Abstract
The ubiquitination of the receptor that mediates signaling induced by the polypeptide pituitary hormone prolactin (PRL) has been shown to lead to the degradation of this receptor and to the ensuing negative regulation of cellular responses to PRL. However, the mechanisms of PRL receptor (PRLr) proteolysis remain largely to be determined. Here we provide evidence that PRLr is internalized and primarily degraded via the lysosomal pathway. Ubiquitination of PRLr is essential for the rapid internalization of PRLr, which proceeds through a pathway dependent on clathrin and the assembly polypeptide 2 (AP2) adaptor complexes. Recruitment of AP2 to PRLr is stimulated by PRLr ubiquitination, which also is required for the targeting of already internalized PRLr to the lysosomal compartment. While mass spectrometry analysis revealed that both monoubiquitination and polyubiquitination (via both K48- and K63-linked chains) occur on PRLr, the results of experiments using forced expression of ubiquitin mutants indicate that PRLr polyubiquitination via K63-linked chains is important for efficient interaction of PRLr with AP2 as well as for efficient internalization, postinternalization sorting, and proteolytic turnover of PRLr. We discuss how specific ubiquitination may regulate early and late stages of endocytosis of PRLr and of related receptors to contribute to the negative regulation of the magnitude and duration of downstream signaling.
Endocytosis of signaling receptors is a major mechanism used by cells to restrict the magnitude and duration of signal transduction induced by extracellular ligands. Ligand-induced endocytosis of cell surface receptors may occur through clathrin-dependent or -independent pathways. The clathrin-dependent pathway links receptors with clathrin-coated vesicles, which are specialized invaginations of the plasma membrane that concentrate receptors that become internalized. This pathway relies on the interaction of the assembly polypeptide 2 (AP2) clathrin adaptor complexes with specific endocytic signals located within the cytoplasmic domain of the receptors (5).
AP2 complexes, which are involved in the assembly of clathrin triskelions at the plasma membrane, are composed of four components, including two adaptin subunits (α and β2) and two smaller subunits (μ2 and σ2); among these subunits, each has different biological functions (34). There are specific endocytic motifs that are essential for receptor clustering on the membrane and clathrin-dependent internalization of receptors. For example, both tyrosine- and leucine-based motifs can be recognized by the AP2 complex through the interaction with its μ2 subunit and with the β2 or α/σ2 hemicomplexes, respectively (5, 8, 13).
The process of receptor endocytosis is not exclusively regulated by the linear motifs located on the cytoplasmic tail of receptors. Posttranslational modification by ubiquitination has also emerged as an important factor in the endocytosis and sorting of surface receptors. Ubiquitin is a 76-amino-acid protein that forms an isopeptide bond between its terminal glycine and a lysine residue of a target substrate. Each individual ubiquitin moiety harbors seven lysine residues, allowing for the formation of ubiquitin chains linked through its internal lysine residues such as K48 and K63, etc. While the overall role of ubiquitination in mediating the internalization of many yeast receptors (6, 21) and mammalian receptors (such as the nerve growth factor receptor TrkA [16], major histocompatibility complex class II proteins [14], growth hormone receptor [42], model fusion proteins [3], and many others) has been reported previously, the requirements of ubiquitination specificity differ substantially between the various receptors.
We have recently demonstrated that linkages of both K48 and K63 types were required for efficient internalization of the alpha/beta interferon receptor chain IFNAR1 (24). We proposed that interaction of specific ubiquitin moieties with diverse linear endocytic motifs within individual receptors is a major mechanism that regulates the rate of receptor endocytosis. However, the specifics of ubiquitin-dependent internalization of given receptors, such as their requirement for a particular type of ubiquitination (mono- versus polyubiquitination) and for different linkages of polyubiquitin chains, await further elucidation. Besides early endocytic steps, cargo receptor ubiquitination also plays an important role in the prevention of the recycling of internalized receptors and in the targeting of these receptors to the lysosome for degradation. Ubiquitinated cargo destined for lysosomal degradation generally remains associated with the endosome as the vesicles mature and decrease their pH, progressing to vesicles of the late endosome and lysosome (11, 41). While any type of polyubiquitination appeared to suffice for the lysosomal targeting of IFNAR1 (24), the role of specific types of ubiquitination for sorting of other signaling receptors is yet to be determined.
Here we aimed to understand how ubiquitination of the receptor for prolactin (PRL), a polypeptide hormone, mediates receptor degradation. PRL is an essential regulator of the growth and maturation of the mammary gland epithelium, which regulates induction and maintenance of lactation. In addition, PRL signaling that is mediated by a cognate receptor (PRL receptor [PRLr]) seems to play an important role in breast cancer; this role is underscored by findings that PRLr is highly expressed in human cancers (reviewed in references 12 and 38). Ubiquitination and degradation of PRLr is mainly mediated by the β-transducin-repeat-containing protein (β-TrCP) E3 ubiquitin ligase that is recruited to PRLr in a manner that requires phosphorylation of Ser349 of the receptor (28). Signaling induced by PRL (but not by a PRLr antagonist) and mediated by catalytic activation of Janus kinase 2 (Jak2) stimulates this Ser349 phosphorylation in addition to ubiquitination, initial internalization, and degradation of PRLr (39). Furthermore, the intracellular localization of Ser349-phosphorylated PRLr in human mammary tissues is consistent with its targeting to the lysosomal compartment (27).
These findings are in line with the hypothesis that ligand-stimulated PRLr ubiquitination leads to PRLr internalization, sorting to the lysosomes, and lysosomal degradation. However, the role of ubiquitination and lysosomes in PRLr internalization and sorting has not been determined. In fact, temperature-sensitive hamster fibroblasts that harbored a functionally deficient ubiquitin-activating E1 enzyme and expressed bovine PRLr did not display any defect in internalization of radiolabeled bovine PRL (29). Furthermore, treatment with proteasomal inhibitors delayed the downregulation of PRLr in a PRL-deficient clone of breast cancer cells. In these cells, ligand treatment led to proteasome-dependent generation of a PRLr fragment that contained an intact extracellular domain (29).
Thus, the goal of our current study was to systematically delineate the mechanisms by which the ubiquitination of PRLr leads to its degradation. We demonstrate that PRLr is degraded primarily through the lysosomal pathway and that efficient clathrin-dependent internalization of PRLr requires both receptor ubiquitination and the function of the AP2 complex. We provide evidence that the ubiquitination of PRLr stimulates its interaction with the AP2 complex. We also demonstrate that polyubiquitination and K63 linkages are required for the internalization, postinternalization sorting, and lysosomal degradation of PRLr.
MATERIALS AND METHODS
Antibodies and immunotechniques.
Commercially available antibodies against Flag (M2 and M1) and clathrin (TD.1; Sigma-Aldrich, St Louis, MO), hemagglutinin (HA) (Y-11 [Santa Cruz Biotechnology, Inc., Santa Cruz, CA], 12CA5 [Roche], and MMS101R [Covance]), ubiquitin (FK2; BIOMOL International LP), PRLr extracellular domain (1A2B1; Invitrogen Corp., Carlsbad, CA), and α-adaptin and β-actin (Affinity BioReagents, Inc., Golden, CO) were purchased. Anti-phospho-β-catenin and anti-β-catenin antibodies were obtained from Cell Signaling (Cambridge, MA). Secondary antibodies conjugated to horseradish peroxidase (HRP) were purchased from Chemicon and Pierce. MG132 and lactacystin were obtained from BIOMOL and methylamine (MA) from Sigma. All the immunoprecipitation and immunoblotting procedures were performed as previously reported (40). Densitometry analysis data were generated and quantitated using Image software (version beta 4.0.2; Scion), and the digital images were prepared using Photoshop 7.0 software (Adobe). Human PRL was purchased from the National Hormone and Peptide program (A. F. Parlow). PRLr antagonist PRLΔ1-9,G129R was produced and purified as previously described (4). Monensin was purchased from Sigma.
Cells and constructs.
293T human embryo kidney cells were propagated and transfected as previously described (28). Descriptions of the plasmids expressing wild-type (WT) Flag-tagged PRLr or the S349A mutant were previously reported (28). The HA-tagged ubiquitin expression constructs were kindly provided by Yosef Yarden (Weizmann Institute, Israel). The open coding region of C-terminally Flag-tagged PRLr (WT or S349A) was amplified by PCR and subcloned into pBABE-puro to generate retroviral vectors pBABE-puro-PRLrWT and pBABE-puro-PRLrS349A. Retroviruses were obtained from 293T cells transfected with these plasmids along with vesicular stomatitis virus-G and polymerase-Gag expression vectors. These retroviruses were used to stably transduce MCF10AΔp53 cells, a human mammary epithelial MCF10A-derivative cell line in which p53 expression is knocked down by RNA interference (a generous gift of Alan Eastman, Dartmouth University) (26). MCF10AΔp53 cells stably expressing PRLr were selected in medium containing puromycin (0.75 μg/ml) and were further maintained in Dulbecco's modified Eagle's medium/F12 medium (DMEM/F12) (1:1) containing 5% fetal bovine serum, 20 ng of epidermal growth factor (EGF)/ml, 0.5 μg of hydrocortisone/ml, 100 ng of cholera toxin/ml, 10 μg of insulin/ml, 0.75 μg of puromycin/ml, and 100 μg of G418/ml.
Transfections were performed with Lipofectamine Plus or Lipofectamine 2000 (Invitrogen Corporation, Carlsbad, CA) or FuGENE 6 (Roche) and analyzed after 48 h according to the manufacturer's recommendations. The knockdown of clathrin heavy chain was performed using a short hairpin obtained from Sigma (MISSION short hairpin RNA [shRNA] plasmid DNA; catalog no. SHDNAC-TRCN0000007982). The efficiency of knockdown was determined by immunoblotting using an anticlathrin antibody in 293T cells that were transfected with the short hairpin construct. The short hairpin constructs directed against β-TrCP (28, 40) and the small interfering RNA against AP2 (3) were previously characterized elsewhere.
Fluorescence-based internalization assay.
This assay measures the loss of cell surface immunoreactivity of epitope-tagged or endogenous receptors by use of an enzyme-linked immunosorbent assay as described previously (3), with the following modifications. Briefly, 293T cells transfected with the various N-terminal HA-epitope-tagged PRLr plasmids and/or pcDNA3 were plated onto 24-well plates. Cells were starved for 1 h in serum-free DMEM, washed twice in 1× phosphate-buffered saline (PBS), starved for an additional 1 h in serum-free DMEM, and chilled on ice for 15 min. Internalization was initiated by incubation of cells with warm (37°C) serum-free DMEM containing PRL (50 ng/ml) for indicated time periods at 37°C and terminated by placing the plates on ice. Cells were washed, blocked, and incubated with anti-HA antibody (or with anti-PRLr antibody when the endogenous receptor was analyzed) for 1 h, washed, and incubated with HRP-conjugated goat anti-mouse secondary antibody (Molecular Probes) for 1.5 h. After extensive washes, cells were incubated with AmplexRed Ultra reagent (10-acetyl-3,7-dihydroxyphenoxazine; Molecular Probes). Aliquots were transferred to black 96-well plates, and fluorescence readings were taken with a Beckman Coulter DTX880 multimode detector using filters (530 nm for excitation and 590 nm for emission). Results were expressed as a percentage of the fluorescence registered prior to internalization after subtraction of the value obtained with mock-transfected cells. Average results of three independent experiments (each experiment was conducted in quadruplicate; results represent means ± standard errors of the means [SEM]) are presented. Where indicated, pretreatment with monensin (100 μM in ethanol for 30 min prior to initiation of internalization) was used to prevent receptor recycling as described elsewhere for EGF receptor (46). In these cases, control cells received equal volumes of vehicle (ethanol).
Reversible cell surface biotinylation.
Cell surface biotinylation was performed as previously described (20, 24, 39). This procedure uses immunoblotting to measure levels of biotinylated proteins that are protected from debiotinylation as a result of being internalized. In brief, MCF10AΔp53 cells (26) that stably express either WT PRLr or the PRLr harboring the S349A mutation were grown in 100-mm-diameter dishes, starved in serum-free DMEM/F12 for 2 h, placed on ice, and washed with ice-cold PBS. Surface biotinylation was performed with 0.25 mg of EZ-Link-Sulfo-NHS-S-S-biotin (Pierce)/ml in 150 mM Na2B4O7 (pH 8.0) for 15 min on ice. The EZ-Link solution was removed, and the excess unreacted biotin was quenched by washing the plates three times with DMEM containing 0.1% bovine serum albumin followed by an additional wash with ice-cold HEPES-buffered saline (HBS; 10 mM HEPES-NaOH [pH 7.4], 150 mM NaCl) containing 0.7 mM CaCl2 and 0.5 mM MgCl2 (HBS+). The input samples and the untreated plates (time point zero) remained on ice, while the other plates were treated with 50 ng of PRL/ml in warm DMEM for the indicated times, allowing for the internalization of PRLr. Following ligand exposure, all the plates were placed on ice, washed with HBS+, and, with the exception of the input sample, subjected to three 30-min incubations with 100 mM sodium 2-mercaptoethanesulfonic acid (MESNA) in 50 mM Tris-HCl (pH 8.6)-100 mM NaCl-1 mM EDTA-0.2% bovine serum albumin to selectively remove biotin remaining at the cell surface. Time point 0 was set using cells that were biotinylated and kept on ice and then treated with MESNA to remove surface biotin. Following the third MESNA treatment, cells were washed two times with HBS+ and any residual MESNA was quenched by a 15-min treatment with ice-cold 120 mM iodoacetamide-HBS+. After two additional washes with ice-cold HBS, cells were lysed in the plate with 60 mM n-octyl β-d-glucopyranoside (Fisher) and 0.1% sodium dodecyl sulfate (SDS) in HBS containing protease inhibitor cocktail. Lysed cells were transferred to a cold labeled tube, incubated on ice for 20 min, and centrifuged at 15,000 rpm for 15 min. Biotinylated proteins were recovered by incubating with immobilized NeutrAvidin resins (Pierce Inc.) overnight. The proteins were washed, boiled in SDS-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, separated by 10% SDS-PAGE, and analyzed by immunoblotting with anti-Flag M2 antibody to detect Flag-tagged PRLr. Densitometry analysis was performed using Image software.
PRLr recycling assay.
To measure the recycling of internalized HA-PRLr expressed in 293T cells, cells were first labeled on ice for 1 h with anti-HA antibody (1/500 dilution). Then, the temperature was shifted to 37°C for 1 h to allow anti-HA PRLr complex internalization in the presence of PRL (50 ng/ml) while control cells were maintained on ice. Cells were then cooled down to 0°C and stripped five to six times with low-pH solution (0.2 M acetic acid, 0.2 M NaCl; pH 2.5) to remove anti-HA antibody remaining at the cell surface. Control experiments showed that 90% of the anti-HA antibody at the cell surface was removed by the acid stripping in both cases (PRLrWT and PRLrS349A). The quantification of the total amount of internalized antibody was done by subtracting the amount of anti-HA antibody at the cell surface before internalization from the amount seen after internalization. The cells were then incubated at 37°C for 1 to 6 h to allow PRLr complexes with anti-HA antibody to recycle to the cell surface. Cell surface levels of anti-HA antibody were quantified by an Amplex Red assay after secondary anti-mouse HRP-conjugated labeling was performed as described above for the protocol for the fluorescence-based internalization assay. The percentages of internalized PRLr that reappeared on the cell surface were calculated and plotted as a graph (see Fig. S4 in the supplemental material).
In vivo ubiquitination assay.
The in vivo ubiquitination assay was performed as previously described (25). Briefly, 293T cells were cotransfected with 1 μg of Flag-tagged PRLr and 4 μg of the indicated ubiquitin vector. As a control, cells were transfected with 5 μg of empty vector. At 40 h posttransfection, plates were placed on ice and washed with ice-cold PBS. Cells were harvested and lysed with two pellet volumes of 2% SDS in Tris-buffered saline. Samples were boiled for 10 min, sonicated for 30 s, and placed on ice. After neutralization of the SDS with 10 volumes of 1% Triton X-100, samples were precleared with protein A agarose (Invitrogen Corporation, Carlsbad, CA) and PRLr was immunoprecipitated using anti-Flag antibody-conjugated beads. After the beads were washed, the samples were boiled in SDS-PAGE loading buffer and a fraction from each sample was analyzed by immunoblotting with the anti-FLAG antibody to normalize receptor amounts. Equal levels of expressed receptor were analyzed using the antiubiquitin antibody.
PRLr-AP2 interactions.
293T cells were cotransfected with 1 μg of the indicated Flag-tagged PRLr construct (WT or S349A) and either 3 μg of shRNA constructs (either a control short hairpin or a short hairpin against β-TrCP) when investigating the role of ubiquitination or with 3 μg of an indicated ubiquitin construct when investigating the role of the type of ubiquitination. As a control for these experiments, cells were transfected with either an equivalent amount of empty vector or a short hairpin control empty vector where applicable. At 48 h posttransfection, cells were starved in serum-free DMEM containing 20 mM MA hydrochloride for 4 h followed by stimulation with PRL at 37°C. The samples were prepared as previously described (24), and equal amounts of lysates were precleared and incubated with anti-Flag antibody-conjugated beads overnight. The beads were washed with lysis buffer, resuspended in 3× SDS-PAGE loading buffer, and boiled. An aliquot from each sample was analyzed by immunoblotting with the anti-Flag antibody to normalize receptor levels. Based on the normalized amounts of PRLr, equivalent amounts from each sample were separated by 6% SDS-PAGE and analyzed by immunoblotting with the anti-α-adaptin antibody and with the anti-Flag antibody to detect PRLr. Also, equal amounts of whole-cell extract (reserved from the immunoprecipitation reaction mixture) were separated by 8% SDS-PAGE and analyzed with the anti-α-adaptin antibody.
Identification of the nature of PRLr ubiquitination by mass spectrometric analysis (AQUA method).
For purification of the ubiquitinated species of PRLr, 50 10-cm-diameter dishes of 293T cells were used for transfection and coexpression of WT ubiquitin and Flag-tagged PRLr or a vector control by the calcium phosphate method. Purification of ubiquitinated PRLr was done similarly to purification of ubiquitinated IFNAR1 as previously described (24). Briefly, cells were treated with PRL (50 ng/ml for 30 min) and lysed using NP-40 lysis buffer containing 200 mM NaCl, protease inhibitors, and 10 mM NEM (N-ethyl maleimide). Total cell lysate was precleared with protein A-agarose for 4 h at 4°C followed by immunoprecipitation of 50 mg of total protein immobilized on beads conjugated to the anti-Flag M2 antibody. The beads were washed sequentially with buffers containing 1% NP-40 with 1 M NaCl and 0.1% SDS. Subsequent washes were carried out using lower concentrations of NaCl (200 mM NaCl and then 100 mM NaCl) and finally using Tris-buffered saline. The precipitated proteins were eluted with 0.1 M glycine (pH 3.0) and resolved by SDS-PAGE. A 1% volume of the eluate was analyzed by immunoblotting with an antiubiquitin antibody (FK2; BIOMOL) to determine the apparent molecular weight of “ubiquitinated” receptor, and with an anti-FLAG M2 antibody, 4% of the eluate was analyzed by silver staining and 70% by colloidal Coomassie (Invitrogen) staining.
Gel slices containing the “ubiquitinated” receptor and the corresponding region in the vector control lane representing the Coomassie-stained gel were used for analysis of polyubiquitin linkages according to previously established methods (23, 47). Briefly, the proteins in the gel slices were digested by trypsin with the addition of eight stable isotope-labeled peptides, including all seven GG peptides and one ubiquitin peptide (Thr55-Lys63), as internal standards. The resulting peptide mixtures were analyzed by reverse-phase liquid chromatography-tandem mass spectrometry (liquid chromatography-MS-MS) on an LTQ-Orbitrap hybrid mass spectrometer (Thermo Electron, San Jose, CA). The instrument was operated to monitor the eight peptides and their related native counterparts by selective reaction monitoring (SRM). The quantification analysis was carried out using Xcalibur software (Thermo Finnigan, San Jose, CA).
Cycloheximide chase assay.
293T cells (transfected with the indicated plasmids for 48 h) or T47D cells were serum starved overnight followed by stimulation with PRL (20 ng/ml or 100 ng/ml, respectively) in the presence of 50 μg of cycloheximide/ml. Cells were harvested at different time points, lysed, and analyzed by immunoblotting using anti-Flag (M2) or anti-PRLr antibodies. Digital images were processed with Adobe Photoshop 7.0 software.
pHv measurement.
To monitor the endocytic trafficking of internalized PRLr, the vesicular pH (pHv) of cargo-containing vesicles was determined by fluorescence ratio image analysis (FRIA) essentially as described for CD4 or IFNAR1 (3, 24). Cell surface PRLr were labeled with anti-HA primary antibody (Covance MMS101R) and fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse secondary Fab (Jackson ImmunoResearch Laboratories, West Grove, PA) by incubating primary (1/500 dilution) and secondary (1/500 dilution) antibodies together routinely for 1 h at 37°C in the presence of 50 ng of PRL (WT or antagonist)/ml. The presence of PRL did not affect binding of the antibodies used in this analysis (data not shown). Cells were then washed (140 mM NaCl, 5 mM KCl, 20 mM HEPES, 10 mM glucose, 0.1 mM CaCl2, 1 mM MgCl2; pH 7.3) and chased for 30 to 240 min at 37°C. Fluid-phase antibody uptake in mock-transfected cells was not detectable by FRIA (data not shown).
FITC-Fab antibody-loaded endolysosomes were imaged at 35°C using an Axiovert 100 inverted fluorescence microscope (Carl Zeiss MicroImaging, Inc., Toronto, Ontario, Canada) (at 35°C) equipped with a Hamamatsu ORCA-ER 1394 (Hamamatsu, Japan) cooled charge-coupled-device camera and a Planachromat (63×, 1.4-numerical-aperture) objective. Fluorescence ratio image acquisition and analysis were performed with MetaFluor software (Molecular Devices, Downingtown, PA). Images were acquired at 490-nm (± 5 nm) and 440-nm (± 10 nm) excitation wavelengths by use of a 535-nm (± 25 nm) emission filter.
Both primary and secondary antibodies remained associated with PRLr throughout the course of the experiments, judging from the inability of buffers with pH of 4.0 and higher to dissociate these antibodies in control experiments (data not shown). Calibration curves, describing the relationship between the fluorescence ratio values and pHv, served to calculate the luminal pH of individual vesicles, following fluorescence background subtraction at both excitation wavelengths. In situ calibration was performed by clamping the pHv between 4.5 and 7.4 in K+-rich medium (135 mM KCl, 10 mM NaCl, 20 mM HEPES or 20 mM MES [morpholineethanesulfonic acid], 1 mM MgCl2, and 0.1 mM CaCl2) in the presence of 10 μM nigericin and 10 μM monensin (Sigma-Aldrich, Oakville, Ontario, Canada) and recording the fluorescence ratio. In each experiment the pHv of 450 to 800 vesicles was determined. As an internal control, one-point calibration was performed for each coverslip by clamping the pHv to 6.5 with monensin and nigericin. Mono- or multipeak Gaussian distributions of pHv values were obtained with Origin 7.0 software (OriginLab Corporation, Northampton, MA). The mean pHv of each vesicle population was calculated as the arithmetic mean of the data and was identical to the Gaussian mean, based on single-peak distribution fitting. At least three independent experiments were performed for each set of conditions. To override the effect of endogenous ubiquitin and to detect the effect of the ubiquitin configuration on endocytic trafficking, 293T cells were cotransfected with ubiquitin variants and the expression plasmid coding for PRLr at a 5:1 ratio. FRIA was performed on cells expressing PRLr while assuming that the twos plasmids were expressed together. The mean pHv values obtained in at least three experiments are shown in the figures.
Immunofluorescence microscopy.
Lysosomes were labeled with FITC-dextran (50 μg/ml; molecular mass, 10 kDa) as described previously for CD4 or IFNAR1 (3, 24). Cells expressing HA-PRLr were allowed to internalize anti-HA antibody complexed with FITC-conjugated goat anti-mouse Fab for 1 h in DMEM and were chased for 4 h. During the last 45 min, cells were labeled with tetramethyl rhodamine isocyanate-transferrin (TRITC-transferrin) (10 μg/ml) to visualize recycling endosomes. Single optical sections were collected by use of a Zeiss LSM510 laser confocal fluorescence microscope, equipped with a Plan-Apochromat 63×/1.4 objective (Carl Zeiss Microimaging, Inc.), as previously described (2). Images were processed with Adobe Photoshop (Adobe Systems Incorporated, San Jose, CA) software.
RESULTS
Degradation of PRLr is primarily a lysosome-dependent process.
Given somewhat controversial data on the importance of lysosomal versus proteasomal pathways in the degradation of PRLr, we treated T47D human breast cancer cells with either the proteasome inhibitor MG132 or MA, an inhibitor of lysosomal delivery, and assessed the degradation of endogenous PRLr by use of a cycloheximide chase assay. Treatment with MA completely abrogated the proteolytic turnover of PRLr, while MG132 treatment only moderately inhibited the degradation of this receptor (Fig. 1A). Intriguingly, lactacystin, a more specific inhibitor of proteasomes, failed to noticeably impair the rate of PRLr degradation (Fig. 1B) while being active in stabilizing β-catenin phosphorylated on Ser33 (see Fig. S1 in the supplemental material), which is a bona fide proteasomal substrate (1). In addition, treatment with MA to inhibit the lysosomal pathway also prevented PRL-stimulated degradation of endogenous PRLr in human embryo kidney 293T cells (see Fig. S2 in the supplemental material). These data, which are in line with our results with respect to the degradation of exogenously expressed PRLr in these cells as obtained by pulse-chase analysis (27), suggest that the proteolytic turnover of PRLr is primarily mediated by the lysosomal pathway.
FIG. 1.
Effect of proteasomal and lysosomal inhibitors on the degradation of endogenous PRLr in human T47D cells. (A) Degradation of endogenous PRLr was analyzed by a cycloheximide (CHX) chase procedure (treatment with CHX [50 μg/ml] and PRL [100 ng/ml]) using T47D cells following 2 h of pretreatment with either the proteasomal inhibitor MG132 (MG [25 μM]) or the lysosomal inhibitor MA (10 mM). Cell lysates were analyzed by immunoblotting with antibodies against PRLr and β-actin (β-act) (loading control). DMSO, dimethyl sulfoxide. (B) Degradation of PRLr following pretreatment with the proteasomal inhibitor lactacystin (LC [10 μM]) or MA (10 mM) or a combination of the two was analyzed as described for panel A.
Ubiquitination is required for the efficient clathrin-dependent internalization of PRLr.
Considering that ubiquitination of PRLr is important for its degradation (27), which appears to proceed via the lysosomal pathway (Fig. 1), we sought to investigate how ubiquitination of PRLr contributes to its lysosomal turnover. Efficient internalization of cell surface receptors is a common first step toward their degradation in the lysosomes. We used two independent methods (fluorescence-based assays and cell surface biotinylation assays; see Materials and Methods and references 24 and 39) to characterize the processes that control the initial rate of PRLr internalization. Use of shRNA that efficiently knocked down the expression of clathrin heavy chain (see Fig. S3 in the supplemental material) noticeably impaired the internalization of both endogenous PRLr in 293T cells (Fig. 2A) and HA-tagged PRLr expressed in these cells (Fig. 2B). These results indicate that PRLr internalization is clathrin dependent and confirm our previous observations that in this experimental system, endocytosis of recombinant HA-tagged PRLr faithfully recapitulates the processes involved in the regulation of the endogenous receptor (39).
FIG. 2.
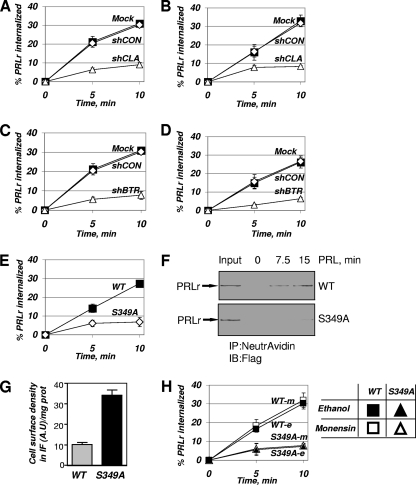
Efficiency of the clathrin-dependent internalization of PRLr is dependent on PRLr ubiquitination. (A) Internalization of endogenous PRLr in 293T cells that were left untransfected (Mock [squares]) or were transfected with shRNA against clathrin heavy chain (shCLA) or green fluorescent protein (shCON [diamonds]) analyzed by the fluorescence-based assay using anti-PRLr antibody as outlined in Materials and Methods. (B) Internalization of HA-tagged PRLrWT exogenously expressed in 293T cells that were left untransfected (Mock [squares]) or were transfected with shRNA against heavy-chain clathrin (shCLA) or green fluorescent protein (shCON [diamonds]) analyzed by the fluorescence-based assay using anti-HA antibody as outlined in Materials and Methods. (C) Internalization of endogenous PRLr in 293T cells that were left untransfected (Mock [squares]) or transfected with shRNA against β-TrCP2 (shBTR) or green fluorescent protein (shCON [diamonds]) analyzed as outlined for panel A. (D) Internalization of HA-tagged PRLrWT exogenously expressed in 293T cells that were left untransfected (Mock [squares]) or transfected with shRNA against β-TrCP2 (shBTR) or green fluorescent protein (shCON [diamonds]) analyzed as outlined for panel B. (E) Internalization of a WT or ubiquitination-deficient S349A mutant of HA-tagged PRLr expressed in 293T cells was analyzed as described for panel B. (F) Internalization of a WT or ubiquitination-deficient S349A mutant of Flag-tagged PRLr stably expressed in MCF10AΔp53 human mammary epithelial cells was analyzed by reversible biotinylation as outlined in Materials and Methods. Results of immunoblotting (IB) analysis of proteins pulled down with Neutravidin beads by use of anti-Flag M2 antibody are shown. “Input” denotes the samples that did not undergo debiotinylation. IP, immunoprecipitation. (G) PRLr cell surface density was determined using anti-HA antibody and secondary anti-mouse HRP-conjugated antibody binding at 0°C in 293T cells expressing either the WT or the S349A mutant. Data (given in arbitrary units [A.U]) were obtained by subtracting the nonspecific background (nonspecific M1 antibody binding to mock-transfected 293T cells), were normalized per total levels of cellular protein (prot), and are presented as means ± SEM [n = 3]. (H) Rates of internalization of WT or ubiquitination-deficient S349A mutant HA-tagged PRLr expressed in 293T cells pretreated with 100 μM monensin (-m) or vehicle (ethanol) (-e) were analyzed as described for panel B.
In line with previous results, the initial rates of internalization of both endogenous PRLr (Fig. 2C) and HA-tagged PRLr (Fig. 2D) were dramatically impaired in cells in which β-TrCP was knocked down. Given that β-TrCP is a major E3 ubiquitin ligase for PRLr, this result may suggest that ubiquitination of PRLr is required for the efficient endocytosis of PRLr. Alternatively, knockdown of β-TrCP may affect PRLr internalization indirectly via the numerous other known β-TrCP targets (15, 48). To distinguish between these possibilities and to directly determine the role of PRLr ubiquitination in its internalization, we compared the initial rates of internalization of WT PRLr and of its ubiquitination-deficient S349A mutant, which does not efficiently recruit β-TrCP (28). Remarkably, this mutant exhibited much slower kinetics of internalization in 293T cells (Fig. 2E). This defect could not be attributed to an artifact of transient expression of PRLr or to a peculiarity of the fluorescence-based assay, because similar data were obtained independently using the reversible cell surface biotinylation assay and MCF10AΔp53 human mammary epithelial cell lines stably transduced with the relevant PRLr expression constructs. In these cells, internalization of the PRLrS349A mutant was noticeably delayed compared to the internalization of WT receptor (Fig. 2F). Accordingly, while we aimed at expressing comparable levels of total PRLr in these experiments (for an example, see Fig. 3A, left panel), we still detected three times more of the ubiquitination-deficient mutant (compared to the WT PRLr level) at the cell surface (Fig. 2G).
FIG. 3.
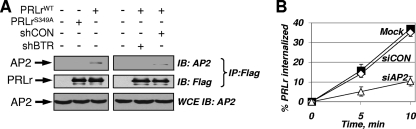
Ubiquitination of PRLr promotes its interaction with components of AP2, which are required for PRLr internalization. (A) Interactions between Flag-tagged PRLr (WT or S349A mutant) and the endogenous α subunit of the AP2 complex in 293T cells (transfected with the indicated plasmids and treated with PRL [50 ng/ml for 5 min]) were analyzed by immunoprecipitation (IP) using M2 anti-Flag antibody followed by immunoblotting (IB) using the indicated antibodies. Material used for immunoprecipitation was normalized to yield comparable levels of PRLr in all lanes. Levels of the α subunit in whole-cell extracts (WCE) were also assessed. (B) Internalization of endogenous PRLr in 293T cells that were left untransfected (Mock [squares]) or were transfected with small interfering RNA against the α-adaptin subunit of AP2 (shAP2) or luciferase (siCON [diamonds]) was analyzed by the fluorescence-based assay using anti-PRLr antibody as outlined for Fig. 2A.
High levels of surface PRLr in the PRLrS349A-expressing cells may result not only from deficient internalization but also from the increased recycling of mutant receptor that we have also observed (albeit at much later time points) (see Fig. S4 in the supplemental material). In order to exclude the possibility that our interpretation of internalization assays could have been affected by receptor recycling, we used monensin, a well-known inhibitor of this process, because this inhibitor has been extensively used to differentiate internalization and recycling phenotypes of various receptors, including EGF receptor (46). Given that pretreatment with monensin did not rescue the endocytic phenotype of PRLrS349A (Fig. 2H), these data together suggest that the ubiquitination of PRLr is required for its efficient initial internalization, which mainly proceeds via a clathrin-dependent pathway.
Ubiquitination of PRLr stimulates its interaction with the AP2 adaptor complex.
How does the ubiquitination of PRLr stimulate the internalization of this receptor via the clathrin-dependent pathway? Clathrin-coated pits associate with cargo through adaptor protein complexes (e.g., AP2) which interact with the cargo and membrane to form an invagination (5). Our previous findings revealed that ubiquitination of IFNAR1 stimulates its endocytosis by promoting the recruitment of the AP2 components. Intriguingly, while interaction of the WT Flag-tagged PRLr with the endogenous α-adaptin subunit of AP2 was detected by coimmunoprecipitation from the lysates of 293T cells, such interaction was much less pronounced when it involved a ubiquitination-deficient PRLrS349A mutant (Fig. 3A).
Furthermore, knockdown of β-TrCP also decreased the interaction between Flag-tagged PRLr and α-adaptin of the AP2 complex (Fig. 3A). Given that knockdown of α-adaptin dramatically suppressed the rate of PRLr internalization (Fig. 3B), these results, taken together, are consistent with the idea that ubiquitination may stimulate PRLr endocytosis by promoting the interaction of PRLr with AP2, which is essential for mediating efficient internalization of PRLr via the clathrin-dependent pathway.
Ubiquitination of PRLr is required for postinternalization sorting.
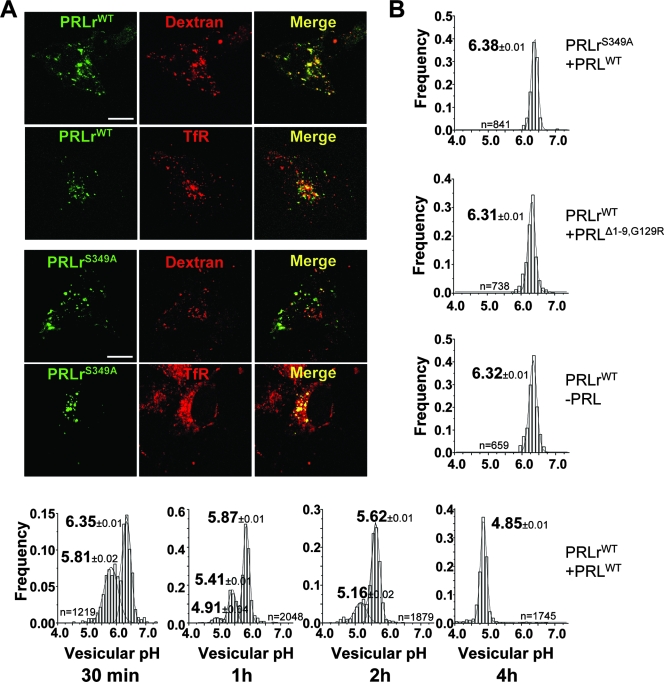
We then sought to investigate whether receptor ubiquitination plays a regulatory role in the postinternalization sorting of PRLr. Immunocytochemical analysis showed that internalized WT PRLr mostly colocalized with fluorophor-conjugated dextran (targeted to the late endosomes/lysosomes; see references 3 and 24). However, PRLr colocalized to a noticeably lesser extent with transferrin (which marks early/recycling endosomes). PRLrS349A, a receptor deficient in ubiquitination, displayed an opposite pattern of colocalization with transferrin but not with dextran (Fig. 4A). These results, along with the previously demonstrated increased ability of the PRLrS349A protein to recycle back to cell surface after internalization (see Fig. S4 in the supplemental material), indicate that ubiquitination of PRLr might be required for efficient targeting of this receptor to the lysosomes.
FIG. 4.
Efficient postinternalization sorting of PRLr into the lysosomes requires PRLr ubiquitination. (A) Localization of internalized HA-tagged WT PRLr or PRLrS349A ubiquitin-deficient mutant. These proteins were internalized for 1 h in the presence of PRL, anti-HA antibody, and FITC-conjugated secondary Fab followed by a 4-h chase procedure. Lysosomes and recycling endosomes were labeled with TRITC-dextran and TRITC-transferrin (TfR), respectively. Dextran completely colocalized with Lamp1 (reference 24 and data not shown). Single optical sections of representative cells obtained by laser confocal fluorescence microscopy are depicted. Bar, 10 μm. (B) pHv of HA-tagged PRLr-containing endosomes/lysosomes at indicated time points following a postinternalization chase procedure using 293T cells transfected with the indicated HA-PRLr constructs (WT or S349A PRLr) and treated (or not) with the indicated ligands (WT PRL or its antagonist, PRLΔ1-9,G129R). Following the internalization and chase procedures described in Materials and Methods, the endosomal pH was measured by FRIA and the frequency distribution of the pHv among the indicated numbers of analyzed vesicles (n) was plotted. Means ± SEM (representing the results obtained with three independent experiments) are also denoted.
To validate these findings we used quantitative FRIA (see Materials and Methods and references 2, 24, and 35). This analysis, which is based on monitoring the luminal pH of cargo-containing vesicles (pHv) and relies on the distinct pHv of early/recycling endosomes (pH of approximately 6.4 to 6.5) and late endosomes/lysosomes (pH < 5.5), was used to determine the efficiency of sorting of internalized PRLr from early endosomes to the lysosomal compartment. As shown in Fig. 4B, the majority of vesicles containing WT PRLr shifted their pH to an acidic (pH 4.85) condition within 4 h of treatment of cells with PRL. Our previous reports indicated that either incubating cells without PRL or substituting WT PRL for the signaling-deficient mutant PRLΔ1-9,G129R (17) decreased the extent of Ser349 phosphorylation (39) and of PRLr ubiquitination. Remarkably, the efficiency of PRLr targeting to the lysosomes was decreased in cells that received no PRL or PRLΔ1-9,G129R (Fig. 4B), indicating that PRLr ubiquitination might be involved in regulating this process. Indeed, under the conditions with which cells were treated with WT PRL, the delivery of ubiquitination-deficient PRLrS349A mutant to the acidic lysosomal compartments was substantially impaired (Fig. 4B). These data indicate that ligand-induced ubiquitination of PRLr plays an important role in the postinternalization sorting of this receptor to the lysosomes.
K63-linked polyubiquitination regulates PRLr internalization, postinternalization sorting, and degradation.
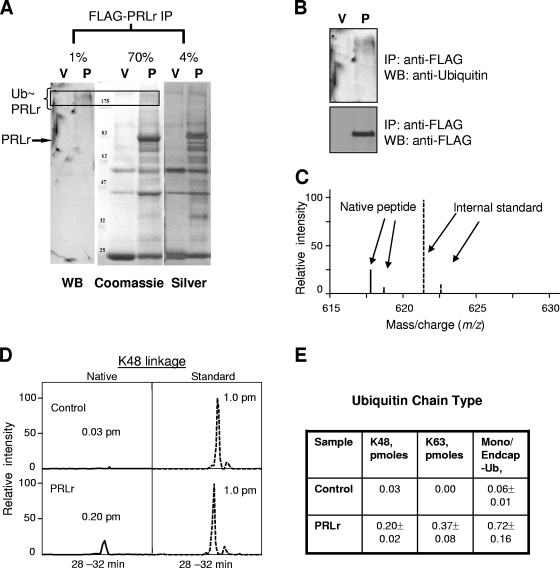
Proteasomal degradation of ubiquitinated proteins usually requires polyubiquitination via K48-linked chains. Given that different types of ubiquitination (mono- versus polyubiquitination via different types of linkages) may play a role in the lysosomal degradation of various receptors, we next sought to investigate what type of ubiquitination occurs with PRLr. To this end, Flag-tagged PRLr was expressed in cells, purified under stringent conditions (Fig. 5A and B), and subjected to direct determination of levels of ubiquitinated species by using a quantitative MS approach (see Materials and Methods and reference 24). While limited coverage of PRLr-derived peptides did not allow the identification of the lysine residues within its intracellular domain that serve as ubiquitin acceptors (data not shown), a sufficient amount of information regarding the peptides which originated from ubiquitin sequences was retrieved (see Fig. 5C and D for an example of characterization and quantification of K48-linked chains). While both mono- and polyubiquitination were revealed with PRLr, among polyubiquitin linkages K48- and K63-linked chains were predominant and the latter linkages constituted a major fraction (Fig. 5E).
FIG. 5.
MS analysis of the nature of ubiquitin chain linkages for PRLr. (A) 293T cells were transfected with Flag-tagged PRLr (P) or pcDNA3 empty vector (V). PRLr was immunoprecipitated (IP) using Flag M2 antibody as described in Materials and Methods. Results represent analysis of 1%, 4%, and 70% of Flag-immunoprecipitated material obtained by immunoblotting (WB) with antiubiquitin antibody, silver staining, and colloidal Coomassie staining, respectively. The position of unmodified PRLr is indicated by an arrow. The region corresponding to ubiquitin-PRLr (Ub∼PRLr) that was used for MS analysis is demarcated. (B) The fraction used to demonstrate the presence of the ubiquitin moiety in the analyzed material was examined by immunoblotting using anti-Flag antibody to detect expression of Flag-PRLr. (C) Representative quantification of K48 linkage based on liquid chromatography-SRM. A spectrum of SRM for the PRLr sample, the internal standard GG peptide, was labeled with a heavy stable isotope, and the results are shown as two peaks at 621.6 and 622.6 m/z (dashed lines). The native peptide corresponding to the K48 linkage was displayed as two additional peaks at 617.9 and 618.9 m/z (solid lines). (D) To quantify the native and standard peptide levels, their elution profiles (intensity versus retention time) were generated for a comparison. The peak area reflected the greatest abundance of the peptides. An example of the results of analysis of K48 linkages is depicted. (E) Results of quantification of types and quantities of ubiquitin chains (given in pmoles) appended to PRLr by the MS-based AQUA method as described in Materials and Methods are presented.
Given that the portions of gel taken for analysis spanned most of the ubiquitinated PRLr (Fig. 5A), it is likely that the detected pattern is representative of the overall population of ubiquitinated PRLr species. However, we cannot exclude the possibility that these results reveal characteristics of a specific subset of modified receptors. To further determine the role of linkage-specific polyubiquitination in PRLr internalization, sorting, and degradation, we used forced expression of ubiquitin mutants in 293T cells to overcome the effect of the presence of endogenous ubiquitin (see Fig. S7 in the supplemental material). Despite obvious shortcomings, this approach is the only one currently available and has become standard for delineating the role of diverse ubiquitin chains in receptor internalization and turnover (14, 19, 32). Expression of the lysine-free (K0) ubiquitin mutant, which is incapable of forming polyubiquitin chains, decreased the efficiency of PRLr ubiquitination (Fig. 6A). We next investigated the effect of the presence of this ubiquitin mutant in the degradation of Flag-tagged PRLr expressed in 293T cells under conditions that clearly allowed for the degradation of ubiquitination-competent WT PRLr while prohibiting the turnover of ubiquitination-deficient PRLrS349A mutant (see Fig. S5 in the supplemental material). Expression of the K0 mutant noticeably impaired the rate of degradation of PRLr (Fig. 6B and C) compared to the results seen with cells expressing WT ubiquitin, which by itself did not stimulate PRLr degradation (see Fig. S6 in the supplemental material) despite being expressed and stimulating the overall extent of ubiquitination in cells (see Fig. S7 in the supplemental material). These results suggest that polyubiquitination may be required for the efficient degradation of PRLr.
FIG. 6.
Polyubiquitination of PRLr promotes its internalization, postinternalization sorting, and lysosomal degradation. (A) Ubiquitination of Flag-tagged PRLr coexpressed with WT ubiquitin (Ub) or K0 ubiquitin mutant in 293T cells analyzed by denaturing immunoprecipitation using M2 antibody followed by immunoblotting (IB) using anti-HA antibody. Levels of PRLr were also analyzed by immunoblotting with M2 antibody (lower panel). (B) Expression of the K0 ubiquitin mutant slows down the degradation of coexpressed Flag-PRLr as measured by a cycloheximide (CHX) chase procedure followed by immunoblotting using M2 antibody. Levels of β-actin (loading control) were also analyzed. (C) Average results from four experiments similar to that represented by panel B are depicted as percentages of PRLr remaining at the indicated time points of a chase procedure. (D) Interaction of Flag-tagged PRLr with an endogenous α-adaptin complex in 293T cells expressing the indicated ubiquitin constructs was analyzed by coimmunoprecipitation (IP) as described for Fig. 3A. The results obtained with a fraction of α-adaptin bound to PRLr (per mille) are depicted in the bottom panel, Vec, vector; IB, immunoblotting; WCE, whole-cell extract. (E) Internalization of HA-tagged PRLr coexpressed with either WT ubiquitin or K0 mutant in 293T cells was analyzed as described for Fig. 2B. (F) pHv of HA-tagged PRLr-containing endosomes/lysosomes at 4 h of a chase procedure using 293T cells coexpressing the indicated ubiquitin constructs was analyzed as described for Fig. 4B.
Given that AP2 is required for mediating PRLr internalization (Fig. 3B), we next assessed the effect of the K0 mutant on the interaction of PRLr with α-adaptin. Expression of this K0 mutant not only decreased the recruitment of α-adaptin to PRLr (Fig. 6D) but also noticeably delayed the initial internalization of PRLr (Fig. 6E). In addition, expression of the K0 mutant prevented efficient sorting of the already internalized PRLr to the acidic lysosomal compartments (Fig. 6F). In all, these data indicate that polyubiquitination promotes the lysosomal degradation of PRLr by stimulating its interaction with AP2 followed by promoting both early and late events in PRLr endocytosis and lysosomal targeting.
We next sought to investigate what types of polyubiquitin linkages are important for PRLr internalization, sorting, and degradation. Consistent with the MS data showing that PRLr possesses both K48- and K63-linked chains (Fig. 5E), expression of either K63R or K48R ubiquitin mutants did not affect the overall levels of PRLr ubiquitination (Fig. 7A). Surprisingly, expression of K63R led to a much more pronounced delay in the degradation of PRLr than of K48R (Fig. 7B and C). These data suggest that K63 linkages may play an important role in PRLr degradation. Indeed, while expression of either R63K or R48K reverse mutants (in which all other lysines are converted to arginines) decreased the overall PRLr ubiquitination to similar extents (Fig. 7A), the availability of K63-linked chains in cells expressing R63K mutant allowed for a higher rate of PRLr degradation (Fig. 7D and E). These data together indicate that K63-linked polyubiquitin chains play an important role in PRLr turnover.
FIG. 7.
K63 polyubiquitin linkages are required for efficient internalization, sorting, and degradation of PRLr. (A) Overall ubiquitination of Flag-tagged PRLr coexpressed with the indicated ubiquitin (Ub) constructs in 293T cells was analyzed by denaturing immunoprecipitation (IP) using M2 antibody followed by immunoblotting (IB) using anti-HA antibody (which recognizes HA-tagged ubiquitin). Amounts of material subjected to immunoprecipitation were normalized in order to load comparable levels of PRLr in all lanes (lower panel). VEC, vector. (B) Degradation of Flag-tagged PRLr coexpressed with the indicated direct ubiquitin mutants in 293T cells was analyzed as described for Fig. 6B. CHX, cycloheximide; β-Act, β-actin. (C) Quantitative analysis of three independent experiments similar to that represented by Fig. 7B. The results of the analysis are presented as percentages of PRLr remaining at indicated time points of a chase procedure. (D) Degradation of Flag-tagged PRLr coexpressed with the indicated reverse ubiquitin mutants in 293T cells was analyzed as in Fig. 6B. (E) Quantitative analysis of four independent experiments similar to that represented by Fig. 7D are presented as percentages of PRLr remaining at indicated time points of a chase procedure. (F) Internalization of HA-PRLr coexpressed with the indicated direct (upper panel) and reverse (lower panel) ubiquitin mutants was measured as described for Fig. 2B. WT ubiquitin, squares; K48R, open triangles; R63K, open circles. (G) pHv of HA-tagged PRLr-containing endosomes/lysosomes at 4 h of chase in 293T cells coexpressing the indicated ubiquitin constructs was analyzed as described for Fig. 4B.
As is consistent with the role of the ubiquitination of PRLr in its interaction with AP2 and in internalization, we have also found that expression of ubiquitin mutants (K0, K63R, and R48K) that disrupt K63 linkages decreased the extent of PRLr-α-adaptin interaction whereas expression of ubiquitin mutants that affect K48 chains (K48R or R63K) did not have this effect (Fig. 6D). Furthermore, expression of K63R or R48K mutants noticeably slowed the initial rate of PRLr internalization (Fig. 7F). Additionally, expression of the R48K mutant (but not expression of the R63K mutant) noticeably decreased the efficacy of targeting of already internalized PRLr to the acidic compartments (Fig. 7G). These results indicate that the formation of K63 (but not K48)-linked polyubiquitin chains is important for PRLr degradation via stimulation of both internalization of this receptor and its postinternalization sorting to the lysosomes.
DISCUSSION
Ubiquitination of a protein substrate may affect the stability of this substrate either by recruiting it to the proteasomes or by triggering the chain of trafficking events that bring a membrane-bound protein into the lysosomal compartment. Here we have delineated the mechanisms by which ubiquitination of a receptor for the polypeptide hormone PRL leads to the downregulation and degradation of this receptor, thereby allowing a ligand-specific mechanism for the negative regulation of PRL signaling. We present biochemical and genetic evidence that suggests that PRLr is mono- and polyubiquitinated (via chains of diverse topology) in cells and that ubiquitination of PRLr leads to its lysosomal degradation not only by stimulating the recruitment of the AP2 complex, as is required for clathrin-dependent initial internalization of PRLr, but also by increasing the targeting efficiency of already internalized PRLr to the lysosomes.
To the best of our knowledge, this report is the first to demonstrate a topology-specific role of polyubiquitination in the internalization of PRLr, a polypeptide hormone receptor. This role has been confirmed here by two different approaches (downregulation of E3 and use of ubiquitination-deficient PRLr mutants) and two different methodologies (fluorescence-based assay and reversible cell surface biotinylation). Although endocytosis of bovine PRLr expressed using the Chinese hamster lung cell line ts20, which contains E1, a thermolabile ubiquitin-activating enzyme, was not affected (29), it is still possible that in these cells, ubiquitination of PRLr was mediated by a recently discovered alternative E1 version (9, 22).
While ubiquitination of closely related erythropoietin receptor and growth hormone receptors has been convincingly demonstrated, available data in the literature question the role of ubiquitination of these receptors in their internalization. Remarkably, in similarity to PRLr, β-TrCP E3 ubiquitin ligase also plays a major role in regulating the stability of erythropoietin receptor (31) and growth hormone receptor (43). Although recruitment of β-TrCP to the latter receptor promotes its downregulation, the role of growth hormone receptor ubiquitination per se in this process remains to be elucidated. Furthermore, while catalytic activation of Jak2 by PRL is required not only for the stimulation of PRLr ubiquitination but also for PRLr internalization and degradation (39), erythropoietin-stimulated Jak2 is required for ubiquitination and degradation but is dispensable for internalization of its receptor (45). Furthermore, here we show that PRLr ubiquitination and stimuli that promote this ubiquitination play an important role in the lysosomal targeting of already internalized PRLr. While a similar role of ubiquitination in postinternalization sorting of erythropoietin and growth hormone receptors has been proposed, it remains to be formally demonstrated.
Our analysis has shown that interfering with K63- but not with K48-linked polyubiquitination impedes the internalization, sorting, and degradation of PRLr. Although one cannot rule out the possibility that K63-linked polyubiquitination might affect these processes indirectly, these data, together with analysis of PRLr degradation in the presence of specific proteasomal inhibitors, raise serious doubts regarding a major role of proteasomes (which usually require K48-linked chains) (10) in the proteolytic fate of PRLr. Interpretation of previously reported effects of proteasome inhibitors on the degradation of either PRLr (29) or erythropoietin/growth hormone receptors (42, 45) should include consideration of the possibility of an indirect effect of these inhibitors on the lysosomal pathway, which could be suppressed via depletion of the ubiquitin pool (37). Interestingly, we have never observed a ligand-stimulated formation of a PRLr fragment either when following the fate of endogenous PRLr in T47D or 293T cells or when studying recombinant N-terminally HA-tagged or C-terminally Flag-tagged proteins. Further studies are required to determine the nature of such a fragment, which has been reported to have appeared in a MCF7 variant clone (29).
Intriguingly, in addition to numerous similarities between the role of ubiquitination in the internalization of IFNAR1 and its role in the internalization of PRLr, there are also apparent differences that relate mainly to the type of ubiquitination. Internalization and postinternalization of both receptors require polyubiquitination (this article and reference 24). However, while IFNAR1 internalization requires both K48 and K63 linkages, PRLr internalization relies mainly on K63-conjugated chains. Furthermore, these chains are also needed for the postinternalization sorting of PRLr (Fig. 7) whereas any type of polyubiquitination suffices for the lysosomal targeting of IFNAR1 (24). These differences underscore the fact that different receptors may require different types of ubiquitin linkages for their downregulation. For example, whereas monoubiquitination is required for downregulation of the EGF receptor (18, 19), K63 chains have been shown to contribute to endocytosis and proteolysis of the TrkA nerve growth factor receptor (16), major histocompatibility complex class I and II proteins (14, 33, 36, 44), and some model chimerical receptors (2, 3). Intriguingly, K29-linked chains have been proven to regulate the degradation of the regulator of the Notch pathway, Deltex (7).
Perhaps the interaction between different types of ubiquitination and various other determinants within the receptors is the key to understanding these discrepancies. It is plausible that for PRLr, K63-linked ubiquitination is important in the context of specific PRLr-interacting proteins and specific linear endocytic/sorting motifs present within the cytoplasmic domain of PRLr. Whereas interaction between ubiquitination of IFNAR1 and its tyrosine-based linear motifs has been demonstrated previously (24), analogous motifs and interactors in PRLr have yet to be identified. Putative dileucine motifs that play an important role in the internalization of radiolabeled bovine PRL in COS cells expressing bovine PRLr (30) may represent attractive candidates for such motifs; the role of analogous motifs within human PRLr is currently under investigation. Subsequent studies aimed at delineating the mechanisms of PRLr downregulation should be important for our understanding of the alterations that constitutively augment PRL signaling in human breast cancer and pinpoint potential targets for therapeutic interference.
Supplementary Material
Acknowledgments
We thank A. Eastman, Y. Yarden, Z. Ronai, and A. F. Parlow for reagents, L. Schuler for discussion of the results, and I. Fernandez for critical comments on the manuscript.
This work was supported by NIH grants CA115281 (to S.Y.F.) and AG025688 (to J.P.), by the Canadian Cystic Fibrosis Foundation (H.B.), by grants from CIHR and NIDDK (to G.L.L.), and by funding from Institut National de la Santé et de la Recherche Médicale (to V.G.).
Footnotes
Published ahead of print on 23 June 2008.
Supplemental material for this article may be found at http://mcb.asm.org.
REFERENCES
- 1.Aberle, H., A. Bauer, J. Stappert, A. Kispert, and R. Kemler. 1997. β-Catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 163797-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barriere, H., C. Nemes, K. Du, and G. L. Lukacs. 2007. Plasticity of polyubiquitin recognition as lysosomal targeting signals by the endosomal sorting machinery. Mol. Biol. Cell 183952-3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barriere, H., C. Nemes, D. Lechardeur, M. Khan-Mohammad, K. Fruh, and G. L. Lukacs. 2006. Molecular basis of oligoubiquitin-dependent internalization of membrane proteins in mammalian cells. Traffic 7282-297. [DOI] [PubMed] [Google Scholar]
- 4.Bernichtein, S., C. Kayser, K. Dillner, S. Moulin, J. J. Kopchick, J. A. Martial, G. Norstedt, O. Isaksson, P. A. Kelly, and V. Goffin. 2003. Development of pure prolactin receptor antagonists. J. Biol. Chem. 27835988-35999. [DOI] [PubMed] [Google Scholar]
- 5.Bonifacino, J. S., and L. M. Traub. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72395-447. [DOI] [PubMed] [Google Scholar]
- 6.Bonifacino, J. S., and A. M. Weissman. 1998. Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu. Rev. Cell Dev. Biol. 1419-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chastagner, P., A. Israel, and C. Brou. 2006. Itch/AIP4 mediates Deltex degradation through the formation of K29-linked polyubiquitin chains. EMBO Rep. 71147-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaudhuri, R., O. W. Lindwasser, W. J. Smith, J. H. Hurley, and J. S. Bonifacino. 2007. Downregulation of CD4 by human immunodeficiency virus type 1 Nef is dependent on clathrin and involves direct interaction of Nef with the AP2 clathrin adaptor. J. Virol. 813877-3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiu, Y. H., Q. Sun, and Z. J. Chen. 2007. E1-L2 activates both ubiquitin and FAT10. Mol. Cell 271014-1023. [DOI] [PubMed] [Google Scholar]
- 10.Ciechanover, A., and K. Iwai. 2004. The ubiquitin system: from basic mechanisms to the patient bed. IUBMB Life 56193-201. [DOI] [PubMed] [Google Scholar]
- 11.Clague, M. J., and D. E. Hammond. 2006. Membrane traffic: catching the lysosome express. Curr. Biol. 16R416-R418. [DOI] [PubMed] [Google Scholar]
- 12.Clevenger, C. V., P. A. Furth, S. E. Hankinson, and L. A. Schuler. 2003. The role of prolactin in mammary carcinoma. Endocr. Rev. 241-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doray, B., I. Lee, J. Knisely, G. Bu, and S. Kornfeld. 2007. The gamma/sigma1 and alpha/sigma2 hemicomplexes of clathrin adaptors AP-1 and AP-2 harbor the dileucine recognition site. Mol. Biol. Cell 181887-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duncan, L. M., S. Piper, R. B. Dodd, M. K. Saville, C. M. Sanderson, J. P. Luzio, and P. J. Lehner. 2006. Lysine-63-linked ubiquitination is required for endolysosomal degradation of class I molecules. EMBO J. 251635-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuchs, S. Y., V. S. Spiegelman, and K. G. Kumar. 2004. The many faces of beta-TrCP E3 ubiquitin ligases: reflections in the magic mirror of cancer. Oncogene 232028-2036. [DOI] [PubMed] [Google Scholar]
- 16.Geetha, T., J. Jiang, and M. W. Wooten. 2005. Lysine 63 polyubiquitination of the nerve growth factor receptor TrkA directs internalization and signaling. Mol. Cell 20301-312. [DOI] [PubMed] [Google Scholar]
- 17.Goffin, V., S. Bernichtein, P. Touraine, and P. A. Kelly. 2005. Development and potential clinical uses of human prolactin receptor antagonists. Endocr. Rev. 26400-422. [DOI] [PubMed] [Google Scholar]
- 18.Haglund, K., P. P. Di Fiore, and I. Dikic. 2003. Distinct monoubiquitin signals in receptor endocytosis. Trends Biochem. Sci. 28598-603. [DOI] [PubMed] [Google Scholar]
- 19.Haglund, K., S. Sigismund, S. Polo, I. Szymkiewicz, P. P. Di Fiore, and I. Dikic. 2003. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat. Cell Biol. 5461-466. [DOI] [PubMed] [Google Scholar]
- 20.Hammond, D. E., S. Carter, J. McCullough, S. Urbe, G. Vande Woude, and M. J. Clague. 2003. Endosomal dynamics of Met determine signaling output. Mol. Biol. Cell 141346-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hicke, L., and R. Dunn. 2003. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 19141-172. [DOI] [PubMed] [Google Scholar]
- 22.Jin, J., X. Li, S. P. Gygi, and J. W. Harper. 2007. Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature 4471135-1138. [DOI] [PubMed] [Google Scholar]
- 23.Kirkpatrick, D. S., N. A. Hathaway, J. Hanna, S. Elsasser, J. Rush, D. Finley, R. W. King, and S. P. Gygi. 2006. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat. Cell Biol. 8700-710. [DOI] [PubMed] [Google Scholar]
- 24.Kumar, K. G., H. Barriere, C. J. Carbone, J. Liu, G. Swaminathan, P. Xu, Y. Li, D. P. Baker, J. Peng, G. L. Lukacs, and S. Y. Fuchs. 2007. Site-specific ubiquitination exposes a linear motif to promote interferon-alpha receptor endocytosis. J. Cell Biol. 179935-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar, K. G., J. J. Krolewski, and S. Y. Fuchs. 2004. Phosphorylation and specific ubiquitin acceptor sites are required for ubiquitination and degradation of the IFNAR1 subunit of type I interferon receptor. J. Biol. Chem. 27946614-46620. [DOI] [PubMed] [Google Scholar]
- 26.Levesque, A. A., E. A. Kohn, E. Bresnick, and A. Eastman. 2005. Distinct roles for p53 transactivation and repression in preventing UCN-01-mediated abrogation of DNA damage-induced arrest at S and G2 cell cycle checkpoints. Oncogene 243786-3796. [DOI] [PubMed] [Google Scholar]
- 27.Li, Y., C. V. Clevenger, N. Minkovsky, K. G. Kumar, P. N. Raghunath, J. E. Tomaszewski, V. S. Spiegelman, and S. Y. Fuchs. 2006. Stabilization of prolactin receptor in breast cancer cells. Oncogene 251896-1902. [DOI] [PubMed] [Google Scholar]
- 28.Li, Y., K. G. Kumar, W. Tang, V. S. Spiegelman, and S. Y. Fuchs. 2004. Negative regulation of prolactin receptor stability and signaling mediated by SCFβ-TrCP E3 ubiquitin ligase. Mol. Cell. Biol. 244038-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu, J. C., T. M. Piazza, and L. A. Schuler. 2005. Proteasomes mediate prolactin-induced receptor down-regulation and fragment generation in breast cancer cells. J. Biol. Chem. 28033909-33916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu, J. C., P. Scott, G. J. Strous, and L. A. Schuler. 2002. Multiple internalization motifs differentially used by prolactin receptor isoforms mediate similar endocytic pathways. Mol. Endocrinol. 162515-2527. [DOI] [PubMed] [Google Scholar]
- 31.Meyer, L., B. Deau, H. Forejtnikova, D. Dumenil, F. Margottin-Goguet, C. Lacombe, P. Mayeux, and F. Verdier. 2007. β-Trcp mediates ubiquitination and degradation of the erythropoietin receptor and controls cell proliferation. Blood 1095215-5222. [DOI] [PubMed] [Google Scholar]
- 32.Mosesson, Y., K. Shtiegman, M. Katz, Y. Zwang, G. Vereb, J. Szollosi, and Y. Yarden. 2003. Endocytosis of receptor tyrosine kinases is driven by monoubiquitylation, not polyubiquitylation. J. Biol. Chem. 27821323-21326. [DOI] [PubMed] [Google Scholar]
- 33.Ohmura-Hoshino, M., Y. Matsuki, M. Aoki, E. Goto, M. Mito, M. Uematsu, T. Kakiuchi, H. Hotta, and S. Ishido. 2006. Inhibition of MHC class II expression and immune responses by c-MIR. J. Immunol. 177341-354. [DOI] [PubMed] [Google Scholar]
- 34.Owen, D. J., B. M. Collins, and P. R. Evans. 2004. Adaptors for clathrin coats: structure and function. Annu. Rev. Cell Dev. Biol. 20153-191. [DOI] [PubMed] [Google Scholar]
- 35.Sharma, M., F. Pampinella, C. Nemes, M. Benharouga, J. So, K. Du, K. G. Bache, B. Papsin, N. Zerangue, H. Stenmark, and G. L. Lukacs. 2004. Misfolding diverts CFTR from recycling to degradation: quality control at early endosomes. J. Cell Biol. 164923-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shin, J. S., M. Ebersold, M. Pypaert, L. Delamarre, A. Hartley, and I. Mellman. 2006. Surface expression of MHC class II in dendritic cells is controlled by regulated ubiquitination. Nature 444115-118. [DOI] [PubMed] [Google Scholar]
- 37.Strack, B., A. Calistri, M. A. Accola, G. Palu, and H. G. Gottlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 9713063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swaminathan, G., B. Varghese, and S. Y. Fuchs. 2008. Regulation of prolactin receptor levels and activity in breast cancer. J. Mammary Gland Biol. Neoplasia 1381-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swaminathan, G., B. Varghese, C. Thangavel, C. J. Carbone, A. Plotnikov, K. G. Kumar, E. M. Jablonski, C. V. Clevenger, V. Goffin, L. Deng, S. J. Frank, and S. Y. Fuchs. 2008. Prolactin stimulates ubiquitination, initial internalization, and degradation of its receptor via catalytic activation of Janus kinase 2. J. Endocrinol. 196R1-R7. [DOI] [PubMed] [Google Scholar]
- 40.Tang, W., Y. Li, D. Yu, A. Thomas-Tikhonenko, V. S. Spiegelman, and S. Y. Fuchs. 2005. Targeting beta-transducin repeat-containing protein E3 ubiquitin ligase augments the effects of antitumor drugs on breast cancer cells. Cancer Res. 651904-1908. [DOI] [PubMed] [Google Scholar]
- 41.Tardy, C., P. Codogno, H. Autefage, T. Levade, and N. Andrieu-Abadie. 2006. Lysosomes and lysosomal proteins in cancer cell death (new players of an old struggle). Biochim. Biophys. Acta 1765101-125. [DOI] [PubMed] [Google Scholar]
- 42.van Kerkhof, P., R. Govers, C. M. Alves dos Santos, and G. J. Strous. 2000. Endocytosis and degradation of the growth hormone receptor are proteasome-dependent. J. Biol. Chem. 2751575-1580. [DOI] [PubMed] [Google Scholar]
- 43.van Kerkhof, P., J. Putters, and G. J. Strous. 2007. The ubiquitin ligase SCF(βTrCP) regulates the degradation of the growth hormone receptor. J. Biol. Chem. 28220475-20483. [DOI] [PubMed] [Google Scholar]
- 44.van Niel, G., R. Wubbolts, T. Ten Broeke, S. I. Buschow, F. A. Ossendorp, C. J. Melief, G. Raposo, B. W. van Balkom, and W. Stoorvogel. 2006. Dendritic cells regulate exposure of MHC class II at their plasma membrane by oligoubiquitination. Immunity 25885-894. [DOI] [PubMed] [Google Scholar]
- 45.Walrafen, P., F. Verdier, Z. Kadri, S. Chretien, C. Lacombe, and P. Mayeux. 2005. Both proteasomes and lysosomes degrade the activated erythropoietin receptor. Blood 105600-608. [DOI] [PubMed] [Google Scholar]
- 46.Wang, Y., S. Pennock, X. Chen, and Z. Wang. 2002. Endosomal signaling of epidermal growth factor receptor stimulates signal transduction pathways leading to cell survival. Mol. Cell. Biol. 227279-7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu, P., and J. Peng. 2006. Dissecting the ubiquitin pathway by mass spectrometry. Biochim. Biophys. Acta 17641940-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamasaki, L., and M. Pagano. 2004. Cell cycle, proteolysis and cancer. Curr. Opin. Cell Biol. 16623-628. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.