Abstract
Unlike other class I viral fusion proteins, spike proteins on severe acute respiratory sydrome coronavirus virions are uncleaved. As we and others have demonstrated, infection by this virus depends on cathepsin proteases present in endosomal compartments of the target cell, suggesting that the spike protein acquires its fusion competence by cleavage during cell entry rather than during virion biogenesis. Here we demonstrate that cathepsin L indeed activates the membrane fusion function of the spike protein. Moreover, cleavage was mapped to the same region where, in coronaviruses carrying furin-activated spikes, the receptor binding subunit of the protein is separated from the membrane-anchored fusion subunit.
The spike (S) protein of the enveloped coronaviruses (CoV) belongs to the class I viral fusion proteins (3), which, upon virion-cell attachment, mediate virus entry through an extensive refolding process that involves the formation of a coiled-coil structure and results in the fusion of the viral and target cell membranes. Crucial to the refolding process of class I fusion proteins is the proteolytic activation of their fusogenic potential by cellular, usually furin-like proteases, encountered during biogenesis in the exocytic pathway. Proteolysis occurs N-proximal to the fusion peptide (Fig. 1), allowing the projection of this peptide toward the target cell membrane in a spring-loaded manner. The S proteins of many CoV, such as that of murine hepatitis virus strain A59 (MHV-A59) (7), are cleaved by furin-like proteases, generating a receptor binding unit (S1) and a C-terminal membrane-anchored fusion unit (S2). However, in contrast to that of class I fusion proteins, this cleavage occurs ∼200 residues upstream of the predicted fusion peptide (2, 5) (Fig. 1). Remarkably, in the case of MHV-A59, this furin-mediated cleavage of the spikes appeared not to be essential for virus entry (7). Moreover, in many CoV, including the CoV responsible for severe acute respiratory syndrome (SARS), furin-mediated cleavage of the spikes appeared not to occur (17), leaving a conundrum as to whether and how these proteins are proteolytically primed for fusion. We and others recently demonstrated that SARS-CoV or SARS-CoV spike protein-pseudotyped retroviruses utilize the enzymatic activity of endosomal cathepsin L protease for viral entry, suggesting the possibility of proteolytic activation of the spike protein in the endosomal route during cell entry rather than during cell exit (10, 16). However, no actual cleavage of the S protein by the cathepsin enzyme has been demonstrated yet. In this note we examine whether, and at which position in the SARS-CoV spike protein, cathepsin L proteolytically activates the fusion function.
FIG. 1.
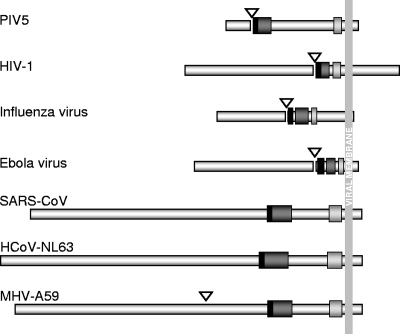
Schematic diagrams of the fusion proteins of parainfluenza virus 5 (PIV5; formerly known as simian virus 5), human immunodeficiency virus type 1 (HIV-1), influenza virus, Ebola virus, SARS-CoV (strain BJ01), MHV-A59, and HCoV-NL63. Filled bars represent the fusion peptide. Dark and light shaded bars, HR1 and HR2 regions, respectively. Open arrowheads indicate the positions of the furin cleavage sites. The fusion proteins are C-terminally anchored in the viral membrane (long vertical shaded bar).
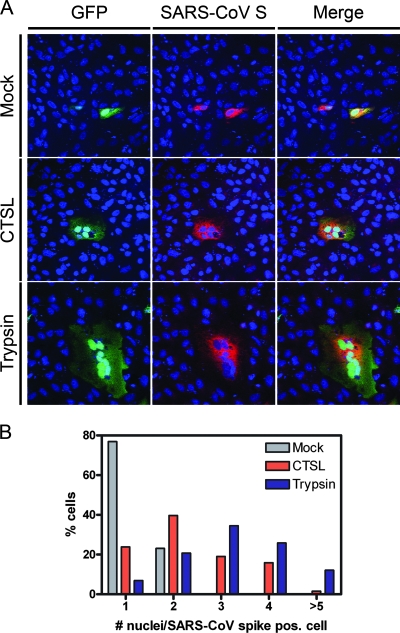
We first investigated whether the fusogenic potential of the SARS-CoV spike protein could be activated by cathepsin L protease in a cell-based system with recombinantly expressed SARS-CoV spike. Vero cells were cotransfected with the pTSh vector containing the human codon-optimized gene encoding the SARS-CoV spike (strain BJ01; a kind gift of H. Deng [13]) and the pEGFP-N1 vector (Clontech). Twenty-four hours after transfection, cells were either mock treated (phosphate-buffered saline [PBS]-Ca2+/Mg2+ [PBS+], pH 6.0) or treated with cathepsin L protease (2 μg/ml in PBS+, pH 6.0; Calbiochem) for 20 min at 37°C, after which the culture medium was replaced with Dulbecco's modified Eagle medium. As a control we used l-1-tosylamide-2-phenylethyl chloromethyl ketone (TPCK)-treated trypsin (2 μg/ml in PBS+, pH 7.4; Sigma), which has been shown to induce SARS-CoV spike-mediated cell-cell fusion (12, 17). Four hours later, cells were fixed, and S protein expression was visualized using a polyclonal antiserum against SARS-CoV S peptide (AZ1; residues 27 to 51; a kind gift of L. A. Babiuk) by confocal laser scanning microscopy (Fig. 2A). Cell-cell fusion was detected in SARS-CoV spike protein-expressing cells treated with cathepsin L protease at pH 6 by the presence of multiple syncytia. Cytoplasmic content mixing among the fused cells was evident by the occurrence of green fluorescent protein (GFP) throughout the syncytia. The extent of syncytium formation was semiquantified by counting the number of nuclei in the S protein-expressing cells (63 cells) (Fig. 2B). Cell-cell fusion was also elicited by trypsin proteolysis at neutral pH. Mock-treated cells remained largely mononucleated, indicating that the cathepsin L protease activity, not the slightly acidic buffer condition, was responsible for the cell fusion. Apparently, cathepsin L proteolysis activates the fusion potential of the spike protein.
FIG. 2.
(A) Confocal fluorescence microscopy of Vero cells transfected with plasmids expressing the SARS-CoV spike or GFP gene and either mock treated or treated with cathepsin L protease (CTSL; 2 μg/ml) or trypsin (2 μg/ml). Nuclei (blue) were labeled with ToPro-3 (Molecular Probes). (B) Semiquantitation of syncytium formation. The number of nuclei in SARS-CoV spike protein-expressing cells/syncytia was counted in mock-treated, CTSL (2 μg/ml)-treated, and trypsin (2 μg/ml)-treated Vero cell cultures. pos., positive.
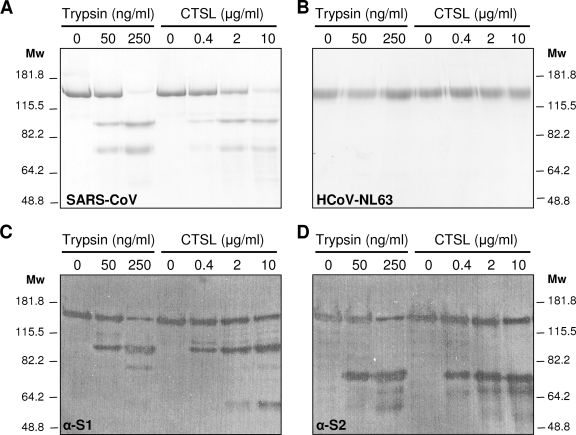
To examine the proteolysis of the SARS-CoV spike protein by the cathepsin L protease, we expressed the S ectodomain (SARS-Se; residues 14 to 1193) in stably transfected Drosophila S2 cell lines (Invitrogen); the protein was flanked by an N-terminal BiP signal sequence, a C-terminal GCN4 domain facilitating its trimerization, and a Strep tag for affinity purification (IBA GmbH). Likewise, we expressed the S protein ectodomain of another human coronavirus, human coronavirus strain NL63 (HCoV-NL63) (NL63-Se; residues 16 to 1294; constructed from pcDNA3.1-HCoV-NL63, a kind gift of H. Choe [10]). Like SARS-CoV, HCoV-NL63 uses the ACE2 receptor for cell entry and carries uncleaved spikes on its virions, but it is independent of cathepsin L activity for its entry into cells (10). SARS-Se and NL63-Se were purified from the culture medium using Strep-Tactin affinity chromatography (IBA GmbH) and gel filtration chromatography. Five micrograms of each protein was incubated with either TPCK-trypsin (0, 50, and 250 ng/ml in PBS [pH 7.4]) or cathepsin L protease (0, 0.4, 2, or 10 μg/ml in PBS [pH 5.5]) for 1 h at room temperature and then analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Treatment of SARS-Se with cathepsin L protease yielded two distinct fragments with apparent molecular masses of 90 kDa and 70 kDa by SDS-PAGE, similar to the two fragments observed after trypsin treatment (Fig. 3A) (11). Cathepsin L cleavage of SARS-Se was also observed at pH 5 and 6 but not upon incubation at a neutral pH (pH 7.4) (data not shown), consistent with the known acid preference of the enzyme's autoactivation (9, 18). NL63-Se was not cleaved by either cathepsin L or trypsin (Fig. 3B), in accordance with the independence of HCoV-NL63 of cathepsin L protease activity during cell entry, which we observed previously (10).
FIG. 3.
Cathepsin L (CTSL) cleavage of the SARS-CoV spike protein. Purified trimeric spike ectodomains of SARS-CoV and HCoV-NL63 were incubated with different concentrations of CTSL or TPCK-treated trypsin for 30 min at room temperature. (A and B) Samples were analyzed by SDS-PAGE and stained with GelCode Blue reagent. (C and D) SARS-CoV samples were also subjected to Western blot analysis using antibodies recognizing the N-terminal part (α-S1) or the C-terminal part (α-S2) of the SARS-CoV spike ectodomain, respectively. The positions of molecular weight (Mw) marker proteins (in thousands) are indicated alongside the gels and blots.
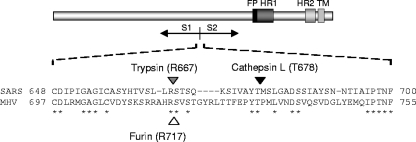
The identities of the two proteolytic SARS-CoV spike fragments were investigated by Western blot analysis using two polyclonal anti-peptide sera specific to sequences within the N-terminal (AZ1; residues 27 to 51) or C-terminal (AZ4; residues 1122 to 1142; a kind gift of L. A. Babiuk) part of the SARS-CoV spike protein. The results confirmed that the 90-kDa and 70-kDa polypeptides correspond to the S1 and S2 domains, respectively (Fig. 3C and D). To precisely map the cathepsin L cleavage site, the proteolytic fragments were electroblotted onto a Sequiblot polyvinylidene difluoride membrane (Bio-Rad) and stained with GelCode Blue (Pierce), and the 70 kDa polypeptide was excised and subjected to N-terminal sequencing using Edman degradation (Cambridge Peptides Ltd.). Interestingly, the cathepsin L cleavage site was found to map to the region of the S protein where in other coronavirus S proteins the furin cleavage site occurs (Fig. 4). Processing of the SARS-CoV S protein by cathepsin L hence gives rise to the characteristic S1 and S2 domains The location of the cathepsin cleavage site was mapped 11 residues downstream of a previously identified trypsin cleavage site (11), consistent with the similar sizes of the products obtained with the two enzymes (Fig. 3).
FIG. 4.
Sequence alignment of the SARS-CoV (strain BJ01; GenBank accession no. AY278488) and MHV-A59 (primary accession no. P11224) spike proteins at the S1-S2 junction. Identical residues are asterisked. The filled, shaded, and open arrowheads indicate the cathepsin L, trypsin, and furin cleavage sites, respectively, with their cleavage positions given in parentheses.
Our observations demonstrate for the first time directly that cathepsin L cleavage activates the SARS-CoV spike protein for membrane fusion. They also demonstrate that this cleavage does not occur at or near the putative fusion peptide but in an apparently exposed protein domain some 200 residues upstream, with no such hydrophobic peptide in the immediate vicinity. Hence, our results seem to rule out the possibility that the fusion activation of coronavirus spikes, not just of SARS-CoV but possibly even of CoV carrying furin-cleaved S proteins, might still be triggered at a very late stage by the proteolytic liberation of the fusion peptide. This feature sets the CoV fusion proteins distinctively apart from the other class I fusion proteins, which generate a fusion peptide at the N terminus of their fusion subunit. It is, however, consistent with the specific properties of the CoV fusion peptides, which share remarkable similarities with the internally located fusion peptides of class II fusion proteins (2).
Cathepsin L cleavage of the SARS-CoV spike protein supposedly occurs at a post-receptor-binding stage during virus entry. It would thus be conceivable that receptor interaction uncovers additional, previously cryptic cathepsin L target sites. However, we did not observe any difference between cathepsin L proteolysis of SARS-CoV spikes bound to the ACE2 receptor and that of unbound spikes (data not shown). This is in agreement with recent cryoelectron microscopic findings showing that ACE2 receptor interaction induces just a minimal conformational change in the SARS-CoV spike (1). Cleavage of the SARS-CoV spike protein brings the protein to a metastable state, thereby allowing the structural transitions that are instrumental to the membrane fusion process. It has been reported that exogenous trypsin cleavage of trimeric spikes at the S1/S2 border resulted in a gradual separation of S1 from the S2 subunit, which in turn resulted in S2 aggregation. The S2 aggregates appeared as rosettes under the electron microscope (8), reminiscent of the postfusion structures of the influenzavirus and paramyxovirus fusion proteins, which form rosettes via interaction of their fusion peptides (4, 6, 15). We therefore favor the hypothesis that cathepsin L cleavage of the SARS-CoV spike protein during cell entry is an essential trigger for the dissociation of S1 from the trimer, with the consequent release of the (internal) fusion peptide toward the target membrane (16). Such a scenario might explain the results of earlier attempts to enhance (pseudo)virion infectivity by exogenous spike cleavage or by engineering a furin cleavage site in the S protein, which may have failed due to premature loss of S1 (8, 12).
The S protein in the SARS-CoV spike apparently exposes a protease-accessible loop between the S1 and S2 subunits that can be targeted by different enzymes, such as cathepsin L, trypsin, and, under physiological conditions, local enzymes such as elastase. Consistent with the presence of such a loop, the syncytium-inducing capacity of expressed SARS-CoV S protein was dramatically enhanced after the introduction of a functional furin cleavage site in the S1/S2 junction region (8, 14). No such loop has been detected yet in spikes of HCoV-NL63 or of most other subgroup 1 coronaviruses. The resistance of the HCoV-NL63 spike ectodomain to cleavage by cathepsin L and trypsin that we observed here is in agreement with our inability to inhibit infection with HCoV-NL63 or an NL63 spike protein-pseudotyped retrovirus by using a broad spectrum of protease inhibitors (10). It is conceivable that during its low-pH-independent cell entry, HCoV-NL63 uses an alternative, possibly nonproteolytic way to activate the fusion function.
Acknowledgments
We thank B. Tummers and V. Bourgonje for technical assistance in the beginning and throughout the project, respectively. R. Wubbolts and A. M. de Graaf are gratefully acknowledged for assistance with the confocal laser scanning microscope. We thank H. Deng (Department of Cell Biology and Genetics, College of Life Sciences, Peking University, Beijing, China) and H. Choe (Pulmonary Division, Children's Hospital, Harvard Medical School, Boston, MA) for providing the plasmids containing the human codon-optimized SARS-CoV and HCoV-NL63 spike genes, respectively, and we thank L. A. Babiuk (Vaccine and Infectious Disease Organization, University of Saskatchewan, Saskatoon, Canada) for providing the polyclonal peptide sera directed to the SARS-CoV S glycoprotein. We thank R. A. P. Romijn (U-Protein Express B.V., Utrecht, The Netherlands) for assistance with the purification and analyses of the spike ectodomains.
B.J.B. is supported by VENI grant 016.062.027 from The Netherlands Organization for Scientific Research (NWO).
Footnotes
Published ahead of print on 18 June 2008.
REFERENCES
- 1.Beniac, D. R., S. L. Devarennes, A. Andonov, R. He, and T. F. Booth. 2007. Conformational reorganization of the SARS coronavirus spike following receptor binding: implications for membrane fusion. PLoS ONE 2e1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bosch, B. J., B. E. Martina, R. Van Der Zee, J. Lepault, B. J. Haijema, C. Versluis, A. J. Heck, R. De Groot, A. D. Osterhaus, and P. J. Rottier. 2004. Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc. Natl. Acad. Sci. USA 1018455-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosch, B. J., R. van der Zee, C. A. de Haan, and P. J. Rottier. 2003. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 778801-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calder, L. J., L. Gonzalez-Reyes, B. Garcia-Barreno, S. A. Wharton, J. J. Skehel, D. C. Wiley, and J. A. Melero. 2000. Electron microscopy of the human respiratory syncytial virus fusion protein and complexes that it forms with monoclonal antibodies. Virology 271122-131. [DOI] [PubMed] [Google Scholar]
- 5.Chambers, P., C. R. Pringle, and A. J. Easton. 1990. Heptad repeat sequences are located adjacent to hydrophobic regions in several types of virus fusion glycoproteins. J. Gen. Virol. 713075-3080. [DOI] [PubMed] [Google Scholar]
- 6.Connolly, S. A., G. P. Leser, H. S. Yin, T. S. Jardetzky, and R. A. Lamb. 2006. Refolding of a paramyxovirus F protein from prefusion to postfusion conformations observed by liposome binding and electron microscopy. Proc. Natl. Acad. Sci. USA 10317903-17908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Haan, C. A., K. Stadler, G. J. Godeke, B. J. Bosch, and P. J. Rottier. 2004. Cleavage inhibition of the murine coronavirus spike protein by a furin-like enzyme affects cell-cell but not virus-cell fusion. J. Virol. 786048-6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Follis, K. E., J. York, and J. H. Nunberg. 2006. Furin cleavage of the SARS coronavirus spike glycoprotein enhances cell-cell fusion but does not affect virion entry. Virology 350358-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Honey, K., and A. Y. Rudensky. 2003. Lysosomal cysteine proteases regulate antigen presentation. Nat. Rev. Immunol. 3472-482. [DOI] [PubMed] [Google Scholar]
- 10.Huang, I. C., B. J. Bosch, F. Li, W. Li, K. H. Lee, S. Ghiran, N. Vasilieva, T. S. Dermody, S. C. Harrison, P. R. Dormitzer, M. Farzan, P. J. Rottier, and H. Choe. 2006. SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J. Biol. Chem. 2813198-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li, F., M. Berardi, W. Li, M. Farzan, P. R. Dormitzer, and S. C. Harrison. 2006. Conformational states of the severe acute respiratory syndrome coronavirus spike protein ectodomain. J. Virol. 806794-6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuyama, S., M. Ujike, S. Morikawa, M. Tashiro, and F. Taguchi. 2005. Protease-mediated enhancement of severe acute respiratory syndrome coronavirus infection. Proc. Natl. Acad. Sci. USA 10212543-12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nie, Y., P. Wang, X. Shi, G. Wang, J. Chen, A. Zheng, W. Wang, Z. Wang, X. Qu, M. Luo, L. Tan, X. Song, X. Yin, M. Ding, and H. Deng. 2004. Highly infectious SARS-CoV pseudotyped virus reveals the cell tropism and its correlation with receptor expression. Biochem. Biophys. Res. Commun. 321994-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiu, Z., S. T. Hingley, G. Simmons, C. Yu, J. Das Sarma, P. Bates, and S. R. Weiss. 2006. Endosomal proteolysis by cathepsins is necessary for murine coronavirus mouse hepatitis virus type 2 spike-mediated entry. J. Virol. 805768-5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruigrok, R. W., A. Aitken, L. J. Calder, S. R. Martin, J. J. Skehel, S. A. Wharton, W. Weis, and D. C. Wiley. 1988. Studies on the structure of the influenza virus haemagglutinin at the pH of membrane fusion. J. Gen. Virol. 692785-2795. [DOI] [PubMed] [Google Scholar]
- 16.Simmons, G., D. N. Gosalia, A. J. Rennekamp, J. D. Reeves, S. L. Diamond, and P. Bates. 2005. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. USA 10211876-11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simmons, G., J. D. Reeves, A. J. Rennekamp, S. M. Amberg, A. J. Piefer, and P. Bates. 2004. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. USA 1014240-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turk, B., D. Turk, and V. Turk. 2000. Lysosomal cysteine proteases: more than scavengers. Biochim. Biophys. Acta 147798-111. [DOI] [PubMed] [Google Scholar]