Abstract
The basic helix-loop-helix (bHLH) transcription factor PTF1a is critical to the development of the embryonic pancreas. It is required early for the formation of the undifferentiated tubular epithelium of the nascent pancreatic rudiment and then becomes restricted to the differentiating acinar cells, where it directs the transcriptional activation of the secretory digestive enzyme genes. Here we report that the complex temporal and spatial expression of Ptf1a is controlled by at least three separable gene-flanking regions. A 14.8-kb control domain immediately downstream of the last Ptf1a exon is highly conserved among mammals and directs expression in the dorsal part of the spinal cord but has very little activity in the embryonic or neonatal pancreas. A 13.4-kb proximal promoter domain initiates limited expression in cells that begin the acinar differentiation program. The activity of the proximal promoter domain is complemented by an adjacent 2.3-kb autoregulatory enhancer that is able to activate a heterologous minimal promoter with high-level penetrance in the pancreases of transgenic mice. During embryonic development, the enhancer initiates expression in the early precursor epithelium and then superinduces expression in acinar cells at the onset of their development. The enhancer contains two evolutionarily conserved binding sites for the active form of PTF1a, a trimeric complex composed of PTF1a, one of the common bHLH E proteins, and either RBPJ or RBPJL. The two sites are essential for acinar cell-specific transcription in transfected cell lines and mice. In mature acinar cells, the enhancer and PTF1a establish an autoregulatory loop that reinforces and maintains Ptf1a expression. Indeed, the trimeric PTF1 complex forms dual autoregulatory loops with the Ptf1a and Rbpjl genes that may maintain the stable phenotype of pancreatic acinar cells.
PTF1a is a basic helix-loop-helix (bHLH) transcription factor with multiple, stage-specific roles during pancreatic organogenesis and is critical to the early development that leads to the formation of the endocrine and acinar compartments. Ptf1a expression begins during the evagination of the dorsal and ventral epithelial buds that arise from the posterior of the foregut endoderm and constitute the early murine pancreatic anlagen (10, 14). It persists during the early period of epithelial growth and morphogenesis that leads to the formation of a branched tubule complex of progenitor cells at mid-pancreatic development. At this time, the epithelium undergoes a transformation that establishes a tissue architecture that produces the islet, acinar, and ductal cell lineages (14, 17). As development continues, islet precursor cells shed from the tubules in the central regions of the epithelium, coalesce into amorphous endocrine cell clusters, and differentiate. Around the periphery of the epithelium, branching tubules form the small connecting ducts, and acinar cell clusters bud from their termini. The early epithelial growth and morphogenesis are blunted in Ptf1a-deficient embryos, so that the epithelial transformation is precluded and the islets, acini, and smaller peripheral ducts do not form (10, 13, 14).
During normal development, Ptf1a expression is extinguished in the central region of the precursor tubular epithelium and superinduced in a population of multipotent progenitors forming at the peripheral tips of the epithelium (14, 26). Subsequently, Ptf1a is inactivated in cells that make a final commitment to the ductal and islet lineages and is maintained at a high level in nascent acinar cells (14). In the new acinar cells, PTF1a partners with one of the common E proteins (e.g., TCF12) and RBPJ to form a trimeric complex (PTF1-J) that binds and activates the gene encoding a pancreas-restricted RBPJ paralogue, RBPJL (14). As RBPJL accumulates during acinar cell differentiation, it replaces RBPJ in the complex to form PTF1-L, which binds and activates the acinar cell-specific digestive enzyme genes such as those for amylase (Amy1), chymotrypsin B (Ctrb), carboxypeptidase A (Cpa1), and elastase 1 (Ela1). In mature acinar cells, RBPJL resides with PTF1a on acinar cell-specific promoters, whereas RBPJ does not (4).
Thus, PTF1a appears to play at least two distinct developmental roles. One is required for the growth and morphogenesis of the early epithelium; without PTF1a, the epithelial foundation required for the secondary developmental transition does not form (10). The later role is for acinar cell differentiation. Because PTF1a is the crucial transcriptional activator for the production of the acinar secretory enzymes (4, 12, 20), the superinduction of Ptf1a expression at the onset of acinar cell formation is an important developmental event. The expression of Ptf1a is maintained in mature acinar cells and remains absent in pancreatic endocrine and ductal cells.
The mechanisms that regulate this critical pancreatic transcription factor are unknown. In this report, we identify three broad transcriptional regulatory domains of the mouse Ptf1a gene: a 13.4-kb proximal promoter region, a 2.3-kb distal autoregulatory enhancer, and a 12.4-kb 3′ regulatory domain. The proximal promoter and 3′ regulatory regions direct only sporadic expression in the embryonic pancreas beginning at middevelopment but drive extensive expression in the dorsal neural tube, another principal site of Ptf1a expression. We show that the distal 5′ enhancer is sufficient to activate Ptf1a expression early and superinduce it at the onset of acinar cell formation. The early and late activities of the enhancer require autoregulation via a trimeric PTF1 complex. Two binding sites for the three-subunit PTF1 complex are present in the autoregulatory enhancer, and the mutation of the two sites abolishes the activity of the enhancer in transfected acinar cell lines and the induction in forming acinar cells in the embryonic pancreases of transgenic mice.
MATERIALS AND METHODS
Cloning of the mouse Ptf1a regulatory regions.
The mouse Ptf1a gene and flanking sequences were previously isolated from a P1 phage library of mouse DNA (4, 20). P1 phage GS#12689 includes the region of mouse chromosome 2 (chr2) encompassing the Ptf1a gene and 5′ and 3′ flanking sequences. Based on the numbering of the February 2006 mouse genome mm8 assembly (NCBI) and the provisional NCBI RefSeq sequence record NM_018809 for the Mus musculus pancreas-specific transcription factor 1a (PTF1a) mRNA, the transcribed region is located at chr2 positions 19363417 to 19365246. For cross-species comparisons to determine sequence conservation, we conducted BLAT analyses (http://genome.ucsc.edu/cgi-bin/hgBlat) (11).
Cell culture and transient transfections.
The 5′ enhancer (chr2 positions 19347844 to 19350141), the promoter region (chr2 positions 19350049 to 19363446), the region comprising the 5′ enhancer plus the promoter (chr2 positions 19347844 to 19363446), and the 3′ control region (chr2 positions 19365786 to 19378222) were ligated upstream of the rat Ela1 minimal promoter (positions −92 to +8) (8) and the luciferase gene in pGL3-basic (Promega). The expression plasmids for mouse PTF1a, mouse RBPJL, human RBPJ, and HEB have been described previously (4).
Human embryonic kidney 293 cells (ATCC CRL-1573) and mouse acinar pancreatic 266-6 cells (ATCC CRL-2151) were grown in Dulbecco's modified Eagle's medium with 10% bovine serum. Rat pancreatic acinar AR4-2J cells (ATCC CRL-1492) were cultured in F12K medium with 20% bovine serum. For transient transfections, plasmid DNA was introduced into the cells with Fugene 6 according to the instructions of the manufacturer (Roche, Basel, Switzerland). Cells were harvested and luciferase assays were performed 48 h after transfection, as described previously (4). Luciferase activity was normalized to the β-galactosidase activity of pCMVβ (Clontech, Mountain View, CA), with which the cells were cotransfected. All reporter gene analyses were performed at least in triplicate, and the data are expressed as the means ± the standard deviations.
EMSAs.
Syrian hamster E12, mouse PTF1a, human RBPJ, and mouse RBPJL proteins were synthesized in vitro using an SP6 and T7 TNT quick-coupled lysate system (Promega, Madison, WI). Mouse nuclear extract was obtained from adult C57BL/6 mouse pancreas with a CelLytic NuCLEAR extraction kit (Sigma, St. Louis, MO). The plasmids encoding the transcription factors, electrophoretic mobility shift assay (EMSA) buffers, and electrophoresis conditions were as described previously (4). The following oligonucleotides were used as labeled probes in EMSAs: for the proximal PTF1 site, CACATGTGTTATGATTCCCACG (top strand); for the distal PTF1 site, CACAAGTGGCGACATTCCCATGG (top strand); for the TC box mutant form of the proximal site, CACATGTGTTATGATTCaatCG (top strand); and for the TC box mutant form of the distal site, CACAAGTGGCGACATTCtatTGG (top strand). Lowercase letters correspond to mutations.
EMSA supershift analyses used the following antibodies: affinity-purified rabbit anti-PTF1a (20), affinity-purified rabbit anti-RBPJL (4), and rat monoclonal anti-RBPJ (code no. 2ARBP1) from the Institute of Immunology Co., Ltd. (Tokyo, Japan).
ChIP.
Mouse pancreatic chromatin was prepared from formaldehyde-cross-linked nuclei as previously described for rat pancreatic chromatin (4) and sheared further by sonication. A total of 100 μl of purified chromatin (1/20 of the total chromatin from one adult pancreas) in 900 μl of chromatin immunoprecipitation (ChIP) dilution buffer (Upstate, Lake Placid, NY) was incubated with 3 to 5 μg of affinity-purified anti-PFT1a, anti-RBPJL, or monoclonal anti-RBPJL for ≥4 h at 4°C and then incubated with blocked protein G-Sepharose beads (Upstate) for ≥1 h at 4°C. Bound chromatin was eluted from the beads, the formaldehyde cross-linking was reversed, and the immunoprecipitated DNA was purified for PCR analysis. The quantification of ChIP enrichment of promoter regions was performed with Sybr green master mix (Applied Biosystems, Foster City, CA) using the ABI Prism 7700 (Applied Biosystems), and quantities were calculated as the amount of the target gene relative to that of the 28S region of the large rRNA gene.
Generation of transgenic embryos and mice and detection of transgene expression.
To test the activities of the various Ptf1a-flanking sequences in transgenic mice, the fragments were placed 5′ of a lacZ reporter gene. The promoter region (chr2 positions 19350049 to 19363608) and the region comprising the 5′ enhancer and the promoter (chr2 positions 19347844 to 19363608) contained the natural promoter and transcriptional start site of the Ptf1a gene, while the 5′ enhancer (chr2 positions 19347844 to 19350141) and the 3′ control region (chr2 positions 19365786 to 19378222) were linked to the lacZ reporter gene through the minimal promoter of the rat Ela1 gene. The lacZ reporter for the 5′ enhancer construct also contained a nuclear localization signal. Each construct was isolated from a recombinant plasmid and microinjected into pronuclei of fertilized eggs by the Transgenic Core Facility of the University of Texas Southwestern, Dallas. Timed embryos were collected between embryonic day 10.5 (E10.5) and E17.5. Whole embryos or dissected embryonic organs, including the pancreas, were prefixed and stained for β-galactosidase activity as described previously (7). Transgenic embryos were identified by PCR with primers specific for the lacZ sequence.
Mouse lines were derived similarly from mouse eggs microinjected with an enhanced green fluorescent protein (EGFP) gene-based transgene containing the 2.3-kb enhancer linked to the minimal beta-globin promoter, the EGFP coding sequence, and the bovine growth hormone gene 3′ untranslated region. EGFP fluorescence was detected and imaged with a Bio-Rad MRC 1-24 confocal microscope.
Immunolocalization.
The immunodetection of CPA1, PTF1a, β-galactosidase, and glucagon was performed with 20-μm frozen sections of 4% paraformaldehyde-fixed samples or with 10-μm frozen sections of 0.2% glutaraldehyde-fixed embryonic tissues. Guinea pig anti-mouse PTF1a (used at 1:2,000) was a gift from Kei Hori (9); anti-CPA1 (1:1,000) was purchased from Chemicon, anti-insulin (1:2,000) and antiglucagon (1:8,000) were obtained from Linco, and anti-β-galactosidase (1:X000, where X000 is any multiple of a thousand) was purchased from MP Biomedicals. Conventional immunofluorescence photomicroscopy was performed with a Leica DMRXE microscope, and confocal imaging was carried out with a Zeiss LSM-510 Meta microscope.
RESULTS
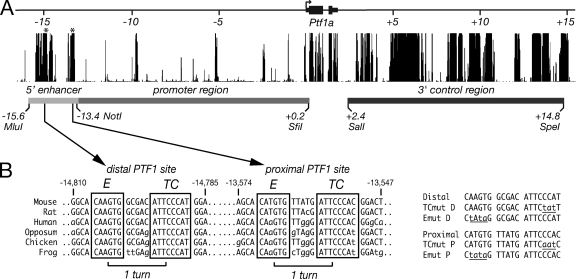
As an initial step to identify transcriptional regulatory regions, we surveyed the nucleotide sequence of the mouse Ptf1a gene 15 kb upstream and downstream of the transcriptional start site for evolutionarily conserved areas (Fig. 1). A highly conserved region (the 5′ enhancer) lies between −15.6 and −13.4 kb relative to the transcriptional start site, whereas the rest of the 5′-end-proximal flanking region (the promoter region) is relatively unconserved. The Ptf1a coding region is followed by a region of extensive conservation (the 3′ control region) that begins after the 3′ untranslated region of the Ptf1a mRNA and continues for 12.4 kb. More than one-third of this 3′ flanking sequence is highly conserved among species from zebrafish to mice.
FIG. 1.
The flanking regions of the Ptf1a gene contain domains highly conserved among vertebrates. (A) Regions of the Ptf1a gene with phylogenetic conservation. (Top line) Schematic of the Ptf1a gene and approximately 15 kb of 5′ and 3′ flanking sequences. (Center) Graphical representation of the sequence homology between the mouse gene and rat, human, opossum, chicken, and frog genes as determined using BLAT (11). Asterisks indicate the positions of the PTF1 complex binding sites. (Bottom line) The gene segments tested in this work were the distal 5′ flanking region from 15.6 to 13.4 kb upstream of the start of transcription (the 5′ enhancer), the proximal 5′ flanking sequence from 13.4 kb upstream to 0.2 kb downstream (the promoter region), and the 3′ flanking sequence from 2.4 to 14.8 kb downstream (the 3′ control region). The locations and identities of restriction sites that separate the regions are shown (the MluI site at kb −15.6 was created by PCR), and the exact coordinates of each of the Ptf1a gene segments on mouse chr2 are given in Materials and Methods. (B) Two PTF1 binding sites in the 5′ enhancer are conserved from amphibians to mammals. (Left panel) Each PTF1 site has an E box and a TC box separated by one helical DNA turn. Lowercase letters indicate divergence from a consensus sequence. (Right panel) Mutations (underlined) in the TC and E boxes of the distal and proximal PTF1 binding sites analyzed in this study.
To determine whether Ptf1a transcription may be controlled by the PTF1 trimeric complex, we searched the 30 kb of the gene and flanking regions for PTF1 binding sites conserved among vertebrates. Only two conserved PTF1 binding sites were found, and both were in the 5′ enhancer region (Fig. 1). Each of these potential binding sites conforms to the known consensus sequence (5, 20), with an E box and the TC box separated by one helical turn of DNA. In the interaction of the PTF1 complex with DNA, the E protein-PTF1a dimer binds the E box while RBPJ or RBPJL binds the TC box (4, 21). The 5′ enhancer together with the promoter region was able to direct the expression of a reporter gene in pancreatic AR4-2J acinar cells but not in nonpancreatic HEK-293 cells (Fig. 2A). Despite the absence of discernible PTF1 binding sites, both the promoter region alone and the 3′ control region also were active in the pancreatic cells but not in the nonpancreatic cells. Thus, all three regions have potential pancreatic transcriptional activity.
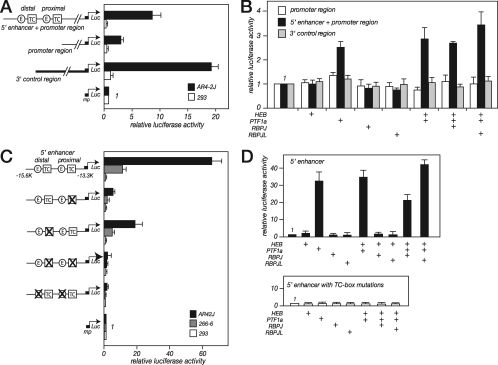
FIG. 2.
The 5′ enhancer, the promoter region, and the 3′ control region are active in pancreatic acinar cell lines, but only the 5′ enhancer is responsive to PTF1. (A) The 5′ enhancer together with the promoter region, the promoter region alone, or the 3′ control region was linked to a minimal promoter (mp) and a luciferase (Luc) reporter gene (see Materials and Methods), and AR4-2J pancreatic acinar cells or 293 nonpancreatic cells were transfected with the constructs. (B) To determine which regions were responsive to the PTF1 complex, 293 cells were cotransfected with the same constructs and expression plasmids for the three components of the PTF1 complex. +, present. (C) To test whether the activity of the enhancer required the PTF1 binding sites, AR4-2J and 266-6 acinar cells and 293 nonacinar cells were transfected with the 5′ enhancer linked to a minimal promoter driving luciferase expression or with the same construct bearing mutations in the proximal, the distal, or both PTF1 binding sites. (D) To verify that the PTF1 complex acted through the known PTF1 binding sites, 293 cells were cotransfected with the 5′ enhancer construct or the 5′ enhancer construct with both TC boxes mutated and plasmids expressing components of the PTF1 trimer. Transfections were normalized according to the expression levels of a pCMVβ plasmid with which the cells were cotransfected. Error bars represent standard deviations.
To determine whether the PTF1 complex controls the transcriptional activities of the three control regions, 293 cells were transfected with expression plasmids for the PTF1 subunits along with reporter genes linked to the Ptf1a-flanking sequences (Fig. 2B). The 5′ enhancer region together with the promoter region was activated 2.5-fold by the ectopic expression of PTF1a. The addition of the other components of the complex (HEB, RBPJ, and RBPJL) did not increase the activity further, likely due to the presence of endogenous RBPJ and E proteins in 293 cells (data not shown). The promoter and 3′ control regions, however, were unresponsive to the PTF1 subunits.
The 5′ enhancer alone induced the expression of a reporter gene with a minimal promoter in the AR4-2J and 266-6 pancreatic acinar cell lines but not in 293 cells (Fig. 2C). To determine whether this activity depended on the presence of the two PTF1 binding sites, the TC box of each site was inactivated by three contiguous base changes (Fig. 1). For each of the sites, the mutation of the TC box blocked the binding of the PTF1 complex in EMSAs (Fig. 3A, lane 15). Nearly all of the enhancer activity in transfected acinar cells was lost when the proximal site was altered, about two-thirds of the activity was lost when the distal site was altered, and the activity was reduced to background levels when both sites were altered (Fig. 2C). In a similar fashion, the mutation of the E box associated with each TC box eliminated PTF1 complex binding in EMSAs (data not shown) and transcriptional activity in both acinar cell lines (Fig. 2C). In this transfection assay, therefore, both the bHLH binding and the RBPJ binding parts of the bipartite PTF1 sites were required for complete enhancer activity.
FIG. 3.
The trimeric PTF1 complex binds PTF1 sites in EMSAs and in vivo. (A) Complexes formed on the proximal or distal PTF1 sites of the 5′ enhancer. Antibodies against PTF1a (αPTF1a), RBPJ, or RBPJL were tested for their ability to supershift the proposed PTF1 complexes formed either with nuclear extract from adult mouse pancreas or with subunits synthesized in vitro. Bands: 1, monomer of RBPJ or RBPJL; 2, E protein-PTF1a heterodimer; 3, PTF1 trimeric complex containing either RBPJ or RBPJL; #, supershifted RBPJ monomer complex; SS and *, supershifted trimeric complexes. +, present; TC mut, TC box mutant form. (B) PTF1 complex subunits bound to the proximal and distal PTF1 sites in nuclear chromatin were analyzed by ChIP with antibodies against PTF1a, RBPL, RBPJ, or control immunoglobulin G (IgG). Error bars represent standard deviations.
To confirm that the 5′ enhancer is activated by the binding of PTF1 to the conserved PTF1 binding sites, reporter genes directed by either the 5′ enhancer or the 5′ enhancer with both TC boxes mutated were introduced into 293 cells, along with expression plasmids for the PTF1 subunits (Fig. 2D). The activity of the enhancer was dependent on the coexpression of PTF1a and the presence of an intact TC box. This requirement for a TC box also confirms that PTF1a is acting as part of a complete trimeric complex of HEB-PTF1a-RBPJ or HEB-PTF1a-RBPJL, because an HEB-PTF1a dimer can bind to an E box in the absence of a TC box but this heterodimer is transcriptionally ineffective (4).
Trimeric PTF1 complexes formed with in vitro-synthesized subunits can bind to oligonucleotides matching the sequence of either the proximal or distal PTF1 binding site of the 5′ enhancer (Fig. 3A). The subunit compositions of the complexes were confirmed by the ability of antibodies against PTF1a, RBPJ, or RBPJL to supershift the EMSA bands. For both the HEB-PTF1a-RBPJ and HEB-PTF1a-RBPJL complexes, the in vitro-synthesized proteins bound the proximal binding site slightly more effectively than the distal binding site (Fig. 3A, compare lanes 3 and 7). When nuclear extract from adult mouse pancreas tissue was incubated with either the proximal or distal PTF1 site, a trimeric PTF1 complex also formed (Fig. 3A, lane 11). The results of antibody supershifts showed that for both sites, most if not all of the complex contained RBPJL (Fig. 3A, compare lanes 13 and 14), even though RBPJ was also present in the nuclear extract (Fig. 3A, lane 11, band 1) (4). Therefore, in adult pancreatic acini, the predominant form of PTF1 capable of binding the PTF1 sites in the enhancer contains RBPJL rather than RBPJ.
To determine whether both sites are bound by the PTF1 complex in acinar nuclei, we performed ChIP of cross-linked, sheared chromatin from adult mouse pancreas tissue with antibodies specific for PTF1a, RBPJL, and RBPJ (Fig. 3B). The region containing the proximal PTF1 site was enriched 6.2- and 3.2-fold in chromatin immunoprecipitated with anti-PTF1a and anti-RBPJL, respectively. No enrichment was seen with anti-RBPJ, which is consistent with the results for PTF1 sites in genes for several pancreatic enzymes that also are bound by the RBPJL form exclusively (4). The distal PTF1 site of the 5′ enhancer was enriched only slightly with antibodies to PTF1a and not enriched with antibodies specific for either RBPJ or RBPJL. Thus, the distal site is less important than the proximal site for activity in transfected acinar cells (Fig. 2C), binds the PTF1 complex to a lesser extent than the proximal site does (Fig. 3A), and is not occupied by a complete complex in vivo.
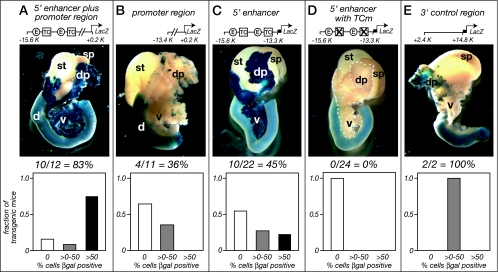
lacZ reporter transgenes were created with the 5′ enhancer, the promoter region, and the 3′ control region to examine the role of each region during pancreatic development. Founder embryos were collected at 17.5 days postcoitum (E17.5) and stained for β-galactosidase activity. When the transgene comprised the 5′ enhancer plus the promoter region, β-galactosidase was detectable in most pancreatic acinar cells of nearly all transgenic embryos (10 of 12) (Fig. 4A). The β-galactosidase activity was restricted to the acinar cells (Fig. 5), where PTF1a is normally present at this stage, and a few scattered islet cells. The promoter region alone also directed acinar cell-specific expression, but only in about one-third of transgenic embryos and then only in a small minority of acinar cells (Fig. 4B). The 5′ enhancer was sufficient to direct acinar cell-specific β-galactosidase activity, although with lower penetrance (10 of 22 embryos) and fewer positive pancreatic cells than were obtained with the 5′ enhancer in combination with the promoter region (Fig. 4C). The dependence of the activity of the 5′ enhancer on the PTF1 binding sites was confirmed when mutations of both TC boxes were introduced into the transgene. None of the 24 transgenic embryos bearing the 5′ enhancer with TC box mutations had detectable β-galactosidase in the pancreas (Fig. 4D). The 3′ control region also directed pancreatic expression, but in very few and widely scattered cells (Fig. 4E).
FIG. 4.
PTF1 sites in the Ptf1a promoter augment transcription in developing acinar cells. Ptf1a regions were ligated upstream of a lacZ reporter and introduced into fertilized mouse eggs. Embryos derived from the implanted oocytes were sacrificed at E17.5 and stained for β-galactosidase (βgal) activity; embryos bearing transgenes were identified by PCR genotyping. A diagram of each transgene is at the top, a photograph shows representative staining, and the bar graph summarizes the results of pancreas staining for all transgenic animals examined for each transgene. dp, dorsal pancreas; v, ventral pancreas; d, duodenum; sp, spleen; st, stomach.
FIG. 5.
Regions of the Ptf1a 5′ flanking sequence direct acinar cell-specific expression at E17.5 but no pancreatic expression at E10.5. (A to C) Transgenic embryos at E17.5 were fixed and stained to detect β-galactosidase (Fig. 4), embedded in paraffin, and sectioned. The staining of sections carrying the transgene comprising the 5′ enhancer and the promoter (A), the promoter region alone (B), and the 5′ enhancer alone (C) is shown. For the construct with the 5′ enhancer plus the promoter and for the promoter region alone (A and B), the β-galactosidase was cytoplasmic. For the 5′ enhancer (C), the β-galactosidase reporter gene contained a nuclear localization signal. The arrowhead (B) points to β-galactosidase-positive cells in an acinus of an animal with the promoter region transgene. Sections in all three panels were stained with hematoxylin; that in the lower panel was also stained with eosin. The bar represents 50 μm and applies to all three panels. a, acinus; e, endocrine cell clusters (preislets). (D to H) The same Ptf1a transgenes diagrammed in Fig. 4 were introduced into fertilized mouse oocytes. Embryos were sacrificed at E10.5 and stained for β-galactosidase activity. The staining of sections carrying the transgene comprising the 5′ enhancer and the promoter (D), the promoter region alone (E), the 5′ enhancer alone (F), and the 3′ control region (G) is shown. (H) Expression of the endogenous Ptf1a locus monitored by β-galactosidase staining of a Cre-activated Rosa26R conditional lacZ reporter in a Ptf1acre/+ heterozygous embryo. Arrows point to the position of the pancreas (stained only in the Ptf1acre/+ control embryo); arrowheads point to the neural tube. The numbers along the side indicate the number of transgenic embryos examined for each construct (D to G), all without pancreatic expression.
None of the lacZ-based transgenes with Ptf1a-flanking regions were active in early pancreatic development, prior to the appearance of acinar cells (Fig. 5D to G). At E10.5, the transgenes directed by the 5′ enhancer and the promoter region, the promoter region alone, the 5′ enhancer alone, or the 3′ control region failed to be expressed in the pancreas, although each was active in the spinal cord, a known site of Ptf1a expression (6). The endogenous Ptf1a locus is active in both the pancreas and the spinal cord at this developmental stage (Fig. 5H).
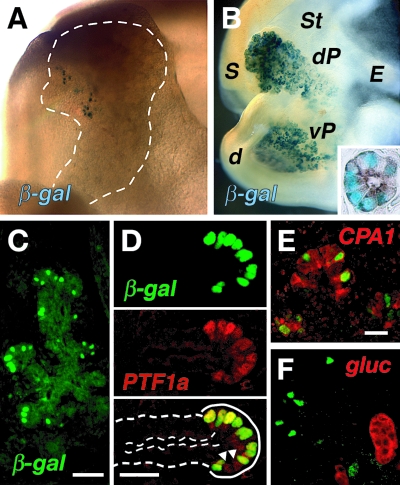
Expression of the transgenic β-galactosidase reporter was first detected in a few scattered cells at E13.5 (Fig. 6A and Table 1) and in many more cells, often in distinct acinar rosettes, by E14.5 (Fig. 6B). As development progresses, a greater fraction of transgenic embryos and acinar cells activate the enhancer. Cells detected by immunofluorescence analysis for β-galactosidase (Fig. 6C) were also positive for PTF1a (Fig. 6D). Many of the β-galactosidase-containing cells at E13.5 also had the early acinar marker CPA1 (Fig. 6E). At E14.5, emerging acinar cells were much more prominent and most β-galactosidase-positive cells were associated with CPA1-positive acini (Fig. 6B, inset). Like PTF1a, transgenic β-galactosidase was excluded from the glucagon-containing cells, which are the prevalent endocrine cells at this developmental stage (see, e.g., Fig. 6F), and from the insulin-containing cells (data not shown). These results indicate that the 5′ enhancer is active in cells beginning the acinar developmental program.
FIG. 6.
Activation of the isolated 5′ enhancer at the onset of acinar cell development. (A) Scattered cells of an E13.5 embryo stained for β-galactosidase (β-gal) are present near the periphery of the pancreatic epithelium (outlined) lying on the surface of the stomach primordium. (B) In this E14.5 embryo, staining for the β-galactosidase activity from the enhancer-driven transgene is more extensive and restricted to the developing acini of the ventral pancreas (vP), positioned in the curvature of the first duodenal loop, and the dorsal pancreas (dP), which lies on the surface of the stomach below the spleen. St, stomach; S, spleen; E, esophagus; d, duodenum. (Inset) Example of a forming acinus with CPA1 immunolocalized to the apical cytoplasmic domain and blue staining indicating transgenic β-galactosidase activity throughout the cytoplasm. (C) At E13.5, immunofluorescence from the transgenic β-galactosidase is present in numerous nuclei of cells around the rounded periphery buds of the pancreatic epithelium, a position consistent with newly forming acini. Bar, 50 μm. (D) Coimmunofluorescence of the transgenic β-galactosidase (green) and endogenous PTF1a (red) showed that the enhancer activity was first detected at E13.5 in nascent acinar cells that contained PTF1a. The overlay shows that β-galactosidase-positive cells in a newly forming acinus (solid line) were also PTF1a positive (yellow), whereas the cells of the connecting tubules (dashed lines) were devoid of β-galactosidase and PTF1a. Arrowheads, PTF1+ nuclei without β-galactosidase. Bar, 20 μm for all images in panel D. (E) β-Galactosidase-positive cells often were also CPA1 positive. (F) β-Galactosidase was not detected in the early endocrine cells, most of which expressed glucagon (gluc). Bar, 20 μm for panels E and F.
TABLE 1.
Summary of the activity of the 5′ enhancer during embryonic pancreatic development
| Age of embryos (dpc)a | No. of transgenic embryos | No. of embryos with CNS stainingb | No. of embryos with pancreas stainingc |
|---|---|---|---|
| 10.5 | 7 | 6 | 0 (0) |
| 13.5 | 23 | 14 | 2 (14) |
| 14.5 | 5 | 4 | 2 (50) |
| 17.5d | 22 | 10 | 10 (100) |
dpc, days postcoitum.
Staining for β-galactosidase activity in the neural tube (central nervous system [CNS]), which is a site of strong endogenous Ptf1a expression, is an indicator that the transgene can be activated in an appropriate developmental context at each of these stages. See also Fig. 5.
Pancreas staining indicated transgenic β-galactosidase activity in pancreatic epithelial tissue. Values in parentheses are the percentages of embryos with CNS staining that also had pancreas staining.
Results from Fig. 4 are summarized.
To investigate more thoroughly the activity of the enhancer during early development, we examined the expression of the 5′ enhancer driving an EGFP reporter in embryos of two independently derived transgenic lines (rather than founder embryos). In contrast to the lacZ reporter construct in founder embryos prior to E13.5, which lacked detectable expression, this construct was active at E10.5 (Fig. 7A and B) and E12.5 (Fig. 7E), prior to the emergence of acinar cells during the secondary transition. At E10.5, EGFP fluorescence was present in the epithelial cells of the ventral and dorsal buds. At E12.5, the activity of the enhancer increased around the periphery of the epithelium (Fig. 7E), in accordance with the accumulation of PTF1a protein in these proacinar crescents. At E14.5, enhancer activity was localized exclusively to PTF1a+ proacinar cells (Fig. 7F). Nonepithelial cells and the epithelial cells of nontransgenic embryos had no fluorescence over the background level. Expression levels in the precursor epithelium at E10.5 and E12.5 were much lower than those in nascent acinar cells at E14.5 (Fig. 7G), which may explain the absence of detectable activity of the lacZ-based enhancer transgene. In contrast to the construct comprising the 5′ enhancer and the green fluorescent protein reporter gene, which was active at E10.5, the 3′ control region with a fluorescent protein reporter gene was inactive at this stage in three transgenic lines, although fluorescence was intense in the neural tube and later in sparse cells of the E13.5 pancreas (data not shown).
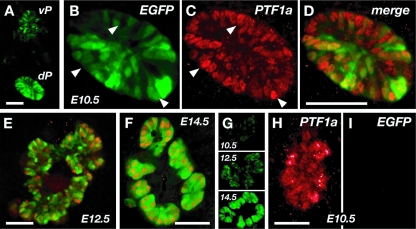
FIG. 7.
Activity of the 5′ enhancer in the epithelium of the early pancreatic buds. EGFP fluorescence is shown in green; PTF1a immunofluorescence is shown in red. (A) Expression of the 5′ enhancer transgene-EGFP reporter in the dorsal (dP) and ventral (vP) pancreatic buds of an E10.5 embryo of an established mouse line. (B to D) Overlapping patterns of transgenic EGFP (B) and the endogenous PTF1a protein (C) and the merging of the two patterns (D). Arrowheads indicate nuclei with high levels of PTF1a but little or no EGFP. (E and F) Enhancer activity (green) in the dorsal pancreas at E12.5 and E14.5, respectively, compared to the distribution of PTF1a (red). (G) Relative levels of enhancer activities in the established transgenic line prior to (E10.5 and E12.5) and during (E14.5) the emergence of acinar cells. (H and I) For embryos with the WA mutant form of PTF1a only, the 5′ enhancer is inactive in the early pancreatic epithelium (I), even though the PTF1a(W298A) protein is present (H). Bars: panels A, D, E, and F, 50 μm; panel H, 30 μm.
The expression patterns for the transgenic enhancer-EGFP gene construct and the endogenous PTF1a detected by immunofluorescence were not congruent: the relative intensities of EGFP and anti-PTF1a fluorescence did not coincide for many of the expressing cells. Although the majority of the epithelial cells expressed both, there were cells with a high level of one but little or none of the other (Fig. 6B and C). The discrepancies may be due to the absence of control sequences outside the 2.3-kb enhancer fragment. As observed for the expression of the endogenous Ptf1a locus, our results show that the 5′ enhancer is active in the early pancreatic epithelium, albeit at low levels, and then superinduced in cells committed to the acinar fate.
To determine whether the activity of the 5′ enhancer in the precursor epithelium requires the early trimeric PTF1-J complex, we examined the expression of the EGFP gene-based enhancer transgene in homozygous Ptf1a(W298A) embryos. The tryptophan-to-alanine substitution at position 298 eliminates the early developmental functions of PTF1a (14) because the altered PTF1a cannot bind and recruit RBPJ into the trimeric complex effectively (4). The effects of the W298A mutation on pancreatic development are identical to those of the Ptf1a null mutation (14): the pancreatic evaginations of the endoderm initiate, but the subsequent early growth and morphogenesis of the epithelium are curtailed beginning at about E10.5, and the secondary transition with its formation of islet and acinar tissues does not occur. At E10.5, the 5′ enhancer was inactive in the pancreatic epithelia of homozygous Ptf1a(W298A) embryos (Fig. 7I), even though the endogenous Ptf1a(W298A) locus was active and the W298A protein was present (Fig. 7H). Therefore, because this early activity of the enhancer requires the interaction between PTF1a and RBPJ, it must be due to autoregulation. Because the initial induction of Ptf1a gene transcription cannot depend on preexisting PTF1a protein, the autoregulation of the enhancer must represent a maintenance function that follows an initial activation event. Furthermore, because the endogenous locus is active at E10.5 in Ptf1a(W298A) pancreas tissue, but the isolated enhancer is not, critical regulatory information for this initial activation of Ptf1a transcription must reside outside the 2.3-kb enhancer fragment.
DISCUSSION
PTF1a is a critical regulator with multiple, temporally distinct functions for pancreatic organogenesis: early functions for growth and morphogenesis (10, 14), functions at middevelopment for the acinar developmental program, and functions thereafter for the maintenance of the acinar cell phenotype (1, 12). To understand better the complex control of Ptf1a during pancreatic development, we identified and analyzed the gene-associated regulatory sequences for pancreatic transcription. With this report, we show that the proximal 13.4-kb 5′ and 12.5-kb 3′ regions of the Ptf1a gene contain little information for pancreas-restricted transcription. The principal control region for pancreatic transcription is within 2.3 kb located 15.6 to 13.4 kb upstream of the transcriptional start site. Because this control region has, minimally, the ability to act adjacent to or at a distance from either its cognate or a heterologous promoter, it has the properties of an enhancer. This 5′ enhancer contains two bipartite binding sites for a trimeric PTF1 bHLH complex. The detrimental effects in vivo of mutating these binding sites and the detection of PTF1a at the gene-proximal site show that PTF1a maintains the transcription of its own gene.
Autoregulation of Ptf1a transcription.
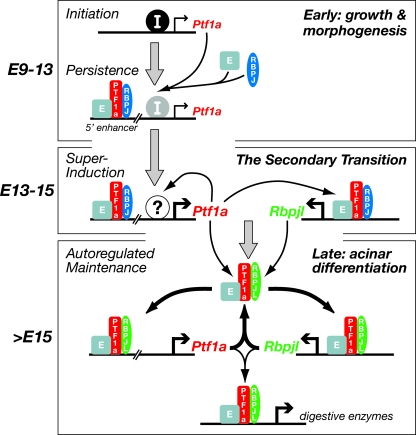
The active form of PTF1a in the mature pancreas is a three-subunit complex of PTF1a, a common E protein such as TCF12/HEB, and RBPJL. RBPJL is restricted to the pancreas, intestine, lung, and parts of the brain (4, 16) (www.brain-map.org) but localizes with PTF1a only in pancreatic acinar cells. The RBPJL form of the PTF1 complex resides on the promoters of the pancreatic secretory digestive enzyme genes and is responsible for the acinar cell-specific activity of most (if not all) of these genes (4, 5). The transcription of Rbpjl in acinar cells is also controlled by the RBPJL form of the PTF1 complex (14). The Rbpjl promoter contains a bipartite PTF1 binding site occupied by PTF1-L in adult acinar nuclei and critical for the acinar activity of the promoter. Thus, the two pancreas-restricted subunits of the PTF1 complex are coordinately autoregulated by the complex (Fig. 8).
FIG. 8.
Autoregulatory functions during pancreatic development. During early growth and morphogenesis, Ptf1a transcription in the nascent epithelial buds at E9 is initiated by unknown factors (I) that bind outside the 5′ enhancer. Once PTF1a is produced, it forms a complex with a common E protein (generally HEB/TCF12) and RBPJ, which binds the 5′ enhancer to autoactivate Ptf1a transcription upon the disappearance of the inducing factors. It is this form of the PTF1 complex (PTF1-J) that is required for the early growth and morphogenesis of the pancreatic epithelium (14). During the secondary transition, in conjunction with the 5′ enhancer, Ptf1a is superinduced in cells beginning the acinar developmental program either by the activation of unknown activators (?) or the loss of repressors. Upon the increase in PTF1a, the transcription of Rbpjl is induced by the PTF1-J complex (14). RBPJL replaces RBPJ in the PTF1 complex, which binds and maintains the activities of the Ptf1a enhancer and the Rbpjl promoter. During acinar differentiation and maintenance, the accumulation of the RBPJL form of the PTF1 complex drives the transcription of downstream regulators of acinar development, as well as the genes encoding the secretory digestive enzymes characteristic of the differentiated acinar phenotype. In mature acinar cells, dual autoregulatory loops (thick arrows) maintain the transcription of Ptf1a and Rbpjl via PTF1-L. PTF1-L continues driving the transcription of the digestive enzyme genes (4), as well as downstream regulatory genes, for the faithful maintenance of the acinar phenotype.
Feedback loops with autoregulation are common motifs in transcription factor networks that control the development and maintain the stability of complex biological systems (24, 25). The transcriptional activation of Ptf1a and Rbpjl by a shared DNA binding complex containing each of their products (Fig. 8) is a variation of a two-node positive feedback loop with autoregulation of each node (see, e.g., reference 2). Such a positive feedback loop of two transcription factors at the top of a regulatory network can lock a cell fate decision by irreversibly activating a developmental program. In differentiated cells, this simple regulatory motif acts as molecular memory to maintain the mature phenotype of a particular cellular lineage.
In this instance with Ptf1a and Rbpjl, an obligatory complex (PTF1-L) between the two transcription factors mediates the feedback activation of both nodes. PTF1-L is composed of a heterodimer of PTF1a and an E protein, which interact through their bHLH domains and bind an E box DNA motif, and a third subunit (RBPJL) that is recruited to the complex through a C-terminal portion of PTF1a and binds a typical RBP (RBPJ or RBPJL) DNA recognition sequence (a TC-rich box). Two unusual attributes of the trimeric complex restrict the action of PTF1a and RBPJL to a highly selective subset of E boxes and RBP binding sites (4).
First, specific DNA binding by the trimeric complex is highly concerted and much more selective than the simple sum of the binding of the individual components. Thus, even though the PTF1a-E protein heterodimer can bind an isolated E box and RBPJL can bind an isolated TC box, DNA binding by the trimeric complex requires both DNA boxes and their proper spacing. By using a consensus binding sequence with permissible variations derived from a compilation of known DNA binding sites for PTF1, potential binding sites can be calculated to occur randomly only once approximately every 15 kb.
Second, without an RBP subunit, PTF1a-E protein heterodimers have little (if any) activity on their own; thus, mutations in the PTF1a C-terminal domain that prevent the recruitment of an RBP into the complex drastically diminish transcriptional activity in cell transfection experiments (4) and disrupt pancreatic development for both mice and humans (14, 22). For Ptf1a and Rbpjl, the binding of the complete trimeric complex is critical to their acinar transcription; alterations of the bipartite PTF1 binding sites in the Ptf1a enhancer and the Rbpjl promoter that permit the binding of the bHLH heterodimer but not the trimer nonetheless abolish the activities of the enhancer (Fig. 4D) and the promoter (14). Due to the high-level binding selectivity of the trimeric complex and the apparent inactivity of the bHLH heterodimer, the feedback activation loops, as well as the control of downstream effectors of acinar development and maintenance, are highly discriminating. The strong cooperative action of PTF1-L may be the basis of the secure phenotype of pancreatic acinar cells. It will be interesting to test whether perturbations of the stability of the complex affect the phenotype of the mature acinar cell or the known transdifferentiation relationships between pancreatic acinar cells and ductal cells (3, 15) and pancreatic acinar cells and hepatocytes (18, 19, 23).
Developmental control of Ptf1a: the 5′ enhancer.
Ptf1a gene transcription has four temporally distinct phases: initiation at the onset of pancreatic bud formation (E9 to E9.5), maintenance at a low level in the early precursor epithelium (E10 to E12), superinduction in multipotent precursor cells at E12.5, and maintenance at the superinduced level in cells committed to acinar differentiation (E13 to adult). The results of this study show that the autoregulated enhancer controls the two maintenance phases.
The detection of activity of the 5′ enhancer with the EGFP transgene in stable transgenic mouse lines showed that the enhancer could direct expression as early as E10.5, during the early phase of growth and morphogenesis, prior to the beginning of acinar cell development at E13. To distinguish whether this activity of the 5′ enhancer represents the initial induction event for the Ptf1a gene or the autoregulatory maintenance of transcription postinduction, we analyzed the effect of the homozygous WA mutation of Ptf1a on transgene expression. PTF1a(W298A) had tryptophan 298 replaced with alanine, and its ability to recruit RBPJ into a functional trimeric PTF1 complex was nearly completely eliminated (4). The inability to form the PTF1-J complex disrupts the early phase of pancreatic development (14). The PTF1a(W298A) protein appeared and then disappeared much more quickly than PTF1a does in normal embryos (14), which suggests that the initial inducers of Ptf1a transcription are transient and that continued expression relies on autoregulation through the 5′ enhancer. The inactivity of the 5′ enhancer-EGFP transgene at E10.5, when the only PTF1a was the WA mutant form, indicates that the postinduction activity is autoregulated through the binding of a trimeric complex containing RBPJ. Although the direct binding of a PTF1 complex is likely required for the acinar activity of the 5′ enhancer, it remains a formal possibility that the requirement for the PTF1-J complex at E10.5 is indirect.
The analysis of the 5′ enhancer in lacZ-based transgenes showed that the switch from the early and broad Ptf1a expression in the precursor epithelium to the more intense and selective expression in newly forming acinar cells also required the action of a PTF1 complex on the 5′ enhancer. In this context, the 5′ enhancer was sufficient for transcriptional activation in these late-forming cells and the PTF1 complex binding sites in the enhancer were necessary. Because the superinduction of Ptf1a is an initial step in the formation of acinar cells (26) and precedes the appearance of RBPJL (14), the acinar PTF1-L complex cannot be the initial activator. Because the PTF1-J complex is present throughout the early pancreatic precursor epithelium, spatially restricted signaling and the induction of new transcriptional regulators must be invoked for the superinduction in collaboration with PTF1-J.
Recently, the results of lineage-tracing analyses by Zhou and colleagues (26) indicated that the cells in the tips of the branching epithelium at E12.5 that first superinduce Ptf1a are not yet restricted to the acinar fate and can contribute to islet and ductal cell populations as well. Subsequent regulatory mechanisms must maintain high levels of Ptf1a expression in acinar cells and suppress expression in islet and ductal cells. The identification of these events and the molecular players is critical to understanding the initiation of the acinar lineage and its resolution from the islet and ductal lineages at pancreatic middevelopment.
Acknowledgments
We thank Robert Hammer and the University of Texas Southwestern Transgenic Core Facility for generating transgenic embryos, Yanjie Chang for analysis of transgenic embryos, Kei Hori for the guinea pig anti-mouse PTF1a, and John Shelton, William Richardson, and the Pathology Research Core Facility for extensive expert advice and histological services.
This work was supported by Public Health Service grants DK61220 (R.J.M.) and HD37032 (J.E.J.).
Footnotes
Published ahead of print on 7 July 2008.
REFERENCES
- 1.Adell, T., A. Skoudy, A. Gomez-Cuadrado, T. Palomero, and F. X. Real. 2000. Role of the basic helix-loop-helix transcription factor p48 in the differentiation phenotype of exocrine pancreas cancer cells. Cell Growth Differ. 11137-147. [PubMed] [Google Scholar]
- 2.Alon, U. 2007. Network motifs: theory and experimental approaches. Nat. Rev. Genet. 8450-461. [DOI] [PubMed] [Google Scholar]
- 3.Arias, A. E., and M. Bendayan. 1993. Differentiation of pancreatic acinar cells into duct-like cells in vitro. Lab. Investig. 69518-530. [PubMed] [Google Scholar]
- 4.Beres, T. M., T. Masui, G. H. Swift, L. Shi, R. M. Henke, and R. J. MacDonald. 2006. PTF1 is an organ-specific and Notch-independent basic helix-loop-helix complex containing the mammalian Suppressor of Hairless (RBP-J) or its paralogue, RBP-L. Mol. Cell. Biol. 26117-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cockell, M., B. J. Stevenson, M. Strubin, O. Hagenbüchle, and P. K. Wellauer. 1989. Identification of a cell-specific DNA-binding activity that interacts with a transcriptional activator of genes expressed in the acinar pancreas. Mol. Cell. Biol. 92464-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glasgow, S. M., R. M. Henke, R. J. MacDonald, C. V. E. Wright, and J. E. Johnson. 2005. Ptf1a determines GABAergic over glutamatergic neuronal cell fate in the spinal cord dorsal horn. Development 1325461-5469. [DOI] [PubMed] [Google Scholar]
- 7.Hale, M. A., H. Kagami, L. Shi, A. M. Holland, H. P. Elsasser, R. E. Hammer, and R. J. MacDonald. 2005. The homeodomain protein PDX1 is required at mid-pancreatic development for the formation of the exocrine pancreas. Dev. Biol. 286225-237. [DOI] [PubMed] [Google Scholar]
- 8.Hammer, R. E., G. H. Swift, D. M. Ornitz, C. J. Quaife, R. D. Palmiter, R. L. Brinster, and R. J. MacDonald. 1987. The rat elastase I regulatory element is an enhancer that directs correct cell specificity and developmental onset of expression in transgenic mice. Mol. Cell. Biol. 72956-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hori, K., J. Cholewa-Waclaw, Y. Nakada, S. M. Glasgow, T. Masui, R. M. Henke, T. M. Beres, H. Wildner, B. Martarelli, J. A. Epstein, M. A. Magnuson, R. J. MacDonald, C. Birchmeier, and J. E. Johnson. 2008. A non-classical bHLH-Rbpj transcription factor complex is required for specification of GABAergic neurons independent of Notch signaling. Genes Dev. 22166-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawaguchi, Y., B. Cooper, M. Gannon, M. Ray, R. J. MacDonald, and C. V. E. Wright. 2002. The role of the transcriptional regulator PTF1a in converting intestinal to pancreatic progenitors. Nat. Genet. 32128-134. [DOI] [PubMed] [Google Scholar]
- 11.Kent, W. J. 2002. BLAT: the BLAST-like alignment tool. Genome Res. 12656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krapp, A., M. Knofler, F. Frutiger, G. J. Hughes, O. Hagenbuchle, and P. K. Wellauer. 1996. The p48 DNA-binding subunit of transcription factor PTF1 is a new exocrine pancreas-specific basic helix-loop-helix protein. EMBO J. 154317-4329. [PMC free article] [PubMed] [Google Scholar]
- 13.Krapp, A., M. Knofler, B. Ledermann, K. Burki, C. Berney, N. Zoerkler, O. Hagenbuchle, and P. K. Wellauer. 1998. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev. 123752-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masui, T., Q. Long, T. M. Beres, M. A. Magnuson, and R. J. MacDonald. 2007. Early pancreatic development requires the vertebrate Suppressor of Hairless (RBPJ) in the PTF1 bHLH complex. Genes Dev. 212629-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Means, A. L., I. M. Meszoely, K. Suzuki, Y. Miyamoto, A. K. Rustgi, R. J. Coffey, C. V. Wright, D. A. Stoffers, and S. D. Leach. 2005. Pancreatic epithelial plasticity mediated by acinar cell transdifferentiation and generation of nestin-positive intermediates. Development 1323767-3776. [DOI] [PubMed] [Google Scholar]
- 16.Minoguchi, S., T. Ikeda, S. Itohara, T. Kaneko, H. Hokaichi, and T. Honjo. 1999. Studies on the cell-type specific expression of RBP-L, a RBP-J family member, by replacement insertion of beta-galactosidase. J. Biochem. 126738-747. [DOI] [PubMed] [Google Scholar]
- 17.Pictet, R. L., W. R. Clark, R. H. Williams, and W. J. Rutter. 1972. An ultrastructural analysis of the developing embryonic pancreas. Dev. Biol. 29436-467. [DOI] [PubMed] [Google Scholar]
- 18.Rao, M. S., M. Bendayan, R. D. Kimbrough, and J. K. Reddy. 1986. Characterization of pancreatic-type tissue in the liver of rat induced by polychlorinated biphenyls. J. Histochem. Cytochem. 34197-201. [DOI] [PubMed] [Google Scholar]
- 19.Rao, M. S., and J. K. Reddy. 1995. Hepatic transdifferentiation in the pancreas. Semin. Cell Biol. 6151-156. [DOI] [PubMed] [Google Scholar]
- 20.Rose, S. D., G. H. Swift, M. J. Peyton, R. E. Hammer, and R. J. MacDonald. 2001. The role of PTF1-P48 in pancreatic acinar gene expression. J. Biol. Chem. 27644018-44026. [DOI] [PubMed] [Google Scholar]
- 21.Roux, E., M. Strubin, O. Hagenbuchle, and P. K. Wellauer. 1989. The cell-specific transcription factor PTF1 contains two different subunits that interact with the DNA. Genes Dev. 31613-1624. [DOI] [PubMed] [Google Scholar]
- 22.Sellick, G. S., K. T. Barker, I. Stolte-Dijkstra, C. Fleischmann, R. J. Coleman, C. Garrett, A. L. Gloyn, E. L. Edghill, A. T. Hattersley, P. K. Wellauer, G. Goodwin, and R. S. Houlston. 2004. Mutations in PTF1A cause pancreatic and cerebellar agenesis. Nat. Genet. 361301-1305. [DOI] [PubMed] [Google Scholar]
- 23.Shen, C. N., J. M. Slack, and D. Tosh. 2000. Molecular basis of transdifferentiation of pancreas to liver. Nat. Cell Biol. 2879-887. [DOI] [PubMed] [Google Scholar]
- 24.Stathopoulos, A., and M. Levine. 2005. Genomic regulatory networks and animal development. Dev. Cell 9449-462. [DOI] [PubMed] [Google Scholar]
- 25.Tapscott, S. J. 2005. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development 1322685-2695. [DOI] [PubMed] [Google Scholar]
- 26.Zhou, Q., A. C. Law, J. Rajagopal, W. J. Anderson, P. A. Gray, and D. A. Melton. 2007. A multipotent progenitor domain guides pancreatic organogenesis. Dev. Cell 13103-114. [DOI] [PubMed] [Google Scholar]