Abstract
Aberrant cell division is a hallmark of cancer, but the molecular circuitries of this process in tumor cells are not well understood. Here, we used a high-throughput proteomics screening to identify novel molecular partners of survivin, an essential regulator of mitosis overexpressed in cancer. We found that survivin associates with the small GTPase Ran in an evolutionarily conserved recognition in mammalian cells and Xenopus laevis extracts. This interaction is regulated during the cell cycle, involves Ran-GTP, requires a discrete binding interface centered on Glu65 in survivin, and is independent of the Ran effector Crm1. Disruption of a survivin-Ran complex does not affect the assembly of survivin within the chromosomal passenger complex or its cytosolic accumulation, but it inhibits the delivery of the Ran effector molecule TPX2 to microtubules. In turn, this results in aberrant mitotic spindle formation and chromosome missegregation in tumor, but not normal, cells. Therefore, survivin is a novel effector of Ran signaling, and this pathway may be preferentially exploited for spindle assembly in tumor cells.
Deregulated cell division is a hallmark of cancer (31), which results in unrestrained cell proliferation, abrogation of cell cycle checkpoints, and propensity to aneuploidy (27). These processes hinge on spatial-temporal assembly of a bipolar mitotic spindle (23), where microtubules nucleating from duplicated centrosomes or assembled in proximity of mitotic chromosomes “search and capture” chromatids and ultimately segregate them between daughter cells (46). Reconstitution experiments in model organisms, particularly cycled Xenopus laevis extracts, uncovered a pivotal role of the small GTPase Ran in the mechanism of spindle formation (36). We now know that a gradient of Ran-GTP assembles on mitotic chromosomes (20), largely through the chromatin-associated activity of the nucleotide exchange factor RCC1. In turn, this releases Ran effector molecules implicated in spindle formation (9, 15), including microtubule-stabilizing TPX2 (14), from an inhibitory interaction with importin α/β receptors (16, 45). Depletion of Ran in Xenopus extracts (9) or mammalian cells (15) or targeting its effector molecules (26, 41, 43) profoundly impairs spindle formation, causing the appearance of flattened mitotic spindles, severely depleted of microtubules, and abnormal chromosomal segregation.
Among the regulators of cell division aberrantly overexpressed in cancer is survivin (3), a member of the inhibitor of apoptosis (IAP) gene family with dual roles in suppression of cell death and control of mitosis (1, 29). The latter has been linked to a multiplicity of functions, including kinetochore targeting of the chromosomal passenger complex (22), enhancement of Aurora B kinase activity (7), control of kinetochore-microtubule interactions for proper chromosomal alignment (39), participation in the spindle assembly checkpoint (11), and regulation of microtubule dynamics for spindle formation (37). Although these functions are essential (32) and some of them have been conserved evolutionarily from yeasts to mammals (1, 29), a unifying model for how survivin orchestrates cell division has not been obtained. This is now a priority because survivin antagonists are being tested as anticancer agents in humans (2), and elucidation of the molecular requirements of survivin regulation of mitosis may lead to better anticancer regimens.
In this study, we took an unbiased approach to identify molecular regulators of survivin in the control of cell division. Using high-throughput proteomics screenings in mammalian cells and Xenopus laevis extracts, we found that survivin physically associates with Ran and that this interaction is required to deliver the Ran effector molecule TPX2 to microtubules for proper spindle formation (36).
MATERIALS AND METHODS
Cells and cell culture.
Mouse NIH 3T3 fibroblasts, primary WS-1 human fibroblasts, and cervical carcinoma HeLa cells were obtained from the American Tissue Type Collection (ATCC) (Manassas, VA) and maintained in culture according to the supplier's specifications. To generate a stable cell line that expresses survivin, HeLa cells were transduced with a survivin cDNA construct carrying a carboxyl-terminal hemagglutinin (HA) tag linked to an interleukin 2 receptor subunit surface marker in the pOZ retroviral vector. Transduced cells were selected by repeated cycles of affinity cell sorting by flow cytometry using reactivity for interleukin 2 receptor expression. Parental HeLa cells or stable HeLa-survivin-HA cells (HeLa-SVV) were maintained in a humidified atmosphere containing 5% CO2 at 37°C in Dulbecco's modified Eagle's medium containing 10% heat-inactivated fetal bovine serum, nonessential amino acids, minimal essential medium, sodium pyruvate, 100 μg/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). For cell cycle synchronization, HeLa cells were arrested at the G1/S transition by treatment with 4 mM thymidine (Sigma) for 16 h at 37°C, released into fresh medium, and analyzed by Western blotting or DNA content by flow cytometry at increasing time intervals for the first 20 h after release.
Antibodies and reagents.
The following antibodies to survivin (Novus Biologicals), Ran (Novus Biologicals, Cell Signaling, Santa Cruz Biotechnology), HA (Santa Cruz Biotechnology), α-tubulin (Sigma-Aldrich), Aurora A (Cell Signaling), Borealin (Abgent), Aurora B (Cell Signaling), TPX2 (Novus Biologicals), RCC1 (Santa Cruz Biotechnology), cyclin B1 (Santa Cruz Biotechnology), or β-actin (Sigma-Aldrich), were used. An antibody to Xenopus survivin was raised against bacterially expressed, recombinant full-length Xenopus survivin, affinity purified, and characterized as described previously (8). Restriction enzymes were purchased from New England BioLabs. Recombinant glutathione S-transferase (GST)-Rac1 was purchased from Cytoskeleton, Inc.
Plasmid construction and transfections.
Full-length cDNAs encoding survivin or Ran were amplified by reverse transcription-PCR from HeLa cells and subcloned into HindIII/BamHI sites of pcDNA3.1 or BamHI/XhoI sites of pGEX-4T-1 vectors carrying an HA tag at the N or C terminus, respectively. Site-directed mutagenesis of survivin or Ran cDNA was carried out by PCR. Individual mutants were confirmed by DNA sequencing and inserted into HindIII/BamHI sites of pcDNA3.1 or BamHI/XhoI sites of pGEX-4T-1 vectors carrying HA or FLAG tag at the N or C terminus, respectively. The various cDNA constructs in pGEX vector were expressed in Escherichia coli BL21(DE3) strain, grown in LB medium containing ampicillin after induction with 0.1 mM isopropyl-1-thio-d-galactopyranoside (IPTG) at an optical density at 600 nm of ∼0.5. Xenopus survivin was expressed in E. coli BL21 and affinity purified as described previously (8). Bacterial cultures were harvested after 3 h, lysed by sonication (10 pulses of 20 s each), and GST fusion proteins were purified by affinity chromatography on glutathione-Sepharose TM4B beads (Amersham Biosciences). Proteins were eluted with free glutathione, and in some experiments, the GST frame was removed by overnight treatment with thrombin followed by immediate neutralization with benzamidine. The following recombinant mutant proteins were generated (numbers in parentheses refer to the amino acid sequence): pGEX-Ran(1-45), pGEX-Ran(1-74), pGEX-Ran(1-125), pGEX-Ran(1-170), pGEX-Ran(171-216), pGEX-Ran-G19V, pGEX-Ran-T24N, pGEX-Ran-Q69L, pGEX-Survivin(15-142)-flag, pGEX-Survivin(38-142)-flag, pGEX-Survivin(55-142)-flag, pGEX-Survivin(71-142)-flag, pGEX-Survivin(81-142)-flag, pGEX-Survivin(88-142), pGEX-flag-Survivin(1-70), pGEX-flag-Survivin(1-80), pGEX-Survivin(1-87), pGEX-Survivin(55-70), pcDNA3-HA-survivin-E65A, pcDNA3-survivin-E65A-HA, pcDNA3-survivin, pcDNA3-survivin-E65A, pcDNA3-HA-survivin(55-142), and pcDNA3-HA-survivin(71-142), and individual Ala substitutions of each residue comprised within the survivin sequence Ala55 and Asp70 (A55 to D70). For transient-transfection experiments, subconfluent cultures of HeLa cells were mixed with various cDNA constructs in pcDNA3.1 by Lipofectamine (Invitrogen), as recommended by the manufacturer, and harvested after 24 to 48 h at 37°C.
Proteomics screening of survivin-associated molecules in tumor cells.
To identify molecules bound to survivin in vivo, a survivin-HA complex was immunoprecipitated from parental HeLa cells or HeLa-SVV cultures using an antibody to HA coupled to agarose beads (Sigma-Aldrich) for 4 h under constant agitation. After washes in 0.1 M KCl-containing buffer (20 mM Tris-HCl [pH 8.0], 5 mM MgCl2, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, 0.1% Tween 20, plus 10 mM β-mercaptoethanol), bound proteins were eluted from HA-agarose beads by the addition of soluble HA peptide (0.2 mg/ml; Sigma-Aldrich) for 60 min in the same binding buffer. Samples were centrifuged at 20,000 rpm for 1 h, and fractions collected from the top of the centrifugation tube were separated by high-resolution sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and silver stained. Selected protein bands were analyzed using an Ettan matrix-assisted laser desorption ionization-time-of-flight system by Genomine, Inc. (South Korea), and candidate sequences were matched to the SWISS-PROT and NCBI databases using the search program ProFound (http://prowl.rockefeller.edu/).
Proteomics screening of survivin-associated molecules in Xenopus egg extracts.
Cytostatic factor (CSF)-arrested egg extracts supplemented with sperm DNA (100 sperm/μl) were activated in the presence of Ca2+ for 45 min at 22°C, and nuclei were analyzed by epifluorescence microscopy to confirm their progression into S phase. GST-His6 or GST-Xenopus-survivin-His6 was added to the S-phase extracts (30 μg/200 μl of extracts), which were cycled back into mitosis by adding fresh CSF extract (20% initial sample volume) for 60 to 90 min at 22°C. The appearance of mitotic DNA in the extracts was confirmed by epifluorescence microscopy. Mitotic egg extracts mixed with recombinant proteins were diluted four times in binding buffer containing 20 mM Tris-HCl (pH 7.9), 500 mM NaCl, and 5 mM imidazole in the presence of phosphatase (50 mM NaF and 1 mM Na3VO4) and protease inhibitors (100 μM phenylmethylsulfonyl fluoride, 10 μg/ml each of leupeptin, pepstatin A, and aprotinin). Survivin-associated proteins were isolated by two-step affinity chromatography as described previously (8). Survivin protein complexes were isolated from 14 batches of 200-μl M-phase egg extracts, pooled together, and separated by 12% SDS-PAGE. Colloidal blue (Invitrogen)-stained bands in the survivin eluate, but not GST, were excised from the gel, and gel slices were washed once in distilled water, followed by two 1-ml washes in 50% acetonitrile. Liquid was removed from the gel slices with a 27-gauge needle, and samples were frozen at −80°C. Protein bands were analyzed by mass spectrometry and peptide sequence analysis (Harvard Microchemistry Facility, Harvard University, Cambridge, MA).
Spindle assembly in Xenopus egg extracts.
CSF-arrested extracts were prepared from unfertilized Xenopus eggs and used to assemble bipolar spindles as described previously (8). GST-Ran mutant proteins were incubated in CSF-arrested extracts (final concentration of 2 μM) for 60 to 90 min at 22°C. Spindles and associated chromosomes in the various samples (2 μl of extracts) were visualized in the presence of 6 μl of Hoechst fixative (60% glycerol, 0.1 mM HEPES [pH 7.5], 10% formaldehyde, and 1 μg/ml bisbenzimide) and fluorescence microscopy.
In vitro protein binding assays.
For analysis of protein interactions by pulldown experiments, GST or various GST fusion proteins (15 to 30 μg) immobilized on glutathione-Sepharose 4B beads were incubated with purified recombinant proteins (20 or 100 ng) or aliquots of HeLa cell extracts (500 μg) for 2 h at 22°C. After five washes (0.5 ml/each) in phosphate-buffered saline (PBS) (pH 7.2), bound proteins were separated by SDS-PAGE and analyzed by Western blotting. In some experiments, recombinant Ran was incubated with 2 μM purified GDP or GTP for 2 h at 22°C and mixed with increasing concentrations (100 to 200 ng) of GST or GST-survivin, and bound proteins were analyzed by Western blotting. For analysis of survivin-Ran protein interactions in Xenopus egg extracts, purified GST-tagged Ran proteins (2 μM) were added to CSF-arrested extracts, and samples were incubated for 60 min at 22°C.
Subcellular fractionations.
Cells were washed in PBS (pH 7.2), suspended in 4 to 5 volumes of cold lysis buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM EDTA, 50 mM NaF, 0.5% Nonidet P-40, plus protease inhibitors (Roche Applied Science). After 30-min incubation at 0°C, cell extracts were cleared by centrifugation at 14,000 rpm for 20 min at 4°C. Nuclear and cytosolic fractions were prepared from 1 × 108 HeLa cells using CelLytic NuCLEAR extraction kit (Sigma-Aldrich), according to the supplier's instructions. Protein content was determined using a BCA-200 protein assay kit (Pierce). TPX2 reactivity was used as marker for the nuclear fraction.
Immunoprecipitation and Western blotting.
Aliquots (800 μg) of cellular or subcellular extracts were incubated with nonbinding immunoglobulin G (IgG) or an antibody to Ran or HA prebound to protein A-Sepharose. In some experiments, HeLa cells were incubated in the presence or absence of the Crm1 inhibitor leptomycin B (LMB) (Sigma-Aldrich) (10 nM) for 2 h, harvested, and incubated with the various primary precipitating antibodies. After 4-h incubation at 4°C, the immune complexes were washed five times in lysis buffer, and proteins were separated by SDS-PAGE and analyzed by Western blotting. For these experiments, proteins (20 to 30 μg) were transferred to nitrocellulose membranes, blocked for 1 h at 22°C in 5% nonfat milk in TBST (20 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.1% Tween 20), and incubated with various primary antibodies for 16 h at 4°C. After washes, samples were incubated with horseradish peroxidase-conjugated secondary reagents for 1 h at 22°C, and protein bands were visualized by enhanced chemiluminescence (Amersham). In another series of experiments, GST or GST-Ran was incubated with extracts from untreated or LMB-treated HeLa cells, and proteins in bound or unbound fractions were analyzed by Western blotting. For a control, untreated or LMB-treated HeLa cells were analyzed for differential subcellular distribution of cyclin B1 by immunofluorescence microscopy.
siRNA transfections.
Gene silencing by small interfering RNA (siRNA) was carried out with control (VIII) or survivin-directed (S4) double-stranded RNA oligonucleotide (50 nmol) as described previously (6). In another series of experiments, WS-1 primary human fibroblasts were transfected with nontargeted (VIII) or Ran-directed siRNA GAAAUUCGGUGGACUGAGAUU (Dharmacon), characterized in recent studies (48). Subconfluent cultures were transfected using Oligofectamine (Invitrogen), harvested after 48 to 96 h at 37°C, and analyzed by Western blotting; cell cycle analysis of the cultures was performed by examining DNA content and flow cytometry or, alternatively, by fluorescence microscopy. For these experiments, cells with disorganized mitotic spindles by tubulin staining and misaligned chromosomes by 4′,6′-diamidino-2-phenylindole (DAPI) reactivity were scored as aberrant mitotic cells.
Microtubule assembly in vitro.
Transfected HeLa cells (1 × 108) were harvested by centrifugation at 5,000 rpm for 5 min at 22°C. Cells were washed in PBS (pH 7.2) and suspended in 2 ml of microtubule stabilization buffer (MSB) at 4°C after the final wash. MSB consisted of 0.1 M 2-(N-morpholino)ethane sulfonic acid, 1 mM MgSO4, and 1 mM EGTA (pH 6.6). The mixture was homogenized with 20 up-and-down strokes in a 1-ml Teflon glass homogenizer at 4°C and then immediately centrifuged at 12,000 rpm for 5 min at 4°C. The resulting supernatant was then centrifuged at 55,000 rpm for 1 h at 4°C and further incubated with 20 μM Taxol and 1 mM GTP for 30 min at 37°C (step 1). The mixture was then transferred to 4°C for 30 min in the presence of GTP (step 2), and steps 1 and 2 were repeated two or three times. Samples were loaded on 1-ml 10% sucrose cushion in MSB containing 2 μM Taxol and centrifuged at 25,000 rpm for 30 min at 30°C. The microtubule pellet was suspended in 1 ml of MSB and analyzed by Western blotting.
Fluorescence microscopy and flow cytometry.
HeLa cells under the various conditions tested were grown on optical-grade glass coverslips, synchronized by thymidine block for 16 h at 37°C, and fixed 12 h after release (corresponding to the mitotic transition, as determined by DNA content analysis and flow cytometry) in ice-cold methanol-acetone (1:1) for 5 min at 22°C. Cells were permeabilized with 0.1% Triton X-100 for 10 min, blocked in PBS containing 3% bovine serum albumin and 0.2% Tween 20 for 1 h at 22°C, and labeled with a primary antibody to α-tubulin, TPX2, or HA (1:2000) for 1 h at 22°C, followed by the addition of Alexa Fluor 594-conjugated secondary reagent (1:1,000) of appropriate specificity (Molecular Probes). Nuclei were stained with DAPI (6.5 μg/ml; Sigma) for 5 min at 22°C. After washes, cells were mounted and analyzed for nuclear morphology using a fluorescence microscope (Axioplan 2; Zeiss) equipped with a charge-coupled device camera (Axiocam; Zeiss). Quantification of TPX2 fluorescence intensity on mitotic spindles and spindle microtubule fibers of transfected cells was carried out using images imported in Adobe Photoshop v.7 and analyzed using ImageJ software. An average of 120 mitotic cells was counted per condition. Spindles in Xenopus extracts were captured with a Roper CoolSNAP HQ charge-coupled device camera mounted to a Nikon E800 fluorescence microscope and analyzed using Metamorph software v5.0r1 (Universal Imaging Corp.).
Statistical analysis.
Data were analyzed using the unpaired t test on a GraphPad Prism 4.0 software package for Windows (GraphPad Software). A P value of 0.05 was considered statistically significant.
RESULTS
Proteomics screening for novel survivin-associated molecules in tumor cells.
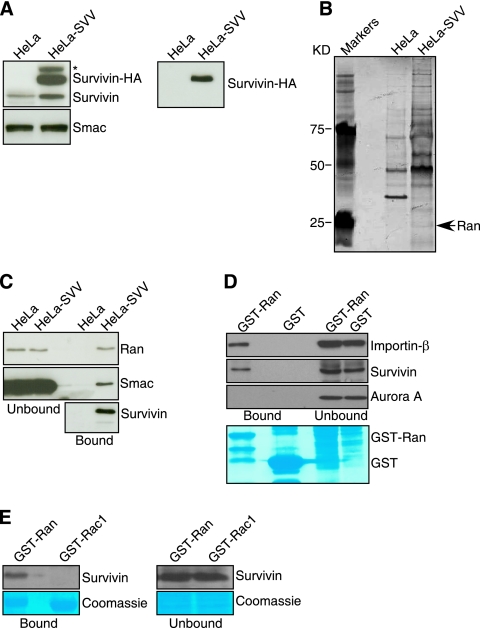
To identify novel proteins that associate with survivin in the regulation of cell division (1, 29), we stably transfected a carboxyl-terminal HA-tagged wild-type (WT) survivin cDNA in HeLa cells and selected survivin-expressing clones using a surface marker by flow cytometry. One cell line, designated HeLa-SVV, expressed endogenous survivin, as well as HA-tagged survivin, as determined by Western blotting, whereas parental HeLa cells expressed only endogenous survivin (Fig. 1A). Next, we affinity purified survivin-HA complexes from HeLa-SVV cells and identified eluted proteins by mass spectrometry and matrix-assisted laser desorption ionization-time-of-flight analysis. Several protein bands were enriched in HeLa SVV eluates compared with parental HeLa cells, and a silver-stained band of ∼26 kDa (Fig. 1B) was identified as the small GTPase, Ran (National Center for Biotechnology Information Protein Database accession no. AAC99400) on the basis of the sequences of the following peptides: SIVFHR, NVPNWHR, LVLVGDGGTGK, and NLQYYDISAK (16% protein coverage). Accordingly, eluates from HeLa-SVV extracts contained Ran, survivin, as well as Smac (Fig. 1C), a molecule known to associate with survivin (42) by Western blotting. In pulldown experiments, recombinant GST-Ran strongly associated with importin-β in HeLa cell extracts, but not Aurora A (Fig. 1D), in agreement with previous observations (9, 15). GST-Ran also formed a complex with survivin in HeLa cell extracts, whereas GST alone was ineffective (Fig. 1D). Under conditions in which a survivin-Ran complex was readily detected in HeLa cell extracts, a related small GTPase, Rac1, did not associate with survivin in pulldown experiments in vivo (Fig. 1E).
FIG. 1.
Proteomics identification of Ran as a survivin-associated molecule in tumor cells. (A) Characterization of HeLa-SVV. Parental cultures or HeLa cells stably transfected with Survivin-HA (HeLa-SVV) were analyzed by Western blotting with an antibody to survivin (left panels) or HA (right panel). Reactivity for Smac was used as a loading control. The position of nonspecific protein bands is indicated by an asterisk. (B) Identification of Ran as a survivin-associated molecule. A ∼26-kDa silver-stained band eluted from survivin-HA beads (arrowhead) was identified as Ran by mass spectrometry. The positions of molecular mass markers are indicated to the left of the gel. (C) Western blotting. Proteins eluted from survivin-HA beads (bound) or supernatant (unbound) were analyzed by Western blotting. (D) Pulldown analysis. GST or GST-Ran was incubated with HeLa cell extracts, and proteins in pellets (bound) or supernatants (unbound) were analyzed by Western blotting. The bottom panel depicts Coomassie blue staining of incubation reaction mixtures. The positions of GST and GST-Ran are indicated. (E) Specificity of a Ran-survivin complex. GST-Ran or GST-Rac1 was mixed with HeLa cell extracts, and proteins in the bound or unbound fractions were analyzed by Western blotting. The bottom panels depict Coomassie blue staining of incubation reaction mixtures.
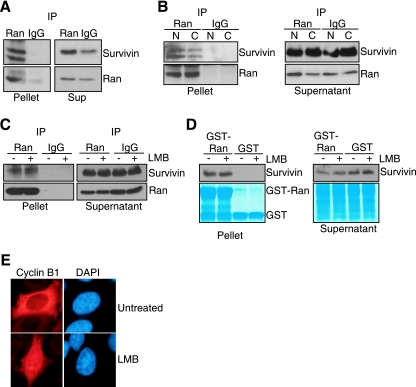
To determine whether a survivin-Ran complex occurred in vivo, we next carried out immunoprecipitation experiments of endogenous proteins. Ran immune complexes precipitated from HeLa cell extracts contained associated survivin, whereas control IgG immunoprecipitates were negative (Fig. 2A). In immunoprecipitation experiments from isolated subcellular fractions, endogenous Ran formed complexes with survivin in both nuclear and cytosol extracts (Fig. 2B). In contrast, IgG immunoprecipitates did not contain survivin in the various subcellular fractions (Fig. 2B). Next, we carried out two experiments to test whether survivin formed a complex with Ran in vivo independently of Crm1, a known Ran effector that interacts with survivin in nucleocytoplasmic shuttling (24). Treatment of HeLa cells with the Crm1 inhibitor LMB did not affect the association of endogenous Ran with endogenous survivin as determined by immunoprecipitation and Western blotting (Fig. 2C). In contrast, control samples immunoprecipitated with nonimmune IgG did not contain Ran or survivin in the presence or absence of LMB (Fig. 2C). Similarly, in pulldown assays in vivo, GST-Ran indistinguishably associated with endogenous survivin in HeLa cell extracts in the presence or absence of LMB, whereas GST was unreactive (Fig. 2D). Confirming its Crm1 inhibitory activity, LMB treatment resulted in prominent nuclear accumulation of cyclin B1 in asynchronous HeLa cells as seen by immunofluorescence microscopy (Fig. 2E).
FIG. 2.
Survivin-Ran interaction in vivo. (A) Immunoprecipitation. HeLa cell extracts were immunoprecipitated (IP) with an antibody to Ran or IgG, and proteins in pellets or supernatants (Sup) were analyzed by Western blotting. (B) Survivin-Ran complexes in isolated subcellular fractions. Nuclear (N) or cytosolic (C) extracts from HeLa cells were immunoprecipitated (IP) with an antibody to Ran or IgG, and proteins in pellets or supernatants were analyzed by Western blotting. (C) Leptomycin B treatment. HeLa cells were incubated in the presence (+) or absence (−) of LMB, immunoprecipitated (IP) with an antibody to Ran or IgG, and proteins in pellets or supernatants were analyzed by Western blotting. (D) In vivo pulldown analysis. GST or GST-Ran was incubated with HeLa cell extracts treated with LMB (+) or not treated with LMB (−), and proteins in pellets or supernatants were analyzed by Western blotting. The bottom panels depict Coomassie blue staining of GST or GST-Ran recombinant proteins. (E) Effect of LMB on nucleocytoplasmic shuttling of cyclin B1. HeLa cells were treated with LMB or left untreated and analyzed by fluorescence microscopy. DNA was stained with DAPI.
Cell cycle requirements of a survivin-Ran complex.
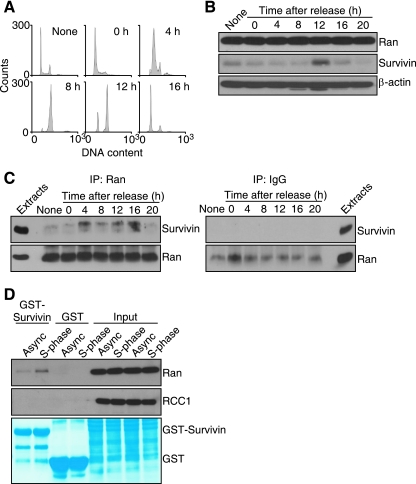
Because survivin expression is regulated by the cell cycle and peaks at mitosis, we next asked whether a survivin-Ran complex assembled in specific cell cycle phases in vivo. For these experiments, we synchronized HeLa cells at the G1/S boundary and harvested aliquots of the culture at various time intervals after thymidine release (Fig. 3A). Present at low levels in interphase cells, survivin expression peaked abruptly at mitosis, 12 h after thymidine release, and was again downregulated with the next G1 phase, 16 to 20 h after release (Fig. 3B), in agreement with previous observations (1). In contrast, Ran expression remained constant throughout the cell cycle in synchronized HeLa cells (Fig. 3B). Ran immunoprecipitated from synchronized cultures exhibited a biphasic interaction with survivin, first at 4 h after thymidine release, thus corresponding to S phase (Fig. 3A), and again at mitosis, 10 to 12 h after release (Fig. 3C, left panel). In contrast, control samples immunoprecipitated with nonimmune IgG did not contain survivin at any cell cycle phase (Fig. 3C, right panel). Consistent with this, GST-survivin associated with endogenous Ran, but not RCC1, in asynchronous HeLa cell extracts, and this interaction was increased in S phase 4 h after thymidine release (Fig. 3D). In contrast, GST alone did not associate with Ran or RCC1 in asynchronous or S-phase extracts (Fig. 3D).
FIG. 3.
Cell cycle-regulated survivin-Ran interaction. (A) DNA content. HeLa cells were synchronized by thymidine block, released in fresh medium, and analyzed for DNA content at the indicated times (in hours) by propidium iodide staining and flow cytometry. (B) Cell cycle-dependent expression of survivin. Synchronized HeLa cell extracts were harvested at the indicated times after thymidine release and analyzed by Western blotting. Asynchronous cells are shown in the lane labeled None. (C) Cell cycle regulation of survivin-Ran interaction. Synchronized HeLa cell extracts harvested at the indicated times (in hours) after thymidine release were immunoprecipitated (IP) with an antibody to Ran (left panel) or with IgG (right panel), and associated proteins were analyzed by Western blotting. Asynchronous cells are shown in the lanes labeled None. (D) In vivo pulldown analysis. HeLa cell extracts from asynchronous (Async) cell cultures or synchronized cultures in S phase (4 h after thymidine release) were mixed with GST or GST-survivin, and bound proteins were analyzed by Western blotting. The bottom panel depicts Coomassie blue staining of incubation reaction mixtures. The positions of GST and GST-Ran are indicated.
Conservation of a survivin-Ran interaction in Xenopus laevis eggs.
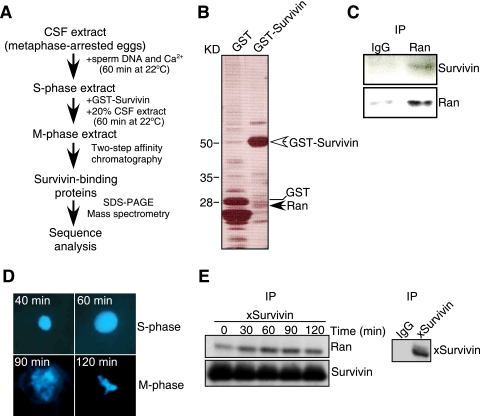
To determine whether survivin-Ran recognition had been conserved evolutionarily, we next carried out a proteomics screening in Xenopus laevis egg extracts (Fig. 4A), a model extensively used to study mitotic functions. For these experiments, we expressed Xenopus survivin fused to GST and His6, mixed it with metaphase-arrested Xenopus egg extracts, and analyzed protein complexes by sequential affinity chromatography and mass spectrometry (Fig. 4B). Using this protocol, Xenopus Ran was identified (22% protein coverage based on peptide sequences) as one of the molecules binding to GST-survivin, but not GST (Fig. 4B). Accordingly, an antibody to Ran precipitated survivin from Xenopus egg extracts, whereas a control IgG was ineffective (Fig. 4C). To determine whether a survivin-Ran complex in Xenopus egg extracts also assembled in various cell cycle phases, we harvested aliquots of cycled extracts during S and M phases, as determined by fluorescence microscopy of Hoechst-stained chromatin (Fig. 4D). Similar to the results obtained in mammalian cells, Xenopus survivin immunoprecipitated from cycled egg extracts was found in complex with Ran in both the S and M phases of the cell cycle (Fig. 4E). In contrast, IgG immune complexes did not associate with Ran (Fig. 4E, right panel).
FIG. 4.
Proteomics screening for survivin-associated molecules in Xenopus laevis. (A) Protocol for identification of survivin-associated proteins in cycled Xenopus egg extracts. CSF, cytostatic factor. (B) Survivin-associated molecules in Xenopus egg extracts. GST-Xenopus-survivin-His6 or GST-His6 was mixed with cycled Xenopus egg extracts, and protein complexes were analyzed by silver staining. The positions of GST, GST-survivin, and Ran are indicated to the right of the gel. The positions of molecular mass markers are indicated to the left of the gel. (C) Survivin-Ran complexes in cycled Xenopus extracts. Protein complexes immunoprecipitated (IP) from CSF extracts with an antibody to Ran or IgG were analyzed by Western blotting. (D) Synchronized Xenopus egg extracts. Aliquots of cycling Xenopus extracts were harvested at the indicated times representative of S or M phase and stained with Hoechst (left panels). (E) Cell cycle regulation of a survivin-Ran complex in Xenopus egg extracts. Cycled Xenopus extracts harvested at the indicated time intervals were immunoprecipitated (IP) with an antibody to Xenopus survivin (xSurvivin), and bound proteins were analyzed by Western blotting. Control immunoprecipitation (IP) reactions were carried out with nonbinding IgG (right panel).
Nucleotide requirement of a survivin-Ran complex.
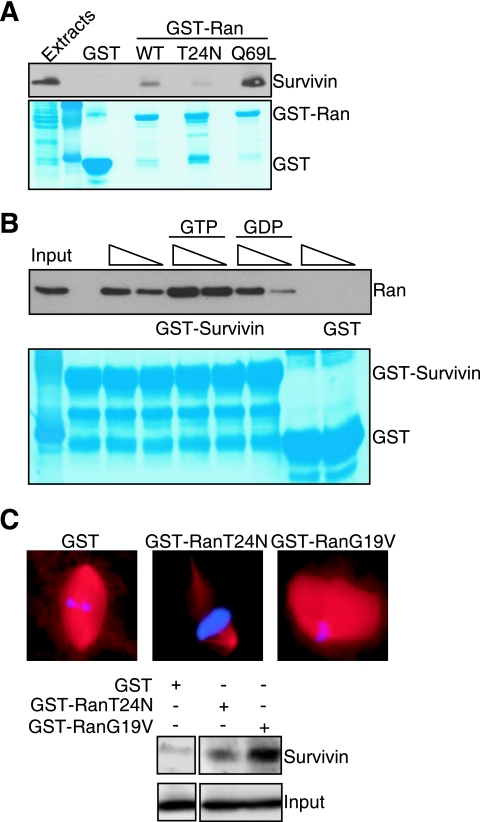
The association of Ran with exportin/importin-bound cargos is regulated by its GDP- or GTP-bound state. To test the nucleotide requirements of a survivin-Ran interaction, we first analyzed two well-characterized Ran mutants for their ability to associate with survivin. For these experiments, we mixed recombinant WT or mutant Ran proteins fused to GST with HeLa cell extracts to allow nucleotide loading and then analyzed their binding to recombinant survivin. WT Ran associated with recombinant survivin, but not GST (Fig. 5A), consistent with the data reported above. A constitutively active Ran Q69L mutant, which is unable to hydrolyze GTP, bound considerably more strongly to survivin than WT Ran did (Fig. 5A). In contrast, a Ran T24N mutant, which has low binding affinity for GTP and GDP, nearly completely lost the ability to bind survivin (Fig. 5A). In a second series of experiments, we loaded recombinant Ran with GDP or GTP in vitro, mixed the reactions with GST or GST-survivin, and analyzed bound proteins. As shown in Fig. 5B, GST-survivin bound GTP-loaded Ran with considerably higher efficiency than GDP-Ran did and in a concentration-dependent manner (Fig. 5B). In contrast, GST did not bind Ran in the presence of GDP or GTP (Fig. 5B). Similar results were obtained in Xenopus extracts. A loss-of-function Ran T24N mutant failed to promote spindle assembly in Xenopus extracts (Fig. 5C, top panels), in agreement with previous observations (36), and weakly associated with survivin (Fig. 5C, bottom panels). Conversely, a constitutively active Ran G19V mutant promoted exaggerated spindle formation in Xenopus extracts (Fig. 5B, top panel), and bound strongly to survivin (Fig. 5C, bottom panels).
FIG. 5.
Survivin recognition of Ran-GTP. (A) In vivo pulldown analysis. WT Ran fused to GST (GST-Ran) or the indicated Ran mutants fused to GST were mixed with HeLa cell extracts, and bound proteins were analyzed by Western blotting. The bottom panel depicts Coomassie blue staining of the incubation reaction mixtures. The positions of GST and GST-Ran are indicated to the right of the gel. (B) Nucleotide requirement of survivin-Ran interaction. Recombinant Ran was incubated with 2 μM GDP or GTP, mixed with increasing concentrations (100 to 200 ng [indicated by the height of the triangle above the gel]) of GST or GST-survivin, and bound proteins were analyzed by Western blotting. The bottom panel depicts Coomassie blue staining of incubation reaction mixtures. The positions of GST and GST-survivin are indicated to the right of the gel. (C) Xenopus extract pulldown analysis. Cycled Xenopus extracts were incubated (+) with GST or the indicated GST-Ran mutants and analyzed for spindle formation by fluorescence microscopy (top panels) or differential binding to survivin (bottom panels). Western blots with comparable amounts of added GST or GST-Ran proteins are labeled Input and are shown at the bottom.
Structure and function of a survivin-Ran complex.
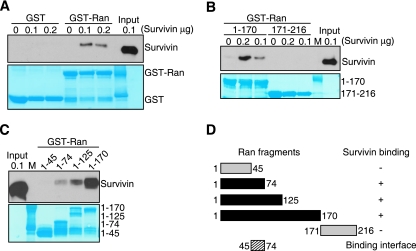
To elucidate whether survivin interacted with Ran directly or as a part of a complex with other Ran-GTP effector molecules, we next carried out binding studies with recombinant proteins in vitro. GST-Ran strongly associated with isolated survivin, whereas GST alone was ineffective (Fig. 6A). This interaction required the amino-terminal region of Ran, because a fragment comprising residues 1 to 170, but not residues 171 to 216 in Ran, bound survivin in a dose-dependent reaction in vitro (Fig. 6B). In addition, fragments spanning Ran residues 1 to 74 and 1 to 125 also bound survivin, whereas the most amino-terminal residues 1 to 45 were ineffective (Fig. 6C). Therefore, amino acids 45 to 74 in Ran contain a binding site for survivin (Fig. 6D).
FIG. 6.
Identification of a survivin binding site in Ran. (A to C) In vitro pulldown analysis. GST or various GST-Ran proteins were incubated with recombinant survivin (0, 0.1, or 0.2 μg), and bound complexes were analyzed by Western blotting. Various fragments of GST-Ran proteins were used in panels B and C (fragments of GST-Ran proteins from residues 1 to 170 and 171 to 216 were used in panel B, and fragments from residues 1 to 45, 1 to 74, 1 to 125, and 1 to 170 were used in panel C). Lanes M, molecular weight markers. The bottom panels depict Coomassie blue staining of GST or GST-Ran recombinant proteins. (D) Ran binding site. A summary of Ran fragments analyzed for differential binding to survivin is shown. The ability of the Ran fragments to bind to survivin are shown to the right as follows: +, bound to survivin; −, did not bind to survivin.
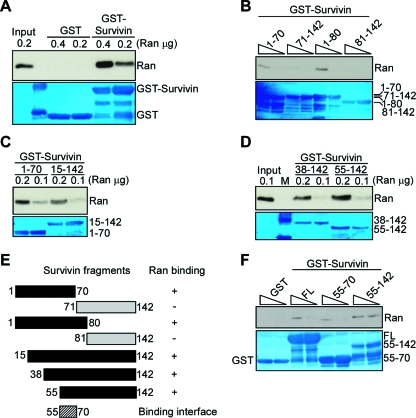
We next used a similar approach to map a Ran binding site in human survivin in vitro. GST-survivin bound Ran in a concentration-dependent manner, whereas GST was unreactive (Fig. 7A). In addition, fragments of survivin containing amino acids 1 to 70, 1 to 80 (Fig. 7B), 15 to 142 (Fig. 7C), 38 to 142, and 55 to 142 (Fig. 7D), also associated with Ran. Conversely, fragments duplicating the COOH terminal α-helix of survivin between amino acids 71 to 142 and 81 to 142, which have been implicated in electrostatic interactions with chromosomal passenger proteins Borealin and INCENP (18), did not bind Ran (Fig. 7B). Therefore, residues 55 to 70 in survivin comprise a unique binding site for Ran (Fig. 7E). Consistent with this prediction, a truncated survivin mutant spanning amino acids 55 to 70 retained the ability to bind Ran indistinguishably from WT survivin or a survivin mutant containing amino acids 55 to 142, whereas GST was ineffective (Fig. 7F).
FIG. 7.
Identification of a Ran binding site in survivin. (A to D) In vitro pulldown analysis. GST or various GST-survivin proteins were incubated with recombinant Ran (0.1, 0.2, or 0.4 μg), and bound complexes were analyzed by Western blotting. Various fragments of GST-survivin proteins were used in panels B, C, and D. Fragments of GST-survivin proteins from residues 1 to 70, 71 to 142, 1 to 80, and 81 to 142 were used in panel B. The amount (0.4 or 0.2 μg) of GST-survivin protein in the lanes is indicated by the height of the triangle above the gel in panel B. Lane M, molecular weight markers. The bottom panels in panels A to D depict Coomassie blue staining of GST or various GST-survivin recombinant proteins. (E) Survivin binding site. A summary of survivin fragments differentially associating with Ran is shown. The ability of the survivin fragments to bind to Ran are shown to the right as follows: +, bound to Ran; −, did not bind to Ran. (F) Survivin A55-D70-Ran interaction. GST or various GST-survivin constructs (0.1 to 0.2 μg [indicated by the height of the triangle above the gel]) were incubated with recombinant Ran, and bound complexes were analyzed by Western blotting. The bottom panel in panel F depicts Coomassie blue staining of various GST-survivin recombinant proteins, including the full-length (FL) protein.
Regulation of TPX2 recognition by a survivin-Ran complex.
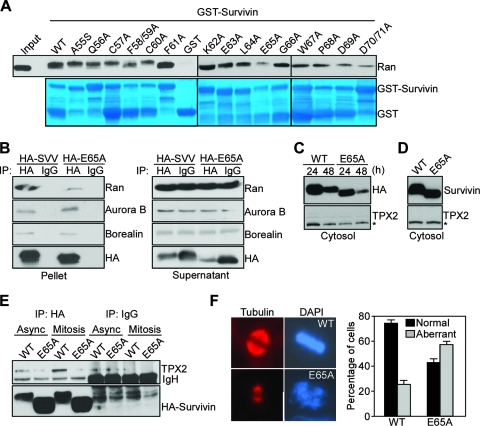
To begin testing a role of a survivin-Ran complex in tumor cells, we first mapped the structural requirements of this interaction in more detail. For these experiments, we mutated to Ala every residue in the survivin sequence containing amino acids 55 to 70 (Fig. 7E), expressed the individual point mutants as GST fusion proteins, and tested their ability to bind Ran in vitro (Fig. 8A). Only a survivin Glu65Ala (E65A) mutant, thus outside of a putative nuclear export sequence in survivin (24), exhibited nearly complete loss of Ran binding in vitro (Fig. 8A). In contrast, Ala mutagenesis of all other amino acids in the survivin region from amino acids 55 to 70 did not significantly affect Ran binding in vitro (Fig. 8A). We next asked whether mutation of Glu65 in survivin affected the recognition of other mitotic regulator(s), in particular the chromosomal passenger proteins (18). For these experiments, we transfected asynchronous HeLa cells with HA-WT survivin or HA-E65A survivin mutant and immunoprecipitated aliquots of these cell extracts with IgG or an antibody to HA. WT survivin immune complexes contained Ran as determined by Western blotting (Fig. 8B), whereas immunoprecipitates of survivin E65A mutant were largely devoid of Ran (Fig. 8B), in agreement with the data presented above. Under these experimental conditions, both WT survivin and survivin E65A mutant associated with chromosomal passenger proteins Aurora B and Borealin to comparable levels in vivo (Fig. 8B). In control experiments, IgG immune complexes did not contain associated molecules, and Western blotting with an antibody to HA confirmed comparable levels of immunoprecipitation of WT or mutant survivin (Fig. 8B).
FIG. 8.
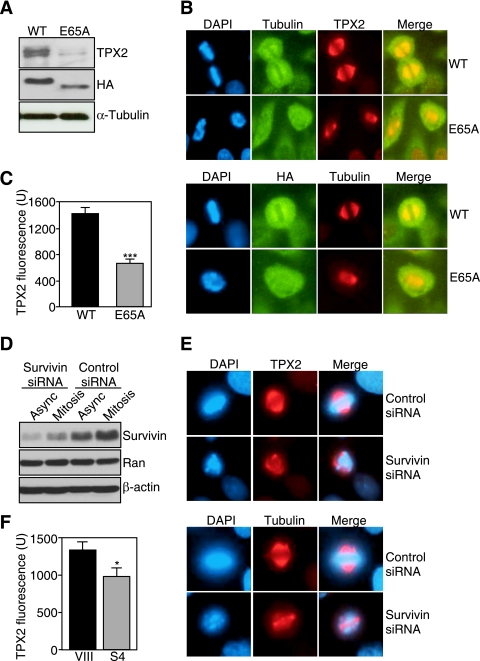
Molecular targeting of a survivin-Ran complex results in aberrant mitotic spindles. (A) Ala-scanning mutagenesis. Each residue in the survivin sequence from A55 to D70 was mutated to Ala, and cDNA constructs encoding the various indicated mutants or wild-type survivin were expressed as GST fusion proteins and incubated with recombinant Ran, and bound proteins were analyzed by Western blotting. Input, 0.2 μg. The bottom panel depicts Coomassie blue staining of GST or GST-survivin recombinant proteins. (B) Immunoprecipitation. Asynchronous HeLa cells were transfected with HA-WT survivin (HA-SVV) or HA-survivin E65A mutant (HA-E65A), immunoprecipitated (IP) with IgG or with an antibody to HA, and proteins in pellets or supernatants were analyzed by Western blotting. (C and D) Cytosolic accumulation. HeLa (C) or NIH 3T3 (D) cells were transfected with the indicated HA-tagged (C) or untagged (D) cDNAs, harvested after 24 or 48 h, and analyzed by Western blotting. The position of nonspecific protein bands is indicated by an asterisk. (E) Survivin-TPX2 interaction. HeLa cells transfected with WT HA-survivin or HA-E65A survivin mutant were maintained asynchronously (Async) or were synchronized at mitosis, immunoprecipitated (IP) with an antibody to HA or with IgG, and the immune complexes were analyzed by Western blotting. IgH, immunoglobulin heavy chain. (F) Spindle defects. Synchronized HeLa cells transfected with WT survivin or survivin E65A mutant were harvested at mitosis and labeled for tubulin or DNA (DAPI) (left panels). Spindle defects were quantified by fluorescence microscopy in 10 high-power fields in the bar graph shown at the right. The means plus standard error of the means (error bars) are shown in the graph.
Because survivin has been reported to interact with the Ran target Crm1 in a pathway required for nucleocytoplasmic shuttling (24), we also tested whether mutation of Glu65 in survivin affected its nuclear export. HA-WT survivin and HA-E65A survivin mutant transfected in HeLa cells exhibited comparable levels of accumulation in isolated cytosolic extracts as determined by Western blotting (Fig. 8C). A potential source of artifacts in survivin overexpression studies is the addition of “tag” moieties, which may affect subcellular trafficking and/or inhibit binding to regulatory molecules (21). To rule out this possibility, we transfected untagged WT survivin or survivin E65A mutant in NIH 3T3 cells, which express very low levels of endogenous survivin, and analyzed isolated subcellular fractions. Similar to the data presented above, both WT and survivin E65A mutant accumulated at comparable levels in cytosolic extracts of NIH 3T3 fibroblasts (Fig. 8D).
Regulation of mitotic spindle assembly by a survivin-Ran complex.
Next, we asked whether a survivin-Ran complex was required to localize Ran effector molecules during mitosis, and we focused on TPX2 for its role in spindle formation. In immunoprecipitation experiments from HeLa cell extracts, immune complexes of WT survivin contained TPX2 in vivo (Fig. 8E). This interaction was readily detected in asynchronous culture, and significantly increased in cells synchronized at mitosis (Fig. 8E). In contrast, immune complexes of survivin E65A mutant, which has lost the ability to bind Ran, did not contain TPX2 in asynchronous or mitotic cell extracts, and IgG immune complexes were unreactive (Fig. 8E). When analyzed by fluorescence microscopy, HeLa cells expressing survivin E65A mutant exhibited multiple mitotic defects, characterized by short, flattened mitotic spindles and grossly abnormal chromosome congression (Fig. 8F).
To determine whether this phenotype was due to mislocalization of TPX2 from the mitotic apparatus, we assembled microtubules from HeLa cells transfected with WT survivin or survivin E65A mutant in vitro and looked for potential differences in TPX2 recruitment. Both WT survivin and survivin E65A mutant associated with polymerized microtubules to comparable levels as determined by Western blotting with an antibody to HA (Fig. 9A). However, while polymerized microtubules from cells expressing WT survivin contained TPX2, transfection of survivin E65A mutant in HeLa cells almost completely abolished the recruitment of TPX2 to microtubules (Fig. 9A). In control experiments, comparable amounts of tubulin were assembled in cells transfected with WT or survivin E65A mutant (Fig. 9A), thus ruling out the possibility that modulation of TPX2 reactivity was due to a decrease in microtubule content. By fluorescence microscopy, synchronized HeLa cells transfected with WT survivin and harvested at mitosis exhibited bipolar mitotic spindles with a full complement of microtubules, proper chromosome congression, and strong reactivity for TPX2 at spindle poles and metaphase spindle fibers (Fig. 9B), in agreement with previous observations (15). In contrast, synchronized mitotic cells expressing survivin E65A mutant exhibited grossly abnormal, flattened mitotic spindles, with considerably attenuated TPX2 reactivity at spindle poles and microtubule fibers (Fig. 9B and C).
FIG. 9.
Survivin-regulated delivery of TPX2 to microtubules. (A) Requirement of survivin for TPX2 delivery to microtubules. Polymerized microtubules were assembled in vitro from HeLa cell extracts transfected with WT survivin or survivin E65A mutant and analyzed by Western blotting. (B) Depletion of TPX2 from mitotic spindles. Synchronized HeLa cells transfected with WT survivin or survivin E65A mutant were harvested at mitosis, stained with the indicated antibodies, and analyzed by fluorescence microscopy. DNA was labeled with DAPI. The merged images are shown in the panels labeled Merge. (C) Quantification of TPX2 fluorescence intensity in transfected cells. The two values were significantly different (P < 0.0001) (n = 120 mitotic cells) as indicated by the three asterisks. (D) Survivin knockdown analysis. Asynchronous (Async) HeLa cells or HeLa cells synchronized at mitosis were transfected with nontargeted (Control) or survivin-directed siRNA and analyzed by Western blotting. (E) TPX2 expression in survivin-depleted cells. Synchronized mitotic HeLa cells transfected with control or survivin-directed siRNA were stained with the indicated antibodies and analyzed by fluorescence microscopy. DNA was stained with DAPI. The merged images are shown in the panels labeled Merge. (F) Quantification of TPX2 fluorescence intensity in transfected cells. The two values were significantly different (P < 0.037) (n = 11 to 14 mitotic cells) as indicated by the asterisk.
Next, we asked whether acute ablation of survivin by siRNA reproduced the defect in microtubule localization of TPX2 observed after expression of survivin E65A mutant. Transfection of a survivin-directed double-stranded RNA oligonucleotide (S4) characterized in previous studies (6) resulted in significant reduction of survivin levels in both asynchronous cultures and HeLa cells synchronized at mitosis compared with nontargeted siRNA (Fig. 9D). In contrast, siRNA targeting of survivin did not affect Ran levels in asynchronous or mitotic HeLa cells (Fig. 9D). Transfection of HeLa cells with control siRNA did not influence the expression or localization of TPX2 to the mitotic apparatus as examined by fluorescence microscopy (Fig. 9E). In contrast, even a partial reduction of survivin levels by siRNA (Fig. 9D) was sufficient to cause severe spindle defects, with reduced expression of TPX2 on spindle poles and metaphase spindle fibers (Fig. 9E and F).
Selectivity of a survivin-Ran functional complex in tumor cells.
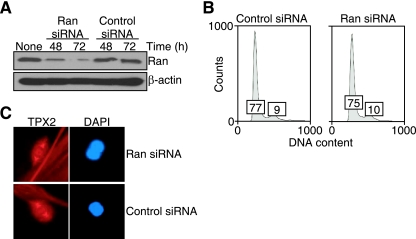
To test whether a survivin-Ran complex in the regulation of spindle formation was preferentially operative in tumor cells, we acutely ablated Ran expression in WS-1 primary human fibroblasts. Transfection of these cells with a Ran-directed siRNA resulted in significant suppression of endogenous Ran expression 48 and 72 h after treatment, whereas a control, nontargeted siRNA was without effect (Fig. 10A), in agreement with recent observations (48). Under these experimental conditions, Ran knockdown in WS-1 fibroblasts was not associated with cell cycle defects compared with control transfectants as ascertained by DNA content analysis and flow cytometry (Fig. 10B). At variance with the results obtained with tumor cell types, normal human fibroblasts acutely silenced for Ran expression did not exhibit formation of aberrant mitotic spindles or changes in the expression of TPX2 on the mitotic apparatus as seen by fluorescence microscopy (Fig. 10C), consistent with a selective role of a Ran-survivin pathway in tumor cell mitosis (48).
FIG. 10.
Effect of Ran targeting on normal cells. (A) Ran knockdown analysis. WS-1 primary human fibroblasts were transfected with control nontargeted or Ran-directed siRNA, harvested at the indicated times, and analyzed by Western blotting. Asynchronous cells are shown in the lane labeled None. (B) Cell cycle analysis. WS-1 fibroblasts transfected with control or Ran-directed siRNA were analyzed after 72 h for DNA content by propidium iodide staining and flow cytometry. The percentages of cells in the G1 and G2/M fractions are indicated in the first and second boxes, respectively, in each graph. (C) Fluorescence microscopy. Mitotic WS-1 fibroblasts transfected with control or Ran-directed siRNA were analyzed for expression of TPX2 or DNA (DAPI) by fluorescence microscopy.
DISCUSSION
In this study, we have shown that survivin associates with the small GTPase Ran (36) and that this interaction is required for mitotic spindle formation selectively in tumor cells. Differently from a canonical model of Ran-GTP signaling centered on the release of spindle assembly factors from importin α/β inhibition (14), a survivin-Ran complex serves as a physical scaffold to help deliver the Ran effector molecule TPX2 (36) to microtubules. Relevant features of the survivin-Ran recognition include evolutionary conservation in Xenopus laevis extracts, biphasic cell cycle requirements in S phase and mitosis, independence from the Ran target (Crm1), preferential recruitment of Ran-GTP, and a discrete binding interface centered on Glu65 in survivin, which is separate from the chromosomal passenger complex-interacting domain.
Although survivin is essential for mitosis (32) and this pathway has been highly conserved during evolution (28), its molecular requirements have long remained elusive. In particular, it has been difficult to reconcile the multiple subcellular localizations of mitotic survivin with a single, unifying mechanism for its role at cell division (1, 29). Conversely, the data presented here position survivin in an evolutionary conserved pathway of mitotic spindle formation. Accordingly, a survivin-Ran complex assembled on microtubules may help increase the local concentration of the “spindle assembly factor” TPX2 (15) on the mitotic apparatus, thus stabilizing microtubules and enhancing spindle formation. This is in agreement with previous observations of a tight interaction of TPX2 with polymerized microtubules (47), and prior evidence linking survivin to increased microtubule stability (13), direct suppression of microtubule dynamics in vivo (37), and an evolutionary conserved role in spindle formation in cycled Xenopus extracts (8). The participation of survivin in spindle assembly is also consistent with its function as a chromosomal passenger protein (29). We now know that not only do these molecules participate in central spindle positioning and cytokinesis but they also contribute to a Ran-independent mechanism of chromatin-associated spindle formation (38). As an indispensable regulator of the core chromosomal passenger complex, responsible for its subcellular localization and biochemical assembly (18), survivin has been shown to enhance the catalytic activity of Aurora B kinase (7), resulting in inhibitory phosphorylation of the microtubule-depolymerizing kinesin MCAK (44), and increased microtubule growth around mitotic chromosomes.
The data presented here also fit well with an expanding involvement of survivin in Ran-GTP signaling. Recent studies have shown that survivin associated with the Ran effector Crm1 and that this interaction contributed to nucleocytoplasmic shuttling of survivin for proper kinetochore targeting of the chromosomal passenger complex (24) and modulation of antiapoptotic survivin levels in the cytosol (25). At variance from this paradigm, we have shown here that survivin interacts with Ran directly in a reaction insensitive to the Crm1 inhibitor LMB, and domain mapping of this recognition using isolated recombinant proteins identified Glu65 as critically required for Ran-GTP binding. This region is physically distinct from an “atypical” nuclear export sequence that has been proposed to span residues 89 to 98 in survivin and has been implicated in Crm1 binding (25), as well as recognition of chromosomal passenger proteins INCENP and Borealin (18). Accordingly, a survivin E65A mutant, which failed to associate with Ran in vitro and in vivo, retained the ability to bind the chromosomal passenger proteins (18) Aurora B and Borealin and the ability to accumulate in the cytosol (24), indistinguishably from WT survivin.
Although Ran signaling provides an essential mechanism of spindle formation in many organisms (46), it has long been appreciated that some of its critical downstream factors, for instance TPX2 and NuMa, have not been conserved during evolution (17). Whether this reflects a cell- and/or context-specific mechanism by which diverse Ran effector molecules are differentially recruited for spindle formation is currently not known, but it is intriguing that many Ran targets in mammalian cells also function as cancer genes, differentially expressed or deregulated in tumors, compared with normal tissues (40). These include an oncogenic kinase with transforming potential, Aurora A (12), a microtubule-associated protein overexpressed in certain cancers, HURP (26, 41), a DNA repair/checkpoint protein complex, BRCA1/BARD1 with critical roles in genomic integrity (19), and now survivin (this study), a general regulator of tumor cell viability and proliferation (1, 29). Here, acute silencing of Ran in primary human fibroblasts was surprisingly well tolerated and did not result in cell cycle defects or abnormalities of mitotic spindle formation, in agreement with recent observations (48). Collectively, these data support a model that a Ran-survivin complex may be preferentially, if not exclusively, exploited in tumor cells, as opposed to normal cells. It remains to be seen whether such differential, “tumor-selective” utilization of oncoproteins in the Ran pathway contributes to tumorigenesis in vivo. However, the results of initial gene profiling experiments with ovarian cancer (33) and experimental breast cancer (34) seem consistent with this possibility, linking increased Ran expression to unfavorable disease outcome. Mechanistically, deregulated Ran signaling may favor increased propensity to aneuploidy (35), in agreement with Ran playing a role in chromosome positioning (17), loading of spindle checkpoint proteins (5), and organization of kinetochore fibers (4). Consistent with this, the Ran targets Aurora A and TPX2 were identified in a 70-gene signature predictive of chromosomal instability and poor outcome in various cancers (10), and expression of survivin has almost invariably been linked to aggressive tumor behavior, metastatic disease, and worse prognosis (2). In this context, exploitation of survivin as a novel effector of Ran signaling may be ideally suited to further deregulate spindle assembly in tumor cells, obliterating mitotic checkpoint(s) (11, 30), promoting aneuploidy, and antagonizing apoptosis (1) in response to cell division defects, or antimitotic therapies (49).
In summary, we have identified a novel, evolutionary conserved mechanism of Ran-GTP signaling (14) centered on a physical association with survivin and contributing to spindle formation selectively in tumor cells. For the differential expression of survivin and other Ran targets in cancer, as opposed to normal tissues (40), molecular or pharmacologic antagonists of this pathway may be envisioned as potential therapeutic approaches to selectively disrupt tumor cell mitosis.
Acknowledgments
This work was supported by Public Health Service grants HL54131, CA78810, and CA90917 (D.C.A.) and GM62542 (T.M.G.).
Footnotes
Published ahead of print on 30 June 2008.
REFERENCES
- 1.Altieri, D. C. 2006. The case for survivin as a regulator of microtubule dynamics and cell-death decisions. Curr. Opin. Cell Biol. 18609-615. [DOI] [PubMed] [Google Scholar]
- 2.Altieri, D. C. 2003. Validating survivin as a cancer therapeutic target. Nat. Rev. Cancer 346-54. [DOI] [PubMed] [Google Scholar]
- 3.Ambrosini, G., C. Adida, and D. C. Altieri. 1997. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat. Med. 3917-921. [DOI] [PubMed] [Google Scholar]
- 4.Arnaoutov, A., Y. Azuma, K. Ribbeck, J. Joseph, Y. Boyarchuk, T. Karpova, J. McNally, and M. Dasso. 2005. Crm1 is a mitotic effector of Ran-GTP in somatic cells. Nat. Cell Biol. 7626-632. [DOI] [PubMed] [Google Scholar]
- 5.Arnaoutov, A., and M. Dasso. 2003. The Ran GTPase regulates kinetochore function. Dev. Cell 599-111. [DOI] [PubMed] [Google Scholar]
- 6.Beltrami, E., J. Plescia, J. C. Wilkinson, C. S. Duckett, and D. C. Altieri. 2004. Acute ablation of survivin uncovers p53-dependent mitotic checkpoint functions and control of mitochondrial apoptosis. J. Biol. Chem. 2792077-2084. [DOI] [PubMed] [Google Scholar]
- 7.Bolton, M. A., W. Lan, S. E. Powers, M. L. McCleland, J. Kuang, and P. T. Stukenberg. 2002. Aurora B kinase exists in a complex with survivin and INCENP and its kinase activity is stimulated by survivin binding and phosphorylation. Mol. Biol. Cell 133064-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Canovas, P. M., and T. M. Guadagno. 2007. Functional analysis of Survivin in spindle assembly in Xenopus egg extracts. J. Cell. Biochem. 100217-229. [DOI] [PubMed] [Google Scholar]
- 9.Carazo-Salas, R. E., O. J. Gruss, I. W. Mattaj, and E. Karsenti. 2001. Ran-GTP coordinates regulation of microtubule nucleation and dynamics during mitotic-spindle assembly. Nat. Cell Biol. 3228-234. [DOI] [PubMed] [Google Scholar]
- 10.Carter, S. L., A. C. Eklund, I. S. Kohane, L. N. Harris, and Z. Szallasi. 2006. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat. Genet. 381043-1048. [DOI] [PubMed] [Google Scholar]
- 11.Carvalho, A., M. Carmena, C. Sambade, W. C. Earnshaw, and S. P. Wheatley. 2003. Survivin is required for stable checkpoint activation in taxol-treated HeLa cells. J. Cell Sci. 1162987-2998. [DOI] [PubMed] [Google Scholar]
- 12.Giet, R., C. Petretti, and C. Prigent. 2005. Aurora kinases, aneuploidy and cancer, a coincidence or a real link? Trends Cell Biol. 15241-250. [DOI] [PubMed] [Google Scholar]
- 13.Giodini, A., M. Kallio, N. R. Wall, G. J. Gorbsky, S. Tognin, P. C. Marchisio, M. Symons, and D. C. Altieri. 2002. Regulation of microtubule stability and mitotic progression by survivin. Cancer Res. 622462-2467. [PubMed] [Google Scholar]
- 14.Gruss, O. J., and I. Vernos. 2004. The mechanism of spindle assembly: functions of Ran and its target TPX2. J. Cell Biol. 166949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gruss, O. J., M. Wittmann, H. Yokoyama, R. Pepperkok, T. Kufer, H. Sillje, E. Karsenti, I. W. Mattaj, and I. Vernos. 2002. Chromosome-induced microtubule assembly mediated by TPX2 is required for spindle formation in HeLa cells. Nat. Cell Biol. 4871-879. [DOI] [PubMed] [Google Scholar]
- 16.Harel, A., and D. J. Forbes. 2004. Importin beta: conducting a much larger cellular symphony. Mol. Cell 16319-330. [DOI] [PubMed] [Google Scholar]
- 17.Hetzer, M., O. J. Gruss, and I. W. Mattaj. 2002. The Ran GTPase as a marker of chromosome position in spindle formation and nuclear envelope assembly. Nat. Cell Biol. 4E177-E184. [DOI] [PubMed] [Google Scholar]
- 18.Jeyaprakash, A. A., U. R. Klein, D. Lindner, J. Ebert, E. A. Nigg, and E. Conti. 2007. Structure of a survivin-borealin-INCENP core complex reveals how chromosomal passengers travel together. Cell 131271-285. [DOI] [PubMed] [Google Scholar]
- 19.Joukov, V., A. C. Groen, T. Prokhorova, R. Gerson, E. White, A. Rodriguez, J. C. Walter, and D. M. Livingston. 2006. The BRCA1/BARD1 heterodimer modulates ran-dependent mitotic spindle assembly. Cell 127539-552. [DOI] [PubMed] [Google Scholar]
- 20.Kalab, P., A. Pralle, E. Y. Isacoff, R. Heald, and K. Weis. 2006. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature 440697-701. [DOI] [PubMed] [Google Scholar]
- 21.Kang, B. H., and D. C. Altieri. 2006. Regulation of survivin stability by the aryl hydrocarbon receptor-interacting protein. J. Biol. Chem. 28124721-24727. [DOI] [PubMed] [Google Scholar]
- 22.Klein, U. R., E. A. Nigg, and U. Gruneberg. 2006. Centromere targeting of the chromosomal passenger complex requires a ternary subcomplex of Borealin, Survivin, and the N-terminal domain of INCENP. Mol. Biol. Cell 172547-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kline-Smith, S. L., and C. E. Walczak. 2004. Mitotic spindle assembly and chromosome segregation: refocusing on microtubule dynamics. Mol. Cell 15317-327. [DOI] [PubMed] [Google Scholar]
- 24.Knauer, S. K., C. Bier, N. Habtemichael, and R. H. Stauber. 2006. The Survivin-Crm1 interaction is essential for chromosomal passenger complex localization and function. EMBO Rep. 71259-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knauer, S. K., O. H. Kramer, T. Knosel, K. Engels, F. Rodel, A. F. Kovacs, W. Dietmaier, L. Klein-Hitpass, N. Habtemichael, A. Schweitzer, J. Brieger, C. Rodel, W. Mann, I. Petersen, T. Heinzel, and R. H. Stauber. 2007. Nuclear export is essential for the tumor-promoting activity of survivin. FASEB J. 21207-216. [DOI] [PubMed] [Google Scholar]
- 26.Koffa, M. D., C. M. Casanova, R. Santarella, T. Kocher, M. Wilm, and I. W. Mattaj. 2006. HURP is part of a Ran-dependent complex involved in spindle formation. Curr. Biol. 16743-754. [DOI] [PubMed] [Google Scholar]
- 27.Kops, G. J., B. A. Weaver, and D. W. Cleveland. 2005. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat. Rev. Cancer 5773-785. [DOI] [PubMed] [Google Scholar]
- 28.Kotwaliwale, C., and S. Biggins. 2006. Microtubule capture: a concerted effort. Cell 1271105-1108. [DOI] [PubMed] [Google Scholar]
- 29.Lens, S. M., G. Vader, and R. H. Medema. 2006. The case for Survivin as mitotic regulator. Curr. Opin. Cell Biol. 18616-622. [DOI] [PubMed] [Google Scholar]
- 30.Lens, S. M., R. M. Wolthuis, R. Klompmaker, J. Kauw, R. Agami, T. Brummelkamp, G. Kops, and R. H. Medema. 2003. Survivin is required for a sustained spindle checkpoint arrest in response to lack of tension. EMBO J. 222934-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malumbres, M., and M. Barbacid. 2001. To cycle or not to cycle: a critical decision in cancer. Nat. Rev. Cancer 1222-231. [DOI] [PubMed] [Google Scholar]
- 32.Okada, H., and T. W. Mak. 2004. Pathways of apoptotic and non-apoptotic death in tumour cells. Nat. Rev. Cancer 4592-603. [DOI] [PubMed] [Google Scholar]
- 33.Ouellet, V., M. C. Guyot, C. Le Page, A. Filali-Mouhim, C. Lussier, P. N. Tonin, D. M. Provencher, and A. M. Mes-Masson. 2006. Tissue array analysis of expression microarray candidates identifies markers associated with tumor grade and outcome in serous epithelial ovarian cancer. Int. J. Cancer 119599-607. [DOI] [PubMed] [Google Scholar]
- 34.Papaconstantinou, A. D., I. Shanmugam, L. Shan, I. S. Schroeder, C. Qiu, M. Yu, and E. G. Snyderwine. 2006. Gene expression profiling in the mammary gland of rats treated with 7,12-dimethylbenz[a]anthracene. Int. J. Cancer 11817-24. [DOI] [PubMed] [Google Scholar]
- 35.Pihan, G., and S. J. Doxsey. 2003. Mutations and aneuploidy: co-conspirators in cancer? Cancer Cell 489-94. [DOI] [PubMed] [Google Scholar]
- 36.Quimby, B. B., and M. Dasso. 2003. The small GTPase Ran: interpreting the signs. Curr. Opin. Cell Biol. 15338-344. [DOI] [PubMed] [Google Scholar]
- 37.Rosa, J., P. Canovas, A. Islam, D. C. Altieri, and S. J. Doxsey. 2006. Survivin modulates microtubule dynamics and nucleation throughout the cell cycle. Mol. Biol. Cell 171483-1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sampath, S. C., R. Ohi, O. Leismann, A. Salic, A. Pozniakovski, and H. Funabiki. 2004. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell 118187-202. [DOI] [PubMed] [Google Scholar]
- 39.Sandall, S., F. Severin, I. X. McLeod, J. R. Yates III, K. Oegema, A. Hyman, and A. Desai. 2006. A Bir1-Sli15 complex connects centromeres to microtubules and is required to sense kinetochore tension. Cell 1271179-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanderson, H. S., and P. R. Clarke. 2006. Cell biology: Ran, mitosis and the cancer connection. Curr. Biol. 16R466-R468. [DOI] [PubMed] [Google Scholar]
- 41.Sillje, H. H., S. Nagel, R. Korner, and E. A. Nigg. 2006. HURP is a Ran-importin beta-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr. Biol. 16731-742. [DOI] [PubMed] [Google Scholar]
- 42.Sun, C., D. Nettesheim, Z. Liu, and E. T. Olejniczak. 2005. Solution structure of human survivin and its binding interface with Smac/Diablo. Biochemistry 4411-17. [DOI] [PubMed] [Google Scholar]
- 43.Tsai, M. Y., C. Wiese, K. Cao, O. Martin, P. Donovan, J. Ruderman, C. Prigent, and Y. Zheng. 2003. A Ran signalling pathway mediated by the mitotic kinase Aurora A in spindle assembly. Nat. Cell Biol. 5242-248. [DOI] [PubMed] [Google Scholar]
- 44.Tulu, U. S., C. Fagerstrom, N. P. Ferenz, and P. Wadsworth. 2006. Molecular requirements for kinetochore-associated microtubule formation in mammalian cells. Curr. Biol. 16536-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weis, K. 2003. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell 112441-451. [DOI] [PubMed] [Google Scholar]
- 46.Wittmann, T., A. Hyman, and A. Desai. 2001. The spindle: a dynamic assembly of microtubules and motors. Nat. Cell Biol. 3E28-E34. [DOI] [PubMed] [Google Scholar]
- 47.Wittmann, T., M. Wilm, E. Karsenti, and I. Vernos. 2000. TPX2, a novel Xenopus MAP involved in spindle pole organization. J. Cell Biol. 1491405-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xia, F., C. W. Lee, and D. C. Altieri. 2008. Tumor cell dependence on Ran-GTP-directed mitosis. Cancer Res. 681826-1833. [DOI] [PubMed] [Google Scholar]
- 49.Zhou, J., A. O'Brate, A. Zelnak, and P. Giannakakou. 2004. Survivin deregulation in beta-tubulin mutant ovarian cancer cells underlies their compromised mitotic response to taxol. Cancer Res. 648708-8714. [DOI] [PubMed] [Google Scholar]