Abstract
Set2 (KMT3)-dependent methylation (me) of histone H3 at lysine 36 (H3K36) promotes deacetylation of transcribed chromatin and represses cryptic promoters within genes. Although Set2 is the only methyltransferase (KMTase) for H3K36 in yeast, it is not known if Set2 is regulated or whether the different methylation states at H3K36 are functionally distinct. Here we show that the N-terminal 261 residues of Set2 (Set21-261), containing the SET KMTase domain, are sufficient for H3K36me2, histone deacetylation, and repression of cryptic promoters at STE11. Set2-catalyzed H3K36me2 does not require either Ctk1-dependent phosphorylation of RNA polymerase II (RNAPII) or the presence of the phospho-C-terminal domain (CTD) interaction (SRI) domain of Set2. This finding is consistent with a known correlation between H3K36me2 and whether a gene is on or off, but not the level of activity of a gene. By contrast, H3K36me3 requires Spt6, proline 38 on histone H3 (H3P38), the CTD of RNAPII, Ctk1, and the C-terminal SRI domain of Set2. We suggest that the C-terminal region of Set2, in conjunction with the phosphorylated CTD of RNAPII, influences the KMTase activity to promote H3K36me3 during transcription elongation.
The packaging of DNA into chromatin plays an important role in the fidelity of DNA-related processes. The position of nucleosomes, covalent histone modifications, and the integrity of individual histone proteins all contribute to the accessibility and function of DNA. Histones and their modifications influence the expression of genes both positively and negatively and act at the initiation, elongation, and termination stages of transcription.
One such modification, methylation of lysine 36 on histone H3 (H3K36), catalyzed by the Set2 histone methyltransferase (KMT3), is associated with the elongation phase of transcription and is found within the coding regions of actively transcribed genes (19, 24, 25, 39, 41, 42, 46). Set2 catalyzes all three states of methylation at H3K36, monomethyl (me1), dimethyl (me2), and trimethyl (me3), and negatively influences gene expression (20), including the repression of cryptic promoters within the coding region of genes such as STE11 (5, 11, 23). Repression is proposed to result from recruitment of the Rpd3S histone deacetylase complex to specific modifications on the chromatin, including an interaction between methylated H3K36 and the chromodomain of Eaf3, leading to deacetylation and stabilization of newly transcribed chromatin, particularly on poorly expressed genes (5, 11, 15, 22, 23). The presence of H3K36me2 correlates with whether a gene is expressed or not (39), while H3K36me3 tends to correlate with highly expressed genes (38), supporting distinct functions for the two modifications.
Analysis of the C-terminal SRI domain of Set2 indicates that it interacts with the phosphorylated carboxy-terminal domain (CTD) of Rbp1(26, 45), the largest subunit of RNA polymerase II (RNAPII), and is required for normal levels of H3K36me (16). This interaction supports cotranscriptional deposition of methyl groups on H3K36 during transcription elongation. Further support for H3K36 methylation being a cotranscriptional modification comes from observations that Ctk1, a subunit of a cyclin-dependent kinase complex (CTD-K) that acts on the CTD of Rbp1 within RNAPII (6, 21, 36, 46), and Spt6, a transcription elongation factor (4, 9, 10, 14), are required for H3K36 methylation (8, 15, 19, 47) and for repression of the cryptic promoters at STE11 (5).
Spt6 is an essential protein that binds to histone H3 and is capable of nucleosome assembly in vitro (4). Spt6 is known to control the initiation (1), elongation (10, 13, 14, 18, 27), and termination (12) phases of transcription, including cotranscriptional pre-mRNA processing and export (48). The protein has several domains, including an EF hand, a resolvase domain, a helix-hairpin-helix DNA binding domain, and an SH2 domain. The SH2 domain of mammalian Spt6 binds to the CTD on RNAPII when phosphorylated at Ser2 (48). In Saccharomyces cerevisiae, the requirement for Spt6 in maintaining H3K36 methylation is allele specific (8). Strains expressing the spt6-140 allele show normal levels of H3K36 methylation, while those expressing the spt6-1004 allele show defects in H3K36 dimethylation and trimethylation.
At present it is not known if the different methylation states at H3K36 are associated with different functions, although there are functions associated with Set2 that are likely to be independent of the Rpd3S complex and cotranscriptional histone deacetylation (3, 43, 44). Moreover, it is not known if Set2 KMTase activity is regulated in any way. Here we show that the region of Set2 (1-261) containing the SET domain is sufficient for H3K36me2, histone deacetylation, and repression of cryptic promoters at STE11 but not for H3K36me3. Moreover, Set2-catalyzed H3K36me2 is largely independent of Ctk1-dependent CTD phosphorylation and the SRI domain of Set2, consistent with the observations that H3K36me2 correlates with whether a gene is “on” or “off,” but not the level of activity of a gene (39). By contrast, H3K36me3 requires the integrity of H3P38, Spt6, the CTD of RNAPII, Ctk1, and the SRI domain of Set2. These data indicate that the different methylation states created by the Set2 KMTase are likely to be regulated and are functionally distinct.
MATERIALS AND METHODS
Strains used in this study are as follows: two strains based on the BY4741 genetic background (Euroscarf) (a set2Δ strain and a ctk1Δ strain); OY 51, 56, 57, and 58 {MATα his4-912δ lys2-128δ suc2Δuas(−1900/−390) his3Δ200 ura3-52 leu2 rpb1Δ187::HIS3 [RPB1 rpb1Δ104 rpb1Δ103 (or rpb1Δ101) URA3 CEN]} (33) (CTD truncations); WZY42 (49) (H3P38V); and FY2181 (MATa leu2δ1 his4-912 lys2-128 SPT6/FLAG) (ctk1Δ) and its derivative FY2180 (MATa leu2δ1 his4-912 lys2-128 spt6-1004/FLAG) or FY119 (MATα his4-912 lys2-128 leu2 1 ura3-52 trp1-63) (Spt6 truncation). Truncations and tagging of Set2 were carried out as described previously (16, 46). FLAG-tagged SET2 and truncated derivatives were expressed from the SET2 genomic locus or from the ADH1 promoter in plasmid PN823 (32). Other strains, including epitope-tagged derivatives, truncations, and gene deletions, were constructed by single-step gene replacement using PCR-generated DNA fragments (29). The truncation of Spt6 (Spt6ΔC) was constructed in the genomic locus by replacing the last 606 bases (202 amino acids) with a TGA stop codon and the ADH1 terminator. As a control the stop codon of wild-type (WT) Spt6 was replaced with a TGA codon followed by the ADH1 terminator. Appropriate targeting of disruption cassettes was confirmed by PCR and, in case of double mutants, by mating single mutant haploids and basic genetic analysis after sporulation and tetrad dissection. Epitope tagging was confirmed by Western blotting. For Northern blotting, chromatin immunoprecipitation (ChIP) experiments, and chromatin preparation, strains were grown in yeast extract-peptone-dextrose (YPD; glucose), YPR (raffinose), or YPG (galactose) medium at 30°C to exponential phase (approximately 1 × 107 to 1.5 × 107 cells/ml). The sequences of all primers used in this work are available on request.
RNA preparation and Northern blotting.
Total RNA was prepared by hot acid-phenol extraction and separated on 1.5% formaldehyde-morpholinepropanesulfonic acid-agarose gels for >5 h in FA buffer, blotted onto nylon. Fixed RNA was sequentially hybridized to radiolabeled probes overnight at 64°C in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Membranes were washed four times for 15 min in 0.1× SSC-0.1% sodium dodecyl sulfate (SDS) at room temperature (twice) and then at 64°C (twice). Bands were detected by autoradiography.
Generation of antiserum specific for Set2.
Bacterially expressed, recombinant Set2 containing N-terminal amino acids 1 to 261 was prepared using induction and purification procedures as previously described (42). This N-terminal Set2 fragment was emulsified in Freund's complete adjuvant and injected into rabbits following the protocol and procedures from Covance, Inc. The resulting blood samples from several injected rabbits were screened for specificity to Set2 via Western blot analysis of yeast wild-type and set2Δ whole-cell extracts. Antisera were compared to the matched preimmune serum for each rabbit as a control, and the blood sample of highest antibody avidity was selected. Data indicate that the antiserum recognizes a major epitope at the C-terminal region of Set2. Thus, epitope tagging was necessary to detect the C-terminally truncated versions of Set2.
Coimmunoprecipitation.
Coimmunoprecipitation experiments were performed essentially as previously described (46). In brief, the indicated strains were grown in YPG to induce Set2 expression, and whole-cell extracts were prepared. Immunoprecipitations were performed in a final volume of 0.8 ml and equalized with extraction buffer containing 1.5 mg of whole-cell extract protein. The extracts were incubated with 4 μl of Set2 antibody or 4 μl of the matched preimmune serum for 2 h at 4°C, after which each sample was incubated for 1 h with 15 μl protein A-Sepharose beads (GE Healthcare). The beads were then washed twice for 10 min in extraction buffer, eluted in SDS-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, and incubated at 100°C for 5 min, and bead-bound proteins were analyzed by immunoblot analysis using antibodies targeted against the Ser5 phospho-CTD (H14; Covance Inc.) or the Set2 antibody.
Preparation of whole-cell or nuclear extracts and immunoblot assays.
Cells were grown in YPD at 30°C to 1.5 × 107 cells/ml. Extracts were prepared in duplicate by breaking the cells with glass beads in 8 M urea for 3 min and boiling in Laemmli buffer. Extracts were separated by SDS-PAGE on 15% gels, transferred to Bio-Rad nitrocellulose membranes, and incubated with the antibodies indicated. Interactions were detected using enhanced chemiluminescence (GE Healthcare or Pierce) and a horseradish peroxidase anti-species secondary antibody. Nuclear extracts were prepared as described previously (17).
Chromatin immunoprecipitation protocol.
Chromatin immunoprecipitation was performed as described elsewhere (30, 31). In summary, ChIP was done using 50-ml cultures fixed with 1% formaldehyde for 15 min followed by addition of glycine to 0.25 mM final concentration. Yeast cells were broken using glass beads on a MagnaLyser (Roche), and fixed chromatin was sheared by sonication using a bioruptor (Diagenode). Average DNA fragment lengths were 150 to 300 bp. After centrifugation (30 min, 10,000 × g, 4°C), the soluble chromatin was incubated with antibody to the following epitopes: 12 μl of H5 (Covance), 10 μl hemagglutinin (HA; Roche), 3 μl of Y80 (Santa Cruz), 3 μl of Set2, 3 μl of FLAG M2 (Sigma), and 10 μl of myc (Sigma) in 1.5-ml siliconized Eppendorf tubes at 4°C for 15 to 20 h and immunoprecipitated with protein A-Sepharose or anti-immunoglobulin M-agarose for H5 antibodies for 90 min at room temperature. After washing, the chromatin was eluted from the beads at 65°C for 30 min. Cross-links were reversed by incubation at 65°C for 6 to 20 h and treated with protease and RNase A. DNA was purified using Qiagen PCR mini-columns and eluted in 100 μl water. IP samples and IP controls (no antibody) were used undiluted, while input DNA samples were diluted accordingly. Samples were subject to real-time PCR using a Corbett Rotorgene and Sybr green mix (Sensymix; Quantace). Real-time PCR was used to amplify regions corresponding to those shown at STE11. Data [(IP − no antibody)/total] were expressed as a percentage of the input. Error bars reflect the standard deviations of the average signals obtained between different experiments (n = 2 to 4). Full details of the procedure are available on request.
HMT assays.
Yeast strains containing SET2 expressed from the GAL1 promoter were grown in raffinose to prevent Set2 expression. Whole-cell extracts were prepared and sonicated as for ChIP, except for the following alteration in buffer components: 150 mM NaCl, 50 mM Tris pH 9.0, 0.5 mM EDTA, and 10% glycerol. Histone methyltransferase (HMT) buffers and the expression of recombinant Set2 fused to calmodulin binding protein (rSet2) were prepared as described previously (16, 42), where the resulting cell lysate was used directly for the in vitro histone methyltransferase assay. HMT reaction mixtures contained 100 μg of sonicated yeast cell extract with HMT buffer, and 50 μM S-adenosylmethionine (SAM), with or without 2 μl of rSet2 lysate. Samples were incubated for 30 min at 30°C, resolved by SDS-PAGE, and analyzed for H3K36me2 and H3K36me3 by Western blotting.
RESULTS
The SRI domain of Set2 is not absolutely required for recruitment of Set2 to chromatin.
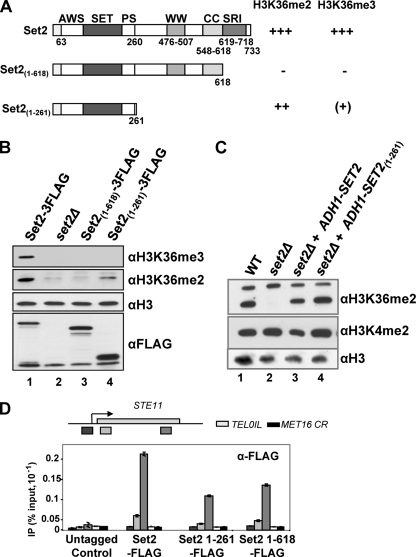
As the SRI domain of Set2 interacts with the phosphorylated CTD of elongating RNAPII, we proposed that Set2 recruitment via the SRI domain to the phosphorylated CTD leads to methylation of the chromatin during transcription (16). Consistent with this, a strain expressing a form of Set2 without the SRI domain, Set21-618, lacks both H3K36me2 and H3K36me3 (Fig. 1A and B) (16). We show here, however, by using ChIP of FLAG-tagged Set21-618, that the SRI domain is not essential for the recruitment of Set2 to chromatin (Fig. 1A and D). Levels of protein associated with STE11 are 50% of those for full-length Set2, and this decrease does not reflect lower Set21-618 protein levels (Fig. 1B). This suggests that the SRI domain-phospho-CTD interaction is not essential for Set2 recruitment to chromatin. In addition, there is no further loss of the ChIP signal at STE11 when an additional 357 residues from Set21-618 are removed, leaving the amino-terminal 261 residues, including the SET domain, on Set21-261 (Fig. 1A and D).
FIG. 1.
The amino-terminal 261 residues of Set2, including the SET domain, are sufficient for H3K36me2. (A) Schematic of the three versions of Set2 analyzed in this work, showing the various domains and their requirement for H3K36me2 and H3K36me3. +++, WT levels; ++, moderate reduction; (+), occasionally seen; −, not detectable. (B and C) Western blots of whole-cell extracts prepared from the indicated strains cultured at 30°C and incubated with antibodies against histone H3, H3K36me3, H3K36me2, the 3FLAG epitope, or H3K4me2. (B) Strains engineered to express full-length Set2-3FLAG, or Set2-3FLAG derivatives containing the indicated regions, from the endogenous locus. (C) Plasmid-borne full-length Set2, or a truncated derivative, expressed from the ADH1 promoter in a set2Δ strain. (D) Chromatin immunoprecipitation to detect Set2-FLAG or derivatives at three positions on STE11, the coding region (CR) of MET16 and telomere 1L (TEL01L) in the strains indicated. Data are expressed as the percentage of input signal.
Residues 1 to 261, including the SET domain, are sufficient for H3K36me2 by Set2.
Although the strain expressing FLAG-tagged Set21-618 lacks global H3K36me3 and H3K36me2 (Fig. 1A and B) (16), H3K36me2 is present in the strain expressing the smaller protein, Set21-261-FLAG, from either the endogenous promoter (Fig. 1B) or from the ADH1 promoter (Fig. 1C). The lower global levels of H3K36me2 in strains expressing Set21-261-FLAG from the endogenous promoter may reflect reduced recruitment to chromatin, observed at STE11 (Fig. 1D). At this gene, however, the distribution and levels of H3K36me2 in the strain expressing Set21-261 are similar to the WT (Fig. 2A). These observations suggest that the region from residues 262 to 618 has a specific inhibitory function on the methyltransferase activity associated with residues 1 to 261. Taken together these data suggest that the Set2 KMTase H3K36me3 versus H3K36me2 activity may be regulated, that the SET domain within residues 1 to 261 of Set2 is sufficient for H3K36me2, and that the SRI domain is not essential for Set2 recruitment to chromatin. We next investigated whether H3K36me2 can direct Rpd3S-mediated chromatin deacetylation at STE11, leading to repression of the cryptic promoters.
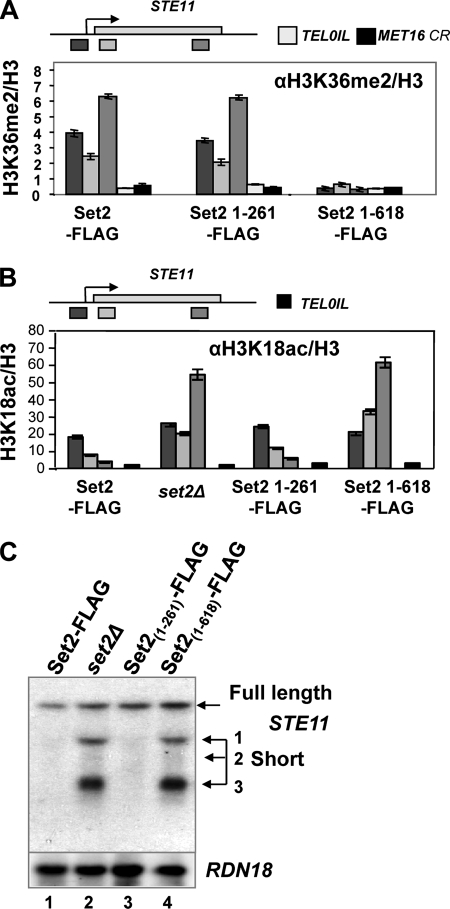
FIG. 2.
H3K26me2 is sufficient for histone deacetylation and repression of internally initiated transcripts at STE11. (A) Chromatin immunoprecipitation to detect H3K36me2 at three positions on STE11, the coding region (CR) of MET16, and telomere 1L (TEL01L) in the strains indicated. Data are presented as a percentage of the input signal and are shown normalized to the histone H3 signals. (B) Chromatin immunoprecipitation to detect H3K18ac at three positions on STE11 and telomere 1L (TEL01L) in the strains indicated. Data are presented as a percentage of input signal and shown normalized to the histone H3 signals. (C) Northern blots of total RNA isolated from the indicated strains, hybridized with a probe from the 3′ region of STE11 or RDN18 (18S rRNA). The full-length transcript and the three (1 to 3) short internally initiated transcripts are indicated.
H3K36me2 is sufficient to direct histone deacetylation and to repress the internal promoters at STE11.
Having a strain capable of dimethylating but not trimethylating H3K36 enabled us to ask if H3K36me2 is sufficient to direct Rpd3S-mediated deacetylation of chromatin at STE11. In the control strain expressing Set2-FLAG, H3K18ac is observed at the promoter and drops off toward the 3′ region of the gene (Fig. 2B). This pattern of distribution for H3K18ac is similar to that observed in microarray analysis of active genes on chromosome III (28). In contrast, strains lacking Set2 or expressing Set21-618-FLAG display high levels of H3K18ac at the promoter and throughout the transcribed region of STE11, consistent with a failure to deacetylate H3K18 toward the 3′ region of STE11 (Fig. 2B). In the strain expressing Set21-261-FLAG, the distribution of H3K18ac resembles the wild-type strain, although the signal is reproducibly higher at the promoter. This suggests that H3K36me2 is sufficient to direct deacetylation of histone H3K18 within the coding region of STE11.
Set2-dependent deacetylation of the chromatin in the transcribed region of STE11 correlates with repression of internal cryptic promoters, as observed in the strain expressing Set2-FLAG (Fig. 2C). In the strain expressing Set21-618-FLAG and the set2Δ strain, initiation at the internal promoters is observed, consistent with the high levels of H3K18ac at STE11. In the strain expressing Set21-261-FLAG, by contrast, initiation of transcription is repressed. This suggests that dimethylation of H3K36 by Set21-261 is sufficient for chromatin deacetylation and repression of the internal cryptic promoters at STE11. To support our observation that H3K36me2 is sufficient to repress internal cryptic promoters at STE11 and to ask if H3K36me2 and H3K36me3 by Set2 might be under different regulation and thus require different factors, we examined mutant strains reported to affect H3K36 methylation, including spt6-1004 (8), ctk1Δ (19, 46), a strain expressing a valine substitution at H3P38 (34), and strains expressing truncations of the CTD on the large subunit of RNAPII (8).
The integrity of H3P38 and the CTD of RNAPII are required for optimal H3K36me3 but not H3K36me2.
The cis-trans isomerization of histone H3 proline 38 by the Fpr4 peptidyl prolyl isomerases influences H3K36me3 (34). The substitution of valine for proline at position 38 is proposed to fix the histone H3 tail in the trans (straight) conformation, resulting in a loss of H3K36me3 (34). This loss of H3K36me3 does not result from the inability of the H3K36me3 antibody to recognize its epitope in the H3P38V strain (34). We show here that in a strain expressing H3P38V, H3K36me2 is present at similar levels to the WT strain, suggesting that proline 38 is not important for dimethylation but does influence trimethylation of H3K36 (Fig. 3A). This suggests that H3P38V is not inherently inhibitory to Set2 methylation but that trimethylation is more sensitive to the loss of proline 38 on histone H3 compared to dimethylation.
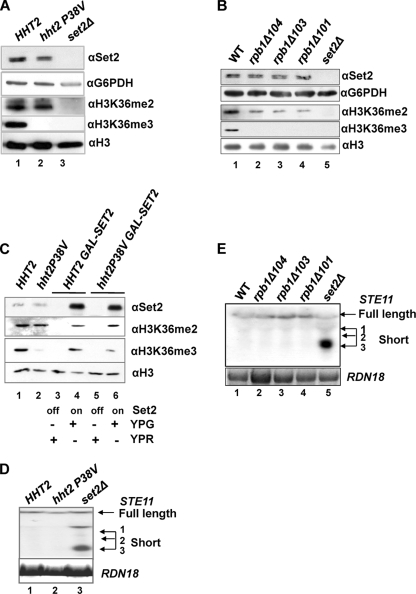
FIG. 3.
Rpb1 CTD- and H3P38-dependent H3K36me3, but not H3K36me2. (A) Western blot of total protein isolated from an hht1-hhf1Δ hht2-hhf2Δ strain expressing HHT2-HHF2 or the P38V derivatives from a plasmid (lane 1, wild-type histone H3; lane 2, H3 with a P38V substitution; lane 3, set2Δ) incubated with the antibodies indicated. Anti-glyceraldehyde-6-phosphate dehydrogenase (αG6PDH) acted as a loading control. (B) Western blot of total protein extracts from strains carrying truncation of the CTD of Rbp1 (33) incubated with the indicated antibodies. (C) Western blots of total protein extracts from strains expressing SET2 from the GAL1 promoter integrated at the SET2 locus cultured in raffinose (lanes 3 and 5; YPR off) or galactose (lanes 4 and 6; YPG on) incubated with the indicated antibodies. Lanes 1 and 2 contain extract from the strains shown in which SET2 is expressed from the endogenous SET2 promoter. (D and E) Northern blots of total RNA isolated from the strains indicated below, hybridized with a probe from the 3′ region of STE11 or RDN18 (18S rRNA). The full-length transcript and the three (1 to 3) short internally initiated transcripts are indicated. (D) H3P38V substitution; (E) Rpb1 derivatives.
In a similar way, strains expressing the RNAPII large subunit Rpb1 with 10 or 11 copies of the CTD, which are just sufficient for viability, rather than the normal 26 (35), also show loss of H3K36me3 while H3K36me2 is present, but at reduced levels compared to WT (Fig. 3B) (8). This reduction is unlikely to reflect Set2 protein levels but may reflect the slow growth associated with these strains.
The levels of Set2 protein in the H3P38V strain are also similar to the WT control, suggesting that the lower levels of H3K36me3 are unlikely to result from insufficient enzyme (Fig. 3A and C). To test whether increasing the amount of Set2 protein would restore trimethylation of H3K36 in the H3P38V strain, we introduced the GAL1 promoter in place of the SET2 promoter at the genomic SET2 locus. As this is the only copy of SET2 in the genome, Set2 protein is only expressed when the cells are cultured on galactose (YPG) (Fig. 3C, lanes 4 and 6) and no Set2 is produced in these strains when grown in raffinose (YPR) (Fig. 3C, lanes 3 and 5). Set2 protein is evident in total protein extracts prepared from the hht2P38V GAL1:SET2 strain and the HHT2 GAL1:SET2 strain when induced with galactose (Fig. 3C, lanes 4 and 6), and this is significantly increased over the levels of Set2 in the equivalent background expressing SET2 from the endogenous SET2 promoter (Fig. 3C, lanes 1 and 2). We examined the levels of H3K36me2 in the strains induced in galactose. Levels were similar to the control strains, suggesting that the increased levels of Set2 protein do not result in an increase in global levels of dimethylated H3K36. Next we looked at trimethylated H3K36. In this experiment there was a very low level of H3K36me3 detectable in the hht2P38V strain, similar to that described previously (34) (Fig. 3C, lane 2). The increased levels of Set2 in the hht2P38V GAL1:SET2 strain results in a small increase in H3K36me3 levels (Fig. 3C, lane 6); however, this is low compared to the levels in the WT strains, especially when the loading is taken into account (Fig. 3C, compare lanes 1, 4, and 6). Thus, increased levels of Set2 protein are not sufficient to restore H3K36me3 to normal levels when H3P38 is replaced with valine. We conclude that H3P38 influences trimethylation of H3K36 even when Set2 levels are increased above normal.
We asked if the presence of H3K36me2 in the strains expressing histone H3 with the valine substitution at P38 (H3P38V) or the Rpb1 CTD truncations was sufficient to repress the cryptic internal promoters at STE11. In all four strains, no internal initiation of transcription was observed at STE11 (Fig. 3D and E), supporting the idea that H3K36me2 is sufficient for repression. In addition, these data suggest specific roles for H3P38 and the CTD of RNAPII in trimethylation of H3K36.
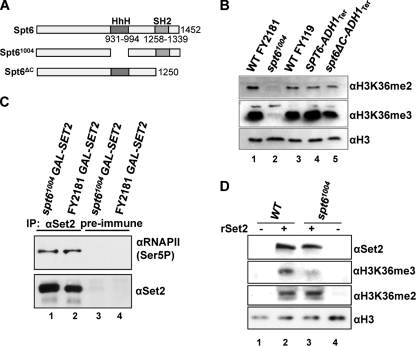
Spt6 influences Set2 protein levels.
A strain expressing the spt6-1004 allele shows a defect in methylation of H3K36 (8) (Fig. 4A). The loss of H3K36 methylation can be partly explained by the low level of Set2 protein present in the spt6-1004 mutant (Fig. 4A, lane 4) compared to the isogenic WT (Fig. 4A, lane 3). Since SET2 mRNA levels are unaffected in the spt6-1004 strain (see Fig. S1 in the supplemental material), this may reflect defects in Set2 protein synthesis or stability. We note that the overall levels of Set2 protein vary in different WT strain backgrounds (Fig. 4A, lanes 1 and 3, compare BY4741 and FY2181), but this does not appear to influence the levels of H3K36me3, which on the Western blot assays correlates with the amount of histone H3 in the extracts, not the levels of Set2 protein. In addition, we noticed that the effect of the spt6-1004 allele on Set2 protein levels varied slightly from experiment to experiment (data not shown). This is similar to the effect of the spt6-1004 allele on Ctr9 protein levels and may reflect inherent instability of the Spt6-1004 protein (12).
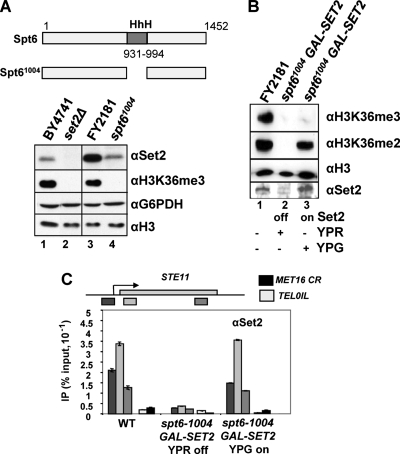
FIG. 4.
Spt6-dependent H3K36me3, but not H3K36me2, when Set2 protein is restored. (A) Spt6 influences the levels of Set2 protein. Western blots of total protein extracts from the strains indicated incubated with the antibodies shown. (B) Western blots of total protein extracts from strains expressing SET2 from the GAL1 promoter integrated at the SET2 locus in the FY2181 background (WT or spt6-1004) cultured in raffinose (lane 2; YPR off) or galactose (lane 3; YPG on) incubated with the indicated antibodies. Lane 1 contains extract from the WT parental background in which SET2 is expressed from the endogenous SET2 promoter. (C) Chromatin immunoprecipitation to detect Set2 at three positions on STE11, the coding region of MET16 (CR), and telomere 01L in the strains indicated, cultured at 30°C in raffinose (YPR off) or galactose (YPG on). Data are expressed as a percentage of the input signal. Note that the profile for Set2 at STE11 detected using the polyclonal antibodies is different from that with the FLAG epitope (see Fig. 1D). These differences are reproducible and have been observed by others using different epitope tags on Set2 (7).
In order to understand more precisely how Spt6 influences H3K36 methylation, we restored Set2 protein levels by expressing SET2 from the GAL1 promoter in a construct integrated into the SET2 locus in the WT and spt6-1004 strains (Fig. 4B). As this is the only copy of SET2 in the genome, Set2 protein is only expressed when the cells are cultured on galactose (Fig. 4B, lane 3), and no Set2 is produced in these strains when grown in raffinose (Fig. 4B, lane 2). Set2 protein is evident in total protein extracts prepared from the spt6-1004 GAL1:SET2 strain when induced with galactose (Fig. 4B, lane 3), and this is increased over the levels of Set2 in the WT isogenic parental strain (Fig. 4B, lane 1).
Next we asked whether the increased levels of Set2 protein facilitate its recruitment to chromatin (Fig. 4C). Although the global levels of Set2 protein are increased, the amount and distribution of Set2 on chromatin, assessed by ChIP at STE11, MET16, and a telomere in the spt6-1004 strain induced with galactose to express GAL1:SET2, resemble what are observed in the control WT strain. Importantly, we observed no association of Set2 with an inactive gene (MET16) or at telomeres in this induced strain. This suggests that the Set2 protein is correctly targeted to chromatin even when expressed from the GAL1 promoter and that Spt6 is not essential for this.
Spt6 is required for H3K36 trimethylation but not dimethylation when Set2 protein levels are restored.
We next asked if the association of Set2 with the chromatin of active genes in the spt6-1004 mutant might be sufficient to restore both di- and trimethylation at H3K36. In the spt6-1004 GAL1:SET2 strain induced with galactose, we observed no signal for H3K36me3, while a robust signal for dimethylated H3K36 was evident at levels similar to WT (Fig. 4B, lane 3). We conclude that under conditions when levels of Set2 are restored at or above those normally observed, Spt6 is required for the ability of Set2 to effectively trimethylate H3K36.
The C-terminal regions of Spt6, including the SH2 domain, are not required for H3K36me3 or H3K36me2.
Our data support a role for the CTD of RNAPII in trimethylation of H3K36. Both yeast and mammalian Spt6 contain a SH2 domain. In mammalian Spt6 the SH2 domain interacts with the serine 2-phosphorylated CTD of RNAPII in vitro (48), but the SH2 domain of yeast Spt6 protein is not characterized. To test whether Spt6 might play a role in H3K36 methylation via an interaction with the CTD, we constructed a strain expressing a C-terminal truncation of Spt6 lacking 202 residues, including the SH2 domain (Fig. 5A). This strain shows a marked slow growth phenotype and a small reduction in H3K36me3 compared to the isogenic parental strain (Fig. 5B, compare lanes 4 and 5). No defect in H3K36me2 was observed (Fig. 5B, compare lanes 4 and 5). This suggests that Spt6 does not influence H3K36 methylation via its SH2 domain.
FIG. 5.
Spt6 influences the ability of Set2 to trimethylate chromatin in vitro but does not influence Set2 association with RNAPII. (A) Schematic of Spt6 showing the regions missing in the two derivatives. spt6-1004-FLAG is in the FY2181 background in which the WT Spt6 is also FLAG tagged, and Spt6ΔC is in the FY118 background in which a stop codon and the ADH1 terminator has replaced the SPT6 sequences at positions 1250 (Spt6ΔC) or 1452 (WT). (B) Western blots of total protein extracts from the indicated strains incubated with the antibodies indicated. Strains were cultured at 30°C, the permissive growth temperature for spt6-1004. (C) Extracts from strains expressing FLAG-tagged Spt6 (WT FY2181) or Spt6-1004 were immunoprecipitated (IP) with anti-Set2 antibodies (lanes 1 and 2) or preimmune serum (lanes 3 and 4), and association with Ser5 P-CTD was assessed using H14 antibodies by Western blotting. Levels of Set2 in the IP were monitored to assess recovery. (D) Spt6 controls trimethylation of H3K36 by recombinant Set2. WT and spt6-1004 strains, with GAL1:SET2 at the SET2 locus, were grown in YPR (raffinose) to ensure repression of SET2 under the GAL1 promoter. Equal amounts of sonicated whole-cell extract were used in a HMT reaction in which the sonicated extracts were incubated with 50 μM SAM (all lanes) and with (lanes 2 and 3) or without (lanes 1 and 4) an equal amount of purified recombinant Set2 (rSet2) (16) at 30°C for 30 min. Aliquots of the reaction mixture were analyzed by Western blotting using antibodies raised against Set2, H3K36me2, H3K36me3, and histone H3. Due to the challenges in the use of the H3K36me2 antibody as described previously (17), representative blots from more than three experiments are shown. In each case, the independent replicates yielded equivalent results. Note that no H3K36me3 or H3K36me2 was detected without the addition of rSet2, despite the presence of SAM.
To explore directly whether the Set2 protein expressed from the GAL1 promoter in the spt6-1004 strain is recruited to RNAPII, we immunoprecipitated Set2 and asked if phosphorylated RNAPII was present in the precipitated material (Fig. 5C). The Set2 protein from the WT and the spt6-1004 strains is able to pull down equal amounts of phosphorylated RNA polymerase II. We conclude that, when expressed from the GAL1 promoter, the Set2 protein is targeted to RNAPII in the spt6-1004 strain. We also confirmed that the spt6-1004 allele does not significantly influence phosphorylation of the CTD at serine 2 or the distribution of RNAPII at STE11 (see Fig. S2 in the supplemental material). Furthermore, the reduced levels of Set2 in the spt6-1004 allele are unlikely to result from a failure to recruit Set2 to the phospho-CTD via the SRI domain, as protein levels in the WT and spt6-1004 backgrounds are not greatly influenced by the presence or absence of the SRI domain (see Fig. S3 in the supplemental material).
Spt6 is required for trimethylation but not dimethylation of H3K36 on a chromatin template by recombinant Set2.
Spt6 is known to bind to the globular domains of histone H3 and H4 and regulate H3K36me3 (4). We therefore designed an experiment to test, in vitro, the role of Spt6 in promoting Set2-dependent methylation of nucleosomes, the preferred substrate for Set2 (42). Cell lysates containing sonicated chromatin from WT and spt6-1004 strains in which SET2 is not expressed were incubated with recombinant Set2 and SAM for 30 min at 30°C (Fig. 5D). These reactions were analyzed by Western blotting using antibodies against H3K36me3 and H3K36me2. Antibodies against H3 and Set2 were used as a loading control or to verify the presence of rSet2, respectively. We observed a significant reduction in the ability of recombinant Set2 to trimethylate chromatin obtained from the spt6-1004 strain compared to the WT strain (Fig. 5D, compare lanes 2 and 3). However, equivalent signals for H3K36me2 were obtained in extracts from both strains. A negligible level of H3K36me2 and H3K36me3 was present in either strain background in the absence of rSet2, despite the addition of SAM. This result recapitulates the data in vivo and suggests that the chromatin in the spt6-1004 strain is a poor substrate for trimethylation of H3K36 by Set2.
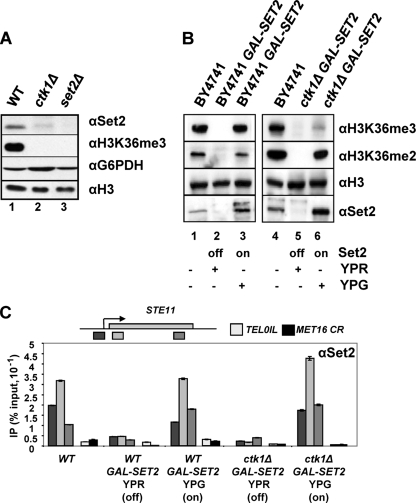
Ctk1 influences the levels of Set2 protein.
Ctk1 is a kinase that phosphorylates the CTD of Rbp1. Unlike the strains expressing CTD truncations, the ctk1Δ strain affects dimethylation and trimethylation at lysine 36 (8, 19, 46), suggesting additional functions for Ctk1 in Set2 function. To resolve this, we examined the levels of Set2 protein and its recruitment to STE11 in the ctk1Δ strain. We observe reduced levels of Set2 in the ctk1Δ strain (Fig. 6A). Since SET2 mRNA levels are unaffected in this strain (46), the loss of H3K36 methylation is likely to be due to a reduction in Set2 protein synthesis, consistent with a role for Ctk1 in translation elongation (40) or a problem with the stability of Set2 (2).
FIG. 6.
Ctk1 is required for H3K36me3 but not H3K36me2 when Set2 levels are restored. (A) Ctk1 influences the levels of Set2 protein. Western blot results shown are of total protein extracts from the strains indicated incubated with antibodies indicated. In this and subsequent experiments set2Δ and ctk1Δ are in the BY4741 background, the Euroscarf reference strain. (B) Western blots of total protein extracts from strains expressing SET2 from the GAL1 promoter in the BY4741 background (left panel) or the same background with ctk1Δ (right panel) cultured in raffinose (YPR off) or galactose (YPG on) incubated with the antibodies indicated. Lanes 1 and 4 contain extracts from the WT parental backgrounds in which SET2 is expressed from the endogenous promoter. (C) Chromatin immunoprecipitation to detect Set2 at three positions on STE11, the coding region of MET16 (CR), and telomere 01L in the strains indicated, cultured at 30°C in raffinose (YPR off) or galactose (YPG on). Data are expressed as a percentage of input signal. Note that the profile for Set2 at STE11 detected using the polyclonal antibodies is different from that with the FLAG epitope (see Fig. 1D). These differences are reproducible and have been observed by others using different epitope tags on Set2 (7).
In order to understand more precisely how Ctk1 influences H3K36 methylation, we restored Set2 protein levels by expressing SET2 from the GAL1 promoter in a construct integrated into the SET2 locus in the WT and ctk1Δ strain (Fig. 6B). Set2 is not expressed when the cells are cultured on raffinose (Fig. 6B, lanes 2 and 5). Set2 protein is evident in total protein extracts prepared from the WT and ctk1Δ GAL1:SET2 strains when induced with galactose (Fig. 6B, lanes 3 and 6), and these are all increased over the levels of Set2 in the WT isogenic parental strains (Fig. 6B, lanes 1 and 4).
Next we asked whether the increased levels of Set2 protein facilitate its recruitment to chromatin (Fig. 6C). The amount and distribution of Set2 on chromatin, assessed by ChIP at STE11, MET16, and a telomere, in the WT strain and the ctk1Δ strain cultured in galactose to induce SET2 expression from the GAL1 promoter resemble those in the control WT strain (Fig. 6C). Importantly, we observed no association of Set2 with an inactive gene (MET16) or at telomeres in these induced strains. This suggests that the Set2 protein was correctly targeted to chromatin in this experiment and that Ctk1 is not essential for this.
Ctk1 is required for H3K36 trimethylation but not dimethylation when Set2 protein levels are restored.
Given the ability to restore Set2 levels in the ctk1Δ strain, we next asked if the association of Set2 with the chromatin of active genes in this mutant might be sufficient to restore both di- and trimethylation at H3K36. In the WT strain expressing GAL1:SET2, the increased Set2 levels lead to a small decrease in global H3K36me2 and no change in H3K36me3 (Fig. 4B, compare lanes 1 and 3). We observed only a trace signal for H3K36me3 in the ctk1Δ strain (Fig. 6B, lane 6). However, a robust signal for dimethylated H3K36 was evident at levels similar to WT (Fig. 6B, compare lanes 4 and 6). We conclude that under conditions when Set2 proteins levels are restored at or above those normally observed, Ctk1 is required for Set2 to effectively trimethylate H3K36. Taken together these data implicate Ctk1, the phosphorylated CTD of RNAPII, and the C-terminal domain of Set2 in trimethylation but not dimethylation of H3K36.
Both Ctk1 and Spt6 are important for H3K36me2 maintenance, but only Ctk1 has a role in repression of the cryptic promoters at STE11.
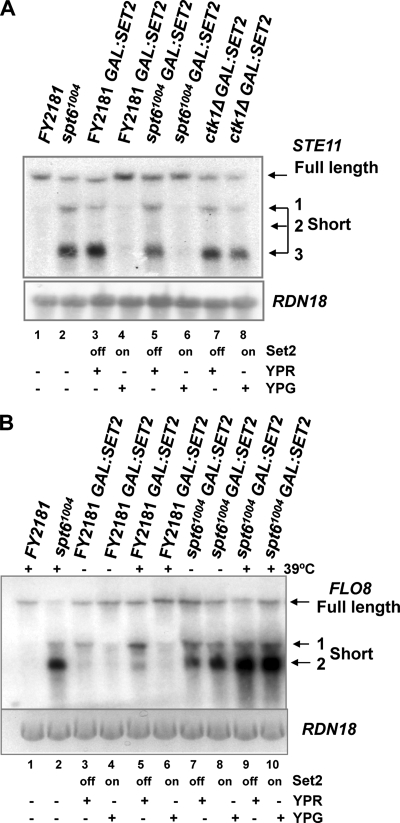
We have shown that dimethylation of H3K36 by Set21-261 is sufficient for chromatin deacetylation and repression of the cryptic promoters at STE11. Next we asked if dimethylation of H3K36 in the spt6-1004 GAL1:SET2 and ctk1Δ GAL1:SET2 strains induced with galactose is sufficient for repression of cryptic promoters at STE11. This might indicate additional functions for Spt6 and Ctk1 beyond H3K36me2.
The presence of H3K36me2 in the induced spt6-1004 GAL-SET2 strain was correlated with efficient repression of the cryptic promoters at STE11 (Fig. 7A, lane 6). This suggests that Spt6 plays no direct role in repression of cryptic internal promoters at STE11 other than maintaining H3K36me2. By contrast, in the ctk1Δ GAL1-SET2 strain, restoration of H3K36me2 was not sufficient to repress the cryptic promoters (Fig. 7A, lane 8). We propose that Ctk1 may have an additional function, independent of Set2, in repression of cryptic initiation at STE11.
FIG. 7.
H3K36me2 is sufficient to repress the cryptic promoters at STE11 and FLO8. Northern blots show total RNA isolated from the WT, spt6-1004, and ctk1Δ strains expressing SET2 from the GAL1 promoter induced with galactose (YPG on) or repressed with raffinose (YPR off), hybridized with a probe from the 3′ region of STE11 (A) or FLO8 (B). RDN18 (18S rRNA) acted as a loading control. For the experiment shown in panel B, WT and spt6-1004 strains were heat shocked at 39°C for 60 min to inactive Spt6-1004 function. The full-length transcript and short internally initiated transcripts at each gene are indicated.
Spt6 represses internal initiation at FLO8 independently of H3K36me2.
Genes such as FLO8 also show a low level of internal initiation of transcription in strains lacking H3K36 methylation (Fig. 7B, lanes 3 and 5) (5). Spt6 is required to repress cryptic initiation at FLO8 (Fig. 7B, lanes 2, 7, and 9) (5, 13). The effect of loss of H3K36 methylation or Spt6 on the amount of internally initiated transcript is enhanced by heat shock (Fig. 7B, lanes 2, 5, and 9, compared to lanes 3 and 7). The low levels of internal initiation that occur when Set2 is absent are repressed when SET2 is expressed from the GAL1 promoter (Fig. 7B, lanes 4 and 6). We asked if restoration of Set2 protein levels in the spt6-1004 strain would repress internal initiation at FLO8, as observed at STE11 (Fig. 7A). No repression was observed in either RNA prepared from yeast grown at 30°C (Fig. 7B, lane 8 [same RNA samples as analyzed in A]) or after a heat shock at 39°C (Fig. 7B, lane 10). We conclude that Spt6 represses internal initiation at FLO8 by a mechanism that is distinct from H3K36me2-mediated histone deacetylation.
DISCUSSION
The current view of methylation of H3K36 is that it is tightly linked to the elongation phase of transcription through the recruitment of Set2 via the SRI domain to the CTD of RNAPII when phosphorylated at serine 2 by Ctk1. In this scenario, deletion of either CTK1 or the region encoding the SRI domain on SET2 leads to loss of Set2-RNAPII association which, in turn, results in a loss of H3K36 dimethylation and trimethylation, a failure to deacetylate transcribed chromatin, and internal initiation of transcription within genes such as STE11. Here we showed that only trimethylation of H3K36 is linked to the elongating form of RNAPII. H3K36me3 requires the C-terminal region of Set2, including the SRI domain, and is dependent on the CTD of RNAPII, the Ctk1 kinase, the integrity of proline 38 on histone H3, and the transcription elongation factor Spt6. In contrast, these factors are not required for dimethylation of H3K36. Moreover, the N-terminal domain of Set2, containing the SET domain, is sufficient for H3K36me2. The discrepancies between our data and the previously published data can be resolved (8, 16, 19, 46). First, mutant strains such as ctk1Δ and spt6-1004 may only show loss of H3K36 dimethylation because levels of Set2 protein are decreased. Set2 protein has a short half-life (S. Fuchs and B. Strahl, unpublished data) (2). In addition, Ctk1 is required for efficient translation elongation in yeast (40). Together these can explain the reduced levels of Set2 protein in the ctk1Δ strain. A possible explanation for how Spt6 influences Set2 function might have been provided if Spt6 could directly associate with, and thereby stabilize, Set2. However, we detected no interaction between Spt6 and Set2 using conventional coimmunoprecipitation experiments (data not shown). Therefore, the ability of Spt6 to regulate Set2 protein levels is likely due to a downstream effect of the HhH domain and its function on transcribed genes. Reduced levels of other proteins, such as Ctr9, have also been observed in the spt6-1004 strain (12), suggesting that the effect is not specific to Set2. We also ruled out a role for Spt6 in recruiting or stabilizing the interaction between Set2 and chromatin, as recruitment of Set2 to chromatin in the spt6-1004 mutant can be restored by overexpression of SET2.
The second discrepancy concerns the role of the SRI domain (16). Our data suggest that an interaction between the CTD and the SRI domain is necessary for trimethylation of H3K36 in the context of the full-length Set2 protein. However, we previously showed that strains expressing a version of Set2 lacking the C-terminal SRI domain lose both H3K36me2 and H3K36me3. We suggest that the sequences between the N-terminal AWS-SET-PS domain and the C-terminal SRI domain may inhibit the activity of the SET domain (especially in the absence of the SRI domain), raising the possibility that Set2 activity is regulated. In this model, the middle region of the WT protein represses Set2 trimethylation activity, and repression is relieved by the SRI interaction with the CTD. Further work is necessary to confirm this. However, our data support tight control over Set2 function. We propose that without the internal conformation changes to Set2, correct recruitment, and an appropriate nucleosomal substrate, Set2 may become unstable or degraded in a regulated way, limiting Set2 action to properly configured nucleosomes. Furthermore, our studies point to the ability of Set2 to recognize and methylate H3 in a context that is CTD independent; however, CTD binding is likely required for optimal or full catalytic activity.
As a result of having strains that lack H3K36me3, we have been able to demonstrate that H3K36me2 is sufficient for histone deacetylation and repression of the cryptic promoters at STE11. This is consistent with the observation that the chromodomain of Eaf3, a component of the Rpd3S histone deacetylase complex, forms a specific interaction with nucleosomes or peptides dimethylated at H3K36 (5, 23). This result suggests another function for H3K36me3 in the transcription process that remains to be defined. We have been unable to show any role for H3K36me3 in functions associated with the expression of telomere proximal genes (43) (K. Kizer, unpublished data) or the PAF1 complex (M. Youdell, unpublished data). For example, Paf1-dependent poly(A) site usage at MAK21 (37) is not affected in the Set21-261 strain, although paf1Δ strains do show reduced levels of Set2 protein. Although we have been unable to define a specific function for H3K36me3, we have defined other factors that are specific to H3K36me3 but not H3K36me2, including the integrity of proline 38 on histone H3 and Spt6.
Proline 38 is reported to undergo cis (bent) to trans (straight) isomerization by Fpr4, a putative peptidyl-prolyl isomerase for H3P38 (34). This change is expected to have a major influence on the local structure of the histone H3 N-terminal tail in the nucleosome. In their paper, Kouzarides and coworkers proposed that cis-proline 38 would facilitate methylation at K36 in the deep cleft at the active site of Set2. Our data suggest that this cannot be the case for H3K36me2, as this is independent of H3P38. However, trimethylation by Set2 is influenced by H3P38. Whether this reflects cis-trans isomerization of P38 is unknown; however, it is clear that the local environment on histone H3 is important for the establishment of H3K36me states.
Our data also suggest that the role for Spt6 in trimethylation of K36 may be related to the chromatin template rather than recruitment of Set2 to RNAPII. Sonicated whole-cell lysates containing fragmented chromatin from WT cells support di- and trimethylation by recombinant Set2, while chromatin from a spt6-1004 strain is only capable of supporting dimethylation. These experiments suggest an Spt6-dependent defect in the chromatin structure that specifically prevents trimethylation. However, we cannot rule out the possibility that Spt6 controls the expression of an as-yet-unknown cofactor that could be required for H3K36me3 by Set2. Future studies using purified chromatin may be able to elucidate these questions. Despite our attempts to do this, we were unable to produce a system where H3K36me3 directed by recombinant Set2 was possible, even from a WT extract. As we are unable to demonstrate experimentally a direct link between Fpr4 and Spt6 (Youdell, unpublished), it is unlikely that Spt6 influences P38 directly. Instead, a role for Spt6 in maintaining a chromatin structure suitable for trimethylation fits very well with its proposed function as an H3/H4 chaperone during transcription elongation. We cannot, however, rule out roles for Spt6 in creating nucleosomal accessibility through an interaction with DNA or antagonizing a chromatin-associated repressor of H3K36me3.
It is clear that from this work that Spt6 does not play a direct role in repressing transcription initiation from the H3K36 methylation-sensitive cryptic promoters at STE11, but rather it is an indirect role, with Spt6 maintaining Set2 protein levels. Once Set2 protein levels are restored, initiation from the cryptic promoter at STE11 is repressed, correlating with the restoration of H3K36me2 in the spt6-1004 background. Based on this, and the absence of transcripts initiated from the cryptic promoter at STE11 in other strains that also have H3K36me2 but not K36me3, we suggest that dimethylation is sufficient for repression at STE11. This is entirely consistent with genome-wide data showing that H3K36me2 correlates with gene expression but not with transcription frequency (39). Moreover, the CTD phosphorylation independence of H3K36me2 is consistent with its establishment during the initial phases of gene activation (31), with subsequent transcription having only a maintenance role.
Longer genes and infrequently transcribed genes tend to be more dependent on the Set2/Rpd3S deacetylation pathway for accurate transcription and repression of internally initiated transcripts (23). FLO8 falls into this category and shows low levels of internal initiation in the absence of Set2 that are repressed when SET2 is expressed. A key question is whether the Set2/Rpd3S pathway is the only mechanism to maintain accurate transcription at this group of genes. At FLO8, H3K36me2 is not sufficient to repress internal initiation in the spt6-1004 mutant, raising the possibility that at this gene H3K36me3 is required or that Spt6 has an additional function unrelated to H3K36 methylation. No internal initiation is observed at FLO8 in the H3P38V strains or the CTD truncations which lack H3K36me3, leading us to conclude that there is a distinct mechanism by which Spt6 functions at FLO8.
In contrast to Spt6, Ctk1 does play a role in repressing the cryptic promoters at STE11, even when H3K36me2 has been restored by Set2 overexpression. This may reflect additional activities associated with Ctk1, for example, its role in limiting H3K4me3 to the 5′ regions of genes, which is independent of its role in maintaining H3K36 methylation (47). Given that H3K4me3 is associated with histone acetylation and unstable chromatin at the 5′ region of genes, the presence of trimethylated K4 toward the 3′ region of open reading frames, together with loss of other Ctk1 functions, may be sufficient to permit transcription from the cryptic promoter even in the presence of H3K36me2. We note that other mutant strains, such as spt16-197, behave like the ctk1Δ strain and fail to repress the internal initiation at STE11, even in the presence of H3K36me2 (8) (data not shown). This strain carries a conditional mutation in SPT16, encoding a putative chaperone function for H2A/H2B during transcription elongation with an additional role in transcription initiation at promoters (3, 8). We suggest that these factors will have additional functions in the repression of transcription initiation at STE11.
Our data strongly support a role for H3K36me2 in creating a repressive chromatin environment on genes that are activated for transcription, as proposed previously (8, 15). Here we show that this is critical to maintain the correct usage of RNA initiation sites. The strains used here that lack H3K36me3, but with H3K36me2, show a slow growth rate, as observed for strains lacking the Bur1/2 kinase that also lack H3K36me3 (7, 8, 15). Slow growth can be overcome by deletion of SET2, supporting the idea that H3K36me2 is a repressive mark (15). In the cell, cotranscription of H3K36me3 would be required to overcome this repression. Our data define a range of conditions that, in addition to the Bur1/2 kinase, are required for the transition from H3K36me2 to H3K36me3 and relief from the repressive chromatin environment. This complex dynamic would ensure that a gene is efficiently expressed while maintaining the integrity of the chromatin and transcription initiation.
Supplementary Material
Acknowledgments
We thank Anitha Nair for excellent technical support, Fred Winston, Greg Prelich, and Karen Arndt for strains and advice during the course of this work, Alexandre Akoulitchev, Laurent Kuras, and Dominico Libri for comments on the manuscript, and all members of the laboratories for their support during this work.
This work was funded by grants to J.M. from The Wellcome Trust, BBSRC, and CRUK and by a grant to B.D.S. from the National Institutes of Health. M.Y. holds a BBSRC studentship. B.D.S. is a Pew Scholar in the Biomedical Sciences.
Footnotes
Published ahead of print on 9 June 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adkins, M. W., and J. K. Tyler. 2006. Transcriptional activators are dispensable for transcription in the absence of spt6-mediated chromatin reassembly of promoter regions. Mol. Cell 21405-416. [DOI] [PubMed] [Google Scholar]
- 2.Belle, A., A. Tanay, L. Bitincka, R. Shamir, and E. K. O'Shea. 2006. Quantification of protein half-lives in the budding yeast proteome. Proc. Natl. Acad. Sci. USA 10313004-13009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biswas, D., R. Dutta-Biswas, D. Mitra, Y. Shibata, B. D. Strahl, T. Formosa, and D. J. Stillman. 2006. Opposing roles for Set2 and yFACT in regulating TBP binding at promoters. EMBO J. 254479-4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bortvin, A., and F. Winston. 1996. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science 2721473-1476. [DOI] [PubMed] [Google Scholar]
- 5.Carrozza, M. J., B. Li, L. Florens, T. Suganuma, S. K. Swanson, K. K. Lee, W. J. Shia, S. Anderson, J. Yates, M. P. Washburn, and J. L. Workman. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123581-592. [DOI] [PubMed] [Google Scholar]
- 6.Cho, E. J., M. S. Kobor, M. Kim, J. Greenblatt, and S. Buratowski. 2001. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 153319-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu, Y., R. Simic, M. H. Warner, K. M. Arndt, and G. Prelich. 2007. Regulation of histone modification and cryptic transcription by the Bur1 and Paf1 complexes. EMBO J. 264646-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu, Y., A. Sutton, R. Sternglanz, and G. Prelich. 2006. The BUR1 cyclin-dependent protein kinase is required for the normal pattern of histone methylation by SET2. Mol. Cell. Biol. 263029-3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark-Adams, C. D., and F. Winston. 1987. The SPT6 gene is essential for growth and is required for delta-mediated transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 7679-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartzog, G. A., T. Wada, H. Handa, and F. Winston. 1998. Evidence that Spt4, Spt5 and Spt6 control transcription elongation by RNA polymerase II. Genes Dev. 12357-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joshi, A. A., and K. Struhl. 2005. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol. Cell 20971-978. [DOI] [PubMed] [Google Scholar]
- 12.Kaplan, C. D., M. J. Holland, and F. Winston. 2005. Interaction between transcription elongation factors and mRNA 3′-end formation at the Saccharomyces cerevisiae GAL10-GAL7 locus. J. Biol. Chem. 280913-922. [DOI] [PubMed] [Google Scholar]
- 13.Kaplan, C. D., L. Laprade, and F. Winston. 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 3011096-1099. [DOI] [PubMed] [Google Scholar]
- 14.Kaplan, C. D., J. R. Morris, C.-T. Wu, and F. Winston. 2000. Spt5 and Spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev. 142623-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keogh, M. C., S. K. Kurdistani, S. A. Morris, S. H. Ahn, V. Podolny, S. R. Collins, M. Schuldiner, K. Chin, T. Punna, N. J. Thompson, C. Boone, A. Emili, J. S. Weissman, T. R. Hughes, B. D. Strahl, M. Grunstein, J. F. Greenblatt, S. Buratowski, and N. J. Krogan. 2005. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123593-605. [DOI] [PubMed] [Google Scholar]
- 16.Kizer, K. O., H. P. Phatnani, Y. Shibata, H. Hall, A. L. Greenleaf, and B. D. Strahl. 2005. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol. Cell. Biol. 253305-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kizer, K. O., T. Xiao, and B. D. Strahl. 2006. Accelerated nuclei preparation and methods for analysis of histone modifications in yeast. Methods 40296-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krogan, N. J., M. Kim, S. H. Ahn, G. Zhong, M. S. Kobor, G. Cagney, A. Emili, A. Shilatifard, S. Buratowski, and J. F. Greenblatt. 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 226979-6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krogan, N. J., M. Kim, A. Tong, A. Golshani, G. Cagney, V. Canadien, D. P. Richards, B. K. Beattie, A. Emili, C. Boone, A. Shilatifard, S. Buratowski, and J. Greenblatt. 2003. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 234207-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landry, J., A. Sutton, T. Hesman, J. Min, R.-M. Xu, M. Johnston, and R. Sternglanz. 2003. Set2-catalyzed methylation of histone H3 represses basal expression of GAL4 in Saccharomyces cerevisiae. Mol. Cell. Biol. 235972-5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, J. M., and A. L. Greenleaf. 1991. CTD kinase large subunit is encoded by CTK1, a gene required for normal growth of Saccharomyces cerevisiae. Gene Expr. 1149-167. [PMC free article] [PubMed] [Google Scholar]
- 22.Li, B., M. Gogol, M. Carey, D. Lee, C. Seidel, and J. L. Workman. 2007. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science 3161050-1054. [DOI] [PubMed] [Google Scholar]
- 23.Li, B., M. Gogol, M. Carey, S. G. Pattenden, C. Seidel, and J. L. Workman. 2007. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev. 211422-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, B., L. Howe, S. Anderson, J. R. Yates III, and J. L. Workman. 2003. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 2788897-8903. [DOI] [PubMed] [Google Scholar]
- 25.Li, J., D. Moazed, and S. P. Gygi. 2002. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J. Biol. Chem. 27749383-49388. [DOI] [PubMed] [Google Scholar]
- 26.Li, M., H. P. Phatnani, Z. Guan, H. Sage, A. L. Greenleaf, and P. Zhou. 2005. Solution structure of the Set2-Rpb1 interacting domain of human Set2 and its interaction with the hyperphosphorylated C-terminal domain of Rpb1. Proc. Natl. Acad. Sci. USA 10217636-17641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindstrom, D. L., S. L. Squazzo, N. Muster, T. A. Burckin, K. C. Wachter, C. A. Emigh, J. A. McCleery, J. R. Yates IIII, and G. A. Hartzog. 2003. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell. Biol. 231368-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, C. L., T. Kaplan, M. Kim, S. Buratowski, S. L. Schreiber, N. Friedman, and O. J. Rando. 2005. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 3e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Longtine, M. S., A. McKenzie, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14953-961. [DOI] [PubMed] [Google Scholar]
- 30.Meluh, P. D., and J. R. Broach. 1999. Immunological analysis of yeast chromatin. Methods Enzymol. 304414-430. [DOI] [PubMed] [Google Scholar]
- 31.Morillon, A., N. Karabetsou, A. Nair, and J. Mellor. 2005. Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol. Cell 18723-734. [DOI] [PubMed] [Google Scholar]
- 32.Mumberg, D., R. Muller, and M. Funk. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156119-122. [DOI] [PubMed] [Google Scholar]
- 33.Murray, S., R. Udupa, S. Yao, G. A. Hartzog, and G. Prelich. 2001. Phosphorylation of the RNA polymerase II carboxy-terminal domain by the Bur1 cyclin dependent kinase. Mol. Cell. Biol. 214089-4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nelson, C. J., H. Santos-Rosa, and T. Kouzarides. 2006. Proline isomerization of histone H3 regulates lysine methylation and gene expression. Cell 126905-916. [DOI] [PubMed] [Google Scholar]
- 35.Nonet, M., D. Sweetser, and R. A. Young. 1987. Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell 50909-915. [DOI] [PubMed] [Google Scholar]
- 36.Patturajan, M., N. K. Conrad, D. B. Bregman, and J. L. Corden. 1999. Yeast carboxyl-terminal domain kinase I positively and negatively regulates RNA polymerase II carboxyl-terminal domain phosphorylation. J. Biol. Chem. 27427823-27828. [DOI] [PubMed] [Google Scholar]
- 37.Penheiter, K. L., T. M. Washburn, S. E. Porter, M. G. Hoffman, and J. A. Jaehning. 2005. A posttranscriptional role for the yeast Paf1-RNA polymerase II complex is revealed by identification of primary targets. Mol. Cell 20213-223. [DOI] [PubMed] [Google Scholar]
- 38.Pokholok, D. K., C. T. Harbison, S. Levine, M. Cole, N. M. Hannett, T. I. Lee, G. W. Bell, K. Walker, P. A. Rolfe, E. Herbolsheimer, J. Zeitlinger, F. Lewitter, D. K. Gifford, and R. A. Young. 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122517-527. [DOI] [PubMed] [Google Scholar]
- 39.Rao, B., Y. Shibata, B. D. Strahl, and J. D. Lieb. 2005. Dimethylation of histone H3 at lysine 36 demarcates regulatory and nonregulatory chromatin genome-wide. Mol. Cell. Biol. 259447-9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rother, S., and K. Strasser. 2007. The RNA polymerase II CTD kinase Ctk1 functions in translation elongation. Genes Dev. 211409-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schaft, D., A. Roguev, K. M. Kotovic, A. Shevchenko, M. Sarov, A. Shevchenko, K. M. Neugebauer, and A. F. Stewart. 2003. The histone 3 lysine 36 methyltransferase, SET2, is involved in transcriptional elongation. Nucleic Acids Res. 312475-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strahl, B. D., P. A. Grant, S. D. Briggs, Z. W. Sun, J. R. Bone, J. A. Caldwell, S. Mollah, R. G. Cook, J. Shabanowitz, D. F. Hunt, and C. D. Allis. 2002. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 221298-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tompa, R., and H. D. Madhani. 2007. Histone H3 lysine 36 methylation antagonizes silencing in Saccharomyces cerevisiae independently of the Rpd3S histone deacetylase complex. Genetics 175585-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tripic, T., D. G. Edmondson, J. K. Davie, B. D. Strahl, and S. Y. Dent. 2006. The Set2 methyltransferase associates with Ssn6 yet Tup1-Ssn6 repression is independent of histone methylation. Biochem. Biophys. Res. Commun. 339905-914. [DOI] [PubMed] [Google Scholar]
- 45.Vojnic, E., B. Simon, B. D. Strahl, M. Sattler, and P. Cramer. 2006. Structure and carboxyl-terminal domain (CTD) binding of the Set2 SRI domain that couples histone H3 Lys36 methylation to transcription. J. Biol. Chem. 28113-15. [DOI] [PubMed] [Google Scholar]
- 46.Xiao, T., H. Hall, K. O. Kizer, Y. Shibata, M. C. Hall, C. H. Borchers, and B. D. Strahl. 2003. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 17654-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao, T., Y. Shibata, B. Rao, R. N. Laribee, R. O'Rourke, M. J. Buck, J. F. Greenblatt, N. J. Krogan, J. D. Lieb, and B. D. Strahl. 2007. The RNA polymerase II kinase Ctk1 regulates positioning of a 5′ histone methylation boundary along genes. Mol. Cell. Biol. 27721-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoh, S. M., H. Cho, L. Pickle, R. M. Evans, and K. A. Jones. 2007. The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes Dev. 21160-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang, W., J. R. Bone, D. G. Edmondson, B. M. Turner, and S. Y. Roth. 1998. Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 173155-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.