Abstract
The maturation of cytosolic iron-sulfur (Fe/S) proteins in mammalian cells requires components of the mitochondrial iron-sulfur cluster assembly and export machineries. Little is known about the cytosolic components that may facilitate the assembly process. Here, we identified the cytosolic soluble P-loop NTPase termed huNbp35 (also known as Nubp1) as an Fe/S protein, and we defined its role in the maturation of Fe/S proteins in HeLa cells. Depletion of huNbp35 by RNA interference decreased cell growth considerably, indicating its essential function. The deficiency in huNbp35 was associated with an impaired maturation of the cytosolic Fe/S proteins glutamine phosphoribosylpyrophosphate amidotransferase and iron regulatory protein 1 (IRP1), while mitochondrial Fe/S proteins remained intact. Consequently, huNbp35 is specifically involved in the formation of extramitochondrial Fe/S proteins. The impaired maturation of IRP1 upon huNbp35 depletion had profound consequences for cellular iron metabolism, leading to decreased cellular H-ferritin, increased transferrin receptor levels, and higher transferrin uptake. These properties clearly distinguished huNbp35 from its yeast counterpart Nbp35, which is essential for cytosolic-nuclear Fe/S protein assembly but plays no role in iron regulation. huNbp35 formed a complex with its close homologue huCfd1 (also known as Nubp2) in vivo, suggesting the existence of a heteromeric P-loop NTPase complex that is required for both cytosolic Fe/S protein assembly and cellular iron homeostasis.
Proteins carrying iron-sulfur (Fe/S) clusters as inorganic cofactors are involved in fundamental cellular processes such as respiration, enzyme catalysis, and regulation of gene expression (6, 28), accounting for their presence in all kingdoms of life. In eukaryotes, known iron-sulfur (Fe/S) proteins are localized within the mitochondria, plastids, cytosol, and nucleus. Biosynthesis of the Fe/S clusters and their insertion into apoproteins require complex machineries with a steadily growing number of known components. Studies mainly performed in yeast have identified three different biogenesis systems: the iron-sulfur cluster (ISC) assembly, the ISC export machineries located within mitochondria, and the cytosolic iron-sulfur protein assembly (CIA) apparatus (for a recent comprehensive review, see reference 40). Relatives of central components of the yeast ISC assembly and export machineries have been identified in higher eukaryotes and found to perform tasks similar to those of their yeast counterparts (8, 12, 50, 64, 70).
The matrix of yeast mitochondria harbors at least 15 ISC assembly proteins, most of which were inherited from bacteria (40). Key events catalyzed by this machinery are common to virtually all eukaryotes investigated thus far. Individual steps of biogenesis comprise the abstraction of sulfur from cysteine by the desulfurase complex Nfs1p-Isd11p, the formation of a transient Fe/S cluster on the scaffold protein Isu1p, presumably requiring the assistance by frataxin (yeast Yfh1p) as an iron donor. Subsequently, the Isu1p-bound Fe/S cluster is transferred to recipient apoproteins with the help of a dedicated Hsp70 chaperone system and the monothiol glutaredoxin Grx5p. Most ISC proteins, in addition to their function in the maturation of mitochondrial Fe/S proteins, are crucial for the maturation of Fe/S proteins outside mitochondria (40). Mitochondria appear to export a still-unknown compound to the cytosol via the ISC export machinery, where it is used for maturation of extramitochondrial Fe/S proteins. The key component of the ISC export machinery is the ABC transporter of the mitochondrial inner membrane termed Atm1p in yeast and ABCB7 in mammals (7, 12, 15, 50). Its ablation specifically impairs the maturation of extra-mitochondrial Fe/S proteins without affecting the biosynthesis of mitochondrial Fe/S proteins.
In contrast to the well-conserved ISC assembly and export machineries, the mechanism and components responsible for the maturation of extramitochondrial Fe/S proteins in lower and higher organisms are just emerging. In yeast, the CIA machinery has been shown to be specifically responsible for the assembly of cytosolic and nuclear, but not of mitochondrial Fe/S proteins (40). Thus far, four CIA components termed Cfd1p, Nbp35p, Nar1p, and Cia1p have been identified in yeast (4, 5, 22, 55). According to a current model, a transient Fe/S cluster is assembled on the heterotetrameric Cfd1p-Nbp35p complex which serves as a scaffold for Fe/S cluster synthesis (47). Cfd1p and Nbp35p are the prototypical members of the Mrp/NBP35 subclass of the large protein family of P-loop NTPases (36). Recently, a mitochondrial member of this protein family (termed Ind1) was identified in Yarrowia lipolytica and shown to perform a specific role in the assembly of respiratory complex I (9). Typically, all Mrp/NBP35 subclass members contain conserved C-terminal cysteine motifs that may be involved in transient Fe/S cluster synthesis and binding (37, 60). Fe/S cluster assembly on Cfd1p-Nbp35p is dependent on the function of the mitochondrial ISC assembly and export machinery. The labile Fe/S cluster is then transferred to target apoproteins, a process which in vivo requires the function of the iron-only hydrogenase-like protein Nar1p and the WD40 repeat protein Cia1p (47).
In human cells, the majority of the ISC assembly components huNfs1, huIsu1, huNfu1, and frataxin are present inside mitochondria, and yet low amounts have been detected in the cytosol and/or nucleus (1, 39, 69). Hence, a spatial duplication of the ISC assembly machinery has been proposed, suggesting a maturation mechanism in the cytosol-nucleus resembling that within mitochondria (54, 71). A direct test of this idea was performed for mammalian Nfs1 and Isu1. The cytosolic version of mammalian Nfs1 did not suffice to assemble Fe/S clusters on the cytosolic Fe/S protein iron regulatory protein 1 (IRP1) in HeLa cells (8). Likewise, specific depletion of the cytosolic isoform of huIsu1 had no profound effect on the maturation of cytosolic Fe/S proteins (70). However, a delay in the repair-regeneration of Fe/S proteins was seen after destruction of the Fe/S cluster of IRP1 by H2O2 or iron limitation (70).
Intriguingly, proteins with sequence similarity to the four known yeast CIA components have been identified in mammals. The mammalian proteins were proposed to participate in rather diverse cellular functions. For instance, the Cia1p relative Ciao1 was shown to interact with the Wilms' tumor protein with a potential role in transcription regulation (29). A task in Fe/S protein biogenesis was suggested by the ability of Ciao1 to complement Cia1p-deficient yeast cells (63). The yeast Nar1p homologue IOP1 was first assumed to modulate the activity of hypoxia-inducible factor 1α (26) but was recently shown to perform an essential function in the formation of the cytosolic Fe/S proteins IRP1 and xanthine oxidase (61). The mouse homologues of yeast Nbp35p and Cfd1p (previously termed nucleotide binding proteins 1 and 2, respectively) were found to interact with kinesin 5A in murine cells and hence were suggested to participate in centrosome duplication (13, 46, 58). The putative involvement of the latter proteins and Ciao1 in Fe/S protein biogenesis has not been experimentally tested in mammals yet.
Here, we present a biochemical and cellular analysis of the functional role of the human counterparts of yeast Nbp35p-Cfd1p (in the following designated huNbp35 and huCfd1, respectively) in the maturation of cellular Fe/S proteins. Since this process is intimately linked to cellular iron homeostasis, we also recorded changes in iron regulation. Using a vector-based RNA interference (RNAi) approach, endogenous huNbp35 was depleted in HeLa cells to critical levels and the physiological consequences were studied. Our findings suggest an essential function of huNbp35, in complex with huCfd1, for cell growth, for the maturation of cytosolic but not mitochondrial Fe/S proteins, and for cellular iron regulation.
MATERIALS AND METHODS
Plasmids.
pOTB7 cloning vectors containing the cDNAs of HUNBP35 (NUBP1; NM_002484) or HUCFD1 (NUBP2; NM_012225) were obtained from the German Resource Center for Genome Research (RZPD, Berlin, Germany). Bacterial overproduction of N-terminally His-tagged huNbp35 was achieved by subcloning the HUNBP35 open reading frame into the bacterial expression vector pETDuet-1 via EcoRI and BsrGI restriction sites. For mammalian expression, HUNBP35 was cloned into the pEGFP-C1-derived (Clontech) mammalian expression vectors pHA-MCS-C1 (kindly provided by G. Suske, Marburg, Germany) or pHis-MCS-C1 (His6 tag instead of hemagglutinin [HA] tag) via EcoRI and BsrGI/Acc65I restriction sites. In order to achieve a C-terminal enhanced green fluorescent protein (EGFP)-tagged version, HUNBP35 was amplified by the Phusion DNA polymerase (Finnzymes, Finland) using the primer pair 5′-CAA CGG GAC TTT CCA AAA TG-3′ and 5′-ATC TCG AGG GAA CTG ATG AGG TTC TCT TC-3′. The PCR product was cloned into the pEGFP-N3 vector (Clontech) via EcoRI and XhoI/SalI sites. Tagging and mammalian expression of HUCFD1 was achieved by insertion into pEGFP-C1 via SacI and PstI sites.
A vector-based RNAi approach was used to deplete huNbp35 in human HeLa cells. Three different gene-specific targeting sequences (si#1, CGT TAG CCT ACA GAA GTA T; si#2, GGT CGA CTA CCT CAT TGT G; and si#3, CGG CTA TAG AGG AAA TCA A) were obtained from Ambion, and respective oligonucleotides were cloned into the pSUPER-derived vector pSUPERIOR.puro (OligoEngine) using the BglII and HindIII restriction sites in order to obtain vector-encoded short hairpin (sh) constructs for RNAi.
Silent mutations corresponding to RNAi sequences si#1 and si#3 were introduced into HUNBP35. The cloning vector pOTB7 containing the wild-type version of HUNBP35 was amplified by the Phusion DNA Polymerase using the mismatch-containing primer pairs sm#1 (5′-GAT TCC CCA GCC ACC CTT GCG TAC CGG TCA ATA ATT CAG AGA ATC-3′ and 5′-GAT TCT CTG AAT TAT TGA CCG GTA CGC AAG GGT GGC TGG GGA ATC-3′) and sm#3 (5′-G GCC ACT CCG GAC ACC GCA ATT GAA GAG ATA AAG GAG AAA ATG AAG AC-3′ and 5′-GT CTT CAT TTT CTC CTT TAT CTC TTC AAT TGC GGT GTC CGG AGT GGC C-3′) (mismatches are underlined). PCR products were subsequently digested and ligated at their newly created AgeI and MunI restriction sites, respectively. Silently mutated HUNBP35 open reading frames (HUNBP35SM#1 and HUNBP35SM#3) were subcloned into pHis-MCS-C1 via EcoRI and BsrGI/Acc65I restriction sites.
A pcDNA3.1 vector encoding a murine GPAT (glutamine phosphoribosylpyrophosphate amidotransferase) version that is not capable of propeptide processing due to a single amino acid exchange in its active site (muGPAT_C1F) was obtained from Martelli et al. (42).
Purification of recombinant His-tagged huNbp35.
Yeast Nbp35p and huNbp35 were produced as described previously (22, 47). Proteins were expressed in Escherichia coli in the presence of the bacterial isc operon using vector pET-15b (Novagen) and purified anaerobically by nickel-nitrilotriacetic acid affinity chromatography. The proteins were immediately desalted on a PD10 column and subsequently chemically reconstituted (47). Incorporation of Fe/S clusters was monitored by UV-Vis spectroscopy. Electron paramagnetic resonance (EPR) spectra were recorded with a Bruker ESP 300E X-band spectrometer, equipped with an Oxford Instruments ESR910 helium flow cryostat (temperature, 10 K; microwave power, 3.2 mW; microwave frequency, 9,460 ± 1 MHz; modulation frequency, 100 kHz; modulation amplitude, 1.25 mT).
Cell culture, transfection, and flow cytometry.
Human cervix carcinoma cells (HeLa) were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 7.5% fetal calf serum, 1 mM glutamine, and 1% penicillin-streptomycin (complete DMEM). All cell culture reagents were obtained from PAA (Germany). After harvesting by trypsin treatment and washing in transfection buffer (21 mM HEPES, 137 mM NaCl, 5 mM KCl, 0.7 mM Na2HPO4, and 6 mM dextrose), 3.5 × 106 cells were resuspended in 525 μl of transfection buffer and supplemented with 2.5 to 25 μg of each of the required plasmids. Transfections were carried out by electroporation (250 V, 1,500 μF, about 30-ms duration) using an EASYJect+ device (64), and cells were immediately cultured in complete DMEM supplemented with 20% conditioned HeLa medium. In experiments where huNbp35 was depleted in the presence of shRNAs, 3.5 × 106 cells were retransfected every third day in order to prolong the time period of depletion. When an EGFP tag was included, transfection efficiency was analyzed by flow cytometry (FACScan; Becton Dickinson). The protein content of the harvested cells was determined by the Bradford method (Bio-Rad, Germany).
Choice of suitable HUNBP35 mRNA-directed shRNAs and of appropriate vector amounts for transfection.
The three shRNAs (shRNA#1 to shRNA#3) directed against HUNBP35 mRNA were analyzed for their efficacy to deplete huNbp35, and the two silently mutated versions of HUNBP35 (sm#1 and sm#3) were tested for complementation of depletion. shRNAi construct #2 was least active and exerted only weak effects on huNbp35 protein levels (not shown). Application of shRNA#3 led to a substantial decrease of huNbp35 protein and resulted in the strongest cellular phenotype of all three shRNAs without showing reversal by His-huNbp35sm#3, suggesting off-target effects exerted by shRNA#3 (not shown). Only shRNA#1 led to a substantial decrease of huNbp35 protein, which was complemented by simultaneous expression of His-huNbp35sm#1 (see Results). Consequently, shRNA#1 was used to investigate the function of huNbp35 in HeLa cells.
The degree of complementation was strongly dependent on the amount of plasmid used. While 25 μg of the shRNAi#1 vector were required to induce a profound RNAi depletion effect, only 2.5 μg of the His-huNbp35sm#1 encoding vector were sufficient to achieve growth complementation. Despite the low amounts of vector the transfection efficiency was at least 80%, as estimated by control transfections with equal amounts of an EGFP-encoding vector.
Labeling with Tf-FITC.
Transferrin receptor (TfR) expression was evaluated by a fluorometric approach (10, 27). Human holotransferrin was dissolved in sodium carbonate buffer (pH 8.0) at a concentration of 30 mg/ml and coupled to dimethyl sulfoxide-solved fluorescein isothiocyanate (FITC) according to the manufacturer's instructions. Tf-FITC was separated by a PD-10 column, eluted with Hanks balanced salt solution (HBSS), sterile filtered, and stored at a concentration of 10 mg/ml in HBSS supplemented with 1% bovine serum albumin.
After each transfection, cells were seeded in 48-well cell culture plates at 25 × 103 cells per well. At the time points indicated cells were washed with HBSS, and Tf-FITC was added for 60 min at a concentration of 100 μg/ml in HBSS. Subsequently, cells were washed with phosphate-buffered saline, and cell-associated fluorescence was analyzed in a Tecan infinite 200 microplate reader (Tecan, Switzerland; excitation wavelength, 482 nm; emission wavelength, 532 nm). Background fluorescence was recorded in wells that contained transfected but unlabeled cells. In some cases, cells were challenged either with 125 μM ferric ammonium citrate in combination with 125 μM sodium ascorbate or with 125 μM deferoxamine 24 h prior to Tf-FITC labeling. Since TfR expression and Tf-FITC binding is cell density dependent (24, 43, 51; our observations), each plate included control-transfected HeLa cells seeded at different densities in order to derive a reference curve. The total protein content of each well was determined by the bicinchoninic acid method (Pierce), including a protein standard in each plate. The three differentially transfected groups were compared by two-way analysis of variance using the “day after first transfection” as the second factor and Tukey's post-hoc test.
Coimmunoprecipitation of huNbp35 and huCfd1 fusion proteins.
HeLa cells were transfected with the respective plasmids, grown for 3 days, harvested, washed twice with phosphate-buffered saline, snap-frozen in liquid nitrogen, and stored at −80°C until use. Coimmunoprecipitation was carried out at 4°C. Dry cell pellets (corresponding to about 3 mg of total protein) were lysed in TNGT buffer (10 mM Tris, 150 mM NaCl, 10% [wt/vol] glycerol, 0.25% [vol/vol] Triton X-100, 1 mM phenylmethylsulfonyl fluoride), and the lysate was clarified by centrifugation at 13,000 × g for 10 min. Protein A-Sepharose (GE Healthcare, Uppsala, Sweden)-conjugated HA probe (monoclonal mouse antibody HA.11; Covance) was added to the clarified samples for 1 h under gentle agitation. Sepharose beads were washed four times with TNGT buffer and resuspended in loading buffer for sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Miscellaneous biochemical methods.
Published methods were used for harvesting and fractionating HeLa cells by digitonin treatment (8, 16), as well as for determination of enzyme activities (65): aconitase activity by a coupled aconitase-isocitrate dehydrogenase assay (16), succinate dehydrogenase activity by the DCIP assay in combination with decyl ubiquinone (21, 64), citrate synthase (CS) and lactate dehydrogenase (LDH) activities (8, 25, 62), and iron-responsive element (IRE) RNA-binding capacity of IRP1 by RNA electrophoretic mobility shift assay (REMSA) (8, 45). The following primary antibodies were used for indirect immunostaining (cf. references 8 and 64): affinity-purified rabbit anti-huNbp35 serum, mouse anti-alpha tubulin (clone DM1α; Sigma, Germany) and anti-actin (clone 2G2) (19) monoclonal antibodies, mouse anti-IRP1 monoclonal antibody (clone 295B) (14), rabbit anti-IRP2 serum (14), affinity-purified rabbit anti-murine GPAT serum (42), rabbit anti-mitochondrial aconitase serum (L. Szweda, Oklahoma), rabbit anti-human ferritin serum (ICN), mouse anti-TfR monoclonal antibody (clone H68.4; Zymed), mouse anti-HA monoclonal antibody (clone HA.11; Covance), and rabbit anti-EGFP serum. An Alexa 488-labeled goat anti-rabbit antibody (Molecular Probes/Invitrogen) or peroxidase-conjugated goat anti-rabbit and anti-mouse antibodies (Bio-Rad, Germany) were used as secondary reagents.
RESULTS
huNbp35 is an Fe/S protein located in the cytosol.
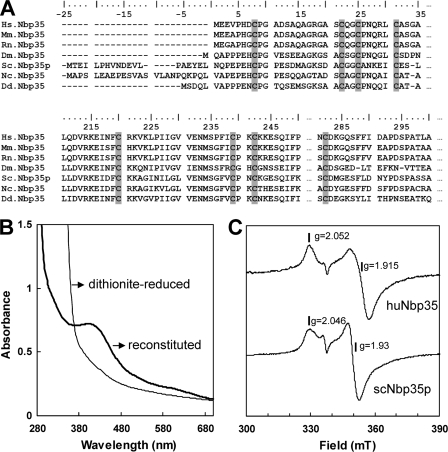
The P-loop NTPase huNbp35 carries, at its N and C termini, several conserved cysteine residues (Fig. 1A) that may form iron-binding sites. In order to analyze the protein's capability of coordinating an Fe/S cluster, it was overproduced in E. coli and purified under anaerobic conditions. Recombinant huNbp35 had a dark brown color, and exhibited a structured UV-visible (UV-Vis) spectrum with an absorption peak at 420 nm that compared well with that of typical Fe/S proteins (not shown). However, cluster occupancy was low (∼0.9 Fe and S per monomer), and only after chemical reconstitution a consistent content of ∼4.5 to 6.0 Fe and S per monomer was present in the protein. UV-Vis (Fig. 1B) and low-temperature EPR spectroscopy (Fig. 1C) of chemically reconstituted, reduced huNbp35 showed signals and average g values similar to yeast Nbp35p (22). These findings were consistent with the presence of a [4Fe-4S] cluster demonstrating that huNbp35 is an Fe/S protein.
FIG. 1.
Purified huNbp35 is an Fe/S protein. (A) Partial sequence alignment of Nbp35 homologues from various species. Conserved cysteine residues are highlighted in gray. The numbering at the top refers to huNbp35, which is 320 amino acid residues long. Abbreviations: Hs, Homo sapiens; Mm, Mus musculus; Rn, Rattus norvegicus; Dm, Drosophila melanogaster; Sc, Saccharomyces cerevisiae; Nc, Neurospora crassa; Dd, Dictyostelium discoideum. (B) UV-VIS spectrum of N-terminally His-tagged huNbp35 (in 50 mM Tris-HCl [pH 8]) after Fe/S cluster reconstitution and after additional reduction with 20 mM sodium dithionite. (C) EPR spectra of recombinant purified and reconstituted huNbp35 and yeast Nbp35p (scNbp35p). Traces and g values indicate the similar spectral properties of the Fe/S clusters present in both proteins.
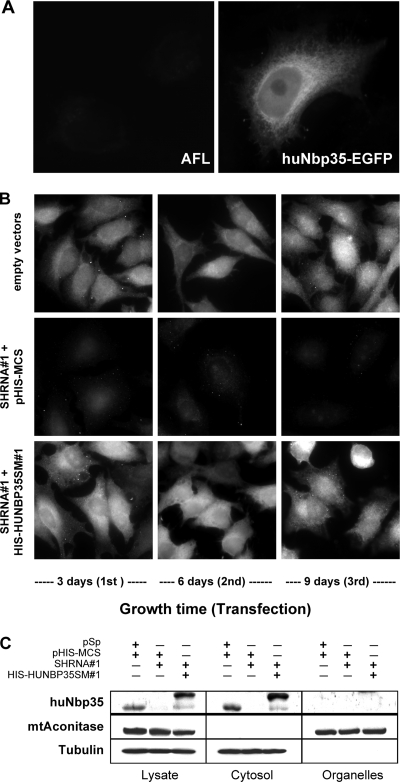
To determine the cellular localization of huNbp35, HeLa cells were transiently transfected with an expression vector encoding a C-terminally EGFP-tagged huNbp35 fusion protein. Three days after transfection EGFP-fluorescence was distributed throughout the entire cell body without any specific organellar staining (Fig. 2A). As an alternative method, endogenous huNbp35 was visualized in HeLa cells by indirect immunofluorescence microscopy using an affinity-purified polyclonal rabbit anti-huNbp35 antibody. The fluorescent stain was distributed throughout the entire cell, a finding consistent with a cytosolic localization of the protein (Fig. 2B, top panels). A vector-based RNAi treatment leading to depletion of huNbp35 (see also below) verified the specificity of the immunofluorescence signal. HeLa cells were consecutively transfected for three times at three day intervals with 25 μg of a vector encoding a huNbp35 mRNA-directed short hairpin RNA (shRNA#1). This treatment strongly diminished the fluorescence staining (Fig. 2B, middle panels), demonstrating the efficacy of the RNAi approach. Cotransfection of 2.5 μg of a vector encoding a RNAi-resistant, N-terminally His-tagged version of huNbp35 (His-huNbp35sm#1) led to complementation of the lack of endogenous huNbp35 and resulted in a strong immunostaining similar to that found in control cells (Fig. 2B, bottom panels).
FIG. 2.
Human Nbp35 is a cytosolic protein. (A) EGFP fluorescence of a HeLa cell transiently transfected with a vector encoding a huNbp35-EGFP fusion protein (right) in comparison to the endogenous autofluorescence (AFL) of control cells (left). (B) Immunofluorescence detection of huNbp35 in HeLa cells cotransfected with expression vectors encoding shRNA#1 (for RNAi treatment) and His-huNbp35sm#1 (containing silent mutations, for complementation) or with the corresponding empty vectors. Every third day, cells were harvested, and a fraction was retransfected. Staining was performed at the indicated time points after the first transfection. (C) Subcellular localization of huNbp35 by cell fractionation. HeLa cells were transfected with the vectors indicated (see panel B). Six days after the first transfection cells were permeabilized with digitonin, and the cell lysate was centrifuged for 10 min at 15,000 × g. Supernatant (cytosol) and pellet (organelles) fractions were analyzed by immunoblotting. HuNbp35 exclusively colocalizes with tubulin in the cytosolic fraction, but not with mitochondrial aconitase (mtAconitase) present in the membrane fraction.
The cytosolic localization of huNbp35 was further verified by cell fractionation (Fig. 2C). HeLa cells were cotransfected twice at a three day interval with expression vectors encoding shRNA#1 and His-huNbp35sm#1, or with the corresponding empty vectors as described above. Six days after the first transfection cells were harvested and permeabilized by digitonin treatment. A cytosolic and a membrane-containing fraction were prepared by centrifugation and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting. HuNbp35 and its His-tagged version were exclusively present in the cytosolic fractions containing α-tubulin (Fig. 2C). None of these proteins was found in the organelle fractions, which harbored the mitochondrial aconitase as a leading enzyme. Upon RNAi treatment with shRNA#1, endogenous huNbp35 was strongly depleted demonstrating the efficacy of the RNAi approach (Fig. 2C). We conclude from these results that huNbp35 is a soluble cytosolic protein.
RNAi-mediated depletion of huNbp35 impairs cell growth.
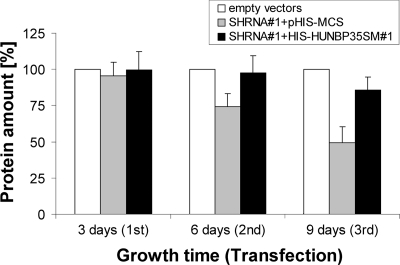
Since many Fe/S proteins are involved in vital processes, we studied the physiological consequences of RNAi-mediated huNbp35 deficiency. HeLa cells were transfected three times as described above, harvested 3 days after each transfection, and analyzed for their protein content. In shRNA#1-expressing cells, protein content was gradually diminished by up to 60% on day 9 after the first transfection (Fig. 3, gray bars). Since no substantial cell death was observed (not shown), this effect could be mainly attributed to reduced proliferation. The effect of shRNA#1 was specific, since protein levels were normalized by the presence of His-huNbp35sm#1 (Fig. 3, black bars). Complementation was most efficient when the average His-huNbp35sm#1 level was comparable to the amount of endogenous huNbp35 (cf. Fig. 2C and Materials and Methods). Cell counting as an alternative measure of cell growth confirmed these observations (data not shown). Taken together, the impaired growth of HeLa cells upon the RNAi-mediated depletion of huNbp35 reveals a crucial role of the protein.
FIG. 3.
Depletion of huNbp35 by RNAi leads to impaired cell growth. HeLa cells were transfected with the vectors indicated as described in Fig. 2B. At the indicated time points, cells were harvested, and their protein content was determined as a measure of cell growth and presented relative to that found in the respective control cells transfected with empty vectors (mean ± the standard deviation; 6 < n < 7).
Depletion of huNbp35 diminishes the levels of the cytosolic Fe/S protein GPAT.
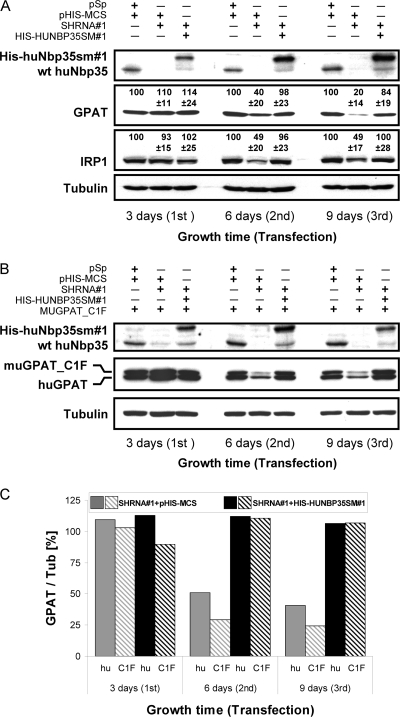
We next examined whether huNbp35 is involved in the maturation of cytosolic Fe/S proteins. GPAT is translated within the cytosol as an inactive proenzyme, and for maturation to the active enzyme it requires both the incorporation of a [4Fe-4S] cluster and the subsequent removal of an 11 residues long propeptide at the N terminus (42, 76). Studies on GPAT in Bacillus subtilis revealed that Fe/S cluster insertion determines protein stability (20). In support of this, iron depletion in HeLa cell culture by addition of the iron chelator deferoxamine at a concentration of 125 μM for 18 h lead to a twofold depletion of GPAT (not shown). We therefore examined whether the RNAi-mediated depletion of huNbp35 affects the steady-state protein levels of endogenous human GPAT (huGPAT) or of a murine GPAT version that is not capable of propeptide processing due to a single amino acid exchange at its active site (muGPAT_C1F) (42). To this end, HeLa cells were transfected as described above with the shRNA#1- and His-huNbp35sm#1-expressing vectors either alone (Fig. 4A) or together with a pcDNA3.1 vector encoding muGPAT_C1F (Fig. 4B). Cell lysates were then analyzed for huNbp35 and GPAT levels.
FIG. 4.
Depletion of huNbp35 diminishes cellular steady-state levels of the cytosolic Fe/S proteins GPAT and IRP1. (A) HeLa cells were repeatedly cotransfected with expression vectors encoding shRNA#1 and His-huNbp35sm#1 or with the corresponding empty vectors in 3-day intervals as described for Fig. 2B. Cells were harvested, and lysates were analyzed by immunostaining for huNbp35, GPAT, IRP1, and tubulin. The signals of the protein-associated chemiluminescence for GPAT and IRP1 relative to tubulin in the various samples are indicated (mean ± the standard deviation; GPAT, n = 7; IRP1, n = 12). (B) HeLa cells were additionally transfected with a vector encoding a murine GPAT version that is not capable of propeptide processing (muGPAT_C1F). Samples were then treated and analyzed for huNbp35, GPAT, and tubulin as in panel A. (C) The ratios of the protein-associated chemiluminescence signals of GPAT and tubulin (Tub) in the various samples from part B were depicted relative to the ratios in control samples (set to 100%; data not shown for simplicity). hu, huGPAT; C1F, muGPAT_C1F.
Immunostaining showed an almost complete depletion of huNbp35 in shRNA#1-expressing cells already 3 days after the first transfection (Fig. 4A and B). Both huGPAT and muGPAT_C1F levels decreased at significantly slower rates resulting in a 60% decrease after six and a 80% decrease of endogenous human GPAT after 9 days of huNbp35 depletion. The amount of vector-encoded, nonprocessed muGPAT_C1F dropped to even lower levels (Fig. 4C), thereby providing an explanation why immature endogenous huGPAT was not detectable in huNbp35-deficient cells. These findings show that huNbp35 is required for maintaining normal levels of both endogenous huGPAT and muGPAT_C1F. Presumably, the lack of Fe/S cluster assembly in GPAT proteins affected protein stability and led to degradation of these proteins. Strikingly, the stability of GPAT was not significantly influenced by the processing of the propeptide, indicating that the increased degradation of GPAT in the absence of huNbp35 was the sole effect of the lack of the Fe/S cluster. Cellular GPAT content could be completely restored to normal levels by coexpression of His-huNbp35sm#1 (Fig. 4), indicating that the decrease in GPAT was a direct consequence of the low amounts of huNbp35 and not due to unspecific effects. The concentrations of the cytosolic control proteins α-tubulin (Fig. 4A and B) and actin (not shown) were not affected by huNbp35 depletion. These results show that huNbp35 is crucial for maintaining normal levels of GPAT thus suggesting an involvement of huNbp35 in the maturation of this cytosolic Fe/S protein.
Depletion of huNbp35 decreases the cellular level and aconitase activity of IRP1.
IRP1 is a well-known cytosolic Fe/S protein with a dual function (53, 73). In the presence of an [4Fe-4S] cluster it functions as a cytosolic aconitase (cytAco). When the cluster is completely missing, it may bind to mRNA stem-loop structures called IREs and interfere with mRNA expression. Extended loss of the Fe/S cluster may affect IRP1 protein stability leading to lower protein levels (14, 74). We therefore studied the consequences of huNbp35 depletion on the cellular levels, aconitase function and IRE binding activity of IRP1 in more detail. As described above HeLa cells were repeatedly transfected with respective vectors, harvested 3 days after each transfection, and cell extracts were prepared. Upon depletion of huNbp35 the levels of IRP1 decreased twofold (Fig. 4A). As for GPAT, this reduction was a specific consequence of the shRNA#1-induced depletion of endogenous huNbp35, since IRP1 levels could be fully restored by coexpression of His-huNbp35 from the silently mutated construct.
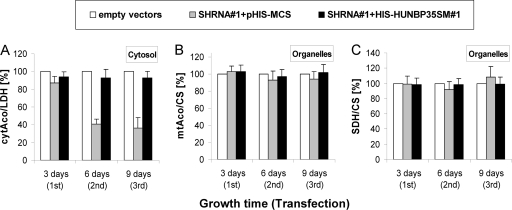
To more directly test whether the decrease in IRP1 levels was a consequence of the loss of the Fe/S cluster, we measured the cytAco activity of IRP1 after fractionating the HeLa cells by treatment with digitonin and centrifugation into a cytosolic fraction (containing IRP1) and a membrane-containing organelle fraction (including mitochondria). The efficiency of this fractionation procedure was evaluated by measuring the specific enzyme activities of cytosolic LDH and mitochondrial CS in both fractions. Routinely, <10% of cytosolic LDH activity was present in the organelle fraction, and <3% of mitochondrial CS activity was measured in the cytosol fraction (data not shown; cf. reference 8). Depletion of huNbp35 resulted in a time-dependent decrease of the specific cytAco activity of IRP1 (Fig. 5A, gray bars). After the third round of transfection its activity was decreased by 70%. The effect was specifically due to depletion of huNbp35, since the expression of His-huNbp35sm#1 restored the specific cytAco activity to control levels (Fig. 5A, black bars). Strikingly, the loss of cytAco activity was somewhat more pronounced than the lowering of IRP1 protein levels (compare Fig. 4A and 5A), suggesting that protein degradation of IRP1 occurred subsequently to the impairment of Fe/S cluster formation on IRP1. In contrast to cytAco, neither the enzyme activity (Fig. 5B) nor the protein levels (cf. Figure 2C) of mtAco of the organelle fractions changed significantly upon huNbp35 depletion. Likewise, the specific activity of mitochondrial Fe/S protein SDH and the non-Fe/S control protein CS remained unchanged (Fig. 5C). Collectively, the data indicate the specific requirement of huNbp35 for Fe/S cluster assembly on cytosolic but not on mitochondrial Fe/S proteins.
FIG. 5.
Depletion of huNbp35 impairs cytAco activity of IRP1. HeLa cells were transfected, harvested, and fractionated as described for Fig. 2B and C. Specific activities of the Fe/S proteins cytAco (IRP1-related) (A), mitochondrial aconitase (mtAco) (B), and succinate dehydrogenase (SDH) (C) were measured in the indicated fractions. The data are presented relative to the specific activities of the non-Fe/S cluster-containing marker enzymes CS or LDH and were normalized to the respective control samples (mean ± the standard deviation; 3 < n < 4).
huNbp35 deficiency promotes IRE-binding of IRP1.
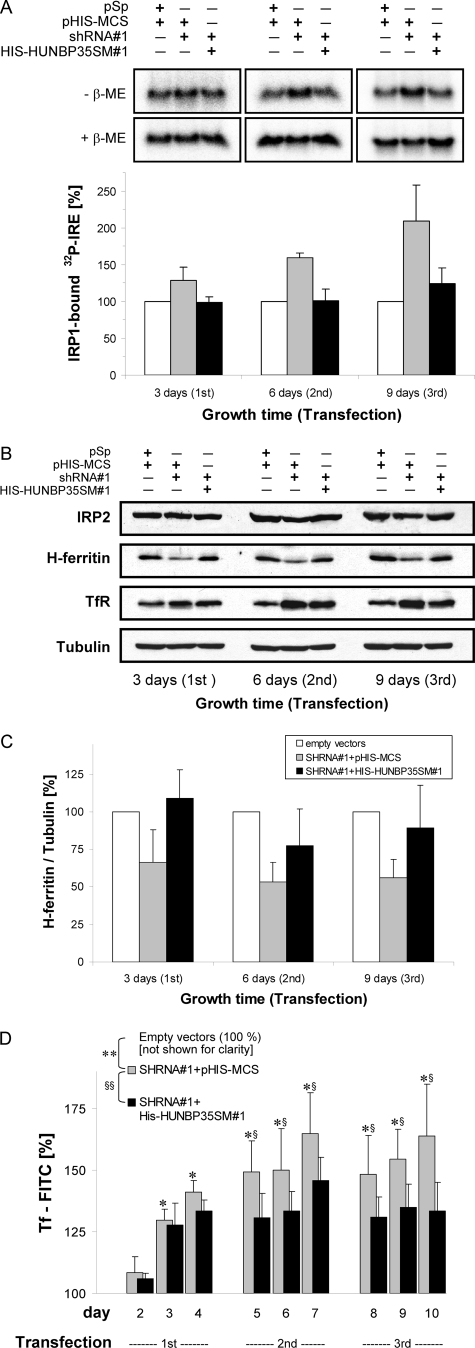
In order to analyze whether the depletion of huNbp35 would increase IRE binding of IRP1, extracts of huNbp35-depleted HeLa cells were analyzed by a REMSA. The capacity of IRP1 to bind to an [α-32P]CTP-labeled IRE of ferritin mRNA was distinguished from that of the non-Fe/S cluster-containing homolog IRP2 by inclusion of an anti-IRP2 antibody in order to induce a supershift on IRP2. The lower IRP1 protein levels in huNbp35-deficient cells were taken into account by following the maximal IRP1 binding capacity of IRP1 after treatment of the samples with β-mercaptoethanol (β-ME). The ratio of IRP1 binding in the absence and presence of β-ME reflects the IRE binding capacity of IRP1. Upon huNbp35 depletion, the β-ME-induced IRE binding activity matched the reduced IRP1 protein level, while the IRE-binding capacity of IRP1 strongly increased in a time-dependent manner (Fig. 6A; cf. Fig. 4A), i.e., in an behavior opposite to the decrease seen in cytAco activity (see Fig. 5A). The effect was almost completely reversed by expression of His-huNbp35sm#1, unequivocally demonstrating the specific involvement of huNbp35 in the maturation of the cytosolic Fe/S protein IRP1.
FIG. 6.
Depletion of huNbp35 increases IRE binding of IRP1 altering the steady-state levels of ferritin and TfR. HeLa cells were transfected with the indicated vectors and harvested at 3-day intervals as described in Fig. 2B. (A) Cell extracts were analyzed for IRP1 binding activity (IRP2 supershift method) to 32P-labeled IRE of human ferritin mRNA by native gel electrophoresis. Binding of IRE to IRP1 was visualized in the absence (−) or presence (+) of β-ME using phosphorimaging (upper panels). The amounts of IRP1-bound IRE in the various transfected cells were normalized to the maximal binding capacity of β-ME-treated samples and presented relative to the amount of IRP1-bound IRE in control vector-transfected cells (lower panel; mean ± the standard deviation; n = 4). (B) Total cell lysates were immunoblotted and stained for IRP2, H-ferritin, TfR, and tubulin. (C) The ratios of the protein-associated chemiluminescence signals of H-ferritin and tubulin in the various samples from part B are depicted relative to the ratios in control samples (mean ± the standard deviation; n = 5). (D) HeLa cells transfected with the indicated vectors were labeled with FITC-conjugated transferrin at the indicated days after the first transfection and were analyzed for cell-associated fluorescence as a measure of TfR expression. The fluorescence intensity was corrected for cell density dependency and is expressed relative to the fluorescence of control cells. The overall statistical significance of the differences between the values for control-, SHRNA#1-, and HIS-HUNBP35SM#1-transfected cells was P < 0.001 (** and §§, two-way analysis of variance). At each individual time point, the significance of the differences between shRNA#1-expressing cells versus control-transfected cells was P < 0.05 (*; Tukey post hoc) and between shRNA#1-expressing cells versus His-huNbp35-expressing cells was P < 0.05 (§; Tukey post hoc).
huNbp35 deficiency affects ferritin and TfR translation in an IRP1-dependent fashion.
The effect of huNbp35 depletion on IRP1 activity predicts physiological consequences on iron homeostasis. However, the in vivo effects of the increased IRE-binding capacity of IRP1 in huNbp35-depleted cells are more difficult to assess than the altered mRNA-binding activity of IRP1. Apart from IRP1, its homologue IRP2 also binds to IREs and posttranscriptionally regulates gene expression (73). IRP2 is not known to carry an Fe/S cluster, and its IRE-binding activity is mainly regulated by iron-dependent degradation. Cellular IRP2 levels, measured by immunostaining, were only slightly (20%) decreased in huNbp35-deficient cells (Fig. 6B, upper panel). We therefore conclude that the posttranslational changes through the IRP-IRE system in huNbp35-depleted cultured HeLa cells are mainly caused through the action of IRP1.
We directly monitored the fate of two target proteins of the IRP-mediated posttranscriptional regulation, i.e., cytosolic ferritin and TfR. Each of the mRNAs of H and L ferritin subunits contains a single IRE in its 5′ noncoding region (68). Binding of IRPs to these IREs inhibits ferritin translation and thus decreases cytosolic iron storage capacity. The amount of H-type ferritin (H-ferritin) in HeLa cells upon huNbp35 depletion was followed by immunoblotting (Fig. 6B). Densitometric analysis of H-ferritin levels relative to tubulin revealed an up to twofold decrease (Fig. 6C), a finding consistent with the increased IRE-binding capacity of IRP1 (Fig. 6A). These changes were an indirect effect of the functional defect of huNbp35 in maturing IRP1, since H-ferritin levels could be almost completely restored by the simultaneous expression of His-huNbp35sm#1 (Fig. 6B and C). The TfR is a cell surface receptor responsible for uptake of iron that is bound to the soluble carrier protein transferrin (51). In contrast to ferritins, the TfR mRNA contains five IREs in its 3′ noncoding region. Binding of IRPs to these IREs stabilizes the mRNA against degradation, leading to increased TfR mRNA translation and hence transferrin uptake. As expected TfR levels were increased in the absence of functional huNbp35 and strongly reversed by coexpression of His-huNbp35sm#1 (Fig. 6B).
In addition to this immunostaining approach, we sought to analyze TfR expression by other means, mainly for two reasons. First, HeLa cells were harvested by trypsin treatment, and TfR, as a transmembrane protein, might have at least partially been degraded. Second, TfR gene expression depends on cell proliferation (24, 43, 51) and thus varies with HeLa cell densities (unpublished data). To account for these potential problems, we monitored the cellular uptake of a fluorescently labeled transferrin (Tf-FITC) as a measure of the changes of TfR expression upon huNbp35 depletion (Fig. 6D). Adherent HeLa cells were incubated with Tf-FITC and analyzed for cell-associated fluorescence with respect to their cell density. Depletion of huNbp35 resulted in a significant (P < 0.001) and time-dependent (P < 0.001) increase of cell-associated fluorescence (Fig. 6D, gray bars). After three transfections cell-associated fluorescence was >1.6-fold higher than that of control cells. The time course of the fluorescence increase was similar to the rise of IRP1 binding to mRNA IREs (cf. Fig. 6A). Simultaneous expression of His-huNbp35sm#1 significantly attenuated the increase of cell-associated fluorescence by about one-half (Fig. 6D, black bars; P < 0.001) substantiating that the increased Tf-FITC uptake was directly linked to the impaired maturation of IRP1 after depletion of endogenous huNbp35. Intriguingly, complementation of Tf-FITC binding in vivo was less pronounced than that of IRP1 binding to IREs and other shRNA#1-induced effects analyzed in vitro, reflecting that regulation of TfR expression is a complex process which, among other things, integrates the availability of iron (73) and oxygen (41, 66), mitogenic signals (51), and Fe/S cluster biosynthesis (40). Despite the fact that shRNA#1-treated cells showed an increased Tf-FITC binding compared to control cells, they were still able to respond to both iron addition and removal (data not shown).
Taken together, the substantial alterations of ferritin and TfR synthesis in huNbp35-deficient cells clearly indicate that huNbp35, via its role in the maturation of IRP1, plays a crucial function in the maintenance of cellular iron homeostasis. Importantly, huNbp35 differs markedly from its yeast counterpart which does not impact on iron uptake and regulation (22, 23).
huNbp35 interacts with huCfd1 in vivo.
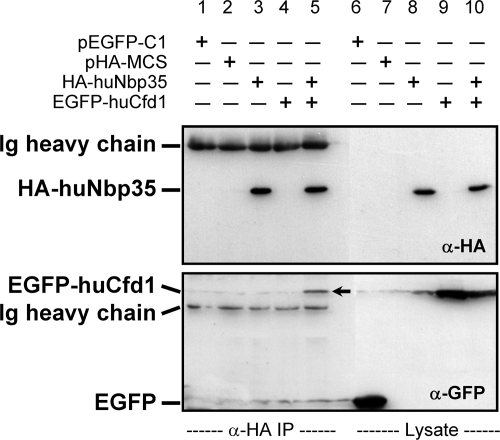
In yeast cells, Nbp35p forms a tight complex with Cfd1p both in vivo and in vitro (47). We therefore tested whether huNbp35 interacts with huCfd1, the human homologue of yeast Cfd1p. To this end, N-terminally HA-tagged huNbp35 and N-terminally EGFP-tagged huCfd1 were coexpressed in HeLa cells. Cell extracts were subjected to anti-HA immunoprecipitation, and both proteins were detected on immunoblots using the respective tags. Specific coimmunoprecipitation of HA-huNbp35 and EGFP-huCfd1 was observed (Fig. 7, lane 5). Single or no expression of HA-huNbp35 or of EGFP-huCfd1 served as specificity controls for the immunoprecipitation assay (Fig. 7, lanes 1 to 4). The coimmunoprecipitation of EGFP-huCfd1 and HA-huNbp35 demonstrated an interaction between both proteins in vivo, suggesting that the two proteins form a functional complex. Together, our observations suggest a highly conserved function of the P-loop NTPases from yeast to humans in the maturation of extramitochondrial Fe/S proteins.
FIG. 7.
HuNbp35 and huCfd1 form a complex in vivo. HeLa cells were transfected with vectors encoding N-terminally HA-tagged huNbp35 and/or N-terminally EGFP-tagged huCfd1 or with respective empty vectors as indicated. After cell harvest, cleared lysates were prepared and subjected to immunoprecipitation by anti-HA-coupled protein A-Sepharose. Immunoprecipitates (α-HA IP) and lysates were immunoblotted and stained for HA tag (top) and EGFP (bottom). Secondary reagents also labeled heavy chains of anti-HA and anti-EGFP immunoglobulins (Ig). α, anti.
DISCUSSION
The components involved in the maturation of cytosolic Fe/S proteins in mammalian cells are ill defined, and the mechanism of biogenesis is a matter of debate. In the present study we characterized huNbp35 as an essential cytosolic component required for the assembly of Fe/S clusters on extramitochondrial proteins. Using a vector-based RNAi approach, endogenous huNbp35 was depleted in HeLa cells, and the resulting phenotypes and biochemical consequences were analyzed. The specificity of the RNAi approach was verified by the simultaneous plasmid-based expression of an RNAi-resistant huNBP35 version that was silently mutated at nine positions within the shRNA target site. Collectively, our study demonstrated an essential and specific function of huNbp35 in cytosolic Fe/S protein biogenesis in the human cell.
HuNbp35 was first identified in human cells as a new member of the large family of P-loop NTPases (58), and sequence analysis revealed a close similarity to yeast Nbp35p (72). Both proteins carry a typical P-loop NTPase motif, in addition to two conserved cysteine motifs located at the N and C termini (36). In yeast Nbp35p, these cysteine motifs are involved in coordinating two Fe/S clusters (22, 47). Analysis of a bacterially expressed His-tagged version of huNbp35 by both UV-Vis and EPR spectroscopy demonstrated the conservation of Fe/S cluster association. The EPR signal of recombinant huNbp35 was highly similar to that of yeast Nbp35, indicating that both proteins carry the same type(s) of [4Fe-4S] clusters.
Fluorescence microscopy as well as digitonin-based cell fractionation demonstrated a cytosolic localization in human HeLa cells, a finding consistent with the lack of any predictable targeting sequence. Depletion of huNbp35 by a vector-based RNAi approach resulted in a substantial decrease in the growth rate, demonstrating a crucial function of the protein. This effect was not attributable to an impaired assembly of Fe/S clusters within mitochondria, because the activities of two mitochondrial Fe/S proteins, aconitase and SDH, were not affected. In contrast, the cellular steady-state protein levels of two cytosolic Fe/S proteins, GPAT and IRP1, were considerably diminished, a finding indicative of an insufficient assembly of Fe/S clusters on these proteins in the absence of huNbp35. The disparate maturation defect of mitochondrial and cytosolic Fe/S proteins was pointing to a direct role of huNbp35 in the formation of extramitochondrial Fe/S clusters rather than an unspecific damage of Fe/S proteins.
In higher eukaryotes, only enzyme and RNA binding assays have been used to follow Fe/S protein biogenesis. Unfortunately, a 55Fe radiolabeling assay for de novo Fe/S protein assembly developed in yeast (32) has not been successfully adapted to mammalian cells thus far, most likely because the iron depletion needed to achieve a sufficiently high specific radioactivity cannot easily be applied to cultured human cells. As cytosolic marker Fe/S proteins we therefore used GPAT and IRP1. GPAT catalyzes the first step of the de novo purine nucleotide synthesis. The function and stability of the protein depend on the insertion of its Fe/S cluster (42, 76). Transgenic expression of avian GPAT in HeLa cells and analysis of the respective homologue in B. subtilis had revealed that an impaired Fe/S cluster assembly may impede proper folding and is eventually leading to protein degradation (20, 76). The present study additionally demonstrated that the iron levels and the presence of huNbp35 are major determinants for protein stability of GPAT independently of propeptide processing. Consequently, the reduced cellular GPAT level is indicative of a direct role of huNbp35 in the formation of Fe/S clusters on GPAT.
IRP1, our second target Fe/S protein, is related to mitochondrial aconitases and may exert two different activities (73). When it harbors a [4Fe-4S] cluster, it functions as an aconitase, but when the cluster is absent, the apoform of IRP1 has the ability to bind to certain mRNA stem-loop structures, so-called IREs. Recent studies revealed that apo-IRP1 is unstable when it lacks its Fe/S cluster over longer time periods, e.g., when proper Fe/S cluster formation is not possible (8, 14, 50, 74). Especially under the latter conditions IRP1 protein stability seems to be iron regulated (14). In the present study, huNbp35 depletion not only resulted in the reduction of IRP1 levels but also in an even more pronounced drop of cytAco activity, and in turn increased the IRE-binding capacity of IRP1. Together, these results demonstrate the essential function of huNbp35 in the maturation of IRP1 to a functional aconitase and support the view that an impaired Fe/S cluster assembly is leading to diminished IRP1 levels.
The impaired maturation of IRP1 upon huNbp35 depletion had profound consequences on cellular iron homeostasis. In mammalian cells, iron uptake and metabolism are mainly regulated by IRP1 and IRP2 in a posttranscriptional fashion (for a comprehensive review, see reference 73). Depletion of huNbp35 in the present study resulted in a strong decrease of H-ferritin levels, as indicated by immunoblotting. This in vivo effect is consistent with a stalled translation due to an increased IRP1 binding, as indicated by the gel shift assay. An additional contribution of IRP2 in altering ferritin expression seems unlikely since huNbp35 depletion did not result in an elevation of IRP2 levels but rather a slight decrease. In contrast to ferritins, IRP binding to 3′ IREs stabilizes TfR mRNA and promotes TfR mRNA translation. As a result of the increased IRE-binding activity of IRP1, huNbp35 deficiency strongly increased the cellular TfR levels which was measured both by immunostaining and by quantification of the cellular uptake of a fluorescently labeled transferrin (Tf-FITC). Increased cellular Tf-FITC uptake may result in elevated cellular iron levels, providing a logical explanation for the slightly decreased levels of IRP2 by degradation in an iron-dependent manner (73). Since apo-IRP1 protein stability is also iron regulated (14), elevated cellular iron levels might have contributed to its degradation. Taken together, these findings demonstrate that the phenotype of huNbp35-deficient HeLa cells with respect to ferritin and TfR expression resembles that of an iron-starved cell (73). Despite their impaired iron metabolism, huNbp35-deficient cells were still able to respond to changes in external iron levels (data not shown), which in part might be due to incomplete depletion of huNbp35 under our RNAi conditions.
The largely altered synthesis of ferritin and TfR in huNbp35-deficient cells reemphasizes the general importance of Fe/S cluster biosynthesis for the maintenance of iron metabolism (40, 73). In fact, the role of frataxin and ABCB7 in cellular iron homeostasis was well established even before their primary involvement in the formation of Fe/S proteins was identified (3, 17, 31). Depletion of other ISC assembly or export components such as Nfs1 or Isu1 in yeast or mammalian cells generally result in similar iron deficiency phenotypes (18, 34, 38, 70). Remarkably, the molecular basis for the regulation of iron homeostasis in yeast and mammals is substantially different. In yeast, the iron metabolism is mainly regulated at the transcriptional level via the transcription factors Aft1p-Aft2p and associated proteins (30, 33, 57), which are not known to directly associate with Fe/S clusters. This might explain why the depletion of yeast CIA components such as Nbp35p and Cfd1p has no effect on iron metabolism (23, 40). The molecular basis of how the mitochondrial ISC machineries modulate the cellular iron metabolism in yeast is still unclear (33, 48). Conversely, in mammalian cells the involvement of the cytosolic Fe/S protein IRP1 in the posttranscriptional regulation of iron metabolism provides a reasonable explanation for the role of the Fe/S protein biogenesis systems, including the CIA machinery in cellular iron homeostasis. The involvement of huNbp35 (the present study) and IOP1 (61) in IRP1 maturation satisfactorily explains the observed huge alterations in mammalian iron metabolism.
The phenotypes observed for huNbp35-deficient HeLa cells are rather similar to those described recently for depletion of the human Nar1p homologue IOP1 (61). As for huNbp35, the impaired iron homeostasis upon IOP1 depletion could be ascribed to a disrupted Fe/S protein biosynthesis within the cytosol. The primary role of both huNbp35 and IOP1 in extramitochondrial Fe/S protein biogenesis establishes the functional conservation of these CIA machinery components from yeasts to humans and suggests that the molecular mechanisms of Fe/S protein biogenesis are similar in higher and lower eukaryotes. This notion is supported by the in vivo interaction of huNbp35 with huCfd1 demonstrated here, which may imply a function of this P-loop NTPase complex as a scaffold for Fe/S cluster assembly, as shown for its yeast counterpart (47). It will therefore be interesting to directly analyze the cellular role of huCfd1 and clarify the relative tasks of the two related proteins. In contrast, a functional involvement of cytosolic duplicates of mitochondrial ISC assembly components in the maturation of extramitochondrial Fe/S proteins was proposed (1, 39, 69). However, in contrast to the strong effects observed for the depletion of huNbp35 and IOP1, specific RNAi knockdown of the cytosolic version of huIsu1 did not impair Fe/S cluster maturation on IRP1 (70). Similarly, exclusive synthesis of huNfs1 in the cytosol-nucleus of HeLa cells did not suffice to generate cytosolic Fe/S proteins (8). Together, these studies document the central importance of human CIA components in cytosolic Fe/S protein biogenesis, underlining the high conservation of Fe/S protein biogenesis pathways in eukaryotes.
The involvement of huNbp35 and huCfd1 in the maturation of extramitochondrial Fe/S proteins might also provide a link to the observation that an RNAi-mediated knockdown of murine Nbp35 and Cfd1 resulted in phenotypes similar to those observed after knockdown of KIFC5A, a mouse kinesin-like protein that is implicated in the control of centrosome duplication (13). Archaebacterial homologues of the two well-known DNA helicases XPD/Ercc2 and FancJ/Bach1 were recently identified as putative Fe/S proteins (56, 75), thereby connecting cell cycle progression with Fe/S cluster biosynthesis. Moreover, its conceivable that huNbp35 plays a dual role in both the maturation of cytosolic Fe/S proteins and in centrosome duplication. In humans, impaired functions of XPD/Ercc2 and FancJ/Bach1 lead to a certain type of xeroderma pigmentosum (35) and to Fanconi's anemia (67), respectively. Since more and more diseases are identified to be related to an impaired assembly of Fe/S proteins, including Friedreich's ataxia (52) and X-linked sideroblastic anemia and cerebellar ataxia (2), as well as some forms of myopathies (44, 49), microcytic anemia (11), and erythropoietic protoporphyria (59), the elucidation of the mechanisms leading to the maturation of Fe/S proteins within various eukaryotic compartments is of utmost medical importance. The identification of huNbp35 as a critical component of the cytosolic CIA machinery for Fe/S protein maturation may help to achieve deeper insights into these mechanisms.
Acknowledgments
We thank L. Szweda and M. Wattenhofer-Donzé for aconitase and GPAT antibodies, E. W. Müllner for pSPT-fer plasmid containing the ferritin IRE, and B. Steiniger for generously supporting the fluorescence-activated cell sorting experiments.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB 593 and TR1, Gottfried-Wilhelm Leibniz program, and GRK 1216) and the Fonds der Chemischen Industrie (R.L.); the French National Agency for Research (ANR-05-MRAR-013-01) and the French Medical Research Foundation Equipe FRM 2005 (DEQ2005-1205774) (H.P.); and National Institutes of Health grant DK66600 and U.S. Department of Agriculture grant 2006-35200-16604 (R.S.E.).
Footnotes
Published ahead of print on 23 June 2008.
REFERENCES
- 1.Acquaviva, F., I. De Biase, L. Nezi, G. Ruggiero, F. Tatangelo, C. Pisano, A. Monticelli, C. Garbi, A. M. Acquaviva, and S. Cocozza. 2005. Extra-mitochondrial localization of frataxin and its association with IscU1 during enterocyte-like differentiation of the human colon adenocarcinoma cell line Caco-2. J. Cell Sci. 1183917-3924. [DOI] [PubMed] [Google Scholar]
- 2.Allikmets, R., W. H. Raskind, A. Hutchinson, N. D. Schueck, M. Dean, and D. M. Koeller. 1999. Mutation of a putative mitochondrial iron transporter gene (ABC7) in X-linked sideroblastic anemia and ataxia (XLSA/A). Hum. Mol. Genet. 8743-749. [DOI] [PubMed] [Google Scholar]
- 3.Babcock, M., D. de Silva, R. Oaks, S. Davis-Kaplan, S. Jiralerspong, L. Montermini, M. Pandolfo, and J. Kaplan. 1997. Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science 2761709-1712. [DOI] [PubMed] [Google Scholar]
- 4.Balk, J., D. J. Aguilar Netz, K. Tepper, A. J. Pierik, and R. Lill. 2005. The essential WD40 protein Cia1 is involved in a late step of cytosolic and nuclear iron-sulfur protein assembly. Mol. Cell. Biol. 2510833-10841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balk, J., A. J. Pierik, D. J. Netz, U. Mühlenhoff, and R. Lill. 2004. The hydrogenase-like Nar1p is essential for maturation of cytosolic and nuclear iron-sulphur proteins. EMBO J. 232105-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beinert, H., R. H. Holm, and E. Munck. 1997. Iron-sulfur clusters: nature's modular, multipurpose structures. Science 277653-659. [DOI] [PubMed] [Google Scholar]
- 7.Bekri, S., G. Kispal, H. Lange, E. Fitzsimons, J. Tolmie, R. Lill, and D. F. Bishop. 2000. Human ABC7 transporter: gene structure and mutation causing X-linked sideroblastic anemia with ataxia with disruption of cytosolic iron-sulfur protein maturation. Blood 963256-3264. [PubMed] [Google Scholar]
- 8.Biederbick, A., O. Stehling, R. Rösser, B. Niggemeyer, Y. Nakai, H. P. Elsasser, and R. Lill. 2006. Role of human mitochondrial Nfs1 in cytosolic iron-sulfur protein biogenesis and iron regulation. Mol. Cell. Biol. 265675-5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bych, K., S. Kerscher, D. J. Netz, A. J. Pierik, K. Zwicker, M. A. Huynen, R. Lill, U. Brandt, and J. Balk. 2008. The iron-sulphur protein Ind1 is required for effective complex I assembly. EMBO J. 271736-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caltagirone, A., G. Weiss, and K. Pantopoulos. 2001. Modulation of cellular iron metabolism by hydrogen peroxide. Effects of H2O2 on the expression and function of iron-responsive element-containing mRNAs in B6 fibroblasts. J. Biol. Chem. 27619738-19745. [DOI] [PubMed] [Google Scholar]
- 11.Camaschella, C., A. Campanella, L. De Falco, L. Boschetto, R. Merlini, L. Silvestri, S. Levi, and A. Iolascon. 2007. The human counterpart of zebrafish shiraz shows sideroblastic-like microcytic anemia and iron overload. Blood 1101353-1358. [DOI] [PubMed] [Google Scholar]
- 12.Cavadini, P., G. Biasiotto, M. Poli, S. Levi, R. Verardi, I. Zanella, M. Derosas, R. Ingrassia, M. Corrado, and P. Arosio. 2007. RNA silencing of the mitochondrial ABCB7 transporter in HeLa cells causes an iron-deficient phenotype with mitochondrial iron overload. Blood 1093552-3559. [DOI] [PubMed] [Google Scholar]
- 13.Christodoulou, A., C. W. Lederer, T. Surrey, I. Vernos, and N. Santama. 2006. Motor protein KIFC5A interacts with Nubp1 and Nubp2, and is implicated in the regulation of centrosome duplication. J. Cell Sci. 1192035-2047. [DOI] [PubMed] [Google Scholar]
- 14.Clarke, S. L., A. Vasanthakumar, S. A. Anderson, C. Pondarre, C. M. Koh, K. M. Deck, J. S. Pitula, C. J. Epstein, M. D. Fleming, and R. S. Eisenstein. 2006. Iron-responsive degradation of iron-regulatory protein 1 does not require the Fe-S cluster. EMBO J. 25544-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Csere, P., R. Lill, and G. Kispal. 1998. Identification of a human mitochondrial ABC transporter, the functional orthologue of yeast Atm1p. FEBS Lett. 441266-270. [DOI] [PubMed] [Google Scholar]
- 16.Drapier, J. C., and J. B. Hibbs, Jr. 1996. Aconitases: a class of metalloproteins highly sensitive to nitric oxide synthesis. Methods Enzymol. 26926-36. [DOI] [PubMed] [Google Scholar]
- 17.Foury, F., and O. Cazzalini. 1997. Deletion of the yeast homologue of the human gene associated with Friedreich's ataxia elicits iron accumulation in mitochondria. FEBS Lett. 411373-377. [DOI] [PubMed] [Google Scholar]
- 18.Gerber, J., K. Neumann, C. Prohl, U. Mühlenhoff, and R. Lill. 2004. The yeast scaffold proteins Isu1p and Isu2p are required inside mitochondria for maturation of cytosolic Fe/S proteins. Mol. Cell. Biol. 244848-4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonsior, S. M., S. Platz, S. Buchmeier, U. Scheer, B. M. Jockusch, and H. Hinssen. 1999. Conformational difference between nuclear and cytoplasmic actin as detected by a monoclonal antibody. J. Cell Sci. 112(Pt. 6)797-809. [DOI] [PubMed] [Google Scholar]
- 20.Grandoni, J. A., R. L. Switzer, C. A. Makaroff, and H. Zalkin. 1989. Evidence that the iron-sulfur cluster of Bacillus subtilis glutamine phosphoribosylpyrophosphate amidotransferase determines stability of the enzyme to degradation in vivo. J. Biol. Chem. 2646058-6064. [PubMed] [Google Scholar]
- 21.Hatefi, Y., and D. L. Stiggall. 1978. Preparation and properties of succinate: ubiquinone oxidoreductase (complex II). Methods Enzymol. 5321-27. [DOI] [PubMed] [Google Scholar]
- 22.Hausmann, A., D. J. Aguilar Netz, J. Balk, A. J. Pierik, U. Mühlenhoff, and R. Lill. 2005. The eukaryotic P loop NTPase Nbp35: an essential component of the cytosolic and nuclear iron-sulfur protein assembly machinery. Proc. Natl. Acad. Sci. USA 1023266-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hausmann, A., B. Samans, R. Lill, and U. Mühlenhoff. 2008. Cellular and mitochondrial remodeling upon defects in iron-sulfur protein biogenesis. J. Biol. Chem. 2838318-8330. [DOI] [PubMed] [Google Scholar]
- 24.Hirsch, S., and W. K. Miskimins. 1995. Mitogen induction of nuclear factors that interact with a delayed responsive region of the transferrin receptor gene promoter. Cell Growth Differ. 6719-726. [PubMed] [Google Scholar]
- 25.Howell, B. F., S. McCune, and R. Schaffer. 1979. Lactate-to-pyruvate or pyruvate-to-lactate assay for lactate dehydrogenase: a re-examination. Clin. Chem. 25269-272. [PubMed] [Google Scholar]
- 26.Huang, J., D. Song, A. Flores, Q. Zhao, S. M. Mooney, L. M. Shaw, and F. S. Lee. 2007. IOP1, a novel hydrogenase-like protein that modulates hypoxia-inducible factor-1alpha activity. Biochem. J. 401341-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang, S. N., M. A. Phelps, and P. W. Swaan. 2003. Involvement of endocytic organelles in the subcellular trafficking and localization of riboflavin. J. Pharmacol. Exp. Ther. 306681-687. [DOI] [PubMed] [Google Scholar]
- 28.Johnson, D. C., D. R. Dean, A. D. Smith, and M. K. Johnson. 2005. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74247-281. [DOI] [PubMed] [Google Scholar]
- 29.Johnstone, R. W., J. Wang, N. Tommerup, H. Vissing, T. Roberts, and Y. Shi. 1998. Ciao 1 is a novel WD40 protein that interacts with the tumor suppressor protein WT1. J. Biol. Chem. 27310880-10887. [DOI] [PubMed] [Google Scholar]
- 30.Kaplan, J., D. McVey Ward, R. J. Crisp, and C. C. Philpott. 2006. Iron-dependent metabolic remodeling in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1763646-651. [DOI] [PubMed] [Google Scholar]
- 31.Kispal, G., P. Csere, B. Guiard, and R. Lill. 1997. The ABC transporter Atm1p is required for mitochondrial iron homeostasis. FEBS Lett. 418346-350. [DOI] [PubMed] [Google Scholar]
- 32.Kispal, G., P. Csere, C. Prohl, and R. Lill. 1999. The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 183981-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumanovics, A., O. Chen, L. Li, D. Bagley, E. Adkins, H. Lin, N. N. Dingra, C. E. Outten, G. Keller, D. Winge, D. M. Ward, and J. Kaplan. 2008. Identification of FRA1 and FRA2 as genes involved in regulating the yeast iron regulon in response to decreased mitochondrial iron-sulfur cluster synthesis. J. Biol. Chem. 28310276-10286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lange, H., T. Lisowsky, J. Gerber, U. Mühlenhoff, G. Kispal, and R. Lill. 2001. An essential function of the mitochondrial sulfhydryl oxidase Erv1p/ALR in the maturation of cytosolic Fe/S proteins. EMBO Rep. 2715-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lehmann, A. R. 2001. The xeroderma pigmentosum group D (XPD) gene: one gene, two functions, three diseases. Genes Dev. 1515-23. [DOI] [PubMed] [Google Scholar]
- 36.Leipe, D. D., Y. I. Wolf, E. V. Koonin, and L. Aravind. 2002. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 31741-72. [DOI] [PubMed] [Google Scholar]
- 37.Lezhneva, L., K. Amann, and J. Meurer. 2004. The universally conserved HCF101 protein is involved in assembly of [4Fe-4S]-cluster-containing complexes in Arabidopsis thaliana chloroplasts. Plant J. 37174-185. [DOI] [PubMed] [Google Scholar]
- 38.Li, J., M. Kogan, S. A. Knight, D. Pain, and A. Dancis. 1999. Yeast mitochondrial protein, Nfs1p, coordinately regulates iron-sulfur cluster proteins, cellular iron uptake, and iron distribution. J. Biol. Chem. 27433025-33034. [DOI] [PubMed] [Google Scholar]
- 39.Li, K., W. H. Tong, R. M. Hughes, and T. A. Rouault. 2006. Roles of the mammalian cytosolic cysteine desulfurase, ISCS, and scaffold protein, ISCU, in iron-sulfur cluster assembly. J. Biol. Chem. 28112344-12351. [DOI] [PubMed] [Google Scholar]
- 40.Lill, R., and U. Mühlenhoff. 2008. Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu. Rev. Biochem. 77669-700. [DOI] [PubMed] [Google Scholar]
- 41.Lok, C. N., and P. Ponka. 1999. Identification of a hypoxia response element in the transferrin receptor gene. J. Biol. Chem. 27424147-24152. [DOI] [PubMed] [Google Scholar]
- 42.Martelli, A., M. Wattenhofer-Donze, S. Schmucker, S. Bouvet, L. Reutenauer, and H. Puccio. 2007. Frataxin is essential for extramitochondrial Fe-S cluster proteins in mammalian tissues. Hum. Mol. Genet. 162651-2658. [DOI] [PubMed] [Google Scholar]
- 43.Miskimins, W. K., A. McClelland, M. P. Roberts, and F. H. Ruddle. 1986. Cell proliferation and expression of the transferrin receptor gene: promoter sequence homologies and protein interactions. J. Cell Biol. 1031781-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mochel, F., M. A. Knight, W. H. Tong, D. Hernandez, K. Ayyad, T. Taivassalo, P. M. Andersen, A. Singleton, T. A. Rouault, K. H. Fischbeck, and R. G. Haller. 2008. Splice mutation in the iron-sulfur cluster scaffold protein ISCU causes myopathy with exercise intolerance. Am. J. Hum. Genet. 82652-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Müllner, E. W., B. Neupert, and L. C. Kühn. 1989. A specific mRNA binding factor regulates the iron-dependent stability of cytoplasmic transferrin receptor mRNA. Cell 58373-382. [DOI] [PubMed] [Google Scholar]
- 46.Nakashima, H., M. J. Grahovac, R. Mazzarella, H. Fujiwara, J. R. Kitchen, T. A. Threat, and M. S. Ko. 1999. Two novel mouse genes—Nubp2, mapped to the t-complex on chromosome 17, and Nubp1, mapped to chromosome 16—establish a new gene family of nucleotide-binding proteins in eukaryotes. Genomics 60152-160. [DOI] [PubMed] [Google Scholar]
- 47.Netz, D. J., A. J. Pierik, M. Stümpfig, U. Mühlenhoff, and R. Lill. 2007. The Cfd1-Nbp35 complex acts as a scaffold for iron-sulfur protein assembly in the yeast cytosol. Nat. Chem. Biol. 3278-286. [DOI] [PubMed] [Google Scholar]
- 48.Ojeda, L., G. Keller, U. Mühlenhoff, J. C. Rutherford, R. Lill, and D. R. Winge. 2006. Role of glutaredoxin-3 and glutaredoxin-4 in the iron regulation of the Aft1 transcriptional activator in Saccharomyces cerevisiae. J. Biol. Chem. 28117661-17669. [DOI] [PubMed] [Google Scholar]
- 49.Olsson, A., L. Lind, L. E. Thornell, and M. Holmberg. 2008. Myopathy with lactic acidosis is linked to chromosome 12q23.3-24.11 and caused by an intron mutation in the ISCU gene resulting in a splicing defect. Hum. Mol. Genet. 171666-1672. [DOI] [PubMed] [Google Scholar]
- 50.Pondarre, C., B. B. Antiochos, D. R. Campagna, S. L. Clarke, E. L. Greer, K. M. Deck, A. McDonald, A. P. Han, A. Medlock, J. L. Kutok, S. A. Anderson, R. S. Eisenstein, and M. D. Fleming. 2006. The mitochondrial ATP-binding cassette transporter Abcb7 is essential in mice and participates in cytosolic iron-sulfur cluster biogenesis. Hum. Mol. Genet. 15953-964. [DOI] [PubMed] [Google Scholar]
- 51.Ponka, P., and C. N. Lok. 1999. The transferrin receptor: role in health and disease. Int. J. Biochem. Cell. Biol. 311111-1137. [DOI] [PubMed] [Google Scholar]
- 52.Puccio, H., and M. Koenig. 2002. Friedreich ataxia: a paradigm for mitochondrial diseases. Curr. Opin. Genet. Dev. 12272-277. [DOI] [PubMed] [Google Scholar]
- 53.Rouault, T. A. 2006. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat. Chem. Biol. 2406-414. [DOI] [PubMed] [Google Scholar]
- 54.Rouault, T. A., and W. H. Tong. 2005. Iron-sulphur cluster biogenesis and mitochondrial iron homeostasis. Nat. Rev. Mol. Cell. Biol. 6345-351. [DOI] [PubMed] [Google Scholar]
- 55.Roy, A., N. Solodovnikova, T. Nicholson, W. Antholine, and W. E. Walden. 2003. A novel eukaryotic factor for cytosolic Fe-S cluster assembly. EMBO J. 224826-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rudolf, J., V. Makrantoni, W. J. Ingledew, M. J. Stark, and M. F. White. 2006. The DNA repair helicases XPD and FancJ have essential iron-sulfur domains. Mol. Cell 23801-808. [DOI] [PubMed] [Google Scholar]
- 57.Rutherford, J. C., L. Ojeda, J. Balk, U. Mühlenhoff, R. Lill, and D. R. Winge. 2005. Activation of the iron regulon by the yeast Aft1/Aft2 transcription factors depends on mitochondrial but not cytosolic iron-sulfur protein biogenesis. J. Biol. Chem. 28010135-10140. [DOI] [PubMed] [Google Scholar]
- 58.Shahrestanifar, M., D. P. Saha, L. A. Scala, A. Basu, and R. D. Howells. 1994. Cloning of a human cDNA encoding a putative nucleotide-binding protein related to Escherichia coli MinD. Gene 147281-285. [DOI] [PubMed] [Google Scholar]
- 59.Shaw, G. C., J. J. Cope, L. Li, K. Corson, C. Hersey, G. E. Ackermann, B. Gwynn, A. J. Lambert, R. A. Wingert, D. Traver, N. S. Trede, B. A. Barut, Y. Zhou, E. Minet, A. Donovan, A. Brownlie, R. Balzan, M. J. Weiss, L. L. Peters, J. Kaplan, L. I. Zon, and B. H. Paw. 2006. Mitoferrin is essential for erythroid iron assimilation. Nature 44096-100. [DOI] [PubMed] [Google Scholar]
- 60.Skovran, E., and D. M. Downs. 2003. Lack of the ApbC or ApbE protein results in a defect in Fe-S cluster metabolism in Salmonella enterica serovar Typhimurium. J. Bacteriol. 18598-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song, D., and F. S. Lee. 2008. A role for Iop1 in mammalian cytosolic iron-sulfur protein biogenesis. J. Biol. Chem. 2839231-9238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Srere, P. A., M. Brazil, and L. Gonen. 1963. The citrate condensing enzyme of pigeon breast muscle and moth flight muscle. Acta Chem. Scand. 17129-134. [Google Scholar]
- 63.Srinivasan, V., D. J. Netz, H. Webert, J. Mascarenhas, A. J. Pierik, H. Michel, and R. Lill. 2007. Structure of the yeast WD40 domain protein Cia1, a component acting late in iron-sulfur protein biogenesis. Structure 151246-1257. [DOI] [PubMed] [Google Scholar]
- 64.Stehling, O., H. P. Elsässer, B. Brückel, U. Mühlenhoff, and R. Lill. 2004. Iron-sulfur protein maturation in human cells: evidence for a function of frataxin. Hum. Mol. Genet. 133007-3015. [DOI] [PubMed] [Google Scholar]
- 65.Stehling, O., P. M. Smith, A. Biederbick, J. Balk, R. Lill, and U. Mühlenhoff. 2007. Investigation of iron-sulfur protein maturation in eukaryotes. Methods Mol. Biol. 372325-342. [DOI] [PubMed] [Google Scholar]
- 66.Tacchini, L., L. Bianchi, A. Bernelli-Zazzera, and G. Cairo. 1999. Transferrin receptor induction by hypoxia. HIF-1-mediated transcriptional activation and cell-specific posttranscriptional regulation. J. Biol. Chem. 27424142-24146. [DOI] [PubMed] [Google Scholar]
- 67.Taniguchi, T., and A. D. D'Andrea. 2006. Molecular pathogenesis of Fanconi anemia: recent progress. Blood 1074223-4233. [DOI] [PubMed] [Google Scholar]
- 68.Theil, E. C., M. Matzapetakis, and X. Liu. 2006. Ferritins: iron/oxygen biominerals in protein nanocages. J. Biol. Inorg. Chem. 11803-810. [DOI] [PubMed] [Google Scholar]
- 69.Tong, W. H., G. N. Jameson, B. H. Huynh, and T. A. Rouault. 2003. Subcellular compartmentalization of human Nfu, an iron-sulfur cluster scaffold protein, and its ability to assemble a [4Fe-4S] cluster. Proc. Natl. Acad. Sci. USA 1009762-9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tong, W. H., and T. A. Rouault. 2006. Functions of mitochondrial ISCU and cytosolic ISCU in mammalian iron-sulfur cluster biogenesis and iron homeostasis. Cell Metab. 3199-210. [DOI] [PubMed] [Google Scholar]
- 71.Tong, W. H., and T. A. Rouault. 2007. Metabolic regulation of citrate and iron by aconitases: role of iron-sulfur cluster biogenesis. Biometals 20549-564. [DOI] [PubMed] [Google Scholar]
- 72.Vitale, G., E. Fabre, and E. C. Hurt. 1996. NBP35 encodes an essential and evolutionary conserved protein in Saccharomyces cerevisiae with homology to a superfamily of bacterial ATPases. Gene 17897-106. [DOI] [PubMed] [Google Scholar]
- 73.Wallander, M. L., E. A. Leibold, and R. S. Eisenstein. 2006. Molecular control of vertebrate iron homeostasis by iron regulatory proteins. Biochim. Biophys. Acta 1763668-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang, J., C. Fillebeen, G. Chen, A. Biederbick, R. Lill, and K. Pantopoulos. 2007. Iron-dependent degradation of apo-IRP1 by the ubiquitin-proteasome pathway. Mol. Cell. Biol. 272423-2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weiner, B. E., H. Huang, B. M. Dattilo, M. J. Nilges, E. Fanning, and W. J. Chazin. 2007. An iron-sulfur cluster in the C-terminal domain of the p58 subunit of human DNA primase. J. Biol. Chem. 28233444-33451. [DOI] [PubMed] [Google Scholar]
- 76.Zhou, G., S. S. Broyles, J. E. Dixon, and H. Zalkin. 1992. Avian glutamine phosphoribosylpyrophosphate amidotransferase propeptide processing and activity are dependent upon essential cysteine residues. J. Biol. Chem. 2677936-7942. [PubMed] [Google Scholar]