Abstract
Genes for pentatricopeptide repeat (PPR) proteins are found in all eukaryotic genomes analyzed but are particularly abundant in land plants. The majority of analyzed PPR proteins play a role in the processing or translation of organellar RNAs. Few PPR proteins have been studied in detail, and the functional repertoire and mechanisms of action of proteins in the PPR family are poorly understood. Here we analyzed a maize ortholog of the embryo-essential Arabidopsis thaliana gene AtPPR5. A genome-wide analysis of chloroplast RNAs that coimmunoprecipitate with Zea mays PPR5 (ZmPPR5) demonstrated that ZmPPR5 is bound in vivo to the unspliced precursor of trnG-UCC. Null and hypomorphic Zmppr5 insertion mutants are embryo viable but are deficient for chloroplast ribosomes and die as seedlings. These mutants show a dramatic decrease in both spliced and unspliced trnG-UCC RNAs, while the transcription of trnG-UCC is unaffected. These results, together with biochemical data documenting the sequence-specific binding of recombinant PPR5 to the trnG-UCC group II intron, suggest that PPR5 stabilizes the trnG-UCC precursor by directly binding and protecting an endonuclease-sensitive site. These findings add to the evidence that chloroplast-localized PPR proteins that are embryo essential in Arabidopsis typically function in the biogenesis of the plastid translation machinery.
As descendants of prokaryotic endosymbionts, chloroplasts and mitochondria retain remnants of bacterial chromosomes that carry essential information for organelle biogenesis and metabolism. Some components of the organellar gene expression systems are encoded by these chromosomes, including rRNAs, most tRNAs, and many ribosomal proteins; however, the vast majority of factors involved in organellar gene expression are encoded by nuclear genes (4). Some such genes are derived from the prokaryotic ancestors of organelles by gene transfer from endosymbionts (29), but recent findings suggest that many are innovations of the host (5, 43). The analysis of such eukaryote-derived proteins is a prerequisite for understanding how endosymbionts integrate into the developmental programs and signaling circuits of hosts.
Despite the simplicity of the organellar genomes, the mechanisms of gene expression in plant organelles are highly complex. Transcripts are generated by several different RNA polymerases; precursor RNAs can be processed by RNA editing, RNA splicing, and various cleavage reactions; and mRNA-specific translational activators are sometimes required (7, 35, 40). Recently, a family of putative RNA binding proteins involved in many posttranscriptional aspects of plant organellar gene expression has come into focus, the pentatricopeptide repeat (PPR) protein family (11, 26). PPR proteins are characterized by the presence of tandem arrays of a degenerate 35-amino-acid (pentatricopeptide) motif, similar to and probably derived from the tetratricopeptide repeat (43). PPR proteins are found in all eukaryotic organisms but are absent in prokaryotes. The family has expanded to a particularly large size in land plants, where more than 450 PPR proteins have been annotated in both rice and Arabidopsis genome sequences (26). Almost all of the PPR proteins analyzed to date are targeted to mitochondria or chloroplasts. This makes the PPR protein family one of the most conspicuous evolutionary novelties in nuclear-organelle interactions. Genetic approaches have demonstrated roles for PPR proteins in various steps in organellar gene expression, including RNA splicing (12, 30, 42), RNA editing (25, 32), intercistronic RNA cleavage (20), and translation (9, 15). Three PPR proteins have been demonstrated to be associated with their target RNAs in vivo (16, 41, 42), and two have been shown to bind in vitro to their genetically determined physiological target RNAs (31, 33). By extrapolation, it is anticipated that many PPR proteins bind directly to the RNAs whose metabolism they influence.
So far, most PPR proteins analyzed in detail are required for the expression of subunits of the photosynthetic or respiratory apparatus and thus for chloroplast or mitochondrial biogenesis. However, a large number of PPR proteins in Arabidopsis thaliana impact early embryo development (10, 26). Embryo lethality caused by the disruption of mitochondrial-localized PPR proteins is not unexpected, as respiration is essential for embryogenesis. However, the molecular basis for the embryo-essential role of plastid-localized PPR proteins has been more enigmatic. This has been elucidated for one chloroplast PPR protein, PPR4. It was demonstrated that the maize PPR4 ortholog is associated with the trans-spliced intron of the chloroplast rps12 gene and is required for splicing this intron (42). The absence of PPR4 leads to the failure to express the ribosomal protein RPS12 and, consequently, to the loss of chloroplast translation. The embryo-lethal phenotype associated with mutations in Arabidopsis PPR4 and in various genes encoding chloroplast tRNA synthetases, translation factors, and ribosomal proteins (6, 39, 42, 50) strongly suggests that chloroplast translation is essential for embryo development in Arabidopsis. It seems likely that this reflects a requirement for the products of several plastid open reading frames (ORFs) whose orthologs have been shown to be essential for cellular survival in tobacco (13, 24).
Here we present a functional analysis of a second plastid-localized PPR protein that is essential for embryo development in Arabidopsis (At4g39620) (10). We named the orthologous group corresponding to this gene PPR5, and we refer to the Arabidopsis ortholog as AtPPR5. We demonstrate that the maize ortholog of AtPPR5 (Zea mays PPR5 [ZmPPR5]) is bound in vivo to the intron of the plastid trnG-UCC precursor and is required for its stabilization; in the absence of PPR5, neither unspliced nor spliced trnG-UCC accumulates. Our results add tRNA biogenesis to the list of functions for PPR proteins. This single direct function for PPR5 can account for the loss of plastid ribosomes and plastid-encoded proteins observed in ppr5 mutants. By extrapolation from these data and those for PPR4, we hypothesize that chloroplast-localized PPR proteins that are essential for embryo development in Arabidopsis directly affect the expression of one or more of the chloroplast genes that are essential for chloroplast translation, or specifically affect the expression of a chloroplast ORF whose ortholog has been shown to be cell essential in tobacco.
MATERIALS AND METHODS
Plant material.
Insertion alleles of ppr5 were identified in a reverse genetic screen of a collection of ∼2,300 Mu transposon-induced nonphotosynthetic maize mutants (http://chloroplast.uoregon.edu). DNA from pooled mutant seedlings was analyzed by PCR with ppr5-specific primers (5′-CGTCTAATCTGTAATCTGCACG-3′ and 5′-CCAAGCACATCAAGTCCATGGC-3′) in conjunction with a Mu transposon terminal inverted repeat primer (5′-GCCTCCATTTCGTCGAATCCCG-3′), using previously established methods (54). The flanking sequences were amplified by PCR and sequenced to locate the insertion sites. Mutations were tested for complementation by crossing plants heterozygous for each allele. Fifty-one ears were recovered from ppr5-1/+ × ppr5-2/+ crosses, 18 of which segregated chlorophyll-deficient mutants. Other maize mutants used in this work include hcf7 mutants, which are pale green and show reduced chloroplast polysome assembly and aberrant metabolism of 16S rRNA (3), and ppr4-1/ppr4-2 mutants, which are pale green and have a plastid ribosome deficiency due to defects in the splicing of the chloroplast rps12 RNA (42). Seedlings were grown in soil in a growth chamber under a 16 h light/8 h dark cycle at 27°C and harvested between 7 and 9 days after planting.
Analysis of RNA in mutant tissue.
Total RNA was extracted from leaf tissue using TRIzol (Invitrogen) following the manufacturer's protocol. A total of 4 μg of RNA was electrophoresed in 1.8% agarose-formaldehyde gels or 20% polyacrylamide gels and then transferred to Hybond N (Amersham) membranes. PCR fragment probes were radiolabeled by random priming (Hexalabel kit, Fermentas). Oligonucleotide probes were radiolabeled with [γ-32P]ATP using T4 polynucleotide kinase (Fermentas). The sequences of the primers used for the PCR, along with their positions within the maize chloroplast DNA, are given in Table S2 in the supplemental material. Prehybridization of the membranes was carried out for 30 min in Church hybridization buffer (0.5 M NaPO4 [pH 7.2], 7% [wt/vol] sodium dodecyl sulfate [SDS]). Hybridization to PCR fragments was carried out in the same buffer overnight at 65°C, followed by three washes at the same temperature for 20 min each in 0.2× SSC/0.1% SDS (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Hybridization to oligonucleotide probes was carried out accordingly but at 50°C during prehybridization, hybridization, and washing and using 0.5× SSC/0.1% SDS for the washes. For Northern analysis of the coimmunoprecipitated RNA, pellet RNA extracts were spiked with total RNA from albino ppr5 mutants to compensate for the ionic differences between the pellet and supernatant fractions that otherwise lead to distorted gel migration behaviors. This added RNA did not yield any signal when probed for the presence of trnG-UCC (Fig. 4 and data not shown). The probe was removed from the hybridized RNA gel blots by boiling in 0.1% SDS for 10 min. The blots were exposed to a phosphorimager screen to verify successful stripping before reprobing.
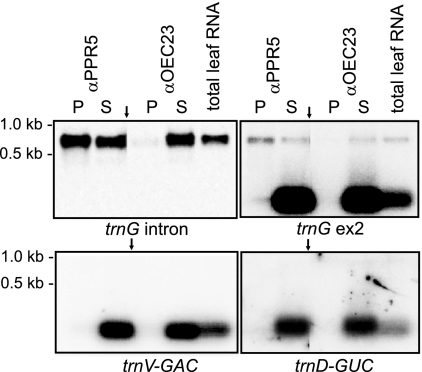
FIG. 4.
Coimmunoprecipitation of unspliced trnG-UCC precursors but not spliced trnG-UCC with PPR5. RNA purified from a PPR5 immunoprecipitation supernatant (S) and pellet (P) was analyzed on duplicate RNA gel blots by hybridization to the indicated exon-specific and intron-specific probes from the trnG-UCC locus (top panels). An immunoprecipitation with antibody to OEC23 served as a negative control. The probes were removed, and the blots were reprobed for the presence of trnD-GUC and trnV-GAC (bottom panels). Arrows point to where irrelevant lanes have been removed from images.
Reverse transcription-PCR (RT-PCR) quantification of the ppr5 mRNA was performed by RT from 2 μg total leaf RNA with the gene-specific primers ppr5-cDNAR3 and Actin-R using the Quantitect reverse transcription kit (Qiagen) according to the manufacturer's recommendations, followed by PCR amplification with these primers in conjunction with the forward primers ppr5-cDNAF3 and Actin-F. PCRs (20 μl) contained 0.4 μl of cDNA. ppr5 cDNA PCRs included 0.5 μM of each primer in PCR buffer (5× Phusion HF buffer; Finnzymes), 0.8 mM deoxynucleotide triphosphates, and 0.4 units of Phusion DNA polymerase (Finnzymes) and were performed with the following PCR profile: 98°C/30 s, followed by 25 to 32 cycles of 98°C/10 s, 53°C/30 s, and 72°C/30 s and a final extension of 72°C/5 min. Actin cDNA PCRs included 0.2 μM of each primer in PCR buffer (10× PCR buffer, Qiagen), 0.8 mM deoxynucleotide triphosphate, and 2.5 units of Taq DNA polymerase (Qiagen) and were performed with the following PCR profile: 94°C/3 min, followed by 25 to 32 cycles of 94°C/30 s, 53°C/30 s, and 72°C/30 s and a final extension of 72°C/5 min.
Antibody production.
A fragment of PPR5 (amino acids 109 to 149) with a C-terminal six-His tag was expressed in Escherichia coli and used for the generation of antibodies in rabbits at the University of Oregon antibody facility. The expression clone pPPR5ex was produced by the PCR amplification of DNA from the inbred line B73 (for primers, see Table S2 in the supplemental material). The PCR product was digested with NcoI and XhoI and cloned into pet28b (Novagen) that had been digested with NcoI and XhoI. For antigen expression, BL21 Star (DE3) cells (Invitrogen) were transformed with pPPR5ex, and a 1-liter culture was grown with Luria-Bertani medium plus kanamycin (30 μg/liter) to an optical density at 600 nm of 0.6; protein expression was then induced by the addition of 1 mM isopropylthio-β-galactoside, and the cells were grown at 37°C for 120 min. After harvesting, the cell pellets were suspended in 60 ml of buffer A (50 mM Na phosphate [pH 8], 300 mM NaCl, 10 mM imidazole, and 5 mM β-mercaptoethanol) and lysed by passage through a French press. Inclusion bodies were pelleted by centrifugation at 15,000 × g for 15 min. and washed with buffer A containing 2 M urea. Antigen was solubilized in buffer A containing 8 M urea. The clarified antigen was purified by affinity chromatography on nickel-nitrilotriacetic acid agarose beads (Qiagen) that had been equilibrated in buffer A plus 8 M urea. Antigen was eluted in 2 ml of elution buffer (100 mM Na phosphate [pH 4.5] and 8 M urea), precipitated with ethanol, suspended in phosphate-buffered saline, and used for immunization. Antibodies were affinity-purified on a column in which the same PPR5 fragment had been covalently attached to cyanogen bromide-activated Sepharose. Other antibodies used were generated by us and described previously (34, 51, 53).
Chloroplast fractionation and protein analyses.
Total leaf protein was extracted and analyzed by immunoblotting as described previously (2). Chloroplast subfractions were those described previously by Williams and Barkan (54). Stromal extracts (0.5 mg of stromal protein per experiment) were fractionated by sedimentation through sucrose gradients according to Jenkins and Barkan (22).
Analysis of RNAs that coimmunoprecipitate with PPR5.
RNA coimmunoprecipitation assays via RNA coimmunoprecipitation and chip analysis (RIP-chip) and slot blot hybridization were performed as described previously (41). Replicate assays used antibodies from two different rabbits immunized with the PPR5 antigen. In brief, stromal protein extract containing ∼1 mg of protein was used for each immunoprecipitation. RNA was purified from the pellet and supernatant fractions and resuspended in 20 μl RNase-free water. Ten microliters of the pellet RNA and 5 μl of the supernatant RNA were coupled to the fluorescent dyes Cy5 and Cy3, respectively, by a direct RNA labeling method (Micromax ASAP RNA labeling kit; Perkin-Elmer). The two fractions were combined and hybridized to a maize chloroplast tiling microarray (fragments described at MIAME Express A-MEXP-164). The array was scanned for Cy3 and Cy5 fluorescence, thus assigning each spot a red (Cy5) and green (Cy3) fluorescent value. The ratios were compared between replicate samples and negative controls, which leads to a value termed “differential enrichment” that was plotted against the genomic position of each PCR product on the array (described in detail in reference 41). For the slot blot hybridizations, 1/6 and 1/12 of the RNAs recovered from the immunoprecipitation pellet and supernatant, respectively, were applied to each slot.
Run-on transcription analysis.
Intact chloroplasts from ∼15 9-day-old maize seedlings were isolated according to Jenkins and Barkan (22). Run-on transcription reactions were performed essentially as described by Zoschke et al. (57). A total of 1.9 × 107 chloroplasts were used in run-on transcription experiments, performed at 25°C for 10 min in 50 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 10 mM β-mercaptoethanol, 20 U RNase inhibitor, and 0.2 mM each of ATP, GTP, and CTP, in the presence of [α-32P]UTP (10 μCi/μl). RNA was extracted and hybridized overnight at 58°C to DNA fragments (1 μg) dot-blotted onto nylon membranes. Signals were detected using a PhosphorImaging system and its associated software (Bio-Rad). The sequences of the primers used for the generation of the DNA probes along with their positions within the maize DNA are given in Table S2 in the supplemental material.
Expression of recombinant PPR5 and RNA binding assays.
A maltose-binding protein (MBP)-PPR5 fusion was generated from a plasmid constructed in the following manner. A PCR product was generated from a ppr5 cDNA (GenBank accession no. EU037901) by PCR amplification with primers 5′-CGGGATCCGCGGCGGAGGGGGTGG-3′ and 5′-CGCGTCGACTTATGTAGCAGCTACATGCCA-3′. The PCR product was digested with BamHI and SalI and cloned into BamHI/SalI-cut pMAL-TEV to generate plasmid pmaltevPPR5. This plasmid encodes an MBP-PPR5 fusion protein in which the PPR moiety starts at PPR5 amino acid 46, which corresponds to the transit peptide cleavage site predicted by ChloroP. The MBP-PPR5 fusion protein was expressed by introducing pmaltevPPR5 into Rosetta 2 (DE3) cells (Novagen). The cultures were grown at 37°C in LB with carbenicillin and chloramphenicol to an optical density at 600 nm of 0.8 and transferred to ice for 20 min. Protein expression was then induced by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to 1 mM and incubation for 4 h at 20°C. The harvested cells were suspended in ice-cold lysis buffer [50 mM Na phosphate (pH 7.5), 300 mM NaCl, 10 mM β-mercaptoethanol, 0.13 mM phenylmethylsulfonyl fluoride, 0.5 μg/ml leupeptin, 0.06 μg/ml pepstatin, 0.01% CHAPS (3-[(3-cholamidopropyl)-dimethylammonio]-1-propranesulfonate)]. The cells were lysed at 4°C by sonication, cooled on ice for 5 min, and then sonicated a second time. The lysate was cleared by centrifugation at 13,000 × g for 20 min. The MBP-PPR5 fusion protein was purified by incubating the cleared lysate with amylose-coupled agarose resin (New England BioLabs) at 4°C for 1 h with rotation. After three washes with lysis buffer, protein was eluted in the same buffer supplemented with 15 mM maltose. The eluted protein was filtered (Amicon Ultrafree-MC, 0.45 μm) and resolved on a 1- by 30-cm Superdex 200 column (GE Healthcare Life Sciences) in 50 mM Na phosphate (pH 7.5), 300 mM NaCl, 5 mM β-mercaptoethanol. The peak fractions, which eluted at a position corresponding to the monomeric molecular weight of MBP-PPR5, were dialyzed against 50 mM Na phosphate (pH 7.5), 300 mM NaCl, 5 mM β-mercaptoethanol, and 50% glycerol and stored at −20°C. The MBP used as a negative control was recovered from a preparation that was initiated in the same manner but that was cleaved with tobacco etch virus protease prior to amylose-affinity chromatography. The MBP was further purified by gel filtration chromatography.
RNAs used in gel mobility shift assays were generated by in vitro transcription from PCR products with a T7 promoter incorporated into the 5′ primer sequence. Transcription reactions (20 μl) included 1 μl T7 RNA polymerase (Promega); 1 μl of [α-32P]UTP (800 Ci/mmol, 10 mCi/ml); 4 μl of a mixture of ATP, CTP, and GTP (2.5 mM each); 1 μl of 1 mM UTP; 1 μl of 100 mM dithiothreitol; 1 μl RNasin (Promega); 0.2 μM DNA template; and the transcription buffer supplied by the manufacturer. The reactions were incubated for 1 h at 37°C and then treated with 1 unit of RQ1 DNase (Promega) for 15 min at 37°C. The radiolabeled RNAs were purified on denaturing polyacrylamide gels.
Immediately prior to the binding reactions, the purified RNAs (10 μl of a 100 pM solution) were heated to 94°C for 2 min and snap-cooled on ice. The RNA binding reactions (25 μl) contained 50 mM Na phosphate (pH 7.5), 200 mM NaCl, 4 mM dithiothreitol, 0.04 mg/ml bovine serum albumin, 10% glycerol, 2 mg/ml heparin, 10 units RNasin, 40 pM radiolabeled RNA, and protein concentrations as indicated in Fig. 6. The reactions were incubated for 30 min at 25°C. The samples were applied to a running 5% polyacrylamide (29:1 acrylamide/bis-acrylamide) gel prepared in 1× THE (66 mM HEPES, 34 mM Tris [pH 7.5], 0.1 mM EDTA). The gels were run at 15 W (constant power) at 4°C with buffer recirculation until the bromophenol blue in an adjacent lane had migrated 5 cm. The gels were fixed, dried, and imaged with a phosphorimager.
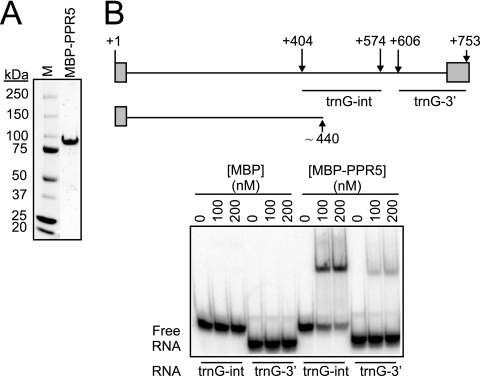
FIG. 6.
RNA binding activity of recombinant PPR5. (A) Purified MBP-PPR5 used in RNA binding assays. The protein recovered after amylose-affinity and gel filtration chromatography was resolved by SDS-polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue. M, molecular mass marker. (B) Gel mobility shift assays demonstrating that MBP-PPR5 binds with high affinity to the central region of the trnG-UCC intron. The trnG-UCC locus is diagrammed at the top, with exons shown as rectangles, the intron as a line, and nucleotide positions with respect to the 5′ end (+1) indicated. The sequences represented in RNAs used for the gel mobility shift assays (trnG-int and trnG-3′) and the truncated RNA of ∼440 nucleotides that accumulates in a PPR5-dependent manner (see Fig. 5) are diagrammed below. MBP was used in parallel binding assays as a negative control.
Sequence comparisons.
Clustal W (48) and the Phylip software package version 3.67 (http://evolution.genetics.washington.edu/phylip/general.html) were used for the protein sequence alignments and for calculating distance trees using the neighbor-joining method.
RESULTS
Null alleles of PPR5 are embryo lethal in Arabidopsis but survive to the seedling stage in maize.
AtPPR5 (At4g39620) is predicted to be targeted to chloroplasts by both the TargetP (14) and Predotar (44) algorithms. Transfer-DNA insertions in this gene have been reported to lead to embryo arrest at the early globular stage (10). We previously recovered germinating mutations in the maize orthologs of two Arabidopsis genes encoding plastid-localized, embryo-essential PPR proteins; null alleles of these genes, ppr2 and ppr4, condition albino, seedling lethal mutants in maize (42, 54). We therefore reasoned that null mutants in the maize ppr5 ortholog might likewise be recoverable. The rice (Oryza sativa) ortholog of AtPPR5, OsPPR5, is predicted to be Os02g51480 (http://pogs.uoregon.edu; see Figure S1A in the supplemental material). OsPPR5 was used to query public maize sequence data using the BlastN program. The most similar maize sequence had 81% nucleotide identity to OsPPR5, whereas the next most closely related maize gene had only 27% identity. When this maize sequence was used to query the complete rice genome, OsPPR5 was its best hit. The phylogenetic tree shown in Figure S1A in the supplemental material supports the orthology among these maize, rice, and Arabidopsis proteins. This maize gene is referred to as Zmppr5. The maize, rice, and Arabidopsis PPR5 orthologs each encode a protein with 10 PPR repeats and show extensive similarity throughout their length except at their N termini, which are predicted to be cleavable chloroplast targeting signals (see Figure S1B in the supplemental material).
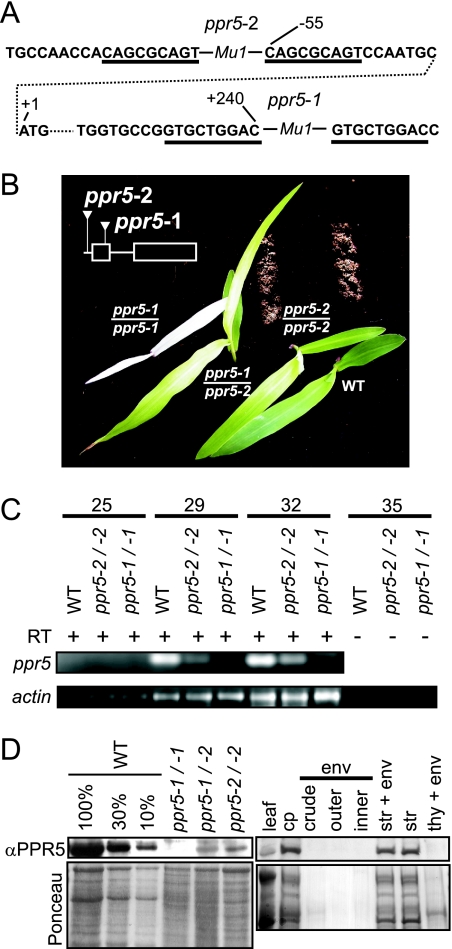
The Zmppr5 sequence was used to identify Zmppr5 mutants in a reverse genetic screen of our large collection of Mu-transposon-induced nonphotosynthetic maize mutants (http://pml.uoregon.edu/). Two insertion alleles were recovered: ppr5-1 has a Mu1 insertion in protein coding sequences in exon 1, and ppr5-2 has a Mu1 insertion 55 nucleotides upstream of the predicted start codon (Fig. 1A). Seeds homozygous for these insertions germinated, giving rise to seedling-lethal albino (ppr5-1) or pale green (ppr5-2) mutants (Fig. 1B). The level of ppr5 mRNA in the mutants correlates with the severity of their chlorophyll deficiencies (Fig. 1C): the ppr5 mRNA is reduced but detectable in pale green ppr5-2 mutants, whereas no ppr5 mRNA was detectable by RT-PCR in albino ppr5-1 mutants. The progeny of crossing ppr5-1/+ plants with ppr5-2/+ plants segregated seedlings with a phenotype intermediate between those of the two parental alleles (Fig. 1B), demonstrating that the insertions in Zmppr5 are responsible for the chlorophyll-deficient phenotypes. The noncomplementing progeny of these crosses were used for most of the following experiments, because it is virtually certain that mutant phenotypes exhibited by these plants result from disruption of Zmppr5 and because albino mutants such as ppr5-1 homozygotes exhibit various pleiotropic defects in chloroplast gene expression (see, e.g., references 53 and 54) and are a poor source for chloroplast isolation.
FIG. 1.
Maize ppr5 mutants. (A) Positions of the Mu transposon insertions in ppr5-1 and ppr5-2 mutants. The DNA sequence flanking the Mu insertions is shown. Residue numbers with respect to the start codon in the genomic sequence are indicated. The start codon is shown in the second row, connected to upstream and downstream noncontiguous sequences by a dotted line. The type of Mu element at each site (Mu1) was deduced from polymorphisms in the terminal inverted repeat sequences. The direct nonamer repeats that were duplicated during the insertion process are underlined. (B) Phenotypes of ppr5 mutant seedlings grown on soil for 14 days. The positions of the Mu insertions with respect to the ppr5 protein coding sequence are diagrammed in the insert. (C) ppr5 mRNA is decreased in ppr5 mutants. RT-PCR was used to assay ppr5 mRNA levels using primers located in exon 1 and exon 2 that flank the Mu1 insertion in ppr5-1 mutants. The number of PCR cycles is indicated at the top. The bottom panel shows the RT-PCR amplification of actin mRNA in the same RNA samples. The 35-cycle PCRs on the same samples without the inclusion of a RT step demonstrate that the signals do not arise from contaminating genomic DNA. Furthermore, no intron-containing amplification products were detected, demonstrating that the bands observed arose from RNA templates. (D) PPR5 localizes to chloroplast stroma. For the left panel, total leaf proteins from wild-type and ppr5 mutants were separated by SDS-polyacrylamide gel electrophoresis, blotted, and probed with the PPR5 antibody. A portion of the same blot stained with Ponceau S is shown below. Dilutions of the wild-type sample were included to aid in quantification. For the right panel, an immunoblot of the subcellular fractions described and verified previously (1) was probed with the PPR5 antibody. The chloroplast and subchloroplast samples are derived from the same quantity of chloroplasts (cp). Replicate blots were probed previously to detect markers for the stroma (str; Cp60), the thylakoid membrane (thy; PsbA), and the inner envelope membrane (env; IM35) (1). WT, wild type.
An antibody raised to a recombinant PPR5 fragment was used to probe immunoblots of total leaf extracts (Fig. 1D). A protein of the expected size (51 kDa after transit peptide cleavage) was found in wild-type leaf, but was undetectable in ppr5-1/ppr5-1 mutants and was strongly reduced in ppr5-1/ppr5-2 and ppr5-2/ppr5-2 mutants. PPR5 is enriched in isolated chloroplasts and in the purified stromal fraction, in comparison with its concentration in total leaf extract (Fig. 1D, right lanes). It was not detected in purified thylakoid or envelope membranes. These results demonstrate that PPR5 localizes to the chloroplast stroma.
Zmppr5 mutants have a reduced content of plastid ribosomes.
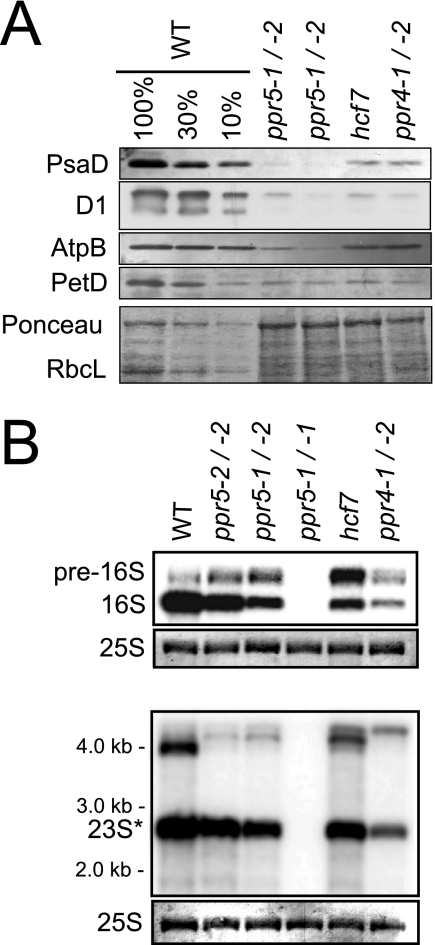
The albino leaf pigmentation of ppr5 null mutants (ppr5-1/-1) resembles that of previously described maize mutants lacking plastid ribosomes (e.g., ppr4, ppr2, crs2-1, rnc1, and iojap; 23, 42, 52, 53, 54), suggesting that ppr5 mutants might be defective in plastid translation. In support of this possibility, ppr5-1/ppr5-2 mutants exhibit a reduction in all photosynthetic enzyme complexes that include plastid-encoded subunits (Rubisco, photosystem II, cytochrome b6f, photosystem I, and ATP synthase; Fig. 2A). None of these proteins were detectable in albino ppr5-1 homozygotes (data not shown), as is typical of albino maize mutants (23, 42). To determine whether a deficiency of plastid ribosomes underlies these protein deficiencies, the accumulation of plastid rRNAs in ppr5 mutants was analyzed. The level of mature 16S rRNA is decreased and a precursor of 16S rRNA overaccumulates in hypomorphic ppr5 mutants (Fig. 2B, top panel), as in the previously described hcf7 and ppr4 mutants that are impaired in plastid translation (3, 42). Analogous results were obtained for the plastid 23S rRNA (Fig. 2B, bottom panel). In albino plants homozygous for the ppr5-1 null allele, no plastid rRNA was detectable. Taken together, these results show that ppr5 is necessary for the accumulation of plastid ribosomes and, therefore, for the accumulation of photosynthetic enzyme complexes harboring plastid-encoded subunits. The additional analyses described below strongly suggest that these reductions in rRNA processing and rRNA accumulation are secondary effects of a primary defect in the biogenesis of a plastid tRNA.
FIG. 2.
Loss of plastid rRNAs and plastid-encoded proteins in ppr5 mutants. (A) Immunoblot analysis of photosynthetic enzyme accumulation in ppr5 mutants. Total leaf proteins (5 μg or the indicated dilution of the wild-type sample) were analyzed by probing immunoblots with antisera to representative subunits of photosystem I (PsaD), photosystem II (D1), ATP synthase (AtpB), and the cytochrome b6f complex (PetD). The same filter was stained with Ponceau S to visualize total proteins (bottom panel). RbcL, the large subunit of Rubisco, is marked. The hcf7 and ppr4 mutants were shown previously to have a global loss of plastid-encoded proteins due to a plastid ribosome deficiency (3, 42). (B) RNA gel blot hybridizations showing plastid rRNA defects in ppr5 mutants. Total leaf RNA (0.5 μg) was analyzed by hybridization to probes for the plastid 16S (top panel) or 23S (bottom panel) rRNA. Excerpts of the same filters stained with methylene blue show the cytosolic 25S rRNA as a loading control. Previously described mutants with lesions in 16S rRNA maturation (hcf7 and ppr4-1/ppr4-2) are shown for comparison (3, 42). The maize chloroplast 23S rRNA exists as two cleavage products in vivo, one of which overlaps the probe used here (23S*). WT, wild type.
PPR5 is associated in vivo with unspliced trnG-UCC RNA.
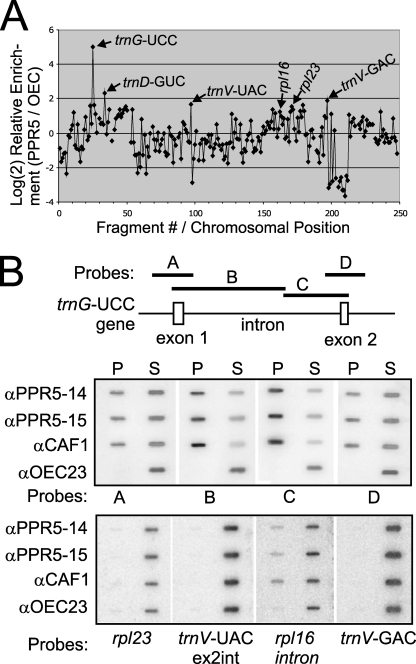
The pleiotropic defects associated with plastid ribosome deficiencies can make it difficult to distinguish the direct from the indirect effects of the underlying mutation (see, for example, reference 54). To gain insight into the direct function of PPR5, RNAs that are associated with ZmPPR5 in chloroplast extract were identified using a RIP-chip assay (41) as an initial screen. ZmPPR5 was immunoprecipitated from maize stromal extract, and RNAs purified separately from the immunoprecipitation pellet and supernatant were labeled with Cy5 and Cy3, respectively. The RNA samples were combined and hybridized to a maize chloroplast genome tiling microarray. The normalized ratio of fluorescence from Cy5 versus Cy3 (F635:F532) for replicate spots across replicate experiments reflects the enrichment of an RNA in the pellet of an immunoprecipitation. Two replicate experiments were performed, using anti-PPR5 antibodies from two different immunized rabbits. In addition, two negative control experiments used antibodies to OEC23 and OEC16; these proteins are part of the oxygen-evolving complex of photosystem II and are not anticipated to bind RNA. Comparison of the F635:F532 ratios in replicate PPR5 assays with those in the negative control assays identified RNAs that are differentially enriched in PPR5 immunoprecipitations. The significance of this differential enrichment for each sequence on the array was scored using Student's t test (see Table S1 in the supplemental material). To highlight the RNAs that are differentially enriched, the log-transformed median enrichment ratio for each array fragment among replicate PPR5 assays was plotted according to chromosomal position, after subtracting the corresponding values for the replicate control assays (Fig. 3A). Only one prominent peak of differential enrichment was observed, which mapped to the intron-containing gene trnG-UCC. A t test analysis confirmed that RNAs corresponding to this array fragment were significantly enriched in PPR5 immunoprecipitations in comparison to the control immunoprecipitations (fragment 25; see Table S1 in the supplemental material). These experiments point to RNA from the trnG-UCC locus as the primary target for PPR5.
FIG. 3.
Identification of RNAs associated with PPR5 in chloroplast stroma. (A) Summary of RIP-chip data. The log2-transformed enrichment ratios (F635:F532) were normalized between two assays involving PPR5 antisera from different immunized rabbits and two control experiments with antibodies to OEC23 and OEC16. The median normalized values for replicate spots from the control assays were subtracted from those from the PPR5 assays and plotted according to fragment number, which reflects their chromosomal position. Fragments for which at least three replicate spots per array did not meet our background cutoff in the supernatant (F532) channel were not used in this plot and appear as gaps in the curve. (B) Validation of RIP-chip data by slot blot hybridization of RNAs that coimmunoprecipitate with PPR5. A schematic map of the trnG-UCC gene and the probes used for slot blot hybridization (A through D) are shown at the top. One-sixth of the RNA recovered from each immunoprecipitation pellet (P) and 1/12 of the RNA recovered from each supernatant (S) were applied to replicate slot blots and hybridized with probes A through D. Antibodies from two immunized rabbits were used in replicate assays (αPPR5-14 and αPPR5-15). An immunoprecipitation with antibody to OEC23 was used as a negative control. An immunoprecipitation with antibody against the splicing factor CAF1, which includes unspliced trnG-UCC among its ligands (34), served as a positive control. The same blots were reprobed after the decay of the previous signal to detect four RNAs for which the RIP-chip data suggested possible minor enrichment (bottom panel).
As an independent verification of the RIP-chip data, RNAs that coimmunoprecipitate with PPR5 were analyzed by slot blot hybridization using probes from the trnG-UCC locus and from loci corresponding to minor RIP-chip peaks. An immunoprecipitation with antibody to CAF1, which is established to bind the trnG-UCC intron in vivo (34, 41), was used as a positive control. Sequences throughout the trnG-UCC locus were strongly enriched by immunoprecipitation with PPR5 and CAF1 antibodies but not with OEC23 antibody (Fig. 3B, top panel). Notably, the strongest enrichment was detected with probes containing predominantly intronic sequences, suggesting that PPR5 is bound to intron sequences, with flanking exons coprecipitated due to their being tethered to the same RNA molecule (Fig. 3B). RNAs from five loci corresponding to minor peaks in the RIP-chip data (rpl23, trnV-UAC, trnV-GAC, rps2, and rpl33) scored negative in the slot blot assay (Fig. 3B, bottom panel, and Figure S2 in the supplemental material). Likewise, RNA gel blot analysis of the immunoprecipitated RNAs did not detect the enrichment of RNA from the trnD-GUC or trnV-GAC locus (Fig. 4, bottom panels). In contrast, rpl16 intron RNA, for which a statistical analysis of the RIP-chip data suggested a possible association with PPR5 (see Table S1 in the supplemental material), was confirmed by slot blot hybridization to coimmunoprecipitate with PPR5, albeit rather weakly. Together, these coimmunoprecipitation data demonstrate that RNA from the trnG-UCC locus is the major ligand of PPR5. They suggest further that rpl16 RNA may be a minor PPR5 ligand and that PPR5 has few, if any, additional RNA ligands.
PPR5 is associated with the unspliced trnG-UCC precursor but not with the mature tRNA.
To test whether PPR5 associates with the unspliced trnG-UCC precursor, the spliced tRNA, or both, we analyzed RNAs that coimmunoprecipitate with PPR5 from stromal extract by RNA gel blot hybridization. Replicate blots were probed with intron-specific or exon-specific trnG-UCC probes (Fig. 4, top panels); the same blots were stripped and then reprobed to detect trnD-GUC and trnV-GAC RNAs, which correspond to minor peaks in the RIP-chip data (Fig. 4, bottom panels). The unspliced trnG-UCC precursor was enriched in the pellets of PPR5 immunoprecipitations relative to the supernatant RNA, whereas the mature trnG-UCC, trnD-GUC, and trnV-GAC RNAs were not detected in the PPR5 immunoprecipitation pellet. These results indicate that PPR5 associates with the unspliced trnG-UCC precursor but not with the mature tRNA.
PPR5 is required for the accumulation of the unspliced trnG-UCC precursor.
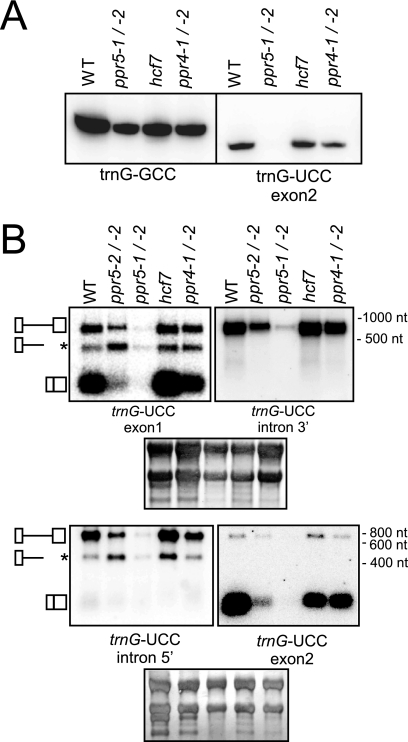
To determine whether PPR5 influences the metabolism of the unspliced trnG-UCC precursor, we analyzed the accumulation of trnG-UCC transcripts in ppr5 mutants. The ppr5-1/ppr5-2 progeny of complementation crosses and ppr5-2 homozygotes were used for these analyses rather than the more severely affected ppr5-1 homozygotes because severe plastid-ribosome deficiencies, as observed in ppr5-1/ppr5-1 seedlings, are known to cause secondary effects on plastid RNA accumulation (17, 18, 21, 23, 54). To control for possible pleiotropic effects of the comparatively mild ribosome deficiencies in ppr5-2/ppr5-2 and ppr5-1/ppr5-2 material, RNAs from ppr4-1/ppr4-2 and hcf7 mutants were analyzed in parallel. Leaf RNAs were fractionated on polyacrylamide gels, blotted, and probed with an oligonucleotide complementary to exon 2 of trnG-UCC and, as a control, with an oligonucleotide specific for a different plastid-encoded tRNA isoacceptor, trnG-GCC. trnG-UCC was strongly reduced in ppr5-1/ppr5-2 mutants, whereas trnG-GCC abundance was similar to that in the control mutants (Fig. 5A). To analyze the accumulation of unspliced precursors, the same RNAs were fractionated on agarose gels and probed with exon-specific or intron-specific trnG-UCC probes (Fig. 5B). The results confirmed the loss of mature trnG-UCC and, in addition, demonstrated that the unspliced precursor is reduced specifically in ppr5 mutants. The loss of both mature trnG-UCC and the unspliced precursor is more severe in ppr5-1/ppr5-2 mutants than in ppr5-2/ppr5-2 mutants, correlating with the degree to which the expression of ppr5 is disrupted in each mutant background (Fig. 1). Neither the precursor nor the mature transcript is detectable in ppr5-1 homozygotes (data not shown), as in other albino maize mutants lacking plastid ribosomes (e.g., rnc1 and iojap; 53). These results indicate that PPR5 is required for the synthesis or stabilization of the trnG-UCC precursor.
FIG. 5.
RNA gel blot hybridizations of trnG-UCC transcripts in ppr5 mutants. (A) Total leaf RNA (4 μg/lane) from seedlings with the indicated genotypes was fractionated on a 20% polyacrylamide gel and transferred to a nylon membrane. The membrane was cut into two strips, which were hybridized with the indicated oligonucleotide probes. (B) Total leaf RNA (4 μg/lane) was fractionated on agarose gels, transferred to nylon membranes, and hybridized to the probes indicated. The filter shown in the upper panels was first hybridized to a trnG-UCC exon 1 probe and, after removal of this probe, reprobed with a probe from the 3′ region of the trnG-UCC intron. The filter shown in the lower panels was first hybridized to a trnG-UCC exon 2 probe and, after removal of this probe, reprobed with a probe from the 5′ region of the trnG-UCC intron. Shown below are the same blots stained with methylene blue to illustrate equal sample loading. The diagrams on the left depict the three transcripts detected (top, unspliced tRNA; middle, truncated precursor tRNA; bottom, spliced tRNA). WT, wild type; nt, nucleotides.
The RNA gel blots in Fig. 5B detected an unanticipated RNA of ∼440 nucleotides that hybridized to an exon 1-specific oligonucleotide and to sequences near the 5′ end of the trnG-UCC intron (see asterisk in Fig. 5B). This band was not detected with probes specific for the 3′ region of the intron or exon 2, indicating that it is a truncated form of the primary transcript that ends within the intron. Intriguingly, the ratio of this truncated transcript to the unspliced precursor is consistently higher in ppr5 mutants than in wild-type siblings or in hcf7 or ppr4 control mutants. These results suggest that PPR5 either inhibits an endonucleolytic cleavage that generates this “dead-end” RNA fragment or inhibits premature transcription termination within the trnG-UCC intron.
To determine whether PPR5 plays a similar role in the metabolism of the rpl16 RNA, which was also enriched in PPR5 immunoprecipitations albeit much less robustly than trnG-UCC RNA (Fig. 3B), rpl16 transcripts in ppr5 mutants were examined on RNA gel blots (Figure S3 in the supplemental material). A large intron-containing precursor RNA is reduced and a smaller intron-containing RNA is increased in ppr5 mutants (see asterisks in Figure S3 in the supplemental material) in a manner that is reminiscent of the results for trnG-UCC transcripts; however, the accumulation of mature rpl16 transcripts is unaffected. RNAs from several other loci corresponding to minor peaks in the RIP-chip experiments were also assayed in this manner, but the transcript patterns were similar in the wild-type and ppr5 mutant samples (see Figure S3 in the supplemental material).
Recombinant PPR5 binds with high affinity to the central region of the trnG-UCC intron.
The coimmunoprecipitation data demonstrated that PPR5 is associated with the unspliced trnG-UCC precursor in vivo. To elucidate whether PPR5 interacts directly with this RNA, recombinant PPR5 was tested for its ability to bind RNA in vitro. PPR5 was expressed in E. coli as a fusion to MBP and purified by sequential amylose-affinity and gel filtration chromatography (Fig. 6A). Under stringent binding conditions (200 mM NaCl, 2 μg/μl heparin), MBP-PPR5 bound with high affinity to an RNA containing an interior segment of the trnG-UCC intron, whereas it bound only weakly to an RNA of similar size from the 3′ region of the intron (Fig. 6B). The residual binding detected to the latter is nonspecific, as several other RNA molecules of this length gave similar results (data not shown). These results show that PPR5 is an RNA binding protein that has a high affinity binding site near the 3′ end of the truncated transcript that accumulates at the expense of the intact precursor in hypomorphic ppr5 mutants (see Fig. 5).
PPR5 is not required for trnG-UCC transcription.
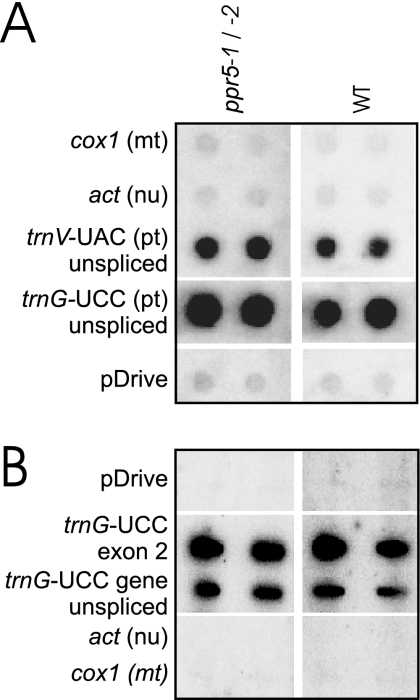
The reduction of trnG-UCC precursor RNA in ppr5 mutants could be explained by a loss of transcription, decreased RNA stability, or both. To distinguish between these possibilities, chloroplast transcription run-on experiments were performed with wild-type and ppr5-1/ppr5-2 mutant chloroplasts. The ratio of run-on transcripts from the trnG-UCC locus with respect to those from the trnV-UAC locus was similar in ppr5-1/5-2 mutant and wild-type chloroplasts (Fig. 7A), suggesting that PPR5 is not needed for normal rates of trnG-UCC transcription initiation. Furthermore, the ratio of run-on transcripts from exon 2 of trnG-UCC relative to those from the complete trnG-UCC gene was similar in the wild-type and ppr5-1/5-2 mutants (Fig. 7B and data not shown), suggesting that transcription does not terminate prematurely within the trnG-UCC intron in the absence of PPR5. These results, together with the RNA gel blot data demonstrating a truncated trnG-UCC precursor transcript, support a role for PPR5 in stabilizing unspliced trnG-UCC by inhibiting an endonucleolytic cleavage event. However, the fidelity of transcription termination in chloroplast transcription run-on assays has not been thoroughly explored, so we cannot eliminate the possibility that transcription through the trnG-UCC locus terminates prematurely in ppr5 mutants in vivo.
FIG. 7.
Run-on transcription assay of trnG-UCC transcription in ppr5 mutants. (A) RNAs purified from run-on transcription assays involving plastids isolated from ppr5-1/ppr5-2 seedlings or from their normal siblings (wild-type) were hybridized to the indicated DNA fragments on nylon membranes. Both the trnV-UAC and trnG-UCC loci include a group II intron, and these probes encompass exon and intron sequences. DNA fragments from the mitochondrial (mt) cox1 gene, nuclear (nu) actin, and the pDrive vector into which these probes were cloned were included as controls for nonspecific hybridization. The signals were quantified and used to calculate the mean trnG-to-trnV ratio of two replicate experiments. This was similar for the wild-type (2.6) and ppr5 mutants (1.7). pt, plastid. (B) Run-on transcripts were hybridized to a single-stranded antisense oligonucleotide specific for trnG-UCC exon 2 and to a PCR product spanning the entire trnG-UCC gene. The enhanced signal observed with the oligonucleotide probe relative to the PCR product is typical for experiments of this nature (data not shown) and likely represents the inefficient denaturation of double-stranded DNAs on the filter. WT, wild type.
To determine whether PPR5 influences the editing of trnG-UCC intron sequences, we amplified the complete intron from wild-type RNA by RT-PCR and determined the nucleotide sequence of bulk PCR products. No C-to-U editing events were detected (data not shown). No amplifications were detected in reactions where reverse transcriptase was omitted, suggesting that the DNase-treated RNA preparations were free of DNA.
PPR5 resides in large complexes that include the plastid splicing factors CAF1 and RNC1.
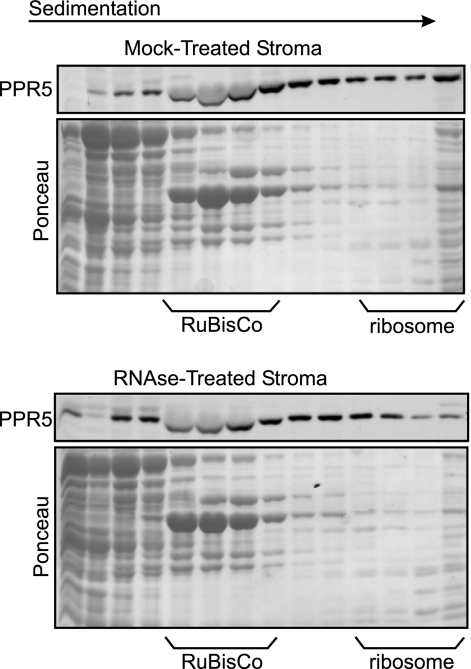
The experiments above established that PPR5 is associated in vivo with trnG-UCC intron-containing RNAs. To estimate the size of these ribonucleoprotein particles and to determine what fraction of PPR5 is associated with them, chloroplast stroma was fractionated on sucrose gradients and the distribution of PPR5 across the gradient was determined by immunoblotting (Fig. 8). PPR5 was found throughout the gradient. The treatment of stroma with RNase A prior to sedimentation had only minor effects, including a possible reduction in its presence in large complexes in the bottom fractions of the gradient (Fig. 8, bottom panel). These results indicate that PPR5 is found in complexes of various sizes and provide additional evidence that this protein is associated with RNA. The resistance of group II introns in chloroplast extract to digestion by RNase has been shown previously (49 and data not shown) and is presumably a consequence of their compact tertiary structure and association with proteins. Thus, the rather small effect of RNase treatment on the sedimentation of the PPR5-containing particles is consistent with PPR5's association with a group II intron RNA.
FIG. 8.
Sucrose gradient sedimentation analysis of PPR5-containing particles in stromal extract. Stromal extract was treated with RNase A or incubated under the same conditions in the absence of RNase and sedimented through sucrose gradients. An equal volume of each gradient fraction was analyzed on immunoblots probed with PPR5 antibody. Blots stained with Ponceau S are shown below. The last lane in each panel contains material that pelleted in the gradients. The position of ribosomes was determined by the pattern of Ponceau S-stained bands and by the appearance of rRNAs in these fractions (data not shown).
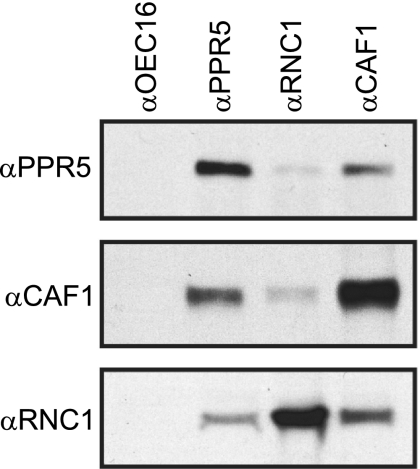
Previously, two proteins that interact with the trnG-UCC intron were identified, CAF1 and RNC1, both of which promote its splicing (34, 53). To test whether PPR5 associates with CAF1 and RNC1 in vivo, antisera against these three proteins were used in coimmunoprecipitation experiments with chloroplast stroma. The CAF1 and RNC1 antisera both coimmunoprecipitated PPR5, and reciprocally, PPR5 antisera coimmunoprecipitated CAF1 and RNC1 (Fig. 9). The coprecipitation of PPR5 with RNC1 is markedly less efficient than its coprecipitation with CAF1, which could be indicative that PPR5 and RNC1 do not always associate simultaneously with the trnG-UCC intron RNA. These results demonstrate that PPR5 is found in large ribonucleoprotein complexes that also contain the trnG-UCC splicing factor CAF1 and possibly also RNC1.
FIG. 9.
Coimmunoprecipitation of PPR5 with CAF1 and RNC1 from chloroplast extract. Chloroplast stroma was incubated with the affinity-purified antiserum indicated above each lane. Immunoprecipitates were analyzed on immunoblots to detect PPR5 (top panel), CAF1 (middle panel), or RNC1 (bottom panel). Antiserum to OEC16 was used as a negative control.
DISCUSSION
PPR proteins as protectors of RNase-sensitive sites.
Most PPR proteins analyzed to date are required for a specific posttranscriptional step in the expression of one or several organellar genes. The specificity of the defects encountered in mutants with lesions in PPR-encoding genes suggested that PPR proteins interact with specific RNA sequences. However, experimental verification of specific interactions either in vivo or in vitro remains sparse. Moreover, although PPR proteins have been implicated in various steps in gene expression, such as the splicing of a specific intron or translation of a specific mRNA, the mechanisms by which PPR proteins act in these processes have not been elucidated. We demonstrate here that the primary ligand of PPR5 in vivo is an unspliced plastid tRNA precursor, pre-trnG-UCC; that recombinant PPR5 binds to this RNA in vitro; and that PPR5 is essential for the accumulation of this precursor and its spliced product. In hypomorphic ppr5 mutants, a truncated precursor molecule overaccumulates relative to the intron-containing pre-trnG-UCC, yet transcription through the trnG-UCC locus is unaffected during transcription run-on experiments. A high-affinity binding site for recombinant MBP-PPR5 maps near the 3′ terminus of this truncated transcript, suggesting that direct interaction between PPR5 and the RNA in this region promotes the accumulation of the full-length precursor. In a parallel study, we fine-mapped the binding site of recombinant untagged PPR5 and found that the 3′ end of the truncated trnG-UCC RNA lies within the ∼45-nucleotide minimal PPR5 binding site (55). Together, these results strongly suggest that in wild-type plants, PPR5 binds to an RNase-hypersensitive site in the intron of pre-trnG-UCC, preventing access by an endonuclease, and that this protection is essential for the accumulation of the unspliced pre-trnG-UCC substrate for splicing. In ppr5 mutants, pre-trnG-UCC is not protected and thus becomes cleaved. The 3′ cleavage product is then degraded, whereas the 5′ product accumulates. As a consequence of precursor degradation, the mature tRNA is lost as well.
The endonucleolytic cleavage of chloroplast RNAs has been recognized as one entry route into the degradation of chloroplast mRNAs (7). Among the previously characterized factors determining the stabilities of specific chloroplast RNAs are two other PPR proteins: MCA1 is required for the regulated accumulation of the petA mRNA in Chlamydomonas spp. chloroplasts (37) and PGR3 of Arabidopsis is required for the accumulation of the chloroplast petL-petG-psaJ precursor (56). PPR proteins in Drosophila spp., yeast, and trypanosomes have also been shown to stabilize specific RNAs (27, 28, 36). The function of the PPR protein PET309 in Saccharomyces cerevisiae mitochondria is particularly reminiscent of that of PPR5, in that its absence causes the loss of group II intron-containing cox1 pre-mRNAs (29). Interestingly, the pentatricopeptide repeats in PET309 are not required for its role in cox1 mRNA accumulation (47). However, in the case of PPR5, the PPR tract makes up almost all of the protein (see Figure S1 in the supplemental material) and is therefore likely to be critical for its effect on pre-trnG-UCC stability.
It has been proposed that the PPR domain functions merely as a sequence-specific RNA adaptor to recruit additional proteins that carry out active processes like RNA editing or endonucleolytic cleavage (26, 43). However, for passive functions like the protection of an RNase-sensitive site, no such additional factor has to be invoked. This mechanistic simplicity and the fact that all PPR proteins known to influence RNA stability belong to the phylogenetically old and widespread P-type subfamily of PPR proteins (26) suggest that protection of RNA against degradation might be an ancient function of PPR proteins. An interaction with an ancestral PPR5 protein could have allowed the maintenance of a mutation in the trnG-UCC intron that, in the absence of PPR5, would result in precursor decay. This would resemble the situation of PPR proteins that restore fertility in certain male sterile backgrounds, which are an evolutionary answer to sterility-causing aberrancies in plant mitochondrial genomes (8, 19).
The finding that PPR5 stabilizes an unspliced tRNA precursor does not rule out that it might also promote the splicing of the trnG-UCC intron. The position of the PPR5 binding site, as established through in vitro assays, suggests that the PPR5/intron interaction may, in fact, promote splicing by enhancing the accessibility of an intron element, EBS1, which must pair with complementary IBS1 sequences in the 5′ exon for accurate splicing (55). In fact, the data presented here show that the interaction of PPR5 with the intron overlaps temporally with the presence of CAF1 and RNC1, two proven splicing factors of trnG-UCC (34, 53). It is unclear at present whether PPR5 impacts the splicing of the trnG-UCC intron and, if so, which function—RNA stabilization or splicing—evolved first.
Embryo-essential Arabidopsis chloroplast PPR proteins may generally function in the biogenesis of the chloroplast translation machinery.
The chloroplast genomes of angiosperms encode 30 tRNA genes that are sufficient to decode all plastid ORFs (46). Among them are two glycine isoacceptors, of which only trnG-UCC contains an intron, which is the site for PPR5 binding. We show here that the loss of PPR5 in maize leads to an albino seedling-lethal defect and to the loss of trnG-UCC. The absence of plastid ribosomes and plastid-encoded proteins in ppr5 mutants can be explained by an essential role for mature trnG-UCC in chloroplast translation and thus for chloroplast development. Strikingly, the tobacco ortholog of trnG-UCC decodes all plastid glycine codons and was recently shown to be essential for cellular viability (38), whereas our results suggest that this tRNA in maize is necessary for chloroplast biogenesis but not for embryo viability. The different impacts of chloroplast gene expression on cell survival and embryo development in dicot versus monocot plants have been noted earlier and have been attributed to differences in chloroplast gene content: whereas the cell-essential genes ycf1, ycf2, and accD are found in dicot plastid genomes, they are absent from plastid chromosomes in the grasses (1, 42, 45, 54). PPR5 and the previously characterized protein PPR4 are both required for the maturation of specific RNAs that are necessary for plastid translation (a tRNA and a ribosomal protein mRNA, respectively). The null alleles of the Arabidopsis PPR5 and PPR4 orthologs are embryo lethal, as are the null alleles of Arabidopsis PPR2, whose maize ortholog is necessary for plastid ribosome accumulation (54). Thus, the need for PPR2, PPR4, and PPR5 during dicot embryo development is presumably due to their essential roles in plastid translation and, consequently, for the expression of ycf1, ycf2, and accD. It seems likely that other plastid-localized PPR proteins that have been shown to be essential for embryo development in Arabidopsis (10, 50) are likewise involved in the expression of specific components of the plastid translation apparatus or function directly in the expression of one of the cell-essential plastid ORFs. The combination of RIP-chip and mutant analysis in maize will be a valuable tool for determining the direct functions of other PPR proteins that are essential for embryo development in Arabidopsis.
Supplementary Material
Acknowledgments
Skillful technical assistance by Susan Belcher, Margarita Rojas, Tiffany Kroeger, and Reik Modrozynski is gratefully acknowledged.
Funding for this project was provided by a DFG Emmy-Noether stipend to C.S.-L. and NSF grant DBI-0421799 to A.B.
Footnotes
Published ahead of print on 30 June 2008.
Supplemental data for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Asakura, Y., and A. Barkan. 2007. A CRM domain protein functions dually in group I and group II intron splicing in land plant chloroplasts. Plant Cell 193864-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barkan, A. 1998. Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Methods Enzymol. 29738-57. [Google Scholar]
- 3.Barkan, A. 1993. Nuclear mutants of maize with defects in chloroplast polysome assembly have altered chloroplast RNA metabolism. Plant Cell 5389-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barkan, A., and M. Goldschmidt-Clermont. 2000. Participation of nuclear genes in chloroplast gene expression. Biochimie 82559-572. [DOI] [PubMed] [Google Scholar]
- 5.Barkan, A., L. Klipcan, O. Ostersetzer, T. Kawamura, Y. Asakura, and K. P. Watkins. 2007. The CRM domain: an RNA binding module derived from an ancient ribosome-associated protein. RNA 1355-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berg, M., R. Rogers, R. Muralla, and D. Meinke. 2005. Requirement of aminoacyl-tRNA synthetases for gametogenesis and embryo development in Arabidopsis. Plant J. 44866-878. [DOI] [PubMed] [Google Scholar]
- 7.Bollenbach, T. J., G. Schuster, V. Portnoy, and D. Stern. 2007. Processing, degradation, and polyadenylation of chloroplast transcripts, p. 175-211. In R. Bock (ed.), Cell and molecular biology of plastids, vol. 19. Springer, Berlin, Heidelberg, Germany. [Google Scholar]
- 8.Chase, C. D. 2007. Cytoplasmic male sterility: a window to the world of plant mitochondrial-nuclear interactions. Trends Genet. 2381-90. [DOI] [PubMed] [Google Scholar]
- 9.Coffin, J. W., R. Dhillon, R. G. Ritzel, and F. E. Nargang. 1997. The Neurospora crassa cya-5 nuclear gene encodes a protein with a region of homology to the Saccharomyces cerevisiae PET309 protein and is required in a post-transcriptional step for the expression of the mitochondrially encoded COXI protein. Curr. Genet. 32273-280. [DOI] [PubMed] [Google Scholar]
- 10.Cushing, D. A., N. R. Forsthoefel, D. R. Gestaut, and D. M. Vernon. 2005. Arabidopsis emb175 and other ppr knockout mutants reveal essential roles for pentatricopeptide repeat (PPR) proteins in plant embryogenesis. Planta 221424-436. [DOI] [PubMed] [Google Scholar]
- 11.Delannoy, E., W. A. Stanley, C. S. Bond, and I. D. Small. 2007. Pentatricopeptide repeat (PPR) proteins as sequence-specificity factors in post-transcriptional processes in organelles. Biochem. Soc. Trans. 351643-1647. [DOI] [PubMed] [Google Scholar]
- 12.de Longevialle, A. F., E. H. Meyer, C. Andres, N. L. Taylor, C. Lurin, A. H. Millar, and I. D. Small. 2007. The pentatricopeptide repeat gene OTP43 is required for trans-splicing of the mitochondrial nad1 intron 1 in Arabidopsis thaliana. Plant Cell 193256-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drescher, A., S. Ruf, T. Calsa, Jr., H. Carrer, and R. Bock. 2000. The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J. 2297-104. [DOI] [PubMed] [Google Scholar]
- 14.Emanuelsson, O., H. Nielsen, S. Brunak, and G. von Heijne. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 3001005-1016. [DOI] [PubMed] [Google Scholar]
- 15.Fisk, D. G., M. B. Walker, and A. Barkan. 1999. Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 182621-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gillman, J. D., S. Bentolila, and M. R. Hanson. 2007. The petunia restorer of fertility protein is part of a large mitochondrial complex that interacts with transcripts of the CMS-associated locus. Plant J. 49217-227. [DOI] [PubMed] [Google Scholar]
- 17.Han, C.-D., R. J. Derby, P. S. Schnable, and R. A. Martienssen. 1995. Characterization of the plastids affected by class II albino mutations of maize at the morphological and transcript levels. Maydica 4013-22. [Google Scholar]
- 18.Han, C.-D., W. Patrie, M. Polacco, and E. H. Coe. 1993. Aberrations in plastid transcripts and deficiency of plastid DNA in striped and albino mutants in maize. Planta 191552-563. [Google Scholar]
- 19.Hanson, M. R., and S. Bentolila. 2004. Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell 16(Suppl.)S154-S169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hashimoto, M., T. Endo, G. Peltier, M. Tasaka, and T. Shikanai. 2003. A nucleus-encoded factor, CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis. Plant J. 36541-549. [DOI] [PubMed] [Google Scholar]
- 21.Hess, W. R., A. Prombona, B. Fieder, A. R. Subramanian, and T. Borner. 1993. Chloroplast rps15 and the rpoB/C1/C2 gene cluster are strongly transcribed in ribosome-deficient plastids: evidence for a functioning non-chloroplast-encoded RNA polymerase. EMBO J. 12563-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenkins, B. D., and A. Barkan. 2001. Recruitment of a peptidyl-tRNA hydrolase as a facilitator of group II intron splicing in chloroplasts. EMBO J. 20872-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenkins, B. D., D. J. Kulhanek, and A. Barkan. 1997. Nuclear mutations that block group II RNA splicing in maize chloroplasts reveal several intron classes with distinct requirements for splicing factors. Plant Cell 9283-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kode, V., E. A. Mudd, S. Iamtham, and A. Day. 2005. The tobacco plastid accD gene is essential and is required for leaf development. Plant J. 44237-244. [DOI] [PubMed] [Google Scholar]
- 25.Kotera, E., M. Tasaka, and T. Shikanai. 2005. A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433326-330. [DOI] [PubMed] [Google Scholar]
- 26.Lurin, C., C. Andres, S. Aubourg, M. Bellaoui, F. Bitton, C. Bruyere, M. Caboche, C. Debast, J. Gualberto, B. Hoffmann, A. Lecharny, M. Le Ret, M. L. Martin-Magniette, H. Mireau, N. Peeters, J. P. Renou, B. Szurek, L. Taconnat, and I. Small. 2004. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 162089-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mancebo, R., X. Zhou, W. Shillinglaw, W. Henzel, and P. Macdonald. 2001. BSF binds specifically to the bicoid mRNA 3′ untranslated region and contributes to stabilization of bicoid mRNA. Mol. Cell. Biol. 213462-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manthey, G. M., and J. E. McEwen. 1995. The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J. 144031-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin, W., T. Rujan, E. Richly, A. Hansen, S. Cornelsen, T. Lins, D. Leister, B. Stoebe, M. Hasegawa, and D. Penny. 2002. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. USA 9912246-12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meierhoff, K., S. Felder, T. Nakamura, N. Bechtold, and G. Schuster. 2003. HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell 151480-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakamura, T., K. Meierhoff, P. Westhoff, and G. Schuster. 2003. RNA-binding properties of HCF152, an Arabidopsis PPR protein involved in the processing of chloroplast RNA. Eur. J. Biochem. 2704070-4081. [DOI] [PubMed] [Google Scholar]
- 32.Okuda, K., F. Myouga, R. Motohashi, K. Shinozaki, and T. Shikanai. 2007. Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing. Proc. Natl. Acad. Sci. USA 1048178-8183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okuda, K., T. Nakamura, M. Sugita, T. Shimizu, and T. Shikanai. 2006. A pentatricopeptide repeat protein is a site recognition factor in chloroplast RNA editing. J. Biol. Chem. 28137661-37667. [DOI] [PubMed] [Google Scholar]
- 34.Ostheimer, G. J., R. Williams-Carrier, S. Belcher, E. Osborne, J. Gierke, and A. Barkan. 2003. Group II intron splicing factors derived by diversification of an ancient RNA-binding domain. EMBO J. 223919-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peled-Zehavi, H., and A. Danon. 2007. Translation and translational regulation in chloroplasts, p. 249-282. In R. Bock (ed.), Cell and molecular biology of plastids, vol. 19. Springer, New York, NY. [Google Scholar]
- 36.Pusnik, M., I. Small, L. K. Read, T. Fabbro, and A. Schneider. 2007. Pentatricopeptide repeat proteins in Trypanosoma brucei function in mitochondrial ribosomes. Mol. Cell. Biol. 276876-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raynaud, C., C. Loiselay, K. Wostrikoff, R. Kuras, J. Girard-Bascou, F. A. Wollman, and Y. Choquet. 2007. Evidence for regulatory function of nucleus-encoded factors on mRNA stabilization and translation in the chloroplast. Proc. Natl. Acad. Sci. USA 1049093-9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rogalski, M., D. Karcher, and R. Bock. 2008. Superwobbling facilitates translation with reduced tRNA sets. Nat. Struct. Mol. Biol. 15192-198. [DOI] [PubMed] [Google Scholar]
- 39.Ruppel, N. J., and R. P. Hangarter. 2007. Mutations in a plastid-localized elongation factor G alter early stages of plastid development in Arabidopsis thaliana. BMC Plant Biol. 737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmitz-Linneweber, C., and A. Barkan. 2007. RNA splicing and RNA editing in chloroplasts, p. 213-248. In R. Bock (ed.), Cell and molecular biology of plastids, vol. 19. Springer, Berlin, Heidelberg, Germany. [Google Scholar]
- 41.Schmitz-Linneweber, C., R. Williams-Carrier, and A. Barkan. 2005. RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. Plant Cell 172791-2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmitz-Linneweber, C., R. Williams-Carrier, P. Williams, T. Kroeger, A. Vichas, and A. Barkan. 2006. A pentatricopeptide repeat protein binds to and facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell 182650-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Small, I., and N. Peeters. 2000. The PPR motif—a TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 2546-47. [DOI] [PubMed] [Google Scholar]
- 44.Small, I., N. Peeters, F. Legeai, and C. Lurin. 2004. Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 41581-1590. [DOI] [PubMed] [Google Scholar]
- 45.Stern, D. B., M. R. Hanson, and A. Barkan. 2004. Genetics and genomics of chloroplast biogenesis: maize as a model system. Trends Plant Sci. 9293-301. [DOI] [PubMed] [Google Scholar]
- 46.Sugiura, M., T. Hirose, and M. Sugita. 1998. Evolution and mechanism of translation in chloroplasts. Annu. Rev. Genet. 32437-459. [DOI] [PubMed] [Google Scholar]
- 47.Tavares-Carreon, F., Y. Camacho-Villasana, A. Zamudio-Ochoa, M. Shingu-Vazquez, A. Torres-Larios, and X. Perez-Martinez. 2007. The pentatricopeptide repeats present in Pet309 are necessary for translation but not for stability of the mitochondrial COX1 mRNA in yeast. J. Biol. Chem. 2831472-1479. [DOI] [PubMed] [Google Scholar]
- 48.Thompson, J., D. Higgins, and T. Gibson. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Till, B., C. Schmitz-Linneweber, R. Williams-Carrier, and A. Barkan. 2001. CRS1 is a novel group II intron splicing factor that was derived from a domain of ancient origin. RNA 71227-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tzafrir, I., R. Pena-Muralla, A. Dickerman, M. Berg, R. Rogers, S. Hutchens, T. C. Sweeney, J. McElver, G. Aux, D. Patton, and D. Meinke. 2004. Identification of genes required for embryo development in Arabidopsis. Plant Physiol. 1351206-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Voelker, R., and A. Barkan. 1995. Two nuclear mutations disrupt distinct pathways for targeting proteins to the chloroplast thylakoid. EMBO J. 143905-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walbot, V., and E. H. Coe. 1979. Nuclear gene iojap conditions a programmed change to ribosome-less plastids in Zea mays. Proc. Natl. Acad. Sci. USA 762760-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watkins, K. P., T. S. Kroeger, A. M. Cooke, R. E. Williams-Carrier, G. Friso, S. E. Belcher, K. J. van Wijk, and A. Barkan. 2007. A ribonuclease III domain protein functions in group II intron splicing in maize chloroplasts. Plant Cell 192606-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams, P. M., and A. Barkan. 2003. A chloroplast-localized PPR protein required for plastid ribosome accumulation. Plant J. 36675-686. [DOI] [PubMed] [Google Scholar]
- 55.Williams-Carrier, R. E., T. Kroeger, and A. Barkan. Sequence-specific binding of a chloroplast pentatricopeptide repeat protein to its native group II intron ligand. RNA, in press. [DOI] [PMC free article] [PubMed]
- 56.Yamazaki, H., M. Tasaka, and T. Shikanai. 2004. PPR motifs of the nucleus-encoded factor, PGR3, function in the selective and distinct steps of chloroplast gene expression in Arabidopsis. Plant J. 38152-163. [DOI] [PubMed] [Google Scholar]
- 57.Zoschke, R., K. Liere, and T. Borner. 2007. From seedling to mature plant: arabidopsis plastidial genome copy number, RNA accumulation and transcription are differentially regulated during leaf development. Plant J. 50710-722. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.