Abstract
The assembly of cytochrome c oxidase (CcO) in yeast mitochondria is dependent on a new assembly factor designated Coa2. Coa2 was identified from its ability to suppress the respiratory deficiency of coa1Δ and shy1Δ cells. Coa1 and Shy1 function at an early step in maturation of the Cox1 subunit of CcO. Coa2 functions downstream of the Mss51-Coa1 step in Cox1 maturation and likely concurrent with the Shy1-related heme a3 insertion into Cox1. Coa2 interacts with Shy1. Cells lacking Coa2 show a rapid degradation of newly synthesized Cox1. Rapid Cox1 proteolysis also occurs in shy1Δ cells, suggesting that in the absence of Coa2 or Shy1, Cox1 forms an unstable conformer. Overexpression of Cox10 or Cox5a and Cox6 or attenuation of the proteolytic activity of the m-AAA protease partially restores respiration in coa2Δ cells. The matrix-localized Coa2 protein may aid in stabilizing an early Cox1 intermediate containing the nuclear subunits Cox5a and Cox6.
Cytochrome c oxidase (CcO) is the terminal enzyme of the energy-transducing respiratory chain in mitochondria. Eukaryotic CcO consists of 12 or 13 subunits, with three mitochondrially encoded subunits (Cox1 to Cox3) forming the core enzyme embedded within the mitochondrial inner membrane (IM). The remaining nucleus-encoded subunits pack on the periphery of the catalytic core (35). The core enzyme contains three copper atoms and two heme a cofactors (34).
The biogenesis of CcO, occurring within the IM, commences with the mitochondrial synthesis of the Cox1 subunit, followed by the insertion of heme a and copper cofactors and the addition of the remaining subunits. The translation of Cox1 on mitoribosomes occurs in juxtaposition to the IM and is mediated in the yeast Saccharomyces cerevisiae by the IM-tethered translational activator Pet309 along with the IM-associated Mss51 protein (23, 24, 28, 30). The translational activators may recruit mitoribosomes and promote cotranslational IM insertion together with the Oxa1 translocase (9, 17).
Insights have also been gleaned on the assembly process of CcO through characterization of stalled intermediate complexes in human cells of patients with mutations in assembly factors. One intermediate that accumulates in Leigh's syndrome patients with SURF1 mutations consists of Cox1, CoxIV, and CoxVa (32, 37, 40). This intermediate fails to accumulate in patient cells with mutations in Cox10 and Cox15, two enzymes that function in heme a biosynthesis. Thus, the prediction is that heme a insertion occurs prior to the addition of CoxIV and CoxVa (corresponding to yeast Cox5a/b and Cox6) (1, 2). Two heme a moieties and a copper ion are buried inside a barrel within Cox1 (34). These cofactors may be inserted through an interface obstructed by the association of the Cox2 subunit. Consistent with this postulate is the fact that Cox2 associates downstream of the Cox1, Cox5a, and Cox6 complex (18).
In addition to translational initiation, Mss51 has a posttranslational function in yeast Cox1 maturation (8, 28). Mss51 and Cox14 form a complex that interacts with Cox1 and regulates Cox1 translation/elongation. In cells lacking the yeast gene SURF1, i.e., shy1Δ cells, Cox1 translation is stated to be attenuated (8). We recently reported that Coa1 functions in an early step of Cox1 assembly in yeast (29). Coa1 is part of the Mss51, Cox14, and Cox1 complex and, in addition, binds to Shy1 in a separate complex (25, 29). Coa1 appears to coordinate the transition of newly synthesized Cox1 from the Mss51 complex to a later intermediate, involving Shy1, that likely functions in heme a3 insertion. We isolated a series of extragenic suppressors of coa1Δ cells (29). The high-copy-number genes MSS51 and COX10 suppress the respiratory defect of coa1Δ cells. In addition, we observed that respiration in both coa1Δ and shy1Δ cells is enhanced when MSS51 and COX10 are coexpressed. Cox10 is a farnesyl transferase involved in the biosynthesis of heme a. The synergistic suppression is consistent with Mss51 and Cox10 affecting Cox1 translation and heme a3 insertion, suggesting that Coa1 links cotranslational insertion and heme addition to Cox1.
A fourth extragenic suppressor of coa1Δ cells was found. High-copy expression of the unannotated yeast open reading frame (ORF) YPL189C-A, designated Coa2 (cytochrome oxidase assembly 2), is shown here to effectively restore respiratory growth to coa1Δ and shy1Δ cells. We report that Coa2 functions in Cox1 maturation downstream from the Mss51 translational elongation step and at the heme a3 insertion step involving Shy1.
MATERIALS AND METHODS
Yeast strains and vectors.
Yeast strains used in this study are described in Table 1. The deletion strains were created by disruption, using homologous recombination of KanMX4 or Candida albicans URA3. Proper integration was confirmed by PCR of the locus, and reversion of the phenotype induced by the deletion was ensured by complementation with a wild-type (WT) copy of the gene on a plasmid. Cells were cultured in either rich medium or synthetic complete (SC) medium lacking the appropriate nutrients for plasmid selection. The carbon source used was either 2% glucose, 2% glycerol-2% lactate, or 2% raffinose. COA2 was cloned into YEp lac181 with 400 bp of its own promoter and terminator, using BamHI and SalI. The COA2 ORF was cloned into pRS416 under the control of the MET25 promoter and the CYC1 terminator (26), using BamHI and SalI. COX5a and COX6 were inserted into pRS426 and pRS423 under the control of their own promoters (600 and 430 bp, respectively) and terminators (500 bp) by restriction with BamHI and XhoI. The vector containing Yta12(E614Q) was a gift from Thomas Langer. The COX10 ORF with the coding sequence for a 3′ six-His peptide tag was cloned into pRS426 under the control of the MET25 promoter and CYC1 terminator. The same promoter and terminator set was used for cloning of the MSS51 ORF into pRS423. The SHY1 ORF with a 13-Myc 3′ tag was cloned into pRS416 under the control of its own promoter and the ADH1 terminator. Sequencing was used to confirm cloning products in all created vectors. Yeast strains were transformed using lithium acetate.
TABLE 1.
Yeast strains used in this studya
| Strain | Genotype | Reference or source |
|---|---|---|
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Invitrogen Corp. |
| BY4741 Δcoa2 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Δcoa2::kanMX4 | This work |
| W303 | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 | |
| W303 Δcoa2 | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δcoa2::kanMX4 | This work |
| W303 Δcoa2 Δcox14 | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δcoa2::kanMX4 Δcox14::CaURA3 | This work |
| W303 Δcoa2 | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δcoa2::CaURA3 | This work |
| W303 Δcoa2 Δyta12 | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δcoa2::CaURA3 Δyta12::kanMX4 | This work |
| W303 Δcoa2 Δcoa1 | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δcoa2::CaURA3 Δcoa1::kanMX4 | This work |
| W303 Δshy1 | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δshy1::URA3 | 7 |
| W303 Δshy1 Δcoa2 | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δshy1::URA3 Δcoa2::kanMX4 | This work |
| W303 Δcox5a | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δcox5a::HIS3 | 16 |
| cox1Δ::ARG8m | MATalys2 leu2-3,112 arg8::hisG ura3-52 [cox1Δ::ARG8m] | 28 |
| cox1Δ::ARG8m Δcoa2 | MATalys2 leu2-3,112 arg8::hisG ura3-52 [cox1Δ::ARG8m] Δcoa2::kanMX4 | This work |
| COX1(1-512)::ARG8 | MATalys2 leu2-3,112 arg8::hisG ura3-52 [COX1(1-512)::ARG8m] | 28 |
| COX1(1-512)::ARG8 Δcoa2 | MATalys2 leu2-3,112 arg8::hisG ura3-52 [COX1(1-512)::ARG8m] Δcoa2::kanMX4 | This work |
| COX1(1-512)::ARG8 Δcox5a | MATalys2 leu2-3,112 arg8::hisG ura3-52 [COX1(1-512)::ARG8m] Δcox5a::kanMX4 | This work |
| DY5113 | MATaade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 | 20 |
| COA2-3HA | MATaade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 COA2-3HA::TRP1 | This work |
| COA2-13Myc | MATaade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 COA2-13Myc::TRP1 | This work |
| COA2-13Myc Δpet309 | MATaade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 COA2-13Myc::TRP1 Δpet309::CaURA3 | This work |
| MSS51-13Myc COA1-3HA | MATaade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 COA1-3HA::TRP1 MSS51-13Myc::HIS3MX6 | 29 |
| MSS51-13Myc COA1-3HA Δcoa2 | MATaade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 COA1-3HA::TRP1 MSS51-13Myc::HIS3MX6 Δcoa2::kanMX4 | This work |
| MSS51-13Myc COA1-3HA Δpet309 | MATaade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 COA1-3HA::TRP1 MSS51-13Myc::HIS3MX6 Δpet309::CaURA3 | This work |
| BY4743 Δshy1 | MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 MET15/met15Δ0 ura3Δ0/ura3Δ0 Δshy1::kanMX4 | Invitrogen Corp. |
| BY4743 Δcoa1 | MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 MET15/met15Δ0 ura3Δ0/ura3Δ0 Δcoa1::kanMX4 | Invitrogen Corp. |
| BY4743 | MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 MET15/met15Δ0 ura3Δ0/ura3Δ0 | Invitrogen Corp. |
| BY4743 Δcox5a | MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 MET15/met15Δ0 ura3Δ0/ura3Δ0 Δcox5a::kanMX4 | Invitrogen Corp. |
| BY4743 Δcox6 | MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 MET15/met15Δ0 ura3Δ0/ura3Δ0 Δcox6::kanMX4 | Invitrogen Corp. |
| BY4743 Δcox7 | MATa/α hs3Δ1/his3Δ1 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 MET15/met15Δ0 ura3Δ0/ura3Δ0 Δcox7::kanMX4 | Invitrogen Corp. |
The strains with the 3HA or 13Myc tag were generated in the DY5113 background by PCR-based gene modification, using the template pFA6a-3HA-TRP1, pFA6a-13MYC-TRP1, pFA6a-3HA-HIS3MX6, or pFA6a-13MYC-HIS3MX6 (22).
Mitochondrial purification and assays.
Intact mitochondria were isolated from yeast as described previously (13, 14). The total mitochondrial protein concentration was determined by Bradford assay (11). Cytochrome c oxidase activity was assessed by monitoring the oxidation of reduced cytochrome c, and reaction rates were normalized to total mitochondrial protein (12). The oxygen consumption of cells grown to stationary phase was determined on a model 5300A biological oxygen monitor (Yellow Springs Instrument Co.). The linear response was considered in calculating the rate of oxygen consumption. Heme analysis was conducted on purified mitochondria (1 to 2 mg of protein) extracted with 0.5 ml of acetone containing 2.5% HCl, as described previously (29). The pH of the extract was adjusted to 4 by addition of 1 μl formic acid and titration with KOH. The sample was clarified by centrifugation at 13,000 rpm for 5 min, and 1 ml was injected onto a 3.9- by 300-mm Bondclone C18 column (Phenomex).
Hydrogen peroxide sensitivity, respiratory growth assay, and arginine auxotrophy.
Sensitivities of the strains and transformants to hydrogen peroxide were assessed as previously described (21). To test respiratory competence, the strains were grown overnight in selective medium containing 2% raffinose-0.2% glucose, serially diluted, and spotted on 2% glucose-containing medium (control) and 2% glycerol-2% lactate-containing rich (YPLG) or SC (SCLG) medium. The cox1Δ::ARG8 COX1(1-512)::ARG8 strains and their derivatives were grown overnight in selective medium containing 2% glucose and 1× arginine (20 mg/liter Arg) or 0.2× Arg, as indicated. The cultures were serially diluted and dropped onto SC medium with 2% glucose containing no Arg or 1× Arg (control).
In vivo mitochondrial protein translation assay.
The cells were grown overnight in selective medium containing 2% raffinose and then reinoculated in YP-2% raffinose to grow to an absorbance of 1 at 600 nm. The labeling and preparation of the samples for 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) or 15% SDS-PAGE (to visualize mp15) were done as described previously (7). The gel was dried, and radiolabeled proteins were visualized by exposing autoradiographic films at −80°C.
BN-PAGE.
Blue native PAGE (BN-PAGE) analysis was performed essentially as described previously (38), except that 1.5% digitonin was used. After incubation for 15 min on ice and centrifugation (20,000 × g for 10 min at 2°C), supernatants were mixed with sample buffer (5% Coomassie brilliant blue G250, 0.5 M 6-aminocaproic acid, pH 7.0) and loaded on a gradient polyacrylamide gel. Separated complexes were detected by immunoblotting on a polyvinylidene difluoride membrane.
Immunoassays.
Protein samples were separated in 15% acrylamide gels and transferred to nitrocellulose. Proteins were visualized using enhanced chemiluminescence (ECL) reagents with horseradish peroxidase-conjugated secondary antibodies. Anti-Myc and antihemagglutinin (anti-HA) antisera were purchased from Santa Cruz, antiporin was purchased from Molecular Probes, and antisera to Cox1 to Cox3 were purchased from Mitosciences. Antiserum to Sod2 was provided by Val Culotta, and antisera to Cyb2 and Cyc1 were provided by Carla Koehler. Alex Tzagoloff provided antiserum to F1 ATP synthase, and Peter Rehling provided antiserum to Cyt1. For immunoprecipitation (IP), purified mitochondria (0.3 mg protein) were solubilized by incubation in 0.5 ml 40 mM potassium phosphate, 200 mM ammonium sulfate, 1 mM phenylmethylsulfonyl fluoride, and 1% digitonin (IP buffer) for 10 min at 4°C. Lysates were centrifuged at 16,000 × g for 30 min at 4°C. IP was performed by incubating the clarified lysates overnight at 4°C with 30 μl anti-HA-agarose conjugate (Santa Cruz Biotechnology). The beads were pelleted by centrifugation at 1,000 × g for 1 min and washed with 0.5 ml of IP buffer for 5 min, and this was repeated four times. The agarose beads were eluted by boiling for 5 min in 50 μl SDS-PAGE loading buffer. The clarified lysate, final wash, and elution fractions were analyzed by immunoblotting.
RESULTS
Coa2 is a multicopy suppressor of coa1Δ and shy1Δ cell respiratory growth defect.
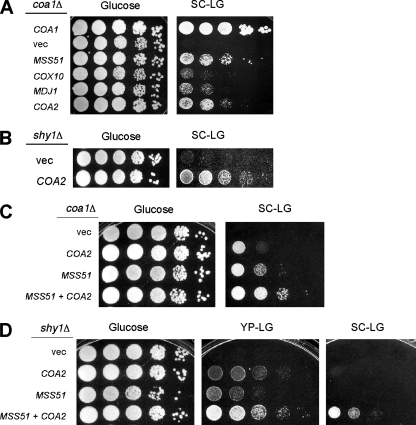
In a search for high-copy-number suppressors of the respiratory growth defect of coa1Δ cells, we recovered MSS51, COX10, and MDJ1 (29). A fourth suppressor, which we designated COA2, was also identified (Fig. 1A). Five plasmids recovered from respiration-competent clones contained a DNA fragment encompassing YPL189c-a, an uncharacterized ORF that was identified by homology to an ORF in Ashbya gossypii (10, 19). Our identification of this gene as an extragenic suppressor and the respiratory deficiency of the deletion of YPL189c-a (10, 19) imply that YPL189c-a is a functional gene. The complementation of the respiratory growth defect of coa1Δ cells by overexpression of COA2 is similar to that observed by overexpression of MSS51 (Fig. 1A). Since Coa1 and Shy1 function at a related step in CcO assembly (29), we tested whether COA2 overexpression rescued the respiratory defect of shy1Δ cells. Coa2 overexpression was able to partially complement the respiratory growth defect of shy1Δ cells (Fig. 1B). A synergistic effect of MSS51 and COA2 on the suppression of the respiratory defect of coa1Δ and shy1Δ strains was found (Fig. 1C and D), consistent with Mss51 and Coa2 affecting distinct aspects of CcO maturation. This is analogous to the synergistic suppression effects we reported previously for MSS51 and COX10 in coa1Δ and shy1Δ strains (29).
FIG. 1.
Overexpression of COA2 suppresses the respiratory growth defect of coa1Δ and shy1Δ cells and is synergistic with MSS51. (A) coa1Δ cells (BY4743 background) transformed with episomal vectors expressing MSS51, MDJ1, COX10, or COA2 were grown in SC-2% raffinose, serially diluted, and spotted on SCLG or 2% glucose. The plates were incubated at 30°C for 2 days (glucose) or 7 days (SCLG). (B) shy1Δ cells (W303 background) transformed with an episomal vector expressing COA2 or an empty vector were grown as described for panel A and spotted on YPLG or 2% glucose. The plates were incubated at 30°C for 2 days (glucose) or 4 days (YPLG). (C) BY4743 coa1Δ cells transformed with episomal vectors expressing MSS51, COA2, or both were grown in SC-2% raffinose, serially diluted, and spotted on SCLG or 2% glucose. The plates were incubated at 30°C for 2 days (glucose) or 6 days (SCLG). (D) BY4743 shy1Δ cells transformed with the same vectors as those described for panel C were grown in SC-2% raffinose, serially diluted, and spotted on SCLG, 2% glucose, or YPLG. The plates were incubated at 30°C for 2 days (glucose) or 6 days.
coa2Δ cells are respiration deficient, with a CcO-specific defect.
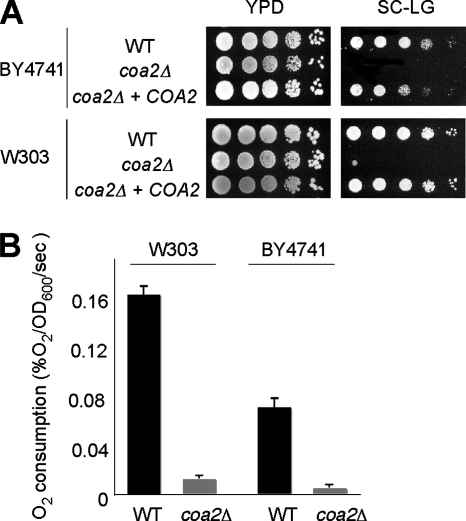
The COA2 ORF was deleted by homologous recombination with a KanMX4 cassette in BY4741 and W303 strain backgrounds. The coa2Δ strains showed normal growth on glucose medium but were unable to propagate on respiratory medium containing glycerol and lactate as carbon sources (Fig. 2A). Introduction of a centromeric plasmid expressing COA2 under the control of the MET25 promoter and CYC1 terminator in the coa2Δ strains restored respiratory growth to the WT level. Consistent with the growth defect observed on glycerol-lactate medium, oxygen consumption was strongly reduced in the coa2Δ strains, compared to that in the isogenic WT strains (Fig. 2B). Overall, these results confirm that Coa2 is necessary for respiration in S. cerevisiae, as reported recently in a high-throughput study (19).
FIG. 2.
coa2Δ cells are respiration deficient. (A) coa2Δ cells, isogenic WT cells, and coa2Δ cells transformed with a centromeric vector expressing the COA2 ORF under the control of the MET25 promoter and the CYC1 terminator were grown in SC-2% glucose, serially diluted, and spotted on yeast extract-peptone-dextrose (YPD) or SCLG. The plates were incubated at 30°C for 2 days (YPD) or 4 days (SCLG). (B) coa2Δ cells and the WT were grown in liquid YPD at 30°C, and oxygen consumption (% O2/s/optical density value at 600 nm [OD600]) was measured. The data represent the averages for four independent experiments, and the error bars indicate the standard errors.
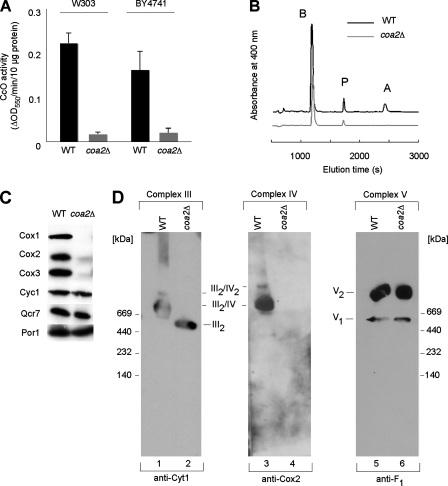
Since Coa1 and Shy1 are part of the CcO assembly machinery, we assessed whether the absence of COA2 led to a CcO defect. CcO activity measured on purified mitochondria was diminished to about 10% of the WT level in the coa2Δ strains (Fig. 3A), whereas bc1 activity (complex III) was reduced to 50 to 60% of the WT level (data not shown). Heme a, a cofactor exclusive to CcO, was undetectable in mitochondrial extracts of coa2Δ strains, and heme b levels in mitochondria were 58% of the WT level, consistent with the partial reduction in bc1 activity (Fig. 3B). Numerous CcO assembly mutants have reduced bc1 activity arising from partial pleiotropy (36). Immunoblotting showed an absence of Cox1 to Cox3 in mitochondria purified from the coa2Δ strain. Despite the reduced bc1 activity, cytochrome c and Qcr7, a subunit of complex III, were present at WT levels (Fig. 3C). CcO was undetectable in the coa2Δ mitochondria by BN-PAGE, whereas the CcO supercomplexes were evident in WT mitochondria (Fig. 3D). The absence of CcO in coa2Δ cells caused complex III to be redistributed from the III2-IV2 and III2-IV supercomplexes seen in WT cells into the dimeric form (III2) (Fig. 3D). The deletion of COA2 had no effect on complex V (Fig. 3D). Overall, these data argue for a specific CcO defect in coa2Δ cells.
FIG. 3.
Deletion of COA2 results in a CcO-specific defect. (A) coa2Δ cells and the isogenic WT were grown in YP-2% raffinose, and mitochondria were purified and assayed for CcO activity (ΔOD550/min/10 μg protein). The data represent the averages for three independent experiments, and the error bars indicate the standard errors. (B) Heme was extracted from mitochondria (2 mg protein) purified from coa2Δ or W303 cells and separated by reverse-phase high-performance liquid chromatography. The peaks corresponding to heme b (B), heme a (A), and protoporphyrin (P) are shown. (C) Immunoblot of mitochondria (30 μg protein) from coa2Δ cells or from WT W303 cells. (D) Mitochondria (200 μg protein) isolated from either the WT or coa2Δ strain were solubilized in buffer containing 1.5% digitonin. Lysates were loaded onto a continuous 4 to 13% gradient gel, and protein complexes were separated by BN-PAGE. The distribution of respiratory complexes was analyzed by immunoblotting with antisera against Cyt1, Cox2, and the F1 subunit.
Coa2 is a mitochondrial matrix soluble protein partially associated with the IM.
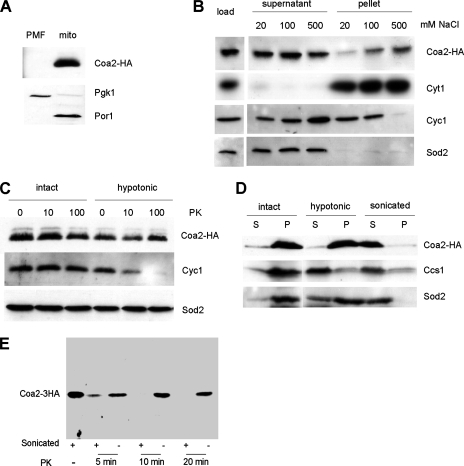
We chromosomally integrated three copies of the HA epitope at the 3′ end of the COA2 ORF. The tag did not affect the function of Coa2, as judged by the WT growth on respiratory medium of the COA2-3HA strain (data not shown). Nycodenz gradient-purified mitochondria isolated from the COA2-3HA strain showed a single band at ∼17 kDa upon immunodetection with HA antiserum (Fig. 4A). This apparent mass is higher than the expected 10.8 kDa, although the basic isoelectric point of Coa2 (10.6) may account for its aberrant migration in SDS-PAGE gels. Coa2 was detected predominantly in the soluble fraction of sonicated mitochondria, like the soluble proteins Cyc1 and Sod2 (Fig. 4B). When the concentration of sodium chloride in the sonication buffer was increased, Cyc1 lost its membrane association, as expected, whereas the fraction of Coa2 recovered in the pellet increased (Fig. 4B). This salt effect suggests that Coa2 may have a hydrophobic interaction with a mitochondrial membrane or with one of its proteins. This candidate interaction may involve a hydrophobic segment between residues 13 and 33. In order to determine the subcompartmentalization of Coa2, we treated intact or hypotonically lysed mitochondria with proteinase K. The intermembrane space (IMS) protein Cyc1 was degraded in mitoplasts, whereas Coa2 was protected, similar to the matrix protein Sod2 (Fig. 4C). Coa2, like Sod2, was retained in the insoluble fraction of mitoplasts, whereas it was liberated in the soluble fraction upon sonication of mitochondria (Fig. 4D). In contrast, Ccs1, an IMS soluble protein, was mostly present in the soluble fraction of mitoplasts. Coa2 was susceptible to proteinase K degradation upon sonication of mitochondria (Fig. 4E) or solubilization of mitochondria with Triton X-100 (data not shown). Taken together, these results show that Coa2 is a soluble protein in the mitochondrial matrix and that Coa2 may have an interaction with an IM-anchored protein or the IM itself.
FIG. 4.
Coa2 is a soluble matrix protein loosely associated with the IM. (A) Immunoblot of mitochondria (mito) and the postmitochondrial fraction (PMF) purified from the COA2-3HA strain. Pgk1 is a cytosolic marker, and Por1 is a mitochondrial outer membrane protein. (B) Purified mitochondria from the COA2-3HA strain were sonicated in 20 mM HEPES, pH 7.4, and the indicated concentration of NaCl. The soluble and insoluble fractions were separated by centrifugation at 100,000 × g for 1 h and analyzed by immunoblotting. Cyt1 is an integral IM protein, Cyc1 is associated with the IM, and Sod2 is a matrix soluble protein. (C) Purified mitochondria from the COA2-3HA strain were incubated in 20 mM HEPES, pH 7.4, with 1.2 M sorbitol (intact) or without it (hypotonic), for 30 min on ice, digested on ice for 40 min with the indicated concentration of proteinase K (PK) (μg/ml), and analyzed by immunoblotting. (D) Purified mitochondria from the COA2-3HA strain were sonicated or incubated in 20 mM HEPES, pH 7.4, with 1.2 M sorbitol (intact) or without it (hypotonic), for 30 min on ice. The soluble (S) and insoluble (P) fractions were separated by centrifugation at 15,000 × g for 20 min and analyzed by immunoblotting. Ccs1 is a soluble IMS protein. (E) Purified mitochondria as described in panel C were either sonicated (+) three times with 30-s pulses to rupture them or left intact (−). The samples were then treated with 100 μg/ml proteinase K (PK) at 24°C. At each time point (5, 10, and 20 min), 10 μg of total protein was removed for immunoblotting.
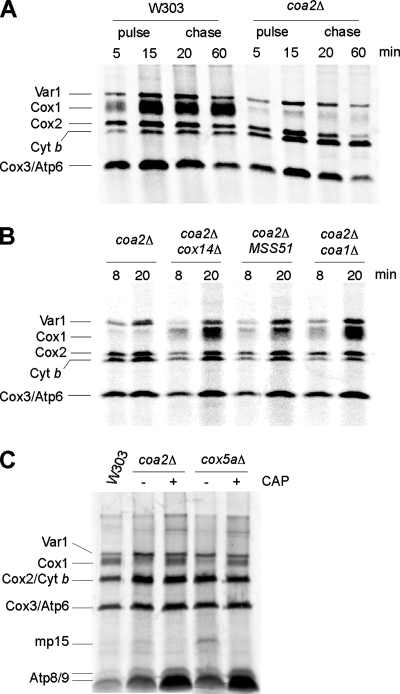
The absence of COA2 leads to a low level of newly synthesized Cox1.
In vivo labeling of mitochondrial translation products revealed that very limited amounts of Cox1 were detected in coa2Δ cells in both W303 and BY4741 backgrounds (Fig. 5A). Cox2, Cox3, and Var1 were translated at WT levels, while Cob (cytochrome b) translation was slightly increased (Fig. 5A; data not shown). During the 60-min chase, the levels of Var1 and cytochrome b remained stable, whereas Cox2 and Cox3 levels diminished markedly in coa2Δ cells, a phenomenon consistent with a CcO assembly defect. A low abundance of Cox1 during the pulse phase of in vivo mitochondrial translation experiments has been reported for most CcO mutants (8). This has been proposed to result from the accumulation of an Mss51-Cox14-Cox1 complex which titrates Mss51 out of its Cox1 translational activator role (8). The synthesis of Cox1 can be restored in certain CcO mutants when Mss51 is overexpressed, when COX14 is deleted (8), or when COA1 is deleted (29). Deletion of COX14 or COA1 in coa2Δ cells restored WT levels of newly synthesized Cox1, whereas overexpression of Mss51 only partially returned the Cox1 level (Fig. 5B). However, none of these strains recovered respiratory growth (see Fig. 9A; data not shown).
FIG. 5.
The absence of COA2 leads to a low level of newly synthesized Cox1. (A) In vivo labeling of mitochondrial translation products. coa2Δ and WT cells were pulsed for 5 and 15 min with [35S]methionine. After 15 min, 20 mM cold methionine (Met) was added, and the reaction was chased for 20 and 60 min at 30°C. The samples were run by 12% SDS-PAGE, and the gel was dried and exposed to autoradiographic film. (B) Cells were labeled for 8 and 20 min with [35S]Met at 30°C. The samples were analyzed as described for panel A. (C) W303 coa2Δ or cox5aΔ cells were treated (+) or not (−) for 3 h with 2 mg/ml chloramphenicol (CAP) and then pulsed for 20 min with [35S]Met. The samples were run by 15% SDS-PAGE, and the gel was dried and exposed to autoradiographic film.
FIG. 9.
Overexpression of Cox10 and Cox5a can partially rescue the respiratory deficiency of coa2Δ cells. (A) coa2Δ cells were transformed with an empty vector, COA2, COX10, or MSS51, grown in SC-2% raffinose, and then serially diluted and spotted on YPLG or YPD. The plates were incubated at 30°C for 2 days (YPD) or 4 days (YPLG). (B) In vivo labeling of mitochondrial translation products in coa2Δ cells overexpressing COX10. Cells were labeled for 8 and 20 min with [35S]Met at 30°C (P). After the 20-min pulse, excess unlabeled Met was added, and the cells were chased for 100 min (C). The samples were run by 12% SDS-PAGE, and the gel was dried and exposed to autoradiographic film. (C) In vivo labeling of mitochondrial translation products in cells lacking CcO subunits. Cells (BY4743 background) were labeled and analyzed as described for panel B. (D) W303 coa2Δ cells transformed with episomal vectors expressing COX5a or COX6 were grown in SC-2% raffinose, serially diluted, and spotted on YPLG or YPD. The plates were incubated at 30°C and 37°C for 2 days (YPD) or 7 days (YPLG). (E) Immunoblot of trichloroacetic acid extracts from W303, coa2Δ, and cox19Δ cells containing a centromeric vector expressing a three-HA C-terminally tagged version of COX5a under the control of its own promoter. Porin (Por1) is shown as a loading control.
Recently, Barrientos and coworkers showed that attenuation of Mss51 levels or titration of Mss51 out of its Cox1 translational activator role leads to the synthesis of mp15, a truncated translation product of COX1 mRNA, in the W303 background (39). mp15 was apparent in cox5aΔ cells, but its abundance was lower in coa2Δ cells (Fig. 5C). Consistent with reported data (39), mp15 synthesis was abrogated when cox5aΔ or coa2Δ cells were treated with low levels of chloramphenicol prior to being labeled, and the synthesis of some Cox1 was restored (Fig. 5C). The low abundance of mp15 in coa2Δ cells suggests that Mss51 is still able to fulfill its translation functions in the absence of COA2.
Cox1 is translated but rapidly degraded in coa2Δ cells.
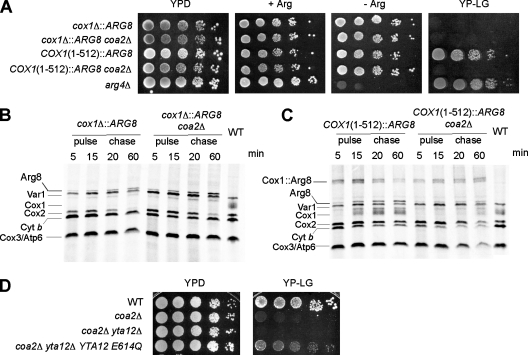
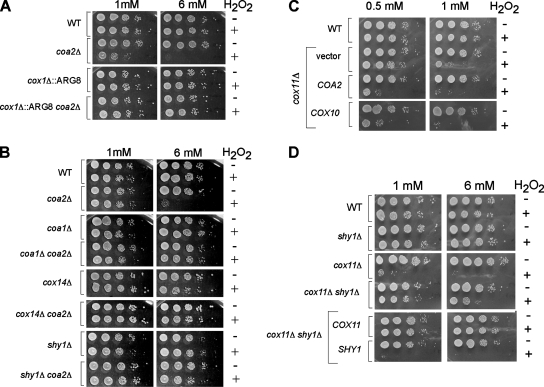
To evaluate the status of translation at the COX1 locus in coa2Δ cells, we used Arg8 reporter strains constructed by Fox and coworkers (28). Arg8 is encoded by a nuclear gene and is imported to the mitochondrial matrix, where it participates in the biosynthesis of arginine. These reporter strains, constructed in an arg8Δ background, contain the ARG8 gene in place of the COX1 ORF (cox1Δ::ARG8) or fused at the 3′ end of the 512-codon intronless form of the COX1 coding sequence [COX1(1-512)::ARG8] (28). The growth of the fusion strains in medium lacking arginine is indicative of translation of COX1(1-512)::ARG8 mRNA. Deletion of COA2 in these two strains did not produce an arginine auxotrophy, implying that translation was occurring at the COX1 locus (Fig. 6A). As expected, the COX1(1-512)::ARG8 strain was unable to respire when COA2 was deleted (Fig. 6A). To ascertain that the arginine prototrophy of the cox1Δ::ARG8 and COX1(1-512)::ARG8 strains lacking COA2 was not sustained only by small amounts of Arg8, we performed in vivo mitochondrial translation assays with these strains. The levels of mature Arg8 detected in the cox1Δ::ARG8 strain were comparable whether COA2 was present or not (Fig. 6B). For the COX1(1-512)::ARG8 strain, a band corresponding to the unprocessed fusion was detected during the pulse phase of the reaction (Fig. 6C). The unprocessed fusion band, although clearly detectable, was reduced in the absence of COA2, as was its processing into mature Cox1 during the chase. The experiments with the Arg8 reporter strains show that efficient translation occurs at the COX1 locus even when the COA2 gene is deleted.
FIG. 6.
Cox1 is translated in the absence of COA2. (A) COA2 was deleted in arg8Δ strains in which ARG8 replaces the COX1 codons (cox1Δ::ARG8) or ARG8 is fused to 512 codons of COX1 [COX1(1-512)::ARG8]. The strains were grown in SC-2% glucose containing 0.2× Arg (4 mg/liter), serially diluted, and spotted on YPD, SC-2% glucose with or without 1× Arg, and YPLG. The plates were incubated at 30°C for 2 days (glucose plates) or 4 days (YPLG). The arg4Δ strain is shown as a control. (B) In vivo labeling of mitochondrial translation products of the cox1Δ::ARG8 strain, with or without the COA2 gene. The cells were treated as described in the legend to Fig. 5A. The additional band is the mature Arg8 protein. WT corresponds to the W303 strain labeled for 20 min. (C) The COX1(1-512)::ARG8 strain, with or without the COA2 gene, was radiolabeled as described in the legend to Fig. 5A. The bands corresponding to the Cox1::Arg8 fusion and to the mature Arg8 protein are marked. WT corresponds to the W303 strain labeled for 20 min. (D) coa2Δ yta12Δ cells transformed with a centromeric vector expressing Yta12(E614Q) were grown in SC-2% raffinose, serially diluted, and spotted on YPLG or YPD. The plates were incubated at 30°C for 2 days (YPD) or 6 days (YPLG).
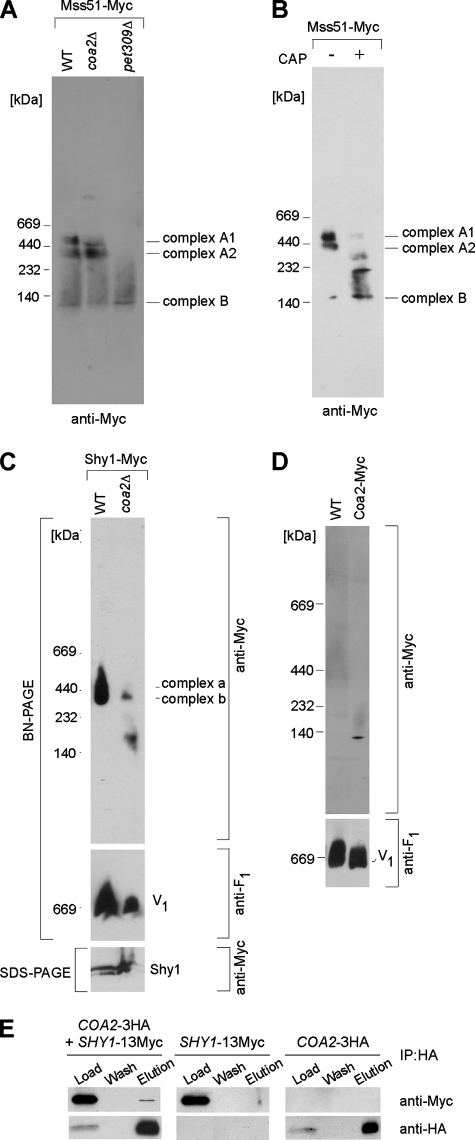
We previously reported that Mss51 is distributed into two complexes upon BN-PAGE on a 4 to 13% gradient gel (29). The largest complex (complex A) has been postulated to contain Cox14, Coa1, and newly synthesized Cox1, based on the observation that this complex is disrupted upon deletion of COX14 and is stabilized in shy1Δ cells (29). Complex A was resolved into two different complexes, A1 and A2, in a 5 to 13% gradient gel (Fig. 7A). In the absence of Coa2, Mss51 reproducibly formed the A2 complex, but the A1 complex was attenuated (Fig. 7A). Neither the A1 nor A2 Mss51 complex was present in pet309Δ cells lacking the Cox1 translational activator (Fig. 7A). Both A1 and A2 Mss51 complexes were also abrogated in cells treated with chloramphenicol at a concentration that completely blocks mitochondrial translation (Fig. 7B). These results suggest either that Cox1 is present in the A1 and A2 complexes or that Cox1 synthesis induces formation of the two Mss51 complexes. The steady-state level of Mss51 was unaffected by chloramphenicol treatment (data not shown). The clear presence of the A2 Mss51 complex in coa2Δ cells is consistent with Cox1 translation occurring in these cells.
FIG. 7.
BN-PAGE analyses of protein complexes. (A) Mitochondria (200 μg protein) isolated from either the WT, coa2Δ, or pet309Δ strain containing a genomically tagged 13-Myc MSS51 gene (Mss51-Myc) were solubilized in buffer containing 1.5% digitonin. Lysates were loaded onto a continuous 5 to 13% gradient gel, and protein complexes were separated by BN-PAGE. The distribution of Mss51 complexes was analyzed by immunoblotting with anti-Myc antibody. (B) WT cells expressing endogenously tagged MSS51 were grown to mid-logarithmic phase and incubated for an additional 3 h in the presence (+) or absence (−) of chloramphenicol (CAP; 8 mg/ml). Isolated mitochondria were lysed, subjected to BN-PAGE, and ana- lyzed as described above. (C) Mitochondria were isolated from the WT or coa2Δ strain containing chromosomally tagged SHY1 (Shy1-Myc) and were analyzed by both native (BN) and denaturing (SDS) PAGE, using anti-Myc antibodies. Anti-F1 antiserum was used to visualize the monomeric (V1) form of complex V, which served as a loading control for BN-PAGE. (D) Mitochondria isolated from either the WT or a strain containing a genomically tagged 13-Myc COA2 gene (Coa2-Myc) were solubilized in buffer containing 1.5% digitonin. Lysates were separated by BN-PAGE (6 to 16% gradient gel). (E) Coa2 interacts with Shy1. Mitochondria from COA2-3HA cells, COA2-3HA cells expressing episomal SHY1-13Myc, or SHY1-13Myc cells were solubilized in IP buffer, and clarified extracts were immunoprecipitated with goat polyclonal anti-HA beads. Load, 1% of the soluble extract; wash, 50% of the last wash concentrated by trichloroacetic acid precipitation; and elution, entire fraction of the bead eluate. All samples were analyzed by immunoblotting as indicated.
The m-AAA protease, composed of Yta10-Yta12, is known to degrade nonassembled mitochondrially encoded proteins within the IM (4). Additionally, the m-AAA protease is essential for mitoribosome assembly through processing of MrpL32 (33). To evaluate whether the m-AAA protease was responsible for the low level of newly synthesized Cox1 in coa2Δ cells, we used a mutant m-AAA containing an E614Q substitution in Yta12 that attenuates its proteolytic activity yet maintains its processing function (3, 33). Respiratory growth was partially restored in coa2Δ cells when YTA12 was deleted and replaced with the Yta12 E614Q allele (Fig. 6D). Therefore, rapid degradation of Cox1 contributes to the CcO deficiency in coa2Δ cells.
Coa2 acts near the Shy1 step in the CcO assembly pathway.
The absence of Coa2 had a dramatic effect on Shy1 complexes visualized by BN-PAGE (Fig. 7C). The Shy1 complexes were attenuated in coa2Δ cells, and a lower-molecular-mass complex was observed. The reduced Shy1 complex level may arise from diminished Cox1 levels, since the Shy1 complexes were reported to contain Cox1 (25). The appearance of an altered mass of Shy1 may be consistent with a physical complex between Shy1 and Coa2. To test whether Coa2 binds Shy1, we carried out immunoprecipitation studies with cells containing chromosomally tagged Coa2-3HA and Shy1-13Myc. Immunoprecipitation of Coa2 reproducibly resulted in coprecipitation of limited amounts of Shy1 (Fig. 7E). Genomically tagged Coa2 migrated in BN-PAGE gels as a single complex whose mass was approximately 100 kDa (Fig. 7D). This is in contrast to the predicted mass of 27 kDa for Coa2-13Myc. The observed Coa2 complex is smaller than any of the Shy1-containing complexes (25), suggesting that Coa2 may not form a complex with Shy1 that can be visualized by BN-PAGE.
Shy1 is postulated to function in the heme a3 insertion step. Rhodobacter cells lacking Shy1 are compromised in heme a3 insertion (31). Deletion of certain CcO assembly factors, such as Cox11 and Sco1, renders yeast cells sensitive to hydrogen peroxide (6), a phenotype generated by a pro-oxidant heme a3-containing Cox1 assembly intermediate (21). Cells lacking Shy1 are peroxide resistant (21). We found that coa2Δ cells are sensitive to hydrogen peroxide in a Cox1-dependent way, as the cox1Δ::ARG8 strain did not exhibit sensitivity in the absence of COA2 (Fig. 8A). Thus, despite low levels of Cox1 observed in the 35S translation assay in coa2Δ cells, sufficient Cox1 was present to cause peroxide sensitivity. This highlights the sensitivity of the hydrogen peroxide assay. As expected, deletion of SHY1 in coa2Δ cells abrogated the peroxide sensitivity (Fig. 8B). The peroxide sensitivity of coa2Δ cells was also suppressed by the deletion of either COA1 or COX14 (Fig. 8B), despite the elevated levels of newly synthesized Cox1 detected in coa2Δ coa1Δ and coa2Δ cox14Δ cells (Fig. 5B). These results suggest that heme a3 insertion is precluded in the absence of COA1 or COX14 and that COA2 acts downstream of these two assembly factors. The interaction between Shy1 and Coa2, the perturbation of the Shy1 complexes in the absence of COA2, and the partial suppression of the respiratory defect of shy1Δ cells by overexpression of COA2 suggest a role for Coa2 in the Shy1-mediated step of CcO maturation.
FIG. 8.
Coa2 acts downstream of Coa1 and Shy1 in the assembly pathway of CcO. (A) Cells were grown to mid-exponential phase and incubated with (+) or without (−) the indicated concentration of H2O2 for 2 h at 30°C. Serial dilutions were spotted onto YPD plates and incubated for 36 to 48 h at 30°C. (B) Cells (W303 background) were treated as described for panel A. (C) cox11Δ cells expressing either an empty vector, COA2, or COX10 were grown to mid-exponential phase, incubated with (+) or without (−) the indicated concentrations of hydrogen peroxide, and spotted onto YPD plates. (D) WT, shy1Δ, and cox11Δ cells as well as shy1Δ cox11Δ cells expressing either COX11 or SHY1 were treated as described for panel A.
Coa2 has a stabilizing role with Cox1.
The peroxide sensitivity of coa2Δ cells suggests that limited quantities of heme a3-Cox1 persist, but Cox1 is highly unstable in these cells. Since the absence of COA2 leads to rapid degradation of newly synthesized Cox1, we hypothesized that Coa2 may participate in stabilizing Cox1. Overexpression of COA2 enhanced the hydrogen peroxide sensitivity of cox11Δ cells (Fig. 8C). This may have arisen from a stabilization of the heme a3-Cox1 pro-oxidant intermediate. As expected, the H2O2 sensitivity of cox11Δ cells is dependent on Shy1, since cox11Δ shy1Δ cells were resistant (Fig. 8D).
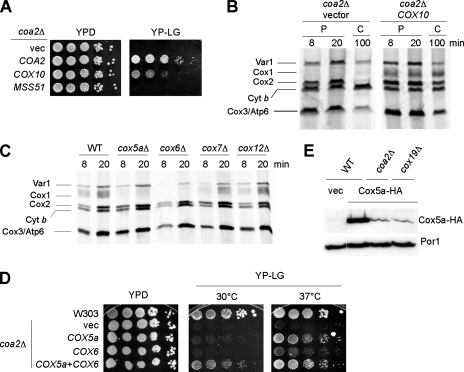
Cox10 is a farnesyl transferase converting heme b to heme o in the heme a biosynthetic pathway. The high-copy-number COX10 gene is a weak suppressor of the respiratory deficiency of coa1Δ cells and enhances the respiration of shy1Δ cells overexpressing Mss51. Cox10 is also partially effective in restoring respiratory growth of coa2Δ cells (Fig. 9A). As expected from the observed respiratory growth, the COX10 transformants showed elevated levels of newly synthesized Cox1 in a mitochondrial translation assay (Fig. 9B). This result further supports the hypothesis that the CcO assembly defect in coa2Δ cells is posttranslational. Like the case for Coa2, Cox10 overexpression exacerbates the peroxide sensitivity of cox11Δ cells (Fig. 8C). Therefore, Cox10 and Coa2 probably participate in stabilizing Cox1 at a stage of assembly downstream of heme a3 insertion.
Cox1 occupies a central position in the mature CcO structure (35). We reasoned that the rapid degradation of Cox1 in coa2Δ cells likely occurs at an early stage in CcO assembly. In humans, CoxIV and CoxVa (homologous to yeast Cox5a and Cox6, respectively) associate with Cox1 as the first subassembly intermediate (27, 37), and these subunits are in close contact with Cox1 in bovine CcO. We addressed whether Cox5a and Cox6 have a significant effect on Cox1 stability at early stages of CcO assembly. We performed in vivo mitochondrial translation experiments with strains carrying deletions of different CcO subunits, early assembled subunits, such as Cox5a and Cox6, and subunits assembled at a later stage, such as Cox7 and Cox12. Only minimal levels of Cox1 were detected in cox5aΔ and cox6Δ cells, whereas cox7Δ and cox12Δ cells showed WT levels of Cox1 during the pulse phase of the reaction (Fig. 9C). This result prompted us to overexpress Cox5a and Cox6 in coa2Δ cells. The two genes, overexpressed individually, showed suppression of the coa2Δ respiratory defect at 37°C (Fig. 9D). However, Cox5a and Cox6 had to be overexpressed together to produce significant growth of coa2Δ cells at 30°C (Fig. 9D). Coa2 is probably not required for Cox5a stability per se, since the same residual level of Cox5a was detected in coa2Δ cells or in other CcO-deficient strains, e.g., cox19Δ cells (Fig. 9E). Coa2 may therefore aid in complex formation between Cox5a and newly synthesized Cox1. However, no interactions were detected between Coa2 and Cox5a by co-IP.
COA2 and MSS51 have nonredundant complementary functions.
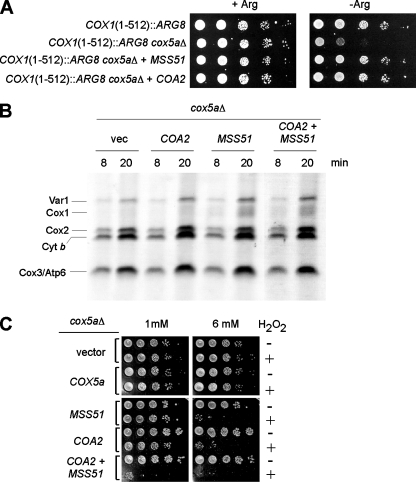
As in coa2Δ cells, Cox1 synthesis is also unusually low in cox5aΔ cells. In order to test whether the low level of Cox1 detected in cox5aΔ cells (Fig. 9C) was a result of a lack of translation or rapid degradation, we disrupted COX5a in the COX1(1-512)::ARG8 strain. This strain showed partial arginine auxotrophy, which was reversed by overexpression of MSS51 or COA2 (Fig. 10A). Consistent with the Cox1 translation block model (8), overexpression of MSS51 in a cox5aΔ strain partially returned the level of newly synthesized Cox1, while COA2 overexpression did not (Fig. 10B). These results suggest that MSS51 probably increases translation of the Cox1-Arg8 fusion to rescue the arginine auxotrophy of the COX1(1-512)::ARG8 cox5aΔ strain, whereas COA2 suppression may arise in a posttranslational step through stabilization of translated Cox1-Arg8. The low intensity of this band by in vivo mitochondrial translation assay of the COX1(1-512)::ARG8 cox5aΔ strain prevented us from directly testing this hypothesis.
FIG. 10.
Coa2 and Mss51 overexpression in cox5aΔ cells results in increased levels of Cox1. (A) The COX1(1-512)::ARG8 strain lacking COX5a, with or without episomal vectors expressing COA2 or MSS51, was grown in SC-2% glucose containing 1× Arg (20 mg/liter), serially diluted, and spotted on SC-2% glucose containing 1× Arg (+Arg) or no Arg (−Arg). The plates were incubated at 30°C for 2 days. (B) W303 cox5aΔ cells containing the same plasmids as those described for panel A were labeled for 8 and 20 min with [35S]Met at 30°C. The samples were run by 12% SDS-PAGE, and the gel was dried and exposed to autoradiographic film. (C) The same cells as those in panel B were grown to mid-exponential phase and incubated with (+) or without (−) the indicated concentration of H2O2 for 2 h at 30°C. Serial dilutions were spotted onto YPD plates and incubated for 36 to 48 h at 30°C.
We subsequently assayed the peroxide sensitivity of the cox5aΔ strain to assess heme a3-Cox1 levels. Cells lacking Cox5a were not sensitive unless MSS51 or COA2 was overexpressed (Fig. 10C). The coexpression of the two proteins further increased the peroxide sensitivity, arguing for nonredundant functions of MSS51 and COA2. High levels of Mss51 and Coa2 appeared to enhance Cox1 levels, via translational and posttranslational effects, respectively.
DISCUSSION
Coa2 is a 68-residue mitochondrial protein shown to be required for respiration and to have a specific role in CcO assembly in stabilizing Cox1 concurrent with the Shy1-dependent step of heme a3 insertion. The most striking aspect of the CcO deficiency of coa2Δ cells is a rapid degradation of newly synthesized Cox1. Only a limited quantity of newly synthesized Cox1 is observed with in vivo mitochondrial translation, and this limitation results from rapid degradation rather than impaired synthesis. Multiple observations support this conclusion. First, deletion of COA2 in a COX1(1-512)::ARG8 fusion strain resulted in normal growth on medium lacking arginine, whereas deletion of COX5a induced Arg auxotrophy, partially due to restricted translation. cox5aΔ cells and coa2Δ cells have equally low levels of Cox1 in the W303 background. Second, the A2 Mss51 complex persists in coa2Δ cells, and this complex is dependent on Cox1 translation. Thus, Cox1 translation is unimpaired in coa2Δ cells. Third, respiration was partially recovered in coa2Δ cells by diminishing the activity of the m-AAA protease, a complex which has been shown to degrade misfolded IM proteins (5). Fourth, the respiratory defect of coa2Δ cells is partially suppressed by Cox10, a protein that functions at a posttranslational step in Cox1 maturation.
COX1 translation is impaired in many CcO assembly mutants due to the titration of Mss51 into a posttranslational complex, preventing its function as a translational activator of COX1 (8). This COX1 translation block is unlikely or at least limited in coa2Δ cells, since Arg8 is translated essentially normally in COX1(1-512)::ARG8 coa2Δ cells. The attenuation of Mss51 as a translational activator is also known to result in synthesis of a truncated Cox1 translation product called mp15 (39). Whereas mp15 synthesis occurs in cox5aΔ cells, only limited quantities of mp15 are detected in coa2Δ cells.
The translation block occurring in most CcO mutants can be obviated by depleting Coa1 or Cox14. The deletion of COX14 leads to a marked enhancement in Cox1 synthesis, but the newly synthesized Cox1 does not progress to assembled CcO (8). Deletion of either COA1 or COX14 in coa2Δ cells leads to a pronounced increase in newly synthesized Cox1 in an in vivo mitochondrial translation assay. The accumulated Cox1 must be in an environment where it is protected from the protease responsible for its rapid degradation in coa2Δ cells. Cox14 and Coa1 are essential to coordinate Mss51's posttranslational function, leading to cofactor insertion in Cox1 (29). Despite the elevation in the level of newly synthesized Cox1, the deletion of COX14 or COA1 in coa2Δ cells reverses the peroxide sensitivity, suggesting that this expanded Cox1 is devoid of heme a3. Therefore, disruption of COX14 or COA1 likely prevents newly synthesized Cox1 from joining the assembly chain and acquiring the heme a3 center.
At what step in CcO assembly is Coa2 important? Two lines of evidence suggest that Coa2 functions downstream of Coa1 and Mss51. First, Coa2 is a suppressor of the respiratory defect of coa1Δ cells. Second, the Coa1-Mss51-Cox14 A2 complex persists in coa2Δ cells. The attenuation of the A1 Mss51 complex in coa2Δ cells may arise if this Mss51 complex contains Cox1. This interpretation is consistent with the absence of this complex in pet309Δ cells and in WT cells treated with chloramphenicol. The presence of the A2 Mss51 complex in coa2Δ cells but not in chloramphenicol-treated WT cells suggests that either the limited amount of Cox1 present in coa2Δ cells is sufficient to stabilize this complex or this complex is indirectly dependent on Cox1 translation. In any case, the presence of the A2 Mss51 complex in coa2Δ cells supports the assertion that Cox1 is translated and that Coa2 functions downstream of Mss51 in a posttranslational step.
Coa2 appears to have a role near the heme a3-related function of Shy1. The Shy1 protein complex observed by BN-PAGE is destabilized and size shifted in coa2Δ cells. Shy1 protein complexes of 250 to 400 kDa contain Cox1 (25), so part of the destabilization of the Shy1 complexes observed in coa2Δ cells may arise from attenuated Cox1 levels. However, the interaction between Coa2 and Shy1 detected by co-IP suggests a direct effect of Coa2 on Shy1. The Coa2-Shy1 complex does not appear to be of sufficient stability for visualization by BN-PAGE with tagged Coa2 cells. This may also account for the low level of co-IP.
The absence of Coa2 does not preclude heme a3 insertion, as coa2Δ cells are peroxide sensitive and this sensitivity is reversed by the deletion of SHY1. The role of Shy1 in heme a3 insertion is corroborated by the observation that the peroxide sensitivity of cox11Δ cells is reversed with depletion of Shy1. Thus, Shy1 appears to participate in heme a3 insertion in yeast as well as in Rhodobacter (31). Heme a3 insertion is not strictly dependent on Shy1, since shy1Δ cells have residual CcO activity and the respiratory defect of the null cells is suppressed by overexpression of either MSS51 or COA2. Although Shy1 appears to be important in the heme a3 insertion step, Shy1 stays associated with CcO until the supercomplex stage (25). It remains unclear whether Shy1 has any function downstream of the proposed heme a3 assembly step.
Proteolytic degradation of Cox1 may be a protective response to the assembly of the pro-oxidant heme a3-Cox1 intermediate. In the absence of heme a3 insertion, Cox1 assembly intermediates, e.g., in cox14Δ cells, are not deleterious and perhaps not a priority for the cell to remove proteolytically. Cox1 degradation appears to be attenuated in coa2Δ cells by the overexpression of COX10 or COX5a/COX6. COX5a and COX6 were recently shown to partially suppress the respiratory defect of shy1Δ cells (15). In this case, it was suggested that Cox5a and Cox6 protect newly synthesized Cox1 from proteolytic degradation. It is possible that at least part of the decrease in the amount of newly synthesized Cox1 observed in shy1Δ cells actually results from rapid degradation of Cox1, not entirely from a translation block. This argument is supported by the observations that a COX1(1-512)::ARG8 shy1Δ strain has no growth defect on medium lacking Arg (28) and that normal levels of Arg8 are present in the COX1(1-512)::ARG8 strain in the absence of SHY1 (28). Therefore, the same rapid Cox1 proteolysis may occur in shy1Δ cells as well as in coa2Δ cells, suggesting that in the absence of these factors, Cox1 forms an unstable conformer. Given the close packing of Cox5a against Cox1, Cox5a may have a stabilizing effect on newly synthesized Cox1.
An early step in Cox1 maturation is formation of the Cox1-Cox5a-Cox6 subassembly complex (32, 37). Cox5a packs against transmembrane helices 11 and 12 in Cox1 and, together with Cox6, docks onto the matrix face of the Cox1 helical barrel, stabilizing loops connecting the 12 membrane helices. This complex accumulates in mammalian cells with mutations in SURF1 (Shy1) (32, 37). This assembly intermediate likely contains heme a, since mutant Cox10 or Cox15 fibroblast lines fail to form this intermediate (1, 2). Heme a is axially coordinated by histidyl residues from Cox1 helices 2 and 10, and its farnesyl tail packs between helices 1, 11, and 12 (34). Thus, heme a insertion may assist in organizing the Cox1 helices creating the Cox5a interface.
It is conceivable that heme a produced by Cox10/Cox15 and inserted into newly synthesized Cox1 creates a Cox1 conformer competent for Cox5a-Cox6 binding. The heme a3-CuB site may be formed in a second step mediated by Shy1 and Cox11. These two steps may occur concurrently rather than sequentially. In support of concurrent steps, a 300-kDa Cox1 assembly intermediate observed by BN-PAGE contains both Shy1 and Cox5a, although distinct 250-kDa complexes exist with either Shy1 or Cox5a (25). Coa2 may also have a chaperone activity in promoting Cox5a/Cox6 association with Cox1. The inability to efficiently form the Cox1-Cox5a-Cox6 intermediate in coa2Δ cells could expose Cox1 to rapid proteolysis. Coa2 likely suppresses the shy1Δ respiratory defect via a posttranslational effect on Cox1. We did not detect any enhancement of the suppression of shy1Δ cells by Coa2 when Cox5a and Cox6 were also overexpressed. Therefore, it is probable that Coa2 suppresses the respiratory defect of shy1Δ cells by a similar mechanism to that for Cox5a and Cox6, which was proposed to be due to increasing the stability of newly synthesized Cox1 (15). Such a role of Coa2 would also explain the H2O2 sensitivity of a cox5aΔ strain overexpressing Coa2 and the enhanced H2O2 sensitivity of cox11Δ cells with high levels of COA2.
Cells lacking Coa2 may stall CcO assembly due to additional factors other than merely Cox1 instability. Details on additional contributions of Coa2 to CcO assembly will await future studies on Coa2.
Acknowledgments
We acknowledge Tom Fox for the generous gift of the ARG8 reporter strains, Alex Tzagoloff for the cox5aΔ strain, and Thomas Langer for the YTA12 mutant plasmid.
This work was supported by grant ES 03817 from the National Institutes of Environmental Health Sciences, NIH, to D.R.W. We acknowledge support from the United Mitochondrial Disease Foundation for support for P.A.C.
Footnotes
Published ahead of print on 9 June 2008.
REFERENCES
- 1.Antonicka, H., S. C. Leary, G. H. Guercin, J. N. Agar, R. Horvath, N. G. Kennaway, C. O. Harding, M. Jaksch, and E. A. Shoubridge. 2003. Mutations in COX10 result in a defect in mitochondrial heme A biosynthesis and account for multiple, early-onset clinical phenotypes associated with isolated COX deficiency. Hum. Mol. Genet. 122693-2702. [DOI] [PubMed] [Google Scholar]
- 2.Antonicka, H., A. Mattman, C. G. Carlson, D. M. Glerum, K. C. Hoffbuhr, S. C. Leary, N. G. Kennaway, and E. A. Shoubridge. 2003. Mutations in COX15 produce a defect in the mitochondrial heme biosynthetic pathway causing early-onset fatal hypertrophic cardiomyopathy. Am. J. Hum. Genet. 72101-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arlt, H., G. Steglich, R. Perryman, B. Guiard, W. Neupert, and T. Langer. 1998. The formation of respiratory chain complexes in mitochondria is under the proteolytic control of the m-AAA protease. EMBO J. 174837-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arlt, H., R. Tauer, H. Feldmann, W. Neupert, and T. Langer. 1996. The YTA10-12 complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell 85875-885. [DOI] [PubMed] [Google Scholar]
- 5.Arnold, I., and T. Langer. 2002. Membrane protein degradation by AAA proteases in mitochondria. Biochim. Biophys. Acta 159289-96. [DOI] [PubMed] [Google Scholar]
- 6.Banting, G. S., and D. M. Glerum. 2006. Mutational analysis of the Saccharomyces cerevisiae cytochrome c oxidase assembly protein Cox11p. Eukaryot. Cell 5568-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barrientos, A., D. Korr, and A. Tzagoloff. 2002. Shy1p is necessary for full expression of mitochondrial COX1 in the yeast model of Leigh's syndrome. EMBO J. 2143-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrientos, A., A. Zambrano, and A. Tzagoloff. 2004. Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 233472-3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonnefoy, N., F. Chalvet, P. Hamel, P. P. Slonimski, and G. Dujardin. 1994. OXA1, a Saccharomyces cerevisiae nuclear gene whose sequence is conserved from prokaryotes to eukaryotes controls cytochrome oxidase biogenesis. J. Mol. Biol. 239201-212. [DOI] [PubMed] [Google Scholar]
- 10.Brachat, S., F. S. Dietrich, S. Voegeli, Z. Zhang, L. Stuart, A. Lerch, K. Gates, T. Gaffney, and P. Philippsen. 2003. Reinvestigation of the Saccharomyces cerevisiae genome annotation by comparison to the genome of a related fungus: Ashbya gossypii. Genome Biol. 4R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bradford, N. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 12.Capaldi, R. A., M. F. Marusich, and J. W. Taanman. 1995. Mammalian cytochrome-c oxidase: characterization of enzyme and immunological detection of subunits in tissue extracts and whole cells. Methods Enzymol. 260117-132. [DOI] [PubMed] [Google Scholar]
- 13.Daum, G., P. C. Bohni, and G. Schatz. 1982. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J. Biol. Chem. 25713028-13033. [PubMed] [Google Scholar]
- 14.Diekert, K., A. I. De Kroon, G. Kispal, and R. Lill. 2001. Isolation and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol. 6537-51. [DOI] [PubMed] [Google Scholar]
- 15.Fontanesi, F., C. Jin, A. Tzagoloff, and A. Barrientos. 2007. Transcriptional activators HAP/NF-Y rescue a cytochrome c oxidase defect in yeast and human cells. Hum. Mol. Genet. 17775-788. [DOI] [PubMed] [Google Scholar]
- 16.Glerum, D. M., and A. Tzagoloff. 1997. Submitochondrial distributions and stabilities of subunits 4, 5, and 6 of yeast cytochrome oxidase in assembly defective mutants. FEBS Lett. 412410-414. [DOI] [PubMed] [Google Scholar]
- 17.Hell, K., W. Neupert, and R. A. Stuart. 2001. Oxa1p acts as a general membrane insertion machinery for proteins encoded by mitochondrial DNA. EMBO J. 201281-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horan, S., I. Bourges, J.-W. Taanman, and B. Meunier. 2005. Analysis of COX2 mutants reveals cytochrome oxidase subassemblies in yeast. Biochem. J. 390703-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kastenmayer, J. P., L. Ni, A. Chu, L. E. Kitchen, W. C. Au, H. Yang, C. D. Carter, D. Wheeler, R. W. Davis, J. D. Boeke, M. A. Snyder, and M. A. Basrai. 2006. Functional genomics of genes with small open reading frames (sORFs) in S. cerevisiae. Genome Res. 16365-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keller, G., A. Bird, and D. R. Winge. 2005. Independent metalloregulation of Ace1 and Mac1 in Saccharomyces cerevisiae. Eukaryot. Cell 41863-1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khalimonchuk, O., A. Bird, and D. R. Winge. 2007. Evidence for a pro-oxidant intermediate in the assembly of cytochrome oxidase. J. Biol. Chem. 28217442-17449. [DOI] [PubMed] [Google Scholar]
- 22.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14953-961. [DOI] [PubMed] [Google Scholar]
- 23.Manthey, G. M., and J. E. McEwen. 1995. The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J. 144031-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manthey, G. M., B. D. Przybyla-Zawislak, and J. E. McEwen. 1998. The Saccharomyces cerevisiae Pet309 protein is embedded in the mitochondrial inner membrane. Eur. J. Biochem. 255156-161. [DOI] [PubMed] [Google Scholar]
- 25.Mick, D. U., K. Wagner, M. van der Laan, A. E. Frazier, I. Perschil, M. Pawlas, H. E. Meyer, B. Warscheid, and P. Rehling. 2007. Shy1 couples Cox1 translational regulation to cytochrome c oxidase assembly. EMBO J. 264347-4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mumberg, D., R. Muller, and M. Funk. 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use of heterologous expression. Nucleic Acids Res. 225767-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nijtmans, L. G., J. W. Taanman, A. O. Muijsers, D. Speijer, and C. Van den Bogert. 1998. Assembly of cytochrome-c oxidase in cultured human cells. Eur. J. Biochem. 254389-394. [DOI] [PubMed] [Google Scholar]
- 28.Perez-Martinez, X., S. A. Broadley, and T. D. Fox. 2003. Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 225951-5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pierrel, F., M. L. Bestwick, P. A. Cobine, O. Khalimonchuk, J. A. Cricco, and D. R. Winge. 2007. Coa1 links the Mss51 post-translational function to Cox1 cofactor insertion in cytochrome c oxidase assembly. EMBO J. 264335-4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siep, M., K. van Oosterum, H. Neufeglise, H. van der Spek, and L. A. Grivell. 2000. Mss51p, a putative translational activator of cytochrome c oxidase subunit-1 (COX1) mRNA, is required for synthesis of Cox1p in Saccharomyces cerevisiae. Curr. Genet. 37213-220. [DOI] [PubMed] [Google Scholar]
- 31.Smith, D., J. Gray, L. Mitchell, W. E. Antholine, and J. P. Hosler. 2005. Assembly of cytochrome c oxidase in the absence of the assembly protein Surf1p leads to loss of the active site heme. J. Biol. Chem. 28017652-17656. [DOI] [PubMed] [Google Scholar]
- 32.Stiburek, L., K. Vesela, H. Hansikova, P. Pecina, M. Tesarova, L. Cerna, J. Houstek, and J. Zeman. 2005. Tissue-specific cytochrome c oxidase assembly defects due to mutations in SCO2 and SURF1. Biochem. J. 392625-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tatsuta, T., S. Augustin, M. Nolden, B. Friedrichs, and T. Langer. 2007. m-AAA protease-driven membrane dislocation allows intramembrane cleavage by rhomboid in mitochondria. EMBO J. 26325-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsukihara, T., H. Aoyama, E. Yamashita, T. Tomizaki, H. Yamaguchi, K. Shinzawa-Itoh, R. Hakashima, R. Yaono, and S. Yoshikawa. 1995. Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8A. Science 2691069-1074. [DOI] [PubMed] [Google Scholar]
- 35.Tsukihara, T., H. Aoyama, E. Yamashita, T. Tomizaki, H. Yamaguichi, K. Shinzawa-Itoh, R. Nakashima, R. Yaono, and S. Yoshikawa. 1996. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science 2721136-1144. [DOI] [PubMed] [Google Scholar]
- 36.Tzagoloff, A., A. Akai, and R. B. Needleman. 1975. Assembly of the mitochondrial membrane system. Characterization of nuclear mutants of Saccharomyces cerevisiae with defects in mitochondrial ATPase and respiratory enzymes. J. Biol. Chem. 2508228-8235. [PubMed] [Google Scholar]
- 37.Williams, S. L., I. Valnot, P. Rustin, and J.-W. Taanman. 2004. Cytochrome c oxidase subassemblies in fibroblast cultures from patients carrying mutations in COX10, SCO1 or SURF1. J. Biol. Chem. 2797462-7469. [DOI] [PubMed] [Google Scholar]
- 38.Wittig, I., H. P. Braun, and H. Schagger. 2006. Blue native PAGE. Nat. Protoc. 1418-428. [DOI] [PubMed] [Google Scholar]
- 39.Zambrano, A., F. Fontanesi, A. Solans, R. Leite de Oliveira, T. D. Fox, A. Tzagoloff, and A. Barrientos. 2007. Aberrant translation of cytochrome c oxidase subunit 1 mRNA species in the absence of Mss51p in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 18523-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu, Z., J. Yao, T. Johns, K. Fu, I. De Bie, C. Macmillan, A. P. Cuthbert, R. F. Newbold, J. Wang, M. Chevrette, G. K. Brown, R. M. Brown, and E. A. Shoubridge. 1998. SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat. Genet. 20337-343. [DOI] [PubMed] [Google Scholar]