Abstract
The monoclonal antibody M1-1A, specific for the hemagglutinin-neuraminidase (HN) protein of human parainfluenza type 2 virus (HPIV2), blocks virus-induced cell-cell fusion without affecting the hemagglutinating and neuraminidase activities. F13 is a neutralization escape variant selected with M1-1A and contains amino acid mutations N83Y and M186I in the HN protein, with no mutation in the fusion protein. Intriguingly, F13 exhibits reduced ability to induce cell-cell fusion despite its multistep replication. To investigate the potential role of HPIV2 HN protein in the regulation of cell-cell fusion, we introduced these mutations individually or in combination to the HN protein in the context of recombinant HPIV2. Following infection at a low multiplicity, Vero cells infected with the mutant virus H-83/186, which carried both the N83Y and M186I mutations, remained as nonfused single cells at least for 24 h, whereas most of the cells infected with wild-type virus mediated prominent cell-cell fusion within 24 h. On the other hand, the cells infected with the mutant virus, carrying either the H-83 or H-186 mutation, mediated cell-cell fusion but less efficiently than those infected with wild-type virus. Irrespective of the ability to cause cell-cell fusion, however, every virus could infect all the cells in the culture within 48 h after the initial infection. These results indicated that both the N83Y and M186I mutations in the HN protein are involved in the regulation of cell-cell fusion. Notably, the limited cell-cell fusion by H-83/186 virus was greatly promoted by lysophosphatidic acid, a stimulator of the Ras and Rho family GTPases.
Human parainfluenza type 2 virus (HPIV2) is one of the major human respiratory pathogens and belongs to genus Rubulavirus in the family Paramyxoviridae (14, 16). The members of genus Rubulavirus, as well as those of genera Respirovirus and Avulavirus in the same family, have two envelope glycoproteins: the hemagglutinin-neuraminidase (HN) and fusion (F) proteins (16). The HN protein is responsible for binding to the viral receptor sialoconjugate (hemagglutinating activity) and for enzymatic destruction of the receptor (neuraminidase activity), while the F protein mediates membrane fusion such as cell-cell fusion or virus-cell fusion. Cleavage of the F precursor (F0) by cellular proteases into subunits F1 and F2 is a prerequisite for its fusion-inducing activity. Interestingly, the F protein requires the fusion-promoting function of its homologous HN protein (10), except for the W3A strain of a rubulavirus parainfluenza virus 5 (PIV5) (29). The stalk region of the HN protein is inferred to contain the site that determines the F protein specificity for promoting fusion (7, 19, 37, 41), while the globular head region carries both the receptor-binding and neuraminidase activities (22, 38).
Although it is not well known how the HN protein promotes the F protein-mediated membrane fusion, it is believed that the fusion is induced through a series of conformational changes of the F protein that is initiated by an interaction between HN and F proteins (15, 24). Interestingly, relationships among the three activities (receptor binding, neuraminidase, and fusion promotion) of the HN protein vary depending on the virus type. On the one hand, mutational analysis of the HN protein of a respirovirus HPIV3 has indicated that high neuraminidase activity reduces the extent of membrane fusion by counteracting the receptor-binding function (31). On the other hand, in the case of the HN protein of an avulavirus Newcastle disease virus (NDV), neither neuraminidase nor receptor-binding activity affects the extent of membrane fusion (5) although receptor binding seems essential for fusion induction (18).
It is thought that the HN and F proteins of paramyxoviruses mediate membrane fusion at neutral pH. Thus, the viruses can fuse with the plasma membrane, and the virus-infected cells mediate cell-cell fusion with the neighboring cells, sometimes resulting in extensive syncytium formation that leads to cell death. Intriguingly, the PIV5 strain SER induces minimal cell-cell fusion (39) even though virus-cell fusion for SER virus occurs at nearly wild-type (WT) levels (6), indicating that there is a difference between virus-cell fusion and cell-cell fusion. However, reports dealing with the molecular mechanism of SER virus-mediated cell-cell fusion are inconsistent with each other (2, 36), and further studies are needed to know what kinds of viral and cellular factors are required for SER virus to induce cell-cell fusion.
We reported previously that the anti-HPIV2 HN monoclonal antibody M1-1A could inhibit cell-cell fusion without affecting hemagglutinating and neuraminidase activities (42). The epitope for M1-1A was mapped to the stalk region of the HN protein, as revealed by analyzing the reactivity of M1-1A to the neutralization escape variants (41, 49). Interestingly, we found that one of the escape variants did not mediate detectable cell-cell fusion of infected HeLa cells up to 72 h after infection, whereas the parent Toshiba strain induced prominent cell-cell fusion within 48 h (49). This mutant, designated F13, has amino acid mutations N83Y and M186I in the HN protein, with no mutation in the F protein. Since F13 could perform multistep replication (49), it was suggested that the mutations in the HN protein could reduce cell-cell fusion without significantly affecting virus-cell fusion. However, we could not rule out the possibility that mutation(s) in other proteins of F13 might also be involved in its reduced ability to induce cell-cell fusion, although protein expression analyses indicated that the cell-cell fusion-promoting activity of the F13 HN protein was significantly lower than that of WT HN protein (49). In order to investigate the potential role of the HN protein in the regulation of cell-cell fusion, we created three recombinant viruses that harbor either or both of the mutations (N83Y and M186I) in the HN protein. The mutant viruses were then analyzed by using Vero cells instead of HeLa cells because the former type of cells was found to be more susceptible to HPIV2-induced cell-cell fusion (49). Furthermore, our preliminary experiment indicated that Vero cells can be cultured for at least 7 days without significant morphological changes and thus are more suitable for observation of extremely slow cell-cell fusion induced by F13. As a result, the revealed properties of the mutant viruses have suggested a novel function of the HN protein that may be involved in the regulation of cell-cell fusion.
MATERIALS AND METHODS
Cells.
Vero cell monolayers were maintained in Eagle's minimum essential medium (MEM) supplemented with 5% calf serum. BSR T7/5 cell monolayers stably expressing T7 RNA polymerase (3) were maintained in MEM supplemented with 10% fetal calf serum and 1 mg/ml of G418 (geneticin; Gibco BRL).
Generation of recombinant HPIV2 with HN protein mutations.
The recombinant plasmid, pPIV2, that harbors full-length cDNA of the HPIV2 genome (Toshiba strain; 15,654 nucleotides [nt]; GenBank accession number AB176531) was described previously (26). In order to construct recombinant HPIV2 plasmids with mutations in the HN gene, restriction enzyme sites for BssHII and AgeI were introduced into the 3′ noncoding region (nt 6779) and the 5′ noncoding region (nt 8669) of the HN gene in plasmid pPIV2, respectively. cDNA fragments, corresponding to the nucleotides (nt 6779 to 8669) in pPIV2, were then produced by PCR using the recombinant SRα plasmids that encode the HN proteins with amino acid mutation(s) N83Y, M186I, or N83Y plus M186I (49) as the templates. Simultaneously, the cDNA fragments were provided with BssHII and AgeI sites at their ends. After being cloned into the plasmid pDrive (Qiagen), each cDNA fragment was cut out by BssHII and AgeI and used to replace the HN protein-encoding region of pPIV2. Recombinant viruses were then rescued by using BSR T7/5 cell monolayers as reported previously (26).
Viral growth kinetics.
Confluent monolayers of Vero cells in 96-well culture plates or those in 24-well culture plates were infected with each recombinant virus at a multiplicity of infection (MOI) of 5 or 0.0001, respectively. The infected cells were incubated in MEM containing 2% calf serum at 37°C at various times postinfection, and the amount of infectious virus in the culture supernatant was quantified by plaque-forming assay on Vero cells, as described previously (42).
Indirect immunofluorescent staining.
Vero cell monolayers on glass coverslips in six-well culture plates were infected with recombinant virus at an MOI of 0.05. After being incubated in MEM at 37°C, the cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100 in phosphate-buffered saline (PBS). The viral nucleoproteins in the infected cells were then immunostained with monoclonal antibody 306-1 (42) and fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulins (ICN Biomedicals), while the nuclei of cells were visualized with 4′,6-diamidino-2-phenylindole (DAPI; Sigma). The results were observed by using a fluorescence microscope (Olympus, Tokyo, Japan), and the digital images were obtained.
Penetration assay.
Penetration rate of the recombinant viruses was measured as described previously (42) with some modifications. Briefly, Vero cell monolayers in 12-well culture plates were inoculated with approximately 75 PFU/well of each virus and incubated at 37°C. Viruses that had not penetrated into the cells were then inactivated by treating with 1 ml/well of citrate buffer (40 mM citric acid [pH 3.0], 10 mM KCl, 135 mM NaCl) at 23°C for 1 min. After two washes with PBS containing 0.1 mM CaCl2 and 1 mM MgCl2, the cells were overlaid with agarose-containing medium and incubated at 37°C for the examination of plaques.
Cell viability assay.
A colorimetric assay for quantification of cell viability was performed as described by Cid-Arregui et al. (4) with some modifications. Briefly, Vero cell monolayers in 12-well culture plates were infected with recombinant virus at an MOI of 5 and incubated at 37°C. At various times after infection, the culture medium in each well was replaced with 1 ml of a 0.5 mg/ml solution of MTT [3(4,5-dimethylthiazol-2-yl) 2,5-diphenyl-tetrazolium bromide; Roche] in phenol red-free MEM and incubated at 37°C for 2 h. The culture supernatant was then carefully removed, and the purple formazan crystals that had been formed in the cells were dissolved in 200 μl/well isopropanol. Absorbance of the formazan solution in isopropanol was measured at 550 nm after removal of the cell debris by centrifugation (10,000 × g for 5 min). Positive control wells with mock-infected cells and negative control wells without cells were run in parallel.
Hemagglutination and neuraminidase assays.
For the hemagglutination assay, virus suspension containing 107 PFU was centrifuged at 15,000 × g for 60 min. The virions in the pellet were resuspended in 25 μl of PBS containing 0.1% bovine serum albumin and 0.01% gelatin (PBS-GB, pH 7.0), and serial twofold dilutions were made in U-type 96-well plates. After addition of 25 μl of 0.4% suspension of guinea pig erythrocyte in PBS-GB to each well, the plates were kept at 4°C or 37°C and the number of hemagglutination units (HAU) was estimated. For the neuraminidase assay, each virus pellet (107 PFU) was resuspended in 20 μl of a 25 mg/ml solution of bovine fetuin in 0.4 M acetate buffer (pH 4.0 to pH 7.0) and incubated at 37°C for 100 min. The released sialic acids were then assayed by the thiobarbituric acid method of Warren as described by Ito et al. (13), and the absorbance value at 550 nm was measured.
Receptor binding assay.
Virus suspension (4 HAU, which was estimated by the hemagglutination assay at 4°C) in 25 μl of PBS-GB (pH 7.0 or pH 5.5) was mixed with an equal volume of 0.4% suspension of guinea pig erythrocytes in PBS-GB (pH 7.0 or pH 5.5) and incubated at 37°C. Digital images of the plates were taken by using an image scanner (Epson, Nagano, Japan).
Quantification of cell-cell fusion.
For analysis of cell-cell fusion by H-83/186 virus, Vero cell monolayers in 96-well culture plates were infected with the virus at an MOI of 0.5 or 5 and incubated at 37°C for 42 h in MEM containing 1% fetal calf serum. After three washes with MEM, the cells were further incubated at 37°C for 8 h in MEM containing 4 mg/ml of fatty acid-free bovine serum albumin (Sigma) in the presence or absence of 50 μM LPA (1-oleoyl-sn-glycero-3-phosphate; Sigma). In the experiment using both LPA and Y-27632 (Calbiochem), treatment of H-83/186 virus-infected cells with 20 μM Y-27632 was started at 24 h postinfection and ended at 50 h postinfection.
For analysis of cell-cell fusion by WT virus, Vero cell monolayers in 96-well culture plates were incubated in MEM with or without 20 μM Y-27632 at 37°C for 24 h. The cells were then inoculated with the virus (MOI of 5) at 37°C for 1 h and further incubated in MEM in the presence or absence of 20 μM Y-27632 at 37°C for 20 h. In the experiment using LPA, WT virus-infected Vero cells were incubated in MEM containing 1% fetal calf serum for 12 h and further incubated for 8 h in MEM containing 4 mg/ml of fatty acid-free bovine serum albumin in the presence or absence of 50 μM LPA.
Finally, the cells were fixed with 4% paraformaldehyde and stained with Giemsa's solution. For each sample, the number of syncytia (with more than four nuclei) was counted microscopically, and the average number of syncytia per well was estimated by observing six wells. Otherwise, digital images of 15 randomly chosen fields were obtained for each sample and subjected to morphometric measurement of syncytia, and the average fusion index was estimated as described previously (41).
Cell surface biotinylation and immunoprecipitation.
Vero cell monolayers in six-well culture plates were infected with H-83/186 or WT virus and treated with LPA or Y-27632 as described above. Biotinylation of cell surface proteins was then carried out as described previously (40) with some modifications. Briefly, the cells were treated with 0.3 mg/ml sulfo-NHS-LC-biotin (sulfosuccinimidyl-6-biotinamido-hexanoate; Pierce) solution in PBS containing 0.1 mM CaCl2 and 1 mM MgCl2 at 23°C for 30 min and lysed with 500 μl/well of lysis buffer (25 mM HEPES [pH 7.6], 1% Triton X-100, 3 mM β-glycerophosphate, 3 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 137 mM NaCl). For the coimmunoprecipitation experiment, another lysis buffer containing lauryl maltoside (50 mM HEPES [pH 7.3], 10 mM lauryl maltoside, 1 mM phenylmethylsulfonyl fluoride, 100 mM NaCl) was used as reported by Yao et al. (47).
The viral glycoproteins were immunoprecipitated from the cell lysate with protein A-Sepharose Fast Flow (GE Healthcare Bio-Sciences AB, Sweden), to which anti-HN (42S1) or anti-F (144-1A) monoclonal antibody (42) had been bound in advance. After the Sepharose beads had been washed with lysis buffer or with buffer containing lauryl maltoside (for coimmunoprecipitation), the proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electroblotted to Hybond-P polyvinylidene difluoride membranes (Amersham Biosciences).
For detection of the biotinylated proteins on the membrane by enhanced chemiluminescence, the membrane was treated with streptavidin-biotin-peroxidase complex (Vector Laboratories) and Western blotting Luminol reagent (Santa Cruz Biotechnology), followed by exposure to X-ray film (Konica, Tokyo, Japan). Quantification of the signals on the developed film was performed with the aid of the graphics software NIH Image, version 1.60.
Molecular modeling of the HN proteins.
Molecular modeling of the WT HN and H-186 HN proteins was performed on the automated comparative protein modeling server SWISS-MODEL (http://swissmodel.expasy.org/SWISS-MODEL.html) by using the PIV5 HN protein structure (Protein Data Bank code 1Z4Y) as the template. The structures of the HN proteins were analyzed with the aid of the Deep View Swiss-Pdb Viewer software program (GlaxoSmithKline R&D and the Swiss Institute of Bioinformatics).
RESULTS
Properties of recombinant viruses in Vero cells after infection at low MOIs.
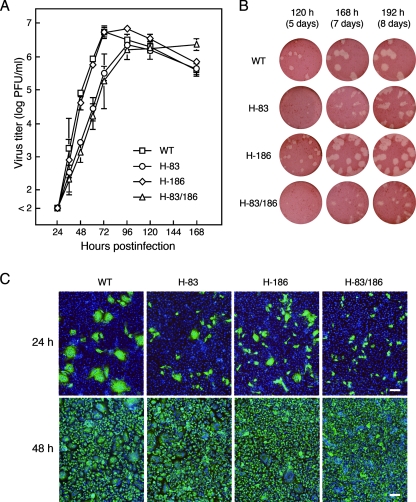
We previously reported that the neutralization escape variant F13, which has N83Y and M186I mutations in the HN protein and no mutation in the F protein, exhibits a markedly reduced ability to induce cell-cell fusion either in HeLa cells or in Vero cells compared to its highly fusogenic parent Toshiba strain (49). Since F13 did perform multistep replication in these cells, we supposed that its ability to mediate virus-cell fusion might not significantly be affected by the mutations. To investigate the potential role of HPIV2 HN protein in the regulation of cell-cell fusion, we generated three recombinant viruses, H-83, H-186, and H-83/186, which harbor the N83Y, M186I, and N83Y plus M186I mutations, respectively. First, in order to see the multistep growth properties of the mutant viruses, Vero cell monolayers were infected with each recombinant virus at a low MOI (0.0001), and the infectious virus released into the culture supernatant was quantified by a plaque-forming assay (Fig. 1A). The production of all the three mutant viruses was as efficient as that of WT virus although the virus production rates of H-83 and H-83/186 viruses were slower than those of WT and H-186 viruses. Intriguingly, the amount of H-83/186 virus in the culture supernatant was increasing even at 168 h postinfection, while that of the other viruses decreased within 120 h postinfection. Figure 1B shows the morphology of representative plaques formed by the recombinant viruses: the plaques formed by either of the viruses were generated after the appearance of extensive cell-cell fusion and subsequent cell death (data not shown). Although WT and H-186 viruses formed clear plaques within 120 h postinfection, the plaques formed by H-83/186 virus were hardly visible at 120 h postinfection and were small and fuzzy even at 192 h postinfection. The plaques by H-83 virus exhibited an intermediate phenotype between those of H-186 and H-83/186 viruses. On the other hand, most of the WT virus-infected cells mediated prominent syncytia (cell-cell fusion) within 24 h postinfection, whereas the H-83/186 virus-infected cells remained as nonfused single cells (Fig. 1C). H-83 and H-186 viruses displayed intermediate phenotypes between those of WT and H-83/186 viruses in terms of the ability to induce cell-cell fusion at 24 h postinfection. Therefore, the onset of cell-cell fusion by H-186 virus was evidently retarded compared to that of WT virus, whereas it mediated extensive cell-cell fusion at a WT level after 120 h, as described above (Fig. 1B). Notably, on the other hand, every recombinant virus could infect almost all cells in the well within 48 h postinfection (Fig. 1C). These results indicate that both the N83Y and M186I mutations in the HN protein are needed to effectively suppress cell-cell fusion as demonstrated by the properties of H-83/186 virus, which can perform multistep replication while inducing very slow cell-cell fusion. It should be pointed out that either of the mutations is deleterious to the viral activity to induce cell-cell fusion.
FIG. 1.
Properties of recombinant viruses in Vero cells after infection at low MOIs. (A) Growth kinetics. Vero cell monolayers in 24-well culture plates were infected with each mutant or WT virus at an MOI of 0.0001 and incubated at 37°C. The amount of infectious virus in the culture supernatant at the time points indicated was quantified by plaque-forming assay on Vero cells. Results are averages of triplicate samples; error bars indicate standard deviations. Representative plaque morphology is presented in panel B. (C) Syncytium formation and spread of infection. Confluent monolayers of Vero cells on glass coverslips in six-well culture plates were infected with each virus at an MOI of 0.05. After incubation at 37°C for the times indicated, the cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100. The infected cells were detected with antinucleoprotein monoclonal antibody and fluorescein isothiocyanate-conjugated secondary antibody while the nuclei of cells were visualized with DAPI. Scale bar, 100 μm.
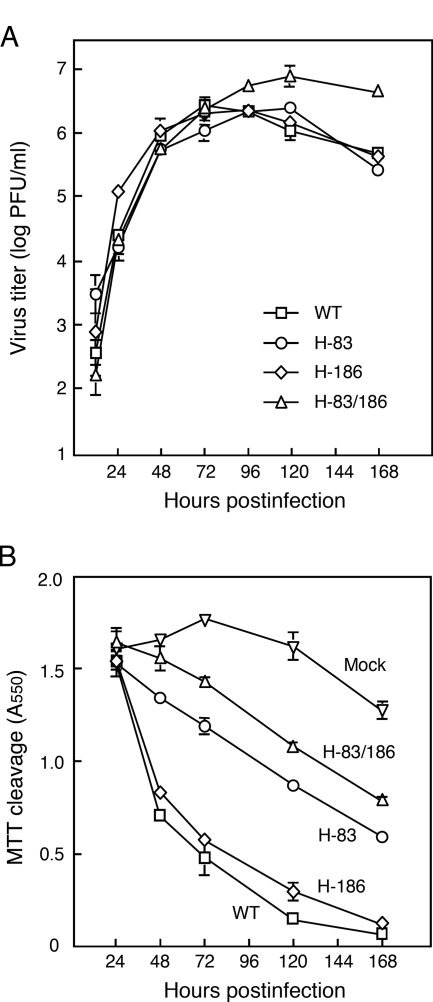
H-83/186 virus exhibits low cytopathicity.
As described above, at least during 24 h after infection, Vero cells that had been infected with H-83/186 virus at a low MOI did not fuse with the surrounding uninfected cells. Therefore, it seemed very likely that the seemingly slow production rate of H-83/186 virus shown in Fig. 1A might reflect the slow rate of cell-to-cell spread of the virus in the early phase of infection. Thus, to see whether there is substantial difference in virus production rates between the recombinant viruses, Vero cells were infected with each recombinant virus at a high multiplicity (MOI of 5), and infectious virus in the culture supernatant was quantified. As shown in Fig. 2A, this experiment proved that the four recombinant viruses displayed nearly identical growth kinetics for 72 h after infection. Interestingly, however, the amount of WT and H-186 viruses reached a peak level at 72 h postinfection, while that of H-83 and H-83/186 viruses reached a peak level at 120 h postinfection. Notably, the amount of H-83/186 virus at 168 h postinfection was about 1 order of magnitude higher than the amounts of other viruses. On the other hand, WT and H-186 viruses induced extensive cell-cell fusion and exhibited severe cytopathicity within 48 h postinfection, whereas H-83/186 virus induced limited cell-cell fusion even at 72 h postinfection (data not shown). Intriguingly, H-83 virus induced more cell-cell fusion at 24 h postinfection but less cell-cell fusion (or cytopathicity) at 48 h or 72 h postinfection than H-186 virus (data not shown). Therefore, in order to compare kinetically the cytopathicity of the recombinant viruses, Vero cells were infected at an MOI of 5, and viability was measured (Fig. 2B). At 48 h postinfection, the viability of cells infected with WT and H-186 viruses became less than 50% that of mock-infected control cells, whereas the viability of H-83/186 virus-infected cells was nearly equal to that of the control cells. Notably, even at 168 h postinfection, the viability of H-83/186 virus-infected cells was more than 50% that of control cells while the cells infected with WT or H-186 viruses exhibited little viability. It should be pointed out, however, that extensive cell-cell fusion was observed in the H-83/186 virus-infected cells at 168 h postinfection (data not shown). The viability of H-83 virus-infected cells was intermediate between that of cells infected with H-186 virus and H-83/186 virus at any time point (Fig. 2B), which is consistent with the above finding that the progression of cell-cell fusion induced by H-83 virus was slower than that induced by H-186 virus (Fig. 1B). These results indicate that the mutant viruses exhibit a marked difference in their abilities to induce cell-cell fusion and to exhibit cytopathicity whereas they replicate as efficiently as the WT virus. Notably, the mutant virus H-83/186 carrying both the N83Y and M186I mutations exhibited the lowest cytopathicity among the recombinant viruses, which is consistent with its having the lowest ability to induce cell-cell fusion.
FIG. 2.
Properties of recombinant viruses in Vero cells after infection at a high MOI. Vero cell monolayers in 96-well culture plates (A) or those in 12-well culture plates (B) were infected with each virus at an MOI of 5 and incubated at 37°C. (A) Growth kinetics. The amount of infectious virus in the culture supernatant was quantified by plaque-forming assay. Results are averages of triplicate samples; error bars indicate standard deviations. (B) Cell viability. The viability of infected cells was measured by the MTT method as described in Materials and Methods. Results are averages of triplicate samples; error bars indicate standard deviations.
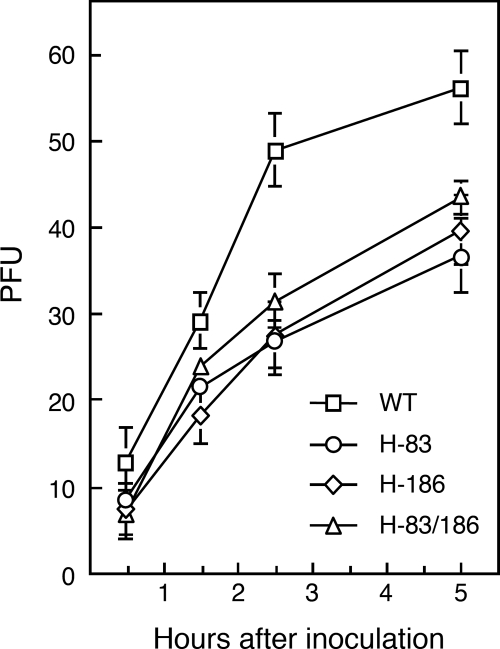
Ability of the recombinant viruses to undergo penetration.
Although H-83/186 virus induced very slow cell-cell fusion, it could perform multistep replication at nearly WT levels. Thus, it seemed as if the ability of H-83/186 virus to mediate virus-cell fusion was not significantly affected by the HN protein mutations. Thus, in order to examine the fusing activity of the recombinant viruses in terms of virus-cell fusion, we carried out the penetration assay. It should be noted that the penetration assay may reflect not only the efficiency of virus-cell fusion but also other factors such as viral attachment efficiency to cells. First, Vero cell monolayers were inoculated with each recombinant virus (approximately 75 PFU/well). Second, the cells were treated with acidic buffer (pH 3.0) at 0.5, 1.5, 2.5, and 5 h after inoculation to inactivate nonfused, and thus unpenetrated, viruses. Finally, the cells were overlaid with agarose-containing medium, and the number of plaques was counted, which would represent the number of the virus particles that had completed penetration (or virus-cell fusion). As shown in Fig. 3, the penetration rates of the mutant viruses were apparently slower than that of WT virus, suggesting that either the N83Y or M186I mutation affects the penetration efficiency. Importantly, H-83/186 virus proved to infect cells with a penetration rate similar to that of H-186 virus, which can induce extensive cell-cell fusion at a WT level as described above.
FIG. 3.
Penetration rate of recombinant viruses. Vero cell monolayers in 24-well culture plates were inoculated with each virus (about 75 PFU/cell), and the number of plaques was counted as described in Materials and Methods. Results are averages of triplicate samples; error bars indicate standard deviations.
The M186I mutation raises neuraminidase activity.
In the case of a rubulavirus mumps virus (20, 45) and a respirovirus HPIV3 (11), it has been shown that a given virus with high neuraminidase activity exhibits reduced ability to induce cell-cell fusion, presumably through depletion of sialoconjugate receptors on the cell surface. If this were also the case with H-83/186 virus, then the neuraminidase activity of the virus under physiological conditions (37°C and pH 7.0) would be much higher than that of H-186 virus or WT virus. As shown in Fig. 4A, every recombinant virus showed higher neuraminidase activity at pH 5.5 than at pH 7.0, which is consistent with the general property of the paramyxovirus neuraminidases which have acidic pH optima (16). Notably, it was proved that the neuraminidase activity of H-83 virus was lower while that of H-186 virus or H-83/186 virus was higher than that of WT virus independently of pH, indicating that the M186I mutation raises neuraminidase activity while the N83Y mutation lowers it. When measured at pH 7.0, interestingly, the neuraminidase activity of H-83/186 virus was the highest among the viruses, suggesting that the reduced ability of the virus to induce cell-cell fusion might be due to its “high” neuraminidase activity. However, since H-83 virus exhibited the lowest neuraminidase activity even though it induced cell-cell fusion less efficiently than the WT virus, as described above, a clear correlation between neuraminidase activity and cell-cell fusion could not be corroborated.
FIG. 4.
Interaction of recombinant viruses with receptor. (A) Neuraminidase activity. Neuraminidase (NA) activity of each virus (107 PFU) was measured as described in Materials and Methods. Results are averages of triplicate samples; error bars indicate standard deviations. (B) Hemagglutinating activity. Hemagglutinating activity of each virus (107 PFU) at pH 7.0 was measured by incubation at 4°C or 37°C as described in Materials and Methods. The HAU values of duplicate samples which gave identical results are shown. (C) Receptor-binding activity at different pH. Virus suspension (4 HAU) in 25 μl of PBS-GB (pH 7.0 or pH 5.5) was mixed with an equal volume of 0.4% suspension of guinea pig erythrocytes in PBS-GB (pH 7.0 or pH 5.5) and incubated at 37°C for the times indicated.
The M186I mutation reduces receptor-binding activity.
Since H-186 and H-83/186 viruses proved to possess significantly high neuraminidase activity at pH 7.0 compared to WT and H-83 viruses, we anticipated that at physiological pH and temperature, receptor-binding activities of the former viruses would be much lower than those of the latter viruses. To test this hypothesis, the receptor-binding activities of the viruses were measured at pH 7.0 under conditions in which the neuraminidase activity is active (37°C) or inactive (4°C). First, 107 PFU of each virus was suspended in PBS-GB (pH 7.0), and serial twofold dilutions were made in U-type 96-well plates at 4°C. Second, guinea pig erythrocyte suspension was added to each well at 4°C. Finally, the plates were incubated at 4°C or 37°C and the number of HAU was estimated. As shown in Fig. 4B, after incubation with erythrocytes at 4°C for 3 h, WT virus showed the highest receptor-binding activity (28 HAU) while H-83/186 virus showed the lowest (26 HAU). The H-83 and H-186 viruses had the same HAU value (27), which was an intermediate between the values of WT and H-83/186 viruses. Thus, it seemed as if the HAU value of a given virus (measured at 4°C) paralleled its ability to induce cell-cell fusion. However, when viruses were incubated with erythrocytes at a physiological temperature (37°C) for 3 h, the HAU values of H-186 and H-83/186 dropped below the detection level, while WT and H83 viruses gave the same HAU as that measured at 4°C. Notably, after incubation at 37°C for 12 h, H-83 virus still retained the same HAU value whereas WT virus showed no detectable hemagglutination. To see more precisely the receptor-binding activities of the recombinant viruses at physiological temperature, each virus (4 HAU) was mixed with erythrocytes, and the time course of the hemagglutination profile was observed (Fig. 4C, left panel). Without virus, sedimentation of erythrocytes terminated within 30 min. When erythrocytes were mixed with H-186 or H-83/186 virus, sedimentation of erythrocytes terminated within 2 h or 4 h, respectively. When erythrocytes were mixed with WT virus, sedimentation of erythrocytes progressed very slowly and did not terminate even after 6 h. As expected, no sedimentation of erythrocytes was observed when they were mixed with H-83 virus, indicating that the binding of the virus to the receptor was remarkably quick and stable compared to that of H-186 and H-83/186 viruses. Therefore, the M186I mutation lowered the receptor-binding activity of the HN protein while the N83Y mutation raised it, most likely by elevating or reducing the neuraminidase activity, respectively. Taken together, under physiological conditions, the receptor-binding activity of H-83 virus was higher, while that of H-186 and H-83/186 was lower, than that of the WT virus. Since the receptor-binding activity of H-83/186 virus was slightly higher than that of H-186 virus, presumably due to the N83Y mutation, the reduced ability of H-83/186 virus to induce cell-cell fusion did not seem to be attributable to its “relatively low” receptor-binding activity. On the other hand, faster sedimentation of erythrocytes was observed at pH 5.5 than at pH 7.0 for all of the recombinant viruses; in the case of H-186 and H-83/186 viruses, the sedimentation terminated within 30 min at pH 5.5. Interestingly, when erythrocytes were mixed with H-83 virus at pH 5.5, sedimentation of erythrocytes could be observed within 2 h, indicating that even the low neuraminidase activity of H-83 virus might be enough to cleave the sialoconjugates in the acidic interior of the trans-Golgi network, which would enable the virus to be efficiently released from the infected cells.
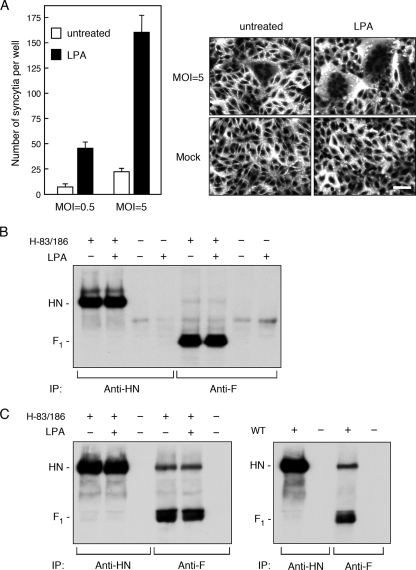
LPA promotes syncytium formation by H-83/186 virus.
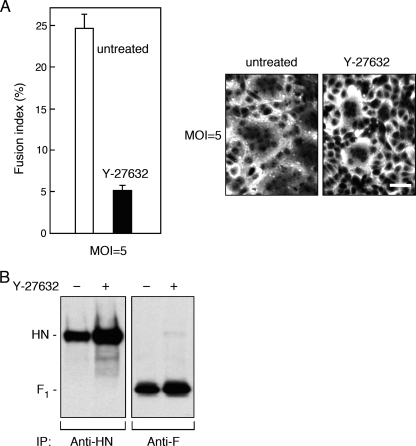
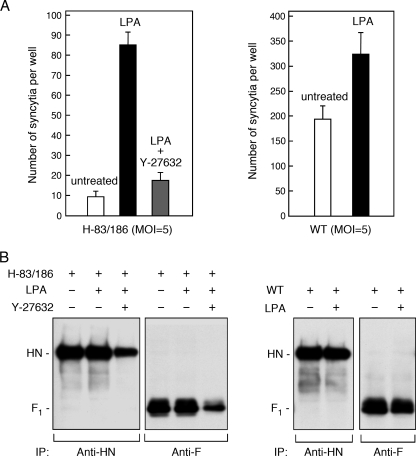
The data presented so far indicated that neither high neuraminidase activity nor low receptor-binding activity of H-83/186 virus confers its reduced ability to induce cell-cell fusion. On the other hand, it has been recently reported for respiratory syncytial virus (RSV), a member of genus Pneumovirus in the family Paramyxoviridae (16), that RhoA signaling is required for an infected cell to fuse with a target cell (8). RhoA is a member of Rho family small GTPases, in which RhoA, Rac1, and Cdc42 have been studied in great detail (46). To see whether activation of RhoA could also facilitate syncytium formation of cells infected with H-83/186 virus, we treated the H-83/186 virus-infected Vero cells with the lipid mediator LPA, which activates multiple signal transduction pathways including those initiated by small GTPases of Rho and Ras families (23). As shown in Fig. 5A and B, LPA greatly promoted syncytium formation induced by H-83/186 virus without affecting the expression levels of viral glycoproteins on the cell surface, indicating that activation of the GTPases of Rho and Ras families could compensate H-83/186 virus for the reduced ability to induce cell-cell fusion. To investigate whether the physical interaction between the HN and F proteins would be affected by the mutations in the HN protein, we carried out a coimmunoprecipitation experiment (Fig. 5C). Notably, it was proved that the H-83/186 HN protein was coprecipitated with the F protein similarly to the WT HN protein although the F protein was only faintly coprecipitated with these HN proteins. Furthermore, treatment with LPA did not affect the coprecipitation efficiency of the H-83/186 HN protein. Thus, it was indicated that the reduced ability of H-83/186 virus to induce cell-cell fusion was not due to a defect in the physical interaction between HN and F proteins on the infected cell surface. These findings suggested, on the other hand, that WT virus may somehow activate RhoA or other small GTPase(s) in the infected cells and thereby facilitate cell-cell fusion. To ascertain whether this assumption is correct, we treated WT virus-infected Vero cells with the Rho kinase inhibitor Y-27632, which specifically blocks RhoA signaling (12, 43), and found that cell-cell fusion by WT virus was effectively inhibited by Y-27632 (Fig. 6A). Intriguingly, the expression level of either the HN or F protein on the surface of Y-27632-treated cells was about 1.6-fold higher than that on the surface of untreated control cells (Fig. 6B), indicating that the reduction of cell-cell fusion by Y-27632 is not due to suppression of viral glycoprotein expression. Similarly, the promoting effect of LPA on H-83/186 virus-mediated cell-cell fusion was effectively suppressed by Y-27632 (Fig. 7A, left panel). However, since the cell surface expression levels of HN and F proteins of H-83/186 virus decreased by about 50% when cells were treated with LPA and Y-27632 compared to cells treated with LPA only (Fig. 7B, left panel), involvement of RhoA signaling in the LPA-promoted cell-cell fusion by H-83/186 virus could not be certified. On the other hand, treatment of WT virus-infected cells with LPA resulted in about a 1.7-fold increase in the extent of cell-cell fusion without significantly affecting the surface expression levels of the viral glycoproteins (Fig. 7A and B, right panels).
FIG. 5.
Effect of LPA on cells infected with H-83/186 virus. (A) Syncytium formation. Vero cell monolayers in 96-well plates were infected with H-83/186 virus at an MOI of 0.5 or 5. After incubation at 37°C for 42 h, the cells were further incubated for 8 h in the presence or absence of 50 μM LPA. After cells were stained with Giemsa's solution, the average number of syncytia per well was estimated as described in Materials and Methods. Error bars indicate standard deviations. Representative digital images are shown in the right panel (scale Bar, 50 μm). (B) Cell surface expression of viral glycoproteins. Vero cell monolayers in six-well culture plates were infected with H-83/186 virus at an MOI of 5, incubated at 37°C for 42 h, and treated with or without LPA as described above. The cell surface-localized proteins were then biotinylated, immunoprecipitated with anti-HN or anti-F monoclonal antibody, and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by transfer to a polyvinylidene difluoride membrane. The biotinylated proteins on the membrane were detected by enhanced chemiluminescence. (C) Interaction between the HN and F proteins. Immunoprecipitation of the glycoproteins of H-83/186 virus was carried out as described in the legend for panel B, except that the lysis buffer containing lauryl maltoside was used (left panel). As the control, Vero cell monolayers were infected with WT virus at an MOI of 5 and incubated at 37°C for 24 h, and the viral glycoproteins were immunoprecipitated (right panel).
FIG. 6.
Effect of Y-27632 on cells infected with WT virus. (A) Syncytium formation. Vero cell monolayers in 96-well plates were incubated with or without 20 μM Y-27632 at 37°C for 24 h. The cells were then inoculated with WT virus at an MOI of 5 at 37°C for 1 h and further incubated at 37°C for 24 h in the presence or absence of 20 μM Y-27632. After cells were stained with Giemsa's solution, the average fusion index was estimated as described in Materials and Methods. Error bars indicate standard deviations. Representative digital images are shown in the right panel (scale bar, 50 μm). (B) Cell surface expression of viral glycoproteins. Vero cell monolayers in six-well culture plates were infected with WT virus at an MOI of 5 and treated with or without Y-27632 as described above. After biotinylation, the viral glycoproteins on the cell surface were immunoprecipitated and visualized as described in the legend of Fig. 5B.
FIG. 7.
Effects of LPA and Y-27632 on cells infected with H-83/186 and WT viruses. (A) Syncytium formation. Vero cell monolayers in 96-well plates were infected with WT or H-83/186 virus at an MOI of 5. After incubation at 37°C for 12 h (WT virus) or 42 h (H-83/186 virus), the cells were further incubated for 8 h in the presence or absence of 50 μM LPA. Treatment of H-83/186 virus-infected cells with Y-27632 was started at 24 h postinfection and ended at 50 h postinfection. After cells were stained with Giemsa's solution, the average number of syncytia per well was estimated as described in Materials and Methods. Error bars indicate standard deviations. (B) Cell surface expression of viral glycoproteins. Vero cell monolayers in six-well plates were infected with WT or H-83/186 virus and treated with LPA (and Y-27632) as described above. After biotinylation, the viral glycoproteins on the cell surface were immunoprecipitated and visualized as described in the legend of Fig. 5B.
Taken together, these results suggest that signaling via RhoA may be required for the induction of cell-cell fusion by HPIV2 and that WT HN protein may somehow trigger this signal transduction pathway while the H-83/186 HN protein may not efficiently do so.
It is worth noting that even the WT HN protein appeared not to be fully active in triggering signal transduction since LPA can promote WT virus-mediated cell-cell fusion to a moderate degree.
The M186I mutation can affect the conformation of the receptor-binding site.
As described above, both the mutations N83Y and M186I in the HN protein proved to be involved in the reduced ability of H-83/186 virus to induce cell-cell fusion. It was also proved that the M186I mutation raises neuraminidase activity and lowers receptor-binding activity. By contrast, the N83Y mutation lowers neuraminidase activity and raises receptor-binding activity. Intriguingly, Met-186 is located in the globular head region of the HN protein while Asn-83 resides in the stalk region, as suggested previously (49). Recently, the three-dimensional (3D) structure of the globular head region of PIV5 HN protein was clarified and indicated that a single site is involved in both the receptor-binding and neuraminidase activities (48). Since HPIV2 and PIV5 are closely related members of genus Rubulavirus (16), we were able to predict the 3D structure of WT HN protein by molecular modeling on the web using the PIV5 HN structure as the template. As shown in Fig. 8A and D, it was suggested that Met-186 is located at the dimer interface and very close to the receptor-binding site. Therefore, in order to see whether the M186I mutation would affect the structure of the receptor-binding site, we predicted the 3D structure of the H-186 HN protein, which has Ile-186 (Fig. 8B and E), and compared it with that of WT HN protein, which has Met-186 (Fig. 8C and F). Notably, we found that there is subtle but clear difference between the two structures in the predicted positions of eight amino acids ([aa] 191 to 198) that are located in the vicinity of Met-186 (or Ile-186). Importantly, Leu-191 is the counterpart of Gln-186 in the PIV5 HN protein, which is directly involved in binding to the sialyllactose, a neuraminidase substrate (48). Interestingly, it was suggested that, due to the M186I mutation, the side chain of Leu-191 might change its position toward the center of the receptor-binding site by about 1.5 Å. Therefore, the difference in neuraminidase and receptor-binding activities between H-186 and WT viruses could be explained by a possible difference in conformation of the receptor-binding sites between their HN proteins.
FIG. 8.
Possible influence of the M186I mutation on the conformation of HN protein. Top views (A, B, and C) and side views (D, E, and F) of the HN monomers. Molecular modeling of WT HN (A and D) and H-186 HN (B and E) proteins was performed as described in Materials and Methods. Shown in panels C and F are superimposed images of WT and H-186 HN proteins. Sixteen amino acids (Arg-168, Ile-169, Leu-191, Tyr-246, Glu-252, Tyr-256, Tyr-311, Tyr-357, His-358, Glu-395, Arg-410, Ser-412, Ile-464, Arg-502, Tyr-530, and Glu-551) are shown in cyan as a space-filling model, except for Leu-191 in the H-186 HN protein, which is shown in red. The PIV5 HN counterparts of these 16 amino acids are involved in binding to sialyllactose, a neuraminidase substrate (48). Met-186 in the WT HN protein and Ile-186 in the H-186 HN protein are shown in yellow and blue, respectively. The carbon and oxygen backbones of the amino acids from Leu-191 to Gln-198 in the H-186 HN protein, whose positions differ from the WT HN counterparts, are shown in red. In panels A and D, the backbones and side chains of the amino acids that would be buried in the dimer interface (48) are shown in purple. The backbones of the remaining amino acids are shown in green. Arrowheads in D, E, and F indicate the positions of Leu-123, which is located at the boundary between the stalk and globular head regions (48).
DISCUSSION
In the present study, we have shown that the recombinant virus H-83/186, which contains amino acid mutations N83Y and M186I in the HN protein, induces considerably slower cell-cell fusion than WT virus. We have also shown that H-83/186 virus undergoes penetration as efficiently as does H-186 virus, which possesses the M186I mutation only and induces extensive cell-cell fusion at a WT level. Accordingly, H-83/186 virus can perform multistep replication while inducing minimal cell-cell fusion, and thus the infected cells continue to produce infectious virus longer than cells infected with H-186 or WT virus.
Interestingly, Seth et al. (36) reported that the F protein of SER virus, a pig isolate of PIV5, has low-pH-dependent membrane-fusing activity, and thus entry of the virus occurs in the endosome. Accordingly, cells infected with SER virus do not display apparent cell-cell fusion, but treating the infected cells with low pH results in induction of cell-cell fusion (36). However, Bissonnette et al. (2) could not confirm the requirement for low-pH triggering of SER virus-mediated fusion. In contrast, it has been reported for Hendra virus, a member of genus Henipavirus in the family Paramyxoviridae (16), that the entry of the virus occurs via an endocytic pathway, in which the uncleaved precursor F0 on the virus is cleaved by the endosomal enzyme cathepsin L and then mediates virus-cell fusion in the endosome (21, 27). Thus, Hendra virus can infect cells and perform multistep replication without inducing cell-cell fusion. With regard to H-83/186 virus, cell-cell fusion of the infected cells was not promoted by low-pH treatment (our unpublished data). Moreover, extensive cell-cell fusion at physiological pH could be observed in the H-83/186 virus-infected cells after a long incubation period such as 8 days (Fig. 1B) (our unpublished observations). It should also be noted that the cell surface-localized F protein of H-83/186 virus was fully cleaved the same as that of WT virus (Fig. 6B and 8B). On the basis of these findings, the reduced ability of H-83/186 virus to induce cell-cell fusion could not be explained by pH-dependent membrane fusion or by a defect in the F protein cleavage.
We have found that the limited cell-cell fusion by H-83/186 virus can be significantly promoted by LPA. Importantly, LPA can also promote WT virus-mediated cell-cell fusion to a moderate degree. LPA is a bioactive phospholipid that activates multiple signal transduction pathways including those initiated by Rho and Ras family GTPases, thereby regulating cell proliferation, migration, and morphogenesis (23, 28). Y-27632, in contrast, is a highly selective inhibitor of Rho kinases that are activated by RhoA, a member of Rho family GTPases, and thus does not inhibit p21-mediated kinases (25, 43) that are activated by other members of the Rho family, namely, Cdc42 and Rac1 (35). Our present finding that the cell-cell fusion caused by WT virus can be effectively inhibited by Y-27632 strongly suggests that activation of RhoA-mediated signaling pathways is involved in the induction of cell-cell fusion caused by HPIV2. In all likelihood, therefore, LPA may promote H-83/186 virus-mediated cell-cell fusion through activation of RhoA-mediated signaling pathways, although the inhibitory effect of Y-27632 on the LPA-promoted cell-cell fusion could not definitely be certified since it also affected surface expression levels of the HN and F proteins. Presumably, blocking Rho kinases by Y-27632 in the H-83/186 virus-infected cells might have resulted in activation of other LPA-stimulated signaling pathway(s), which might affect surface expression levels of the viral glycoproteins as well as cell-cell fusion. It should be pointed out in this context that LPA does promote H-83/186 virus-mediated cell-cell fusion without affecting either the surface expression levels of the viral glycoproteins or the physical interaction between HN and F proteins.
Notably, it was reported that activation of RhoA in Swiss/3T3 cells results in formation of vinculin-mediated focal adhesions and assembly of actin stress fibers (33), and it has been suggested that RhoA-mediated actin polymerization in HEp-2 cells is associated with the syncytium-inducing phenotype of RSV (8). However, typical actin stress fibers were not observed in Vero cells even after treatment with LPA or infection with WT virus, while they were abundantly formed in HeLa cells or LLC-MK2 cells irrespective of LPA treatment or virus infection (our unpublished observations). Moreover, we could not observe any definite effect of Y-27632 on the actin distribution in Vero cells whereas the actin stress fibers in HeLa or LLC-MK2 cells were severely disrupted by Y-27632 (our unpublished observations). Thus, further investigations are needed to clarify the mechanism by which the WT HN protein triggers the putative RhoA-mediated signal transduction and to elucidate what kind(s) of cellular molecule functions in promoting cell-cell fusion when the infected cells are activated by this signal transduction.
It has been reported that RhoA and its downstream signaling cascades are activated during RSV infection of HEp-2 cells (9), and the Rho kinase inhibitor Y-27632 effectively reduces RSV-induced cell-cell fusion (8). Schowalter et al. (34) have shown, however, that the effects of Rho family GTPases on cell-cell fusion mediated by paramyxovirus glycoproteins are dependent on both the viral glycoproteins and the cell type. For example, constitutively active RhoA inhibits cell-cell fusion of BHK cells induced by Hendra virus glycoproteins but does not affect that induced by PIV5 glycoproteins. For HPIV2, the expression of the WT HN protein in the infected Vero cells seems to activate signal transduction via RhoA, leading to promotion of cell-cell fusion. Thus, the cell-cell fusion-promoting function of HPIV2 HN protein may include not only the interaction with the F protein but also a putative new role played in the signal transduction which is apparently not required for virus-cell fusion. Although it seems likely that receptor binding of the HN protein triggers the activation of RhoA, stable binding may not be required for the triggering since H-186 virus can induce cell-cell fusion at a WT level despite its extremely low receptor-binding activity.
Several amino acid mutations in the stalk region of the NDV HN protein are known to affect the neuraminidase activity (17, 44), whose active site is located in the globular head region (22). In addition, it has been reported that severe loss of neuraminidase activity of the HPIV3 C28a variant is due to synergistic effect of two mutations in the HN protein: P111S in the stalk region and D216N in the globular head region (30, 32). Our present observation that the N83Y mutation lowers neuraminidase activity also seems consistent with the involvement of the stalk region in the neuraminidase function. In the absence of information on the 3D structure of the stalk region, however, it is not clear whether the stalk region can sterically influence the neuraminidase active site in the globular head region. Interestingly, the mutant virus H-83/186, which harbors mutations in the stalk (N83Y) and globular head (M186I) regions of the HN protein, exhibits the highest neuraminidase activity at physiological pH among the viruses used in the present study. Furthermore, the receptor-binding activity of H-83/186 virus is very low, most likely due to its high neuraminidase activity. However, the low receptor-binding activity of the virus cannot explain its reduced ability to promote cell-cell fusion. This is consistent with the result of mutational analysis of the NDV HN protein, indicating that neither neuraminidase activity nor receptor-binding activity correlates with the fusion-promoting activity of the HN protein (5) although HN protein receptor binding is required for cell-cell fusion (18). In the case of mumps virus (20, 45) and HPIV3 (11, 32), by contrast, it has been shown that either high neuraminidase activity or low receptor-binding activity of the HN protein counteracts the fusion promoting function. Interestingly, Ah-Tye et al. (1) have reported that HPIV3 does not induce cell-cell fusion of CV1 cells after infection at a high multiplicity (MOI of 5), presumably due to depletion of receptors on the cell surface by neuraminidase, whereas HPIV1 and HPIV2 induce cell-cell fusion under the same conditions. Consistently, our present study has shown that HPIV2 (WT and H-186 viruses) can induce extensive cell-cell fusion in Vero cells when the cells are infected at an MOI of 5. Thus, there is an interesting discrepancy in receptor requirement for cell-cell fusion between HPIV2 and HPIV3. Whether the difference in biological properties and pathogenicity between these viruses reflects this discrepancy remains to be examined.
We have previously suggested that an amino acid domain (aa 37 to 94) in the stalk region and another amino acid domain (aa 148 to 209) in the globular head region work cooperatively when HPIV2 HN protein promotes cell-cell fusion through its interaction with the F protein (41). Moreover, we found that the N83Y and M186I mutations in the HN protein of F13, which exhibits an aberrant fusion phenotype, had taken place in these domains (41, 49). However, our present study has indicated that these mutations do not affect the HN-F interaction per se. Rather, they seem to hamper the presumptive role of HN protein in triggering the RhoA-mediated signal transduction pathways, leading to effective suppression of cell-cell fusion. We also found that the N83Y mutation reduces cell-cell fusion to some extent and that the M186I mutation brings a delay to the onset of cell-cell fusion. Therefore, either of the mutations seems to have a deleterious, but not particularly strong, effect on the HN protein's activity in promoting cell-cell fusion. It is likely that the elevated neuraminidase activity due to M186I mutation suppresses cell-cell fusion by reducing the receptor-binding efficiency of the HN protein, but this suppression, if it occurs at all, seems to be temporary. With regard to the N83Y mutation, we assume that it suppresses cell-cell fusion to some extent by moderately inhibiting the putative signal-transducing activity of the HN protein. If these assumptions are correct, then combination of the N83Y and M186I mutations may result in more effective inhibition of cell-cell fusion. Another possibility is that there exists cooperation between the two mutations in terms of the inhibition of the signal transduction. Since Met-186 is predicted to be located at the dimer interface of the HN protein, the M186I mutation may affect the integrity of the dimer interface, possibly resulting in a conformational change in the stalk region. This putative conformational change may not greatly influence the signal-transducing activity of the HN protein by itself but may effectively inhibit it when combined with the N83Y mutation in the stalk region.
As discussed so far, we believe that HPIV2 HN protein has a novel function that is involved in RhoA-mediated signal transduction, thereby regulating cell-cell fusion and cytopathicity. Although this notion may not be applicable to other paramyxovirus HN proteins, this putative fourth function of the HN protein would be an effective viral strategy to cope with the host cells, leading to immediate spread of infection by promoting cell-cell fusion or otherwise leading to prolonged virus replication by suppressing cell-cell fusion.
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research (grant 18590447) from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Footnotes
Published ahead of print on 18 June 2008.
REFERENCES
- 1.Ah-Tye, C., S. Schwartz, K. Huberman, E. Carlin, and A. Moscona. 1999. Virus-receptor interactions of human parainfluenza viruses types 1, 2 and 3. Microb. Pathog. 27329-336. [DOI] [PubMed] [Google Scholar]
- 2.Bissonnette, M. L. Z., S. A. Connolly, D. F. Young, R. E. Randall, R. G. Paterson, and R. A. Lamb. 2006. Analysis of the pH requirement for membrane fusion of different isolates of the paramyxovirus parainfluenza virus 5. J. Virol. 803071-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cid-Arregui, A., V. Juárez, and H. zur Hausen. 2003. A synthetic E7 gene of human papillomavirus type 16 that yields enhanced expression of the protein in mammalian cells and is useful for DNA immunization studies. J. Virol. 774928-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Connaris, H., T. Takimoto, R. Russell, S. Crennell, I. Moustafa, A. Portner, and G. Taylor. 2002. Probing the sialic acid binding site of the hemagglutinin-neuraminidase of Newcastle disease virus: identification of key amino acids involved in cell binding, catalysis, and fusion. J. Virol. 761816-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connolly, S. A., and R. A. Lamb. 2006. Paramyxovirus fusion: real-time measurement of parainfluenza virus 5 virus-cell fusion. Virology 355203-212. [DOI] [PubMed] [Google Scholar]
- 7.Deng, R., A. M. Mirza, P. J. Mahon, and R. M. Iorio. 1997. Functional chimeric HN glycoproteins derived from Newcastle disease virus and human parainfluenza virus-3. Arch. Virol. 13(Suppl.)115-130. [DOI] [PubMed] [Google Scholar]
- 8.Gower, T. L., M. K. Pastey, M. E. Peeples, P. L. Collins, L. H. McCurdy, T. K. Hart, A. Guth, T. R. Johnson, and B. S. Graham. 2005. RhoA signaling is required for respiratory syncytial virus-induced syncytium formation and filamentous virion morphology. J. Virol. 795326-5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gower, T. L., M. E. Peeples, P. L. Collins, and B. S. Graham. 2001. RhoA is activated during respiratory syncytial virus infection. Virology 283188-196. [DOI] [PubMed] [Google Scholar]
- 10.Hu, X., R. Ray, and R. W. Compans. 1992. Functional interactions between the fusion protein and hemagglutinin-neuraminidase of human parainfluenza viruses. J. Virol. 661528-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huberman, K., R. W. Peluso, and A. Moscona. 1995. Hemagglutinin-neuraminidase of human parainfluenza 3: role of the neuraminidase in the viral life cycle. Virology 214294-300. [DOI] [PubMed] [Google Scholar]
- 12.Ishizaki, T., M. Uehata, I. Tamechika, J. Keel, K. Nonomura, M. Maekawa, and S. Narumiya. 2000. Pharmacological properties of Y-27632, a specific inhibitor of Rho-associated kinases. Mol. Pharmacol. 57976-983. [PubMed] [Google Scholar]
- 13.Ito, Y., Y. Kimura, I. Nagata, and A. Kunii. 1974. Effects of l-glutamine deprivation on growth of HVJ (Sendai virus) in BHK cells. J. Virol. 13557-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karron, R. A., and P. L. Collins. 2007. Parainfluenza viruses, p. 1497-1526. In D. M. Knipe, P. M. Howley, D. E. Griffin, M. A. Martin, R. A. Lamb, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 15.Lamb, R. A., and T. S. Jardetzky. 2007. Structural basis of viral invasion: lessons from paramyxovirus F. Curr. Opin. Struct. Biol. 17427-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamb, R. A., and G. D. Parks. 2007. Paramyxoviridae: the viruses and their replication, p. 1449-1496. In D. M. Knipe, P. M. Howley, D. E. Griffin, M. A. Martin, R. A. Lamb, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 17.Li, J., E. Quinlan, A. Mirza, and R. M. Iorio. 2004. Mutated form of the Newcastle disease virus hemagglutinin-neuraminidase interacts with the homologous fusion protein despite deficiencies in both receptor recognition and fusion promotion. J. Virol. 785299-5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGinnes, L. W., and T. G. Morrison. 2006. Inhibition of receptor binding stabilizes Newcastle disease virus HN and F protein-containing complexes. J. Virol. 802894-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melanson, V. R., and R. M. Iorio. 2006. Addition of N-glycans in the stalk of the Newcastle disease virus HN protein blocks its interaction with the F protein and prevents fusion. J. Virol. 80623-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merz, D. C., and J. S. Wolinsky. 1981. Biochemical features of mumps virus neuraminidases and their relationship with pathogenicity. Virology 114218-227. [DOI] [PubMed] [Google Scholar]
- 21.Meulendyke, K. A., M. A. Wurth, R. O. McCann, and R. E. Dutch. 2005. Endocytosis plays a critical role in proteolytic processing of the Hendra virus fusion protein. J. Virol. 7912643-12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mirza, A. M., J. P. Sheehan, L. W. Hardy, R. L. Glickman, and R. M. Iorio. 1993. Structure and function of a membrane anchor-less form of the hemagglutinin-neuraminidase glycoprotein of Newcastle disease virus. J. Biol. Chem. 26821425-21431. [PubMed] [Google Scholar]
- 23.Moolenaar, W. H., L. A. van Meeteren, and B. N. Giepmans. 2004. The ins and outs of lysophosphatidic acid signaling. Bioessays 26870-881. [DOI] [PubMed] [Google Scholar]
- 24.Morrison, T. G. 2003. Structure and function of a paramyxovirus fusion protein. Biochim. Biophys. Acta 161473-84. [DOI] [PubMed] [Google Scholar]
- 25.Narumiya, S., T. Ishizaki, and M. Uehata. 2000. Use and properties of ROCK-specific inhibitor Y-27632. Methods Enzymol. 325273-284. [DOI] [PubMed] [Google Scholar]
- 26.Nishio, M., M. Tsurudome, M. Ito, D. Garcin, D. Kolakofsky, and Y. Ito. 2005. Identification of paramyxovirus V protein residues essential for STAT protein degradation and promotion of virus replication. J. Virol. 798591-8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pager, C. T., and R. E. Dutch. 2005. Cathepsin L is involved in proteolytic processing of the Hendra virus fusion protein. J. Virol. 7912714-12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panetti, T. S. 2002. Differential effects of sphingosine 1-phosphate and lysophosphatidic acid on endothelial cells. Biochim. Biophys. Acta 158190-196. [DOI] [PubMed] [Google Scholar]
- 29.Paterson, R. G., S. W. Hiebert, and R. A. Lamb. 1985. Expression at the cell surface of biologically active fusion and hemagglutinin/neuraminidase proteins of the paramyxovirus simian virus 5 from cloned cDNA. Proc. Natl. Acad. Sci. USA 827520-7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porotto, M., O. Greengard, N. Poltoratskaia, M. A. Horga, and A. Moscona. 2001. Human parainfluenza virus type 3 HN-receptor interaction: effect of 4-guanidino-Neu5Ac2en on a neuraminidase-deficient variant. J. Virol. 757481-7488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Porotto, M., M. Murrell, O. Greengard, L. Doctor, and A. Moscona. 2005. Influence of the human parainfluenza virus 3 attachment protein's neuraminidase activity on its capacity to activate the fusion protein. J. Virol. 792383-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Porotto, M., M. Murrell, O. Greengard, and A. Moscona. 2003. Triggering of human parainfluenza virus 3 fusion protein (F) by the hemagglutinin-neuraminidase (HN) protein: an HN mutation diminishes the rate of F activation and fusion. J. Virol. 773647-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ridley, A. J., and A. Hall. 1992. The small GTP-binding protein Rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell 70389-399. [DOI] [PubMed] [Google Scholar]
- 34.Schowalter, R. M., M. A. Wurth, H. C. Aguilar, B. Lee, C. L. Moncman, R. O. McCann, and R. E. Dutch. 2006. Rho GTPase activity modulates paramyxovirus fusion protein-mediated cell-cell fusion. Virology 350323-334. [DOI] [PubMed] [Google Scholar]
- 35.Schwartz, M. 2004. Rho signalling at a glance. J. Cell Sci. 1175457-5458. [DOI] [PubMed] [Google Scholar]
- 36.Seth, S., A. Vincent, and R. W. Compans. 2003. Activation of fusion by the SER virus F protein: a low-pH-dependent paramyxovirus entry process. J. Virol. 776520-6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanabayashi, K., and R. W. Compans. 1996. Functional interaction of paramyxovirus glycoproteins: identification of a domain in Sendai virus HN which promotes cell fusion. J. Virol. 706112-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson, S. D., and A. Portner. 1987. Localization of functional sites on the hemagglutinin-neuraminidase glycoprotein of Sendai virus by sequence analysis of antigenic and temperature-sensitive mutants. Virology 1601-8. [DOI] [PubMed] [Google Scholar]
- 39.Tong, S., M. Li, A. Vincent, R. W. Compans, E. Fritsch, R. Beier, C. Klenk, M. Ohuchi, and H.-D. Klenk. 2002. Regulation of fusion activity by the cytoplasmic domain of a paramyxovirus F protein. Virology 301322-333. [DOI] [PubMed] [Google Scholar]
- 40.Tsurudome, M., M. Ito, M. Nishio, M. Kawano, H. Komada, and Y. Ito. 2006. A mutant fusion (F) protein of simian virus 5 induces hemagglutinin-neuraminidase-independent syncytium formation despite the internalization of the F protein. Virology 34711-27. [DOI] [PubMed] [Google Scholar]
- 41.Tsurudome, M., M. Kawano, T. Yuasa, N. Tabata, M. Nishio, H. Komada, and Y. Ito. 1995. Identification of regions on the hemagglutinin-neuraminidase protein of human parainfluenza virus type 2 important for promoting cell fusion. Virology 213190-203. [DOI] [PubMed] [Google Scholar]
- 42.Tsurudome, M., M. Nishio, H. Komada, H. Bando, and Y. Ito. 1989. Extensive antigenic diversity among human parainfluenza type 2 virus isolates and immunological relationships among paramyxoviruses revealed by monoclonal antibodies. Virology 17138-48. [DOI] [PubMed] [Google Scholar]
- 43.Uehata, M., T. Ishizaki, H. Satoh, T. Ono, T. Kawahara, T. Morishita, H. Tamakawa, K. Yamagami, J. Inui, M. Maekawa, and S. Narumiya. 1997. Calcium sensitization of smooth muscle mediated by a Rho-associated protein kinase in hypertension. Nature 1389990-994. [DOI] [PubMed] [Google Scholar]
- 44.Wang, Z., and R. M. Iorio. 1999. Amino acid substitutions in a conserved region in the stalk of the Newcastle disease virus HN glycoprotein spike impair its neuraminidase activity in the globular domain. J. Gen. Virol. 80749-753. [DOI] [PubMed] [Google Scholar]
- 45.Waxham, M. N., and J. Aronowski. 1988. Identification of amino acids involved in sialidase activity of the mumps virus hemagglutinin-neuraminidase protein. Virology 167226-232. [DOI] [PubMed] [Google Scholar]
- 46.Wennerberg, K., and C. J. Der. 2004. Rho-family GTPases: it's not only Rac and Rho (and I like it). J. Cell Sci. 1171301-1312. [DOI] [PubMed] [Google Scholar]
- 47.Yao, Q., X. Hu, and R. W. Compans. 1997. Association of the parainfluenza virus fusion and hemagglutinin-neuraminidase glycoproteins on cell surfaces. J. Virol. 71650-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuan, P., T. B. Thompson, B. A. Wurzburg, R. G. Paterson, R. A. Lamb, and T. S. Jardetzky. 2005. Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure 13803-815. [DOI] [PubMed] [Google Scholar]
- 49.Yuasa, T., M. Kawano, N. Tabata, M. Nishio, S. Kusagawa, H. Komada, H. Matsumura, Y. Ito, and M. Tsurudome. 1995. A cell fusion-inhibiting monoclonal antibody binds to the presumed stalk domain of the human parainfluenza type 2 virus hemagglutinin-neuraminidase protein. Virology 2061117-1125. [DOI] [PubMed] [Google Scholar]