- 9.McCormick, C. J., L. Challinor, A. Macdonald, D. J. Rowlands, and M. Harris. 2004. Introduction of replication-competent hepatitis C virus transcripts using a tetracycline-regulable baculovirus delivery system. J. Gen. Virol. 85429-439. [DOI] [PubMed] [Google Scholar]
- 10.McCormick, C. J., D. J. Rowlands, and M. Harris. 2002. Efficient delivery and regulable expression of hepatitis C virus full-length and minigenome constructs in hepatocyte-derived cell lines using baculovirus vectors. J. Gen. Virol. 83383-394. [DOI] [PubMed] [Google Scholar]
- 11.Meyers, G. 2003. Translation of the minor capsid protein of a calicivirus is initiated by a novel termination-dependent reinitiation mechanism. J. Biol. Chem. 27834051-34060. [DOI] [PubMed] [Google Scholar]
- 12.Meyers, G. 2007. Characterization of the sequence element directing translation reinitiation in RNA of the calicivirus rabbit hemorrhagic disease virus. J. Virol. 819623-9632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rohayem, J., I. Robel, K. Jager, U. Scheffler, and W. Rudolph. 2006. Protein-primed and de novo initiation of RNA synthesis by norovirus 3Dpol. J. Virol. 807060-7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vázquez, A. L., J. M. Alonso, and F. Parra. 2000. Mutation analysis of the GDD sequence motif of a calicivirus RNA-dependent RNA polymerase. J. Virol. 743888-3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ward, V. K., C. J. McCormick, I. N. Clarke, O. Salim, C. E. Wobus, L. B. Thackray, H. W. Virgin, and P. R. Lambden. 2007. Recovery of infectious murine norovirus using pol II-driven expression of full-length cDNA. Proc. Natl. Acad. Sci. USA 10411050-11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wirblich, C., H. J. Thiel, and G. Meyers. 1996. Genetic map of the calicivirus rabbit hemorrhagic disease virus as deduced from in vitro translation studies. J. Virol. 707974-7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wobus, C. E., S. M. Karst, L. B. Thackray, K. O. Chang, S. V. Sosnovtsev, G. Belliot, A. Krug, J. M. Mackenzie, K. Y. Green, and H. W. Virgin. 2004. Replication of Norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2e432. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.