Abstract
Saccharomyces cerevisiae chromosome III encodes 11 autonomously replicating sequence (ARS) elements that function as chromosomal replicators. The essential 11-bp ARS consensus sequence (ACS) that binds the origin recognition complex (ORC) has been experimentally defined for most of these replicators but not for ARS318 (HMR-I), which is one of the HMR silencers. In this study, we performed a comprehensive linker scan analysis of ARS318. Unexpectedly, this replicator depends on a 9/11-bp match to the ACS that positions the ORC binding site only 6 bp away from an Abf1p binding site. Although a largely inactive replicator on the chromosome, ARS318 becomes active if the nearby HMR-E silencer is deleted. We also performed a multiple sequence alignment of confirmed replicators on chromosomes III, VI, and VII. This analysis revealed a highly conserved WTW motif 17 to 19 bp from the ACS that is functionally important and is apparent in the 228 phylogenetically conserved ARS elements among the six sensu stricto Saccharomyces species.
Chromosomal origins of DNA replication in budding yeast are called autonomously replicating sequence (ARS) elements and were identified about 30 years ago by their ability to confer autonomous replication to originless plasmids (30). A conserved 11-bp sequence called the ARS consensus sequence (ACS) was initially identified by the analysis of the DNA sequences of four ARS elements (8). The ACS is now known to comprise a binding site for the origin recognition complex (ORC), the essential initiator protein in all eukaryotes (reviewed in reference 2). The consensus sequence of the AT-rich ACS element (WTTTAYRTTTW) is degenerate, and bona fide ARSs sometimes contain only a 10/11- or 9/11-bp match to this sequence. A W indicates the base A or T. When additional ACSs were identified experimentally, a 17-bp extended ACS (EACS) was defined and reflected that the bases flanking the ACS were often A's or T's (WWW-ACS-[G/T]WW) (47). Although the ACS is essential for replicator activity, it cannot be the sole determinant of ORC binding and/or origin specification. By pattern matching, there are 860 exact matches to the 11-bp ACS and 13,978 ACSs, allowing one mismatch in the yeast nuclear genome (http://seq.yeastgenome.org/cgi-bin/PATMATCH/nph-patmatch). Since there are only ∼350 functional ARS elements in Saccharomyces cerevisiae, excluding the ribosomal DNA locus (25), additional sequences, chromatin environment, active transcription units, higher-order nucleosome structure, and/or nuclear organization might further restrict the locations of functional ARS elements. In agreement with one of these predictions, genome-wide studies of ORC and MCM binding sites revealed that >90% of putative ARSs are intergenic (50, 51). Furthermore, additional sequences in the B region adjacent to the ACS (Fig. 1) are known to contribute to ORC binding and origin activity in S. cerevisiae (26, 37, 41, 46).
FIG. 1.
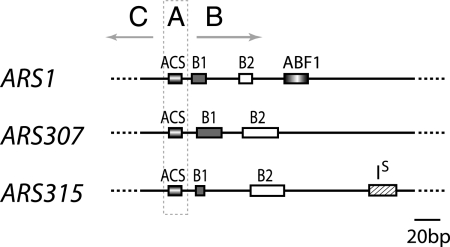
Structure of yeast replicators. ARS1 (26), ARS307 (37, 46), and ARS315 (12) are diagrammed, highlighting the A, B, and C regions important for replicator activity. Region A contains the ACS, and region B is further defined by modular B1, B2, and B3 (Abf1) elements. ACS and B1 together comprise the ORC binding site. IS depicts an inhibitory element within a positioned nucleosome. Some replicators contain transcription factor binding sites in region C that stimulate their activity (not shown).
Detailed analysis of ARS1 (26) and ARS307 (37, 46) showed that they have modular structures (Fig. 1). In addition to the ACS (A element), they also contain B1 and B2 elements 3′ to the T-rich strand of the ACS. The ARS1 B region additionally contains an Abf1p binding site, also called the B3 element. ARS302 (43), ARS305 (12, 22), and ARS315 (12) contain B1 and B2 elements as well, and studies of several chromosome VI ARS elements also defined domain B elements that were important for origin activity (39). The B1 sequences at ARS1 and ARS307 include sequences important for ORC binding (38, 41), and it is now recognized that ORC contacts a bipartite DNA sequence consisting of nucleotides in the ACS and B1 elements (21). Some ARS elements also contain stimulatory sequences 5′ to the T-rich strand of the ACS in domain C. For several of these ARS elements, domain C contains transcription factor binding sites for Abf1p (49), Rap1p (43), Mcm1p (11), or Sum1p (20) that stimulate, but are not required for, origin activity. Recently, an ARS inhibitory element that likely influences local chromatin structure was discovered 3′ to the B2 element in ARS305 and ARS315 (12).
Because the sequence characteristics of the B1 and B2 elements have not been experimentally defined at multiple replicators and because there may be as-yet-undefined regulatory elements, we are analyzing additional replicators on chromosome III to identify their functional sequences. Chromosomes III and VI were the first entire chromosomes to be analyzed systematically for the presence of ARS elements. ARS elements were identified by their ability to confer autonomous plasmid replication and then confirmed as chromosomal replicators by neutral-neutral two-dimensional (2D) gel analysis to detect origin activity at their endogenous chromosomal locations (18, 29, 36, 44, 52). Chromosome III contains 11 active replicators, and chromosome VI contains 10.
Here, we have completed the identification of the essential ACS(s) of the replicators on chromosome III by analyzing ARS313, ARS316, ARS317, ARS318, and ARS319 and have determined the detailed structure of the silent mating-type replicator ARS318 (HMR-I). ARS318 has an A-B1-B2 structure but also contains an Abf1p binding site 6 bp 5′ to the ACS that contributes to its activity. Although ARS318 is largely inactive on the chromosome, we found that ARS318 becomes active when the adjacent HMR-E silencer is deleted. Furthermore, although the 11/11-bp match to the ACS of ARS317 is a well-studied ORC binding site, we identified a redundant ACS that functions in its absence. ARS313, ARS316, and ARS319 each contained a single essential ACS. Analysis of multiple active origins on chromosomes III, VI, and VII revealed a conserved and highly significant WTW motif present within the B1 element that is very important for replicator function. This sequence is also conserved (P ≤ 1 × 10−10) within the 228 ARS elements phylogenetically conserved among the six sensu stricto Saccharomyces species (32). Previous studies have noted a bias toward AT base pairs in this region (6, 30, 51). Therefore, when comparative analysis was confined to active or phylogenetically conserved replicators, the conserved WTW motif was revealed to be highly significant. This conserved sequence within B1 likely makes important ORC-DNA contacts at many replicators based on previous ORC-ARS1 and ORC-ARS307 binding studies (21, 38, 41) and our phenotypic analysis of WTW mutants. The ACSs for all active replicators on chromosome III have now been defined experimentally, with the exception of ARS308, which overlaps the CEN3 centromere. In addition, the detailed structures of ARS1 (26), ARS305 (12, 22), ARS307 (37, 46), ARS315 (12), and ARS318 are now known, allowing for some generalization about replicator structure.
MATERIALS AND METHODS
Yeast strains and methods.
The yeast strains used in this study were W303-1A (MATa ade2-1 trp1-1 ura3-1 leu2-3,112 his3-11,15 can1-100), W303-1B (MATα ade2-1 trp1-1 ura3-1 leu2-3,112 his3-11,15 can1-100) (48), and CFY2133, which is W303-1B containing an 800-bp deletion of the HMR-E locus (10, 28). Yeast transformation and culturing were performed according to standard methods. YPD medium contains 20 g yeast extract, 10 g peptone, and 20 g dextrose per liter. Synthetic complete medium (SCM) contains 50 g dextrose and 6.7 g yeast nitrogen base without amino acids (Difco) per liter and is supplemented with 20 μg/ml uracil and adenine and the 20 amino acids.
Plasmid construction.
All wild-type chromosome III ARS elements were cloned into the pARS1-WT (CEN4 URA3) backbone (26), replacing ARS1, and were described previously (12), with the exception of pFJ23 and pRF23. A 318-bp ARS316 fragment (chromosomal coordinates 272768 to 273085) was amplified with EcoRI-HindIII sites and cloned into the same sites of pARS1-WT to give pFJ23. pRF23 similarly has ARS318 chromosomal coordinates 294567 to 295148 cloned on an EcoRI-HindIII fragment into pARS1-WT. The deletion derivatives within ARS318 (Fig. 2) were made by QuikChange mutagenesis as follows: pFJ10 (Δ1) deletes 94 bp from the EcoRI site of pRF23, pVMH40 (Δ2) deletes 194 bp from the EcoRI site of pRF23, pVMH42 (Δ3) deletes 252 bp from the EcoRI site of pRF23, pFJ12 (Δ4) deletes 294 bp from the EcoRI site of pRF23, pFJ14 (Δ5) deletes 143 bp from the HindIII site of pVMH40, and pFJ13 (Δ6) deletes 185 bp from the HindIII site of pVMH40. pFJ157 contains chromosomal coordinates 292134 to 292652 directly to the left (centromere proximal region) of HMR-E on an EcoRI-HindIII fragment in pARS1-WT. pFJ166 is pFJ157 containing an XhoI linker (CCTCGAG) that eliminates the two overlapping 9/11-bp matches to the ACS in site A of Fig. 5B. All other plasmid derivatives used in the study deviate from the wild-type sequence as shown in the figures.
FIG. 2.
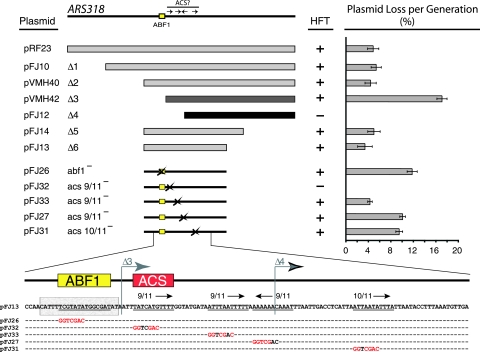
Identifying the ACS for ARS318. Various deletions and point mutations within ARS318 sequences were constructed and tested for their ability to confer ARS activity by transformation into W303-1A. HFT indicates high frequency transformation. The loss rates of these same plasmids were quantitated using a plasmid loss assay (right). The sequence at bottom indicates the positions of the SalI linker mutations as well as deletions Δ3 and Δ4 within ARS318. The shaded sequence represents the DNase I Abf1p footprint from reference 9, and the arrows indicate the 9/11- and 10/11-bp matches to the ACS.
FIG. 5.
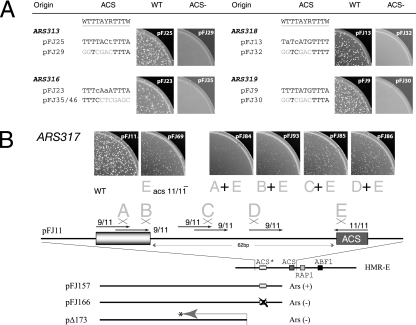
Identifying the ACS for ARS313, ARS316, ARS317, ARS318, and ARS319. (A) ARS elements were cloned into a URA3 CEN4 plasmid (Table 1) and then tested for wild-type (WT) ARS activity by transformation of W303-1A and selection for Ura+ transformants. The same plasmids containing the indicated SalI linker mutations within the essential ACS (described in the text) eliminated ARS activity by the same ACS− transformation assay. (B) ARS317 (HMR-E) contains a redundant ACS 3′ to the silencer. pFJ11 (URA3 CEN4) containing the HMR-E silencer or various mutants (diagrammed at bottom) were transformed into W303-1A and assayed for growth at 30°C. The 11/11-bp match to the ACS (site E) is defined as the ACS element within the silencer. The five 9/11-bp matches to the ACS within the B-region of the ARS317 replicator are indicated as arrows, and mutations A to D disrupt these sequences (see the text for additional details).
Plasmid stability assay.
We performed the plasmid stability assays essentially as described previously (35). At least six independent cultures were inoculated from single colonies and grown overnight at 25°C in SCM without uracil for ∼18 h. The cultures were diluted 1:2,000 into SCM and grown for 24 h at 25°C, during which time they underwent approximately 10 doublings in the absence of selection for the ARS plasmid. Dilutions of both overnight cultures were plated onto SCM and SCM-Ura plates, and colonies were counted 72 h later. Generational loss rates were calculated, and standard errors of the mean (SEM) are reported for each assay.
Alignment of chromosome sequences and statistical analysis.
Active origins on chromosomes III, VI, and VII were compiled with the ACS elements aligned, and the sequence logos were generated using a web-based application, WebLogo (13) (http://weblogo.berkeley.edu/). The WebLogo sequences of 228 confirmed ARS elements identified by Nieduszynski et al. (32) were also generated using the aligned ACS elements. The statistical significance of the proportion of WTW hits in the 228 ARS elements at positions 19 to 21 distal to the ACS was estimated by first creating 1 million samples, each containing 228 random 31-mer sequences selected from the whole genome. For each sample, the proportion of the 228 sequences containing WTW in the equivalent position was calculated. The mean and standard deviation for these million samples approximates the mean and standard error for the expected proportion. The P value was estimated based on a normal approximation for the proportions, which was reasonable based on a visual inspection of the distribution for the random samples.
Determining conservation of WTW motif across the sensu stricto species.
Forty-two phylogenetically conserved and confirmed S. cerevisiae replicators that had at least 12 out of 15 base identities within the proposed 15-bp ACS motif (proACS) (32) as determined by comparing Saccharomyces paradoxus, S. mikatae, S. kudriavzevii, and S. bayanus to S. cerevisiae were selected (see Fig. S2 in the supplemental material). The alignments among the sensu stricto species for each of these ACS elements can be seen at OriDB (Origin Database) (31). For each replicator and for each species, the proACS motif was retrieved with the addition of 30 bp 5′ and 30 bp 3′. For each species, the resulting 42 sequences were used to generate the WebLogo diagrams in Fig. S3 in the supplemental material.
Analysis of replication intermediates.
DNA was extracted from log-phase cultures of W303-1B and CFY2133 as described previously (7). Sixty micrograms of DNA was then digested with XbaI-EcoRV and subjected to BND-cellulose (Sigma) chromatography as described in reference 15, except that volumes were cut in half. Neutral-neutral 2D agarose gels were blotted to Nytran SPC (Whatman) and hybridized as described previously (46). Membranes were probed with the 1-kb XbaI-BglII fragment of pYND70 (36), which was labeled with [α-32P]dATP (Perkin Elmer), using the Megaprime DNA-labeling system (GE Healthcare).
RESULTS
Identifying the ACS for ARS318.
We cloned 14 ARS elements from chromosome III into the same CEN4 URA3 reporter plasmid (12) and determined their loss rates in W303-1A (Table 1). Most loss rates varied from ∼3 to 8% per generation except that ARS302, ARS306, ARS314, and ARS316 were lost at higher rates, from 12 to 30% per generation. ARS318, which coincides with the HMR-I silencer near the right end of chromosome III, is largely inactive as a chromosomal replicator (40). However, a plasmid carrying a 583-bp insert containing ARS318 was lost at ∼4.5% per generation (Fig. 2), indicating that ARS318 is an efficient replicator when removed from its native chromosomal context (1, 8). Highly efficient chromosomal replicators, such as ARS1, which are active in nearly every cell cycle, have similar low plasmid loss rates. Successive ∼50-bp deletions from the left (telomere distal) end of the 583-bp ARS318 fragment did not alter plasmid stability (Fig. 2, Δ1 and Δ2). However, further deletion of the Abf1p binding site (9) caused a fourfold increase in the plasmid loss rate (Fig. 2, Δ3). Removal of an additional 41 bp eliminated replicator activity (Fig. 2, Δ4), indicating that sequences within this 41-bp region were essential for ARS318 function. We then deleted telomere-proximal sequences and defined a ∼200-bp fragment that was maintained as well as the original 583-bp ARS318 fragment (Fig. 2, Δ6).
TABLE 1.
Loss rates for chromosome III ARS plasmids
| Replicator | Plasmid | Loss rate/generation (mean % ± SEM) |
|---|---|---|
| ARS301 | pRF12 | 5.8 ± 1.5 |
| ARS302 | pAC28 | 28.3 ± 1.6 |
| ARS305 | pRF4 | 3.7 ± 0.8 |
| ARS306 | pDP153 | 15.0 ± 0.7 |
| ARS307 | pRF11 | 7.4 ± 0.6 |
| ARS309 | pDP166 | 3.0 ± 2.3 |
| ARS310 | pDP172 | 4.8 ± 2.9 |
| ARS313 | pFJ25 | 4.3 ± 0.5 |
| ARS314 | pMW561 | 12.6 ± 1.0 |
| ARS315 | pAC25 | 5.5 ± 0.4 |
| ARS316 | pRF32 | 13.0 ± 0.6 |
| pFJ23 | 16.8 ± 0.5 | |
| ARS317 | pFJ11 | 5.4 ± 1.4 |
| ARS318 | pFJ10 | 5.4 ± 0.9 |
| ARS319 | pFJ9 | 4.3 ± 1.4 |
There are three 9/11-bp matches and one 10/11-bp match to the ACS within the smaller ARS318 plasmid pFJ13. To determine sequences important for ARS activity, we made 7-bp SalI linker substitution mutations (GGTCGAC) within each potential ACS and in the Abf1p binding site (Fig. 2). Mutation of the Abf1p site increased the plasmid loss rate approximately threefold, indicating that this site, which is known to bind Abf1p, contributes significantly to ARS activity. The 9/11-bp match directly adjacent to the Abf1p site was essential for ARS function, as shown by the failure of plasmids carrying this mutation to produce Ura+ transformants. In contrast, mutation of the three remaining putative ACSs caused at most a twofold increase in the plasmid loss rate. Thus, the 10/11-bp match to the ACS is not essential for ARS activity, as was suggested based on its higher degree of similarity to the ACS (8). However, sequences within the 10/11-bp match to the ACS likely contribute to ARS function by encoding the B2 element (next section).
ARS318 has a modular Abf1-A-B1-B2 structure.
Since the organization of the ARS318 replicator appeared to be different from that of known ARS elements, we performed a linker scan analysis of ARS318 in pFJ13 to determine the positions of its functional elements. We created a series of 29 plasmids that contained ordered 7-bp SalI linker mutations and tested their ability to transform W303-1A to Ura+ (Fig. 3). All of the plasmids transformed the wild-type strain with high frequencies except for pFJ32 and pFJ42, which carry mutations in the 9/11-bp match to the ACS described above. We conclude that the TATCATGTTTT sequence corresponds to the functional ACS at ARS318. Another sequence located between 16 and 26 bp 3′ to the ACS was also important for ARS function (Fig. 3). However, mutation of sequences from base pairs +16 to +22 had a much greater effect on ARS activity (41% loss rate) than mutation of base pairs +23 to +26, which increased the loss rate about twofold, to 10%. The positions of the sequences 16 to 26 bp from the ACS correspond to the B1 elements of ARS1, ARS305, ARS307, and ARS315. Two additional linker scan mutations 23 bp and 30 bp distal to B1 caused a twofold decrease in plasmid stability. These overlap the 10/11-bp match to the ACS and correspond to the B2 elements defined in the four replicators cited above. Finally, mutations in the Abf1p binding site increased the plasmid loss rate two- to threefold (Fig. 3). There were no other sequences surrounding ARS318 that promoted or inhibited ARS activity.
FIG. 3.
Structure of ARS318. The structure of ARS318 was determined by assaying 29 derivatives of pFJ13 carrying 7-bp SalI linker scan mutations for their ability to transform W303-1A and also for their plasmid loss rate. Wild-type (WT) ARS318 was lost at a rate of about 4.5% per generation. The boxed regions within the DNA sequence indicate the functional regions delimited by the linker scan mutations and correspond to the Abf1p binding site, the ACS (A), and the B1 and B2 elements. Plasmids containing mutations in the ACS could not be assayed, since they fail to yield transformants. Plasmid loss rates represent the averages ± SEM of results from at least six independent measurements.
ARS318 is an active chromosomal replicator in the absence of the HMR-E silencer.
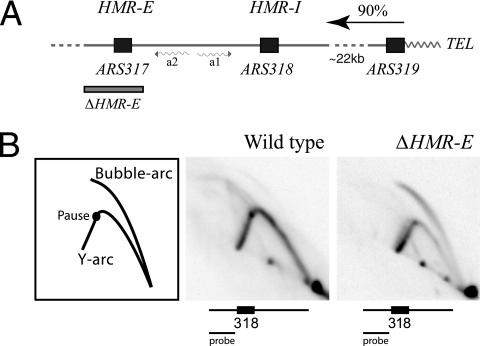
Since ARS318 was an efficient replicator in a plasmid context but largely inactive on the chromosome, we thought its chromosomal position might render it inactive. ARS317 and ARS318 flank the transcriptionally silent a-type mating genes on the right end of chromosome III about 22 kb from the telomere. ARS318 is proximal to the telomere, and the entire HMR region is replicated in ∼90% of cell cycles from leftward-moving forks that originate from the subtelomeric replicator ARS319 (36) (Fig. 4A). ARS317 replicates this region in the remaining 10% of cell cycles. Therefore, proximity to the telomere, the nearby highly active ARS319 replicator, the local heterochromatin at the HMR silencer, or the adjacent ARS317 replicator could potentially inhibit ARS318 activity. We tested the latter two possibilities by deleting HMR-E on the chromosome and examining ARS318 activity by 2D gel electrophoresis. Under this condition, both the adjacent ARS317 replicator and the heterochromatic environment are eliminated, since HMR-E is essential for heterochromatin formation at the HMR locus (42). In the wild type, HMR-I was largely inactive, as indicated by the strong Y arc (passive replication from ARS319) and the very faint bubble arc (Fig. 4B), as previously shown (40). However, when HMR-E was deleted, ARS318 then became an active replicator, as evidenced by the strong bubble arc. We estimate that ARS318 is active in more than 50% of cell cycles. These data suggest that ARS317 or the heterochromatic environment at HMR (or both) inhibits ARS318 activity.
FIG. 4.
ARS318 is an active replicator in the absence of ARS317. (A) The right end of chromosome III is diagrammed, highlighting the ARS317, ARS318, and ARS319 replicators. The arrow at the right indicates the direction of forks that replicate this region emanating from ARS319, and the shaded box indicates the extent of the 800-bp HMR-E deletion. (B) Southern blots of 2D gels showing ARS318 replicator activity in the wild-type (W303-1B) and ΔHMR-E (CFY2133) strains.
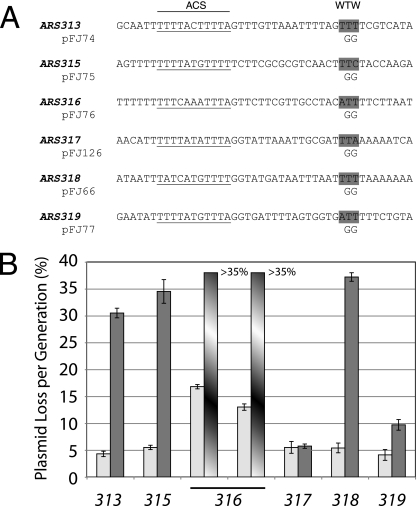
Determining the ACSs for ARS313, ARS316, and ARS319.
We cloned ARS313 and ARS319 into the CEN4 URA3 plasmid on 287-bp and 393-bp fragments, respectively. ARS313 and ARS319 fragments contain single 10/11-bp and 11/11-bp matches to the ACS, respectively, and both plasmids were maintained very efficiently (Table 1). A 7-bp SalI substitution mutation within each putative ACS resulted in a loss of ARS activity (Fig. 5A). Thus, these sequences are essential for ARS activity and likely represent the ACS for these replicators. The situation for ARS316 was more complex because its sequence had not been narrowly defined; ARS316 had been mapped to an ∼1,185-bp fragment (36). We cloned ARS316 on a 502-bp fragment that contained five 9/11-bp matches to the ACS (but no 10/11- or 11/11-bp matches) (see Fig. S1 in the supplemental material). The pFJ23 plasmid, carrying a deletion of 184 bp from one end of this clone, removed three potential ACSs but retained ARS activity, indicating that pFJ23 contained the essential ACS. Sequences within this ∼184-bp fragment stimulated ARS activity since the loss rate increased from 13% to 16.8%. Mutation of one 9/11-bp-match ACS (TTTCAAATTTA) abolished origin activity within pFJ23 (Fig. 5A) and in the context of the larger, 502-bp fragment in pRF32 (see Fig. S1 in the supplemental material). Mutation of the other 9/11-bp-match ACS allowed high efficiency transformation of yeast and, therefore, was not essential for ARS activity (see Fig. S1 in the supplemental material). Thus, this single 9/11-bp match to the ACS is required for ARS316 replicator activity. Our data for ARS313, ARS316, and ARS319 are in agreement with the results of a recent genome-wide study of phylogenetically conserved ARS (EACS) elements (32).
Redundancy in the ARS317 replicator.
The HMR-E silencer on the right end of chromosome III has replicator activity corresponding to ARS317. ARS317 is active in ≤10% of cell cycles in its chromosomal context (40) but is an efficient replicator in a plasmid context (Table 1) (1). Therefore, its chromosome position or additional chromosomal sequences contribute to its relative inactivity within its native context. An 11/11-bp match to the ACS in ARS317 binds ORC (3, 34). However, mutation of the ACS abolishes ARS activity only within a minimal 138-bp fragment containing the silencer elements, i.e., the ORC, Rap1p, and Abf1p binding sites (Fig. 5B) (19, 28, 33). In other words, if natural sequences flanking this 138-bp minimal fragment are present, the ACS− mutant retains replicator activity both in the plasmid and on the chromosome. The residual origin activity is due to activation of redundant nearby origins, which are normally suppressed by the HMR-E silencer (19, 33).
We cloned a 345-bp fragment containing HMR-E and determined the sequences required for ARS activity, using SalI linker mutagenesis as described above. Confirming previous studies, mutation of the 11/11-bp match to the ACS reduced but did not eliminate ARS activity, as evidenced by the appearance of slower-growing Ura+ transformants (Fig. 5B, mutant E). There are five 9/11-bp matches to the ACS within 80 bp of the ORC binding site, corresponding to the B region of ARS317. We made SalI mutations that eliminated the 9/11-bp matches; however, none of these mutations (A to D) affected the transformation frequencies of the plasmids or the sizes of the resulting Ura+ colonies (data not shown). This indicated that sites A to E were not essential for ARS activity. In contrast, mutations in sites A or B when combined with the site E mutation caused the formation of tiny Ura+ transformants that could not be propagated (Fig. 5B). Two overlapping 9/11-bp matches to the ACS were affected by the A and B mutations; mutation A affects both potential ACSs but mutation B only the second. Therefore, the second 9/11-bp match to the ACS probably confers ARS activity in the absence of the primary ORC binding site.
A weak ARS activity that functions only in the absence of ARS317 has been mapped within ∼500 bp to the left of HMR-E (40). Our analogous plasmid clone pFJ157 also had weak ARS activity, which was eliminated by mutation A, affecting the overlapping 9/11-bp-match ACSs (Fig. 5B, pFJ166). Therefore, these 9/11-bp ACS matches are required for this ARS activity. Since plasmid pΔ173 begins 40 base pairs to the left of the 9/11-bp matches and lacks ARS activity (1) (Fig. 5B), this also confirms that the essential sequences for the left ARS activity map to this region.
Alignment of known chromosomal ARS elements reveals a WTW motif within B1.
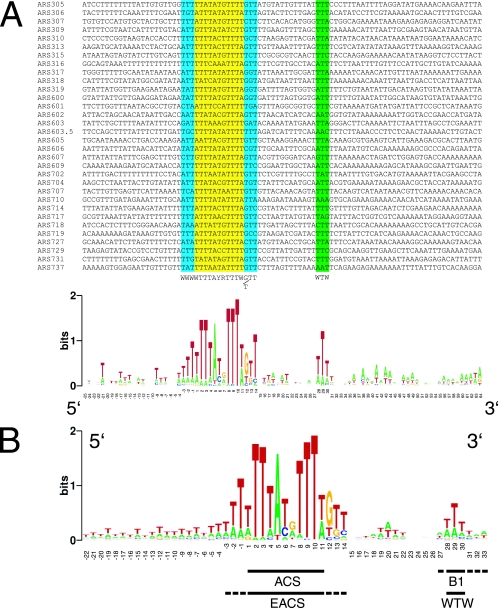
Since we determined the ACSs for four more ARS elements on chromosome III in this study and also recently analyzed the structures of ARS305 and ARS315 (12), we aligned the active chromosome III, VI, and VII ACSs and looked for additional conserved sequences flanking the ACS by using WebLogo (13) (Fig. 6A). We numbered the 11-bp ACS from +1 to +11. As expected, we found a selection for WWW at residues −1, −2, and −3 from the ACS and (G/T)TT at positions +12, +13, and +14, which matched the EACS (47). In addition, we saw a strong selection for WTW beginning 17 bp 3′ to the ACS (positions +28 to +30). This sequence falls within the B1 elements of ARS1, ARS302, ARS305, ARS307, ARS315, and ARS318. Interestingly, these nucleotides were conserved at levels similar to key nucleotides within the EACS. A bias toward AT base pairs in this region was previously noted based on alignment of known origins (47) and also computational prediction and evaluation of ARS elements (6). However, the enrichment that we observed in these 32 active chromosomal origins was more highly significant.
FIG. 6.
(A) Alignment of 32 ARS elements on chromosomes III, VI, and VII by use of WebLogo reveals the EACS and a conserved WTW motif within B1. The ACS is numbered from +1 to +11. See the text for details. (B) WTW is conserved among the 228 phylogenetically conserved ARS elements. The 228 ARS elements conserved in the six sensu stricto Saccharomyces species (32) were aligned using WebLogo (13) and shown to conserve a WTW motif within the B1 element.
This result prompted us to look for the WTW sequence in the 228 evolutionarily conserved ARS elements within the sensu stricto Saccharomyces species (32). These six species diverged within the last 35 million years but retain the ability to mate with each other, yielding sterile diploids (24). The conservation of these ARS elements is a strong indicator that they are functional replicators across species, and this supposition was directly demonstrated using a plasmid ARS assay with S. cerevisiae for the 228 replicators (32). Alignment of the 228 S. cerevisiae ARS elements showed once again a highly significant enrichment of WTW 17 to 19 bp distal to the ACS (Fig. 6B), with no further conserved sequences at least 30 bp 3′ to the WTW sequence (data not shown). The P value for the chance occurrence of the WTW sequence was ≤1 × 10−10 in comparison to a similar enrichment of this sequence within the genome. Interestingly, the WTW motif was also apparent among the 42 most highly conserved ACS elements across the sensu stricto species (see Fig. S2 and S3 in the supplemental material).
WTW is important for ARS activity but is not required at the silencer origin ARS317.
The conservation of the WTW sequence near the ACS, and the fact that nucleotides within this region affect ORC binding at ARS1 and ARS307, strongly suggested that these nucleotides would bind ORC at other replicators and influence ARS activity in vivo. We tested this directly at six chromosome III ARSs by mutating WTW to WGG (Fig. 7A) and then measuring plasmid loss rates for the resulting mutants compared to the wild-type levels. For ARS313, ARS315, ARS316, and ARS318, mutation of this sequence dramatically increased the loss rate to 30% per generation or higher (Fig. 7B). We could not accurately measure the loss rates for ARS316 mutants, because they grew very poorly in liquid medium, but the loss rates had to be greater than 35% since that rate is measurable. The WGG mutation at the subtelomeric ARS319 mutant increased plasmid loss rate twofold (Fig. 7B). In contrast, mutation of WTW at ARS317 did not affect its stability. Interestingly, ARS317 binds ORC 10-fold more tightly than efficient origins such as ARS1 (34). Our data therefore suggest the possibility that mutation of WTW at ARS317 does not sufficiently disrupt ORC binding (or subsequent ORC-dependent steps in origin activation) to alter origin activity. At the other origins tested, mutation of this site strongly destabilized origin function, indicating that the conservation of this sequence is functionally relevant.
FIG. 7.
The WTW sequence is important for origin activity at multiple ARS elements. (A) The DNA sequence surrounding the ACS and B1 elements are shown for six ARS elements. The essential ACS is underlined, and the WTW sequence within B1 is shaded. (B) The wild-type ARS plasmids (light gray bars) and the corresponding WTW→WGG derivatives (dark gray bars) were transformed into W303-1A and quantitated for their plasmid loss rates. Two wild-type ARS316 plasmids (pRF32 and pFJ23) were assayed together with their respective WGG derivatives. Plasmid loss rates represent the averages ± SEM of results from at least six independent measurements.
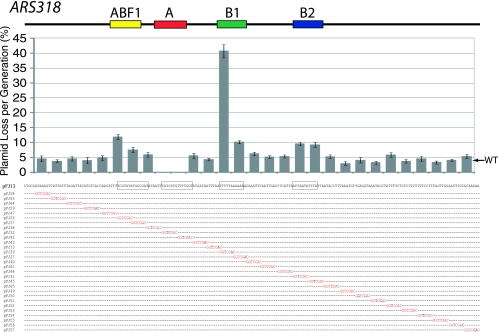
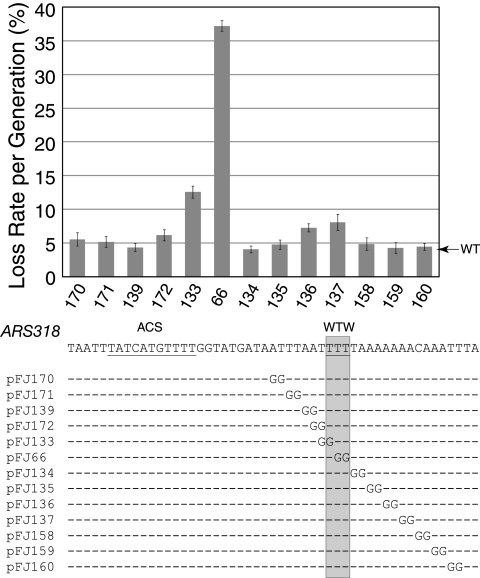
We performed a 2-bp linker scan over the ARS318 B1 region to determine if there were sequences other than WTW that were important for replicator activity (Fig. 8). Mutation of the WTW nucleotides decreased ARS activity from three- to eightfold; however, mutation of nucleotides directly adjacent to WTW had little effect. Mutation of nucleotides 5 to 8 bp distal to WTW decreased ARS activity modestly (∼70 to 80%), but no other nucleotides were important for ARS activity. Thus, at ARS318 the WTW sequence is the most important determinant of the B1 element.
FIG. 8.
WTW is the critical sequence within the ARS318 B1 element. A series of 2-bp linker scan mutations spanning the B1 region of ARS318 defined by the 7-bp SalI linkers in Fig. 3 were constructed. Mutation of the WTW base pairs but not adjacent nucleotides substantially decreased ARS activity as determined by a plasmid stability assay, as in Fig. 7. WT, wild type.
ACS plus WTW is a better predictor of ARS element location than the ACS alone.
We found that the ACS plus the WTW sequence was a better search string for identifying known ARSs. We searched chromosome X for matches to the ACS (including 1 bp on either side) with or without the WTW sequence in B1 and identified 3,577 matches to the 13-bp ACS (up to 2-bp mismatches) and 994 matches by including the WTW sequence. Both sets included 15 of the 16 known chromosome X ARS elements with conserved ACSs (32), indicating that the search strings had good sensitivity but ACS plus WTW improved the stringency since it eliminated about three-fourths of the false hits. However, even the ACS-plus-WTW search string was not stringent enough, because it identified 979 additional matches, most of which were probably not ARSs. Limiting the search to intergenic sequences did not improve the stringency (data not shown); however, it reduced the number of hits, as expected. Lastly, searching with a 15-bp ACS (including 2 bp on either side of the ACS) with or without WTW did not significantly alter the above-described outcome. Therefore, the ACS plus the WTW sequence is a better predictor of known replicators but additional genetic or epigenetic information must determine the positions of ARS elements in budding yeast.
DISCUSSION
The ACS is conserved at yeast replicators because it is a binding site for the initiator complex ORC (2). Point mutations within the ACS or B1 element that disrupt ORC binding also impair ARS activity. In addition, the spacing of the B1 element and the ACS is critical for ORC binding. Deletion of one or more base pairs between the ARS603 ACS and B1 elements causes impaired ORC binding in vitro (4). Therefore, it is not surprising that multiple replicators have defined B1 elements similarly positioned with respect to the ACS. The B1 element is located within 10 to 31 bp 3′ to the ACS, as defined by 7- or 8-bp linker scan mutations (12, 26, 37, 46). By use of 3-bp linkers at ARS307, the B1 sequence has been more narrowly defined and contains critical residues positioned 17 to 19 bp and 26 to 31 bp from the ACS (38). Similarly, at the ARS1 B1 element, single nucleotides 17 to 20 bp distal to the ACS are important for ARS activity (37). Finally, several studies have noted a bias toward AT base pairs within the B1 region at many ARS elements (6, 47, 51).
We showed, using multiple sequence alignment of active or phylogenetically conserved origins, that a WTW sequence is conserved within the B1 region at positions 17 to 19 bp from the ACS (P ≤ 1 × 10−10). We found no other DNA sequence conservation within multiple ARS elements apart from the EACS and the WTW sequence; however, there was a small bias for AT-rich sequences 8 to 10 bp from the ACS (Fig. 6B). The WTW sequence is likely to be conserved at multiple replicators because it makes an important contribution to ARS activity. A double point mutation from WTW to WGG at ARS313, ARS315, ARS316, and ARS318 caused a six- to eightfold increase in the plasmid loss rate, indicating that ARS function had been substantially impaired. A similar mutation within ARS319 (a subtelomeric X-element ARS) caused a twofold increase in the loss rate. The corresponding nucleotides at ARS307 are TTA, and mutation of this sequence to GGG impaired ORC binding in vitro and impairs ARS activity in vivo (38). Furthermore, in vitro ORC DNA footprinting studies showed that ORC contacts nucleotides overlapping this region (21). Thus, we infer that mutations within the WTW sequence generally impair ORC binding in vivo.
Mutation of the WTW sequence has different effects on ARS activity that might depend on the strength of the ORC-DNA interaction at each particular origin. In support of this idea, mutation of the WTW motif within ARS317, which binds ORC 10-fold more tightly than at the efficient ARS1 origin (34), had no effect on ARS activity. One explanation for this finding is that additional nucleotides within ARS317 besides EACS and WTW contribute to tight ORC binding and, therefore, ORC binding is less affected by mutations in the WTW sequence. In contrast, the principal stabilizing ORC-DNA contacts at ARS313 or ARS315 (for example) are perhaps mediated by the EACS and WTW alone.
We also performed a detailed linker scan analysis of the HMR-I silencer origin ARS318. Although this ARS element has little chromosomal replicator activity, it is important for transcriptional silencing at the HMR silent mating-type locus (reviewed in reference 42). Deletion of HMR-I has a modest effect on HMR silencing in the wild-type strain but substantially impairs silencing when combined with mutations in HMR-E (1, 5, 40). Thus, a detailed analysis of the functional components of this silencer element provide a starting point for further studies to test how this element cooperates to determine the epigenetic state of the silencer. The first surprise was that a 9/11-bp match to the ACS was required for origin activity and not the 10/11-bp match. This finding highlights the necessity of empirically determining the ACS at each ARS element, because these elements still cannot be predicted accurately. Although ARS318 conforms to the A-B1-B2 structure seen for several origins, it is unusual in that it has an Afb1p binding site 6 bp 5′ to the ACS. The Abf1p DNase I footprint at this origin shows that Abf1p protects 21 nucleotides surrounding the Abf1p site (9) (Fig. 2). ORC recognizes the ACS at multiple origins and protects ∼10 to 12 bp 5′ to the ACS from DNase I digestion (3, 14). ORC subunits can also be cross-linked to DNA within this same interval (21). A DNase I footprint of ORC at HMR-I revealed a pattern resembling two ORC binding sites, with the stronger protection adjacent to and overlapping the Abf1p binding site (3). This agrees well with our molecular characterization of ARS318, indicating that the 9/11-bp match adjacent to the Abf1p binding site is the essential ACS that likely binds ORC in vivo. The second region of DNase I protection seen in vitro is consistent with an ORC-B2 interaction. Furthermore, the Abf1p and ORC proteins could conceivably contact each other at ARS318 or they could bind to opposite faces of the DNA helix.
How does Abf1p enhance origin or silencer activities? Although Abf1p stimulates silencing at HMR-E and HML-E, it is thought to do this by its ability to position nucleosomes flanking the silencer and not through recruitment of silencer proteins (17). Abf1p has not been shown to make protein-protein contacts with known heterochromatin proteins. Abf1p similarly stimulates ARS1 origin activity through its ability to exclude nucleosomes from the B region, which is where prereplicative complex assembly occurs (23, 45). Interestingly, ORC has also been shown to position nucleosomes adjacent to the silencer and at origins of replication (23). Since ORC can also position nucleosomes, it is unclear why Abf1p would stimulate origin activity at ARS318 two- to threefold through a similar activity. We have now defined the functional cis elements at ARS318 for replicator activity, which will allow the contribution of the Abf1p and ORC binding sites to be determined for transcriptional silencing. We propose that the ability of Abf1p to bend DNA (27) may alter ORC-DNA interactions to stimulate ORC binding to the origin or, alternatively, stimulate subsequent steps in origin activation. Abf1p-mediated DNA bending might also facilitate the activity of the silencer. In any event, defining the functional sequences of ARS318 allows further investigation of these sites to determine their precise roles in origin and silencer function. The ARS318 replicator is inhibited by its chromosomal context since deleting HMR-E (which includes ARS317) results in substantial ARS318 replicator activity. Since ARS317 is active in only ∼10% of cell cycles, it seems unlikely that the infrequent firing of this replicator inactivates ARS318. It is more likely that the heterochromatic environment at HMR inhibits ARS318 activity. This contrasts with the ARS301 replicator at HML-E, which is also active in a plasmid context but inactive on the chromosome, since the loss of transcriptional silencing does not lead to its activation (16). Therefore, other positional effects likely inhibit ARS301.
Supplementary Material
Acknowledgments
We thank Catherine Fox for helpful discussions, Ryan Frisch and Don Pappas for several plasmids, and Craig Webb for supporting J.M. in the Laboratory of Metastasis and Angiogenesis at the Van Andel Research Institute.
C.S.N. and J.F.T. were supported by NIH grant GM35679, and M.W. was supported by the Van Andel Research Institute.
Footnotes
Published ahead of print on 23 June 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abraham, J., K. A. Nasmyth, J. N. Strathern, A. J. Klar, and J. B. Hicks. 1984. Regulation of mating-type information in yeast. Negative control requiring sequences both 5′ and 3′ to the regulated region. J. Mol. Biol. 176307-331. [DOI] [PubMed] [Google Scholar]
- 2.Bell, S. P. 2002. The origin recognition complex: from simple origins to complex functions. Genes Dev. 16659-672. [DOI] [PubMed] [Google Scholar]
- 3.Bell, S. P., R. Kobayashi, and B. Stillman. 1993. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science 2621844-1849. [DOI] [PubMed] [Google Scholar]
- 4.Bolon, Y. T., and A. K. Bielinsky. 2006. The spatial arrangement of ORC binding modules determines the functionality of replication origins in budding yeast. Nucleic Acids Res. 345069-5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brand, A. H., L. Breeden, J. Abraham, R. Sternglanz, and K. Nasmyth. 1985. Characterization of a “silencer” in yeast: a DNA sequence with properties opposite to those of a transcriptional enhancer. Cell 4141-48. [DOI] [PubMed] [Google Scholar]
- 6.Breier, A. M., S. Chatterji, and N. R. Cozzarelli. 2004. Prediction of Saccharomyces cerevisiae replication origins. Genome Biol. 5R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brewer, B. J., D. Lockshon, and W. L. Fangman. 1992. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell 71267-276. [DOI] [PubMed] [Google Scholar]
- 8.Broach, J. R., Y. Y. Li, J. Feldman, M. Jayaram, J. Abraham, K. A. Nasmyth, and J. B. Hicks. 1983. Localization and sequence analysis of yeast origins of DNA replication. Cold Spring Harbor Symp. Quant. Biol. 471165-1173. [DOI] [PubMed] [Google Scholar]
- 9.Buchman, A. R., W. J. Kimmerly, J. Rine, and R. D. Kornberg. 1988. Two DNA-binding factors recognize specific sequences at silencers, upstream activating sequences, autonomously replicating sequences, and telomeres in Saccharomyces cerevisiae. Mol. Cell. Biol. 8210-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casey, L., E. E. Patterson, U. Muller, and C. A. Fox. 2008. Conversion of a replication origin to a silencer through a pathway shared by a forkhead transcription factor and an S phase cyclin. Mol. Biol. Cell 19608-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang, V. K., J. J. Donato, C. S. Chan, and B. K. Tye. 2004. Mcm1 promotes replication initiation by binding specific elements at replication origins. Mol. Cell. Biol. 246514-6524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crampton, A., F. Chang, D. L. Pappas, Jr., R. L. Frisch, and M. Weinreich. 2008. An ARS element inhibits DNA replication through a SIR2-dependent mechanism. Mol. Cell 30156-166. [DOI] [PubMed] [Google Scholar]
- 13.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 141188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diffley, J. F., and J. H. Cocker. 1992. Protein-DNA interactions at a yeast replication origin. Nature 357169-172. [DOI] [PubMed] [Google Scholar]
- 15.Dijkwel, P. A., and J. L. Hamlin. 1995. The Chinese hamster dihydrofolate reductase origin consists of multiple potential nascent-strand start sites. Mol. Cell. Biol. 153023-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubey, D. D., L. R. Davis, S. A. Greenfeder, L. Y. Ong, J. G. Zhu, J. R. Broach, C. S. Newlon, and J. A. Huberman. 1991. Evidence suggesting that the ARS elements associated with silencers of the yeast mating-type locus HML do not function as chromosomal DNA replication origins. Mol. Cell. Biol. 115346-5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fox, C. A., and K. H. McConnell. 2005. Toward biochemical understanding of a transcriptionally silenced chromosomal domain in Saccharomyces cerevisiae. J. Biol. Chem. 2808629-8632. [DOI] [PubMed] [Google Scholar]
- 18.Friedman, K. L., B. J. Brewer, and W. L. Fangman. 1997. Replication profile of Saccharomyces cerevisiae chromosome VI. Genes Cells 2667-678. [DOI] [PubMed] [Google Scholar]
- 19.Hurst, S. T., and D. H. Rivier. 1999. Identification of a compound origin of replication at the HMR-E locus in Saccharomyces cerevisiae. J. Biol. Chem. 2744155-4159. [DOI] [PubMed] [Google Scholar]
- 20.Irlbacher, H., J. Franke, T. Manke, M. Vingron, and A. E. Ehrenhofer-Murray. 2005. Control of replication initiation and heterochromatin formation in Saccharomyces cerevisiae by a regulator of meiotic gene expression. Genes Dev. 191811-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, D. G., and S. P. Bell. 1997. Architecture of the yeast origin recognition complex bound to origins of DNA replication. Mol. Cell. Biol. 177159-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin, S., and D. Kowalski. 1997. Functional equivalency and diversity of cis-acting elements among yeast replication origins. Mol. Cell. Biol. 175473-5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lipford, J. R., and S. P. Bell. 2001. Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol. Cell 721-30. [DOI] [PubMed] [Google Scholar]
- 24.Liti, G., and E. J. Louis. 2005. Yeast evolution and comparative genomics. Annu. Rev. Microbiol. 59135-153. [DOI] [PubMed] [Google Scholar]
- 25.MacAlpine, D. M., and S. P. Bell. 2005. A genomic view of eukaryotic DNA replication. Chromosome Res. 13309-326. [DOI] [PubMed] [Google Scholar]
- 26.Marahrens, Y., and B. Stillman. 1992. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science 255817-823. [DOI] [PubMed] [Google Scholar]
- 27.McBroom, L. D., and P. D. Sadowski. 1994. DNA bending by Saccharomyces cerevisiae ABF1 and its proteolytic fragments. J. Biol. Chem. 26916461-16468. [PubMed] [Google Scholar]
- 28.McNally, F. J., and J. Rine. 1991. A synthetic silencer mediates SIR-dependent functions in Saccharomyces cerevisiae. Mol. Cell. Biol. 115648-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newlon, C. S., L. R. Lipchitz, I. Collins, A. Deshpande, R. J. Devenish, R. P. Green, H. L. Klein, T. G. Palzkill, R. B. Ren, S. Synn, and S. T. Woody. 1991. Analysis of a circular derivative of Saccharomyces cerevisiae chromosome III: a physical map and identification and location of ARS elements. Genetics 129343-357. (Erratum, 130:235.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newlon, C. S., and J. F. Theis. 1993. The structure and function of yeast ARS elements. Curr. Opin. Genet. Dev. 3752-758. [DOI] [PubMed] [Google Scholar]
- 31.Nieduszynski, C. A., S. Hiraga, P. Ak, C. J. Benham, and A. D. Donaldson. 2007. OriDB: a DNA replication origin database. Nucleic Acids Res. 35D40-D46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nieduszynski, C. A., Y. Knox, and A. D. Donaldson. 2006. Genome-wide identification of replication origins in yeast by comparative genomics. Genes Dev. 201874-1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palacios DeBeer, M. A., and C. A. Fox. 1999. A role for a replicator dominance mechanism in silencing. EMBO J. 183808-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palacios DeBeer, M. A., U. Muller, and C. A. Fox. 2003. Differential DNA affinity specifies roles for the origin recognition complex in budding yeast heterochromatin. Genes Dev. 171817-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pappas, D. L., Jr., R. Frisch, and M. Weinreich. 2004. The NAD(+)-dependent Sir2p histone deacetylase is a negative regulator of chromosomal DNA replication. Genes Dev. 18769-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poloumienko, A., A. Dershowitz, J. De, and C. S. Newlon. 2001. Completion of replication map of Saccharomyces cerevisiae chromosome III. Mol. Biol. Cell 123317-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rao, H., Y. Marahrens, and B. Stillman. 1994. Functional conservation of multiple elements in yeast chromosomal replicators. Mol. Cell. Biol. 147643-7651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rao, H., and B. Stillman. 1995. The origin recognition complex interacts with a bipartite DNA binding site within yeast replicators. Proc. Natl. Acad. Sci. USA 922224-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rashid, M. B., K. Shirahige, N. Ogasawara, and H. Yoshikawa. 1994. Anatomy of the stimulative sequences flanking the ARS consensus sequence of chromosome VI in Saccharomyces cerevisiae. Gene 150213-220. [DOI] [PubMed] [Google Scholar]
- 40.Rivier, D. H., J. L. Ekena, and J. Rine. 1999. HMR-I is an origin of replication and a silencer in Saccharomyces cerevisiae. Genetics 151521-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rowley, A., J. H. Cocker, J. Harwood, and J. F. Diffley. 1995. Initiation complex assembly at budding yeast replication origins begins with the recognition of a bipartite sequence by limiting amounts of the initiator, ORC. EMBO J. 142631-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rusche, L. N., A. L. Kirchmaier, and J. Rine. 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72481-516. [DOI] [PubMed] [Google Scholar]
- 43.Sharma, K., M. Weinberger, and J. A. Huberman. 2001. Roles for internal and flanking sequences in regulating the activity of mating-type-silencer-associated replication origins in Saccharomyces cerevisiae. Genetics 15935-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shirahige, K., T. Iwasaki, M. B. Rashid, N. Ogasawara, and H. Yoshikawa. 1993. Location and characterization of autonomously replicating sequences from chromosome VI of Saccharomyces cerevisiae. Mol. Cell. Biol. 135043-5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simpson, R. T. 1990. Nucleosome positioning can affect the function of a cis-acting DNA element in vivo. Nature 343387-389. [DOI] [PubMed] [Google Scholar]
- 46.Theis, J. F., and C. S. Newlon. 1994. Domain B of ARS307 contains two functional elements and contributes to chromosomal replication origin function. Mol. Cell. Biol. 147652-7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Theis, J. F., and C. S. Newlon. 1997. The ARS309 chromosomal replicator of Saccharomyces cerevisiae depends on an exceptional ARS consensus sequence. Proc. Natl. Acad. Sci. USA 9410786-10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas, B. J., and R. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56619-630. [DOI] [PubMed] [Google Scholar]
- 49.Walker, S. S., S. C. Francesconi, and S. Eisenberg. 1990. A DNA replication enhancer in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 874665-4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wyrick, J. J., J. G. Aparicio, T. Chen, J. D. Barnett, E. G. Jennings, R. A. Young, S. P. Bell, and O. M. Aparicio. 2001. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins. Science 2942357-2360. [DOI] [PubMed] [Google Scholar]
- 51.Xu, W., J. G. Aparicio, O. M. Aparicio, and S. Tavare. 2006. Genome-wide mapping of ORC and Mcm2p binding sites on tiling arrays and identification of essential ARS consensus sequences in S. cerevisiae. BMC Genomics 7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamashita, M., Y. Hori, T. Shinomiya, C. Obuse, T. Tsurimoto, H. Yoshikawa, and K. Shirahige. 1997. The efficiency and timing of initiation of replication of multiple replicons of Saccharomyces cerevisiae chromosome VI. Genes Cells 2655-665. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.