Abstract
Insulin controls glucose homeostasis and lipid metabolism, and insulin impairment plays a critical role in the pathogenesis of diabetes mellitus. Human skeletal muscle and kidney enriched inositol polyphosphate phosphatase (SKIP) is a member of the phosphatidylinositol 3,4,5-trisphosphate phosphatase family (T. Ijuin et al. J. Biol. Chem. 275:10870-10875, 2000; T. Ijuin and T. Takenawa, Mol. Cell. Biol. 23:1209-1220, 2003). Previous studies showed that SKIP negatively regulates insulin-induced phosphatidylinositol 3-kinase signaling (Ijuin and Takenawa, Mol. Cell. Biol. 23:1209-1220, 2003). We now have generated mice with a targeted mutation of the mouse ortholog of the human SKIP gene, Pps. Adult heterozygous Pps mutant mice show increased insulin sensitivity and reduced diet-induced obesity with increased Akt/protein kinase B (PKB) phosphorylation in skeletal muscle but not in adipose tissue. The insulin-induced uptake of 2-deoxyglucose into the isolated soleus muscle was significantly enhanced in Pps mutant mice. A hyperinsulinemic-euglycemic clamp study also revealed a significant increase in the rate of systemic glucose disposal in Pps mutant mice without any abnormalities in hepatic glucose production. Furthermore, in vitro knockdown studies in L6 myoblast cells revealed that reduction of SKIP expression level increased insulin-stimulated Akt/PKB phosphorylation and 2-deoxyglucose uptake. These results imply that SKIP regulates insulin signaling in skeletal muscle. Thus, SKIP may be a promising pharmacologic target for the treatment of insulin resistance and diabetes.
Glucose homeostasis is controlled by insulin, which stimulates glucose transport in skeletal muscle, liver, and adipose tissues (1, 23, 28). One of the metabolic responses to insulin stimulation is glucose incorporation and glycogen synthesis in these tissues. Defects in insulin action in these tissues lead to insulin resistance and type II diabetes (23, 28). Type II diabetes is a polygenic disease affecting a large and growing population worldwide. Insulin signaling requires the generation of phosphatidylinositol 3,4,5-trisphosphate [PtdIns(3,4,5)P3] mediated by phosphatidylinositol 3 (PI3)-kinase and the subsequent activation of Akt/protein kinase B (PKB), which acts on various downstream effectors of the metabolic actions of insulin (15, 19). PtdIns(3,4,5)P3 phosphatases have similar biochemical functions, and they are potent modulators of the PI3-kinase signaling pathway. Among PtdIns(3,4,5)P3 phosphatases, PTEN (phosphatase and tensin homologue deleted on chromosome 10) and SHIP2 (Src homology domain 2 [SH2]-containing inositol 5-phosphatase 2) are reported to participate in negative regulation of the insulin signaling pathway (9, 19, 20, 21). PTEN is a negative regulator of the PI3-kinase signaling pathway and was first described as a tumor suppressor. A null mutation of Pten in mice resulted in embryonic lethality, and hepatocyte-specific deletion in mice caused tumor progression (11). Type II diabetes-associated PTEN polymorphisms have been reported (14). Skeletal muscle-specific deletion of Pten in mice protects against diet-induced insulin resistance and diabetes (30). Adipose tissue-specific deletion of Pten in mice resulted in enhanced insulin sensitivity and resistance to streptozotocin-induced diabetes (17). SHIP2 is a lipid phosphatase that hydrolyzes the 5′ phosphate from PtdIns(3,4,5)P3. Overexpression of SHIP2 inhibited insulin signaling, leading to glucose uptake and glycogen synthesis in 3T3-L1 adipocytes and L6 myotube cells (24, 25). SHIP2-deficient mice are physically normal, have normal glucose and serum insulin levels, and have normal insulin and glucose tolerances on a chow diet but are highly resistant to diet-induced obesity (4, 26).
Skeletal muscle and kidney enriched inositol polyphosphate phosphatase (SKIP)is a phosphoinositide phosphatase that hydrolyzes PtdIns(3,4,5)P3 and plays a negative role in insulin signaling in L6 myoblast cells (13). To understand the precise role of SKIP in insulin signaling in vivo, we mutated the mouse ortholog of the SKIP gene, Pps, using embryonic stem (ES) cell technology (3, 22).
MATERIALS AND METHODS
Generation of PpsBrdm1/+ mice.
A pair of Pps-specific PCR primers (5′-CCA GTC TCT GGC TAC TGA ACT G-3′ and 5′-AGA TCC CAG CAC CCA CAT AGA A-3′) was used to screen an arrayed library of targeting vectors (32). A prototype vector was isolated from a 5′ Hprt vector mouse genomic library with homology extending from nucleotide position 75361049 to 75367519. The final targeting vector, pTVPps, was prepared by deleting a 1,040-bp HpaI-BstBI fragment from the genomic homology region.
ES cell culture, electroporation, G418 selection, Southern blot screening of ES cell clones, and generation of germ line chimeras were performed as described previously (3, 31). Targeted clones were identified by Southern blotting of EcoRI digests with probe A. Potentially heterozygous progeny from the chimeras were identified visually by coat color and confirmed by Southern blot analysis of tail DNAs.
Southern blot screens with EcoRI digests of tail DNAs and probe A were used to distinguish the wild-type from the PpsBrdm1 allele; BclI digests and probe B (Fig. 1A) were used to differentiate between heterozygous and homozygous mutants. Probe A is the HpaI-BstBI fragment of the targeting vector; probe B is a PCR product amplified with primers 5′-CTG CCC AGC ATG AAA TCT CTT GAG TG-3′ and 5′-ATG CCC CCT GTG ATA TCA GCA GGT TA-3′ from the targeting vector and is located external to the homology region of the targeting vector. For Northern blot analysis, 10 μg per lane of total RNA from quadriceps was used, and mouse cDNA fragments coding for Pps and Gapdh were used as probes.
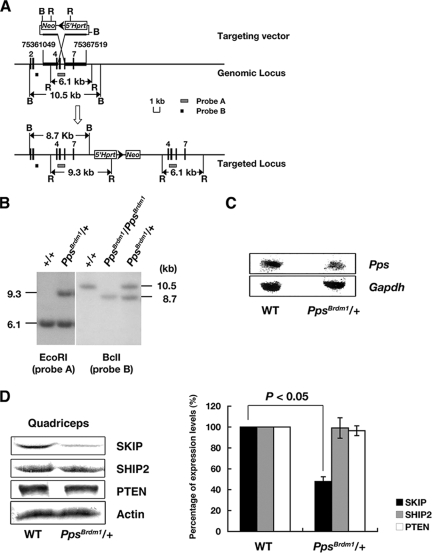
FIG. 1.
The PpsBrdm1 allele and the phenotypic analysis of PpsBrdm1/+ mice. (A) Schematic of the genomic Pps locus, the insertional targeting vector (pTVPps), and the targeted allele PpsBrdm1. Restriction sites are as follows: B, BclI; R, EcoRI. Triangle, loxP; Neo, neomycin-resistance gene cassette; 5′-Hprt, nonfunctional 5′-Hprt cassette. The probes and diagnostic restriction fragments are indicated. (B) Southern blot genotyping of agouti offspring of chimeras was performed with EcoRI-digested tail DNA and probe A. Southern blot genotyping of embryonic day 7.5 embryos, generated by intercrossing of PpsBrdm1/+ mice, was done with BclI-digested DNA and probe B. (C) Northern blot analysis of total RNA from quadriceps of wild-type and PpsBrdm1/+ mice. Pps and Gapdh cDNA fragments were used as probes. (D) Western blot analysis of SKIP, SHIP2, and PTEN using quadriceps lysates of PpsBrdm1/+ mice (n = 6) and wild-type mice (n = 6). Quantification of the Western blotting assay was analyzed by densitometry. WT, wild type.
Metabolic studies of mice and histology of mouse tissues.
PpsBrdm1/+ mice were backcrossed to C57BL/6J mice for at least five generations. Mice were fed with CA-1 (CLEA Japan Inc.) for the normal chow diet and with Quick Fat (CLEA) for the high-fat diet (HFD). Beginning at 4 weeks of age, male mice were fed either the normal chow diet or the high-fat diet and monitored for 6 months. Daily food intake and body length were measured at 12 weeks of age. Serum samples were obtained from male mice between 15 to 18 weeks of age and analyzed for the serum chemistry parameters. Serum triglycerides, nonesterified fatty acid (NEFA), and cholesterol were determined with commercially available diagnostic kits (WAKO). Serum leptin, adiponectin, and insulin were measured by enzyme-linked immunosorbent assay (leptin, Linco, Inc.; adiponectin, R&D Systems; insulin, Morinaga, Inc.).
Hyperinsulinemic-euglycemic clamp.
Hyperinsulinemic-euglycemic clamp analysis was performed as described previously (16), with minor modifications. Briefly, 5 to 7 days before the clamp, mice were anesthetized with sodium pentobarbital (80 to 100 mg/kg of body weight, intraperitoneally), and a catheter was inserted into the right internal jugular vein for infusion. The analysis was performed under nonstressful conditions with conscious mice that had been deprived of food overnight for 4 h. [3-3H]glucose was infused for 2 h at a rate of 0.05 μCi/min before initiation of the clamp, and a blood sample was collected at the end of this period to estimate basal glucose turnover. After a bolus injection of [3-3H]glucose (10 μCi; NEN Life Science) and the onset of subsequent continuous infusion of [3-3H]glucose (0.1 μCi/min), a hyperinsulinemic-euglycemic clamp was applied for 120 min with continuous infusion of insulin at a rate of 2.5 mU/kg of body weight per min. Plasma glucose concentration was monitored every 10 min, and 30% glucose was infused at a variable rate to maintain plasma glucose at basal concentrations. Blood samples were collected 80, 90, 100, 110, and 120 min after the onset of the clamp for determination of the plasma concentrations of [3-3H]glucose and 3H2O. The rates of glucose disposal and hepatic glucose production were calculated as described previously (16). Soleus muscle and epididymal white adipose tissue (EWAT) were isolated 130 min after the onset of the clamp for determination of Akt/PKB phosphorylation.
Glucose and insulin tolerance tests.
Wild-type and PpsBrdm1/+ male mice (15 to 18 weeks of age) were fasted overnight before intraperitoneal injection with either d-glucose (2.0 g/kg of body weight) or insulin (0.5 U/kg of body weight). Blood samples were collected at the time points indicated in Fig. 3A and B and in Fig. S2B in the supplemental material. Blood glucose concentrations were analyzed with a blood glucose test meter (SKK).
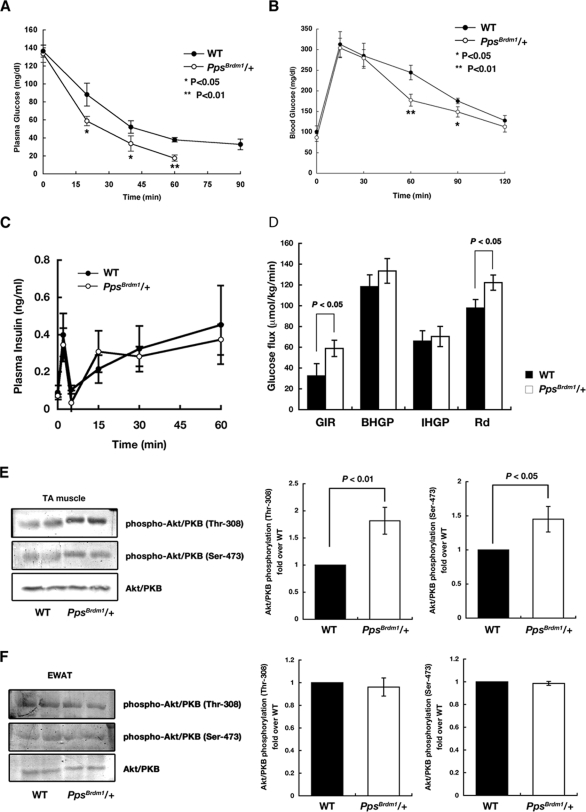
FIG. 3.
Enhanced glucose homeostasis in PpsBrdm1/+ mice. An insulin tolerance test (A) and an oral glucose tolerance test (B) of wild-type (n = 6) and PpsBrdm1/+ (n = 6) male mice were performed. Filled columns indicate wild-type mice; open columns indicate PpsBrdm1/+ mice. (C) Plasma insulin levels of wild-type (n = 3) and PpsBrdm1/+ (n = 3) male mice in response to glucose injection. Glucose-induced insulin secretion was unchanged in PpsBrdm1/+ mice. (D) Hyperinsulinemic-euglycemic clamp analysis in wild-type (n = 3) and PpsBrdm1/+ (n = 3) mice from 17 weeks of age. Glucose infusion rate (GIR), rate of insulin-stimulated glucose disposal (Rd), and basal or insulin-induced hepatic glucose production (BHGP or IHGP, respectively) are indicated. (E and F) Clamp-based insulin-induced Akt/PKB phosphorylation in isolated soleus muscle (TA, for tibialis anterior) and EWAT in wild-type and PpsBrdm1/+ mice. All values are given as means ± standard errors of the means. WT, wild type.
2-Deoxyglucose incorporation analysis.
3H-labeled 2-deoxyglucose and [14C]mannitol (Amersham Biosciences) were used to measure incorporation of 2-deoxyglucose in L6 myoblast cells expressing GLUT4 and soleus muscle isolated from mice. Insulin-induced incorporation of 2-deoxyglucose was measured as described previously (11, 23). Briefly, L6 cells or muscles were preincubated with 2-[3H]deoxyglucose and mannitol. The cells were then stimulated with insulin (0 to 100 nM) for 60 min. Cells were washed, harvested, and solubilized with Soluene 300 (Wako). Incorporation of 2-[3H]deoxyglucose was measured by liquid scintillation counting.
Measurement of insulin signaling in vivo.
Wild-type and PpsBrdm1/+ mice were fasted overnight and injected intravenously with insulin (1 U/kg of body weight). Quadriceps muscle and EWATs were isolated from wild-type and PpsBrdm1/+ mice and homogenized to prepare the lysates. Protein extract (50 μg) was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis for Western blot analysis. Antibodies specific for Akt/PKB, phospho-Akt/PKB, p70 S6 kinase, phospho-p70 S6 kinase, glycogen synthase kinase 3β (GSK3β), phospho-GSK3β, insulin receptor, actin, SKIP, PTEN, and SHIP2 were used. Anti-Akt/PKB and anti-p70 S6 kinase antibodies were from Santa Cruz Biotechnology. Anti-insulin receptor, anti-GSK3β, anti-Myc, and anti-phosphospecific antibodies were from Cell Signaling Technology. SHIP2 antibody was from Upstate Biotechnology. Anti-SKIP antibodies were generated as described previously (13). The protein levels were quantified by densitometry.
Transfection and RNA interference.
RNA interference oligonucleotides corresponding to rat SKIP were subcloned into the pSIREN-DNR vector (BD Clontech). Human SKIP, human PTEN, or rat SHIP2 were cloned into pCMV-3B vector (Stratagene). The full-length or N-terminal truncated isoform gene of SKIP in pCAGGS vector was generated as previously reported (12). These constructs were cotransfected into L6 cells with a positive selection marker. Cells were serum deprived for 20 h and stimulated with insulin (0 to 100 nM). Lysates were analyzed by immunoblotting.
Phosphoinositide phosphatase activity assay.
PtdIns(3,4,5)P3 and phosphatidylinositol 4,5-biphosphate [PtdIns(4,5)P2] were purchased from Cell Signals, and [3H]PtdIns(4,5)P2 was purchased from PerkinElmer, Inc. [32P]PtdIns(3,4,5)P3 was generated with the PI3-kinase p110α subunit as described previously (12). Glutathione S-transferase or glutathione S-transferase-conjugated full-length SKIP or an N-terminal truncated isoform of SKIP was expressed with a baculovirus expression system and incubated with the substrate phosphoinositides for 10 min at 37°C. Lipids were separated by thin-layer chromatography and visualized by autoradiography (12).
GLUT4 translocation assay.
Rat L6 myocytes were cultured in Dulbecco's modified Eagle's medium. A GLUT4 reporter containing Myc epitope tags in the first extracellular loop as well as green fluorescent protein fused at the carboxyl terminus was used (kindly provided by Harvey F. Lodish). Expression was visualized by immunofluorescence with anti-Myc antibodies. GLUT4 translocation was calculated as described previously (2).
RESULTS
Selective expression of full-length SKIP in insulin-sensitive tissues.
There are two alternative splicing isoforms of SKIP, full-length SKIP and a shorter isoform that is truncated by 76 amino acids at the N terminus which eliminates a region conserved among type II inositol polyphosphate phosphatases (SKIP ΔN76) (see Fig. S1A in the supplemental material) (12). In the present study, full-length SKIP was expressed at high levels in insulin-sensitive organs such as skeletal muscle, heart, and brain (see Fig. S1B in the supplemental material). In contrast, the smaller isoform was expressed predominantly in adipose tissue. We compared phosphatase activities of full-length SKIP and SKIP ΔN76 on PtdIns(4,5)P2 and PtdIns(3,4,5)P3. Full-length SKIP efficiently hydrolyzed these phosphoinositides (Km values of 1.80 μM and 0.89 μM, respectively), whereas SKIP ΔN76 had approximately 100-fold lower phosphatase activity (Km of 180 μM and 115 μM, respectively) (see Fig. S1C and D in the supplemental material). As previously reported, expression of full-length SKIP suppressed insulin-induced phosphorylation of Akt/PKB (see Fig. S1E in the supplemental material) (13). In contrast, expression of SKIP ΔN76 did not inhibit these phosphorylations (see Fig. S1E in the supplemental material). These results indicate that SKIP with phosphatase activity is expressed in skeletal muscle, heart, and brain. Skeletal muscle is a major insulin-responsive tissue, and abnormal insulin signaling in skeletal muscle is an early defect in the pathogenesis of obesity and diabetes.
Targeted disruption of the SKIP ortholog Pps in the mouse germ line.
An insertional targeting vector, pTVPps, was used which duplicated exons 4 to 7 of the mouse ortholog of SKIP, Pps, causing a frameshift mutation. The allele, designated PpsBrdm1, was established in mice by standard procedures (Fig. 1A and B). Because Pps lies between p53 and Wnt3 on mouse chromosome 11, the PpsBrdm1 allele was maintained using the balancer chromosome Inv(11)(p53-Wnt3)Brdm (31). We crossed PpsBrdm1/+ mice with Inv(11)(p53-Wnt3)Brdm/+ mice and subsequently intercrossed Inv(11)(p53-Wnt3) Brdm/PpsBrdm1 mice. All of the 176 progeny generated showed the balancer-specific light tail color, and Southern blot analysis confirmed that all progeny were heterozygous for PpsBrdm1, suggesting that PpsBrdm1 is a homozygous lethal allele. Timed matings were established, and embryos were isolated and genotyped at various stages of development. No PpsBrdm1/PpsBrdm1 embryos were identified after embryonic day 10.5, indicating that the homozygous mutation caused embryonic lethality. In PpsBrdm1/+ mice, Northern blot analysis revealed a significant decrease in Pps mRNA in quadriceps muscle (Fig. 1C), and Western blot analysis showed that SKIP protein levels in quadriceps were decreased approximately 55.4% (Fig. 1D). The protein levels of other PtdIns(3,4,5)P3 phosphatases, PTEN, and SHIP2 did not differ from levels in wild-type littermates (Fig. 1D).
PpsBrdm1/+ mice have normal body weight, fat content, and feeding efficiency.
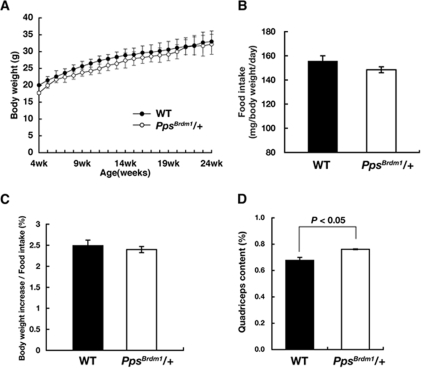
When PpsBrdm1/+ male mice were fed a normal chow diet, they appeared normal, with body weights similar to wild-type male mice (Fig. 2A) and with a normal growth rate (28.4% ± 5.4% increase in body weight from 6 to 18 weeks of age in PpsBrdm1/+ male mice versus 28.2% ± 8.8% increase in wild-type mice; P = 0.95 by Student's t test). PpsBrdm1/+ female mice also have normal body weight (see Fig. S2A in the supplemental material). PpsBrdm1/+ male mice showed normal food intake measured over a 24-h period when normalized by body weight (155.5 ± 4.4 mg/g of body weight in wild-type mice versus 148. 4 ± 2.5 mg/g of body weight in PpsBrdm1/+ mice; P = 0.59) (Fig. 2B). In addition, the increase in feeding efficiency (gain in body weight/food intake) was not observed in PpsBrdm1/+ mice (2.49% ± 0.13% in wild-type mice versus 2.39% ± 0.07% in PpsBrdm1/+ mice; P = 0.81) (Fig. 2C). The weights of major organs such as liver and brain were proportional to body weight, but the weight of quadriceps muscle was increased in PpsBrdm1/+ mice (0.678% ± 0.022% of total body weight in wild-type mice versus 0.761% ± 0.003% of body weight in PpsBrdm1/+ mice; P = 0.045) (Fig. 2D). The weight of soleus muscle also has a tendency to increase in PpsBrdm1/+ mice (0.027% ± 0.002% of body weight in wild-type mice versus 0.032% ± 0.002% in PpsBrdm1/+ mice; P = 0.32). The EWAT content was similar in PpsBrdm1/+ mice to wild-type mice at 18 weeks of age (1.59% ± 0.07% in wild-type mice versus 1.61% ± 0.09% in PpsBrdm1/+ mice; P = 0.80) (see Fig. S3 in the supplemental material). Brown adipose tissue content was also unaffected in PpsBrdm1/+ mice (0.39% ± 0.07% in wild-type mice versus 0.39% ± 0.01% in PpsBrdm1/+ mice; P = 0.27).
FIG. 2.
SKIP mutant mice exhibited normal food intake and adiposity. (A) Growth curves of wild-type (n = 21) and PpsBrdm1/+ (n = 27) mice fed a normal chow diet. (B) Food intake of wild-type (n = 12) and PpsBrdm1/+ (n = 12) male mice fed a normal chow diet at 15 weeks of age. All the parameters are normalized by total body weight. (C) Feeding efficiency of wild-type (n = 6) and PpsBrdm1/+ (n = 6) male mice fed a normal chow diet at 15 weeks of age. Feeding efficiency was calculated by gain in body weight normalized by food intake. (D) Percentage of quadriceps content isolated from wild-type mice (n = 18) and PpsBrdm1/+ (n = 18) male mice at 15 weeks of age. All values are given as means ± standard errors of the means. WT, wild type.
Parameters for glucose and insulin homeostasis were analyzed in PpsBrdm1/+ male mice and in wild-type male mice at 15 to 16 weeks of age. Fasting glucose levels were found to be significantly lower (P = 0.04) in PpsBrdm1/+ male mice than in wild-type male mice (Table 1). PpsBrdm1/+ female mice also showed lower blood glucose levels compared to wild-type mice (115.1 ± 18.8 mg/dl in PpsBrdm1/+ mice versus 129.2 ± 20.4 mg/dl in wild-type; n = 18; P = 0.036). Serum chemistry profiles revealed that PpsBrdm1/+ male mice had significantly lower serum cholesterol, triglycerides, and NEFA levels than wild-type mice (P < 0.05 in all cases) (Table 1). However, serum insulin levels and leptin levels were similar between PpsBrdm1/+ male mice and wild-type mice, whereas blood glucose levels were significantly lower in PpsBrdm1/+ male mice. In contrast, serum cholesterol and NEFAs were comparable in PpsBrdm1/+ female mice and wild-type mice (data not shown).
TABLE 1.
Serum chemistry parameters of wild-type and PpsBrdm1/+ male mice
| Substance(s) | Value in serum under the indicated conditiona
|
|||
|---|---|---|---|---|
| Wild-type mice
|
PpsBrdm1/+ mice
|
|||
| Nonfasted | Fasted | Nonfasted | Fastedb | |
| Glucose (mg/dl) | 241.5 ± 7.5 | 116.8 ± 8.1 | 213.8 ± 3.8 | 92.6 ± 6.3* |
| Triglycerides (mg/dl) | 145.8 ± 1.6 | 142.3 ± 4.7 | 96.9 ± 0.4 | 100.5 ± 2.0** |
| Cholesterol (mg/dl) | 78.5 ± 0.2 | 80.1 ± 4.4 | 62.4 ± 3.9 | 64.6 ± 12.4* |
| NEFA (meq/liter) | 0.89 ± 0.01 | 0.80 ± 0.06 | 0.58 ± 0.03 | 0.52 ± 0.04* |
| Insulin (ng/ml) | 2.71 ± 0.12 | 1.05 ± 0.07 | 2.26 ± 0.06 | 1.04 ± 0.09 |
| Adiponectin (mg/ml) | 7.49 ± 0.18 | 8.30 ± 0.47 | 7.80 ± 0.45 | 8.27 ± 0.24 |
| Leptin (ng/ml) | 2.94 ± 0.28 | 2.69 ± 0.68 | ||
All serum samples were prepared from 15- to 16-week-old male mice fed a normal chow diet. All data are given as means ± standard errors of the means. For diponectin and leptin, n = 6; for all others, n = 12.
*, P < 0.05; **, P < 0.01 (compared to wild-type mice; Student's t test).
Glucose homeostasis and insulin sensitivities are enhanced in PpsBrdm1/+ mice.
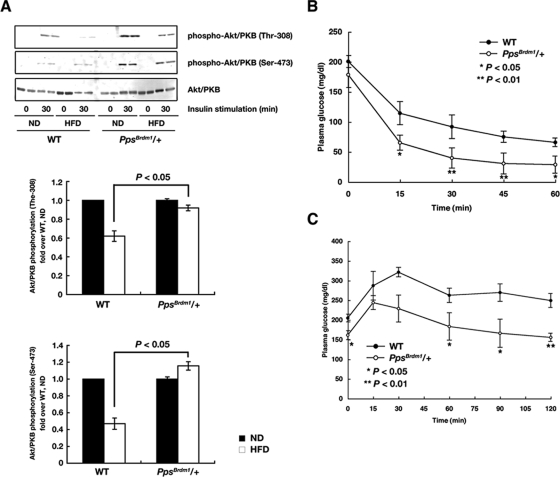
To examine glucose homeostasis, insulin tolerance tests and glucose tolerance tests were performed. Enhanced insulin sensitivity was observed in PpsBrdm1/+ male mice, i.e., an accelerated lowering of plasma glucose after injection of insulin (0.5 U/Kg body weight) (Fig. 3A). PpsBrdm1/+ mice exhibited 53.8% lower blood glucose at 60 min after injection than wild-type mice. Similarly, glucose tolerance tests showed increases in the rapid elimination of blood glucose in PpsBrdm1/+ male mice (Fig. 3B) without change in insulin secretion (Fig. 3C). A similar increase in the blood glucose elimination was also observed in PpsBrdm1/+ female mice (see Fig. S2B in the supplemental material). The average blood glucose of PpsBrdm1/+ mice was 28.5% lower than that of wild-type mice at 60 min after injection (P = 1.1 × 10−4). In order to analyze the systemic insulin sensitivities definitively, we carried out a hyperinsulinemic-euglycemic clamp study (16) and found that the glucose infusion rate and the rate of insulin-stimulated glucose disposal were significantly increased in PpsBrdm1/+ mice (Fig. 3D). On the other hand, PpsBrdm1/+ mice and wild-type mice had similar rates of basal or insulin-induced hepatic glucose production (Fig. 3D). To confirm increased insulin sensitivities in skeletal muscle but not in adipose tissue, we measured Akt/PKB phosphorylations in isolated soleus muscle and EWAT 130 min after the onset of the clamp and found that, in comparison to wild-type mice, Akt/PKB phosphorylations were increased in soleus muscle but not in EWAT in PpsBrdm1/+ male mice (Fig. 3E and F), indicating that SKIP regulates insulin sensitivities in skeletal muscle where the full-length SKIP is abundantly expressed.
PpsBrdm1/+ mice showed enhanced insulin action in skeletal muscle.
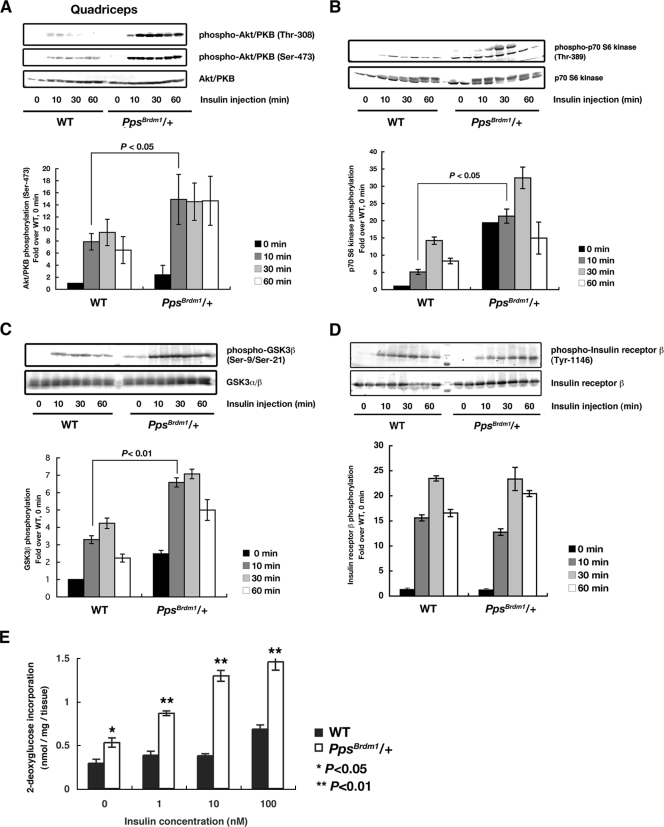
Because SKIP acts negatively on PI3-kinase signaling, this raised the possibility that PpsBrdm1/+ mice have enhanced activation of insulin signaling in skeletal muscle, where full-length SKIP is abundantly expressed (13). Therefore, we examined insulin-induced Akt/PKB phosphorylation. In the absence of insulin stimulation, there was little phosphorylation of Akt/PKB in quadriceps muscle and EWAT of PpsBrdm1/+ mice. However, administration of insulin resulted in higher levels of phosphorylation of Akt/PKB in quadriceps muscle in PpsBrdm1/+ mice than in wild-type mice (Fig. 4A). Such an enhanced phosphorylation of Akt/PKB was not observed in EWAT of PpsBrdm1/+ mice, where full-length SKIP is not expressed (see Fig. S4 in the supplemental material). To confirm these results, we examined phosphorylation of two downstream targets of Akt/PKB, GSK3β, and p70 S6 kinase. Phosphorylation of p70 S6 kinase and GSK3β was also significantly enhanced in PpsBrdm1/+ mice (Fig. 4B and C). Next, insulin-induced incorporation of 2-deoxyglucose was examined using isolated soleus muscle from PpsBrdm1/+ mice. Incorporation of 2-deoxyglucose after stimulation with a submaximal concentration of insulin in soleus muscle (10 nM) was also dramatically enhanced in PpsBrdm1/+ mice (P = 1.4 × 10−3) (Fig. 4D). These results, together with the unchanged level of tyrosine autophosphorylation of the insulin receptor in quadriceps in PpsBrdm1/+ and wild-type mice (Fig. 4E), suggest that reduction of SKIP enhanced insulin signaling by elevating Akt/PKB activities in skeletal muscle.
FIG. 4.
Enhanced insulin signaling in skeletal muscle of PpsBrdm1/+ mice. (A) Phosphorylation of Akt/PKB in quadriceps in response to insulin. Mice were fasted overnight and injected intraperitoneally with insulin (1U/kg of body weight) for the indicated times. Levels of Akt/PKB phosphorylation on Ser-473 and Thr-308 were quantified, and results are shown in the graphs. (B and C) Activation of insulin signaling in quadriceps of PpsBrdm1/+ mice. Phosphorylation of p70 S6 kinase (B) and GSK3β (C) are analyzed. (D) Enhanced insulin-induced glucose uptake in soleus muscle from PpsBrdm1/+ (n = 6) male mice in comparison with wild-type (n = 6) mice at 12 weeks of age. (E) Tyrosine phosphorylation of insulin receptor was not changed between PpsBrdm1/+ mice and wild-type mice. All values are given as means ± standard errors of the means. *, P < 0.05; **, P < 0.01. All values are normalized relative to basal phosphorylation of each protein in wild-type mice. WT, wild-type.
Since insulin signaling in skeletal muscle has been reported to be important for systemic glucose sensitivities (6), we next measured whether insulin-induced Akt/PKB phosphorylation is altered in PpsBrdm1/+ mice fed a HFD. In wild-type mice, a 4-month exposure to a HFD impaired insulin-induced Akt/PKB phosphorylation in skeletal muscle. In contrast, PpsBrdm1/+ mice showed a significantly stronger ability to phosphorylate Akt/PKB (Fig. 5A). Furthermore, elevated insulin sensitivity and glucose tolerance were observed in PpsBrdm1/+ mice (Fig. 5B and C). These results strongly suggest that whole-body glucose uptake improved in PpsBrdm1/+ mice fed a HFD.
FIG. 5.
PpsBrdm1/+ mice are resistant to diet-induced insulin resistance. (A) Western blot analysis of insulin-induced Akt/PKB phosphorylation on Thr-308 and Ser-473 in quadriceps isolated from PpsBrdm1/+ mice fed a normal chow diet (ND) or an HFD. The quantifications of the Western blot analysis are shown. Phosphorylation levels were quantified and normalized by values for wild-type mice fed a normal diet. An insulin tolerance test (B) and an oral glucose tolerance test (C) were performed on wild-type (n = 6) and PpsBrdm1/+ (n = 6) mice fed an HFD for 4 months. All values are given as means ± standard errors of the means. *, P < 0.05; **, P < 0.01. WT, wild type.
Because muscle-specific deletion of Pten protected mice from diet-induced insulin resistance or null-mutation of SHIP2 resulted in resistance to diet-induced obesity in mice, we raised a cohort of PpsBrdm1/+ mice on an HFD. After 6 weeks on the HFD, PpsBrdm1/+ mice gained 30.9% of their body weight, whereas wild-type mice fed on the HFD gained 41.4% of their body weight (see Fig. S5 in the supplemental material). Thus, the PpsBrdm1/+ genotype protects against diet-induced obesity. The HFD caused hyperglycemia in both wild-type and PpsBrdm1/+ mice (Fig. 5B; see also Table S1 in the supplemental material); however, its effect was substantially lower in PpsBrdm1/+ mice. Although PpsBrdm1/+ mice showed normal food intake (126.7 ± 19.7 mg/g of body weight in wild-type mice versus 133.0 ± 7.7 mg/g of body weight in PpsBrdm1/+ mice; P = 0.31), the feeding efficiency had a tendency to decrease in PpsBrdm1/+ mice in comparison with wild-type mice (2.90% ± 0.52% in PpsBrdm1/+ mice versus 6.10% ± 0.17% in wild-type mice; P = 0.092). Cholesterol, insulin, and leptin levels were also lower in PpsBrdm1/+ mice than in wild-type mice (see Table S1 in the supplemental material). In contrast, wild-type mice developed hyperinsulinemia, with the insulin level approximately threefold higher than that of PpsBrdm1/+ mice (see Table S1 in the supplemental material). Therefore, resistance to dietary-induced insulin resistance in PpsBrdm1/+ mice might be affected by adiposity as well as insulin sensitivities in skeletal muscle.
Reduced levels of SKIP increased insulin-induced glucose homeostasis in L6 cells.
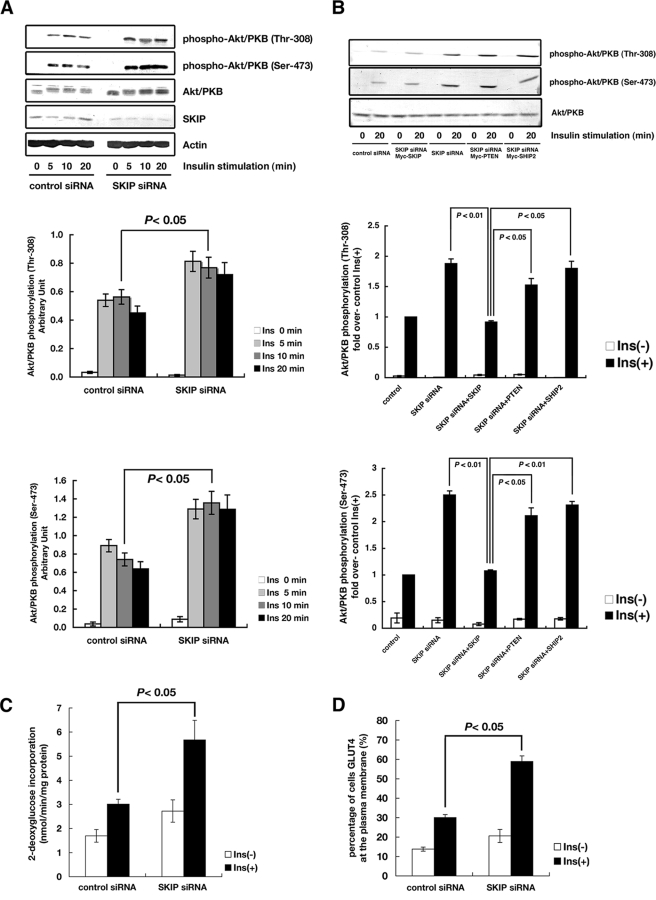
Analyses of SHIP2- and Pten-knockout mice revealed that these proteins negatively regulate insulin signaling (17, 26, 29, 30). However, unlike the targeted mutation of PpsBrdm1/+ in mice, null deletion of SHIP2 or muscle-specific deletion of Pten in mice did not cause the enhanced insulin signaling in skeletal muscle of mice receiving a normal chow diet (26, 30). To clarify the role of SKIP in insulin signaling, small interfering RNA (siRNA) knockdown of SKIP was performed in L6 myoblast cells. An approximately 70% reduction was achieved. Reductions in SKIP did not affect phosphorylation of Akt/PKB without insulin stimulation. However, reductions of SKIP markedly enhanced insulin-induced phosphorylation of Akt/PKB on Ser-473 and Thr-308 (Fig. 6A). To test whether these enhanced Akt/PKB phosphorylations were suppressed by the expression of PtdIns(3,4,5)P3 phosphatases, SKIP, SHIP2, and PTEN were expressed in SKIP knockdown cells. Expression of SKIP but not SHIP2 or PTEN suppressed insulin-induced Akt/PKB phosphorylation on Ser-473 and Thr-308 (Fig. 6B; see also Fig. S6 in the supplemental material). These results indicate that SKIP plays distinct roles from PTEN and SHIP2 in insulin signaling.
FIG. 6.
Enhanced insulin signaling by SKIP suppression in L6 myoblast cells. (A) Reduction of SKIP enhances insulin-induced Akt/PKB phosphorylation. L6 cells were transfected with control or SKIP siRNA and stimulated with insulin. Cells lysates were used for the detection for Akt/PKB and phosphorylated Ser-473 or Thr-308 of Akt/PKB. A time course of insulin-induced Akt phosphorylation is shown. (B) SKIP plays distinct roles from PTEN and SHIP2 in insulin signaling. L6 cells transfected with SKIP siRNA and PtdIns(3,4,5)P3 phosphatases (PTEN, SHIP2, or SKIP) were stimulated with insulin for 20 min. Phosphorylation of Ser-473 and Thr-308 of Akt/PKB is shown. The enhanced effect of insulin on 2-deoxyglucose incorporation (C) and GLUT4 translocation (D) by reduction of SKIP is shown. Insulin-induced (100 nM) 2-deoxyglucose uptake and GLUT4 translocation are shown. All values are given as means ± standard errors of the means. GLUT4 reporter was expressed in L6 cells or L6 cells in conjunction with siRNA vectors. Ins, insulin.
To test whether the increase in PI3-kinase signaling by attenuation of SKIP was correlated with glucose homeostasis, insulin-induced GLUT4 translocation and glucose uptake were measured (Fig. 6C and D). Insulin-induced translocation of GLUT4 was examined with a fluorescent GLUT4 reporter (2). Suppression of SKIP only slightly increased basal glucose incorporation compared to control cells (P = 0.22); however, it markedly enhanced insulin-induced glucose incorporation (P = 0.04) (Fig. 6C). Knockdown of SKIP markedly increased insulin-induced GLUT4 translocation in response to insulin stimulation (P = 0.01), although in the absence of insulin stimulation, membrane-associated GLUT4 levels on the cell surface did not differ significantly from those of controls (Fig. 6D). These results are consistent with the in vivo data from the knockout mice. Therefore, SKIP apparently plays a preeminent role in the regulation of insulin-induced GLUT4 transport to the plasma membrane and glucose incorporation under normal conditions.
DISCUSSION
SKIP is a PtdIns(3,4,5)P3 phosphatase that is abundantly expressed in skeletal muscle, a major insulin-responsive tissue. In a previous study, we have showed that SKIP negatively regulated insulin-induced GLUT4 transport and membrane ruffle formation (13). These regulations require the PtdIns (3,4,5)P3 phosphatase activity of SKIP that decreases intracellular PtdIns(3,4,5)P3 levels. However, at least two other PtdIns(3,4,5)P3 phosphatases, PTEN and SHIP2, also negatively regulate insulin signaling.
In the present study, we generated and characterized a targeted mutation of the Pps gene, which encodes SKIP in mice, and showed enhanced insulin signaling and insulin-induced glucose uptake in skeletal muscle. Because Pps-deficient mice exhibited embryonic lethality, insulin actions were analyzed using PpsBrdm1/+ mice. Insulin tolerance tests, glucose tolerance tests, and hyperinsulinemic-euglycemic clamp assays clearly showed increased systemic insulin sensitivity in PpsBrdm1/+ mice. In agreement with these results, skeletal muscle isolated from PpsBrdm1/+ mice showed a significant increase in insulin-induced Akt/PKB phosphorylation and a twofold greater increase in glucose incorporation than skeletal muscle from wild-type mice. Thus, these results indicate that SKIP is a major regulator of insulin action in skeletal muscle. Although we also found approximately 12% increase in weight of quadriceps, such an increase in muscle mass is not a main contributor to the increase (about twofold) in systemic glucose homeostasis.
Alteration in insulin signaling may affect body weight, food intake, and adiposity, and vice versa. However, PpsBrdm1/+ mice fed a normal chow diet have normal growth rates, leptin levels, adiposity, and feeding efficiency as well as food intake (Fig. 2 and Table 1). These observations may reflect the fact that the full-length SKIP is not expressed in adipose tissue and that an increase in insulin signaling is not detected in adipose tissue in PpsBrdm1/+ mice. A truncated isoform of SKIP that is expressed in adipose tissue exhibited very weak phosphatase activities and did not affect insulin signaling. Together, these results suggest that SKIP is a regulator of insulin signaling in skeletal muscle but not in adipose tissue.
SHIP2-deficient mice and muscle-specific Pten-knockout mice exhibit an enhancement in insulin signaling in skeletal muscle (26, 30). Like PpsBrdm1/+ mice, the reported SHIP2-deficient and muscle-specific Pten-knockout mice also show resistance to diet-induced obesity. However, a major difference among SHIP2-deficient and muscle-specific Pten-knockout mice and PpsBrdm1/+ mice is that PpsBrdm1/+ is the only mutation that led to increased insulin sensitivity and glucose uptake when mice were fed a normal chow diet. Like PpsBrdm1/+ mice, SHIP2-deficient mice showed increases in insulin-induced phosphorylations in Akt/PKB and p70 S6 kinase. However, since SHIP2-deficient male mice fed a normal chow diet showed normal insulin and glucose tolerance, SHIP2 was not implicated in the dysregulation of glucose homeostasis (26). It was also reported that in 3T3-L1 adipocytes, suppression of SHIP2 did not increase insulin-induced glucose uptake or phosphorylation of Akt/PKB (27). Muscle-specific deletion of Pten in mice resulted in protection against insulin resistance and obesity when mice were fed an HFD, but the mutant mice did not show increased insulin sensitivities and insulin-induced glucose uptake when fed a normal chow diet (30). If SHIP2 or PTEN regulated insulin-induced glucose incorporation, these mice would have exhibited increased insulin signaling even under normal conditions.
PpsBrdm1/+ mice fed an HFD also showed increased insulin sensitivity in skeletal muscle and were resistant to HFD-induced insulin resistance, supporting the possibility that SKIP affects insulin signaling in skeletal muscle of mice fed an HFD in addition to normal chow. Taken together, resistance to diet-induced insulin resistance in PpsBrdm1/+ mice may partly be explained by the increased insulin sensitivities in skeletal muscle through direct or indirect effects because skeletal muscle accounts for approximately 75% of whole body glucose homeostasis. However, we could not rule out the possibilities of other underlying changes in PpsBrdm1/+ mice, such as alterations in insulin signaling in the hypothalamus or adipokine secretion, that may play more important roles in causing the phenotypes, including resistance to diet-induced obesity in PpsBrdm1/+ mice fed an HFD. Like muscle-specific Pten and SHIP2 knockout mice, PpsBrdm1/+ mice fed an HFD showed a decrease in the leptin level and a tendency toward decreased feeding efficiency without a change in food intake. These phenotypic features may not be fully explained by the alteration of insulin sensitivities in skeletal muscle. Such ambiguity is well illustrated in recent studies of protein-tyrosine phosphatase 1B (PTP1B), which negatively regulates insulin signaling via inhibiting insulin-induced phosphorylation of insulin receptor and insulin receptor substrate 1 (8). Constitutionally homozygous PTP1B knockout mice (PTP1B−/−) showed increased insulin sensitivity (8) and enhanced glucose uptake in skeletal muscle. In addition, these mutant mice failed to increase their adiposity or body weight when fed an HFD. These phenotypes are very similar to that of PpsBrdm1/+ mice. It has recently been reported that muscle-specific PTP1B knockout in mice did not lead to resistance to HFD-induced obesity, even though insulin signaling in skeletal muscle was enhanced (6), suggesting that the alteration of insulin signaling in skeletal muscle alone may not be sufficient to slow down the weight gain when mice are fed an HFD.
PpsBrdm1/+ mice fed an HFD showed slight increases in body weight and plasma insulin levels compared to those fed a normal chow diet; these effects might be due to the decrease in insulin sensitivities in adipose tissue, where the shorter isoform of SKIP was expressed. Furthermore, full-length SKIP is also expressed highly in the brain, suggesting the possibility that SKIP may also regulate PI3-kinase signaling in brain. Thus, further studies are needed to unravel the precise mechanism of SKIP's regulation of whole-body insulin sensitivity. Generation and characterization of mutant mice carrying muscle-specific deletion of the Pps gene may lead to a better understanding of the relationship between insulin signaling in skeletal muscle and whole-body insulin sensitivity in mice fed an HFD.
We previously showed that the expression of SKIP negatively regulated insulin-induced glucose incorporation and glycogen synthesis in rat L6 myoblast cells (13); thus, the increase in PI3-kinase signaling by the suppression of SKIP may be important for protection against insulin resistance. Our in vitro analyses of L6 myoblast cells also showed that a decrease in SKIP resulted in a significant increase in the insulin-induced activation of downstream targets of Akt/PKB, and such an increase in insulin signaling was not compensated by the expression of SHIP2 and PTEN. In the absence of insulin stimulation, a decrease in SKIP resulted in only a slight increase in glucose uptake in L6 myoblast cells. In response to insulin stimulation, suppression of SKIP resulted in a significant increase in insulin-dependent glucose uptake as well as GLUT4 translocation to the cell surface in myoblast cells. SKIP may be the only PtdIns(3,4,5)P3 phosphatase that controls insulin signaling under normal conditions. Therefore, SKIP is functionally distinct from other PtdIns(3,4,5)P3 phosphatases in insulin signaling. Recently, distinct roles for SHIP2 and PTEN in differentiating L6 cells have been reported. PTEN mainly hydrolyzed a PtdIns(3,4,5)P3 pool that is capable of activating Akt/PKB. In contrast, SHIP2 works on a different pool that is unable to activate Akt/PKB, indicating that SHIP2 and PTEN have distinct roles (18). Together with our in vitro results, these results suggest that SKIP, PTEN, and SHIP2 may have distinct roles in insulin signaling.
The mutational analysis of SKIP, SHIP2, and PTEN has revealed that these major phosphatases do not functionally compensate for each other even though the deficiency of any one of them enhances insulin sensitivity in muscle. Although SKIP and SHIP2 belong to the same family, the type II inositol polyphosphate 5-phosphatases, each of them contains some unique domains. SKIP possesses a C-terminal SKICH domain responsible for the localization at the plasma membrane (10). SKIP is localized in endoplasmic reticulum and the Golgi compartment under resting conditions and is translocated to the plasma membrane when the cells are exposed to insulin (10, 12). On the other hand, SHIP2 possesses an SH2 domain at the N terminus that interacts with other signaling molecules such as Shc and p130cas and a proline-rich domain at the C terminus that interacts with Abl and Shc (7). Unlike SKIP and SHIP2, PTEN is a PtdIns(3,4,5)P3 3-phosphatase with a C-terminal C2 domain for binding to phospholipids (5). SHIP2 and PTEN are mainly localized in cytosol and the plasma membrane through interaction with their specific interaction molecules. Therefore, it is possible that these unique domains may play critical roles in determining the spatial and temporal differences among these phosphatases, which may control different PtdIns(3,4,5)P3 pools in muscle. Therefore, they may regulate different PtdIns(3,4,5)P3 binding molecules located downstream of insulin-mediated PI3-kinase signaling. These differences may underlie the inability of SHIP2 and PTEN to compensate for the function of SKIP in the Pps mutant mice.
These in vitro and in vivo results show that endogenous SKIP may be one of the key regulators of insulin signaling in skeletal muscle for glucose homeostasis. Thus, SKIP may be a potential therapeutic target for obesity and type II diabetes. Although PTEN and SHIP2 may also be good candidates, targeting these proteins may be problematic because PTEN is a tumor suppressor, and a SHIP2 deficiency causes severe growth retardation (7, 22). In contrast, PpsBrdm1/+ mice did not appear to show increased cancer susceptibility over a 1-year observation period or to demonstrate severe growth retardation. A 50% reduction in SKIP expression significantly enhanced insulin sensitivity in mice, suggesting that SKIP may be a more desirable target for modulation of insulin sensitivities. Further analyses of SKIP signaling pathways will improve our understanding of abnormalities in glucose homeostasis and lipogenesis and will lead to new therapeutic interventions for obesity and diabetes.
Supplementary Material
Acknowledgments
We thank H. F. Lodish for the Glut4 cDNA; H. Sakoda and T. Asano for instructions regarding animal experiments; and K. Kawanaka, K. Koshinaka, Z. Li, and T. Kwong for the technical assistance. We are also grateful to A. Suzuki, Y. Horie, and T. Sasaki for helpful discussions and to M. Kamiya for assistance with animal experiments.
This work was supported by grants-in-aid from the MEXT/JST to T. Takenawa, from the NIH/National Cancer Institute to A. Bradley, NIH grant R01HL91519 to Y. E. Yu, and in part by a Roswell Park Cancer Institute Cancer Center Support Grant from the National Cancer Institute.
Footnotes
Published ahead of print on 23 June 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abel, E. D., O. Peroni, J. K. Kim, Y. B. Kim, O. Boss, E. Hadre, T. Minnemann, G. I. Shulman, and B. B. Kahn. 2001. Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver. Nature 409729-733. [DOI] [PubMed] [Google Scholar]
- 2.Bogan, J. S., A. E. McKee, and H. F. Lodish. 2001. Insulin-responsive compartments containing GLUT4 in 3T3-L1 and CHO cells: regulation by amino acid concentrations. Mol. Cell. Biol. 214785-4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradley, A. 1987. Production and analysis of chimeric mice, p. 113-151. In E. Robertson (ed.), Teratocarcinomas and embryonic stem cells—a practical approach. IRL Press, Oxford, United Kingdom.
- 4.Clement, S., U. Krause, F. Desmedt, J. F. Tanti, J. Befrends, X. Pesesse, T. Sasaki, J. Penninger, M. Doherty, W. Malaisse, J. E. Dumont, Y. Le Marchand-Brustel, C. Euneux, L. Hue, and S. Shurmans. 2001. The lipid phosphatase SHIP2 controls insulin sensitivity. Nature 40992-97. [DOI] [PubMed] [Google Scholar]
- 5.Das, S., J. E. Dixon, and C. Wonhwa. 2003. Membrane-binding and activation mechanism of PTEN. Proc. Natl. Acad. Sci. USA 1007491-7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delibegovic, M., K. K. Bence, N. Mody, E.-G. Hong, H. J. Ko, J. K. Kim, B. B. Kahn, and B. G. Neel. 2007. Improved glucose homeostasis in mice with muscle-specific deletion of protein-tyrosine phosphatase 1B. Mol. Cell. Biol. 277727-7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dyson, J. M., A. M. Kong, F. Wiradjaja, M. V. Astle, R. Gurung, and C. A. Mitchell. 2005. The SH2 domain containing inositol polyphosphate 5-phosphatase-2: SHIP2. Int. J. Biochem. Cell Biol. 372260-2265. [DOI] [PubMed] [Google Scholar]
- 8.Elchebly, M., P. Payette, E. Michaliszyn, W. Cromlish, S. Collins, A. L. Loy, D. Normandin, A. Cheng, J. Himms-Hagen, C. C. Chan, C. Ramachandran, M. J. Gresser, M. L. Tremblay, and B. P. Kennedy. 1999. Increased insulin sensitivity and obesity in mice lacking the protein tyrosine phosphatase-1B gene. Science 2831544-1548. [DOI] [PubMed] [Google Scholar]
- 9.Fukui, K., T. Wada, S. Kagawa, K. Nagira, M. Ikubo, H. Ishihara, M. Kobayashi, and T. Sasaoka. 2005. Impact of the liver-specific expression of SHIP2 (SH2-containing inositol 5′-phosphatase 2) on insulin signaling and glucose metabolism in mice. Diabetes 541958-1967. [DOI] [PubMed] [Google Scholar]
- 10.Gurung, R., A. Tan, L. M. Ooms, M. J. McGrath, R. D. Huysmans, A. D. Munday, M. Prescott, J. C. Whisstock, and C. A. Mitchell. 2003. Identification of a novel domain in two mammalian inositol-polyphosphate 5-phosphatases that mediates membrane ruffle localization. The inositol 5-phosphatase skip localizes to the endoplasmic reticulum and translocates to membrane ruffles following epidermal growth factor stimulation. J. Biol. Chem. 27811376-11385. [DOI] [PubMed] [Google Scholar]
- 11.Horie, Y., A. Suzuki, E. Kataoka, T. Sasaki, K. Hamada, J. Sasaki, K. Mizuno, G. Hasegawa, H. Kishimoto, M. Iizuka, M. Naito, K. Enomoto, S. Watanabe, T. W. Mak, and T. Nakano. 2004. Hepatocyte-specific Pten deficiency results in steatohepatitis and hepatocellular carcinomas. J. Clin. Investig. 1131774-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ijuin, T., Y. Mochizuki, K. Fukami, M. Funaki, T. Asano, and T. Takenawa. 2000. Identification and characterization of a novel inositol polyphosphate 5-phosphatase. J. Biol. Chem. 27510870-10875. [DOI] [PubMed] [Google Scholar]
- 13.Ijuin, T., and T. Takenawa. 2003. SKIP negatively regulates insulin-induced GLUT4 translocation and membrane ruffle formation. Mol. Cell. Biol. 231209-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishihara, H., T. Sasaoka, S. Kagawa, S. Murakami, K. Fukui, Y. Kawagishi, K. Yamazaki, A. Sato, M. Iwata, M. Urakaze, M. Ishiki, T. Wada, S. Yaguchi, H. Tsuneki, I. Kimura, and M. Kobayshi. 2003. Association of the polymorphisms in the 5′-untranslated region of PTEN gene with type 2 diabetes in a Japanese population. FEBS Lett. 554450-454. [DOI] [PubMed] [Google Scholar]
- 15.Jiang, G., and B. B. Zhang. 2002. Pi 3-kinase and its up- and down-stream modulators as potential targets for the treatment of a type II diabetes. Front. Biosci. 7d903-d907. [DOI] [PubMed] [Google Scholar]
- 16.Kanda, H., S. Tateya, Y Tamori, K. Kotani, K. Hiasa, R. Kitazawa, S. Kitazawa, H. Miyachi, S. Maeda, K. Egashira, and M. Kasuga. 2006. MCP-1 contributes to macrophage infiltration into adipose tissue, insulin resistance, and hepatic steatosis in obesity. J. Clin. Investig. 1161494-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurlawalla-Martinez, C., B. Stiles, Y. Wang, S. U. Devaskar, B. B. Kahn, and H. Wu. 2005. Insulin hypersensitivity and resistance to streptozotocin-induced diabetes in mice lacking PTEN in adipose tissue. Mol. Cell. Biol. 252498-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandl, A., D. Sarkes, V. Carricaburu, V. Jung, and L. Rameh. 2007. Serum withdrawal-induced accumulation of phosphoinositide 3-kinase lipids in differentiating 3T3-L6 myoblasts: distinct roles for Ship2 and PTEN. Mol. Cell. Biol. 278098-8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin, S. S., T. Haruta, A. J. Morris, A. Klippel, L. T. Williams, and J. M. Olefsky. 1996. Activated phosphatidylinositol 3-kinase is sufficient to mediate actin rearrangement and GLUT4 translocation in 3T3-L1 adipocytes. J. Biol. Chem. 27117605-17608. [DOI] [PubMed] [Google Scholar]
- 20.Nakashima, N., P. M. Sharma, T. Imamura, R. Bookstein, and J. M. Olefsky. 2000. The tumor suppressor PTEN negatively regulates insulin signaling in 3T3-L1 adipocytes. J. Biol. Chem. 27512889-12895. [DOI] [PubMed] [Google Scholar]
- 21.Ono, H., H. Katagiri, M. Funaki, M. Anai, K. Inukai, Y. Fukushima, H. Sakoda, T. Ogihara, Y. Onishi, M. Fujishiro, M. Kikuchi, Y. Oka, and T. Asano. 2001. Regulation of phosphoinositide metabolism, Akt phosphorylation, and glucose transport by PTEN (phosphatase and tensin homolog deleted on chromosome 10) in 3T3-L1 adipocytes. Mol. Endocrinol. 151411-1422. [DOI] [PubMed] [Google Scholar]
- 22.Ramirez-Solis, R., A. C. Davis, and A. Bradley. 1993. Gene targeting in embryonic stem cells. Methods Enzymol. 225855-878. [DOI] [PubMed] [Google Scholar]
- 23.Saltiel, A. R. 2001. New perspectives into the molecular pathogenesis and treatment of type 2 diabetes. Cell 104517-529. [DOI] [PubMed] [Google Scholar]
- 24.Sasaoka, T., T. Hori, T. Wada, M. Ishiki, T. Haruta, H. Ishihara, and M. Kobayashi. 2001. SH2-containing inositol phosphatase 2 negatively regulates insulin-induced glycogen synthesis in L6 myotubes. Diabetologia 441258-1267. [DOI] [PubMed] [Google Scholar]
- 25.Sasaoka, T., K. Fukui, T. Wada, S. Murakami, J. Kawahara, H. Ishihara, M. Funaki, T. Asano, and M. Kobayashi. 2005. Inhibition of endogenous SHIP2 ameliorates insulin resistance caused by chronic insulin treatment in 3T3-L1 adipocytes. Diabetologia 48336-344. [DOI] [PubMed] [Google Scholar]
- 26.Sleeman, M. W., K. E. Wortley, K.-M. V. Lai, L. C. Gowen, J. Kintner, W. O. Kline, K. Garcia, T. N. Stitt, G. D. Yancopoulos, S. J. Wiegand, and D. J. Glass. 2005. Absence of the lipid phosphatase SHIP2 confers resistance to dietary obesity. Nat. Med. 11199-205. [DOI] [PubMed] [Google Scholar]
- 27.Tang, X., A. M. Powelka, N. A. Soriano, M. P. Czech, and A. Guilherme. 2005. PTEN, but not SHIP2, suppresses insulin signaling through the phosphatidylinositol 3-kinase/Akt pathway in 3T3-L1 adipocytes. J. Biol. Chem. 28022523-22529. [DOI] [PubMed] [Google Scholar]
- 28.Virkamaki, A., K. Ueki, and C. R. Kahn. 1999. Protein-protein interaction in insulin signaling and the molecular mechanisms of insulin resistance. J. Clin. Investig. 103931-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wada, T., T. Sasaoka, M. Funaki, H. Hori, S. Murakami, M. Ishiki, T. Asano, W. Ogawa, H. Ishihara, and M. Kobayashi. 2001. Overexpression of SH2-containing inositol phosphatase 2 results in negative regulation of insulin-induced metabolic actions in 3T3-L1 adipocytes via its 5′-phosphatase catalytic activity. Mol. Cell. Biol. 211633-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wijesekara, N., D. Konrad, M. Eweida, C. Jefferies, M. Liadis, A. Giacca, M. Crachower, A. Suzuki, T. W. Mak, C. R. Kahn, A. Klip, and M. Woo. 2005. Muscle-specific Pten deletion protects against insulin resistance and diabetes. Mol. Cell. Biol. 251135-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng, B., M. Sage, W. W. Cai, D. M. Thompson, B. C. Tavsanli, Y. C. Cheah, and A. Bradley. 1999. Engineering a mouse balancer chromosome. Nat. Genet. 22375-378. [DOI] [PubMed] [Google Scholar]
- 32.Zheng, B., A. A. Mills, and A. Bradley. 1999. System for rapid generation of coat color-tagged knockouts and defined chromosomal rearrangements in mice. Nucleic Acids Res. 272354-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.