Abstract
The Sry-related high-mobility-group box transcription factor Sox9 recruits the redundant L-Sox5 and Sox6 proteins to effect chondrogenesis, but the mode of action of the trio remains unclear. We identify here a highly conserved 359-bp sequence 10 kb upstream of the Agc1 gene for aggrecan, a most essential cartilage proteoglycan and key marker of chondrocyte differentiation. This sequence directs expression of a minimal promoter in both embryonic and adult cartilage in transgenic mice, in a manner that matches Agc1 expression. The chondrogenic trio is required and sufficient to mediate the activity of this enhancer. It acts directly, Sox9 binding to a critical cis-acting element and L-Sox5/Sox6 binding to three additional elements, which are cooperatively needed. Upon binding to their specific sites, L-Sox5/Sox6 increases the efficiency of Sox9 binding to its own recognition site and thereby robustly potentiates the ability of Sox9 to activate the enhancer. L-Sox5/Sox6 similarly secures Sox9 binding to Col2a1 (encoding collagen-2) and other cartilage-specific enhancers. This study thus uncovers critical cis-acting elements and transcription factors driving Agc1 expression in cartilage and increases understanding of the mode of action of the chondrogenic Sox trio.
Cartilage is an essential connective tissue. It constitutes the primary skeleton of the vertebrate embryo and drives body growth in highly organized growth plates. Most cartilage is progressively replaced by bone during development, but some is kept through adulthood to support the respiratory and auditory tracts and to ensure deformation-free and friction-free movement of bones in synovial joints. A very abundant extracellular matrix characterizes cartilage. Its tensile strength is provided by a fibrillar scaffold made of up to 95% collagen-2 (14), and its remarkable resiliency is ensured by a gel of proteoglycans, of which aggrecan is the most profuse and specific type (40). Chondrocytes produce and maintain this matrix. Their differentiation from mesenchymal cells is recognized early by formation of condensations that turn on the collagen-2 gene (Col2a1) (28). Upregulation of Col2a1 and activation of the aggrecan gene (Agc1) and other cartilage matrix genes typify overt chondrogenesis. As chondrocytes proceed to terminal maturation in growth plates, they downregulate Col2a1 and Agc1, activate new cartilage matrix genes, and undergo hypertrophy. Permanent chondrocytes, in contrast, express a fairly stable phenotype through life, maintaining high expression of Agc1 but hardly expressing Col2a1.
Deciphering the transcriptional mechanisms underlying chondrocyte differentiation is key to understanding many types of cartilage diseases, such as chondrodysplasias, characterized by malformations of skeletal elements (38), and osteoarthritis, characterized by progressive, irreversible loss of adult articular cartilage (17). Disease-causing mutations have been found in the genes for cartilage regulatory factors and matrix components, including Agc1. For instance, mouse cartilage matrix deficiency is caused by a nonsense mutation in Agc1 (21, 50). While heterozygotes develop mild dwarfism and spinal degeneration (51), homozygotes die at birth with severe dwarfism (39). AGC1 mutations and polymorphisms have been associated in humans with spondyloepiphyseal dysplasia, osteoarthritis, and intervertebral disc degeneration (16, 19, 41). Moreover, most forms of arthritis feature early and severe loss of aggrecan, due to both increased degradation and decreased production (17).
L-Sox5, Sox6, and Sox9 form a trio of transcription factors needed and sufficient for chondrogenesis (1, 28). The Sox family specifies cell fate and differentiation in many lineages (24). Its signature is an Sry-related high-mobility-group (HMG) box DNA-binding domain. This domain preferentially binds the C A/T T T G A/T A/T sequence in vitro (8, 32). It contacts and bends the minor groove of DNA and may thereby fulfill a key role in the organization of transcriptional complexes. L-Sox5, a long product of the Sox5 gene, is highly similar to Sox6. The two proteins share 50% identity with Sox9 in the Sox domain but no other identity. They feature a coiled-coil DNA-independent homodimerization domain but no other known functional domain, whereas Sox9 features a DNA-dependent homodimerization domain and a potent transactivation domain. Sox9 is turned on in chondrogenic cells at a pluripotent mesenchymal stage. It remains expressed in growth plate chondrocytes until prehypertrophy and in permanent chondrocytes through life (35, 43, 53). Sox5 and Sox6 are expressed downstream of Sox9 from the prechondrocyte stage and later remain coexpressed with Sox9 (27). Knockout studies have shown that Sox9 is required for chondrogenesis and that L-Sox5 and Sox6 promote chondrogenesis (2, 3, 5, 45, 46). Sox9 ensures cell survival in precartilaginous condensations and is needed to activate Col2a1, Agc1, and other cartilage genes. L-Sox5 and Sox6 act redundantly and are therefore referred to as L-Sox5/Sox6 hereafter. They allow precartilaginous condensations to undergo overt chondrogenesis and are needed to upregulate Col2a1 and significantly express Agc1 and other cartilage genes. While these studies proved that the Sox trio is needed for chondrogenesis, a forced-expression study has proven that it is also sufficient (20).
Col2a1 was the first gene proposed as a direct target of the trio. Sox9 powerfully activates a 48-bp cartilage-specific enhancer located in its first intron (4, 23, 24, 29, 33, 35, 54), and L-Sox5/Sox6 potentiates the activity of Sox9 (27). The enhancer has no consensus Sox site but has four sites with five or six consensus nucleotides that are all needed for enhancer activity. Sox9 binds the most distal pair of them, and L-Sox5/Sox6 contacts each of them in vitro. It remains unknown, however, whether the proteins bind the enhancer in vivo and how they cooperatively activate it. Sox9 binds and activates enhancers in other cartilage genes, such as Col11a2, CD-RAP, Col9a1, Comp, Cmp, and Crtl1, but a role for L-Sox5/Sox6 in the activation of these enhancers has been suggested only for Cmp and Crtl1 (6, 7, 15, 22, 31, 37, 52).
Despite the crucial roles of aggrecan, little is known about control of its gene. Agc1 has 18 exons spread over 60 kb in the rat genome, and this structure is evolutionarily conserved (10, 30, 47, 48, 49). Sekiya and colleagues showed that Sox9 overexpression results in threefold upregulation of a reporter containing Agc1 sequences between 3 kb upstream and 5 kb downstream of the transcription start site in transiently transfected chondrocytes, but they did not show whether this gene segment binds Sox9 and is sufficient to drive cartilage expression in vivo (44). Doege and colleagues showed that a 4.7-kb segment located 10 kb upstream of the Agc1 transcription start site could activate a 3.4-kb promoter in transiently transfected chondrocytes (9). This segment could drive cartilage-specific expression in transgenic mouse embryos, but weakly and only in hypertrophic chondrocytes in a few cartilage elements. Hence, this 4.7-kb region did not appear sufficient to confer on Agc1 its characteristic expression in cartilage.
We show here that this 4.7-kb region contains a highly conserved enhancer that directs the robust, specific expression of Agc1 in embryonic and adult cartilage and that this enhancer is a direct target of the Sox trio. Interestingly, Sox9 binding to this enhancer occurs only in the presence of L-Sox5/Sox6, and a similar mechanism applies to other cartilage enhancers. Our study therefore identifies key cis-acting elements in Agc1 and provides novel insights into the mechanism of action of the trio.
MATERIALS AND METHODS
Genome sequence analysis.
The rat genome sequence encompassing Agc1 was downloaded from the NCBI website (NW 047560; gene identification: 58968). Sequences conserved in multiple genomes were identified through discontiguous Megablast searches in the whole-genome shotgun sequence database. Sequences were aligned with ClustalX software (http://www.ebi.ac.uk/clustalw/).
Reporter genes.
Agc1 elements were amplified by PCR of 129 mouse genomic DNA. Forward and reverse PCR primers were designed with BamHI and BglII restriction sites, respectively, at the 5′ ends. PCR products were cloned into pCR4-TOPO (Invitrogen), and sequences were verified. Mutations were introduced by PCR of wild-type DNA with appropriate primers. Tandem copies were obtained by ligation of BamHI/BglII-restricted monomers. Monomers and multimers were cloned into a reporter plasmid featuring a −89 to +6 Col2a1 promoter (pCol2-Luc) (30) or a −596 to +90 Agc1 promoter (pAgc1-Luc). The Agc1-lacZ transgene was generated by cloning four copies of A1 into a lacZ reporter containing the −309 to +308 Col2a1 sequences (54). It was excised from the plasmid backbone by digestion with Asp718.
Mice.
Mice were used according to federal guidelines and as approved by the Cleveland Clinic Institutional Animal Care and Use Committee. Agc1-lacZ transgenic founders were generated using standard procedures and identified by lacZ PCR. They were bred to wild-type 129 × B6 mice to generate transgenic lines and to mice harboring Sox5 and Sox6 conditional null alleles (11, 13).
X-Gal staining.
Whole embryos were stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) as described previously (18). They were then cleared in methanol followed by 1:1 benzylbenzoate-benzylalcohol or were embedded in paraffin, sectioned at 20 μm, and counterstained with nuclear fast red. As indicated in the figures, some embryos were stained with X-Gal on frozen sections. They were fixed in 4% paraformaldehyde (PFA) for 1 h, and frozen sections were postfixed with 0.2% PFA. X-Gal staining of knees, ribs, and vertebrae of adult mice was performed upon fixation in 2% PFA, 0.2% glutaraldehyde, and 0.1% Tween 20 in phosphate-buffered saline for 2 h, followed by three washes in phosphate-buffered saline-Tween. Stained samples were demineralized in 0.5 M EDTA, pH 8.0, at 4°C for 3 weeks before processing for paraffin sections.
Cell culture, adenovirus infection, and transfection.
Mouse primary chondrocytes (25) were infected with an adenoviral vector expressing Cre recombinase (AdeCre) or green fluorescent protein (AdeGFP) at 100 PFU/cell (University of Iowa) 12 h after plating. Primary skin fibroblasts (25) and rat chondrosarcoma (RCS) (33), BALB/3T3, HEK-293, and Cos-1 cells were cultured under standard conditions. Fugene6 (Roche) was used for transfection after treatment of RCS cells with collagenase D (33) and of other cells with trypsin-EDTA (12). Unless otherwise indicated, DNA included 300 ng pSV2-betaGal or pGL3 (Promega) control plasmid, 600 ng reporter plasmid, and 200 ng expression plasmid for mouse L-Sox5 or Sox6 or for human SOX9 (27). Cell extracts were assayed 24 h after transfection or 24 to 96 h after adenovirus infection. Transient reporter activities were normalized for transfection efficiency. Transgene activities were normalized for protein concentration in chondrocytes (Dc protein assay kit; Bio-Rad) and for transfection efficiency in fibroblasts. Reporter and transgene activities are presented in relative light units as the averages with standard deviations of measurements obtained for triplicate cultures in one representative experiment.
Antibodies and Western blotting.
Rabbit polyclonal antibodies against Sox proteins were custom-made using peptides in the protein N-terminal region and were purified by peptide affinity chromatography (Invitrogen). Specificity was ascertained by Western blotting (13) (data not shown). Western blots were prepared with cell lysates made in sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer. They were hybridized with custom-made antibodies, Sox9 rabbit polyclonal antibodies (AB5535; Chemicon), or a beta-actin mouse monoclonal antibody (sc-47778; Santa Cruz Biotechnology). Signals were detected by enhanced chemiluminescence (Amersham Biosciences). Protein migration levels were ascertained with Mr standards (Bio-Rad).
ChIP.
Chromatin immunoprecipitation (ChIP) was performed using an EZ-ChIP kit (Upstate Biotechnology). Antibodies consisted of 2 μg Sox antibodies, RNA polymerase II antibodies (Upstate Biotechnology), or normal rabbit immunoglobulin G (IgG; Santa Cruz Biotechnology). PCR primers were designed to match the mouse and rat sequences. They flanked the Agc1 enhancer (forward, ATGTGACCTGGGTCCAGAATGGAA; reverse, GAAATTCCTTTCCCTGCAACGCCT), Col2a1 enhancer (forward, AGGCTTGTTTGCGTTGAGGGATTG; reverse, GCCGTGCGGCATAAGTGATTCTTT), Agc1 promoter (forward, CACACACACACACACTGGGTTTTG; reverse, ACTGGGTTTGGATTTTTCCCTG), or Col11a2 enhancer (forward, AGATAACTATATGGGCGGTGGGCA; reverse, AAGCCAGGGTGAAAGCTGAAGTGA). Qualitative PCR was done with 1 cycle at 94°C for 3 min; 35 cycles at 94°C for 30 s, 57°C for 1 min, and 72°C for 1 min; and 1 cycle at 72°C for 7 min. Products were electrophoresed on a 2% agarose gel. Quantitative PCR was performed with 10-fold serial dilutions of DNA on an ABI Prism 7900HT sequence detection system (Applied Biosystems) with PCR Master Mix for Sybr green assays (Applied Biosystems). Parameters were 1 cycle at 95°C for 10 min and 40 cycles at 95°C for 15 s and 60°C for 1 min. Data were calculated as the percentage of DNA input. They are presented as the averages with standard deviations of triplicate assays for one representative experiment.
EMSA.
RCS nuclear extracts, HEK-293 cell extracts, and the 2HMG probe were prepared and tested in electrophoretic mobility shift assays (EMSA) as described previously (12, 26). Briefly, probes were double-stranded oligonucleotides labeled with [32P]dCTP by Klenow fragment fill-in. All probe-specific activities were equalized by dilution with unlabeled probe. EMSA reaction mixtures contained 10 fmol probe, 1 to 3 μl extracts, and 1 μg poly(dG-dC)·poly(dG-dC) (Amersham) in 20 μl buffer. Supershifts were obtained using 1 μl (0.1 to 0.4 μg) Sox antibodies.
Image acquisition and manipulation.
Embryos were photographed using a Nikon SMZ1500 microscope equipped with C-W10×20 lenses and a Spot Insight digital camera. Sections were analyzed with an Olympus BX50 microscope equipped with Olympus Uplanapo 10×, 0.40-numerical-aperture (NA) and Olympus Uplanapo 20×, 0.70-NA infinity-corrected (effective NA, 0.17) lenses. Images were captured with a Polaroid DMC2 digital camera. Figures were composed using Adobe Photoshop 7.0 software.
RESULTS
Agc1 features three highly conserved nonexonic regions: A1, A2, and A3.
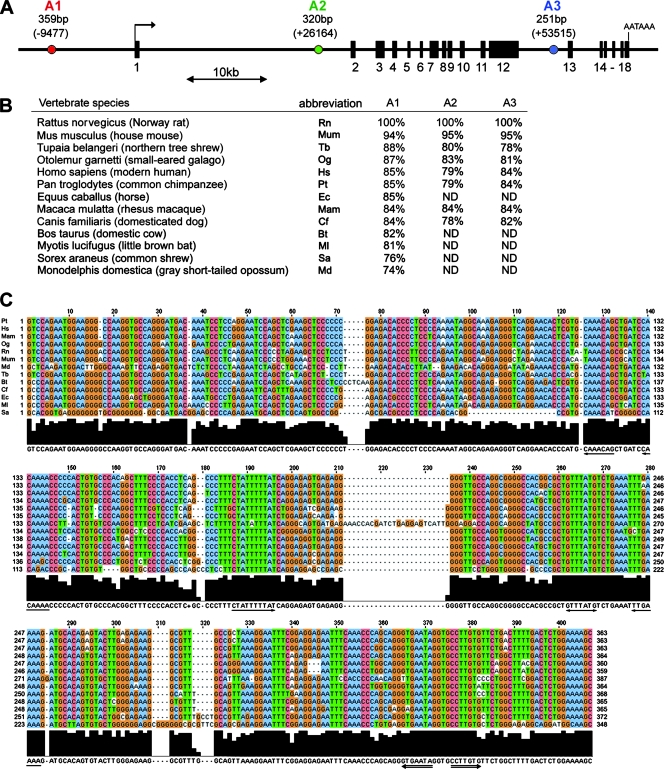
We started a search for Agc1 cis-acting elements based on the hypothesis that these elements have been conserved evolutionarily. We downloaded 91 kb of rat genome sequence, from 20 kb 5′ of the transcription start site to 10 kb 3′ of the stop site. We split this DNA into 5-kb overlapping segments and used these segments to probe the NCBI whole-genome shotgun sequence database. We found three sequences that were nonexonic, ≥100 bp, and ≥75% identical in many mammalian genomes (Fig. 1A and B). We called them A1, A2, and A3. A1 is the most conserved and longest sequence. Its 359 bp start 9,477 bp upstream of the Agc1 transcription start site. A2 is 320 bp and starts at +26164 in the first intron. A3 is 251 bp and starts at +53515 in the 12th intron. Besides its high conservation, each sequence also showed stretches of 100% identity (Fig. 1C and data not shown). Agc1 also showed ≥73% conservation in the −596 to +90 promoter region but much less anywhere else (data not shown).
FIG. 1.
Identification of highly conserved nonexonic sequences in Agc1. (A) Diagram of the rat Agc1 gene showing the locations (in parentheses) and sizes of A1, A2, and A3. The horizontal line represents nonexonic sequences. Black boxes with numbers underneath represent exons. The arrow symbolizes the start of transcription, and AATAAA indicates the polyadenylation site. (B) Percentages of identity of A elements in mammalian genomes. ND, not determined. (C) ClustalW alignment of A1 sequences. The black histogram schematizes the degree of conservation of each nucleotide. The consensus sequence derived from the alignment is shown below the histogram. The L-Sox5/Sox6 and L-Sox5/Sox6/Sox9 binding sites identified in Fig. 7 are underlined with single-shaft and double-shaft arrows, respectively.
A1 is a potent enhancer in transiently transfected chondrocytes.
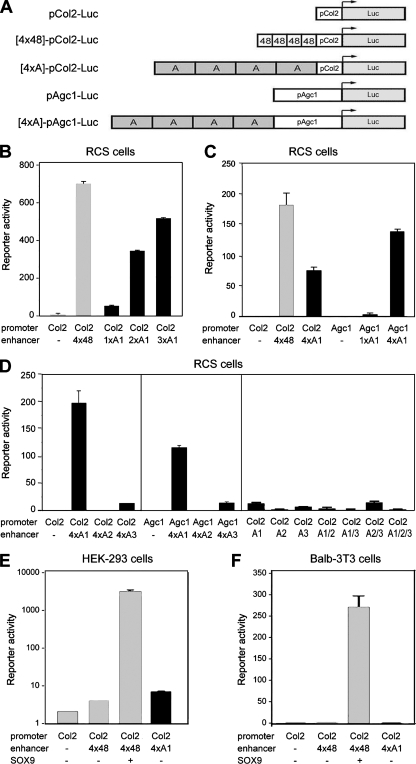
We next determined whether the mouse A1, A2, and A3 elements have transcriptional activity. We constructed luciferase reporters featuring up to four copies of these elements (1×A to 4×A) upstream of an Agc1 −596 to +90 promoter (pAgc1) or upstream of a minimal, exogenous promoter (Fig. 2A). We chose the Col2a1 −89 to +6 promoter (pCol2) as the latter because it contains only a TATA box and a GC-rich element as recognizable motifs and it is virtually silent, with no cell type specificity, in transient transfection of various cell types and in transgenic mice (26, 29). We first tested these constructs in transient transfection of well-differentiated RCS cells (33). The Col2a1 minimal promoter was activated weakly but significantly by 1×A1, and it was activated by 2×A1, 3×A1, and 4×A1 almost as powerfully as by four copies of the 48-bp Col2a1 enhancer (4×48) (Fig. 2B and C). The Agc1 promoter was inactive when tested alone but significantly activated by 1×A1 and powerfully activated by 4×A1 (Fig. 2C). Like typical enhancers, A1 was similarly active in forward and reverse orientations (data not shown). A2 and A3 were inactive in RCS cells, whether tested as four like copies or in any combination of each with A1 (Fig. 2D). We then tested various types of nonchondrocytic cells, including human embryonic kidney HEK-293 cells and mouse mesenchymal BALB/3T3 cells, and found that none of the A elements was active in any of these cells (Fig. 2E and F and data not shown). We concluded that A1, but neither A2 nor A3, might be a chondrocyte-specific enhancer.
FIG. 2.
Enhancer activity of A1 in transiently transfected cells. (A) Schematics of reporter constructs. Each construct contains a −89 to +6 Col2a1 promoter (pCol2) or −596 to +90 Agc1 promoter (pAgc1) driving the firefly luciferase gene (Luc). Some constructs contain up to four tandem copies of the Col2a1 48-bp enhancer (4×48) or Agc1 A element (4×A). (B to F) Transient transfection of RCS, HEK-293, and BALB/3T3 cells with reporter constructs containing the indicated promoter and enhancer elements. Activation of the Col2a1 enhancer construct by SOX9 is shown as positive control in panels E and F.
A1 drives gene expression in transgenic mice with the characteristic spatial and temporal pattern of Agc1.
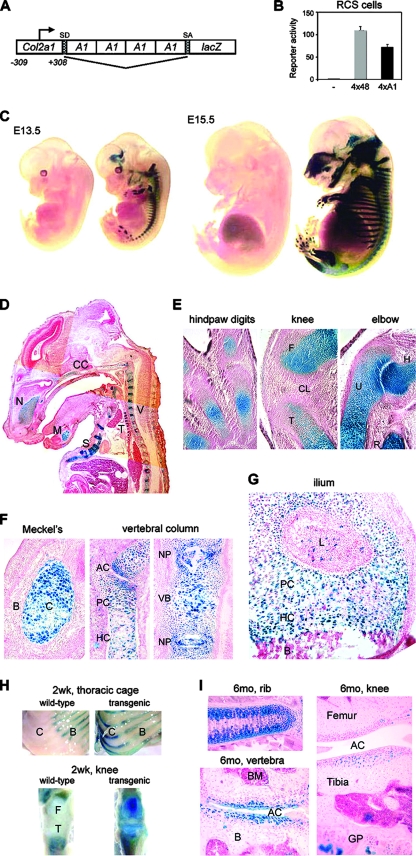
We then tested the activity of A1 in vivo. We made an Agc1-lacZ transgene by cloning 4×A1 into the intron of p309Col2a1lacZ (Fig. 3A). The latter reporter contained the Col2a1 sequence from −309 to +308, i.e., the proximal promoter, first exon (with the translation initiation site mutated into CTG), and 70 bp of the first intron (not including enhancer sequences) (54). It also featured a splice acceptor downstream of the intron segment, followed by lacZ and a polyadenylation site. It was chosen because it is inactive in transgenic mice. When tested in transient transfection, Agc1-lacZ was almost as active in RCS cells as a similar reporter containing the 4×48 Col2a1 enhancer (Fig. 3B), but it was inactive in nonchondrocytic cells (data not shown). We obtained nine Agc1-lacZ transgenic mouse founders and derived a mouse line from each one. One line failed to express the transgene, but all others showed the same expression pattern (data not shown). Embryos stained with the chromogenic beta-galactosidase substrate X-Gal revealed transgene expression in all craniofacial, axial, and appendicular cartilage elements (Fig. 3C). Embryo sections showed expression in differentiated articular and growth plate chondrocytes and in notochord-derived nucleus pulposus cells (which also express Agc1; Fig. 3D to G). As expected, no staining derived from the lacZ product was seen in noncartilaginous tissues and in prechondrocytes, the perichondrium, and presumptive joints. Hypertrophic chondrocytes were still X-Gal positive, even though they were expected to turn off the transgene concomitantly with Agc1. This result is likely due to the high stability of the Escherichia coli beta-galactosidase. We also tested the activity of the Agc1-lacZ transgene postnatally. Interestingly, we found that it was highly expressed in all growth plate and articular cartilage structures of actively growing 2-week-old pups (Fig. 3H) and that it was still as active in the same structures in 6-month-old adult mice (Fig. 3I). We concluded that A1 is sufficient to confer on a transgene the typical expression pattern of Agc1 in both developing and adult cartilage.
FIG. 3.
Agc1-lacZ expression in transgenic mice. (A) Schematic of Agc1-lacZ. Four tandem copies of A1 were inserted in an intron downstream of the −309 to +308 Col2a1 sequence and upstream of lacZ. The intron (bent line) is flanked with splice donor (SD) and acceptor (SA) sites. (B) Transient transfection of RCS cells to compare the activity of Agc1-lacZ to those of similar reporters featuring no enhancer (−) or the 4×48 Col2a1 enhancer. (C) X-Gal staining of wild-type (left) and Agc1-lacZ (right) littermates at embryonic day 13.5 (E13.5) and E15.5. (D) Frozen section through an E15.5 Agc1-lacZ embryo stained with X-Gal. CC, chondrocranium; N, nasal cartilage; M, Meckel's cartilage; S, sternal cartilage; T, tracheal cartilage; V, vertebral cartilage. (E) Paraffin sections of various skeletal elements in E14.5 Agc1-lacZ embryos stained with X-Gal. The digits, which are the least developed, show chondrocytes well differentiated and positive for X-Gal only in the cartilage core regions. The knee, which is more advanced, shows positive staining in most chondrocytes of the femur (F) and tibia (T) but not in the cruciate ligament (CL) and in other cells. The elbow, which is the most advanced, shows positive staining throughout humerus (H), ulna (U), and radius (R) cartilage. (F) Frozen sections of an E15.5 Agc1-lacZ embryo stained with X-Gal. (Left) Positive cells in the Meckel's cartilage but not in surrounding bone (B) and other tissues. (Middle) X-Gal-positive articular (AC), proliferating (PC), and hypertrophic (HC) chondrocytes in vertebral arches. (Right) X-Gal-positive cells in vertebral body (VB) cartilage and intervertebral disc nucleus pulposus (NP). (G) Frozen sections of the distal ilium growth plate of a newborn Agc1-lacZ mouse showing positive X-Gal staining in most chondrocytes. A few bone (B) and ligament (L) cells are X-Gal positive as a result of endogenous beta-galactosidase, as proven by the fact that the same result was obtained with nontransgenic littermates (data not shown). (H) Thoracic cage and knees of 2-week-old wild-type and Agc1-lacZ pups showing X-Gal staining derived from the lacZ transgene in the cartilaginous, ventral portion of the ribs (C) and endogenous beta-galactosidase activity in the ossified, dorsal portion of the ribs (B). F, femur; T, tibia. (I) Histology sections through a rib, vertebral joint, and knee of a 6-month-old Agc1-lacZ mouse, demonstrating positive X-Gal staining in articular (AC) and growth plate (GP) chondrocytes but not in bone marrow (BM).
The Sox trio is needed and sufficient to activate A1.
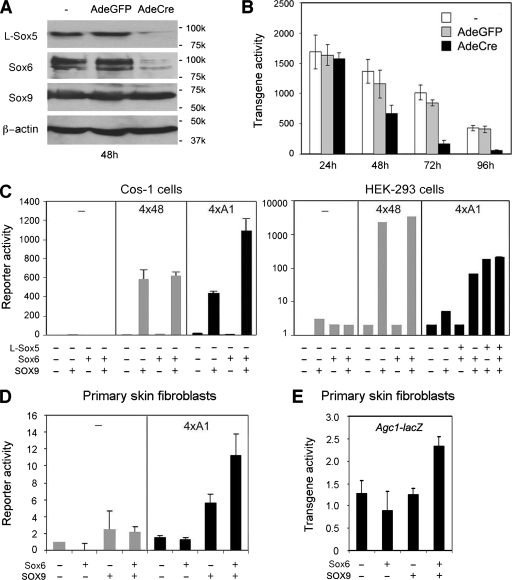
We next asked whether the Sox trio controls A1. We first isolated primary chondrocytes from Sox5fl/fl Sox6fl/fl pups, i.e., pups harboring Sox5 and Sox6 conditional null alleles, and from Sox5fl/fl Sox6fl/f Agc1-lacZ littermates. We left the cells untreated or infected them with AdeCre or AdeGFP. As expected, AdeCre-treated cells lost most of the L-Sox5 and Sox6 proteins in 48 h but maintained a normal level of Sox9 protein for at least 96 h (Fig. 4A and data not shown). All cultures actively expressed Agc1-lacZ 24 h after infection (Fig. 4B). They progressively decreased their transgene activity later on, likely reflecting their typical dedifferentiation in monolayer culture (25). This decrease occurred, however, much faster in mutant cells, indicating that L-Sox5/Sox6 is required for A1 activity.
FIG. 4.
A1 activation by the Sox trio. (A) Western blot of Sox5fl/fl Sox6fl/fl primary chondrocytes cultured for 48 h with no adenovirus (−), AdeGFP, or AdeCre. Primary antibodies and the Mrs of protein markers are indicated. (B) Beta-galactosidase activity in Sox5fl/fl Sox6fl/fl Agc1-lacZ primary chondrocytes cultured as described for panel A and harvested 24 h to 96 h after adenovirus infection. (C) Transfection of Cos-1 and HEK-293 cells with pCol2-Luc reporters harboring no enhancer (−), 4×48, or 4×A1 and Sox expression plasmids, as indicated. (D) Transfection of primary skin fibroblasts with pCol2-Luc reporters harboring no enhancer (−) or 4×A1 and Sox expression plasmids, as indicated. (E) Beta-galactosidase activity in Agc1-lacZ primary skin fibroblasts transfected for 36 h with Sox6 and/or SOX9 expression plasmids, as indicated.
We also tested whether L-Sox5/Sox6 and SOX9 are sufficient to activate the A1 enhancer by forcing their expression in nonchondrocytic cells. As previously described (26, 27), L-Sox5/Sox6 was unable to activate the 4×48-pCol2-Luc reporter in monkey kidney Cos-1 fibroblasts and in HEK-293 cells, whereas SOX9 activated it (Fig. 4C). Similarly, L-Sox5/Sox6 was unable to activate the 4×A1-pCol2-Luc reporter, whereas SOX9 significantly activated it. Interestingly, however, coexpression of L-Sox5/Sox6 and SOX9 did not affect the ability of SOX9 to activate 4×48-pCol2-Luc but drastically increased the ability of SOX9 to activate 4×A1-pCol2-Luc. Similarly, SOX9 activated 4×A1-pCol2-Luc in transiently transfected primary skin fibroblasts, and Sox6 increased its activity (Fig. 4D). In contrast, SOX9 required coexpression of Sox6 to activate the Agc1-lacZ transgene in these cells (Fig. 4E).
Together, these results thus demonstrated that the Sox trio is both needed and sufficient to activate A1, with L-Sox5/Sox6 strongly increasing activation by Sox9.
The Sox trio binds the endogenous A1 region in chondrocytes.
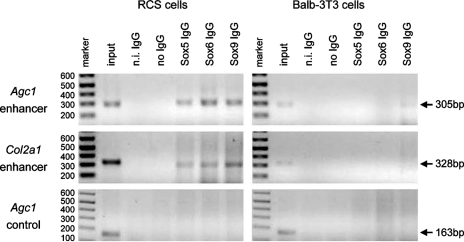
We performed ChIP in RCS and nonchondrocytic cells to test whether the Sox trio binds the endogenous A1 enhancer in live cells. PCR products of the expected sizes were obtained for the Agc1 and Col2a1 enhancers when RCS cell chromatin was precipitated with L-Sox5, Sox6, or Sox9 antibodies (Fig. 5). As expected, no PCR product was obtained for these enhancers when no IgG or nonimmune IgG was used, and no PCR product was obtained either for a control sequence located 2 kb downstream of A1 regardless of whether Sox antibodies or control antibodies were used. No PCR product was obtained either for the A2 and A3 elements in any immunoprecipitation condition (data not shown). Chromatin from BALB/3T3 cells, HEK-293 cells, and skin fibroblasts, which all weakly express Sox9 but not Sox5 and Sox6, occasionally gave a faint signal for Sox9 on the Agc1 and Col2a1 enhancers (Fig. 5 and data not shown). These data thus demonstrated that the Sox trio directly and specifically binds the endogenous Agc1 and Col2a1 enhancers in chondrocytes.
FIG. 5.
A1 binding by the Sox trio in RCS cells. Chromatin immunoprecipitation in RCS and BALB/3T3 cells using Sox and control antibodies, as indicated. Shown are the products obtained upon PCR of immunoprecipitated chromatin with primers for the Agc1 and Col2a1 enhancers and a nonconserved control sequence located 2 kb downstream of A1. The sizes of the products are indicated. The first lane in each panel shows DNA markers, with their sizes indicated, and the second lane shows the PCR product obtained with chromatin input. n.i., nonimmune IgG. The data are representative of three experiments.
The Sox trio contacts several regions in A1.
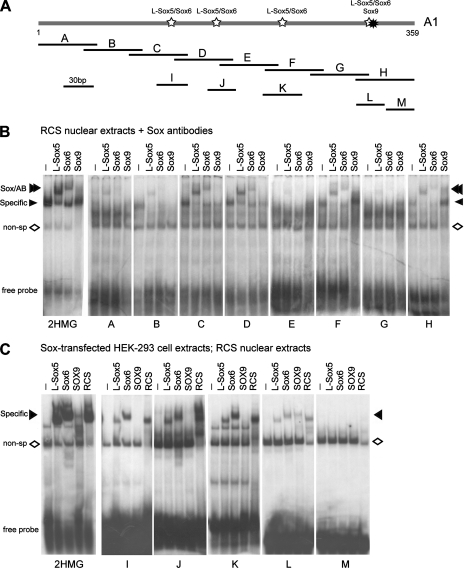
Examination of the A1 sequence revealed no perfect Sox consensus site but many Sox-like sites, i.e., heptamers with five or six consensus nucleotides (data not shown). To find the sequences bound by the Sox trio, we performed EMSA. We first prepared a series of eight probes, which we named A to H (Fig. 6A). These probes encompassed the entire enhancer and significantly overlapped each other. We used a 2HMG probe, which harbors two Sox consensus sites, as a positive control (27).
FIG. 6.
Delineation of A1 regions bound by the Sox trio. (A) Schematic of EMSA probes. Probes A to H are about 60 bp and cover the entire A1 sequence, with 15-bp overlaps. Probes I to M are 23 to 40 bp. Stars represent the regions bound by L-Sox5/Sox6 and Sox9, as identified in panels B and C. (B) EMSA of RCS nuclear extracts with 2HMG and A to H probes and with antibodies, as indicated. The migration levels of free probes, antibody supershifts (Sox/AB), specific complexes (specific), and nonspecific complexes (non-sp) are indicated. (C) EMSA of HEK-293 and RCS extracts with 2HMG and I to M probes. HEK-293 cell extracts were prepared following transfection with Sox expression plasmids, as indicated.
RCS nuclear extracts formed two complexes with the 2HMG probe (Fig. 6B). The higher-mobility complex was nonspecific since it was also seen with extracts from other cells and with other probes (Fig. 6B and C). The other complex, in contrast, was specific. It formed only with chondrocyte extracts and could be supershifted with L-Sox5, Sox6, and Sox9 antibodies. Furthermore, it migrated like complexes formed with extracts from HEK-293 cells forced to express L-Sox5, Sox6, or SOX9.
RCS proteins formed no specific complex or only a weak complex with the A, B, E, and G probes (Fig. 6B). In contrast, they formed a specific complex with the C, D, F, and H probes. L-Sox5 and Sox6 antibodies supershifted or abolished formation of this complex, but Sox9 antibodies supershifted only the specific complex formed with the H probe. The latter supershift was always partial, even when more antibodies were used (data not shown). Its mobility and that of the supershifts were consistent with the notion that each molecule of the probe was binding either L-Sox5/Sox6 or Sox9 but not both types of Sox proteins simultaneously. In agreement with these results, extracts from HEK-293 cells forced to express L-Sox5 or Sox6 formed a similar specific complex only with probes C, D, F, and H, and extracts from cells forced to express SOX9 formed a similar complex only with probe H (data not shown). We thus concluded that L-Sox5/Sox6 specifically binds probes C, D, F, and H and that Sox9 specifically binds the H probe.
We next designed smaller probes, which we called I to M, to further delineate the Sox binding regions (Fig. 6A). These probes were segments of the C, D, F, and H probes and, with the exception of M, used as a negative control, they all featured a Sox-like site(s). Consistent with the results obtained with the longer probes, L-Sox5 and Sox6 made in HEK-293 cells efficiently bound probes I, J, K, and L and SOX9 bound only probe L (Fig. 6C). None of the proteins bound M. In agreement, RCS nuclear extracts formed a specific complex with probes I to L but not with probe M. Moreover, L-Sox5 and Sox6 antibodies supershifted the complex formed with probes I to L, and Sox9 antibodies supershifted only the complex formed with probe L (data not shown). We concluded that L-Sox5 and Sox6 are able to contact the A1 enhancer in four distinct regions, whereas Sox9 is able to bind only in one region.
Sox9 binds A1 to a characteristic Sox-like pair of sites, and L-Sox5/Sox6 binds A1 to several atypical Sox-like elements.
Despite their common ability to bind L-Sox5/Sox6, the I, J, K, and L sequences were fairly different from each other and from the Sox consensus. We therefore used mutant probes to delineate the exact sequences bound by these Sox proteins. We changed AT pairs into GC pairs, since Sox proteins mainly contact AT pairs. Mutations in the I probe suggested that Sox6 binds two Sox-like sites oriented in the same direction and separated by 6 nucleotides (Fig. 7A). Mutations in the J probe suggested that Sox6 binds an AT-rich core featuring overlapping Sox-like sites (Fig. 7B). Mutations in the K probe suggested that Sox6 binds two Sox-like sites with opposite orientations and separated by 8 nucleotides (Fig. 7C). Mutations in the L probe suggested that SOX9 binds to two Sox-like sites arranged in opposite orientations and separated by several nucleotides (Fig. 7D). SOX9 bound these sites as a dimer but could still bind the 3′ site as a monomer when the 5′ site was destroyed. Mutations that varied the spacing between the two sites from 2 to 7 nucleotides did not significantly affect SOX9 or Sox6 binding (Fig. 7E). Interestingly, mutations that further altered spacing abolished SOX9 binding but not Sox6 binding. Also, mutations that changed the relative orientation of the two sites abolished SOX9 binding but not Sox6 binding (Fig. 7F). L-Sox5 bound all probes with efficiency and specificity similar to those for Sox6 (data not shown). These data, together with previous reports, demonstrated that Sox9 binds cartilage enhancers to pairs of variable Sox-like sites oriented back to back and separated by 3 to 5 bp (Fig. 7G). In contrast, L-Sox5/Sox6 has less-stringent requirements for the DNA sequence and the configuration of Sox-like sites and binds cartilage enhancers to several atypical Sox-like sites (Fig. 7H).
FIG. 7.
Delineation of Sox binding sites in A1. (A to F) EMSA of extracts from HEK-293 cells forced to express Sox6 or SOX9 incubated with probes carrying mutations, as indicated. Probes are named according to the positions of mutated nucleotides (A to D), number of nucleotides separating the Sox-like sites (E), and orientation of the sites (F). Shown are the Sox/DNA complexes. Arrows with single and double shafts designate SOX9 monomers and dimers, respectively (D and E). (G) Comparison of Sox9 binding sites in cartilage enhancers. The upper strand of the Sox consensus site is shown in reverse and forward orientations, as indicated with arrows and numbers. The Sox-like sites in enhancers are boxed, with boldface letters for consensus nucleotides and underlined letters for nucleotides present in most enhancers. (H) Comparison of L-Sox5/Sox6 binding sites in the Agc1 and Col2a1 enhancers. All 48 bp of the Col2a1 enhancer and flanking nucleotides are shown on two lanes.
The L-Sox5/Sox6 and Sox9 binding sites in A1 are required for enhancer activity.
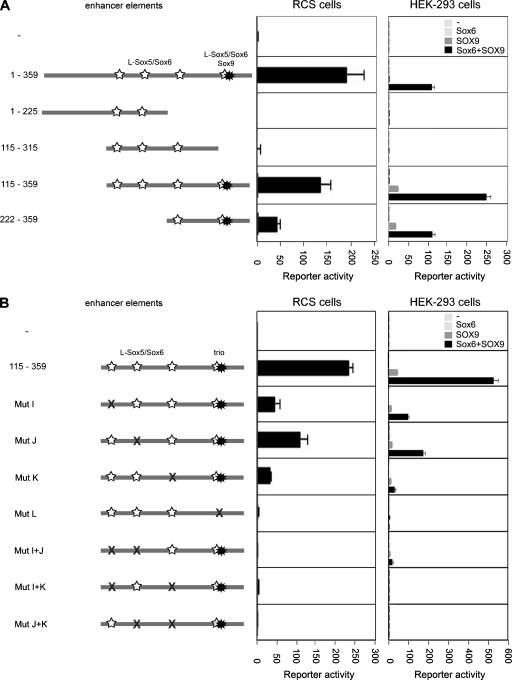
To determine whether the binding sites of the Sox trio on A1 are needed for enhancer activity, we generated mutant reporters consisting of two copies of wild-type or mutant A1 in the minimum promoter pCol2-Luc reporter. Reporters harboring a 1 to 225 fragment (consisting of nucleotides 1 to 225 in the 359-bp A1 enhancer) or 115 to 315 fragment were inactive in RCS cells, and they were not activated by Sox6 and/or SOX9 in HEK-293 cells (Fig. 8A). These fragments contained several L-Sox5/Sox6 sites but lacked the Sox9 site. A 115 to 359 reporter, featuring all Sox sites, was almost as active as full-length A1 in RCS cells, and it was activated by Sox6/SOX9 in HEK-293 cells more efficiently than the full-length A1 enhancer. A 222 to 359 reporter, featuring the Sox9 site and the distal L-Sox5/Sox6 site, was less active in RCS cells than the full-length A1 enhancer, but it was efficiently activated by Sox6/SOX9 in HEK-293 cells. We concluded that the activity of enhancer fragments closely correlated with the presence of the Sox binding sites.
FIG. 8.
Test of the functional importance of the Sox trio binding sites. (A) Transient transfection of RCS cells and HEK-293 cells with pCol2-Luc constructs containing two copies of A1 fragments. Fragments are schematized with numbers indicating nucleotide boundaries. The HEK-293 cells were cotransfected with Sox expression plasmids, as indicated. (B) Similar transfection as in panel A but with pCol2-Luc constructs harboring point mutations in the Sox sites of the 115 to 359 A1 fragment. The mutations corresponded to those described as m7-11-20-24 for segment I (Mut I), m5-9-12-16 for segment J (Mut J), m7-11-15-19-22-26 for segment K (Mut K), and m7-10-14-16 for segment L (Mut L) in Fig. 7.
Point mutations in any L-Sox5/Sox6 site reduced the activity of the 115 to 359 enhancer in RCS cells (Fig. 8B). It also reduced the ability of Sox6/SOX9 to activate the enhancer in HEK-293 cells. Point mutations in the Sox9 site or in any combination of two L-Sox5/Sox6 sites totally abolished enhancer activity in RCS cells and SOX9-mediated activation in HEK-293 cells. We concluded that the Sox9 site is required to mediate the ability of Sox9 to activate the enhancer and that each L-Sox5/Sox6 site contributes to allow L-Sox5/Sox6 to boost the activity of Sox9.
L-Sox5/Sox6 secures binding of Sox9 to chondrocyte-specific enhancers.
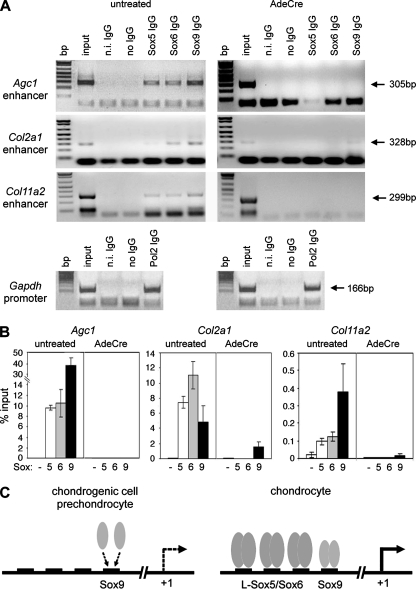
L-Sox5/Sox6 could potentiate the activity of Sox9 in several ways. One of them could be to increase the ability of Sox9 to bind A1. To test this possibility, we prepared Sox5fl/fl Sox6fl/fl chondrocytes and treated them with AdeCre. As expected, binding of each Sox protein was detected in untreated cells by both qualitative and quantitative ChIP assays, and binding of L-Sox5/Sox6 was not detected in AdeCre-treated cells (Fig. 9A and B). Interestingly, while Sox9 was still present in AdeCre-treated cells (Fig. 4A), its binding to A1 was no longer detected. A similar result was obtained for the Col2a1 and Col11a2 enhancers, although weak binding of Sox9 often remained on the Col2a1 enhancer in cells lacking L-Sox5/Sox6. We concluded that L-Sox5/Sox6 cooperates with Sox9 in chondrocytes at least in part by securing Sox9 binding to cartilage-specific enhancers.
FIG. 9.
Test of the importance of L-Sox5/Sox6 for Sox9 binding to cartilage enhancers. (A) ChIP in Sox5fl/fl Sox6fl/fl chondrocytes 48 h after infection with no adenovirus or AdeCre. Cartilage enhancers were detected by PCR of chromatin precipitated with antibodies as indicated. Binding of RNA polymerase II (Pol2) to the Gapdh promoter was used as positive control. (B) Quantitative ChIP assay of the same samples as in panel A, precipitated with IgG control (−) and Sox antibodies, as indicated. (C) Model for the mode of transactivation of Agc1 by the Sox trio. See text for details.
DISCUSSION
This study has uncovered mechanisms underlying transcription of Agc1, the gene for a major cartilage component and chondrocyte marker, and has provided novel insights into the mechanisms whereby the trio the L-Sox5/Sox6 and Sox9 transcription factors drives chondrogenesis.
We identified the Agc1 enhancer based on the hypothesis, proven correct for many genes, that essential cis-acting elements are evolutionarily conserved. The high-stringency criteria that we used in our search allowed us to extract only three conserved nonexonic regions within and around Agc1. The fact that A1 is the longest and most conserved of these regions supports our experimental conclusion that it is a major, if not the only, Agc1 enhancer. Although the other two elements failed to exhibit transcriptional activity, one can nevertheless expect that they have an important function, e.g., in RNA splicing. Interestingly, even less-stringent criteria did not allow us to identify nonmammalian orthologues of A1. The low sequence specificity of the DNA sites bound by L-Sox5/Sox6 and Sox9 may provide an explanation. A1 may indeed feature Sox-like sites in both mammalian and nonmammalian genomes, but these sites may have significantly diverged during evolution.
We showed that A1 drives gene expression with a pattern matching that of Agc1. This pattern is different from that of endogenous Col2a1 and Col2a1 reporters (36, 42, 54) in that it is not expressed in skeletogenic mesenchymal cells, the perichondrium, and presumptive joint cells and is thus strictly specific for differentiated chondrocytes. Moreover, it remains active in growth plate and articular cartilage through adulthood, whereas Col2a1 and Col2a1 transgenes are turned off by weaning age (34). The latter finding is important considering that the specific transcriptional mechanisms operating in adult articular cartilage remain unknown. It is possible that A1 contains cis-acting elements, ensuring its lifelong activation by the Sox trio, whereas the Col2a1 enhancer does not. A1 therefore constitutes a new tool to investigate mechanisms that control gene expression differentially according to cartilage type, age, and physiological or pathological conditions, and it will also be instrumental for expressing transgenes in differentiated chondrocytes only and in adult cartilage.
Doege and colleagues (9) showed that an Agc1 4.7-kb upstream region was able to activate an Agc1 3.4-kb promoter in transgenic embryos, but weakly and only in hypertrophic chondrocytes in a subset of cartilage elements. Since this region contained A1, it is surprising that it did not reproduce Agc1 expression as Agc1-lacZ did. One possibility is that the transgene featured a single copy of the enhancer, whereas we used four copies. Another possibility is that the activity of A1 was weakened by other elements in the 4.7-kb region or promoter. The fact that the 4.7-kb segment was the only Agc1 region having chondrocyte-specific activity nevertheless further strengthens our conclusion that A1 is likely the major driver of Agc1 in cartilage.
We showed that L-Sox5/Sox6 is needed for A1 activity by recording a rapid loss of Agc1-lacZ activity upon inactivating Sox5 and Sox6 in primary chondrocytes. These data also proved that Sox9 is needed, at least indirectly, since Sox5 and Sox6 expression depends on Sox9. They also proved that Sox9 is not sufficient, since Sox5- and Sox6-deficient chondrocytes still express Sox9. These data thus fit with the virtual silencing of Agc1 in mice lacking L-Sox5/Sox6, despite normal expression of Sox9 (45).
By using a ChIP assay, we provided the first report, to our knowledge, of direct binding of the Sox trio to target genes in chondrocytes. We then used a comprehensive EMSA approach to delineate the binding sites of the proteins in A1. We found that Sox9 binds a single element and demonstrated that this site is needed for enhancer activity. This element is a pair of Sox-like sites separated by 4 nucleotides and oriented back to back. We proved that this configuration is critical. Since Sox9 binds similar pairs of sites on other cartilage enhancers, our data add weight to the notion that the consensus Sox9 site in chondrocytes is a pair of Sox-like sites positioned tail-to-tail and separated by 3 to 5 nucleotides. Interestingly, the Sox9 site sequences greatly vary among enhancers but not across species for any gene. This suggests that site variations may affect the efficiency of Sox9 binding to DNA or its interaction with other proteins and may thereby contribute to differences in expression levels and patterns between cartilage-specific genes.
We identified three elements in A1 specifically bound by L-Sox5/Sox6 and needed to mediate enhancer activation by the Sox trio. These sites greatly vary in sequence from each other and from the Sox consensus, and we showed for the first time that L-Sox5/Sox6 binds with similar efficiencies to highly variable Sox-like sequences. However, since the A1 sites are highly conserved evolutionarily, we postulate that L-Sox5/Sox6 uses criteria besides the DNA sequence itself to bind target genes. This finding is a reminder that Sox proteins belong to the superfamily of HMG box proteins, which includes such members as HMG1 and HMG2, which bind DNA with little or no sequence specificity. Like these relatives, L-Sox5/Sox6 may primarily act as architectural organizers of transcriptional complexes, while Sox9 binds DNA with higher sequence specificity and is capable of transactivation.
Ever since it was suggested that L-Sox5/Sox6 cooperates with Sox9 in activating Col2a1 (27), the mode of operation of this trio has remained elusive. This study has provided significant insights into this mode of operation by showing by ChIP that L-Sox5/Sox6 is needed to secure Sox9 binding to the endogenous Agc1, Col2a1, and Col11a2 enhancers. The fact that the L-Sox5/Sox6 binding sites were required for optimal activation of A1 by the Sox trio strongly suggests that L-Sox5/Sox6 secured Sox9 binding to this enhancer by directly binding to this enhancer rather than through indirect mechanisms, such as activation of other genes. L-Sox5 and Sox6 may act in several ways. They may physically interact with Sox9, but no evidence was found of such interaction in solution or in EMSA (27). They may help Sox9 interact with transcriptional partners. A1, however, does not feature consensus sites for other transcription factors and was not found in EMSA to form complexes other than Sox/DNA complexes. Putative partners would thus not be likely to bind DNA, and, since the Sox trio is sufficient for A1 activation in nonchondrocytic cells, they would more likely be ubiquitous than chondrocyte specific. L-Sox5/Sox6 may also secure Sox9 binding to DNA by inducing chromatin remodeling. This possibility would explain why Sox9 is more dependent on L-Sox5/Sox6 to activate Agc1, Col2a1 and Agc1-lacZ than to activate transient reporters.
In conclusion, our data suggest a new model for the chondrogenic action of the Sox trio. Mesenchymal cells and prechondrocytes express Agc1 and other chondrocyte markers at low or undetectable levels because Sox9 has a limited ability to bind to the cartilage enhancers of these genes in the absence of L-Sox5/Sox6 (Fig. 9C). By inducing Sox5 and Sox6 expression in overtly differentiating chondrocytes, Sox9 gives itself the potential to upregulate its own activity as L-Sox5/Sox6 now binds to distinct sites on the enhancers and thereby secures Sox9 binding to its recognition site.
Acknowledgments
This work was supported by NIH/NIAMS grant AR46249 (V.L.).
We are grateful to Josephine Adams for advice on ChIP; Benoit de Crombrugghe for Col2a1 reporters; the Case Transgenic Core for transgenic mouse founders; and Shunichi Murakami, Guang Zhou, Peter Dy, Alfredo Penzo-Méndez, and Pallavi Bhattaram for precious advice with experiments and the manuscript.
Footnotes
Published ahead of print on 16 June 2008.
REFERENCES
- 1.Akiyama, H. 20 March 2008. Control of chondrogenesis by the transcription factor Sox9. Mod. Rheumatol. 18213-219. [DOI] [PubMed] [Google Scholar]
- 2.Akiyama, H., M. C. Chaboissier, J. E. Martin, A. Schedl, and B. de Crombrugghe. 2002. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 162813-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akiyama, H., J. P. Lyons, Y. Mori-Akiyama, X. Yang, R. Zhang, Z. Zhang, J. M. Deng, M. M. Taketo, T. Nakamura, R. R. Behringer, P. D. McCrea, and B. de Crombrugghe. 2004. Interactions between Sox9 and beta-catenin control chondrocyte differentiation. Genes Dev. 181072-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell, D. M., K. K. Leung, S. C. Wheatley, L. J. Ng, S. Zhou, K. W. Ling, M. H. Sham, P. Koopman, P. P. Tam, and K. S. Cheah. 1997. SOX9 directly regulates the type-II collagen gene. Nat. Genet. 16174-178. [DOI] [PubMed] [Google Scholar]
- 5.Bi, W., J. M. Deng, Z. Zhang, R. R. Behringer, and B. de Crombrugghe. 1999. Sox9 is required for cartilage formation. Nat. Genet. 2285-89. [DOI] [PubMed] [Google Scholar]
- 6.Bridgewater, L. C., V. Lefebvre, and B. de Crombrugghe. 1998. Chondrocyte-specific enhancer elements in the Col11a2 gene resemble the Col2a1 tissue-specific enhancer. J. Biol. Chem. 27314998-15006. [DOI] [PubMed] [Google Scholar]
- 7.Bridgewater, L. C., M. D. Walker, G. C. Miller, T. A. Ellison, L. D. Holsinger, J. L. Potter, T. L. Jackson, R. K. Chen, V. L. Winkel, Z. Zhang, S. McKinney, and B. de Crombrugghe. 2003. Adjacent DNA sequences modulate Sox9 transcriptional activation at paired Sox sites in three chondrocyte-specific enhancer elements. Nucleic Acids Res. 311541-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denny, P., S. Swift, F. Connor, and A. Ashworth. 1992. An SRY-related gene expressed during spermatogenesis in the mouse encodes a sequence-specific DNA-binding protein. EMBO J. 113705-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doege, K., L. B. Hall, W. McKinnon, L. Chen, D. T. Stephens, and K. A. Garrison. 2002. A remote upstream element regulates tissue-specific expression of the rat aggrecan gene. J. Biol. Chem. 27713989-13997. [DOI] [PubMed] [Google Scholar]
- 10.Doege, K. J., K. Garrison, S. N. Coulter, and Y. Yamada. 1994. The structure of the rat aggrecan gene and preliminary characterization of its promoter. J. Biol. Chem. 26929232-29240. [PubMed] [Google Scholar]
- 11.Dumitriu, B., P. Dy, P. Smits, and V. Lefebvre. 2006. Generation of mice harboring a Sox6 conditional null allele. Genesis 44219-224. [DOI] [PubMed] [Google Scholar]
- 12.Dy, P., A. Penzo-Méndez, H. Wang, C. E. Pedraza, W. B. Macklin, and V. Lefebvre. 2008. The three SoxC proteins—Sox4, Sox11, and Sox12—exhibit overlapping expressing patterns and molecular properties. Nucleic Acids Res. 363101-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dy, P., Y. Han, and V. Lefebvre. 2008. Generation of mice harboring a Sox5 conditional null allele. Genesis 46294-299. [DOI] [PubMed] [Google Scholar]
- 14.Eyre, D. R. 2004. Collagens and cartilage matrix homeostasis. Clin. Orthop. Relat. Res. 427118-122. [DOI] [PubMed] [Google Scholar]
- 15.Genzer, M. A., and L. C. Bridgewater. 2007. A Col9a1 enhancer element activated by two interdependent SOX9 dimers. Nucleic Acids Res. 351178-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gleghorn, L., R. Ramesar, P. Beighton, and G. Wallis. 2005. A mutation in the variable repeat region of the aggrecan gene (AGC1) causes a form of spondyloepiphyseal dysplasia associated with severe, premature osteoarthritis. Am. J. Hum. Genet. 77484-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldring, M. B., and S. R. Goldring. 2007. Osteoarthritis. J. Cell. Physiol. 213626-634. [DOI] [PubMed] [Google Scholar]
- 18.Hogan, B., R. Beddington, F. Costantini, and E. Lacy. 1994. Manipulating the mouse embryo, p. 373-375. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 19.Horton, W. E., Jr., M. Lethbridge-Cejku, M. C. Hochberg, R. Balakir, P. Precht, C. C. Plato, J. D. Tobin, L. Meek, and K. Doege. 1998. An association between an aggrecan polymorphic allele and bilateral hand osteoarthritis in elderly white men: data from the Baltimore Longitudinal Study of Aging (BLSA). Osteoarthritis Cartilage 6245-251. [DOI] [PubMed] [Google Scholar]
- 20.Ikeda, T., S. Kamekura, A. Mabuchi, I. Kou, S. Seki, T. Takato, K. Nakamura, H. Kawaguchi, S. Ikegawa, and U. I. Chung. 2004. The combination of SOX5, SOX6, and SOX9 (the SOX trio) provides signals sufficient for induction of permanent cartilage. Arthritis Rheum. 503561-3573. [DOI] [PubMed] [Google Scholar]
- 21.Kimata, K., H. J. Barrach, K. S. Brown, and J. P. Pennypacker. 1981. Absence of proteoglycan core protein in cartilage from the cmd/cmd (cartilage matrix deficiency) mouse. J. Biol. Chem. 2566961-6968. [PubMed] [Google Scholar]
- 22.Kou, I., and S. Ikegawa. 2004. SOX9-dependent and -independent transcriptional regulation of human cartilage link protein. J. Biol. Chem. 27950942-50948. [DOI] [PubMed] [Google Scholar]
- 23.Krebsbach, P. H., K. Nakata, S. M. Bernier, O. Hatano, T. Miyashita, C. S. Rhodes, and Y. Yamada. 1996. Identification of a minimum enhancer sequence for the type II collagen gene reveals several core sequence motifs in common with the link protein gene. J. Biol. Chem. 2714298-4303. [DOI] [PubMed] [Google Scholar]
- 24.Lefebvre, V., B. Dumitriu, A. Penzo-Mendez, Y. Han, and B. Pallavi. 2007. Control of cell fate and differentiation by Sry-related high-mobility-group box (Sox) transcription factors. Int. J. Biochem. Cell Biol. 392195-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lefebvre, V., S. Garofalo, G. Zhou, M. Metsäranta, E. Vuorio, and B. De Crombrugghe. 1994. Characterization of primary cultures of chondrocytes from type II collagen/beta-galactosidase transgenic mice. Matrix Biol. 14329-335. [DOI] [PubMed] [Google Scholar]
- 26.Lefebvre, V., W. Huang, V. R. Harley, P. N. Goodfellow, and B. de Crombrugghe. 1997. SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro-alpha 1(II) collagen gene. Mol. Cell. Biol. 172336-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lefebvre, V., P. Li, and B. de Crombrugghe. 1998. A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are co-expressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J. 175718-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lefebvre, V., and P. Smits. 2005. Transcriptional control of chondrocyte fate and differentiation. Birth Defects Res. Part C Embryo Today Rev. 75200-212. [DOI] [PubMed] [Google Scholar]
- 29.Lefebvre, V., G. Zhou, K. Mukhopadhyay, C. N. Smith, Z. Zhang, H. Eberspaecher, X. Zhou, S. Sinha, S. N. Maity, and B. de Crombrugghe. 1996. An 18-base-pair sequence in the mouse proα1(II) collagen gene is sufficient for expression in cartilage and binds nuclear proteins that are selectively expressed in chondrocytes. Mol. Cell Biol. 164512-4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, H., and N. B. Schwartz. 1995. Gene structure of chick cartilage chondroitin sulfate proteoglycan (aggrecan) core protein. J. Mol. Evol. 41878-885. [DOI] [PubMed] [Google Scholar]
- 31.Liu, C. J., Y. Zhang, K. Xu, D. Parsons, D. Alfonso, and P. E. Di Cesare. 2007. Transcriptional activation of cartilage oligomeric matrix protein by Sox9, Sox5, and Sox6 transcription factors and CBP/p300 coactivators. Front. Biosci. 123899-3910. [DOI] [PubMed] [Google Scholar]
- 32.Mertin, S., S. G. McDowall, and V. R. Harley. 1999. The DNA-binding specificity of SOX9 and other SOX proteins. Nucleic Acids Res. 271359-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mukhopadhyay, K., V. Lefebvre, G. Zhou, S. Garofalo, J. H. Kimura, and B. de Crombrugghe. 1995. Use of a new rat chondrosarcoma cell line to delineate a 119-bp chondrocyte-specific enhancer element and to define active promoter segments in the mouse pro-alpha 1(II) collagen gene. J. Biol. Chem. 27027711-27729. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura, E., M.-T. Nguyen, and S. Mackem. 2006. Kinetics of tamoxifen-regulated Cre activity in mice using a cartilage-specific CreER™ to assay temporal activity windows along the proximo-distal limb skeleton. Dev. Dyn. 2352603-2612. [DOI] [PubMed] [Google Scholar]
- 35.Ng, L.-J., S. Wheatley, G. E. O. Muscat, J. Conway-Campbell, J. Bowles, E. Wright, D. M. Bell, P. P. L. Tam, K. S. E. Cheah, and P. Koopman. 1997. SOX9 binds DNA, activates transcription, and coexpressed with type II collagen during chondrogenesis in the mouse. Dev. Biol. 183108-121. [DOI] [PubMed] [Google Scholar]
- 36.Ovchinnikov, D. A., J. M. Deng, G. Ogunrinu, and R. R. Behringer. 2000. Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis 26145-146. [PubMed] [Google Scholar]
- 37.Rentsendorj, O., A. Nagy, I. Sinkó, A. Daraba, E. Barta, and I. Kiss. 2005. Highly conserved proximal promoter element harbouring paired Sox9-binding sites contributes to the tissue- and developmental stage-specific activity of the matrilin-1 gene. Biochem. J. 389705-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rimoin, D. L., D. Cohn, D. Krakow, W. Wilcox, R. S. Lachman, and Y. Alanay. 2007. The skeletal dysplasias: clinical-molecular correlations. Ann. N. Y. Acad. Sci. 1117302-309. [DOI] [PubMed] [Google Scholar]
- 39.Rittenhouse, E., L. C. Dunn, J. Cookingham, C. Calo, M. Spiegelman, G. B. Dooher, and D. Bennett. 1978. Cartilage matrix deficiency (cmd): a new autosomal recessive lethal mutation in the mouse. J. Embryol. Exp. Morphol. 4371-84. [PubMed] [Google Scholar]
- 40.Roughley, P. 2006. The structure and function of cartilage proteoglycans. Eur. Cells Mater. 301292-101. [DOI] [PubMed] [Google Scholar]
- 41.Roughley, P., D. Martens, J. Rantakokko, M. Alini, F. Mwale, and J. Antoniou. 2006. The involvement of aggrecan polymorphism in degeneration of human intervertebral disc and articular cartilage. Eur. Cells Mater. 111-7. [PubMed] [Google Scholar]
- 42.Sakai, K., L. Hiripi, V. Glumoff, O. Brandau, R. Eerola, E. Vuorio, Z. Bosze, R. Fassler, and A. Aszodi. 2001. Stage- and tissue-specific expression of a Col2a1-Cre fusion gene in transgenic mice. Matrix Biol. 19761-767. [DOI] [PubMed] [Google Scholar]
- 43.Salminen, H., E. Vuorio, and A. M. Säämänen. 2001. Expression of Sox9 and type IIA procollagen during attempted repair of articular cartilage damage in a transgenic mouse model of osteoarthritis. Arthritis Rheum. 44947-955. [DOI] [PubMed] [Google Scholar]
- 44.Sekiya, I., K. Tsuji, P. Koopman, H. Watanabe, Y. Yamada, K. Shinomiya, A. Nifuji, and M. Noda. 2000. SOX9 enhances aggrecan gene promoter/enhancer activity and is up-regulated by retinoic acid in a cartilage-derived cell line, TC6. J. Biol. Chem. 27510738-10744. [DOI] [PubMed] [Google Scholar]
- 45.Smits, P., P. Li, J. Mandel, Z. Zhang, J. M. Deng, R. R. Behringer, B. de Crombrugghe, and V. Lefebvre. 2001. The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev. Cell 1277-290. [DOI] [PubMed] [Google Scholar]
- 46.Smits, P., S. Mitra, and V. Lefebvre. 2004. Sox5 and Sox6 are needed to develop and maintain source, columnar and hypertrophic chondrocytes in the cartilage growth plate. J. Cell Biol. 164747-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valhmu, W. B., G. D. Palmer, J. Dobson, S. G. Fischer, and A. Ratcliffe. 1998. Regulatory activities of the 5′- and 3′-untranslated regions and promoter of the human aggrecan gene. J. Biol. Chem. 2736196-6202. [DOI] [PubMed] [Google Scholar]
- 48.Valhmu, W. B., G. D. Palmer, P. A. Rivers, S. Ebara, J. F. Cheng, S. Fischer, and A. Ratcliffe. 1995. Structure of the human aggrecan gene: exon-intron organization and association with the protein domains. Biochem. J. 309535-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watanabe, H., L. Gao, S. Sugiyama, K. Doege, K. Kimata, and Y. Yamada. 1995. Mouse aggrecan, a large cartilage proteoglycan: protein sequence, gene structure and promoter sequence. Biochem. J. 308433-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watanabe, H., K. Kimata, S. Line, D. Strong, L. Y. Gao, C. A. Kozak, and Y. Yamada. 1994. Mouse cartilage matrix deficiency (cmd) caused by a 7 bp deletion in the aggrecan gene. Nat. Genet. 7154-157. [DOI] [PubMed] [Google Scholar]
- 51.Watanabe, H., K. Nakata, K. Kimata, I. Nakanishi, and Y. Yamada. 1997. Dwarfism and age-associated spinal degeneration of heterozygote cmd mice defective in aggrecan. Proc. Natl. Acad. Sci. USA 946943-6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xie, W. F., X. Zhang, S. Sakano, V. Lefebvre, and L. J. Sandell. 1999. Trans-activation of the mouse cartilage-derived retinoic acid-sensitive protein gene by Sox9. J. Bone Miner. Res. 14757-763. [DOI] [PubMed] [Google Scholar]
- 53.Zhao, Q., H. Eberspaecher, V. Lefebvre, and B. de Crombrugghe. 1997. Parallel expression of Sox9 and Col2a1 in cells undergoing chondrogenesis. Dev. Dyn. 209377-386. [DOI] [PubMed] [Google Scholar]
- 54.Zhou, G., G. Garofalo, K. Mukhopadhyay, V. Lefebvre, C. N. Smith, H. Eberspaecher, and B. de Crombrugghe. 1995. A 182 bp fragment of the mouse pro-alpha 1(II) collagen gene is sufficient to direct chondrocyte expression in transgenic mice. J. Cell Sci. 1083677-3684. [DOI] [PubMed] [Google Scholar]