Abstract
Exposure of phosphatidylserine (PS) on the cell surface occurs early during apoptosis and serves as a recognition signal for phagocytes. Clearance of apoptotic cells by a membrane PS receptor is one of the critical anti-inflammatory functions of macrophages. However, the PS binding receptors and their recognition mechanisms have not been fully investigated. Recently, we reported that stabilin-2 is a PS receptor that mediates the clearance of apoptotic cells, thus releasing the anti-inflammatory cytokine, transforming growth factor β. In this study, we showed that epidermal growth factor (EGF)-like domain repeats (EGFrp) in stabilin-2 can directly and specifically recognize PS. The EGFrps also competitively impaired apoptotic cell uptake by macrophages in in vivo models. We also showed that calcium ions are required for stabilin-2 to mediate phagocytosis via EGFrp. Interestingly, at least four tandem repeats of EGF-like domains were required to recognize PS, and the second atypical EGF-like domain in EGFrp was critical for calcium-dependent PS recognition. Considering that PS itself is an important target molecule for both apoptotic cells and nonapoptotic cells during various cellular processes, our results should help elucidate the molecular mechanism by which apoptotic cell clearance in the human body occurs and also have implications for targeting PS externalization of nonapoptotic cells.
Rapid and efficient cell corpse clearance protects normal healthy cells from cytotoxic and antigenic compounds, thereby reducing the amount of tissue damage that occurs as a result of inappropriate inflammation or autoimmune responses (29). The central element of the recognition process is the cell surface presentation of repulsive ligands (also termed “don't eat me” signals) and adhesive engulfment ligands (also termed “eat me” signals) by apoptotic cells (10, 13, 20). Although accumulating data identified several engulfment ligands, such as phosphatidylserine (PS), calrecticulin, and modified surface sugars (8), the best-characterized marker of apoptotic cells is the exposure of PS on the outer leaflet of the plasma membrane, which is associated with a loss of phospholipid asymmetry (30). The underlying mechanisms mediating the process of PS externalization seem to involve inhibition of the ATP-dependent aminophospholipid translocase and activation of a Ca2+-dependent phospholipid scramblase (33, 38).
Phagocytes recognize the PS on the apoptotic cell surface through either the membrane PS receptor or secreted bridging molecules. The former is able to directly recognize PS and subsequently engulf apoptotic cells and release anti-inflammatory cytokines, whereas the latter directly bind to exposed PS and enhance the engulfment of apoptotic cells by bridging PS with its receptors on the phagocytes, including protein S, Gas6, and MFG-E8 (36). However, the above-mentioned bridging molecules do not explain all of the observed responses to PS, including the attendant anti-inflammatory and anti-immunogenic effects (8). We recently reported that stabilin-2 is a membrane receptor that recognizes aged and apoptotic cells and subsequently mediates the engulfment of apoptotic cells and the release of an anti-inflammatory cytokine, transforming growth factor β, by macrophages (25). PS liposomes and its structural analogues specifically inhibited the binding and engulfment of aged and apoptotic cells, suggesting that stabilin-2 stereospecifically recognizes PS on the apoptotic cell surface. We therefore proposed to define the domain structure and molecular mechanism of stabilin-2-mediated PS-recognition during the phagocytosis of apoptotic cells.
Stabilin-2 contains a large extracellular domain that consists of seven FAS1 domains, one X-link domain, and four epidermal growth factor (EGF)-like domain repeats (EGFrps). The FAS1 domain was originally described in fasciclin I, which is expressed on subsets of the axon pathways during neuronal development in the grasshopper (3), and FAS1-containing molecules, such as βig-h3, have been reported to function as adhesion molecules (18). The Link domain is a hyaluronan-binding region that is found in vertebrate proteins that are involved in the assembly of the extracellular matrix, cell adhesion, and migration (19). The EGF-like domain includes six or eight cysteine residues that are known to be involved in disulfide bond formation. Although the functional significance of these EGF-like domains is still unknown, a certain EGF-like domain was shown to mediate homophilic or heterophilic protein-protein interaction (2, 14). Therefore, it is possible that the stabilin-2 receptor can interact with various ligands through its multidomain structure. In the present study, we demonstrate that EGFrp is responsible for the interaction of stabilin-2 with PS. In addition, the potential for the second atypical EGF-like domain to serve as a domain for calcium-dependent PS-recognition is addressed and supported by the evidence reported herein.
MATERIALS AND METHODS
Reagents and antibodies.
Monoclonal antibody (MAb) to c-Myc (clone 9E10) was obtained from Santa Cruz. PerCP-Cy5 conjugated anti-mouse CD11b antibody (M1/70) was purchased from BD Pharmingen. MAb to CD68 (clone KP1) was purchased from Abcam. Horseradish peroxidase-conjugated goat anti-His antibody was purchased from Invitrogen. MAb to human stabilin-2 (clone 5G3) was previously described (25). NBD-PC {1-oleoyl-2-[6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl]-sn-glycero-3-phosphocholine}, NBD-PS {1-oleoyl-2-[6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl]-sn-glycero-3-phospho-l-serine}, and all phospholipids were obtained from Avanti-Polar Lipids.
Cell culture.
L cells that stably expressed stabilin-2 (L/Stab-2 cells) and parental L cells were grown in Dulbecco modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS), as previously described (25). U937 and Jurkat T cells were maintained in RPMI 1640 medium containing 10% FBS. HEK293 cells were maintained in DMEM (high glucose) medium containing 10% FBS. Human monocytes were obtained from normal donors, isolated using monocyte isolation kit II systems (Milteny Biotec), and then cultured in X-Vivo 10 (BioWhittaker) containing 10% human serum. The differentiated macrophages, human monocyte-derived macrophages (HMDMs), were utilized at 7 to 10 days of culture.
Preparation of recombinant protein in bacteria.
A Nus tag was fused to each recombinant protein to enhance the solubility of proteins corresponding to the repeated unit. Nus-tagged recombinant proteins corresponding to repeated units, EGFrps, or fifth FAS1 domain were expressed and purified as described previously (15). To produce recombinant proteins corresponding to the C-terminal deletion mutants of EGFrp (E3), fragments of stabilin-2 cDNA encoding amino acids 1333 to 1596, 1333 to 1554, 1333 to 1512, 1333 to 1470, 1333 to 1431, and 1333 to 1383 were generated by PCR; cloned into the BamHI and XhoI sites of pET-28a (Novagen); and designated E3-16, -15, -14, -13, -12, and -1, respectively. To produce recombinant proteins corresponding to the N-terminal deletion mutants of EGFrp (E3), fragments of stabilin-2 cDNA encoding amino acids 1384 to 1596, 1432 to 1596, 1471 to 1596, 1513 to 1596, and 1555 to 1596 were generated by PCR; cloned into the BamHI and XhoI sites of pET-28a (Novagen); and then designated E3-26, -36, -46, -56, and -6, respectively. To produce recombinant proteins corresponding to the EGFrp mutant that lacked the first atypical EGF-like domain (E3-25), the coding regions of stabilin-2 amino acids 1384 to 1554 were generated by PCR and then cloned into the BamHI and XhoI sites of pET-28a. To produce recombinant proteins corresponding to the EGFrp lacking the second atypical EGF-like domain (E3-15Δ2), the coding regions of stabilin-2 amino acids 1333 to 1383 were cloned into BamHI and EcoRI sites of pET28a, and subsequently the coding regions of stabilin-2 amino acids 1432 to 1554 were cloned into EcoRI and XhoI sites. These His-tagged recombinant proteins were then expressed in BL-21cells, harvested, and purified by using Ni-NTA resin (Qiagen) according to the manufacturer's instructions.
To produce thioredoxin-His6-EGFrp from Escherichia coli K-12 strain Origami (DE3) (Novagen), fragments of stabilin-2 cDNA encoding amino acids 1333 to 1596 were generated by PCR and cloned into the BamHI and XhoI sites of pET32a (Novagen). Positive clones were cultured overnight at 37°C in LB medium (with 50 mg of kanamycin/ml, 50 mg of ampicillin/ml, and 15 mg of tetracycline/ml), diluted 1:100, and grown until the optical density at 600 nm reached 0.5. At this point, 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added to induce expression of the fusion protein, and cells were incubated overnight at 18°C. Protein was purified from the cell lysate as previously described (15). EGFrp was cleaved from the fusion protein by using thrombin (Amersham Pharmacia) in accordance with the manufacturer's instructions. Contaminating endotoxins were removed with Detoxi-gel (Pierce).
Preparation of recombinant protein in mammalian cells.
To generate an expression vector for EGFrp-Fc fusion protein, cDNA encoding immunoglobulin Fc region was amplified by PCR, cloned into pcDNA3.1(+) (Invitrogen), and then designated pcDNA-Fc. Next, fragments of stabilin-2 cDNA encoding signal peptide and third EGFrp were amplified from full-length stabilin-2 cDNA by PCR, cloned into pcDNA-Fc vector, and then designated pcDNA-E3-Fc. HEK293 cells were then transfected with pcDNA-E3-Fc vector for 6 h by using Lipofectamine (Invitrogen), in accordance with the manufacturer's instructions. At 24 h after transfection, the culture medium was exchanged with serum-free DMEM. The supernatant was collected after 48 h, and E3-Fc proteins were purified by using protein A-conjugated agarose (Amersham Pharmacia) in accordance with the manufacturer's instructions.
Stable transfection.
To generate an expression vector for mutant stabilin-2, in which the extracellular domain was replaced with the third EGFrp, fragments of stabilin-2 cDNA encoding signal peptide, the third EGFrp, and the transmembrane and cytoplasmic region were amplified from full-length stabilin-2 cDNA by PCR; cloned into pcDNA3.1(−)/Myc-His vector (Invitrogen); and then designated pcDNA-EGF3. To generate an expression vector for mutant stabilin-2, in which the extracellular domain was replaced with the fourth repeated unit, fragments of stabilin-2 cDNA encoding signal peptide, the third EGFrp, and the transmembrane and cytoplasmic region were amplified from full-length stabilin-2 cDNA by PCR; cloned pcDNA3.1(−)/Myc-His vector (Invitrogen); and then designated pcDNA-U4. Mouse fibroblast L cells were transfected with pcDNA-EGF3 or pcDNA-U4 vector using Lipofectamine (Invitrogen) in accordance with the manufacturer's instructions. For stable transfection, L cells were selected in G418 (400 μg/ml), and then individual G418-resistant colonies were isolated after 10 to 12 days of culture. The final clones are designated L/EGF3-# or L/U4-#. Negative control clones were selected randomly by transfection with the empty vector (L/Mock). The expression of stabilin-2 was assessed via flow cytometry.
Fluorescence-activated cell sorting (FACS) analysis.
For the analysis of surface staining, the cells were incubated with 1 μg of MAb 5G3/ml on ice for 30 min with gentle agitation every 5 min. Next, the cells were washed twice, resuspended in phosphate-buffered saline (PBS), and incubated with 4 μg of fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse immunoglobulin G antibody/ml for 45 min on ice. After three washes with PBS, the cells were resuspended in PBS and then analyzed via flow cytometry on a FACScalibur cytometer (BD Biosciences).
Liposome preparation and liposome incorporation assay.
Liposomes containing the indicated phospholipids and phosphatidylcholines in a molar ratio of 50:50 were prepared. The lipids were mixed in chloroform and then dried under nitrogen gas. The dried lipids were resuspended in PBS at final concentration of 5 mM and then sonicated for 10 min on ice. Fluorescence-labeled liposomes were prepared in the presence of N-(lissamin rhodamine B sulfonyl)-l-α-phosphatidylethanolamine (Avanti-Polar Lipids) at a concentration of 1% of the total phospholipids, as previously described (17). The affinity of fluorescent liposomes to L cell transfectants was then quantified by flow cytometry.
Binding and phagocytosis of aged RBCs.
Aged red blood cells (RBCs) were prepared via the incubation of cells in PBS (20% hematocrit) at 37°C, as previously described (24). Exposure of PS on the cell surface was detected via annexin V-FITC using an annexin V apoptotic detection kit (Santa Cruz). Next, aged RBCs were added to the transfected L cells or HMDMs and then incubated for 1 h to assess binding and engulfment in the presence of various recombinant stabilin-2 proteins (10 μΜ). Recombinant stabilin-2 proteins were preincubated with aged RBCs for 30 min at 37°C in medium (containing 1.8 mM CaCl2). After the unbound RBCs were washed away, the uningested RBCs were lysed via addition of deionized H2O for 10 s, which was followed by immediate replacement with DMEM as described previously (23). The cells were then fixed with methanol and stained by using a DiffQuick staining kit (IMEB, Inc.). The binding and engulfment of aged RBCs was then quantified via light microscopy prior to and after hypotonic lysis, respectively. The percentages of binding and phagocytosis were determined as the percentages of phagocytes that were positive for binding and engulfment, respectively, as described previously (25). At least 100 cells were scored per well, and all experiments were repeated at least three times. In certain experiments, the phagocytosis assay was performed in the presence of MAb 5G3 (10 μg/ml), isotype controls (immunoglobulin G1, 10 μg/ml), or 10 μM liposomes containing phosphatidylcholine (PC) or PS.
Binding and phagocytosis of apoptotic cells.
Apoptosis was induced in Jurkat T cells by anti-Fas antibody (CH-11 [Upstate], 100 ng/ml, 6 h) as described previously (24). Loss of phospholipid asymmetry and exposure of PS were evaluated by annexin V-FITC binding. Next, apoptotic cells were added to the transfected L cells, which were then incubated for 2 h to assess the uptake that occurred in the presence of recombinant stabilin-2 proteins. After extensive washing away of the uningested cells, the uptake of apoptotic cells was evaluated by using an in situ cell death detection kit (Roche) and quantified under a fluorescence microscope (Carl Zeiss).
Solid-phase ELISA and cell adhesion assay.
A solid phase enzyme-linked immunosorbent assay (ELISA) for the binding of stabilin-2 recombinant proteins to PS was conducted as previously described (12). Briefly, a phospholipid solution (3 μg/ml, 100 μl) in methanol was added to ELISA plates, which were then air dried. Next, the wells were treated with PBS containing 2% bovine serum albumin to block nonspecific bindings. Stabilin-2 derivatives were then added to the wells, which were subsequently incubated at room temperature for 1 h in binding buffer (10 mM HEPES [pH 7.4], 140 mM NaCl, and 2.5 mM CaCl2). After a washing step with PBS containing 0.05% Tween 20, protein bound to the wells was quantified by ELISA with peroxidase-conjugated anti-His antibody.
A cell adhesion assay was then performed as previously described (18). Briefly, ELISA plates were coated with PC or PS as described above and then blocked for 1 h at room temperature with PBS containing 2% bovine serum albumin. Next, the cells were suspended in medium (containing 1.8 mM CaCl2) at a density of 3 × 105 cells/ml, and 0.1 ml of the cell suspension was added to each well. After incubation for 1 h at 37°C, unattached cells were removed by two rinses with PBS. The attached cells were then incubated for 1 h at 37°C in 50 mM citrate buffer (pH 5.0) containing 3.75 mM p-nitrophenyl-N-acetyl-β-d-glycosaminide and 0.25% Triton X-100. The enzyme activity was then blocked by adding 50 mM glycine buffer (pH 10.4) containing 5 mM EDTA, and the absorbance was measured at 405 nm in a microplate reader (Bio-Rad).
Fluorescence binding assays and data analysis.
NBD-labeled liposome was prepared from PC and PC:PS (1:1) as previously described (16) with the addition of NBD-PC and NBD-PS, respectively (ratio, 0.5 mol%). EGFrp or its deletion mutants were added to NBD-liposome containing binding buffer (50 mM HEPES [pH 7.4], 100 mM NaCl, 25 mM KCl, 2.5 mM CaCl2). Quenching of the NBD-liposome fluorescence was then monitored by using a Perkin-Elmer L55 fluorescence spectrometer with an excitation wavelength of 460 nm and excitation and emission slits of 5 and 10 nm, respectively.
The quenching data can be described by the Stern-Volmer law as Fo/(Fo − F) = 1/fa + 1/(faK[Mt]), where K is a dynamic quenching constant and fa is the fraction of total fluorophore. The extent of quenching is normalized by subtraction of the buffer to eliminate possible quenching phenomena due to dilution or liposome flexibility. After normalization, the quenching constant, K, is then directly related to the binding affinity of the complex, which can be determined by plotting the quenching extent [Fo/(Fo − F)] versus the inverse protein concentration (/[protein]).
In vivo phagocytosis of apoptotic cells.
The in vivo phagocytic clearance of apoptotic cells was examined according to a previously described method (34). Briefly, mice were injected intraperitoneally with 2 ml of sterile 3% Brewer's thioglycolate to induce peritonitis. Four days later, 2 × 107 apoptotic thymocytes prepared as previously described (28) were incubated with 1 μM EGFrp proteins for 30 min at 37°C in binding buffer (10 mM HEPES [pH 7.4], 140 mM NaCl, and 2.5 mM CaCl2) and then injected into the peritoneum in a volume of 200 μl. Control apoptotic thymocytes were incubated for the same period with control Nus protein. After 45 min, the mice were sacrificed, and the peritoneal cells were collected by lavaging the cavity with ice-cold HBSS containing 5 mM EDTA. FACS analysis of the phagocytic uptake was then performed by fluorescent labeling of the apoptotic cells prior to injection using a Vybrant cell tracer kit (Molecular Probes).
Gel filtration chromatography.
Purified bacterial EGFrp was loaded onto Superdex 75 HR column that was equilibrated with the binding buffer (20 mM HEPES [pH 7.4], 100 mM NaCl) with the flow rate of 12 ml/h and a fraction size of 0.4 ml using an AKTA purifier (Amersham Biosciences). The monomer fractions were collected for further analysis.
SDS-PAGE.
The 20-μl aliquot of monomer fraction was mixed with dithiothreitol (DTT)-containing sample buffer, whereas other aliquot mixed with non-DTT sample buffer. Each sample was boiled for 5 min and subjected to sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (SDS-12% PAGE). At the end of electrophoresis, the gel was stained with Coomassie brilliant blue.
MALDI-TOF MS.
Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (MS) analysis was performed by using a Voyager-DE STR (Perseptive Biosystems) mass spectrometer. For reducing conditions, aliquots of the monomer fraction were incubated with 10 mM DTT for 30 min at room temperature. For nonreducing condition, the aliquots of the monomer fractions were prepared without DTT treatment. Then, 1 μl of desalted sample was mixed with 1 μl of 3,5-dimethoxy-4-hydroxycinnamic acid (Aldrich) solution in 50% acetonitrile and 0.5% trifluoroacetic acid. A total of 1,000 shots from 20 different spots were scanned to acquire an MS spectrum. The intensity of each ion was obtained by integrating from the first to the third isotopic peaks of the ion. Explorer software (Applied Biosystems) was used to calculate the peak area. The relative percentage was the ratio of the intensity of each ion to the summated intensity of the detected total ions.
Statistical analysis.
The statistical significance was assessed by using an analysis of variance (ANOVA) test, and a P of <0.05 was considered statistically significant.
RESULTS
EGFrps of stabilin-2 competitively inhibit the uptake of aged RBCs and apoptotic cells via direct and preferential interaction with PS.
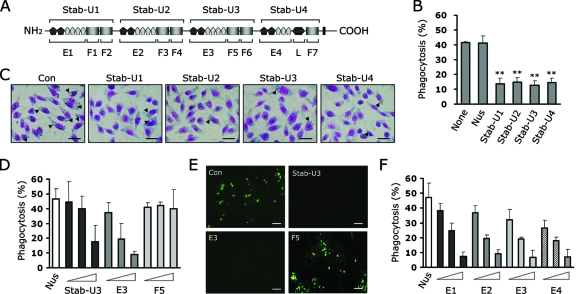
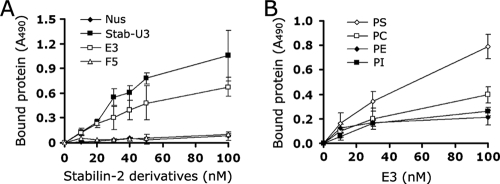
Our recent studies on stabilin-2 and its potential role in cell corpse clearance prompted us to investigate the molecular mechanism by which PS recognition occurs. To determine which regions of stabilin-2 are involved in PS recognition, we dissected the extracellular region into four repeated units (Stab-U1, -U2, -U3, and -U4) (Fig. 1A) and then examined the inhibitory effect of each unit protein on the phagocytic activity of stabilin-2-expressing cells (L/Stab-2 cells). Each repeated unit shares 48% sequence homology and consists of one EGFrp and two FAS1 domains (the fourth unit has a single FAS1 domain and a Link domain). Pretreatment of aged RBCs with each unit protein significantly inhibited their ingestion (Fig. 1B and C), indicating that each repeated unit contains a PS-binding domain in parallel and therefore competitively inhibits the uptake of aged RBCs. In order to further evaluate the PS-binding domain within the repeated unit, we next generated recombinant proteins of the FAS1 domain (F5) and EGFrp (E3) from the third unit (Stab-U3) of stabilin-2 and then examined their effects on the phagocytic activity of L/Stab-2 cells. The stab-U3 and E3 proteins impeded the engulfment of aged RBCs in a dose-dependent manner, whereas the F5 and control Nus proteins had no effect (Fig. 1D). Similar results were observed when apoptotic Jurkat T cells were used as target cells (Fig. 1E). For further experiments, we selected aged RBCs as target cells because they do not bind to phagocytes without apoptotic signals and allow the distinction between binding and engulfment via hypotonic lysis (9). EGFrps obtained from different units of stabilin-2 also showed a comparable inhibitory effect on the engulfment of aged cells, and this effect also occurred in a dose-dependent manner (Fig. 1F). These results suggest that EGFrps are responsible for the recognition of PS during engulfment of aged RBCs or apoptotic cells. To further demonstrate that EGFrps directly interact with PS, ELISA plates were coated with PS and then incubated with stabilin-2 derivatives. The stab-U3 and E3 proteins bound efficiently to the PS-coated plates, whereas the F5 and control Nus proteins did not (Fig. 2A). To evaluate the binding preference of EGFrp for PS among other plasma membrane phospholipids, ELISA plates were coated with various phospholipids and then incubated with the E3 protein (Fig. 2B). E3 showed a strong preference for PS, but not for other phospholipids, which is in agreement with the previous results that showed stabilin-2-mediated phagocytosis to be specifically inhibited by PS liposomes.
FIG. 1.
EGF-like domain repeats (EGFrp) of stabilin-2 competitively inhibit the uptake of aged RBCs and apoptotic cells. (A) Schematic diagram of stabilin-2 protein and deletion mutant> proteins. A Nus tag was fused to each recombinant protein to enhance the solubility of the proteins. Stab-U1, -U2, and -U3 are composed of one EGFrp and two FAS1 domains. Stab-U4 is composed of an EGFrp, a FAS1 domain, and a Link domain. (B) Effect of four repeated units on engulfment of aged RBCs by L/Stab-2 cells. Aged RBCs were preincubated with 10 μM concentrations of the indicated stabilin-2 proteins and then added to L/Stab-2 cells. After incubation for 1 h, the percentages of cells engulfing the aged RBCs were determined. **, P < 0.01 (ANOVA). (C) Representative images of stabilin-2-mediated phagocytosis in the presence of each repeated unit. Scale bar, 50 μm. (D) Effect of EGFrp-containing proteins on the engulfment of aged RBCs by L/Stab-2 cells. Aged RBCs were preincubated with three different concentrations (0.1, 1.0, or 10 μM) of the indicated stabilin-2 proteins and then added to L/Stab-2 cells. After incubation for 1 h, the percentages of cells engulfing the aged RBCs were determined. (E) Representative images of apoptotic cell engulfment by L/Stab-2 cells in the presence of recombinant stabilin-2 proteins. Apoptotic Jurkat T cells were preincubated with the indicated stabilin-2 proteins (10 μM) and then added to L/Stab-2 cells. After incubation for 1 h, the cells were extensively washed, stained using an in situ cell-death detection kit (Roche), and then observed under a fluorescence microscope. Scale bar, 100 μm. (F) Effect of EGFrps on the engulfment of aged RBCs by L/Stab-2 cells. Aged RBCs were preincubated with three different concentrations (0.1, 1.0, or 10 μM) of the indicated proteins and then added to L/Stab-2 cells. The percentages of cells engulfing the aged RBCs were determined. The results are expressed as the means ± the standard deviation (SD) from at least three experiments.
FIG. 2.
EGFrps directly and preferentially interact with PS. (A) Affinity of recombinant stabilin-2 proteins to PS. ELISA plates were coated with PS and then incubated with the indicated stabilin-2 proteins. After 1 h, proteins bound to the wells were quantified using anti-His antibody. (B) ELISA plates were coated with PS, PC, phosphatidylethanolamine (PE), or phosphatidylinositol (PI) and then incubated with the EGFrp protein (E3). After 1 h, proteins bound to the wells were quantified using anti-His antibody. The results are expressed as the means ± the SD from at least three experiments.
EGFrp-expressing cells mediate the binding of aged RBCs via the recognition of PS.
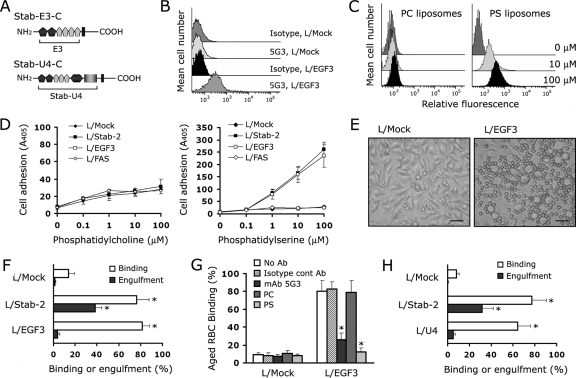
We next investigated whether the EGFrp on the cell surface is sufficient to recognize the PS of apoptotic cells. We established L-cell transfectants (L/EGF3 cells) that stably express mutant stabilin-2, in which the extracellular domain was replaced with the third EGFrp (Fig. 3A). Surface exposure of EGFrp was confirmed by flow cytometry using a monoclonal anti-stabilin-2 antibody (MAb 5G3) (25) that recognizes the third EGFrp of stabilin-2 (Fig. 3B). As expected, L/EGF3 cells were able to bind to fluorescence-labeled PS liposomes but not to PC liposomes, and this occurred in a dose-dependent manner (Fig. 3C). In addition, L/EGF3 cells were also able to specifically bind to the PS-coated plate as efficiently as the L/Stab-2 cells that express the full-length stabilin-2 (Fig. 3D). However, L/FAS7 cells that express the seventh FAS1 domain on the cell surface showed only basal binding affinity to both the PS- and the PC-coated plates (Fig. 3D). Although L/EGF3 cells were able to bind to aged RBCs in a similar manner to what was observed in L/Stab-2 cells (Fig. 3E and F), the L/EGF3 cells showed no engulfment of aged RBCs (Fig. 3F). The EGFrp-mediated recognition was demonstrated to be valid because binding was disrupted by either PS liposome or MAb 5G3 (Fig. 3G). These findings imply that, although EGFrp is critical for the tethering of apoptotic cells, EGFrp alone is not sufficient for the tickling process to mediate the engulfment of aged or apoptotic cells. One possible explanation for this would be that an unidentified additional interaction between apoptotic cells and full-length stabilin-2 is required for the EGFrp-tethered cell corpse to become internalized. To next investigate whether the FAS1 domain is required for unidentified additional interaction, we established L-cell transfectants (L/U4 cells) that stably express mutant stabilin-2, in which the extracellular domain was replaced with the fourth repeated unit. Although L/U4 cells were able to bind to aged RBCs in a manner similar to what was observed in L/Stab-2 cells, the L/U4 cells also showed no engulfment of aged RBCs (Fig. 3H). The results with L/Stab-2 cells and L/U4 could eliminate the possibility that the inability of L/EGF3 cells to engulf is not due to lack of unidentified additional interaction through the FAS1 domain. A previous study suggests that a multimeric form of HARE (which is identical to stabilin-2) may be required to mediate effective removal of hyaluronan molecules from the blood (37). Thus, we propose that a single EGFrp or repeated unit is not sufficient to convert stabilin-2 into a functional conformation.
FIG. 3.
Binding of aged RBCs by L cells expressing a third EGFrp of stabilin-2 (L/EGF3). (A) Schematic representation of mutant stabilin-2 protein (Stab-E3-C and Stab-U4-C). (B) Surface expression of EGFrp on L cells expressing a third EGFrp of stabilin-2 (L/EGF3). Isotype-matched control antibody was used as a control for anti-stabilin-2 antibody (5G3). A representative result of three independent experiments is shown. (C) Dose-dependent binding of PS liposome to the surface of L/EGF3 cells. L/EGF3 cells were incubated with two different concentrations (10 or 100 μM) of fluorescence-labeled PS or PC liposomes for 1 h at 4°C. After extensive washing, the amount of fluorescence-labeled liposome associated with the cells was determined via flow cytometry. (D) Adhesion of L/EGF3 cells to PS. ELISA plates were coated with PS (right) or PC (left) and then incubated with L/Mock, L/Stab-2, L/EGF3, or L/FAS7 cells at 37°C for 1 h. After extensive washing, cells attached to the surfaces were quantified by hexosaminidase assay. (E) Representative images of aged RBC binding in L/EGF3 (right) and L/Mock cells (left). Scale bar, 50 μm. (F) Microscopic quantification of the binding and engulfment of aged RBCs in L/Mock, L/Stab-2, and L/EGF3 cells. The results are expressed as the means ± the SD from at least three experiments. **, P < 0.01 (ANOVA). (G) Binding of the aged RBCs by L/EGF3 cells in the presence of liposomes containing PC or PS (10 μM), anti-stabilin-2 antibody, or isotype-matched control antibody. L/EGF3 cells were preincubated with liposomes containing PC or PS (10 μM), anti-stabilin-2 antibody, or isotype-matched control antibody, and then aged RBCs were added. After incubation for 1 h, the percentages of cells binding the aged RBCs were determined. The results are expressed as the means ± the SD from at least three experiments. **, P < 0.01 (ANOVA). (H) Binding and engulfment of aged RBCs in L/Mock, L/Stab-2, and L/U4 cells. The results are expressed as the means ± the SD from at least three experiments. **, P < 0.01 (ANOVA).
EGFrp impairs the engulfment of aged and apoptotic cells by macrophages in vitro and in vivo.
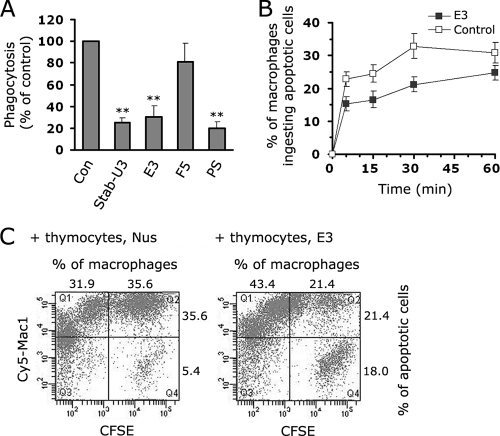
Next, to determine whether EGFrp could inhibit the phagocytosis of aged or apoptotic cells by macrophages in vitro and in vivo, we first tested whether the EGFrp protein inhibits the phagocytosis of aged RBCs by HMDMs. Pretreatment of aged RBCs with Stab-U3 or E3 inhibited their ingestion to the same degree as was seen with PS liposome (Fig. 4A). To confirm whether EGFrp plays a role in the interaction with PS on the apoptotic cell surface in vivo, we injected fluorescence-labeled apoptotic thymocytes into the peritoneal cavity of mice whose peritoneal macrophages had been induced with thioglycolate (34). When the apoptotic thymocytes were pretreated with the EGFrp protein (E3), the percentage of macrophages that ingested apoptotic cells was 67.1% of the Nus-treated control group after 5 min, and this decrease continued for up to 30 min (Fig. 4B). In the E3-pretreated group, the impaired phagocytosis caused a delay in the clearance of the apoptotic thymocytes from the peritoneum, resulting in 18% of the apoptotic cells remaining after 30 min, whereas only 5.4% of the apoptotic cells were sustained in the Nus-treated control group (Fig. 4C). These results strongly suggest that EGFrp is responsible for binding to PS on the apoptotic cell surface, indicating that it has a critical role in the tethering process during phagocytosis.
FIG. 4.
EGFrp inhibits the phagocytosis of apoptotic cells in vitro and in vivo. (A) Effect of EGFrp on the engulfment of aged RBCs by HMDMs. Aged RBCs were preincubated with 10 μM EGFrp and then added to HMDMs. The percentages of cells engulfing apoptotic cells were then determined. The results are expressed as the means ± the SD from at least three experiments. **, P < 0.01 (ANOVA). (B) EGFrp (E3) inhibits the phagocytic clearance of apoptotic thymocytes during sterile peritonitis. A graph shows the percentages of inflammatory macrophages from C57BL/6 mice engulfing apoptotic thymocytes at different time points after injection. Apoptotic thymocytes were preincubated with a 1 μM concentration of the third EGFrp (E3, ▪) or control Nus protein (□) prior to injection. The results are expressed as the means ± the SD from at least three experiments. (C) FACS analysis of the phagocytic uptake of carboxyfluorescein diacetate succinimidyl ester (CFSE)-labeled apoptotic cells by Mac-1+ peritoneal macrophages 30 min after the injection of apoptotic cells in C57BL/c mice. A representative result of three independent experiments is shown.
At least four EGF-like domains containing an atypical EGF-like domain and calcium are required for PS recognition.
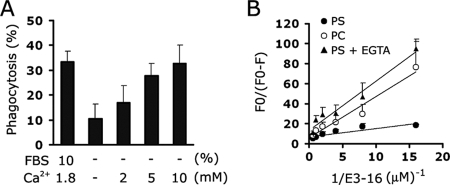
Although the biochemical mechanisms by which PS recognition by phagocytic receptors occurs are unclear, previous studies on several interfacial proteins have shown that PS-binding domains used Ca2+ to bridge the head group of the PS and protein, resulting in PS specificity (32, 35). Therefore, we tested the calcium ion to determine whether it is also involved in stabilin-2-mediated phagocytosis. As shown in Fig. 5A, stabilin-2-expressing cells (L/Stab-2 cells) showed a minimal level of phagocytosis in the absence of calcium under serum-free conditions. The addition of calcium enhanced the stabilin-2-mediated phagocytosis in a dose-dependent manner (Fig. 5A). To further evaluate the calcium-dependent PS recognition of EGFrp, the binding of E3 to PS liposomes was evaluated by observing the quenching of NBD fluorescence incorporated into the liposomes as previously described (5). NBD fluorescence is extremely sensitive to alterations in the local environment; therefore, changes in fluorescence caused by lipid segregation or interactions with amino acid groups can be detected upon protein attachment to the NBD fluorescence liposome. As shown in Fig. 5B, a considerable change in fluorescence occurred upon addition of E3 to PS liposomes compared to the change that occurred when E3 was added to PC liposomes. However, this PS-specific binding was significantly inhibited when the calcium ion was removed by a calcium chelator, EGTA. These results further indicate that EGFrp binds preferentially and directly to PS in a calcium-dependent manner and also suggest that extracellular calcium plays a regulatory role in the engulfment of aged and apoptotic cells through modulation of PS binding.
FIG. 5.
Stabilin-2 mediated phagocytosis via calcium-dependent PS recognition. (A) Effect of calcium ions on stabilin-2-mediated phagocytosis. L/Stab-2 cells were incubated with aged RBCs in the presence of the indicated concentrations of calcium ion under serum-free conditions. After incubation for 1 h, the percentages of cells engulfing the aged RBCs were determined. The results are expressed as the means ± the SD from at least three experiments. (B) Dependence of NBD-PC or NBD-PS fluorescence quenching on the concentration of EGFrp (E3). EGFrp protein was added to NBD-liposome containing binding buffer. Quenching of the NBD-liposome fluorescence was then monitored by using a fluorescence spectrometer. The extent of quenching is plotted versus the inverse protein concentration as previously described (5). The results are expressed as the means ± the SD from at least three experiments.
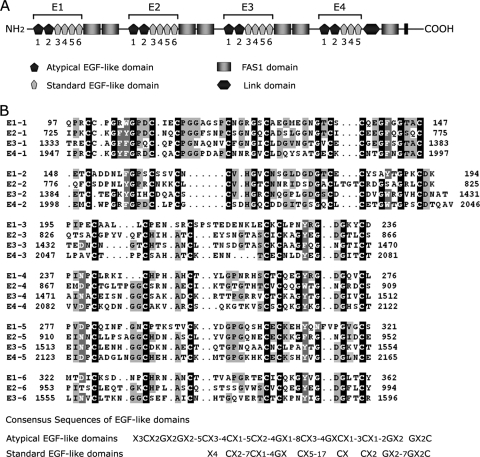
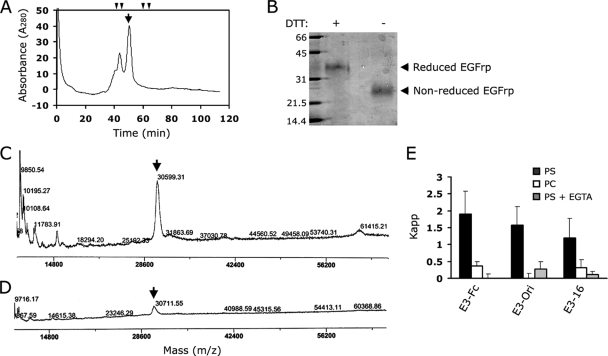
Each EGFrp in stabilin-2 consists of six tandem repeats of the EGF-like domain, with the exception of the fourth EGFrp, which has five EGF-like domains (Fig. 6). The first and second EGF-like domains in each EGFrp can be distinguished from the standard EGF-like domain by the presence of two extra cysteine residues that form an additional disulfide bond (Fig. 6). This type of EGF-like domain has been found in several proteins, including the MEGF family of proteins, SREC, and CED-1, and has therefore been described as the atypical EGF-like domain (22). The EGFrp of stabilin-2 carries 41 cysteine residues, and 40 cysteine residues are supposed to be involved in disulfide bonding. Due to the unpaired Cys residue in the first atypical EGF-like domain (Cys1337 in E3), we observed fractions of dimeric or multimeric forms in gel filtration chromatogram, but ca. 70% of recombinant proteins was purified as monomer (Fig. 7A). This multimerization was eliminated by addition of 5 mM DTT or mutation of unpaired cysteine to serine (C1337S mutant, data not shown). When this monomeric fraction was subjected to the SDS-PAGE with or without reducing agent (DTT) (Fig. 7B), the nonreducing protein ran faster than the reducing protein due to its conformational difference by internal disulfide bonds. We next tried to verify the disulfide bonds of the purified monomer by using MALDI-TOF MS. Since 40 Cys residues are supposed to be involved in the disulfide bond in EGFrp, it was expected that a 40-Da mass would be decreased in EGFrp protein by disulfide bonding. Although the assay was not simple because of the error range of MALDI-TOF MS analysis (estimated as ±0.1% and increased with an increase in the molecular mass of sample), we consistently observed a decrease in the molecular mass for the nonreducing protein sample compared to that of the reducing protein in multiple experiments (Fig. 7C and D).
FIG. 6.
Protein structure of stabilin-2 and alignments of the EGF-like domains. (A) Domain structure of stabilin-2. Stabiln-2 contains four EGF-like domain repeats (EGRrps, E1 to E4). Each EGFrp consists of two atypical EGF-like domains and four standard EGF-like domains (the fourth EGFrp [E4] has three EGF-like domains). (B) Alignment of the EGF-like domains in stabilin-2. The consensus sequences of atypical and standard EGF-like domains are derived from previous studies (6, 22, 39).
FIG. 7.
Characterization of bacterial EGFrp. (A) Gel filtration chromatography of bacterial EGFrp. Fast-performance liquid chromatography-gel filtration was carried out by using a Superdex 75 HR 10/30 column that had been calibrated with molecular mass standards. The molecular mass standards were indicated in the chromatogram (arrowheads indicate 67, 43, 25, and 13.7 kDa). Arrow, monomeric fraction. (B) SDS-PAGE of the purified monomer of EGFrp. Lane 1, molecular weight markers (kDa); lane 2, purified EGFrp monomer with DTT; lane 3, purified EGFrp monomer without DTT. (C and D) MALDI-TOF MS analysis of purified EGFrp monomer under nonreducing (DTT−, C) or reducing (DTT+, D) conditions. Arrows, monomeric EGFrp. (E) E3-Fc, E3-ori, or E3-16 was added to NBD-liposome containing binding buffer. Quenching of NBD-liposome fluorescence was then monitored by using a fluorescence spectrometer. The binding affinities of EGFrps (E3-Fc, E3-Ori, and E3-16) were determined by using the Kapp (apparent association constant, which is given by 1/K, using the Stern-Volmer law, which is as follows: Fo/(Fo − F) = 1/fa + 1/(faK[Mt]). The quenching constant, K, after buffer normalization is directly related to the binding affinity of the complex, which can be determined by plotting the quenching extent [Fo/(Fo − F)] versus the inverse protein concentration (1/[protein]).
To further assure correct folding with appropriate disulfide bonds of recombinant proteins in E. coli, we expressed Fc-fusion EGFrp in mammalian cells and compared binding activity with bacterial expressed EGFrp. Furthermore, we also expressed EGFrp in a modified E. coli trxB gor (Origami) mutant strain, which is deficient in glutathione and thioredoxin reductase systems and allows the formation of disulfide bonds in cytoplasmic proteins (31). We compared the binding activity among mammalian EGFrp (E3-Fc), Origami-origin EGFrp (E3-Ori), and bacterial EGFrp (E3-16) by using a fluorescence quenching assay. The Origami-origin EGFrp (E3-Ori) showed comparable binding activity with mammalian EGFrp, indicating soluble EGFrp from Origami strain formed authentic conformation. The bacterial EGFrp showed ca. 63 or 76% binding activity compared to that of the mammalian or Origami-origin EGFrp, respectively (Fig. 7E). Since all EGFrps from different sources showed identical binding characteristics—specific binding to phosphatidylserine and a calcium requirement for binding— these data suggest that more than 60% of the bacterial EGFrp have formed active conformation with appropriate disulfide bonds, thereby showing 63 to 76% binding activity.
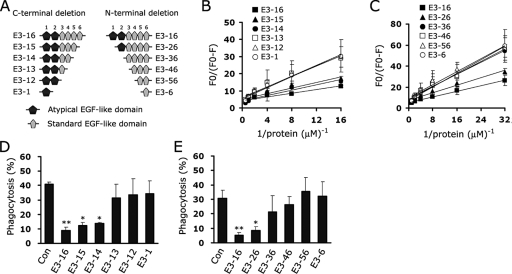
To further analyze the biochemical mechanism by which EGFrp binds to PS, we generated a series of N- and C-terminal deletion mutants that contained different subsets of EGF-like domains within the EGFrp (Fig. 8A) and then evaluated their ability to bind to PS using fluorescence quenching of the NBD-PS that had been incorporated into the liposome. Experiments with serial C-terminal deletion mutants of EGFrp showed that E3-16, E3-15, and E3-14 significantly bind to PS liposomes, indicating that at least four tandem repeats of the EGF-like domain are required for binding to PS (Fig. 8B). In addition, when a series of N-terminal deletion mutants were tested, only deletion mutants (E3-16 and E3-26) including more than one atypical EGF-like domain were able to bind to PS (Fig. 8C). Furthermore, only background binding was observed for PC-containing liposomes, which demonstrates that the binding was specific to PS (data not shown). The results of the cell-based phagocytosis assays also support the fluorescence binding experiments. When aged RBCs were pretreated with EGFrp deletion mutants, more than four repeats of the EGF-like domains, including atypical EGF-like domains, were able to inhibit the uptake of aged RBCs in L/Stab-2 cells (Fig. 8D and E). These results indicate that a certain conformation assembled by at least four EGF-like domains, including more than one atypical EGF-like domain, is necessary for EGFrp to bind to PS.
FIG. 8.
EGFrps containing atypical EGF-like domains mediate the recognition of PS. (A) Schematic representation of deletion mutant stabilin-2 proteins. (B and C) NBD-PS-labeled PS liposomes were successively incubated with C-terminal deletion (B) or N-terminal deletion (C) mutants of EGFrp, and the proteins bound to the liposomes were quantified by fluorescence quenching assays. The extent of quenching is plotted versus the inverse protein concentration. (D and E) Aged RBCs were preincubated with C-terminal deletion (D) or N-terminal deletion (E) mutants of stabilin-2 and then added to L/Stab-2 cells. After incubation for 1 h, the percentages of cells engulfing aged RBCs were determined. The results are expressed as the means ± the SD from at least three experiments. *, P < 0.05; **, P < 0.01 (ANOVA).
A second atypical EGF-like domain plays a key role in calcium-dependent PS-recognition.
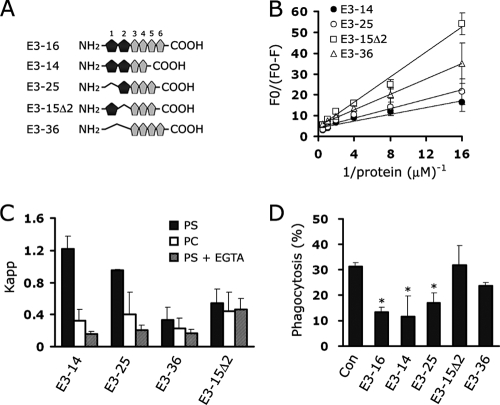
To determine which atypical EGF-like domain is most essential for PS binding, we generated EGFrp mutants lacking single or double atypical EGF-like domains (Fig. 9A). All of the mutants contained four repeats of the EGF-like domain to rule out any possible effect on binding due to the length of the proteins. Deletion mutants containing a second atypical EGF-like domain (E3-14 and E3-25) showed PS-binding activity comparable to that of E3-16, whereas deletion mutants lacking the second atypical EGF-like domain (E3-15Δ2 and E3-36) showed a significant decrease in the ability to bind PS, indicating that the second atypical EGF-like domain plays a pivotal role in specific PS recognition (Fig. 9B). Furthermore, a calcium chelator significantly inhibited the interaction between proteins E3-25 and PS similar to what is observed in the interaction between E3-16 and PS, whereas the binding of PS to proteins E3-15Δ2 was almost identical to that of PC irrespective of the presence of EGTA (Fig. 9C). In addition, mutants lacking both atypical EGF-like domains (E3-36) showed only background binding activity to PS, and slightly less activity was observed in the presence of EGTA; however, this difference was not statistically significant. We also tested the inhibitory effect of mutant proteins on the uptake of aged RBC in L/Stab-2 cells. In agreement with the biochemical studies, E3-14 and E3-25 were able to efficiently inhibit phagocytosis, but E3-15Δ2 was unable to do so (Fig. 9D). These results suggest that the second atypical EGF-like domain plays a central role in calcium-dependent PS-specific binding.
FIG. 9.
The presence of a second atypical EGF-like domain is a key role in calcium-dependent PS recognition. (A) Schematic representation of EGFrp deletion mutants containing four EGF-like domains. (B) EGFrp deletion mutants were added to NBD-PS liposome containing binding buffer. Quenching of the NBD-PS liposome fluorescence was then monitored by using a fluorescence spectrometer. The extent of quenching is plotted versus the inverse protein concentration. (C) The binding affinities of EGFrp mutants were determined by using the Kapp [apparent association constant, which is given by 1/K, using the Stern-Volmer law, which is as follows: Fo/(Fo − F) = 1/fa + 1/(faK[Mt])]. The quenching constant, K, after buffer normalization is directly related to the binding affinity of the complex, which can be determined by plotting the quenching extent [Fo/(Fo − F)] versus the inverse protein concentration (1/[protein]). (D) The inhibitory effects of each EGFrp mutant were tested by using a phagocytosis assay as described in Materials and Methods, and the percentages of cells engulfing aged RBCs were determined. The results are expressed as the means ± the SD from at least three experiments. *, P < 0.05 (ANOVA).
DISCUSSION
Although several proteins involved in apoptotic cell engulfment have recently been identified, the domain structures that recognize the PS of apoptotic cells during engulfment have not been investigated. Recently, we demonstrated that stabilin-2 is capable of ingesting apoptotic cells quickly via PS recognition (25), and the present study provides the first evidence that EGFrp in stabilin-2 specifically recognizes PS in a calcium-dependent manner and is involved in the clearance of apoptotic cells. Multiple repeats of the EGF-like domain (EGFrp) are frequently found in the extracellular domains of various cell surface receptors and are known to be involved in the ligand-receptor interaction (6). These domains can be categorized into four distinct groups: (i) the standard EGF-like domain, which contains only six conserved cysteine residues and is subdivided into three major groups—type I, II, and III (1); (ii) the calcium-binding EGF-like domain; (iii) the laminin type EGF-like domain, which includes eight conserved cysteine residues over a larger (>60 amino acids) region; and (iv) the atypical EGF-like domain, which consists of 40 to 45 amino acids regions containing eight conserved cysteine residues (22). Each EGFrp in stabilin-2 contains two copies of the atypical EGF-like domain and four copies of the standard EGF-like domain (the fourth EGF-like repeat has three standard EGF-like domains). The atypical EGF-like repeat was first identified in MEGF6 and then found in potential scavenger receptors, such as SREC, CED-1, and MEGF10 (22). Among these, CED-1 (39) and its human homolog MEGF-10 (11), which are well known for their ability to recognize apoptotic cells, contain multiple copies of the atypical EGF-like domain in their extracellular domains, suggesting that the atypical EGF-like domain plays a role in the clearance of cell corpses. The results of the present study showed that EGFrps containing the second atypical EGF domain specifically recognized PS during cell corpse clearance. In addition, EGFrps composed of only standard EGF-like domains showed only minimal PS binding activity, which is consistent with the previous observation that, although protein S contained four tandem repeats of the EGF-like domain, its N-terminal Gla domain is responsible for binding to the appropriate phospholipid membranes in the presence of Ca2+ ions (27). However, a recent study showed that EGF-like domains are also important for protein S activity and suggested that EGF modules may be able to constrain the protein S molecule in an appropriate configuration (21). This is consistent with our observation that at least four tandem repeats of the EGF-like domain were required for binding to PS, which suggests that at least four repeats of the EGF-like domains may be necessary to form a conformation that presents the second atypical EGF-like domain in the proper orientation to target PS.
The results in the present study demonstrated that EGFrp specifically bind to PS in a calcium-dependent manner. However, the several binding experiments indicated that the interaction of EGFrp (even mammalian expressed EGFrp) to PS showed low-affinity binding compared to that of MFG-E8 proteins, which is known as soluble bridging molecule with high PS-binding affinity (5, 12). To eliminate possibility that this low affinity of EGFrp might be caused by improper folding in a bacterial expressing system, we examined disulfide bonds of EGFrp using SDS-PAGE and MALDI-TOF MS and compared the binding affinity with mammalian EGFrp (Fc-EGFrp). Only 1.5-fold-higher activity was observed in mammalian EGFrp compared to the bacterial EGFrp implying the binding activity of EGFrp is generically low. Although the generic binding affinity of EGFrp is weaker than C2 domain (PS recognition domain) of MFG-E8, it is possible that relatively low-affinity EGFrp interactions with PS are markedly enhanced by localized exposure of PS on the apoptotic cell and by cooperative binding by other recognition receptor. Furthermore, if the EGFrp in each unit forms a PS recognition conformation like a snap-button to bind the PS patch on the apoptotic cell surface, the sporadic distribution of EGFrps could tether apoptotic cells efficiently with even higher affinity through multimer formation. This distribution of EGFrps is a distinctive feature of stabilin-2 compared to the continuous distribution of multiple repeats of atypical EGF-like domains in CED-1 and MEGF10.
Although the domain structures for PS recognition in phagocytic receptors are unknown, several domains in interfacial proteins are known to mediate the recognition of PS, including the annexin domain in the annexin family, the C2 domain in protein kinase C, and the Gla domain in blood coagulation proteins. The results of a previous study showed that the Ca2+-bridges are key features in the membrane attachment exhibited by annexin V (32). In addition, the structural information on the PKCα-C2 domain also suggested a membrane-binding mechanism in which the calcium ion directly mediates the PS-specific recognition (35). In the present study, we found that calcium ions are required for stabilin-2-mediated phagocytosis and EGFrp-mediated PS recognition. Interestingly, the deletion of the second EGF-like domain in the EGFrp caused a complete loss of calcium dependence for PS recognition, thereby leading to a significant decrease of inhibitory effect on aged-cell clearance, suggesting that the second atypical EGF-like domain acts as a noncanonical calcium-binding EGF-like domain. Therefore, EGFrp containing the second atypical EGF-like domain may interact with PS through a Ca2+-bridging mechanism similar to those that have been observed with other calcium-binding interfacial proteins (32, 35). Future structural studies will provide a clear view of the PS-binding mechanism, however, the present study provides a better understanding of the mechanism by which Ca2+-dependent PS-binding occurs, which may be extended to other EGFrp-containing proteins.
Although PS is an important flag molecule for phagocytosis, the externalization of PS also occurs in nonapoptotic cells during various cellular processes, including platelet activation, cell fusion during myotube formation, syncytiotrophoblast formation, platelet release from progenitor cells, immunoglobulin E-dependent stimulation of mast cells, T-cell migration, and the formation of tumor vasculature (7, 26, 30). Furthermore, a recent study reported that inhibition of PS recognition heightens the immunogenicity of irradiated lymphoma cells (4). Therefore, our in vivo data showing that apoptotic cell clearance was delayed by EGFrp suggest that EGFrp treatment could also be used to increase the efficiency of apoptotic cell-based vaccines by interfering with the PS-dependent recognition and immunosuppressive clearance of dying cells.
In summary, the results of the present study provide evidence that the four EGFrps of stabilin-2 play a key role in the clearance of aged and apoptotic cells by directly binding to exposed PS on the apoptotic cell surface. The identification of the EGFrps that contain an atypical EGF-like domain as a critical domain that directly recognizes PS would help to elucidate the molecular mechanism by which PS is recognized during apoptotic cell clearance and would have broad implications as a PS marker in many physiological and pathological conditions.
Acknowledgments
This study was supported by the grant RTI04-01-01 from the Regional Technology Innovation Program of the Ministry of Knowledge Economy; grant 0720550-2 from the National R&D Program for Cancer Control, Ministry of Health and Welfare & Family Affairs, Republic of Korea; and the Brain Korea 21 Project in 2008. The MALDI-TOF experiment was performed by Korea Basic Science Institute.
Footnotes
Published ahead of print on 23 June 2008.
REFERENCES
- 1.Appella, E., I. T. Weber, and F. Blasi. 1988. Structure and function of epidermal growth factor-like regions in proteins. FEBS Lett. 2311-4. [DOI] [PubMed] [Google Scholar]
- 2.Balzar, M., I. H. Briaire-de Bruijn, H. A. Rees-Bakker, F. A. Prins, W. Helfrich, L. de Leij, G. Riethmuller, S. Alberti, S. O. Warnaar, G. J. Fleuren, and S. V. Litvinov. 2001. Epidermal growth factor-like repeats mediate lateral and reciprocal interactions of Ep-CAM molecules in homophilic adhesions. Mol. Cell. Biol. 212570-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bastiani, M. J., A. L. Harrelson, P. M. Snow, and C. S. Goodman. 1987. Expression of fasciclin I and II glycoproteins on subsets of axon pathways during neuronal development in the grasshopper. Cell 48745-755. [DOI] [PubMed] [Google Scholar]
- 4.Bondanza, A., V. S. Zimmermann, P. Rovere-Querini, J. Turnay, I. E. Dumitriu, C. M. Stach, R. E. Voll, U. S. Gaipl, W. Bertling, E. Poschl, J. R. Kalden, A. A. Manfredi, and M. Herrmann. 2004. Inhibition of phosphatidylserine recognition heightens the immunogenicity of irradiated lymphoma cells in vivo. J. Exp. Med. 2001157-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borisenko, G. G., S. L. Iverson, S. Ahlberg, V. E. Kagan, and B. Fadeel. 2004. Milk fat globule epidermal growth factor 8 (MFG-E8) binds to oxidized phosphatidylserine: implications for macrophage clearance of apoptotic cells. Cell Death Differ. 11943-945. [DOI] [PubMed] [Google Scholar]
- 6.Campbell, I. D., and P. Bork. 1993. Epidermal growth factor-like modules. Curr. Opin. Struct. Biol. 3385-392. [Google Scholar]
- 7.Fadeel, B., and D. Xue. 2006. PS externalization: from corpse clearance to drug delivery. Cell Death Differ. 13360-362. [DOI] [PubMed] [Google Scholar]
- 8.Gardai, S. J., D. L. Bratton, C. A. Ogden, and P. M. Henson. 2006. Recognition ligands on apoptotic cells: a perspective. J. Leukoc. Biol. 79896-903. [DOI] [PubMed] [Google Scholar]
- 9.Gigli, I., and R. A. Nelson, Jr. 1968. Complement dependent immune phagocytosis. I. Requirements for C′1, C′4, C′2, C′3. Exp. Cell Res. 5145-67. [DOI] [PubMed] [Google Scholar]
- 10.Grimsley, C., and K. S. Ravichandran. 2003. Cues for apoptotic cell engulfment: eat-me, don't eat-me and come-get-me signals. Trends Cell Biol. 13648-656. [DOI] [PubMed] [Google Scholar]
- 11.Hamon, Y., D. Trompier, Z. Ma, V. Venegas, M. Pophillat, V. Mignotte, Z. Zhou, and G. Chimini. 2006. Cooperation between engulfment receptors: the case of ABCA1 and MEGF10. PLoS ONE 1e120.17205124 [Google Scholar]
- 12.Hanayama, R., M. Tanaka, K. Miwa, A. Shinohara, A. Iwamatsu, and S. Nagata. 2002. Identification of a factor that links apoptotic cells to phagocytes. Nature 417182-187. [DOI] [PubMed] [Google Scholar]
- 13.Henson, P. M., D. L. Bratton, and V. A. Fadok. 2001. Apoptotic cell removal. Curr. Biol. 11R795-R805. [DOI] [PubMed] [Google Scholar]
- 14.Ishii, J., H. Adachi, J. Aoki, H. Koizumi, S. Tomita, T. Suzuki, M. Tsujimoto, K. Inoue, and H. Arai. 2002. SREC-II, a new member of the scavenger receptor type F family, trans-interacts with SREC-I through its extracellular domain. J. Biol. Chem. 27739696-39702. [DOI] [PubMed] [Google Scholar]
- 15.Jung, M. Y., S. Y. Park, and I. S. Kim. 2007. Stabilin-2 is involved in lymphocyte adhesion to the hepatic sinusoidal endothelium via the interaction with αMβ2 integrin. J. Leukoc. Biol. 821156-1165. [DOI] [PubMed] [Google Scholar]
- 16.Kagan, V. E., B. Gleiss, Y. Y. Tyurina, V. A. Tyurin, C. Elenstrom-Magnusson, S. X. Liu, F. B. Serinkan, A. Arroyo, J. Chandra, S. Orrenius, and B. Fadeel. 2002. A role for oxidative stress in apoptosis: oxidation and externalization of phosphatidylserine is required for macrophage clearance of cells undergoing Fas-mediated apoptosis. J. Immunol. 169487-499. [DOI] [PubMed] [Google Scholar]
- 17.Kawasaki, Y., A. Nakagawa, K. Nagaosa, A. Shiratsuchi, and Y. Nakanishi. 2002. Phosphatidylserine binding of class B scavenger receptor type I, a phagocytosis receptor of testicular Sertoli cells. J. Biol. Chem. 27727559-27566. [DOI] [PubMed] [Google Scholar]
- 18.Kim, J. E., S. J. Kim, B. H. Lee, R. W. Park, K. S. Kim, and I. S. Kim. 2000. Identification of motifs for cell adhesion within the repeated domains of transforming growth factor-beta-induced gene, betaig-h3. J. Biol. Chem. 27530907-30915. [DOI] [PubMed] [Google Scholar]
- 19.Kohda, D., C. J. Morton, A. A. Parkar, H. Hatanaka, F. M. Inagaki, I. D. Campbell, and A. J. Day. 1996. Solution structure of the link module: a hyaluronan-binding domain involved in extracellular matrix stability and cell migration. Cell 86767-775. [DOI] [PubMed] [Google Scholar]
- 20.Lauber, K., S. G. Blumenthal, M. Waibel, and S. Wesselborg. 2004. Clearance of apoptotic cells: getting rid of the corpses. Mol. Cell 14277-287. [DOI] [PubMed] [Google Scholar]
- 21.Mille-Baker, B., S. M. Rezende, R. E. Simmonds, P. J. Mason, D. A. Lane, and M. A. Laffan. 2003. Deletion or replacement of the second EGF-like domain of protein S results in loss of APC cofactor activity. Blood 1011416-1418. [DOI] [PubMed] [Google Scholar]
- 22.Nakayama, M., D. Nakajima, T. Nagase, N. Nomura, N. Seki, and O. Ohara. 1998. Identification of high-molecular-weight proteins with multiple EGF-like motifs by motif-trap screening. Genomics 5127-34. [DOI] [PubMed] [Google Scholar]
- 23.Ogden, C. A., A. deCathelineau, P. R. Hoffmann, D. Bratton, B. Ghebrehiwet, V. A. Fadok, and P. M. Henson. 2001. C1q and mannose binding lectin engagement of cell surface calreticulin and CD91 initiates macropinocytosis and uptake of apoptotic cells. J. Exp. Med. 194781-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oka, K., T. Sawamura, K. Kikuta, S. Itokawa, N. Kume, T. Kita, and T. Masaki. 1998. Lectin-like oxidized low-density lipoprotein receptor 1 mediates phagocytosis of aged/apoptotic cells in endothelial cells. Proc. Natl. Acad. Sci. USA 959535-9540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park, S. Y., M. Y. Jung, H. J. Kim, S. J. Lee, S. Y. Kim, B. H. Lee, T. H. Kwon, R. W. Park, and I. S. Kim. 2007. Rapid cell corpse clearance by stabilin-2, a membrane phosphatidylserine receptor. Cell Death Differ. doi: 10.1038/sj.cdd. 4402242. [DOI] [PubMed]
- 26.Ran, S., and P. E. Thorpe. 2002. Phosphatidylserine is a marker of tumor vasculature and a potential target for cancer imaging and therapy. Int. J. Radiat. Oncol. Biol. Phys. 541479-1484. [DOI] [PubMed] [Google Scholar]
- 27.Rezende, S. M., D. A. Lane, B. Mille-Baker, M. M. Samama, J. Conard, and R. E. Simmonds. 2002. Protein S Gla-domain mutations causing impaired Ca2+-induced phospholipid binding and severe functional protein S deficiency. Blood 1002812-2819. [DOI] [PubMed] [Google Scholar]
- 28.Sano, H., D. K. Hsu, J. R. Apgar, L. Yu, B. B. Sharma, I. Kuwabara, S. Izui, and F. T. Liu. 2003. Critical role of galectin-3 in phagocytosis by macrophages. J. Clin. Investig. 112389-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Savill, J., and V. Fadok. 2000. Corpse clearance defines the meaning of cell death. Nature 407784-788. [DOI] [PubMed] [Google Scholar]
- 30.Schlegel, R. A., and P. Williamson. 2001. Phosphatidylserine, a death knell. Cell Death Differ. 8551-563. [DOI] [PubMed] [Google Scholar]
- 31.Su, Z., Y. Huang, Q. Zhou, Z. Wu, X. Wu, Q. Zheng, C. Ding, and X. Li. 2006. High-level expression and purification of human epidermal growth factor with SUMO fusion in Escherichia coli. Protein Peptide Lett. 13785-792. [DOI] [PubMed] [Google Scholar]
- 32.Swairjo, M. A., N. O. Concha, M. A. Kaetzel, J. R. Dedman, and B. A. Seaton. 1995. Ca2+-bridging mechanism and phospholipid head group recognition in the membrane-binding protein annexin V. Nat. Struct. Biol. 2968-974. [DOI] [PubMed] [Google Scholar]
- 33.Tang, X., M. S. Halleck, R. A. Schlegel, and P. Williamson. 1996. A subfamily of P-type ATPases with aminophospholipid transporting activity. Science 2721495-1497. [DOI] [PubMed] [Google Scholar]
- 34.Taylor, P. R., A. Carugati, V. A. Fadok, H. T. Cook, M. Andrews, M. C. Carroll, J. S. Savill, P. M. Henson, M. Botto, and M. J. Walport. 2000. A hierarchical role for classical pathway complement proteins in the clearance of apoptotic cells in vivo. J. Exp. Med. 192359-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verdaguer, N., S. Corbalan-Garcia, W. F. Ochoa, I. Fita, and J. C. Gomez-Fernandez. 1999. Ca2+ bridges the C2 membrane-binding domain of protein kinase Cα directly to phosphatidylserine. EMBO J. 186329-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu, Y., N. Tibrewal, and R. B. Birge. 2006. Phosphatidylserine recognition by phagocytes: a view to a kill. Trends Cell Biol. 16189-197. [DOI] [PubMed] [Google Scholar]
- 37.Zhou, B., J. A. Oka, A. Singh, and P. H. Weigel. 1999. Purification and subunit characterization of the rat liver endocytic hyaluronan receptor. J. Biol. Chem. 27433831-33834. [DOI] [PubMed] [Google Scholar]
- 38.Zhou, Q., J. Zhao, J. G. Stout, R. A. Luhm, T. Wiedmer, and P. J. Sims. 1997. Molecular cloning of human plasma membrane phospholipid scramblase: a protein mediating transbilayer movement of plasma membrane phospholipids. J. Biol. Chem. 27218240-18244. [DOI] [PubMed] [Google Scholar]
- 39.Zhou, Z., E. Hartwieg, and H. R. Horvitz. 2001. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in Caenorhabditis elegans. Cell 10443-56. [DOI] [PubMed] [Google Scholar]