Abstract
The human polynucleotide cytidine deaminases APOBEC3G (hA3G) and APOBEC3F (hA3F) are antiviral restriction factors capable of inducing extensive plus-strand guanine-to-adenine (G-to-A) hypermutation in a variety of retroviruses and retroelements, including human immunodeficiency virus type 1 (HIV-1). They differ in target specificity, favoring plus-strand 5′GG and 5′GA dinucleotide motifs, respectively. To characterize their mutational preferences in detail, we analyzed single-copy, near-full-length HIV-1 proviruses which had been hypermutated in vitro by hA3G or hA3F. hA3-induced G-to-A mutation rates were significantly influenced by the wider sequence context of the target G. Moreover, hA3G, and to a lesser extent hA3F, displayed clear tetranucleotide preference hierarchies, irrespective of the genomic region examined and overall hypermutation rate. We similarly analyzed patient-derived hypermutated HIV-1 genomes using a new method for estimating reference sequences. The majority of these, regardless of subtype, carried signatures of hypermutation that strongly correlated with those induced in vitro by hA3G. Analysis of genome-wide hA3-induced mutational profiles confirmed that hypermutation levels were reduced downstream of the polypurine tracts. Additionally, while hA3G mutations were found throughout the genome, hA3F often intensely mutated shorter regions, the locations of which varied between proviruses. We extended our analysis to human endogenous retroviruses (HERVs) from the HERV-K(HML2) family, finding two elements that carried clear footprints of hA3G activity. This constitutes the most direct evidence to date for hA3G activity in the context of natural HERV infections, demonstrating the involvement of this restriction factor in defense against retroviral attacks over millions of years of human evolution.
Human immunodeficiency virus type 1 (HIV-1) infection is characterized by the development of considerable genetic variation in the viral population and continuous evolution and adaptation of the virus to its host (4, 9). This variation results from a combination of high viral replication rates, large viral population sizes, and the inherent infidelity of the viral reverse transcriptase (RT), as well as recombination, and is driven by various selective pressures in the infected host (62). Mutations may additionally be induced in HIV-1 proviruses by members of the APOBEC3 (apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3, or hA3) family of human cytidine deaminases, which form part of the innate antiviral defense system and are capable of specifically inducing plus-strand guanine-to-adenine (G-to-A) mutations (6, 26, 44, 47, 52, 66, 90, 92). As these mutations usually occur at a very high frequency in affected sequences, they are collectively termed hypermutation and typically result in viral inactivation (79).
hA3G and hA3F are the most thoroughly investigated members of the hA3 family; both exhibit potent anti-HIV-1 activity (6, 47, 66, 83, 88, 92) and are expressed at high levels in lymphocytes, the major target cells for HIV-1 infection (47, 83). The activity of these proteins is counteracted by the HIV-1 accessory protein Vif, which prevents hA3 incorporation into virions during assembly by targeting them for degradation through the ubiquitin-proteasome pathway (15, 35, 48, 53, 67, 87). In the absence of Vif, hA3 proteins become incorporated into progeny virions in an infected cell, and when such a virion subsequently infects another cell, they act to restrict viral replication (26, 66).
The hA3 proteins target the single-stranded minus-strand DNA intermediate of the HIV-1 reverse transcription reaction, which results in extensive cytosine-to-uracil (C-to-U) deamination (26, 44, 52, 73, 86, 90). Minus-strand C-to-U mutations subsequently become fixed as plus-strand G-to-A changes; reporting of hypermutation conventionally makes reference to changes occurring on the plus strand. In addition, hA3 proteins might also restrict HIV-1 replication through hypermutation-independent mechanisms (5, 24, 25, 28, 31, 46, 50, 54, 57, 59, 64, 85). The presence of proviruses carrying G-to-A hypermutations in sequence sets derived from natural infections suggests that hA3 proteins occasionally circumvent HIV-1 Vif in vivo (32, 38, 40, 78).
Previous in vitro analyses of both hypermutated subgenomic HIV-1 fragments and non-HIV-1 sequences have identified some sequence preferences for hA3G and hA3F cytidine deamination; they preferentially cause G-to-A mutations at plus-strand 5′GG or 5′GA dinucleotide motifs, respectively (target nucleotide underlined) (1, 6, 13, 26, 47, 73, 86, 90). Furthermore, hA3G has been shown to favor 5′TGGG and disfavor 5′nGGC contexts, while hA3F preferentially causes mutations at 5′WGAA (W equals A/T) motifs (1, 6, 13, 47, 73, 86). The preference of hA3G for 5′TGG-to-5′TAG (tryptophan to stop codon) mutations explains why hypermutation commonly results in premature truncation of viral proteins. In addition, several recent studies have presented results suggesting that, at the genome-wide level, hA3G induces twin gradients of hypermutation, increasing from the central and 3′ polypurine tracts (cPPT and 3′PPT) (60, 72, 84, 86). Second-strand synthesis during reverse transcription is initiated from these motifs, and hypermutation is thought to be most intense in the regions furthest from them, which are exposed as single-stranded DNA substrates for the longest times (72, 86).
To characterize hA3G and hA3F mutational preferences in greater detail, we analyzed sets of near-full-length HIV-1 sequences that had been hypermutated by either hA3G or hA3F in single infection cycles in vitro and evaluated the local and genome-wide context preferences for each deaminase (1, 47, 86). We show that hA3G- and hA3F-induced G-to-A mutation rates are significantly influenced by the wider nucleotide context of the target G. Then, through analyzing mutation rates at different types of overlapping G-containing tetranucleotide motifs, we demonstrate that hA3G and, to a lesser extent, hA3F display clear hierarchies of tetranucleotide preferences, which are manifested irrespective of the genomic region examined and the overall hypermutation rate. By analyzing hypermutated sequences from HIV-positive patients using a novel method to generate reference sequences, we show that the majority of these carry signatures strongly correlating with those induced by hA3G in vitro. Moreover, we confirm the influence of the PPTs on the genome-wide hypermutation profiles and demonstrate that the profiles induced by hA3G and hA3F are distinct.
The hA3 family has also been demonstrated to restrict replication of other viruses and retroelements (e.g., hepatitis B virus [75]), endogenous long terminal repeat (LTR) retroelements (e.g., the murine MusD and IAP [19, 21], and yeast Ty1 retroelements [17, 65]), and non-LTR endogenous retrotransposons (e.g., Alu [8, 14] and L1 [58, 70]). Here, we analyzed whether there was evidence of hA3 activity in HERV infections. HERVs constitute approximately 5 to 8% of the human genome (41) and are assumed to have become fixed in the population following infection of germ cells and transmission to offspring (3). The most recently active HERVs belong to the HERV-K(HML2) family, of which many elements are unique to humans (2, 56). No replication-competent HERV-K(HML2) elements have been isolated; most carry multiple frameshift mutations, premature stop codons, or have undergone recombinational deletion between the two viral LTRs (76). However, active HERV-K(HML2) elements may still circulate at low frequencies in human populations (2, 3).
Several lines of evidence are consistent with a role for the A3 proteins in the innate defense against attacks by endogenous retroviruses (27, 36, 63). First, G-to-A mutations consistent with murine A3 (mA3) activity are present in proviruses from the Pmv and Mpnv subgroups of endogenous nonecotropic murine leukemia viruses (MLVs) that are fixed in the mouse genome, suggesting this deaminase may have contributed to their inactivation (34). Second, phylogenetic analysis has demonstrated that the hA3 family has been subject to extremely strong positive selection throughout primate evolution (63, 91), predating the oldest known lentiviruses (37). Third, hA3G and hA3F are expressed at high levels in testes and ovaries, where infection of germ line cells must take place for fixation of endogenous retroelements to occur (33, 77). Furthermore, recent results demonstrated that a reconstituted HERV-K(HML2) element could be inhibited by hA3F in vitro (45). Here, we find mutational footprints strongly correlating with those induced by hA3G on HIV-1 in vitro and in vivo and in two naturally occurring hypermutated HERV-K(HML2) elements. Our analysis provides the most direct evidence to date of hA3G-mediated restriction of HERVs during human evolution and may also highlight novel features of the HERV-K(HML2) replication strategy.
MATERIALS AND METHODS
PCR amplification and sequencing of proviruses hypermutated by hA3G or hA3F in vitro.
Total DNA was extracted from 293T cells infected for 24 h with the G protein of vesicular stomatitis virus (VSV-G)-pseudotyped vif-deficient HIV-1IIIB viruses produced in the presence of hA3G or hA3F, as previously described (6). Following DpnI treatment to eliminate carry-over transfection mixture, near-full-length single HIV-1 proviruses were amplified by limiting dilution nested PCR using the Advantage 2 polymerase mix (TakaraBio/Clontech, Paris, France). All primers used were designed where possible to anneal to sites lacking 5′GG or 5′GA (forward primers) or 5′CC or 5′TC (reverse primers) motifs (the preferred contexts for hA3G and hA3F activity, respectively), to reduce the potential for inefficient amplification of hypermutated viruses. When it was not possible to design a suitable primer lacking these motifs, primers were designed with the motifs restricted to the 5′ end. All PCR primer sequences are given in Table S1 of the supplemental material. First-round PCR resulted in the amplification of an 8.5-kb fragment spanning the gag-to-3′LTR region; this amplicon was used as a template for four second-round PCRs amplifying gag-to-pol, pol-to-vif, vif-to-env, and env-to-3′LTR fragments. The PCR conditions were identical for both first- and second-round PCRs: 95°C (1 min) hot start, followed by 15 cycles of 95°C denaturation (30 s), 60°C annealing (30 s), and 68°C extension (10 min), and then 20 cycles consisting of 95°C denaturation (30 s) and 68°C annealing/extension (10 min), with a final cycle of extension at 68°C (extra 10 min). Amplicons were visualized on 1% agarose gels in Tris-acetate-EDTA containing 0.4 ng/μl ethidium bromide and purified using the QIAquick PCR purification kit (Qiagen, CA); they were sequenced from both directions using the primers listed in Table S1 in the supplemental material by using the Dyedeoxy terminator sequencing system (Applied Biosystems, CA) on an Applied Biosystems 3730xl DNA analyzer. DNA reads were assembled and proofread using Pregap4 and Gap4 within the Staden package (69); sequences with multiple peaks at the same nucleotide position were assumed to represent multiple proviruses within the starting PCR mix and so were discarded. Sequences lacking a G-to-A mutation in the 3′LTR, copied from the engineered G-to-A mutation at HXB2 position 571 during reverse transcription (6), were assumed to be carry-over transfection mixture and were therefore also discarded. Sequences were aligned using a pairwise alignment algorithm with the MacClade software (51), followed by manual adjustment. The alignments generated are given in Fasta format in Fig. S1 of the supplemental material.
Analysis of hypermutated sequences.
To analyze the local nucleotide substrate preferences of hA3G/hA3F activity in a given query sequence, the numbers of (i) guanine bases, (ii) dinucleotide contexts containing guanine (5′Gn [target guanine underlined; n represents any nucleotide]), and (iii) tetranucleotide contexts containing guanine (5′Gnnn, 5′nGnn, and 5′nnGn) were determined for a relevant reference sequence. The number of these contexts carrying guanine-to-adenine (G-to-A) mutations in the query sequence were then counted, such that the proportion of each type of context carrying G-to-A mutations could be calculated. In our analysis, each G-to-A mutation was considered independently and its context was defined by the index nucleotides in the parental virus sequence. C-to-T, CC-to-CT, and TC-to-TT mutation rates were assessed in some cases to give an indication of the noise associated with certain analyses. In cases where more than a single G-to-A mutation occurred within a particular tetranucleotide (e.g., 5′GnGn to 5′AnAn), misreporting of the context of one or the other mutation was likely (but not definite), depending on which guanine was mutated first, the separation of the mutated Gs in the tetranucleotide (i.e., 5′nGGn, 5′nGnG, or 5′GnnG) and the particular tetranucleotide analysis being employed (i.e., 5′Gnnn, 5′nGnn, or 5′nnGn).
To assess the extent of potential misreporting of the contexts of G-to-A mutations in these data sets, we determined the number of mutations occurring within three nucleotides of other mutations (data not shown). The analysis showed that a maximum of approximately 12.6% of the hA3G-induced G-to-A mutations and 20.9% of hA3F-induced G-to-A mutations were potentially misreported. Eliminating tetranucleotides carrying multiple G-to-A mutations from the analysis might remove this potentially confounding factor but would create a new one, since these sites clearly constitute prime targets for hA3 activity. There is some evidence that the 5′G in a poly(G) motif is most likely to be mutated first by hA3G in vitro (13), and the apparent preference of this deaminase for 5′TGGG over 5′TGGG in our data set is consistent with this notion. This effect could potentially be modeled into the analysis, but this approach would still depend on assumptions, which may thwart the results as mentioned above, and therefore has not been carried out here. Furthermore, this discussion still assumes that each mutation does occur independently, but it is possible that a cooperative effect may operate. A second mutation may be more likely in the vicinity of a recently induced mutation.
In some experiments, data for individual sequences were pooled to summarize results and to increase statistical power. Profiles of G-to-A mutational burden across individual hypermutated genomes were generated by first counting the number of target (GG and GA) motifs within a 400-bp sliding window to the 3′ of a given base of a reference sequence (advancing in single nucleotide steps), and second, counting the number of these target motifs carrying a GG-to-AG or GA-to-AA mutation. Using these data, plots of the proportion of target motifs across hypermutated genomes were constructed.
Statistical analyses.
To assess the influence of the wider nucleotide context on G-to-A mutation rates, chi-square tests were performed. For each individual near-full-length provirus hypermutated in vitro by hA3G or hA3F, the independence of G-to-A mutation rates on the nucleotide at each position spanning the region from 100 bp upstream of the target G to 100 bp downstream was determined (chi-square test, three degrees of freedom). To identify the nucleotides in each entire data set that influenced mutation rates, the P values derived from the chi-square analyses of individual proviruses were combined using Fisher's method for combining independent tests (22). To investigate which particular nucleotides contributed to the effects, observed nucleotide frequencies relative to those expected under independence were plotted.
To determine whether the hypermutation preferences observed in one sequence or set of sequences predicted those observed in a second sequence or set of sequences, the relationship between the arrays of observed mutation rates at each relevant context in the two data sets was tested. This was assessed in two ways. First, we used Poisson regression with an identity link function, weighting errors to take into account the different number of contexts available for mutation under the response conditions (55). The goodness of fit of these regression lines was assessed using McFadden's pseudo-R2 [defined as 1 − (log likelihood of the linear model)/(log likelihood for the null model)], which accounts for the number of available target contexts. However, the P values of these regressions, as determined from a likelihood ratio test, were liberal due to the stronger influence of points where the observed mutation rate in the predictor variable was very small. Accordingly, we also tested the strength of correlation using Spearman's rank correlation test, a conservative nonparametric statistic that is robust to the misspecification of errors. For both tests, contexts where the observed mutation rate was zero (i.e., where no contexts were mutated) were excluded because such data are unsuitable for the Poisson regression analysis and since a large number of tied ranks can compromise Spearman's test.
Analysis of hypermutation in sequences derived from HIV-1-infected patients for which no parental sequence is available.
For an ideal hypermutation analysis, hypermutated sequences should be compared with their parental sequence (i.e., the sequence from the previous replication cycle). This is possible in vitro; however, in natural infections, the exact parental sequence is invariably unknown. Some previous studies have used consensus sequences derived from nonhypermutated sequences from the same patient, but no such sequences were available for the majority of hypermutated near-full-length HIV genomes in the Los Alamos Sequence database (http://www.hiv.lanl.gov/hiv-db) (32, 38, 40, 72). We therefore developed a method to improve the generation of reference sequence estimates for analysis of hypermutated sequences. Phylogenetic trees are useful for identifying closely related taxa; unfortunately, hypermutated sequences skew trees, often clustering together (due to common G-to-A mutations) and bearing long branches (due to larger numbers of mutations). To remove the skewing effect of hypermutation, sites in sequence alignments where the hA3 proteins may have recently acted (i.e., sites represented by both GG and AG or by GA and AA dinucleotide motifs) were “repaired”: at such sites, AG and AA were repaired to NG and NA, respectively. Phylogenetic trees reconstructed from such repaired sequence alignments were presumed to be minimally influenced by recent hA3 activity, since N makes no contribution to the construction of the tree, and therefore depict more genuine phylogenetic relationships. Thus, sequences closely related to the hypermutated sequence can be identified, without the skewing effect of hypermutation. This is a conservative approach for removing the influence of hA3-type mutations, yet it will also remove the signal of variation caused by other means, such as reverse transcription; however, typically no more than 20% of sequence information was lost through this approach, leaving a large amount of sequence data from which phylogenetic relationships could be inferred.
We downloaded all hypermutated and nonhypermutated sequences from a given subtype from the database, having carried out a search for complete genomes, including problematic sequences. We aligned and “repaired” the sequences as described above; neighbor-joining trees were constructed using the “repaired” alignments according to the Felstenstein 84 (F84) model of nucleotide substitution using the PAUP* software (74). A subset of sequences clustering with the hypermutated sequence was then identified and reextracted from the database. The hypermutated sequence was removed from this alignment, and the consensus nucleotide at each position was derived from the remaining nonhypermutated sequences (using a 50% majority rule as implemented by the Se-Al software [http://tree.bio.ed.ac.uk/software/seal/]) to give an estimate of a reference sequence against which the hypermutated isolate could be analyzed. The hypermutated sequence was realigned to this reference for analysis as described above. The method is limited by the genetic distance from the available neighbor taxa, which will be minimized when sequences are available from the same patient, or at least the same local epidemic. In several cases, only one or a few sequences of the same subtype are present in the database, and consequently the level of noise in such analyses may be higher.
We generated reference sequences specific for each of the hypermutated proviruses present in the Los Alamos database at the time of writing, which belonged to subtypes also represented by nonhypermutated sequences (accession numbers are listed in Table S2 in the supplemental material).
Analysis of HERV-K (HML2) sequences.
For a preliminary screen of HERV-K(HML2) proviral elements for evidence of hA3-mediated hypermutation, each proviral sequence was aligned to a consensus sequence of the one of the two major HERV-K(HML2) lineages to which it belonged, which was used as a reference sequence (3). Near-full-length proviruses spanning gag to the 3′LTR were analyzed; the 292-bp sequence at the pol-env boundary of type 2 HERV-K(HML2) isolates was omitted from the analysis (49). For each provirus, GN-to-AN mutation rates were determined, relative to the appropriate consensus sequence. Two-by-two chi-square tests for the independence of G-to-A mutation rates with respect to the presence of a purine (R = A or G) or a pyrimidine (Y = C or T) at the +1 position were carried out. HERV-K(HML2) elements for which there was evidence of dependence of mutation rates on the type of downstream nucleotide after Bonferroni correction for multiple testing (P < [0.05/n], where n = number of independent tests) were analyzed further, using the method described above for analysis of hypermutated HIV sequences from the Los Alamos database (hypermutation of HIV-1 sequences by hA3 proteins in vitro showed a marked bias for inducing mutation at GR dinucleotides, compared with GY). Elements 79c12, 74c19, 154c11, 102c6, 8c8, 2c7, K113, K103, 5c22, 172c1, 196c5, 140c3, 84c1, 3q27, 39c5, and 110c10 were used to generate a reference sequence estimate for elements 11c21 and 158c3; elements 119c9, 88c11, 83c19, and 30c19 were used to generate a reference estimate for 103c19. The chromosomal locations of the 44 HERV-K(HML2) elements included in this analysis are given in Table S3 of the supplemental material, and the alignment used in the analysis is presented in Fig. S3 of the supplemental material.
The HERV-K(HML2) tree was constructed by maximum likelihood using PAUP* 4.0b10 (74) and the GTR+Γ model of nucleotide substitution, based on an alignment of the protein-coding regions (gag to env) of the HERV-K(HML2) elements. We employed a heuristic search, starting with a neighbor-joining tree, followed by two successive rounds of branch swapping (TBR and NNI) and parameter optimization. hA3-type mutations within the hypermutated elements 11c21 and 158c3 were repaired prior to construction of the tree.
RESULTS
Sequence preferences for hA3-mediated hypermutation of HIV-1 proviruses in vitro.
To characterize the mutational preferences of hA3G and hA3F, we carried out infections of 293T cells in vitro with vif-deficient VSV-G-pseudotyped HIV-1IIIB produced in the presence of either hA3G or hA3F (6). Near-full-length HIV-1 sequences extending from gag to the 3′LTR were amplified from cell lysates using limiting dilution PCR (hA3G, 10 sequences, 83.7 kb total; hA3F, 9 sequences, 6 of which contained short gaps, 68.8 kb total). The local sequence preferences for hA3-induced mutations were determined through comparison with the known sequence of the parental virus. The vast majority of mutations observed in each sequence set were plus-strand G-to-A changes, with hA3G and hA3F preferentially mutating 5′ GG and 5′ GA dinucleotide motifs (minus-strand 5′ CC and 5′ TC), respectively (Tables 1 and 2), as previously described (1, 26, 47).
TABLE 1.
Summary of mutations induced in HIV-1 proviruses in vitro by hA3G and hA3F
| Parental base | Mutated base
|
Total available | |||
|---|---|---|---|---|---|
| A | C | G | T | ||
| hA3G | |||||
| A | 0 | 0 | 0 | 30,030 | |
| C | 0 | 0 | 4 | 14,833 | |
| G | 1,500 | 1 | 1 | 20,019 | |
| T | 1 | 2 | 0 | 18,816 | |
| hA3F | |||||
| A | 0 | 0 | 0 | 24,710 | |
| C | 0 | 0 | 2 | 12,189 | |
| G | 953 | 0 | 0 | 16,436 | |
| T | 1 | 2 | 0 | 15,492 | |
TABLE 2.
Summary of dinucleotide preferences of G-to-A mutations induced in HIV-1 proviruses in vitro by hA3G and hA3F
| Gn dinucleotide type and mutation | No. of mutations | No. of available contexts | % Contexts mutated | 95% confidence interval |
|---|---|---|---|---|
| hA3G | ||||
| Gn-to-An | 1,499 | 19,998 | 7.50 | 7.13-7.87 |
| GA-to-AA | 127 | 6,924 | 1.83 | 1.53-2.18 |
| GC-to-AC | 2 | 3,717 | 0.05 | 0.01-0.19 |
| GG-to-AG | 1,359 | 5,610 | 24.22 | 23.11-25.37 |
| GT-to-AT | 11 | 3,747 | 0.29 | 0.15-0.52 |
| hA3F | ||||
| Gn-to-An | 951 | 16,422 | 5.79 | 5.44-6.16 |
| GA-to-AA | 718 | 5,701 | 12.59 | 11.74-13.48 |
| GC-to-AC | 77 | 3,037 | 2.54 | 2.01-3.16 |
| GG-to-AG | 136 | 4,597 | 2.96 | 2.49-3.49 |
| GT-to-AT | 20 | 3,087 | 0.65 | 0.40-1.00 |
Influence of surrounding nucleotides on hA3-mediated mutation rates.
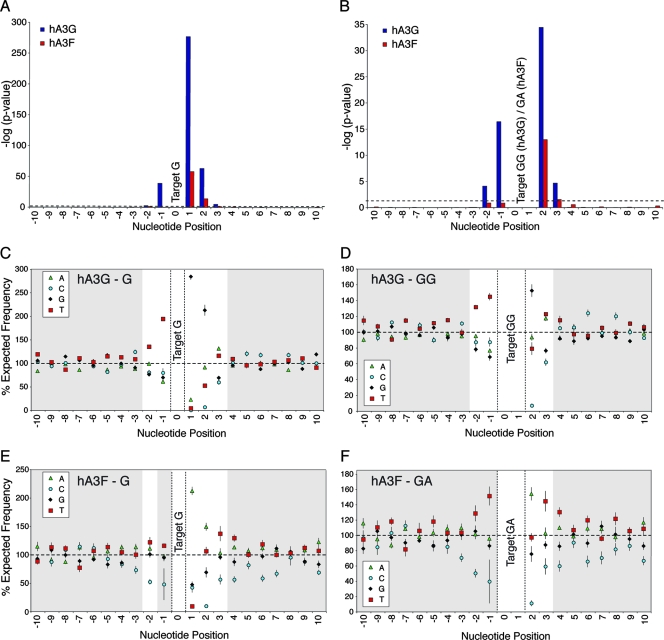
While previous studies have suggested various preferred and disfavored wider nucleotide contexts for hA3 activity, we systematically analyzed how the likelihood of observing a G-to-A mutation depended on the wider context around the target G nucleotide. We performed chi-square analyses, testing the independence of mutation frequencies on the nucleotide at each position ranging from 100 bases upstream to 100 bases downstream of the target G. Each individual hypermutated provirus was first analyzed separately; P values were subsequently combined using Fisher's method for combining independent tests (22) to obtain the overall probability of independence for each nucleotide in both the hA3G and hA3F data sets (Fig. 1A and B).
FIG. 1.
The influence of surrounding nucleotides on hA3G- and hA3F-induced G-to-A mutation rates in HIV-1 proviruses in vitro. (A and B) To analyze the influence of the surrounding nucleotides on G-to-A mutation rates, a series of chi-square analyses (with three degrees of freedom) examining the independence of mutation rates on the nucleotide at each consecutive position from 100 nucleotides downstream to 100 nucleotides upstream of the target plus-strand G (panel A), GG, or GA (panel B) motif were carried out (i.e., N(−99X)G, N(−98X)G… G(+98X)N, G(+99X)N; or N(−99X)GG… GG(+98X)N, where the target G = position 0). Only nucleotides spanning positions −10 to +10 are shown here for clarity; no significant deviations from independence were observed outside of this region in any data set. Each individual sequence mutated by hA3G or hA3F was analyzed independently; P values for each nucleotide position from each sequence were then combined using Fisher's method for combining independent tests to assess the influence of each nucleotide position for the hA3G (blue bars) and hA3F (red bars) data sets, respectively. Data represent the negative log10 of the P value; the dashed line indicates the value corresponding to P < 0.05. (C to F) The frequency of each nucleotide, relative to its expected frequency, at each position with respect to target G (C [hA3G] and E [hA3F]), target GG (D, hA3G) or target GA (F, hA3F) motifs. Data points represent the mean percentages of the expected nucleotide frequency; error bars depict the standard errors of the means; significant data are highlighted with a white background.
For hA3G, the observed mutation rates were dependent on the nucleotides spanning positions −2 to +3 relative to the target G (position 0); the most significant effect was exerted by the nucleotide at position +1, reflecting the extreme preference for 5′GG motifs (Fig. 1C). The nucleotides at positions +2 and −1 were also strong determinants of mutation frequencies, while those at +3 and −2 mediated lesser, yet still significant, effects (Fig. 1A). Similarly, GG-to-AG mutation rates were found to be dependent on the nucleotides occupying positions −2 to +3, demonstrating the importance of the wider context of the target dinucleotide on hA3G-induced mutation frequencies (Fig. 1B). hA3F-induced mutation rates depended most on the nucleotide at position +1, reflecting the preference for 5′GA motifs (Fig. 1E), and were also highly influenced by the nucleotide at position +2 (Fig. 1A). The data were less conclusive regarding the influence of the nucleotides at positions −2, −1, and +3 but were suggestive of an effect (Fig. 1A and B).
To investigate which particular nucleotides were favored or disfavored, the observed frequency of each nucleotide at each of these positions was compared to its expected frequency if mutation rates were independent of the wider nucleotide context (Fig. 1C to F). These analyses indicated that the presence of T at positions −2 and −1, G at +1 and +2, and T or A at +3 were associated with increased hA3G-induced mutation rates; in contrast, the presence of C at positions +2 and +3 and, to a lesser extent, T at +2, was associated with lower mutation rates (Fig. 1C and D). For hA3F, T at positions −2 and −1 (for which the chi-square test tended toward significance), A at +1 and +2, and T at +3 were associated with increased mutation rates, while C at positions −2, −1, +2, and +3 was associated with reduced mutation rates (Fig. 1E and F).
Local sequence preferences for hA3G- and hA3F-mediated mutation of HIV-1 proviruses in vitro.
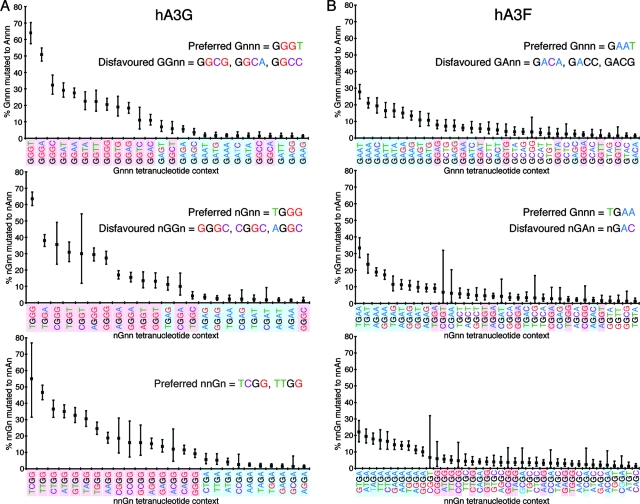
To evaluate the influence of specific combinations of nucleotides on hA3-induced deamination, we determined the mutation rates associated with overlapping G-containing tetranucleotide contexts; analysis of overlapping tetranucleotides was used to ensure the important −2 to +3 region was covered (i.e., Gnnn-to-Annn [0 to +3], nGnn-to-nAnn [−1 to +2], and nnGn-to-nnAn [−2 to +1] analysis), while retaining wide representation of different types of motif (Fig. 2A and B). Raw data for these tetranucleotide analyses (both for individual hypermutated proviruses and for the pooled data sets) are presented in Fig. S2 of the supplemental material.
FIG. 2.
Nucleotide context preferences of G-to-A mutations induced in HIV-1 proviruses by hA3G and hA3F in vitro. The tetranucleotide mutational preferences in proviral sequences, spanning gag-3′LTR, isolated from 293T cells infected with VSV-G-pseudotyped vif-deficient HIV-1IIIB generated in the presence of hA3G (A) or hA3F (B), were analyzed relative to their known parental sequence. The proportion of each type of available G-containing tetranucleotide context carrying G-to-A mutations (Gnnn-to-Annn, nGnn-to-nAnn, and nnGn-to-nnAn, respectively) was determined; these overlapping tetranucleotides covered the region spanning positions −2 to +3 relative to the target G (position 0). Data from the hA3G (10 sequences, 83.7kb) and hA3F (9 sequences, 68.8kb) sequence sets were pooled. Only tetranucleotide contexts with mutation rates greater than 1% are shown; tetranucleotides highlighted in pink and blue contain the hA3G 5′GG and hA3F 5′GA preferred dinucleotides, respectively; the target G nucleotide is black; the surrounding nucleotides are colored differently for clarity; error bars represent 95% confidence intervals based on a binomial distribution.
For both hA3G and hA3F, the most highly mutated tetranucleotide contexts contained the known target 5′GG and 5′GA dinucleotide motifs, respectively; hA3G targeted 5′TGGG motifs almost twice as frequently as any other context, and hA3F most often mutated 5′TGAA motifs. For both hA3G and hA3F, 5′GNC contexts were rarely mutated, demonstrating the marked inhibitory effect of a C at position +2; this effect was the strongest effect observed, overriding the observed beneficial effect for mutation of T at position −1 (data not shown). For hA3G, mutation frequencies at the preferred GGG motifs, and also at GGA, were enhanced by the presence of T or A at +3; the presence of a T at −2 was also favored by hA3G. For hA3F, T at −1 was generally associated with increased mutation frequencies. Together, these effects make hierarchies of nucleotide substrate preferences apparent for both hA3G and hA3F (Fig. 2A and B).
Conservation of nucleotide preference hierarchies in individual hypermutated proviruses and subgenomic fragments.
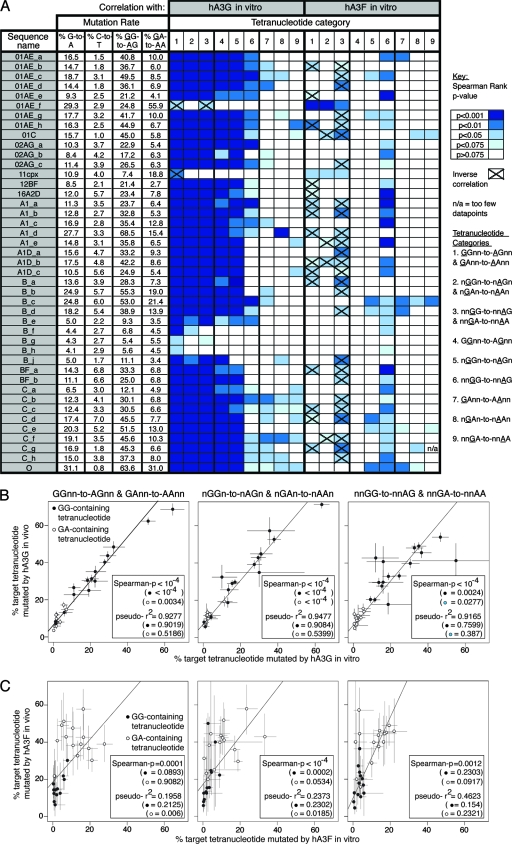
To assess whether the tetranucleotide preference hierarchies observed across the pooled in vitro data sets were highly influenced by subsets of individual proviruses, we compared the mutation preferences in the pooled data sets with those in each individual provirus. The pooled hA3G data strongly predicted the nucleotide preference hierarchy in the majority of individual sequences, even when only GG-containing tetranucleotide contexts were considered (Fig. 3A, categories 4, 5, and 6 [hA3G]). Similarly, although the association was less strong, the pooled hA3F data set predicted the mutational preference hierarchy in individual viruses mutated by hA3F, and most sequences carried analogous mutation signatures even when considering only GA-containing tetranucleotide contexts (Fig. 3A, categories 7, 8, and 9 [hA3F]). This suggests that for both deaminases, considerable substrate specificity exists beyond their preferred dinucleotide targets. Furthermore, the substrate preference hierarchies existed irrespective of the level of hypermutation in the individual sequences, although the sequences least representative of the pooled data sets tended to be those with the lowest overall levels of mutation; however, this may simply reflect a reduction in statistical power in these cases.
FIG. 3.
Conservation of tetranucleotide preference hierarchies in individual hypermutated proviruses and subgenomic fragments. (A) Correlation between the tetranucleotide preference hierarchies observed in the pooled hA3G and hA3F data sets and in each individual provirus comprising the pooled data sets. Spearman rank correlations between the arrays of mutation rates observed in individual sequences and the pooled data sets were determined, considering mutation within different categories of tetranucleotide contexts; darker shades of blue indicate more highly significant correlations; contexts with zero mutation rates were excluded, since a large number of tied ranks can compromise the Spearman's rank test. For each individual provirus, G-to-A, GG-to-AG, and GA-to-AA mutation rates are indicated. Weighted Poisson regression analyses were also carried out, yielding similar results (data not shown). (B) Correlation of tetranucleotide mutational preferences observed in different subgenomic regions of HIV-1 sequences by hA3G and hA3F in vitro. Near-full-length proviruses mutated by hA3G or hA3F were divided arbitrarily into four 2.1-kb fragments (spanning gag-pol (HXB2 1200-3325), pol-vif (HXB2 3326-5450), vif-env (HXB2 5451-7575), and env-3′LTR (HXB2 7576-9680); four additional non-full-length (env-3′LTR) sequences from the hA3G experiment and six additional non-full-length (env-3′LTR) sequences for the hA3F experiment, derived from the same infections, were added to this analysis. The correlation between the tetranucleotide substrate preferences in each fragment with that in each other fragment, for the categories of tetranucleotide context shown, was assessed using Spearman rank correlations, color coded as described above. Weighted Poisson regression analyses were also carried out, yielding similar results (data not shown). Contexts with zero mutation rates were excluded.
To elucidate whether the apparent conservation of hA3G and hA3F tetranucleotide preference hierarchies reflected general features of the deaminase activities or was an artifact of investigating hypermutation in the context of a particular viral sequence, we determined whether the hierarchies were conserved across different subgenomic regions. The hypermutated proviral sequences were arbitrarily divided into four 2.1-kb fragments spanning gag-pol, pol-vif, vif-env, and env-3′LTR, and the mutation preferences were reanalyzed in each case. For hA3G, the tetranucleotide preference hierarchy in any given fragment was still significantly correlated with those of any other when only nGGn contexts were considered, and there was a significant correlation in the majority of cases when GGnn motifs were analyzed alone (Fig. 3B). When GA-containing contexts were considered alone for hA3F, the correlations were less strong but still significant in most cases. These data demonstrate that for hA3G, and to a lesser extent hA3F, hierarchies of tetranucleotide substrate preferences exist irrespective of the sequence investigated and the overall level of mutation.
hA3 footprints in naturally occurring hypermutated HIV-1 sequences.
To determine the correlation between the tetranucleotide preferences found in vitro with those present in hypermutated sequences isolated from natural infections, we analyzed the majority of patient-derived near-full-length hypermutated proviruses in the Los Alamos database (www.hiv.lanl.gov). These belonged to an array of subtypes. A precise characterization of the nucleotide mutation preferences in these sequences is limited by the absence of relevant reference sequences for most of them. Ideally, references generated from nonhypermutated sequences isolated from the same infected individual should be used for analyzing a hypermutated variant, as in several studies of hypermutated subgenomic fragments (32, 38, 40) and a single study of a full-length O-group hypermutant (72). In the absence of such sequences, previous studies of near-full-length genomes have either used references generated from arbitrarily chosen nonhypermutated sequences from the same subtype or measures of G-to-A mutational burden which were nonspecific at the nucleotide level (39, 60, 72).
To optimize our analysis of the naturally occurring near-full-length hypermutated sequences, we developed a method to improve reference sequence estimates; briefly, we used a combination of “repairing” potential hA3-induced mutation in each sequence alignment and subsequent phylogenetic tree analysis to identify the most closely related nonhypermutated sequences, from which a consensus sequence was generated for use as a reference. Using these optimized reference sequences, we determined the genome-wide in vivo hypermutation characteristics of all near-full-length hypermutated proviruses for which nonhypermutated genomes from the same subtype were available.
The in vivo G-to-A mutation rates determined were typically higher than those observed in vitro, yet the C-to-T mutation rates were also notable. This indicates that, even after improving estimates of reference sequences as described, considerable genetic distance was still present between these reference estimates and the genuine, but unknown, parental sequences; thus, not all of the G-to-A mutations recorded were likely accounted for by hA3 activity (Fig. 4A). Nevertheless, the hierarchy of tetranucleotide preferences defined for hA3G-induced mutations in vitro strongly predicted the mutation characteristics, even when only GGnn or nGGn motifs were included in the analysis, in 38/43 (88%) of the database sequences, irrespective of the overall level of hypermutation (Fig. 4A, categories 1 to 5 [hA3G]). The tetranucleotide mutation preferences of 2/43 sequences (5%) were not predicted by the in vitro hA3G data (see Table S2, 01AE_f and 11cpx, in the supplemental material). However, the preferences in these two sequences did correlate with the hA3F in vitro preferences when considering data combined for GG- and GA-containing motifs (Fig. 4A, categories 1 to 3 [hA3F]), but not when GA-containing tetranucleotide contexts were assessed alone (Fig. 4A, categories 7 to 9 [hA3F]). Thus, the significant correlation was solely attributable to GG-containing contexts having low mutation rates in both data sets. Consequently, these two proviruses were more likely to have been mutated by hA3F than hA3G.
FIG. 4.
Correlation of tetranucleotide mutational preferences observed in hypermutated HIV-1 sequences in vivo with those observed in proviruses hypermutated by hA3G or hA3F in vitro. (A) The tetranucleotide preferences hierarchies in 43 near-full-length HIV-1 sequences marked as hypermutated in the Los Alamos HIV sequence database were determined using reference sequences estimated as described. Each sequence was assigned a name according to its subtype. Spearman rank correlations between the arrays of mutation rates observed in each individual in vivo sequence and those in the pooled data sets for proviruses hypermutated in vitro by hA3G or hA3F were determined, considering mutation within different categories of tetranucleotide contexts; darker shades of blue indicate more significant correlations. Contexts with zero mutation rates were excluded since a large number of tied ranks can compromise the Spearman's rank test; pairs of data for which a significant inverse correlation was found are indicated. For each individual provirus, G-to-A, GG-to-AG, and GA-to-AA mutation rates are indicated; C-to-T mutation rates are shown to give an indication of the noise associated with each analysis. Weighted Poisson regression analyses were also carried out, yielding similar results (data not shown). (B) The tetranucleotide preference data (with the target guanine at either position 1, 2, or 3 of the tetranucleotide) from the 38 in vivo proviruses carrying strong evidence of hA3G activity were pooled and correlated with the pooled tetranucleotide mutational preferences for proviruses hypermutated by hA3G in vitro. Each point represents a particular tetranucleotide context; GG- and GA-containing tetranucleotide contexts are represented by black filled and unfilled circles, respectively. Spearman rank correlation P values are indicated and take into consideration both the GG- and GA-containing contexts together, with the P values determined when only GG- or GA-containing tetranucleotide contexts were considered (shown in parentheses); similarly, the McFadden Pseudo-R2 statistic, a measure of the goodness of fit of the regression which accounts for the availability of each target context, is indicated. Contexts with zero mutation rates were excluded. Error bars correspond to binomial 95% confidence intervals. (C) As for panel B, the tetranucleotide preference data (with the target guanine either at position 1, 2, or 3 of the tetranucleotide) from the two in vivo proviruses potentially mutated by hA3F were pooled and correlated with the pooled tetranucleotide mutational preferences for proviruses hypermutated by hA3F in vitro.
Three subtype B proviruses, which all originated from the same patient, carried unusual hypermutation profiles and preferences (see Table S2, sequences B_f, B_g, and B_h in the supplemental material) (81). The sequences were very similar and highly hypermutated in gag and pol, but not elsewhere in the genome, in contrast to most hA3G-mutated proviruses which were hypermutated throughout the viral genome (see Fig. S5 in the supplemental material). Moreover, the tetranucleotide mutation preferences only ever correlated with those induced by hA3G in vitro when GG- and GA-containing tetranucleotide contexts were considered together and never when GG-containing contexts were analyzed alone (Fig. 4A). It is therefore less clear whether hypermutation in these sequences was hA3G mediated, and consequently they were excluded from later analyses.
We collated the tetranucleotide preference data for the 38 sequences carrying hA3G-like mutations; the in vivo hierarchies correlated strongly with those observed for hA3G activity in vitro, even when only contexts containing the hA3G GG target dinucleotide were considered (Fig. 4B). When we looked at the combined hA3F-like in vivo data set, the in vitro hA3F tetranucleotide preferences predicted the in vivo hierarchies only when both GA- and GG-containing contexts were considered, and the significance was lost when only contexts containing the hA3F GA target dinucleotide were analyzed (Fig. 4C). Thus, while the association between the in vitro and in vivo hA3F data sets was ambiguous, the hierarchy of tetranucleotide substrate preferences for hA3G activity appeared highly conserved in vitro and in vivo.
Distinct genome-wide hA3G and hA3F hypermutation profiles.
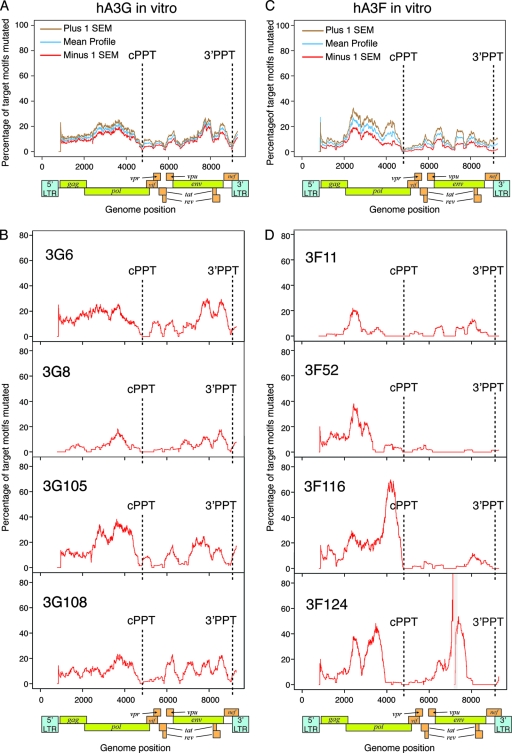
We next examined the distribution of hA3G- and hA3F-induced hypermutation across the near-full-length HIV-1 proviruses, accounting for the distribution of target dinucleotide motifs (Fig. 5; see also Fig. S4 in the supplemental material). We observed high mutation frequencies in the pol and gp41-nef regions, with lower levels of hypermutation induced downstream of both PPTs, consistent with previous studies (72, 84, 86) (Fig. 5A and C). The levels of hA3G- and hA3F-induced hypermutation typically remained low for 1 to 2 kb downstream of the cPPT; however, the level of mutation induced by hA3G rapidly increased to levels similar to those observed in the gp41-env region within 500 bp of the 3′ PPT. In contrast, all of the sequences mutated by hA3F displayed low levels of G-to-A mutation throughout this region (Fig. 5C), except one that carried a high mutational burden (3F117 [see Fig. S4 in the supplemental material]).
FIG. 5.
Genome-wide hypermutation profiles induced in vitro by hA3G and hA3F. Genome-wide hypermutation profiles were generated by calculating the proportion of GG and GA dinucleotide targets mutated to AG and AA, respectively, in 400-bp sliding windows to the 3′ of the base under consideration, advancing in 1-bp steps across the genome. Consequently, the influence of a particular position on the profile commences 400 bp upstream of the position on the plot, and aberrant effects on the profiles may be observed within 400 bp of the ends of the sequences or gaps. The exact locations of the cPPT and 3′PPT in each sequence are indicated. (A) Mean profile (blue line) for proviruses hypermutated by hA3G in vitro (representative of 10 near-full-length proviruses); brown and red lines represent plus and minus 1 standard error of the mean. For each sequence, data for positions where less than 100 bases of actual sequence data were present in the 400-bp window (such as at the start of the sequence or around a gap) were omitted to avoid potential skewing of the mean profiles. (B) Representative profiles of hypermutation in four individual proviruses mutated by hA3G. The remaining profiles are shown in Fig. S4A of the supplemental material. (C) Mean profile (blue line) for proviruses hypermutated by hA3F in vitro (representative of nine near-full-length proviruses, six of which contained short gaps); brown and red lines represent plus and minus 1 standard error of the mean. For each sequence, data for positions where less than 100 bases of actual sequence data were present in the 400-bp window (such as at the start of the sequence or around a gap) were omitted to avoid potential skewing of the mean profiles. (D) Representative profiles of hypermutation in four individual proviruses mutated by hA3F. The positions of gaps in sequences are marked with gray boxes. The remaining profiles are shown in Fig. S4B of the supplemental material.
The hA3G-induced hypermutation profiles were distinct from those induced by hA3F (Fig. 5B and D; see also Fig. S1 in the supplemental material). While hA3G-hypermutated proviruses carried quite conserved genome-wide hypermutation profiles, those mutated by hA3F often contained some intensely mutated regions, while the rest of the genome contained little or no hypermutation; the boundaries of the intensely hypermutated regions did not necessarily coincide with the PPTs and varied between proviruses. Some harbored intensely hypermutated regions only in the 5′ half of the genome; some were only hypermutated significantly in the 3′ half; others were highly hypermutated in both halves of the genome (Fig. 5D; see also Fig. S4 in the supplemental material). Regions of intense hypermutation frequently contained runs of guanine bases followed by an adenine (GnA motifs; n > 1) in which several of the Gs preceding the conventional GA target dinucleotide also were mutated; indeed, for over 70% of the hA3F-mediated mutations classified as GG-to-AG mutations (Table 2), the following G was also mutated (i.e., equivalent to the GGA-to-AAA mutation [data not shown]), which is consistent with hA3F creating new GA target dinucleotides for itself (i.e., GGA-to-GAA-to-AAA).
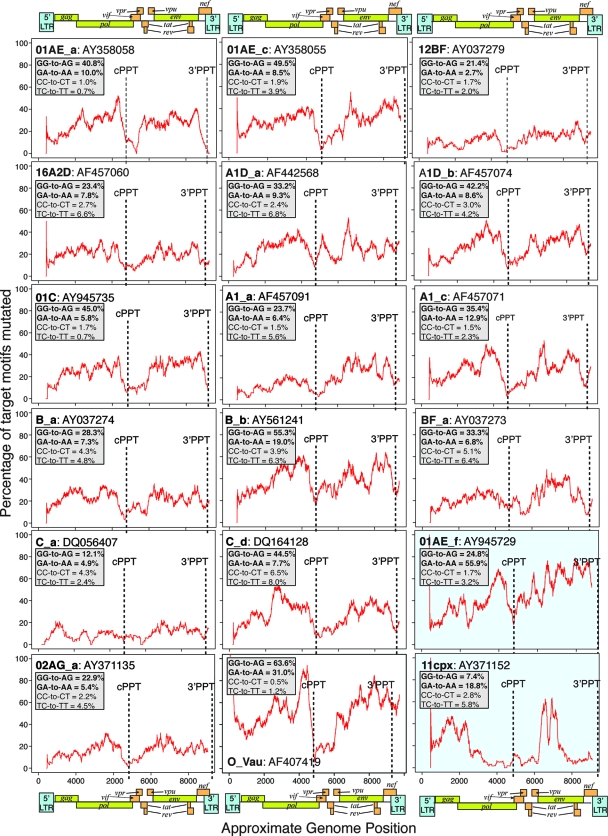
We did similar profile analysis of individual hA3G-hypermutated genomes derived from natural infections. In most cases, regardless of subtype, mutational minima existed at positions corresponding to the PPTs, with levels of hypermutation increasing toward pol and in the gp41-nef region (Fig. 6; see also Fig. S5 in the supplemental material). Analogous to the patterns of mutation induced in vitro by hA3G, the level of hypermutation frequently remained low 1 to 2 kb downstream of the cPPT while increasing to higher levels within 500 bp of the 3′ PPT. Of the two proviruses potentially hypermutated by hA3F in vivo (Fig. 4A and C), one (11cpx) displayed a mutational profile similar to that induced by hA3F in vitro, with short regions of intense hypermutation, while the other (01AE_f) displayed high levels of hypermutation throughout the genome (Fig. 6).
FIG. 6.
Representative profiles of hypermutation in proviruses derived from natural infections. Profiles of hypermutation across each near-full-length in vivo genome investigated were generated by calculating the proportion of target GG and GA dinucleotides mutated to AG and AA, respectively, in 400-bp sliding windows to the 3′ of the base under consideration, advancing in 1-bp steps across the genome. Consequently, the influence of a particular position on the profile commences 400 bp upstream of the position on the plot, and aberrant effects on the profiles may be observed within 400 bp of the end of the sequence. Sequence names according to subtype, as given in Table S2 of the supplemental material, are shown, together with the GenBank accession number of the sequence. The marked locations of the cPPT and 3′PPT indicated for each sequence are exact; these do not necessarily align with the approximate genome maps shown, as the lengths of the hypermutated sequences analyzed were variable. GG-to-AG and GA-to-AA mutation rates are indicated, together with the equivalent minus-strand mutations (plus-strand CC-to-CT and TC-to-TT) to give an indication of the noise associated with each analysis. The panels highlighted in blue indicate the proviruses carrying predominantly hA3F-type 5′GA-to-AA mutations; the remainder carried predominantly hA3G-type 5′GG-to-AG mutations. The remaining profiles are shown in Fig. S5 of the supplemental material.
Two HERV-K(HML2) variants carry footprints of hA3G activity.
The A3 proteins have been under strong positive selection throughout primate evolution, suggesting they have been important in defense against pathogens or mobile genetic elements for millions of years (63, 91). Many proviruses from the Pmv and Mpmv subgroups of endogenous nonecotropic MLVs carry signatures of mA3 activity, which may have contributed to their inactivation (34), and the ability of the hA3 proteins to restrict other types of endogenous retroelements has been demonstrated (8, 14, 17, 20, 21, 58, 65, 70).
The HERV sequences in the human genome provide a large archive of ancestral retroviral infections that were conceivably targets for the hA3 proteins. To analyze whether any HERV sequences carried footprints of hA3 activity in the same manner as the in vivo hypermutated HIV-1 proviruses, we determined the mutational preferences in members of the HERV-K(HML2) family, the most recently active lineage in humans. Each element was initially aligned to the consensus sequence of the major lineage to which it belonged (shown in Fig. 3 in Belshaw et al. [3]), and GR-to-AR and GY-to-AY mutation rates were determined (R = a purine, A or G; Y = a pyrimidine, C or T). The HIV-1 proviruses hypermutated by hA3G or hA3F in vitro displayed a marked bias toward plus-strand GR-to-AR (R = purine, A or G) mutation over GY-to-AY (Y = pyrimidine, C or T) mutations (chi-square test for independence of GR-to-AR and GY-to-AY mutation rates, P < 10−200 for hA3G and P < 10−70 for hA3F). Chi-square tests were therefore carried out for each HERV-K(HML2) element to screen for potential hA3-mediated hypermutation.
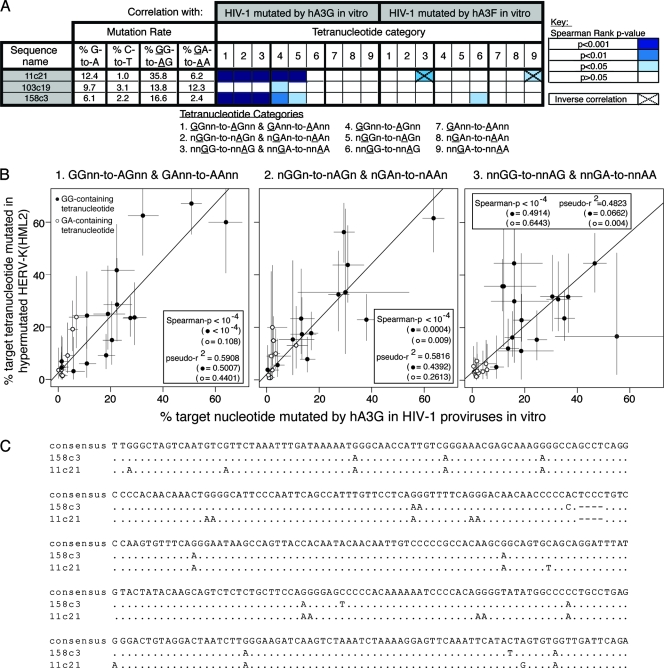
After Bonferroni correction for multiple testing, 3 out of 44 elements displayed significantly different mutation rates at GR and GY dinucleotides. These included two elements, 11c21 (P < 10−24) and 158c3 (P < 10−9), which had previously been shown to carry 11 of the 16 stop codons on internal branches of a HERV-K(HML2) phylogenetic tree; moreover, their branch lengths were longer than those of the surrounding elements (3). These characteristics were initially presumed to reflect the use of complementation in trans as a second mode of replication in the HERV-K(HML2) family (3). However, we noticed that, unlike in other HERV-K(HML2) elements with long branch lengths and multiple stop codons, a high proportion of the stop codons occurred as Trp-to-stop mutations (>75% in each case [data not shown]). Thus, these elements displayed several features of hA3-induced hypermutation: long branch lengths on a phylogenetic tree, abundant common Trp-to-stop mutations, and an excessive burden of GR-to-AR mutations. The third element identified was 103c19 (P = 0.00025). An apparent bias for mutation of GR over GY motifs was also observed in a few other elements, but these correlations were not significant after Bonferroni correction (data not shown).
We generated improved reference sequence estimates for each of these three elements and characterized the tetranucleotide preference hierarchies in each. The preference hierarchies for elements 11c21 and 158c3 correlated strongly with the hA3G tetranucleotide hierarchies determined by analyzing hypermutation in HIV-1 in vitro; the correlations were highly significant even when only GG-containing tetranucleotides were considered (target G at positions 1 or 2 of the tetranucleotide) and when the data were pooled (Fig. 7A, categories 4 and 5, and B). In contrast, the mutational preferences of 103c19 did not correlate significantly with the preferences observed in the hypermutated HIV-1 sequences (Fig. 7A). A short region of the genome demonstrating hypermutation in elements 11c21 and 158c3 is shown in Fig. 7C.
FIG. 7.
Correlation of tetranucleotide mutational preferences in naturally occurring hypermutated HERV-K(HML2) sequences with those observed in proviruses hypermutated in vitro by hA3G or hA3F. (A) HERV-K(HML2) proviruses were screened for hypermutation as described elsewhere. The tetranucleotide mutational preference hierarchies in the HERV-K(HML2) elements carrying evidence of potential hA3 activity (11c21, 103c19, and 158c3) were determined using improved reference sequences estimated as described elsewhere. Spearman rank correlations between the arrays of mutation rates observed in each element and those in the pooled data sets for proviruses hypermutated in vitro by hA3G or hA3F were determined, considering mutation within different categories of tetranucleotide contexts. Darker shades of blue indicate more significant correlations; contexts with zero mutation rates were excluded, since a large number of tied ranks can compromise the Spearman's rank test. Pairs of data for which a significant inverse correlation was found are indicated. For each HERV-K(HML2) element, G-to-A, GG-to-AG, and GA-to-AA mutation rates are indicated; C-to-T mutation rates are shown to give an indication of the noise associated with each analysis. Weighted Poisson regression analyses were also carried out, yielding similar results (data not shown). (B) The tetranucleotide preference data (with the target G either at position 1, 2, or 3 of the tetranucleotide) from the two HERV-K(HML2) elements that showed strong evidence of hA3G activity (11c21 and 158c3) were pooled and correlated with the pooled tetranucleotide mutational preferences for proviruses hypermutated by hA3G in vitro. Each point represents a particular tetranucleotide context; GG- and GA-containing tetranucleotide contexts are represented by black-filled and unfilled circles, respectively. Spearman rank correlation P values are indicated and were generated considering both the GG- and GA-containing contexts together, with the P values determined when only GG- or GA-containing tetranucleotide contexts were considered shown in parentheses; similarly, the McFadden Pseudo-R2 statistic, a measure of the goodness of fit of the regression, is indicated. Contexts with zero mutation rates were excluded. Error bars correspond to binomial 95% confidence intervals. (C) Section of HERV-K(HML2) gag sequence from hypermutated elements 11c21 and 158c3.
Thus, our results strongly suggest that the hypermutation found in elements 11c21 and 158c3 was induced by hA3G, while it remains unknown whether the apparent bias for mutation at GR motifs in element 103c19, and in the other sequences tending toward such a bias, was due to the activity of one or more hA3 proteins, or not.
Hypermutation profiles in HERV-K (HML2) reveal putative cPPT and CTS regions.
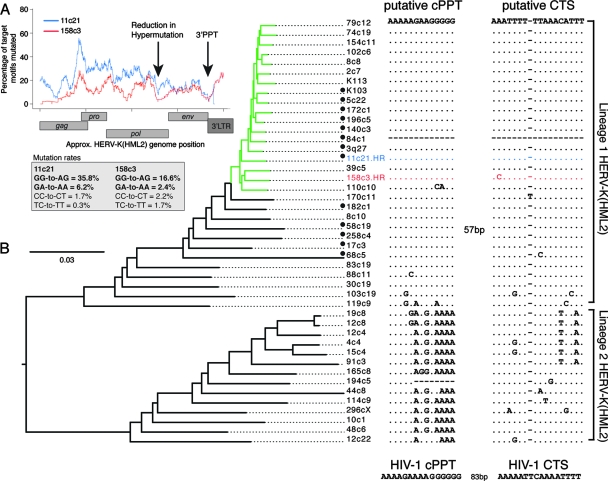
We analyzed the mutational profiles across the two hypermutated HERV-K(HML2) elements (Fig. 8A). As in hypermutated HIV-1 sequences, mutation levels decreased at the 3′ PPT; however, while the data were ambiguous with regard to the existence of hypermutational gradients, an additional reduction in hypermutation levels was found in both proviruses near the 3′ end of the pol gene at a position corresponding to a putative PPT-like sequence (5′-AAAAAGAAGGGGGAG-3′). A central termination site (CTS)-like sequence, characterized by a dA3-dT6 motif (12), occurred 57 bp downstream of the putative cPPT. These sequence motifs are analogous to those present in the HIV-1 genome, which permit initiation of plus-strand cDNA synthesis from a second site and formation of the central DNA flap (12, 89).
FIG. 8.
Conservation of putative cPPT and CTS motifs in a group of HERV-K(HML2) elements. (A) Hypermutation profiles across the HERV-K(HML2) elements 11c21 (blue line) and 158c3 (red line) were generated by calculating the proportion of target GG and GA dinucleotides mutated to AG and AA, respectively, in 400-bp sliding windows to the 3′ of the base under consideration, advancing in 1-bp steps across the genome. Consequently, the influence of a particular position on the profile commences 400 bp upstream of the position on the plot, and aberrant effects on the profiles may be observed within 400 bp of the ends of the sequences. The position of a common reduction in hypermutational burden in the two sequences is indicated. GG-to-AG and GA-to-AA mutation rates are indicated, together with the equivalent minus-strand mutations (plus-strand CC-to-CT and TC-to-TT) to give an indication of the noise associated with each analysis. (B) Maximum likelihood tree generated from 44 near-full-length HERV-K elements; the hA3-type mutations within the hypermutated elements 11c21 (blue) and 158c3 (red), denoted HR, were repaired prior to construction of the tree. The human-specific subgroup of HERV-K(HML2) elements is indicated in green. An alignment of the putative cPPT and CTS regions for each HERV-K(HML2) element in the tree is shown, with the two major lineages designated lineage 1 and lineage 2. The two regions are separated by 57 bp. No sequence for this region is present in element 84c1. Type 1 HERV-K(HML2) sequences, characterized by a 292-bp deletion at the pol-env boundary, are indicated with a black circle; all others are type 2 sequences. The HIV-1 cPPT and CTS sequences are shown for comparison.
We hypothesized that if the putative cPPT and CTS sequences were functionally significant for HERV-K(HML2) replication in general, they should be conserved in other HERV-K(HML2) sequences. While the CTS sequence was found in 43 of the 44 near-full-length HERV-K(HML2) genomes (element 84c1 carried a deletion in this region), the specific PPT-like motif was not so conserved. When we superimposed the putative cPPT region on a phylogenetic tree based on these sequences, it was apparent that the presence or absence of the cPPT motif correlated with the separation of the two major HERV-K(HML2) lineages, close to the root of the tree; the putative cPPT motif was only conserved in lineage 1. However, the sequence present at this location in lineage 2 was also composed entirely of purines, so a similar functional role cannot be excluded (Fig. 8B) (3). Removing the putative cPPT and CTS motifs from the alignment had no effect on the overall topology of the tree (data not shown). Furthermore, the previous nonphylogenetic designation of HERV-K(HML2) into type 1 and type 2 subgroups, based on the presence or absence of a 292-bp deletion at the pol-env boundary, did not correlate with these lineages (Fig. 8B) (3, 49).
DISCUSSION
Here, we demonstrate that hA3G and, to a lesser extent, hA3F leave well-defined footprints of mutational activity on retroviral sequences, beyond their known dinucleotide signatures (1, 47, 90). While some wider nucleotide motifs have been previously reported as preferred or disfavored substrates for hA3G and hA3F (1, 6, 13, 47, 73, 86), we show that the nucleotides spanning the region 2 nucleotides upstream to 3 nucleotides downstream of a target plus-strand G significantly influence the likelihood of a G-to-A mutation occurring; in addition, we present detailed tetranucleotide preference hierarchies for both deaminases. Furthermore, we show that the hA3G preference hierarchies are conserved not only in hypermutated HIV-1 proviruses in vitro and in vivo but also in two hypermutated members of the HERV-K(HML2) family of human endogenous retroviruses.
The highly significant correlation between the hA3G tetranucleotide preferences in vitro and in vivo suggests this deaminase was responsible for the hypermutation observed in vivo. The substrate preference hierarchies were apparent even when we analyzed only those tetranucleotide contexts that contained the preferred hA3G dinucleotide 5′GG target (i.e., GGnn and nGGn), further demonstrating that the target context wider than the dinucleotide strongly influences the likelihood of hA3G inducing a mutation. These hierarchies were typically maintained irrespective of the overall level of hypermutation both in vitro and in vivo.
In contrast, although the hA3F-induced mutation preferences appeared consistent in most hypermutated proviruses in vitro, they did not correlate significantly with those from the sequences carrying predominantly hA3F-type GA-to-AA mutations in vivo when we considered GA-containing tetranucleotide motifs alone (i.e., GAnn and nGAn). This may reflect either the smaller hA3F sample size, that the nucleotide preferences for hA3F activity are less conserved beyond the dinucleotide level, or that one or both of these two sequences were mutated by an hA3F-independent mechanism (e.g., other hA3 family members, such as hA3B [6]).
Irrespective of subtype, over 85% of the in vivo hypermutated HIV-1 proviruses carried clear signatures of hA3G activity and no more than 5% carried footprints of hA3F activity, although we cannot exclude the existence of low-level hA3F mutation in sequences carrying large amounts of hA3G-like mutations. The overrepresentation of proviruses carrying hA3G-like mutations appears to contradict previous suggestions that hA3F is the major contributor to hypermutation in natural HIV infections (47). This proposal was based in part on the observation that hA3F is partially resistant to HIV-1 Vif in vitro, as well as on the predominance of GA-to-AA mutations in a short fragment of the HIV-1 protease gene from one set of patients (32, 47). Assuming no significant biases in sampling or amplification of hA3G- and hA3F-hypermutated sequences within the database samples, which are derived from several independent studies, our data are consistent with hA3G being the major contributor to hypermutation in vivo. However, while hypermutation provides a useful diagnostic marker of hA3G and hA3F activity, we emphasize that it cannot be used to conclude that one or the other deaminase is more significant in terms of the overall hA3-mediated antiviral effect. More specifically, there is evidence that hA3 proteins may exert antiviral phenotypes in the absence of DNA editing in vitro (5, 24, 25, 28, 29, 31, 46, 50, 54, 57, 59, 64, 85).
Our data are consistent with earlier reports demonstrating the influence of the PPTs on the genome-wide hypermutation profiles (72, 84, 86). In the majority of sequences hypermutated in vitro and in vivo by hA3G, and in vitro by hA3F, reductions in mutation frequencies were observed in the genomic regions immediately downstream from the PPTs, which are exposed as single-stranded DNA for the shortest times during reverse transcription. However, since high levels of mutation were observed relatively close to the 3′ PPT in sequences hypermutated by hA3G, factors other than time exposed as single-stranded DNA may modify the hA3G-substrate interactions.
In contrast to the quite conserved genome-wide hypermutation profiles induced by hA3G, hA3F activity resulted in sporadic regions of intense hypermutation and other regions with little or no hypermutation, despite the availability of hA3F target motifs throughout the HIV-1 genome. The intensely hypermutated regions often included mutation of several consecutive guanines in plus-strand 5′ GnA (n > 1) motifs, which is consistent with hA3F creating novel target dinucleotides for itself. However, it is unknown whether these multiple mutations are caused by a single hA3F unit, processively mutating, creating, and itself mutating the newly created targets, or by multiple deaminases subsequently encountering newly created minus-strand 5′TC substrates. If these multiple mutations were catalyzed by a single hA3F unit, it would imply hA3F processed in a minus-strand 5′-to-3′ direction, in contrast to hA3G, which has been shown to act processively on target oligonucleotides in a minus-strand 3′-to-5′ direction in vitro (13). For both hA3G- and hA3F-mediated mutation, the time that DNA is exposed as a single strand, together with the distribution of preferred target motifs, and other as-yet-undefined factors, likely combine to determine the observed hypermutation profiles.
The hA3 proteins have been shown to be under strong positive selection throughout primate evolution (63, 91) and are expressed at high levels in testis, specifically in the ductus seminiferous (where spermatozoa are generated), and in the ovaries; the retrotransposition events that lead to endogenization must occur in these tissues (33, 77). Consequently, they have been suggested to play a role in protection against potentially detrimental transmission of functional retroelements (27, 63). Here, we present evidence that hA3G activity has influenced the natural history of HERVs, as 2 out of 44 HERV-K(HML2) elements were found to carry mutational signatures that correlated strongly with the footprints of hA3G activity observed in hypermutated HIV-1 genomes. These elements, 11c21 and 158c3, are unique to humans and occur near the base of the human-specific HERV-K(HML2) subgroup, which suggests that they are several million years old (2). Other HERV-K(HML2) family members also harbored higher numbers of GR-to-AR than GY-to-AY mutations and were therefore also potentially influenced by lower-level hA3 activity. For hypermutation to have occurred in these HERV-K(HML2) elements, we presume that hA3G became incorporated into HERV-K(HML2) virions that subsequently infected germ line cells, where it induced deamination of nascent viral DNA, prior to integration. The presence of these hypermutated elements in the human genome reveals that hA3G activity did not prevent transmission to offspring of HERV genetic material but may have reduced potential detrimental effects associated with transmission of functional, nonhypermutated retroviruses.
However, since only 2 out of 44 HERV-K(HML2) elements carried footprints of hA3G activity, the extent of its protective effect against these retroviruses may be limited. Proviruses of the Pmv and Mpmv subgroups of nonecotropic MLVs are proposed to have been inactivated, at least in part, by mA3-induced deamination (34); consistent with this proposition is the lack of purifying selection within these subgroups of murine ERVs. In contrast, the HERV-K(HML2) family has been under continuous purifying selection (like the Xmv subgroup of nonecotropic MLVs) and therefore largely has not been inactivated by hA3 proteins (3, 34). Nevertheless, the presence of hA3G-type hypermutation in two HERV-K(HML2) elements illustrates that these retroviruses have some susceptibility to this restriction factor in vivo. It may be of note that the proportion of the HERV-K(HML2) family carrying hA3G-type hypermutation is similar in magnitude to the proportion of HIV-1 proviruses bearing hypermutation in natural HIV-1 infections (38). HERV-K(HML2) may therefore have employed a means of hA3 evasion, functionally analogous to that mediated by Vif in HIV-1 infection, possibly explaining the lack of hA3G footprints in the majority of family members.
Our results may appear to contradict those of Lee and Bieniasz who, using an in vitro infectivity assay, demonstrated that a reconstituted HERV-K(HML2) virus was resistant to hA3G but sensitive to inhibition by hA3F (45). However, our data demonstrate that in vivo hypermutated HIV-1 sequences frequently carry footprints of hA3G activity, even though in vitro infectivity assays have suggested that, owing to Vif, wild-type HIV-1 is resistant to hA3G in the virus' natural target cells (23, 66, 68, 80). Therefore, the ability of the cytidine deaminases to reduce infectivity in an in vitro assay does not necessarily correlate with the presence of hypermutation in vivo. In spite of this, we would like to highlight that the apparent absence of hA3F-type mutations does not exclude that hA3F may also have influenced the natural history of these viruses.
As with hypermutated HIV-1 sequences, the mutational profiles of the HERV-K(HML2) elements showed reductions in mutation levels at the 3′ PPT. Furthermore, they allowed identification of a putative cPPT for priming plus-strand DNA synthesis in the HERV-K(HML2) family, as a decrease in hypermutation levels was observed toward the 3′ end of the pol gene, where a PPT-like motif was located. In hypermutated HIV-1 sequences, such reductions were seen downstream of the cPPT, which is also located toward the 3′ end of pol. This effect was lost in an HIV-1 variant carrying a mutated, nonfunctional cPPT motif (84). Consequently, the observed reduction in hypermutation in the HERV-K(HML2) elements are consistent with this putative cPPT being functional. Moreover, a CTS-like sequence (dA3-dT6) (12, 43) was present 57 bp downstream from the putative cPPT; similar motifs, located 88 and 98 bp downstream from the HIV-1 cPPT, mediate termination of plus-strand synthesis and formation of the central DNA flap (12, 18, 89). The importance of the combination of the cPPT and CTS-like sequences is suggested by their conservation over millions of years across one of the two HERV-K(HML2) lineages. Several of the more complex genera of retroviruses have been reported to possess cPPTs, including the lentiviruses (e.g., HIV-1 [10, 11], visna virus [7], feline immunodefiency virus [82], and equine infectious anemia virus [71]), spumaviruses (82), and piscine epsilon-retroviruses (e.g., walleye dermal sarcoma virus [30], walleye epidermal hyperplasia virus [42], and Atlantic salmon swim sarcoma viruses, phylogenetically placed between gamma- and epsilon-retroviral genera [61]). The functional relevance of these sequence signatures could be examined through site-directed mutagenesis of the motifs in the recently reconstituted HERV-K(HML2)-like viruses (16, 45).
It would be interesting to investigate whether members of other HERV families carry evidence of hA3 activity. However, many HERV families exhibit extreme “star-like” phylogenies, characterized by short internal and long terminal branch lengths, most likely due to accumulation of a large number of neutral mutations, induced postintegration (36). These would greatly increase the noise in similar analyses and would consequently make detection of hA3 activity more difficult than for HERV-K(HML2).
In summary, our study defines detailed and conserved nucleotide preferences for hA3G-mediated hypermutation and suggests different genome-wide mutational profiles for hA3G and hA3F. Such data will prove useful in assessing the contributions of the various hA3 proteins, particularly hA3G, to the generation of genetic diversity observed in natural retroviral infections. Moreover, this analysis provides the most direct evidence to date that hA3G has been in conflict with retroviruses over millions of years of human evolution.
Supplementary Material
Acknowledgments
We thank Michael Malim for reagents and helpful discussions, and we acknowledge the Computational Biology Research Group, Medical Sciences Division, Oxford, for use of their services in this project.
This work was supported by the Medical Research Council (MRC), United Kingdom, the Royal Society, and the Elizabeth Glaser Pediatric AIDS Foundation. A.E.A. is a holder of an MRC studentship; A.K. was funded by an MRC fellowship; K.N.B. is a Royal Society Dorothy Hodgkin Research Fellow.
Footnotes
Published ahead of print on 18 June 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Beale, R. C., S. K. Petersen-Mahrt, I. N. Watt, R. S. Harris, C. Rada, and M. S. Neuberger. 2004. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J. Mol. Biol. 337585-596. [DOI] [PubMed] [Google Scholar]
- 2.Belshaw, R., A. L. Dawson, J. Woolven-Allen, J. Redding, A. Burt, and M. Tristem. 2005. Genomewide screening reveals high levels of insertional polymorphism in the human endogenous retrovirus family HERV-K(HML2): implications for present-day activity. J. Virol. 7912507-12514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belshaw, R., V. Pereira, A. Katzourakis, G. Talbot, J. Paces, A. Burt, and M. Tristem. 2004. Long-term reinfection of the human genome by endogenous retroviruses. Proc. Natl. Acad. Sci. USA 1014894-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhattacharya, T., M. Daniels, D. Heckerman, B. Foley, N. Frahm, C. Kadie, J. Carlson, K. Yusim, B. McMahon, B. Gaschen, S. Mallal, J. I. Mullins, D. C. Nickle, J. Herbeck, C. Rousseau, G. H. Learn, T. Miura, C. Brander, B. Walker, and B. Korber. 2007. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science 3151583-1586. [DOI] [PubMed] [Google Scholar]
- 5.Bishop, K. N., R. K. Holmes, and M. H. Malim. 2006. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 808450-8458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bishop, K. N., R. K. Holmes, A. M. Sheehy, N. O. Davidson, S. J. Cho, and M. H. Malim. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 141392-1396. [DOI] [PubMed] [Google Scholar]
- 7.Blum, H. E., J. D. Harris, P. Ventura, D. Walker, K. Staskus, E. Retzel, and A. T. Haase. 1985. Synthesis in cell culture of the gapped linear duplex DNA of the slow virus visna. Virology 142270-277. [DOI] [PubMed] [Google Scholar]
- 8.Bogerd, H. P., H. L. Wiegand, A. E. Hulme, J. L. Garcia-Perez, K. S. O'Shea, J. V. Moran, and B. R. Cullen. 2006. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc. Natl. Acad. Sci. USA 1038780-8785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brander, C., and B. D. Walker. 2003. Gradual adaptation of HIV to human host populations: good or bad news? Nat. Med. 91359-1362. [DOI] [PubMed] [Google Scholar]
- 10.Charneau, P., M. Alizon, and F. Clavel. 1992. A second origin of DNA plus-strand synthesis is required for optimal human immunodeficiency virus replication. J. Virol. 662814-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charneau, P., and F. Clavel. 1991. A single-stranded gap in human immunodeficiency virus unintegrated linear DNA defined by a central copy of the polypurine tract. J. Virol. 652415-2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charneau, P., G. Mirambeau, P. Roux, S. Paulous, H. Buc, and F. Clavel. 1994. HIV-1 reverse transcription. A termination step at the center of the genome. J. Mol. Biol. 241651-662. [DOI] [PubMed] [Google Scholar]
- 13.Chelico, L., P. Pham, P. Calabrese, and M. F. Goodman. 2006. APOBEC3G DNA deaminase acts processively 3′→5′ on single-stranded DNA. Nat. Struct. Mol. Biol. 13392-399. [DOI] [PubMed] [Google Scholar]
- 14.Chiu, Y. L., H. E. Witkowska, S. C. Hall, M. Santiago, V. B. Soros, C. Esnault, T. Heidmann, and W. C. Greene. 2006. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc. Natl. Acad. Sci. USA 10315588-15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conticello, S. G., R. S. Harris, and M. S. Neuberger. 2003. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 132009-2013. [DOI] [PubMed] [Google Scholar]
- 16.Dewannieux, M., F. Harper, A. Richaud, C. Letzelter, D. Ribet, G. Pierron, and T. Heidmann. 2006. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 161548-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dutko, J. A., A. Schafer, A. E. Kenny, B. R. Cullen, and M. J. Curcio. 2005. Inhibition of a yeast LTR retrotransposon by human APOBEC3 cytidine deaminases. Curr. Biol. 15661-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dvorin, J. D., P. Bell, G. G. Maul, M. Yamashita, M. Emerman, and M. H. Malim. 2002. Reassessment of the roles of integrase and the central DNA flap in human immunodeficiency virus type 1 nuclear import. J. Virol. 7612087-12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esnault, C., O. Heidmann, F. Delebecque, M. Dewannieux, D. Ribet, A. J. Hance, T. Heidmann, and O. Schwartz. 2005. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature 433430-433. [DOI] [PubMed] [Google Scholar]
- 20.Esnault, C., J. Maestre, and T. Heidmann. 2000. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 24363-367. [DOI] [PubMed] [Google Scholar]
- 21.Esnault, C., J. Millet, O. Schwartz, and T. Heidmann. 2006. Dual inhibitory effects of APOBEC family proteins on retrotransposition of mammalian endogenous retroviruses. Nucleic Acids Res. 341522-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fisher, R. A. 1948. Combining independent tests of significance. Am. Stat. 230. [Google Scholar]
- 23.Gabuzda, D. H., K. Lawrence, E. Langhoff, E. Terwilliger, T. Dorfman, W. A. Haseltine, and J. Sodroski. 1992. Role of vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J. Virol. 666489-6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo, F., S. Cen, M. Niu, J. Saadatmand, and L. Kleiman. 2006. Inhibition of tRNA3Lys-primed reverse transcription by human APOBEC3G during human immunodeficiency virus type 1 replication. J. Virol. 8011710-11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo, F., S. Cen, M. Niu, Y. Yang, R. J. Gorelick, and L. Kleiman. 2007. The interaction of APOBEC3G with human immunodeficiency virus type 1 nucleocapsid inhibits tRNA3Lys annealing to viral RNA. J. Virol. 8111322-11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113803-809. [DOI] [PubMed] [Google Scholar]
- 27.Holmes, E. C. 2004. Adaptation and immunity. PLoS Biol. 2e307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holmes, R. K., F. A. Koning, K. N. Bishop, and M. H. Malim. 2007. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J. Biol. Chem. 2822587-2595. [DOI] [PubMed] [Google Scholar]
- 29.Holmes, R. K., M. H. Malim, and K. N. Bishop. 2007. APOBEC-mediated viral restriction: not simply editing? Trends Biochem. Sci. 32118-128. [DOI] [PubMed] [Google Scholar]
- 30.Holzschu, D. L., D. Martineau, S. K. Fodor, V. M. Vogt, P. R. Bowser, and J. W. Casey. 1995. Nucleotide sequence and protein analysis of a complex piscine retrovirus, walleye dermal sarcoma virus. J. Virol. 695320-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iwatani, Y., D. S. Chan, F. Wang, K. S. Maynard, W. Sugiura, A. M. Gronenborn, I. Rouzina, M. C. Williams, K. Musier-Forsyth, and J. G. Levin. 2007. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 357096-7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janini, M., M. Rogers, D. R. Birx, and F. E. McCutchan. 2001. Human immunodeficiency virus type 1 DNA sequences genetically damaged by hypermutation are often abundant in patient peripheral blood mononuclear cells and may be generated during near-simultaneous infection and activation of CD4+ T cells. J. Virol. 757973-7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jarmuz, A., A. Chester, J. Bayliss, J. Gisbourne, I. Dunham, J. Scott, and N. Navaratnam. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79285-296. [DOI] [PubMed] [Google Scholar]
- 34.Jern, P., J. P. Stoye, and J. M. Coffin. 2007. Role of APOBEC3 in genetic diversity among endogenous murine leukemia viruses. PLoS Genet. 32014-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kao, S., M. A. Khan, E. Miyagi, R. Plishka, A. Buckler-White, and K. Strebel. 2003. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J. Virol. 7711398-11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katzourakis, A., A. Rambaut, and O. G. Pybus. 2005. The evolutionary dynamics of endogenous retroviruses. Trends Microbiol. 13463-468. [DOI] [PubMed] [Google Scholar]
- 37.Katzourakis, A., M. Tristem, O. G. Pybus, and R. J. Gifford. 2007. Discovery and analysis of the first endogenous lentivirus. Proc. Natl. Acad. Sci. USA 1046261-6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kieffer, T. L., P. Kwon, R. E. Nettles, Y. Han, S. C. Ray, and R. F. Siliciano. 2005. G→A hypermutation in protease and reverse transcriptase regions of human immunodeficiency virus type 1 residing in resting CD4+ T cells in vivo. J. Virol. 791975-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kijak, G. H., M. Janini, S. Tovanabutra, E. E. Sanders-Buell, D. L. Birx, M. L. Robb, N. L. Michael, and F. E. McCutchan. 2007. HyperPack: a software package for the study of levels, contexts, and patterns of APOBEC-mediated hypermutation in HIV. AIDS Res. Hum. Retrovir. 23554-557. [DOI] [PubMed] [Google Scholar]
- 40.Koulinska, I. N., B. Chaplin, D. Mwakagile, M. Essex, and B. Renjifo. 2003. Hypermutation of HIV type 1 genomes isolated from infants soon after vertical infection. AIDS Res. Hum. Retrovir. 191115-1123. [DOI] [PubMed] [Google Scholar]
- 41.Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody, J. Baldwin, K. Devon, K. Dewar, M. Doyle, W. FitzHugh, R. Funke, D. Gage, K. Harris, A. Heaford, J. Howland, L. Kann, J. Lehoczky, R. LeVine, P. McEwan, K. McKernan, J. Meldrim, J. P. Mesirov, C. Miranda, W. Morris, J. Naylor, C. Raymond, M. Rosetti, R. Santos, A. Sheridan, C. Sougnez, N. Stange-Thomann, N. Stojanovic, A. Subramanian, D. Wyman, J. Rogers, J. Sulston, R. Ainscough, S. Beck, D. Bentley, J. Burton, C. Clee, N. Carter, A. Coulson, R. Deadman, P. Deloukas, A. Dunham, I. Dunham, R. Durbin, L. French, D. Grafham, S. Gregory, T. Hubbard, S. Humphray, A. Hunt, M. Jones, C. Lloyd, A. McMurray, L. Matthews, S. Mercer, S. Milne, J. C. Mullikin, A. Mungall, R. Plumb, M. Ross, R. Shownkeen, S. Sims, R. H. Waterston, R. K. Wilson, L. W. Hillier, J. D. McPherson, M. A. Marra, E. R. Mardis, L. A. Fulton, A. T. Chinwalla, K. H. Pepin, W. R. Gish, S. L. Chissoe, M. C. Wendl, K. D. Delehaunty, T. L. Miner, A. Delehaunty, J. B. Kramer, L. L. Cook, R. S. Fulton, D. L. Johnson, P. J. Minx, S. W. Clifton, T. Hawkins, E. Branscomb, P. Predki, P. Richardson, S. Wenning, T. Slezak, N. Doggett, J. F. Cheng, A. Olsen, S. Lucas, C. Elkin, E. Uberbacher, M. Frazier, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409860-921. [DOI] [PubMed] [Google Scholar]
- 42.LaPierre, L. A., D. L. Holzschu, P. R. Bowser, and J. W. Casey. 1999. Sequence and transcriptional analyses of the fish retroviruses walleye epidermal hyperplasia virus types 1 and 2: evidence for a gene duplication. J. Virol. 739393-9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lavigne, M., P. Roux, H. Buc, and F. Schaeffer. 1997. DNA curvature controls termination of plus strand DNA synthesis at the centre of HIV-1 genome. J. Mol. Biol. 266507-524. [DOI] [PubMed] [Google Scholar]
- 44.Lecossier, D., F. Bouchonnet, F. Clavel, and A. J. Hance. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 3001112. [DOI] [PubMed] [Google Scholar]
- 45.Lee, Y. N., and P. D. Bieniasz. 2007. Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog. 3e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li, X. Y., F. Guo, L. Zhang, L. Kleiman, and S. Cen. 2007. APOBEC3G inhibits DNA strand transfer during HIV-1 reverse transcription. J. Biol. Chem. 28232065-32074. [DOI] [PubMed] [Google Scholar]
- 47.Liddament, M. T., W. L. Brown, A. J. Schumacher, and R. S. Harris. 2004. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 141385-1391. [DOI] [PubMed] [Google Scholar]
- 48.Liu, B., P. T. Sarkis, K. Luo, Y. Yu, and X. F. Yu. 2005. Regulation of Apobec3F and human immunodeficiency virus type 1 Vif by Vif-Cul5-ElonB/C E3 ubiquitin ligase. J. Virol. 799579-9587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lower, R., R. R. Tonjes, C. Korbmacher, R. Kurth, and J. Lower. 1995. Identification of a Rev-related protein by analysis of spliced transcripts of the human endogenous retroviruses HTDV/HERV-K. J. Virol. 69141-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo, K., T. Wang, B. Liu, C. Tian, Z. Xiao, J. Kappes, and X. F. Yu. 2007. Cytidine deaminases APOBEC3G and APOBEC3F interact with human immunodeficiency virus type 1 integrase and inhibit proviral DNA formation. J. Virol. 817238-7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maddison, D. R., and W. P. Maddison. 2003. MacClade, ed. 4.06. Sinauer, Sunderland, MA.
- 52.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 42499-103. [DOI] [PubMed] [Google Scholar]
- 53.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 91398-1403. [DOI] [PubMed] [Google Scholar]
- 54.Mbisa, J. L., R. Barr, J. A. Thomas, N. Vandegraaff, I. J. Dorweiler, E. S. Svarovskaia, W. L. Brown, L. M. Mansky, R. J. Gorelick, R. S. Harris, A. Engelman, and V. K. Pathak. 2007. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J. Virol. 817099-7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCullagh, P., and J. Nelder. 1989. Generalized linear models. Chapman & Hall, London, England.
- 56.Medstrand, P., and D. L. Mager. 1998. Human-specific integrations of the HERV-K endogenous retrovirus family. J. Virol. 729782-9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miyagi, E., S. Opi, H. Takeuchi, M. Khan, R. Goila-Gaur, S. Kao, and K. Strebel. 2007. Enzymatically active APOBEC3G is required for efficient inhibition of human immunodeficiency virus type 1. J. Virol. 8113346-13353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muckenfuss, H., M. Hamdorf, U. Held, M. Perkovic, J. Lower, K. Cichutek, E. Flory, G. G. Schumann, and C. Munk. 2006. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J. Biol. Chem. 28122161-22172. [DOI] [PubMed] [Google Scholar]
- 59.Newman, E. N., R. K. Holmes, H. M. Craig, K. C. Klein, J. R. Lingappa, M. H. Malim, and A. M. Sheehy. 2005. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 15166-170. [DOI] [PubMed] [Google Scholar]
- 60.Pace, C., J. Keller, D. Nolan, I. James, S. Gaudieri, C. Moore, and S. Mallal. 2006. Population level analysis of human immunodeficiency virus type 1 hypermutation and its relationship with APOBEC3G and vif genetic variation. J. Virol. 809259-9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Paul, T. A., S. L. Quackenbush, C. Sutton, R. N. Casey, P. R. Bowser, and J. W. Casey. 2006. Identification and characterization of an exogenous retrovirus from Atlantic salmon swim bladder sarcomas. J. Virol. 802941-2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rambaut, A., D. Posada, K. A. Crandall, and E. C. Holmes. 2004. The causes and consequences of HIV evolution. Nat. Rev. Genet. 552-61. [DOI] [PubMed] [Google Scholar]
- 63.Sawyer, S. L., M. Emerman, and H. S. Malik. 2004. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2e275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schumacher, A. J., G. Hache, D. A. Macduff, W. L. Brown, and R. S. Harris. 2008. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD, and human immunodeficiency virus type 1 restriction. J. Virol. 822652-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schumacher, A. J., D. V. Nissley, and R. S. Harris. 2005. APOBEC3G hypermutates genomic DNA and inhibits Ty1 retrotransposition in yeast. Proc. Natl. Acad. Sci. USA 1029854-9859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418646-650. [DOI] [PubMed] [Google Scholar]
- 67.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 91404-1407. [DOI] [PubMed] [Google Scholar]
- 68.Simon, J. H., N. C. Gaddis, R. A. Fouchier, and M. H. Malim. 1998. Evidence for a newly discovered cellular anti-HIV-1 phenotype. Nat. Med. 41397-1400. [DOI] [PubMed] [Google Scholar]
- 69.Staden, R., K. F. Beal, and J. K. Bonfield. 2000. The Staden package, 1998. Methods Mol. Biol. 132115-130. [DOI] [PubMed] [Google Scholar]
- 70.Stenglein, M. D., and R. S. Harris. 2006. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J. Biol. Chem. 28116837-16841. [DOI] [PubMed] [Google Scholar]
- 71.Stetor, S. R., J. W. Rausch, M. J. Guo, J. P. Burnham, L. R. Boone, M. J. Waring, and S. F. Le Grice. 1999. Characterization of (+) strand initiation and termination sequences located at the center of the equine infectious anemia virus genome. Biochemistry 383656-3667. [DOI] [PubMed] [Google Scholar]
- 72.Suspene, R., C. Rusniok, J. P. Vartanian, and S. Wain-Hobson. 2006. Twin gradients in APOBEC3 edited HIV-1 DNA reflect the dynamics of lentiviral replication. Nucleic Acids Res. 344677-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Suspene, R., P. Sommer, M. Henry, S. Ferris, D. Guetard, S. Pochet, A. Chester, N. Navaratnam, S. Wain-Hobson, and J. P. Vartanian. 2004. APOBEC3G is a single-stranded DNA cytidine deaminase and functions independently of HIV reverse transcriptase. Nucleic Acids Res. 322421-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Swofford, D. L. 2003. PAUP*: phylogenetic analysis using parsimony (*and other methods), 4.0 b10 ed. Sinauer Associates, Sunderland, MA.
- 75.Turelli, P., B. Mangeat, S. Jost, S. Vianin, and D. Trono. 2004. Inhibition of hepatitis B virus replication by APOBEC3G. Science 3031829. [DOI] [PubMed] [Google Scholar]
- 76.Turner, G., M. Barbulescu, M. Su, M. I. Jensen-Seaman, K. K. Kidd, and J. Lenz. 2001. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr. Biol. 111531-1535. [DOI] [PubMed] [Google Scholar]
- 77.Uhlen, M., E. Bjorling, C. Agaton, C. A. Szigyarto, B. Amini, E. Andersen, A. C. Andersson, P. Angelidou, A. Asplund, C. Asplund, L. Berglund, K. Bergstrom, H. Brumer, D. Cerjan, M. Ekstrom, A. Elobeid, C. Eriksson, L. Fagerberg, R. Falk, J. Fall, M. Forsberg, M. G. Bjorklund, K. Gumbel, A. Halimi, I. Hallin, C. Hamsten, M. Hansson, M. Hedhammar, G. Hercules, C. Kampf, K. Larsson, M. Lindskog, W. Lodewyckx, J. Lund, J. Lundeberg, K. Magnusson, E. Malm, P. Nilsson, J. Odling, P. Oksvold, I. Olsson, E. Oster, J. Ottosson, L. Paavilainen, A. Persson, R. Rimini, J. Rockberg, M. Runeson, A. Sivertsson, A. Skollermo, J. Steen, M. Stenvall, F. Sterky, S. Stromberg, M. Sundberg, H. Tegel, S. Tourle, E. Wahlund, A. Walden, J. Wan, H. Wernerus, J. Westberg, K. Wester, U. Wrethagen, L. L. Xu, S. Hober, and F. Ponten. 2005. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol. Cell. Proteomics 41920-1932. [DOI] [PubMed] [Google Scholar]
- 78.Vartanian, J. P., M. Henry, and S. Wain-Hobson. 2002. Sustained G→A hypermutation during reverse transcription of an entire human immunodeficiency virus type 1 strain Vau group O genome. J. Gen. Virol. 83801-805. [DOI] [PubMed] [Google Scholar]
- 79.Vartanian, J. P., A. Meyerhans, B. Asjo, and S. Wain-Hobson. 1991. Selection, recombination, and G→A hypermutation of human immunodeficiency virus type 1 genomes. J. Virol. 651779-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.von Schwedler, U., J. Song, C. Aiken, and D. Trono. 1993. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J. Virol. 674945-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang, B., M. Mikhail, W. B. Dyer, J. J. Zaunders, A. D. Kelleher, and N. K. Saksena. 2003. First demonstration of a lack of viral sequence evolution in a nonprogressor, defining replication-incompetent HIV-1 infection. Virology 312135-150. [DOI] [PubMed] [Google Scholar]
- 82.Whitwam, T., M. Peretz, and E. Poeschla. 2001. Identification of a central DNA flap in feline immunodeficiency virus. J. Virol. 759407-9414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wiegand, H. L., B. P. Doehle, H. P. Bogerd, and B. R. Cullen. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 232451-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wurtzer, S., A. Goubard, F. Mammano, S. Saragosti, D. Lecossier, A. J. Hance, and F. Clavel. 2006. Functional central polypurine tract provides downstream protection of the human immunodeficiency virus type 1 genome from editing by APOBEC3G and APOBEC3B. J. Virol. 803679-3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang, Y., F. Guo, S. Cen, and L. Kleiman. 2007. Inhibition of initiation of reverse transcription in HIV-1 by human APOBEC3F. Virology 36592-100. [DOI] [PubMed] [Google Scholar]
- 86.Yu, Q., R. Konig, S. Pillai, K. Chiles, M. Kearney, S. Palmer, D. Richman, J. M. Coffin, and N. R. Landau. 2004. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 11435-442. [DOI] [PubMed] [Google Scholar]
- 87.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 3021056-1060. [DOI] [PubMed] [Google Scholar]
- 88.Zennou, V., and P. D. Bieniasz. 2006. Comparative analysis of the antiretroviral activity of APOBEC3G and APOBEC3F from primates. Virology 34931-40. [DOI] [PubMed] [Google Scholar]
- 89.Zennou, V., C. Petit, D. Guetard, U. Nerhbass, L. Montagnier, and P. Charneau. 2000. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell 101173-185. [DOI] [PubMed] [Google Scholar]
- 90.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 42494-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang, J., and D. M. Webb. 2004. Rapid evolution of primate antiviral enzyme APOBEC3G. Hum. Mol. Genet. 131785-1791. [DOI] [PubMed] [Google Scholar]
- 92.Zheng, Y. H., D. Irwin, T. Kurosu, K. Tokunaga, T. Sata, and B. M. Peterlin. 2004. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 786073-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.