Abstract
Precise and robust regulation of alternative splicing provides cells with an essential means of gene expression control. However, the mechanisms that ensure the tight control of tissue-specific alternative splicing are not well understood. It has been demonstrated that robust regulation often results from the contributions of multiple factors to one particular splicing pathway. We report here a novel strategy used by a single splicing regulator that blocks the formation of two distinct prespliceosome complexes to achieve efficient regulation. Fox-1/Fox-2 proteins, potent regulators of alternative splicing in the heart, skeletal muscle, and brain, repress calcitonin-specific splicing of the calcitonin/CGRP pre-mRNA. Using biochemical analysis, we found that Fox-1/Fox-2 proteins block prespliceosome complex formation at two distinct steps through binding to two functionally important UGCAUG elements. First, Fox-1/Fox-2 proteins bind to the intronic site to inhibit SF1-dependent E′ complex formation. Second, these proteins bind to the exonic site to block the transition of E′ complex that escaped the control of the intronic site to E complex. These studies provide evidence for the first example of regulated E′ complex formation. The two-step repression of presplicing complexes by a single regulator provides a powerful and accurate regulatory strategy.
Alternative splicing is an important mechanism for regulating gene expression in higher eukaryotes. Through alternative splicing, one pre-mRNA generates several mRNAs, leading to synthesis of multiple proteins with distinct biological functions (6, 7, 33, 34). Alternative splicing plays a particularly important role in regulating neuronal gene expression (27, 49). Alternative splicing is often tightly regulated, leading to the expression of specific isoforms in different tissues or developmental stages. Misregulation of alternative splicing has been linked to several serious diseases (53).
Regulation of alternative splicing builds upon the basic splicing machinery that joins both constitutive and regulated exons within nuclear pre-mRNA molecules. Removal of introns from pre-mRNAs is carried out by a large macromolecular machine known as the spliceosome, which includes five snRNPs (U1, U2, U4, U5, and U6) and several hundred associated proteins (21). The spliceosome forms de novo on the pre-mRNA molecule in a stepwise fashion that can be detected by in vitro splicing analysis using synthetic pre-mRNAs (8, 46). The first complex to form, the H complex, contains a number of heterogeneous nuclear ribonucleoproteins (4). Next to form are the E′ and E complexes, the earliest complexes committed to the splicing pathway (11, 18, 23, 37). The ATP-independent E′ complex assembles in U2 snRNP auxiliary factor (U2AF)-depleted HeLa nuclear extract and contains U1 snRNP bound to the 5′ splice site and splicing factor 1 (SF1) bound to the branch point. The E′ complex can be chased into E complex through recruitment of U2AF to the polypyrimidine tract and 3′ splice site (23). E complex formation is followed by formation of the A complex, the first ATP-dependent step in assembly, which involves U2 snRNP replacing SF1 at the branch point (10, 36). Subsequently, recruitment of the U4/U5/U6 tri-snRNP results in formation of the B complex. Next, the catalytically competent C complex forms by recruitment of additional protein factors, along with significant structural rearrangements during which the U1 and U4 snRNPs dissociate and the U6 snRNP base pairs with the 5′ splice site and U2 snRNA during catalytic activation (24).
Alternative splicing has been shown to be regulated at different points throughout the assembly pathway (17). Splicing can be regulated during formation of the prespliceosome E or A complex or at the transition from the A to the B complex (6, 13, 14, 16, 28). Regulation of splicing can also occur between the first and second catalytic steps (25, 41). The mechanisms that ensure the tight control of tissue-specific alternative splicing are not well understood. Several well-studied examples suggest that robust regulation results from the contributions of multiple factors to individual splicing pathways (6, 15, 27, 35).
Recently, Fox-1/Fox-2 proteins have emerged as tissue-specific splicing regulators that are enriched in the heart, skeletal muscle, and brain (19). Although it is clear that Fox-1/Fox-2 proteins regulate inclusion of several alternative exons through binding to UGCAUG elements, the underlying mechanisms have not been explored extensively (3, 19, 39, 40, 50, 56). Recently, we showed that Fox-1/Fox-2 proteins are major regulators of the neuron-specific alternative RNA processing pathway of the calcitonin/CGRP pre-mRNA (56). These proteins interact with two UGCAUG elements surrounding the 3′ splice site of the calcitonin-specific exon 4 and promote skipping of this exon (56). Of several groups of splicing factors that contribute to regulation of exon 4 inclusion, the Fox-1/Fox-2 proteins provide the most robust regulation (30, 31, 56, 58, 59). However, the precise role of Fox-1/Fox-2 proteins in this regulation is not known.
Here, we present the results of biochemical analysis demonstrating that Fox-1/Fox-2 proteins directly inhibit splicing of the intron preceding exon 4. More importantly, they block prespliceosome complex formation at two distinct steps, the prespliceosome E′ and E complexes, by binding to two different recognition elements. Furthermore, we demonstrate that association of SF1 with the E′ complex and U2AF with the E complex is blocked by Fox-1/Fox-2 proteins, also in a position-dependent manner. These studies reveal a novel strategy of splicing regulation and provide evidence for the first example of regulated E′ complex formation.
MATERIALS AND METHODS
Plasmids.
The human calcitonin/CGRP reporter constructs used in transfection experiments consist of calcitonin/CGRP gene exons 4 to 6 fused to a heterologous first exon from adenovirus (59). To construct the plasmids pFBEwt, pFBE-34, pFBE+45, pFBEdbl, and pFBEese used for in vitro transcription, the pCTA-wt, pCTA-34, pCTA+45, pCTA-dbl, and pCT-ESEs plasmids (56) were digested with XbaI and NsiI and then subcloned into the pGEM-4 vector between the XbaI and PstI sites. A fragment containing three MS2 hairpins was generated by annealing the oligonucleotides 5′-GATCCCGTACACCATCAGGGTACGAGCTAGCCCATGGCGTACACCATCAGGGTACGACTAGTAGATCTCGTACACCATCAGGGTACGG-3′ and 5′-AATTCCGTACCCTGATG GTGTACGAGATCTACTAGTCGTACCCTGATGGTGTACGCCATGGGC TAGCTCGTACCCTGATGGTGTACGG-3′ (57). The double-stranded DNA products were subcloned into the pGEM-3 plasmid between BamHI and EcoRI sites. The fragments amplified by PCR from the templates pCT-wt, pCT-34, pCT+45, and pCT-34+45+177+252 were inserted into the 5′ of MS2 hairpins to generate plasmids pFBEwt-M, pFBE-34-M, pFBE+45-M, and pFBEdbl-M used for MS2-MBP affinity purification.
Expression of His-U2AF65, His-Tra2β, and His-SRp55 proteins.
His-tagged U2AF65 was overexpressed in Escherichia coli and purified by HisPur cobalt resin (Pierce). The recombinant viruses expressing His-Tra2β were generated by using the BAC-to-BAC baculovirus expression system (Invitrogen). The His-SRp55-expressing viruses, as well as His-SRp35-expressing viruses, were provided by James Patton (Vanderbilt University). His-Tra2β, His-SRp55, and His-SRp35 were purified from Sf9 insect cells.
Cell transfection and RNA and protein analysis.
HeLa cell transfection was carried out as previously described (56). For each transfection, 2 μg of the calcitonin/CGRP reporter plasmid, 50 ng of plasmids expressing F011 (Fox-2), and 400 ng or 800 ng of plasmids expressing Tra2β, SRp55, SF1, or CUGBP1 were used. Procedures for total RNA and protein isolation, reverse transcription-PCR (RT-PCR) analysis, and Western blot were described previously (56).
Nuclear extracts.
Nuclear extracts from HeLa and CA77 cells were prepared as described previously (59). Extracts depleted of poly(U)-binding proteins including U2AF65 were carried out as described previously (23, 32). The U2AF65 level in the depleted extracts was analyzed by Western blotting.
In vitro assays.
Pre-mRNAs were transcribed in vitro from the plasmid templates. The in vitro splicing reaction was carried out as previously described (54). The E and H complex assembly assay was carried out as described previously (11). Pre-mRNAs (5 × 104 cpm) were incubated in ATP-deleted nuclear extract in the absence of ATP, creatine phosphate, and heparin. The spliceosome complexes were separated by 1.5% native agarose gel. E′ and E complex formation in Fig. 5 was carried out as described previously (23). The reactions were performed in a volume of 10 μl containing 25% (vol/vol) of either normal or U2AF65-depleted HeLa or CA77 nuclear extract, 60 mM KCl, 10 U of RNaseOUT, and 5 × 104 cpm of 32P-labeled RNA substrates. After incubation at 30°C for 60 min, the reaction was terminated by the addition of 2 μl of termination buffer (1× Tris-borate-EDTA [TBE], 0.1% bromophenol blue, 0.1% xylene cyanol blue, 20% glycerol, and 2.5 mM EDTA) and immediately loaded onto a 1.2% horizontal agarose-0.5× TBE gel. The gels were run for 10 h at 4°C. Add-back experiments were carried out in U2AF65-depleted HeLa or CA77 extract with 60 μM recombinant U2AF65. The UV cross-linking/immunoprecipitation (IP) and psoralen cross-linking reactions were carried out as previously described (56, 59).
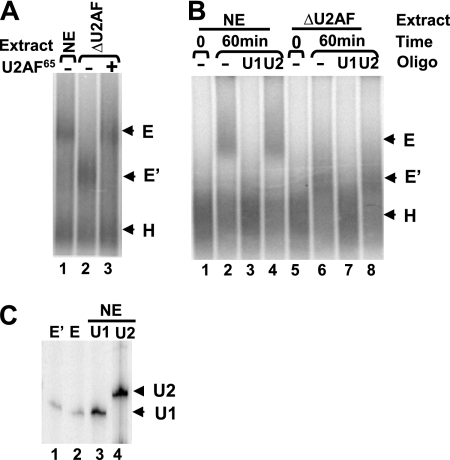
FIG. 5.
Formation of the prespliceosome complex E′ on the FBEdbl pre-mRNA. (A) Native agarose gel analysis of complexes formed in HeLa nuclear extract (NE), U2AF-depleted nuclear extract (ΔU2AF), or ΔU2AF extract supplemented with the recombinant His-U2AF65. The migration positions of the complexes are shown on the right. (B) Complex formation was analyzed in the absence or presence of anti-U1 or anti-U2 2′-O-methyl oligonucleotides. (C) Primer-extension analysis of RNA eluted from E and E′ complexes assembled on FBEdbl RNA. The E′ (lane 1) and E (lane 2) complexes were excised from agarose gels, and RNAs were extracted and analyzed with primers specific for U1 and U2 snRNA. RNAs isolated from HeLa nuclear extract were used as controls (lane 3 for U1 and lane 4 for U2 snRNA).
MS2-MBP affinity purification.
MS2-MBP fusion protein (the construct of which was a gift from Douglas L. Black) was expressed in E. coli. The fusion protein was purified as described previously (20, 57). For purification of all spliceosome complexes, MS2 binding site-containing pre-mRNAs were preincubated with 50-fold excess of recombinant MS2-MBP fusion protein prior to assembly. The assembly reactions (1.0 ml) contained 10 nM pre-mRNA, 500 nM MS2-MBP, 20 mM creatine phosphate, 2 mM ATP, 1.5 mM MgCl2, 1.5% polyethylene glycol, 0.15 mM dithiothreitol, 50 U of RNaseOUT, and 44% (vol/vol) HeLa or CA77 nuclear extract. Reaction mixtures were incubated at 30°C for 30 min and then treated with 0.5 mg of heparin/ml for 5 min at 30°C. Complex purification was carried out as described previously (20). The reaction mix was loaded onto a 5-ml S-500 sizing column (GE) equilibrated with sizing column buffer SCB1 (250 mM KCl, 5 mM EDTA, 1 mM dithiothreitol, 20 mM Tris [pH 7.9]) plus 0.05% NP-40. Pooled solution from the S-500 sizing column was fractionated into 18 aliquots, and the radioactivity was determined by scintillation counting. The peak fractions were pooled and passed twice through a column of 500-μl amylose beads preequilibrated in buffer SCB1. The column was washed with 10 column volumes of buffer SCB1. Bound complexes were eluted with 20 mM maltose in buffer SCB1. Proteins in the final eluate were concentrated by the addition of sodium deoxycholate. The pelleted proteins were resuspended in 2× sample buffer. H and E complexes were purified under the same conditions except that neither ATP nor creatine phosphate was present and the nuclear extract was depleted of ATP by preincubation at room temperature for 30 min. Western blot analysis was carried out after MS2-MBP purification with anti-Fox-2 for Fox-2 (1:1,000), SF1-24D1 for SF1 (1:500), anti-tra2β for tra2β (1:2,000), G-15 for SRp55 (1:200), MC3 for U2AF65 (1:2,500), and 9H10 for hnRNPA1 (1:2,000).
Primer extension.
Primer extension analysis of snRNA was performed by using reverse transcriptase. RNAs purified from the complexes were hybridized with 32P-labeled primers, which were complementary to nucleotides 142 to 159 of human U1 snRNA, nucleotides 150 to 170 of U2 snRNA, nucleotides 64 to 83 of U4 snRNA, nucleotides 94 to 109 of U5 snRNA, or nucleotides 77 to 92 of U6 snRNA. Products were analyzed on a denaturing 10% polyacrylamide gel.
RESULTS
Fox-1/Fox-2 proteins block splicing before formation of the prespliceosome E complex.
We demonstrated previously that Fox-1/Fox-2 proteins repress inclusion of the calcitonin-specific exon 4 (56). We suspected that Fox-1/Fox-2 proteins might block splicing of intron 3 of the calcitonin/CGRP pre-mRNA because two functionally important Fox-1/Fox-2 binding sites lie on either side of the 3′ splice site of this intron. To test whether and how Fox-1/Fox-2 proteins affect splicing, we carried out in vitro splicing analysis with nuclear extracts prepared from HeLa or CA77 cells and the in vitro-transcribed pre-mRNA substrates shown in Fig. 1A. Our previous transfection studies using a calcitonin/CGRP minigene reporter showed that HeLa and CA77 cells process the reporter pre-mRNA via the calcitonin- and CGRP-specific pathways, respectively (see Fig. S1A to C in the supplemental material). CA77 is a cell line derived from a rat medullary thyroid carcinoma and exhibits a number of neuronal features (43). High expression levels of Fox-1/Fox-2 proteins in CA77 cells correlate well with the CGRP-specific splicing pattern (see Fig. S1D in the supplemental material). We demonstrated previously that Fox-1 and Fox-2 proteins are interchangeable as splicing regulators in the calcitonin/CGRP system (56).
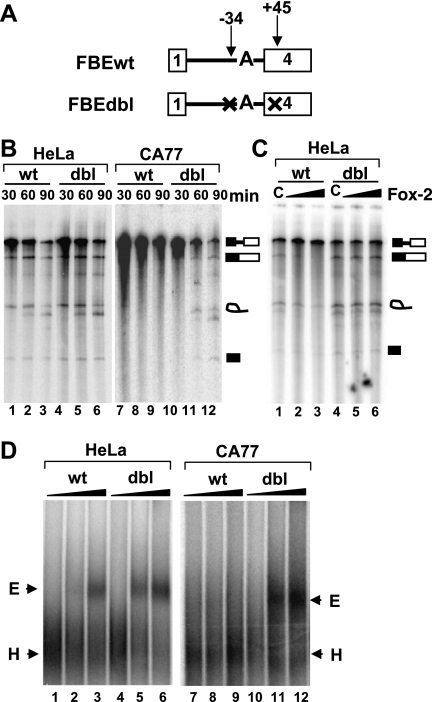
FIG. 1.
Splicing and spliceosome assembly of the calcitonin splicing substrate in HeLa or CA77 nuclear extract. (A) Diagram of splicing substrates used in these experiments. Both substrates contain the branch point mutation from UACUGUC to UACUGAC. FBEdbl also contains FBE mutations. An “X” indicates disruption of the wild-type UGCAUG to UGACUG. (B) Comparison of splicing of FBEwt to FBEdbl substrate in either HeLa (lanes 1 to 6) or CA77 nuclear extract (lanes 7 to 12). Splicing was carried out for 30, 60, or 90 min. The diagrams to the right represent the migration position of the pre-RNA processing products. (C) In vitro splicing of FBEwt and FBEdbl substrates in HeLa extract containing truncated GST-Fox-2 (100 ng/μl) (lane 1) or increasing amounts of GST-Fox-2 (50 and 100 ng/μl) (lanes 2 to 3) for 60 min. Lanes C, truncated GST-Fox-2 control (56). (D) Formation of prespliceosome E complex on the FBEwt or FBEdbl substrate in HeLa (lanes 1 to 6) or CA77 (lanes 7 to 12) nuclear extract. The pre-mRNAs were incubated in nuclear extract in the absence of ATP for 0 min (lanes 1, 4, 7, and 10), 15 min (lanes 2, 5, 8, and 11), or 30 min (lanes 3, 6, 9, and 12). Complexes were separated by 1.5% native agarose gel, and the position of each complex is indicated.
The calcitonin splicing substrate, which was derived from the minigene reporter used in transfection experiments, contains sequences extending from adenovirus exon 1 to the middle of calcitonin-specific exon 4 (Fig. 1A). In particular, this splicing substrate contains the −34 and +45 UGCAUG silencer elements (designated Fox-1/Fox-2 protein binding element [or FBE in the present study]), which were shown to be bound by Fox-1/Fox-2 proteins and repress the calcitonin-specific exon 4 inclusion (56). The wild-type calcitonin splicing substrate has a suboptimal uridine branch point (see Fig. S1B in the supplemental material) and thus is a very poor splicing substrate in vitro (data not shown) (2). To increase splicing efficiency, the branch point was changed to the canonical adenosine to create all of the splicing substrates used in Fig. 1, 5, and 6 (Fig. 1A). Our previous cell transfection studies demonstrated that Fox-1/Fox-2 proteins could still efficiently inhibit exon 4 inclusion when this strong branch point adenosine was present (56).
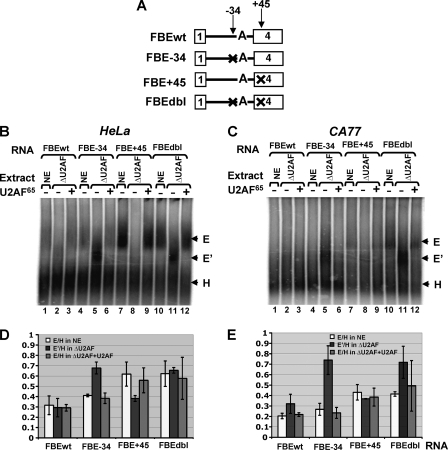
FIG. 6.
The −34 silencer element blocks prespliceosome E′ complex formation. (A) Diagram of pre-mRNA substrates. Formation of E′ or E complex on different RNA substrates in the untreated extract (NE), U2AF65-depleted extracts (ΔU2AF), or ΔU2AF extract supplemented with recombinant His-U2AF65. The migration positions of the complexes are shown on the right. (B) HeLa extract. (C) CA77 extract. (D) Quantitative evaluation of the E′ or E complex formation shown in panel B. Ratio of the E′ to H or of E to H (amount of E′ or E complex divided by amount of the H complex) was calculated based on three independent experiments. (E) Quantitative evaluation of the E′ or E complex formation shown in the panel C.
As shown in Fig. 1B, splicing intermediates and products of the FBEwt substrate were readily detected in HeLa extract but not in CA77 extract (Fig. 1B, lanes 1 to 3 and lanes 7 to 9). Importantly, the two nuclear extracts support the splicing reaction and spliceosome assembly of the Minx pre-mRNA with similar efficiencies (see Fig. S2 in the supplemental material). To determine whether splicing repression is dependent on the two FBEs, we tested splicing of the substrate FBEdbl, in which both FBEs are mutated. In HeLa extract, the FBEdbl substrate was spliced slightly more efficiently than the FBEwt substrate (Fig. 1B, lanes 1 to 6), a finding consistent with the presence of low levels of Fox-2 proteins in HeLa cells (see Fig. S1D in the supplemental material). In CA77 extract, which contains high levels of Fox-1 and Fox-2, and where the FBEwt substrate could not be spliced, splicing was readily detected with the FBEdbl substrate (Fig. 1B, lanes 7 to 12). Furthermore, addition of glutathione S-transferase (GST)-Fox-2 significantly repressed splicing of the FBEwt substrate in a dose-dependent manner but had no effect on the FBEdbl substrate (Fig. 1C). Collectively, these results demonstrate that Fox-1/Fox-2 proteins repress splicing of the calcitonin splicing substrate in an FBE-dependent manner.
We next investigated how Fox-1/Fox-2 proteins block splicing. We reasoned that Fox-1/Fox-2 proteins most likely block formation of the prespliceosome E complex, because we demonstrated previously that these proteins reduced binding of U2AF65 to the polypyrimidine tract of the 3′ splice site (56). We therefore examined E complex formation by native gel analysis. Incubation of FBEwt pre-mRNA in HeLa extract in the absence of ATP and heparin at 30°C led to formation of the E complex (Fig. 1D, lanes 1 to 3), which has a slower mobility than the H complex. However, in CA77 extract, no E complex could be detected on the FBEwt pre-mRNA (Fig. 1D, lanes 7 to 9). This result suggests that the transition from H to E complex on the FBEwt pre-mRNA is blocked in CA77 cells.
We next examined complex formation on the FBEdbl substrate in which the two functionally important FBEs are mutated. In HeLa extract, moderately more E complex formed on the FBEdbl substrate than on the FBEwt substrate (Fig. 1D, lanes 1 to 6). The difference in complex formation between these substrates was more dramatic in CA77 extract, in which significantly more E complex was formed on FBEdbl than FBEwt substrate (Fig. 1D, lanes 7 to 12). Collectively, the results shown in Fig. 1B and D demonstrate that mutation of the FBEs results in the loss of splicing repression and allows formation of spliceosome complexes in CA77 extract.
Fox-1/Fox-2 proteins prevent binding of U2AF65 to the polypyrimidine tract of the 3′ splice site.
The large subunit of U2AF, U2AF65, plays a critical role in the formation of the prespliceosome E complex (36). Previously, using a UV cross-linking/IP assay, we demonstrated that Fox-1/Fox-2 proteins reduced U2AF65 binding to the polypyrimidine tract sequence of the 3′ splice site upstream of exon 4 in an FBE-dependent manner (56). To understand the mechanism by which Fox-1/Fox-2 proteins inhibit U2AF65 binding, we examined the components of the presplicing complexes by the well-established MS2-MBP affinity purification approach. For this experiment, three MS2 hairpins that bind to the MS2-MBP fusion protein were ligated to the 3′ end of four different calcitonin splicing substrates containing the wild-type branch point, as well as wild-type or mutated UGCAUG at −34, +45, or all of the four positions (−34, +45, +177, and +252) (Fig. 2A). The FBEwt construct without the MS2 hairpins was used as a negative control. In our previous cell transfection experiments, the quadruple mutant had a similar splicing pattern as the double mutant −34+45, indicating that the UGCAUG elements at +177 and +252 contribute little to Fox-1/Fox-2 protein-mediated regulation of exon 4 inclusion (56). The addition of the hairpins did not alter the in vitro splicing pattern of the pre-mRNAs (data not shown). The RNA substrates were incubated with recombinant MS2-MBP fusion protein and HeLa extract, and the reaction mix was subsequently separated by a sizing column. The radioactivity peaked in fractions 6 and 7 (see Fig. S3A in the supplemental material), which thus were pooled and passed through a column of amylose beads. The bound complexes were eluted with maltose and analyzed for their RNA and protein content. We first analyzed the RNA components of the eluted complex by primer extension. As shown in Fig. S3B, U1, U2, U4, U5, and U6 snRNA were detected in the complex purified from either FBEwt-M or FBEqua-M (see Fig. S3B in the supplemental material). This result indicates that the purified complexes contain all of the major snRNPs.
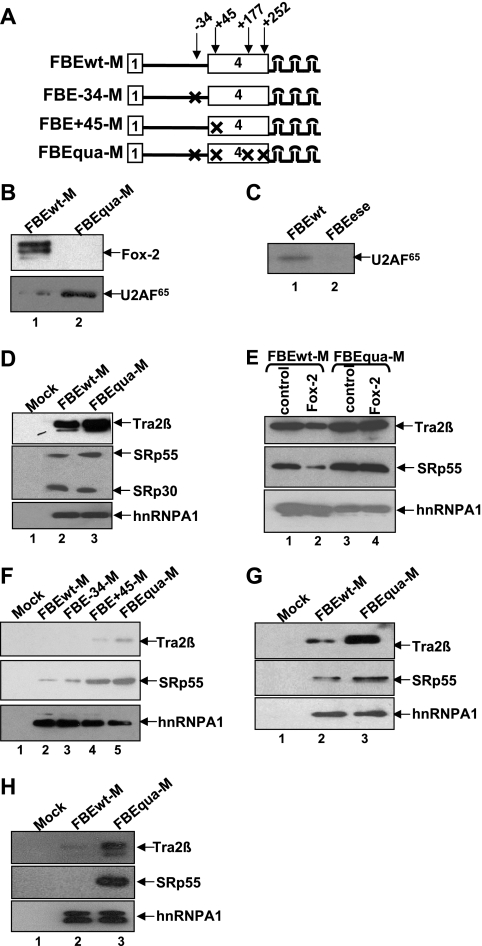
FIG. 2.
The +45 silencer element blocks U2AF65 binding by interfering with association of Tra2β and SRp55 with ESEs. (A) Diagram of RNA substrates. All of the substrates contain three MS2 hairpins at the 3′ end. An “X” indicates disruption of FBE elements. HeLa cell nuclear extracts were used in panels B to G, and CA77 cell nuclear extracts were used in panel H. (B) Western blot showing association of Fox-2 or U2AF65 with the FBEwt-M or FBEqua-M RNA substrates. (C) UV cross-linking/IP of U2AF65 with wild-type (FBEwt) or ESE mutant (FBEese) substrates. These two substrates do not contain MS2 hairpins. (D) Western blot showing association of Tra2β and SRp55 with FBEwt-M or FBEqua-M. Wild-type RNA substrate without MS2 hairpin is used as a negative mock control in panels D, F, G, and H. HnRNPA1 is used as a loading control in panels D to H. (E) Western blot showing association of Tra2β and SRp55 with the FBEwt-M RNA substrate in the presence of GST-Fox-2 protein (10 ng/μl). A truncated Fox-2 protein (10 ng/μl) is used as a control (lanes 1 and 3). (F) Western blot showing that mutation of the +45 but not the −34 silencer element increases association of Tra2β and SRp55 with the RNA substrate. (G) Western blot showing that the increase of the Tra2β and SRp55 association with the RNA substrates as a result of mutation of +45 silencer element occurs prior to the E complex formation. FBEwt-M or FBEqua-M substrate was incubated with HeLa extract in the absence of ATP and heparin. (H) Western blot showing that the increase of the Tra2β and SRp55 association with the RNA substrates as a result of mutation of +45 silencer element occurs in CA77 nuclear extracts.
We next examined the protein components of the eluted complexes by Western blot analysis. First, we determined whether Fox-1/Fox-2 proteins and U2AF65 were differentially associated with the complexes formed on the wild-type (FBEwt-M) and mutant (FBEqua-M) RNA. As expected, a significant amount of Fox-2 protein was detected in the complexes formed on FBEwt-M, while no Fox-2 was detected in the complexes formed on FBEqua-M (Fig. 2B). Conversely, there was dramatically more U2AF65 in the FBEqua-M than in the FBEwt-M complexes (Fig. 2B). These results are consistent with our previous UV cross-linking/IP analysis (56). More importantly, they demonstrate that the MS2-MBP purification approach provides a very useful means to investigate the mechanisms of Fox-1/Fox-2-mediated splicing inhibition.
Fox-1/Fox-2 proteins interfere with binding of Tra2β and SRp55 to the ESE element.
Previous studies demonstrated that two exon splicing enhancer (ESE) elements A and B located on exon 4 are required for inclusion of this exon (51). Mutation of these ESE elements dramatically decreased exon 4 inclusion (56). The UGCAUG element at +45 is 18 and 62 nucleotides upstream of the A and B elements, respectively (see Fig. S1B in the supplemental material). Our previous cell transfection results indicated that the +45 UGCAUG silencer element function to suppress ESE-dependent exon 4 inclusion (56). Previous studies by others also showed that Tra2β and SRp55 bound to the A and B on exon 4, respectively, and promoted exon 4 inclusion (47, 48, 51). Therefore, we sought to determine whether Fox-1/Fox-2 proteins interfere with Tra2β and/or SRp55 association with these ESE elements in a +45 UGCAUG silencer element-dependent manner.
As shown in a UV-cross-linking/IP experiment, mutation of the ESEs abolishes U2AF65 binding (Fig. 2C). Addition of recombinant His-Tra2β or His-SRp55 slightly increased cross-linking of U2AF65 to the wild-type RNA substrate (data not shown). These results suggest that the Tra2β and SRp55 promote U2AF65 binding by interacting with the ESE elements.
We next used the MS2-MBP affinity purification approach to determine whether the Fox-1/Fox-2 proteins block binding of Tra2β/SRp55. Note that in the biochemical purification and analysis experiments described in Fig. 2 and 3, RNA substrates that contain wild-type uridine branch point were used (Fig. 2A). Increased association of both proteins with the FBEqua-M complexes compared to FBEwt-M complexes was observed (Fig. 2D). SRp30 and hnRNPA1 in the FBEqua-M and FBEwt-M complexes serve as loading controls (Fig. 2D). The addition of GST-Fox-2 reduced the association of Tra2β or SRp55 with the FBEwt-M substrate (Fig. 2E, lanes 1 to 2). Importantly, however, association of Tra2β/SRp55 with the FBEqua-M substrate was not affected by GST-Fox-2 (Fig. 2E, lanes 3 to 4). Interestingly, as shown in Fig. 2F, mutation of the +45 UGCAUG element increased Tra2β/SRp55 association (compare lanes 2 and 4), while mutation of the −34 UGCAUG element did not affect Tra2β/SRp55 association (compare lanes 2 and 3). These results demonstrate that the Fox-1/Fox-2 proteins interfere with Tra2β and SRp55 binding to ESE elements in a +45 UGCAUG silencer element-dependent manner.
FIG. 3.
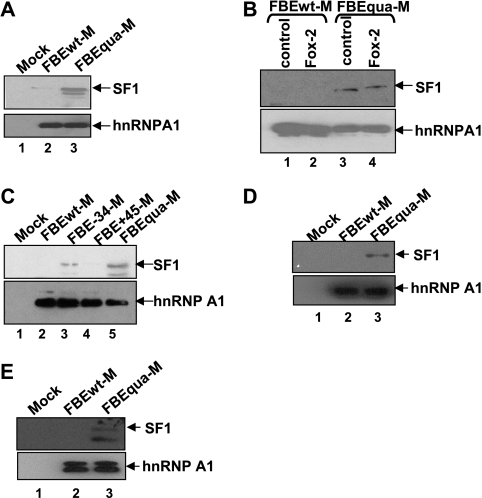
The −34 silencer element interferes with the association of SF1 with the branch point sequence. HeLa cell nuclear extracts were used in panels A to D, and CA77 cell nuclear extracts were used in panel E. (A) Western blot showing association of SF1with RNA substrates FBEwt-M or FBEqua-M. The wild-type RNA substrate without MS2 hairpins is used as a negative mock control in panels A, C, and D. hnRNPA1 is used as a loading control in panels A to D. (B) The addition of GST-Fox-2 (10 ng/μl) reduces the association of SF1 with the FBEwt-M RNA substrate. A truncated GST-Fox-2 (10 ng/μl) is used as a negative control (lanes 1 and 3). (C) Western blot showing that mutation of the −34 but not the +45 silencer element increases SF1 association with the RNA substrate. (D) Western blot showing that the increase of SF1 association with the RNA substrates as a result of silencer element mutations occurs prior to the E complex formation. FBEwt-M or FBEqua-M substrate was incubated with HeLa extract in the absence of ATP and heparin. (E) Western blot showing that the increase of SF1 association with the RNA substrates as a result of silencer element mutations occurs in CA77 nuclear extracts.
To test whether blocking of Tra2β/SRp55 association occurs before E complex assembly, the FBEwt-M or FBEqua-M substrate was incubated with HeLa extract in the absence of ATP and heparin at 30°C to allow formation of the nonspecific H complex and prespliceosome E complex. The assembly process was monitored by native gel analysis (data not shown). These H and E complexes were purified by MS2-MBP affinity. Again, Tra2β and SRp55 were detected in the FBEqua-M complex but to a lesser extent in the FBEwt-M complex, suggesting that blocking of Tra2β and SRp55 occurs before E complex formation (Fig. 2G).
To determine whether Fox-1/Fox-2 proteins block Tra2β/SRp55 association with the calcitonin exon 4 in CA77 cell nuclear extract, where these proteins are expressed at significantly higher levels than in HeLa cells, we carried out similar complex purification and analysis using CA77 nuclear extracts. Similar to HeLa extracts, increased association of Tra2β and SRp55 proteins with the FBEqua-M complexes compared to FBEwt-M complexes was observed in CA77 extracts (Fig. 2H). Again, hnRNPA1 serves as a loading control (Fig. 2H). The double bands detected by the anti-hnRNP A1 antibody most likely represent alternatively spliced isoforms of hnRNP A1. This result indicates that Fox-1/Fox-2 proteins interfere with binding of Tra2β and SRp55 to the ESE element in CA77 cells.
Fox-1/Fox-2 proteins block SF1 binding to the branch point sequence.
SF1, like its yeast ortholog branch point binding protein (BBP), has been shown to interact specifically with the pre-mRNA branch point sequence (5). Since the UGCAUG element at −34 is only six nucleotides upstream from the branch point sequence, we tested whether Fox-1/Fox-2 proteins disrupt this early step of splice site recognition using the same MS2-MBP affinity purification approach. Western blot analysis using anti-SF1 antibody indicates minimal amounts of the SF1 protein in the complex formed on FBEwt-M and significantly increased amounts in the complex formed on FBEqua-M (Fig. 3A). We also added GST-Fox-2 fusion protein into the HeLa extract before MS2-MBP affinity purification. Because the SF1 interaction with FBEwt-M was undetectable in this particular experiment (likely due to the use of the noncanonical branch point-containing substrates), we could not examine whether GST-Fox-2 protein blocks SF1 binding (Fig. 3B, lanes 1 and 2). However, the presence of GST-Fox-2 protein could not repress SF1 binding to the FBEqua-M substrate (Fig. 3B, lanes 3 and 4).
In contrast to Tra2β/SRp55, mutation of the −34 UGCAUG element but not the +45 UGCAUG element increased SF1 association (Fig. 3C). Similar to Tra2β and SRp55, blocking of SF1 association by Fox-1/Fox-2 proteins occurs before E complex assembly (Fig. 3D).
We next used the MS2-MBP affinity purification approach to determine whether the Fox-1/Fox-2 proteins block binding of SF1 in CA77 nuclear extracts. Association of SF1 protein with the FBEqua-M complexes was also increased compared to FBEwt-M complexes in this extract (Fig. 3E).
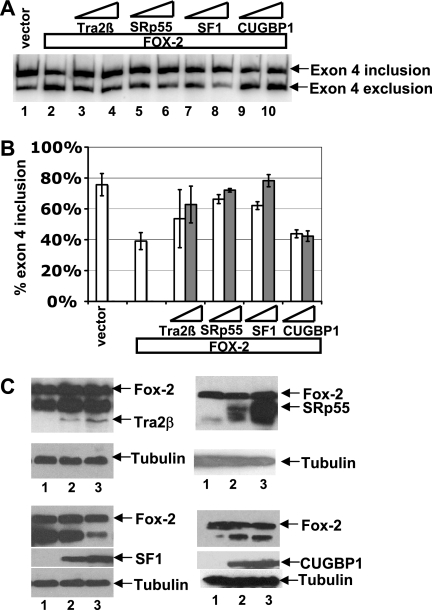
Overexpression of Tra2β/SRp55/SF1 reverses the Fox-1/Fox-2-mediated reduction of calcitonin exon 4 inclusion.
Since Fox-1/Fox-2 proteins interfered with Tra2β/SRp55/SF1 association through binding to the −34 and +45 UGCAUG silencer elements, we hypothesized that increasing the level of Tra2β/SRp55/SF1 may reverse the effect of Fox-1/Fox-2 proteins on the alternative splicing of calcitonin-specific exon 4 in transfected cells. Indeed, in HeLa cells, overexpression of Tra2β, SRp55, or SF1 reversed the effect of Fox-2, thereby restoring calcitonin-specific splicing pattern of the calcitonin/CRGP reporter pre-mRNA shown in Fig. S1B in the supplemental material (Fig. 4). In contrast, overexpression of CUGBP1, another RNA-binding protein, had no effect on Fox-1/Fox-2-mediated regulation of exon 4 (Fig. 4). These results indicate that the antagonism between Fox-2 protein and basic splicing factors observed in vitro can be duplicated inside cells.
FIG. 4.
Overexpression of Tra2β, SRp55, or SF1 reverses the Fox-2 protein-mediated repression of calcitonin-specific exon 4. (A) RT-PCR assay showing that Tra2β, SRp55, or SF1 counteracts the effect of overexpressed Fox-2. HeLa cells were cotransfected with the calcitonin/CGRP minigene reporter shown in Fig. S1B in the supplemental material and vector DNA (lane 1) and 50 ng of the Fox-2 expression plasmid F011 (lanes 2 to 10) in the absence (lane 2) or presence of increasing amounts (400 or 800 ng) of expression plasmid encoding Tra2β (lanes 3 and 4), SRp55 (lanes 5 and 6), SF1 (lanes 7 and 8), or CUGBP1 (lanes 9 and 10). (B) PhosphorImager quantification of the RT-PCR results shown in panel A. For each analysis, the percentage of exon 4 inclusion [exon 4 inclusion/(exon 4 inclusion + exon 4 exclusion)] is calculated based on at least three independent transfections. (C) Western blot analysis of expression of the cotransfected Fox-2, Tra2β, SRp55, SF1, and CUGBP1 proteins in HeLa cells. In each blot, Fox-2 proteins is present at constant levels in all of the three lanes. Lane 1 has no additional cotransfected proteins, while lanes 2 and 3 show increasing levels of indicated proteins. The blot was probed with anti-tubulin for loading control, anti-Myc for Tra2β or SRp55, and anti-His for SF1 or CUGBP1.
The −34 and +45 UGCAUG silencer elements blocks formation of the E′ and E complexes, respectively.
It was shown that formation of the E′ complex is dependent on SF1 interacting with the branch point region, as well as the U1 snRNA-5′ splice site base pairing interaction (23). As described above, Fox-1/Fox-2 proteins interfere with SF1 binding to the branch point sequence, and this effect depends on the −34 but not the +45 silencer element. We therefore suspected that the −34 silencer element bound by Fox-1/Fox-2 proteins blocks formation of the E′ complex. To examine E′ complex formation, we depleted nuclear extract of U2AF65 (see Fig. S4 in the supplemental material) (23). After incubation of the FBEdbl pre-mRNA in HeLa extract in the absence of ATP for 60 min at 30°C, E complex was formed (Fig. 5A, lane 1). However, in the U2AF65-depleted HeLa extract, a complex that moves faster than the E complex was detected (Fig. 5A, lane 2). This complex is most likely the previously described E′ complex (23) based on the results of the following experiments. First, addition of recombinant U2AF65 efficiently chased the more rapidly migrating complex into the E complex (Fig. 5A, lanes 3). Second, formation of this, as well as the E complex, requires a base pairing interaction between the U1 snRNA and the 5′ splice site (Fig. 5B, lanes 3 and 7). Interaction of the U2 snRNA with the branch point sequence is not required (Fig. 5B, lanes 4 and 8). Furthermore, we eluted both E and E′ complexes from agarose gels and assayed for the presence of U1 and U2 snRNAs by primer extension. U1 snRNA, but not U2 snRNA was detected in both the E and the E′ complexes (Fig. 5C). These results strongly suggest that the rapidly migrating complex is the previously described E′ complex (23).
To investigate how the −34 and +45 silencer elements affect the formation of E′ and E complexes, four RNA substrates were examined in prespliceosome complex analysis (Fig. 6A). We used a PhosphorImager to detect the radioactivity in each complex and calculated the E′/H or E/H ratio. The FBE-34 mutant, which did not significantly affect E complex formation, resulted in a significant increase of E′/H ratio compared to FBEwt (0.68 versus 0.30) (Fig. 6B, lanes 1 to 6, and Fig. 6D). Conversely, the E′/H ratio was not significantly increased on the FBE+45 compared to the FBEwt substrate (0.38 versus 0.30) (Fig. 6B, lanes 1 to 3 and lanes 7 to 9). The FBE+45 substrate promoted the E complex formation compared to the wild-type substrate (E/H ratio of 0.62 versus 0.32) (Fig. 6B lanes 1 to 3 and lanes 7 to 9, and Fig. 6D). The formation of both E′ complex and the E complex were dramatically increased on the FBEdbl substrate (Fig. 6B lanes 1 to 3 and lanes 10 to 12, and Fig. 6D). The results demonstrate that the −34 silencer element blocks E′ complex formation, while the +45 silencer element inhibits the transition from E′ complex to E complex.
In CA77 extract, which contains significantly more endogenous Fox-1/Fox-2 proteins than HeLa extract, no E complex could be detected, and minimal E′ complex was formed on the FBEwt pre-mRNA (Fig. 6C, lanes 1 to 3), a finding consistent with the extremely low splicing activity of this extract on the calcitonin substrate (Fig. 1). As observed in HeLa extract, mutation of the −34 silencer element dramatically increased E′ complex formation and did not significantly affect E complex formation (Fig. 6C, lanes 1 to 6, and Fig. 6E), while mutation of the +45 silencer element increased E complex formation and had little effect on E′ complex formation (Fig. 6C, lanes 1 to 3 and lanes 7 to 9, and Fig. 6E).
The results of the CA77 nuclear extract indicate that Fox-1/Fox-2 proteins block the E′ as well as E complex formation in a position-dependent manner.
DISCUSSION
Fox-1/Fox-2 proteins are tissue-specific splicing regulators that control the inclusion of a number of alternative exons through binding to UGCAUG elements. Very little is known about the molecular mechanisms through which Fox-1/Fox-2 proteins regulate alternative splicing. Fukumura et al. showed that Fox-1 binds to intron 8 of the hF1γ pre-mRNA to interfere with E complex formation on intron 9 (12). This cross-exon repression by Fox-1 protein is different from the mechanism by which most regulatory factors repress splicing of the intron containing their binding sites (9, 52). In the calcitonin/CGRP system, Fox-1/Fox-2 proteins inhibit inclusion of the calcitonin-specific exon 4 by binding to two UGCAUG silencer elements, which are located on exon 4 and its upstream intron 3. We demonstrate in this report that Fox-1/Fox-2 proteins repress exon inclusion on a single substrate through multiple mechanisms.
Multistep regulation by a single splicing regulator as a robust mechanism to ensure proper alternative splicing.
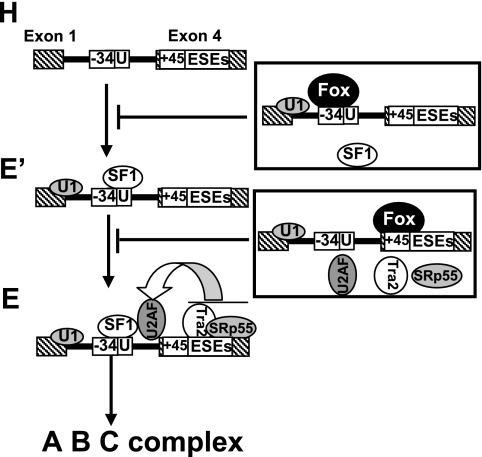
The two-step repression of calcitonin-specific splicing described here reveals a novel strategy for splicing regulation. Although the idea that splice site selection involves the integrated actions of multiple positive and negative factors is well established, regulation is usually directed toward the formation of a single subspliceosome complex. Our observation provides a novel example of how one regulatory factor represses splicing through blocking the formation of different prespliceosome complexes. In this example, both steps play an important role in the repression of calcitonin-specific exon 4 inclusion. In the first step, the −34 silencer element strongly blocks formation of prespliceosome E′ complex (Fig. 7). However, the formation of a small amount of prespliceosome E′ complex in wild-type or mutant +45 silencer element RNA substrate indicates that the −34 silencer element does not entirely prevent prespliceosome E′ complex formation, allowing some prespliceosome assembly through the E′ complex (Fig. 5 and 6). In the second step, the +45 silencer element blocks the transition of E′ complex that escaped the −34 silencer element control to E complex by interfering with U2AF65 binding (Fig. 7). Thus, blocking of E complex formation can be considered a fail-safe mechanism that provides an additional barrier to splicing, thus ensuring highly robust regulation. Importantly, overexpression of Fox-2 protein with wild-type or different mutant calcitonin/CGRP reporter in transfected cells also indicated that both silencer elements played an important role in regulation of calcitonin-specific exon 4 inclusion (see Fig. S5 in the supplemental material).
FIG. 7.
Model for two-step repression of prespliceosome complex formation. The production of CGRP in neuronal cells regulated by Fox-1/Fox-2 proteins involves two distinct regulatory events. First, the −34 silencer element prevents E′ complex formation through repressing SF1 binding. Second, +45 silencer element blocks the transition from E′ complex that is escaped from −34 silencer element control to E complex via inhibiting U2AF65 binding to polypyrimidine tract.
The −34 silencer element represses SF1 binding and E′ complex formation.
Spliceosome assembly has been described as passing through three intermediates (E-A-B) before formation of the final catalytically active C complex (8, 46). Recently, Kent et al. showed that the prespliceosome complex E′ is formed in U2AF-depleted HeLa nuclear extracts prior to E complex formation (23). The E′ complex is analogous to the yeast commitment complex CC1 (26, 44, 55). Formation of the E′ complex is dependent on interaction of SF1 with the branch point region and the U1 snRNA with the 5′ splice site. Because the E′ complex can be chased into E complex and commits the pre-mRNA to the splicing pathway, it functions as a precursor to E complex in spliceosome assembly (23). A number of previous studies have shown that, in most mammalian systems, regulated selection of splice sites can occur at the step of formation of the early ATP-independent E complex or the ATP-dependent association of U2 snRNP during A complex formation (6, 15, 17, 34). In the present study, we show that the −34 UGCAUG silencer element represses splicing of calcitonin-specific exon 4 by preventing the formation of the prespliceosome E′ complex. The Fox-1/Fox-2 proteins, by interacting with the −34 UGCAUG silencer element, block SF1 binding to the branch point without affecting U1 snRNP binding (data not shown). To our knowledge, this is the first example of regulated E′ complex formation. It is, however, very likely that other regulatory factors capable of affecting U1 snRNP or SF1 binding may also regulate E′ complex formation. For example, of the 20 computationally identified Fox-1/Fox-2 binding sites that are phylogenetically and spatially conserved, 8 are within 35 nucleotides of a 5′ splice site and 1 is 4 nucleotides from a 3′ splice site (38).
SF1 is a basic component of the earliest presplicing complexes; however, a role of SF1 in alternative splicing has not been reported. Via its KH-QUA2 domain, SF1 specifically recognizes branch point sequence in the pre-mRNA transcripts during initial spliceosome assembly (29). The yeast CC1 and mammalian E′ complex can be formed in ΔMud2p/U2AF-depleted extract but not in ΔBBP/SF1-depleted extract, suggesting that BBP/SF1 is more important to this particular commitment complex formation than Mud2p/U2AF (23). In the present study, our results demonstrate that interference of association of SF1 by Fox-1/Fox-2 proteins is dependent on the −34 not on the +45 UGCAUG silencer element (Fig. 3). The −34 UGCAUG silencer element is only six nucleotides upstream from the branch point sequence. It is therefore highly likely that the −34 silencer element bound by Fox-1/Fox-2 proteins inhibit prespliceosome E′ complex formation through physically interfering with SF1 binding (Fig. 7).
The 3′ splice site upstream of calcitonin-specific exon 4 contains a noncanonical branch point (uridine in human and cytosine in the rat and mouse), which was demonstrated to function as a negative element. Mutation of uridine to the canonical branch point adenosine resulted in a significant increase in calcitonin exon 4 inclusion in all cell lines tested (1, 2). In our previous transfection studies using splicing reporters, we also showed that mutation of the uridine branch point to adenosine increased exon 4 inclusion in both HeLa and CA77 cells, although exon 4 inclusion of the mutant pre-mRNA in CA77 cells was significantly lower than HeLa cells, suggesting that other proteins present in CA77 cells function to lower inclusion of this exon (56). Furthermore, overexpression of Fox-1 or Fox-2 protein in HeLa cells strongly repressed exon 4 inclusion from the mutant RNA (56). In the current study, we showed that in vitro splicing of RNA substrates containing the canonical adenosine branch point was significantly blocked by Fox-2 protein. Collectively, these data demonstrate that Fox-mediated repression of exon 4 inclusion can override the presence of a strong branch point.
The +45 silencer element prevents U2AF65 binding and E complex formation.
The large subunit of U2AF, U2AF65, plays a critical role in the formation of the prespliceosome E complex. It binds to the polypyrimidine tract and promotes the ATP-dependent binding of U2 snRNP to the branch point sequence of a 3′ splice site (42). A number of splicing regulatory factors modulate the formation of E complex by blocking or promoting U2AF65 binding (6, 45). In the present study, we show that Fox-1/Fox-2 proteins reduce U2AF65 binding and block E complex formation on the calcitonin splicing substrate in a +45 UGCAUG element-dependent manner (Fig. 7).
Repression by the +45 silencer element requires the existence of ESE elements A and B (56); thus, the repression is more complex than simple competition between Fox-1/Fox-2 proteins and U2AF65. The ESEs are located 18 nucleotides upstream from the +45 UGCAUG silencer element (see Fig. S1B in the supplemental material). Tra2β and SRp55 are recruited to the A and B elements, respectively, and activate exon 4 inclusion (47, 48, 51). In the present study, we show that the A and B elements dramatically promote U2AF65 binding to weak polypyrimidine tracts (Fig. 2C), a finding consistent with the model for ESE-dependent 3′ splice site activation by SR proteins (22). Interestingly, Fox-1/Fox-2 proteins interfere with binding of Tra2β and SRp55 to the ESEs, which is dependent on the +45 silencer element but not the −34 silencer element. It is therefore possible that repression of U2AF65 binding by the +45 silencer element occurs by antagonizing the binding of Tra2β and SRp55 to ESEs.
In summary, our studies demonstrate that Fox-1/Fox-2 proteins block splicing of intron 3 of the calcitonin/CGRP pre-mRNA by repressing two distinct steps of prespliceosome assembly. This built-in redundant double block mechanism provides for robust regulation of alternative splicing. Furthermore, our studies illustrate the diversity of mechanisms by which spliceosome assembly can be modulated to achieve differential splicing patterns within a given pre-mRNA.
Supplementary Material
Acknowledgments
We thank the following individuals for providing antibodies and plasmids: Angela Krämer at the University of Geneva (anti-SF1 antibody), Stefan Stamm at the University of Erlangen (anti-Tra2β antibody), Douglas Black at the University of California at Los Angeles (MS2-MBP plasmid and anti-Fox-1 antibody), and James Patton at Vanderbilt University (His-SRp55-expressing and His-SRp35-expressing viruses). We thank Helen Salz, Jo Ann Wise, and Melissa Hinman for critical reading and editing of the manuscript.
This study was supported by an NIH grant (NS-049103-01) to H.L. H.-L.Z. was supported by a postdoctoral fellowship from the American Heart Association.
Footnotes
Published ahead of print on 23 June 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adema, G. J., R. A. Bovenberg, H. S. Jansz, and P. D. Baas. 1988. Unusual branch point selection involved in splicing of the alternatively processed calcitonin/CGRP-I pre-mRNA. Nucleic Acids Res. 169513-9526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adema, G. J., K. L. van Hulst, and P. D. Baas. 1990. Uridine branch acceptor is a cis-acting element involved in regulation of the alternative processing of calcitonin/CGRP-l pre-mRNA. Nucleic Acids Res. 185365-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baraniak, A. P., J. R. Chen, and M. A. Garcia-Blanco. 2006. Fox-2 mediates epithelial cell-specific fibroblast growth factor receptor 2 exon choice. Mol. Cell. Biol. 261209-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett, M., S. Pinol-Roma, D. Staknis, G. Dreyfuss, and R. Reed. 1992. Differential binding of heterogeneous nuclear ribonucleoproteins to mRNA precursors prior to spliceosome assembly in vitro. Mol. Cell. Biol. 123165-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berglund, J. A., M. L. Fleming, and M. Rosbash. 1998. The KH domain of the branch point sequence binding protein determines specificity for the pre-mRNA branchpoint sequence. RNA. 4998-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black, D. L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72291-336. [DOI] [PubMed] [Google Scholar]
- 7.Blencowe, B. J. 2006. Alternative splicing: new insights from global analyses. Cell 12637-47. [DOI] [PubMed] [Google Scholar]
- 8.Brow, D. A. 2002. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 36333-360. [DOI] [PubMed] [Google Scholar]
- 9.Caputi, M., A. Mayeda, A. R. Krainer, and A. M. Zahler. 1999. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 184060-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Champion-Arnaud, P., O. Gozani, L. Palandjian, and R. Reed. 1995. Accumulation of a novel spliceosomal complex on pre-mRNAs containing branch site mutations. Mol. Cell. Biol. 155750-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das, R., and R. Reed. 1999. Resolution of the mammalian E complex and the ATP-dependent spliceosomal complexes on native agarose mini-gels. RNA. 51504-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukumura, K., A. Kato, Y. Jin, T. Ideue, T. Hirose, N. Kataoka, T. Fujiwara, H. Sakamoto, and K. Inoue. 2007. Tissue-specific splicing regulator Fox-1 induces exon skipping by interfering E complex formation on the downstream intron of human F1γ gene. Nucleic Acids Res. 355303-5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graveley, B. R. 2001. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 17100-107. [DOI] [PubMed] [Google Scholar]
- 14.Graveley, B. R. 2000. Sorting out the complexity of SR protein functions. RNA 61197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hertel, K. J. 2008. Combinatorial control of exon recognition. J. Biol. Chem. 2831211-1215. [DOI] [PubMed] [Google Scholar]
- 16.House, A. E., and K. W. Lynch. 2006. An exonic splicing silencer represses spliceosome assembly after ATP-dependent exon recognition. Nat. Struct. Mol. Biol. 13937-944. [DOI] [PubMed] [Google Scholar]
- 17.House, A. E., and K. W. Lynch. 2007. Regulation of alternative splicing: more than just the ABCs. J. Biol. Chem. 2831217-1221. [DOI] [PubMed] [Google Scholar]
- 18.Jamison, S. F., A. Crow, and M. A. Garcia-Blanco. 1992. The spliceosome assembly pathway in mammalian extracts. Mol. Cell. Biol. 124279-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin, Y., H. Suzuki, S. Maegawa, H. Endo, S. Sugano, K. Hashimoto, K. Yasuda, and K. Inoue. 2003. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 22905-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jurica, M. S., L. J. Licklider, S. R. Gygi, N. Grigorieff, and M. J. Moore. 2002. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA 8426-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jurica, M. S., and M. J. Moore. 2003. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 125-14. [DOI] [PubMed] [Google Scholar]
- 22.Kan, J. L., and M. R. Green. 1999. Pre-mRNA splicing of IgM exons M1 and M2 is directed by a juxtaposed splicing enhancer and inhibitor. Genes Dev. 13462-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kent, O. A., D. B. Ritchie, and A. M. Macmillan. 2005. Characterization of a U2AF-independent commitment complex (E′) in the mammalian spliceosome assembly pathway. Mol. Cell. Biol. 25233-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konarska, M. M., and P. A. Sharp. 1986. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell 46845-855. [DOI] [PubMed] [Google Scholar]
- 25.Lallena, M. J., K. J. Chalmers, S. Llamazares, A. I. Lamond, and J. Valcarcel. 2002. Splicing regulation at the second catalytic step by Sex-lethal involves 3′ splice site recognition by SPF45. Cell 109285-296. [DOI] [PubMed] [Google Scholar]
- 26.Legrain, P., B. Seraphin, and M. Rosbash. 1988. Early commitment of yeast pre-mRNA to the spliceosome pathway. Mol. Cell. Biol. 83755-3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, Q., J. A. Lee, and D. L. Black. 2007. Neuronal regulation of alternative pre-mRNA splicing. Nat. Rev. Neurosci. 8819-831. [DOI] [PubMed] [Google Scholar]
- 28.Lim, S. R., and K. J. Hertel. 2004. Commitment to splice site pairing coincides with A complex formation. Mol. Cell 15477-483. [DOI] [PubMed] [Google Scholar]
- 29.Liu, Z., I. Luyten, M. J. Bottomley, A. C. Messias, S. Houngninou-Molango, R. Sprangers, K. Zanier, A. Kramer, and M. Sattler. 2001. Structural basis for recognition of the intron branch site RNA by splicing factor 1. Science 2941098-1102. [DOI] [PubMed] [Google Scholar]
- 30.Lou, H., D. M. Helfman, R. F. Gagel, and S. M. Berget. 1999. Polypyrimidine tract-binding protein positively regulates inclusion of an alternative 3′-terminal exon. Mol. Cell. Biol. 1978-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lou, H., K. M. Neugebauer, R. F. Gagel, and S. M. Berget. 1998. Regulation of alternative polyadenylation by U1 snRNPs and SRp20. Mol. Cell. Biol. 184977-4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacMillan, A. M., P. S. McCaw, J. D. Crispino, and P. A. Sharp. 1997. SC35-mediated reconstitution of splicing in U2AF-depleted nuclear extract. Proc. Natl. Acad. Sci. USA 94133-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maniatis, T., and B. Tasic. 2002. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature 418236-243. [DOI] [PubMed] [Google Scholar]
- 34.Matlin, A. J., F. Clark, and C. W. Smith. 2005. Understanding alternative splicing: toward a cellular code. Nat. Rev. Mol. Cell. Biol. 6386-398. [DOI] [PubMed] [Google Scholar]
- 35.Melton, A. A., J. Jackson, J. Wang, and K. W. Lynch. 2007. Combinatorial control of signal-induced exon repression by hnRNP L and PSF. Mol. Cell. Biol. 276972-6984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michaud, S., and R. Reed. 1993. A functional association between the 5′ and 3′ splice site is established in the earliest prespliceosome complex (E) in mammals. Genes Dev. 71008-1020. [DOI] [PubMed] [Google Scholar]
- 37.Michaud, S., and R. Reed. 1991. An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes Dev. 52534-2546. [DOI] [PubMed] [Google Scholar]
- 38.Minovitsky, S., S. L. Gee, S. Schokrpur, I. Dubchak, and J. G. Conboy. 2005. The splicing regulatory element, UGCAUG, is phylogenetically and spatially conserved in introns that flank tissue-specific alternative exons. Nucleic Acids Res. 33714-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakahata, S., and S. Kawamoto. 2005. Tissue-dependent isoforms of mammalian Fox-1 homologs are associated with tissue-specific splicing activities. Nucleic Acids Res. 332078-2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ponthier, J. L., C. Schluepen, W. Chen, R. A. Lersch, S. L. Gee, V. C. Hou, A. J. Lo, S. A. Short, J. A. Chasis, J. C. Winkelmann, and J. G. Conboy. 2006. Fox-2 splicing factor binds to a conserved intron motif to promote inclusion of protein 4.1R alternative exon 16. J. Biol. Chem. 28112468-12474. [DOI] [PubMed] [Google Scholar]
- 41.Query, C. C., and M. M. Konarska. 2006. Splicing fidelity revisited. Nat. Struct. Mol. Biol. 13472-474. [DOI] [PubMed] [Google Scholar]
- 42.Ruskin, B., P. D. Zamore, and M. R. Green. 1988. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell 52207-219. [DOI] [PubMed] [Google Scholar]
- 43.Russo, A. F., T. M. Lanigan, and B. E. Sullivan. 1992. Neuronal properties of a thyroid C-cell line: partial repression by dexamethasone and retinoic acid. Mol. Endocrinol. 6207-218. [DOI] [PubMed] [Google Scholar]
- 44.Seraphin, B., and M. Rosbash. 1989. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell 59349-358. [DOI] [PubMed] [Google Scholar]
- 45.Sharma, S., A. M. Falick, and D. L. Black. 2005. Polypyrimidine tract binding protein blocks the 5′ splice site-dependent assembly of U2AF and the prespliceosomal E complex. Mol. Cell 19485-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Staley, J. P., and C. Guthrie. 1998. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92315-326. [DOI] [PubMed] [Google Scholar]
- 47.Tran, Q., T. P. Coleman, and J. R. Roesser. 2003. Human transformer 2β and SRp55 interact with a calcitonin-specific splice enhancer. Biochim. Biophys. Acta 1625141-152. [DOI] [PubMed] [Google Scholar]
- 48.Tran, Q., and J. R. Roesser. 2003. SRp55 is a regulator of calcitonin/CGRP alternative RNA splicing. Biochemistry 42951-957. [DOI] [PubMed] [Google Scholar]
- 49.Ule, J., and R. B. Darnell. 2006. RNA-binding proteins and the regulation of neuronal synaptic plasticity. Curr. Opin. Neurobiol. 16102-110. [DOI] [PubMed] [Google Scholar]
- 50.Underwood, J. G., P. L. Boutz, J. D. Dougherty, P. Stoilov, and D. L. Black. 2005. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol. Cell. Biol. 2510005-10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Oers, C. C., G. J. Adema, H. Zandberg, T. C. Moen, and P. D. Baas. 1994. Two different sequence elements within exon 4 are necessary for calcitonin-specific splicing of the human calcitonin/calcitonin gene-related peptide I pre-mRNA. Mol. Cell. Biol. 14951-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wagner, E. J., and M. A. Garcia-Blanco. 2001. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 213281-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, G. S., and T. A. Cooper. 2007. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat. Rev. Genet. 8749-761. [DOI] [PubMed] [Google Scholar]
- 54.Zeng, C., and S. M. Berget. 2000. Participation of the C-terminal domain of RNA polymerase II in exon definition during pre-mRNA splicing. Mol. Cell. Biol. 208290-8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, D., and M. Rosbash. 1999. Identification of eight proteins that cross-link to pre-mRNA in the yeast commitment complex. Genes Dev. 13581-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou, H. L., A. P. Baraniak, and H. Lou. 2007. Role for Fox-1/Fox-2 in mediating the neuronal pathway of calcitonin/calcitonin gene-related peptide alternative RNA processing. Mol. Cell. Biol. 27830-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou, Z., J. Sim, J. Griffith, and R. Reed. 2002. Purification and electron microscopic visualization of functional human spliceosomes. Proc. Natl. Acad. Sci. USA 9912203-12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu, H., R. A. Hasman, V. A. Barron, G. Luo, and H. Lou. 2006. A nuclear function of Hu proteins as neuron-specific alternative RNA processing regulators. Mol. Biol. Cell 175105-5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu, H., R. A. Hasman, K. M. Young, N. L. Kedersha, and H. Lou. 2003. U1 snRNP-dependent function of TIAR in the regulation of alternative RNA processing of the human calcitonin/CGRP pre-mRNA. Mol. Cell. Biol. 235959-5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.