Abstract
The p53 tumor suppressor pathway limits oncogenesis by inducing cell cycle arrest or apoptosis. A key p53 target gene is PUMA, which encodes a BH3-only proapoptotic protein. Here we demonstrate that Puma deletion in the Eμ-Myc mouse model of Burkitt lymphoma accelerates lymphomagenesis and that ∼75% of Eμ-Myc lymphomas naturally select against Puma protein expression. Furthermore, approximately 40% of primary human Burkitt lymphomas fail to express detectable levels of PUMA and in some tumors this is associated with DNA methylation. Burkitt lymphoma cell lines phenocopy the primary tumors with respect to DNA methylation and diminished PUMA expression, which can be reactivated following inhibition of DNA methyltransferases. These findings establish that PUMA is silenced in human malignancies, and they suggest PUMA as a target for the development of novel chemotherapeutics.
The p53 tumor suppressor blocks cancer cell growth and impairs survival by functioning within signaling pathways that respond to various forms of stress, including oncogene activation, DNA damage, and hypoxia (for a review, see reference 47). Upon cellular stress, the p53 protein becomes stabilized and activates the transcription of multiple target genes that negatively regulate cell proliferation (e.g., p21CIP1 and PTPRV) (9, 14) and survival (e.g., Bax, Noxa, and Puma) (18, 34, 35, 37, 52). Thus, disruption of the p53 gene in the mouse results in aberrant cell cycle control and the acquisition of resistance to stress-induced death stimuli in a broad range of cell types. Human tumors frequently bypass these growth restrictions by inactivating p53 directly through mutation. This pathway can also be disrupted through alterations in upstream regulators (e.g., ATM, Mdm2, and p14ARF) and downstream effectors (e.g., Caspase-9 and APAF1) (4).
Puma (p53-upregulated modulator of apoptosis) encodes a proapoptotic BH3-only protein belonging to the Bcl-2 family of proteins (18, 35, 52). Upon induction, Puma initiates programmed cell death as a sensitizer and/or direct activator of Bax and Bak, resulting in cytochrome c release and caspase activation (28, 50). Indeed, deletion of Puma efficiently protects against apoptosis induced by either DNA damage or oncogene activation, rivaling the degree of protection when p53 is deleted (25, 46). Therefore, Puma plays a significant role in p53-dependent cell death. Further, Puma activates apoptotic pathways independently of p53, such as when murine myeloid progenitors are deprived of growth factors or during glucocorticoid treatment of primary thymocytes (25). Thus, Puma is a critical mediator of both p53-dependent and p53-independent cell death.
Human Burkitt lymphomas (BLs) arise through the translocation of the c-MYC proto-oncogene into the immunoglobulin heavy- or light-chain promoter-enhancers in B lymphocytes, resulting in the constitutive overproduction of c-MYC (3). The Eμ-Myc transgenic mouse model resembles the human disease, and these mice develop fully penetrant, lethal B-cell lymphomas (1). Premalignant Eμ-Myc B cells undergo p53-dependent apoptosis, which serves as a rate-limiting step in lymphomagenesis (12, 42, 43). Consequently, 75% of the resulting B-cell lymphomas express mutant p53, delete or silence p19Arf (the murine homolog of human p14ARF), and/or overexpress Mdm2 (12), similar to reported findings on human BL (29, 49). Therefore, the Arf-p53-Mdm2 axis serves as a guardian that prevents tumorigenesis in c-Myc-driven B-cell malignancies in both mice and humans.
Previous studies implicated the apoptotic arm of the p53 signaling pathway as its primary tumor suppressor function (30, 40, 43, 45). Since Puma functions as a critical mediator of p53-dependent cell death, we explored the role of Puma in Myc-driven lymphomagenesis in (i) the Eμ-Myc transgenic mouse model, (ii) established human BL cell lines, and (iii) primary human BLs. We report that Puma restricts B-cell lymphomagenesis in the mouse and that Puma protein is undetectable in most Eμ-Myc lymphomas and, most importantly, demonstrate that PUMA expression is silenced in 40% of human BLs. Further, we present evidence for the epigenetic silencing of PUMA through DNA and histone methylation, providing mechanistic insight into how PUMA expression can be repressed in BL and possibly other tumor types. These findings demonstrate PUMA inactivation in primary human cancer and highlight PUMA as a potential target for the development of novel therapeutics.
MATERIALS AND METHODS
Cell culture.
KemI and KemIII are established human BL cell lines that maintain either a type I or a type III Epstein-Barr virus (EBV) latency, respectively, and were maintained in RPMI 1640 medium supplemented with 10% defined fetal bovine serum (HyClone), 2 mM l-glutamine, penicillin (100 IU/ml), and streptomycin (100 μg/ml) at 37°C in 5% CO2 (39). DNA methyltransferase activity was inhibited by the addition of 5 μM 5-aza-2′-deoxycytidine (5-Aza; Sigma-Aldrich, St. Louis, MO) to the tissue culture medium, which was replenished every 24 h. At the indicated intervals, the cells were harvested and lysed and total RNA was isolated for real-time PCR analysis (see below).
Animals.
Puma−/− (C57BL/6/129) (25) mice were interbred with transgenic Eμ-Myc mice (C57BL/6) (11). Puma+/− and Puma+/−; Eμ-Myc littermates (F1 generation) were intercrossed to obtain nontransgenic and Eμ-Myc transgenic Puma+/+, Puma+/−, and Puma−/− animals (F2 generation). Both male and virgin female mice were then observed daily for signs of morbidity and tumor development. Animals were sacrificed, and tumors were harvested for RNA and protein analysis. Spleen and bone marrow samples were also harvested at 7 weeks of age from each genotype to characterize disease progression. All animal-related procedures were approved by the St. Jude Children's Research Hospital (St. Jude) Institutional Animal Care and Use Committee.
B-cell proliferation and apoptosis assays.
Proliferation of B220+/IgM+ B cells was measured in vivo with bromodeoxyuridine (BrdU), followed by cell sorting with a Flow Cytometry kit (BD Biosciences PharMingen, San Diego, CA). Briefly, nontransgenic and transgenic wild-type and Puma-deficient mice were injected intraperitoneally with 100 μl BrdU (10 mg/ml) in sterile phosphate-buffered saline. Animals were sacrificed at 12 h postinjection, and spleens were harvested. Splenic B cells (1 × 106) were isolated with a FACScalibur cell sorter (Becton Dickinson, Franklin Lakes, NJ) and antibodies specific for B220, immunoglobulin M (IgM), and BrdU. Apoptosis was measured in splenocytes (1 × 105) with antibodies against B220 and IgM, followed by staining with annexin V and propidium iodide and fluorescence-activated cell sorter (FACS) analysis (36). Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling assays were performed with an ApopTag Fluorescein In Situ Apoptosis Detection kit (Chemicon) by following the provided protocol.
Real-time PCR analysis.
Analysis of primary human BL samples was approved by the St. Jude Institutional Review Board. Total cellular RNA was isolated from primary tumor samples and BL cell lines with RNA/DNA kits (Qiagen, Valencia, CA) and iMACS mRNA isolation columns (Miltenyi Biotec, Auburn, CA), respectively, according to the manufacturers’ recommendations. Complementary DNA was prepared from 1 μg of RNA with the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). A two-step real-time PCR assay was performed with the IQ Sybr green Supermix and iCycler PCR instrument (Bio-Rad) and oligonucleotide primers PUMA-F (5′ACGACCTCAACGCACAGTACGA) and PUMA-R (5′GTAAGGGCAGGAGTCCCATGATGA). The samples were denatured at 95°C, followed by an annealing and extension step at 60.2°C. Relative expression was quantified by the ΔCT method and normalized to human ubiquitin expression, which was determined with primers UB-F (5′ACCTGACCAGCAGCGTCTGATATT) and UB-R (5′TCGCAGTTGTATTTCTGGGCAAGC).
Microarray analysis.
cRNA was synthesized from total cellular RNA with One-Cycle Target Labeling and Control Reagent kits (Affymetrix) and hybridized to 430A mouse chips (Affymetrix). Samples were analyzed with an Affymetrix GeneChip Scanner 3000 and Spotfire software (36).
Methylation-specific PCR analysis.
Genomic DNA was isolated with a DNA extraction kit (Chemicon International Inc., Temecula, CA). As a positive control for methylation, 100 ng of nonmethylated DNA was treated in vitro with SssI methylase (New England BioLabs, Ipswich, MA) to convert all CpG dinucleotides to methylated cytosines. Bisulfite DNA modification was performed with the CpGenome DNA modification kit according to the manufacturer's recommendations (Chemicon). Two sequential PCRs were then performed on the modified DNA with the following degenerate oligonucleotides corresponding to PUMA exon 2: PUMA-exon2F1 (5′GGGTAGGTTGGAGGTATAGTGGGT), PUMA-exon2F2 (5′GGAGTTYGTAGAGGGTTTGGTTYG), and PUMA-exon2R1 CCRATCTCCAACCCTCTCTCTTCC) (R = A/G; Y = C/T). The first amplification step was performed with PUMAexon2F1 and PUMAexon2R1 by denaturing at 95°C for 30 s, annealing at 55°C for 1 min, and extension at 72°C for 1 min. The PCR product was purified and used as a template for a second round of PCR with the PUMAexon2F2 and PUMAexon2R1 primers under the same conditions. The PCR fragments were purified by gel electrophoresis and sequenced by the St. Jude Hartwell Center for Biotechnology. Additional details regarding primer sequences and conditions for examining CpG sites 1 and 3 are available upon request.
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were performed with anti-dimethyl-histone H3 (Lys9) antibody (Upstate, Lake Placid, NY) as described by the manufacturer, with minor modifications. Human BL cells (3 × 106/sample) were treated with 2% paraformaldehyde for 15 min to cross-link proteins and then 1.25 M glycine to terminate the reaction. Cells were harvested and resuspended in piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES)-NP-40 buffer (5 mM PIPES [pH 8.0], 85 mM KCl, 0.5% NP-40), incubated on ice for 10 min, and centrifuged at 5,000 rpm for 5 min. The pellet was resuspended in sodium dodecyl sulfate lysis buffer and processed according to the manufacturer's protocol. The DNA isolated by ChIP was then amplified with the PUMA forward (5′AGTACATCCTCTGGGCTCTGC) and reverse (5′CGGACAAGTCAGGACTTGCAGG) primers by 31 cycles of denaturation at 95°C for 30 s, annealing at 57°C for 30 s, and extension at 72°C for 1 min.
Southern blot analysis.
High-molecular-weight genomic DNA was purified from mouse B-cell lymphoma samples and digested with AflII and BamHI for Arf and p53 analysis, respectively. DNA fragments were separated by agarose gel electrophoresis, transferred to nitrocellulose membranes, and probed with radiolabeled cDNAs coding for Arf (exon 1β) and p53 (exons 2 to 10) as previously described (12).
Western blot analysis.
Mouse tumor tissue and BL cell lines were lysed in NTNE buffer (1% NP-40, 0.01 M triethanolamine-HCl [pH 7.8], 0.15 M NaCl, 5 mM EDTA, protease inhibitor cocktail tablet [Roche], 1 mM phenylmethylsulfonyl fluoride). Lysates were cleared by centrifugation at 13,000 × g for 10 min, and the protein concentration was determined with a bicinchoninic acid protein assay kit (Pierce). Equal quantities of proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis under reducing conditions, transferred to nitrocellulose or polyvinylidene difluoride membrane (Millipore), and blocked in 5% nonfat skim milk in TBS-T (10 mM Tris, 150 mM NaCl, 0.01% Tween 20). Blots were probed with antibodies to the following diluted in a 1% milk TBS-T solution: Puma N terminus (Sigma), p53 (pAb122; Becton Dickinson [BD]), AB7 (Calbiochem), β-actin (AC-15; Sigma), Bax (N20; Santa Cruz), Bcl-XL (clone 44; BD), Bcl-2 (clone 7; BD), Bim-EL (22-40; Calbiochem), Mcl-1 (Rockland Labs), and p19Arf (SC32748; Santa Cruz). Membranes were washed with TBS-T and hybridized with horseradish peroxidase-linked secondary antibodies for 1 h at room temperature. The membranes were washed with TBS-T and developed with Supersignal West Dura Extended Substrate according to the manufacturer's recommendations (Pierce).
RESULTS
Puma deficiency accelerates Eμ-Myc lymphomagenesis.
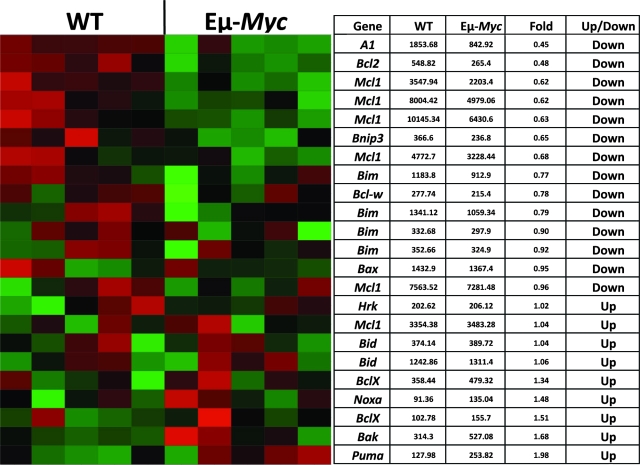
Previous studies established that the p53 tumor suppressor signaling pathway is rate limiting for B-cell tumorigenesis in the Eμ-Myc transgenic mouse model of BL (2, 12, 19, 23, 42, 43). Furthermore, an Eμ-Myc transplant model suggested that p53-mediated apoptosis serves as its principal tumor suppressor activity, as the selection against p53 is abolished by coexpression of the Bcl-2 antiapoptotic protein (43). As shown here, microarray analysis identified Puma as the most highly induced Bcl-2 family member in premalignant Eμ-Myc B cells compared to normal B cells (Fig. 1). Conversely, several survival factors, such as Bcl-2, Mcl-1, and A1, were concomitantly downregulated in the premalignant transgenic B cells. Elevated Puma mRNA levels in premalignant Eμ-Myc B cells and lymphomas were subsequently verified by quantitative real-time PCR (qRT-PCR; see Fig. 4B). We therefore interrogated the contribution of Puma to B-lymphocyte tumor development by crossing Puma−/− and Eμ-Myc transgenic mice. As expected, Eμ-Myc wild-type mice developed B-cell lymphomas at a median age of 15 weeks (Fig. 2) (2, 12). Transgenic Puma+/− mice developed tumors at an intermediate rate (13 weeks; P = 0.08). Notably, loss of Puma accelerated lymphomagenesis, as Eμ-Myc; Puma−/− littermates developed tumors with an average latency of 11 weeks (P ≤ 0.002). Tumor onset in these mice is similar to that previously reported for Eμ-Myc; Bax−/− mice (11).
FIG. 1.
Expression profiles of Bcl-2 family members in normal and premalignant Eμ-Myc B cells. Splenic B cells (B220+/IgM+) from aged-matched wild-type (WT) and Eμ-Myc transgenic mice were analyzed for the expression of Bcl-2-related genes by Affymetrix as described in Materials and Methods. In some cases, multiple probe sets for a particular gene are represented. Puma is the most highly induced Bcl-2 family member in premalignant Eμ-Myc B cells compared to nontransgenic normal B lymphocytes.
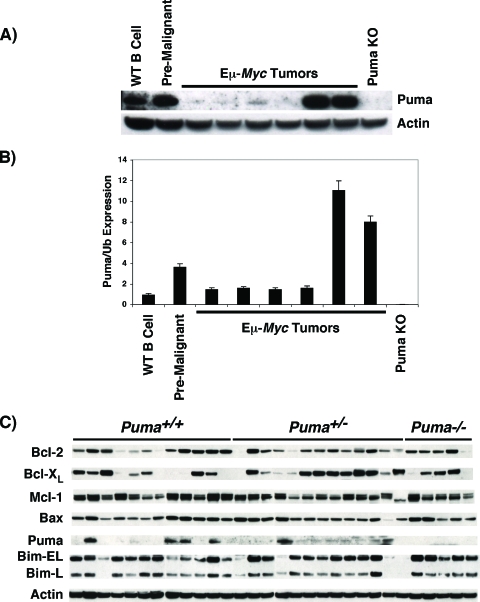
FIG. 4.
Selection against Puma protein expression in Eμ-Myc lymphomas. Normal splenic B lymphocytes (pooled B220+ B cells isolated from four wild-type mice, WT B cell), premalignant Eμ-Myc B cells (pooled B220+ B cells from four transgenic mice, Premalignant), and Eμ-Myc lymphomas (independent tumor samples) were analyzed for Puma expression by (A) Western blotting and (B) qRT-PCR. Samples in panel A are identical to and arranged in the same order as those in panel B. The Puma mRNA level was normalized to the Ubiquitin mRNA level and compared to that of WT B cells, which was arbitrarily set to a value of 1. (C) Expression of Bcl-2 family members in primary Eμ-Myc lymphomas from Puma+/+ (n = 12), Puma+/− (n = 13), and Puma−/− (n = 5) mice was determined by Western blot analysis as described in Materials and Methods.
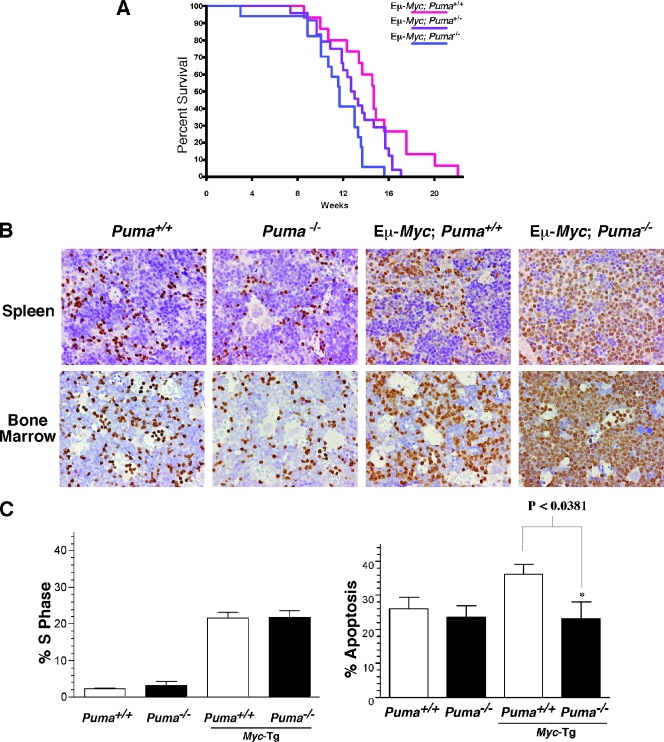
FIG. 2.
Puma deficiency accelerates Myc-induced lymphomagenesis. (A) Tumor-free survival of Eμ-Myc; Puma−/− (n = 17, blue), Eμ-Myc; Puma+/− (n = 24, purple), and Eμ-Myc; Puma+/+ (n = 15, red) animals. (B) Early infiltration of premalignant B cells in the spleens and bone marrow of Eμ-Myc; Puma−/− mice. Histology of spleen tissue (upper) and bone marrow (lower) from Puma+/+, Puma−/−, Eμ-Myc; Puma+/+, and Eμ-Myc; Puma−/− mice. Specimens were stained with the B-cell marker Pax-5 (data shown are representative of three independent experiments). (C) Puma deficiency protects against Eμ-Myc transgenic B-cell apoptosis without altering Myc-induced cell proliferation. Splenic B220+/IgM+ B cells from 7-week-old mice were analyzed by FACS for cell proliferation (left) and apoptosis (right) (n = 3 individual animals of each genotype).
To assess the possible effects of Puma loss on the precancerous phase of this malignancy, we analyzed spleen and bone marrow specimens from 7-week-old Puma+/+, Puma−/−, Eμ-Myc, and Eμ-Myc; Puma−/− littermates. Histopathological analysis of B-lymphocyte markers Pax5 and B220 demonstrated a significant increase in the numbers of B cells within both the spleens and bone marrow of Eμ-Myc; Puma−/− mice compared to those of Eμ-Myc mice (Fig. 2B and data not shown), consistent with the shortened survival of Puma-deficient Myc transgenic mice (Fig. 2A). Therefore, Puma plays a physiological role in suppressing Myc-induced murine B-cell lymphomagenesis.
Puma deficiency alleviates c-Myc-induced apoptosis of B lymphocytes.
Activation of p53 by c-Myc induces the expression of genes involved in cell cycle arrest and cell death (12, 48, 53). To determine the consequence of Puma loss on the proliferative capacity of premalignant Eμ-Myc B cells, 7-week-old littermate Eμ-Myc control and Eμ-Myc; Puma−/− mice were injected with BrdU and 12 h later spleens were harvested for the isolation of B220+/IgM+ B lymphocytes. As expected (36), Eμ-Myc transgenic mouse B lymphocytes displayed a markedly higher number of cells in S phase in comparison to those of their nontransgenic counterparts (Fig. 2C, left panel). However, deletion of Puma had no overt effect on this process, indicating that the cooperation between Puma loss and c-Myc overexpression in B-cell lymphomagenesis is not associated with alterations in cell proliferation.
Apoptosis is an early event that restricts the development of B-cell lymphomas in response to c-Myc overexpression. We previously demonstrated that Puma−/− mouse embryo fibroblasts are highly resistant to c-Myc-induced cell death (25). We therefore reasoned that Puma loss would accelerate lymphomagenesis in Eμ-Myc transgenic animals by reducing the propensity of B cells to undergo apoptosis in response to oncogenic stress. To test this hypothesis, we measured the degree of apoptosis in primary B cells freshly isolated from 7-week-old Eμ-Myc and Eμ-Myc; Puma−/− littermate mice by annexin V and propidium iodide staining, followed by FACS analysis (Fig. 2C, right panel). The levels of apoptosis were similar between wild-type and Puma knockout B lymphocytes, indicating that Puma does not contribute to lymphocyte death during normal B-cell homeostasis. As expected, Eμ-Myc B cells displayed a marked increase in apoptosis over nontransgenic controls. By contrast, the level of apoptosis in Eμ-Myc; Puma−/− B cells was significantly less than that of wild-type Eμ-Myc B cells (P < 0.0381) and indeed was roughly equivalent to that in nontransgenic lymphocytes (Fig. 2C). Therefore, inactivation of Puma impairs apoptosis in premalignant c-Myc transgenic B cells, a scenario that intuitively would foster lymphomagenesis.
Selection against the p53 tumor suppressor pathway in Eμ-Myc/Puma-deficient lymphomas.
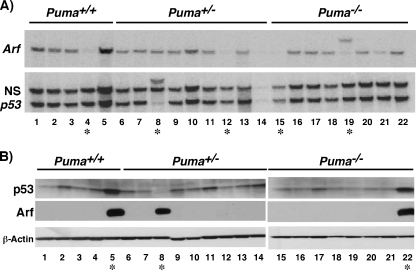
The attenuation of c-Myc-induced apoptosis in Puma knockout B cells would be predicted to bypass events that inactivate the p53 tumor suppressor pathway during lymphomagenesis. To test this hypothesis, the integrity of the p53 tumor suppressor signaling pathway in Eμ-Myc transgenic Puma+/+, Puma+/−, and Puma−/− lymphomas was examined by monitoring the status of p53, Arf, and Mdm2 by Southern and Western blot analyses (Fig. 3A and B and data not shown). Lymphomas expressing mutant p53 and consequently high levels of p19Arf (12) or those harboring Arf deletions were observed in each of the three Eμ-Myc; Puma genotypes, including the Puma knockouts, to various degrees. Therefore, Puma deficiency is not sufficient to eliminate the selective pressure against the p53 tumor suppressor pathway during Myc-induced lymphoma development.
FIG. 3.
The p53 tumor suppressor pathway is bypassed in Puma-deficient lymphomas. Eμ-Myc transgenic lymphomas from Puma+/+ (n = 5), Puma+/− (n = 9), and Puma−/− (n = 8) mice were analyzed for changes in p53 and p19Arf gene expression. (A) Southern blot analysis. (B) Western blot analysis. Lymphoma samples with chromosomal alterations in Arf (no. 4, 12, 15, and 19) or mutations in p53 (no. 5, 8, and 22) are highlighted by asterisks.
Myc suppresses the expression of Bcl-2 and Bcl-XL and induces Puma mRNA and protein expression in premalignant Eμ-Myc B cells, resulting in cell death (Fig. 1, 2, and 4A and B) (13, 32). It is also well established that the overexpression of Bcl-2 or the targeted deletion of Bax or Bim accelerates Eμ-Myc lymphomagenesis (10, 11, 43, 44). Therefore, B cells must overcome the apoptotic response elicited by c-Myc for lymphomagenesis to proceed. Remarkably, B-cell lymphomas arising in Eμ-Myc; Puma+/+ and Eμ-Myc; Puma+/− mice naturally select against Puma protein expression. Indeed, Puma levels are significantly reduced in the majority of lymphomas compared to premalignant Eμ-Myc; Puma+/+ B cells and nontransgenic normal B cells (Fig. 4A and B). Loss of Puma protein expression does not appear to be completely accounted for by a reduction in mRNA levels, implying a posttranscriptional mechanism of regulation (Fig. 4A and B). Analysis of an extended panel of Eμ-Myc lymphomas from Puma+/+ and Puma+/− mice demonstrates that approximately 75% (19/25) of the B-cell lymphomas are deficient in Puma protein. In contrast, the BH3-only family member Bim (both the Bim-EL and Bim-L isoforms), which has also been shown to harness lymphoma development in Eμ-Myc transgenics (10), was expressed in nearly all Eμ-Myc lymphomas (26/30), regardless of their Puma status (Fig. 4C). Puma deficiency also had little or no effect on the expression of other Bcl-2 family members in Eμ-Myc lymphomas, indicating that there were no obvious compensatory effects of Puma loss (Fig. 4C). Similarly, the few Eμ-Myc lymphomas that expressed high levels of Puma protein showed no obvious differences in the expression of other Bcl-2 family members. Finally, there was no selection for loss of the remaining wild-type allele in Eμ-Myc; Puma+/− B-cell lymphomas, and most, if not all, tumors expressed Puma mRNA based on genomic PCR, DNA sequencing, and qRT-PCR (data not shown). Therefore, by these criteria, Puma functions as a tumor modifier rather than a classic tumor suppressor gene and loss of Puma protein expression is a selective event in Eμ-Myc lymphoma.
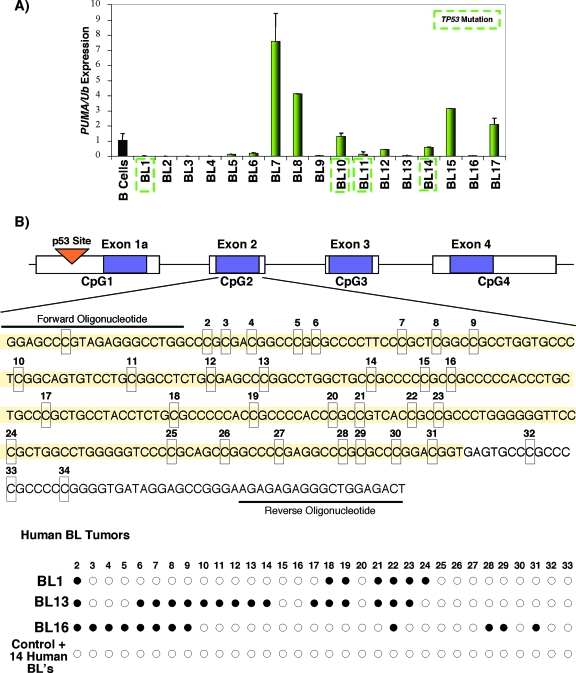
Frequent loss of PUMA expression in primary human BL.
To determine whether PUMA may also limit the development of human BL, we screened a panel of primary tumors for PUMA expression by qRT-PCR. All samples were from subjects with extensive bone marrow involvement and are therefore associated with a less favorable prognosis. Analysis of total cellular RNA isolated from BL samples revealed significantly diminished expression of PUMA in 12 of 17 primary lymphomas compared to levels expressed in a pool of normal B lymphocytes (70% reduction), and of these 12 samples, seven tumors (41%) failed to express any detectable PUMA mRNA (Fig. 5A). The integrity and quantification of each RNA sample was validated by determining the level of Ubiquitin mRNA by qRT-PCR, which was roughly equivalent throughout all 17 BL samples and normal B-cell controls (data not shown). Mutations in TP53 were observed in only 3 of the 12 BL samples with decreased PUMA mRNA levels (and in 1 BL sample with normal PUMA expression) (Fig. 5A), indicating that the loss of PUMA expression likely occurs via a p53-independent mechanism(s) (21).
FIG. 5.
Loss of PUMA expression in human BL correlates with DNA methylation. (A) Primary human BLs (n = 17) were analyzed by real-time PCR for PUMA and Ubiquitin expression and normalized to nontransformed human B-cell transcript levels. TP53 status was previously determined and is boxed in green (21). Equal relative levels of Ubiquitin mRNA across all samples serve as an internal loading control and an indicator of RNA integrity. Representative results of two independent assays performed in triplicate are shown. (B) Genomic structure of PUMA with exons (purple) and predicted CpG islands (white). DNA was subjected to methylation and DNA sequence analysis as described in Materials and Methods. Primary sequence analysis of normal B-cell DNA and DNA modified in vitro with SssI methylase served as negative and positive controls, respectively. Modified sites within exon 2 observed in the primary tumor samples are highlighted by the filled circles. Lymphoma samples B1, B13, and B16 show considerable overlap in methylated CpG sequences within exon 2, and all three tumors have undetectable PUMA mRNA transcripts.
PUMA harbors an unusually high CpG dinucleotide content (ranging from 55 to 76%) throughout its promoter and coding regions. This observation led us to assess whether epigenetic mechanisms, such as DNA methylation, might contribute to the silencing of PUMA expression in human BL. Using established algorithm programs (CpG Island Searcher at http://www.uscnorris.com/cpgislands/cpg.cgi and the European Bioinformatics Institute at http://www.ebi.ac.uk) that identify CpG islands and potential sites of DNA methylation, we defined multiple regions within PUMA that might be susceptible to this modification (Fig. 5B). Both programs identified the same domains, showing that (i) CpG1 spans the PUMA promoter through exon 1a into intron 1; (ii) CpG2 encompasses all of exon 2, which encodes the translation start codon; (iii) CpG3 covers exon 3, which encodes the entire BH3 domain; and (iv) CpG4 contains exon 4 and a portion of the 3′ downstream region. Intronic sequences were not recognized as potential DNA methylation sites, demonstrating specificity for particular regions within the PUMA gene that may be modified.
To determine whether BL tumors with low or no PUMA expression were associated with increased DNA methylation, we analyzed the methylation status of the first three CpG regions of PUMA by using a sodium bisulfite modification and a PCR-based approach (17). Interestingly, three of the six primary BL tumors that lack detectable PUMA expression (samples B1, B13, and B16) exhibited significant DNA methylation within exon 2 (Fig. 5B). The other three tumors and the remaining 11 BL samples, as well as the normal B-cell control, which expressed low, normal, and high levels of PUMA, showed no evidence of methylation in exon 2 or throughout any other region examined. These findings identify discrete sites of DNA methylation in PUMA in a subset of primary human BL tumors that lack detectable levels of PUMA expression. Therefore, DNA methylation may negatively regulate PUMA expression in human cancers.
Activation of PUMA expression by 5-Aza in human BL cell lines.
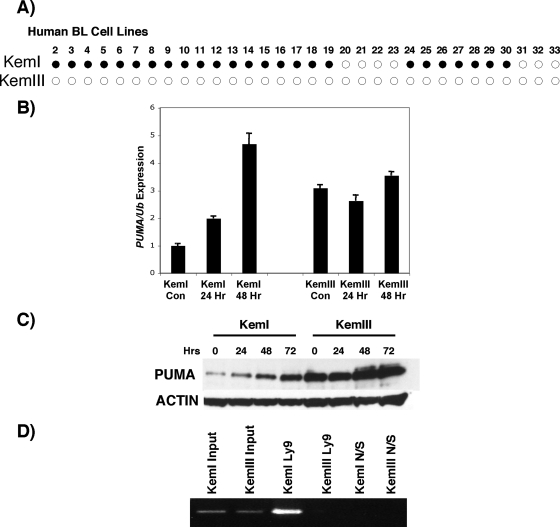
Since the primary human BL tumors are limiting in size and not viable, we further explored the regulation of PUMA expression in a matched set of established human BL cell lines, KemI and KemIII, that were cloned from the same tumor. In culture, KemI cells retain the phenotype of the parental tumor and maintain an epigenetically silenced EBV genome characteristic of EBV-positive BLs. By contrast, EBV latency gene expression within KemIII cells has converted to the viral transcriptional program maintained by EBV-immortalized B-lymphoblastoid cell lines that express the full complement of EBV latent-infection genes. Reactivation of EBV gene expression in KemIII cells, relative to their KemI counterparts and other BL cell lines that epigenetically restrict EBV gene expression, has been attributed to lower levels of the chromatin boundary protein CTCF (5) and may also be a consequence of reduced expression of maintenance and de novo DNA methyltransferases (J. T. Sample, unpublished observation). When we examined the methylation status of exon 2 of the PUMA gene within KemI cells, we observed a pattern similar to that seen in primary tumors that express low levels of PUMA mRNA (Fig. 6). No evidence of DNA methylation was detected in KemIII cells, which express high levels of PUMA mRNA and protein (Fig. 6). Thus, the PUMA gene, like the EBV genome, is subject to DNA methylation in BL.
FIG. 6.
Association of DNA and histone methylation with low PUMA expression in human BL cell lines. (A) Established BL cell lines were screened for DNA methylated sites. KemIII was negative for methylation at all sites, whereas KemI was modified at identical CpG sites detected in primary tumors (Fig. 5). (B) Inhibition of DNA methyltransferases with 5-Aza induces PUMA mRNA expression in KemI but not KemIII cells. Con, control. (C) PUMA protein levels are also induced by 5-Aza in KemI but not KemIII cells. (D) ChIP assay with lysine 9-dimethylated histone H3-specific antibody selectively pulls down the PUMA promoter region flanking the p53 binding site in KemI cells. The input was 1% of the immunoprecipitated material. Normal serum (N/S) served as a negative control. Representative results of three independent experiments are shown.
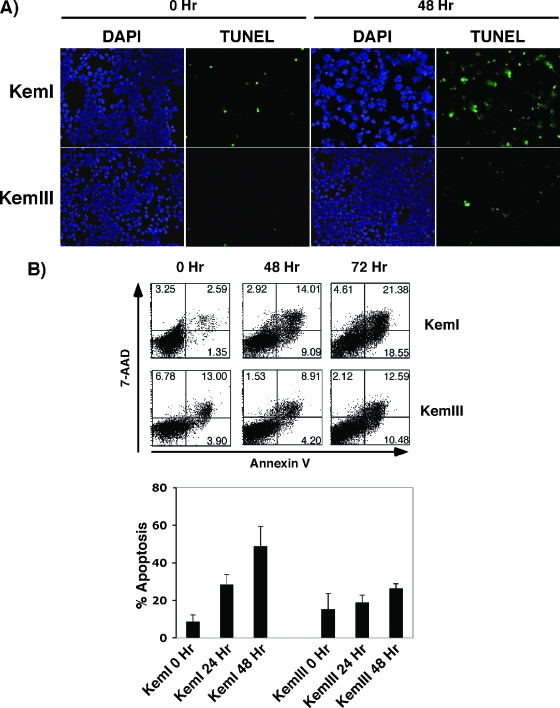
To test the functional significance of DNA methylation to PUMA gene regulation, we treated KemI and KemIII cells with the DNA methyltransferase inhibitor 5-Aza, which derepresses silenced, methylated genes (6). The steady-state levels of PUMA mRNA in control and 5-Aza-treated KemI and KemIII cells were determined by qRT-PCR (Fig. 6B). Treatment of KemI BL cells with 5-Aza resulted in a marked induction of PUMA mRNA, which reached maximal levels by 48 h (about fivefold). By contrast, 5-Aza treatment of KemIII cells had little effect on PUMA expression, as expected for a cell line that displayed no evidence of methylation within PUMA (Fig. 6B). Western blot analysis revealed a similar response for PUMA protein expression, where 5-Aza induced PUMA levels in KemI but not in KemIII cells (Fig. 6C). The induction of PUMA in KemI cells was associated with the acquisition of hallmark features of apoptosis, such as membrane blebbing, nuclear condensation, and annexin V staining (Fig. 7 and data not shown). This response, including the induction of PUMA expression, is p53 independent, as the KemI cells encode mutant p53 (R282W [data not shown]). Collectively, these results are consistent with the silencing of PUMA expression in BL through DNA methylation, which can be relieved by the inhibition of DNA methyltransferases.
FIG. 7.
5-Aza induces apoptosis in epigenetically altered human BL cells. (A) Human BL cells were treated with 5 μM 5-Aza for 48 h. Cell viability was monitored by staining with 4′,6′-diamidino-2-phenylindole (DAPI) and terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL). (B) Apoptosis was quantitated by annexin V-7-amino-actinomycin D (7-AAD), followed by FACS analysis as described in Materials and Methods. Treatment with 5-Aza induced substantial apoptosis in the PUMA DNA-methylated cell line (KemI) but not the nonmethylated cells (KemIII). The averages of three independent experiments are shown.
Although DNA methylation of PUMA does not occur directly within the 5′ promoter regulatory region, previous studies have demonstrated that downstream DNA methylation can serve to nucleate additional 5′ and 3′ epigenetic alterations, including the methylation of chromatin-associated histone proteins (7, 8, 22, 26, 33). Gene silencing also occurs through the dimethylation of histone H3 at lysine 9 (me2-H3-Ly9), which often accompanies DNA methylation (for a review, see reference 41). We therefore examined the PUMA promoter region in KemI and KemIII BL cells for patterns of histone methylation by using a ChIP assay (Fig. 6D). Notably, ChIP analyses of chromatin from KemI and KemIII cells with a me2-H3-Ly9 antibody, followed by PCR with oligonucleotide primers specific for the PUMA promoter region spanning the p53 consensus site to exon 1a (−150 to +50), demonstrated that this promoter fragment was only immunoprecipitated from the KemI BL DNA samples. By contrast, no product was immunoprecipitated with an isotype-matched rabbit polyclonal antibody negative control. Therefore, both DNA and histone H3 methylations occur within the PUMA locus, which correlates with a marked suppression of PUMA gene expression in BL cells.
DISCUSSION
The Puma knockout mouse provides compelling evidence that this BH3-only protein contributes to nearly all of the proapoptotic activity ascribed to the p53 tumor suppressor (25, 46). Puma−/− thymocytes and neuronal cells throughout the developing central nervous system are essentially as resistant to DNA damage as p53-null cells. Puma knockout mice can also withstand doses of ionizing radiation that are sufficient to kill 100% of wild-type animals, which either rivals or exceeds the short-term survival observed for young, p53-deficient mice (J. R. Jeffers and G. P. Zambetti, unpublished results). Moreover, Puma knockout mouse embryo fibroblasts are as resistant to oncogenic stress induced by c-Myc activation as are p53−/− fibroblasts (25). In light of previous findings implicating the apoptotic response as p53's primary tumor suppressor function, one would logically expect that the Puma-deficient mice would be inherently tumor prone. However, this is clearly not the case, as Puma−/− mice do not spontaneously form tumors (15, 25, 46). However, as shown here, Puma deficiency cooperates with c-Myc in accelerating tumor development in a mouse model of BL.
The p53 signaling pathway functions to suppress Eμ-Myc lymphomagenesis. The majority of tumors arising in c-Myc-transgenic mice express mutant p53, lack Arf (required for p53 activation during oncogenic stress), and/or overexpress Mdm2 (the negative regulator of p53) (12, 13). Consistent with these observations, the loss of a single allele of Mdm2 delays tumor formation (2) whereas Arf deletion accelerates the onset of lymphomagenesis (12, 42). Similarly, the loss of Bax or Bim in the context of Eμ-Myc mice shortens the latency of lymphomagenesis to an extent that is similar to that manifested by loss of Puma, indicating that cell death actively limits tumor development (10, 11). However, Bax and Bim do not appear to be naturally targeted in Eμ-Myc lymphomas (Fig. 4) or most types of human B-cell lymphoma (38) Therefore, other mechanisms for the inactivation of the p53 tumor suppressor pathway likely occur. Our findings demonstrating that the loss of Puma accelerates lymphomagenesis in the mouse, which is consistent with previous work by Lowe and coworkers (20); that Puma protein is not detectable in most Eμ-Myc lymphomas; and that PUMA is silenced in a high percentage of human BL highlight PUMA as one such cooperating factor.
The loss of Puma, Bim (10), or Bax (11) accelerates lymphoma onset, but the effect of this loss on tumorigenesis is not as striking as the effects of Bcl-2 overexpression, where Eμ-Myc; Eμ-Bcl-2 double transgenic mice are often born with lympholeukemia and die by 5 to 6 weeks of age (44; M. A. Hall and J. L. Cleveland, unpublished data). Thus, although Puma, Bim, and Bax play important roles in restricting lymphomagenesis, there also appears to be some redundancy in their regulation of apoptosis, which is overcome by Bcl-2 overexpression. Further, the effects of Puma loss on lymphomagenesis do not match those manifested by Arf loss, where no Eμ-Myc; Arf−/− mice survive past 7 to 8 weeks (12). Interestingly, mutations in p53 and loss of Arf were detected in a subset of Eμ-Myc; Puma−/− lymphomas. These results suggest that although Puma deficiency can enhance tumorigenesis, there remains a selective pressure against other activities imparted by Arf or p53, such as cell cycle arrest and senescence, which also contribute to tumor suppression (16, 43, 51). Conversely, the inactivation of the p53 pathway through the deletion of Arf relieves the selection against Puma protein expression in Eμ-Myc; Arf−/− lymphomas (see Fig. S1 in the supplemental material). Based upon these findings, we speculate that in the absence of Puma, p53 still becomes activated in response to deregulated c-Myc expression and suppresses the development of emerging lymphoma cells, no longer principally through the regulation of apoptosis but by blocking cell proliferation and/or inducing cell senescence (16, 45, 51). Therefore, for lymphomas to become fully established, they must likely overcome not only p53-mediated apoptotic responses but also cell cycle inhibition and cell senescence activities, which may explain why the Puma-null mice are not inherently prone to spontaneous tumors (12, 25, 46). The coexistence of p53 mutations in human BL tumors with no or low PUMA expression (Fig. 5A) is also consistent with this hypothesis.
Only 5 of the 12 primary human BL tumors with decreased levels of PUMA transcripts can be accounted for by p53 mutations and/or DNA methylation, and the mechanism(s) contributing to diminished PUMA expression in the remaining 7 tumors is currently unknown. The fact that all BL tumors were sequenced and found to be wild type for PUMA within both the promoter and coding regions (data not shown) eliminates the likelihood of mutations as a cause of the loss of PUMA gene expression. For those human lymphomas with elevated PUMA mRNA levels (e.g., B7 and B8 [Fig. 5]), these tumors may lack PUMA protein, a scenario that we have shown to occur in ∼75% of Eμ-Myc lymphomas, or they may have acquired downstream alterations within the apoptotic pathway. Alternatively, these tumors may tolerate the overexpression of PUMA by upregulating the expression of antiapoptotic factors, in particular, BCL-2, BCL-XL, and/or MCL-1. However, at least within the context of mouse Eμ-Myc B-cell lymphomas, compensation by Bcl-2 family members in response to deregulated Puma expression seems unlikely, as the 25% of tumors that expressed Puma did not show obvious alterations in the expression of other Bcl-2 family members (Fig. 4C). Regardless, the fact that PUMA is suppressed at the mRNA level in ∼70% of human BLs by epigenetic or other means and in 75% of murine Eμ-Myc lymphomas at the protein level underscores the importance of understanding pathways that control Puma expression in tumorigenesis.
Our studies establish Puma as a tumor modifier gene that limits lymphomagenesis, and presumably this property applies to other tumor types as well. More importantly, PUMA is undetectable in approximately 40% of primary human BLs and here the loss of PUMA expression is often associated with specific sites of DNA hypermethylation. Further, analyses of BL cell lines confirmed and extended these findings by demonstrating that the 5′ promoter region of PUMA harbors transcriptionally inactive histone H3 marks (me2-H3-Ly9) and that inhibition of DNA methyltransferases significantly induces PUMA expression. These findings provide compelling evidence that PUMA is inactivated in BL and that this may occur through epigenetic mechanisms.
The application of DNA methyltransferase inhibitors such as 5-Aza (decitabine) for the treatment of human hematopoietic malignancies and dysplasias is currently being tested in clinical trials with promising results (24, 27, 31). PUMA is one such target of 5-Aza that limits tumorigenesis. We speculate that the restoration of PUMA expression in high-grade BL may hold therapeutic potential.
Supplementary Material
Acknowledgments
We thank the Hartwell Center and Flow Cytometry and Cell Sorting Facility at St. Jude Children's Research Hospital for their technical assistance, John T. Sandlund for providing the BL samples, Guangchun Son for bioinformatic analyses, and Jeremy Graff for critically reading the manuscript.
This work was supported in part by NIH grants (CA63230, CA71907, CA76379, and CA73544) and NIH/NCI Cancer Center Support CORE grant CA21765. We are also grateful to the American Lebanese Syrian Associated Charities of St. Jude Children's Research Hospital for their generous support.
Footnotes
Published ahead of print on 23 June 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adams, J. M., A. W. Harris, C. A. Pinkert, L. M. Corcoran, W. S. Alexander, S. Cory, R. D. Palmiter, and R. L. Brinster. 1985. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 318533-538. [DOI] [PubMed] [Google Scholar]
- 2.Alt, J. R., T. C. Greiner, J. L. Cleveland, and C. M. Eischen. 2003. Mdm2 haploinsufficiency profoundly inhibits Myc-induced lymphomagenesis. EMBO J. 221442-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boxer, L. M., and C. V. Dang. 2001. Translocations involving c-myc and c-myc function. Oncogene 205595-5610. [DOI] [PubMed] [Google Scholar]
- 4.Cadwell, C., and G. P. Zambetti. 2001. The effects of wild-type p53 tumor suppressor activity and mutant p53 gain-of-function on cell growth. Gene 27715-30. [DOI] [PubMed] [Google Scholar]
- 5.Chau, C. M., X. Y. Zhang, S. B. McMahon, and P. M. Lieberman. 2006. Regulation of Epstein-Barr virus latency type by the chromatin boundary factor CTCF. J. Virol. 805723-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christman, J. K. 2002. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene 215483-5495. [DOI] [PubMed] [Google Scholar]
- 7.Curradi, M., A. Izzo, G. Badaracco, and N. Landsberger. 2002. Molecular mechanisms of gene silencing mediated by DNA methylation. Mol. Cell. Biol. 223157-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Douglas, D. B., Y. Akiyama, H. Carraway, S. A. Belinsky, M. Esteller, E. Gabrielson, S. Weitzman, T. Williams, J. G. Herman, and S. B. Baylin. 2004. Hypermethylation of a small CpGuanine-rich region correlates with loss of activator protein-2α expression during progression of breast cancer. Cancer Res. 641611-1620. [DOI] [PubMed] [Google Scholar]
- 9.Doumont, G., A. Martoriati, C. Beekman, S. Bogaerts, P. J. Mee, F. Bureau, E. Colombo, M. Alcalay, E. Bellefroid, F. Marchesi, E. Scanziani, P. G. Pelicci, and J.-C. Marine. 2005. G1 checkpoint failure and increased tumor susceptibility in mice lacking the novel p53 target Ptprv. EMBO J. 243093-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egle, A., A. W. Harris, P. Bouillet, and S. Cory. 2004. Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc. Natl. Acad. Sci. USA 1016164-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eischen, C. M., M. F. Roussel, S. J. Korsmeyer, and J. L. Cleveland. 2001. Bax loss impairs Myc-induced apoptosis and circumvents the selection of p53 mutations during Myc-mediated lymphomagenesis. Mol. Cell. Biol. 217653-7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eischen, C. M., J. D. Weber, M. F. Roussel, C. J. Sherr, and J. L. Cleveland. 1999. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 132658-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eischen, C. M., D. Woo, M. F. Roussel, and J. L. Cleveland. 2001. Apoptosis triggered by Myc-induced suppression of Bcl-XL or Bcl-2 is bypassed during lymphomagenesis. Mol. Cell. Biol. 215063-5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.el-Deiry, W. S., T. Tokino, V. E. Velculescu, D. B. Levy, R. Parsons, J. M. Trent, D. Lin, W. E. Mercer, K. W. Kinzler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75817-825. [DOI] [PubMed] [Google Scholar]
- 15.Erlacher, M., V. Labi, C. Manzl, G. Böck, A. Tzankov, G. Häcker, E. Michalak, A. Strasser, and A. Villunger. 2006. Puma cooperates with Bim, the rate-limiting BH3-only protein in cell death during lymphocyte development, in apoptosis induction. J. Exp. Med. 2032939-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finch, A., J. Prescott, K. Shchors, A. Hunt, L. Soucek, T. B. Dansen, L. B. Swigart, and G. I. Evan. 2006. Bcl-xL gain of function and p19 ARF loss of function cooperate oncogenically with Myc in vivo by distinct mechanisms. Cancer Cell 10113-120. [DOI] [PubMed] [Google Scholar]
- 17.Hajkova, P., O. El-Maarri, S. Engemann, J. Oswald, A. Olek, and J. Walter. 2002. DNA-methylation analysis by the bisulfite-assisted genomic sequencing method, p. 143-154. In K. I. Mills and B. H. Ramsahoye (ed.), DNA methylation protocols. Human Press, Totowa, NJ. [DOI] [PubMed]
- 18.Han, J., C. Flemington, A. B. Houghton, Z. Gu, G. P. Zambetti, R. J. Lutz, L. Zhu, and T. Chittenden. 2001. Expression of bbc3, a pro-apoptotic BH3-only gene, is regulated by diverse cell death and survival signals. Proc. Natl. Acad. Sci. USA 9811318-11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hemann, M. T., J. S. Fridman, J. T. Zilfou, E. Hernando, P. J. Paddison, C. Cordon-Cardo, G. J. Hannon, and S. W. Lowe. 2003. An epi-allelic series of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nat. Genet. 33396-400. [DOI] [PubMed] [Google Scholar]
- 20.Hemann, M. T., J. T. Zilfou, Z. Zhao, D. J. Burgess, G. J. Hannon, and S. W. Lowe. 2004. Suppression of tumorigenesis by the p53 target PUMA. Proc. Natl. Acad. Sci. USA 1019333-9338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hemann, M. T., A. Bric, A. Herbst, J. Felstein, C. Cordon-Cardo, J. L. Cleveland, W. P. Tansey, and S. W. Lowe. 2005. Evasion of the p53 tumor surveillance network by tumor-derived myc mutants. Nature 436807-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsieh, C.-L. 1997. Stability of patch methylation and its impact in regions of transcriptional initiation and elongation. Mol. Cell. Biol. 175897-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu, B., M. C. Marin, A. K. el-Naggar, L. C. Stephens, S. Brisbay, and T. J. McDonnell. 1995. Evidence that c-myc mediated apoptosis does not require wild-type p53 during lymphomagenesis. Oncogene 11175-179. [PubMed] [Google Scholar]
- 24.Issa, J. P., G. Garcia-Manero, F. J. Giles, R. Mannari, D. Thomas, S. Faderl, E. Bayar, J. Lyons, C. S. Rosenfeld, J. Cortes, and H. M. Kantarjian. 2004. Phase 1 study of low-dose prolonged exposure schedules of the hypomethylating agent 5-aza-2′-deoxycytidine (decitabine) in hematopoietic malignancies. Blood 1031635-1640. [DOI] [PubMed] [Google Scholar]
- 25.Jeffers, J. R., E. Parganas, Y. Lee, C. Yang, J. Wang, J. Brennan, K. H. MacLean, J. Han, T. Chittenden, J. N. Ihle, P. J. McKinnon, J. L. Cleveland, and G. P. Zambetti. 2003. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell 4321-328. [DOI] [PubMed] [Google Scholar]
- 26.Jones, P. A., and S. B. Baylin. 2002. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 3415-428. [DOI] [PubMed] [Google Scholar]
- 27.Kantarjian, H., J. P. Issa, C. S. Rosenfeld, J. M. Bennett, M. Albitar, J. DiPersio, V. Klimek, J. Slack, C. de Castro, F. Ravandi, R. Helmer III, L. Shen, S. D. Nimer, R. Leavitt, A. Raza, and H. Saba. 2006. Decitabine improves patient outcomes in myelodysplastic syndromes: results of a phase III randomized study. Cancer 1061794-1803. [DOI] [PubMed] [Google Scholar]
- 28.Kim, H., M. Rafiuddin-Shah, H. C. Tu, J. R. Jeffers, G. P. Zambetti, J. J. Hsieh, and E. H. Cheng. 2006. Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat. Cell Biol. 81348-1358. [DOI] [PubMed] [Google Scholar]
- 29.Lindström, M. S., U. Klangby, and K. G. Wiman. 2001. p14ARF homozygous deletion or MDM2 overexpression in Burkitt lymphoma lines carrying wild type p53. Oncogene 202171-2177. [DOI] [PubMed] [Google Scholar]
- 30.Liu, G., J. M. Parant, G. Lang, P. Chau, A. Chavez-Reyes, A. K. El-Naggar, A. Multani, S. Chang, and G. Lozano. 2004. Chromosome stability, in the absence of apoptosis, is critical for suppression of tumorigenesis in Trp53 mutant mice. Nat. Genet. 3663-68. [DOI] [PubMed] [Google Scholar]
- 31.Lyko, F., and R. Brown. 2005. DNA methyltransferase inhibitors and the development of epigenetic cancer therapies. J. Natl. Cancer Inst. 971498-1506. [DOI] [PubMed] [Google Scholar]
- 32.Maclean, K. H., U. B. Keller, C. Rodriguez-Galindo, J. A. Nilsson, and J. L. Cleveland. 2003. c-Myc augments gamma irradiation-induced apoptosis by suppressing Bcl-XL. Mol. Cell. Biol. 237256-7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malumbres, M., I. Perez de Castro, J. Santos, J. Fernandez Piqueras, and A. Pellicer. 1999. Hypermethylation of the cell cycle inhibitor p15INK4b 3′-untranslated region interferes with its transcriptional regulation in primary lymphomas. Oncogene 18385-396. [DOI] [PubMed] [Google Scholar]
- 34.Miyashita, T., and J. C. Reed. 1995. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 80293-299. [DOI] [PubMed] [Google Scholar]
- 35.Nakano, K., and K. H. Vousden. 2001. PUMA, a novel pro-apoptotic gene, is induced by p53. Mol. Cell 7683-694. [DOI] [PubMed] [Google Scholar]
- 36.Nilsson, J. A., U. B. Keller, T. A. Baudino, C. Yang, S. Norton, J. A. Old, L. M. Nilsson, G. Neale, D. L. Kramer, C. W. Porter, and J. L. Cleveland. 2005. Targeting ornithine decarboxylase in Myc-induced lymphomagenesis prevents tumor formation. Cancer Cell 7433-444. [DOI] [PubMed] [Google Scholar]
- 37.Oda, E., R. Ohki, H. Murasawa, J. Nemoto, T. Shibue, T. Yamashita, T. Tokino, T. Taniguchi, and N. Tanaka. 2000. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science 2881053-1058. [DOI] [PubMed] [Google Scholar]
- 38.Peng, H., A. Aiello, G. Packham, P. G. Isaacson, and L. Pan. 1998. Infrequent bax gene mutations in B-cell lymphomas. J. Pathol. 186378-382. [DOI] [PubMed] [Google Scholar]
- 39.Ruf, I. K., P. W. Rhyne, H. Yang, C. M. Borza, L. M. Hutt-Fletcher, J. L. Cleveland, and J. T. Sample. 1999. Epstein-Barr virus regulates c-MYC, apoptosis, and tumorigenicity in Burkitt lymphoma. Mol. Cell. Biol. 191651-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryan, K. M., and K. H. Vousden. 1998. Characterization of structural p53 mutants which show selective defects in apoptosis but not cell cycle arrest. Mol. Cell. Biol. 183692-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Santos-Rosa, H., and C. Caldas. 2005. Chromatin modifier enzymes, the histone code and cancer. Eur. J. Cancer. 412381-2402. [DOI] [PubMed] [Google Scholar]
- 42.Schmitt, C. A., M. E. McCurrach, E. de Stanchina, R. R. Wallace-Brodeur, and S. W. Lowe. 1999. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev. 132670-2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmitt, C. A., J. S. Fridman, M. Yang, E. Baranov, R. M. Hoffman, and S. W. Lowe. 2002. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell 1289-298. [DOI] [PubMed] [Google Scholar]
- 44.Strasser, A., A. W. Harris, M. L. Bath, and S. Cory. 1990. Novel primitive lymphoid tumours induced in transgenic mice by cooperation between myc and bcl-2. Nature 348331-333. [DOI] [PubMed] [Google Scholar]
- 45.Ventura, A., D. G. Kirsch, M. E. McLaughlin, D. A. Tuveson, J. Grimm, L. Lintault, J. Newman, E. E. Reczek, R. Weissleder, and T. Jacks. 2007. Restoration of p53 function leads to tumour regression in vivo. Nature 445661-665. [DOI] [PubMed] [Google Scholar]
- 46.Villunger, A., E. M. Michalak, L. Coultas, F. Mullauer, G. Bock, M. J. Ausserlechner, J. M. Adams, and A. Strasser. 2003. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science 3021036-1038. [DOI] [PubMed] [Google Scholar]
- 47.Vogelstein, B., D. Lane, and A. J. Levine. 2000. Surfing the p53 network. Nature 408307-310. [DOI] [PubMed] [Google Scholar]
- 48.Wagner, A. J., J. M. Kokontis, and N. Hay. 1994. Myc-mediated apoptosis requires wild-type p53 in a manner independent of cell cycle arrest and the ability of p53 to induce p21waf1/cip1. Genes Dev. 82817-2830. [DOI] [PubMed] [Google Scholar]
- 49.Wilda, M., J. Bruch, L. Harder, D. Rawer, A. Reiter, A. Borkhardt, and W. Woessmann. 2004. Inactivation of the ARF-MDM-2-p53 pathway in sporadic Burkitt lymphoma in children. Leukemia 18584-588. [DOI] [PubMed] [Google Scholar]
- 50.Willis, S. N., J. I. Fletcher, T. Kaufmann, M. F. van Delft, L. Chen, P. E. Czabotar, H. Ierino, E. F. Lee, W. D. Fairlie, P. Bouillet, A. Strasser, R. M. Kluck, J. M. Adams, and D. C. Huang. 2007. Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science 315856-859. [DOI] [PubMed] [Google Scholar]
- 51.Xue, W., L. Zender, C. Miething, R. A. Dickins, E. Hernando, V. Krizhanovsky, C. Cordon-Cardo, and S. W. Lowe. 2007. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445656-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu, J., L. Zhang, P. M. Hwang, K. W. Kinzler, and B. Vogelstein. 2001. PUMA induces the rapid apoptosis of colorectal cancer cells. Mol. Cell 7673-682. [DOI] [PubMed] [Google Scholar]
- 53.Zindy, F., C. M. Eischen, D. H. Randle, T. Kamijo, J. L. Cleveland, C. J. Sherr, and M. F. Roussel. 1998. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 122424-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.