Abstract
Specific regions of genomes (fragile sites) are hot spots for the chromosome rearrangements that are associated with many types of cancer cells. Understanding the molecular mechanisms regulating the stability of chromosome fragile sites, therefore, has important implications in cancer biology. We previously identified two chromosome fragile sites in Saccharomyces cerevisiae that were induced in response to the reduced expression of Pol1p, the catalytic subunit of DNA polymerase α. In the study presented here, we show that reduced levels of Pol3p, the catalytic subunit of DNA polymerase δ, induce instability at these same sites and lead to the generation of a variety of chromosomal aberrations. These findings demonstrate that a change in the stoichiometry of replicative DNA polymerases results in recombinogenic DNA lesions, presumably double-strand DNA breaks.
Maintaining intact chromosomes is essential for normal cellular growth and viability. If cell division is initiated in the presence of partially replicated or broken chromosomes, or if broken chromosomes are repaired by recombination events involving nonhomologous chromosomes, the resulting daughter cells will have abnormal numbers of chromosomes or chromosome rearrangements (deletions, duplications, and translocations) (13). Such genomic alterations are commonly observed in many types of cancers (3, 7, 8, 11, 17, 21, 29). Therefore, understanding the molecular mechanisms that lead to chromosome breakage has important implications in cancer biology.
The chromosome rearrangements observed in some types of cancers are associated with regions known as chromosome fragile sites (3, 7, 8, 11, 17, 21, 29). These sites are chromosomal regions that are prone to breakage under DNA replication stresses. There are two types of fragile sites. Rare fragile sites are small regions associated with long tandem arrays of certain trinucleotide repeats that can form unusual DNA structures that block replication (27). In contrast, common fragile sites are large regions not associated with a specific DNA sequence. The common fragile sites are replicated late in the S phase (1, 19) and are induced by drugs, such as aphidicolin, that inhibit DNA replication (9). Thus, both rare and common fragile sites are preferred sites for double-strand break (DSB) formation under conditions of altered DNA replication.
We have previously identified two common chromosome fragile sites in Saccharomyces cerevisiae (20). These fragile sites were hot spots for chromosome rearrangements in cells that had reduced expression of the catalytic subunit of DNA polymerase α. To determine whether other forms of replication stress might induce the expression of these chromosome fragile sites in yeast, we examined the genomic stability of a strain with reduced expression of Pol3p, the catalytic subunit of DNA polymerase δ.
As described below, we found that low levels of Pol3p led to an increased sensitivity to DNA-damaging agents. In addition, reduced levels of Pol3p resulted in a mutator phenotype (an elevated rate of forward mutations at the CAN1 locus) and increases in the rate of chromosome loss and mitotic recombination. Another important phenotype induced by low levels of Pol3p was an increase in the efficiency of illegitimate mating, an assay of the rate of chromosome loss and rearrangements involving chromosome III. Most of these rearrangements occurred at the same fragile site (FS2; an inverted pair of retrotransposons) identified in our study of genomic instability induced by low levels of DNA polymerase α.
MATERIALS AND METHODS
Genetic analysis and media.
Genetic procedures (transformation, tetrad analysis, etc.) and media used were standard (12). Media containing high (0.05%) and low (0.005%) levels of galactose also contained 3% raffinose and the standard supplements for rich growth media (with the exception of glucose). Strains were grown at 30°C.
Strain construction.
All of the strains used in this study (with the exception of the mating-type testers) were isogenic with MS71, a LEU2 derivative of AMY125 (MATα ade5-1 leu2-3 trp1-289 ura3-52 his7-2) (16); EAS18 is a MATa derivative of AMY125 (18). All alterations from this genotype were generated by transformation or by crosses with isogenic strains. The details of the strain construction are described in the supplemental material.
Microarray analysis of chromosome rearrangements.
Comparative genomic hybridization (CGH) microarray analysis of illegitimate diploids was performed as previously described (20). In brief, total genomic DNA was isolated from Zymolyase-treated cells. The purified genomic DNA was sonicated to approximately 500-bp fragments. Five micrograms of this sheared DNA was labeled with either Cy3-dUTP (for the reference sample from strain MS71) or Cy5-dUTP (for the experimental sample from the illegitimate diploids) (Amersham Biosciences). The samples were then purified, mixed, and used to probe open reading frame (ORF)-containing microarrays prepared as described by DeRisi et al. (4). The arrays were washed and dried, then scanned using a GenePix 4000B scanner (Axon Instruments). The patterns were analyzed using GenePix Pro 4.1 (Axon Instruments) and CGH-Miner for Excel 1.0 (http://www-stat.stanford.edu/∼wp57/CGH-Miner).
Measurement of mutation rates, chromosome loss rates, and mitotic recombination rates.
The methods for measuring the rate of mutation at CAN1 were as described previously (18); this analysis is fully discussed in the supplemental material. The procedure for the sequence analysis of can1 mutant genes is also described in the supplemental material. Chromosome loss and mitotic recombination rates were determined for diploid strains (DNPD1 and DNPD13) that were heterozygous for markers on the left (can1) and right (his1) arms of chromosome V as described previously (18). The details of this analysis are in the supplemental material.
Measurement of legitimate and illegitimate mating frequencies.
The frequencies of legitimate and illegitimate mating for the wild-type (MS71) and GAL-POL3 MATα (RJK341) haploid strains grown in the presence of high (0.05%) or low (0.005%) levels of galactose were measured as previously described (20). In brief, we grew the wild-type and GAL-POL3 cells for about 24 h in medium containing either high or low levels of galactose. These strains were then mixed with a fivefold excess of W303a (MATa tester to measure legitimate mating) or W303α (MATα tester to measure illegitimate mating). The mixture was concentrated on a filter paper that was placed on a plate containing medium with high (0.05%) levels of galactose. After 6 h, the cells were washed off the filter and plated onto medium that selected for the growth of diploids (medium lacking histidine and adenine).
We isolated multiple, independent illegitimate diploids derived from the mating of either the wild-type or the GAL-POL3 strain (pregrown in the presence of either high or low levels of galactose) with the 1225 mating-type tester strain. MS71 and RJK341 are MATα HIS4 THR4, whereas 1225 is MATα his4 thr4. The matings were performed in the presence of high (0.05%) levels of galactose, and the resulting illegitimate diploids were selected on synthetic defined complete medium lacking adenine and lysine. From each independent mating, one diploid was purified, and the ability of the strain to grow in the absence of histidine or threonine was examined.
We measured the relative frequencies of the legitimate and illegitimate mating of FJL043 (a Trp+ GAL-POL3 derivative with an intact FS2) and FJL016 (a Trp− GAL-POL3 derivative in which the centromere-proximal Ty element of FS2 was replaced with the hygromycin resistance gene [HYGR]). Independent single colonies of these two strains were mixed and grown in the presence of either high (0.05%) or low (0.005%) levels of galactose. The mixture was then mated with 1225 in the presence of high (0.05%) levels of galactose. A single diploid was purified (selected by growth in medium lacking adenine and lysine), and its Trp phenotype was determined. Diploids that were Trp+ were assumed to be derived from the FJL043 parent, and diploids that were Trp− were assumed to be derived from the FJL016 parent. Approximately 100 independent matings were performed for each experiment.
Analysis of chromosome rearrangements by gel electrophoresis.
We analyzed chromosome rearrangements using two types of gels. CHEF (contour-clamped homogeneous electric field) gels were used to examine intact chromosomes. The sample preparation and DNA analysis were done as previously described (20). To determine the size of chromosome III, we did Southern analysis using a PCR fragment encompassing the CHA1 gene on the left arm of chromosome III (Saccharomyces Genome Database coordinates 15838 to 16857) as a hybridization probe. For five of the illegitimate diploids, we did a detailed analysis of the breakpoints by standard gel electrophoresis, followed by Southern analysis. Mapping of the comparable genomic regions of the two parental haploid strains (RJK341 and 1225) was also done. These analyses are presented in detail in the supplemental material.
RESULTS
Low levels of Pol3p result in increased sensitivity to DNA-damaging agents.
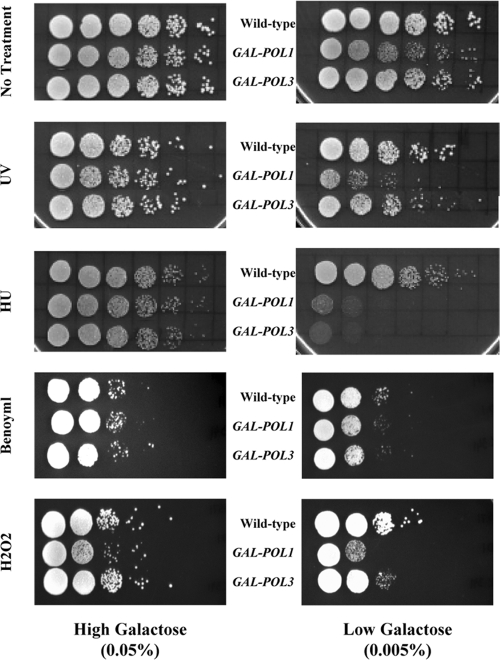
We demonstrated previously that the reduced expression of Pol1p, the catalytic subunit of DNA polymerase α, causes increased sensitivity to various types of DNA-damaging agents (20). To determine if this phenotype is associated with the reduced expression of other DNA polymerases, we examined a strain that expresses Pol3p (the catalytic subunit of DNA polymerase δ) under the control of the galactose-inducible GAL1,10 promoter. This strain, RJK341, has been described previously (18). Compared to the expression level from its native promoter, the expression of Pol3p in this GAL-POL3 strain is 8-fold higher in the presence of high (0.05%) levels of galactose and 14-fold lower in the presence of low (0.005%) levels of galactose (18). As shown in Fig. 1, elevated Pol3p expression had no effect on cell growth in either the absence or the presence of two DNA-damaging agents, UV irradiation and hydroxyurea (HU). Reduced Pol3p expression increased the sensitivity of cells to HU but did not alter their sensitivity to UV (Fig. 1); we previously showed that low levels of DNA polymerase δ or α increased the sensitivity of cells to methyl methanesulfonate (18, 20). Strains with low levels of DNA polymerase δ are slightly more sensitive than wild-type cells to the oxidizing agent hydrogen peroxide but have the same sensitivity as wild-type cells to the spindle-depolymerizing drug benomyl (Fig. 1). In addition, low levels of DNA polymerase δ had no effect on the growth of cells in the absence of DNA-damaging agents.
FIG. 1.
Low levels of Pol3p increase sensitivity to DNA-damaging agents. Wild-type, GAL-POL1, and GAL-POL3 strains, pregrown in medium containing high (0.05%) levels of galactose, were serially diluted and spotted onto rich growth medium in the presence of either high (0.05%; left) or low (0.005%; right) levels of galactose. The various panels show cells plated on rich growth medium in the absence of any mutagen, rich growth medium followed by exposure to UV irradiation (35 J/m2), or rich growth medium containing HU (50 mM), benomyl (40 μg/ml), or hydrogen peroxide (1 mM). The depictions of the wild-type and GAL-POL1 strains are from Figure S3 of Lemoine et al. (reprinted from Cell [20] with permission of the publisher).
Low levels of Pol3p induce a mutator phenotype.
We also examined the effects of reduced Pol3p expression on the forward mutation rate of the CAN1 locus. The mutation rates (95% confidence limits) for the wild-type strain were 1.1 (0.9 to 1.5) × 10−7 and 2 (1.7 to 2.6) × 10−7 when the cells were grown in high (0.05%) and low (0.005%) levels of galactose, respectively. The comparable rates for the GAL-POL3 strain were 1.9 (1.8 to 2.2) × 10−7 and 22 (6 to 30) × 10−7. Although elevated Pol3p expression results in a very modest (2-fold) increase in mutations, reduced Pol3p expression has a substantially increased effect (12-fold increase). In the same genetic background, a mutation that eliminates DNA mismatch repair elevates can1 mutations about 30-fold (26). We have previously measured the mutator phenotype associated with low levels of DNA polymerase δ using somewhat different conditions and a different control strain (18). The results do not differ significantly from those above.
We previously did a limited amount of sequence analysis of the can1 mutants in the wild-type strain and in the GAL-POL3 strain (18). Of the 20 can1 mutants from the wild-type strain that were examined, 11 had single base changes, 4 had small insertions/duplications, 3 had deletions or additions of a single base in mononucleotide runs, 1 had a single base deletion that was not in a mononucleotide run, and 1 had a deletion of 18 bp in which the deletion was flanked by short direct repeats. A similar distribution of types of mutations was observed in the GAL-POL3 strain grown in high levels of galactose. In striking contrast, 58% of the can1 mutants derived from the GAL-POL3 strain grown in low levels of galactose were deletions that were greater than 10 bp and were flanked by direct repeats (18).
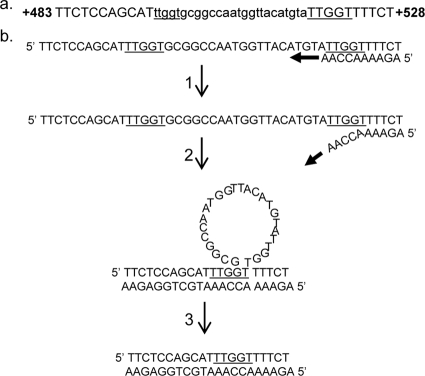
There are two explanations for the high frequency of deletions in strains with low levels of DNA polymerase δ. First, it is possible that low levels of polymerase lead to elevated rates of DNA slippage between short repeats within the CAN1 gene mediated by DNA polymerase δ. Alternatively, the deletions could reflect the increased activity of error-prone DNA polymerases that could have an increased rate of slippage (25). The second possibility predicts that mutations in the error-prone DNA polymerases should reduce or eliminate the can1 deletion class. Consequently, we constructed a GAL-POL3 strain (RJK456) that lacked both RAD30 (encoding DNA polymerase eta) and REV3 (encoding a subunit of DNA polymerase zeta). The mutation rate (95% confidence interval) at the CAN1 locus in this strain grown in low levels of galactose was 7.3 (6 to 10) × 10−7. This rate is less than that observed in the GAL-POL3 strain without the rev3 and rad30 mutations, although the difference is not statistically significant. In order to determine whether the mutation spectrum was affected by the rev3 and rad30 mutations, we measured the frequency of deletions greater than 8 bp (see details in Materials and Methods). Of the 14 isolates examined, 11 had a deletion greater than or equal to 8 bp. We sequenced seven of the deletions and found that all were flanked by short direct repeats. An example of one such deletion is shown in Fig. 2a. DNA polymerase slippage could produce such a deletion, as shown in Fig. 2b, although other mechanisms (such as single-strand annealing) could generate similar deletions (16). The sizes of the sequenced deletions and the flanking repeat sizes (shown in parentheses) in base pairs were 8 (4), 68 (7), 25 (5), 30 (4), 198 (3), 32 (6), and 63 (8). In summary, we found that low levels of DNA polymerase δ result in an elevation in the deletions between short direct repeats, and the generation of these deletions is independent of the error-prone DNA polymerases zeta and eta.
FIG. 2.
Deletions in the can1 gene between short direct repeats. (a) A common mutation in strains with low levels of DNA polymerase δ is a small deletion with short direct repeats at the deletion breakpoints. The sequence in this figure is from a can1 mutant derived from the RJK456 (rev3 rad30 GAL-POL3) strain grown in a low level of galactose. The coordinates are shown relative to the first base of CAN1. The mutation is a 25-bp deletion (shown in small letters), and the small direct repeat is underlined. Because of the repeat, we cannot determine whether the deletion removes bases 494 to 518 (as shown) or 499 to 523. (b) Deletions resulting from DNA polymerase slippage. A replicating DNA molecule is shown with the primer strand shown on the bottom and an arrow showing the direction of replication. In step 1, the primer strand partially dissociates from the template. In step 2, the primer strand reassociates with the AACCA sequence pairing with a noncontiguous TTGGT sequence; the intervening 25 bases is displaced as a DNA loop. In step 3, the loop is removed and the strands are ligated, resulting in the 25-bp deletion.
Low levels of Pol3p increase chromosome V loss and mitotic recombination.
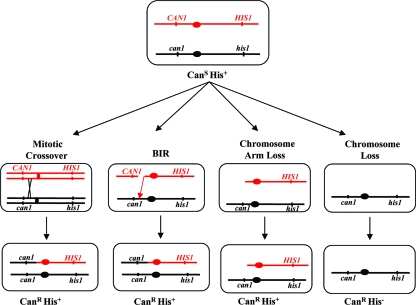
We examined the effects of low levels of DNA polymerase δ on chromosome loss and mitotic exchange in diploid strains using a standard assay (13). This assay, illustrated in Fig. 3, uses a diploid strain that has wild-type alleles (CAN1 and HIS1) on the left and right arms, respectively, of one copy of chromosome V and recessive alleles (can1 and his1) on the other copy of chromosome V. Since both mutations are recessive, the diploid strain is canavanine-sensitive (CanS) and His+. Derivatives of this strain that lose the wild-type allele-carrying copy of chromosome V are CanR and His−. Several different mechanisms can result in CanR His+ cells (Fig. 3): reciprocal mitotic crossing-over, break-induced replication (BIR), and chromosome arm loss followed by telomere capping. In addition, a mutation in the wild-type CAN1 allele can result in a CanR His+ derivative. As discussed below, based on the relatively low rate of CAN1 mutations measured in the haploid strain with low DNA polymerase δ (2.2 × 10−6) compared to the level of CanR derivatives in the diploid (Table 1), mutations are not likely to contribute substantially to the rate of CanR derivatives in the diploid.
FIG. 3.
Assay for mitotic recombination and loss of chromosome V. This commonly used assay employs a diploid cell that is heterozygous for the CAN1 and HIS1 loci on the left and right arms, respectively, of chromosome V. Cells of this genotype are CanS His+. Canavanine-resistant derivatives of this strain can be generated by a mitotic crossing-over event, BIR, loss of a portion of the left arm of chromosome V followed by telomere capping, or loss of one copy of chromosome V. The first three classes of events result in CanR His+ cells, and the chromosome loss events result in CanR His− cells.
TABLE 1.
Rates of chromosome V loss and mitotic recombination in wild-type (DNPD1) and GAL-POL3 (DNPD13) diploids grown in high (0.05%) and low (0.005%) levels of galactose
| Level of galactose and strain | Ratea
|
|
|---|---|---|
| Chromosome V loss (10−6) | Mitotic recombination (10−5) | |
| High | ||
| DNPD1 (wild type) | 4 (3-5) [1] | 1.1 (0.8-2) [1] |
| DNPD13 (GAL-POL3) | 6.1 (5-7) [1.5] | 2.8 (2-3) [2.5] |
| Low | ||
| DNPD1 (wild type) | 7.4 (5-12) [1] | 1.9 (1-6) [1] |
| DNPD13 (GAL-POL3) | 58 (40-70) [7.8] | 67 (10-70) [35] |
Using strains with the markers shown in Fig. 3, we measured the frequencies of CanR His− and CanR His+ derivatives in about 20 independent cultures for each strain in each experimental condition. The rates and 95% confidence limits of the rates (shown in parentheses) were calculated from the frequency data (26). The rates in brackets are normalized to the wild-type strain under the same growth condition.
Using the system shown in Fig. 3, we determined the chromosome loss rates for the wild-type (DNPD1) and GAL-POL3 (DNPD13) diploids grown in the presence of high (0.05%) or low (0.005%) levels of galactose. We found that low levels of DNA polymerase δ result in an eightfold increase in the rate of chromosome V loss (Table 1). Elevated levels of polymerase δ had no significant effect on chromosome loss rates.
We also used this assay to determine the mitotic recombination rates for wild-type (DNPD1) and GAL-POL3 (DNPD13) diploids grown in the presence of high (0.05%) or low (0.005%) levels of galactose. The results (Table 1) show that, following growth in medium containing low levels of galactose, there is a 35-fold increase in the rate of mitotic recombination in the GAL-POL3 strain relative to that in the wild-type strain; low levels of DNA polymerase α resulted in a 22-fold elevation of the rates of mitotic recombination (20). Our experiments do not distinguish among the several classes of mitotic recombination events illustrated in Fig. 3.
Although the effect of low levels of DNA polymerase δ on chromosome V loss and mitotic recombination are slightly greater than observed in strains with low levels of DNA polymerase α, the significance of these differences is not clear. The increase is calculated as a ratio of two rates, each associated with confidence intervals. The confidence interval of the ratio is greater than that associated with either of the rates. For example, the chromosome V loss rates (95% confidence limits) for the wild-type strain and the strain with low levels of DNA polymerase δ were 7.4 (5 to 12) × 10−6 and 58 (40 to 70) × 10−6, respectively (Table 1). If we calculate the confidence limits for the ratios by dividing 70 × 10−6 by 5 × 10−6 and 40 × 10−6 by 12 × 10−6, we estimate that low DNA polymerase δ elevates chromosome loss between 3- and 14-fold.
Low levels of Pol3p increase the efficiency of illegitimate mating.
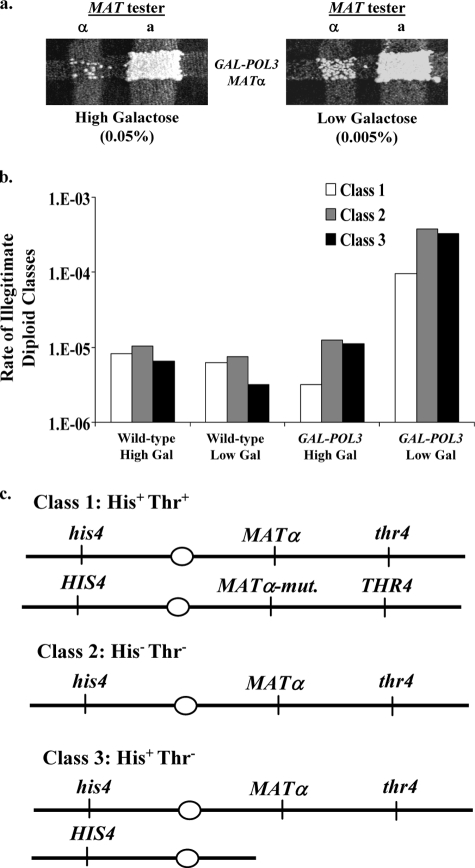
The reduced expression of Pol1p has been shown to increase the efficiency of mating between two MATα haploids (20). This “illegitimate” mating is usually a consequence of the deletion or mutation of the MATα locus in one of the two mated haploids (10, 20, 23). To determine whether a reduced expression of Pol3p elevated illegitimate mating, we first used a streak mating assay (Fig. 4a). From this analysis, it was evident that the rate of illegitimate mating was increased in the GAL-POL3 strain grown in the presence of low galactose (0.005%) relative to the rate in the presence of high (0.05%) galactose. In a more quantitative assay of illegitimate mating (described in Materials and Methods), we found that growing cells in low levels of galactose, which results in low levels of DNA polymerase δ, elevated illegitimate mating by about 50-fold relative to a wild-type strain examined under the same conditions (Table 2). In the same quantitative assay performed with GAL-POL1 strains, illegitimate mating was elevated about 200-fold relative to that of a wild-type strain (20).
FIG. 4.
Low levels of Pol3p increase the efficiency of illegitimate mating. (a) Frequency of illegitimate mating in the GAL-POL3 strain grown in the presence of high (0.05%; left) or low (0.005%; right) levels of galactose. After the strains grew overnight, they were mated (by replica plating) to both MATa and MATα tester strains. Diploids were selected on omission medium. (b) Rates of class 1 to 3 illegitimate diploids. We determined the relative frequencies of class 1 (His+ Thr+), 2 (His− Thr−), and 3 (His+ Thr−) illegitimate diploids by examining the phenotypes of about 100 independent diploids for each strain and experimental condition. These frequencies were multiplied by the rates of illegitimate mating to determine the rates of class 1 to 3 events. (c) Chromosome III composition of class 1 to 3 illegitimate diploids. Class I illegitimate diploids have a mutation in the MATα locus of the GAL-POL3 haploid but retain all other markers, making these cells phenotypically His+ Thr+. Class 2 illegitimate diploids have lost the entire chromosome III molecule from the GAL-POL3 haploid, making these cells phenotypically His− Thr−. Class 3 illegitimate diploids have lost sequences from the right arm of chromosome III from the GAL-POL3 haploid at some position centromere proximal to the MATα locus but have retained the other markers, making these cells phenotypically His+ Thr−.
TABLE 2.
Rates of illegitimate mating in wild-type (MS71) and GAL-POL3 (RJK341) haploids grown in medium with high (0.05%) and low (0.005%) levels of galactose
| Strain | Rate of illegitimate mating (10−5) for indicated levels of galactosea
|
|
|---|---|---|
| High | Low | |
| MS71 (wild type) | 2.5 (2-5.8) [1] | 1.7 (0.5-3.3) [1] |
| RJK341 (GAL-POL3) | 2.7 (2.4-3.2) [1.1] | 80 (59-170) [47] |
Rates of illegitimate mating were calculated as described in Materials and Methods. The numbers in parentheses represent 95% confidence intervals, and the numbers in brackets show the normalized rates (normalized to a wild-type rate of 1). The wild-type data were in the supplemental materials of Lemoine et al. (20).
Diploids resulting from illegitimate mating often have chromosome rearrangements of several different types (20). We employed a previously described assay that allows for the identification of the various types of illegitimate diploids (20). We used a MATα GAL-POL3 experimental strain (RJK341) with wild-type HIS4 and THR4 alleles on the left and right arms, respectively, of chromosome III and a MATα tester strain (1225) with recessive his4 and thr4 alleles; the MATα locus is centromere proximal to THR4, which is on the right arm of chromosome III. When these strains are forced to mate illegitimately by selecting for complementing mutations in the two strains, the phenotype of the resulting diploids indicates the mechanism responsible for the illegitimate mating (20). Class 1 diploids, which have the His+ Thr+ phenotype, are likely to be the result of a point mutation in the MATα locus, a mating-type switch, or a rare cell fusion event. Class 2 diploids (His− Thr− phenotype) reflect the loss of chromosome III from the GAL-POL3 haploid. Class 3 diploids (His+ Thr− diploids) represent the loss of sequences from the right arm of chromosome III in the GAL-POL3 haploid, such that these diploids have lost the MATα locus and the THR4 locus but retain the HIS4 locus (on the left arm of chromosome III). As will be discussed below, there are several different types of class 3 events.
Using this illegitimate mating assay, we examined the rates of each class of events in wild-type cells and in cells with reduced expression of Pol3p. From each strain and for each experimental condition, we isolated about 100 independent illegitimate diploids and determined their ability to grow in the absence of histidine and threonine. We then multiplied the ratios of each phenotypic class by the rate of illegitimate mating to determine the rate of appearance of each phenotypic class. As depicted in Fig. 4b, the distribution of classes 1, 2, and 3 in wild-type cells was not affected by galactose concentration; class 3 was the smallest class. The relative ratios of the three classes in the GAL-POL3 strain, however, were different from those observed in the wild-type strains. Specifically, chromosome loss (class 2) and chromosome arm loss (class 3) events together comprised about 90% of the GAL-POL3 illegitimate diploids examined, and class 1 was the smallest. Additionally, the rates with which all three illegitimate diploid classes arose were about 30-fold greater in the GAL-POL3 strain grown in the presence of low levels of galactose than in the same strain grown in the presence of high levels of galactose. In summary, the altered expression of Pol3p substantially induces the rate of appearance of two classes of diploids with the genotypes expected for chromosome III loss or the loss of sequences from the right arm of chromosome III.
In our previous analysis of the effects of low DNA polymerase α on illegitimate mating, we observed an approximately 200-fold elevation compared to the 50-fold elevation observed in strains with low DNA polymerase δ. As discussed above, estimates of fold increases tend to have large error bars. To obtain a more accurate measurement of the relative effects of low DNA polymerase, we examined 100 legitimate matings with mixtures of two strains (TRP1 GAL-POL3 strain FJL0430 and trp1 GAL-POL1 strain NPD1), grown on medium containing low levels of galactose, to the MATa derivative of 1225. We also analyzed 100 illegitimate matings with the same mixture of two strains, grown on medium with low levels of galactose, to the MATα 1225 strain. By examining whether the resulting diploids were Trp+ or Trp−, we could determine whether the mating involved the GAL-POL3 or the GAL-POL1 strain. The ratio of Trp+ to Trp− diploids for the legitimate mating was 71:29; this difference is likely to reflect the slower growth of the GAL-POL1 strain, relative to the GAL-POL3 strain, in medium with low levels of galactose. The ratio of Trp+ to Trp− diploids for the illegitimate mating was 64:36. Since this ratio is not significantly different from that observed for the legitimate diploids, we conclude that the low levels of the two DNA polymerases have similar effects on the rates of illegitimate mating.
We also determined whether the relative effects of low levels of the two DNA polymerases on illegitimate mating were affected by FS2. We previously showed that the inverted pair of Ty elements at FS2 was a hot spot for chromosome rearrangements associated with illegitimate mating; the replacement of either of the Ty elements at FS2 caused the loss of this hot spot (20). To determine the relative effects of low levels of the two polymerases on illegitimate mating in the absence of a functional FS2, we did competitive matings with FJL014 (trp1 GAL-POL1 FS2Δty::HYG) and MD497 (TRP1 GAL-POL3 FS2Δty::HYG) as described above. The ratio of Trp+:Trp− diploids was 66:34 for the legitimate diploids and 51:49 for the illegitimate matings. The result demonstrates that the effects of low levels of the two DNA polymerases are similar even in the absence of a functional FS2.
Illegitimate mating induced by low levels of Pol3p leads to chromosome rearrangements.
To confirm our conclusions about the nature of the illegitimate diploid classes 1, 2, and 3, we performed CGH microarray analysis and various other studies of multiple, independent illegitimate diploids. For the single class 1 (His+ Thr+) strain examined, no alterations in gene dosage were observed. Since (as discussed above) mutations in the MATα locus result in strains that mate as MATa strains, class 1 strains are likely to have a point mutation or small deletion within the MAT locus. Alternatively, class 1 diploids could be the result of a mating-type switch event in which the parental GAL-POL3 strain used a recombination-based mechanism to “switch” its mating type from MATα to MATa (15). Such a switch would allow the resulting MATa GAL-POL3 strain to legitimately mate with the MATα tester strain, and the resulting diploid would be a legitimate (MATa/MATα) rather than an illegitimate (matα/MATα or matα-Δ/MATα) diploid.
Several lines of evidence suggest that the GAL-POL3-derived class 1 diploids are mostly illegitimate diploids. First, of 43 His+ Thr+ diploids derived from the GAL-POL3 strain pregrown in low (0.005%) levels of galactose, none were able to sporulate. Since MATa/MATα diploids are sporulation proficient, this finding suggests that the diploids are not MATa/MATα. Second, each of the 43 class 1 diploids was able to mate subsequently with a MATa strain but not with a MATα strain (data not shown). This result is also consistent with the diploids being matα/MATα or matα-Δ/MATα diploids. Taken together, these findings suggest that class 1 diploids derived from the GAL-POL3 strain pregrown in low levels of galactose are the product of illegitimate mating resulting from a mutation (or silencing) of the MATα locus, or a rare fusion event between two MATα cells.
We also examined one class 2 (His− Thr−) illegitimate diploid (DNPD353) derived from the GAL-POL3 strain pregrown in low (0.005%) levels of galactose. CGH analysis demonstrated that this strain was monosomic for chromosome III (Fig. 5a). This finding supports the prediction that class 2 illegitimate diploids are the product of illegitimate mating induced by the loss of chromosome III from the GAL-POL3 parent. It should be noted that the loss of chromosome III in a haploid should be a lethal event, as chromosome III contains many essential genes. However, this lethality is rescued by the subsequent illegitimate mating because the resulting illegitimate diploids contain at least one copy of each of these essential genes. Therefore, this system allows for the selection of specific chromosome loss events that would otherwise be undetectable.
FIG. 5.
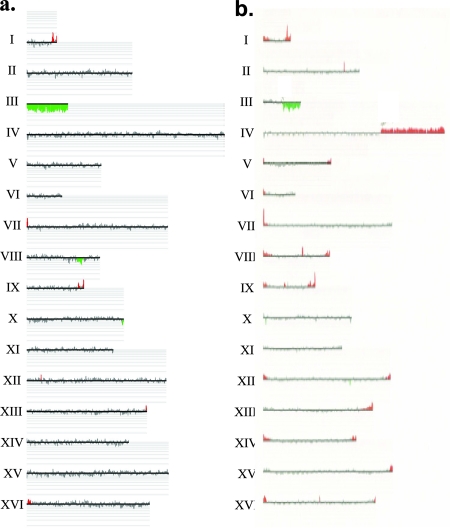
CGH analysis of class 2 and class 3A illegitimate diploids generated by crossing the GAL-POL3 haploid to the wild-type haploid 1225. (a) Class 2 illegitimate diploid (DNPD353). As discussed in the text, class 2 illegitimate diploids, based on their His− Thr− phenotype, are expected to be monosomic for chromosome III. CHG analysis was performed as described in Materials and Methods, and the data are depicted with CGH-Miner software. Each horizontal line represents a chromosome (labeled on the left). The vertical lines represent each individual ORF on that chromosome. The height of the vertical line is proportional to the copy number of that ORF. Green vertical lines below the horizontal line indicate a deletion relative to the wild-type diploid, and red vertical lines above the horizontal line indicate a duplication relative to the wild-type diploid. This class 2 diploid is monosomic for chromosome III. (b) Class 3A illegitimate diploid (DNPD328). In this diploid, a deletion of sequences on the right arm of chromosome III and a duplication on the right arm of chromosome IV are evident. A higher resolution analysis of the breakpoints indicates that the deletion breakpoint is the centromere proximal Ty of FS2 and that the duplication breakpoint is at a Ty element (YDRCTy1-3) on the right arm of chromosome IV.
From our previous analysis of class 3 (His+ Thr−) illegitimate diploids derived from strains with low levels of DNA polymerase α, we predicted three subclasses of events. These subclasses were distinguished using a combination of Southern (using both CHEF and standard agarose gels) and CGH analyses. In class 3A strains, there is a novel chromosome III-hybridizing chromosome (shown by Southern analysis of a CHEF gel), and by CGH analysis, the strains also contain both a deletion of part of the right arm of chromosome III and an amplification of a genomic region from a nonhomologous chromosome (Fig. 5b). Class 3B strains have a large deletion of chromosome III but no other alterations in gene dosage. Class 3C strains have a chromosome III of normal size and no evident changes in gene dosage. In our previous study (20), we found the ratio of class 3A:3B:3C of 3:1:4 in illegitimate diploids resulting from the growth of the GAL-POL1 haploid in low levels of galactose; the 3A:3B:3C ratio in illegitimate diploids derived from wild-type strains grown in low levels of galactose was 3:1:7. In our present study, examining 49 independent class 3 (His+ Thr−) illegitimate diploids derived from the GAL-POL3 strain pregrown in low levels of galactose, we found the ratio of these classes to be 22:0:27. The ratios of the three subclasses in the GAL-POL1 and GAL-POL3 strains were not significantly different (P = 0.12 by the Fisher exact test).
Our distinctions between class 3A, 3B, and 3C strains were initially made by CHEF gel analysis and standard Southern analysis. We subsequently analyzed 16 of these strains in more detail using DNA microarrays. For 14 of these strains, we have a definitive description of the chromosome rearrangement (Table 3). In our study of chromosome rearrangements in strains with low levels of DNA polymerase α, we showed that class 3A strains usually had a translocation that included sequences from the left end of chromosome III to a region between CEN3 and the mating-type locus fused to a region derived from a nonhomologous chromosome. Using CGH analysis, we found that 12 of 14 class 3A strains had a gene dosage pattern consistent with such a translocation. In two of the strains, however, we found a different type of alteration. In these strains, there was an interstitial deletion of chromosome III with breakpoints within the mating-type locus and HMR, a region of homology located about 90 kb away. This deletion, commonly described as the Hawthorne deletion (14), is the product expected as a consequence of unequal crossing-over between MAT and HMR.
TABLE 3.
Rearranged chromosomes in class 3A illegitimate diploids
| Straina | Type of chromosome rearrangement | Chromosomes involved | Size of altered chromosome (kb) | Breakpoints of translocation or deletion | Sequences used as hybridization probes in CHEF analysis |
|---|---|---|---|---|---|
| DNPD302b | Translocation | III/XVI | 320 | Centromere-proximal Ty1 of FS2; YPRCTy1-2 | 15838-16857 (III); 812439-812922 (XVI) |
| DNPD303 | Translocation | I/III | 210 | YARWdelta6; FS1 | 15838-16857 (III) |
| DNPD304b | Translocation | III/V | 260 | Centromere-proximal Ty1 of FS2; YERCTy1-2 | 15838-16857 (III) |
| DNPD307b | Interstitial deletion | III | 250 | MATα and HMR | 15838-16857 |
| DNPD317b | Translocation | III/X | 420 | Centromere-proximal Ty1 of FS1; YJRWTy1-2 | 15838-16857 (III); 485018-485931 (X) |
| DNPD318 | Translocation | II/III | 360 | YBLCdelta7; FS1 | 15838-16857 (III) |
| DNPD319b | Isochromosome | III | 260 | Centromere-distal Ty1 of FS2; YCRCdelta1 | 15838-16857 |
| DNDP324 | Translocation | III/XIII | 355 | FS2; YMRWdelta19c | 15838-16857 (III) |
| DNPD328b | Translocation | III/IV | 730 | Centromere-proximal Ty1 of FS2; YDRCTy1-3 | 15838-16857 (III); 994323-995436 (IV) |
| DNPD336 | Translocation | III/XII | 380 | FS2; YLRWTy1-3 | 15838-16857 (III) |
| DNPD344 | Translocation | III/V | 270 | FS2; YERCTy1-1 | 15838-16857 (III) |
| DNPD350 | Translocation | III/XII | 530 | FS2; YLRCdelta19 | 15838-16857 (III) |
| DNPD351 | Interstitial deletion | III | 250 | MATα and HMR | 15838-16857 |
| DNPD352 | Translocation | II/III | 700 | YBRWTy1-2; FS1 | 15838-16857 (III) |
All strains were examined by microarrays and CHEF gel electrophoresis.
Additional confirmatory Southern analysis was performed (as described in the supplemental material).
In some yeast strains, there is a Ty1 element integrated at SGD coordinate 748521 on chromosome XIII (A. Gabriel, personal communication), a position very close to YMRWdelta19.
The remaining 12 class 3A diploids had a terminal deletion of the right arm of one of the chromosome III molecules and a terminal amplification, usually (11 of 12 diploids) involving sequences of a nonhomologous chromosome. For example, in the class 3A strain DNPD328, CGH analysis revealed a terminal deletion of the right arm of chromosome III and a terminal amplification of the right arm of chromosome IV (Fig. 5b). The microarray analysis allowed us to determine the breakpoints of the deletions and the amplifications within the resolution of a single ORF. The breakpoints had several interesting properties. First, all of the terminal deletions of the right arm of chromosome III originated at one of two specific regions, termed fragile sites 1 and 2 (FS1 and FS2) in our previous study (20). Both FS1 and FS2 are composed of two closely linked Ty1 retrotransposable elements (20). In FS1, these elements are arranged in tandem and share a single long terminal repeat, or delta element. In FS2, these two elements are arranged in an inverted orientation with 283 bp separating the long terminal repeats of the Ty1 elements. Similar to the previously described results observed with the GAL-POL1 strain, more of the deletions began at FS2 (8/12) than at FS1 (4/12) in the GAL-POL3-derived illegitimate diploids. The second interesting feature of the breakpoints is that all of the detected terminal chromosome arm amplifications began at either a Ty or delta element and did not include the centromere. Thus, most class 3A strains have a monocentric translocation composed of a portion of chromosome III and a portion of another chromosome generated by homologous recombination between the Ty elements.
To confirm this conclusion, we did Southern blots of CHEF gels to measure the size of the rearranged chromosome and to demonstrate that the translocated chromosome hybridized to probes derived from two different homologues. Each of the 12 class 3A diploids with a putative translocation had the size predicted from the microarray analysis, and this translocation hybridized to the predicted probes (Table 3). For example, in the DNPD328 strain, we detected a novel 730-kb chromosome band that hybridized with probes to both the left arm of chromosome III and the right arm of chromosome IV.
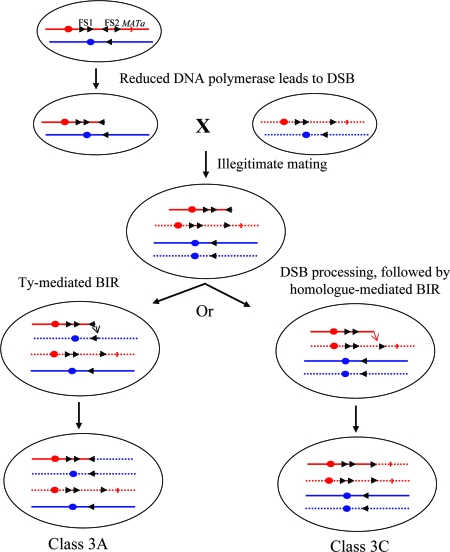
For five of the class 3A strains (Table 3), we did a detailed restriction mapping of the breakpoints of the translocations (described in Materials and Methods). This mapping confirmed that the deletion breakpoint was within FS1 or FS2 on the right arm of chromosome III and the amplification breakpoint involved a Ty element or delta element on a different chromosome. The most likely mechanism for the formation of these strains is that reduced levels of DNA polymerase δ in the GAL-POL3 haploids result in elevated frequencies of DSBs at FS1 and FS2 with a subsequent loss of the DNA sequences distal to the break (Fig. 6). Since these sequences include the MATα locus, the haploids mate illegitimately to the other haploid parent. The broken chromosome is repaired by a BIR event, utilizing a Ty element on a nonhomologous chromosome as a template (left side of Fig. 6).
FIG. 6.
Generation of class 3A and class 3C strains as a consequence of BIR. Chromosome III is shown in red. Arrowheads indicate the positions and orientations of Ty elements, and circles indicate the centromeres; mating-type loci are indicated by the short vertical lines. Nonhomologous chromosomes with Ty elements are shown in blue. The solid lines indicate the chromosomes derived from the GAL-POL3 strain, and the dotted lines indicate the chromosome derived from the MATα tester strain. Reduced DNA polymerase δ in the GAL-POL3 strain results in elevated rates of DSBs at FS2. The chromosome fragment located distal to the DSB, which includes the mating-type locus, is lost, allowing the diploid to mate to the MATα tester strain. We envisage two pathways of repair. In one pathway (left side), the broken chromosome with a terminal Ty element is repaired by a BIR event using a Ty element on the nonhomologous chromosome, resulting in a class 3A event (deletion on chromosome III plus duplication of a segment of a nonhomologous chromosome). In the second pathway (right side), the terminal Ty element is removed by exonucleases, and the processed broken end is repaired using a single-copy sequence of chromosome III derived from the MATα tester strain. This pathway results in a class 3C strain.
Although most of the chromosome rearrangements in Table 3 are translocations, DNPD319 involves a different type of chromosome rearrangement. In this strain, the deletion breakpoint was at FS2, and the duplication breakpoint was on the right arm of chromosome III at a solo delta element (YCRCdelta1). The resulting chromosome rearrangement has the sequences centromere distal to YCRCdelta1 duplicated on both arms; such rearrangements are called isochromosomes. Such a chromosome could be formed by a DSB in FS2 that is repaired by a BIR event initiated at YCRCdelta1.
As discussed above, simple terminal deletions of chromosome III (class 3B), although observed in our study of fragile sites in strains with low levels of DNA polymerase α, were not observed in the present study. Class 3C strains were the most abundant subclass (27 of 49). In these diploids, as discussed above, the wild-type THR4 allele derived from the GAL-POL3 strain has been lost, but there is no deletion or duplication of the chromosome III sequence and chromosome III is of the normal size. As in our previous study, we suggest that these events reflect breaks that occur in chromosome III centromere proximal to the MATα locus in the GAL-POL3 haploid. The loss of the sequences distal to the break allows the haploid to mate illegitimately. We suggest that the broken chromosome is repaired in the resulting illegitimate diploid by a BIR event utilizing the intact chromosome III from the other haploid as a substrate (right side of Fig. 6). Thus, classes 3A and 3C may be initiated by similar events, but differences in the substrates chosen for the repair of the initiating breaks result in different outcomes.
Efficient illegitimate mating induced by low levels of Pol3p requires FS2.
Two-thirds of the class 3A chromosome rearrangements involve FS2 and one-third involve FS1. We next examined whether the elevated level of illegitimate mating (an indicator of chromosome rearrangements) in the GAL-POL3 strains grown in the presence of low levels of galactose required the presence of an intact FS2 (two inverted Ty1 elements separated by 283 bp). For this analysis, we generated two strains: (i) FJL043, a TRP1 derivative of the GAL-POL3 haploid strain RJK341, and (ii) FJL016, a derivative of RJK341 (mutant trp1 gene) in which the HYGR gene replaces the centromere-proximal Ty1 element of FS2. We mixed equal numbers of FJL043 and FJL016 cells and grew the mixture for about 24 h on solid growth medium with either high (0.05%) or low (0.005%) levels of galactose. The resulting cells were then mated both legitimately (to trp1 strains of the opposite mating type) and illegitimately (to trp1 strains of the same mating type), and the phenotypes of the resulting diploids were analyzed. Trp+ diploids reflected mating of the FJL043 parent (intact FS2) and Trp− diploids reflected mating of the FJL016 parent (disrupted FS2).
We examined the Trp phenotype of 96 independent legitimate and 94 independent illegitimate diploids derived from these strain mixtures pregrown in high galactose-containing medium. For the legitimate diploids, we found that 60% (58/96) were Trp+ and 40% (38/96) were Trp−. For the illegitimate diploids, we found that 70% (66/94) were Trp+ and 30% (28/94) were Trp−. Although the strain with the disrupted FS2 has a somewhat reduced efficiency of illegitimate mating relative to the strain with the intact FS2 (normalizing for the efficiency of legitimate mating), the difference is not statistically significant. We also examined the Trp phenotype of 99 independent legitimate and 98 independent illegitimate diploids derived from these strain mixtures pregrown in low galactose-containing medium. For the legitimate diploids, we found that 82% (81/99) were Trp+ and 18% (18/99) were Trp−. For the illegitimate diploids, we found that 97% (95/98) were Trp+ and 3% (3/99) were Trp−. The strain with the disrupted FS2 has a significantly reduced ability to mate illegitimately compared to the strain with the intact FS2 (P < 0.001 by the Fisher exact test). One interpretation of this result (discussed further below) is that when the level of DNA polymerase δ is low, FS2 is a hot spot for DNA breaks that initiate the chromosome rearrangements resulting in illegitimate mating. This result is also consistent with the observation that one can physically detect (by CHEF gels) chromosome III molecules that are broken at FS2 in both the GAL-POL1 and GAL-POL3 strains, incubated in medium containing glucose but not galactose (20). The frequency of legitimate diploids was also greater for the strain with the intact FS2 than that for the strain with the disrupted FS2 (82% versus 18%). It is likely that this difference simply reflects the relative frequencies of the two strains after 24 h of cocultivation rather than an effect of FS2 on legitimate mating.
DISCUSSION
We previously reported the identification and characterization of two regions of chromosome III (FS1 and FS2) that are hot spots for chromosome rearrangements in yeast strains with low levels of DNA polymerase α (20). We argued that these sites were analogous to fragile sites in mammalian cells, which are hot spots for chromosome breakage in cells incubated in drugs that inhibit DNA replication (3, 7, 8, 11, 17, 21, 29). In this study, we examine the effects of low levels of DNA polymerase δ in several different assays of genome stability: sensitivity to DNA-damaging agents, rate of chromosome loss and mitotic recombination, and rate of illegitimate mating. In many of these assays, reduced levels of DNA polymerase δ and DNA polymerase α have similar effects. Given the quite different roles of the two DNA polymerases at the replication fork and in DNA repair, this result is somewhat surprising.
We found that the reduced expression of Pol3p results in a 12-fold increase in the forward mutation rate of the CAN1 locus, an 8-fold increase in the rate of chromosome V loss, and a 35-fold increase in the rate of mitotic recombination (Table 1). Interestingly, each of these rates are considerably higher than those observed following the reduced expression of Pol1p (20). One interpretation of this result is that the reduction in the level of DNA polymerase δ may reduce the efficiency of both replicative DNA synthesis (leading to elevated DSBs) and DNA synthesis associated with DNA repair, whereas reduced levels of DNA polymerase α may affect primarily replicative DNA synthesis (2, 16). The reduced expression of DNA polymerase δ also resulted in sensitivity to HU (Fig. 1).
The mutator phenotype associated with low levels of DNA polymerase δ was also observed in our previous study (18). Sequence analysis of the can1 mutations obtained in the strains with low levels of DNA polymerase δ indicated a somewhat different spectrum of mutations than that observed in the wild-type strain (18). In the wild-type strain or the GAL-POL3 strain grown in high (0.05%) levels of galactose, most can1 mutations are single base changes, whereas in the GAL-POL3 strain grown in low (0.005%) levels of galactose, about half of the can1 mutations are small deletions with end points in short direct repeats (18). Reduced levels of DNA polymerase δ may result in elevated levels of DNA polymerase slippage or an increased amount of synthesis by error-prone DNA polymerases. Since the spectrum of mutations in the strain with low levels of DNA polymerase and with mutations in rev3 and rev30 does not differ significantly from the strain with low levels of DNA polymerase and wild-type REV3 and RAD30, we argue in favor of the first possibility.
Reduced levels of DNA polymerase δ also substantially elevated (47-fold) illegitimate mating (Table 2), although the degree of elevation is 4-fold less than that observed in strains with low levels of DNA polymerase α (20). We previously proposed that reducing the expression of DNA polymerase α induces DSBs at fragile sites by stalling replication forks that then allow single-stranded DNA to form hairpin-like structures (20); such structures would be readily formed in genomic regions such as FS2 that have inverted repeats (22). A reduction in DNA polymerase δ might also be expected to have a destabilizing effect by uncoupling the replication of the lagging and leading strands. The observation that reduced levels of DNA polymerase α result in a stronger destabilization than that observed for DNA polymerase δ can be explained two ways. First, since DNA polymerase δ is a processive enzyme involved in the elongation stage of DNA replication (2, 16), whereas DNA polymerase α is the primase that is responsible for the synthesis of Okazaki fragments on the lagging strand template (2), the cell may simply require more DNA polymerase α. Alternatively, DNA polymerase ɛ may be able to partially substitute for reduced levels of DNA polymerase δ but not for reduced levels of DNA polymerase α. Finally, we note that the mechanism proposed for FS2-associated chromosome translocations in yeast is strikingly similar to that proposed for the frequently observed t(11,22) translocation in humans (6).
Although there is a strong rationale for the mechanism by which FS2 functions as a fragile site, the mechanism by which FS1 acts is less clear. It is possible that DSBs initiated at FS2 are sometimes processed by nucleases, resulting in a recombinogenic structure ending in FS1. Consistent with this possibility, VanHulle et al. (28) recently showed that a homothallism-induced DSB located 30 kb centromere distal to FS2 could stimulate chromosome rearrangements involving FS2. There are two other alternative possibilities. First, Ty elements, which are transcribed at high levels, may be prone to replication fork pausing, and this tendency could be stronger in conditions of perturbed DNA replication. Finally, it is possible that DSBs, other than the DSB at FS2, are random along the chromosome, but only those DSBs that occur in repetitive DNA elements result in chromosome rearrangements. DSBs in nonrepetitive DNA sequences could be repaired by sister chromatid recombination or by recombination with the homologous chromosome (class 3C events).
All of the chromosomal rearrangements observed in our analysis are consistent with the repair of a DSB in a Ty element of FS2 or FS1 by recombination with a Ty or delta element at an ectopic location (Table 3). These results are similar to those that were previously reported in strains with reduced Pol1p expression (20), and these observations argue that hindering DNA replication by different mechanisms can lead to the same type of chromosome rearrangements. Our results also reinforce the conclusions, from a variety of studies, that many of the chromosome rearrangements that occur in the lab and during the evolution of the yeast genome reflect Ty-Ty recombination (24).
Chromosome fragile sites in human cells are hot spots for the chromosome rearrangements observed in cells derived from human cancers (3, 7, 8, 11, 17, 21, 29). Although the DNA lesions responsible for fragile sites in mammalian cells have not yet been defined, one clear difference between mammalian and yeast fragile sites is the manner by which the initiating DNA lesion is repaired. In our studies, the translocations observed in strains with low DNA polymerase α or δ reflect homologous recombination between dispersed repeats. In contrast, the breakpoints of deletions associated with the mammalian fragile site FHIT indicate that the recombinogenic lesions are repaired by nonhomologous end-joining (5). Despite this difference, the identification of fragile sites in S. cerevisiae may be useful in modeling some aspects of fragile sites in mammalian cells. For example, it would be interesting to determine whether the transient downregulation of DNA polymerase or a DNA replication cofactor could be responsible for some of the genetic instability associated with cancer cells.
Supplementary Material
Acknowledgments
We thank Malgorzata Gawel for assistance with the Southern blots and CGH microarray analysis; Jennifer Gerton, Phoebe Lee, and Jeremy DeMai for strain constructions; and all members of the Petes lab for useful discussions. We also thank Abram Gabriel for sharing unpublished information.
The research was supported by an NIH grant to T.D.P. (GM52319) and a grant from the Leukemia and Lymphoma Society to F.J.L. (3427-07).
Footnotes
Published ahead of print on 30 June 2008.
REFERENCES
- 1.Arlt, M. F., A. M. Casper, and T. W. Glover. 2003. Common fragile sites. Cytogenet. Genome Res. 10092-100. [DOI] [PubMed] [Google Scholar]
- 2.Burgers, P. M. 1998. Eukaryotic DNA polymerases in DNA replication and DNA repair. Chromosoma 107218-227. [DOI] [PubMed] [Google Scholar]
- 3.Castro, P. D., J. Fairman, and L. Nagarajan. 1998. The unexplored 5q13 locus: a role in hematopoietic malignancies. Leuk. Lymphoma 30443-448. [DOI] [PubMed] [Google Scholar]
- 4.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278680-686. [DOI] [PubMed] [Google Scholar]
- 5.Durkin, S. G., R. L. Ragland, M. F. Arlt, J. G. Mulle, S. T. Warren, and T. W. Glover. 2008. Replication stress induces tumor-like microdeletions in FHIT/FRA3B. Proc. Natl. Acad. Sci. USA 105246-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emanuel, B. S., and S. C. Saitta. 2007. From microscopes to microarrays: dissecting recurrent chromosomal rearrangements. Nat. Rev. Genet. 8869-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fundia, A., N. Gorla, and I. Larripa. 1994. Spontaneous chromosome aberrations in Fanconi's anemia patients are located at fragile sites and acute myeloid leukemia breakpoints. Hereditas 12047-50. [DOI] [PubMed] [Google Scholar]
- 8.Fundia, A. F., and I. B. Larripa. 1989. Coincidence in fragile site expression with fluorodeoxyuridine and bromodeoxyuridine. Cancer Genet. Cytogenet. 4141-48. [DOI] [PubMed] [Google Scholar]
- 9.Glover, T. W., C. Berger, J. Coyle, and B. Echo. 1984. DNA polymerase alpha inhibition by aphidicolin induces gaps and breaks at common fragile sites in human chromosomes. Hum. Genet. 67136-142. [DOI] [PubMed] [Google Scholar]
- 10.Gordenin, D. A., Y. Y. Proscyavichus, A. L. Malkova, M. V. Trofimova, and A. Peterzen. 1991. Yeast mutants with increased bacterial transposon Tn5 excision. Yeast 737-50. [DOI] [PubMed] [Google Scholar]
- 11.Gumus, G., A. Sunguroglu, A. Tukun, D. B. Sayin, and I. Bokesoy. 2002. Common fragile sites associated with the breakpoints of chromosomal aberrations in hematologic neoplasms. Cancer Genet. Cytogenet. 133168-171. [DOI] [PubMed] [Google Scholar]
- 12.Guthrie, C., and G. R. Fink. 1991. Guide to yeast genetics and molecular biology. Academic Press, San Diego, CA.
- 13.Hartwell, L. H., and D. Smith. 1985. Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S. cerevisiae. Genetics 110381-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hawthorne, D. C. 1963. A deletion in yeast and its bearing on the structure of the mating type locus. Genetics 481727-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hicks, J. B., and I. Herskowitz. 1976. Interconversion of yeast mating types I. Direct observations of the action of the homothallism (HO) gene. Genetics 83245-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hindges, R., and U. Hubscher. 1997. DNA polymerase delta, an essential enzyme for DNA transactions. Biol. Chem. 378345-362. [DOI] [PubMed] [Google Scholar]
- 17.Ishii, H., and Y. Furukawa. 2004. Alterations of common chromosome fragile sites in hematopoietic malignancies. Int. J. Hematol. 79238-242. [DOI] [PubMed] [Google Scholar]
- 18.Kokoska, R. J., L. Stefanovic, J. DeMai, and T. D. Petes. 2000. Increased rates of genomic deletions generated by mutations in the yeast gene encoding DNA polymerase delta or by decreases in the cellular levels of DNA polymerase delta. Mol. Cell. Biol. 207490-7504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Beau, M. M., F. V. Rassool, M. E. Neilly, R. Espinosa III, T. W. Glover, D. I. Smith, and T. W. McKeithan. 1998. Replication of a common fragile site, FRA3B, occurs late in S phase and is delayed further upon induction: implications for the mechanism of fragile site induction. Hum. Mol. Genet. 7755-761. [DOI] [PubMed] [Google Scholar]
- 20.Lemoine, F. J., N. P. Degtyareva, K. Lobachev, and T. D. Petes. 2005. Chromosomal translocations in yeast induced by low levels of DNA polymerase: a model for chromosome fragile sites. Cell 120587-598. [DOI] [PubMed] [Google Scholar]
- 21.Lengauer, C., K. W. Kinzler, and B. Vogelstein. 1998. Genetic instabilities in human cancers. Nature 396643-649. [DOI] [PubMed] [Google Scholar]
- 22.Lobachev, K. S., D. A. Gordenin, and M. A. Resnick. 2002. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108183-193. [DOI] [PubMed] [Google Scholar]
- 23.McCusker, J. H., and J. E. Haber. 1981. Evidence of chromosomal breaks near the mating-type locus of Saccharomyces cerevisiae that accompany MATα × MATα matings. Genetics 99383-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mieczkowski, P. A., F. J. Lemoine, and T. D. Petes. 2006. Recombination between retrotransposons as a source of chromosome rearrangements in the yeast Saccharomyces cerevisiae. DNA Repair (Amsterdam) 51010-1020. [DOI] [PubMed] [Google Scholar]
- 25.Rattray, A. J., and J. N. Strathern. 2003. Error-prone DNA polymerases: when making a mistake is the only way to get ahead. Annu. Rev. Genet. 3731-66. [DOI] [PubMed] [Google Scholar]
- 26.Sia, E. A., R. J. Kokoska, M. Dominska, P. Greenwell, and T. D. Petes. 1997. Microsatellite instability in yeast: dependence on repeat unit size and DNA mismatch repair genes. Mol. Cell. Biol. 172851-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sutherland, G. R. 2003. Rare fragile sites. Cytogenet. Genome Res. 10077-84. [DOI] [PubMed] [Google Scholar]
- 28.VanHulle, K., F. J. Lemoine, V. Narayanan, B. Downing, K. Hull, C. McCullough, M. Bellinger, K. Lobachev, T. D. Petes, and A. Malkova. 2007. Inverted DNA repeats channel repair of distant double-strand breaks into chromatid fusions and chromosomal rearrangements. Mol. Cell. Biol. 272601-2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yunis, J. J., and A. L. Soreng. 1984. Constitutive fragile sites and cancer. Science 2261199-1204. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.