Abstract
A genome-wide screen for synthetic lethal (SL) interactions with loss of the nuclear exosome cofactors Rrp47/Lrp1 or Air1 identified 3′→5′ exonucleases, the THO complex required for mRNP assembly, and Ynr024w (Mpp6). SL interactions with mpp6Δ were confirmed for rrp47Δ and nuclear exosome component Rrp6. The results of bioinformatic analyses revealed homology between Mpp6 and a human exosome cofactor, underlining the high conservation of the RNA surveillance system. Mpp6 is an RNA binding protein that physically associates with the exosome and was localized throughout the nucleus. The results of functional analyses demonstrated roles for Mpp6 in the surveillance of both pre-rRNA and pre-mRNAs and in the degradation of “cryptic” noncoding RNAs (ncRNAs) derived from intergenic regions and the ribosomal DNA spacer heterochromatin. Strikingly, these ncRNAs are also targeted by other exosome cofactors, including Rrp47, the TRAMP complex (which includes Air1), and the Nrd1/Nab3 complex, and are degraded by both Rrp6 and the core exosome. Heterochromatic transcripts and other ncRNAs are characterized by very rapid degradation, and we predict that functional redundancy is an important feature of ncRNA metabolism.
The results of genetic and biochemical analyses indicate that the exosome 3′-to-5′ exonuclease complex has potent in vivo RNA degradation activity, even on highly structured RNAs and large RNA-protein (RNP) complexes. However, the purified eukaryotic exosome shows limited activity in vitro, and this has been interpreted as showing its dependence on cofactors for in vivo activity (reviewed in reference 21). Consistent with this idea, several exosome cofactors have been identified. The Ski2/3/8 complex functions with the cytoplasmic exosome and Ski7 during mRNA turnover and surveillance (3, 53). Known nuclear cofactors include the RNA binding proteins Rrp47 (Lrp1) (31, 34), the Nrd1/Nab3 heterodimer (54), and the TRAMP polyadenylation complexes (16, 23, 27, 51, 60). However, analyses of mutations in the exosome complex components have revealed nuclear RNA degradation phenotypes that are not shown by mutations in the TRAMP complexes, Rrp47, or Nrd1/Nab3, strongly suggesting that additional exosome cofactors remained to be identified.
Rrp47 is believed to act as a specific cofactor for the nuclear 3′ exonuclease Rrp6 (31, 44). Although Rrp6 is associated with the nuclear exosome, strains lacking the activity of Rrp6 show RNA processing defects that are distinctly different from those seen following the loss or inactivation of any other exosome component. In contrast, very similar phenotypes are seen in strains with mutations in any of the “core” exosome components that are common to the nuclear and cytoplasmic forms of the complex (reviewed in reference 21). This suggests that the activity of the core exosome requires the intact structure of the complex and is functionally distinct from the activity of Rrp6. The TRAMP complexes appear to function together with both Rrp6 and the core exosome. Saccharomyces cerevisiae has two forms of the TRAMP complex; both contain the essential DEVH box, putative helicase Mtr4, but differ in the presence of the poly(A) polymerase Trf4 in the TRAMP4 complex or the homologous Trf5 in TRAMP5 (23). In addition, each TRAMP complex contains a zinc knuckle protein, either Air1 or Air2; these proteins are ∼60% identical.
Here we have made use of a recently developed screen for the identification of synthetic lethal (SL) genetic interactions termed genetic interaction mapping (GIM) (13) to identify genes that become essential for cell growth in the absence of Rrp47 or Air1. These screens identified several potential SL interactions, including a novel yeast exosome cofactor that we designate Mpp6.
MATERIALS AND METHODS
Strains and microbiological techniques.
Standard procedures were used for the propagation and maintenance of yeast. A full list of strains used in this study can be found in Table 1. All deletion and GAL1-regulated strains were constructed by using a one-step PCR strategy. Transformants were selected for resistance to G418 or nourseothricin (NAT) and screened by PCR and immunoblotting. For depletion of GAL1-regulated Mpp6 and Rrp47, cells were pregrown on permissive medium containing 2% galactose and harvested immediately prior to transfer (zero-hour samples) and 6 h after transfer to nonpermissive medium containing 2% glucose.
TABLE 1.
Strains used in this study
| Strain | Genotypea | Reference or source |
|---|---|---|
| BY4741 | MATahis3D1 leu2D0 met15D0 ura3D0 | 9 |
| D658 | ade2 arg4 leu2-3,112 trp1-289 ura3-52 Csl4-TAP::KlURA3 | Euroscarf |
| YLM151 | As BY4741 but Mpp6-TAP::HIS | 18 |
| D270 | MATaade2 his3 leu2 trp1 ura3 | 57 |
| D271 | MATaade2 his3 leu2 trp1 ura3 | 57 |
| YLM140 | As D270 but mpp6D::NATR | This study |
| YLM142 | As D270 but HIS::GAL1-3HA-mpp6 | This study |
| YLM144 | As D270 but rrp47D::KANR | This study |
| YLM146 | As D270 but HIS::GAL1-3HA-rrp47 | This study |
| YLM148 | AS YLM140 but HIS::GAL1-3HA-rrp47 | This study |
| YLM150 | As YLM144 but HIS::GAL1-3HA-mpp6 | This study |
| YLM152 bob-GFP | As D270 but mpp6-eGFP::KANR | This study |
| YLM155 | As D270 but KANR::GAL1-3HA-rrp41 | This study |
| D342 | ura3-52 mtr3-1 | 26 |
| YLM156 | As D271 but rrp47D::NATR | This study |
| rna14.1 | MATaade2-1 his3-11 leu2-3,112 trp1-1 ura3-1 rna14.1 | 30 |
| YLM160 | As rna41.1 but rrp6D::KANR | This study |
| YLM161 | As rna14.1 but mpp6D::NATR | This study |
| prp2-1 | MATaura3-52 prp2-1 | 35 |
| YLM163 | As prp2-1 but rrp6D::KANR | This study |
| YLM165 | As prp2-1 but mpp6D::NATR | This study |
| BMA38a | MATaade2-1 his3 Δ200 leu2-3,112 trp13-1 ura3-1 can1-100 | 5 |
| YDC40 trf4Δ | As BMA38a but trf4Δ::KANRMX4 | 11a |
| YDC46 air1Δ/air2Δ | As BMA38a but air1Δ::KANRMX4 air2Δ::NATRMX6 | 11a |
| NRD1 | MATα ura3Δ0 his3Δ1 leu2Δ0 met15Δ0 | YJC1163 45 |
| nrd1.102 | MATα ura3Δ0 his3Δ1 leu2Δ0 met15Δ0 nrd1-102 | YJC1166 45 |
| nab3.11 | MATα ade2 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 nab3-11 | 12 |
3HA, triple-hemagglutinin tag; KlURA3, Kluveromyces lactis URA3.
Glycerol gradient analysis.
Glycerol gradient analysis was performed on a 10 to 30% gradient as previously described (31). Protein was precipitated from each fraction by using trichloroacetic acid and resolved by using standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) techniques. The sedimentation profile of Mpp6-TAP was detected by immunoblotting with peroxidase-conjugated rabbit immunoglobulin G (IgG) (Sigma). A polyclonal, antipeptide antibody was raised against Rrp43 (data not shown) and used to indicate the sedimentation profile of the core exosome.
Affinity purification of Mpp6-TAP.
Affinity purification of Mpp6-TAP was performed as previously described (38) with the following modifications. Frozen yeast pellet obtained from exponentially growing cells (optical density of 0.6 at 600 nm) was ground in liquid nitrogen and thawed in extraction buffer (50 mM HEPES, pH 7.4, 50 mM KCl, 5 mM MgCl2, 10% glycerol). Clarified extract was incubated with IgG-Sepharose for 1.5 h at 4°C. Washes and TEV cleavage were performed in buffer containing 50 mM HEPES, 100 mM KCl, 5 mM MgCl2, 0.1% NP-40, and 1 mM dithiothreitol. Calmodulin binding and release was performed as previously described (38).
SDS-PAGE analysis and in-gel digestion.
The entire lane of the silver-stained gel was chopped into small pieces and placed in a 96-well plate for trypsin digestion as described elsewhere (43). In brief, proteins were reduced in 20 mM dithiothreitol for 30 min at 37°C, alkylated in 50 mM iodoacetamide for 30 min at room temperature in the dark, and digested at 37°C overnight with 12.5 ng/μl trypsin (Proteomics; Grade; Sigma). The digestion medium was then acidified to 0.1% of trifluoroacetic acid and spun onto StageTips as described elsewhere (36). Peptides were eluted in 20 μl 80% acetonitrile, 0.5% acetic acid and were concentrated to 2 μl (concentrator 5301; Eppendorf AG, Hamburg, Germany). They were then diluted to 5 μl with 0.1% trifluoroacetic acid for liquid chromatography-tandem mass spectroscopy (LC-MS-MS) analysis.
MS analysis.
An LTQ-Orbitrap MS (ThermoElectron, Bremen, Germany) was coupled online to an Agilent 1100 binary nanopump (Palo Alto, CA) and an HTC PAL autosampler (CTC, Zwingen, Switzerland). To prepare an analytical column with a self-assembled particle frit (24), C18 material (3-μm ReproSil-Pur C18-AQ; Maisch GmbH, Ammerbuch-Entringen, Germany) was packed into a spray emitter (75-μm inner diameter, 8-μm opening, 70-mm length; New Objectives, United States) by using an air pressure pump (Proxeon Biosystems, Odense, Denmark). Mobile phase A consisted of water, 5% acetonitrile, and 0.5% acetic acid, and mobile phase B of acetonitrile and 0.5% acetic acid. The gradient went from 0% to 20% buffer B in 75 min and then to 80% B in 13 min at a 300-nl/min flow speed. The six most-intense peaks of the MS scan were selected in the ion trap for MS-MS (normal scan, wideband activation, filling 5e−5 ions for MS scan and 104 ions for MS-MS, 100-ms maximum fill time, dynamic exclusion for 180 s). The raw files were processed by using DTAsupercharge 0.62 (a kind gift from Matthias Mann, Munich, Germany). The lists of peaks generated were searched against the Saccharomyces Genome Database (version 11.05.2007) by using Mascot 2.0 with the following parameters: 8-ppm peptide tolerance and 0.6-Da MS-MS tolerance for monoisotopic masses and electrospray ionization TRAP parameters of fully tryptic specificity, cysteine carbamidomethylation as fixed modification, oxidation on methionine as variable modification, and two missed cleavage sites allowed. The results were parsed through MSQuant (http://msquant.sourceforge.net/), and a cutoff 5-ppm peptide tolerance applied to the recalibrated list. Peptides with scores of 25 and higher were reported and in individual cases manually validated.
RNA extraction and Northern hybridization.
RNA was extracted as previously described (49). For high-molecular-weight RNA analysis, 4 μg of total RNA was glyoxyl denatured and resolved on a 1.2% agarose gel as previously described (41). Low-molecular-weight RNAs were resolved on standard 6% acrylamide-8.3 M urea gels. The sequences of oligonucleotides used for probes are shown in Table 2. The quantification was carried out by using a Fujifilm FL-5100. The means of the results from three independent experiments are shown, and standard error was used throughout.
TABLE 2.
Oligonucleotides used in this study
| Name | Identification no. | Sequence (5′ to 3′) |
|---|---|---|
| RPS26a | 995 | GTTTGACGTGACCTCTACCT |
| SCR1 | 250 | ATCCCGGCCGCCTCCATCAC |
| MFA2 | 402 | GGCGGGATCCCAGAAGAGGCCCTTGATTAT |
| CYH2 | 405 | GTGCTTTCTGTGCTTACCGATACGACCTTTACCG |
| ITS2 5′B | 020 | TGAGAAGGAAATGACGCT |
| 27SA3 | 003 | TGTTACCTCTGGGCCC |
| 20S | 004 | CGGTTTTAATTGTCCTA |
Immunofluorescence.
For Mpp6-green fluorescent protein (GFP) localization studies, cells were fixed in 3.7% paraformaldehyde at room temperature, spheroplasted by using zymolase, and dehydrated in 70% ethanol overnight at −20°C. To stain nuclear DNA, 4′,6′-diamidino-2-phenylindole (DAPI) was included in the mounting medium (Vectashield; Vector laboratories). Images were captured by using a Coolsnap CCD camera fitted to a DeltaVision RT restoration imaging system. The images captured by using the DeltaVision system were subjected to real-time two-dimensional deconvolution algorithms. Single optical sections were selected following deconvolution and assembled by using Adobe Photoshop software.
RNA binding assays.
Binding of [35S]Mpp6 to homopolymeric nucleotides and biotinylated, in vitro-transcribed RNA was performed as previously described (32, 37). Equivalent amounts of protein recovered from bound, unbound, and total fractions were separated by SDS-PAGE on a 4 to 12% Bis-Tris gel. Following migration, the gel was dried and protein was visualized by autoradiography.
RESULTS
Mpp6 is a novel exosome-associated protein.
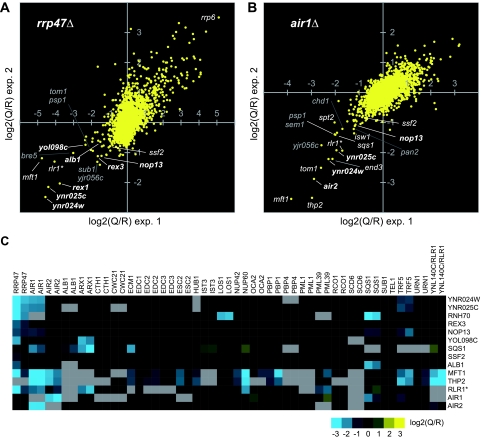
To identify novel factors that functionally interact with the nuclear exosome cofactor Rrp47 or the TRAMP complex, we performed genetic screens with strains lacking Rrp47 or the Air1 component of the TRAMP complex by using the GIM strategy (13). Briefly, query strains were constructed by replacing the KANR marker in the rrp47Δ and air1Δ MATα strains from the systematic gene knock-out collection (19) with a haploid-specific NAT resistance cassette (prMFα2-NAT). These strains were each crossed by being mated en masse to a pool of 4,800 MATa strains from the knockout collection, each of which has a single open reading frame (ORF) replaced by a KANR cassette containing unique “barcode” sequences. The diploids obtained were sporulated en masse, and the resulting haploid double-mutant spores were selected in rich medium containing Geneticin and NAT and grown in competition for about 18 generations. The change in the relative level of the cells bearing both the query mutation and a given deletion in the final population was estimated with comparative barcode-specific microarrays. Ratios between query results and the results for a reference ORF (obtained in experiments using the deletion of a dubious ORF, considered neutral, instead of the deletion of RRP47 or AIR1) were around 1 for no effect and less than 1 for synthetic growth defects (negative values when the logarithms of the query/reference ratios are calculated). Each GIM screen was performed twice and gave reproducible results (Fig. 1A and B). In the data presented in Fig. 1A and B, genes indicated in the lower-left quadrant showed reduced recovery in two independent experiments. The results of the two screens analyzed here were also compared with the results obtained by applying the GIM strategy to other gene deletions (13). Some genes were identified in multiple different screens and may, therefore, be nonspecific (Fig. 1A and B).
FIG. 1.
Identification of YNR024w by genetic screens using the rrp47Δ and air1Δ mutations. (A) The values obtained in two independent GIM screens for the relative growth rate of double-deletion strains using as query the deletion of rrp47 were represented as a scatter plot to assess the reproducibility of the assay and to identify strong interactions. Each point represents log2(Q/R) values {the logarithm in base 2 of the ratio between the average signal measured for a given double mutant xΔ rrp47Δ [query (Q) mutation] and the signal measured for the same deletion when combined with a reference mutation considered to be neutral, xΔ referenceΔ [reference (R) mutation]}. The genes whose deletion was SL or caused synthetic growth inhibition when combined with rrp47Δ are found in the lower-left quadrant and are indicated and shaded differently based on the presumed specificity of the observed genetic interaction, as estimated by the synthetic growth defect scores after performing 41 screens (13). Deletions that appeared to be specific to the screen are indicated in bold. Deletions identified in multiple different GIM screens, which may be nonspecific despite high negative values, are in gray. (B) As described for panel A, but with air1Δ as the query mutation. (C) The genetic interaction profiles for selected genes that showed synergistic growth defects with rrp47Δ and/or air1Δ were retrieved from the published GIM data (13) to assess the specificity of the effects. Query gene deletions are indicated above. The genes showing the largest number of synthetic growth defects, Mft1, Thp2, and Ynl140c, were all components of the THO complex (an ORF that overlaps Rlr1 and is indicated as Rlr1*).
The GIM screens identified several mutant combinations that showed putative SL or synergistic growth inhibition and appeared to be biologically relevant. For example, gene deletions that are underrepresented in the progeny recovered with the rrp47Δ mutation (Fig. 1A) encode two known 3′-to-5′ exonucleases, Rex1 (Rnh70) and Rex3 (52). Other genes identified also participate in RNA metabolism, including factors implicated in RNA polymerase II transcription elongation and mRNA export; the THO complex (11) components Mft1, Thp2, and Rlr1 (represented by the overlapping hypothetical ORF YNL140C); and the ribosome synthesis factors Nop13 (59), Ssf2 (17), and Alb1 (28). The deletion of the largely uncharacterized ORF YOL098C, encoding a putative metalloprotease (25), also yielded a synergistic growth defect with the rrp47Δ mutation. The screen with the air1Δ mutation identified the homologous Air2 protein, as expected, but also the components of the THO complex (Mft1, Thp2, and Rlr1) and ribosome synthesis factors, including Nop13, Ssf2, and Sqs1 (Fig. 1B). Rrp47 functions as a cofactor for the nuclear exonuclease Rrp6, and the rrp6Δ mutation conferred improved relative growth in the presence of the rrp47Δ mutation. In other GIM screens, such improved relative growth was also seen for other pairs of mutations affecting the same process, and this was attributed to epistasis by Decourty et al. (13).
Two gene deletions were identified in both screens and appeared to be highly specific to the rrp47Δ and air1Δ mutations (Fig. 1C shows profiles obtained with various factors in a number of subsequent GIM screens [data extracted from reference 13]). These were YNR024W and the overlapping hypothetical ORF YNR025C, which is effectively an independent mutation of YNR024W, since YNR025C is probably not a bona fide gene. These interactions suggested that Ynr024w could be a novel cofactor for the nuclear exosome.
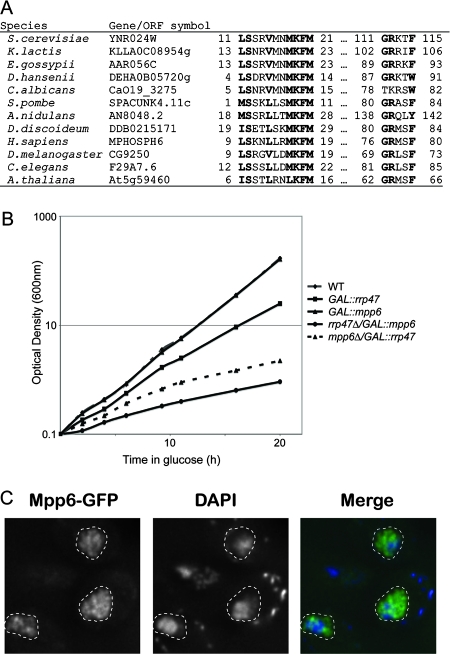
Ynr024w is a nonessential, 21-kDa protein, with potential exosome interactions from high-throughput proteomic data (22). BLAST searches with Ynr024w only yielded homologues in other Saccharomycetaceae. However, the results of bioinformatic analyses identified it as a homologue of the mammalian exosome-associated protein M-phase phosphoprotein 6 (Mpp6 or Mphosph6). Using mammalian and fission yeast (Schizosaccharomyces pombe) Mpp6 homologues (42) as starting points for consecutive PSI-BLAST searches and HMMer searches based on truncated multiple sequence alignments, we found several additional fungal homologues. This enabled us to identify two consensus motifs for Mpp6 homologues (Fig. 2A), both of which occur in a single Saccharomyces cerevisiae sequence, namely, Ynr024w. The P value for random cooccurrence of these motifs in a single sequence is less than 0.0015. Human Mpp6 is similar in size to Ynr024w and was reported to physically interact with the human exosome (42). YNR024W was therefore designated Mpp6 and selected for detailed analysis.
FIG. 2.
Ynr024w is the yeast homologue of the human exosome cofactor Mpp6. (A) The alignment shows the two conserved Mpp6 motifs from model eukaryotes, with consensus residues in bold. Start and end positions are indicated. K. lactis, Kluyveromyces lactis; E. gossypii, Eremothecium gossypii; D. hansenii, Debaryomyces hansenii; C. albicans, Candida albicans; A. nidulans, Aspergillus nidulans; D. discoideum, Dictyostelium discoideum; H. sapiens, Homo sapiens; D. melanogaster, Drosophila melanogaster; C. elegans, Caenorhabditis elegans; and A. thaliana, Arabidopsis thaliana. (B) Double-mutant strains carrying either the mpp6Δ PGAL-RRP47 or the rrp47Δ PGAL-MPP6 mutations are inhibited in growth 6 h after transfer to glucose medium. The wild-type (WT) strain and mutant strains carrying the PGAL-RRP47, PGAL-MPP6, mpp6Δ PGAL-RRP47, or rrp47Δ PGAL-MPP6 mutations were grown at 25°C in rich medium containing 2% galactose and then transferred for the time indicated on the x axis to rich medium containing 2% glucose. (C) Mpp6-GFP is restricted to the nucleus. In the merged image, Mpp6-GFP is shown in green and DAPI in blue. The nucleus is outlined with a dotted line.
To confirm the predicted genetic interaction, the rrp47::KAN strain was crossed individually to an mpp6::NAT strain. Among 52 haploid progeny tested, no NATR KANR strains were recovered, indicating that the mutations are indeed SL. Rrp47 functions as a cofactor for the exonuclease Rrp6, and we also tested the mpp6::NAT strain in combination with the rrp6::KAN strain. No double-mutant progeny were recovered from 71 haploids tested, showing the mpp6Δ and rrp6Δ mutations to be SL for spore viability in combination. In order to allow phenotypic analyses and to test the phenotype of the double mutant in mitotic cells, we constructed double-mutant strains carrying either the mpp6Δ PGAL-RRP47 or the rrp47Δ PGAL-MPP6 mutations. On plates, both strains were viable for growth on permissive galactose medium but unable to grow on glucose medium (data not shown). In liquid medium, the growth of either double mutant was detectably inhibited 6 h following transfer to glucose medium (Fig. 2B) and both GAL-regulated proteins were undetectable by Western blot (data not shown). We conclude that the absence of Mpp6 is lethal in combination with the absence of either Rrp6 or its cofactor Rrp47.
To determine the subcellular localization of Mpp6, a C-terminal GFP fusion was constructed in the genome under the control of the endogenous MPP6 promoter (Fig. 2C). Comparison with the results of DAPI staining of the DNA showed the Mpp6-GFP fusion to be restricted to the nucleus, but no clear subnuclear localization was evident. The nuclear localization of Mpp6 is consistent with the SL interactions with Rrp47 and Rrp6, which are also nuclear specific in yeast.
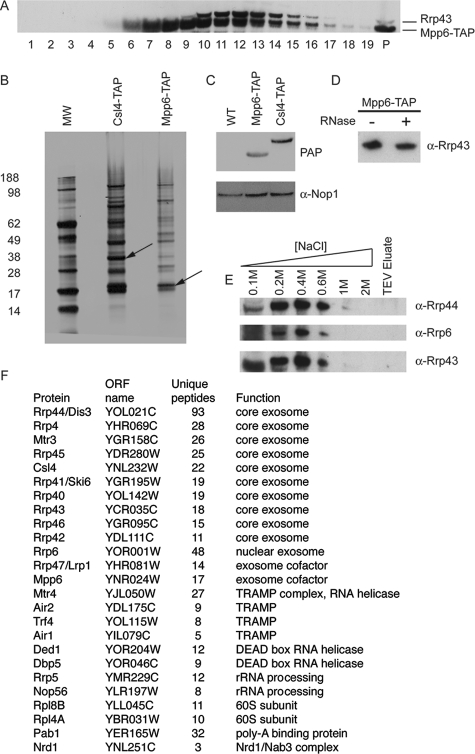
To assess the physical association between Mpp6 and the exosome, a C-terminal TAP fusion was constructed in the genome under the control of the endogenous MPP6 promoter. The results of glycerol gradient analysis followed by Western blotting showed good cosedimentation of Mpp6-TAP with the core exosome component Rrp43, and both proteins sedimented with the velocity expected for the exosome from the results of previous analyses (Fig. 3A). To confirm this association, we compared the proteins that copurified with the TAP-tagged core exosome component Csl4-TAP and with Mpp6-TAP (Fig. 3B). A similar band pattern was visible in each case, but the amount of protein recovered with Mpp6-TAP was substantially lower. Comparisons of total protein preparations revealed that Mpp6-TAP is approximately threefold less abundant than Csl4-TAP (Fig. 3C). The results of previous analyses indicated that approximately one-third of the total exosome population is associated with Rrp6, and this is assumed to represent the nuclear exosome fraction (2). To test whether the association of Mpp6 with the exosome was mediated by substrate RNAs, Mpp6-TAP bound to IgG was treated with RNase A prior to elution by TEV cleavage (Fig. 3D). RNase A treatment did not affect the association of Mpp6 with the exosome component Rrp43. To assess the stability of the Mpp6-exosome interaction, Mpp6-TAP bound to IgG was washed with increasing salt concentrations (Fig. 3E). The association of the core exosome components Rrp43 and Rrp44 with Mpp6-TAP was largely resistant to 0.1 M NaCl and progressively lost at salt concentrations of from 0.2 M to 0.6 M NaCl. The retention of a significant fraction of the association after washing with 0.4 M NaCl indicates that the Mpp6-exosome interaction is relatively robust.
FIG. 3.
Mpp6 physically associates with the exosome. (A) Glycerol gradient analysis of a C-terminal Mpp6-TAP fusion protein. Cell lysate from a strain expressing Mpp6-TAP was loaded on to a 10-to-30% glycerol gradient. Lane 1 is the top of the gradient, and P the pellet. Western blotting was done using peroxidase antiperoxidase, which recognizes the protein A region of the TAP tag, and a polyclonal anti-Rrp43 antibody. (B) Silver-stained gel of affinity purification of Csl4-TAP- and Mpp6-TAP-containing complexes. Arrows indicate the positions of the tagged proteins. MW, molecular weight in thousands. (C) Western analysis of total protein extracts from a wild-type (WT) strain and strains expressing either Mpp6-TAP or Csl4-TAP using peroxidase antiperoxidase (PAP) and an anti-Nop1 antibody. (D) Western blot showing the recovery of the core exosome component Rrp43 with TAP-purified Mpp6 following (+) or without (−) treatment of the IgG-bound complex with RNase A. (E) Western blot showing the elution of the exosome components Rrp43, Rrp44, and Rrp6 from Mpp6-TAP bound to IgG in the presence of increasing salt concentrations. (F) Proteins identified as coprecipitated with Mpp6 by LC-MS. α, anti.
Proteins coprecipitated with Mpp6-TAP were identified by LC-MS (Fig. 3F). All 11 components of the nuclear exosome complex were identified (Rrp4, Rrp40 to Rrp46, Mtr3, Csl4, and Rrp6), as were its characterized cofactors, including both TRAMP complexes (Trf4, Trf5, Air1, Air2, and Mtr4), Nrd1, and Rrp47. A number of other nuclear RNA binding proteins were also identified, including Pab1 and Ded1.
The data are consistent with the association of most or all of the nuclear exosome population with Mpp6 and with quantitative association of Mpp6 with the nuclear exosome.
The potential RNA binding activity of Mpp6 was tested in vitro (Fig. 4). MPP6 was transcribed in vitro, and Mpp6 was then 35S labeled during translation. The binding substrates were poly(U) Sepharose and biotinylated poly(dT), poly(A), and poly(A/U) bound to streptavidin agarose. In addition, RNAs corresponding to the D-A2 region of the pre-rRNA internal transcribed spacer 1 (ITS1) or tRNAimet were biotinylated during in vitro transcription. Mpp6 bound efficiently to poly(U) with substantially weaker binding to oligo(dT) and was not recovered above the level of the background control (protein A Sepharose or streptavidin agarose without bound RNA) with the other RNAs tested. We conclude that Mpp6 has robust in vitro binding activity on a single-stranded poly(U) RNA. However, it seems likely that additional factors influence binding-site selection in vivo.
FIG. 4.
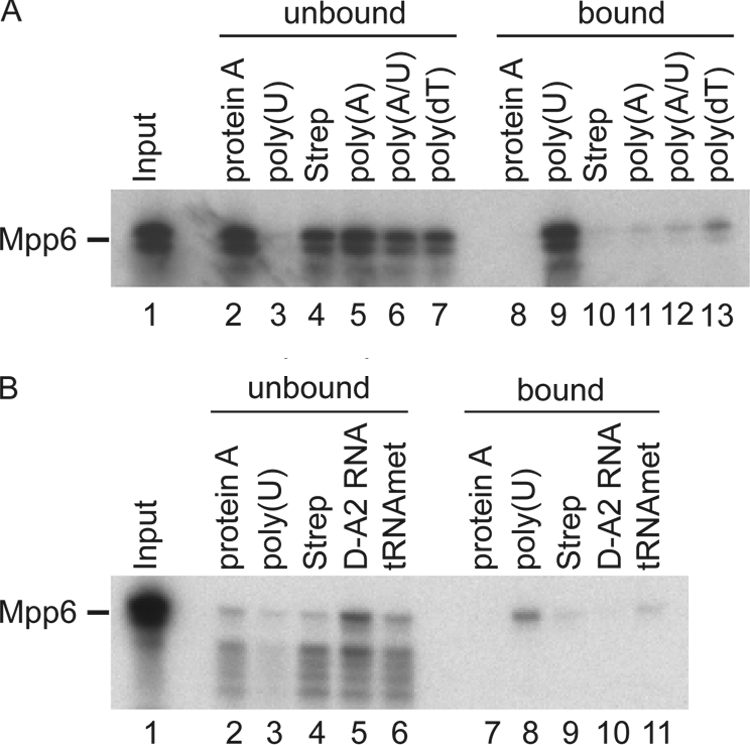
Mpp6 displays in vitro RNA binding activity. 35S-labeled Mpp6 was generated from a TNT reaction and incubated with resins carrying the factors indicated. Following incubation, proteins were recovered from bound and unbound fractions. Equivalent amounts were run along with the corresponding input (lane 1) on a 4 to 12% Bis-Tris gradient acrylamide gel and visualized by autoradiography. Resins used were protein A-Sepharose (protein A), poly(U)-Sepharose [poly(U)], streptavidin agarose alone (Strep), and streptavidin-agarose loaded with either biotinylated A20 [poly(A)], biotinylated A20/U20 [poly(A/U)], biotinylated dT20 [poly(dt)], or an in vitro-transcribed and biotinylated pre-rRNA fragment (D-A2 RNA) or tRNAimet (tRNAmet).
Mpp6 functions together with the nuclear exosome.
To assess the potential role of Mpp6 in nuclear RNA processing and degradation, RNA was extracted from the mpp6Δ GAL::rrp47 and rrp47Δ GAL::mpp6 strains and the corresponding single mutants during growth on permissive galactose medium and following transfer to glucose medium for 6 h. Strains lacking Rrp47 have a pronounced defect in 3′ maturation of the 5.8S rRNA (31), with very strong accumulation of a species with a 30-nt extension, 5.8S+30 (Fig. 5A, lane 3). The mpp6Δ strain and GAL::mpp6 strain grown on glucose showed mild accumulation of 5.8S+30; however, this accumulation appears low partly because 5.8S+30 is highly abundant in the rrp47Δ strain, in which it can readily be seen by ethidium staining of total RNA. Most yeast mutants with defects in ribosome synthesis show low levels of pre-rRNA accumulation, consistent with rapid surveillance (reviewed in reference 56). The double-mutant strains with rrp47 were also not more impaired in 5.8S maturation than the corresponding rrp47Δ and GAL::rrp47 strains (Fig. 5A). The loss of Mpp6 had little effect on the levels of other pre-rRNA species examined.
FIG. 5.
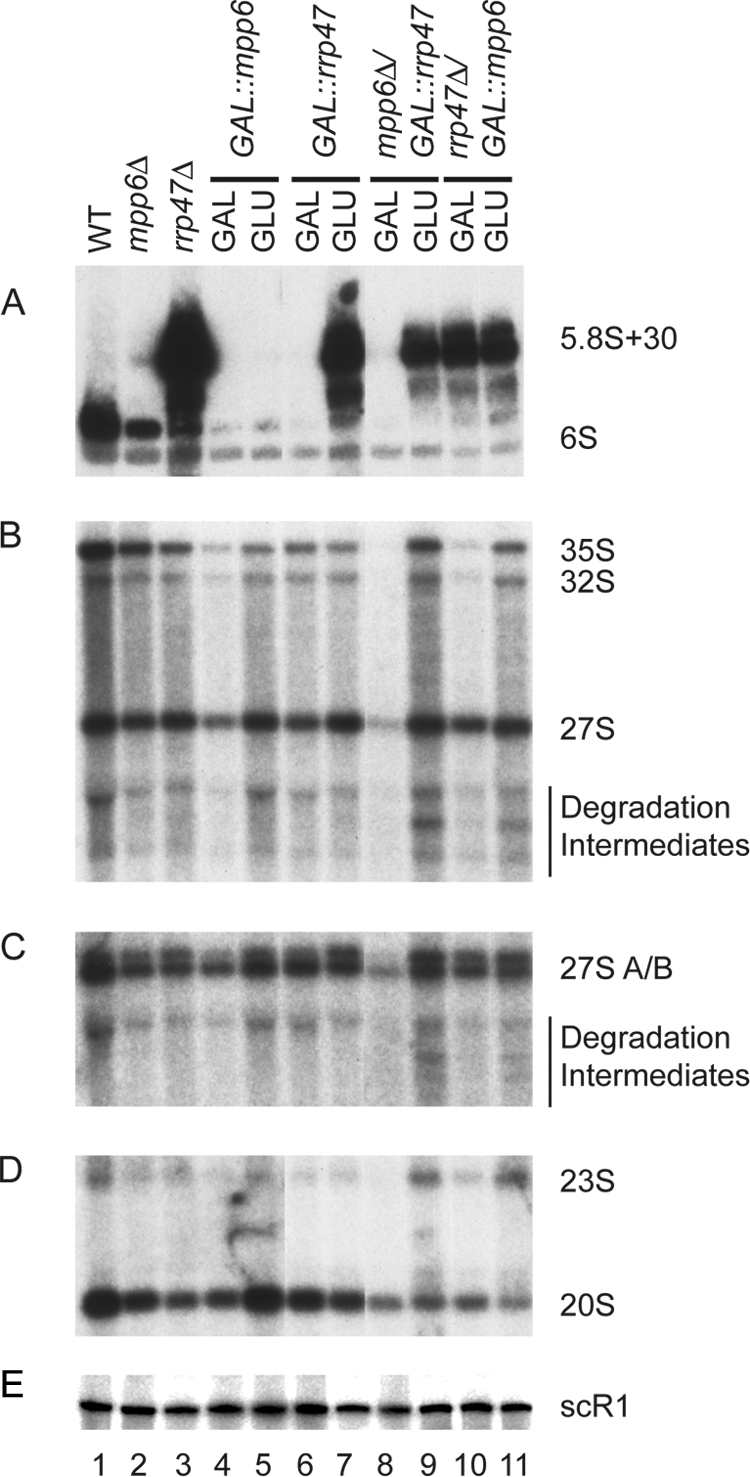
Results of analyses of pre-rRNA processing in Mpp6 mutants. Total RNA was extracted from the wild-type (WT) strain (lane 1) and strains carrying the mpp6Δ (lane 2) or rrp47Δ (lane 3) mutation during growth on glucose medium and strains carrying PGAL-MPP6 (lanes 4 and 5), PGAL-RRP47 (lanes 6 and 7), mpp6Δ PGAL-RRP47 (lanes 8 and 9), or rrp47Δ PGAL-MPP6 (lanes 10 and 11) during growth on galactose medium (GAL) and 6 h after transfer to glucose medium (GLU). (A) Northern analyses of low-molecular-weight RNA species resolved on a 6% polyacrylamide-8.3 M urea gel. (B, C, D, and E) Northern analyses of high-molecular-weight RNA species resolved on a 1.2% agarose gel following glyoxyl denaturation. The same RNA preparations were used for each analysis. Eight micrograms of total RNA was used for each lane. Probes used were ITS2 5′B (A), 27SA3 (B), ITS2 5′B (C), 20S (E), and scR1 (E).
However, the aberrant 23S RNA, which is a characterized substrate for RNA surveillance by the TRAMP and exosome complexes, accumulated mildly (Fig. 5D, lanes 9 and 11). In addition, a set of bands accumulated markedly in both the mpp6Δ GAL::rrp47 and the rrp47Δ GAL::mpp6 double mutant on glucose medium (Fig. 5B and C, lanes 9 and 11). These do not correspond to known pre-rRNA species and likely represent intermediates in pre-rRNA degradation which have been stabilized by the loss of both Mpp6 and Rrp47. We propose that the efficient degradation by the exosome of pre-rRNAs that have been targeted by surveillance pathways requires the partially redundant activities of Mpp6 and Rrp47.
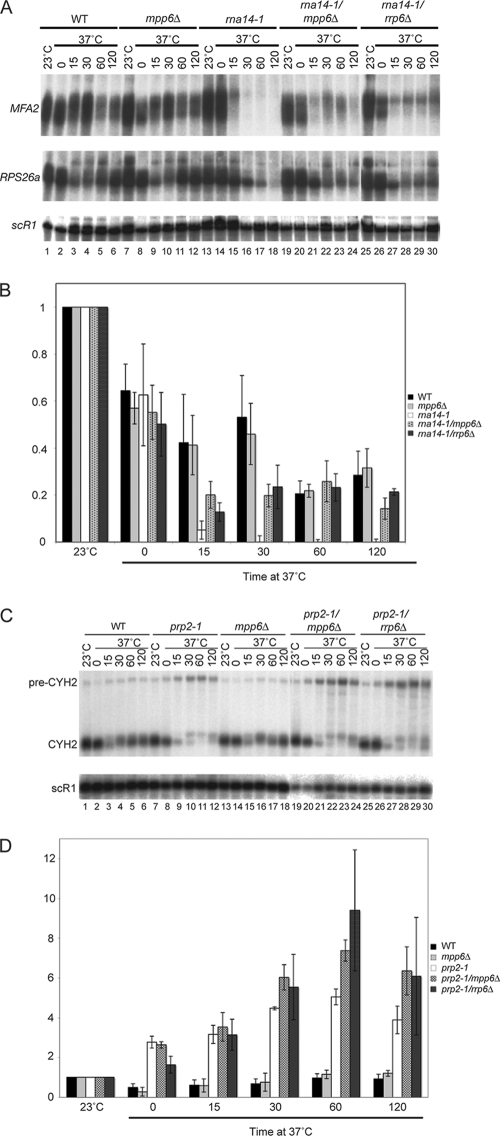
The requirement for Mpp6 in surveillance of nuclear pre-mRNAs was also tested. Strains carrying the rna14-1 mutation are defective in 3′ cleavage and polyadenylation of mRNA precursors, leading to rapid loss of mRNA synthesis following transfer to 37°C (Fig. 6A, lanes 13 to 18) (30). The absence of the nuclear exosome component Rrp6 partially restores mRNA synthesis in rna14-1 strains, probably by suppressing pre-mRNA surveillance activity (Fig. 6A, lanes 25 to 30) (50). In the rna14-1 mpp6Δ strain, mRNA levels were elevated relative to the level in the rna14-1 single mutant following transfer to 37°C (Fig. 6A, compare lanes 13 to 18 with lanes 19 to 24). The data are quantified for MFA2 in Fig. 6B; quantification of the data for RPS26 also showed statistically significant stabilization in the rna14-1 mpp6Δ strain (data not shown).
FIG. 6.
Mpp6 is involved in surveillance and degradation of nuclear pre-mRNAs. (A) Northern analyses of the RNA species resolved on a 6% polyacrylamide-8.3 M urea gel. Total RNA was extracted from wild-type cells (lanes 1 to 6) and strains carrying the single or double mutations mpp6Δ (lanes 7 to 12), rna14-1 (lanes 13 to 18), rna14-1 mpp6Δ (lanes 19 to 24), and rna14-1 rrp6Δ (lanes 25 to 30) during growth at 23°C and for the times indicated (minutes) after shift to 37°C. (B) Quantification of MFA2 mRNA. The graph shows mean values of the results from three independent experiments ± 1 standard error for MFA2 mRNA normalized to the level of the stable, cytoplasmic RNA scR1. (C) Northern analyses of RNA species resolved on a 2% agarose gel. Total RNA was extracted from wild-type cells (lanes 1 to 6) and strains carrying the single or double mutations prp2-1 (lanes 7 to 12), mpp6Δ (lanes 13 to 18), prp2-1 mpp6Δ (lanes 19 to 24), and prp2-1 rrp6Δ (lanes 25 to 30) during growth at 23°C and for the times indicated (minutes) after shift to 37°C. (D) Quantification of CYH2 mRNA. The graph shows mean values of the results from three independent experiments ± 1 standard error for CYH2 mRNA normalized to the level of the stable, cytoplasmic RNA scR1. WT, wild type.
The exosome was also implicated in the degradation of unspliced pre-mRNAs that have been targeted for splicing but have failed to complete the process (8). At 37°C the prp2-1 mutation allows commitment complex formation but blocks splicing prior to the first catalytic step (48). In the prp2-1 single-mutant strain, the spliced mRNA product is lost following transfer to 37°C but the unspliced pre-mRNA does not accumulate to a corresponding degree, due to its rapid degradation (Fig. 6C, lanes 7 to 12). In prp2-1 strains lacking Rrp6 (Fig. 6C, lanes 25 to 30) or Mpp6 (Fig. 6C, lanes 19 to 24), unspliced pre-CYH2 RNA was accumulated relative to the level in the prp2-1 single mutant and, although modest, the effects were statistically significant (quantification shown in Fig. 6D). These effects were, however, much less marked than those previously observed in prp2-1 strains with defects in the core exosome (8).
We conclude that Mpp6 participates in surveillance of defective nuclear pre-mRNAs, presumably helping to target the exosome to these RNAs.
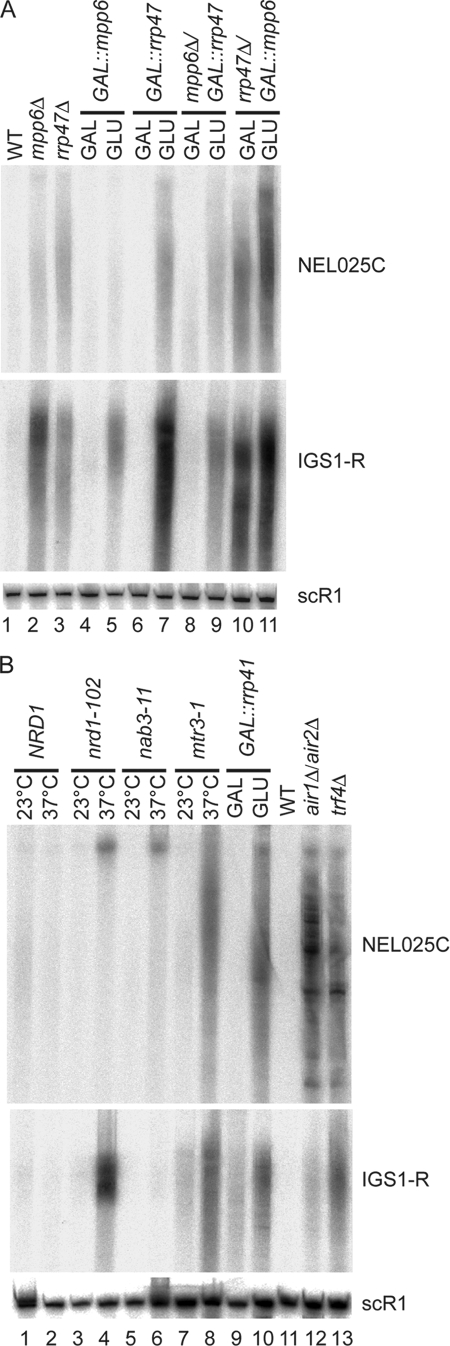
Rrp47 is implicated in the degradation of a class of non-protein-coding RNAs (ncRNAs), termed cryptic unstable transcripts (CUTs), which are generated from intergenic spacer regions at many sites in the yeast genome (4, 60). To assess whether Mpp6 also participates in CUT degradation, we examined the well-characterized species NEL025C (Fig. 7A). The NEL025C CUT shows substantial 3′ heterogeneity and therefore appears as a smear in Northern hybridization. NEL025C is almost undetectable in wild-type cells but was accumulated in the rrp47Δ strain (Fig. 7A, lane 3) and in the GAL::rrp47 strain on glucose medium (Fig. 7A, lane 7). Similar increases in NEL025C abundance were seen in the mpp6Δ strain and in the GAL::mpp6 strain on glucose medium, although the effect was milder in this strain, probably due to residual expression. Both the mpp6Δ GAL::rrp47 and the rrp47Δ GAL::mpp6 double-mutant strain showed higher NEL025C accumulation on glucose medium than any single mutant, indicating that Mpp6 and Rrp47 each participate in NEL025C degradation. Notably, when grown on galactose medium, the mpp6Δ GAL::rrp47 strain did not show the NEL025C accumulation seen in the mpp6Δ single mutant (Fig. 7A, lane 8). This suppression of the phenotype for mpp6Δ for NEL025C degradation probably results from overexpression of Rrp47, since GAL-regulated constructs are generally overexpressed on galactose medium.
FIG. 7.
Exosome cofactors involved in surveillance of ncRNAs. Northern analyses of RNA species resolved on a 6% polyacrylamide-8.3 M urea gel probed for the CUT NEL025C, the rDNA intergenic transcript IGS1-R, or cytoplasmic scR1 RNA. (A) Total RNA was extracted from the wild-type strain (lane 1); strains carrying either the mpp6Δ (lane 2) or rrp47Δ (lane 3) mutation during growth on glucose medium; and strains carrying the single or double mutations PGAL-MPP6 (lanes 4 and 5), PGAL-RRP47 (lanes 6 and 7), mpp6Δ PGAL-RRP47 (lanes 8 and 9), and rrp47Δ PGAL-MPP6 (lanes 10 and 11) during growth on galactose medium (GAL) and 6 h after transfer to glucose medium (GLU). (B) Total RNA was extracted from the NRD1 mutant strain (lanes 1 and 2); from nrd1-102 (lanes 3 and 4), nab3-11 (lanes 5 and 6), and mtr3-1 (lanes 7 and 8) mutant strains during growth at 23°C or 1 h after transfer to 37°C; from a PGAL-RRP41 mutant strain (lanes 9 and 10) during growth in galactose medium (GAL) or after transfer to medium containing glucose for 20 h (GLU); from an isogenic wild-type strain (lane 11); and from strains carrying either the air1Δ air2Δ (lane 12) or trf4Δ (lane 13) mutation. WT, wild type.
Another class of ncRNA is transcribed from the telomeres and ribosomal DNA (rDNA) spacer regions in S. cerevisiae, which are generally described as heterochromatic (20, 22, 55). The most-studied rDNA spacer transcript is designated IGS1-R and is very rapidly degraded following transcription. Like NEL025C, the IGS1-R transcripts show great 3′ heterogeneity (20). The levels of IGS1-R transcripts were elevated in the mpp6Δ and rrp47Δ single-mutant strains and in GAL::mpp6 and GAL::rrp47 strains on glucose medium (Fig. 7A). In the mpp6Δ GAL::rrp47 and the rrp47Δ GAL::mpp6 double mutant, accumulation on glucose medium was not clearly higher than in the single mutants. However, as for NEL025C, growth of the mpp6Δ GAL::rrp47 strain on galactose medium suppressed the mpp6Δ phenotype.
We conclude that Mpp6 and Rrp47 both participate in the rapid degradation of ncRNAs. The apparent suppression of the mpp6Δ phenotype by overexpression of Rrp47 is consistent with direct interactions or closely related functions.
Both ncRNAs, NEL025C and IGS1-R, were stabilized by the loss of components of the TRAMP complex, either the poly(A) polymerase Trf4 or Air1 plus Air2 (Fig. 7B) (20, 55, 60). Stabilization of the ncRNAs was also seen in strains depleted of the core exosome component Rrp41 or carrying a temperature-sensitive-lethal point mutation in the core exosome component Mtr3 (Fig. 7B), and these RNAs are also stabilized in strains lacking the nuclear-specific exonuclease Rrp6 (data not shown) (20, 60). The NEL025C RNA accumulated in the exosome appeared to be longer and more heterogeneous than in the TRAMP mutants, probably due to hyperadenylation by TRAMP in the absence of exosome activity. In strains with temperature-sensitive-lethal point mutations in the Nrd1/Nab3 complex, nrd1-102 and nab3-11, stabilization of longer forms of NEL025C was seen (Fig. 7B), consistent with the reported dual roles of Nrd1/Nab3 in transcription termination and subsequent degradation. The IGS1-R transcripts were strongly stabilized by nrd1-102 but, as previously observed (20), there was little stabilization in the nab3-11 strain, probably reflecting allele specificity.
Together these observations reveal a remarkable degree of apparent redundancy in ncRNA degradation, with at least four cofactor complexes and two nucleases participating in their degradation.
DISCUSSION
Here we report the use of genome-wide screens for genetic interactions to identify a new nuclear cofactor for the yeast exosome.
Gene deletions underrepresented in the collection of double mutants with the rrp47Δ mutation included the 3′ to 5′ exonucleases, Rex1 (Rnh70) and Rex3. Rrp47 is believed to function as a cofactor for the nuclear exosome component Rrp6, and deletion of RRP6 was previously shown to be SL with rex1Δ (52). The identification of these apparently biologically relevant putative SL interactions provided a strong indication that the screen had identified bona fide interacting genes. These interactions also suggested that common substrates are shared by the exosome and other, individually nonessential, 3′ exonucleases present in yeast cells.
The rrp47Δ screen also identified genes encoding the ribosome synthesis factors Ssf2, Nop13, and Alb1. Since the exosome and its cofactors are involved in surveillance of defective preribosomes (1, 14, 15), the identification of synergistic growth inhibition resulting from the combination of defects in ribosome synthesis and degradation was not entirely unexpected, but it is less clear why this particular set of mutations were identified. By definition, only the few ribosome synthesis factors that are not essential for viability could have been identified in the screen, which used deletion mutations. One possibility is that the encoded proteins have important functions during preribosome surveillance, conceivably signaling defects in ribosome synthesis to the degradation machinery.
GIM screens with both rrp47Δ and air1Δ identified strong negative interactions with components of the THO complex, which is probably required for packaging of newly synthesized mRNPs and also interacts genetically with Rrp6 (11, 29, 33, 40, 61). These interactions suggest roles for both Rrp47 and Air1 in the surveillance of pre-mRNAs. In addition to their roles in the TRAMP complexes, Air1 and Air2 were reported to bind the arginine methyltransferase Hmt1 and inhibit its activity. Hmt1 methylates the glycine-arginine repeat tracts on several RNA binding proteins, including the mRNP proteins Npl3 and Hrp1. Whether this is related to the SL interactions of air1Δ with THO complex mRNP proteins remains to be determined. There are no previously reported functional distinctions between Air1 and Air2; the single mutants show no detectable growth inhibition and no clear defects in RNA metabolism, whereas double mutants are severely impaired in growth and show strong defects in TRAMP activity. However, the GIM screens indicated that mpp6Δ is SL with air1Δ, but not with air2Δ (Fig. 1C), apparently showing that there are indeed clear functional distinctions.
Strong negative interactions with the mpp6Δ mutation were identified for both the rrp47Δ and air1Δ mutation, and the SL interaction between the rrp47Δ and mpp6Δ mutations was subsequently confirmed by crossing. The mpp6Δ mutation was also SL in combination with the rrp6Δ mutation, making it unlikely that Mpp6 functions in parallel with Rrp47 in targeting RNA substrates to Rrp6. Rather, we predict that Mpp6 targets RNAs to the core exosome, possibly to the active 3′ exonuclease Rrp44. Mpp6 bound RNA in vitro, showing substantially higher binding to poly(U) than to poly(dT), with only background binding to poly(A), poly(A/U), or natural RNAs with more structure. The lack of binding to poly(A) may be significant, since many exosome substrates are polyadenylated and a high affinity of Mpp6 for poly(A) might well interfere with their degradation. Yeast Rrp47, in contrast, specifically binds structured RNAs (44), indicating that Mpp6 and Rrp47 will have distinctly different preferred binding sites in vivo.
The results of detailed bioinformatics analyses starting from the human exosome-binding protein Mhosph6 show that Mpp6 is the only budding yeast (S. cerevisiae) protein with the two signature motifs for Mphosph6 homologues and that the occurrence of both motifs in a protein is statistically significant (P < 0.0015). Human MPHOSPH6 was reported to associate with the human exosome and also bound single-stranded RNA (42).
Strains lacking Mpp6 showed weak but detectable stabilization of unspliced pre-mRNAs, and stronger stabilization was seen for mRNAs with defects in 3′ cleavage and polyadenylation. These data show that Mpp6 participates in the surveillance of defective mRNA precursors. We predict that the effects of loss of Mpp6 on the degradation of these exosome substrates are relatively mild because of functional redundancy with other exosome cofactors and, possibly, other nucleases.
The role of redundancy in RNA degradation.
The results of recent analyses for yeast and humans suggest that ncRNAs are very wide-spread and may, indeed, dominate total Pol II transcription (7, 46, 47). Moreover, regions of the genome that are embedded in heterochromatin, including the rDNA spacers, were long considered to be transcriptionally inert since transcripts derived from these regions could not be detected, whereas it is now clear that, at least in yeast, heterochromatic regions are actively transcribed (10, 20, 55, 58). This raises the obvious question of why the apparently almost ubiquitous ncRNA transcripts largely escaped detection in previous analyses.
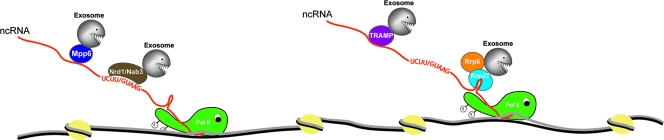
At least part of the answer is that many ncRNAs, including those derived from the rDNA heterochromatin described here, are subject to very rapid degradation, keeping their steady-state levels very low. This, in turn, raises the question of how such rapid degradation can be achieved. One feature that likely contributes to the rapid degradation of ncRNAs is the cotranscriptional binding of at least some exosome cofactors (12, 20), potentially allowing “pretargeting” of the nascent transcript and rapid degradation as soon as transcription termination makes the 3′ end available (Fig. 8) (reviewed in reference 22). A second important feature is likely to be the functional redundancy that is manifested among the components of the degradation machinery (Fig. 8). The ncRNAs analyzed here appear to be targeted by at least four exosome cofactors: Mpp6, Rrp47, the Nrd1-Nab3 heterodimer, and the TRAMP4 polyadenylation complex. Moreover, inhibition of the activity of either the nuclear-specific exonuclease Rrp6 or the core exosome also stabilized the ncRNAs. This indicates that multiple cofactors can target the ncRNAs to different exonucleases for degradation. Such competition would presumably avoid any bottlenecks during degradation—if any factor fails to bind, another will do the job. The suppression of the mpp6Δ phenotype on the CUT ncRNA observed in strains overexpressing Rrp47 supports the idea of competition for the substrate.
FIG. 8.
Multiple cofactors and exonucleases target ncRNAs. The figure highlights two aspects of ncRNA metabolism that are likely to contribute to very rapid degradation. (i) At least some of the exosome cofactors bind the nascent RNA cotranscriptionally (12, 20), potentially “pretargeting” it for degradation as soon as the 3′ end is released from the transcribing polymerase. (ii) Multiple, partially redundant cofactors can target the ncRNA for degradation (see text).
The results of recent analyses indicate that at least some ncRNAs exert their influence on gene expression at the level of chromatin structure and act in trans (6, 39). Since there can only be a very small number of target genes per cell for each ncRNA, maintaining accurate, and very low, levels of the ncRNAs may be crucial for the regulation of gene expression. More generally, such “genetic buffering” may protect the cell against the failure of any specific component.
Functional redundancy may also be important for the efficient degradation of “difficult” substrates, a notable example of which may be the preribosomes. The predominant phenotype of most ribosome synthesis mutants in yeast is not the accumulation of defective preribosomes but the rapid and complete degradation of the pre-rRNAs (reviewed in reference 56). These RNAs are kilobases in length, highly structured, and packaged with many different proteins but can be completely degraded without the appearance of clear intermediates. Consistent with the importance of redundancy in pre-rRNA surveillance, strains lacking both Mpp6 and Rrp47 accumulated heterogeneous pre-rRNA fragments, which we predict represent degradation intermediates.
Redundancy is believed to be important for robustness in many biological systems, and this probably includes nuclear RNA surveillance.
Acknowledgments
We thank Emma Thomson, Kim Kotovic, and Jon Houseley for critical reading of the manuscript and Martin Kos for assistance with Fig. 8.
L.M. was supported by the BBSRC. This work was supported by the Wellcome Trust.
Footnotes
Published ahead of print on 30 June 2008.
REFERENCES
- 1.Allmang, C., P. Mitchell, E. Petfalski, and D. Tollervey. 2000. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 281684-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allmang, C., E. Petfalski, A. Podtelejnikov, M. Mann, D. Tollervey, and P. Mitchell. 1999. The yeast exosome and human PM-Scl are related complexes of 3′→5′ exonucleases. Genes Dev. 132148-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Araki, Y., S. Takahashi, T. Kobayashi, H. Kajiho, S. Hoshino, and T. Katada. 2001. Ski7p G protein interacts with the exosome and the Ski complex for 3′-to-5′ mRNA decay in yeast. EMBO J. 204684-4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arigo, J. T., D. E. Eyler, K. L. Carroll, and J. L. Corden. 2006. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol. Cell 23841-851. [DOI] [PubMed] [Google Scholar]
- 5.Baudin, A., O. Ozier-Kalogeropoulos, A. Denouel, F. Lacroute, and C. Cullin. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 213329-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berretta, J., M. Pinskaya, and A. Morillon. 2008. A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Genes Dev. 22615-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birney, E., J. A. Stamatoyannopoulos, A. Dutta, R. Guigo, T. R. Gingeras, E. H. Margulies, Z. Weng, M. Snyder, E. T. Dermitzakis, R. E. Thurman, M. S. Kuehn, C. M. Taylor, S. Neph, C. M. Koch, S. Asthana, A. Malhotra, I. Adzhubei, J. A. Greenbaum, R. M. Andrews, P. Flicek, P. J. Boyle, H. Cao, N. P. Carter, G. K. Clelland, S. Davis, N. Day, P. Dhami, S. C. Dillon, M. O. Dorschner, H. Fiegler, P. G. Giresi, J. Goldy, M. Hawrylycz, A. Haydock, R. Humbert, K. D. James, B. E. Johnson, E. M. Johnson, T. T. Frum, E. R. Rosenzweig, N. Karnani, K. Lee, G. C. Lefebvre, P. A. Navas, F. Neri, S. C. Parker, P. J. Sabo, R. Sandstrom, A. Shafer, D. Vetrie, M. Weaver, S. Wilcox, M. Yu, F. S. Collins, J. Dekker, J. D. Lieb, T. D. Tullius, G. E. Crawford, S. Sunyaev, W. S. Noble, I. Dunham, F. Denoeud, A. Reymond, P. Kapranov, J. Rozowsky, D. Zheng, R. Castelo, A. Frankish, J. Harrow, S. Ghosh, A. Sandelin, I. L. Hofacker, R. Baertsch, D. Keefe, S. Dike, J. Cheng, H. A. Hirsch, E. A. Sekinger, J. Lagarde, J. F. Abril, A. Shahab, C. Flamm, C. Fried, J. Hackermuller, J. Hertel, M. Lindemeyer, K. Missal, A. Tanzer, S. Washietl, J. Korbel, O. Emanuelsson, J. S. Pedersen, N. Holroyd, R. Taylor, D. Swarbreck, N. Matthews, M. C. Dickson, D. J. Thomas, M. T. Weirauch, J. Gilbert, et al. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447799-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bousquet-Antonelli, C., C. Presutti, and D. Tollervey. 2000. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102765-775. [DOI] [PubMed] [Google Scholar]
- 9.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14115-132. [DOI] [PubMed] [Google Scholar]
- 10.Buhler, M., W. Haas, S. P. Gygi, and D. Moazed. 2007. RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell 129707-721. [DOI] [PubMed] [Google Scholar]
- 11.Chavez, S., T. Beilharz, A. G. Rondon, H. Erdjument-Bromage, P. Tempst, J. Q. Svejstrup, T. Lithgow, and A. Aguilera. 2000. A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J. 195824-5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Ciais, D., M. Bohnsack, and D. Tollervey. 2008. The mRNA encoding the yeast ARE-binding protein Cth2 is generated by a novel 3′ processing pathway. Nucleic Acids Res. 363075-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conrad, N. K., S. M. Wilson, E. J. Steinmetz, M. Patturajan, D. A. Brow, M. S. Swanson, and J. L. Corden. 2000. A yeast heterogeneous nuclear ribonucleoprotein complex associated with RNA polymerase II. Genetics 154557-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Decourty, L., C. Saveanu, K. Zemam, F. Hantraye, E. Frachon, J.-C. Rousselle, M. Fromont-Racine, and A. Jacquier. 2008. Linking functionally related genes by sensitive and quantitative characterization of genetic interaction profiles. Proc. Natl. Acad. Sci. USA 1055816-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dez, C., M. Dlakic, and D. Tollervey. 2007. Roles of the HEAT repeat proteins Utp10 and Utp20 in 40S ribosome maturation. RNA 131516-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dez, C., J. Houseley, and D. Tollervey. 2006. Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae. EMBO J. 251534-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egecioglu, D. E., A. K. Henras, and G. F. Chanfreau. 2006. Contributions of Trf4p- and Trf5p-dependent polyadenylation to the processing and degradative functions of the yeast nuclear exosome. RNA 1226-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fatica, A., A. D. Cronshaw, M. Dlakic, and D. Tollervey. 2002. Ssf1p prevents premature processing of an early pre-60S ribosomal particle. Mol. Cell 9341-351. [DOI] [PubMed] [Google Scholar]
- 18.Ghaemmaghami, S., W. K. Huh, K. Bower, R. W. Howson, A. Belle, N. Dephoure, E. K. O'Shea, and J. S. Weissman. 2003. Global analysis of protein expression in yeast. Nature 425737-741. [DOI] [PubMed] [Google Scholar]
- 19.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K.-D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C.-Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418387-391. [DOI] [PubMed] [Google Scholar]
- 20.Houseley, J., K. Kotovic, A. El Hage, and D. Tollervey. 2007. Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. EMBO J. 264996-5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Houseley, J., J. LaCava, and D. Tollervey. 2006. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 7529-539. [DOI] [PubMed] [Google Scholar]
- 22.Houseley, J., and D. Tollervey. 2008. The nuclear RNA surveillance machinery: the link between ncRNAs and genome structure in budding yeast. Biochim. Biophys. Acta 1779239-246. [DOI] [PubMed] [Google Scholar]
- 23.Houseley, J., and D. Tollervey. 2006. Yeast Trf5p is a nuclear poly(A) polymerase. EMBO Rep. 7205-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishihama, Y., J. Rappsilber, J. S. Andersen, and M. Mann. 2002. Microcolumns with self-assembled particle frits for proteomics. J. Chromatogr. A 979233-239. [DOI] [PubMed] [Google Scholar]
- 25.Jonson, L., J. F. Rehfeld, and A. H. Johnsen. 2004. Enhanced peptide secretion by gene disruption of CYM1, a novel protease in Saccharomyces cerevisiae. Eur. J. Biochem. 2714788-4797. [DOI] [PubMed] [Google Scholar]
- 26.Kadowaki, T., R. Schneiter, M. Hitomi, and A. M. Tartakoff. 1995. Mutations in nucleolar proteins lead to nucleolar accumulation of polyA+ RNA in Saccharomyces cerevisiae. Mol. Biol. Cell 61103-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LaCava, J., J. Houseley, C. Saveanu, E. Petfalski, E. Thompson, A. Jacquier, and D. Tollervey. 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 21713-724. [DOI] [PubMed] [Google Scholar]
- 28.Lebreton, A., C. Saveanu, L. Decourty, J. C. Rain, A. Jacquier, and M. Fromont-Racine. 2006. A functional network involved in the recycling of nucleocytoplasmic pre-60S factors. J. Cell Biol. 173349-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Libri, D., K. Dower, J. Boulay, R. Thomsen, M. Rosbash, and T. H. Jensen. 2002. Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol. Cell. Biol. 228254-8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minvielle-Sebastia, L., P. J. Preker, and W. Keller. 1994. RNA14 and RNA15 proteins as components of a yeast pre-mRNA 3′-end processing factor. Science 2661702-1705. [DOI] [PubMed] [Google Scholar]
- 31.Mitchell, P., E. Petfalski, R. Houalla, A. Podtelejnikov, M. Mann, and D. Tollervey. 2003. Rrp47p is an exosome-associated protein required for the 3′ processing of stable RNAs. Mol. Cell. Biol. 236982-6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oeffinger, M., M. Dlakic, and D. Tollervey. 2004. A pre-ribosome-associated HEAT-repeat protein is required for export of both ribosomal subunits. Genes Dev. 18196-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olesen, J. R., D. Libri, and T. H. Jensen. 2005. A link between transcription and mRNP quality in Saccharomyces cerevisiae. RNA Biol. 245-48. [DOI] [PubMed] [Google Scholar]
- 34.Peng, W. T., M. D. Robinson, S. Mnaimneh, N. J. Krogan, G. Cagney, Q. Morris, A. P. Davierwala, J. Grigull, X. Yang, W. Zhang, N. Mitsakakis, O. W. Ryan, N. Datta, V. Jojic, C. Pal, V. Canadien, D. Richards, B. Beattie, L. F. Wu, S. J. Altschuler, S. Roweis, B. J. Frey, A. Emili, J. F. Greenblatt, and T. R. Hughes. 2003. A panoramic view of yeast noncoding RNA processing. Cell 113919-933. [DOI] [PubMed] [Google Scholar]
- 35.Plumpton, M., M. McGarvey, and J. D. Beggs. 1994. A dominant negative mutation in the conserved RNA helicase motif “SAT” causes splicing factor PRP2 to stall in spliceosomes. EMBO J. 13879-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rappsilber, J., Y. Ishihama, and M. Mann. 2003. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75663-670. [DOI] [PubMed] [Google Scholar]
- 37.Rexach, M., and G. Blobel. 1995. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell 83683-692. [DOI] [PubMed] [Google Scholar]
- 38.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 171030-1032. [DOI] [PubMed] [Google Scholar]
- 39.Rinn, J. L., M. Kertesz, J. K. Wang, S. L. Squazzo, X. Xu, S. A. Brugmann, L. H. Goodnough, J. A. Helms, P. J. Farnham, E. Segal, and H. Y. Chang. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 1291311-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rougemaille, M., R. K. Gudipati, J. R. Olesen, R. Thomsen, B. Seraphin, D. Libri, and T. H. Jensen. 2007. Dissecting mechanisms of nuclear mRNA surveillance in THO/sub2 complex mutants. EMBO J. 262317-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 42.Schilders, G., R. Raijmakers, J. M. Raats, and G. J. Pruijn. 2005. MPP6 is an exosome-associated RNA-binding protein involved in 5.8S rRNA maturation. Nucleic Acids Res. 336795-6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68850-858. [DOI] [PubMed] [Google Scholar]
- 44.Stead, J. A., J. L. Costello, M. J. Livingstone, and P. Mitchell. 2007. The PMC2NT domain of the catalytic exosome subunit Rrp6p provides the interface for binding with its cofactor Rrp47p, a nucleic acid-binding protein. Nucleic Acids Res. 355556-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steinmetz, E. J., N. K. Conrad, D. A. Brow, and J. L. Corden. 2001. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature 413327-331. [DOI] [PubMed] [Google Scholar]
- 46.Steinmetz, E. J., C. L. Warren, J. N. Kuehner, B. Panbehi, A. Z. Ansari, and D. A. Brow. 2006. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol. Cell 24735-746. [DOI] [PubMed] [Google Scholar]
- 47.Struhl, K. 2007. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat. Struct. Mol. Biol. 14103-105. [DOI] [PubMed] [Google Scholar]
- 48.Teigelkamp, S., M. McGarvey, M. Plumpton, and J. D. Beggs. 1994. The splicing factor PRP2, a putative RNA helicase, interacts directly with pre-mRNA. EMBO J. 13888-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tollervey, D., and I. W. Mattaj. 1987. Fungal small nuclear ribonucleoproteins share properties with plant and vertebrate U-snRNPs. EMBO J. 6469-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Torchet, C., C. Bousquet-Antonelli, L. Milligan, E. Thompson, J. Kufel, and D. Tollervey. 2002. Processing of 3′ extended read-through transcripts by the exosome can generate functional mRNAs. Mol. Cell 91285-1296. [DOI] [PubMed] [Google Scholar]
- 51.Vanacova, S., J. Wolf, G. Martin, D. Blank, S. Dettwiler, A. Friedlein, H. Langen, G. Keith, and W. Keller. 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 3e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Hoof, A., P. Lennertz, and R. Parker. 2000. Three conserved members of the RNase D family have unique and overlapping functions in the processing of 5S, 5.8S, U4, U5, RNase MRP and RNase P RNAs in yeast. EMBO J. 191357-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Hoof, A., R. R. Staples, R. E. Baker, and R. Parker. 2000. Function of the Ski4p (Csl4p) and Ski7p proteins in 3′-to-5′ degradation of mRNA. Mol. Cell. Biol. 208230-8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vasiljeva, L., and S. Buratowski. 2006. Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol. Cell 21239-248. [DOI] [PubMed] [Google Scholar]
- 55.Vasiljeva, L., M. Kim, N. Terzi, L. M. Soares, and S. Buratowski. 2008. Transcription termination and RNA degradation contribute to silencing of RNA polymerase II transcription within heterochromatin. Mol. Cell 29313-323. [DOI] [PubMed]
- 56.Venema, J., and D. Tollervey. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33261-311. [DOI] [PubMed] [Google Scholar]
- 57.Venema, J., and D. Tollervey. 1996. RRP5 is required for formation of both 18S and 5.8S rRNA in yeast. EMBO J. 155701-5714. [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, S.-W., A. L. Stevenson, S. E. Kearsey, S. Watt, and J. Bähler. 2008. Global role for polyadenylation-assisted nuclear RNA degradation in posttranscriptional gene silencing. Mol. Cell. Biol. 28656-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu, K., P. Wu, and J. P. Aris. 2001. Nucleolar protein Nop12p participates in synthesis of 25S rRNA in Saccharomyces cerevisiae. Nucleic Acids Res. 292938-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wyers, F., M. Rougemaille, G. Badis, J.-C. Rousselle, M.-E. Dufour, J. Boulay, B. Régnault, F. Devaux, A. Namane, B. Séraphin, D. Libri, and A. Jacquier. 2005. Cryptic Pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121725-737. [DOI] [PubMed] [Google Scholar]
- 61.Zenklusen, D., P. Vinciguerra, J. C. Wyss, and F. Stutz. 2002. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell. Biol. 228241-8253. [DOI] [PMC free article] [PubMed] [Google Scholar]