Abstract
ABCG2 is recognized as an important efflux transporter in clinical pharmacology and is potentially important in resistance to chemotherapeutic drugs. To identify epigenetic mechanisms regulating ABCG2 mRNA expression at its 3′ untranslated region (3′UTR), we performed 3′ rapid amplification of cDNA ends with the S1 parental colon cancer cell line and its drug-resistant ABCG2-overexpressing counterpart. We found that the 3′UTR is >1,500 bp longer in parental cells and, using the miRBase TARGETs database, identified a putative microRNA (miRNA) binding site, distinct from the recently reported hsa-miR520h site, in the portion of the 3′UTR missing from ABCG2 mRNA in the resistant cells. We hypothesized that the binding of a putative miRNA at the 3′UTR of ABCG2 suppresses the expression of ABCG2. In resistant S1MI80 cells, the miRNA cannot bind to ABCG2 mRNA because of the shorter 3′UTR, and thus, mRNA degradation and/or repression on protein translation is relieved, contributing to overexpression of ABCG2. This hypothesis was rigorously tested by reporter gene assays, mutational analysis at the miRNA binding sites, and forced expression of miRNA inhibitors or mimics. The removal of this epigenetic regulation by miRNA could be involved in the overexpression of ABCG2 in drug-resistant cancer cells.
ABCG2, a ubiquitous ATP-binding cassette (ABC) transporter, besides playing a significant role in absorption, distribution, and elimination of its substrate drugs, may also confer multidrug resistance in cancer cells (1). ABCG2 overexpression is frequently observed in human cancer cell lines selected with various anticancer drugs (12, 34, 38, 44, 52).
The molecular mechanisms regulating ABCG2 expression are not well understood. Like most TATA-less gene promoters, the ABCG2 promoter contains numerous Sp1, AP1, and AP2 sites and a CCAAT box. ABCG2 also has a putative CpG island located upstream of the gene (50). To date, most studies examining the regulation of ABCG2 are focused at the transcriptional level. Two functional cis elements in the ABCG2 promoter, namely, the estrogen (15) and hypoxia (28) response elements, and a peroxisome proliferator-activated receptor response element upstream of the ABCG2 gene (48) have been reported. An aryl hydrocarbon receptor response element has been proposed, but the exact sequence has not been identified (13). In drug-resistant MCF-7 cells, alternative promoter use due to differential expression of splice variants at the 5′ untranslated region (5′UTR) of ABCG2 mRNA has been observed (39). We recently reported that DNA methylation and histone modifications play important roles in the regulation of human ABCG2 in renal carcinoma cell lines (50) and in some drug-selected resistant cell lines (49), respectively.
Less is known about the 3′UTR of ABCG2. In eukaryotic mRNAs, the 3′UTR plays an important role in regulating gene expression at the posttranscriptional level by modulating nucleocytoplasmic mRNA transport, polyadenylation status, subcellular targeting, translation efficiency, stability, and rates of degradation (11, 40, 45, 47, 54). The 3′UTR of ABCG2 mRNA reported in GenBank (accession no. NM_004827) and in the UTR database (UTR database entry UTR:3HSA117529) is about 2 kb in length, which is considerably longer than the average 770 bp observed for human mRNAs (24, 30), suggesting that it may have one or more important roles in the regulation of gene expression.
MicroRNAs (miRNAs) represent a large class of gene regulatory molecules that control fundamental cellular processes in animals and plants (2, 16, 21, 36). Gene regulation by miRNAs is typically mediated by the formation of imperfect hybrids with the 3′UTR sequences of target mRNAs, inducing translational repression and/or mRNA degradation (10, 29, 42).
In this study, we demonstrate that ABCG2 mRNA adopts a longer 3′UTR in the parental S1 colon cancer cell line than in its drug-resistant counterpart and that a miRNA (hsa-miR-519c) decreases endogenous ABCG2 mRNA and protein levels by acting through a putative hsa-miR-519c binding site located within the longer 3′UTR region found only in parental cells. These findings suggest that escape from miRNA-mediated translational repression and mRNA degradation could lead to overexpression of ABCG2 in drug-resistant cancer cells.
MATERIALS AND METHODS
Tissue culture.
The human colon cancer cell line S1 and its resistant subline S1MI80 have been described previously (38). SW620, MCF-7, H460, and SF295 cells were chosen from the NCI Tumor Drug Screen for this study. The cell lines were maintained in Iscove modified Eagle medium (S1, S1MI80, and MCF-7 cells) or RPMI medium (SW620, SF295, and H460 cells) supplemented with 10% fetal bovine serum, 100 units/ml streptomycin sulfate, and 100 units/ml penicillin G sulfate and incubated at 37°C in 5% CO2.
RNA degradation analysis.
Parental S1 and resistant S1MI80 cells in 10-cm tissue culture dishes were grown to subconfluence, and then actinomycin D (Sigma, St. Louis, MO) was added to a final concentration of 5 μg/ml to arrest de novo RNA synthesis. Four, 8, and 16 h after actinomycin D treatment, the cells were harvested and ABCG2 mRNA was quantified by semiquantitative reverse transcription-PCR (RT-PCR) as described below. c-myc and glyceraldeyde-3-phosphate (GAPDH) mRNA levels were also monitored as controls. For HEK293 cells transfected with pcDNA3.1-based vectors, actinomycin D was added 48 h after transfection, and the cells were harvested at the same time points thereafter. The expression level of cotransfected green fluorescent protein (GFP) (pEGFP-C1; BD Bioscience Clontech) was evaluated for normalization purposes. The value recorded was the percentage of mRNA remaining compared with the amount before the addition of actinomycin D. All experiments were repeated three times.
Semiquantitative RT-PCR.
Total RNA was isolated using Trizol reagent (Invitrogen, Carlsbad, CA). RNA (1 μg) was reverse transcribed using a commercially available cDNA synthesis kit (Bioline, Randolph, MA). Amplification of cDNA was done using primers specific for ABCG2 (5′-CAATGGGATCATGAAACCTG-3′ [forward] and 5′-GAGGCTGATGAATGGAGAA-3′ [reverse]) and GAPDH (5′ACCACAGTCCATGCCATCAC-3′ [forward] and 5′TCCACCACCCTGTTGCTGTA-3′ [reverse]). The primers were designed in such a way that they span introns 3 to 6 in ABCG2 and intron 8 in GAPDH. Amplification of GAPDH cDNA served as an internal control. PCR amplification was performed at an annealing temperature of 55°C for 28 cycles (ABCG2) or 25 cycles (GAPDH), to yield a 584- or 330-bp product, respectively. No PCR product was obtained when reverse transcriptase was omitted from cDNA synthesis mixtures. The PCR products were resolved in 2% agarose gels and stained with ethidium bromide. Gel images were captured, and band intensities were quantitated by using the APP_Collage PPC4.0 analysis software program.
Real-time PCR.
For the mRNA degradation assay, measurement of ABCG2 mRNA levels was repeated by a real-time quantitative PCR method, using a LightCycler Fast Start DNA Master Taqman kit (Universal ProbeLibrary; Roche Molecular Biochemicals, Indianapolis, IN) with a LightCycler 2.0 system according to the manufacturer's recommendations. Primers were designed for ABCG2 and GAPDH (for normalization), using the Roche UPL custom design service, and the appropriate fluorescent probes were chosen (probes 56 and 60 for ABCG2 and GAPDH, respectively). The sequences of the primers used for the analysis of ABCG2 and GAPDH expression were as follows: ABCG2 forward, 5′-TGGCTTAGACTCAAGCACAGC-3′; ABCG2 reverse, 5′-TCGTCCCTGCTTAGACATCC-3′; GAPDH forward, 5′-AGCCACATCGCTCAGACAC-3′; and GAPDH reverse, 5′-GCCCAATACGACCAAATCC-3′. The relative ABCG2 and GADPH expression levels were determined by reading from a standard curve generated by five fourfold serial dilutions of cDNA from MCF-7 cells, which plotted the threshold cycle against the log input amount of template.
Analysis of the relative stability of the ABCG2 3′UTR was repeated by quantitative real-time RT-PCR, using a LightCycler Fast Start DNA Master Plus Sybr green I kit (Roche Applied Science). Quantitative PCR was performed using a LightCycler system for 35 cycles at 90°C for 10 s, the primer-specific annealing temperature for 10 s, and 72°C for 18 s. After each cycle, the temperature was raised to the primer's respective acquisition temperature, and the fluorescence of Sybr green bound to double-stranded DNA was measured at 530 nm (Light Cycler fluorescence channel F1). The cycle number at which the fluorescence of the sample exceeded that of the background was determined by the LightCycler software (version 3.5), using the second derivative method. A melting curve analysis was performed after the amplification phase to eliminate the possibility of nonspecific amplification or primer dimer formation.
3′RACE assay.
Total RNAs were extracted from parental S1 and resistant S1MI80 cells by use of Trizol reagent (Invitrogen). First-strand cDNA synthesis was performed with a 3′ rapid amplification of cDNA ends (3′RACE) cDNA amplification kit (Bioline) according to the manufacturer's protocol. 3′RACE uses an adapter primer (AP) (Table 1; see Fig. S3A in the supplemental material), an oligo(dT) primer with a defined (non-dT) sequence at its 5′ end, to prime first-strand cDNA synthesis. The oligo(dT) portion of AP hybridizes to the mRNA poly(A) tail to prime an RT reaction. The defined sequence at the 5′ end of AP is thus incorporated into all cDNA molecules synthesized. PCR was then carried out on the first-strand cDNA with Bio-X-ACT Long DNA polymerase (Bioline). An outer reverse primer (Table 1; see Fig. S3A in the supplemental material) complementary to the defined sequence in AP can then be used in conjunction with an upstream primer which binds to a known ABCG2 sequence to specifically amplify from the upstream primer to sites of mRNA polyadenylation. Primer ABCG2 ex16 (Table 1; see Fig. S3A in the supplemental material) was used as an upstream primer for the 3′RACE reactions reported in our study. To increase the specificity of the first PCR, a nested PCR (PCR 1) was performed by using a nested N1 forward primer, the outer reverse primer, and the first PCR mixture as the DNA template. DNA from the PCR was subcloned as a mixture into a pCR2.1 vector (Invitrogen). Multiple clones were then sequenced by the DNA sequencing core facility at the Laboratory of Experimental Carcinogenesis, NCI. All sequences were aligned against the reported mRNA sequence for ABCG2 (GenBank accession no. NM_004827). To confirm the RACE findings for clones with a long 3′UTR, nested PCR 2 was performed, using the nested N1 forward primer (ABCG2 nucleotides [nt] 2401 to 2429) and an ABCG2-specific nested N2 reverse primer annealing to near the 3′ end of the reported mRNA sequence (nt 4240 to 4263) (Table 1; see Fig. S3A in the supplemental material). For S1 cells, 3′RACE analysis was repeated by using total RNA harvested from cells after transfection with an hsa-miR519c-specific inhibitor (Dharmacon).
TABLE 1.
Oligonucleotide primers used in this study
| Use and primer | Sequence (5′-3′) (nt) |
|---|---|
| 3′RACE | |
| ABCG2 ex16 | AAAGCAGGGCATCGATCTCTCAC (2338-2360) |
| Outer reverse primer | CAGTCGGTCCTGCAGGGTTCAAGCGCATCTGAGG |
| Nested N1 forward primer | GATTGTTATTTTCCTCACAATTGCCTACC (2401-2429) |
| Nested N2 reverse primer | TATACCAAGAACAGGAAAAACTGA (4263-4240) |
| 3′RACE AP | CCCTGTTCAAGCGCATCTGAGGTGAACCATGAACCGTGC(T)18 |
| 3′UTR RT-PCR | |
| A-Fw | TTCCCCTTAATTCAGTATGATTTATCCTCACA (2464-3030) |
| A-R | TACCACTAGACAGGAATATCAGTAAGTTC |
| B-Fw | GAGAATTGAATTCTGGAAACTCCTGA (2731-3291) |
| B-R | ATCCAGGAGTGGTCAGATTCCTTTATGA |
| C-FW | GAACTTACTGATATTCCTGTCTAGTGGTA (3002-3558) |
| C-R | CTGAGTAGGCTTTTGTGTGCTACAATCTTCTAT |
| D-Fw | TCATAAAGGAATCTGACCACTCCTGGAT (3264-3812) |
| D-R | CATTAAGGGAAGCTGAGCAAAGAGTAGACAGGAACTTT |
| E-Fw | ATAGAAGATTGTAGCACACAAAAGCCTACTCAG (3526-4024) |
| E-R | CTCAGTTAACTCCTGTAAGTGCTGA |
| F-Fw | AAAGTTCCTGTCTACTCTTTGCTCAGCTTCCCTTAATG (3775-4388) |
| F-R | GTGGCTGTTTTGTTTCTTTTGAAGTATATTTTGTG |
| Mutagenesis for preparing luciferase reporter constructs | |
| 3′UTR (3820/3841) | |
| miR519c site F | CGGAAAGATCGCCGTGTAATTTTATATAGCAAGATGCATTTGTCTAGAGTCGGGGCGGC |
| miR519c site R | GCCGCCCCGACTCTAGACAAATGCATCTTGCTATATAAAATTACACGGCGATCTTTCCG |
| 3′UTR scramble (3820/3841) | |
| Scramble (3820-3841) F | CGGAAAGATCGCCGTGTAATTACTGTCGTTAAAGTTTAAGATTCTAGAGTCGGGGCGGC |
| Scramble (3820-3841) R | GCCGCCCCGACTCTAGAATCTTAAACTTTAACGACAGTAATTACACGGCGATCTTTCCG |
| 3′UTR FL Δ(3820/3841) | |
| Deletion (Δ3820-3841) F | CTACTCTTTGCTCAGCTTCCCTTAATGTTAACATTCAAAGCTAACAAGTTAACATTGGTAC |
| Deletion (Δ3820-3841) R | GTACCAATGTTAACTTGTTAGCTTTGAATGTTAACATTAAGGGAAGCTGAGCAAAGAGTAG |
Bioinformatics.
miRNA target site prediction of hsa-miR-519c was performed by using miRBase TARGETS (19) and RNAHybrid (43) software. The ABCG2 3′UTR was analyzed using the UTRScan program (http://bighost.area.ba.cnr.it/BIG/UTRHome/) to search for the presence of functional elements (41).
Analysis of RNA secondary structure.
RNA secondary structure analysis was performed using the mfold program (56) (available at http://www.bioinfo.rpi.edu/applications/mfold). The folding temperature was fixed at 37°C. The miR-519c binding sequence, with or without the neighboring nucleotides ± 100 bp, was used for analysis. Sequences were folded with mfold in a locally automated manner. The most thermodynamically stable structure is shown in Fig. S8 in the supplemental material.
Cell transfection with miRNA inhibitor or mimic.
A miRNA inhibitor and mimic were purchased from Dharmacon. Cells were transfected with up to 120 nM of the specific hsa-miR519c inhibitor or mimic in a total volume of 10 ml by using Oligofectamine (Invitrogen). The mimic is a double-stranded RNA that contains the mature miRNA sequence. When the mimic is transfected into the cell, the result is similar to adding an exogenous, processed miRNA that functions comparably to endogenous miRNA. The inhibitor contains a sequence that is complementary to the mature miRNA sequence and acts as a decoy. The endogenous miRNA binds tightly to the inhibitor and is sequestered, so there is less available for binding of endogenous targets. The expression of miRNAs was assessed by stem-loop RT-PCR as described below.
Stem-loop RT-PCR for miRNAs.
Stem-loop RT for mature hsa-miR-519c and hsa-miR-520h was performed as previously described (6, 46). All reagents for miRNA stem-loop RT were obtained from Applied Biosystems (Foster City, CA), using an hsa-miR-519c (P/N:4373251)- or hsa-miR-520h (P/N:4373258)-specific stem-loop RT primer. The RT reaction mixture was assembled and incubated for 10 min at 25°C, 60 min at 42°C, and 5 min at 70°C and then held at 4°C. PCR products were then amplified with the following primers: miR-519c forward, 5′-GCGGCAAAGTGCATCTTTTTAGAGGAT-3′; miR-519c reverse, 5′-GTGCAGGGTCCGAGGT-3′; miR-520h forward, 5′-GCGGCACAAAGTGCTTCCCTTTAGAGT-3′; miR-520h reverse, 5′-GTGCAGGGTCCGAGGT-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′; and U6 reverse, 5′-AACGCTTCACGAATTTGCGT-3′. PCR products were analyzed in 2% agarose gels. U6 small nuclear RNA was used as an internal control.
Luciferase reporter assays.
Luciferase reporter constructs were prepared using a pGL3-control vector (Promega, Madison, WI) carrying a simian virus 40 (SV40) promoter-luciferase expression unit. A 5′UTR fragment of ABCG2 (nt −1289 to +396) was amplified by PCR from a previously described promoter construct (50), and a 3′UTR (nt +2462 to +4430) fragment was generated by RT-PCR from H460 cell total RNA. They were inserted downstream of the firefly luciferase gene, into an XbaI site located between the luciferase coding sequence and the poly(A) signal in the pGL3-control vector. Proper orientation of the constructs was verified by restriction mapping and sequence analysis. From the full-length (FL) 3′UTR construct (bases +2462 to + 4430), a series of 3′ deletion constructs were made, with the 3′ ends terminating at bases +4060, +3028, and +2761. The numbering of bases was designated relative to nucleotide 1, defined in GenBank accession no. NM_004827. Another construct (3′UTR FL Δ3820/3841) was prepared by deleting a putative hsa-miR519c binding site from the FL construct. A short sequence corresponding to the putative hsa-miR519c binding site was also cloned downstream of the luciferase gene, resulting in the 3′UTR (nt 3820 to 3841) construct. As a control, a vector with a scrambled sequence from nt 3820 to 3841 was prepared similarly (see Fig. 4A; also see Fig. S9 in the supplemental material).
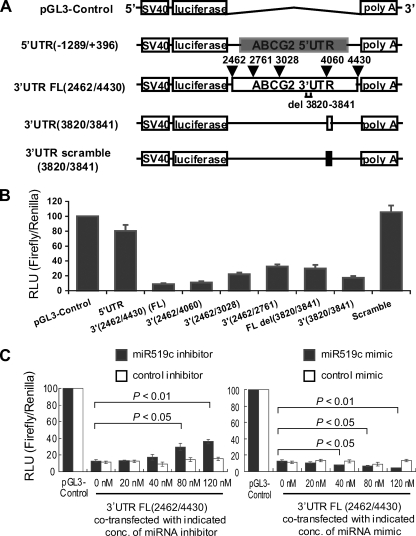
FIG. 4.
ABCG2 luciferase reporter assay. (A) Schematic diagram showing the luciferase reporter constructs used to study the 3′UTR of ABCG2 mRNA. The parental vector, pGL3-control, contains the coding region of firefly luciferase, whose expression is driven by an SV40 promoter (Promega). Nucleotide “1” refers to the first nucleotide in the ABCG2 transcript, as defined in sequence NM_004827. The following constructs were generated: 3′UTR FL (2462/4430); three truncated 3′UTR constructs (based on the FL 3′UTR construct, with progressive deletions from the 3′ end of the 3′UTR, whose 3′ ends are marked at the top of the FL construct, i.e., nt 2761, 3028, and 4060); 3′UTR(3820/3841), which contains only the 22-nt putative miRNA binding site for hsa-miR-519c; 3′UTR FL(Δ3820-3841), which contains the FL 3′UTR lacking the hsa-miR-519c binding site; 3′UTR (scramble 3820/3841), which contains the same 22 nt of the miRNA binding site, but in random order; and a 5′UTR construct (−1289/+396), prepared by inserting a fragment (−1289 to +396) from the ABCG2 5′UTR downstream of the luciferase coding sequence (serves as a control to demonstrate the specific effect of the 3′UTR on the luciferase activity of the reporter assay). The diagram is not drawn to scale. A more detailed diagram illustrating the reporter constructs can be found in Fig. S9 in the supplemental material. (B) Luciferase reporter activities of various reporter constructs, as indicated. Mean reporter activities ± SD (firefly/Renilla luciferase units) from three independent experiments are presented. pGL3-control represents the vector backbone without the ABCG2 3′UTR. (C) Luciferase reporter activity of the FL ABCG2 3′UTR FL(2462/4430) construct in cells cotransfected with either miRNA inhibitor (left) or mimic (right). Mean reporter activities ± SD were obtained as described for panel B. Statistical analysis was performed by the Student t test.
The firefly luciferase constructs (200 ng) were transfected into S1 cells in a 12-well plate, using Fugene 6 transfection reagent (Roche) following the manufacturer's protocol. To monitor transfection efficiency, cells were cotransfected with 50 ng of the phRG-Basic plasmid (Promega), encoding Renilla luciferase. Luminescence was measured 48 h after transfection, using a dual-luciferase reporter assay system (Promega). All transfections were performed in duplicate, and data were analyzed by normalizing firefly luciferase activity to Renilla luciferase activity for each sample. Each construct was tested in three independent transfections. For those experiments testing the effect of miRNA inhibitor or mimic on the reporter constructs, the DharmacFECT Duo transfection reagent (Dharmacon) was used for transfection. Results are expressed as means ± standard deviations (SD) for three independent experiments. Data sets were compared by unpaired Student's t test, and significant differences were considered to be those with P values of <0.05.
Western blot analysis.
Whole-cell lysates prepared from transfected HEK293 or A549 cells were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and subjected to immunoblot analysis with the respective antibodies for ABCG2 (Kamiya Biomedical Company, Seattle, WA), GAPDH (American Research Products, Belmont, MA), and GFP (Cell Signaling Technology, Danvers, MA). The blot was analyzed with an Odyssey (Li-Cor, Licolin, NE) infrared imaging system after incubation with a 1:10,000 dilution of goat anti-mouse (IRDye800CW) or anti-rabbit (IRDye680) secondary antibody (Li-Cor).
RESULTS
Stability of ABCG2 mRNA in parental S1 and resistant S1MI80 cell lines.
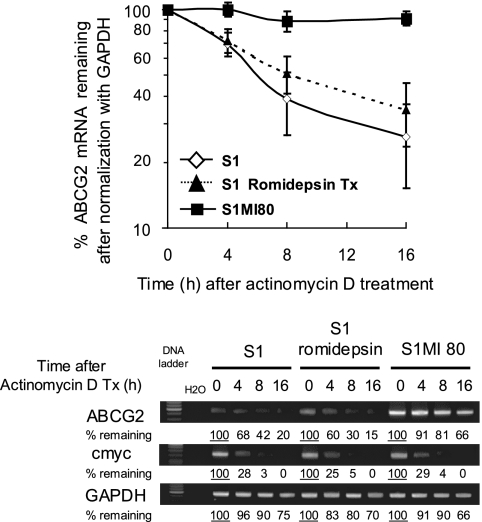
The half-life of ABCG2 mRNA in the parental S1 colon cancer cell line and its drug-selected resistant subline S1MI80 was measured after the cells were treated with actinomycin D to inhibit RNA synthesis. Total RNA was isolated at various time intervals after actinomycin D treatment and analyzed for the ABCG2 mRNA level by RT-PCR. Results are presented as the percentages of ABCG2 mRNA remaining after normalization with GAPDH (Fig. 1). While ABCG2 mRNA in S1MI80 cells was fairly stable for over 16 h after actinomycin D treatment, ABCG2 mRNA in parental S1 cells had a half-life of about 6 h. Upregulation of ABCG2 by romidepsin in parental S1 cells (49) did not alter its mRNA stability (Fig. 1). To ensure that transcriptional shutoff by actinomycin D was efficient in our experiments, RT-PCR was carried out to detect c-myc and GAPDH mRNAs as controls for short (15 to 40 min) and long (more than 24 h) half-lives, respectively (see Fig. S1 in the supplemental material). Importantly, the degradation profiles for GAPDH and c-myc mRNAs in untreated and romidepsin-treated S1 and S1MI80 cells were similar (see Fig. S1 in the supplemental material). Analysis of ABCG2 and GAPDH levels in the mRNA stability study was repeated by quantitative real-time PCR (universal probe library gene assay; Roche Applied Science, Indianapolis, IN) (see Fig. S2A in the supplemental material), and the results were comparable to those described above. We postulated that the reduced degradation rate of ABCG2 mRNA may contribute to the upregulation of ABCG2 in resistant S1MI80 cells. Differential ABCG2 mRNA stability was also observed in several other pairs of parental and ABCG2-overexpressing resistant cell lines (data not shown).
FIG. 1.
(Top) ABCG2 mRNA is more stable in resistant S1MI80 cells than in parental S1 cells. Although ABCG2 is also upregulated by romidepsin treatment (2 ng/ml for 24 h), its mRNA stability is not affected. Nascent RNA synthesis was inhibited with actinomycin D (5 μg/ml), and RNAs were harvested at 0, 4, 8, and 16 h posttreatment. RT-PCR analysis of ABCG2 mRNA was carried out to trace the remaining amount of ABCG2 mRNA with time. c-myc and GAPDH mRNA levels were also monitored as controls for fast-degrading and stable mRNAs, respectively. The value recorded was the percentage of mRNA remaining compared with the amount before the addition of actinomycin D, after normalization with GAPDH. The data shown represent the means ± SD for three independent experiments. (Bottom) Representative gel image showing RT-PCR analysis of ABCG2, c-myc, and GAPDH in cells pretreated with actinomycin D.
Identification of the 3′ end of the human ABCG2 mRNA by 3′RACE assay.
3′RACE reactions were performed to examine the 3′ end of the ABCG2 mRNA and to define the locations of poly(A) sites (17). First-strand cDNA synthesis was performed using AP hybridized to template mRNAs isolated from parental S1 and resistant S1MI80 cells. PCR (first PCR) was then performed on the first-strand cDNA, using the outer reverse primer, complementary to the unique sequence in AP, in conjunction with an ABCG2-specific forward primer, ABCG2 ex16, complementary to nt 2338 to 2360 at exon 16 of ABCG2, upstream of the stop codon (see Fig. S3A in the supplemental material). This PCR yielded multiple small products (200 to 400 bp) for the resistant S1MI80 subline (first PCR) (see Fig. S3B in the supplemental material). However, no PCR product could be obtained from parental S1 cells, probably because of the extremely low level of ABCG2 expression in this cell line. There was no significant reaction with samples primed without RT (data not shown). To increase the specificity of the first PCR, nested PCR 1 was performed using an ABCG2-specific nested N1 forward primer (complementary to nt 2401 to 2429 at exon 16 of ABCG2, still upstream of the stop codon) and the outer reverse primer (data not shown). Nested PCR 1 gave sharper PCR bands but essentially the same pattern as in the first PCR. Since the length of the ABGC2 3′UTR obtained from our 3′RACE assay (about 200 to 400 bp long for S1MI80 cells) is much shorter than that reported in GenBank and in the UTR database, we evaluated other sensitive parental cell lines (SW620, MCF7, SF295, and H460) to see if this was a cell type-specific observation. To our surprise, 3′RACE assay yielded a long product (∼2 kb) and multiple small products (200 to 400 bp) for other parental cell lines as well (see Fig. S3B in the supplemental material) (no PCR product was obtained from SW620 parental cells, probably because of the low level of ABCG2 expression in this cell line).
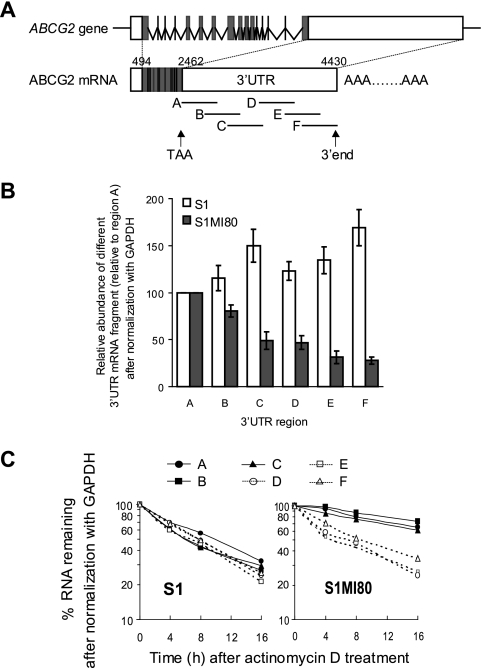
It should be noted that in PCRs containing multiple templates with various sequence lengths between primer binding sites (i.e., first-strand cDNAs from 3′UTRs of different lengths), the shorter templates are usually preferentially amplified (4). This could lead to bias in determining the 3′UTR length by the 3′RACE assay, by underrepresenting the long 3′UTR PCR product. To overcome this problem and to rule out alternative splicing in the ABCG2 3′UTR, we conducted RT-PCR experiments by using total RNAs from pairs of parental and resistant cell lines and primers complementary to different regions in the ABCG2 3′UTR cDNA (Fig. 2A; Table 1). Six separate and overlapping PCR fragments (A to F) that covered the reported ABCG2 3′UTR sequence (NM_004827) were amplified (Fig. 2A). All six PCR fragments were obtained, with the predicted sizes, for all RT samples tested, eliminating the possibility of alternative splicing at the ABCG2 3′UTR. No PCR product was obtained when reverse transcriptase was omitted from the cDNA synthesis mixture. Calibration curves were constructed using a range of input genomic DNAs from S1 cells (see Fig. S4 in the supplemental material). The relative abundances of the different 3′UTR fragments compared to the most upstream region, A, after normalization with GAPDH, were estimated by reading the calibration curves. For parental S1 cells, the relative abundances of the six 3′UTR regions were found to be similar (Fig. 2B). However, for resistant S1MI80 cells, the relative abundances of the more downstream 3′UTR regions became lower and lower, from regions A to F, inferring a larger proportion of truncated 3′UTR in the resistant cells (Fig. 2B).
FIG. 2.
RT-PCR analysis investigating the relative abundances of different fragments of ABCG2 3′UTR in parental S1 and resistant S1MI80 cells. (A) Schematic showing the approximate positions of the six different overlapping fragments within the ABCG2 3′UTR analyzed by RT-PCR. Each fragment is about 500 bp long. (B) Relative abundances of different 3′UTR fragments were plotted relative to the amount in region A after normalization with GAPDH. Open bars, parental S1 cells; closed bars, resistant S1MI80 cells. The data shown represent the means ± SD for three independent experiments. Since the binding site for hsa-miR-519c is located at nt 3820 to 3841 within the ABCG2 3′UTR, only the long ABCG2 transcripts that can be regulated by hsa-miR-519c gave a PCR product for fragment F (nt 3775 to 4388). However, both the long and short ABCG2 transcripts produced PCR products for fragment A (nt 2462 to 3030). By comparing the PCR products for fragments A and F, we estimated that about 77% of ABCG2 transcripts in the parental S1 cells can be regulated by hsa-miR-519c. (C) Relative mRNA stabilities of the six different 3′UTR fragments. Total RNA was isolated at various time intervals after actinomycin D (5 μg/ml) treatment and analyzed for mRNA levels of the six 3′UTR regions by RT-PCR as described for panel A. The data from a representative experiment were plotted as the percentage of mRNA remaining for each 3′UTR fragment compared with the amount before the addition of actinomycin D, after normalization with GAPDH.
Next, the relative mRNA stabilities of the six different 3′UTR regions were evaluated by RT-PCR with total RNAs isolated from S1 and S1MI80 cells at various time intervals after actinomycin D treatment (Fig. 2C; see Fig. S5 in the supplemental material). While all six regions were found to degrade at similar rates in the parental S1 cells, the more upstream regions (i.e., A to C) were more stable than the downstream regions (i.e., D to F) in the resistant S1MI80 cells (Fig. 2C; see Fig. S5 in the supplemental material). This 3′UTR PCR analysis was repeated with quantitative real-time PCR using SYBR green detection (Roche Applied Sciences), and comparable results were obtained (see Fig. S6 in the supplemental material). Together with the 3′RACE result, this suggests that shorter 3′UTR transcripts exist in resistant S1MI80 cells and that they are more stable than their longer counterparts in the parental S1 cells.
Sequence analysis of ABCG2 3′UTRs with different lengths.
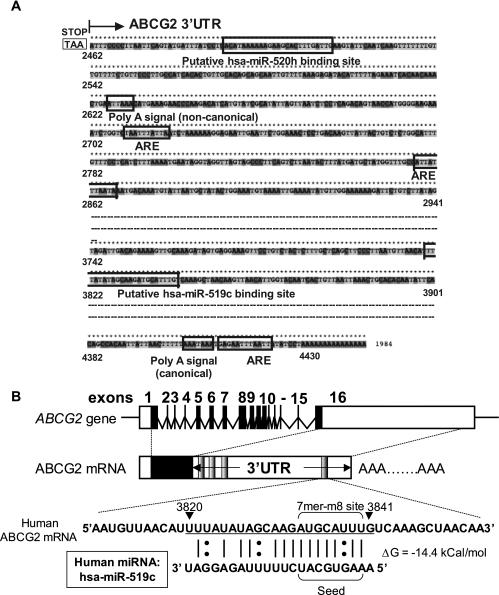
Working first with the 3′RACE product from parental H460 cells and later with those from parental S1 cells and S1MI80 cells, we cloned the 3′RACE PCR mixture as a whole (first PCR) into a TOPO vector and subjected the resulting clones to DNA sequencing. Figure 3A illustrates the relative locations of some putative functional elements within the 3′UTR of the ABCG2 mRNA transcript reported in GenBank (accession no. NM_004827) [including the poly(A) signals, putative AU-rich elements (AREs), and several putative binding sites for miRNAs (see below)]. From our sequencing results, five ABCG2 3′UTR transcripts of different lengths were found. Consistent with the 3′RACE assay, while there were a long (∼2 kb; ends at nt 4419) and four much shorter (100 to 400 bp; ends at nt 2471, 2525, 2644, and 3013) 3′UTR transcripts in the parental cells, only the four shorter ones were found in the resistant cells. The numbering of nucleotides at the 3′UTR is relative to the first nucleotide in the ABCG2 transcript designated in sequence NM_004827, which is used throughout this whole report. Sequence alignment by Clustal X 2.0 multiple sequence comparison software (ftp://ftp.ebi.ac.uk/pub/software/clustalw2) indicated that the five 3′UTR sequences are highly similar, except for the different lengths at the 3′ end (see Fig. S7 in the supplemental material). A 3′UTR sequence of similar length to the long one identified in parental cells in our study has also been reported for ABCG2 (GenBank accession no. BC092408; UTR database entry UTR:3HSA117529). This observation suggests that the different lengths were a result of multiple-site polyadenylation of the 3′UTR, not alternative splicing.
FIG. 3.
(A) Putative functional elements in the reported GenBank sequence (NM_004827) for the 3′UTR of ABCG2 mRNA. Locations of the stop codon (TAA), the canonical (AAUAAA) and noncanonical (AUUAAA) poly(A) signals, three putative AREs (UAAUUUAUUA, AUUAUUUAAUA, and GAGAAUUUAAUU), and the putative hsa-miR-519c and miR-520h binding sites within the 3′UTR of ABCG2 are marked. (B) Diagram illustrating the four clusters of putative miRNA target sites (shaded regions), as predicted by miRBase TARGETS, version 4 (19), within the 3′UTR of ABCG2. Detailed information about these putative miRNAs and their binding site sequences on the ABCG2 3′UTR are summarized in Table 2. The most upstream cluster can putatively be bound by 13 different human miRNAs (hsa-miR-518c, hsa-miR-519d, hsa-miR-302a, hsa-miR-302d, and hsa-miR-520a-h), the second site can be bound by hsa-miR-142-5p, the third site can be bound by hsa-miR-100, and the most downstream site can be bound by hsa-miR-519c. RNA hybrid free energy (ΔG) calculations and the theoretical miRNA-mRNA duplex pairing are also shown for hsa-miR-519c. The “seed” region within the hsa-miR-519c sequence, which determines the specificity of a putative miRNA, is also marked, as are locations of G-U wobbles within the putative binding site for hsa-miR-519c which may destabilize miRNA-mRNA interactions. The G-U wobble found at position 4 from the 5′ end of the hsa-miR-519c sequence may be detrimental to miRNA target recognition, but it has been demonstrated that such a G-U base pairing can usually be tolerated in the seed region (9).
A closer look at the sequences shown in Fig. S7 in the supplemental material identified two poly(A) signal sequences preceding the five alternative poly(A) ends and one or more copies of putative AREs. Apparently, the poly(A) signal at nt 2626 (AUUAAA; a noncanonical signal), downstream from the stop codon TAA (at nt 2459), gives rise to the shorter 3′UTRs, while the other one, at nt 4405 (AAUAAA; a canonical signal), facilitates the formation of the transcript with the long 3′UTR (Fig. 3A; see Fig. S7 in the supplemental material). Moreover, except for the transcripts with the shorter 3′UTRs (ending at nt 2471, 2525, and 2644), all other transcripts of ABGC2 had one or more copies of putative AREs or their variants (UAAUUUAUUA, AUUAUUUAAUA, and GAGAAUUUAAUU, found at nt 2710, 2857, and 4412, respectively). The role of AREs is to degrade targeted mRNA, so the presence of different lengths of ABCG2 3′UTRs with or without AREs may differentiate their mRNA stabilities.
The ABCG2 3′UTR contains four putative clusters of miRNA binding sites.
To look for miRNA binding sites that might be involved in ABCG2 mRNA stability, in silico analysis was performed using the miRBase TARGETS database (http://microrna.sanger.ac.uk) (19) and RNAHybrid (43) to look for putative miRNA binding sites at the ABCG2 3′UTR. The RNAHybrid program uses a free energy (ΔG) algorithm to calculate a favorable binding interaction between a miRNA and potential sites within the mRNA of interest. These analyses identified four putative miRNA binding sites (nt 2491 to 2512, 2823 to 2841, 3052 to 3073, and 3820 to 3841) (Fig. 3B) and several potential miRNAs (summarized in Table 2). Among the four putative miRNA binding sites on the ABCG2 3′UTR, the binding sequence specific to hsa-miR-519c was located at the far 3′ end of the longer 3′UTR and was the only one missing from the shorter 3′UTRs in the resistant cells (position 3820). Note that this site was found only in humans.
TABLE 2.
Putative miRNAs and their binding site sequences on ABCG2 3′UTR
| Positiona | miRNA binding site sequence at 3′UTR | Putative miRNA | ΔG (kcal/mol) |
|---|---|---|---|
| 2492-2513 | CACAUAAAAAAGAGCACUUU | hsa-miR-302a | −13.24 |
| UCACAUAAAAAAGAAGCACUUU | hsa-miR-302d | −18.53 | |
| 2493-2514 | CACAUAAAAAAGAAGCACUUUG | hsa-miR-518c | −17.27 |
| 2492-2514 | ACAUAAAAAAGAGCACUUUG | hsa-miR-519d | −16.28 |
| 2492-2513 | ACAUAAAAAAGAGCACUUU | hsa-miR-520a | −15.10 |
| 2493-2513 | UCACAUAAAAAAGAAGCACUUU | hsa-miR-520b | −15.26 |
| 2491-2513 | CUCACAUAAAAAAGAAGCACUUU | hsa-miR-520c | −16.59 |
| CACAUAAAAAAGAAGCACUUU | hsa-miR-520d | −23.26 | |
| 2493-2513 | ACAUAAAAAAGAAGCACUUU | hsa-miR-520e | −17.96 |
| 2491-2512 | CUCACAUAAAAAAGAAGCACUU | hsa-miR-520f | −15.56 |
| 2492-2515 | ACAUAAAAAAGAAGCACUUUGA | hsa-miR-520g | −20.43 |
| 2494-2515 | AUAAAAAAGAAGACUUUGA | hsa-miR-520h | −17.43 |
| 2493-2513 | CACAUAAAAAAGAAGCACUUU | hsa-miR-526b | −15.43 |
| 2824-2842 | UUCAGUCUUAAUACUUUAUG | hsa-miR-142-5p | −15.43 |
| 2953-2974 | GAGGGUUUGGAACUGUGGGU | hsa-miR-100 | −24.71 |
| 3820-3842 | AGUAAAUACUUUUCAGCACUUAC | hsa-miR-519c | −14.48 |
Nucleotide “1” refers to the first nucleotide in the ABCG2 transcript, as defined in the NM_004827 sequence.
We asked whether hsa-miR-519c could be involved in ABCG2 repression. An important determinant for miRNA repression is the stability of the miRNA-mRNA duplex within the “seed” sequence, from nt 2 to 8 at the 5′ end of the miRNA (5, 10, 20, 23). A so-called “7-mer-m8 site” seed match (containing the seed match augmented by a match to miRNA nucleotide 8) (20) is predicted for hsa-miR-519c (Fig. 3B), with a favorable free energy of hybridization (−14.4 kcal/mol).
Target accessibility has long been established as an important factor for effective antisense oligonucleotide- and siRNA-mediated silencing (31). We therefore also analyzed the stability of the predicted miRNA binding site sequence to determine whether we could find RNA structural features that might affect accessibility and enhance the specificity of target prediction (55). Therefore, the presence of these stabilizing or destabilizing elements could be useful to support the in silico screen for putative miRNA targets. Using mFold (56), we found that the putative hsa-miR-519c binding site was located in an unstable region at the ABCG2 3′UTR, based on free energy prediction (ΔG) (see above) and secondary RNA structure. An unstable hairpin loop was predicted for the secondary structure of the hsa-miR-519c binding site sequence (Fig. S8 in the supplemental material shows the secondary RNA folding structures created by mFold for the putative hsa-miR-519c binding site alone or in the context of ±100-bp neighboring nucleotides), thereby strengthening our in silico prediction for miRNA targeting.
Validation of the putative hsa-miR-519c target site in the 3′UTR of ABCG2 mRNA in S1 cells.
Previous studies have indicated that miRNA binding sites are sufficient to confer miRNA-dependent gene silencing in reporter constructs (10, 53). We inserted ABCG2 3′UTRs of different lengths immediately downstream of a luciferase open reading frame under the control of a constitutively active SV40 promoter (Fig. 4A; full details of all the reporter constructs can be found in Fig. S9 in the supplemental material). In this assay, levels of luciferase activity correlate with translational efficiency and/or stability of the luciferase mRNA. The FL ABCG2 3′UTR (nt 2462 to 4430) dramatically decreased the luciferase activity, by 90%, compared with the pGL3-control without any 3′UTR insert (Fig. 4B). Specific repression of luciferase activity by the 3′UTR was also demonstrated by the minimal inhibition of reporter activity (<20% decrease) by a construct with the 5′UTR of ABCG2 (nt −1289 to +396) inserted downstream of the luciferase open reading frame. Reporter constructs with progressive deletions from the 3′ end of the ABCG2 3′UTR resulted in less and less inhibition of luciferase activity, suggesting a repressive element at the far 3′ end of the 3′UTR (Fig. 4B). Importantly, the FL ABCG2 3′UTR construct with a deletion of the putative hsa-miR-519c binding site (Δ3820-3841) restored the luciferase activity to about 40% of the pGL3-control level. Moreover, the hsa-miR-519c binding site alone could also repress luciferase activity to about 20% of the pGL3-control level. However, a scramble sequence of the putative hsa-miR-519c binding site did not have any effect on luciferase activity.
To confirm that hsa-miR-519c was the specific miRNA repressing the activity of the ABCG2 3′UTR luciferase reporter constructs, we performed experiments in which the endogenous pool of hsa-miR-519c was altered in S1 cells by using a synthetic miRIDIAN hsa-miR-519c inhibitor or mimic (Dharmacon, Lafayette, CO). The specific hsa-miR-519c inhibitor used in our study is a chemically modified antisense oligonucleotide, perfectly complementary to the mature miRNA, which binds tightly to the endogenous miRNA, resulting in its sequestration (8). The specific hsa-miR-519c inhibitor restored luciferase activity in the FL 3′UTR reporter construct by approximately fourfold at the highest dose, 120 nM (Fig. 4C). On the other hand, the hsa-miR-519c mimic is a double-stranded RNA that contains the mature miRNA sequence. When added to cells, the mimic functions in the same way as the endogenous miRNA. When the specific hsa-miR-519c mimic was cotransfected together with the reporter plasmid containing the FL 3′UTR sequence of ABCG2 in S1 cells, a further dose-dependent reduction in luciferase activity was observed compared with that of the reporter construct alone (Fig. 4C). As controls, universal negative controls for miRNA mimic and inhibitor based on a Caenorhabditis elegans miRNA sequence (cel-miR-67 [Dharmacon]; this miRNA has been confirmed to have minimal sequence identity with miRNAs in humans, mice, and rats) did not have any effect on the luciferase activity (Fig. 4C). Moreover, neither hsa-miR-519c mimic nor inhibitor had an effect on the hsa-miR-519c site-deleted FL ABCG2 3′UTR reporter construct (see Fig. S10 in the supplemental material). These results indicate that hsa-miR-519c specifically interacts with the target binding site identified in the human ABCG2 3′UTR, which in turn represses the ABCG2 mRNA and protein expression.
Since the putative hsa-miR-519c site has been validated for ABCG2 3′UTR in S1 cells, we repeated the 3′RACE assay, using S1 cells transfected with the specific hsa-miR-519c inhibitor. We used the specific hsa-miR-519c inhibitor to increase the level of ABCG2 3′UTR in S1 cells (see Fig. S11A in the supplemental material) and subsequently confirmed the long sequence in S1 cells and its absence in S1MI80 cells with a nested PCR 1 and nested PCR 2 (see Fig. S11B in the supplemental material).
hsa-miR-519c regulates ABCG2 expression by both translation repression and mRNA degradation.
Regulation by miRNAs can be due to translation repression and/or mRNA degradation. To assess whether hsa-miR-519c has a functional role in the downregulation of ABCG2 expression by either of these mechanisms, an ABCG2 expression vector (pcDNA3-ABCG2) and two of its derivatives, one with a tandem repeat of three copies of hsa-miR-519c binding site sequence (pcDNA3-ABCG2-3×miRNA binding site) and the other with a tandem repeat of three copies of a scramble sequence of the hsa-miR-519c site (pcDNA3-ABCG2-3×xscramble binding site) fused immediately after the stop codon of the ABCG2 coding sequence in the vector, were transfected into HEK293 cells and ABCG2 expression levels measured. As an internal control, the level of cotransfected GFP (pEGFP-C1; BD Bioscience Clontech, Mountain View, CA) was assessed as well and was used to normalize the ABCG2 mRNA levels. ABCG2 expression was found to be lower in cells transfected with the 3x×miRNA binding site vector than in those with the original ABCG2 expression vector without any 3′UTR insert, at both the mRNA (−24%) and protein (even lower, at −49%) levels (see Fig. S12A in the supplemental material). The scramble binding site vector gave similar ABCG2 expression to that of the plain ABCG2 expression vector. An hsa-miR-519c inhibitor was cotransfected with the pcDNA3-ABCG2-3×miR519c binding site vector to verify whether hsa-miR-519c inhibits ABCG2 expression specifically. Both ABCG2 mRNA and protein expression levels were restored to the levels in cells transfected with the plain pcDNA3-ABCG2 vector by the specific hsa-miR-519c inhibitor but not by a universal negative control miRNA inhibitor (see Fig. S12A in the supplemental material).
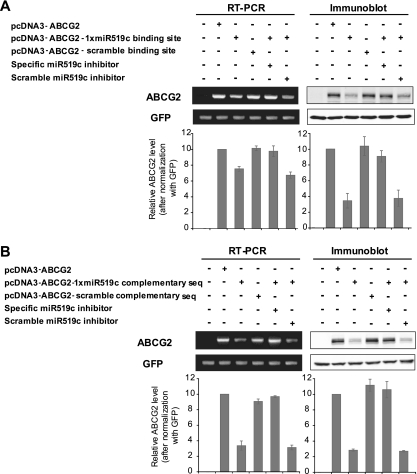
The differential effect on ABCG2 mRNA and protein expression by the hsa-miR-519c inhibitor was quite reproducible and led us to postulate that protein translation blockade may play a more important role than mRNA degradation in the repression of ABCG2 by hsa-miR-519c. The degree of miRNA-mRNA pairing has been proposed to affect the mechanism of repression, with perfect pairing leading to mRNA cleavage but imperfect pairing leading to protein translation block (23). We prepared an ABCG2 expression vector as described above, but immediately following the stop codon we placed either a tandem repeat of three copies of a 22-nt sequence perfectly complementary to the miR519c sequence (pcDNA3-ABCG2-3×miRNA complementary sequence) or a tandem repeat of three copies of scramble sequence (pcDNA3-ABCG2-3×scramble complementary sequence). ABCG2 expression was then examined in HEK293 cells transfected with these new vectors. As with the previous vectors, ABCG2 expression was found to be lower in cells transfected with the 3×miRNA complementary sequence vector than in cells with the plain ABCG2 expression vector without anything after the stop codon, but similarly at both the mRNA (−61%) and protein (−70%) levels (see Fig. S12B in the supplemental material), suggesting that while affecting both levels, the effect of hsa-miR-519c on protein translation predominates in the wild-type 3′UTR. The 3×scramble complementary sequence vector gave similar ABCG2 expression to that with the original ABCG2 expression vector. Interestingly, similar observations were obtained when the vectors contained only a single copy of either the hsa-miR-519c binding site or complementary sequence (Fig. 5A and B).
FIG. 5.
hsa-miR-519c binding site facilitates regulation of ABCG2 expression by both mRNA degradation and protein translation blockade. (A) HEK293 cells were transfected with an ABCG2 expression vector (pcDNA3-ABCG2) or one of two of its derivatives, one with a copy of the hsa-miR-519c binding sequence (pcDNA3-ABCG2-1×miRNA binding site) and the other with a scramble sequence of the hsa-miR-519c site (pcDNA3-ABCG2-scramble binding site) fused immediately after the stop codon of the ABCG2 coding sequence in the vector, and ABCG2 mRNA and protein expression was evaluated 48 h after transfection. A specific hsa-miR-519c or negative control miRNA inhibitor was also cotransfected into the cells to see if it could increase the ABCG2 expression level. ABCG2 expression levels were normalized with GFP (pEGFP-C1; DB Bioscience Clontech) and plotted in the bar graph relative to the level obtained for cells transfected with the pcDNA3-ABCG2 vector. The data shown represent the means ± SD for three independent experiments. (B) Perfect Watson-Crick pairing between hsa-miR-519c and 3′UTR sequences promotes predominantly mRNA degradation in the repression of ABCG2. HEK293 cells were transfected with similar ABCG2 expression vectors to those described for panel A. However, in the two derivative vectors, instead of the hsa-miR-519c binding site, either one copy of a 22-nt sequence complementary to the hsa-miR-519c binding site (pcDNA3-ABCG2-1×miRNA complementary sequence) or its scramble sequence (pcDNA3-ABCG2-scramble complementary sequence) was fused immediately downstream of the ABCG2 coding sequence. ABCG2 expression was found to be lower in cells transfected with the 1×miRNA complementary sequence vector than in those transfected with the plain ABCG2 expression vector without anything after the stop codon, but expression was similar at both the mRNA (−65%) and protein (−70%) levels. The scramble complementary sequence vector gave similar ABCG2 expression to that of the original ABCG2 expression vector. hsa-miR-519c was also found to be responsible for repression, because an hsa-miR-519c inhibitor, but not the scramble control miRNA inhibitor, could restore ABCG2 expression in cells transfected with the pcDNA3-ABCG2-1×miRNA complementary sequence. ABCG2 expression levels were normalized with GFP and plotted relative to the level obtained in cells transfected with the pcDNA3-ABCG2 vector.
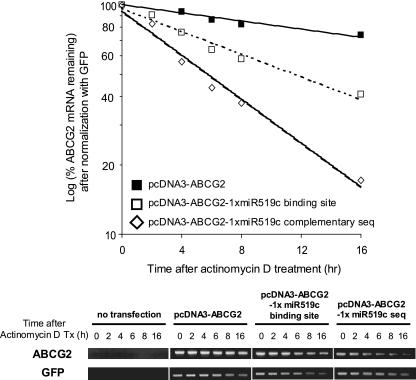
ABCG2 mRNA stability was also measured in HEK293 cells transfected with the various vectors (having a single copy of either the hsa-miR-519c binding site or complementary sequence). Actinomycin D was added at 48 h posttransfection to block transcription, and total RNA was isolated at various time intervals and analyzed for ABCG2 mRNA levels by using RT-PCR. As an internal control, the level of cotransfected GFP was measured for normalization. As shown in Fig. 6, ABCG2 mRNA expressed from the vector with the hsa-miR-519c complementary sequence downstream of the ABCG2 coding sequence was the least stable (half-life [t1/2], ∼6 h), followed by the vector with the wild-type hsa-miR-519c binding site (t1/2, >16 h). In contrast, the ABCG2 without any 3′UTR was fairly stable for up to 24 h after actinomycin D treatment. The assay was repeated by real-time PCR, using the Universal ProbeLibrary gene assay, and similar results were obtained. These results underscore the destabilizing effects in vivo of the hsa-miR-519c binding site in the ABCG2 3′UTR (nt 3820 to 3841), but only partially by promoting mRNA degradation. Interestingly, the perfect pairing of hsa-miR-519c with the hsa-miR-519c complementary sequence led to the most substantial degradation of ABCG2 mRNA.
FIG. 6.
hsa-miR-519c binding site decreases ABCG2 expression by promoting mRNA degradation. (Top) An ABCG2 expression vector (pcDNA3-ABCG2) and two of its derivatives, one with a copy of the hsa-miR-519c binding sequence (pcDNA3-ABCG2-1×miRNA binding site) and the other with a sequence complementary to that of hsa-miR-519c (pcDNA3-ABCG2-1×miRNA complementary sequence) fused immediately after the stop codon of the ABCG2 coding sequence in the vector, were transfected into HEK293 cells, and ABCG2 mRNA stability was monitored. Actinomycin D was added at 48 h posttransfection to block transcription, and total RNA was isolated at various time intervals and analyzed for ABCG2 mRNA levels by using RT-PCR. As an internal control, the level of GFP mRNA (from cotransfection of pEGFP-C1 [BD Bioscience Clontech]) was assessed and was used to normalize the ABCG2 mRNA levels. The graph shows the mean results for three independent and reproducible experiments. ABCG2 mRNA expressed from the vector with the hsa-miR-519c complementary sequence downstream of the ABCG2 coding sequence was the least stable (t1/2, ∼6 h), followed by the vector with the hsa-miR-519c binding site (t1/2, >16 h). In contrast, the ABCG2 mRNA without any 3′UTR was fairly stable for up to 24 h after actinomycin D treatment. (Bottom) Representative gel image showing the RT-PCR analysis summarized in the top panel.
hsa-miR-519c regulates endogenous ABCG2 mRNA and protein expression.
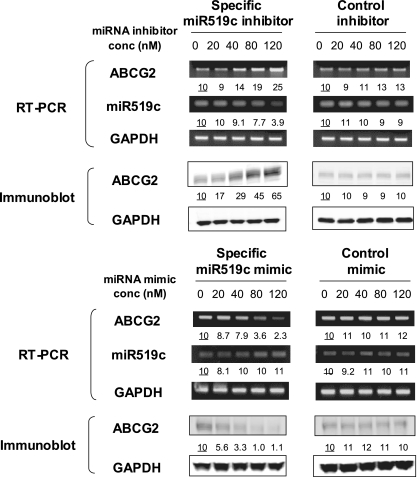
Finally, we evaluated whether hsa-miR-519c has a functional role in the downregulation of endogenous ABCG2 expression. A549 cells were chosen for our study because endogenous ABCG2 protein expression is readily detectable in these cells by Western blot analysis, and similar to other parental cell lines used in our study, A549 cells have both long and short ABCG2 3′UTRs, as determined by 3′RACE assay (data not shown). Transfection of A549 cells with various concentrations of hsa-miR-519c inhibitor, a single-stranded modified RNA that has a complementary sequence to the mature miRNA, led to a dose-dependent induction of ABCG2 at both the mRNA and, albeit more pronounced, protein levels. ABCG2 mRNA and protein were upregulated over twofold and sixfold, respectively, with 120 nM of miR519c inhibitor, while no change was noted in GAPDH (Fig. 7, top panel). The negative control miRNA inhibitor did not alter the ABCG2, GAPDH, or hsa-miR-519c level. Although miRNA inhibitors principally function by sequestering endogenous miRNAs, the level of the latter was also decreased at a higher dose of miRNA inhibitor (Fig. 7, top panel). In fact, antisense oligonucleotides have been shown to reduce the cellular concentration of their cognate miRNAs (8, 29).
FIG. 7.
hsa-miR-519c regulates endogenous ABCG2 expression in A549 cells. A549 cells were transfected with 0 to 120 nM of either specific hsa-miR-519c inhibitor, hsa-miR-519c mimic, or the respective negative control (based on the cel-miR-67 sequence [Dharmacon]; this miRNA has been confirmed to have minimal sequence homology with miRNAs in humans, mice, and rats) for 48 h. Total RNA and whole-cell lysates were harvested for subsequent RT-PCR and Western blot analysis of ABCG2 expression. ABCG2 expression was normalized with GAPDH and reported relative to that of mock-treated cells. The transcript for hsa-miR-519c was also measured by stem-loop RT-PCR. The figure shows a representative result for three independent and reproducible experiments. The universal miRNA negative control inhibitor or mimic did not affect ABCG2, hsa-miR-519c, or GAPDH expression.
In parallel with these studies, A549 cells were transfected with a specific hsa-miR-519c mimic, a synthetic RNA duplex that mimics endogenous hsa-miR-519c. ABCG2 mRNA and protein levels were repressed approximately 70% and 90%, respectively, with 120 nM of the specific hsa-miR-519c mimic, but GAPDH was not affected (Fig. 7, bottom panel). The negative control miRNA mimic also did not alter the ABCG2, GAPDH, or hsa-miR-519c level.
Another repressive miRNA(s) located at the 3′UTR of ABCG2.
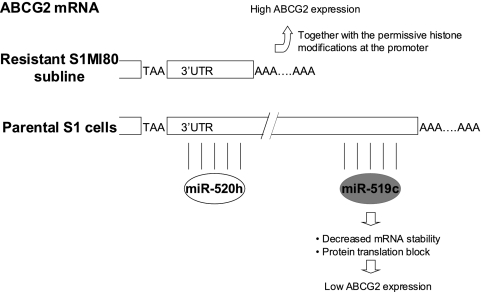
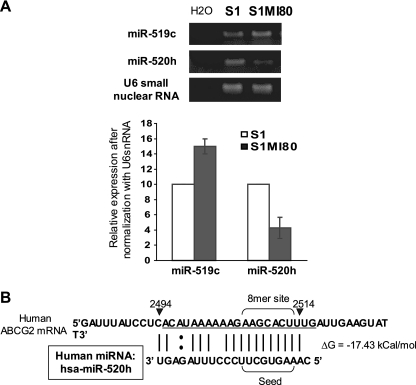
As indicated by the luciferase reporter assay (Fig. 4B), the substantial inhibition of reporter activity by the shortest ABCG2 3′UTR construct (nt 2462 to 2761) suggests that some other unknown repressive elements exist at the more upstream region of the ABCG2 3′UTR. While this report was under preparation, Liao et al. (34) reported that hsa-miR-520h promotes differentiation of hematopoietic stem cells into progenitor cells by inhibiting ABCG2 expression. According to miRNA target prediction by the miRBase TARGETS database (19) and RNAHybrid (43), the putative binding sequence for hsa-miR-520h is located much more upstream (nt 2494 to 2514, relative to the first nucleotide defined in sequence NM_004827) (Fig. 3A; see Fig. 9) at the ABCG2 3′UTR than that for hsa-miR-519c. More importantly, hsa-miR-520h binds to both long and short ABCG2 3′UTRs in parental S1 and resistant S1MI80 cells. We therefore examined the expression level of hsa-miR-520h by stem-loop RT-PCR with S1 and S1MI80 cells to try to elucidate its role in regulating ABCG2. Interestingly, the hsa-miR-520h level in S1MI80 was found to be only about half of that in S1 cells (43% ± 13%) (Fig. 8A).
FIG. 9.
Proposed model for involvement of miRNA in the regulation of ABCG2 at the 3′UTR. A putative miRNA, hsa-miR-519c, binds to the 3′UTR of ABCG2 and suppresses the expression of ABCG2 in the parental cell lines. In the resistant cell lines, hsa-miR-519c cannot bind to ABCG2 mRNA because of the shorter 3′UTR, and thus mRNA degradation and/or repression of protein translation is relieved, contributing to the overexpression of ABCG2 by other mechanisms, such as permissive histone modifications at its promoter (49). hsa-miR-520h is another putative miRNA targeting ABCG2 mRNA, which was reported while the manuscript was under preparation (34). Unlike hsa-miR-519c, identified in our study, hsa-miR-520h binds to the 3′UTR of ABCG2 in both S1 and S1MI80 cells. Interestingly, S1MI80 cells express less hsa-miR-520h than do S1 cells, presumably allowing less repression of ABCG2 in the former.
FIG. 8.
(A) Differential hsa-miR-520h expression level between parental S1 and resistant S1MI80 cells. hsa-miR-519c and hsa-miR-520h expression was measured by stem-loop RT-PCR (7, 46). U6 small nuclear RNA served as the loading control. The figure shows representative results for three independent and reproducible experiments, with means (± SD) plotted in the lower panel. (B) Location of the putative hsa-miR-520h binding site at ABCG2 3′UTR, RNA hybrid free energy (ΔG) calculations, and the theoretical miRNA-mRNA duplex pairing for hsa-miR-520h.
DISCUSSION
Overexpression of the ABCG2 gene is frequently observed in cancer cell lines selected with a number of chemotherapeutic drugs (1, 12, 35, 38, 44, 52). However, little is known about the mechanisms underlying its upregulation. Gene amplification and chromosome translocation have been found to play a role in the increased expression of the ABCG2 gene in some drug-resistant cell lines (27). The use of alternative 5′ promoters at the ABCG2 gene in drug-selected cells may offer another novel mechanism of ABCG2 upregulation (39), a finding similar to observations for MDR-1, where rearrangement of the 5′ region of MDR-1 resulted in capture of that gene by other promoters (22, 26, 37). Recently, we reported that a set of permissive histone modifications and a chromatin remodeling factor, Brg-1, associated with the ABCG2 proximal promoter are required for the upregulation of ABCG2 in resistant cells (49). In an effort to further understand the regulation of ABCG2 in resistant cancer cells, we found that the ABCG2 mRNA is less stable in a number of sensitive parental cancer cell lines than in their drug-selected and ABCG2-overexpressing resistant counterparts. We also observed that parental cells produce ABCG2 mRNA with both long and short 3′UTRs, whereas resistant cells produce only the shorter ones.
Since the 3′UTR of eukaryotic mRNA contains regulatory elements affecting mRNA stability, translation, and transport, it is logical to suspect that the absence of some regulatory elements in the short 3′UTR compared with the long 3′UTR may lead to differential regulation of ABCG2 in sensitive and resistant cells. We hypothesized that a putative miRNA binds to the 3′UTR of ABCG2 in parental cells, thereby suppressing ABCG2 expression, but that the binding site for this miRNA is lost in the shorter 3′UTR in the resistant cells. miRNAs have a direct role in regulating the processes of development and differentiation, and more than 4,000 have been identified in plant and animal species (miRBase Registry [http://microrna.sanger.ac.uk/sequences]). miRNA dysregulation has been correlated with the progression and aggressiveness of several forms of cancer (3, 25). In silico analysis revealed a putative target site for hsa-miR-519c at the far 3′ end of the long ABCG2 3′UTR transcript identified in sensitive cells.
In fact, the differential repression of alternative transcripts with 3′UTRs of different lengths by miRNA was proposed recently (32). When a miRNA target is located between two variant sites of polyadenylation, the shorter transcript will be target-free and can escape miRNA-mediated inhibition, while longer transcripts will be inhibited. ABCG2 mRNA contains both a canonical AAUAAA poly(A) signal and its noncanonical variant AUUAAA. The canonical and noncanonical poly(A) signals are usually processed differently. This provides a means for mRNAs with multiple poly(A) sites to be regulated by synthesis of specific mRNA forms, thereby regulating gene expression (14, 18). Based on our 3′RACE assay and the subsequent DNA sequencing of ABCG2, it seems that the noncanonical AUUAAA poly(A) signal is preferentially used in resistant cells, giving only the short 3′UTR transcripts. In parental cells, both the canonical AAUAAA and the noncanonical AUUAAA signals are used (Fig. 3; see Fig. S7 in the supplemental material), so that long and short 3′UTR transcripts are generated.
Our hypothesis that ABCG2 is a true target for hsa-miR-519c was rigorously tested. Based on a current prediction algorithm (19), the targeting is likely because hsa-miR-519c binds to the 3′UTR of ABCG2 mRNA with a “7-mer-m8 site” complementary to the hsa-miR-519c seed sequence (Fig. 3B). The presence of a destabilizing element (hairpin loop) in the target ABCG2 3′UTR region further strengthens the likelihood that it is a direct target (see Fig. S8 in the supplemental material). Using reporter gene assays, the FL ABCG2 3′UTR significantly inhibited the reporter activity, while the 3′-truncated reporter construct did so to a lesser extent. Moreover, the inhibition of reporter activity was also significantly decreased when the hsa-miR-519c binding site was deleted from the FL ABCG2 3′UTR. The hsa-miR-519c binding sequence fused immediately downstream of the luciferase reporter coding sequence could alone confer a decrease in reporter activity, though this type of result typically requires three copies of the miRNA binding sequence and a short linker sequence (20, 33, 55). Furthermore, the FL ABCG2 3′UTR luciferase construct, but not the one with a truncation at the 3′ end of the ABCG2 3′UTR, is specifically responsive to hsa-miR-519c inhibitor or mimic (Fig. 4C; see Fig. S10 in the supplemental material). In transient transfection experiments, the repressive effect of hsa-miR-519c was demonstrated by comparing an expression vector containing the hsa-miR-519c binding site downstream of the ABCG2 coding sequence to one containing a scramble sequence. Mutation of the hsa-miR-519c site in the expression vector abolished the repressive effect. Finally, endogenous ABCG2 was found to be upregulated by the specific hsa-miR-519c inhibitor and downregulated by the specific hsa-miR-519c mimic in A549 human lung cancer cells (Fig. 7). Thus, multiple tests confirmed that ABCG2 is a true target of hsa-miR-519c. These tests have also been used to confirm multiple other miRNA-target pairs, including let-7-RAS (25), miR-1-Hand2 (55), miR-127-BCL6 (46), miR-130a-GAX (7), and miR-196-HOXB8 (53).
Although repression of gene expression by miRNAs can be mediated by mRNA degradation and/or inhibition of translation from existing mRNA (51), our results suggest that the effect of hsa-miR-519c inhibiting ABCG2 protein translation is more pronounced than that promoting mRNA degradation, which could be predicted by the suboptimal degree of pairing between hsa-miR-519c and the human ABCG2 3′UTR. When the hsa-miR-519c binding site was replaced with a sequence complementary to hsa-miR-519c in the same ABCG2 expression vector, ABCG2 was repressed predominantly by mRNA degradation (Fig. 5A and B and 6). This perfect Watson-Crick complementarity between miRNA and the target mRNA allows mRNA cleavage according to the same mechanisms of gene silencing occurring with the experimental use of small interfering RNA (siRNA) (23).
The resistant S1MI80 subline used in this study was derived from the S1 clone of the LS174 human colon cancer cell line by selection in mitoxantrone (38). Interestingly, no amplification of the chromosome band 4q21-q22 but a balanced t(4,7) translocation downstream of the ABCG2 gene was observed in S1MI80 cells (27). In this study, we observed the absence of the long 3′UTR, leaving behind only the shorter ones, in S1MI80 cells, which is coincident with this translocation. Since the breakpoint for this translocation is believed to be 3′ of the ABCG2 gene, it remains to be determined if there is any correlation between the translocation and the differential 3′UTR length adopted in resistant cells.
In summary, we have shown that ABCG2 adopts different polyadenylation sites (and thus different 3′UTR lengths) at its 3′UTR in parental and resistant cells and that there is an interplay between alternative polyadenylation and hsa-miR-519c targeting. Figure 9 presents a model where hsa-miR-519c targets the 3′UTR of ABCG2 only in parental cells, not in resistant cells, thereby allowing the overexpression of ABCG2 in the latter by permissive histone modifications and chromatin remodeling events at the gene promoter (49). ABCG2 mRNA that is highly expressed in the presence of hsa-miR-519c by other mechanisms, such as gene amplification, translocation, or distinct histone modifications, is under selection pressure to avoid binding sites for this miRNA. While the manuscript was under preparation, another putative miRNA (hsa-miR-520h) was reported to target ABCG2 in hematopoietic stem cells during their differentiation into progenitor cells (34). Unlike hsa-miR-519c, identified in our study, hsa-miR-520h binds to the 3′UTR of ABCG2 in both S1 and S1MI80 cells. Interestingly, S1MI80 cells express less hsa-miR-520h (∼40%) than do S1 cells.
Two implications of this work can be considered. If other repressive elements or other inhibitory miRNA binding sites in the 3′UTR could be identified, vectors containing such a miRNA could be generated for delivery to patient tumors to overcome limitations imposed by ABCG2 overexpression on drug accumulation, provided that problems with translation of gene therapy to the clinic are overcome. Second, genetic variations (particularly single-nucleotide polymorphisms [SNPs]) in drug transporter genes have been implicated in altering cancer drug pharmacology and, presumably, drug effects. To date, most SNPs identified in ABCG2 lie within its coding region. Since we have identified a miRNA binding site at the ABCG2 3′UTR, it follows that SNP variations (if found at the 3′UTR) could interact with the binding of miRNA(s), thereby affecting the overall ABCG2 expression. The effect of these SNPs is therefore predictable and opens up another avenue for the study of interindividual variation in transporter activity. Undoubtedly, continued efforts to delineate miRNA function in regulating ABCG2 could eventually provide a new way of altering ABCG2 expression to manipulate the disposition of ABCG2 substrate drugs.
Supplementary Material
Acknowledgments
This research was supported by the intramural research program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Published ahead of print on 23 June 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Allikmets, R., L. M. Schriml, A. Hutchinson, V. Romano-Spica, and M. Dean. 1998. A human placenta-specific ATP-binding cassette gene (ABCP) on chromosome 4q22 that is involved in multidrug resistance. Cancer Res. 585337-5339. [PubMed] [Google Scholar]
- 2.Ambros, V. 2004. The functions of animal microRNAs. Nature 431350-355. [DOI] [PubMed] [Google Scholar]
- 3.Bandres, E., E. Cubedo, X. Agirre, R. Malumbres, R. Zarate, N. Ramirez, A. Abajo, A. Navarro, I. Moreno, M. Monzo, and J. Garcia-Foncillas. 2006. Identification by real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol. Cancer 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basler, C. F., and M. S. Horwitz. 1996. Subgroup B adenovirus type 35 early region 3 mRNAs differ from those of the subgroup C adenoviruses. Virology 215165-177. [DOI] [PubMed] [Google Scholar]
- 5.Brennecke, J., A. Stark, R. B. Russell, and S. M. Cohen. 2005. Principles of microRNA-target recognition. PLoS Biol. 3e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, C., D. A. Ridzon, A. J. Broomer, Z. Zhou, D. H. Lee, J. T. Nguyen, M. Barbisin, N. L. Xu, V. R. Mahuvakar, M. R. Andersen, K. Q. Lao, K. J. Livak, and K. J. Guegler. 2005. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 33e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, Y., and D. H. Gorski. 2008. Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood 1111217-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis, S., B. Lollo, S. Freier, and C. Esau. 2006. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res. 342294-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Didiano, D., and O. Hobert. 2006. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat. Struct. Mol. Biol. 13849-851. [DOI] [PubMed] [Google Scholar]
- 10.Doench, J. G., and P. A. Sharp. 2004. Specificity of microRNA target selection in translation repression. Genes Dev. 18504-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donnini, M., A. Lapucci, L. Papucci, E. Witort, A. Jacquier, G. Brewer, A. Nicolin, S. Capaccioli, and N. Schiavone. 2004. Identification of TINO: a new evolutionarily conserved BCL-2 AU-rich element RNA-binding protein. J. Biol. Chem. 27920154-20166. [DOI] [PubMed] [Google Scholar]
- 12.Doyle, L. A., W. Yang, L. V. Abruzzo, T. Krogmann, Y. Gao, A. K. Rishi, and D. D. Ross. 1998. A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc. Natl. Acad. Sci. USA 9515665-15670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ebert, B., A. Seidel, and A. Lampen. 2005. Identification of BCRP as transporter of benz[a]pyrene conjugates metabolically formed in Caco-2 cells and its induction by Ah-receptor agonists. Carcinogenesis 261754-1763. [DOI] [PubMed] [Google Scholar]
- 14.Edwalds-Gilbert, G., K. L. Veraldi, and C. Milcarek. 1997. Alternative poly(A) site selection in complex transcription units: mean to an end? Nucleic Acids Res. 252547-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ee, P. L., S. Kamalakaran, D. Tonetti, X. He, D. D. Ross, and W. T. Beck. 2004. Identification of a novel estrogen response element in the breast cancer resistance protein (ABCG2) gene. Cancer Res. 641247-1251. [DOI] [PubMed] [Google Scholar]
- 16.Farh, K. K., A. Grimson, C. Jan, B. P. Lewis, W. K. Johnston, L. P. Lim, C. B. Burge, and D. P. Bartel. 2005. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science 3101817-1821. [DOI] [PubMed] [Google Scholar]
- 17.Frohman, M. A. 1990. RACE: rapid amplification of cDNA ends, p. 28-38. In M. A. Inns, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, San Diego, CA.
- 18.Graber, J. H., C. R. Cantor, S. C. Mohr, and T. F. Smith. 1999. In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc. Natl. Acad. Sci. USA 9614055-14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffiths-Jones, S. 2006. miRBase: the microRNA sequence database. Methods Mol. Biol. 342129-138. [DOI] [PubMed] [Google Scholar]
- 20.Grimson, A., K. K. Farh, W. K. Johnston, P. Garrett-Engele, L. P. Lim, and D. P. Bartel. 2007. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 2791-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He, L., and G. J. Hannon. 2004. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5522-531. [DOI] [PubMed] [Google Scholar]
- 22.Huff, L. M., J. S. Lee, R. W. Robey, and T. Fojo. 2006. Characterization of gene rearrangements leading to activation of MDR-1. J. Biol. Chem. 28136501-36509. [DOI] [PubMed] [Google Scholar]
- 23.Hutvagner, G., and P. D. Zamore. 2002. A microRNA in a multiple-turnover RNAi enzyme complex. Science 2972056-2060. [DOI] [PubMed] [Google Scholar]
- 24.Jensen, L. E., and A. S. Whitehead. 2004. The 3′untranslated region of the membrane-bound IL-1R accessory protein mRNA confers tissue-specific destabilization. J. Immunol. 1736248-6258. [DOI] [PubMed] [Google Scholar]
- 25.Johnson, S. M., H. Grosshans, J. Shingara, M. Byrom, R. Jarvis, A. Cheng, E. Labourier, K. L. Reinert, D. Brown, and F. J. Slack. 2005. RAS is regulated by the let-7 microRNA family. Cell 120635-647. [DOI] [PubMed] [Google Scholar]
- 26.Knutsen, T., L. A. Mickley, T. Ried, E. D. Green, S. du Manoir, E. Schrock, M. Macville, Y. Ming, R. Robey, M. Polymeropoulous, R. Torres, and T. Fojo. 1998. Cytogenetic and molecular characterization of random chromosomal rearrangements activating the drug resistance gene, MDR1/P-glycoprotein in drug-selected cell lines and patients with drug refractory ALL. Genes Chromosomes Cancer 2344-54. [DOI] [PubMed] [Google Scholar]
- 27.Knutsen, T., V. K. Rao, T. Ried, L. Mickey, E. Schneider, K. Miyake, B. M. Ghadimi, H. Padilla-Nash, S. Pack, L. Greenberger, K. Cowan, M. Dean, T. Fojo, and S. Bates. 2000. Amplification of 4q21-q22 and the MXR gene in independently derived mitoxantrone-resistant cell lines. Genes Chromosomes Cancer 27110-116. [PubMed] [Google Scholar]
- 28.Krishnamurthy, P., D. D. Ross, T. Nakanishi, K. Bailey-Dell, S. Zhou, K. E. Mercer, B. Sarkadi, B. P. Sorrentino, and J. D. Schuetz. 2004. The stem cell marker Bcrp/ABCG2 enhances hypoxic cell survival through interactions with heme. J. Biol. Chem. 27924218-24225. [DOI] [PubMed] [Google Scholar]
- 29.Krutzfeldt, J., N. Rajewky, R. Braich, K. G. Rajeev, T. Tuschl, M. Manoharan, and M. Stoffel. 2005. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438685-689. [DOI] [PubMed] [Google Scholar]
- 30.Lander, E. S., L. M. Linton, B. Birren, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409860-921. [DOI] [PubMed] [Google Scholar]
- 31.Lee, N. S., T. Dihjima, G. Bauer, H. Li, M. J. Li, A. Ehsani, P. Salvaterra, and J. Rossi. 2002. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 20500-505. [DOI] [PubMed] [Google Scholar]
- 32.Legendre, M., W. Ritchie, F. Lopez, and D. Gautheret. 2006. Differential repression of alternative transcripts: a screen for miRNA targets. PLoS Comput. Biol. 2e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis, B. P., C. B. Burge, and D. P. Bartel. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 12015-20. [DOI] [PubMed] [Google Scholar]
- 34.Liao, R., J. Sun, L. Zhang, G. Lou, M. Chen, D. Zhou, Z. Chen, and S. Zhang. 2008. MicroRNAs play a role in the development of human hematopoietic stem cells. J. Cell Biochem. 104805-817. [DOI] [PubMed] [Google Scholar]
- 35.Maliepaard, M., M. A. van Gastelen, L. A. de Jang, D. Pluim, R. C. van Waardenburg, M. C. Ruevekamp-Helmers, B. G. Floot, and J. H. Schellens. 1999. Overexpression of the BCRP/MXR/ABCP gene in a topotecan-selected tumor cell line. Cancer Res. 594559-4563. [PubMed] [Google Scholar]
- 36.Meister, G., M. Landthaler, Y. Dorsett, and T. Tuschi. 2004. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA 10544-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mickley, L. A., B. A. Spengler, T. A. Knutsen, J. L. Biedler, and T. Fojo. 1997. Gene rearrangement: a novel mechanism for MDR-1 gene activation. J. Clin. Investig. 991947-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyake, K., L. Mickley, T. Litman, Z. Zhan, R. Robey, B. Cristensen, M. Brangi, L. Greenburger, M. Dean, T. Fojo, and S. E. Bates. 1999. Molecular cloning of cDNAs which are highly overexpressed in mitoxantrone-resistance cells: demonstration of homology to ABC transport genes. Cancer Res. 598-13. [PubMed] [Google Scholar]
- 39.Nakanishi, T., K. J. Bailer-Dell, B. A. Hassel, K. Shiozawa, D. M. Sullivan, J. Turner, and D. D. Ross. 2006. Novel 5′ untranslated region variants of BCRP mRNA are differentially expressed in drug-selected cancer cells and in normal human tissues: implications for drug resistance, tissue-specific expression, and alternative promoter usage. Cancer Res. 665007-5011. [DOI] [PubMed] [Google Scholar]
- 40.Nishimori, T., H. Inoue, and Y. Hirata. 2004. Involvement of the 3′-untranslated region of cyclooxygenase-2 gene in its post-transcriptional regulation through the glucocorticoid receptor. Life Sci. 742505-2513. [DOI] [PubMed] [Google Scholar]
- 41.Pesole, G., and S. Liuni. 1999. Internet resources for the functional analysis of 5′ and 3′ untranslated regions of eukaryotic mRNA. Trends Genet. 15378. [DOI] [PubMed] [Google Scholar]
- 42.Pillai, R. S. 2005. MicroRNA function: multiple mechanisms for a tiny RNA? RNA 111753-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rehmsmeier, M., P. Steffen, M. Hochsmann, and R. Giegerich. 2004. Fast and effective prediction of microRNA/target duplexes. RNA 101507-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robey, R., W. Y. Medina-Perez, K. Nishiyama, T. Lahusen, K. Miyake, T. Litman, A. M. Senderowicz, D. D. Ross, and S. E. Bates. 2001. Overexpression of the ATP-binding cassette half transporter, ABCG2 (Mxr/BCrp/ABCP1) in flavopiridol-resistant human breast cancer cells. Clin. Cancer Res. 7145-152. [PubMed] [Google Scholar]
- 45.Roe, D. F., G. L. Craviso, and J. C. Waymire. 2004. Nicotinic stimulation modulates tyrosine hydroxylase mRNA half-life and protein binding to the 3′UTR in a manner that requires transcription. Brain Res. Mol. Brain Res. 12091-102. [DOI] [PubMed] [Google Scholar]
- 46.Saito, Y., G. Liang, G. Egger, J. M. Friedman, J. C. Chuang, G. A. Coetzee, and P. A. Jones. 2006. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell 8435-443. [DOI] [PubMed] [Google Scholar]
- 47.Schiavone, N., P. Rosini, A. Quattrone, M. Donnini, A. Lapucci, L. Citti, A. Bevilacqua, A. Nicolin, and S. Capaccioli. 2000. A conserved AU-rich element in the 3′ untranslated region of bcl-2 mRNA is endowed with a destabilizing function that is involved in bcl-2 down-regulation during apoptosis. FASEB J. 14174-184. [DOI] [PubMed] [Google Scholar]
- 48.Szatmari, I., G. Vamosi, P. Brazda, B. L. Balint, S. Benko, L. Szeles, V. Jeney, C. Ozvegy-Laczka, A. Szanto, E. Barta, J. Balla, B. Sarkadi, and L. Nagy. 2006. Peroxisome proliferator-activated receptor gamma-regulated ABCG2 expression confers cytoprotection to human dendritic cells. J. Biol. Chem. 28123812-23823. [DOI] [PubMed] [Google Scholar]
- 49.To, K. K. W., O. Polgar, L. Huff, M. Kuniaki, and S. E. Bates. 2008. Histone modifications at the ABCG2 promoter following treatment with HDAC inhibitor mirror those in multidrug-resistant cells. Mol. Cancer Res. 6151-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.To, K. K. W., Z. Zhan, and S. E. Bates. 2006. Aberrant promoter methylation of the ABCG2 gene in renal carcinoma. Mol. Cell. Biol. 268572-8585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Valencia-Sanchez, M. A., J. Liu, G. J. Hannon, and R. Parker. 2006. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 20515-524. [DOI] [PubMed] [Google Scholar]
- 52.Volk, E. L., K. M. Farley, Y. Wu, F. Li, R. W. Robey, and E. Schneider. 2002. Overexpression of wild-type breast cancer resistance protein mediates methotrexate resistance. Cancer Res. 625035-5040. [PubMed] [Google Scholar]
- 53.Yekta, S., I. H. Shih, and D. P. Bartel. 2004. MicroRNA-directed cleavage of HOXB8 mRNA. Science 304594-596. [DOI] [PubMed] [Google Scholar]
- 54.Yu, H., S. Stasinopoulos, P. Leedman, and R. L. Medcalf. 2003. Inherent instability of plasminogen activator inhibitor type 2 mRNA is regulated by tristetraprolin. J. Biol. Chem. 27813912-13918. [DOI] [PubMed] [Google Scholar]
- 55.Zhao, Y., E. Samal, and D. Srivastava. 2005. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 436214-220. [DOI] [PubMed] [Google Scholar]
- 56.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 313406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.