Abstract
A common feature of tumor cells is the aberrant expression of ion channels on their plasma membrane. The molecular mechanisms regulating ion channel expression in cancer cells are still poorly known. K+ channels that belong to the human ether-a-go-go-related gene 1 (herg1) family are frequently misexpressed in cancer cells compared to their healthy counterparts. We describe here a posttranslational mechanism for the regulation of hERG1 channel surface expression in cancer cells. This mechanism is based on the activity of hERG1 isoforms containing the USO exon. These isoforms (i) are frequently overexpressed in human cancers, (ii) are retained in the endoplasmic reticulum, and (iii) form heterotetramers with different proteins of the hERG family. (iv) The USO-containing heterotetramers are retained intracellularly and undergo ubiquitin-dependent degradation. This process results in decreased hERG1 current (IhERG1) density. We detailed such a mechanism in heterologous systems and confirmed its functioning in tumor cells that endogenously express hERG1 proteins. The silencing of USO-containing hERG1 isoforms induces a higher IhERG1 density in tumors, an effect that apparently regulates neurite outgrowth in neuroblastoma cells and apoptosis in leukemia cells.
Ion channels control several cellular functions from neuronal signaling and hormone secretion to the regulation of cell volume and salt and water fluxes across epithelia (19). Recent evidence indicates that ion channels are also relevant in the regulation of tumor establishment and progression (15). The contribution of ion channels to the neoplastic phenotype is as diverse as the ion channel families themselves: some types of K+ and Cl− channels are necessary for cell proliferation (6) or are involved in the control of apoptosis (10). K+ and Na+ channels have also been found to modulate the invasive phenotype and metastatic potential of cancer cells (24). Sometimes, these effects can be exerted through the regulation of neoangiogenesis in tumors (31, 34).
The number and biophysical features of ion channels expressed on the plasma membrane are crucial to these physiological functions (19). Several mechanisms control the supply of ion channels to the plasma membrane, including gene transcription and RNA processing, posttranslational modifications (glycosylation and phosphorylation, etc.), and protein degradation (29).
We studied the regulation of the surface expression of K+ channels encoded by human eag (ether-à-go-go)-related gene 1 (herg1) in cancer cells. hERG1 channels are often aberrantly expressed in cancer cells, and their activity is involved in different aspects of tumor establishment and progression (5). The biophysical features of hERG1 current (IhERG1) can be regulated by different mechanisms. First, hERG channels are tetramers, and both homo- and heterotetramers can occur. The coassembly of different isoforms of the hERG1 protein, as well as of channel proteins encoded by different herg genes, occurs. For example, the two full-length hERG1 isoforms (hERG1A and hERG1B) coassemble in tumor cells (14) as well as in the heart (18, 22). The heterotetrameric interaction of proteins encoded by different erg genes has also been reported (42, 43) and gives rise to functional channels. A relevant level of control of IhERG1 is represented by the amount of hERG1 protein expressed on the cell surface. This control is mainly exerted by regulation of hERG1 protein folding and subsequent regulation of forward trafficking. This mechanism also accounts for some forms of long QT syndrome (41). Sequences relevant to hERG1 trafficking to the plasma membrane have been identified in the C terminus of the protein (1, 2, 8, 25). In particular, the 104-amino-acid stretch located immediately downstream of the CNB domain has the function of masking an RGR sequence, which is located downstream and behaves as an endoplasmic reticulum (ER) retention signal. This RGR retention signal becomes exposed when mutations truncate the hERG1 C terminus (26). A physiological example of this mechanism is represented by a herg1 splice variant named herg1USO (25). herg1USO encodes a protein, hERG1USO, which lacks most of the domain, to be substituted by a specific domain (88 amino acids), encoded by the USO exon. hERG1USO does not give rise to any current when transfected in mammalian cells; nevertheless, it can modify the final IhERG1 when coexpressed with the full-length herg1 (25).
We report here evidence for a posttranslational control mechanism for the regulation of hERG1 channel expression on the plasma membranes of tumor cells.
MATERIALS AND METHODS
Cell culture and transfection.
Human embryonic kidney (HEK) 293 and SH-SY5Y human neuroblastoma cells were cultured in Dulbecco's modified Eagle medium containing 4.5 g/liter of glucose and 10% fetal calf serum (complete medium; HyClone). Clones of HEK 293 cells stably expressing the empty vector (HEK-MOCK), herg1a (HEK-hERG1A), herg1b (HEK-hERG1B), herg1USO (HEK-hERG1USO), and herg1bUSO (HEK-hERG1BUSO) genes were selected by limiting dilution and growing them in complete medium supplemented with Geneticin (0.8 mg/ml). Cells were routinely tested for the corresponding channel expression by means of RNase protection assay, Western blot (WB), and patch clamp. Human acute myeloid leukemia cells (FLG 29.1, NB4, and HL60), and human lymphoblastic leukemia cell lines (697, REH, and RS) were cultured in RPMI 1640 medium with 10% fetal calf serum. Cells were incubated at 37°C in a humidified atmosphere with 5% CO2. Cells were transfected or cotransfected with the various herg1 constructs by Lipofectamine 2000 reagent (Invitrogen), following the instructions provided by the manufacturer. In experiments where different amounts of pcDNA3.1-herg1USO and pcDNA3.1-herg1bUSO were transfected with an equal amount of pcDNA3.1-herg1 and pcDNA3.1-herg1b, the total amount of plasmids was kept constant so that the same transfection efficiency was maintained; this was achieved by adding the empty pcDNA3.1 vector to make up to the desired amount.
RNA extraction, RNase protection assay, and Northern blotting.
RNA extraction and RNase protection assay were performed as previously reported (14). Commercially available human heart, brain, and testis RNAs (Ambion) were used for both Northern blotting and real-time quantitative PCR (RQ-PCR). Northern blotting was performed either by electrophoresing 20 μg of total RNA from each sample on a 1% agarose-formaldehyde gel and transferring the RNA onto Hybond-N+ membranes (Amersham Biosciences) or by probing the FirstChoice human blot 1 (Ambion). In both protocols, blots were hybridized using the USO exon fragment encompassing nucleotides 2427 to 2661 of the herg1USO sequence (GenBank accession number NM_172056) according to reference 9.
RQ-PCR.
mRNA quantification of herg1 isoforms by RQ-PCR was performed using the ABI Prism 7700 sequence detection system and the Sybr green master mix kit (both from Applied Biosystems) according to reference 34. The primers were used at a final concentration of 50 nM for herg1a, 100 nM for herg1b, and 300 nM for herg1USO/herg1bUSO and for gapdh. The primer sequences for herg1USO/herg1bUSO were as follows: sense, 5′-CGGAATTCCGGGCACTGAACTGGAAATG-3′; antisense, 5′-AGGCGGCCGCCTACTTTAAGGAAGCAAAAA-3′. The primer sequences for herg1a and herg1b were reported previously in reference 34. The levels of the various transcripts reported throughout the manuscript are normalized to the level of the corresponding transcript detected in HEK 293 cells.
Production of anti-USO polyclonal antibody.
Rabbit polyclonal antibodies against hERG1 (pan-hERG1; Alexis) (27) and hERG1B (Alexis) (12) have been described previously. A specific peptide (CRIRHKQTLFASLK) located within the USO exon-encoded region was synthesized by PRIMM (Milan, Italy), coupled to ovalbumin, and used for immunization of adult male rabbits (Charles River). The antiserum was further immunopurified on a column packed with CNBr-Sepharose beads (Amersham Biosciences) covalently bound to the antigenic peptides. The specificity of the antibody was tested by enzyme-linked immunosorbent assay and with preabsorption experiments (see Results). The specificity of the three anti-hERG1 antibodies toward each isoform is illustrated in Table 1.
TABLE 1.
Antibodies used for the experiments in this study
| hERG1 isoform | Ability of the following antibody to recognize hERG1 isoforma:
|
||
|---|---|---|---|
| Anti-pan-hERG1b | Anti-hERG1Bc | Anti-USOd | |
| hERG1 | + | − | − |
| hERG1B | + | + | − |
| hERG1USO | − | − | + |
| hERG1BUSO | − | + | + |
The ability (+) or inability (−) of the three antibodies to recognize hERG1 isoforms is shown.
Residues 1106 to 1158 of the sequence deposited in GenBank under accession number NP_000229.
Residues 16 to 28 of the sequence deposited in GenBank under accession number CAD54447.
Residues 36 to 548 of of the sequence deposited in GenBank under accession number CAE82156.
Protein extraction, IP, and immunoblotting.
Total cell lysates were prepared as follows: cells were collected by centrifugation, washed three times with ice-cold phosphate-buffered saline (PBS), and lysed on ice with lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% Nonidet P-40, 5 mM EDTA, 100 mM NaF, 0.4 mM Na3VO4, 100 mM Na4P2O7, protease inhibitors [Complete mini; Roche]). The supernatant was then cleared by centrifugation at 16,000 × g for 10 min at 4°C. Proteinase K treatment of cells was performed as reported previously (14). For N-glycosidase F treatment, proteins (50 μg/sample) that had been resuspended in 100 μl of 20 mM sodium phosphate (pH 7.5) containing 0.1% sodium dodecyl sulfate (SDS), 0.75% Nonidet P-40, 50 mM β-mercaptoethanol, and protease inhibitors (Complete mini; Roche) were incubated with 10 mU of N-glycosidase F (Roche) for 17 h or without N-glycosidase F. For endoglycosidase H treatment, proteins (50 μg/sample) that had been resuspended in 100 μl of 50 mM sodium acetate (pH 5.5), 25 mM EDTA, 1% SDS, 0.1% Nonidet P-40, and protease inhibitors were incubated with 10 mU of endoglycosidase H (Roche) for 17 h or without endoglycosidase H. Reactions were stopped by adding reducing Laemmli buffer. For immunoprecipitation (IP) experiments, cell proteins (1 mg) were first precleared with 40 μl of protein A (Sigma) (33% gel slurry) and then incubated with purified anti-USO antibodies overnight at 4°C. Protein A (30 μl) was added for another 2 hours of incubation. Bound proteins were washed three times in lysis buffer and three times in PBS before heating the samples in reducing Laemmli buffer.
WB was performed by using anti-pan-hERG1 (Alexis) (1:1,000), anti-hERG1B (Alexis) (1:500), anti-USO (Alexis) (1:500), antitubulin (Sigma) (1:500), and antiubiquitin (clone P4G7, Stressgene) (1:250), diluted in PBS with 0.1% Tween 20, supplemented with 5% bovine serum albumin (BSA). For preabsorption experiments, anti-USO antibodies (5 μl) were first incubated for 2 h at room temperature with 10 μg of antigenic peptide before probing the WBs.
When required, quantitative measurement of the pixel density of fully and core-glycosylated hERG1A and/or hERG1B bands, as well as ubiquitinated hERG proteins, was performed with Scion image software (Scion Corporation). Values were normalized to the corresponding tubulin band (for hERG1A and hERG1B) or to the immunoprecipitated hERG1A band (for ubiquitin experiments).
Immunofluorescence (IF) laser confocal microscopy.
HEK 293 and SH-SY5Y cells were plated onto polylysine- or laminin-coated coverslips, respectively. Forty-eight hours after transfection, cells were fixed with 3.5% paraformaldehyde in PBS for 15 min at room temperature and processed as reported in reference 12. The following primary antibodies were used: polyclonal antibodies anti-pan-hERG1 (Alexis) (1:100), anti-hERG1B (Alexis) (1:400), and anti-USO (1:400) and monoclonal antibodies anti-KDEL (Stressgene) (1:400) and anti-β1 integrin (clone TS2/16, American Type Culture Collection) (1:50). All the primary antibodies were diluted in PBS containing 10% BSA and incubated overnight at 4°C. The following secondary antibodies were used: tetramethyl rhodamine isothiocyanate-conjugated goat anti-rabbit immunoglobulin G (Southern Biotechnology Associates) and fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse immunoglobulin G (ICN Biomedical), goat anti-mouse antibody-Alexa Fluor 488 (Invitrogen), and goat anti-rabbit antibody-Alexa Fluor 635 (Invitrogen). The former was used for HEK 293 cells, while the latter was used for SH-SY5Y cells. All the antibodies were diluted 1:500 in PBS containing 10% BSA and incubated for 1 h at room temperature. In experiments where two rabbit polyclonal antibodies were used (see Fig. 4C, panels d and e), pilot experiments were performed in order to assess that complete saturation was achieved after incubation with the first antibody, before adding the other primary antibody. Coverslips were mounted in ProLong antifade reagent (Invitrogen) and imaged with a Leica SP2-AOBS confocal microscope. Images were acquired using either a 63× HCX PL APO 1.4-numerical-aperture oil immersion objective or a 40× HCX PL APO 1.25 oil immersion, using a 1,024- by 1,024-pixel image format and adjusting the zoom level to match the voxel size to the Nyquist criterion. The pinhole size was always set at 1 airy unit (airy disk), and each plane was Kalman averaged three times to reduce noise. In each experiment, the same instrumental settings were used for all image acquisitions. All images were Gaussian filtered to eliminate single-pixel noise before analysis.
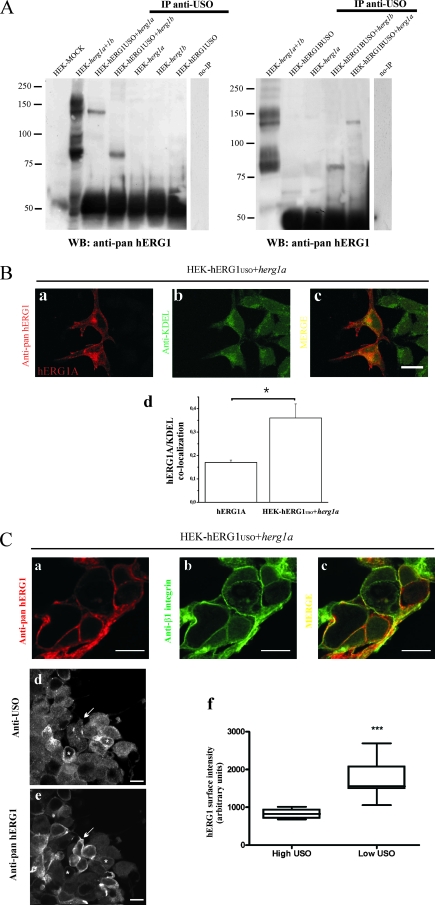
FIG. 4.
hERG1A and hERG1B coassemble with hERG1USO and hERG1BUSO. (A) (Right) Coimmunoprecipitation of hERG1USO with hERG1A and hERG1B. Total lysates (1 mg) from HEK-hERG1USO cells transfected with herg1a (HEK-hERG1USO+herg1a) or herg1b (HEK-hERG1USO+herg1b) and HEK 293 cells transiently transfected with herg1a (HEK-herg1a) and herg1b (HEK-herg1b) were immunoprecipitated with anti-USO antibody and run through a 7.5% SDS-polyacrylamide gel, and the following WB was revealed with anti pan-hERG1 antibody. Total lysates (5 μg each) from mock-transfected HEK 293 cells (HEK-MOCK), cells cotransfected with herg1a and herg1b (HEK-herg1a+1b) and from HEK-hERG1USO cells (HEK-hERG1USO) were run. The no-IP lane contains samples in which no antibody was added to cell lysates during immunoprecipitation. (Left) Coimmunoprecipitation of hERG1BUSO with hERG1A and hERG1B. Total lysates (1 mg) from HEK-hERG1BUSO transfected with herg1b (HEK-hERG1BUSO+herg1b), herg1a (HEK-hERG1BUSO+herg1a), and HEK 293 cells transiently transfected with herg1a (HEK-herg1a) and herg1b (HEK-herg1b), were treated as described above for the right gel in panel A. Total lysates (5 μg each) from HEK-hERG1BUSO cells and from HEK 293 cells cotransfected with herg1a and herg1b (HEK-herg1a+1b), were also run. Lane no-IP, no antibody was added during immunoprecipitation. The positions of molecular mass markers (in kilodaltons) are indicated to the left of the gels. (B) Cellular localization of hERG1 in cotransfected cells. HEK-hERG1USO cells were transiently transfected with herg1a and stained with anti-pan-hERG1 (a) or anti-KDEL (b) antibodies. The merged image of panels a and b is shown in panel c. (d) hERG1A/KDEL colocalization, determined through the calculation of the Manders' coefficient (see Materials and Methods). To do this, images from HEK-hERG1USO cells cotransfected with herg1a (from the same dish as the pictures in panels a to c), as well as the images as those reported in Fig. 2F, panel b, were used. The values calculated by the Manders' coefficient are as follows: 0.17 ± 0.01 for 10 cells transfected with herg1a and 0.36 ± 0.06 for 10 HEK-hERG1USO cells cotransfected with herg1a (P < 0.01, Student's t test). (C) Cellular localization of hERG1 in cotransfected cells. HEK-hERG1USO cells, transiently transfected with herg1a, were stained with anti-pan-hERG1 (a) or anti-β1 integrin (b) antibodies. The merged image of panels a and b is shown in panel c. HEK-hERG1USO cells transiently transfected with herg1a and stained with anti-USO (d) and pan-hERG1 (e) antibodies. To quantify the level of hERG1A expression on the plasma membranes of hERG1USO cells cotransfected with herg1a, a sequential labeling protocol was used, after optimizing the blocking step and diluting the secondary antibodies to avoid any aspecific signal. hERG1A protein expression on the plasma membrane was quantified according to reference 13 (see Materials and Methods). Bar, 10 μm (applies to all panels). (f) Distribution of hERG1 intensity in HEK-hERG1USO cells, displaying high and low USO expression. ImageJ and statistical analyses were performed as described in Materials and Methods. Statistical significance for the Mann-Whitney test (two-tailed) is P < 0.0001. The sum of ranks and median value for high-USO-expressing cells are 45 and 1,557, respectively, and the sum of ranks and median value for low-USO-expressing cells are 108 and 822, respectively (n = 20).
Quantitative colocalization analysis of antigens shown below (see Fig. 4B) was performed on acquired images analyzed as individual channels. The Manders' overlap coefficients (M1 and M2) (30) were calculated using the intensity correlation analysis plugin of the open-source software WCIF-ImageJ (28). Manders' overlap coefficients indicate an overlap of the signals and thus represent the degree of colocalization between the red and green pixels: their values range from 0 (no overlap) to 1 (complete overlap). Briefly, the background signal on each image was initially corrected using the ImageJ background subtraction function, and whenever possible, single cells on the images were selected using the lasso tool (which defines a so-called region of interest). Colocalization was then calculated, after choosing the threshold values for the green and red channels, with the above cited plugin on the regions of interest previously defined. Zero/zero pixel was excluded. We reported only the Manders' coefficient relative to the hERG1A/KDEL overlap.
In order to quantify the plasma membrane expression of the hERG1A protein, we followed the procedure described in reference 13 with minor modifications. Briefly, the quantization of hERG1 fluorescence was conducted on z stacks of 15 sections corresponding to a thickness of approximately 2.5 μm passing through the middle of the cells. Regions of interest were manually drawn around each cell, and the integrated intensity after background correction was measured in hERG1 and USO channels (Alexa Fluor 635 and Alexa Fluor 488, respectively). To account for differences in cell size, the integrated intensity in each region of interest was divided by its measured perimeter. The median value of USO intensity was calculated and used to divide the population of measured cells into two groups: low-USO-expressing cells (all cells below the median value) and high-USO-expressing cells (all cells above the median value). The distribution of the normalized integrated intensity of hERG1 in the two groups of cells was then compared and statistically tested for significance with the nonparametric two-tailed Mann-Whitney test. ImageJ software was used for fluorescence quantization, and GraphPad Prism 4.0 was used for statistical analysis. Quantitative colocalization analysis of antigens was performed on acquired images analyzed as individual channels.
Electrophysiology.
HEK 293 cells were cultured as described above. Approximately 2 × 104 cells were transfected (see above) with 1 to 5 μg of the appropriate plasmid, along with 0.2 μg of a plasmid containing the gene for enhanced green fluorescent protein (pEGFP-C1; Clontech). Currents were recorded 24 to 72 h after transfection.
(i) Solutions and drugs.
During the patch clamp experiments, cells were maintained in standard extracellular solution (130 mM NaCl, 5 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 10 mM HEPES-NaOH, and 5 mM d-glucose) (pH 7.4). During the specific biophysical tests, cells were perfused with a high K+ external solution (the concentration of K+ outside the cell was 20 mM), where NaCl was replaced by an equimolar amount of KCl. The pipette solution contained 130 mM K+-aspartate, 10 mM NaCl, 2 mM MgCl2, 10 mM EGTA-KOH, 10 mM HEPES-KOH, had a pH of 7.30, and a nominal intracellular [Ca2+] of ∼50 nM. When needed, the specific ERG blocker WAY-123,398 (way, 40) was used at 2 μM, and the resulting traces were subtracted from control traces to obtain the way-sensitive currents. The extracellular solutions were delivered through a 9-hole (0.6-mm) remote-controlled linear positioner with an average response time of 1 or 2 seconds that was placed near the cell under study. Experiments aimed at determining whether the specific ERG blocker E4031 could rescue USO isoforms that encoded ERG1 currents were performed according to reference 45, except for the incubation time extended to 48 h.
(ii) Patch clamp recordings and data analysis.
ERG currents were always elicited under conditions of relatively high extracellular K+ concentrations (20 mM) in order to measure currents under optimal signal-to-noise conditions. The currents were recorded at room temperature by using the MultiClamp 700A (Axon Instruments) as previously described (35). Pipette resistances were about 1.5 to 2.2 MΩ. Cell capacitance and series resistance errors were compensated (85 to 90%) before each voltage clamp protocol was run, in order to reduce the voltage errors to less than 5% of the protocol pulse. pClamp 8.2 (Axon Instruments) and Origin 7 (Microcal Inc.) software were routinely used during data acquisition and analysis. The steady-state activation curves were obtained by plotting the normalized peak tail currents at −120 mV versus the preconditioning potential (−80 to +20 mV for 15 seconds), as reported previously (37).
RNA interference.
The small interfering RNA (siRNA) specific for the USO exon of the herg1USO gene (sense, 5′-ACA GAC ACU UUU UGC UUC CTT-3′) was custom synthesized by Ambion. The siRNA negative control 1 (Ambion) was used as a negative control. siRNAs were transfected using the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. siRNA specific for USO-containing transcripts was used at a final concentration of 100 nM, while siRNA negative control 1 was used at a final concentration of 50 nM as recommended. In some cases, siRNA were Cy3 labeled using the Cy3 labeling kit (Ambion) following the manufacturer's instructions. Sequential excitation with a 514-nm laser line was used when imaging cells transfected with Cy3-labeled siRNA. The same approach was used to determine the percentage of siRNA-transfected cells to evaluate the effect of USO silencing.
Flow cytometry. (i) hERG1 labeling.
To quantify hERG1 expression on the plasma membranes of SH-SY5Y cells transfected with siRNA USO or siRNA negative control, cells were labeled with an Alexa Fluor 488-conjugated anti-human hERG1 monoclonal antibody (1:50) produced in our laboratory and available commercially (Alexis). This antibody is specific for a 14-amino-acid peptide located in the extracellular S5-P loop of hERG1 protein, and it is able to recognize its epitope in flow cytometry in living cells (for details, see reference 33). Cells that had been previously washed with PBS were stained for 15 min at room temperature in the dark. After a careful wash to remove any aspecific background, samples were analyzed by flow cytometry on a BD FACSCalibur flow cytometer using the Diva software (BD Pharmingen).
(ii) Annexin V and PI labeling.
Double staining of leukemia cells with annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) (annexin V-FLUOS staining kit; Roche) was used to analyze the apoptotic rate. Cells were resuspended in 100 μl buffer (provided by the kit) and incubated with FITC-conjugated annexin V and PI for 15 min at room temperature in the dark. Cells were washed once and then used for analysis by flow cytometry on a BDLSRII cytofluorimeter using the Diva software (BD Biosciences). We defined three main different cell populations, viable (annexin V negative and PI negative), early apoptotic (only annexin V positive), and late apoptotic or necrotic (annexin V positive and PI positive), according to the manufacturer's instructions.
Nucleotide sequence accession number.
The sequence of herg1buso has been submitted to GenBank under accession no. AJ 609614.
RESULTS
Cloning of an alternative transcript of the herg1 gene, herg1bUSO.
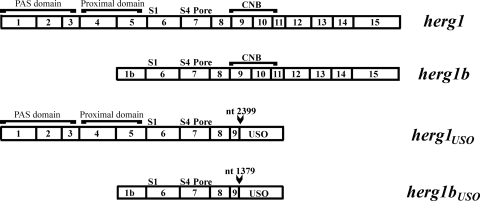
Three variants of the herg1 gene (KCNH2) have been cloned so far (Fig. 1). We cloned a fourth variant by 3′ rapid amplification of cDNA ends and PCR, starting from RNA extracted from neuroblastoma and leukemia cells. This variant bears the 1b exon at the 5′ end and the USO exon at the 3′ end. Therefore, we named this new clone herg1bUSO. The structures of the four herg1 cDNAs and a list of the antibodies we developed and used to discriminate the four encoded proteins are shown in Fig. 1 and Table 1, respectively. When the herg1USO and herg1bUSO genes and the corresponding proteins exhibit common features, they will be collectively called “USO-containing transcripts” and “USO-containing isoforms,” respectively.
FIG. 1.
Schematic representations of herg1, herg1b, herg1USO, and herg1bUSO cDNAs. Some structural features are also indicated. herg1 and herg1USO sequences are identical up to nucleotide (nt) 2399, while herg1b and herg1bUSO sequences are identical up to nt 1379 (both indicated by arrowheads). Boxed numbers indicate the number of exons relative to the herg1 gene.
The hERG1BUSO protein does not give rise to detectable IhERG1 in transfected cells: evidence for intracellular retention.
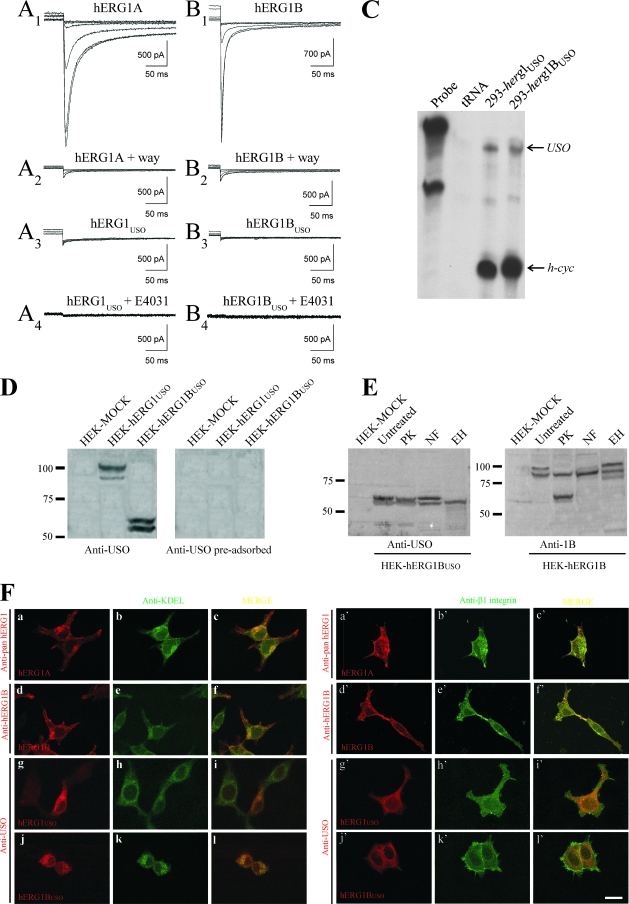
It has been reported that hERG1USO is unable to produce macroscopic IhERG1 when expressed in mammalian cells (25). We verified whether this lack of current also occurred for hERG1BUSO. As shown in Fig. 2, a typical IhERG1 can be recorded from HEK 293 cells transfected with herg1a (panel A1) and herg1b (panel B1). The use of the specific hERG1 blocker WAY-123,398 (way) confirms the presence of these currents (Fig. 2, panels A2 and B2) with the expected biophysical properties. In contrast, no IhERG1 is detectable in HEK 293 cells transfected either with herg1USO (panel A3) or herg1bUSO (panel B3). No currents were detected even after long-term incubation with the hERG1 blocker E4031, a pharmacological tool commonly used to rescue misfolded LQT2-hERG1 mutants (45) (Fig. 2, panels A4 and B4, and the legend to Fig. 2). The proper expression of the transfected herg1USO and herg1bUSO constructs in the cells used for patch clamping was checked by RNase protection assay (Fig. 2C).
FIG. 2.
hERG1A/hERG1B and hERG1USO/hERG1BUSO encoded currents in HEK 293 cells. (A1 and B1) Superimposed traces (elicited at −120 mV from 15-second-long preconditionings at +20, 0, −20, −40, −60, and −80 mV) from two representative cells transfected with herg1a (A1) or herg1b (B1). (A2 and B2) Same cells as for traces A1 and B1, treated for 2 min with the specific ERG blocker WAY-123,398 (way) (2 μM). (A3 and B3) Same as in traces A1 and B1 but from two representative cells transfected with herg1USO or herg1bUSO, respectively. Scale bars are identical in panels A and B. The voltage clamp protocol is described in Materials and Methods. (A4 and B4) Experiments aimed at determining whether the ERG blocker E4031 could rescue IhERG1 as reported previously (41). Two representative cells transfected with herg1USO (n = 9) (A4) or herg1bUSO (B4) (n = 11) exposed for 48 h to 5 μM E4031. Scale bars are identical in panels A and B. The voltage clamp protocol is described in Materials and Methods. (C) RNase protection assay showing the proper expression of herg1USO and herg1bUSO mRNAs in stably transfected clones of HEK 293 cells. The top arrow labeled USO points to the USO exon-protected band, while the bottom arrow (h-cyc) points to the human cyclophilin gene-protected band used as an internal loading control. Saccharomyces cerevisiae tRNA was used as a negative control to test for the presence of probe self-protection bands. (D to F) hERG1USO and hERG1BUSO proteins are retained in the ER. (D) Characterization of the anti-USO polyclonal antibody. Protein lysates (50 μg/lane) from mock-transfected HEK cells (HEK-MOCK), HEK-hERG1USO, and HEK-hERG1BUSO cells were separated by 7.5% SDS-polyacrylamide gel electrophoresis (PAGE) and blotted, and the following WB was revealed with a purified anti-USO antibody. In the right panel, the same blot was revealed with the anti-USO antibody preadsorbed with an excess antigenic peptide (Anti-USO pre-adsorbed). (E) WBs of proteins extracted from hERG1B and hERG1BUSO cells after enzymatic treatments. Total lysates from HEK-hERG1BUSO (left panel) and HEK-hERG1B (right panel) cells were subjected to SDS-PAGE as described above for panel D, and the following WBs were revealed with anti-USO antibodies and anti-hERG1B (anti-1B). Lanes: PK, proteinase K (cleavage of proteins expressed on the plasma membranes of intact cells); NF, N-glycosidase F (removal of all glycans from glycosylated proteins); EH, endoglycosidase H (removal of high-mannose oligosaccharides added during core glycosylation of newly synthesized proteins in the ER, before being sorted to the Golgi apparatus). Note that the band of higher Mr (∼100 kDa) disappeared in HEK-hERG1B cells treated with PK, while the 85-kDa band was unaffected by PK treatment as expected. Moreover, when hERG1B protein was treated with NF, a shift of the 100-kDa band toward the 85-kDa band was observed. Furthermore, EH digestion shifted the 85-kDa band to lower Mr, while leaving the 100-kDa band unaltered. The same experiments performed on HEK-hERG1A gave the expected protein band pattern (46; also data not shown). The positions of molecular mass markers (in kilodaltons) are indicated to the left of the gels in panels D and E. (F) Cellular localization of hERG1 isoforms. HEK 293 cells, transiently transfected with herg1a (panels a to c and a' to c'), herg1b (d to f and d' to f'), herg1USO (g to i and g' to i') and herg1bUSO (j to l and j' to l'), were double labeled with the polyclonal anti-hERG(s) antibodies indicated to the left of panels a, d, g, j and a', d', g', and j' and with monoclonal antibodies anti-KDEL (panels b, e, h, and k) or anti-β1 integrin (panels b', e', h', and k') antibodies. Merged images are in panels c, f, i, and l (for KDEL) and c', f', i', and l' (for β1-integrin). Bar, 10 μm (applies to all panels).
The lack of expression of detectable IhERG1 in cells expressing the USO-containing isoforms could be traced back to a shortcoming of plasma membrane localization of the two isoforms, due to missing C-terminal sequences. This possibility was tested by performing WB and immunolocalization experiments (Fig. 2D to F). For these studies, we raised a new rabbit polyclonal antibody against a peptide located in the USO region (anti-USO antibody). The immunoreactivity and specificity of this novel antibody were first tested by WB performed on total lysates from HEK 293 cells stably transfected with herg1USO (HEK-hERG1USO) and herg1bUSO (HEK-hERG1BUSO) (Fig. 2D, anti-USO antibody). The antibody recognized doublets of 97 and 99 kDa in HEK-hERG1USO cell extracts and 60 and 62 kDa in HEK-hERG1BUSO extracts. These results are in agreement with the predicted molecular mass (Mr) of the two USO-containing proteins (Table 2). The WB signals were absent when the antibody was preadsorbed with an excess of antigenic peptide (Fig. 2D, preadsorbed anti-USO antibody).
TABLE 2.
Characteristics of herg1, herg1b, herg1USO, and herg1bUSO cDNAs and the proteins encoded by these cDNAs
| herg1 cDNA/hERG1 protein | No. of nucleotides in coding sequence | No. of amino acids in protein | Predicted molecular mass (kDa)a | GenBank accession no. (cDNA/protein) |
|---|---|---|---|---|
| herg1/hERG1 | 3,480 | 1,159 | 126.6 | NM_000238/NP_000229 |
| herg1b/hERG1B | 2,460 | 819 | 90 | AJ512214/CAD54447 |
| herg1USO/hERG1USO | 2,667 | 888 | 97.5 | NM_172056/NP_742053 |
| herg1bUSO/hERG1BUSO | 1,647 | 548 | 60 | AJ609614/CAE82156 |
The expected molecular mass was determined using BioEdit version 5.0.6 software.
Specific enzymatic treatments of total intact cells and protein extracts were performed to better define hERG1BUSO membrane localization and glycosylation state. An example is shown in Fig. 2E. The doublet representing hERG1BUSO in HEK-hERG1BUSO cells (Fig. 2E, HEK-hERG1BUSO) was both proteinase K and N-glycosidase F resistant. Only endoglycosidase H digested the band of higher Mr. For comparison, the effects of the same enzymes on the hERG1B protein extracted from HEK 293 cells stably transfected with herg1b (HEK-hERG1B) were evaluated, giving the expected results (14) (Fig. 2E, HEK-hERG1B). We can conclude that the lower hERG1BUSO band likely represents the unglycosylated protein, while the upper band represents the core-glycosylated, ER-resident form of the protein. Hence, hERG1BUSO is trapped and core glycosylated in the ER and does not undergo any further glycosylation process.
We also performed IF experiments to confirm these results (Fig. 2F). HEK 293 cells were transiently transfected with the four herg1 isoforms, and after the cells were permeabilized, they were probed with the corresponding antibodies (Table 1), along with either an anti-KDEL antibody (that recognizes a motif shared by a subset of ER-resident chaperones) (Fig. 2F, panels a to l), or an anti-β1 integrin antibody (that mainly stains cell surface integrins) (Fig. 2F, panels a' to l'). In herg1a- and herg1b-transfected cells, the encoded proteins showed both intracellular and plasma membrane distribution (Fig. 2F, panels a to f and a' to f'). On the other hand, the hERG1USO and hERG1BUSO proteins showed only intracellular staining that mostly merged with the anti-KDEL staining (Fig. 2F, panels g to l) and did not colocalize with the β1 integrin (panels g' to l').
hERG1BUSO decreases IhERG1density when coexpressed with current-encoding hERG1 isoforms.
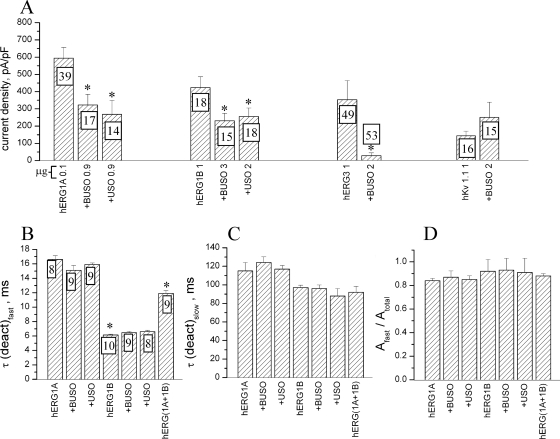
It has been previously reported that expression of hERG1USO in hERG1A-transfected cells induced a decrease in IhERG1 density (25). We tested whether the same occurred for hERG1BUSO and whether there was a selectivity of the USO-containing isoforms for the corresponding current-encoding counterparts. Hence, we measured the average current density of HEK 293 cells cotransfected with herg1a or herg1b as a control, plus either herg1USO or herg1bUSO. The results of these experiments are collectively shown in Fig. 3A. Cotransfection with the USO-containing cDNAs produced a significant inhibition of the currents compared to controls. No selectivity of the two USO-containing isoforms (hERG1USO and hERG1BUSO) for the corresponding current-encoding proteins (hERG1A and hERG1B, respectively) was observed.
FIG. 3.
Biophysical properties of currents obtained in herg1a- and herg1b-transfected HEK 293 cells. The effects of cotransfection with USO-containing cDNAs are shown. The voltage clamp protocols used in all the experiments shown in this figure are described in Materials and Methods. (A) Average maximal current density, recorded at −120 mV from HEK 293 cells, transfected with the different herg1a, herg1b, herg3, and Kv1.1 cDNAs as controls plus herg1USO (+USO) or herg1bUSO (+BUSO). The numbers in the columns indicate the numbers of recorded cells. The numbers on the x axis after the transfected cDNA indicate the amount (in micrograms) of the different cDNAs used to transfect the cells. Values that are significantly different (P < 0.05, two-tailed t test) in the controls (cells transfected with a single cDNA) and cells cotransfected with one USO-containing cDNA are indicated by an asterisk. (B to D) Fast (B) and slow (C) time constants of deactivation (deact) kinetics and the ratio of the fast time constant to the total amplitude (D) measured under control conditions and under USO-containing cDNA cotransfection. The numbers in the columns indicate the numbers of recorded cells. Values that are significantly different (P < 0.05, two-tailed t test) in the controls and cells transfected with the USO-containing cDNAs are indicated by an asterisk.
The USO-induced inhibition of the average current density could also in turn alter the biophysical characteristics of the functional channels, hERG1A and hERG1B. To test this possibility, we analyzed one of the most sensitive biophysical parameters, namely, the deactivation time constants of the current elicited at −120 mV. Various mixtures of the cDNAs used for transfections were tested. All data are reported in Fig. 3B to D. We combined either herg1USO or herg1bUSO cDNAs with herg1a or herg1b. Data obtained from cells cotransfected with herg1a and herg1b are also reported and represent the controls relative to the effects of coassembly of two different hERG1 isoforms (23). In Fig. 3B, C, and D are reported the values of the fast and slow time constants and the ratio of the fast time constant to the total amplitude, respectively. As expected, the fast time constants of IhERG1 in cells cotransfected with herg1a and herg1b [Fig. 3B, hERG(1A+1B)] are quite different from those registered in cells transfected with the single cDNAs. This result indicates that different hERG1 monomers can coassemble to form functional heterotetramers with biophysical properties which are intermediate between those of the corresponding homotetramers (23). No difference was found between controls and HEK 293 cells cotransfected with the USO-containing cDNAs. The same analysis performed on the slow component (Fig. 3C) of the deactivation current gave identical results. Also, the amplitude ratio, shown in Fig. 3D, did not result in significant differences.
Finally, we tested whether USO-containing isoforms exerted the same effects on channels other than the hERG1A and hERG1B channels. We chose one channel, hERG3, that belongs to the same family and another channel, Kv 1.1, that belongs to an unrelated family of voltage-dependent K+ channels. It emerged that cotransfection of herg1bUSO along with herg3 strongly (>90%) reduced the hERG3 current compared to herg3-transfected cells (Fig. 3A, hERG3 and +BUSO 2 bars). On the other hand, cotransfection of herg1bUSO along with Kv 1.1 did not significantly alter the amount of Kv current (Fig. 3A, hKv 1.1 1 and +BUSO 2 bars). These results indicate that the regulatory role of USO-containing isoforms on the amount of the detectable IhERG is family specific, with no effect on K+ channels belonging to unrelated families.
The USO-containing isoforms coassemble with and regulate the trafficking of full-length hERG1 isoforms to the plasma membrane in transfected cells.
The results reported above were interpreted on the assumption that both hERG1USO and hERG1BUSO proteins coassembled and hence formed heterotetramers with hERG1A and/or hERG1B. We then experimentally verified this hypothesis. For this purpose, protein extracts from HEK-hERG1USO or HEK-hERG1BUSO cells and cotransfected with either herg1a or herg1b were immunoprecipitated with anti-USO antibody, and the following WB was revealed with the anti-pan-hERG1 antibody (Fig. 4A). As evident in the left gel of Fig. 4A, the anti-USO antibody was able to immunoprecipitate only one protein band from HEK 293 cells coexpressing herg1a and herg1USO (lane HEK-hERG1USO+herg1a). The Mr of this band corresponds to the Mr of the core-glycosylated, ER-resident, form of the hERG1A protein (135 kDa). Similarly, also in HEK 293 cells coexpressing herg1b and herg1USO (lane HEK-hERG1USO+herg1b) and in HEK 293 cells coexpressing either herg1b or herg1a and herg1bUSO (Fig. 4A, right gel, lanes HEK-hERG1USO+herg1b and HEK-hERG1USO+herg1a), the anti-USO antibody immunoprecipitated only the core-glycosylated forms of hERG1B and hERG1A, respectively.
To better localize the full-length hERG1 proteins in cells expressing the USO isoforms, we performed double IF experiments on HEK-hERG1USO and HEK-hERG1BUSO cells cotransfected with either herg1a or herg1b. Hence, all the cells expressed the stably transfected cDNA, although at different intensities, while only some of them expressed the cotransfected isoform. The latter could be visualized by using the antibody specific for the full-length hERG1 proteins (anti-pan-hERG1). In any case, the latter antibodies were used in conjunction with either anti-KDEL or anti-β1 integrin antibodies. Representative images relative to HEK-hERG1USO cells cotransfected with herg1a are shown in Fig. 4B and C. The hERG1A protein colocalized with KDEL and hence was present in the ER, when coexpressed with an USO transcript (Fig. 4B, panel c). The localization of hERG1A in the ER also occurs in cells transfected with only herg1a (Fig. 2F, panel a). We quantified the colocalization of hERG1A with KDEL in cells expressing only hERG1A and in cells coexpressing hERG1USO and hERG1A through the calculation of the Manders' coefficient (see Materials and Methods and the legend to Fig. 4). It emerged that hERG1A is more localized in the ER when expressed with hERG1USO (Fig. 4B, panel d). These results suggest that hERG1 heterotetramers are retained in the ER, if a USO-containing subunit is present in the tetramer.
We also evaluated whether hERG1A colocalized with a plasma membrane protein, when coexpressed with hERG1USO. To do this, HEK-hERG1USO cells cotransfected with herg1a were labeled with both anti-pan-hERG1 and anti-β1 integrin antibodies. Only a partial merge of the two signals was observed (Fig. 4C, panel c). Finally, we quantified the plasma membrane expression of hERG1A when coexpressed with an USO isoform through the following approach: HEK-hERG1USO cells cotransfected with herg1a were labeled with anti-USO (Fig. 4C, panel d) and anti pan-hERG1 (Fig. 4C, panel e) antibodies. As expected, all the cells expressed the hERG1USO protein, although at different intensities. Two groups of cells were identified, those expressing low levels (see the white arrow in Fig. 4C, panel d) and those expressing high levels of the USO protein (see the white asterisks in Fig. 4D, panel d). In the two groups of cells, the plasma membrane expression of the hERG1A protein was quantified as described previously (13). It emerged that cells displaying a low expression of hERG1USO had the highest level of hERG1A expressed on the plasma membrane (see the white arrow in Fig. 4C, panel c), and vice versa (see the white asterisks in Fig. 4C, panel e). The different plasma membrane expression of the hERG1A protein in the two USO-expressing cell populations was statistically significant (P < 0.0001) (Fig. 4C, panel f).
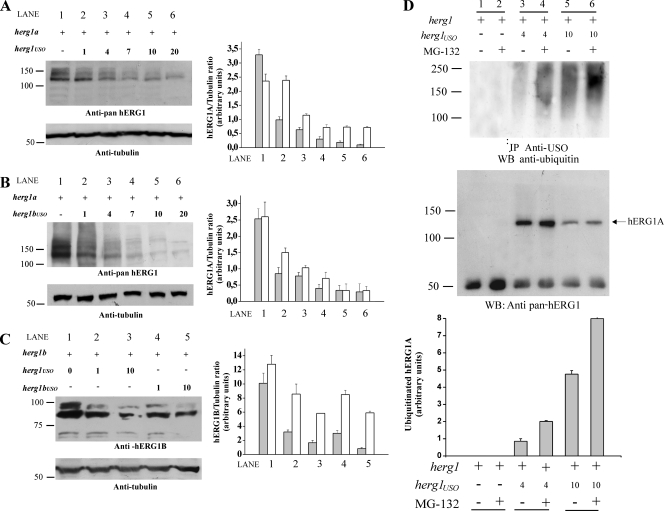
To better define this point, we performed cotransfection experiments in which the amount of herg1a or herg1b was kept constant, while the amount of cotransfected herg1USO or herg1bUSO was progressively increased from an equimolar concentration to a 20-fold excess. Cellular proteins were then extracted, and the following blot was revealed with anti-pan-hERG1 antibody. Results are reported in Fig. 5A to C. When herg1USO (Fig. 5A) or herg1bUSO (Fig. 5B) was coexpressed with herg1a, a significant decrease of mature (155-kDa) hERG1 proteins was observed. Similar results were obtained for the hERG1B protein (Fig. 5C). When amounts of USO-containing cDNAs above fourfold excess were used, a significant decrease in the core-glycosylated forms of both hERG1A and hERG1B proteins was also observed. Quantitative data are reported in the bar graphs in Fig. 5A, B, and C. These results suggest that the USO-containing subunits cause strong retention of heterotetramers in the ER, thus inducing an overall decrease in channel protein expression, with a subsequent impairment of their insertion into the plasma membrane. This effect could rely on increased channel degradation, possibly through ubiquitination and subsequent clearing through the proteasomal pathway. We tested this latter possibility by cotransfecting different amounts of herg1USO along with a constant amount of herg1a, both in the absence and presence of the proteasome inhibitor MG-132. Proteins were immunoprecipitated with the anti-USO antibody, and the following WB was revealed with antiubiquitin antibody which recognizes both mono- and polyubiquitinated proteins. Results are reported in Fig. 5D. It is evident that the level of ubiquitination increases when higher amounts of herg1USO (4 and 10 times) are cotransfected with herg1a (Fig. 5D, top gel, lanes 3 and 5). In addition, the block of the proteasome through the specific inhibitor MG-132 significantly increased the level of ubiquitination (Fig. 5D, top gel, lanes 4 and 6). Reprobing the blot with anti-pan-hERG1 antibodies (Fig. 5D, bottom gel) shows the presence of the core-glycosylated, 135-kDa hERG1A protein in the immunoprecipitate. Hence, comparing the bottom and top gels in Fig. 5D, we can argue that the hERG1A protein undergoes ubiquitination when it coassembles with hERG1USO. Reprobing the blot also shows that the immunoprecipitated hERG1A protein increases in MG-132-treated cells, as expected. The graph at the bottom of Fig. 5D shows the results of densitometric analysis of the level of ubiquitinated hERG1A protein. The data clearly show that hERG1A undergoes a significant ubiquitination when expressed with hERG1USO and that hERG1A channels are likely to be cleared by the proteasome.
FIG. 5.
Modulation of hERG1A and hERG1B expression by hERG1USO and hERG1BUSO. HEK 293 cells were transiently transfected (+) with herg1a or herg1b alone or with herg1USO and herg1bUSO. Total lysates (5 μg) were run through a 7.5% SDS-polyacrylamide gel, and the following WB was revealed with anti-pan-hERG1 (A and B) or anti-hERG1B antibody (C). Equal loading of proteins in each lane was confirmed by reprobing the membranes with antitubulin antibody (bottom gels). Densitometric analysis was performed as described in Materials and Methods. The positions of molecular mass markers (in kilodaltons) are indicated to the left of the gels. (A) Modulation of hERG1A expression by hERG1USO. Lane 1, cells transfected with herg1a alone; lane 2, cells transfected with herg1a/herg1USO at a 1:1 ratio; lane 3, cells transfected with herg1a/herg1USO at a 1:4 ratio; lane 4, cells transfected with herg1a/herg1USO at a 1:7 ratio; lane 5, cells transfected with herg1a/herg1USO at a 1:10 ratio; lane 6, cells transfected with herg1a/herg1USO at a 1:20 ratio. (Right) Densitometric analysis of mature (gray bars) and immature (white bars) hERG1A protein. (B) Modulation of hERG1A expression by hERG1BUSO. Lane 1, cells transfected with herg1a alone; lane 2, cells transfected with herg1a/herg1bUSO at a 1:1 ratio; lane 3, cells transfected with herg1a/herg1bUSO at a 1:4 ratio; lane 4, cells transfected with herg1a/herg1bUSO at a 1:7 ratio; lane 5, cells transfected with herg1a/herg1USO at a 1:10 ratio; lane 6, cells transfected with herg1a/herg1bUSO at a 1:20 ratio. (Right) Densitometric analysis of mature (gray bars) and immature (white bars) hERG1A protein. (C) Modulation of hERG1B expression by hERG1USO/hERG1BUSO. Lane 1, cells transfected with herg1b alone; lane 2, cells transfected with herg1b/herg1USO at a 1:1 ratio; lane 3, cells transfected with herg1b/herg1USO at a 1:10 ratio; lane 4, cells transfected with herg1b/herg1bUSO at a 1:1 ratio; lane 5, cells transfected with herg1b/herg1bUSO at a 1:10 ratio. (Right) Densitometric analysis of mature (gray bars) and immature (white bars) hERG1 protein. (D) hERG1A ubiquitination after coexpression with hERG1USO and effect of proteasome inhibition. HEK 293 cells were transiently transfected with herg1a alone or with herg1USO at a ratio of 1:4 or 1:10. When needed, transfected cells were exposed to the proteasome inhibitor MG-132 for 60 min (+). Total lysates (1 mg) were immunoprecipitated with anti-USO antibody and run through a 7.5% SDS-polyacrylamide gel, and the following WB was revealed either with an antiubiquitin antibody (top gel) or an anti-pan-hERG1 antibody (bottom gel). The results of densitometric analysis of the ratio between ubiquitinated hERG1A proteins and immunoprecipitated (core-glycosylated) hERG1 are shown in the graph at the bottom of panel D.
Expression of herg1bUSO in healthy and neoplastic human tissues.
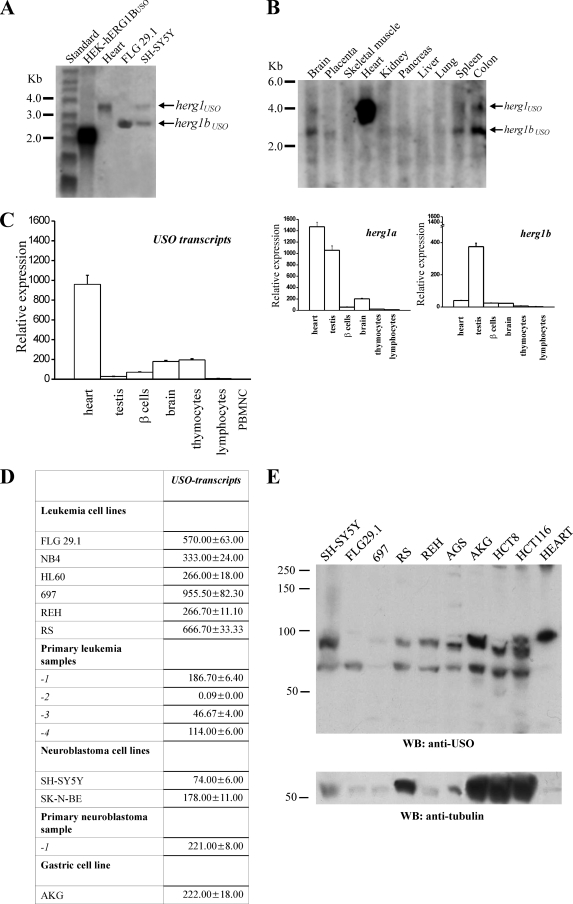
The expression of herg1bUSO transcript in selected human tissues and cell lines was analyzed by Northern blotting using an USO-specific probe. The probe also recognized herg1USO in tissues where it was expressed. The two transcripts can be discriminated on the basis of their length. Figure 6A shows a Northern blot of total RNA extracted from HEK-hERG1BUSO, human heart, FLG 29.1 leukemia, and SH-SY5Y neuroblastoma cells. A larger band (3.8 kb) can be detected in the heart, corresponding to the herg1USO transcript, which is abundantly expressed in this tissue (25). A band of 2.1 kb, corresponding to the herg1bUSO isoform, is present in HEK-hERG1BUSO cells. The bands corresponding to both herg1USO and herg1bUSO can be detected in SH-SY5Y cells, and only the band corresponding to herg1bUSO can be found in FLG 29.1 cells. In these cells, the herg1bUSO transcript showed a slightly greater length (2.6 kb) compared to that detected in transfected cells; this is attributable to the lack of the 5′-untranslated region in the clone used for transfection. The same probe was used to detect transcripts in a commercially available blot containing poly(A) RNAs from different human tissues (Fig. 6B). Apart from the high-level expression of herg1USO in the heart, both herg1USO and herg1bUSO were expressed at very low levels in the tissues examined. We detected faint bands relative to both herg1USO and herg1bUSO in brain, spleen, and colon, while herg1bUSO could be barely detected in placenta, pancreas, and kidney.
FIG. 6.
herg1USO and herg1bUSO mRNA expression in healthy tissues and cancer cell lines. (A) Northern blot of total RNA extracted from HEK-hERG1BUSO, human heart, FLG 29.1, and SH-SY5Y tumor cell lines, hybridized with a USO exon-specific probe (see Materials and Methods). A single band relative to herg1USO is visible in the heart. The herg1bUSO mRNA in HEK-hERG1BUSO cells is shorter than that in FLG 29.1 and SH-SY5Y cells because the former lacks most of the 5′ untranslated region (see primers used for herg1bUSO cloning, 14). (B) Multiple-tissue Northern blot on poly(A) RNA extracted from different human tissues (Ambion) hybridized with the same USO exon-specific probe. The positions of molecular size standards (in kilobases) are indicated to the left of the gels in panels A and B. (C and D) Quantitative expression of USO-containing transcripts in human healthy cells and tissues (C) and tumor cells and tissues (D) by RQ-PCR. The Sybr green method was used (see Materials and Methods). PBMNC, peripheral blood mononuclear cells. (Right) RQ-PCR of herg1a and herg1b expression on the same RNAs as in panel C. The levels of the various transcripts reported on the y axis are normalized to the level of the corresponding glyceraldehyde-3-phosphate dehydrogenase gene. (D) Quantitative expression of USO-containing transcripts in different human tumor cell lines: acute myeloid leukemias (FLG 29.1, NB4, and HL60), acute lymphoblastic leukemias (RS, 697, and REH), neuroblastomas (SH-SY5Y and SK-N-BE), and gastric cancer (AKG). The quantitative expression of USO transcripts in four different primary leukemias (primary leukemia samples 1 to 4) and in a primary neuroblastoma (primary leukemia sample 1) is also reported. (E) WB of proteins extracted from the tumor cell lines listed above plus a gastric cancer cell line (AGS) and two different colon cancer cell lines (HCT8 and HCT116). The membrane was probed with anti-USO antibody (top gel) and with antitubulin antibody (bottom gel). Note that human heart expressed only the hERG1USO isoform, confirming the Northern blot data. The positions of molecular mass markers (in kilodaltons) are indicated to the left of the gels.
The expression of USO-containing transcripts was also quantified by Sybr green RQ-PCR in human tissues. Some of the transcripts were the same as those present in the blot, while others were chosen on the basis of their level of expression of the current-encoding herg1 transcripts (herg1a and herg1b) in humans or rats (38, 44). Results are reported in Fig. 6C (USO transcripts). The quantitative expression of herg1a and herg1b in the same tissues are also reported (herg1a and herg1b graphs). Note that RQ-PCR data do not allow discriminating whether the USO signal can be attributed to herg1USO or herg1bUSO, except in those cases where only herg1a or herg1b transcripts are present. A high level of USO-containing mRNA was confirmed in human heart where the signal is attributable to herg1USO, as indicated by Northern blot data. Moderate levels of USO-containing transcripts were also found in brain and thymocytes. No expression of any of the herg1 transcripts could be detected in peripheral blood mononuclear cells and adherent cell-depleted peripheral blood lymphocytes. The USO-containing transcripts were also detected at very low levels in testis and pancreatic β-cells.
Figure 6D shows the quantitative expression of USO-containing transcripts in tumor cell lines and primary tumors, in which the current-encoding transcripts (herg1a and herg1b) were previously shown to be expressed (14, 27, 34). The USO-containing transcripts are always expressed at high levels in tumor cell lines and primary tumors.
Finally, we tested whether the USO-containing transcripts were properly translated into USO-containing proteins in tumor cells. WB experiments exploiting the anti-USO antibody were therefore performed on several tumor cell lines, including those used for RQ-PCR analysis. Figure 6E shows that, in tumor cell protein extracts, the anti-USO antibody revealed two groups of bands; bands around 97 kDa correspond to hERG1USO, while those around 60 kDa correspond to hERG1BUSO. All tumor cell lines examined expressed both hERG1USO and hERG1BUSO, although at different intensities. FLG 29.1 cells expressed exclusively bands corresponding to hERG1BUSO, while colon carcinoma cell lines expressed bands of slightly different molecular weights (see Discussion). On the whole, USO-containing proteins are expressed in tumor cells, in accordance with Northern blot and RQ-PCR data.
hERG1USO and hERG1BUSO are retained intracellularly and coassemble with current-encoding hERG1 isoforms in tumor cells.
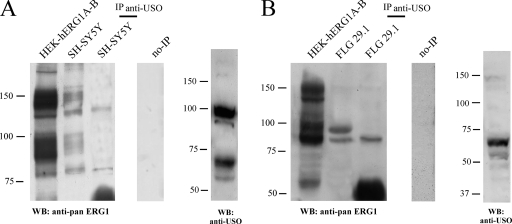
We then examined whether the USO-containing isoforms were retained intracellularly also in cells where they are physiologically expressed. We tested this possibility by evaluating whether the USO-containing isoforms coimmunoprecipitated with the current-encoding hERG1 proteins in tumors, as occurs in transfected cells. SH-SY5Y neuroblastoma cells and FLG 29.1 leukemia cells were used as a model. Total lysates were immunoprecipitated with the anti-USO antibody, and the following WB was revealed with anti-pan-hERG1. In SH-SY5Y cells (Fig. 7A), the anti-USO antibody was able to immunoprecipitate only the immature, ER-resident, hERG1A and hERG1B (compare Fig. 7A with Fig. 4A, HEK-hERG1USO+herg1a and HEK-hERG1USO+herg1b lanes). Correspondingly, in FLG 29.1 cells (Fig. 7B), which express only hERG1B and hERG1BUSO, the anti-USO antibody only immunoprecipitated the immature, ER-resident, hERG1B protein. Total lysates of both SH-SY5Y and FLG 29.1 cells, probed with the anti-USO antibody are shown in the rightmost gels in Fig. 7A and B, respectively.
FIG. 7.
Interaction of hERG1 isoforms in tumor cells. Coprecipitation of USO isoforms with hERG1A and/or hERG1B in SH-SY5Y cells (A) and FLG 29.1 cells (B). Proteins (2 mg) were immunoprecipitated with anti-USO antibody (IP anti-USO) and run through a 7.5% SDS-polyacrylamide gel, and the following WB was revealed with anti-pan-hERG1 antibody. Total lysates from each cell line and from HEK-hERG1A and -hERG1B-expressing cells (HEK-hERG1A-B) were also loaded. The results of WB analysis of protein extracts (50 μg/lane) from SH-SY5Y (A) and FLG 29.1 (B) cells probed with anti-USO antibody are shown in the rightmost gels (WB: anti-USO). The positions of molecular mass standards (in kilodaltons) are indicated to the left of the gels. The no-IP gels contain samples to which no antibody was added to cell lysates during immunoprecipitation.
Collectively, these data suggest that the functional role of USO-containing isoforms that was demonstrated in transfected cells likely occurs in physiological systems.
Physiological roles of hERG1USO and hERG1BUSO in tumor cells: regulation of hERG1 trafficking and IhERG1 density.
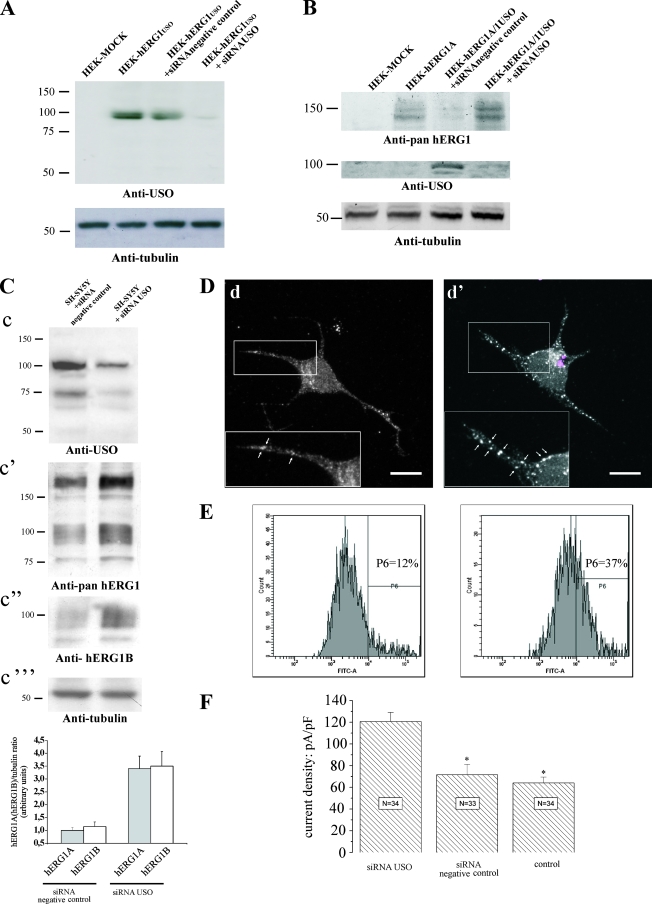
We further confirmed this conclusion by knocking down hERG1USO and hERG1BUSO in tumor cells through RNA interference (Fig. 8). An siRNA against the USO sequence (siRNA USO) was designed and synthesized. The efficiency of this siRNA in silencing hERG1USO and hERG1BUSO was first tested on HEK-hERG1USO and HEK-hERG1BUSO cells. Figure 8A shows that the siRNA USO efficiently impairs the translation of hERG1USO in transfected cells (a similar effect was exerted on hERG1BUSO [not shown]). To simulate the physiological situation, HEK 293 cells were then cotransfected with herg1 and herg1USO; a significant downregulation of the hERG1A protein was observed (Fig. 8B, lane HEK-hERG1A/1USO+siRNA negative control), as already reported (Fig. 5A). The siRNA-mediated hERG1USO silencing restored hERG1A expression (Fig. 8B, lane HEK-hERG1A/1USO+siRNAUSO) to levels comparable to those detected in HEK 293 cells transfected with only herg1a (lane HEK-hERG1A). The same effects were obtained in HEK 293 cells cotransfected with herg1b and herg1bUSO (not shown).
FIG. 8.
Effects of silencing USO-containing isoforms in tumor cells. (A) Effects of siRNA USO on the expression of hERG1USO. HEK 293 cells were transfected with herg1USO and cotransfected with an anti-USO siRNA (siRNA USO) and a scrambled siRNA (siRNA negative control) for 48 h. WB was performed as described in the legend to Fig. 2D. The positions of molecular mass markers (in kilodaltons) are indicated to the left of the gels. (B) Effects of siRNA USO on hERG1A expression. HEK 293 cells were transfected with herg1a (HEK-hERG1A), herg1a/herg1USO at a 1:10 ratio plus siRNA negative control (HEK-hERG1A/1USO+siRNA negative control), and herg1a/herg1USO at a 1:10 ratio plus siRNA USO (HEK-hERG1A/1USO+siRNAUSO). Total lysates (5 μg) were run through a 7.5% SDS-polyacrylamide gel, and the following WB was revealed with anti-pan-hERG1, anti-USO, and antitubulin antibodies. Mock-transfected HEK 293 cells (HEK-MOCK) are shown in the leftmost lanes of panels A and B. (C) Effects of siRNA USO on hERG1A and hERG1B expression in SH-SY5Y cells. SH-SY5Y cells were treated with siRNA USO and siRNA negative control. Total lysates (40 μg) were run through a 7.5% SDS-polyacrylamide gel, and the following WB was revealed with anti-USO, anti-pan-hERG1, anti-hERG1B, and antitubulin antibodies. Densitometric analysis was performed on hERG1A and hERG1B bands (n = 3), and the resulting data are shown in the graph at the bottom of panel C. (D) Localization of hERG1 proteins in SH-SY5Y cells not transfected (d) or transfected with siRNA USO (d'). Cells transfected with Cy3-labeled siRNA USO were labeled with anti-pan-hERG1 antibodies. Images were acquired from both transfected cells (see the siRNA USO visualized as colored dots in panel d') and neighboring nontransfected cells. Highlighted are plasma membrane details where white arrows mark the different hERG1 expression and distribution in nonsilenced (d) and silenced (d') cells. (E) Cytofluorimetric analysis of hERG1 surface expression in SH-SY5Y cells. Cells were labeled with a monoclonal antibody specific for an extracellular epitope of the hERG1 proteins. The antibody was used without previous permeabilization, hence the fluorescent signal marks only the proteins expressed on the cell surface. The cytofluorimetric analysis was performed using Diva software (BD Pharmingen). The percentage of cells expressing high levels of hERG1 (P6 area) is reported inside the panels. (F) IhERG1 density in SH-SY5Y cells transfected with siRNA USO, transfected with siRNA negative control, and not transfected (control). The number of cells analyzed is reported inside each column.
Finally, we tested the effect of the siRNA USO on tumor cells. In this case, transfected cells turned out to be about 40% of the cells (see Materials and Methods and the legend to Fig. 8). Figure 8C shows that the siRNA USO was efficient in decreasing the expression of both hERG1USO and hERG1BUSO in SH-SY5Y cells (panel c); this effect was accompanied by a significant increase in both hERG1A (panel c') and hERG1B (panel c' and c”) proteins. The results are more evident in the densitometric analysis in the graph at the bottom of Fig. 8C. To analyze whether the siRNA USO produced an increase of hERG1A expression on the cell surface, we performed IF experiments (Fig. 8D). SH-SY5Y cells were transfected with the siRNA USO and labeled with anti-pan-hERG1 antibodies. Images were taken from the same culture dish. It is evident that cells properly transfected with the siRNA USO (Fig. 8D, panel d') show numerous discrete spots on the plasma membrane neurites, an aspect not detectable in neighboring, nontransfected cells (panel d) (see also the insets in panels d' and d at higher magnifications) or in cells transfected with the siRNA negative control (not shown). This feature is suggestive of an increased expression of the hERG1 proteins on the plasma membrane, a hypothesis confirmed by flow cytometry (Fig. 8E). To examine this hypothesis, SH-SY5Y cells were labeled with a monoclonal antibody raised in our laboratory that recognizes the S5-S6 extracellular loop of the hERG1 protein. Hence, when used without cell permeabilization, this antibody marks only the hERG1 proteins expressed on the plasma membrane. Figure 8E shows that a greater percentage (37%) of neuroblastoma cells silenced with the siRNA USO (right graph) express high levels of plasma membrane hERG1 proteins compared to the nonsilenced cells (left graph) (12%).
Strikingly, a significant increase in IhERG1 density recorded from SH-SY5Y cells transfected with the siRNA USO compared to either siRNA-transfected cells (negative control) or nontransfected cells accompanied these findings (Fig. 8F).
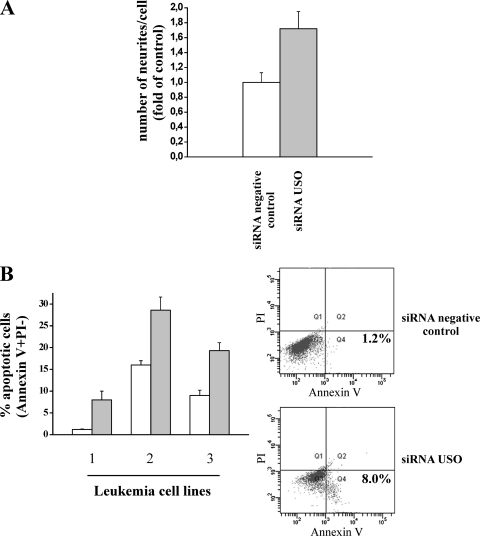
Finally, we asked whether the USO-mediated control of hERG1 channel expression could have relevance in any biological aspect of tumor cells. On the basis of previous observations for neuroblastoma cells (3, 4), we tested whether the USO isoforms could play a role in modulating neurite outgrowth in these cells. In Fig. 9A, it is evident that the number of neurites per cell significantly increases in SH-SY5Y cells transfected with the siRNA USO compared to cells transfected with the siRNA negative control (P = 0.02).
FIG. 9.
Effects of silencing USO-containing isoforms on neurite outgrowth and apoptosis in tumor cells. (A) Number of neurites per cell in SH-SY5Y cells treated with siRNA USO. The number of neurites was evaluated in three separate experiments performed on SH-SY5Y cells transfected with siRNA USO or siRNA negative control. Cells were seeded on polylysine-coated slides (20 μg/ml) and fixed 24 h after transfection with 2.5% glutaraldehyde. Values are reported as changes in the numbers of neurites per cell for the control, and each determination represents the average of three individual fields (P < 0.05). (B) Percentage of apoptotic cells in leukemia cell lines (1, RS; 2, REH; 3, 697) treated with the siRNA negative control (white bars) or with siRNA USO (gray bars). The percentage of apoptotic cells was measured by cytofluorimetric analysis using annexin V and propidium iodide (PI) staining. Data are means plus standard errors of the mean (error bars) of two different experiments. (Right) Representative examples of the flow cytometric density plot of the annexin V test relative to leukemia cell line 1 treated for 24 h with the siRNA negative control or the siRNA USO. Early apoptotic cells are in quadrant 4 (Q4) in the lower part of each plot on the right side (annexin V positive and PI negative). The percentage of apoptotic cells is indicated in Q4. Annexin V binding to the cell surface and surface membrane permeability to PI are shown on the x and y axes, respectively.
Another relevant function of hERG1 channels is their regulatory role in the growth of leukemia cells (32, 39). Hence, we tested the effects of siRNA USO on both cell cycle progression and induction of apoptosis on leukemia cell lines. While cell cycle progression was not affected by siRNA USO treatment (not shown), such treatment induced a significant increase in the percentage of apoptotic cells in all the leukemia cell lines analyzed (Fig. 9B). Two representative examples of the flow cytometry data relative to the annexin V test in the RS cell line are shown (Fig. 9B, graphs to the right) (see the legend to Fig. 9B for details).
DISCUSSION
In this study we provide evidence of a novel mechanism that regulates the surface expression of K+ channels of the hERG1 family in tumor cells. This mechanism is based on the expression of isoforms that are retained in the ER and form heterotetramers with isoforms that are properly targeted to the cell surface and thus modulate their expression level on the plasma membrane. Specifically, two hERG1 proteins that contain the USO sequence at the C terminus (hERG1USO and hERG1BUSO) modulate forward trafficking of the two full-length hERG1 isoforms (hERG1A and hERG1B). This modulation leads to a decreased IhERG1 density. The USO-based mechanism of hERG1 channel regulation was demonstrated in both reconstituted and physiological systems, mainly consisting of tumor cells. In tumors, the decrease in IhERG1 density operated by USO-containing isoforms is apparently relevant in maintaining some aspects of the neoplastic phenotype, such as blocking cell differentiation and escape from apoptotic death.
Two USO-containing transcripts have been cloned so far, herg1USO (25) and a novel transcript we cloned from tumor cells. The novel transcript presents the USO exon at the C terminus and the 1b exon at the N terminus and was hence named herg1bUSO. From various healthy human tissues, we detected a high expression of herg1USO only in the heart. Both herg1USO and herg1bUSO transcripts were expressed at high levels in human tumors. Interestingly, a significant correlation was found between the levels of expression of USO-containing transcripts and those of current-encoding isoforms in leukemias, where the latter data are published (see Table 3 in reference 34). Moreover, USO-containing transcripts were properly translated in tumor cells. A leukemia cell line, FLG 29.1, expressed only hERG1BUSO, while colon carcinoma cells expressed USO proteins of lower molecular weight. This result is suggestive of the presence of specific USO variants at least in some tumor cells.
The functioning and physiological role of USO-containing isoforms in both heterologous systems and tumor cells was defined. First, we showed that the two USO-containing isoforms do not give rise to functional channels when heterologously expressed in mammalian cells, because they are retained intracellularly at the ER level (Fig. 2 and 3). The ER retention of the two USO-containing isoforms also occurs in tumor cells (Fig. 7). The ER entrapment of USO-containing isoforms could be predicted by the absence in these proteins of the part of the hERG1 C terminus implicated in channel maturation (8, 21, 26), in particular the RGR sequence (26). We obtained experimental demonstration of these predictions through the development and use of a novel anti-USO antibody. In addition, we showed that the ER retention of the USO-containing isoforms is not due to protein misfolding, as occurs for some long QT syndrome mutants (36). In fact, neither the addition of the hERG1 blocker E4031 nor lowering the temperature (not shown), two different procedures commonly used to rescue the proper plasma membrane expression of various long LQT syndrome 2 mutants (45), ensues in significant IhERG (Fig. 2, panels A4 and B4).
A novel finding emerging from our data is the demonstration that the USO-containing isoforms can form heterotetramers with the full-length hERG1 proteins. In particular, the USO-containing isoforms coassembled with the core-glycosylated, ER-resident hERG1 full-length proteins. This finding was supported by biochemical and IF data gathered on transfected cells (Fig. 4) and in tumor cells (Fig. 7). This implies that heterotetramers comprising USO-containing isoforms are entrapped in the ER. We also showed that this entrapment produces a decrease in the expression of mature hERG1A and hERG1B proteins and hence in the number of channels that reach the plasma membrane. This occurred in both heterologous systems (Fig. 5) and tumor cells. We indirectly showed this latter point using RNA interference (Fig. 8). In heterologous systems, when the USO-containing isoforms were expressed at higher levels than the full-length hERG1 proteins were, an overall decrease in channel protein was also observed (Fig. 6). Hence, USO-containing isoforms could both entrap current-encoding hERG1 proteins in the ER and also interfere with the trafficking of the proteins toward the plasma membrane, when the levels of expression of the USO isoforms strongly exceed those of the full-length hERG1 proteins. The USO-dependent decrease in full-length hERG1 protein expression is apparently due to an increase of protein ubiquitination and hence to the induction of protein degradation in the proteasome. In other words, hERG1USO or hERG1BUSO might exert a “chaperone-like” function, similar to, although with slight differences with, that reported for Hsp90 (16), arsenic trioxide (17), or ceramide (11).
Finally, we demonstrated that the USO-containing hERG1 isoforms are physiologically relevant in regulating the amount of functional IhERG1 expressed on the plasma membrane. In fact, cotransfection of herg1USO and herg1bUSO with full-length, current-encoding isoforms lead to a significant decrease in IhERG1 density (Fig. 3). This decrease occurred without any modification of IhERG1 biophysical properties. This fact confirms that USO-containing isoforms simply decrease the fraction of hERG1 channels reaching the plasma membrane. The IhERG1-decreasing effect was exerted by hERG1USO or hERG1BUSO on hERG channels encoded by different genes of this family (such as herg3). On the other hand, this mechanism is completely absent for K+ channels belonging to different families, such as the Kv 1.1 channels presented in this paper.
The relevant aspect of our data is the demonstration that in addition to heterologous systems, the USO-dependent modulation of hERG1 proteins and of IhERG1 also occurs in tumor cells. We used RNA interference to demonstrate this point, mainly using neuroblastoma and leukemia cells as a model. siRNA USO was able to increase the expression of current-encoding hERG1 isoforms and their localization onto the plasma membranes of tumor cells (Fig. 8C to E). This effect results in a doubling of IhERG1 in tumor cells (Fig. 8F). In addition, siRNA USO treatment induces an increase of neurite outgrowth in neuroblastoma cells and of cellular apoptosis in leukemia cells.
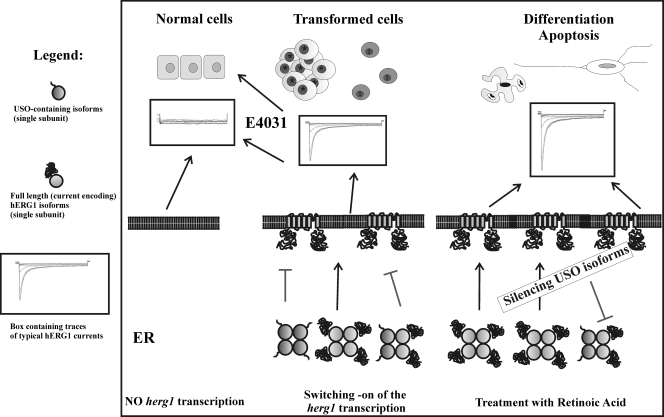
We can speculate that the significance of the USO-mediated regulation of hERG1 channel surface expression in the heart could be that of regulating IKr in pathological conditions. For example, during heart failure, a decrease in K+ currents, including IKr, has been reported. This decrease is not accompanied by a concomitant decrease in RNA transcription of K+ current-encoding genes. This apparent paradox could be interpreted with an upregulation of hERG1USO in these conditions. However, the main question we have tentatively addressed in the present paper is the significance of the USO-mediated mechanism in tumor cells. On the basis of data presented herein and according to reference 5, we hypothesize that oncogenic transformation is accompanied by a herg1 gene(s) switching on. Both full-length and USO-containing isoforms are expressed and assemble in the ER. Only tetramers composed by full-length, current-encoding hERG1 isoforms can reach the plasma membrane. A definite number of functional hERG1 channels on the plasma membrane can in turn control various behavioral features of tumor cells, such as cell proliferation and invasion (6). This can occur either through the clamping of the voltage resting membrane potential to values compatible with cell proliferation or through the well-established modulatory role of hERG1 channels in cell signaling (7, 34). When IhERG1 activity is blocked through specific hERG1 inhibitors, cell proliferation and invasion are concurrently impaired (34), leading to the restoration of a healthy, hERG1-devoid, biophysical profile. When a hERG1 channel density on the cell surfaces of tumor cells that is too high is produced (by either silencing USO-containing isoforms or increasing hERG1 expression through retinoic acid treatment [4]), impairment of cell proliferation through the induction of either cell differentiation or apoptosis ensues. On the whole, proper control of hERG1 protein surface expression is necessary in tumor cells, and the USO-containing isoforms could help in providing the proper number of functional hERG1 channels on the plasma membranes of tumor cells. This hypothesis is outlined in Fig. 10. In this light, the block of the USO-based mechanism of hERG1 channel trapping, through RNA interference, could have therapeutic significance in cancer cells.
FIG. 10.
Proposed model of regulation of hERG1 channel expression on the plasma membrane by USO isoforms. Correlation with different aspects of tumor cell behavior is shown.
Acknowledgments
This work was supported in part by funds from the Italian Association for Cancer Research (AIRC) to A.A., the Association for International Cancer Research (AICR) to A.A., the Italian Ministero dell'Università e della Ricerca Scientifica e Tecnologica (MIUR-PRIN-2003054500_006 and MIUR-PRIN-2006053378_002 to A.A.; MIUR-PRIN-2001055320, 2003052919, MIUR-FIRB2001-RBNE01XMP4-002, and MIUR-FISR2001 0300179 to E.W.), the Università di Milano-Bicocca to E.W., and the Ente Cassa di Risparmio di Firenze to A.A. L.G. is a recipient of a fellowship from the Italian Fulbright Commission. E.R. is a postdoc student of physiology at the Department of Biotechnologies and Biosciences of the University of Milan-Bicocca.
Footnotes
Published ahead of print on 16 June 2008.
REFERENCES
- 1.Akhavan, A., R. Atanasiu, and A. Shrier. 2003. Identification of a COOH-terminal segment involved in maturation and stability of human ether-a-go-go-related gene potassium channels. J. Biol. Chem. 27840105-40112. [DOI] [PubMed] [Google Scholar]
- 2.Akhavan, A., R. Atanasiu, T. Noguchi, W. Han, N. Holder, and A. Shrier. 2005. Identification of the cyclic-nucleotide-binding domain as a conserved determinant of ion-channel cell-surface localization. J. Cell Sci. 1182803-2812. [DOI] [PubMed] [Google Scholar]
- 3.Arcangeli, A., L. Faravelli, L. Bianchi, B. Rosati, A. Gritti, A. Vescovi, E. Wanke, and M. Olivotto. 1996. Soluble or bound laminin elicit in human NB cells short- or long-term potentiation of a K+ inwardly rectifying current: relevance to neuritogenesis. Cell Adhes. Commun. 4369-385. [DOI] [PubMed] [Google Scholar]
- 4.Arcangeli, A., B. Rosati, O. Crociani, A. Cherubini, L. Fontana, B. Passani, E. Wanke, and M. Olivotto. 1999. Modulation of HERG current and herg gene expression during retinoic acid treatment of human neuroblastoma cells: potentiating effects of BDNF. J. Neurobiol. 40214-225. [PubMed] [Google Scholar]
- 5.Arcangeli, A. 2005. Expression and role of hERG channels in cancer cells. Novartis Found. Symp. 266225-232. [PubMed] [Google Scholar]
- 6.Arcangeli, A., and A. Becchetti. 2006. Ion channels and cell cycle, p. 81-94. In D. Janigro (ed.), Cell cycle and the central nervous system. Humana Press Inc., Totowa, NJ.
- 7.Arcangeli, A., and A. Becchetti. 2006. Complex functional interaction between integrin receptors and ion channels. Trends Cell Biol. 16631-639. [DOI] [PubMed] [Google Scholar]
- 8.Aydar, E., and C. Palmer. 2001. Functional characterization of the C-terminus of the human ether-a-go-go-related gene K+ channel (HERG). J. Physiol. 5341-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bianchi, L., B. Wible, A Arcangeli, M. Taglialatela, F. Morra, P. Castaldo, O. Crociani, B. Rosati, L. Faravelli, M. Olivotto, and E. Wanke. 1998. herg encodes a K+ current highly conserved in tumors of different histogenesis: a selective advantage for cancer cells? Cancer Res. 58815-822. [PubMed] [Google Scholar]
- 10.Burg, E. D., C. V. Remillard, and J. X. Yuan. 2006. K+ channels in apoptosis. J. Membr. Biol. 2093-20. [DOI] [PubMed] [Google Scholar]
- 11.Chapman, H., C. Ramstrom, L. Korhonen, M. Laine, K. T. Wann, D. Lindholm, M. Pasternack, and K. Törnquist. 2005. Downregulation of the HERG (KCNH2) K+ channel by ceramide: evidence for ubiquitin-mediated lysosomal degradation. J. Cell Sci. 1185325-5334. [DOI] [PubMed] [Google Scholar]
- 12.Cherubini, A., G. Hofmann, S. Pillozzi, L. Guasti, O. Crociani, E. Cilia, P. Di Stefano, S. Degani, M. Balzi, M. Olivotto, E. Wanke, A. Becchetti, P. Defilippi, R. Wymore, and A. Arcangeli. 2005. Human ether-a-go-go-related gene 1 channels are physically linked to β1 integrins and modulate adhesion-dependent signaling. Mol. Biol. Cell 62972-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Claassen, S., S. Schwarzer, J. Ludwig, and B. J. Zunkler. 2008. Electrophysiological and fluorescence microscopy studies with HERG channel/EGFP fusion proteins. J. Membrane Biol. 22231-41. [DOI] [PubMed] [Google Scholar]
- 14.Crociani, O., L. Guasti, M. Balzi, A. Becchetti, E. Wanke, M. Olivotto, R. S. Wymore, and A. Arcangeli. 2003. Cell cycle-dependent expression of HERG1 and HERG1B isoforms in tumor cells. J. Biol. Chem. 2782947-2955. [DOI] [PubMed] [Google Scholar]
- 15.Felipe, A., R. Vicente, N. Villalonga, M. Roura-Ferrer, R. Martínez-Mármol, L. Solé, J. Ferreres, and E. Condom. 2006. Potassium channels: new targets in cancer therapy. Cancer Detect. Prev. 30375-385. [DOI] [PubMed] [Google Scholar]
- 16.Ficker, E., A. T. Dennis, L. Wang, and A. M. Brown. 2003. Role of the cytosolic chaperones Hsp70 and Hsp90 in maturation of the cardiac potassium channel HERG. Circ. Res. 9287-100. [DOI] [PubMed] [Google Scholar]
- 17.Ficker, E., Y. A. Kuryshev, A. T. Dennis, C. Obejero-Paz, L. Wang, P. Hawryluk, B. A. Wible, and A. M. Brown. 2004. Mechanisms of arsenic-induced prolongation of cardiac repolarization. Mol. Pharmacol. 6633-44. [DOI] [PubMed] [Google Scholar]
- 18.Guasti, L., E. Cilia, O. Crociani, G. Hofmann, S. Polvani, A. Becchetti, E. Wanke, F. Tempia, and A. Arcangeli. 2005. Expression pattern of the ether-a-go-go-related (ERG) family proteins in the adult central nervous system: evidence for coassembly of different subunits. J. Comp. Neurol. 17157-174. [DOI] [PubMed] [Google Scholar]
- 19.Hille, B. 2001. Ionic channels of excitable membranes, 3rd ed. Sinauer Associates, Sunderland, MA.
- 20.Reference deleted.
- 21.Jenke, M., A. Sanchez, F. Monje, W. Stuhmer, R. M. Weseloh, and L. A. Pardo. 2003. C-terminal domains implicated in the functional surface expression of potassium channels. EMBO J. 22395-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones, E. M., E. C. Roti, J. Wang, S. A. Delfosse, and G. A. Robertson. 2004. Cardiac Ikr channels minimally comprise hERG 1a and hERG 1b subunits. J. Biol. Chem. 27944690-44694. [DOI] [PubMed] [Google Scholar]
- 23.Kirchberger, N. M., I. Wulfsen, J. R. Schwarz, and C. K. Bauer. 2006. Effects of TRH on heteromeric rat erg1a/1b K+ channels are dominated by the rerg1b subunit. J. Physiol. 57127-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kunzelmann, K. 2005. Ion channels and cancer. J. Membr. Biol. 205159-173. [DOI] [PubMed] [Google Scholar]
- 25.Kupershmidt, S., D. J. Snyders, A. Raes, and D. M. Roden. 1998. A K+ channel splice variant common in human heart lacks a C-terminal domain required for expression of rapidly activating delayed rectifier current. J. Biol. Chem. 27327231-27235. [DOI] [PubMed] [Google Scholar]
- 26.Kupershmidt, S., T. Yang, S. Chanthaphaychith, Z. Wang, J. A. Towbin, and D. M. Roden. 2002. Defective human Ether-a-go-go-related gene trafficking linked to an endoplasmic reticulum retention signal in the C terminus. J. Biol. Chem. 2627442-27448. [DOI] [PubMed] [Google Scholar]
- 27.Lastraioli, E., L. Guasti, O. Crociani, S. Polvani, G. Hofmann, H. Witchel, L. Bencini, M. Calistri, L. Messerini, M. Scatizzi, R. Moretti, E. Wanke, M. Olivotto, G. Mugnai, and A. Arcangeli. 2004. herg1 gene and HERG1 protein are overexpressed in colorectal cancers and regulate cell invasion of tumor cells. Cancer Res. 64606-611. [DOI] [PubMed] [Google Scholar]
- 28.Li, Q., A. Lau, T. J. Morris, L. Guo, C. B. Fordyce, and E. F. Stanley. 2004. A syntaxin 1, Gαo, and N-type calcium channel complex at a presynaptic nerve terminal: analysis by quantitative immunocolocalization. J. Neurosci. 244070-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma, D., N. Zerangue, Y. F. Lin, A. Collins, M. Yu, Y. N. Jan, and L. Y. Jan. 2001. Role of ER export signals in controlling surface potassium channel numbers. Science 291316-319. [DOI] [PubMed] [Google Scholar]
- 30.Manders, E. M. M., F. J. Verbeek, and J. A. Aten. 1993. Measurement of co-localization of objects in dual-color confocal images. J. Microsc. 169375-382. [DOI] [PubMed] [Google Scholar]
- 31.Masi, A., A. Becchetti, R. Restano-Cassulini, S. Polvani, G. Hofmann, A. M. Buccoliero, M. Paglierani, B. Pollo, G. L. Taddei, P. Gallina, N. Di Lorenzo, S. Franceschetti, E. Wanke, and A. Arcangeli. 2005. hERG1 channels are overexpressed in glioblastoma multiforme and modulate VEGF secretion in glioblastoma cell lines. Br. J. Cancer 93781-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pillozzi, S., M. F. Brizzi, M. Balzi, O. Crociani, A. Cherubini, L. Guasti, B. Bartolozzi, A. Becchetti, E. Wanke, P. A. Bernabei, M. Olivotto, L. Pegoraro, and A. Arcangeli. 2002. herg potassium channels are constitutively expressed in primary human acute myeloid leukemias and regulate cell proliferation of normal and leukemic haemopoietic progenitors. Leukemia 161791-1798. [DOI] [PubMed] [Google Scholar]
- 33.Pillozzi, S., E. De Lorenzo, M. Masselli, O. Crociani, E. Cilia, E. Torre, A. Mugnai, A. Becchetti, and A. Arcangeli. 2007. Treatment with hERG1 K+ channel inhibitors reduces acute myeloid leukemia cell lines engraftment into nonobese diabetic/severe combined immunodeficient mice and prolongs survival of injected mice. Blood (Am. Soc. Hematol. Annu. Meet. Abstr.) 110877. [Google Scholar]
- 34.Pillozzi, S., M. F. Brizzi, P. A. Bernabei, B. Bartolozzi, R. Caporale, V. Basile, V. Boddi, L. Pegoraro, A. Becchetti, and A. Arcangeli. 2007. VEGFR-1 (FLT-1), beta-1 integrin and hERG K+ channel form a macromolecular signalling complex in acute myeloid leukemia: role in cell migration and clinical outcome. Blood 1101238-1250. [DOI] [PubMed] [Google Scholar]
- 35.Restano-Cassulini, R., Y. V. Korolkova, S. Diochot, G. Gurrola, L. Guasti, L. D. Possani, M. Lazdunski, E. V. Grishin, A. Arcangeli, and E. Wanke. 2006. Species diversity and peptide toxins blocking selectivity of ether-a-go-go-related gene subfamily K+ channels in the central nervous system. Mol. Pharmacol. 691673-1683. [DOI] [PubMed] [Google Scholar]
- 36.Robertson, G. A., and C. T. January. 2006. HERG trafficking and pharmacological rescue of LQTS-2 mutant channels. Handb. Exp. Pharmacol. 171349-355. [DOI] [PubMed] [Google Scholar]
- 37.Schonherr, R., B. Rosati, S. Hehl, V. G. Rao, A. Arcangeli, M. Olivotto, S. H. Heinemann, and E. Wanke. 1999. Functional role of the slow activation property of ERG K+ channels. Eur. J. Neurosci. 11753-760. [DOI] [PubMed] [Google Scholar]
- 38.Shi, W., R. S. Wymore, H. S. Wang, Z. Pan, I. S. Cohen, D. McKinnon, and J. E. Dixon. 1997. Identification of two nervous system-specific members of the erg potassium channel gene family. J. Neurosci. 179423-9432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith, G. A., H. W. Tsui, E. W. Newell, X. Jiang, X.-P. Zhu, F. W. L. Tsui, and L. C. Schlichter. 2002. Functional up-regulation of HERG K+ channels in neoplastic hematopoietic cells. J. Biol. Chem. 27718528-18534. [DOI] [PubMed] [Google Scholar]
- 40.Spinelli, W., I. F. Moubarak, R. W. Parsons, and T. J. Colatsky. 1993. Cellular electrophysiology of WAY-123,398, a new class III antiarrhythmic agent: specificity of IK block and lack of reverse use dependence in cat ventricular myocytes. Cardiovasc. Res. 91580-1591. [DOI] [PubMed] [Google Scholar]
- 41.Thomas, D., J. Kiehn, H. A. Katus, and C. A. Karle. 2003. Defective protein trafficking in hERG-associated hereditary long QT syndrome (LQT2): molecular mechanisms and restoration of intracellular protein processing. Cardiovasc. Res. 60235-241. [DOI] [PubMed] [Google Scholar]
- 42.Wimmers, S., I. Wulfsen, C. K. Bauer, and J. R. Schwarz. 2001. erg1, erg2 and erg3 K+ channel subunits are able to form heteromultimers. Pfluegers Arch. 441450-455. [DOI] [PubMed] [Google Scholar]
- 43.Wimmers, S., C. K. Bauer, and J. R. Schwarz. 2002. Biophysical properties of heteromultimeric erg K+ channels. Pflugers Arch. 445423-430. [DOI] [PubMed] [Google Scholar]
- 44.Wymore, R. S., G. A. Gintant, R. T. Wymore, J. E. Dixon, D. McKinnon, and I. S. Cohen. 1997. Tissue and species distribution of mRNA for the IKr-like K+ channel, erg. Circ. Res. 80261-268. [DOI] [PubMed] [Google Scholar]
- 45.Zhou, Z., Q. Gong, and C. T. January. 1999. Correction and defective protein trafficking of a mutant HERG potassium channel in human long QT syndrome. Pharmacological and temperature effects. J. Biol. Chem. 27431123-31126. [DOI] [PubMed] [Google Scholar]
- 46.Zhou, Z., Q. Gong, M. L. Epstein, and C. T. January. 1998. HERG channel dysfunction in human long QT syndrome. Intracellular transport and functional defects. J. Biol. Chem. 27321061-21066. [DOI] [PubMed] [Google Scholar]