Abstract
Cyclin D1 is required at high levels for passage through G1 phase but must be reduced to low levels during S phase to avoid the inhibition of DNA synthesis. This suppression requires the phosphorylation of Thr286, which is induced directly by DNA synthesis. Because the checkpoint kinase ATR is activated by normal replication as well as by DNA damage, its potential role in regulating cyclin D1 phosphorylation was tested. We found that ATR, activated by either UV irradiation or the topoisomerase IIβ binding protein 1 activator, promoted cyclin D1 phosphorylation. Small interfering RNA against ATR inhibited UV-induced Thr286 phosphorylation, together with that seen in normally cycling cells, indicating that ATR regulates cyclin D1 phosphorylation in normal as well as stressed cells. Following double-stranded DNA (dsDNA) breakage, the related checkpoint kinase ATM was also able to promote the phosphorylation of cyclin D1 Thr286. The relationship between these checkpoint kinases and cyclin D1 was extended when we found that normal cell cycle blockage in G1 phase observed following dsDNA damage was efficiently overcome when exogenous cyclin D1 was expressed within the cells. These results indicate that checkpoint kinases play a critical role in regulating cell cycle progression in normal and stressed cells by directing the phosphorylation of cyclin D1.
Cell cycle progression is regulated by the timely production and destruction of cyclins. Among these, cyclin D1 plays the unique role of responding to the extracellular mitogenic environment and then regulating the cell cycle machinery accordingly (29). After its expression is stimulated by mitogenic signaling, such as by activation of the Ras pathway, cyclin D1 binds to and activates cyclin-dependent kinase 4 or 6 (CDK4/6) to generate a kinase for the retinoblastoma protein. Upon phosphorylation, the retinoblastoma protein loses its ability to inhibit E2F transcription factors, resulting in the expression of E2F target genes, which are required for entry into S phase, DNA synthesis, and progression through the later cell cycle phases (30).
Based upon recent studies from this and other labs, it is clear that the expression of cyclin D1 is highly regulated throughout the cell cycle and that its expression level in each cell cycle phase helps determine the overall proliferative characteristics of the cell. Cyclin D1 expression must be high for passage through G1 phase and the initiation of DNA synthesis but must decline rapidly during S phase for efficient DNA synthesis (11, 14). Cyclin D1 inhibits DNA synthesis due to its binding of PCNA, an essential component of the replication machinery (23, 39). At the completion of DNA synthesis, cyclin D1 levels must once again increase during G2 phase if the cell is to continue active cell cycle progression. This G2 phase increase is absolutely dependent upon the stabilization of cyclin D1 mRNA by proliferative signaling (10). In this way, continuing cell cycle progression requires positive growth conditions able to stimulate proliferative signaling. This fact helps explain the importance of the suppression of cyclin D1 levels during S phase. This suppression effectively erases any consequences of proliferative signaling from previous cell cycle phases and requires that the cell reassess its proliferative environment at the beginning of each G2 phase for cell cycle progression to continue (34).
These considerations emphasize the importance of understanding the mechanism of cyclin D1 suppression during S phase. This suppression requires the phosphorylation of Thr286, leading to proteasomal degradation, since cyclin D1 harboring a mutation at position 286 is expressed at constant levels through S phase (11). Previous reports suggested that glycogen synthase kinase 3 (GSK3) is the kinase responsible for this phosphorylation (6, 7). Our extensive studies, however, failed to detect any alteration in GSK3 activity, or in the activity of the phosphatidylinositol-3 kinase/AKT pathway that regulates GSK3, during S phase or any other cell cycle period. Moreover, direct analyses of GSK3 activity conclusively demonstrated that it is not responsible for regulating cyclin D1 levels in any of the cultured cells we analyzed (11). In fact, we were unable to find any signaling pathway responsible for promoting the phosphorylation of Thr286 during S phase. Rather, our studies suggested that this phosphorylation takes place automatically during each S phase regardless of the growth environment of the cell.
In this study, therefore, we began with the assumption that the suppression of cyclin D1 during S phase is due to a kinase activated directly by DNA synthesis. In this way, cyclin D1 levels would always decline during DNA synthesis regardless of the signaling environment of the cell, and the cell would invariably be forced to reassess its growth conditions prior to the initiation of each new cell cycle. The checkpoint kinase ATR (ATM and Rad 3 related) is widely believed to be specifically activated following UV irradiation when single-stranded DNA becomes coated by replication protein A (40). Recent studies, however, indicate that similar structures formed during normal DNA synthesis are also able to stimulate ATR activity (16, 28, 33). In fact, it appears that the activation of ATR during normal DNA synthesis is vital to the overall regulation of the course of replication (33, 37). Thus, ATR not only is activated during DNA synthesis but has been proposed to perform a critical function to regulate the rate of normal DNA synthesis. These data made ATR an attractive candidate for the S phase-specific regulator of the cyclin D1 kinase. In these studies, we have activated ATR in a number of ways and confirmed that it does regulate cyclin D1 phosphorylation on Thr286. This was the case following UV irradiation and also during normal DNA synthesis. Interestingly, the related checkpoint kinase ATM also directs the phosphorylation of cyclin D1. These observations raised the possibility that the phosphorylation of cyclin D1 might also be involved in the checkpoint functions of ATM and ATR.
MATERIALS AND METHODS
Materials.
Neocarzinostatin (NCS), rabbit polyclonal antiactin antibody (Ab), alkaline phosphatase-conjugated secondary antibodies, and bovine serum for cell culture were obtained from Sigma (St. Louis, MO). 2-Morpholin-4-yl-6-thianthren-1-yl-pyran-4-one, an ATM inhibitor, was purchased from EMD Chemicals (San Diego, CA). Mouse monoclonal anti-phospho-histone H2AX (Ser139) Ab (clone JBW301) was purchased from Upstate Cell Signaling Solutions (Lake Placid, NY). Rabbit polyclonal anti-phosho-Thr286 cyclin D1 and anti-phospho-chk1 (Ser317) antibodies were products of Cell Signaling Technology (Danvers, MA) and were originally prepared by and presented as gifts by M. Garrett (11). Mouse monoclonal anti-cyclin D1 Ab (72-13G, sc-450), mouse monoclonal anti-green fluorescent protein (GFP) (B-2, Sc9996), goat polyclonal anti-ATR Ab (N-19, sc-1887), and small interfering RNA (siRNA) against mouse ATR (sc-29764) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Sheep polyclonal antibromodeoxyuridine (anti-BrdU) Ab was obtained from Fitzgerald Industries International (Concord, MA). Rabbit polyclonal anti-phospho-p53 Ab (Ser 15; ab1431) was from Abcam, and rabbit anti-p53 (MCL-p53-CM5p) was from Novacastra. Fluorochrome-labeled secondary antibodies of multilabeling grade were the products of Jackson ImmunoResearch (West Grove, PA). The 5-ethynyl-2′-deoxyuridine (EdU) labeling kit was obtained from Invitrogen. We found that the cross-reactivity between EdU and BrdU during labeling was minimized by staining BrdU prior to EdU.
Western blotting.
NIH 3T3 cells were maintained in 10% bovine serum containing Dulbecco modified Eagle's medium supplemented with penicillin and streptomycin at 37°C under humidified atmosphere with 5% CO2. In order to suppress ATR expression, siRNA was transfected using calcium phosphate coprecipitation overnight. After removal of the transfection solution, the monolayers were rinsed once with phosphate-buffered saline, and then the cells were serum starved (0.5% serum) for 2 days to make them quiescent. The cells were restimulated with serum for 15 h to induce them to progress into S phase, after which they were UV irradiated. One hour following irradiation, the cells were harvested and lysed by sonication in Laemmli sample buffer. The resulting lysates were boiled and applied to a precast sodium dodecyl sulfate-gradient polyacrylamide gel (∼4 to 15%; Bio-Rad, Richmond, CA). The separated proteins were electrotransferred onto a nitrocellulose membrane. To determine ATM involvement in cyclin D1 phosphorylation, proliferating NIH 3T3 cells were treated with 10 μM of the ATM inhibitor, or the same amount of solvent, dimethyl sulfoxide (DMSO), for 30 to 60 min. The cultures were then treated with 5 ng/ml of NCS for 1 h. The cells were scraped from the plates, and the lysates were prepared as described previously. The proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (T = 10%) and blotted onto a nitrocellulose membrane. To detect the proteins or phosphoproteins of interest, the blots were incubated with specific antibodies followed by incubation with alkaline phosphatase-conjugated secondary antibodies. The immunoreactive bands were visualized by StromImager scanning following incubation with enzyme-chemifluorescence substrate (GE Healthcare).
Statistical analyses.
In general, statistical analyses relied upon two-tailed, paired Student's t tests to determine the probability that two data sets differ from one another. In addition, two sample t tests were used with unpaired data, and one-tailed tests were used to test single-tailed hypotheses. Standard errors (SE) displayed graphically were determined by standard methods, as well as by the Kaleidagraph software program. For an analysis of quantitated Western analyses and image analysis results, significance levels were determined using analysis of variance tests with the R software package. In the case of Western analyses of double-stranded breakage induced by NCS in the absence of the proteasomal inhibitor MG132, however, the trends were highly consistent over four analyses, but the base numerical value of the pD1/total D1 ratio varied between analyses due to variations in experimental conditions and due to the reduction in the overall cyclin D1 levels. Consequently, it was necessary to utilize nonparametric methods. These methods are based on a ranking system where each data point in an experiment is ranked from largest to smallest. The analysis is completed by testing the probability that the rankings, not the output seen in each replication, were due to chance alone. Using the R software, we utilized the Wilcoxon signed-rank test to determine the statistical differences between the treatment groups. The Wilcoxon signed-rank test demonstrated that the NCS-treated cells have a slightly larger pD1/total D1 ratio than each of the other three treatments (P = 0.06). The same level of significance was determined when the value of the NCS treatment in the presence of the ATM inhibitor was tested against the NCS value alone (P = 0.06). When the control treatments with DMSO alone, the ATM inhibitor alone (P = 0.44), or the ATM inhibitor together with NCS treatment was compared to either of these, there was no significant difference.
Microinjection and immunofluorescence.
To perform quantitative immunofluorescence and microinjection, NIH 3T3 cells were grown on glass coverslips. To activate ATR, the pEGFP-TopBP1 (978-1286; wild type) expression plasmid (10 μg/ml) was microinjected into the nuclei of the 200 to 300 cells growing on an area of the coverslip designated by a circle scribed on the back of the coverslip. As a control, the inactive pEGFP-TopBP1 (978-1286) W1145R mutant expression vector was injected (prepared by Akiko Kumagai [17]). Three hours following the injection, the cells were fixed with methanol. After blocking with 0.3% bovine serum albumin in phosphate-buffered saline, cyclin D1, phospho-Thr286-D1, and/or phospho-H2AX was stained with specific antibodies and Cy3- or Cy5-labeled secondary antibodies. DNA was stained with DAPI (4′,6′-diamidino-2-phenylindole). Sixteen-bit grayscale digital images of each fluorescent stain and GFP were collected using a Leica DMRB fluorescent microscope (Wetzlar, Germany) equipped with a charge-coupled-device (CCD) camera (PI-MAX; Princeton Instruments, Trenton, NJ). The fluorescent intensity of each staining was determined using an image analysis program, Metamorph, as described previously (14). In order to down-modulate ATR expression, siRNA against ATR (2 μM) was microinjected into the cytoplasm of NIH 3T3 cells of very sparse cultures. As a control, we injected a nonspecific siRNA against cyclin which failed to down-modulate any protein we have been able to identify (data not shown) and has no identifiable effects upon cells. Two days following injection, the effect of ATR down-modulation on cyclin D1 phosphorylation was determined by quantitative image analysis with or without UV irradiation.
To determine the average levels of a fluorochrome in a culture, the fluorescence of that marker and of the DAPI-stained DNA was determined by image analysis for each cell. The cells in each culture were then separated into groups according to DNA content, and the average level of fluorescence of the designated marker was determined for cells in each DNA grouping. This average was plotted against the DNA content for the group of cells. When separate cultures were to be compared, such as a comparison between ATR siRNA and control siRNA injections, the average of each culture was determined, and the ATR siRNA results for each DNA grouping were subtracted from the control results with the same DNA content. The difference values for all cultures within each DNA range were then analyzed to determine the means and SE with Kaleidagraph. The average levels of phosphorylation among different determinations were determined as a simple mean value for each DNA range.
Microinjected cells were identified by their content of GFP in the case of mutant cyclin D1 and topoisomerase IIβ binding protein 1 (TopBP1) plasmids (which contained proteins linked to the GFP sequence). Direct staining of the GFP protein with a fluorescent antibody stain was also utilized. The mutant cyclin D1 containing Ala in the place of Thr286, linked to the GFP sequence, has been described (11). In some cases, these plasmids were also microinjected together with a plasmid expressing GFP alone. In most cases, the cells within an area designated by a circle scribed on the back of the coverslip were all injected. Of the 150 to 200 cells in this area, nearly 90% expressed consistent levels of the introduced material. Wild-type and mutant cyclin D1 expression plasmid DNAs were each injected at a concentration of 1.0 μg/ml, while the GFP-cytomegalovirus plasmid was injected at 10 μg/ml. Extensive previous studies indicate that at this concentration, the amount of exogenous cyclin D1 expressed is approximately equal to the level of endogenous cyclin D1 and remained exclusively in the nucleus as determined by associated GFP fluorescence or antibody staining. Protein expression can be seen beginning within 60 min of the injection and continues for at least 18 h.
RESULTS
Activation of ATR by UV promotes cyclin D1 phosphorylation.
Our first goal was to determine if ATR has the ability to promote the phosphorylation of cyclin D1 Thr286, either directly or indirectly, following the induction of activity by DNA damage. It has been reported that ATR is specifically activated by UV irradiation. NIH 3T3 cells were irradiated with UV light (10 J/m2), and the cells were then fixed and stained for cyclin D1, cyclin D1 phosphorylated on Thr286, DNA, and phosphorylated histone H2AX (γH2AX; a marker of DNA damage). Images taken with a CCD camera of each fluorochrome were then analyzed to determine the level of each marker in each cell. This image analysis technique has been extensively utilized in this laboratory and allows the extraction of quantitative data from digital images (14). To visualize the results, the DNA content for each cell was plotted versus the level of one of the other three markers to yield an expression profile of that marker through the cell cycle (Fig. 1).
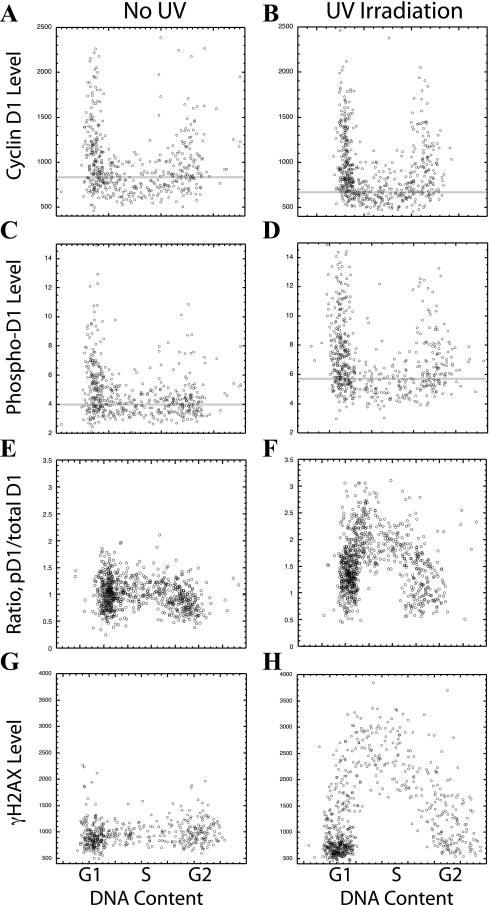
FIG. 1.
UV irradiation induces cyclin D1 phosphorylation. NIH 3T3 cells were UV irradiated (10 J/m2). Sixty minutes later, the cells were fixed and stained for cyclin D1 (A, B) and cyclin D1 phosphorylated on Thr286 (pD1) (C, D) or were stained for phosphorylated histone H2AX (γH2AX) (G, H). The fluorescent intensity of each fluorochrome in each cell was determined by photography with a CCD camera and image analysis and plotted versus the level of DNA determined following DAPI staining (as indicated by the cell cycle stages listed). (E, F) As an indication of the actual cyclin D1 Thr286 kinase activity within each cell, the level of phosphorylated cyclin D1 was divided by the level of total cyclin D1 (pD1/total D1 ratio). The faint horizontal lines indicate the average level of cyclin D1, or phosphorylated cyclin D1, for all S-phase cells in each frame (A to D).
Prior to UV irradiation, cyclin D1 levels were low in S phase and high in the G1 and G2 phases, as previously reported (14; Fig. 1A). These total cyclin D1 levels were slightly decreased by UV irradiation (Fig. 1B). At the same time, phosphorylated Thr286 cyclin D1 (pD1) levels, detected by an antibody able to specifically recognize the phosphorylation of Thr286, were increased by UV irradiation (Fig. 1C and D). To determine a change in the level of cyclin D1 Thr286 kinase activity within the cell following UV treatment, however, it was necessary to consider both the amount of phosphorylated cyclin D1 and the total amount of cyclin D1 available for phosphorylation. This was accomplished by dividing the amount of phosphorylated cyclin D1 by the level of total cyclin D1 within each cell (pD1/total D1), and plotting this value against the DNA content of the cell. This ratio, and therefore the total cyclin D1 kinase activity, was strongly induced by UV irradiation (Fig. 1E and F). For comparison, the general marker for DNA damage within a cell, γH2AX, was also strongly induced following UV irradiation (Fig. 1G and H). Interestingly, both cyclin D1 and H2AX appeared to be phosphorylated most actively during the S phase (Fig. 1F and H). These experiments provide the first indication that ATR activity leads to the phosphorylation of cyclin D1 Thr286. The studies to follow extend this conclusion.
siRNA confirms the ability of ATR to promote cyclin D1 phosphorylation.
To gain conclusive evidence that it is indeed ATR which directs the phosphorylation of cyclin D1 following UV irradiation, we microinjected siRNA against ATR into NIH 3T3 cells 48 h prior to UV irradiation. As before, the change in cyclin D1 Thr286 kinase activity in each cell was measured by determining the ratio of pD1 to total cyclin D1 (pD1/total D1) by image analysis. When individual cells of an untreated culture (Fig. 2A) were compared to a UV-irradiated culture treated with an unrelated siRNA (Fig. 2B), this ratio increased, indicating that kinase activity had been stimulated. On the other hand, the treatment of a separate culture with siRNA against ATR (Fig. 2C) followed by UV irradiation resulted in a much smaller increase in the pD1/total D1 ratio, indicating that the ablation of ATR had limited the increase in cyclin D1 kinase activity induced by UV irradiation. For comparison, the phosphorylation of histone H2AX and of Chk1 (two well-characterized targets of ATR activity) were also reduced by the injection of ATR siRNA followed by UV irradiation (Fig. 2D to I).
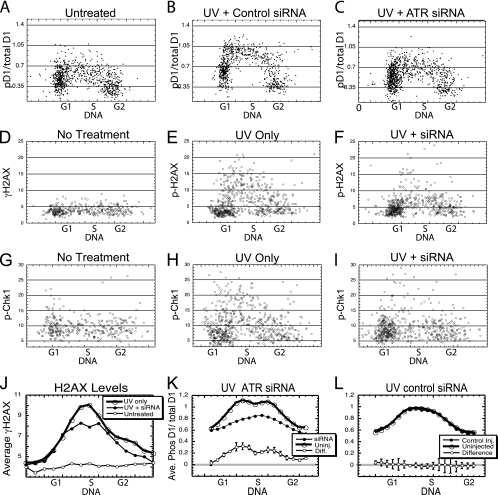
FIG. 2.
Cyclin D1 phosphorylation following UV irradiation depends upon ATR activity. (A to I) NIH 3T3 cells were microinjected with siRNA (2 μM) against ATR, or an unrelated siRNA. Forty-eight hours later, the injected cultures were UV irradiated, while control cultures were not irradiated. All cultures were then fixed and stained for phosphorylated histone H2AX, cyclin D1 and phosphorylated cyclin D1, or for phosphorylated Chk1. The stains were quantitated and plotted as indicated. (J to L) These results were then summarized by determining the average reading for cells of increasing DNA contents to generate a line indicating the average levels of each marker through the cell cycle. (J) The suppression of average levels of H2AX phosphorylation by ATR siRNA is shown. (K, L) The pD1/total D1 ratios for siRNA-injected and for neighboring uninjected cells were determined from six separate injection experiments and average results reported. The differences between injected and uninjected cells for ATR siRNA and control siRNA are also presented (±SE).
To accurately quantitate the reduction in cyclin D1 kinase activity resulting from the ablation of ATR, this experiment was repeated six times and the results averaged. In each experiment, the pD1/total D1 ratio was determined for each injected cell and for an equal number of neighboring uninjected cells. To obtain an average result, the cells were separated into groups according to increasing DNA content and the average pD1/total D1 ratio for each group determined. These values for individual DNA groups in separate experiments were then averaged over all six experiments, and this average value plotted against the DNA level to yield the average profile. When this average profile for ATR siRNA-injected cells was compared to the profile for neighboring uninjected cells (Fig. 2K), it was apparent that cyclin D1 kinase activity (the pD1/total D1 ratio) had been reduced in each of the separate groups with increasing DNA. On the other hand, the control siRNA did not reduce cyclin D1 kinase activity compared to uninjected cells in any of the DNA ranges analyzed (Fig. 2L). To accurately determine how much the pD1/total D1 ratio was suppressed by siRNA, the average pD1/total D1 value for injected cells was subtracted from this value for uninjected cells in each DNA grouping and the differences across all six experiments compared to determine the mean reduction in the pD1/total D1 ratio (±SE). When these differences were plotted versus DNA content, it was apparent that ATR plays a critical role in the induction of cyclin D1 kinase activity following UV irradiation (Fig. 2K and L). As a comparison, the average level of H2AX phosphorylation was similarly determined and plotted versus DNA content for ATR siRNA-injected cells, neighboring uninjected cells, and untreated cells (Fig. 2J). This result makes it clear that the extent of the reduction in cyclin D1 phosphorylation was quite similar to the reduction in H2AX phosphorylation following ATR siRNA treatment.
As a final analysis of these data, the cells of each analysis were separated into separate cell cycle phases based upon labeling with BrdU and DNA content. The combined data from all six experiments were analyzed statistically and indicated that siRNA against ATR reduced the phosphorylation of cyclin D1 significantly (P < 0.001), while the control siRNA failed to significantly reduce phosphorylation. The inhibition of cyclin D1 phosphorylation for S-phase cells was significantly greater than that for cells in either G1 or G2 phase (P < 0.01), while the inhibition during the G1 phase was indistinguishable from the inhibition during the G2 phase (P = 0.5). No significant inhibition was observed during any cell cycle phase following the injection of control siRNA. We conclude that ATR plays a critical role in the UV-induced phosphorylation of cyclin D1 and that this phosphorylation is primarily localized in the S phase.
Activation of ATR by TopBP1 stimulates cyclin D1 phosphorylation.
To provide additional support for the conclusion that ATR is able to direct the phosphorylation of cyclin D1 Thr286, we made use of the fact that ATR is specifically activated by TopBP1 (3, 17). A plasmid expressing an active fragment of this protein linked to the GFP sequence, along with a similar plasmid expressing an inactive TopBP1 mutant (prepared by A. Kumagai), was separately microinjected into NIH 3T3 cells. After 4 h, the level of GFP fluorescence, along with the cellular levels of pD1, cyclin D1, and DNA, was determined by image analysis. It was clear from the ratio of pD1 to total cyclin D1 that the wild-type TopBP1 had effectively induced cyclin D1 phosphorylation in all cell cycle phases (Fig. 3A and B). For comparison, TopBP1 injection also induced γH2AX in all cell cycle phases (Fig. 3D and E). Neither cyclin D1 nor H2AX phosphorylation was observed following the microinjection of the mutant protein (Fig. 3C and F). As observed following UV irradiation, the introduction of the TopBP1 protein resulted in a reduction in total cyclin D1 levels. This was apparent from the cyclin D1 levels in individual injected and uninjected cells (Fig. 3G and H) and for a calculation of the average levels following these injections (Fig. 3I). The reduction in total cyclin D1 levels is also apparent from photographic images taken of an area of cells containing TopBP1-injected cells together with uninjected cells (Fig. 3J). These data support the contention that ATR is able to play a critical role in the phosphorylation of cyclin D1 following activation, although it is not known if this is a direct or an indirect effect.
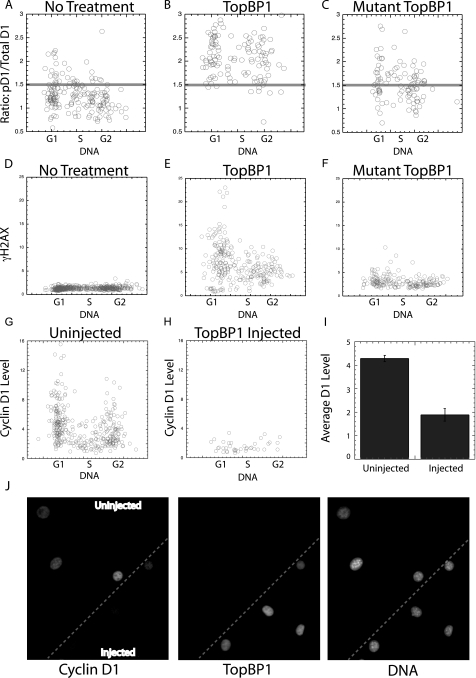
FIG. 3.
TopBP1 induces cyclin D1 phosphorylation. Plasmids expressing the wild-type or an inactive mutant TopBP1 sequence attached to the GFP sequence were injected into NIH 3T3 cells. After 3 h, the cells were fixed, stained, and subjected to image analysis. These experiments were performed in the presence of the proteasomal inhibitor MG132 to block the degradation of total cyclin D1 during the experiment. The ratio of phosphorylated to total cyclin D1 is presented (A to C), as is the level of γH2AX following each injection (D to F). (G, H) To determine the extent to which total cyclin D1 is reduced following TopBP1 expression, the above experiments were repeated in the absence of MG132, and the level of cyclin D1 determined and plotted for injected and neighboring uninjected cells versus DNA content. (I) The average reduction in total cyclin D1 expression was determined for injected and uninjected cells. (J) Images from such an experiment are also presented. Cells at the bottom right of the panel were injected with the TopBP1 plasmid as indicated by the resulting GFP fluorescence (middle panel). Cells containing TopBP1 expressed dramatically reduced levels of cyclin D1 compared to uninjected cells as shown following cyclin D1 staining (see upper left of the left panel). For comparison, the DAPI-stained DNA image is presented to localize all cells (right panel). Injected cells were identified by immunofluorescence staining of GFP.
ATR activation and cyclin D1 phosphorylation are localized in the S phase.
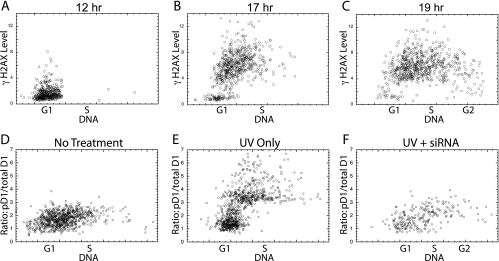
The above results, together with other published studies (12), indicate that ATR is activated by UV irradiation primarily during the S phase as indicated by γH2AX levels. Evidence that cyclin D1 Thr286 is also phosphorylated during the S phase would provide added support for the role of ATR in this event. For this determination, NIH 3T3 cells were synchronized by serum removal, followed by serum addition for various periods to promote cell cycle progression into differing cell cycle phases. Following the UV irradiation of cells at various times following serum stimulation, the level of γH2AX was determined by image analysis. The cells stimulated for 12 h were almost exclusively in the G1 phase and showed little γH2AX (Fig. 4A), while serum stimulation for 17 or 19 h resulted in increasing numbers of S-phase cells with progressively increasing DNA content (Fig. 4B and C). In these cultures, the S-phase cells, but not the remaining G1-phase cells, showed a dramatic increase in H2AX phosphorylation following UV irradiation. This procedure was next utilized to confirm that cyclin D1 phosphorylation was also induced during the S phase by ATR. NIH 3T3 cells were serum deprived and stimulated with serum for 15 h to produce a culture in which approximately half the cells were in the S phase (Fig. 4D). UV irradiation stimulated the pD1-to-total-cyclin-D1 ratio only in cells with S-phase DNA content (Fig. 4E), exactly as seen for γH2AX. In a culture that had received an injection of siRNA against ATR prior to serum removal, however, the phosphorylation of cyclin D1 was low, even in UV-irradiated S-phase cells (Fig. 4F). Note that following TopBP1 expression, both H2AX and cyclin D1 were phosphorylated in all cell cycle phases (Fig. 3B and E). Thus, with these two distinct means of inducing ATR, the cell cycle characteristics of H2AX phosphorylation (and therefore of ATR activation) directly mirrored the cell cycle characteristics of cyclin D1 phosphorylation. This serves as additional support for the notion that ATR is responsible for promoting cyclin D1 phosphorylation. It should be emphasized, however, that while these results confirm that cyclin D1 phosphorylation is primarily localized in S phase, there is evidence that some induction in G1 phase also takes place (see Fig. 1C and D and Fig. 2A to C).
FIG. 4.
UV irradiation induces H2AX and cyclin D1 phosphorylation only during the S phase. (A to C) NIH 3T3 cells were rendered quiescent by serum deprivation, followed by serum addition for the times indicated, UV irradiation, fixation, and determination of the γH2AX content of each cell, which is plotted versus DNA content. (D to F) NIH 3T3 cells treated as above for 15 h with serum were analyzed without UV irradiation (D) or following UV irradiation with (F) or without (E) a prior injection of siRNA against ATR. Again, the ratio of phosphorylated cyclin D1 divided by total cyclin D1 levels is presented.
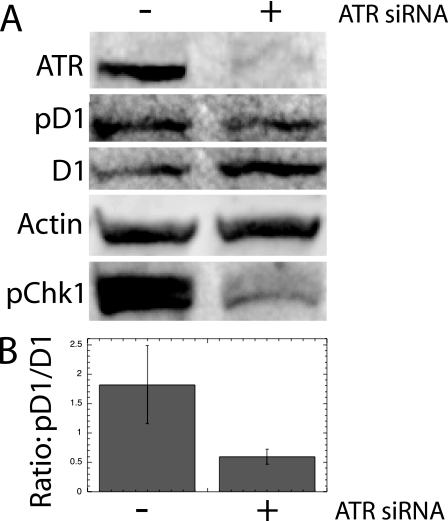
NIH 3T3 cultures synchronized as described above were next studied by Western analysis to confirm the cytometric results described above. A culture transfected with siRNA against ATR was deprived of serum for 48 h and then stimulated with serum for 15 h (to promote the progression of the culture into S phase) prior to UV irradiation and Western analysis. In these UV-irradiated cells, the transfected siRNA against ATR promoted slightly higher levels of total cyclin D1, while lowering the amount of phosphorylated cyclin D1 compared to UV-irradiated cultures without ATR siRNA (Fig. 5A). While neither of these individual changes was great in itself, the overall change in cyclin D1 kinase activity indicated by dividing pD1/total D1 was significantly reduced by ATR siRNA treatment. This was apparent following the quantitation of all three such analyses (Fig. 5B). Both ATR protein and the phosphorylation of its target, Chk1, were efficiently reduced by siRNA treatment, confirming the efficiency of ATR ablation in this analysis. Each of the separate studies reported above provides independent support for the conclusions reached above that ATR was able to promote the phosphorylation of cyclin D1 following UV stimulation.
FIG. 5.
Western analysis following ATR ablation. (A) NIH 3T3 cells were either transfected with siRNA against ATR or mock transfected. After 48 h, they were stimulated with serum for 15 h, UV irradiated, and subjected to Western analysis for the indicated markers. (B) To determine the changes in cyclin D1 Thr286 kinase activity, the ratio of phosphorylated cyclin D1 was divided by the level of total cyclin D1 for three totally independent analyses (±SE). Total Chk1 levels (not shown) were indistinguishable between the two lanes.
ATR promotes cyclin D1 phosphorylation in normal S-phase cells.
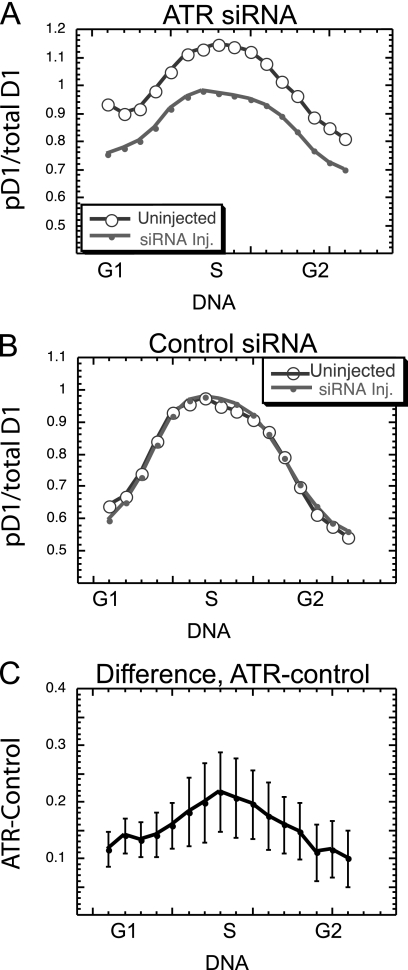
Our model of cell cycle control initially prompted us to predict that ATR might induce the phosphorylation of cyclin D1 in normal cells during the S phase. Since it is clear that ATR is able to induce cyclin D1 phosphorylation, and since it is known that ATR is normally activated during S phase (16, 28, 33), there appears to be good support for this original assumption. To directly confirm this idea, and to determine the extent of ATR phosphorylation of cyclin D1 during S phase, we microinjected siRNA into sparse, actively cycling NIH 3T3 cultures. Forty-eight hours later, the level of cyclin D1 phosphorylation was determined by staining and image analysis. The results from nine such experiments were averaged through the cell cycle as described above (for Fig. 2J to L). siRNA against ATR promoted a consistent suppression of normal cyclin D1 phosphorylation (Fig. 6A), while the control siRNA had no observable effect (Fig. 6B). To quantitate and compare these results, the phosphorylation of cyclin D1 following ATR siRNA injection was subtracted from the suppression with control siRNA for each grouping of cells with increasing DNA content in each experiment. The results of such analysis from all nine experiments were then compared to determine the mean ATR-induced suppression of cyclin D1 phosphorylation (±SE) throughout the cell cycle. In each DNA range, there was a definite reduction in cyclin D1 phosphorylation resulting from the ablation of ATR (Fig. 6D).
FIG. 6.
ATR promotes the phosphorylation of cyclin D1 during normal S phase. ATR siRNA (A) or the control siRNA (B) was microinjected into sparse cultures of NIH 3T3 cells 48 h prior to fixation, staining, and analysis. The phosphorylated to total cyclin D1 ratio was determined for injected and neighboring uninjected cells from nine separate analyses, and average values were determined for groups of cells with increasing DNA content. (C) The pD1/total D1 ratio for siRNA-injected cells was subtracted from this ratio for neighboring uninjected cells in each DNA range to yield the extent to which the ATR siRNA had suppressed normal cell cyclin D1 phosphorylation. The mean (±SE) of this difference for nine separate experiments is presented.
The natural variations in this experiment make it difficult to determine the extent to which ATR is able to promote cyclin D1 phosphorylation in individual cell cycle stages. To address this question, cells from each experiment were separated into cell cycle phases according to BrdU labeling and DNA content, and the results of all experiments were combined and compared statistically. When all cell cycle phases were considered together, the siRNA against ATR suppressed cyclin D1 phosphorylation highly significantly (P < 0.001). The suppression during S phase was also highly significant compared to the control siRNA (P = 0.01), while the significance for G1 phase was slightly reduced (P = 0.03). There appeared to be little evidence that ATR was involved in cyclin D1 phosphorylation during the G2 phase (P = 0.18). These results not only support our initial assumption regarding the role of ATR in the normal suppression of cyclin D1 during the S phase but raise the possibility that it plays some role during G1 phase, a result consistent with the limited but definite increase in cyclin D1 phosphorylation seen during the G1 phase after UV irradiation (Fig. 1F).
ATM is also able to induce cyclin D1 phosphorylation.
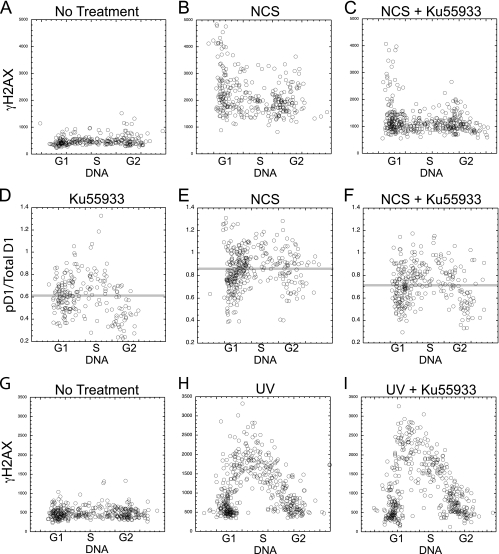
Our results with ATR raised the possibility that a related checkpoint kinase, ATM (ataxia-telangiectasia mutated), might also be able to direct cyclin D1 phosphorylation. ATM is induced by double-stranded DNA (dsDNA) breaks. To specifically induce ATM, therefore, we utilized the bacterial toxin NCS, which is reported to specifically induce dsDNA breakage within living cells and thus to be a specific inducer of ATM (15, 24). This toxin induced γH2AX in all cell cycle phases within 1 h (Fig. 7A and B). As evidence that this was the result of ATM activity, this H2AX phosphorylation was efficiently blocked by treatment with the specific ATM inhibitor Ku55933 (Fig. 7C). Importantly, Ku55933 did not interfere with H2AX phosphorylation following UV irradiation of these same cells (Fig. 7G to I), indicating that it is unable to block ATR activity.
FIG. 7.
ATM induces cyclin D1 phosphorylation. NIH 3T3 cells were treated with NCS, in the presence or absence of the ATM inhibitor Ku55933. (A to C) Treated and untreated cells as indicated were stained for γH2AX, and the levels plotted versus DNA content for each cell. (D to F) The ratio of pD1 to total cyclin D1 is plotted versus DNA for similarly treated cultures. Faint horizontal lines indicate the average cyclin D1 phosphorylation ratio of all cells in the panel. (G to I) The phosphorylation of H2AX was also determined following UV irradiation in the presence or absence of the ATM inhibitor.
To determine if ATM is also able to promote cyclin D1 phosphorylation, NIH 3T3 cells were treated with NCS and analyzed for cyclin D1 Thr286 phosphorylation by image analysis. The ratio of pD1 to total cyclin D1 was increased by NCS treatment (Fig. 7D and E), while the level of total cyclin D1 was reduced significantly (not shown; P = 0.01). This cyclin D1 phosphorylation was efficiently inhibited by Ku55933 (Fig. 7F). Careful analyses (not shown) indicate that the phosphorylation of cyclin D1 following NCS treatment took place in all cell cycle phases but was actually more pronounced during G1 and G2 phases than during S phase.
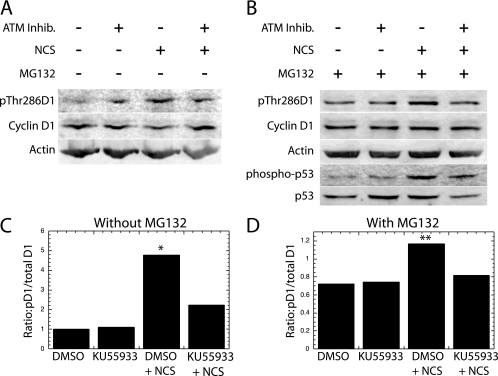
To confirm these observations made with image analysis, bulk cultures were treated with the NCS toxin in the presence or absence of Ku55933 and subjected to Western analysis for Thr286-phosphorylated and total cyclin D1. The results from each of four individual experiments confirmed that NCS treatment induced a slight but clear increase in cyclin D1 Thr286 phosphorylation, while promoting a decrease in the cyclin D1 level (Fig. 8A). As described above, the change in overall cyclin D1 kinase activity must take into consideration each of these alterations. When the pD1/total D1 level obtained by the quantitation of these Western analyses was calculated for all four experiments and averaged, it was clear that the cyclin D1 kinase activity (pD1/total D1 ratio) was increased by NCS treatment and decreased by the ATM inhibitor (Fig. 8C).
FIG. 8.
Western analysis of the ATM-induced phosphorylation of cyclin D1. (A) NIH 3T3 cultures were treated with NCS in the presence (+) or absence (−) of the ATM inhibitor Ku55933 as indicated. Lysates of each culture were then subjected to Western analysis with antibodies specific for cyclin D1 phosphorylated on Thr286 or the other indicated antigens. (B) A similar analysis was performed in the presence (+) of the proteasome inhibitor MG132. The average pD1/total D1 ratio is presented for all four experiments without (C) and for the three experiments with (D) MG132. *, P of 0.06 that NCS induced an increased phosphorylation compared to all other lanes; **, P of <0.01 in the presence of MG132.
The above Western analysis is complicated by the fact that dsDNA breaks reduce cyclin D1 levels while inducing its phosphorylation. This decrease in total cyclin D1 level complicates the calculations described above. Consequently, these analyses were repeated in the presence of the proteasomal inhibitor MG132 to block the degradation of phosphorylated cyclin D1. Since the level of cyclin D1 remained consistent throughout the analysis, it was possible to focus only upon the increase in phosphorylated cyclin D1 (Fig. 8B, lane 3). In each of three separate analyses, there was a definite increase in cyclin D1 phosphorylation following NCS treatment, which was reduced by treatment with Ku55933 (Fig. 8C). The average of all three such analyses indicates that the increase in phosphorylation observed following NCS treatment is significantly blocked by the ATM inhibitor (P = 0.008). We conclude that ATM, in addition to ATR, is able to promote the phosphorylation of cyclin D1 Thr286, although our data do not address the role of ATM in normal cell cycle progression. The efficiency with which the ATM inhibitor functioned in this experiment is evidenced by its ability to block the phosphorylation of p53 on Ser15 (Fig. 8B).
Cyclin D1 regulates the G1-phase checkpoint.
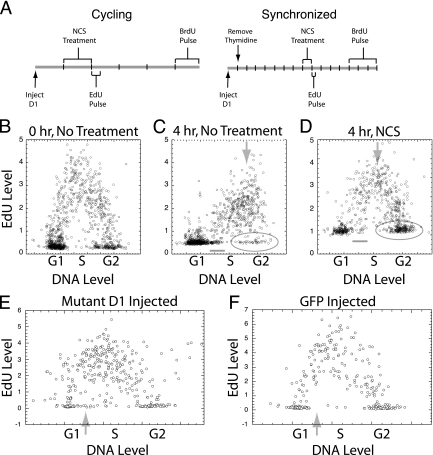
The fact that cyclin D1 phosphorylation was induced by either ATM or ATR raised the possibility that this critical cell cycle regulatory molecule might be involved in the checkpoint control mechanism induced by these two kinase molecules following DNA damage. To directly test this possibility, NIH 3T3 cells were microinjected with either the T286A mutant or the wild-type cyclin D1 sequence linked to the GFP epitope (1.0 μg/ml). Since the T286A mutant could not be phosphorylated at position 286, only this mutant would resist degradation during the S phase (11). The expression of the microinjected plasmids began within 1 to 2 h following injection and continued for at least 18 h. During this time, the expressed cyclin D1 protein remained exclusively nuclear and was expressed at levels at or below the levels of endogenous cyclin D1 (11). The first analysis of the checkpoint function of cyclin D1 was performed in actively cycling cells. These cultures were treated for 1 h with NCS beginning 2 to 4 h after injection of the cyclin D1 expression plasmids. Immediately following the NCS treatment, the cultures were labeled with a 15-min pulse of EdU, which was incorporated only into actively synthesized DNA and served to identify cells in S phase at the time of DNA damage. Following a 3-h incubation, a final pulse of BrdU identified cells in the S phase at the end of the experiment (Fig. 9A).
FIG. 9.
Analysis of cell cycle progression following DNA damage. (A) The experimental timelines for the treatment of cycling cells (left) and cells synchronized by thymidine treatment (right) are presented. Timelines are broken into hours. (B to D) The profile of EdU labeling is shown immediately (B) or 4 h following a 15-min EdU pulse without (C) or with (D) NCS treatment. (C and D) The short, horizontal lines indicate cells that had passed from G1 to S phase during incubation, the downward arrows indicate progression through S phase, and the elongated circles enclose G2-phase cells. (E and F) Cycling cells were injected with the T286A mutant cyclin D1 or the control GFP expression plasmid and treated as described. The profile of EdU labeling versus DNA content for the cells injected with mutant cyclin D1 (E) and with GFP (F) is shown. The faint upward arrows indicate cells which passed from G1 to S phase during this experiment.
The ability of dsDNA breakage to block cell cycle progression in each cell cycle phase could be determined from the EdU versus DNA profiles (Fig. 9B to D). The progression from G1 to S phase was significantly reduced following NCS treatment as indicated by the number of cells that had left G1 phase after EdU labeling. These cells (Fig. 9C and D) had DNA levels of early S phase but no EdU labeling. Extensive analysis indicated that within 6 h, only approximately 25% of the G1-phase cells passed into S phase following NCS treatment, whereas almost all of the G1-phase cells entered S phase in untreated cultures (not shown). Slowed progression through S phase following dsDNA damage was revealed by the failure of the most highly EdU-labeled cells to increase in DNA content during the 4-h incubation following the NCS treatment (Fig. 9C and D). Finally, a blockage in the passage through mitosis was revealed by the failure of the cells initially in G2 phase to divide and enter G1 phase. Thus, most cells in the G2 phase immediately following DNA damage (without EdU labeling and with G2/M DNA content) remained in the G2 phase during the 4 h after NCS treatment, while most G2-phase cells divided in untreated cultures (Fig. 9C and D). Moreover, mitotic figures were not observed in treated cultures (not shown).
Expression of the T286A mutant cyclin D1 did not affect the blockage in cell cycle progression during G2/M phase, since no mitotic cells were seen following the injection (data not shown). The progression of cells through S phase was also not altered by the injected cyclin D1 protein, since the overall profile of EdU labeling at the end of the experiment was not changed by the exogenous cyclin D1 protein (Fig. 9E and F). On the other hand, the cyclin D1 mutant protein was able to promote passage through the G1/S phase block induced by dsDNA breakage, as shown by the EdU labeling. Following the control GFP injection, the cells in G1 phase immediately following DNA damage (those cells with G1 DNA content and without EdU labeling) retained their G1 DNA content to the end of the experiment (Fig. 9F). On the other hand, among the mutant cyclin D1-injected cells, a significant proportion of the EdU-unlabeled cells that were in G1 phase immediately after DNA damage had progressed into early S phase, as evidenced by cells with S-phase DNA content but without EdU labeling (Fig. 9E). At the same time, the total number of cells remaining in the G1 phase throughout the experiment appeared to decrease following cyclin D1 expression.
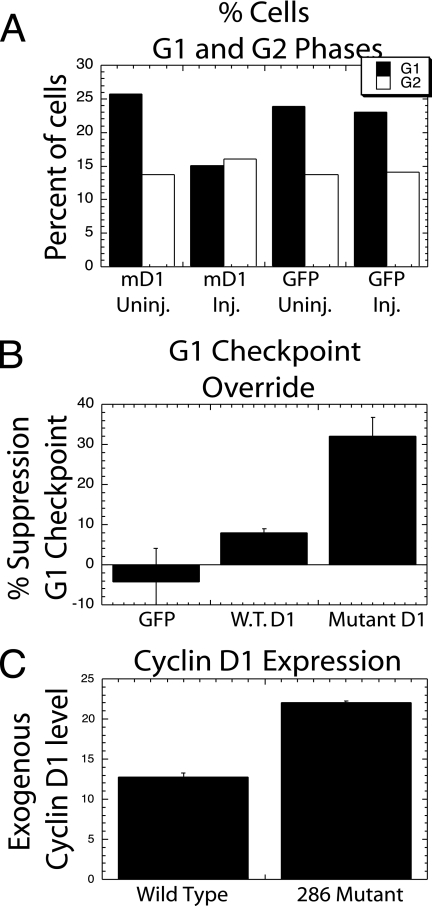
This initial indication that the G1/S-phase checkpoint block had been overridden by the mutant cyclin D1 protein was extended by an analysis of cells labeled by BrdU at the end of the experiment described above. The numbers of cells in G1 phase compared to G2 phase were determined by DNA content and BrdU labeling. The injection of the mutant cyclin D1 expression plasmid into NCS-treated cells resulted in the reduction of G1-phase cells by almost half, compared to either uninjected cells or control GFP-injected cells (Fig. 10A). This reduction in G1-phase cells took place without a major change in the number of G2-phase cells, indicating that the DNA damage had blocked mitosis.
FIG. 10.
Microinjected cyclin D1 overcomes the G1 phase DNA damage checkpoint. (A) Cycling cells were injected with either the T286A cyclin D1-GFP expression plasmid or the control GFP plasmid. The cells were then treated with NCS as described in the legend for Fig. 9A. Based upon BrdU labeling, the proportion of all cells in G1 or G2 phase at the end of each experiment is shown for injected (Inj.) and neighboring uninjected (Uninj.) cells in a single representative experiment. (B) This experiment was repeated multiple times following the injection of mutant cyclin D1, wild-type cyclin D1 (W.T.D1), or control GFP expression plasmids. The number of cells initially in G1 phase that entered S phase following DNA damage (EdU-unlabeled and BrdU-labeled cells with G1/S-phase DNA content) was compared to the number of cells that remained in G1 phase throughout the experiment (BrdU/EdU-unlabeled cells with G1-phase DNA). The proportion of cells entering S phase following each injection along with this proportion for neighboring uninjected cells was determined. The number plotted represents the increase in the G1/S-phase transition induced by the injection over the number observed in uninjected cells (±SE) (n = 7 for control GFP, n = 11 for mutant cyclin D1, and n = 4 for wild-type cyclin D1). (C) The average expression level of the cyclin D1-GFP in microinjected cells was determined following the injection of mutant and wild-type cyclin D1-GFP expression plasmids, followed by determination of the average amount of GFP per cell. Average expression levels from multiple experiments are reported (±SE).
To provide final evidence that cyclin D1 had overcome the G1/S-phase checkpoint block induced by dsDNA damage, and to measure the overall efficiency of this event, the proportion of cells initially in G1 phase (with G1/S DNA content and no EdU labeling) was compared to the proportion of these cells remaining in G1 phase at the end of the experiment (with G1 DNA content and no BrdU labeling). This analysis was made possible by the fact that since no cells passed through mitosis following DNA damage, all cells in G1 phase at the end of the experiment must have been in G1 at the beginning of the experiment. In NCS-treated, DNA-damaged cells, the expression of the mutant cyclin D1 in multiple experiments induced approximately one-third of the cells that would have remained in G1 phase to progress into S phase, while the control GFP injection had no apparent effect (Fig. 10B). To confirm this result, the experiment was performed upon NIH 3T3 cells synchronized in S phase by thymidine treatment (15 h) as diagrammed (Fig. 9A, right). The injection of the GFP control plasmid had no effect in overriding the G1/S cell cycle blockage in these synchronized cells, while mutant cyclin D1 expression induced almost 50% of the otherwise blocked cells to enter S phase following NCS treatment (data not shown). In numerous experiments with cycling or with thymidine-synchronized cells, the probability that cyclin D1 had overcome the G1-phase checkpoint blockage induced by dsDNA breakage was P = 0.007 and P = 0.006, respectively. When cycling cells were treated with UV irradiation in the place of NCS treatment, however, the blockage of cell cycle progression during G1 and G2 phases was slight and variable even in the absence of injection. It is not surprising, therefore, that no consistent evidence for the override of this blockage following cyclin D1 expression was observed (data not shown).
The T286A cyclin D1 mutant was utilized in the above experiments because it would remain stable even in the presence of a Thr286 kinase. To directly demonstrate that the ability to avoid Thr286 phosphorylation was important in the action of the cyclin D1 mutant, the mutant expression plasmid was directly compared to a plasmid expressing the wild-type cyclin D1 protein. Repeated injections of plasmid DNA (1.0 μg/ml) for the two cyclin D1 constructs were performed into cycling cells and analyzed as described above. The wild-type cyclin D1 construct was far less effective than the mutant construct in overcoming G1/S-phase checkpoint blockage, although it might have had some limited effect (Fig. 10B). When the overall expression levels of the two cyclin D1 constructs were compared based upon expression of the linked GFP sequence, it was clear that the wild-type protein was expressed at reduced levels (Fig. 10C). This most likely explained the difference in the activity of the two constructs and confirms the importance of Thr286 phosphorylation in cell cycle blockage induced by checkpoint kinases. Based upon all these considerations, we conclude not only that checkpoint kinases are able to direct the phosphorylation of cyclin D1 Thr286 but that this action plays a critical role in checkpoint cell cycle blockage during G1 phase.
DISCUSSION
Cyclin D1 expression declines during S phase as indicated by cytometric as well as biochemical analyses (2, 10, 14, 18, 19, 27). Without this suppression, DNA synthesis would be inhibited because cyclin D1 binds to and inhibits PCNA (20, 23, 26). Because of its suppression during S phase, the cell must resynthesize cyclin D1 during G2 phase if active cell cycle progression is to continue. This requires the assessment of cellular growth conditions and the activation of proliferative signaling proteins immediately prior to the commitment to continued proliferation during G2 phase (14, 34). Our results indicating that S-phase cyclin D1 suppression requires the phosphorylation of Thr286 by a kinase activated directly by DNA synthesis (11) lead to the prediction tested here that ATR might be involved in this phosphorylation. While identified as a DNA-damage response kinase, this essential protein has recently been shown to become activated by, and to then regulate, normal DNA synthesis (16, 25, 28, 33). We present evidence that ATR is indeed able to promote Thr286 phosphorylation. UV irradiation, which has been shown to specifically activate ATR in S-phase cells (12, 38), induces cyclin D1 phosphorylation together with histone H2AX phosphorylation during S phase. Both were inhibited by pretreatment with siRNA against ATR. The specific ATR activator, TopBP1, stimulates both cyclin D1 Thr286 and H2AX phosphorylation in all cell cycle phases, but an inactive TopBP1 mutant does not. In direct support of the predictions of our cell cycle model, ATR was also shown to promote cyclin D1 Thr286 phosphorylation during S phase of normal cell cycle progression. The overall conclusion that ATR is able to direct cyclin D1 phosphorylation was extended by demonstrating that the related DNA-damage response kinase, ATM, has a similar and perhaps even more robust activity. Our combined data would suggest that while ATR is more likely to direct cyclin D1 phosphorylation in normal cell cycle progression, ATM is more active in cyclin D1 phosphorylation following DNA damage. The validation of our initial hypothesis serves to confirm the previous cell cycle studies upon which it was based.
These data not only confirm the predictions of our cell cycle model but also have interesting implications for the DNA-damage response. The three DNA-damage response kinases, ATM, ATR, and DNA-PK, have the ability to halt cell cycle progression at various stages of the cell cycle, allowing the cell the opportunity to correct the damage before a decision is made to continue proliferation or initiate apoptosis. Upon DNA damage, these kinases stabilize p53, resulting in p21Waf1 synthesis, or destabilize CDC25, leading to elevated CDK Thr14 and Tyr15 phosphorylation, both of which directly inhibit CDK2 or CDK1 activity and block cell cycle progression at G1/S, S, or G2/M phase (13, 31). It has been shown, however, that ionizing irradiation can block cell cycle progression during G1 phase even in p53−/− cells. To explain this observation, it was found that cyclin D1 was suppressed during this cell cycle period by ionizing radiation, although the phosphorylation of Thr286 was not involved (1). Our results directly link DNA-damage response kinases, cyclin D1 phosphorylation, and DNA damage together. From past studies, we know that the suppression of cyclin D1 blocks cell cycle progression during G1 phase, leading to entry into quiescence. The suppression of cyclin D1 by the DNA-damage response kinases, therefore, might provide a means for a cell to exit the cell cycle and enter quiescence, thus allowing the time needed to completely repair all DNA lesions prior to reentering the cell cycle. This could provide an advantage over a temporary halt in cell cycle progression, as many lesions are not rapidly repaired (36). In support of this contention, we were able to directly demonstrate that the expression of a cyclin D1 mutant unable to be phosphorylated at position 286 was able to efficiently override the G1/S-phase blockage observed following dsDNA breakage, even though it had no effect upon the G2/M- or S-phase checkpoints. Thus, by the suppression of cyclin D1 levels, ATM is able to promote a G1-phase arrest to allow the repair of damaged DNA. It is important to note that despite the fact that our cells contain active p53 (Fig. 8B), from 30 to 50% of the G1-phase blockage was apparently due to the suppression of cyclin D1 protein levels following ATM activation (Fig. 10).
The suppression of cyclin D1 by DNA-damage response kinases also has the potential to directly aid DNA repair synthesis. While PCNA plays a central role in DNA replication, it is also known to play an essential role in several types of DNA repair synthesis, including mismatch repair, base excision repair, and nucleotide excision repair (22). Cyclin D1, as indicated above, binds PCNA and blocks its activity. The fact that this binding can actually block repair DNA synthesis has been shown following the UV irradiation of cells in G1 phase. The resulting repair synthesis was greatly enhanced in these cells if the level of cyclin D1 was artificially reduced (23). It appears likely, therefore, that the ability of DNA-damage response kinases to suppress cyclin D1 might aid in the overall process of DNA-damage repair by making PCNA more readily available to the DNA repair machinery.
These observations make an interesting connection between cell cycle progression and the DNA-damage response. ATR is known to be vital for the repair of lesions induced by UV irradiation. Recent studies indicate that ATR is also required, along with ATM and DNA-PK, for the repair of dsDNA lesions (5, 15, 41) and that ATM can be activated by ATR (35). Clearly, ATR serves as a central component of the DNA-damage response. The fact that ATR also plays a necessary role in normal S-phase progression ensures that its activity is always present in actively cycling cells; otherwise, high levels of cyclin D1 would block DNA synthesis. Thus, if a mutation were to eliminate ATR activity, the cell would stop proliferation before it could sustain DNA damage due to the absence of ATR.
These studies leave several unanswered questions. First, we are not certain that ATM and ATR directly phosphorylate cyclin D1. These kinases generally target a serine or threonine followed by a glutamine, making the direct phosphorylation of cyclin D1 unlikely, although perhaps not out of the question (32). It would, therefore, appear more likely that targets of these checkpoint kinases directly catalyze cyclin D1 phosphorylation on Thr286. In this light, it is critical to emphasize that the phosphorylation of cyclin D1 on Thr286, leading to its degradation, takes place continuously throughout the cell cycle (11). This phosphorylation, particularly in G1 and G2 phases, is most likely regulated by a kinase unrelated to ATM or ATR. It remains possible, however, that a single cyclin D1 kinase is responsible for cyclin D1 phosphorylation throughout the cell but that it responds to multiple regulatory signals, including ATM and ATR. Second, it is not clear if ATR possesses a mechanism to promote cyclin D1 suppression in addition to Thr286 phosphorylation. It is reported that at least at a high dose, UV rapidly suppresses cyclin D1 mRNA expression (21). Finally, our preliminary evidence suggests DNA-PK might not promote robust cyclin D1 phosphorylation following dsDNA breakage induced by NCS treatment, but further study will be required to confirm this observation (4).
Acknowledgments
We thank Akiko Kumagai, Division of Biology, California Institute of Technology, for generously providing the TopBP1 plasmids used in this study and for valuable review of the manuscript. We also thank David R. Mason and Michelle D. Garrett for providing antibodies against phosphorylated cyclin D1 early in these studies. We are grateful for the able technical assistance of Yang Guo and Jyoti Harwalkar.
These studies were supported by NIH grant GM02271.
Footnotes
Published ahead of print on 7 July 2008.
REFERENCES
- 1.Agami, R., and R. Bernards. 2000. Distinct initiation and maintenance mechanisms cooperate to induce G1 cell cycle arrest in response to DNA damage. Cell 10255-66. [DOI] [PubMed] [Google Scholar]
- 2.Baldin, V., J. Lukas, M. J. Marcote, M. Pagano, and G. Draetta. 1993. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 7812-821. [DOI] [PubMed] [Google Scholar]
- 3.Bartek, J., and N. Mailand. 2006. TOPping up ATR activity. Cell 124888-890. [DOI] [PubMed] [Google Scholar]
- 4.Collis, S. J., T. L. DeWeese, P. A. Jeggo, and A. R. Parker. 2005. The life and death of DNA-PK. Oncogene 24949-961. [DOI] [PubMed] [Google Scholar]
- 5.Cuadrado, M., B. Martinez-Pastor, M. Murga, L. I. Toledo, P. Gutierrez-Martinez, E. Lopez, and O. Fernandez-Capetillo. 2006. ATM regulates ATR chromatin loading in response to DNA double-strand breaks. J. Exp. Med. 203297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diehl, J. A., M. Cheng, M. F. Roussel, and C. J. Sherr. 1998. Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 123499-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diehl, J. A., F. Zindy, and C. J. Sherr. 1997. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev. 11957-972. [DOI] [PubMed] [Google Scholar]
- 8.Reference deleted.
- 9.Reference deleted.
- 10.Guo, Y., J. Harwalkar, D. W. Stacey, and M. Hitomi. 2005. Destabilization of cyclin D1 message plays a critical role in cell cycle exit upon mitogen withdrawal. Oncogene 241032-1042. [DOI] [PubMed] [Google Scholar]
- 11.Guo, Y., K. Yang, J. Harwalkar, J. M. Nye, D. R. Mason, M. D. Garrett, M. Hitomi, and D. W. Stacey. 2005. Phosphorylation of cyclin D1 at Thr 286 during S phase leads to its proteasomal degradation and allows efficient DNA synthesis. Oncogene 242599-2612. [DOI] [PubMed] [Google Scholar]
- 12.Halicka, H. D., X. Huang, F. Traganos, M. A. King, W. Dai, and Z. Darzynkiewicz. 2005. Histone H2AX phosphorylation after cell irradiation with UV-B: relationship to cell cycle phase and induction of apoptosis. Cell Cycle 4339-345. [PubMed] [Google Scholar]
- 13.Harrison, J. C., and J. E. Haber. 2006. Surviving the breakup: the DNA damage checkpoint. Annu. Rev. Genet. 40209-235. [DOI] [PubMed] [Google Scholar]
- 14.Hitomi, M., and D. W. Stacey. 1999. Cyclin D1 production in cycling cells depends on ras in a cell-cycle-specific manner. Curr. Biol. 91075-1084. [DOI] [PubMed] [Google Scholar]
- 15.Jazayeri, A., J. Falck, C. Lukas, J. Bartek, G. C. Smith, J. Lukas, and S. P. Jackson. 2006. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 837-45. [DOI] [PubMed] [Google Scholar]
- 16.Jirmanova, L., D. V. Bulavin, and A. J. Fornace, Jr. 2005. Inhibition of the ATR/Chk1 pathway induces a p38-dependent S-phase delay in mouse embryonic stem cells. Cell Cycle 41428-1434. [DOI] [PubMed] [Google Scholar]
- 17.Kumagai, A., J. Lee, H. Y. Yoo, and W. G. Dunphy. 2006. TopBP1 activates the ATR-ATRIP complex. Cell 124943-955. [DOI] [PubMed] [Google Scholar]
- 18.Lukas, J., D. Jadayel, J. Bartkova, E. Nacheva, M. J. Dyer, M. Strauss, and J. Bartek. 1994. BCL-1/cyclin D1 oncoprotein oscillates and subverts the G1 phase control in B-cell neoplasms carrying the t(11;14) translocation. Oncogene 92159-2167. [PubMed] [Google Scholar]
- 19.Lukas, J., M. Pagano, Z. Staskova, G. Draetta, and J. Bartek. 1994. Cyclin D1 protein oscillates and is essential for cell cycle progression in human tumour cell lines. Oncogene 9707-718. [PubMed] [Google Scholar]
- 20.Matsuoka, S., M. Yamaguchi, and A. Matsukage. 1994. D-type cyclin-binding regions of proliferating cell nuclear antigen. J. Biol. Chem. 26911030-11036. [PubMed] [Google Scholar]
- 21.Miyakawa, Y., and H. Matsushime. 2001. Rapid downregulation of cyclin D1 mRNA and protein levels by ultraviolet irradiation in murine macrophage cells. Biochem. Biophys. Res. Commun. 28471-76. [DOI] [PubMed] [Google Scholar]
- 22.Moldovan, G. L., B. Pfander, and S. Jentsch. 2007. PCNA, the maestro of the replication fork. Cell 129665-679. [DOI] [PubMed] [Google Scholar]
- 23.Pagano, M., A. M. Theodoras, S. W. Tam, and G. F. Draetta. 1994. Cyclin D1-mediated inhibition of repair and replicative DNA synthesis in human fibroblasts. Genes Dev. 81627-1639. [DOI] [PubMed] [Google Scholar]
- 24.Pereg, Y., D. Shkedy, P. de Graaf, E. Meulmeester, M. Edelson-Averbukh, M. Salek, S. Biton, A. F. Teunisse, W. D. Lehmann, A. G. Jochemsen, and Y. Shiloh. 2005. Phosphorylation of Hdmx mediates its Hdm2- and ATM-dependent degradation in response to DNA damage. Proc. Natl. Acad. Sci. USA 1025056-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petermann, E., and K. W. Caldecott. 2006. Evidence that the ATR/Chk1 pathway maintains normal replication fork progression during unperturbed S phase. Cell Cycle 52203-2209. [DOI] [PubMed] [Google Scholar]
- 26.Prosperi, E., A. I. Scovassi, L. A. Stivala, and L. Bianchi. 1994. Proliferating cell nuclear antigen bound to DNA synthesis sites: phosphorylation and association with cyclin D1 and cyclin A. Exp. Cell Res. 215257-262. [DOI] [PubMed] [Google Scholar]
- 27.Scovassi, A. I., L. A. Stivala, L. Rossi, L. Bianchi, and E. Prosperi. 1997. Nuclear association of cyclin D1 in human fibroblasts: tight binding to nuclear structures and modulation by protein kinase inhibitors. Exp. Cell Res. 237127-134. [DOI] [PubMed] [Google Scholar]
- 28.Shechter, D., V. Costanzo, and J. Gautier. 2004. ATR and ATM regulate the timing of DNA replication origin firing. Nat. Cell Biol. 6648-655. [DOI] [PubMed] [Google Scholar]
- 29.Sherr, C. J. 1995. D-type cyclins. Trends Biochem. Sci. 20187-190. [DOI] [PubMed] [Google Scholar]
- 30.Sherr, C. J., and J. M. Roberts. 1999. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 131501-1512. [DOI] [PubMed] [Google Scholar]
- 31.Shiloh, Y. 2006. The ATM-mediated DNA-damage response: taking shape. Trends Biochem. Sci. 31402-410. [DOI] [PubMed] [Google Scholar]
- 32.Smolka, M. B., C. P. Albuquerque, S. H. Chen, and H. Zhou. 2007. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc. Natl. Acad. Sci. USA 10410364-10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sorensen, C. S., R. G. Syljuasen, J. Lukas, and J. Bartek. 2004. ATR, Claspin and the Rad9-Rad1-Hus1 complex regulate Chk1 and Cdc25A in the absence of DNA damage. Cell Cycle 3941-945. [PubMed] [Google Scholar]
- 34.Stacey, D. W. 2003. Cyclin D1 serves as a cell cycle regulatory switch in actively proliferating cells. Curr. Opin. Cell Biol. 15158-163. [DOI] [PubMed] [Google Scholar]
- 35.Stiff, T., S. A. Walker, K. Cerosaletti, A. A. Goodarzi, E. Petermann, P. Concannon, M. O'Driscoll, and P. A. Jeggo. 2006. ATR-dependent phosphorylation and activation of ATM in response to UV treatment or replication fork stalling. EMBO J. 255775-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vilenchik, M. M., and A. G. Knudson. 2003. Endogenous DNA double-strand breaks: production, fidelity of repair, and induction of cancer. Proc. Natl. Acad. Sci. USA 10012871-12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, X., L. Zou, T. Lu, S. Bao, K. E. Hurov, W. N. Hittelman, S. J. Elledge, and L. Li. 2006. Rad17 phosphorylation is required for Claspin recruitment and Chk1 activation in response to replication stress. Mol. Cell 23331-341. [DOI] [PubMed] [Google Scholar]
- 38.Ward, I. M., K. Minn, and J. Chen. 2004. UV-induced ataxia-telangiectasia-mutated and Rad3-related (ATR) activation requires replication stress. J. Biol. Chem. 2799677-9680. [DOI] [PubMed] [Google Scholar]
- 39.Xiong, Y., H. Zhang, and D. Beach. 1992. D type cyclins associate with multiple protein kinases and the DNA replication and repair factor PCNA. Cell 71505-514. [DOI] [PubMed] [Google Scholar]
- 40.Yajima, H., K. J. Lee, and B. P. Chen. 2006. ATR-dependent phosphorylation of DNA-dependent protein kinase catalytic subunit in response to UV-induced replication stress. Mol. Cell. Biol. 267520-7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoo, H. Y., A. Kumagai, A. Shevchenko, and W. G. Dunphy. 2007. Ataxia-telangiectasia mutated (ATM)-dependent activation of ATR occurs through phosphorylation of TopBP1 by ATM. J. Biol. Chem. 28217501-17506. [DOI] [PubMed] [Google Scholar]