Abstract
In organelles, the posttranscriptional steps of gene expression are tightly controlled by nucleus-encoded factors, most often acting in a gene-specific manner. Despite the molecular identification of a growing number of factors, their mode of action remains largely unknown. In the green alga Chlamydomonas reinhardtii, expression of the chloroplast petA gene, which codes for cytochrome f, depends on two specific nucleus-encoded factors. MCA1 controls the accumulation of the transcript, while TCA1 is required for its translation. We report here the cloning of MCA1, the first pentatricopeptide repeat protein functionally identified in this organism. By chloroplast transformation with modified petA genes, we investigated the function of MCA1 in vivo. We demonstrate that MCA1 acts on the very first 21 nucleotides of the petA 5′ untranslated region to protect the whole transcript from 5′→3′ degradation but does not process the 5′ end of the petA mRNA. MCA1 and TCA1 recognize adjacent targets and probably interact together for efficient expression of petA mRNA. MCA1, although not strictly required for translation, shows features of a translational enhancer, presumably by assisting the binding of TCA1 to its own target. Conversely, TCA1 participates to the full stabilization of the transcript through its interaction with MCA1.
Organelle genomes have retained a limited set of genes from their prokaryotic ancestor, the expression of which is tightly controlled by the nucleus. Within organelles, mRNAs may undergo cis or trans splicing, editing, endo- and exonucleolytic cleavage, and 5′- and 3′-end processing. Their stabilization, translation, and degradation are highly regulated (reviewed in references 3, 5, 23, 26, and 63). Each of these posttranscriptional steps depends on nucleus-encoded factors. Strikingly, most are gene specific, one factor being required for the expression of one or a few organelle mRNAs. Altogether, several hundreds of nucleus-encoded factors may be required for the proper expression of the organelle genome (3, 65).
The green alga Chlamydomonas reinhardtii and the yeast Saccharomyces cerevisiae have been instrumental in the identification of these factors. Genetic analyses of nuclear mutants, defective for the expression of single organelle genes, have emphasized the existence of two major classes of trans-acting factors. Some are required for the proper maturation and stabilization of specific organellar transcripts (reviewed in references 3, 44, and 48), while others are required for their translation (reviewed in references 3, 23, and 78). Among the numerous nucleus-encoded factors involved in the stabilization of specific chloroplast transcripts identified in Chlamydomonas (14, 16, 28, 33, 35, 36, 39, 45, 66), only three have been cloned up to now—NAC2, MCD1, and MBB1—that, respectively, control psbD, petD, and psbB transcript stability (6, 46, 71). They act on the 5′ untranslated regions (5′UTRs) of their target transcripts and protect them from 5′→3′ ribonucleolytic degradation (14, 50, 71), but the molecular basis for their specific RNA recognition is still poorly understood. MBB1 and NAC2 promote the cleavage of a precursor form to generate the stable, mature form of their target mRNA (49, 50, 72). A 5′-end cleavage is also involved in the formation of petD mRNA (58, 70). Understanding whether these 5′ processing events are mandatory for the production of stable and translatable mRNAs in the chloroplast is critical for our knowledge of the basic rules of chloroplast gene expression. Furthermore, involvement of MBB1, NAC2, and MCD1 in the translation of their target mRNA has been hypothesized (15, 49, 72) but has proved difficult to test since template mRNAs fail to accumulate in the absence of these specific stabilization factors.
Here, in an attempt to address these issues, we studied the expression of the chloroplast petA gene, encoding cytochrome f, a major subunit of the cytochrome b6f complex. Previously, we characterized nuclear mutant strains of C. reinhardtii that accumulated the petA mRNA but did not synthesize cytochrome f (76). We subsequently cloned the TCA1 gene coding for a petA-specific translational activator (53). In the present study we focus on another set of nuclear mutants that are defective for the accumulation of the petA mRNA. We present the molecular characterization of MCA1, the nucleus-encoded factor responsible for petA mRNA stabilization. MCA1 turned out to code for the first pentatricopeptide repeat (PPR) protein functionally characterized in Chlamydomonas. Using chimeric versions of the petA gene, we mapped the nucleotide target of MCA1 to the very 5′ end of the petA mRNA and demonstrate that the most upstream 21-nucleotide motif is sufficient to confer MCA1-dependent stability to an unrelated mRNA. Finally, we provide evidence for an enhancer function of MCA1 in translation.
MATERIALS AND METHODS
Strains and growth conditions.
Wild-type, mutant, and transformed strains of C. reinhardtii were grown in Tris-acetate-phosphate (TAP) medium (pH 7.2) (29) under continuous low light (5 to 10 μE m−2 s−1).
mca1 mutant strains were isolated either by UV mutagenesis (CF-792, CF-152, CF-6, and CF-813 hereafter referred to as the mca1-2, mca1-3, mca1-4, and mca1-5 strains) (24) or insertional mutagenesis (strain mca1-1, previously called MΦ11 in reference 28). Cell-wall-less mca1 strains were obtained by crossing the mca1-1 and mca1-2 strains with mutant strain cw15. The nuclear mutant mcd1-1 (14, 46), tca1-1, tca1-2, and tca1-8 (53, 76) strains, as well as the ΔpetA strain, a chloroplast deletion strain (37), were also used in the present study.
Plasmid constructions. (i) Construction of modified petA and petD genes.
Plasmid pF5mut2 was created by cloning the 841-bp AatII-BstEII fragment from plasmid pWF (37) into vector pBKS, digested by SacI and XhoI, both fragments first blunt ended by treatment with Klenow and T4 DNA polymerase.
To delete nucleotides 22 to 63 from the petA 5′UTR, a PCR product was amplified with primers delcod and delrev from template plasmid pF5mut2 (see Table 1 for the sequence of the primers used in the present study), digested with PstI (a unique restriction site introduced in primers [Table 1]), and religated on itself to yield plasmid pF5mut2-del. The 364-bp BglII-HindIII fragment excised from this plasmid was cloned into plasmid pWF digested with the same enzymes to create plasmid pWF-del.
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′→3′)a | Restriction site |
|---|---|---|
| petAcod | GCGAGATCTTCCATCGATGAACTATGCTTTATTTGCTAAAAAAAAGA | |
| petArev | ACAGCTTGTGGTACTTCGATTTCAACTGCTTTTTGC | |
| pGcod | ATTTGGGGGGGGGGGGGGGGGGAAATAGTAATTATGAATGTAAATTACTTATGCTTACT | |
| pGrev | ATTTCCCCCCCCCCCCCCCCCCAAATTTTATTTTTTTTCTTCTCAACATATATTATATCT | |
| DAcod | ATTTTTAGCATGTAAACATTAGAAATACAGCAAGTAATTATGAATGTAAATTACTTATGCTTACT | |
| DArev | TGCTGTATTTCTAATGTTTACATGCTAAAAATTTTTAATATATTATTATATCTTTTTTTTAGCAAATAAAGCATAGTTCATG | |
| delcod | CGCCTGCAGCTAGAATAAACTTGTTGAGGCTGCTTA | PstI |
| delrev | GCGCTGCAGAATTTTATTTTTTTTCTTCTCAACATA | PstI |
| petDcod | CGCCCTAGGACTCACTAAAATTCATTTGCCCGAAGGGACGTCCA | HindIII |
| petDrev | CGTTAGGCCAAGCAGGTTCACCGTAAGTG | |
| ADcod | GATATAATATATGTTGAGAAGAAAAAAAATAAAATTTAAATAGTAATTGGAGTAAAAGAAAAATAT | |
| ADrev | TATTTTTTTTCTTCTCAACATATATTATATCTTTTTTTTAGCATTATATATTTTGTGCGGGATTTA | |
| RevpetA | TGCTGCGCGTAAAGTAGTAAATACTTGGTT | |
| RevpsaB | CAGGTGCAAAGCAAACTTTGCATAC |
Restriction sites are indicated by underscoring.
pG-petA construct was created by a two-step PCR procedure (31). Two pairs of primers (petAcod/pGrev and pGcod/petArev) allowed amplification from template plasmid pWF of two partially overlapping fragments that were mixed and used as templates in a third PCR with the external primers petAcod and petArev. The final amplicon, carrying the poly(G) tract, was digested with BglII and HindIII, two restriction sites on both sides of the mutation, and cloned into plasmid pWF digested with the same enzymes to create plasmid pWF-pG.
Plasmid pWF-DA carrying the modified petA gene, D-petA (described in Fig. 6A), was similarly created by using petAcod/DArev and DAcod/petArev primer pairs.
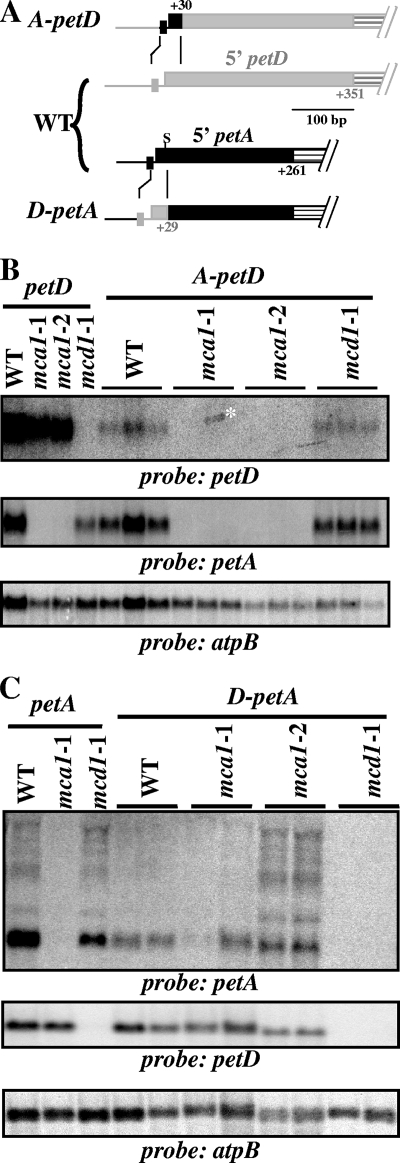
FIG. 6.
The stability of chimeric transcripts depends on their very 5′ end. (A) Schematic representation of the chimeric genes used for these experiments. The 5′ regions of the petA and petD genes are shown in the middle. Filled boxes represent the respective 5′UTR (in black for petA and gray for petD), while dashed boxes symbolize the beginning of the coding sequences. Rectangles immediately upstream of the 5′UTRs depict the −10 boxes of the two genes. In the 5′UTR of the petA gene, “S” indicates a SwaI restriction site, lost in the D-petA chimera, used for RFLP analysis of transformants. The top and bottom lines show, respectively, the structure of the A-petD and D-petA chimeras, with the origin of the sequence elements indicated by the color code. (B) The 30 first nucleotides of the petA 5′UTR confer an MCA1-dependent stability to the A-petD transcript. The A-petD chimera was introduced in the chloroplast of wild-type, mca1-1, mca1-2, and mcd1-1 strains. The accumulation of petA, petD (and atpB as a loading control) transcripts was monitored in three independent transformed strains for each genetic background, as well as in recipient strains. The asterisk points to unspecific stain overlapping two lanes. (C) D-petA transcripts are stabilized through interactions with MCD1. The D-petA chimera was introduced in the chloroplasts of wild-type, mca1-1, mca1-2, and mcd1-1 strains. The accumulation of petA and petD (and atpB as a loading control) mRNAs was monitored in two independent transformants, as well as in recipient strains.
In the three plasmids pWF-del, pWF-pG, and pWF-DA, the SwaI site present in petA 5′UTR was destroyed, creating a restriction fragment length polymorphism (RFLP) marker used to assess the homoplasmy of transformed strains.
Plasmid pWQ-AD carrying a modified petD gene (A-petD, see Fig. 6A) was similarly constructed from template plasmid pWQ, encompassing the petD gene (37), using the two-step PCR strategy with the primer pairs petDcod-ADrev and ADcod-petDrev. The 780-bp amplicon resulting from the third PCR was digested with HindIII and AvrII (a restriction site introduced within the petDcod primer) and cloned into vector pWQ digested by the same enzymes to create plasmid pWQ-AD. In addition to the mutation introduced in the petD 5′UTR, this plasmid contained a 381-bp deletion downstream of AvrII. A control plasmid pWQ-ΔAvrII, bearing the same deletion but no modification in the petD gene, was constructed by cloning the PCR product amplified with the primers petDcod and petDrev from plasmid pWQ into the vector pWQ, both being digested with HindIII and AvrII.
Plasmids pWF-pG, pWF-DA, and pWQ-AD were sequenced to confirm the presence of the appropriate mutations. The 5′psaA-aadA cassette (77), conferring resistance to spectinomycin, was then introduced into the unique HincII site of plasmids pWF-del, pWF-pG, and pWF-DA or into the unique AvrII site from plasmids pWQ-AD and pWQ-ΔAvrII, in reverse orientation with respect to petA or petD genes, to yield plasmids paAKWF-del, paAKWF-pG, and paAKWF-DA or plasmids paAKWQ-AD and paAKWQ-ΔAvrII, respectively.
(ii) Construction of a chimeric psbB gene.
Plasmid pcA-CP47, containing the chimeric gene 5′petA-psbB, was constructed by cloning a 695-bp DNA fragment containing the petA promoter and 5′UTR regions, obtained by digestion of plasmid p5F (11) with NcoI and EcoRV, into vector p38.Nco (72), digested with NcoI and StuI. The recycling aadA cassette (21) was then introduced into the unique restriction site AvrII of plasmid pcA-CP47 filled in with Klenow, in opposite orientation with respect to the psbB gene, to yield plasmid pKrcA-CP47.
Transformation experiments.
Chloroplast transformations were performed by tungsten particle bombardment (7) as described earlier (37). Transformants were selected either on TAP medium supplemented with spectinomycin (100 μg ml−1) under low light (5 to 6 μE m−2 s−1) or on minimal medium under moderate light (100 μE m−2 s−1). They were then subcloned on selective medium until they reached homoplasmy. At least three independent transformants were analyzed for each transformation.
Proper insertion of transforming DNA and homoplasmy were checked by RFLP analysis of PCR products encompassing the petA 5′UTR, amplified with primers petAcod and petArev, since all chimeric petA genes used in that study lacked the SwaI site present in the regular 5′UTR or led to amplicons of different size depending on whether the template contained wild-type or chimeric petA genes.
Nuclear transformation was performed by electroporation, as described previously (64), using the following parameters: 25 μF/720 V cm−1. Transformants were selected for phototrophy on minimal medium (29) under high light (200 μE m−2 s−1).
RNA isolation and analysis.
RNA extraction and Northern blot analysis were performed as described previously (17) with probes derived from coding sequences (18).
For primer extension analysis, 15 μg of total RNA, mixed with 1.5 pmol of the 33P-labeled 5′-phosphorylated oligonucleotides RevpetA and RevpsaB, were denatured at 75°C for 4 min and rapidly cooled down in ethanol-dry ice. Reverse transcription was performed at 54°C for 30 min with avian myeloblastosis virus reverse transcriptase (Finzyme). After LiCl precipitation, half the reaction was run on 5% sequencing gel alongside a sequencing reaction of pWF.
Discrimination between transcription start sites and processing sites of precursor RNAs was done by RNA ligase-mediated rapid amplification of cDNA ends (RLM-RACE), using a Generacer kit (Invitrogen) according to the supplier's protocol, except that we omitted the removal of free 5′-phosphates by calf intestine alkaline phosphatase. RLM-RACE was performed on 5 μg of total RNA from wild-type Chlamydomonas, pretreated or not with tobacco acid pyrophosphatase. Reverse transcription was done with oligonucleotide RevpetA, and the subsequent PCR amplification was performed using the oligonucleotides RevpetA and Generacer 5′ as primers. PCR products were analyzed on agarose gels and scanned for ethidium bromide fluorescence on a Typhoon Trio+ (GE Healthcare Life Sciences).
Protein isolation and analysis.
Protein isolation, separation, and immunoblot analysis were carried out on exponentially growing cells (2 × 106 cells ml−1) as described previously (37). Cell extracts were loaded on an equal chlorophyll basis. Cytochrome f accumulation, normalized to that of the oxygen evolving enhancer 2 (OEE2) subunit from Photosystem II as an internal standard, was quantified from PhosphorImager scans of immunoblots revealed with 125I-labeled protein A, using ImageQuant software (Molecular Dynamics, Sunnyvale, CA), as described previously (11).
RESULTS
Stabilization of petA mRNA is impaired in mca1 mutants.
In Chlamydomonas, petA is the first cistron of a dicistronic unit (Fig. 1A) that also includes the petD gene, encoding subunit IV from the cytochrome b6f complex (70). The petA transcription pattern, schematized in Fig. 1A, can be observed in Fig. 1B. Large transcripts corresponding to processing intermediates of the petA-petD transcription unit (55) are clearly visible, together with the shorter mature petA mRNA in the wild-type lane.
FIG. 1.
mca1 mutant strains lack stable accumulation of petA mRNA. (A) Schematic map of the petA-petD transcription unit. At the top is shown the relevant chloroplast genomic region, with the transcripts shown below. Bent arrows indicate transcription start sites. The dashed wavy line depicts the transient petA-petD cotranscript. This cotranscript is not observed in the wild type (WT, panel B) because of its rapid processing (58, 70). Symbols (diamond, star, and circle) refer to the transcripts visible in panel B. (B and C) Accumulation of petA mRNA (B) and cytochrome f (C) in wild-type and mca1-2 strains and in an mca1-2 cw15 strain complemented by transformation with a MCA1 cDNA clone. Accumulations of atpB mRNA and of the OEE2 subunit from Photosystem II provide the respective loading controls. Sizes of the best-characterized petA transcripts are indicated on the right side of panel B.
Previously, we isolated several mutants of C. reinhardtii that were incapable of phototrophic growth because they lacked stable accumulation of the petA mRNA (24, 28). As can be seen in Fig. 1B, where we overloaded the mutant lane with respect to the control, the petA mature mRNA is fully missing in the mutant mca1-2 strain, while trace amounts of larger petA-containing transcripts are still slightly visible. Accumulation of the petD mRNA is not affected (data not shown).
Genetic analysis demonstrated that all mutations are nuclear, recessive and allelic (see Table S1 in the supplemental material). Two strains were chosen for further study: MΦ11, isolated after insertional mutagenesis using a modified ARG7 cassette (28), now referred to as the mca1-1 strain, and CF792, generated by UV mutagenesis (24), referred to here as the mca1-2 strain. Since the mca1-1 strain still showed active transcription of petA (28), its failure to accumulate the transcript was attributed to increased nucleolytic degradation. The gene mutated in those strains was thus called MCA1 (for maturation/stabilization of cytochrome b6f complex petA mRNA).
MCA1 encodes a PPR protein.
As we reported previously, phototrophic growth of the mca1-1 mutant is restored by complementation with a cosmid containing a 31-kb fragment of wild-type genomic DNA (28), which is also able to complement the mca1-2 strain (data not shown). An 18-kb AvrII-NdeI subfragment, complementing both mca1-1 and mca1-2 mutations, was further restricted to a HindIII 7.7-kb region, which upon transformation, restored the photosynthetic capability of the mca1-2 strain but not of the mca1-1 strain (data not shown). This suggested that, in addition to MCA1, at least one other gene required for phototrophic growth had been disrupted during insertion of the modified ARG7 cassette into the mca1-1 strain. This other gene, TDA1, controlling translation of the α subunit of the ATP synthase complex, is characterized in a separate study (C. Loiselay et al., unpublished data). To avoid any uncontrolled effect of this second mutation on cytochrome f expression, we chose to continue our experimental investigations primarily with the mca1-2 strain. However, as illustrated below, we consistently obtained similar results when we compared the mca1-1 and mca1-2 strains.
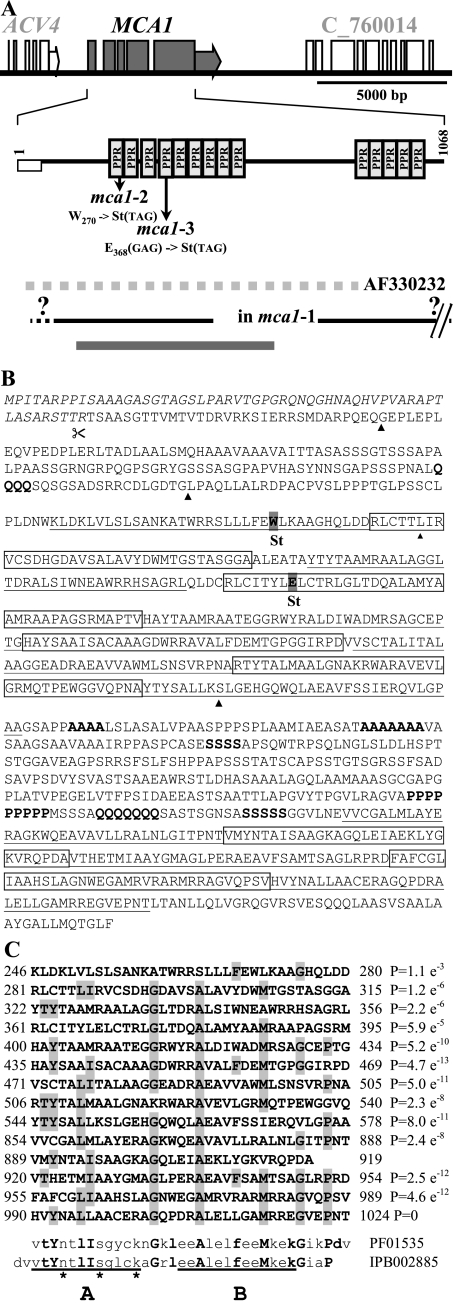
The sequence of the 7.7-kb insert and its surrounding regions was deposited in the EMBL database under accession no. AF330231 and corresponds to nucleotides 335254 to 347320 of contig 75 from the C. reinhardtii draft genome (v3.0) (http://genome.jgi-psf.org/Chlre3/Chlre3.home.html/). The sequence of the CL01e04 cDNA clone from this region, obtained from Kazusa DNA Research Institute (http://www.kazusa.or.jp/en/plant/), supported gene model estExt_fgenesh2_kg.C_750005. The polyadenylation consensus signal typical of C. reinhardtii, TGTAA (67), is found 15 bp upstream of the poly(A) tail. Sequence comparison between the cDNA and the genomic scaffold showed that MCA1 gene is composed of five exons (Fig. 2A). The cDNA lacks the 5′UTR and the first two codons of MCA1. However, it was able to complement the mca1-2 mutation, as demonstrated by the restoration of petA mRNA and cytochrome f in transformed strains (Fig. 1B and C). This confirmed that we actually cloned MCA1. Furthermore, sequence analysis of MCA1 in the mca1-2 and mca1-3 strains revealed that they each harbored single point mutations within the MCA1 coding sequence, changing, respectively, codons UGG(W270) and GAG(E368) to an amber codon (Fig. 2A).
FIG. 2.
MCA1 gene. (A) Structure of the MCA1 gene. The top line shows a schematic map of the MCA1 genomic region. MCA1 exons and 3′UTR are shown as gray boxes and a gray arrow. Adjacent gene models (ACV4 and C_760014) are also drawn as white boxes. A diagram of the protein is shown below with the position of the PPR motifs and the predicted transit peptide depicted as a white rectangle. The positions of mutations in mca1-2 and mca1-3 strains with respect to the protein sequence are indicated. From top to bottom are shown: the localization of the genomic fragment whose sequence is in the database (GenBank accession no. AF330232, dashed gray line), the approximate extend of the deletion in the mca1-1 strain (black line; note that the right border of the deletion has not been determined precisely but lies outside of the figure), and the location of the 7.7-kb DNA fragment able to restore the phototrophic growth of the mca1-2 strain upon transformation (thick gray line). (B) Sequence of the MCA1 protein. The predicted transit peptide is written in italics. PPR motifs are alternatively underlined and boxed. Stretches of four or more consecutive identical residues are in boldface. Arrowheads indicate the position of introns with respect to the coding sequence. Amino acids substituted by an amber codon in the mca1-2 and mca1-3 mutant strains are highlighted. (C) Alignment of the PPR motifs identified by program TPRpred, compared to the consensus sequences PFAM:PF01535 and InterPro:IPB002885. P values, measuring the statistical significance of the occurrence, are indicated for all repeats except repeat XI that was not detected by TPRpred (http://toolkit.tuebingen.mpg.de/tprpred) but was detected by program SMART (http://smart.embl-heidelberg.de/). Residues forming the antiparallel helices A and B are underlined. Asterisks point to the residues exposed on the external surface of helix A that could be involved in specific contacts with nucleotides (see Discussion).
MCA1 codes for a 1,068-residue protein (Fig. 2B), predicted to be targeted to the chloroplast by the programs Wolf PSORT (http://wolfpsort.seq.cbrc.jp/), Predotar (http://urgi.infobiogen.fr/predotar/predotar.html), TargetP (http://www.cbs.dtu.dk/services/TargetP/), and ChloroP (http://www.cbs.dtu.dk/services/ChloroP/). This predicted localization was confirmed since a tagged version of the MCA1 protein was enriched in chloroplast fractions (53). MCA1 displays 14 occurrences of a 35-amino-acid degenerated motif (PPRs) split into two blocks of nine and five PPR repeats. Eleven repeats are canonical PPR motifs (P < 1 e−5), whereas the other three (repeats I, III, and XI) are more degenerated (Fig. 2B and C). The protein also contains tracts of residues A, S, Q, or P (Fig. 2B), as previously reported for other nucleus-encoded factors in Chlamydomonas (6).
BLAST searches of protein databases identified several homologs whose similarity was restricted to the PPR repeats. Only one ortholog showing identity spread all along the sequence was found in the closely related green alga, Volvox carteri (see Fig. S1 in the supplemental material).
The target of MCA1 is located in the 5′UTR of petA mRNA.
We first tested whether the target(s) of MCA1 were localized within the 261-nucleotide petA 5′UTR. To this end, we substituted by chloroplast transformation the regular petA gene of the mca1-2 strain by the chimera 5′psbA-petA, comprising the sequence coding for cytochrome f translated under the control of the unrelated 5′UTR from the psbA gene (43) (Fig. 3A). Phototrophic transformants recovered on minimal medium were undistinguishable from the wild type in terms of growth rate or fluorescence induction kinetics (data not shown). Restoration of cytochrome f expression was confirmed by detection of the chimeric petA transcript in mca1 genetic backgrounds (Fig. 3B), at levels similar to those observed when the 5′psbA-petA gene was expressed in a wild-type nuclear background (43) (Fig. 3B, last lane). Since the mca1 transformed strains are capable of photoautotrophic growth and have a wild-type phenotype, we concluded that petA is the sole gene involved in photosynthesis whose expression is altered in the absence of MCA1.
FIG. 3.
The petA 5′UTR is the target of the MCA1 factor. (A) Schematic map of the chimera used for these experiments. The chloroplast genomic region encompassing the petA gene is shown in the middle line. 5′psbA-petA and 5′petA-psbB chimera, whose expression is analyzed in panels B and C, are, respectively, shown above and below the petA gene. Bent arrows indicate transcription start sites, while “K” stands for the aadA cassette, inserted in the opposite orientation with respect to petA to allow the selection of transformed strains. (B) A chimeric petA mRNA, expressed from the unrelated psbA 5′UTR, accumulates in the absence of MCA1. The accumulation of petA mRNA (either regular or the smaller chimeric 5′psbA-petA mRNA) in wild-type and mca1-2 nuclear backgrounds is shown. Two independent transformants expressing the chimeric 5′psbA-petA gene in the mca1 nuclear context are shown. The accumulation of psbB transcripts in the same strains provides a loading control. (B) Chimeric transcripts, driven by the petA 5′UTR, fail to accumulate in an mca1 mutant strain. The accumulation of psbB mRNAs (either regular or the slightly larger chimeric 5′petA-psbB mRNA) in wild-type and mca1-2 nuclear backgrounds is shown. Three independent transformants expressing the chimeric 5′petA-psbB gene are shown for each nuclear background. Accumulation of the atpB transcript in the same strains provides a loading control.
That the target for MCA1 was fully comprised within the petA 5′UTR was confirmed by using another chimera, 5′petA-psbB, made of the petA 5′UTR fused to the psbB coding sequence and 3′UTR (Fig. 3A). This chimera, associated with a spectinomycin resistance cassette to allow selection of transformed strains (25), was introduced by transformation in place of the endogenous psbB gene in the chloroplast genome of wild-type and mca1-2 strains. The chimeric transcript turned out to be fully MCA1 sensitive since it accumulated in a wild-type genetic context but was absent in mca1 mutants (Fig. 3C).
The petA mRNA displays a single 5′ end, which is MCA1 independent.
In chloroplasts, nucleus-encoded factors often stabilize a processed form of their target transcript (50, 72). This prompted us to map the petA mRNA 5′ end(s) by primer extension in wild-type and mca1 strains with the expectation that the 5′ end of the sole precursor transcript would be observed in mca1 mutant strains, whereas both mature and precursor ends would be detected in the wild type. Figure 4 (left panel) shows that a single 5′ end was detected in the wild type. The same single 5′ terminus was also detected to a much lower extent, in mca1-1 or mca1-2 mutant strains. This terminus, located 13 bp downstream of the first nucleotide of the TATAATAT consensus sequence for chloroplast promoters in Chlamydomonas (−10 box), probably corresponds to the actual start site for petA transcription. When carrying out RLM-RACE on Chlamydomonas RNA treated with tobacco acid pyrophosphatasse (taP), we observed an amplification product of the expected size (Fig. 4, right panel). In contrast, we did not observe this amplicon when the experiment was performed on RNA not treated with taP, but rather several faint unspecific amplification products of various sizes. The petA mRNA is thus almost exclusively found with a triphosphorylated 5′ end, which is not a substrate for ligation with an RNA oligonucleotide. The petA transcript therefore is an unprocessed transcription product, the abundance of which is controlled by MCA1 (53), through a mechanism that does not involve the maturation of its 5′ end.
FIG. 4.
MCA1 controls the abundance, but not the localization of the 5′ end of petA transcript. The left panel shows mapping of the petA 5′ends by extension of primer RevpetA in wild-type, mca1-1, and mca1-2 strains and in the control strain ΔpetA. Mapping of the psaB 5′ end by extension of the primer RevpsaB in the same reaction mixture provides a loading control. The RevpetA primer was used to generate a sequencing ladder for comparison. Arrow points to the 5′ end of the petA transcript on the sequence shown on the left, while the promoter consensus sequence is highlighted by a gray box. Asterisks stress two 5′ termini described by Matsuda and coworkers (41). We did not find these 5′ ends in our experiments and concluded that they probably originated from nonspecific extensions. The right panel shows the 5′ end of the petA transcript analyzed by RLM-RACE after (lane +) and without (lane −) taP treatment of mRNAs. First lane, 100-bp DNA ladder.
MCA1 protects the petA mRNA from 5′→3′ RNases.
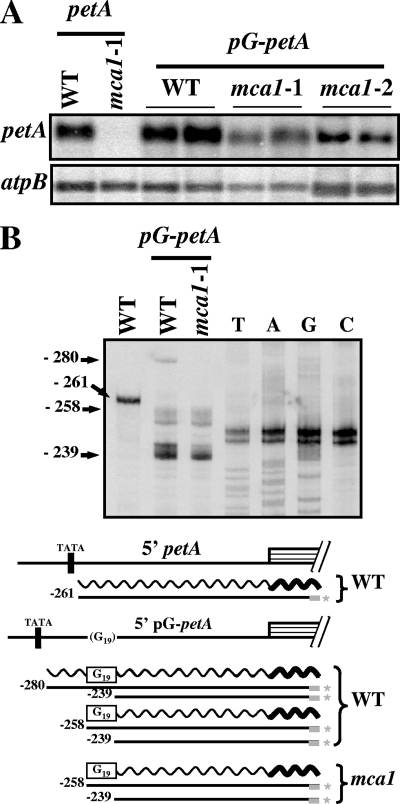
Stabilization factors previously studied in Chlamydomonas chloroplasts protect their target transcript from a 5′→3′ degradation process (14, 49, 72). This conclusion was reached by using poly(G) tracts that form very stable secondary structures impeding the progression of 5′→3′ exoribonucleases. We inserted 19 consecutive G residues in the petA 5′UTR at position +22 (+1 being the 5′ end of the mRNA), which also corresponds to position −239 with respect to the A residue from the initiation codon. This construct was introduced in place of the endogenous petA gene in the chloroplast genome of wild-type and mca1 strains. In a wild-type nuclear context, the pG-petA mRNA accumulated twice as much as the endogenous petA transcript (Fig. 5A). Thus, the accumulation of petA mRNA is limited by 5′→3′ degradation processes even in the presence of MCA1. This conclusion is consistent with MCA1 being in limiting amounts in the chloroplast, as we previously demonstrated (18, 53). In mca1 mutant strains that do not accumulate the petA transcript, the pG-petA mRNA accumulated to a level close to that of the regular transcript in the wild type (Fig. 5A). If the poly(G) tract impedes the progression of 5′→3′ RNases, then the 5′ end of this transcript should map at the 5′ border of the poly(G) tract in mca1 mutant strains. We mapped the 5′ ends of pG-petA transcripts by primer extension (Fig. 5B). Prominent bands in the −239/244 and −252/258 regions map at the beginning and the end of the poly(G) tract in both wild-type and mca1 nuclear backgrounds. The group of shorter bands originates from arrested extension at the beginning of the strongly structured poly(G) tract, as often observed in similar experiments (49, 72). The group of longer bands corresponds to the expected 5′ end of a transcript protected from 5′→3′ RNase degradation by the poly(G) tract. Accumulation of these bands in the wild-type nuclear context is in agreement with the overaccumulation of the pG-petA mRNA. In wild-type strains, but not in mca1 strains expressing the pG-petA chimera, a larger band at position −280 maps to the genuine 5′ end of the pG-petA transcript, just downstream of the −10 box. Its intensity is lower than that of the 5′ end of the regular petA mRNA in the wild type, since most extensions are arrested at the level of the poly(G) tract.
FIG. 5.
Insertion of a poly(G) tract at the beginning of the 5′UTR stabilizes the petA mRNA. (A) Accumulation of transcripts expressed from the pG-petA chimera in two independent transformed clones derived from wild-type, mca1-1, or mca1-2 recipient strains. The accumulation of the endogenous petA transcript in wild-type and mca1-1 strains is also shown. atpB mRNA provides a loading control. (B) 5′ ends of petA transcripts in the wild type and in a transformed strain expressing the pG-petA chimera, determined by primer extension. The labeled primer RevpetA was also used to generate a sequencing ladder from template plasmid pWF-pG. The length (numbered from the initiation codon) of extension products is indicated on the left. Below are schematically shown the structure of the 5′ region of the petA gene, as well as the transcripts (wavy lines) and extension products (straight lines) found in each strain. The position of the −10 box is indicated. The thick gray bar depicts the oligonucleotide primers, labeled at the 5′ end (*).
Thus, in the absence of MCA1, the petA mRNA is degraded by a 5′→3′ ribonucleolytic activity which is blocked by the poly(G) tract. In the wild type, MCA1 protects only a fraction of neosynthesized petA mRNAs from this ribonucleolytic activity.
The MCA1 target is located within the first 30 nucleotides of the 5′UTR of petA mRNA.
The MCA1-dependent protection of the petA mRNA suggests that MCA1 interacts, directly or indirectly, with the very 5′ end of the petA 5′UTR to prevent access of 5′→3′ RNases. In order to better define the target of MCA1 within the petA 5′ UTR, we constructed a reciprocal pair of chimeric genes, each linked to a spectinomycin resistance cassette for selection of transformants.
In the first one, hereafter referred to as A-petD (Fig. 6A), the −10 box and the first 31 transcribed nucleotides from petD were replaced by the −10 box and 30 first transcribed nucleotides from the petA gene. In the second construct, D-petA (Fig. 6A), we substituted the −10 box up to the 26th transcribed nucleotide of petA by the −10 box and first 29 transcribed nucleotides from petD. This construct includes stabilization element I, which is crucial for petD mRNA stability (57) that comprises the nine most upstream nucleotides of the petD transcript (30) and could contribute to the target of MCD1, the petD specific stabilization factor (14).
By chloroplast transformation, we substituted the endogenous petA or petD genes with their chimeric versions D-petA or A-petD in the wild type and in the mca1 and mcd1 strains lacking, respectively, the MCA1 and MCD1 stabilizing functions. When introduced in the wild type, the A-petD transcript accumulated significantly, although to a 10-fold-decreased level than the regular petD mRNA (Fig. 6B). Unlike the petD transcript, its A-petD chimeric version accumulated to similar levels in wild-type and mcd1 strains but failed to accumulate when expressed in mca1 genetic background (Fig. 6B). Thus, the A-petD transcript behaved as the original petA transcript: it is stabilized by MCA1 but insensitive to MCD1. Therefore, an essential target sequence for MCA1 is comprised within the most upstream 30 nucleotides in the 5′UTR of petA.
In the reciprocal experiments, the D-petA transcript was stabilized in a wild-type nuclear background, although to a lower level than the regular petA mRNA (20 to 35%, Fig. 6C). At variance with this, D-petA mRNA accumulated to similar levels in mca1 and wild-type strains but failed to accumulate in mcd1 mutants (Fig. 6C). Thus, the D-petA transcript behaved as the original petD transcript, being stabilized by MCD1 but insensitive to MCA1. The target for MCD1 is comprised within the 30 most upstream nucleotides of the petD 5′UTR.
This domain-swapping assay demonstrated that the specificities of MCA1 and MCD1 reside within the most upstream regions of the 5′UTRs of their respective target mRNAs.
TCA1 does not promote translation through the MCA1 target.
Translation of the petA mRNA is strictly dependent on the presence of TCA1, the petA specific translational activator that acts on its 5′UTR (53, 76). We wondered whether MCA1 and TCA1 shared the same target on the petA 5′UTR. We assessed for translation the transformed strains harboring the MCA1-sensitive A-petD construct. These strains were incapable of photoautotrophic growth because they completely lack accumulation of subunit IV. This defective expression cannot be accounted for by the 10-fold-decreased content in chimeric petD mRNA since strains with 10% of the wild-type level of petD mRNA accumulate more than 50% of subunit IV (20), whereas 5 to 10% of subunit IV is sufficient to sustain phototrophic growth (8, 56). Higgs et al. have proposed that “MCD1 itself appears to be a translation factor, in addition to promoting RNA stability” (30). Most probably, absence of the crucial stability element I within the A-petD mRNA prevents MCD1 to bind the chimeric transcript and to activate its translation.
Thus, stabilization of the A-petD chimeric transcript by MCA1 allowed its accumulation but not its translation. We conclude that part, if not all, of the target for TCA1 is located downstream the first 30 nucleotides from the petA 5′UTR.
TCA1 hampers translation of the D-petA mRNA.
We next examined whether these target elements for TCA1 were preserved in the D-petA construct. To study its translation, we relied on the fact that, in exponentially growing cells, cytochrome f is a stable protein whose accumulation faithfully reflects the rate of translation, a property particularly useful at low expression levels (9, 11, 37). The D-petA mRNA turned out to be translatable since cytochrome f could be detected in transformed cells with a wild-type nuclear background (Fig. 7A). As expected from the absence of the MCA1 target in the D-petA construct, translation of cytochrome f proved quite independent of the presence of MCA1, similar low levels of accumulation being observed in a wild-type or mca1 nuclear background (Fig. 7A).
FIG. 7.
The D-petA chimera is still translated in the absence of TCA1. (A and B) Accumulation of cytochrome f (A) and petA transcript (B) expressed from the D-petA chimera introduced by transformation in the chloroplast genomes of tca1-8 (two independent transformants), mca1-1, and wild-type recipient strains. The accumulation of cytochrome f and of the regular petA mRNA in wild-type and tca1-8 strains is also shown. OEE2 and psbB mRNA provide the respective loading controls.
We then tested whether translation of the D-petA mRNA still required TCA1 by introducing the D-petA chimera in the chloroplast genome of a tca1 mutant strain. In this tca1 nuclear context, the D-petA mRNA accumulated to the same extent as in the wild-type and mca1 nuclear contexts (Fig. 7B). Surprisingly, cytochrome f was better expressed in the absence of TCA1 than in a wild-type nuclear background (Fig. 7A). Thus, the first 29 nucleotides from the petD 5′UTR, which confer MCD1-dependent stability to the D-petA chimera, allow its translation in the absence of TCA1. Moreover, that translation is enhanced in the absence of TCA1 suggests an interaction between TCA1 and the chimeric transcript that hampers MCD1-mediated translation.
MCA1, or its mRNA target, enhances petA translation.
That MCA1 cannot promote translation of the A-petD chimeric transcript by no means excludes its contribution to the translation of a regular petA mRNA. The pG-petA mRNA that still accumulates in the absence of MCA1 gave us the opportunity to address this point by comparing the abundance of cytochrome f translated from this transcript in tca1, mca1, and wild-type nuclear backgrounds (Fig. 8).
FIG. 8.
Cytochrome f can be translated from the pG-petA transcript in the absence of MCA1 but not in the absence of TCA1. (A) Accumulation of cytochrome f (and OEE2 as a loading control) detected by a specific antibody in independent transformants expressing the pG-petA chimera derived from wild-type, mca1-1, or mca1-2 recipient strains. The accumulation of cytochrome f in the recipient strains is also shown for comparison. (B) Expression of the pG-petA chimera, introduced by transformation in the chloroplast genome of the tca1-2 recipient strain. The accumulations of petA mRNA and cytochrome f are shown in the wild-type and tca1-2 recipient strains and in two independent transformants. psbB mRNA and the OEE2 protein provide the respective loading controls.
In a wild-type nuclear background cytochrome f was slightly more expressed from the overaccumulated pG-petA transcript than from the regular petA mRNA (Fig. 8A): hence, the poly(G) tract inserted at position +22 did not impair translation. Despite the high accumulation of the pG-petA transcript in a tca1 nuclear context, cytochrome f was nevertheless not translated from the pG-petA mRNA in the absence of TCA1 (Fig. 8B): translation of the pG-petA mRNA, like that of the endogenous petA transcript, still requires TCA1.
In the absence of MCA1, cytochrome f was still expressed from the pG-petA transcript, but much less efficiently than in a wild-type nuclear context (Fig. 8A). The threefold drop in pG-petA mRNA accumulation in mca1 mutant strains could not account for the ∼10-fold decrease in cytochrome f expression. Thus, MCA1, even if not strictly required for translation of the petA mRNA, could behave as a translational enhancer. Alternatively, translational enhancement may involve nucleotides 1 to 21 that are not protected by the poly(G) tract in the absence of MCA1 (Fig. 5B).
We also noted that in tca1 mutant strains, the accumulation of the pG-petA mRNA remained unchanged, at variance with the regular petA transcript, whose level is reduced fivefold (76) (Fig. 8B). In the absence of TCA1, the petA mRNA is thus degraded by a 5′→3′ ribonucleolytic activity, from which it is normally protected by MCA1. The lower content in petA transcripts in tca1 mutants thus suggests a weakened interaction between the petA transcript and MCA1 in the absence of TCA1.
MCA1 and TCA1 could act on neighboring targets within the petA 5′UTR.
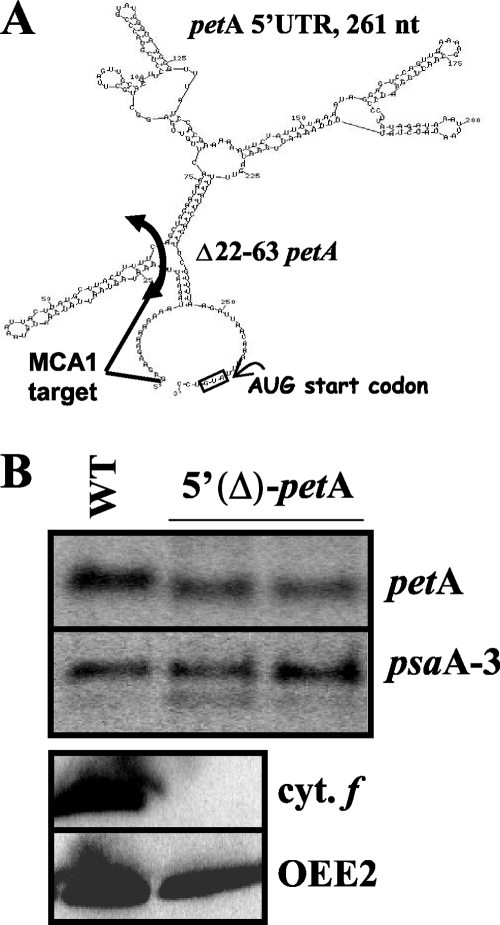
The experiments described above (Fig. 7 and 8) suggest that TCA1 interacts with sequences downstream of the MCA1 binding domain and that the presence of MCA1 and/or its nucleotide targets enhances TCA1-driven translation, while TCA1 improves the MCA1-dependent protection of petA transcripts from 5′→3′ degradation. To check whether the target of TCA1 includes sequences just downstream of those recognized by MCA1, we constructed a modified petA gene that still contained the target of MCA1 (the first 21 transcribed nucleotides from petA mRNA) but lacked nucleotides 22 to 63, predicted by RNA folding programs (Mfold) (80) to form a stem-loop structure (Fig. 9A). We then substituted the endogenous petA gene in wild-type and mca1 strains with this 5′(Δ)petA gene. The 5′(Δ)petA RNA accumulated to ca. 75% of the level observed for the endogenous petA transcript, when expressed in a wild-type nuclear background (Fig. 9B). In contrast, it did not accumulate in mca1 mutant strains (data not shown), indicating that it still contains the major target of MCA1, which could thus be restricted to positions 1 to 21.
FIG. 9.
Nucleotides 22 to 63 from the petA 5′UTR are required for the translation but not for the accumulation of the mRNA. (A) In silico prediction of the possible secondary structure of the petA 5′UTR, using the program RNAfold. Note that the proposed structure lacks experimental support. (B) Transcript accumulation from the regular petA gene in the wild type and from 5′(Δ)petA in transformed strains. psaA provides a loading control. Below is shown the accumulation of cytochrome f (and OEE2 as a loading control) in the same strains.
Cytochrome f was undetectable in transformed cells, either with the wild-type or the mca1 nuclear genome (Fig. 9B): the 5′(Δ)petA mRNA was not translatable despite its significant accumulation. Nucleotides 22 to 63 are critical for translation, probably as part of the TCA1 target. Thus, the targets for MCA1 and TCA1 comprise neighboring, if not overlapping, sequence elements in the petA 5′UTR.
DISCUSSION
MCA1 codes for a PPR protein.
Here, we report the characterization of MCA1, a nucleus-encoded factor specifically required for stabilization of the chloroplast petA mRNA in Chlamydomonas, since mca1 mutant strains still transcribe but do not accumulate the petA mRNA (28). MCA1 turned out to encode the first PPR protein functionally characterized in C. reinhardtii.
PPR proteins are presumably involved in specific interactions with organelle RNAs. They form a large protein family, present in all eukaryotes but greatly expanded in land plants (68). More than 450 PPR proteins were identified in a systematic search on the Arabidopsis genome, most of them predicted as targeted to organelles (38). This is also true for the PPR protein MCA1, since we previously reported that it was enriched in chloroplast fractions (53).
Only a few PPR proteins have been extensively studied up to now, but they most often act on one (or a few) specific target mRNA(s) and participate in any of the various posttranscriptional steps of organelle gene expression (reviewed in reference 1): cis and trans splicing (61), RNA editing (51), RNA processing (42, 47), RNA stabilization (the present study), and RNA translation (22, 40, 60). Other PPR proteins targeted to plant mitochondria are fertility restorers: they prevent expression of mRNAs encoding cytotoxic peptides (13, 75). How these proteins mediate such widely different functions within organelles is still poorly understood. In vascular plants, additional conserved motifs present at the C terminus of some PPR proteins may carry catalytic activity (38, 59). However, PPR proteins from nonplant eukaryotes, including green algae, lack these domains and most probably just recognize and/or bind RNAs. PPR proteins are thus believed to perform their function by recruiting catalytic factors (38).
MCA1 specifically recognizes the very 5′ end of the petA mRNA.
Understanding the basis and specificity of PPR/RNA interactions is thus crucial to elucidate the function of this wide and yet largely unknown protein family. This requires the precise identification of PPR protein targets in vivo. In higher plant chloroplasts, interactions between the PPR protein HCF152 and its target RNA—the transcript of the polycistronic unit psbB-psbT-psbH-petB-petD—have been demonstrated in vitro by UV cross-linking (42, 47). More recently, targets of PPR proteins in the chloroplast of maize were identified in vivo by microarray analysis of RNA immunoprecipitated with antibodies against the PPR proteins CRP1 or PPR4, respectively required for translation of petA and psaC mRNAs or for trans splicing of rps12 mRNA (60, 62).
In Chlamydomonas, short nucleotide sequences required for the stable accumulation of several transcripts have been identified by site-directed mutagenesis of their 5′UTRs (2, 30, 49), such as the stability element I (nucleotides 1 to 9) that is required for stable accumulation of the petD mRNA (30). Here, we show that the first 29 nucleotides from the petD mRNA are sufficient to confer an MCD1-dependent stability to a chimeric transcript. Probably, this major target sequence of MCD1 can further be restricted to stability element I because substitutions of nucleotides 10 to 24 had no effect on petD gene expression (30).
We also identified the sequence elements sufficient to confer in vivo MCA1-dependent stability to a chimeric transcript. We first demonstrated that the target of MCA1 was located within the petA 5′ UTR, a feature shared by most stabilization factors studied up to now in Chlamydomonas (14, 50, 72). Our 5′ end swapping experiments then pointed to the first 30 nucleotides from the petA transcript as the main target of MCA1, which can be further restricted to nucleotides 1 to 21 (it may even be shorter), as it is conserved in the 5′(Δ)petA construct that bears a deletion from nucleotides 22 to 63. This short nucleotide stretch confers MCA1-dependent stability to the A-petD reporter transcript, demonstrating that it is recognized by MCA1. Furthermore, we found no evidence for other critical sequence elements that would allow an independent binding of MCA1 to the petA mRNA: the presence of MCA1 in mcd1 strains expressing the D-petA chimera did not prevent the degradation of the chimeric mRNA, despite it contained the whole petA sequence, only deprived from the very first 26 first nucleotides (Fig. 6B). Conversely, this D-petA transcript proved insensitive to the absence of MCA1: it accumulates to identical levels in wild-type and mca1 genetic contexts (Fig. 6C). These experiments, however, do not exclude a marginal participation of other petA sequences to the interaction with MCA1, which would explain the limited accumulation of the A-petD transcript in a wild-type nuclear context.
PPR motifs are predicted to fold into pairs of antiparallel α helices (helices A and B, see Fig. 2C) (68), and tandem PPR repeats would form a superhelical binding surface. Three polar or charged amino acids exposed on the external side of helix A would make specific contact with target nucleotides (68). Although it has been proposed that each PPR repeat could bind three nucleotides (60), the number of PPR repeats in MCA1 (14), close to the maximal size of its mRNA target (21 nucleotides), rather supports an alternative model where each repeat would bind a single nucleotide, as established from the crystal structure of the analogous PUF repeats (19, 73, 74). We have here an unique opportunity for a better understanding of the structural interactions between PPR motifs and their nucleotide sequence target since both interacting partners (MCA1 and the petA 5′UTR) can exquisitely be modified by site-directed mutagenesis.
To address the specificity of the MCA1/petA 5′UTR interaction, we BLAST searched for other occurrences of the major MCA1 target in chloroplast genomes. This sequence was found only once in the chloroplast genome of Scenedesmus oblicus, a closely related unicellular green alga, 205 nucleotides upstream of the initiation codon of the petA gene (see Fig. S2 in the supplemental material). It was also found 214 nucleotides upstream of the petA translation start codon from another Chlamydomonas species, C. raudensis (27), 37 nucleotides downstream of a putative −10 box (see Fig. S2 in the supplemental material). Once available, the chloroplast genome sequence of Volvox should also prove informative in that respect. In contrast, this target was not found elsewhere in the chloroplast genome of C. reinhardtii, even when the query sequence was restricted to the first 14 nucleotides of the petA transcript. We only found one occurrence of the related sequence GAGAAGATAAAA in the transcribed intergenic region between psaC and petL (see Fig. S2 in the supplemental material), but it does not correspond to the 5′ end of a transcript. Moreover, the accumulation of psaC and petL transcripts is unaltered in mca1 mutant strains (C. Loiselay, unpublished results).
MCA1 protects the petA transcript from 5′→3′ degradation.
MCA1 protects its target RNA from 5′→3′ ribonucleolytic degradation since insertion of a poly(G) tract rescues the accumulation of the downstream sequence of the petA transcript in the absence of MCA1. A single 5′ end of petA mRNA was found in both wild-type and mca1 strains that corresponds to the transcription initiation site. In that respect, MCA1 differs from MBB1, NAC2, or MCD1, whose mRNA stabilizing function is associated with a 5′ end cleavage that converts an unstable mRNA precursor molecule to a stable and mature form (49, 50, 72). Our previous study of the MCA1 dependency of petA mRNA abundance also suggested a stable interaction between the MCA1 factor and its target mRNA, rather than a catalytic role of this factor in the conversion of petA transcripts from an unstable to a stable form (53). In agreement with this, MCA1 has no predicted catalytic domain. Non-PPR sequences within MCA1 are unlikely to carry enzymatic activity since, apart from the N and C termini, they are poorly conserved in size and sequence in the closely related species V. carteri. MCA1 thus probably recognizes and binds the very 5′ end of the petA 5′UTR, thereby preventing the access of RNases to the petA transcript.
In Chlamydomonas, the petA expression system comprises one stabilization factor and one translational activator.
Several chloroplast genes require specific factors for both mRNA stabilization and translation. This was documented for the expression of psbC (33, 54, 79), psaB (12, 69), and psbD (34-36, 61) genes, although the precise number of specific factors of each type remains unknown. In the case of the chloroplast gene petA, we identified only two distinct nuclear loci, encoding MCA1 and TCA1, respectively, that control mRNA accumulation and translation (24, 28, 53, 76). The number of mutant alleles identified thus far for each locus—13 and 9, respectively—argues that these two factors are the sole specific partners in petA gene expression. Since cytochrome f, the product of the petA gene, is highly conserved among organisms performing oxygenic photosynthesis, we looked for orthologs of MCA1 and TCA1 in other photosynthetic eukaryotes. Like TCA1 (53), MCA1 has an ortholog in V. carteri (66% identity), but none was found in higher plants or diatoms. This may reflect the rapid evolution of nucleus-encoded factors in green algae, as we suggested for TCA1 (53). Alternatively, an MCA1 homolog may not be required in vascular plants because of a different gene organization. In C. reinhardtii, petA is the first gene of the petA-petD dicistronic unit, and MCA1 binds to the very 5′ end of the transcribed mRNA. In contrast, petA is the last cistron of a ∼3.5-kb tetracistronic transcript (comprising psaI, ycf4, cemA, and petA) in land plants (4), which may not require petA-specific factors for its stable accumulation. Interestingly, expression of petA in maize does involve another PPR protein, CRP1, that is required for the translation of the petA and psaC mRNAs (4, 22, 60). Like MCA1, CRP1 comprises 14 PPR repeats, but these are organized in a continuous block. Whether these two proteins shared a common origin but diverged in the different lineages cannot be currently excluded. In Chlamydomonas, however, translation of petA mRNA relies on a distinct nucleus-encoded factor, TCA1, a pioneer protein devoid of PPRs (53, 76).
Are some nuclear factors that stabilize chloroplast mRNAs also required for their translation?
For a number of chloroplast genes only mRNA stabilization factors or translation factors have been identified to date. In some cases, it is likely that additional trans-acting factors still await identification. However, some chloroplast genes may actually require a single factor that would both stabilize their mRNA and allow its translation. Insertion of poly(G) tracts into petD, psbB, or psbD 5′UTRs restores the accumulation of these transcripts in the absence of their respective stabilization factor, but not their translation (14, 49, 50, 72), suggesting that these stabilization factors could be required for translation as well. Similarly, in the mitochondria of the yeast S. cerevisiae, mutations that restored the accumulation of the COB RNA coding for cytochrome b, in the absence of its specific stabilization factor, CBP1p, did not restore the synthesis of cytochrome b, which, again, suggests a role for CBP1p in the translation of its target transcript (32).
Our results strongly support a role for the stabilization factor MCD1 in translation. That element I, the putative nucleotide target of MCD1 (see above), is required for efficient petD mRNA translation has been abundantly documented (30, 56, 57). Here, we demonstrate that this MCD1 target allows the translation of a chimeric petA mRNA in the absence of its regular translation activator, TCA1. This argues for a role of MCD1 in translation activation, either by itself or by recruiting a translation activator, which would, however, be devoid of sequence specificity since petA and petD 5′UTRs have no common motif.
Recruitment prevails for the expression of the psbD mRNA, whose translation relies on the binding of a 40-kDa protein to a U-rich region of the psbD 5′UTR (52, 62). Once bound, this protein activates translation by destabilizing a secondary structure that sequesters the initiation codon (34). RNA binding of this protein shows poor sequence specificity, and its interaction with the psbD 5′UTR depends on the presence of NAC2, the psbD-specific stabilization factor (52), with which it interacts (62). Thus, sequence specificity is rather born by NAC2, which, nevertheless, is not able alone to activate translation.
Interaction of MCA1 and TCA1 in the expression of petA mRNAs.
Our work shows that MCA1 does not activate translation by itself since the A-petD chimeric transcript is not translated in vivo. Either it cannot efficiently recruit TCA1 or, if it does, downstream sequences from the petA 5′UTR are subsequently required for translation activation. In both hypotheses, TCA1 should have its own target(s), not restricted to the first 30 nucleotides of the petA 5′UTR. Deletion of nucleotides 22 to 63 prevented translation but not accumulation of the petA mRNA, suggesting that they contribute to the target of TCA1, adjacent to that of MCA1. This organization in tandem of the two target sites is further supported by the higher translation of the D-petA mRNA in the absence of TCA1. Probably, the target of TCA1 is preserved in the D-petA mRNA, and the interaction of TCA1 with the D-petA transcript hampers translation driven by MCD1/petD elements.
MCA1 is dispensable for translation since the pG-petA transcript is still translated in its absence, but not in the absence of TCA1. However, the absence of MCA1 results in poor translation, suggesting that MCA1 acts as an enhancer of translation. Alternatively, the target of TCA1 may extend upstream of position +22 and be degraded in the absence of MCA1 because it remains unprotected by the poly(G) tract. Even if not strictly required for TCA1 recruitment, the interaction of MCA1 with its mRNA target proved sensitive to the presence of TCA1 on its neighboring target (Fig. 10): in the absence of TCA1, much less petA transcript accumulates, due to the same poly(G)-sensitive 5′→3′ nucleolytic degradation that fully destroys petA mRNAs in the absence of MCA1. Thus, the decreased translation efficiency in the absence of MCA1 and the decreased mRNA stability in the absence of TCA1 argue for an interaction between these two trans-acting factors for efficient expression of the petA gene.
FIG. 10.
Working model for the expression of the petA genes used in the present study. At the top is shown the expression of the endogenous petA gene in the wild type. The MCA1, TCA1, and MCD1 factors are shown with their respective targets, respectively, drawn in black, white, and gray. The interaction between the initiation codon and TCA1 is suggested. The expression of the pG-petA, on the left, and D-petA, on the right, transcripts are schematically depicted below in (from top to bottom) wild-type, mca1, and tca1 strains. For each situation, we indicated in parentheses the efficiency of the translation: +++, robust, +, reduced but significant (∼10% of the wild-type level); +/−, low (<5%); −, no synthesis of cytochrome f.
Preliminary biochemical analyses suggest a physical interaction between MCA1 and TCA1 within a high-molecular-weight protein complex, the characterization of which is now under progress (C. Raynaud et al., unpublished data). We note also that a cooperative binding of MCA1 and TCA1 to the petA 5′UTR could account, at least in part, for the low accumulation of the A-petD transcript, since this latter does not contain the TCA1 target.
Being most likely restricted to three components—MCA1, TCA1, and their chloroplast gene target—the petA gene expression system now represents a unique model for an in-depth characterization of the regulation of the expression of a chloroplast gene. Further biochemical analysis of the behavior of TCA1 and MCA1 combined with site-directed mutations of the three partners should provide critical information on the physiological significance of their molecular interactions.
Supplementary Material
Acknowledgments
We thank D. Drapier and C. Raynaud for critical reading of the manuscript, N. Choonee and L. Zig for help with primer extension experiments, S. Bujaldon for constructing the double mutant mca1-2 cw15 strain, K. Wostrikoff for the kind gift of the 5′petA-psbB construct, A. Boulouis for help with the characterization of pG-petA-containing strains, and R. Kuras and all of the members of UMR7141 for stimulating discussions.
This study was supported by the CNRS and the Université Pierre et Marie Curie-Paris6. C.L. was ATER at the Université Pierre et Marie Curie-Paris6.
Footnotes
Published ahead of print on 23 June 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Andres, C., C. Lurin, and I. D. Small. 2007. The multifarious roles of PPR proteins in plant mitochondrial gene expression. Physiol. Plant 12914-22. [Google Scholar]
- 2.Anthonisen, I. L., M. L. Salvador, and U. Klein. 2001. Specific sequence elements in the untranslated regions of rbcL and atpB gene mRNAs stabilize transcripts in the chloroplast of Chlamydomonas reinhardtii. RNA 71024-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barkan, A., and M. Goldschmidt-Clermont. 2000. Participation of nuclear gene in chloroplast gene expression. Biochimie 82559-572. [DOI] [PubMed] [Google Scholar]
- 4.Barkan, A., M. Walker, M. Nolasco, and D. Johnson. 1994. A nuclear mutation in maize blocks the processing and translation of several chloroplast mRNAs and provides evidence for the differential translation of alternative mRNA forms. EMBO J. 133170-3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bollenbach, T. J., G. Schuster, and D. B. Stern. 2004. Cooperation of endo- and exoribonucleases in chloroplast mRNA turnover. Prog. Nucleic Acids Res. Mol. Biol. 78305-337. [DOI] [PubMed] [Google Scholar]
- 6.Boudreau, E., J. Nickelsen, S. D. Lemaire, F. Ossenbuhl, and J.-D. Rochaix. 2000. The Nac2 gene of Chlamydomonas encodes a chloroplast TPR-like protein involved in psbD mRNA stability. EMBO J. 193366-3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boynton, J. E., and N. W. Gillham. 1993. Chloroplast transformation in Chlamydomonas. Methods Enzymol. 217510-536. [DOI] [PubMed] [Google Scholar]
- 8.Chen, X., K. Kindle, and D. B. Stern. 1993. Initiation codon mutations in the Chlamydomonas chloroplast petD gene result in temperature-sensitive photosynthetic growth. EMBO J. 123627-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choquet, Y., D. B. Stern, K. Wostrikoff, R. Kuras, J. Girard-Bascou, and F.-A. Wollman. 1998. Translation of cytochrome f is autoregulated through the 5′ untranslated region of petA mRNA in Chlamydomonas chloroplasts. Proc. Natl. Acad. Sci. USA 954380-4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choquet, Y., and F.-A. Wollman. 2002. Translational regulations as specific traits of chloroplast gene expression. FEBS Lett. 52939-42. [DOI] [PubMed] [Google Scholar]
- 11.Choquet, Y., F. Zito, K. Wostrikoff, and F.-A. Wollman. 2003. Cytochrome f translation in Chlamydomonas chloroplast is autoregulated by its carboxyl-terminal domain. Plant Cell 151443-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dauvillee, D., O. Stampacchia, J. Girard-Bascou, and J.-D. Rochaix. 2003. Tab2 is a novel conserved RNA binding protein required for translation of the chloroplast psaB mRNA. EMBO J. 226378-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desloire, S., H. Gherbi, W. Laloui, S. Marhadour, V. Clouet, L. Cattolico, C. Falentin, S. Giancola, M. Renard, F. Budar, I. Small, M. Caboche, R. Delourme, and A. Bendahmane. 2003. Identification of the fertility restoration locus, Rfo, in radish, as a member of the pentatricopeptide-repeat protein family. EMBO Rep. 4588-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drager, R. G., J. Girard-Bascou, Y. Choquet, K. L. Kindle, and D. B. Stern. 1998. In vivo evidence for 5′→3′ exoribonuclease degradation of an unstable chloroplast mRNA. Plant J. 1385-96. [DOI] [PubMed] [Google Scholar]
- 15.Drager, R. G., D. C. Higgs, K. L. Kindle, and D. B. Stern. 1999. 5′ to 3′ exoribonucleolytic activity is a normal component of chloroplast mRNA decay pathways. Plant J. 19521-531. [DOI] [PubMed] [Google Scholar]
- 16.Drapier, D., J. Girard-Bascou, and F.-A. Wollman. 1992. Evidence for nuclear control of the expression of the atpA and atpB chloroplast genes in Chlamydomonas. Plant Cell 4283-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drapier, D., H. Suzuki, H. Levy, B. Rimbault, K. L. Kindle, D. B. Stern, and F.-A. Wollman. 1998. The chloroplast atpA gene cluster in Chlamydomonas reinhardtii: functional analysis of a polycistronic transcription unit. Plant Physiol. 117629-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eberhard, S., D. Drapier, and F.-A. Wollman. 2002. Searching limiting steps in the expression of chloroplast encoded proteins: relations between gene copy number, transcription, transcript abundance, and translation rate in the chloroplast of Chlamydomonas reinhardtii. Plant J. 311-14. [DOI] [PubMed] [Google Scholar]
- 19.Edwards, T. A., J. Trincao, C. R. Escalante, R. P. Wharton, and A. K. Aggarwal. 2000. Crystallization and characterization of Pumilo: a novel RNA binding protein. J. Struct. Biol. 132251-254. [DOI] [PubMed] [Google Scholar]
- 20.Esposito, D., D. C. Higgs, R. G. Drager, D. B. Stern, and J. Girard-Bascou. 2001. A nucleus-encoded suppressor defines a new factor which can promote petD mRNA stability in the chloroplast of Chlamydomonas reinhardtii. Curr. Genet. 3940-48. [DOI] [PubMed] [Google Scholar]
- 21.Fischer, N., O. Stampacchia, K. Redding, and J.-D. Rochaix. 1996. Selectable marker recycling in the chloroplast. Mol. Gen. Genet. 251373-380. [DOI] [PubMed] [Google Scholar]
- 22.Fisk, D. G., M. B. Walker, and A. Barkan. 1999. Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 182621-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fox, T. D. 1996. Genetics of mitochondrial translation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 24.Girard-Bascou, J., Y. Choquet, N. J. Gumpel, D. Culler, S. Purton, S. Merchant, F. Laquerriere, and F.-A. Wollman. 1995. Nuclear control of the expression of the chloroplast pet genes in Chlamydomonas reinhardtii, p. 683-686. In P. Mathis (ed.), Photosynthesis: from light to biosphere, vol. III. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 25.Goldschmidt-Clermont, M. 1991. Transgenic expression of aminoglycoside adenine transferase in the chloroplast: a selectable marker of site-directed transformation of Chlamydomonas. Nucleic Acids Res. 194083-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grivell, L. A. 1995. Nucleo-mitochondrial interactions in mitochondrial gene expression. Crit. Rev. Biochem. Mol. Biol., 30121-164. [DOI] [PubMed] [Google Scholar]
- 27.Gudynaite-Savitch, L., M. Gretes, R. M. Morgan-Kiss, L. V. Savitch, J. Simmonds, S. E. Kohalmi, and N. P. Huner. 2006. Cytochrome f from the Antarctic psychrophile, Chlamydomonas raudensis UWO 241: structure, sequence, and complementation in the mesophile, Chlamydomonas reinhardtii. Mol. Genet. Genomics 275387-398. [DOI] [PubMed] [Google Scholar]
- 28.Gumpel, N. J., L. Ralley, J. Girard-Bascou, F.-A. Wollman, J. H. Nugent, and S. Purton. 1995. Nuclear mutants of Chlamydomonas reinhardtii defective in the biogenesis of the cytochrome b6f complex. Plant Mol. Biol. 29921-932. [DOI] [PubMed] [Google Scholar]
- 29.Harris, E. H. 1989. The Chlamydomonas source book: a comprehensive guide to biology and laboratory use. Academic Press, Inc., San Diego, CA. [DOI] [PubMed]
- 30.Higgs, D. C., R. Kuras, K. L. Kindle, F.-A. Wollman, and D. B. Stern. 1998. Inversions in the Chlamydomonas chloroplast genome suppress a petD 5′ untranslated region deletion by creating functional chimeric mRNAs. Plant J. 14663-671. [DOI] [PubMed] [Google Scholar]
- 31.Higuchi, R. 1990. Recombinant PCR, p. 177-183. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, Inc., New York, NY.
- 32.Islas-Osuna, M. A., T. P. Ellis, L. L. Marnell, T. M. Mittelmeier, and C. L. Dieckmann. 2002. Cbp1 is required for translation of the mitochondrial cytochrome b mRNA of Saccharomyces cerevisiae. J. Biol. Chem. 27737987-37990. [DOI] [PubMed] [Google Scholar]
- 33.Jensen, K. H., D. L. Herrin, F. G. Plumley, and G. W. Schmidt. 1986. Biogenesis of photosystem II complexes: transcriptional, translational, and posttranslational regulation. J. Cell Biol. 1031315-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klinkert, B., I. Elles, and J. Nickelsen. 2006. Translation of chloroplast psbD mRNA in Chlamydomonas is controlled by a secondary RNA structure blocking the AUG start codon. Nucleic Acids Res. 34386-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuchka, M., S. Mayfield, and J.-D. Rochaix. 1988. Nuclear mutations specifically affect the synthesis and/or degradation of the chloroplast-encoded D2 polypeptide of photosystem II in Chlamydomonas reinhardtii. EMBO J. 7319-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuchka, M. R., M. Goldschmidt-Clermont, J. J. van Dillewijn, and J.-D. Rochaix. 1989. Mutation at the Chlamydomonas nuclear NAC2 locus specifically affects stability of the chloroplast psbD transcript encoding polypeptide D2 of PS II. Cell 58869-876. [DOI] [PubMed] [Google Scholar]
- 37.Kuras, R., and F.-A. Wollman. 1994. The assembly of cytochrome b6f complexes: an approach using genetic transformation of the green alga Chlamydomonas reinhardtii. EMBO J. 131019-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lurin, C., C. Andres, S. Aubourg, M. Bellaoui, F. Bitton, C. Bruyere, M. Caboche, C. Debast, J. Gualberto, B. Hoffmann, A. Lecharny, M. Le Ret, M. L. Martin-Magniette, H. Mireau, N. Peeters, J. P. Renou, B. Szurek, L. Taconnat, and I. Small. 2004. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 162089-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Majeran, W., J. Olive, D. Drapier, O. Vallon, and F.-A. Wollman. 2001. The light sensitivity of ATP synthase mutants of Chlamydomonas reinhardtii. Plant Physiol. 126421-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manthey, G. M., and J. E. McEwen. 1995. The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J. 144031-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsumoto, T., M. Matsuo, and Y. Matsuda. 1991. Structural analysis and expression during dark-light transitions of a gene for cytochrome f in Chlamydomonas reinhardtii. Plant Cell Physiol. 32863-872. [Google Scholar]
- 42.Meierhoff, K., S. Felder, T. Nakamura, N. Bechtold, and G. Schuster. 2003. HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell 151480-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Minai, L., K. Wostrikoff, F.-A. Wollman, and Y. Choquet. 2006. Chloroplast biogenesis of photosystem II cores involves a series of assembly-controlled steps that regulate translation. Plant Cell 18159-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monde, R. A., G. Schuster, and D. B. Stern. 2000. Processing and degradation of chloroplast mRNA. Biochimie 82573-582. [DOI] [PubMed] [Google Scholar]
- 45.Monod, C., M. Goldschmidt-Clermont, and J.-D. Rochaix. 1992. Accumulation of chloroplast psbB RNA requires a nuclear factor in Chlamydomonas reinhardtii. Mol. Gen. Genet. 231449-459. [DOI] [PubMed] [Google Scholar]
- 46.Murakami, S., K. Kuehnle, and D. B. Stern. 2005. A spontaneous tRNA suppressor of a mutation in the Chlamydomonas reinhardtii nuclear MCD1 gene required for stability of the chloroplast petD mRNA. Nucleic Acids Res. 333372-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakamura, T., K. Meierhoff, P. Westhoff, and G. Schuster. 2003. RNA-binding properties of HCF152, an Arabidopsis PPR protein involved in the processing of chloroplast RNA. Eur. J. Biochem. 2704070-4081. [DOI] [PubMed] [Google Scholar]
- 48.Nickelsen, J. 1998. Chloroplast RNA stability, p. 151-163. In J.-D. Rochaix, M. Goldschmidt-Clermont, and S. Merchant (ed.), The molecular biology of chloroplasts and mitochondria in Chlamydomonas. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 49.Nickelsen, J., M. Fleischmann, E. Boudreau, M. Rahire, and J.-D. Rochaix. 1999. Identification of cis-acting RNA leader elements required for chloroplast psbD gene expression in Chlamydomonas. Plant Cell 11957-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nickelsen, J., J. van Dillewijn, M. Rahire, and J.-D. Rochaix. 1994. Determinants for stability of the chloroplast psbD RNA are located within its short leader region in Chlamydomonas reinhardtii. EMBO J. 133182-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okuda, K., F. Myouga, R. Motohashi, K. Shinozaki, and T. Shikanai. 2007. Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing. Proc. Natl. Acad. Sci. USA 1048178-8183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ossenbuhl, F., and J. Nickelsen. 2000. cis- and trans-acting determinants for translation of psbD mRNA in Chlamydomonas reinhardtii. Mol. Cell. Biol. 208134-8142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raynaud, C., C. Loiselay, K. Wostrikoff, R. Kuras, J. Girard-Bascou, F.-A. Wollman, and Y. Choquet. 2007. Evidence for regulatory function of nucleus-encoded factors on mRNA stabilization and translation in the chloroplast. Proc. Natl. Acad. Sci. USA 1049093-9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rochaix, J.-D., M. Kuchka, S. P. Mayfield, M. Schirmer-Rahire, J. Girard-Bascou, and P. Bennoun. 1989. Nuclear and chloroplast mutations affect the synthesis or stability of the chloroplast psbC gene product in Chlamydomonas reinhardtii. EMBO J. 81013-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rymarquis, L. A., D. C. Higgs, and D. B. Stern. 2006. Nuclear suppressors define three factors that participate in both 5′ and 3′ end processing of mRNAs in Chlamydomonas chloroplasts. Plant J. 46448-461. [DOI] [PubMed] [Google Scholar]
- 56.Sakamoto, W., X. Chen, K. L. Kindle, and D. B. Stern. 1994. Function of the Chlamydomonas reinhardtii petD 5′ untranslated region in regulating the accumulation of subunit IV of the cytochrome b6f complex. Plant J. 6503-512. [DOI] [PubMed] [Google Scholar]
- 57.Sakamoto, W., K. L. Kindle, and D. B. Stern. 1993. In vivo analysis of Chlamydomonas chloroplast petD gene expression using stable transformation of β-glucuronidase translational fusions. Proc. Natl. Acad. Sci. USA 90497-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sakamoto, W., N. R. Sturm, K. L. Kindle, and D. B. Stern. 1994. petD mRNA maturation in Chlamydomonas reinhardtii chloroplasts: role of 5′ endonucleolytic processing. Mol. Cell. Biol. 146180-6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salone, V., M. Rudinger, M. Polsakiewicz, B. Hoffmann, M. Groth-Malonek, B. Szurek, I. Small, V. Knoop, and C. Lurin. 2007. A hypothesis on the identification of the editing enzyme in plant organelles. FEBS Lett. 5814132-4138. [DOI] [PubMed] [Google Scholar]
- 60.Schmitz-Linneweber, C., R. Williams-Carrier, and A. Barkan. 2005. RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. Plant Cell 172791-2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmitz-Linneweber, C., R. E. Williams-Carrier, P. M. Williams-Voelker, T. S. Kroeger, A. Vichas, and A. Barkan. 2006. A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell 182650-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schwarz, C., I. Elles, J. Kortmann, M. Piotrowski, and J. Nickelsen. 2007. Synthesis of the D2 protein of photosystem II in Chlamydomonas is controlled by a high molecular mass complex containing the RNA stabilization factor Nac2 and the translational activator RBP40. Plant Cell 193627-3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shikanai, T. 2006. RNA editing in plant organelles: machinery, physiological function, and evolution. Cell. Mol. Life Sci. 63698-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shimogawara, K., S. Fujiwara, A. Grossman, and H. Usuda. 1998. High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics 1481821-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sickmann, A., J. Reinders, Y. Wagner, C. Joppich, R. Zahedi, H. E. Meyer, B. Schonfisch, I. Perschil, A. Chacinska, B. Guiard, P. Rehling, N. Pfanner, and C. Meisinger. 2003. The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. USA 10013207-13212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sieburth, L. E., S. Berry-Lowe, and G. W. Schmidt. 1991. Chloroplast RNA stability in Chlamydomonas: rapid degradation of psbB and psbC transcripts in two nuclear mutants. Plant Cell 3175-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Silflow, C. D. 1998. Organization of the nuclear genome, p. 25-40. In J.-D. Rochaix, M. Goldschmidt-Clermont, and S. Merchant (ed.), The molecular biology of chloroplasts and mitochondria in Chlamydomonas. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 68.Small, I. D., and N. Peeters. 2000. The PPR motif: a TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 2546-47. [DOI] [PubMed] [Google Scholar]
- 69.Stampacchia, O. 1997. Photosystem I biogenesis in Chlamydomonas reinhardtii. Ph.D. thesis. University of Geneva, Geneva, Switzerland.
- 70.Sturm, N. R., R. Kuras, S. Buschlen, W. Sakamoto, K. L. Kindle, D. B. Stern, and F.-A. Wollman. 1994. The petD gene is transcribed by functionally redundant promoters in Chlamydomonas reinhardtii chloroplasts. Mol. Cell. Biol. 146171-6179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vaistij, F. E., E. Boudreau, S. D. Lemaire, M. Goldschmidt-Clermont, and J.-D. Rochaix. 2000. Characterization of mbb1, a nucleus-encoded tetratricopeptide-like repeat protein required for expression of the chloroplast psbB/psbT/psbH gene cluster in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 9714813-14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vaistij, F. E., M. Goldschmidt-Clermont, K. Wostrikoff, and J.-D. Rochaix. 2000. Stability determinants in the chloroplast psbB/T/H mRNAs of Chlamydomonas reinhardtii. Plant J. 21469-482. [DOI] [PubMed] [Google Scholar]
- 73.Wang, X., J. McLachlan, P. D. Zamore, and T. M. Hall. 2002. Modular recognition of RNA by a human pumilio-homology domain. Cell 110501-512. [DOI] [PubMed] [Google Scholar]
- 74.Wang, X., P. D. Zamore, and T. M. Hall. 2001. Crystal structure of a Pumilio homology domain. Mol. Cell 7855-865. [DOI] [PubMed] [Google Scholar]
- 75.Wang, Z., Y. Zou, X. Li, Q. Zhang, L. Chen, H. Wu, D. Su, Y. Chen, J. Guo, D. Luo, Y. Long, Y. Zhong, and Y. G. Liu. 2006. Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell 18676-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wostrikoff, K., Y. Choquet, F.-A. Wollman, and J. Girard-Bascou. 2001. TCA1, a single nuclear-encoded translational activator specific for petA mRNA in Chlamydomonas reinhardtii chloroplast. Genetics 159119-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wostrikoff, K., J. Girard-Bascou, F.-A. Wollman, and Y. Choquet. 2004. Biogenesis of PSI involves a cascade of translational autoregulation in the chloroplast of Chlamydomonas. EMBO J. 222696-2705.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zerges, W. 2000. Translation of mRNAs encoded by chloroplast genomes. Biochimie 82583-601. [DOI] [PubMed] [Google Scholar]
- 79.Zerges, W., and J.-D. Rochaix. 1994. The 5′ leader of a chloroplast mRNA mediates the translational requirements for two nucleus-encoded functions in Chlamydomonas reinhardtii. Mol. Cell. Biol. 145268-5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 313406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.