Abstract
Repair of oxidative DNA damage in mitochondria was thought limited to short-patch base excision repair (SP-BER) replacing a single nucleotide. However, certain oxidative lesions cannot be processed by SP-BER. Here we report that 2-deoxyribonolactone (dL), a major type of oxidized abasic site, inhibits replication by mitochondrial DNA (mtDNA) polymerase γ and interferes with SP-BER by covalently trapping polymerase γ during attempted dL excision. However, repair of dL was detected in human mitochondrial extracts, and we show that this repair is via long-patch BER (LP-BER) dependent on flap endonuclease 1 (FEN1), not previously known to be present in mitochondria. FEN1 was retained in protease-treated mitochondria and detected in mitochondrial nucleoids that contain known mitochondrial replication and transcription proteins. Results of immunofluorescence and subcellular fractionation studies were also consistent with the presence of FEN1 in the mitochondria of intact cells. Immunodepletion experiments showed that the LP-BER activity of mitochondrial extracts was strongly diminished in parallel with the removal of FEN1, although some activity remained, suggesting the presence of an additional flap-removing enzyme. Biological evidence for a FEN1 role in repairing mitochondrial oxidative DNA damage was provided by RNA interference experiments, with the extent of damage greater and the recovery slower in FEN1-depleted cells than in control cells. The mitochondrial LP-BER pathway likely plays important roles in repairing dL lesions and other oxidative lesions and perhaps in normal mtDNA replication.
Mitochondria are critically involved in aging and age-associated diseases (56). Reactive oxygen species (ROS), generated as by-products of oxidative phosphorylation in mitochondria, are believed to be key players in the normal aging process (18, 19, 47). Although some ROS may diffuse out of mitochondria and react with other cellular macromolecules, mitochondria and especially mitochondrial DNA (mtDNA) appear to be the primary targets. The accumulation of mtDNA damage is positively correlated with aging and age-related diseases. Oxidative damage to mtDNA in the human brain markedly increases with age (33, 36), and the frequencies of mtDNA mutations have been shown to be increased in a variety of age-related degenerative disorders (35, 39).
The lack of the protective nucleosome structure and a more limited DNA repair network is generally believed to contribute to the higher mutation rate of mtDNA (40). For the known DNA repair pathways, there is no convincing evidence for nucleotide excision or mismatch repair operating in mammalian mitochondria (3, 13, 62). Although some homologous recombination activities have been reported (54), the question of whether or not a recombination repair mechanism exists in mammalian mitochondria is still controversial (11, 62). At the moment, base excision DNA repair (BER) is the only confirmed pathway in mitochondria (3, 13), and of the two BER subpathways, only single-nucleotide short-patch BER (SP-BER) has been established experimentally (48). In view of the numerous and diverse DNA lesions generated by ROS, efficient mitochondrial BER would likely be important for cell function and survival. Indeed, artificially boosting BER protected neuronal cells from ROS- and cytokine-induced apoptosis (31), while a decline in BER efficiency is a common correlate of aging and age-associated neurodegenerative diseases (5, 10).
ROS generate a variety of oxidative DNA lesions by reacting with most atoms of the DNA bases and the deoxyribose sugars (21). Some of the base lesions are confirmed to be repaired in mitochondria by BER (5). However, the mechanism(s) for repair of oxidative lesions derived from deoxyribose damage has not been extensively investigated, and some may not be handled effectively by single-nucleotide BER. One such lesion is 2-deoxyribonolactone (dL), a major form of oxidized abasic site, which comprises several percent of the total (15, 42). Of note is the observation that dL in DNA can act as a suicide inhibitor for several important DNA repair proteins with intrinsic apurinic/apyrimidinic (AP) lyase or 5′ deoxyribose-5-phosphate (dRP) lyase activity (14, 15, 28). The attempted repair of dL by such activities results in the formation of stable covalent DNA-protein cross-links that are resistant to simple nuclease or protease processing (51). To prevent the covalent trapping of repair proteins, dL in nuclear DNA is repaired almost exclusively via the long-patch subpathway of BER (LP-BER), in which the FEN1 nuclease removes dL as part of a displaced flap (51). In the absence of such a LP-BER pathway in mitochondria, formation of dL in mtDNA might irreversibly inactivate mitochondrial DNA polymerase γ (Polγ), both the repair enzyme and the replicative enzyme for this organelle, by forming cross-links with the protein through its dRP lyase activity. In addition to this problem, dL in the template strand is likely to block DNA synthesis or cause miscoding, so its correction would be important.
Here we report that dL, when present in the template strand, strongly inhibits human mitochondrial Polγ-mediated DNA synthesis in vitro. Although a dA residue can be incorporated opposite a template dL, bypass beyond the dL residue is completely blocked, leading to DNA degradation through the proofreading activity of Polγ. Attempted repair of AP endonuclease-incised dL by the dRP lyase activity of Polγ resulted in the formation of a covalent protein-DNA cross-link, irreversibly inactivating Polγ. However, repair of dL was detected in mitochondrial extracts (ME) of human lymphoblasts, HeLa cells, and mouse liver; this repair was further characterized as LP-BER dependent on flap endonuclease activity that could largely be ascribed to the FEN1 enzyme. Mitochondrial localization of FEN1 was demonstrated by using fractionation experiments, the preparation of mitochondrial nucleoids, and high-resolution immunofluorescence microscopy. Immunodepletion experiments showed that the LP-BER observed in ME was largely dependent on FEN1, and RNA interference studies indicated that repair of oxidative damage in mtDNA is delayed in FEN1-depleted cells.
MATERIALS AND METHODS
Oligonucleotides containing a dL precursor residue (43) were provided by Marc Greenberg (Johns Hopkins University). Other oligonucleotides were synthesized by Operon Technologies or The Midland Reagent Company and denaturing gel purified before use. Radionucleotides were purchased from Perkin Elmer Life Sciences. Reconstituted Polγ holoenzyme (58) and hexahistidine-tagged human DNA Polβ (14) were prepared as previously described. Recombinant human FEN1 was expressed and purified to apparent homogeneity (51).
Preparation of whole-cell extracts and ME from GM1310 lymphoblasts.
GM1310 normal human lymphoblast cells (Coriell Cell Repositories) were grown in a humidified 5% CO2 atmosphere in RPMI medium supplemented with 10% fetal bovine serum, 50 U/ml penicillin, 50 μg/ml streptomycin, and 2 mM glutamine. Whole-cell extracts were prepared as described previously (55), and mitochondria were isolated by differential centrifugation with Percoll gradient as previously described (50).
Formaldehyde cross-linking and preparation of nucleoids.
Purification of formaldehyde cross-linked mtDNA nucleotides and treatment of purified mitochondria with proteinase K were performed exactly as described previously by Bogenhagen et al. (4).
DNA Pol assays.
“Running-start” assays were carried out as described previously (12). The primer-template DNA was generated by annealing a 32P-5′-end-labeled 14-mer (P14) (Table 1) with a lesion-containing 30-mer (30U, 30 dL, or 30A) (Table 1) at a molar ratio of 1:2 in 20 mM HEPES-KOH, pH 7.8, and 10 mM MgCl2. Hydrolytic AP sites were generated in the P14-30U primer-template immediately before use by treatment with a catalytic amount of uracil-DNA glycosylase (NEB). The dL residue was generated by photolysis of the precursor at 350 nm for 45 min (43). Pol reactions were carried out at 30°C for 20 min and contained 50 nM DNA, 50 μM of each deoxynucleotide triphosphate, and various concentrations of DNA Pol (0, 2, 20, and 200 nM) in 20 mM HEPES-KOH, pH 7.8, 10 mM MgCl2, 10 mM dithiothreitol (DTT), 5% glycerol, and 0.1 mg/ml bovine serum albumin. An equal volume of formamide dye (98% formamide, 0.01 M EDTA, 1 mg/ml xylene cyanol, 1 mg/ml bromphenol blue) was added to terminate the reactions. After heat denaturation at 95°C for 5 min followed by a 1-min incubation on ice, primers and extension products were resolved on a 15% urea-denaturing gel and visualized on a PhosphorImager.
TABLE 1.
Oligonucleotides used for substrate constructions
| DNA | Nucleotide sequencea |
|---|---|
| 30U | 5′-GTCACGTGCTGCAUACGACGTGCTGAGCCT |
| 30pre-dL | 5′-GTCACGTGCTGCA-pre-dL-ACGACGTGCTGAGCCT |
| 30A | 5′-GTCACGTGCTGCAAACGACGTGCTGAGCCT |
| 31G | 5′-GAGGCTCAGCACGTCGTGTGCAGCACGTGAC |
| P14 | 5′-AGGCTCAGCACGTC |
| P16 | 5′-AGGCTCAGCACGTCGT |
| S-APnick2cd | 5′-THF-TCTAGAGGATCCCCGGGTACCGAGCT*C*G*A |
| Nick1 | 5′-GCTTGCATGCCTGCAGGTCGA |
| 46G | 5′-TCGAGCTCGGTACCCGGGGATCCTCTCGACCTGCAGGCATGCAAGC |
U, uracil; 30pre-dL, photosensitive precursor of dL; *, a phosphorothioate bond.
Cross-linking reactions.
Standard cross-linking reactions (51) contained 50 mM HEPES-KOH (pH 7.5), 20 mM NaCl, 0.5 mM DTT, 2 mM EDTA, 5% (vol/vol) glycerol, 0.1 mg/ml bovine serum albumin, 10 nM 32P-3′-end-labeled 31-mer DNA substrate, and protein concentrations as indicated in the figure legends. Duplex 31-mer DNA substrates were prepared by hybridization of the 30pre-dL oligonucleotide or the 30U oligonucleotide to the complementary 31G oligonucleotide (Table 1) and then 3′ end labeled by incorporation of [α-32P]dCTP using the exonuclease-free Klenow fragment of DNA Pol I. Following incubation at 30°C for the specified times, the reactions were terminated by the addition of sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis loading buffer and heating at 100°C for 5 min. DNA-protein cross-links and free DNA were resolved by 8% SDS-polyacrylamide gel electrophoresis, and 32P radioactivity associated with cross-links was quantified using a PhosphorImager and the ImageQuant program.
In vitro BER assays.
BER assays with closed circular plasmid DNA were conducted as described previously (51) with slight modifications. Briefly, BER reactions were performed at 30°C using a standard reaction buffer containing 100 mM HEPES-KOH (pH 7.5), 55 mM KCl, 5 mM MgCl2, 1 mM DTT, 0.1 mM EDTA, 2 mM ATP, 0.5 mM β-NAD, 20 μM of each deoxynucleoside triphosphate, 5 mM phosphocreatine, 200 U/ml phosphocreatine kinase, 60 fmol of the appropriate pGEM DNA substrate, and 20 μg of whole-cell mitochondrial extract in a 24-μl volume. When [α-32P]dTTP or [α-32P]dCTP (10 μCi per reaction) was included in the reaction buffer to monitor the SP- or LP-BER subpathway, the corresponding dTTP or dCTP concentration was reduced to 2 μM. After incubation for 60 min at 30°C, the reactions were terminated, and the DNA products were recovered and analyzed (51).
Flap endonuclease reactions.
Flap-containing DNA substrates were generated by annealing an upstream DNA (Nick1) and a 32P-5′-end-labeled downstream DNA (S-APnick2) with a template DNA (46G) (Table 1) at a molar ratio of 2:1:2. The S-APnick2 oligonucleotide contains a tetrahydrofuran (THF) residue at the 5′ end, and three phosphorothioate bonds at the 3′ end to inhibit exonuclease activities. Flap endonuclease reactions were conducted as described previously (20). For the reactions depicted in Fig. 3B and C, the nuclease activities of FEN1 were assayed with Flap substrates by following published protocols (32, 46).
FIG. 3.
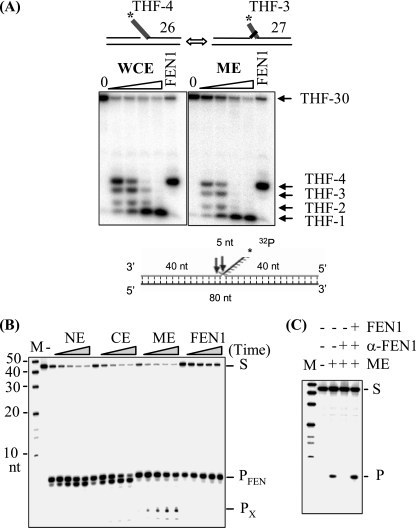
Flap endonuclease activity in human ME. (A) Flap endonuclease activities in GM1310 whole-cell extracts (WCE) and ME. A flap-containing DNA duplex was used to mimic a repair intermediate of LP-BER. The 4-nt flap (with a 5′-terminal THF residue) substrate equilibrates between two structures: a 5′ THF-4-nt single flap and a double flap of 1 nt on the 3′ side and THF-3 nt on the 5′ side. The numbers along the right side indicate the product oligonucleotide sizes (nt). Note that the presence of a 5′ THF residue at the flap ends contributes mobility of <1-nt equivalent. (B) Flap endonuclease activity in HeLa ME. NE, CE, or ME (100 ng each) or 0.1 ng purified FEN1 was incubated with 1 pmol flap substrate at 30°C for 5, 10, 20, 40, and 60 min as indicated by the wedges; the second lane (left panel) corresponds to incubation of the substrate for 60 min without added protein. S indicates the uncleaved substrate, and PFEN and PX indicate the products of FEN1 and nonspecific exonuclease, respectively. M, molecular size marker. (C) FEN1 was depleted from ME with agarose beads conjugated with antibody against FEN1 (α-FEN1). Flap endonuclease activity was assayed with 100 ng ME, FEN1-depeleted ME, or FEN1-depleted ME supplemented with 0.1 ng purified FEN1. M, molecular size markers; S, uncleaved substrate; P, cleavage product of FEN1.
FEN1 immunodepletion and Western blotting.
To immunodeplete FEN1, 24 μg of GM1310 ME was incubated with 5 μg of rabbit polyclonal anti-FEN1 antibody (Novus) in a 15-μl volume at 0°C for 4 h, followed by a 2.5-h incubation after the addition of protein A/G plus agarose beads (Santa Cruz). After a brief centrifugation at 4,000 × g, the supernatant was used directly for repair assays or Western blot analysis. Western blotting was performed using standard protocols with the following antibodies: mouse monoclonal anti-FEN1 antibody (Genetex catalog no. GTX70185), mouse monoclonal antibody (JG1) to mitochondrial heat shock protein 70 (mtHsp70; Genetex catalog no. GTX22799), and mouse monoclonal anti-β-actin (Sigma).
Immunofluorescence microscopy.
To detect endogenous FEN1, HeLa cells were grown overnight on coverslips. To observe exogenous FEN1, HeLa cells were transfected with vectors encoding wild-type or Y83H mutant FEN1 by using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). After being cultured for 24 to 48 h, cells were washed with phosphate-buffered saline and fixed with 3.7% paraformaldehyde for 30 min, permeabilized, and blocked at room temperature in Image-iT FX signal enhancer (Invitrogen, Carlsbad, CA). Antibodies were diluted in phosphate-buffered saline and incubated with samples at room temperature for 2 h (primary antibodies) or 1 h (secondary antibodies). After incubation with secondary antibodies, the cells were stained with 200 ng/ml of DAPI (4′,6-diamidino-2-phenylindole) for 10 min. Samples were mounted with SlowFade Gold (Invitrogen, Carlsbad, CA) antifade reagent, and images were acquired with an LSM510 confocal microscope (Carl Zeiss, Thornwood, NY). The images were processed using Photoshop 7.0 (Adobe, San Jose, CA).
siRNAs.
Stealth small interfering RNAs (siRNAs) against FEN1 or Stealth RNA interference negative control (Invitrogen) were transfected into HeLa cells using Lipofectamine 2000 (Invitrogen) at a final concentration of 75 nM total siRNA. A combination of three siRNAs against FEN1 was used, and the sequences are as follows: FEN1 #1, AUCAAAGACAUACACGGGCUUGAUG; FEN1 #2, UGAGGCUCAGCAGAUGUUUGCACUC; and FEN1 #3, UUUGGCUUCACUGGCAGUCAGCUGU.
H2O2 treatment of siRNA-transfected cells.
siRNA-transfected HeLa cells were seeded at 106 cells per 60-mm culture plate 96 h after transfection and incubated overnight at 37°C. The next morning, cells were washed once with Dulbecco's modified Eagle's medium and treated with H2O2 (0, 0.5, 1, 1.5, 2, and 2.5 mM) at 37°C for 30 min. Cells were either immediately harvested or allowed to recover in complete Dulbecco's modified Eagle's medium for 1, 2, 6, or 21 h. Cells were then collected, and total cellular DNA was extracted using a blood and cell culture DNA minikit (Qiagen).
QPCR assay of DNA damage.
Measurement of nuclear and mitochondrial DNA damage and repair was performed using quantitative PCR (QPCR) that amplifies long DNA targets. The assay was conducted essentially as described previously (44), and primers were used to amplify a 13.5-kb fragment of the β-globin gene (sense primer 5′-CGAGTAAGAGACCATTGTGGCAG and antisense primer 5′-GCACTGGCTTAGGAGTTGGACT) and a 8.9-kb fragment of mtDNA (sense primer 5′-TCTAAGCCTCCTTATTCGAGCCGA and antisense primer 5′-TTTCATCATGCGGAGATGTTGGATGG).
A 221-bp fragment of mtDNA was also amplified to monitor the copy number of the mitochondrial genome and to normalize the data obtained with the 8.9-kb fragment. The small mtDNA fragment was amplified using the above antisense primer with the sense primer 5′-CCCCACAAACCCCATTACTAAACCCA. To ensure that the product amplification is in the exponential phase, a quality control in which only 50% of the template was added to the QPCR reaction was routinely run. Relative amplification of 40 to 60% product was considered acceptable.
RESULTS
dL blocks Polγ-catalyzed DNA synthesis and traps Polγ during attempted repair.
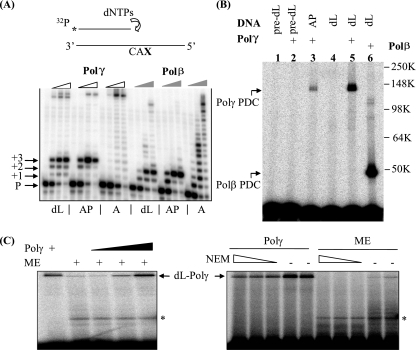
Oxidative damage to DNA generates not only base damage that would be processed into AP sites by DNA glycosylases present in mitochondria (5) but also various oxidized abasic lesions whose effects are just beginning to be assessed (52). A previous study showed that the activity of the mitochondrial DNA Polγ is strongly inhibited by hydrolytic AP sites in the DNA template (38), and we tested the effect of unrepaired dL on Polγ-mediated DNA synthesis. An AP site, a dL residue, or a dA nucleotide was placed on the template strand three bases downstream from the primer terminus. The substrates were verified using the Klenow fragment of Escherichia coli DNA PolI (lacking the 5′-3′ exonuclease), which bypassed the dL and AP sites with slight stalling (data not shown). In contrast, the template dL and AP residues strongly inhibited DNA synthesis by a proofreading-deficient human Polγ holoenzyme (58) and, included for the sake of comparison, by the nuclear DNA repair enzyme Polβ, with different effects for the two enzymes (Fig. 1A). No deoxynucleoside monophosphate residue was incorporated opposite the template dL or AP sites by Polβ. However, Polγ inserted a residue opposite these lesions without efficiently bypassing them (Fig. 1A). In a “standing-start” assay (with the primer terminus immediately before either a dL lesion or an AP lesion), purine nucleotides were incorporated (dAMP more than dGMP) by Polγ much more effectively than pyrimidine nucleotides (see Fig. S1A in the supplemental material). Similar inhibitory effects of dL and AP lesions were observed with a proofreading-competent Polγ catalytic subunit, although stalling at the dL and AP sites led eventually to degradation of the radiolabeled primers by its 3′-5′ exonuclease (see Fig. S1B in the supplemental material).
FIG. 1.
The oxidative DNA lesion dL blocks DNA synthesis by Polγ and inactivates the enzyme by forming a covalent DNA-protein cross-link during attempted SP-BER. (A) A running-start experiment was performed to investigate the effect of dL or an AP site on Polγ-mediated DNA synthesis. The reactions were performed with proofreading-deficient Polγ holoenzyme (indicated by open wedges) and Polβ (indicated by filled wedges). In the schematic drawing, CAX in the template strand indicates the sequence: C and A nucleotides followed by an abasic residue (dL or AP) or an undamaged adenylate nucleotide (A). P, +1, +2, and +3 indicate, respectively, the labeled primer and the extension products with one, two, or three additional nucleotides. The full-length extension products appear near the top of the images and are most readily seen with the undamaged A substrate (see designations below the panel). dNTPs, dinucleotide triphosphates. (B) dL traps purified Polγ. A 3′-end-labeled, nicked-DNA duplex with a 5′ dL in the gap was used to mimic the repair intermediate after Ape1 incision. The protein-DNA complex (PDC) of Polγ formed with an incised AP site and reductive trapping with NaBH4 (37) is shown in lane 3. pre-dL, the dL precursor. Molecular weights in thousands (K) are listed on the right of the gel. (C) dL traps Polγ in ME. A protein-DNA cross-link band was detected with ME of HeLa cells and migrated identically to the recombinant Polγ marker, which was also titrated into the extract for the right three lanes of the left panel. Upon treatment with N-ethylmaleimide (NEM; 5, 10, and 20 mM), this high-molecular-weight band disappeared, consistent with the sensitivity of Polγ to this reagent. The identity of the lower band seen with ME (indicated by an asterisk) is unknown, and it may result from dL cross-linking with contaminating Polβ, a proteolytic fragment of Polγ, or other unidentified mitochondrial proteins with dRP lyase activity.
Once generated in duplex DNA, dL in mammalian cells is recognized and cleaved by AP endonuclease 1 (Ape1) efficiently (57), leading to the formation of a single-strand break with a 5′ dL phosphate residue and a 3′ hydroxyl terminus capable of priming DNA repair synthesis. Ape1 is present both in the nucleus and in mitochondria (9) to drive this reaction. For the nuclear repair enzyme Polβ, attempted excision of 5′ dL by its dRP lyase activity results in a stable protein-DNA cross-link, irreversibly inactivating the enzyme (51). In mitochondria, both replication and DNA repair DNA synthesis are performed by Polγ, which also possesses dRP lyase activity (37). We tested whether human Polγ forms cross-links with 5′ dL at an AP endonuclease-generated strand break. Incubation of purified Polγ with 5′ dL in a 31-mer duplex oligonucleotide pretreated with E. coli endonuclease IV (which cleaves dL in the same way as Ape1) (57) resulted in the appearance of a band with an Mr of ∼145,000 (on SDS-polyacrylamide gels) that was not present in control reactions with dL precursor-containing DNA (Fig. 1B). This new species migrated much more slowly than the Polβ-dL cross-link species (shown for comparison), and the observed Mr was ∼5,000 greater than that of free Polγ, consistent with the covalent addition of the oligonucleotide to form a Polγ-dL cross-link (Fig. 1B). The identity of the Polγ-dL protein-DNA complex was further verified by treatment with proteinase K or DNase I (see Fig. S2 in the supplemental material), which characteristically reduces the mobility of protein-dL complexes (14). The relative amount of cross-linked complex formed with Polβ and Polγ was directly correlated with the dRP lyase activity of each enzyme under the same conditions (data not shown), as expected for a cross-linking reaction that depends on this enzyme mechanism. Furthermore, when the dL-containing oligonucleotide was incubated with ME, a high-Mr band that migrated identically to the species formed with isolated Polγ was detected (Fig. 1C, left panel). This slow-migrating species formed with 5′ dL in ME disappeared upon N-ethylmaleimide treatment (Fig. 1C, right panel), consistent with the sensitivity of Polγ to this reagent (48).
dL is repaired in mitochondria through the LP-BER pathway.
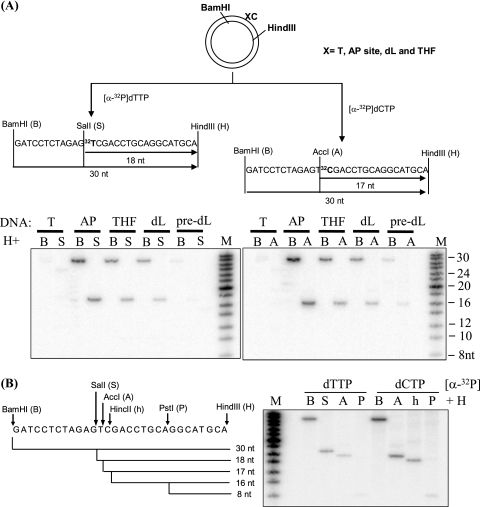
To prevent the formation of dL-Polβ cross-links that constitute a significant obstacle to repair, dL in nuclear DNA is processed exclusively via the LP-BER subpathway (51). However, existing studies have detected only the single-nucleotide SP-BER pathway in mitochondria (48). We examined whether ME can repair dL using a closed circular plasmid DNA substrate with a dL residue, an abasic site analog, a hydrolytic AP site, or a T residue at a defined position, as previously described (51). Regular (hydrolytic) AP sites are predominantly repaired by single-nucleotide BER, while dL and the abasic site analog THF are processed by the multinucleotide LP-BER pathway in nuclear extracts (NE). Lesion-specific and control plasmids were incubated with purified ME of GM1310 human lymphoblasts, and repair was first monitored by the incorporation of [α-32P]dTMP, corresponding to the first residue incorporated during DNA repair synthesis. Repaired DNA was then digested with BamHI and HindIII or with SalI and HindIII to release a 30-nucleotide (nt) fragment or 18-nt fragment, respectively, containing the labeled nucleotide (Fig. 2A). Consistent with the outcomes described in published data (48), the hydrolytic AP site was effectively repaired by the ME, as indicated by the appearance of both 18- and 30-nt fragments (Fig. 2A, left panel). Notably, for both the dL and THF substrates, repaired DNA was also detected after incubation with the ME, indicative of the presence of a functional LP-BER pathway. The substrates with unmodified T or the dL precursor (which is not abasic [43]) at the target site directed only background incorporation of labeled nucleotide.
FIG. 2.
Repair of dL in ME via LP-BER. (A) Plasmid DNA containing a site-specific unmodified base or abasic lesion (X) was constructed and used for repair as described previously (51) (designated by X followed by an unmodified C nucleotide). (Left panel) Repair (via both SP-BER and LP-BER) was monitored by the incorporation of [α-32P]dTMP into the plasmid, and DNA fragments were revealed after digestion with different restriction endonuclease sets (BamHI plus HindIII and SalI plus HindIII). (Right panel) LP-BER was monitored exclusively by the incorporation of [α-32P]dCMP, and the repaired DNA fragments were revealed by treatment with restriction endonuclease sets of BamHI plus HindIII and AccI plus HindIII. In both panels, lanes showing incubations with the dL precursor (pre-dL) confirm a lack of repair in the absence of a BER substrate. (B) LP-BER was confirmed by treatment with different combinations of restriction enzymes. Strand displacement was ruled out as the mechanism of radioactive nucleotide incorporation because, in such a case, an equal incorporation would be expected for every C position downstream of the abasic residue.
To test more directly whether the repair of dL and THF by the ME is by a long patch-type mechanism, we repeated the repair assays with [α-32P]dCTP as the labeled nucleotide. Incorporation of this label indicates repair synthesis of ≥2 nt, as expected for LP-BER. Indeed, incubation of the AP, dL, and THF substrates with ME resulted in the appearance of 32P-labeled repair products (Fig. 2A, right panel), consistent with LP-BER in this organelle. However, the 32P-labeled DNA fragment might also be generated by extensive strand displacement DNA synthesis catalyzed by Polγ (27). To verify that the observed incorporation of [α-32P]dCMP was due to LP-BER rather than strand displacement, we treated the repaired THF plasmid with other restriction endonucleases recognizing sites downstream of the lesion. As shown in Fig. 2B, the radioactive signals of the released DNA fragments became weaker as the cutting sites were moved further from the THF site. This observation was consistent with the repair synthesis of LP-BER proximal to the lesion but not with strand displacement, which instead would generate a more uniform distribution of the radioactive signal throughout the fragment. Repair of dL and THF was also detected by using a mouse liver ME (see Fig. S3 in the supplemental material), indicating that LP-BER is a general repair mechanism in mammalian mitochondria.
Flap endonuclease activity in mitochondria.
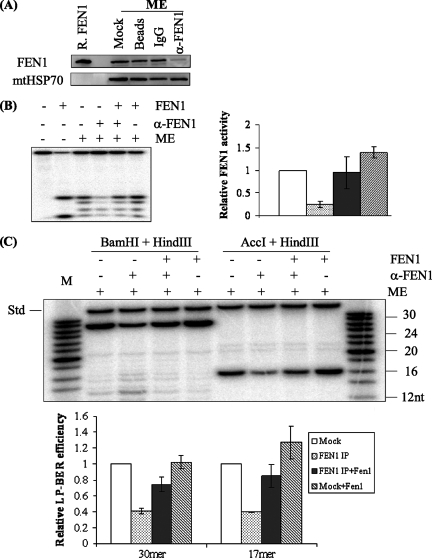
Incorporation of multiple nucleotides during LP-BER generates a displaced 5′ DNA flap downstream of the repair site, which must be removed in order for DNA ligases to complete the process (26). We therefore used a flap-containing DNA substrate that mimics an LP-BER intermediate to test for flap endonuclease activity in mitochondria; this substrate contained a THF residue at the 5′ end of the displaced flap. Annealing of the oligonucleotides Nick1 and S-APnick2 with template 46G (see Materials and Methods) generated a DNA substrate with flaps that equilibrate between two structures: a single THF-4-nt 5′ flap and a THF-3-nt 5′ flap adjacent to a 1-nt 3′ flap (Fig. 3A), which closely mimics an in vivo FEN1 substrate (24). Incubation of this molecule with a GM1310 whole-cell extract led to cleavage of the flaps at or near their junctions with duplex DNA, although there was additional degradation of the liberated product with increasing amounts of extract (Fig. 3A). Purified FEN1 protein generated a single main product by cleaving the THF-4-nt flap at the nick junction or the double flap 1 nt into the annealed region (24). The ME produced a digestion pattern very similar to that generated by the whole-cell extract (Fig. 3A), consistent with the action of a FEN1-like activity but also indicating that other nucleases are present.
FEN1-type activity was also observed in ME of HeLa cells prepared by a slightly different method (see Materials and Methods). These assays employed substrates lacking the THF residue, which are more frequently used to assess FEN1 activity. As shown in Fig. 3B, cleavage at the junction in a double-flap substrate was observed with the HeLa ME, with an incision pattern identical to that of purified FEN1. Results similar to those found with the GM1310 ME were obtained for HeLa ME with the THF-4-nt flap substrate (data not shown). We further demonstrated that the observed flap endonuclease activity was diminished in ME immunodepleted by treatment with FEN1-specific antibody and that the activity could be restored by the addition of recombinant FEN1 (Fig. 3C). Thus, ME of both nontumor human lymphoblastoid cells (hematopoietic) and HeLa cells (epithelial) contain activity very similar to that of authentic FEN1 enzyme. For both cell types, however, there may be additional nucleases that can contribute to end-processing reactions supporting repair.
Mitochondrial localization of FEN1.
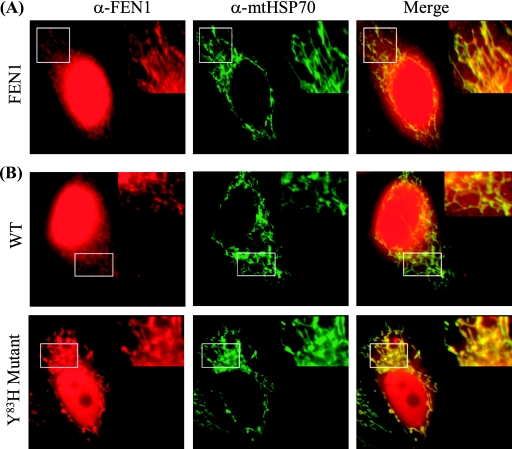
To obtain cytological evidence for mitochondrial localization of FEN1, we stained HeLa cells with polyclonal antibody against human FEN1. In HeLa cells, FEN1 had strong nuclear staining (Fig. 4A), consistent with previous observations and the major functions of this protein in nuclear DNA metabolism (45, 60, 61). However, we also detected FEN1-reactive material colocalized with the mtHsp70 under high-resolution imaging conditions, suggesting that FEN1 translocates into mitochondria (Fig. 4A). Recently, we identified a FEN1 mutant (with the Y83H mutation) that is partially defective in nuclear localization (L. Qian and B. Shen, unpublished data). We hypothesized that decreased nuclear localization of the Y83H form of FEN1 might increase the amount of the protein localized to mitochondria or that the lower nuclear background level might enhance the detection of a mitochondrial form of the protein. To test this hypothesis, we expressed c-myc-tagged versions of wild-type and Y83H mutant FEN1 in HeLa cells and probed with anti-FEN1 antibodies. Similar to the case for endogenous FEN1 (Fig. 4A), a strong nuclear staining was observed for c-myc-tagged wild-type FEN1, with a weaker signal detected in mitochondria (Fig. 4B). However, in cells expressing the mutant Y83H, the decreased amount of the nuclear form enhanced the detection of FEN1 in the mitochondria colocalizing with mtHsp70 (Fig. 4B). This observation was confirmed by costaining of the exogenous FEN1 with anti-c-myc antibody and Mitotracker, a mitochondrion-specific dye (see Fig. S4A in the supplemental material). We also fractionated HeLa cells into an NE, a cytoplasmic extract (CE), and an ME. The nucleus-specific lamin A, mtHsp70, and actin were used as compartment-specific controls. FEN1 was detected in all three fractions (NE, CE, and ME) by immunoblotting with anti-FEN1 antibody (see Fig. S4B in the supplemental material). As expected, lamin A was detected only in the NE fraction; actin was detected in both the NE and CE fractions but not in the ME fraction, and Hsp70 was detected only in the ME.
FIG. 4.
Immunolocalization of FEN1 to mitochondria. (A) Representative images of colocalization of FEN1 and mitochondrion-specific heat shock protein (mtHSP70) in HeLa cells. Fixed HeLa cells were costained with anti-FEN1 (α-FEN1) and anti-mtHsp70 (α-mtHSP70) antibodies. In the merged views, the yellow spots indicate colocalization of FEN1 and mtHsp70. The boxed area in each frame is shown magnified in the upper right; this better illustrates the extranuclear staining of FEN1 and mtHSP70. (B) Representative images of the localization of FEN1 and mtHsp70 in HeLa cells expressing wild-type (WT) and Y83H mutant forms of the nuclease.
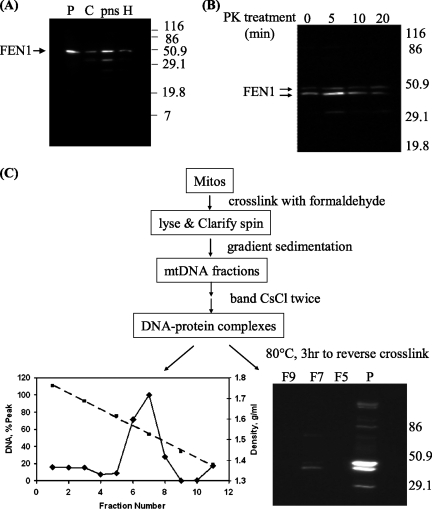
For a more rigorous test of the mitochondrial localization of FEN1, we investigated the effect of protease treatment of mitochondria prior to extraction and tested whether FEN1 is colocalized in nucleoids together with mtDNA replication and transcription proteins (4). As shown in Fig. 5A, a significant amount of FEN1 could be detected in purified mitochondria from HeLa cells by using a monoclonal antibody. Moreover, treatment of these mitochondria with proteinase K prior to extraction showed that much of the FEN1 signal was retained even after extensive digestion (Fig. 5B). Note that known mitochondrial proteins may also be subject to some degradation at the longest digestion times used here (4). We note that two FEN1-cross-reacting bands were identified in this experiment, a result that is further considered below.
FIG. 5.
FEN1 in protease-treated mitochondria and in nucleoids. Molecular weights (in thousands) are given to the side of the gels. (A) FEN1 was detected in purified mitochondria. H, cell homogenate; pns, postnuclear supernatant; C, cytosol fraction; P: purified mitochondria. (B) FEN1 in mitochondria is resistant to proteinase K (PK) treatment. Pure mitochondria were treated with proteinase K for 0, 5, 10, and 20 min before lysis, and 5 μg of mitochondrial lysate was loaded onto each lane for WB. (C) FEN1 is localized in mitochondrial DNA nucleoids. Formaldehyde cross-linked mitochondrial DNA nucleoids were purified as shown in the flow chart. Mitos, mitochondria. After cross-links were reversed, DNA and FEN1 protein in each CsCl fraction were detected by fluorescence and immunoblotting, respectively. F5, F7, and F9, CsCl fractions 5, 7, and 9, respectively; P, purified mitochondria.
Mitochondrial replication and transcription proteins, and perhaps those of DNA repair, are organized within organellar structures called nucleoids (4). The assessment of nucleoids is performed by cross-linking treatment of purified mitochondria, extraction, rigorous isolation of cross-linked protein-DNA complexes on CsCl gradients, and finally reversal of the cross-links and analysis of the recovered proteins. Analysis of such purified nucleoid material by using FEN1 monoclonal antibodies showed that the protein is recovered in nucleoid fractions (Fig. 5C), consistent with the presence of this flap endonuclease in these important organellar substructures. Taken together, our data constitute strong evidence that FEN1 translocates into both nuclei and mitochondria in human cells.
As also seen in higher-resolution gels from the proteinase K experiment (Fig. 5B), the anti-FEN1 monoclonal antibody detected two polypeptides in the purified mitochondria used for the nucleoid experiment (Fig. 5C), with the faster-migrating form (with a Mr ∼4,000 less than that of full-length FEN1) enriched in nucleoids. It seems possible that both forms are present in mitochondria, as both are retained following proteinase K treatment, but understanding the difference between the two forms is outside the scope of the present work.
Dependence of mitochondrial LP-BER on FEN1.
To test whether FEN1 participates in the observed mitochondrial LP-BER (Fig. 2), we immunodepleted FEN1 from GM1310 lymphoblast ME. Levels of repair of THF from mock-treated and FEN1-depleted ME were compared under conditions that permit monitoring of LP-BER exclusively (labeling with [α-32P]dCTP as described for Fig. 2A). Removal of FEN1 from the extracts was confirmed by immunoblotting (Fig. 6A) accompanied by loss of ∼80% of the flap endonuclease activity (Fig. 6B). Most critically, FEN1 depletion from the ME also resulted in removal of most of the LP-BER activity, as measured using a THF substrate (∼60% decrease in LP-BER) (Fig. 6C). Supplementing the depleted ME with purified FEN1 restored LP-BER to a level comparable to that of the mock-treated control, confirming that FEN1 was the only protein whose depletion caused diminished LP-BER activity. Thus, FEN1 is required for most of the LP-BER detected in ME of GM1310 lymphoblasts. The presence of FEN1 in epithelial cells (HeLa) and a similar result with FEN1-depleted HeLa ME (data not shown) further indicate that this is a general phenomenon.
FIG. 6.
LP-BER in mitochondria dependent on FEN1. α-FEN1, FEN1 antibody. (A) Western blot showing immunodepletion of FEN1 from GM1310 ME, with mtHsp70 as a loading control. ME (20 μg) was loaded onto each lane. R. FEN1, purified recombinant FEN1; IgG, immunoglobulin G. (B) Diminished flap endonuclease activity following FEN1 immunodepletion. The substrate used was the THF-4-nt flap (see Fig. 3A). The strong band at the bottom of lane 2 is likely due to the 5′-3′ exonuclease activity of FEN1. (C) LP-BER assay with a THF substrate was performed in the presence of [α-32P]dCTP at 30°C for 30 min. Repair products were revealed after digestion with restriction endonuclease sets BamHI plus HindIII and AccI plus HindIII, which released a 30-nt fragment and a 17-nt fragment, respectively, from the plasmid DNA. The relative LP-BER efficiency of each sample was calculated by normalizing the radioactivity of the 30-nt or the 17-nt fragment to that of the 35-nt internal standard (Std). The internal standard was added after reactions were terminated to control for material loss during phenol extraction and ethanol precipitation. For reactions supplemented with exogenous FEN1, 20 ng of purified FEN1 was added to 12 μg of ME. The experiments were repeated three times. The average values and standard deviations are shown in a bar diagram. M, molecular size marker. IP, immunoprecipitation.
FEN1 is required for efficient repair of mtDNA oxidative damage in vivo.
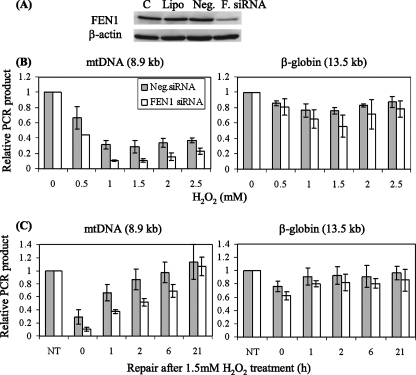
We sought to test the in vivo role of FEN1 in mtDNA repair, for which we employed an approach utilizing RNA interference. This method is necessitated by the fact that FEN1 is an essential protein also involved in nuclear DNA replication through the processing of Okazaki fragments and other reactions (45). Thus, deletion of the FEN1 gene causes embryonic lethality in mice (30). We were able to use siRNA transfection to suppress FEN1 levels significantly in HeLa cells without detectably compromising cell proliferation. In these experiments, the level of FEN1 protein could be reduced to about 15% of the normal amount 4 days after transfection with siRNA specific for FEN1, while a control sequence had no effect (Fig. 7A). The transfected cells were then challenged with H2O2 to introduce oxidative DNA lesions, followed by incubation to allow DNA repair. Lesions in mtDNA or the nucleus were assessed by a gene-specific, QPCR-based assay in which base lesions, abasic sites, or strand breaks interfere with the amplification of long DNA targets (44). This assay has proven particularly useful in examining mtDNA damage and repair kinetics after an oxidative challenge (44). Relatively long PCR products (8.9 kb for mtDNA and 13.5 kb for the β-globin gene) were used diagnostically, and the copy numbers of mtDNA from different samples were normalized to that of a shorter control PCR product (221 bp). As a function of the H2O2 dose (Fig. 7B), the level of mtDNA signal recovered was significantly less in cells transfected with FEN1-specific siRNA than in control cells. The dose-response curve for this effect was not monotonic, perhaps reflecting the two modes of production of lethal H2O2 damage proposed for bacteria (23).
FIG. 7.
Suppression of FEN1 interferes with efficient repair of oxidative damage in mtDNA. (A) FEN1 was knocked down in HeLa cells by using siRNA. C, controls; Lipo, cells treated with Lipofectamine 2000; Neg., negative control siRNA; F. siRNA, FEN1-specific siRNA. (B and C) DNA damage and repair were assayed by long-range QPCR generating an 8.9-kb product from mtDNA and a 13.5-kb product from the β-globin gene for nuclear DNA. These were normalized to the total DNA in the sample for the nuclear product or a 221-bp PCR product for the mtDNA signal. (B) Damage was induced by various doses of H2O2 for 30 min. (C) Repair of mtDNA and nuclear DNA was monitored by QPCR after 30 min of 1.5 mM H2O2 treatment. NT, cells not treated with H2O2. Results shown are averages of three independent siRNA transfections; error bars represent standard deviations.
Time course experiments also supported an important contribution of FEN1 to the repair of oxidative damage in mtDNA. In FEN1-suppressed cells, the amount of oxidative mtDNA damage was more pronounced immediately after the H2O2 treatment, and the recovery of repaired mtDNA was delayed in cells treated with FEN1-specific siRNA (Fig. 7C). These results are consistent with an important role for FEN1 in LP-BER in mitochondria, with possible contributions by other flap endonucleases. It is worth noting that there was also a smaller effect of FEN1 depletion on repair of oxidative DNA damage in nuclear DNA, which reached statistical significance under only one set of conditions (30-min treatment with 1.5 mM H2O2) (Fig. 7B, right panel). The observed greater sensitivity to H2O2 damage of mtDNA compared to nuclear DNA is consistent with the lower degree of protection and more limited repair options thought to exist in mitochondria.
DISCUSSION
How oxidative damage to mtDNA contributes to mitochondrial mutation and dysfunction is not well understood. Various mutagenic oxidative lesions have been measured in mtDNA (21), but most studies have focused on 8-oxoguanine. Despite a 20-fold increase in the level of this lesion in the mtDNA of OGG1-null mice (16), no evidence for mitochondrial dysfunction was found in these animals, suggesting that the functional impact of this lesion in mtDNA is low (49). Contributions from other oxidative lesions, especially those derived from deoxyribose damage, have not been extensively investigated. In this paper, we provide evidence that dL, a common oxidized abasic lesion, has considerable potential to interfere with mtDNA integrity. Unrepaired dL strongly blocks Polγ-catalyzed DNA synthesis. Although a dA residue was incorporated most frequently opposite a template dL, there was an almost complete stalling of Polγ at the lesion site, which eventually led to DNA degradation by the Polγ-associated exonuclease activity. On the other hand, attempted excision of dL following Ape1 cleavage results in covalent trapping and irreversible inactivation of Polγ, the only mitochondrial DNA Pol, which is essential for both replication and repair. Once formed, Polγ-DNA cross-links might become an especially difficult problem if the appropriate DNA repair systems are lacking in mitochondria. This action of dL may contribute to the age-associated decline in the level of mitochondrial Polγ (10). Direct or indirect inhibition of mtDNA replication by dL may lead to mtDNA deletion and depletion, a phenomenon commonly associated with multiple mitochondrial autosomal disorders (22) and age-related neurodegenerative diseases (2, 53).
Mammalian mitochondria have a limited capacity to repair damage incurred by mtDNA (3, 5). Single-nucleotide BER in mitochondria has been demonstrated in multiple studies (5, 31). Using the naturally occurring oxidative lesion dL, which in the nucleus can be processed only by LP-BER (51), we provide here evidence for an LP-BER pathway in mitochondria. This mechanism allows for the removal of dL and presumably other oxidative DNA lesions that otherwise would not be processed. The presence of LP-BER in mitochondria was further confirmed by repair of THF, an artificial analog of abasic sites that in NE is processed exclusively by LP-BER (25, 26, 34). Repair of both dL and THF was clearly detected in extracts of mitochondria prepared from several mammalian sources, and the reactions generated multinucleotide incorporation products.
FEN1 is an essential enzyme participating in nuclear DNA replication, and it is crucial for nuclear LP-BER (29, 30, 45). Our work provides multiple lines of evidence that FEN1 also localizes to mitochondria and that the enzyme plays an important role in mtDNA repair. A flap endonuclease activity similar to that of FEN1 was detected in ME from various sources by using two different DNA substrates. Immunofluorescence microscopy experiments localized FEN1 in mitochondria as well as in the nucleus. FEN1 was retained during proteolytic treatment of mitochondria prior to extraction, supporting the conclusion that the enzyme found in ME is not a contaminant. The protein was also isolated as a component of mitochondrial nucleoids, loci of replication, and other DNA processing reactions. Immunodepletion of FEN1 from ME removed the majority of the observed flap endonuclease and LP-BER activities (85% and 60%, respectively), which demonstrated that LP-BER in ME is strongly dependent on FEN1. The residual LP-BER observed following FEN1 depletion could be due to another flap-digesting nuclease, which remains to be established. Finally, depletion of FEN1 in intact cells led to defects in processing oxidative lesions in mtDNA, supporting an important in vivo contribution of the enzyme.
FEN1 in mitochondria may also contribute to general mtDNA replication. Two models were proposed to explain how mtDNA replicates: the asymmetric strand-displacement model (8, 17, 41) and the symmetric strand-coupled model (59). In the asymmetric strand-displacement model, heavy-strand synthesis starts first and extends through about two-thirds of the genome before light-strand synthesis initiates (while heavy-strand synthesis continues). In the strand-coupled model, mtDNA replicates symmetrically, with leading- and lagging-strand syntheses progressing bidirectionally from multiple origins in a manner similar to nuclear DNA replication. Both models have several lines of evidence to support them, but there is still a lack of consensus in the field. Mitochondrial FEN1 could contribute to either mechanism by removing primers formed during lagging-strand synthesis. Moreover, FEN1 could contribute to the resolution of certain types of stalled replication forks (45). A possible role of FEN1 in mtDNA replication should be investigated in future studies.
A key question remains how FEN1 is translocated into mitochondria. We noted in some experiments (Fig. 5) the presence of a shorter form of the protein cross-reacting with a highly specific monoclonal antibody. Such a slightly truncated form would be consistent with the proteolytic processing involved in the import of many mitochondrial proteins, although internal sequences may also be employed for uptake into this organelle (6). It is interesting that the shorter form appeared to be cross-linked preferentially with the DNA of nucleoids, while both forms remained present in protease-treated mitochondria. Further work will be required to establish the difference between the two forms and their possible differential localizations within mitochondria.
Although LP-BER should generally overcome the threat of lyase-mediated cross-linking of Polγ to incised dL residues, under oxidative stress the formation of numerous lesions might promote such cross-linking. As noted earlier, there is no obvious repair pathway in mitochondria to handle such damage, and even for nuclear DNA, the mechanism is uncertain. The detection of oxidative Polγ-DNA cross-links might therefore provide a useful means to monitor the accumulation of cellular damage involving mitochondria.
Mitochondria are vital organelles of nucleated cells, and the mitochondrial genome is under constant assault by ROS and hydrolytic decay. These sources produce many lesions that can block replication and lead to loss of function. Inherited mutations in mtDNA cause many genetic disorders, and acquired mutations are associated with common diseases, including neurodegenerative disorders and cancer (7, 53, 56). Despite the importance of mtDNA in human health, our understanding of the basic mitochondrial processes that maintain its overall integrity is far from adequate. Studies of pathways involved in mtDNA replication and repair will provide novel information on the relationship of mtDNA damage, mutations, and human diseases and will help focus efforts on finding new preventive and treatment strategies.
Just as our manuscript was completed, a paper indicating the presence of LP-BER operating in parallel with SP-BER in mammalian mitochondria was published (1). However, those authors were unable to find FEN1 in their ME, and they ascribe the necessary flap endonuclease activity to an as-yet-unknown enzyme. The difference between that study and this one could be attributed to the different FEN1 antibodies used: we found that antibodies from different vendors exhibit considerable variation in the sensitivity of FEN1 detection, which is not surprising (P. Liu, D. F. Bogenhagen, and B. Demple, data not shown). The existence of additional enzymes to support LP-BER would certainly be consistent with the work presented here, and studies to identify additional flap endonuclease activities in mitochondria are under way. Moreover, we have provided cellular, biochemical, and in vivo evidence that FEN1 is involved in the LP-BER of oxidative damage in human mitochondria.
Supplementary Material
Acknowledgments
This research was supported by National Institutes of Health (NIH) grants R01GM040000 to B.D., R01CA073764 to B.S., and R01GM029681 to D.F.B. and by the Intramural Research Program of the National Institute on Aging, NIH. P.L. was partially supported by the Center for Medical Countermeasures against Radiation, Education CORE 5U19-A1067751-2 (NIAID, NIH), NCI Radiation Biology training grant T32 CA09078, and a pilot grant from Harvard School of Public Health NIEHS Center Facilities Core (NIEHS-Bio-022).
We are grateful to H. Fung and N. Saydam in the Demple laboratory for useful suggestions and help with the siRNA experiments, M. M. Greenberg (Johns Hopkins University) for providing the dL precursor oligonucleotide, and A. Imrich of Harvard School of Public Health Facility Core for her help with the fluorescence QPCR measurements.
Footnotes
Published ahead of print on 9 June 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Akbari, M., T. Visnes, H. E. Krokan, and M. Otterlei. 2008. Mitochondrial base excision repair of uracil and AP sites takes place by single-nucleotide insertion and long-patch DNA synthesis. DNA Repair (Amsterdam) 7605-616. [DOI] [PubMed] [Google Scholar]
- 2.Bender, A., K. J. Krishnan, C. M. Morris, G. A. Taylor, A. K. Reeve, R. H. Perry, E. Jaros, J. S. Hersheson, J. Betts, T. Klopstock, R. W. Taylor, and D. M. Turnbull. 2006. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat. Genet. 38515-517. [DOI] [PubMed] [Google Scholar]
- 3.Bogenhagen, D. F. 1999. Repair of mtDNA in vertebrates. Am. J. Hum. Genet. 641276-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogenhagen, D. F., D. Rousseau, and S. Burke. 2008. The layered structure of human mitochondrial DNA nucleoids. J. Biol. Chem. 2833665-3675. [DOI] [PubMed] [Google Scholar]
- 5.Bohr, V. A. 2002. Repair of oxidative DNA damage in nuclear and mitochondrial DNA, and some changes with aging in mammalian cells. Free Radic. Biol. Med. 32804-812. [DOI] [PubMed] [Google Scholar]
- 6.Bolender, N., A. Sickmann, R. Wagner, C. Meisinger, and N. Pfanner. 2008. Multiple pathways for sorting mitochondrial precursor proteins. EMBO Rep. 942-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandon, M., P. Baldi, and D. C. Wallace. 2006. Mitochondrial mutations in cancer. Oncogene 254647-4662. [DOI] [PubMed] [Google Scholar]
- 8.Brown, T. A., C. Cecconi, A. N. Tkachuk, C. Bustamante, and D. A. Clayton. 2005. Replication of mitochondrial DNA occurs by strand displacement with alternative light-strand origins, not via a strand-coupled mechanism. Genes Dev. 192466-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chattopadhyay, R., L. Wiederhold, B. Szczesny, I. Boldogh, T. K. Hazra, T. Izumi, and S. Mitra. 2006. Identification and characterization of mitochondrial abasic (AP)-endonuclease in mammalian cells. Nucleic Acids Res. 342067-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, D., G. Cao, T. Hastings, Y. Feng, W. Pei, C. O'Horo, and J. Chen. 2002. Age-dependent decline of DNA repair activity for oxidative lesions in rat brain mitochondria. J. Neurochem. 811273-1284. [DOI] [PubMed] [Google Scholar]
- 11.Clayton, D. A., J. N. Doda, and E. C. Friedberg. 1974. The absence of a pyrimidine dimer repair mechanism in mammalian mitochondria. Proc. Natl. Acad. Sci. USA 712777-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Creighton, S., L. B. Bloom, and M. F. Goodman. 1995. Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading, and lesion bypass efficiencies. Methods Enzymol. 262232-256. [DOI] [PubMed] [Google Scholar]
- 13.Croteau, D. L., R. H. Stierum, and V. A. Bohr. 1999. Mitochondrial DNA repair pathways. Mutat. Res. 434137-148. [DOI] [PubMed] [Google Scholar]
- 14.DeMott, M. S., E. Beyret, D. Wong, B. C. Bales, J. T. Hwang, M. M. Greenberg, and B. Demple. 2002. Covalent trapping of human DNA polymerase beta by the oxidative DNA lesion 2-deoxyribonolactone. J. Biol. Chem. 2777637-7640. [DOI] [PubMed] [Google Scholar]
- 15.Demple, B., and M. S. DeMott. 2002. Dynamics and diversions in base excision DNA repair of oxidized abasic lesions. Oncogene 218926-8934. [DOI] [PubMed] [Google Scholar]
- 16.de Souza-Pinto, N. C., L. Eide, B. A. Hogue, T. Thybo, T. Stevnsner, E. Seeberg, A. Klungland, and V. A. Bohr. 2001. Repair of 8-oxodeoxyguanosine lesions in mitochondrial DNA depends on the oxoguanine DNA glycosylase (OGG1) gene and 8-oxoguanine accumulates in the mitochondrial DNA of OGG1-defective mice. Cancer Res. 615378-5381. [PubMed] [Google Scholar]
- 17.Fish, J., N. Raule, and G. Attardi. 2004. Discovery of a major D-loop replication origin reveals two modes of human mtDNA synthesis. Science 3062098-2101. [DOI] [PubMed] [Google Scholar]
- 18.Harman, D. 1956. Aging: a theory based on free radical and radiation chemistry. J. Gerontol. 11298-300. [DOI] [PubMed] [Google Scholar]
- 19.Harman, D. 1972. The biologic clock: the mitochondria? J. Am. Geriatr. Soc. 20145-147. [DOI] [PubMed] [Google Scholar]
- 20.Harrington, J. J., and M. R. Lieber. 1994. The characterization of a mammalian DNA structure-specific endonuclease. EMBO J. 131235-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hegler, J., D. Bittner, S. Boiteux, and B. Epe. 1993. Quantification of oxidative DNA modifications in mitochondria. Carcinogenesis 142309-2312. [DOI] [PubMed] [Google Scholar]
- 22.Hirano, M., R. Marti, C. Ferreiro-Barros, M. R. Vila, S. Tadesse, Y. Nishigaki, I. Nishino, and T. H. Vu. 2001. Defects of intergenomic communication: autosomal disorders that cause multiple deletions and depletion of mitochondrial DNA. Semin. Cell Dev. Biol. 12417-427. [DOI] [PubMed] [Google Scholar]
- 23.Imlay, J. A., and S. Linn. 1986. Bimodal pattern of killing of DNA-repair-defective or anoxically grown Escherichia coli by hydrogen peroxide. J. Bacteriol. 166519-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kao, H. I., L. A. Henricksen, Y. Liu, and R. A. Bambara. 2002. Cleavage specificity of Saccharomyces cerevisiae flap endonuclease 1 suggests a double-flap structure as the cellular substrate. J. Biol. Chem. 27714379-14389. [DOI] [PubMed] [Google Scholar]
- 25.Kim, K., S. Biade, and Y. Matsumoto. 1998. Involvement of flap endonuclease 1 in base excision DNA repair. J. Biol. Chem. 2738842-8848. [DOI] [PubMed] [Google Scholar]
- 26.Klungland, A., and T. Lindahl. 1997. Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1). EMBO J. 163341-3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korhonen, J. A., X. H. Pham, M. Pellegrini, and M. Falkenberg. 2004. Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 232423-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kroeger, K. M., M. Hashimoto, Y. W. Kow, and M. M. Greenberg. 2003. Cross-linking of 2-deoxyribonolactone and its beta-elimination product by base excision repair enzymes. Biochemistry 422449-2455. [DOI] [PubMed] [Google Scholar]
- 29.Kucherlapati, M., K. Yang, M. Kuraguchi, J. Zhao, M. Lia, J. Heyer, M. F. Kane, K. Fan, R. Russell, A. M. Brown, B. Kneitz, W. Edelmann, R. D. Kolodner, M. Lipkin, and R. Kucherlapati. 2002. Haploinsufficiency of Flap endonuclease (Fen1) leads to rapid tumor progression. Proc. Natl. Acad. Sci. USA 999924-9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larsen, E., C. Gran, B. E. Saether, E. Seeberg, and A. Klungland. 2003. Proliferation failure and gamma radiation sensitivity of Fen1 null mutant mice at the blastocyst stage. Mol. Cell. Biol. 235346-5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LeDoux, S. P., N. M. Druzhyna, S. B. Hollensworth, J. F. Harrison, and G. L. Wilson. 2007. Mitochondrial DNA repair: a critical player in the response of cells of the CNS to genotoxic insults. Neuroscience 1451249-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, R., J. Qiu, L. D. Finger, L. Zheng, and B. Shen. 2006. The DNA-protein interaction modes of FEN-1 with gap substrates and their implication in preventing duplication mutations. Nucleic Acids Res. 341772-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu, T., Y. Pan, S. Y. Kao, C. Li, I. Kohane, J. Chan, and B. A. Yankner. 2004. Gene regulation and DNA damage in the ageing human brain. Nature 429883-891. [DOI] [PubMed] [Google Scholar]
- 34.Matsumoto, Y., K. Kim, and D. F. Bogenhagen. 1994. Proliferating cell nuclear antigen-dependent abasic site repair in Xenopus laevis oocytes: an alternative pathway of base excision DNA repair. Mol. Cell. Biol. 146187-6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mecocci, P., U. MacGarvey, and M. F. Beal. 1994. Oxidative damage to mitochondrial DNA is increased in Alzheimer's disease. Ann. Neurol. 36747-751. [DOI] [PubMed] [Google Scholar]
- 36.Mecocci, P., U. MacGarvey, A. E. Kaufman, D. Koontz, J. M. Shoffner, D. C. Wallace, and M. F. Beal. 1993. Oxidative damage to mitochondrial DNA shows marked age-dependent increases in human brain. Ann. Neurol. 34609-616. [DOI] [PubMed] [Google Scholar]
- 37.Pinz, K. G., and D. F. Bogenhagen. 2000. Characterization of a catalytically slow AP lyase activity in DNA polymerase gamma and other family A DNA polymerases. J. Biol. Chem. 27512509-12514. [DOI] [PubMed] [Google Scholar]
- 38.Pinz, K. G., S. Shibutani, and D. F. Bogenhagen. 1995. Action of mitochondrial DNA polymerase gamma at sites of base loss or oxidative damage. J. Biol. Chem. 2709202-9206. [DOI] [PubMed] [Google Scholar]
- 39.Polidori, M. C., P. Mecocci, S. E. Browne, U. Senin, and M. F. Beal. 1999. Oxidative damage to mitochondrial DNA in Huntington's disease parietal cortex. Neurosci. Lett. 27253-56. [DOI] [PubMed] [Google Scholar]
- 40.Richter, C., J. W. Park, and B. N. Ames. 1988. Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc. Natl. Acad. Sci. USA 856465-6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robberson, D. L., H. Kasamatsu, and J. Vinograd. 1972. Replication of mitochondrial DNA. Circular replicative intermediates in mouse L cells. Proc. Natl. Acad. Sci. USA 69737-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roginskaya, M., W. A. Bernhard, R. T. Marion, and Y. Razskazovskiy. 2005. The release of 5-methylene-2-furanone from irradiated DNA catalyzed by cationic polyamines and divalent metal cations. Radiat. Res. 16385-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roupioz, Y., J. Lhomme, and M. Kotera. 2002. Chemistry of the 2-deoxyribonolactone lesion in oligonucleotides: cleavage kinetics and products analysis. J. Am. Chem. Soc. 1249129-9135. [DOI] [PubMed] [Google Scholar]
- 44.Santos, J. H., J. N. Meyer, B. S. Mandavilli, and B. Van Houten. 2006. Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol. Biol. 314183-199. [DOI] [PubMed] [Google Scholar]
- 45.Shen, B., P. Singh, R. Liu, J. Qiu, L. Zheng, L. D. Finger, and S. Alas. 2005. Multiple but dissectible functions of FEN-1 nucleases in nucleic acid processing, genome stability and diseases. Bioessays 27717-729. [DOI] [PubMed] [Google Scholar]
- 46.Singh, P., L. Zheng, V. Chavez, J. Qiu, and B. Shen. 2007. Concerted action of exonuclease and Gap-dependent endonuclease activities of FEN-1 contributes to the resolution of triplet repeat sequences (CTG)n- and (GAA)n-derived secondary structures formed during maturation of Okazaki fragments. J. Biol. Chem. 2823465-3477. [DOI] [PubMed] [Google Scholar]
- 47.Sohal, R. S., and R. Weindruch. 1996. Oxidative stress, caloric restriction, and aging. Science 27359-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stierum, R. H., G. L. Dianov, and V. A. Bohr. 1999. Single-nucleotide patch base excision repair of uracil in DNA by mitochondrial protein extracts. Nucleic Acids Res. 273712-3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stuart, J. A., B. M. Bourque, N. C. de Souza-Pinto, and V. A. Bohr. 2005. No evidence of mitochondrial respiratory dysfunction in OGG1-null mice deficient in removal of 8-oxodeoxyguanine from mitochondrial DNA. Free Radic. Biol. Med. 38737-745. [DOI] [PubMed] [Google Scholar]
- 50.Stuart, J. A., K. Hashiguchi, D. M. Wilson III, W. C. Copeland, N. C. Souza-Pinto, and V. A. Bohr. 2004. DNA base excision repair activities and pathway function in mitochondrial and cellular lysates from cells lacking mitochondrial DNA. Nucleic Acids Res. 322181-2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sung, J. S., M. S. DeMott, and B. Demple. 2005. Long-patch base excision DNA repair of 2-deoxyribonolactone prevents the formation of DNA-protein cross-links with DNA polymerase beta. J. Biol. Chem. 28039095-39103. [DOI] [PubMed] [Google Scholar]
- 52.Sung, J. S., and B. Demple. 2006. Roles of base excision repair subpathways in correcting oxidized abasic sites in DNA. FEBS J. 2731620-1629. [DOI] [PubMed] [Google Scholar]
- 53.Taylor, R. W., and D. M. Turnbull. 2005. Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 6389-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thyagarajan, B., R. A. Padua, and C. Campbell. 1996. Mammalian mitochondria possess homologous DNA recombination activity. J. Biol. Chem. 27127536-27543. [DOI] [PubMed] [Google Scholar]
- 55.Vodenicharov, M. D., F. R. Sallmann, M. S. Satoh, and G. G. Poirier. 2000. Base excision repair is efficient in cells lacking poly(ADP-ribose) polymerase 1. Nucleic Acids Res. 283887-3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wallace, D. C. 2005. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 39359-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu, Y. J., M. S. DeMott, J. T. Hwang, M. M. Greenberg, and B. Demple. 2003. Action of human apurinic endonuclease (Ape1) on C1′-oxidized deoxyribose damage in DNA. DNA Repair (Amsterdam) 2175-185. [DOI] [PubMed] [Google Scholar]
- 58.Yakubovskaya, E., Z. Chen, J. A. Carrodeguas, C. Kisker, and D. F. Bogenhagen. 2006. Functional human mitochondrial DNA polymerase gamma forms a heterotrimer. J. Biol. Chem. 281374-382. [DOI] [PubMed] [Google Scholar]
- 59.Yang, M. Y., M. Bowmaker, A. Reyes, L. Vergani, P. Angeli, E. Gringeri, H. T. Jacobs, and I. J. Holt. 2002. Biased incorporation of ribonucleotides on the mitochondrial L-strand accounts for apparent strand-asymmetric DNA replication. Cell 111495-505. [DOI] [PubMed] [Google Scholar]
- 60.Zheng, L., H. Dai, J. Qiu, Q. Huang, and B. Shen. 2007. Disruption of the FEN-1/PCNA interaction results in DNA replication defects, pulmonary hypoplasia, pancytopenia, and newborn lethality in mice. Mol. Cell. Biol. 273176-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng, L., H. Dai, M. Zhou, M. Li, P. Singh, J. Qiu, W. Tsark, Q. Huang, K. Kernstine, X. Zhang, D. Lin, and B. Shen. 2007. Fen1 mutations result in autoimmunity, chronic inflammation and cancers. Nat. Med. 13812-819. [DOI] [PubMed] [Google Scholar]
- 62.Zuckerman, S. H., J. F. Solus, F. P. Gillespie, and J. M. Eisenstadt. 1984. Retention of both parental mitochondrial DNA species in mouse-Chinese hamster somatic cell hybrids. Somat. Cell Mol. Genet. 1085-91. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.