Abstract
ERCC1-XPF endonuclease is required for nucleotide excision repair (NER) of helix-distorting DNA lesions. However, mutations in ERCC1 or XPF in humans or mice cause a more severe phenotype than absence of NER, prompting a search for novel repair activities of the nuclease. In Saccharomyces cerevisiae, orthologs of ERCC1-XPF (Rad10-Rad1) participate in the repair of double-strand breaks (DSBs). Rad10-Rad1 contributes to two error-prone DSB repair pathways: microhomology-mediated end joining (a Ku86-independent mechanism) and single-strand annealing. To determine if ERCC1-XPF participates in DSB repair in mammals, mutant cells and mice were screened for sensitivity to gamma irradiation. ERCC1-XPF-deficient fibroblasts were hypersensitive to gamma irradiation, and γH2AX foci, a marker of DSBs, persisted in irradiated mutant cells, consistent with a defect in DSB repair. Mutant mice were also hypersensitive to irradiation, establishing an essential role for ERCC1-XPF in protecting against DSBs in vivo. Mice defective in both ERCC1-XPF and Ku86 were not viable. However, Ercc1−/− Ku86−/− fibroblasts were hypersensitive to gamma irradiation compared to single mutants and accumulated significantly greater chromosomal aberrations. Finally, in vitro repair of DSBs with 3′ overhangs led to large deletions in the absence of ERCC1-XPF. These data support the conclusion that, as in yeast, ERCC1-XPF facilitates DSB repair via an end-joining mechanism that is Ku86 independent.
ERCC1-XPF is a highly conserved endonuclease identified for its essential role in nucleotide excision repair (NER) of helix-distorting DNA lesions, in particular, UV-induced damage (4, 74). Defects in NER cause xeroderma pigmentosum (XP), a rare disorder characterized by photosensitivity, a dramatically increased risk of skin cancer, and neurodegeneration in severe cases. In contrast, the only reported patient with a mutation in ERCC1 had severe congenital anomalies (cerebro-oculo-facial-skeletal syndrome) (33). Patients with subtle mutations in XPF have a mild form of XP (46), consistent with only a partial defect in NER. However, a mutation in XPF that severely compromises protein levels causes dramatically accelerated aging (53). This observation implies additional functions for mammalian ERCC1-XPF distinct from NER. Consistent with that, ERCC1- and XPF-deficient mice have a much more severe phenotype than mice defective in NER. Xpa−/− mice with undetectable NER are indistinguishable from wild-type (WT) mice until challenged with carcinogens (12). In contrast, Ercc1−/− and Xpf−/− mice have a constellation of progeroid symptoms affecting the musculoskeletal, dermatologic, hepatobiliary, renal, and hematopoietic systems (48, 53, 79, 84) and die of liver failure before sexual maturation (72).
XPF contains the catalytic domain of the nuclease (18), whereas ERCC1 is required for DNA binding and stabilization of XPF (51, 80). The endonuclease is structure specific, incising double-stranded DNA 5′ to a junction with single-stranded DNA. Thus, ERCC1-XPF can remove 3′ single-stranded flaps from DNA ends (11) and cleaves the 5′ side of a bubble in NER to excise the lesion (74). Incision by ERCC1-XPF creates a 3′ OH group that is used to prime DNA synthesis to replace excised bases (74). Neither ERCC1 nor XPF has structural domains that suggest that the protein functions other than as a nuclease (73). Thus, novel functions of ERCC1-XPF that protect against rapid aging are likely contributions to other DNA repair mechanisms.
Indeed, ERCC1-XPF is required for the repair of DNA interstrand cross-links (ICLs) via a mechanism distinct from NER (47, 54) and ICLs are implicated in contributing to the dramatic premature aging phenotype caused by ERCC1-XPF deficiency (53). However, orthologs of ERCC1-XPF, AtErcc1-AtRad1 in Arabidopsis thaliana (29), DmERCC1-MEI-9 in Drosophila melanogaster (3), and Rad10-Rad1 in Saccharomyces cerevisiae (22, 32) are also implicated in double-strand break (DSB) repair. DNA DSBs are extremely cytotoxic lesions because both strands of the double helix are affected. DSBs are caused by both environmental and endogenous processes, including ionizing radiation (IR), radiomimetic drugs, programmed cleavage by endonucleases during meiotic recombination and V(D)J recombination, and replication of DNA containing single-strand breaks, ICLs, or topoisomerase I-induced lesions (6, 30). Failure to repair DSBs can lead to the accumulation of chromosomal aberrations or cell death (6, 30, 37). Thus, humans with genetic defects in DSB recognition or repair, or model organisms mimicking these human syndromes, are prone to cancer and segmental premature aging (21, 30).
There are two major mechanisms of DSB repair in eukaryotes: homologous recombination (HR)-mediated repair and nonhomologous end joining (NHEJ) (6). HR is an error-free mechanism in which sequence information lost at a broken end is recovered from the sister chromatid. Therefore, HR is primarily restricted to S and G2 phases of the cell cycle, when a sister chromatid is available. NHEJ, on the other hand, rejoins two broken ends via ligation. Thus, it is not restricted to proliferating cells and this repair mechanism is frequently used in mammals (36). Since NHEJ does not require sequence homology, inappropriate ends can potentially be joined, leading to chromosomal translocations (6, 30). In addition, bases may be lost at broken ends, resulting in deletions. However, NHEJ appears to be primarily error free (27, 81).
There are several well-defined error-prone mechanisms of DSB repair in yeast (28). Both single-strand annealing (SSA) and microhomology-mediated end-joining (MMEJ) pathways align two broken DNA ends by pairing homologous sequences at or near the DSB. In both mechanisms, if the homology is not immediately at the broken end, Rad10-Rad1 endonuclease, the ortholog of ERCC1-XPF, is required to remove the 3′ flap of nonhomologous sequence from the end, permitting DNA synthesis and ligation and thereby creating a deletion. Despite their similarity, SSA and MMEJ appear to have distinct genetic requirements. SSA requires Rad52 and Rad10-Rad1, while for short patches of homology, Rad59, Msh2, and Msh3 are also implicated (28). In contrast, MMEJ is not dependent on Rad52 or Ku86 but requires Rad10-Rad1, Mre11-Rad50-Xrs2 (28), the flap endonuclease Sae2 (40), and mismatch repair proteins Msh2 and Pms1 (10). DSB repair events that utilize short patches of sequence homology were recognized in mammalian cells for quite some time (65). Evidence exists for SSA in mammalian cells (34, 42), particularly when HR or NHEJ is defective (76). NHEJ mutant mammalian cells can still support end joining of DSBs (35) by utilizing short sequences of microhomology at the broken ends (38), demonstrating that MMEJ also occurs in mammalian cells. Recently, this alternative end-joining mechanism of microhomology-mediated DSB repair was shown to support class switch recombination in NHEJ-deficient B cells (87). The genetic requirements for SSA and MMEJ and the biological significance of these pathways in mammals remain unknown.
In this study, we investigated if the 3′ flap endonuclease ERCC1-XPF is involved in DSB repair in mammalian cells. ERCC1-XPF-deficient Chinese hamster ovary cell lines were reported to be moderately hypersensitive to IR, particularly under hypoxic conditions (50, 86). In contrast, human XP-F fibroblasts and murine Ercc1−/− embryonic stem (ES) cells are not hypersensitive to IR (50, 52). To look more systematically for a role of the mammalian nuclease in DSB repair, we screened Ercc1−/− mouse embryonic fibroblasts (MEFs), XPF-deficient human fibroblasts, and ERCC1-deficient mice for sensitivity to IR. We also used a genetic approach to determine if ERCC1-XPF is epistatic with NHEJ proteins with respect to sensitivity to IR.
MATERIALS AND METHODS
Generation of cell lines and mice.
Ercc1−/− mouse ES cells were generated and cultured as previously described (52). Ercc1−/−, DNA-Pkcs−/−, Ku86−/−, and Csb−/− primary MEFs were developed from embryonic day 12 to 15 embryos derived from crossing inbred C57BL/6 mice heterozygous for each null allele, as previously described (13, 41, 43, 84). Ercc1−/− Ku86−/− and Ercc1−/− DNA-Pkcs−/− double-knockout primary MEFs were created from embryos derived from crossing double-heterozygous mice in an inbred C57BL/6 background. Genomic DNA was isolated from a tissue sample of each embryo with the NucleoSpin DNA extraction system (Macherey-Nagel, Inc.). Genotyping of the Ercc1 allele was done by PCR coamplification of the 3′ end of exon 7 from the WT allele and the neomycin resistance marker cloned into exon 7 of the targeted allele with primers specific for exon 7, neor, and intron 7 (5′-AGCCGACCTCCTTATGGAAA, 5′-TCGCCTTCTTGACGAGTTCT, and 5′-ACAGATGCTGAGGGCAGACT, respectively). WT (0.25-kb) and mutant (0.4-kb) products were separated by electrophoresis on a 2% agarose gel. Genotyping of the Ku86 alleles by PCR was done as previously described (82). Genotyping of DNA-Pkcs alleles was done by PCR with forward (5′-GCATCGCCTTCTATCGCCTT) and reverse (5′-GCTGAGACATCCTGGACTGAA) primers for the null allele (0.4 kb) and the forward primer 5′-GGAATTGACTTTGGACATGCG with the same reverse primer as for the WT allele.
MEFs were cultured in a 1:1 mixture of Dulbecco's modified Eagle's medium and Ham's F10 with 10% fetal calf serum and antibiotics and incubated at 3% oxygen (59). Each experimental replica was done with a new MEF line, created from a unique embryo, in its second or third passage. Cell lines derived from WT littermate embryos were used as controls in all experiments. A spontaneously transformed Ercc1−/− MEF line was stably transfected with a plasmid expressing human ERCC1 cDNA fused in frame with enhanced yellow fluorescent protein (YFP) (52). Human fibroblasts immortalized with hTert derived from a normal individual (C5RO) and a patient with severe progeria caused by a mutation in XPF (XFE) (53) were cultured in Ham's F10 with 10% fetal calf serum and antibiotics and incubated at 3% oxygen.
Double-heterozygous (Ercc1+/Δ Ku86+/− and Ercc1+/− Ku86+/−) mice in a mixed genetic background (50:50 C57BL/6 and FVB/n) were bred to recover Ercc1−/Δ Ku86−/− mice. DNA was extracted from an ear plug of 2-week-old pups and genotyped by PCR as described above. The mutant allele of Ercc1 (Δ) was amplified by adding a fourth primer (5′-CTAGGTGGCAGCAGGTCATC) to amplify the neo cassette in the Δ allele (0.5 kb).
Clonogenic survival assays.
Early-passage primary MEFs were seeded in 6-cm dishes in triplicate at 103 to 104 per plate, depending on the dose of genotoxin. After 16 h, cells were irradiated with a 137Cs source. Seven to 10 days later, cultures were fixed and stained with 50% methanol, 7% acetic acid, and 0.1% Coomassie blue. Colonies (defined as ≥10 cells) were counted with a Nikon SMZ 2B stereomicroscope with a 10× eyepiece. The data were plotted as the number of colonies that grew on the treated plates relative to untreated plates ± the standard error of the mean for at least three independent experiments each with a unique cell line.
Immunofluorescence.
Cells were trypsinized and seeded at 25% confluence on glass coverslips. Sixteen hours later, the cells were irradiated with 2 Gy of gamma rays. The cells were incubated in fresh medium at 37°C for the indicated amount of time and then fixed with 2% paraformaldehyde in phosphate-buffered saline, pH 7.4, for 15 min. Cells were permeabilized with 0.1% Triton X-100 in phosphate-buffered saline, and the phosphorylated form of H2AX (γH2AX) was detected with polyclonal anti-γH2AX (1:1,000; Upstate Biotechnology) and Alexa 488-conjugated goat anti-rabbit immunoglobulin (1:500; Molecular Probes) in phosphate-buffered saline with 0.15% glycine and 0.5% bovine serum albumin. γH2AX foci were counted with an Olympus BX51 fluorescent microscope with a 60× to 100× objective.
Animal survival.
Six-week-old WT and mutant animals (six per genotype) were exposed to 6 Gy of whole-body IR from a 137Cs source, and their health was monitored daily. Animals were euthanized when deemed terminal in accordance with the University of Pittsburgh IACUC standards. Survival data were recorded as a Kaplan-Meier curve.
Histological analysis and immunohistochemistry.
Tissues (liver, femur, and bowel) were collected from terminal gamma-irradiated Ercc1−/Δ mice or 2 days postirradiation. Age-matched untreated and treated WT and untreated Ercc1−/Δ mice were sacrificed simultaneously as controls. Tissues were fixed with 10% formalin and embedded in paraffin. Five-micrometer sections were cut and stained with hematoxylin and eosin. Proliferation of cells within the crypts of the small intestine was visualized by immunostaining with a rat anti-mouse Ki67 monoclonal antibody (clone TEC-3; Dako North America, Inc., Carpinteria, CA) as recommended by the manufacturer.
Cytogenetics.
MEFs were transformed by stable transfection with a plasmid expressing the simian virus 40 large T antigen and exposed to 2 Gy (WT and Ercc1−/−) or 0.4 Gy (Ku86−/− and Ercc1−/− Ku86−/−) of IR. Bromodeoxyuridine (10 μM) and colcemid (0.1 μg/10 ml) were added to the medium 48 and 36 h, respectively, prior to harvesting of the cells. The cells were harvested by trypsinization, and metaphase spreads were prepared as previously described (15). The numbers of fragments, breaks, fusions, radials, and marker chromosomes per diploid metaphase spread were determined.
Calculation of population doubling number.
WT and mutant MEFs were plated at a density of 0.5 × 106 per 6-cm dish. Cells were trypsinized at confluence, counted, and replated at the same density until double-knockout cells stopped growing. The total number of cells at each passage was calculated as follows: (no. of cells at previous passage/no. of cells plated) × no. of cells at current passage. The total cell number was plotted as the log cell number.
In vitro DNA repair assay.
WT and Ercc1−/− MEFs were grown on 6-cm dishes and transfected by using either a Nucleofector MEF II kit (Amaxa Biosystems, Gaithersburg, MD) or Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) and following the manufacturer's instructions. pEYFP-N1 (Clontech Laboratories Inc., Mountain View, CA) was linearized by digesting between the promoter and coding sequence of YFP with SmaI (blunt ends), HindIII (5′ complementary), or KpnI-SacI (3′ noncomplementary ends). The linear products were gel purified and transfected into WT or mutant MEFs. After 48 h, cells were sorted and the fraction of cells expressing YFP was determined by a DakoCytomation MoFLo high-speed cell sorter (Dako North America, Carpinteria, CA). The efficiency of repair was calculated as the percentage of YFP-positive cells after transfection with a linear plasmid, relative to transfection with the circular plasmid, from at least three independent experiments. In parallel, transfected plasmid DNA was recovered from MEFs by alkaline lysis (31). After phenol-chloroform extraction, DNA was ethanol precipitated and treated with lambda exonuclease to remove any linear plasmid DNA (16, 44). The DNA was then amplified in Escherichia coli, and individual plasmids were isolated. The plasmids were screened by digestion with SmaI (blunt ends), HindIII (5′ complementary ends), or EcoRI (3′ noncomplementary ends) to distinguish between error-free (sensitive to digestion) and error-prone (resistant to digestion) DSB repair. The plasmids were next screened for deletions by digestion with ApaLI and ClaI, which yields two products of 2.9 and 1.7 kb, the latter of which will be diminished in size if a deletion occurred at the site of the DSB. For plasmids containing small deletions, a 475-bp region flanking the site of the DSB was PCR amplified with the forward primer 5′-TACATCAATGGGCGTGGATA and the reverse primer 5′-GAACTTCAGGGTCAGCTTGC. The amplified fragments were run on a 3% agarose gel to estimate the size of small deletions; the lower limit of detection was approximately 25 bp. Finally, the junction where DSB repair occurred was sequenced for 20 to 30 plasmids from each group (blunt or 5′ or 3′ overhangs) to determine the frequency with which microhomology was utilized and bases were inserted.
RESULTS
Sensitivity of ERCC1-XPF-deficient cells to IR.
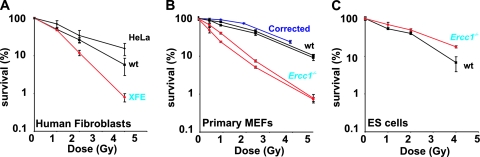
As a first screen to determine if ERCC1-XPF participates in DSB repair, cells harboring mutations in either protein subunit were tested for sensitivity to IR by clonogenic survival assay. Telomerase-immortalized XPF-deficient human fibroblasts were significantly more sensitive to IR than were WT fibroblasts or HeLa cells (Fig. 1A), even though these cells are not completely devoid of XPF (53). Similarly, Ercc1−/− primary MEFs, in which XPF protein is also undetectable (53), were 2.5-fold more sensitive to IR relative to congenic WT MEFs (Fig. 1B). The hypersensitivity was rescued by stable transfection of the Ercc1−/− cells with human ERCC1 cDNA. In contrast, Ercc1−/− mouse ES cells were not sensitive to IR relative to a congenic WT cell line (Fig. 1C), as previously reported (52). These data reveal heterogeneity in sensitivity to genotoxic stress between different types of cells with defects in ERCC1-XPF and indicate that this endonuclease is required to protect differentiated mammalian cells from IR.
FIG. 1.
Clonogenic survival assays after exposure of cells to increasing doses of IR. Error bars represent the standard error of the mean for three or more independent experiments. (A) WT immortalized human fibroblasts (C5RO), HeLa cells, and immortalized fibroblasts derived from an XPF-deficient patient (XFE). (B) Two independent, early-passage, primary WT and Ercc1−/− MEF lines and transformed Ercc1−/− MEFs stably corrected with human ERCC1 cDNA. (C) WT and Ercc1−/− murine ES cells.
Because IR induces numerous types of DNA lesions in addition to DSBs (e.g., single-strand breaks and oxidative base damage [23]), Ercc1−/− cells were also screened for sensitivity to the oxidizing agent paraquat (see Fig. S1 in the supplemental material). Ercc1−/− ES cells were hypersensitive to paraquat and H2O2 (53), whereas Ercc1−/− primary MEFs were not. The cell type-specific pattern of sensitivity is contrary to that of IR, indicating that the hypersensitivity of ERCC1-XPF-deficient MEFs to IR is not due to a failure to repair oxidative base damage.
Quantitation of IR-induced DSBs in ERCC1-XPF-deficient cells.
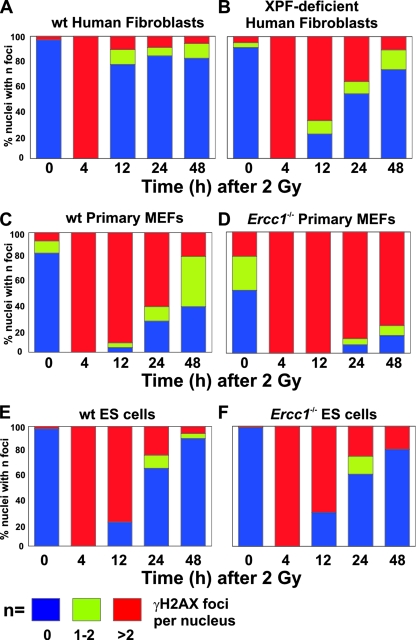
To further probe the cause of the IR sensitivity of ERCC1-XPF-deficient cells, we measured phosphorylated histone variant H2AX, a marker of DSBs (63), in WT and ERCC1-XPF-deficient cells at multiple time points following IR exposure. γH2AX foci provide a quantitative measurement of DSB induction and repair at low doses of irradiation (66). Cells were exposed to 2 Gy of IR, and γH2AX foci were detected by immunofluorescence (see Fig. S2 in the supplemental material). Foci were counted at 0, 4, 12, 24, and 48 h following irradiation. Cells were categorized as having no foci, one or two foci, or more than two foci (Fig. 2). A 2-Gy dose induced multiple γH2AX foci in 100% of WT and XPF mutant fibroblasts. By 12 h postirradiation, 75% of the WT cells no longer had γH2AX foci (Fig. 2A) whereas >60% of the XPF-deficient cells still had multiple foci, suggesting continued accumulation or persistence of DSBs (Fig. 2B). Similar results were obtained with Ercc1−/− primary MEFs (Fig. 2C and D). In both human and murine mutant fibroblasts, the fraction of cells with γH2AX foci decreased at 24 and 48 h postirradiation, consistent with repair of DSBs, albeit substantially delayed relative to WT controls. There was no difference in the number of γH2AX foci in WT and Ercc1−/− ES cells at any time point following irradiation (Fig. 2E and F), correlating with the lack of sensitivity of these cells to IR. These data support a role for the ERCC1-XPF nuclease in facilitating DSB repair in mammalian fibroblasts.
FIG. 2.
Quantitation of γH2AX foci in WT and ERCC1-XPF-deficient cells exposed to IR. Histograms indicate the fractions of cells with no foci (blue), one or two foci (green), or more than two foci (red) at 0, 4, 12, 24, and 48 h postirradiation. Panels: A, immortalized WT human fibroblasts (C5RO); B, immortalized XPF-deficient human fibroblasts (XFE); C, WT primary MEFs; D, Ercc1−/− primary MEFs; E, WT ES cells; F, Ercc1−/− ES cells.
Sensitivity of ERCC1-deficient mice to IR.
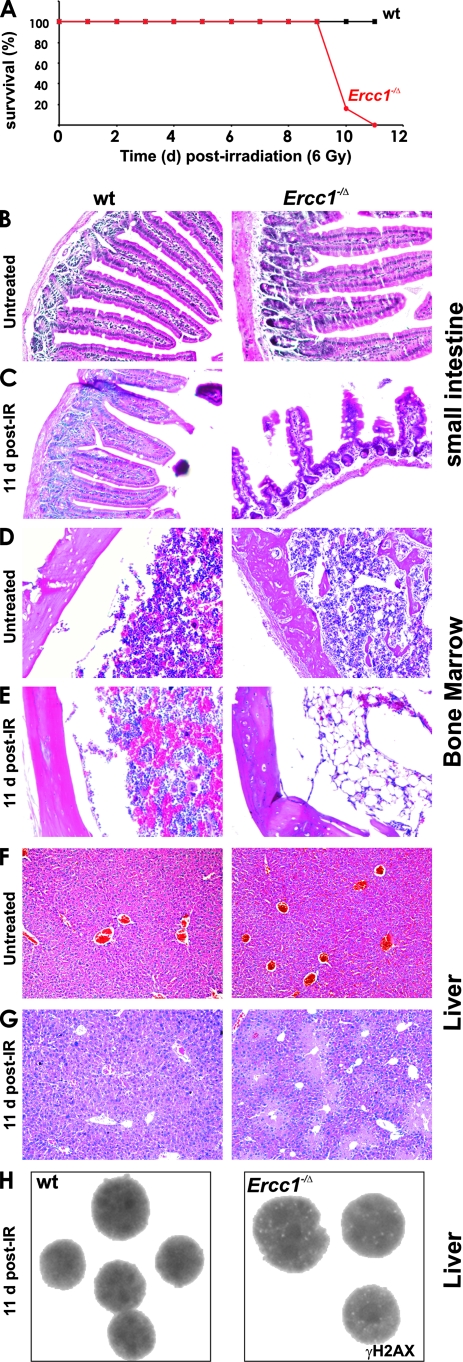
To determine if ERCC1-XPF contributes to protection against IR in vivo, 6-week-old Ercc1−/Δ mice, which are hypomorphic for ERCC1-XPF (14, 84), and WT littermates (n = 6) were exposed to a single dose of 6 Gy of total-body irradiation. At this age, Ercc1−/Δ mice are healthy and have no obvious abnormal phenotype other than growth retardation (unpublished data). A 6-Gy dose of radiation did not acutely affect the survival of WT mice (Fig. 3A and reference 41) but caused 100% lethality of Ercc1−/Δ mice by 11 days postirradiation. Although difficult to compare because of genetic background differences, the sensitivity of Ercc1−/Δ mice is similar to that of DNA-Pkcs−/− mice (41) but less than that of Ku86−/− mice (56), both of which lack a protein required for NHEJ. Therefore, ERCC1-XPF makes a substantial contribution to protecting mammals from radiation-induced DNA damage in vivo.
FIG. 3.
Sensitivity of ERCC1-deficient mice to IR. (A) Six-week-old Ercc1−/Δ mice and WT littermates (n = 6 per group) were exposed to 6 Gy of IR, and survival was recorded in days after exposure. (B to G) Tissue sections from 7- to 8-week-old WT and Ercc1−/Δ mice ± exposure to 6 Gy of IR. (B) Small intestines of untreated mice. (C) Small intestines of mice exposed to IR demonstrating loss of villi in Ercc1−/Δ mice. (D) BM of untreated mice. (E) BM of mice exposed to IR demonstrating fatty replacement in Ercc1−/Δ mice. (F) Livers of untreated mice. (G) Livers of mice exposed to IR demonstrating centrilobular necrosis in Ercc1−/Δ mice. (H) Hepatocyte nuclei demonstrating γH2AX foci in Ercc1−/Δ mice 11 days after IR.
To determine the cause of death of the Ercc1−/Δ mice, gastrointestinal tract, bone marrow (BM), and liver tissue sections from WT and Ercc1−/Δ mice 11 days postirradiation were examined. These tissues were indistinguishable between untreated Ercc1−/Δ and WT littermates (Fig. 3B, D, and F). However, following IR, Ercc1−/Δ mice had dramatically fewer intestinal villi than WT mice (Fig. 3C). Immunostaining for the proliferation marker Ki67 (69) revealed considerably fewer positive cells in Ercc1−/Δ crypts compared to WT mice at 11 days postirradiation, but not at 2 days post-IR or in unexposed animals (see Fig. S3 in the supplemental material). Postirradiation, the femoral BM of WT mice displayed hypercellularity of all hematopoietic lineages consistent with reactive hyperplasia (Fig. 3D and E). In contrast, the BM of Ercc1−/Δ mice was markedly hypocellular with residual cells displaying dysplastic changes including internuclear bridging, multinuclearity, and megablastoid changes, most prominently affecting erythropoiesis. Granulopoiesis displayed a shift to immature cell types. In the livers of Ercc1−/Δ but not WT mice, this dose of IR induced centrilobular necrosis (Fig. 3F and G). Immunostaining of liver sections for γH2AX revealed nuclear foci in hepatocytes of Ercc1−/Δ mice but not WT littermates (Fig. 3H), indicative of persistent DSBs. Collectively, these data indicate that proliferative tissues of Ercc1−/Δ mice are vulnerable to IR-induced genotoxic stress and have diminished regenerative capacity following damage.
ERCC1 deficiency causes embryonic lethality in a Ku86−/− background.
Very recent evidence indicates that ERCC1-XPF facilitates SSA between direct repeats in mammalian cells (2) as do the orthologs Rad10-Rad1 in S. cerevisiae (28). In yeast, Rad10-Rad1 also participates in MMEJ, a Ku-independent mechanism of DSB repair (45). This pathway was recently confirmed in mammals (87). To test if this function of ERCC1-XPF is conserved in mammals, we attempted to breed mice that are doubly deficient in ERCC1 and Ku86. Surprisingly, no live Ercc1−/Δ Ku86−/− mice were recovered (see Table S1 in the supplemental material; 0 of 220 offspring, 14 expected; P < 0.001). Genotyping of pups that died within 24 h of birth also failed to identify double-mutant mice. This was unexpected since both Ku86−/− and Ercc1−/Δ mice are born with Mendelian frequency and live into adulthood, although they have reduced life spans (88 and 30 weeks, respectively [82, 84; unpublished results]). In mammals, combined loss of Ku86 and ERCC1 thus results in lethality, similar to the effect of deleting Ku and Rad1 in budding yeast (45). This supports the conclusion that ERCC1-XPF contributes to a DSB repair pathway that is Ku independent, distinct from NHEJ and other Ku-dependent mechanisms.
Ercc1−/− Ku86−/− MEFs senesce prematurely.
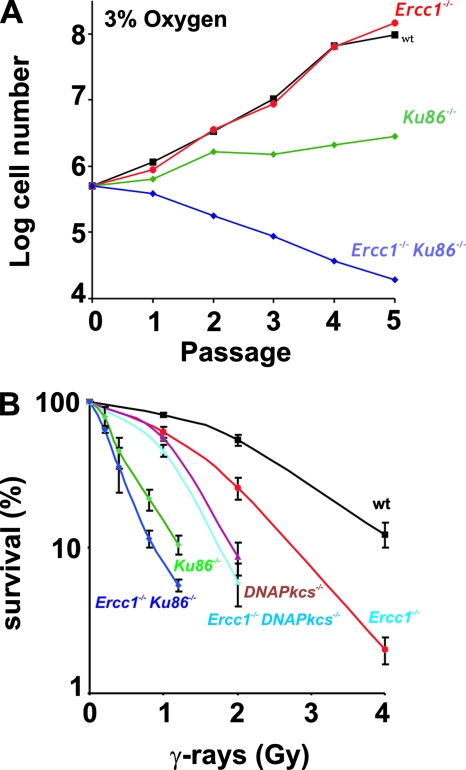
To generate Ercc1−/− Ku86−/− MEFs, double-heterozygous Ercc1+/− Ku86+/− mice were crossed. Ercc1−/− Ku86−/− embryos (embryonic days 13 to 16) were recovered at Mendelian frequency (see Table S2 in the supplemental material), indicating that deficiency of both ERCC1 and Ku86 causes lethality late in embryonic development. MEFs derived from Ercc1−/− Ku86−/− embryos grew extremely poorly in culture compared to single-mutant cell lines derived from littermates (Fig. 4A; see Fig. S4 in the supplemental material). Even when the MEFs were cultured at 3% O2, which alleviates premature senescence due to oxidative stress (59), Ercc1−/− Ku86−/− MEFs did not proliferate beyond passage 5. This indicates that simultaneous deletion of Ercc1 and Ku86 causes premature senescence of primary cells and that cellular proliferation in an oxidative environment requires either Ku-dependent or ERCC1-dependent DNA repair.
FIG. 4.
Growth and IR sensitivity of cells in which the ERCC1 and NHEJ proteins have been deleted. (A) Cell number with each passage of WT, Ercc1−/−, Ku86−/−, and Ercc1−/− Ku86−/− primary MEFs cultured at 3% oxygen. (B) Clonogenic survival of transformed WT, Ercc1−/−, DNA-Pkcs−/−, Ku86−/−, Ercc1−/− Ku86−/−, and Ercc1−/− DNA-Pkcs−/− MEFS after exposure to IR.
Ercc1−/− Ku86−/− MEFs are hypersensitive to IR.
To determine if Ercc1 is epistatic with Ku86 or DNA-Pkcs with respect to DSB repair, Ercc1−/− Ku86−/− and Ercc1−/− DNA-Pkcs−/− cells were screened for sensitivity to IR. Since primary Ercc1−/− Ku86−/− MEFs rapidly senesce, one primary cell line of each genotype was transformed by stable transfection with a plasmid expressing simian virus 40 large T antigen (83). Transformed Ercc1−/−, Ku86−/−, and DNA-Pkcs−/− single-mutant MEFs were hypersensitive to IR (Fig. 4B), as shown in Fig. 1B and as previously reported (8, 41, 43). Unexpectedly, Ercc1−/− DNA-Pkcs−/− MEFs appear to be equally as sensitive to IR as DNA-Pkcs−/− cells. However, Ercc1−/− Ku86−/− MEFs were significantly more sensitive to IR than either Ercc1−/− or Ku86−/− cells, consistent with the additive effect of both mutations in vivo. These results provide further evidence that ERCC1-XPF participates in a DSB repair mechanism that provides a critical back-up for Ku-dependent DSB repair, as do the yeast orthologs Rad10-Rad1.
Genomic instability in Ercc1−/− MEFs after IR.
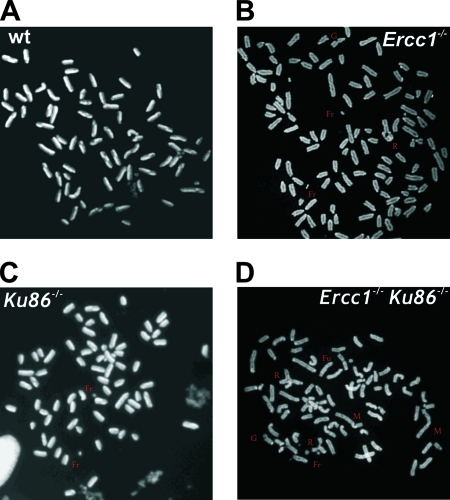
To further investigate whether the hypersensitivity of ERCC1-XPF-deficient cells to IR is caused by DSBs, cells were examined for chromosomal abnormalities after exposure to low-dose IR. Transformed WT and Ercc1−/− MEFs were treated with 2 Gy, while Ku86−/− and Ercc1−/− Ku86−/− MEFs were treated with 0.4 Gy due to their extreme hypersensitivity to IR (Fig. 4A and reference 43). Sister chromatid exchanges (SCEs), which occur via HR of DSBs (75), were significantly increased in Ercc1−/− MEFs relative to WT cells (Table 1). These data demonstrate that homologous recombination is not affected by loss of ERCC1-XPF nuclease. In contrast, the frequency of SCEs was not increased in irradiated Ercc1−/− Ku86−/− cells relative to Ku86−/− cells. This may reflect inefficient processing of radiation-induced breaks lacking 3′ OH groups, which are necessary for HR, when both Ku and ERCC1-XPF are present. Chromosomal aberrations were significantly increased (about fivefold) in Ercc1−/− MEFs treated with IR compared to WT cells (Table 1 and Fig. 5A and B). These abnormalities include gaps, fragments, breaks, fusions, and radials, all characteristic of unrepaired or misrepaired DSBs. Gaps, breaks, fusions, and radials were also significantly increased in Ercc1−/− Ku86−/− MEFs relative to Ku86−/− cells (Table 1 and Fig. 5C and D), providing further evidence that ERCC1-XPF participates in a DSB repair mechanism that functions as a back-up for Ku86-dependent repair.
TABLE 1.
Frequency of chromosomal aberrations in transformed MEFs exposed to IR
| Genotype | Dose (Gy) | No. of cells examined | SCEa | Gapsa | Breaks and fragmentsa | Fusion and radialsa |
|---|---|---|---|---|---|---|
| WT | 2.0 | 25 | 25 | 0.3 | 0.8 | 0 |
| Ercc1−/− | 2.0 | 24 | 42b | 2.6b | 2.6b | 0.5b |
| Ku86−/− | 0.4 | 36 | 25 | 1.2 | 1.2 | 0 |
| Ercc1−/−Ku86−/− | 0.4 | 34 | 18 | 2.8c | 2.8c | 0.5c |
The number of events per diploid genome is reported.
P < 0.05 for WT versus Ercc1−/− cells.
P < 0.05 for Ercc1−/− Ku86−/− versus Ku86−/− cells.
FIG. 5.
Chromosomal aberrations in transformed MEFs after IR. Subconfluent cultures of WT and Ercc1−/− cells were exposed to 2 Gy of IR, subconfluent cultures of Ku86−/− and Ercc1−/− Ku86−/− cells were exposed to 0.4 Gy, and the cells were analyzed 48 h later. (A) Representative metaphase spread from WT MEFs exposed to IR. (B) Metaphase spread from an Ercc1−/− cell demonstrating IR-induced gaps (G), fragments (Fr), and radials (R). (C) Ku86−/− cell demonstrating IR-induced fragments. (D) Ercc1−/− Ku86−/− cell demonstrating IR-induced fragments, gaps, fusions (Fu), radials, and marker chromosomes (M).
ERCC1-XPF facilitates removal of noncomplementary 3′ overhangs.
To determine if ERCC1-XPF participates in an end-joining mechanism of DSB repair similar to Rad10-Rad1, we used an in vitro DNA end-joining assay (67). A plasmid expressing YFP (pYFP-N1) was linearized by digestion with restriction enzymes that produce blunt, complementary 5′ overhangs or noncomplementary 3′ overhangs. These linear plasmids were transfected into WT, Ercc1−/−, Ku86−/−, and Ercc1−/− Ku86−/− cells, and repair events were scored as the percentage of YFP-expressing cells (see Fig. S5 in the supplemental material). Creation of a DSB between the promoter and YFP cDNA reduced the recovery of YFP+ cells by threefold. There was no difference in the frequency of repair of DSBs with either blunt or single-strand overhanging ends. Furthermore, there was no difference in the frequency of repair events in WT, Ercc1−/−, or Ku86−/− cells transfected with any of the repair substrates. The only significant difference observed was decreased recovery of YFP+ Ercc1−/− Ku86−/− cells after transfection with a linear plasmid containing single-strand overhangs. This indicates that, in the absence of both ERCC1-XPF and Ku86, DSBs that cannot be directly ligated are inefficiently repaired.
Repaired pYFP-N1 plasmid DNA was recovered from WT and Ercc1−/− YFP+ cells 48 h after transfection. The plasmid DNA was digested with the same enzyme that was used for linearization prior to transfection to score for error-free repair events. Approximately half of the repair events were error free for DNA with blunt or complementary single-strand overhanging ends in WT and Ercc1−/− cells (Table 2), while all of the repair events were error prone for the DNA with noncomplementary single-strand overhangs, as expected. Error-prone events were divided into three categories based on the size of the deletion, as determined by agarose gel electrophoresis: <100 bp, 100 to 500 bp, and >500 bp. There was no difference in the size of the deletions resulting from repair of blunt ends or 5′ complementary overhangs between WT and Ercc1−/− cells. However, there was a significant increase in very large deletions in substrates with 3′ noncomplementary overhangs repaired in Ercc1−/− cells relative to WT cells. This indicates that, in the absence of ERCC1-XPF, repair of DSBs with 3′ ends that cannot be directly ligated is impaired, leading to increased resection and larger deletions. This is consistent with the substrate specificity of ERCC1-XPF as a 3′ flap endonuclease (11, 74).
TABLE 2.
Size of linear plasmid with blunt or overhanging DNA ends after repair in WT or Ercc1−/− cells
| DNA ends and genotype | n | % Error free | % Error prone | % with <100-bp deletion | % with 101- to 500-bp deletion | % with >500-bp deletion |
|---|---|---|---|---|---|---|
| Blunt | ||||||
| WT | 81 | 50 | 50 | 88 | 10 | 2 |
| Ercc1−/− | 78 | 58 | 42 | 90 | 10 | 0 |
| 5′ Overhangs, complementary | ||||||
| WT | 65 | 43 | 57 | 60 | 40 | 0 |
| Ercc1−/− | 66 | 42 | 58 | 60 | 35 | 5 |
| 3′ Overhangs, noncomplementary | ||||||
| WT | 94 | 0 | 100 | 63 | 31 | 6 |
| Ercc1−/− | 82 | 0 | 100 | 65 | 14a | 21a |
P < 0.05.
A subset of the plasmids with deletions were sequenced to determine the frequency with which DSB repair occurred via a mechanism that utilizes sequence homology between the broken ends (see Tables S3 to S5 in the supplemental material). Short stretches of microhomology (one to five nucleotides) were identified at the broken ends in 70 to 90% of the error-prone DSB repair events, regardless of whether the ends were blunt or had overhangs (Table 3). There was not a significant difference in the utilization of microhomology to repair DSBs in WT and Ercc1−/− cells. However, significantly fewer of the repair events in Ercc1−/− cells resulted in the insertion of bases relative to the WT. This observation is consistent with ERCC1-XPF being required to remove 3′ nonhomologous flaps to create an end that can be used to prime DNA synthesis (74).
TABLE 3.
Sequence analysis of blunt, complementary, and noncomplementary DNA ends after repair in WT or Ercc1−/− cells
| DNA ends and genotype | n | % with nucleotide added | % Microhomology mediated |
|---|---|---|---|
| Blunt | |||
| WT | 18 | 0 | 89 |
| Ercc1−/− | 21 | 0 | 81 |
| 5′ Overhangs, complementary | |||
| WT | 18 | 6 | 72 |
| Ercc1−/− | 17 | 6 | 71 |
| 3′ Overhangs, noncomplementary | |||
| WT | 30 | 37 | 80 |
| Ercc1−/− | 31 | 10a | 71 |
P < 0.05.
DISCUSSION
Mammalian ERCC1-XPF participates in DSB repair.
ERCC1-XPF is a structure-specific endonuclease that incises double-stranded DNA at a junction with single-stranded DNA (3′), nicking bubble structures and 3′ single-strand overhangs (11). This activity is essential for excising damaged DNA in NER (74) and for creating a 3′ end that can be used to prime DNA synthesis to replace the excised bases. In addition to the bubble substrates in NER, ERCC1-XPF incises stem-loop structures (11), cross-linked Y structures (39), and R loops in vitro (78) and removes 3′ single-strand overhangs of nonhomologous sequence that otherwise would prevent HR (1, 52, 68). It has also been proposed that ERCC1-XPF incises D loops (52), G-rich telomeric overhangs (88), and intermediates of DNA ICL repair (54). Such a flap endonuclease activity could facilitate DSB repair, specifically, of ends with 3′ single-strand noncomplementary overhangs or without a 3′ hydroxyl group necessary for DNA synthesis and/or ligation. Indeed, orthologs of ERCC1-XPF in S. cerevisiae (Rad10-Rad1) (22, 32, 58), Arabidopsis thaliana (AtErcc1-AtRad1p) (17, 29), and Drosophila melanogaster (DmERCC1-MEI-9) (24, 61) do facilitate DSB repair and confer resistance to IR. Herein, we provide in vitro, genetic and in vivo evidence that this function is conserved in murine and human cells.
Role of ERCC1-XPF in DSB repair.
ERCC1-XPF-deficient cells are exquisitely sensitive to cross-linking agents and accumulate DSBs in response to cross-link damage (54). We postulated that in ICL repair, ERCC1-XPF is required to incise the cross-link lesion after a DSB is created by stalled replication, before the DSB can be repaired. Here we provide evidence for a distinct role for ERCC1-XPF in DSB repair. ERCC1-XPF-deficient murine and human cells are hypersensitive to IR (Fig. 1), similar to cells defective in HR-mediated or NHEJ DSB repair (77). Murine fibroblasts, but not ES cells, are IR sensitive (Fig. 1). This pattern of cell type-specific sensitivity is similar to that of DNA-Pkcs mutant lines defective in NHEJ (25) but the opposite of that of Rad54−/− cells defective in HR (19). This suggests that ERCC1-XPF may participate in an end-joining mechanism of DSB repair that is more frequently utilized in differentiated cells than in ES cells. In further support of this, SCEs are significantly elevated in irradiated Ercc1−/− MEFs (Table 1), indicating that HR-mediated DSB repair is possible in the absence of ERCC1-XPF. This increase in SCEs might reflect preferential repair of DSBs by HR when end-joining mechanisms are impaired (85). ERCC1-XPF, however, is clearly not part of the core NHEJ machinery, as there is no evidence of severe combined immunodeficiency, a pathognomonic feature of NHEJ deficiency (87), in Ercc1−/− or Xpf−/− mice (70, 79) or humans with XPF deficiency (53). Furthermore, Ercc1−/− MEFs are less sensitive to IR than are DNA-Pkcs−/− or Ku86−/− MEFs, in which NHEJ is absent (Fig. 4B).
IR induces a variety of lesions in addition to DSBs (23). Thus, to confirm that the IR sensitivity of ERCC1-XPF-deficient cells is due, at least in part, to a defect in DSB repair, we quantitated γH2AX foci, a marker of DSBs (63), in cells following IR. γH2AX foci persist in irradiated ERCC1-XPF-deficient fibroblasts (Fig. 2) and in hepatocytes of irradiated ERCC1-deficient mice (Fig. 3H) relative to congenic WT controls, consistent with impaired DSB repair in the absence of ERCC1-XPF both in vitro and in vivo. Furthermore, irradiated Ercc1−/− MEFs had increased gaps, breaks, fragments, and radial structures compared to WT cells (Table 1). These chromosome abnormalities are a consequence of DSBs (49) and thus demonstrate a defect in DSB repair in Ercc1−/− cells. Interestingly, there is no difference in the number of γH2AX foci in Ercc1−/− and WT ES cells at any time point following exposure to IR, correlating with their lack of sensitivity to IR. There is increasing evidence that homologous recombination is essential for genome stability in ES cells (6, 26, 62), further suggesting that HR occurs in ERCC1-deficient cells.
Biological significance of ERCC1-XPF in DSB repair.
A single dose of 6 Gy of IR is universally lethal to mice hypomorphic for ERCC1 (Fig. 3A), whereas 2 Gy is sublethal (data not shown). This level of sensitivity is similar to that of mice in which DNA-PKcs, an essential component of NHEJ, is deleted (41), even though ERCC1-XPF expression is reduced only 85% in the hypomorphic strain (84). This establishes an important role for ERCC1-XPF in the radioprotection of mammals. IR causes loss of intestinal villi and BM hypocellularity in ERCC1-deficient mice, as anticipated (Fig. 3). In addition, ERCC1-deficient mice display centrilobular necrosis of the liver, which is caused by occlusion of small venules with cellular debris, following IR (20). Liver pathology is life limiting for Ercc1−/− mice (72), explaining the particular hypersensitivity of Ercc1−/Δ mouse liver to IR. Mice defective in DSB repair have impaired hematopoietic stem cell function and regenerative capacity after stress (55, 64). We observed significantly reduced proliferative reserve in the BM of Ercc1−/− mice (60) and in the intestine following IR (see Fig. S3 in the supplemental material). Thus, the hypersensitivity of Ercc1−/Δ mice to IR likely is a consequence not only of increased cell death/senescence in response to unrepaired DNA damage but also reduced capacity to regenerate damaged tissues.
Although ERCC1 hypomorphic mice are equally as sensitive to IR as DNA-Pkcs−/− mice are, they are less sensitive than Ku86−/− mice (9, 56). DNA-Pkcs−/− mice do not have a spontaneous phenotype other than severe combined immunodeficiency (25, 41), whereas Ku86−/− mice display growth retardation and progeroid symptoms in addition to immunodeficiency (82). Ercc1−/− mice have a more severe phenotype than either DNA-Pkcs−/− or Ku86−/− mice, including growth retardation, liver dysfunction, renal insufficiency, epidermal atrophy, impaired hematopoiesis, sarcopenia, kyphosis, and neurodegeneration (48, 53, 60, 84), making it unlikely that the phenotype of Ercc1−/− mice can be ascribed in its entirety to a defect in DSB repair. More plausibly, it is the combined defects in multiple DNA repair pathways including global-genome and transcription-coupled NER, ICL repair, and DSB repair, documented here, that contribute to the severe phenotype of Ercc1−/− mice, XFE progeria (53), and a recently identified ERCC1 patient (33).
ERCC1-XPF participates in error-prone, Ku-independent end joining of DSBs.
ERCC1-XPF endonuclease facilitates DSB repair and protects against IR in vivo. Yet the two major mechanisms of DSB repair in mammalian cells, HR and NHEJ, are intact in ERCC1-XPF-deficient cells. Thus, the following question remains: how does this nuclease contribute to DSB repair? In yeast, the orthologs of ERCC1-XPF, Rad10-Rad1, are required for two error-prone DSB repair mechanisms: SSA (22, 32, 57) and MMEJ (45). Rad10-Rad1 are required to remove nonhomologous sequence from the 3′ ends of the break to permit DNA synthesis and/or ligation. MMEJ in yeast is independent of the HR protein Rad52 and the NHEJ protein Ku86 and critical for protection against IR (45). In rad52; yku70 strains, DSB repair is error prone, leading to large deletions and utilizing short homologies between ends (5). Recently, an alternative mechanism of end joining, distinct from NHEJ, was identified in mammals (87). This mechanism supports class switch recombination in the absence of NHEJ, is error prone, and utilizes microhomology to join broken ends, analogous to MMEJ in yeast. Linearized plasmids and I-Sce-I-induced genomic DSBs are repaired in cells deficient in NHEJ, most notably Ku86, leading to deletions and insertions and utilizing microhomology (27, 35, 71). However, the biological significance of this pathway in mammals, when NHEJ is present, remains uncertain.
Our data provide several lines of evidence that support the conclusion that ERCC1-XPF contributes to this alternative mechanism of end joining in mammalian cells. First, Ercc1−/− Ku86−/− MEFs are hypersensitive to IR relative to single-mutant cells (Fig. 4B), similar to yeast (45). In addition, Ercc1−/− Ku86−/− cells accumulate significantly more chromosomal aberrations in response to low-dose IR compared to cells defective in NHEJ only (Ku86−/− cells; Table 1). Furthermore, Ercc1−/− Ku86−/− primary MEFs proliferate poorly, surviving only to passage 5, even when cultured at 3% oxygen (Fig. 4A). These data are consistent with the conclusion that ERCC1-XPF participates in a mechanism of DSB repair that is Ku86 independent and important for protection against IR and oxidative stress, analogous to the role of Rad10-Rad1 in yeast.
The second line of evidence that supports a role for ERCC1-XPF in MMEJ comes from the in vitro end-joining assay. Joining of blunt 5′ or 3′ overhangs is not significantly reduced in Ercc1−/− or Ku86−/− cells compared to WT cells (see Fig. S5 in the supplemental material). However, joining of ends with overhangs is significantly impaired in Ercc1−/− Ku86−/− cells, suggesting loss of two distinct mechanisms of end joining in these double-mutant cells. Ercc1−/− cells are defective in joining ends that cannot be directly ligated, i.e., ends with 3′ noncomplementary overhangs but not blunt ends or ends with 5′ complementary overhangs. In the absence of ERCC1-XPF, joining of nonhomologous 3′ overhangs leads to very large deletions (Table 2), similar to yeast mutants (45). Furthermore, in Ercc1−/− cells, insertion of nucleotides at the joints is significantly reduced (Table 3), consistent with the conserved role of this nuclease in creating a 3′ end that can prime DNA synthesis (58). In addition, there is a trend toward decreased use of microhomology in end joining in Ercc1−/− cells relative to WT cells (Table 3). Deletion of Rad1 diminishes MMEJ to a greater extent in yeast but does not abrogate MMEJ, indicating the presence of redundant 3′ flap endonucleases in yeast (45) and therefore likely also in mammals. Recently, it was demonstrated that telomere fusion likely occurs via a Ku-independent end-joining mechanism (7). Thus, the participation of ERCC1-XPF in this alternative end-joining mechanism is also supported by the fact that ERCC1-XPF trims unprotected telomeric overhangs, allowing telomeric fusion via end joining (88).
In total, these data demonstrate a novel function of ERCC1-XPF nuclease in a Ku86-independent mechanism of DSB repair. Further, our data, in combination with a recent report establishing a role for ERCC1-XPF in SSA (2), indicate that Rad10-Rad1 activities in DSB repair are conserved in mammals. The data also suggest that alternative end joining of DSBs is important for protection against DSBs even when NHEJ and HR repair can occur.
Supplementary Material
Acknowledgments
This work was supported by the University of Pittsburgh Cancer Institute, NCI and NIA CA103730 and CA111525, the Pennsylvania Department of Health, and The Ellison Medical Foundation (AG-NS-0303). J.H.J.H. is supported by the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NOW), Cancer Genomics Center, the Association for International Cancer Research, and the European Commission (IP 512113), RISC-RAD contract F16R-CT-2003-508842.
We thank Richard D. Wood for careful reading of the manuscript.
Footnotes
Published ahead of print on 9 June 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adair, G. M., R. L. Rolig, D. Moore-Faver, M. Zabelshansky, J. H. Wilson, and R. S. Nairn. 2000. Role of ERCC1 in removal of long non-homologous tails during targeted homologous recombination. EMBO J. 195552-5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Minawi, A. Z., N. Saleh-Gohari, and T. Helleday. 2008. The ERCC1/XPF endonuclease is required for efficient single-strand annealing and gene conversion in mammalian cells. Nucleic Acids Res. 361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, B. S., A. T. Carpenter, and P. Ripoll. 1978. The utilization during mitotic cell division of loci controlling meiotic recombination and disjunction in Drosophila melanogaster. Genetics 90531-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biggerstaff, M., D. E. Szymkowski, and R. D. Wood. 1993. Co-correction of the ERCC1, ERCC4 and xeroderma pigmentosum group F DNA repair defects in vitro. EMBO J. 123685-3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boulton, S. J., and S. P. Jackson. 1996. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 155093-5103. [PMC free article] [PubMed] [Google Scholar]
- 6.Brugmans, L., R. Kanaar, and J. Essers. 2007. Analysis of DNA double-strand break repair pathways in mice. Mutat. Res. 61495-108. [DOI] [PubMed] [Google Scholar]
- 7.Capper, R., B. Britt-Compton, M. Tankimanova, J. Rowson, B. Letsolo, S. Man, M. Haughton, and D. M. Baird. 2007. The nature of telomere fusion and a definition of the critical telomere length in human cells. Genes Dev. 212495-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan, D. W., B. P. Chen, S. Prithivirajsingh, A. Kurimasa, M. D. Story, J. Qin, and D. J. Chen. 2002. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 162333-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Couëdel, C., K. D. Mills, M. Barchi, L. Shen, A. Olshen, R. D. Johnson, A. Nussenzweig, J. Essers, R. Kanaar, G. C. Li, F. W. Alt, and M. Jasin. 2004. Collaboration of homologous recombination and nonhomologous end-joining factors for the survival and integrity of mice and cells. Genes Dev. 181293-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Decottignies, A. 2007. Microhomology-mediated end joining in fission yeast is repressed by pku70 and relies on genes involved in homologous recombination. Genetics 1761403-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Laat, W. L., E. Appeldoorn, N. G. Jaspers, and J. H. Hoeijmakers. 1998. DNA structural elements required for ERCC1-XPF endonuclease activity. J. Biol. Chem. 2737835-7842. [DOI] [PubMed] [Google Scholar]
- 12.de Vries, A., C. T. van Oostrom, F. M. Hofhuis, P. M. Dortant, R. J. Berg, F. R. de Gruijl, P. W. Wester, C. F. van Kreijl, P. J. Capel, H. van Steeg, et al. 1995. Increased susceptibility to ultraviolet-B and carcinogens of mice lacking the DNA excision repair gene XPA. Nature 377169-173. [DOI] [PubMed] [Google Scholar]
- 13.de Waard, H., J. de Wit, J. O. Andressoo, C. T. van Oostrom, B. Riis, A. Weimann, H. E. Poulsen, H. van Steeg, J. H. Hoeijmakers, and G. T. van der Horst. 2004. Different effects of CSA and CSB deficiency on sensitivity to oxidative DNA damage. Mol. Cell. Biol. 247941-7948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dollé, M. E., R. A. Busuttil, A. M. Garcia, S. Wijnhoven, E. van Drunen, L. J. Niedernhofer, G. van der Horst, J. H. Hoeijmakers, H. van Steeg, and J. Vijg. 2006. Increased genomic instability is not a prerequisite for shortened lifespan in DNA repair deficient mice. Mutat. Res. 59622-35. [DOI] [PubMed] [Google Scholar]
- 15.Dronkert, M. L. G., H. B. Beverloo, R. D. Johnson, J. H. J. Hoeijmakers, M. Jasin, and R. Kanaar. 2000. Mouse RAD54 affects DNA double-strand break repair and sister chromatid exchange. Mol. Cell. Biol. 203147-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duan, D., Y. Yue, and J. F. Engelhardt. 2003. Consequences of DNA-dependent protein kinase catalytic subunit deficiency on recombinant adeno-associated virus genome circularization and heterodimerization in muscle tissue. J. Virol. 774751-4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dubest, S., M. E. Gallego, and C. I. White. 2002. Role of the AtRad1p endonuclease in homologous recombination in plants. EMBO Rep. 31049-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enzlin, J. H., and O. D. Scharer. 2002. The active site of the DNA repair endonuclease XPF-ERCC1 forms a highly conserved nuclease motif. EMBO J. 212045-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Essers, J., R. W. Hendriks, S. M. Swagemakers, C. Troelstra, J. de Wit, D. Bootsma, J. H. Hoeijmakers, and R. Kanaar. 1997. Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell 89195-204. [DOI] [PubMed] [Google Scholar]
- 20.Fajardo, L. F. 2005. The pathology of ionizing radiation as defined by morphologic patterns. Acta Oncol. 4413-22. [DOI] [PubMed] [Google Scholar]
- 21.Finkel, T., M. Serrano, and M. A. Blasco. 2007. The common biology of cancer and ageing. Nature 448767-774. [DOI] [PubMed] [Google Scholar]
- 22.Fishman-Lobell, J., and J. E. Haber. 1992. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science 258480-484. [DOI] [PubMed] [Google Scholar]
- 23.Friedberg, E. C., G. C. Walker, W. Siede, R. D. Wood, R. A. Schultz, and T. Ellenberger. 2006. DNA repair and mutagenesis, 2nd ed. ASM Press, Washington, DC.
- 24.Gamo, S., T. Megumi, and E. Nakashima-Tanaka. 1990. Sensitivity to ether anesthesia and to gamma-rays in mutagen-sensitive strains of Drosophila melanogaster. Mutat. Res. 2359-13. [DOI] [PubMed] [Google Scholar]
- 25.Gao, Y., J. Chaudhuri, C. Zhu, L. Davidson, D. T. Weaver, and F. W. Alt. 1998. A targeted DNA-PKcs-null mutation reveals DNA-PK-independent functions for KU in V(D)J. recombination. Immunity 9367-376. [DOI] [PubMed] [Google Scholar]
- 26.Griffin, C., H. Waard, B. Deans, and J. Thacker. 2005. The involvement of key DNA repair pathways in the formation of chromosome rearrangements in embryonic stem cells. DNA Repair 41019-1027. [DOI] [PubMed] [Google Scholar]
- 27.Guirouilh-Barbat, J., S. Huck, P. Bertrand, L. Pirzio, C. Desmaze, L. Sabatier, and B. S. Lopez. 2004. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol. Cell 14611-623. [DOI] [PubMed] [Google Scholar]
- 28.Haber, J. E. 2006. Transpositions and translocations induced by site-specific double-strand breaks in budding yeast. DNA Repair 5998-1009. [DOI] [PubMed] [Google Scholar]
- 29.Hefner, E., S. B. Preuss, and A. B. Britt. 2003. Arabidopsis mutants sensitive to gamma radiation include the homologue of the human repair gene ERCC1. J. Exp. Bot. 54669-680. [DOI] [PubMed] [Google Scholar]
- 30.Helleday, T., J. Lo, D. C. van Gent, and B. P. Engelward. 2007. DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA Repair 6923-935. [DOI] [PubMed] [Google Scholar]
- 31.Hesse, J. E., M. R. Lieber, M. Gellert, and K. Mizuuchi. 1987. Extrachromosomal DNA substrates in pre-B cells undergo inversion or deletion at immunoglobulin V-(D)-J. joining signals. Cell 49775-783. [DOI] [PubMed] [Google Scholar]
- 32.Ivanov, E. L., and J. E. Haber. 1995. RAD1 and RAD10, but not other excision repair genes, are required for double-strand break-induced recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 152245-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaspers, N. G., A. Raams, M. C. Silengo, N. Wijgers, L. J. Niedernhofer, A. R. Robinson, G. Giglia-Mari, D. Hoogstraten, W. J. Kleijer, J. H. Hoeijmakers, and W. Vermeulen. 2007. First reported patient with human ERCC1 deficiency has cerebro-oculo-facio-skeletal syndrome with a mild defect in nucleotide excision repair and severe developmental failure. Am. J. Hum. Genet. 80457-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jonnalagadda, V. S., T. Matsuguchi, and B. P. Engelward. 2005. Interstrand crosslink-induced homologous recombination carries an increased risk of deletions and insertions. DNA Repair 4594-605. [DOI] [PubMed] [Google Scholar]
- 35.Kabotyanski, E. B., L. Gomelsky, J. O. Han, T. D. Stamato, and D. B. Roth. 1998. Double-strand break repair in Ku86- and XRCC4-deficient cells. Nucleic Acids Res. 265333-5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karran, P. 2000. DNA double strand break repair in mammalian cells. Curr. Opin. Genet. Dev. 10144-150. [DOI] [PubMed] [Google Scholar]
- 37.Khanna, K. K., and S. P. Jackson. 2001. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 27247-254. [DOI] [PubMed] [Google Scholar]
- 38.Kuhfittig-Kulle, S., E. Feldmann, A. Odersky, A. Kuliczkowska, W. Goedecke, A. Eggert, and P. Pfeiffer. 2007. The mutagenic potential of non-homologous end joining in the absence of the NHEJ core factors Ku70/80, DNA-PKcs and XRCC4-LigIV. Mutagenesis 22217-233. [DOI] [PubMed] [Google Scholar]
- 39.Kuraoka, I., W. R. Kobertz, R. R. Ariza, M. Biggerstaff, J. M. Essigmann, and R. D. Wood. 2000. Repair of an interstrand DNA cross-link initiated by ERCC1-XPF repair/recombination nuclease. J. Biol. Chem. 27526632-26636. [DOI] [PubMed] [Google Scholar]
- 40.Lee, K., and S. E. Lee. 2007. Saccharomyces cerevisiae Sae2- and Tel1-dependent single-strand DNA formation at DNA break promotes microhomology-mediated end joining. Genetics 1762003-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li, X.-L., S.-R. Shen, S. Wang, H.-H. Ouyang, and G. C. Li. 2002. Restoration of T cell-specific V(D)J. recombination in DNA-PKcs−/− mice by ionizing radiation: the effects on survival, development, and tumorigenesis. Acta Biochim. Biophys. Sin. 34149-157. [PubMed] [Google Scholar]
- 42.Liang, F., M. Han, P. J. Romanienko, and M. Jasin. 1998. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc. Natl. Acad. Sci. USA 955172-5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lim, D.-S., H. Vogel, D. M. Willerford, A. T. Sands, K. A. Platt, and P. Hasty. 2000. Analysis of ku80-mutant mice and cells with deficient levels of p53. Mol. Cell. Biol. 203772-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Little, J. W. 1981. Lambda exonuclease. Gene Amplif. Anal. 2135-145. [PubMed] [Google Scholar]
- 45.Ma, J.-L., E. M. Kim, J. E. Haber, and S. E. Lee. 2003. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 238820-8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsumura, Y., C. Nishigori, T. Yagi, S. Imamura, and H. Takebe. 1998. Characterization of molecular defects in xeroderma pigmentosum group F in relation to its clinically mild symptoms. Hum. Mol. Genet. 7969-974. [DOI] [PubMed] [Google Scholar]
- 47.McHugh, P. J., V. J. Spanswick, and J. A. Hartley. 2001. Repair of DNA interstrand crosslinks: molecular mechanisms and clinical relevance. Lancet Oncol. 2483-490. [DOI] [PubMed] [Google Scholar]
- 48.McWhir, J., J. Selfridge, D. J. Harrison, S. Squires, and D. W. Melton. 1993. Mice with DNA repair gene (ERCC-1) deficiency have elevated levels of p53, liver nuclear abnormalities and die before weaning. Nat. Genet. 5217-224. [DOI] [PubMed] [Google Scholar]
- 49.Mills, K. D., D. O. Ferguson, J. Essers, M. Eckersdorff, R. Kanaar, and F. W. Alt. 2004. Rad54 and DNA ligase IV cooperate to maintain mammalian chromatid stability. Genes Dev. 181283-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murray, D., L. Vallee-Lucic, E. Rosenberg, and B. Andersson. 2002. Sensitivity of nucleotide excision repair-deficient human cells to ionizing radiation and cyclophosphamide. Anticancer Res. 2221-26. [PubMed] [Google Scholar]
- 51.Newman, M., J. Murray-Rust, J. Lally, J. Rudolf, A. Fadden, P. P. Knowles, M. F. White, and N. Q. McDonald. 2005. Structure of an XPF endonuclease with and without DNA suggests a model for substrate recognition. EMBO J. 24895-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Niedernhofer, L. J., J. Essers, G. Weeda, B. Beverloo, J. de Wit, M. Muijtjens, H. Odijk, J. H. Hoeijmakers, and R. Kanaar. 2001. The structure-specific endonuclease Ercc1-Xpf is required for targeted gene replacement in embryonic stem cells. EMBO J. 206540-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Niedernhofer, L. J., G. A. Garinis, A. Raams, A. S. Lalai, A. R. Robinson, E. Appeldoorn, H. Odijk, R. Oostendorp, A. Ahmad, W. van Leeuwen, A. F. Theil, W. Vermeulen, G. T. van der Horst, P. Meinecke, W. J. Kleijer, J. Vijg, N. G. Jaspers, and J. H. Hoeijmakers. 2006. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature 4441038-1043. [DOI] [PubMed] [Google Scholar]
- 54.Niedernhofer, L. J., H. Odijk, M. Budzowska, E. van Drunen, A. Maas, A. F. Theil, J. de Wit, N. G. Jaspers, H. B. Beverloo, J. H. Hoeijmakers, and R. Kanaar. 2004. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol. Cell. Biol. 245776-5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nijnik, A., L. Woodbine, C. Marchetti, S. Dawson, T. Lambe, C. Liu, N. P. Rodrigues, T. L. Crockford, E. Cabuy, A. Vindigni, T. Enver, J. I. Bell, P. Slijepcevic, C. C. Goodnow, P. A. Jeggo, and R. J. Cornall. 2007. DNA repair is limiting for haematopoietic stem cells during ageing. Nature 447686-690. [DOI] [PubMed] [Google Scholar]
- 56.Nussenzweig, A., K. Sokol, P. Burgman, L. Li, and G. C. Li. 1997. Hypersensitivity of Ku80-deficient cell lines and mice to DNA damage: the effects of ionizing radiation on growth, survival, and development. Proc. Natl. Acad. Sci. USA 9413588-13593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pâques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pâques, F., and J. E. Haber. 1997. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 176765-6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parrinello, S., E. Samper, A. Krtolica, J. Goldstein, S. Melov, and J. Campisi. 2003. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat. Cell Biol. 5741-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prasher, J. M., A. S. Lalai, C. Heijmans-Antonissen, R. E. Ploemacher, J. H. Hoeijmakers, I. P. Touw, and L. J. Niedernhofer. 2005. Reduced hematopoietic reserves in DNA interstrand crosslink repair-deficient Ercc1−/− mice. EMBO J. 24861-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Radford, S. J., E. Goley, K. Baxter, S. McMahan, and J. Sekelsky. 2005. Drosophila ERCC1 is required for a subset of MEI-9-dependent meiotic crossovers. Genetics 1701737-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Richardson, C., M. E. Moynahan, and M. Jasin. 1998. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 123831-3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rogakou, E. P., C. Boon, C. Redon, and W. M. Bonner. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146905-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rossi, D. J., D. Bryder, J. Seita, A. Nussenzweig, J. Hoeijmakers, and I. L. Weissman. 2007. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature 447725-729. [DOI] [PubMed] [Google Scholar]
- 65.Roth, D. B., and J. H. Wilson. 1986. Nonhomologous recombination in mammalian cells: role for short sequence homologies in the joining reaction. Mol. Cell. Biol. 64295-4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rothkamm, K., I. Kruger, L. H. Thompson, and M. Lobrich. 2003. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell. Biol. 235706-5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rünger, T. M., and K. H. Kraemer. 1989. Joining of linear plasmid DNA is reduced and error-prone in Bloom's syndrome cells. EMBO J. 81419-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sargent, R. G., J. L. Meservy, B. D. Perkins, A. E. Kilburn, Z. Intody, G. M. Adair, R. S. Nairn, and J. H. Wilson. 2000. Role of the nucleotide excision repair gene ERCC1 in formation of recombination-dependent rearrangements in mammalian cells. Nucleic Acids Res. 283771-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Scholzen, T., and J. Gerdes. 2000. The Ki-67 protein: from the known and the unknown. J. Cell Physiol. 182311-322. [DOI] [PubMed] [Google Scholar]
- 70.Schrader, C. E., J. Vardo, E. Linehan, M. Z. Twarog, L. J. Niedernhofer, J. H. Hoeijmakers, and J. Stavnezer. 2004. Deletion of the nucleotide excision repair gene Ercc1 reduces immunoglobulin class switching and alters mutations near switch recombination junctions. J. Exp. Med. 200321-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Secretan, M. B., Z. Scuric, J. Oshima, A. J. Bishop, N. G. Howlett, D. Yau, and R. H. Schiestl. 2004. Effect of Ku86 and DNA-PKcs deficiency on non-homologous end-joining and homologous recombination using a transient transfection assay. Mutat. Res. 554351-364. [DOI] [PubMed] [Google Scholar]
- 72.Selfridge, J., K. T. Hsia, N. J. Redhead, and D. W. Melton. 2001. Correction of liver dysfunction in DNA repair-deficient mice with an ERCC1 transgene. Nucleic Acids Res. 294541-4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sgouros, J., P. H. Gaillard, and R. D. Wood. 1999. A relationship between a DNA-repair/recombination nuclease family and archaeal helicases. Trends Biochem. Sci. 2495-97. [DOI] [PubMed] [Google Scholar]
- 74.Sijbers, A. M., W. L. de Laat, R. R. Ariza, M. Biggerstaff, Y. F. Wei, J. G. Moggs, K. C. Carter, B. K. Shell, E. Evans, M. C. de Jong, S. Rademakers, J. de Rooij, N. G. Jaspers, J. H. Hoeijmakers, and R. D. Wood. 1996. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell 86811-822. [DOI] [PubMed] [Google Scholar]
- 75.Sonoda, E., M. S. Sasaki, C. Morrison, Y. Yamaguchi-Iwai, M. Takata, and S. Takeda. 1999. Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol. Cell. Biol. 195166-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stark, J. M., A. J. Pierce, J. Oh, A. Pastink, and M. Jasin. 2004. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol. Cell. Biol. 249305-9316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Takata, M., M. S. Sasaki, E. Sonoda, C. Morrison, M. Hashimoto, H. Utsumi, Y. Yamaguchi-Iwai, A. Shinohara, and S. Takeda. 1998. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 175497-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tian, M., and F. W. Alt. 2000. Transcription-induced cleavage of immunoglobulin switch regions by nucleotide excision repair nucleases in vitro. J. Biol. Chem. 27524163-24172. [DOI] [PubMed] [Google Scholar]
- 79.Tian, M., R. Shinkura, N. Shinkura, and F. W. Alt. 2004. Growth retardation, early death, and DNA repair defects in mice deficient for the nucleotide excision repair enzyme XPF. Mol. Cell. Biol. 241200-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tsodikov, O. V., J. H. Enzlin, O. D. Scharer, and T. Ellenberger. 2005. Crystal structure and DNA binding functions of ERCC1, a subunit of the DNA structure-specific endonuclease XPF-ERCC1. Proc. Natl. Acad. Sci. USA 10211236-11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.van Heemst, D., L. Brugmans, N. S. Verkaik, and D. C. van Gent. 2004. End-joining of blunt DNA double-strand breaks in mammalian fibroblasts is precise and requires DNA-PK and XRCC4. DNA Repair 343-50. [DOI] [PubMed] [Google Scholar]
- 82.Vogel, H., D. S. Lim, G. Karsenty, M. Finegold, and P. Hasty. 1999. Deletion of Ku86 causes early onset of senescence in mice. Proc. Natl. Acad. Sci. USA 9610770-10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Watanabe, N., H. Odagiri, E. Totsuka, and M. Sasaki. 2004. A new method to immortalize primary cultured rat hepatocytes. Transplant Proc. 362457-2461. [DOI] [PubMed] [Google Scholar]
- 84.Weeda, G., I. Donker, J. de Wit, H. Morreau, R. Janssens, C. J. Vissers, A. Nigg, H. van Steeg, D. Bootsma, and J. H. Hoeijmakers. 1997. Disruption of mouse ERCC1 results in a novel repair syndrome with growth failure, nuclear abnormalities and senescence. Curr. Biol. 7427-439. [DOI] [PubMed] [Google Scholar]
- 85.Weinstock, D. M., and M. Jasin. 2006. Alternative pathways for the repair of RAG-induced DNA breaks. Mol. Cell. Biol. 26131-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wood, R. D., H. J. Burki, M. Hughes, and A. Poley. 1983. Radiation-induced lethality and mutation in a repair-deficient CHO cell line. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 43207-213. [DOI] [PubMed] [Google Scholar]
- 87.Yan, C. T., C. Boboila, E. K. Souza, S. Franco, T. R. Hickernell, M. Murphy, S. Gumaste, M. Geyer, A. A. Zarrin, J. P. Manis, K. Rajewsky, and F. W. Alt. 2007. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature 449478-482. [DOI] [PubMed] [Google Scholar]
- 88.Zhu, X. D., L. Niedernhofer, B. Kuster, M. Mann, J. H. Hoeijmakers, and T. de Lange. 2003. ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol. Cell 121489-1498. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.