Abstract
The histone demethylase lysine demethylase 5b (KDM5b) specifically demethylates lysine 4 of histone H3 (meH3K4), thereby repressing gene transcription. KDM5b regulates cell cycle control genes in cancer and is expressed in the early epiblast. This suggests that KDM5b plays a developmental role by maintaining uncommitted progenitors. Here we show that transient overexpression of KDM5b in embryonic stem cells decreases the expression of at least three different modulators of cell fate decisions, Egr1, p27KIP1, and BMI1, by demethylation of their promoters. Constitutively increased KDM5b expression results in an increased mitotic rate and a decreased global 3meH3K4 but no change in cell identity. Results of two separate differentiation assays, neural differentiation and embryoid body EB (EB) formation, showed that KDM5b reduced the terminally differentiated cells and increased proliferating progenitors. These were achieved by two mechanisms, blocking of the upregulation of cell lineage markers and maintenance of cyclins, that allowed cells to escape differentiation and remain uncommitted. Additionally, EBs maintain high levels of Oct4 and Nanog and can be dissociated to reestablish highly proliferative cultures. The persistence of uncommitted progenitors may be due to the direct regulation of the Tcf/Lef family member mTcf3/hTcf7L1, an upstream regulator of Nanog expression. These findings demonstrate a role for KDM5b in the choice between proliferation and differentiation during development.
Transcriptional control is a dynamic process, during which several different histone residues are modified to change RNA polymerase's ability to access the transcriptional start site (19, 42). A key component in this process is the methylation of histone H3 lysine 4 (H3K4). Methylation of H3K4 is a key regulator of RNA polymerase binding to active genes (41) and of transcription factor binding within promoter elements (43). The ability of this epigenetic mark to control multiple points in transcription suggests that modulation of H3K4 methylation plays a role in both the activation and the repression of genes.
A key aspect of H3K4 methylation is how this epigenetic mark is removed, thereby reducing RNA polymerase's localization to the specific genes. This loss of methyl H3K4 (meH3K4) appears to be an important part of differentiation (30). A group of proteins that may govern the developmental loss of meH3K4 are those of the histone demethylase, lysine (K) demethylase 5 (KDM5)/JARID1 family, members of which have been characterized as di- and trimethyl H3K4 demethylases (9, 18, 25, 48). The KDM5/JARID1 family is composed of four genes, each containing a jumonji (J) C domain (the enzymatic domain), two DNA binding domains, an A/T-rich domain (ARID), a PHD domain, and a jumonji N domain. This family may be critical to many aspects of development, since members have been found to have biologically important roles in cell cycle control (e.g., KDM5a and KDM5b) and neural development through the repression of early differentiation markers (KDM5c and KDM5d). KDM5a and -b appear to have opposing functions, where KDM5a promotes differentiation (3), and KDM5b promotes proliferation (48). Additionally, KDM5b acts as a corepressor of BF/FoxG1b, a proto-oncogene product that regulates neural development, and Pax9, a proto-oncogene product that regulates neural crest development (44). The combination of cell cycle control and developmental target genes suggests that KDM5b may play an important role in cell fate decisions.
Recent evidence suggests that histone modifications perform a key role in the repression of both prodifferentiation genes and cell cycle inhibitors in uncommitted cells, allowing them to maintain their pluripotency and proliferative capabilities (6). The choice of the stem cell identity is an active process maintained at the level of the epigenome (20) through the repression of prodifferentiation genes required for the cell to maintain multilineage potential. Histone modifications can make genes transcriptionally available but not readily transcribed (14). Their transcriptional availability in embryonic stem cells (ESCs), particularly of cell fate genes, is directed by bivalent marks (meH3K4 and meH3K27) on histone H3 in the genes' promoters (4).
The recent identification of histone demethylases changes the paradigm by which developmental gene regulation can be studied, since it opens the possibility that all histone modifications are reversible (39). This suggests a simple model, where epigenetic modifications are dynamic regulators of transcription rather than permanent/static determinants of transcriptional accessibility (40). This element of dynamic control is particularly important for uncommitted cells, where cell lineage genes must be transcriptionally dormant but available for activation in response to the proper differentiation signal(s). This is especially true in mammals, where the correct modulation of H3K4 methylation is crucial to the timing and progression of development (11).
In the earliest phases of cell fate decisions, maintaining the proper control of H3K4 methylation may be of the utmost importance. At every division, cells choose between proliferation and differentiation either by continuing the repression of cell lineage markers or by relieving that repression. KDM5b falls into a class of proteins that may be critical to this choice. KDM5b/PLU1 (29) is highly expressed in the day 5.5 epiblast (12) and regulates G0-to-G1 progression (48) through the repression of cell cycle checkpoint genes. This division represents the key check point at which uncommitted cells choose between proliferation and commitment. The combination of early expression and cell cycle involvement suggests a role for KDM5b in the proliferation of progenitor populations. This is further supported by the upregulation of this gene in prostate (47) and breast cancer (28, 48). The role of KDM5b in multiple cancer types and its early developmental expression suggest that this histone demethylase functions to limit the number of stem or progenitor cells that differentiate, presumably by blocking the cells' ability to exit the cell cycle. We wanted to test this hypothesis directly during early development, using mouse ESCs (mESCs). Using this model, we show that KDM5b plays a role in both the proliferation of stem cells and in the repression of cell lineage genes, allowing cells to remain uncommitted.
MATERIALS AND METHODS
Generation of constructs.
Enhanced green fluorescent protein (EGFP) cDNA was cloned into the pCMV-Flag2 vector (Sigma) to generate the Flag-EGFP construct. The gateway cassette from Invitrogen was further cloned into the Flag-EGFP vector to generate a gateway Flag-EGFP destination (DEST) vector. This vector was then used to generate Flag-KDM5b (Flag-EGFP-KDM5b), using the pENTR clone (KIAA4034, from the KOTC collection) in an LR clonase reaction (Invitrogen). Similarly, the pEF5-KDM5b-V5 construct was generated by using the same pENTR clone and the pEF5/FRT/V5-DEST vector from Invitrogen. While all data from transient transfections shown in Fig. 1 to 7 were taken from experiments using the Flag-EGFP-KDM5b construct, the pEFα-KDM5b-V5 construct was used to confirm all results.
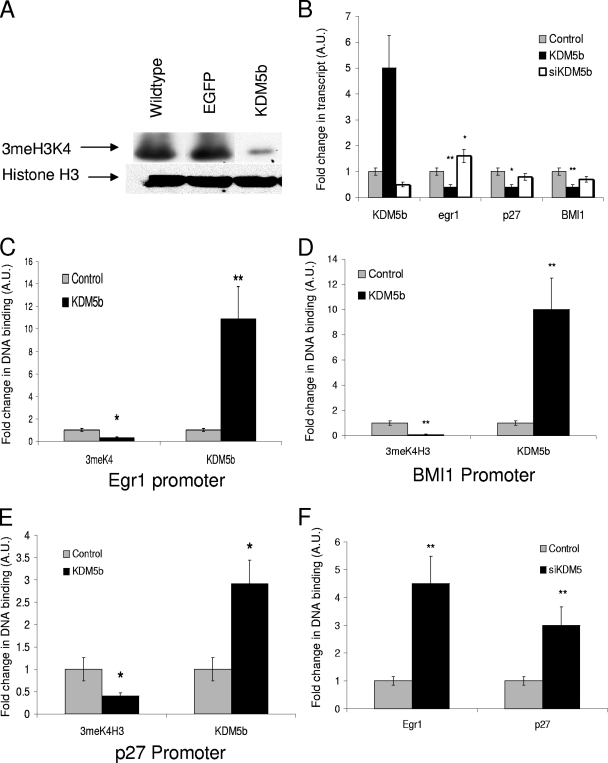
FIG. 1.
KDM5b represses target genes through demethylation of 3meH3K4. (A) Change in 3meH3K4 after 24 h of transient transfection of mESCs with either Flag-EGFP or Flag-KDM5b as measured by Western blotting with either anti-3meH3K4 (Upstate Biotechnology) or anti-Pan H3 (Upstate Biotechnology). (B) qRT-PCR of KDM5b, Egr1, p27, and BMI1 (primers are listed in Table S1 in the supplemental material) after 24 h transfection of either the control (Flag-EGFP), Flag-KDM5b, or siKDM5b (IDTdna). (C, D, and E) qChIP analysis of 3meH3K4 binding or KDM5b binding to Egr1, BMI1, or p27 promoters (primers are listed in Table S2 in the supplemental material). (F) qChIP analysis of 3meH3K4 in Egr1 and p27 promoters after cells were treated with siKDM5b. *, P = 0.05; **, P < 0.01. All data are in triplicate except where noted.
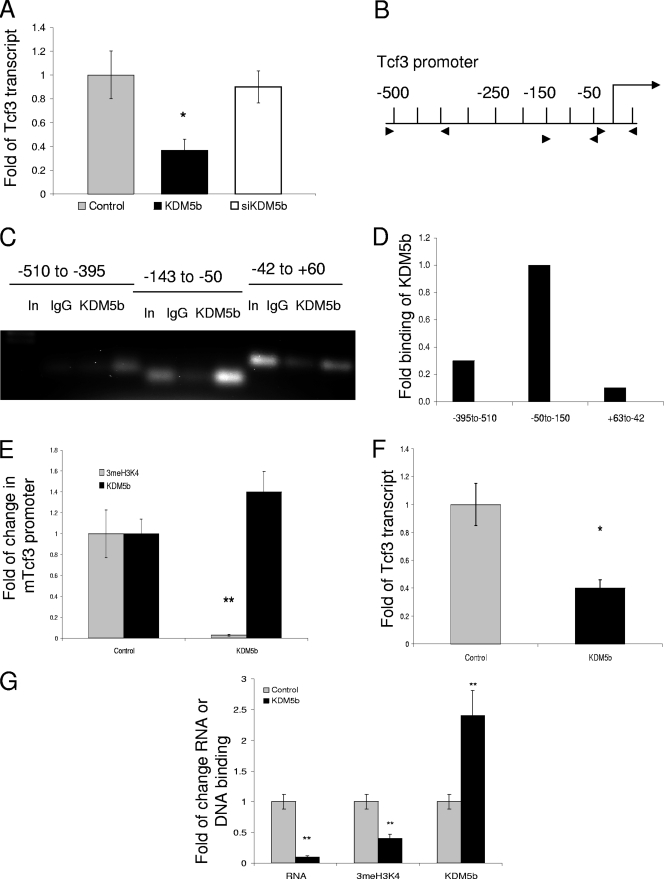
FIG. 7.
KDM5b represses mTcf3 in both mESCs and EBs. (A) qRT-PCR of mTcf3 (primers are listed in Table S1 in the supplemental material) after 24 h transfection of either control (Flag-EGFP), Flag-KDM5b, or siKDM5b. (B) Graphical representation of mTcf3 promoter in base pairs; arrowheads represent PCR primers for qChIP (primers are listed in Table S2 in the supplemental material). (C) ChIP for KDM5b binding within the mTcf3 promoter from normal mESCs. (D) qChIP for KDM5b binding within the mTcf3 promoter from normal mESCs. (E) Comparison of qChIP for 3meH3K4 binding and KDM5b binding at the −143-to-−50 element with either the control (Flag-EGFP) or the Flag-KDM5b transfection. (F) qRT-PCR of mTcf3 in mESCPURO and mESCKDM5b (G) qPCR of RT and svChIP for mTcf3 for day 9 EBs from control (mESCs) and mESCKDM5b of transcript, 3meH3K4 binding, or KDM5b binding. *, P = 0.05; **, P < 0.01. All data in triplicate except where noted.
Cell culture and transient transfections.
E14Tg ESCs, including KDM5b heterozygous (CSA022) cells, were obtained from BayGenomics and maintained in ESC medium consisting of Glasgow minimal essential medium (GMEM) with 4,500 mg/liter d-glucose and l-glutamine (Sigma-Aldrich), 10% fetal bovine serum (FBS) (VWR), 2 mM glutamine (Gibco-BRL), 0.1 mM MEM-nonessential amino acid solution (Gibco-BRL), 0.1 mM β-mercaptoethanol (Sigma-Aldrich), and 1,000 U/ml ESGRO medium (Millipore). Cells were grown at 37°C in a humidified 6% CO2 incubator. The medium was changed on alternate days, and cells were passaged every 2 days.
Transient transfections were carried out in either six-well plates or 100-mm tissue culture dishes, using Lipofectamine LTX with PLUS reagent (Invitrogen). The protocol was performed as per the supplier's directions. Briefly, 1 to 2 μg of plasmid DNA was mixed with 1 to 2 μl of PLUS reagent and 3 to 5 μl of Lipofectamine LTX in a total volume of 50 μl of GMEM. The transfection mixture was then added to a single well of a six-well plate. For 100-mm dishes, 5 to 10 μg of DNA, 2.5 to 5 μl of PLUS reagent, and 10 μl of Lipofectamine LTX were mixed in a total volume of 100 μl of GMEM. The mixture was added to the preplated mESCs and incubated for 4 to 18 h. The cells were harvested after 24 h or 48 h for reverse transcription-PCR (RT-PCR) and chromatin immunoprecipitation (ChIP) analyses.
siRNA.
mESCs at 50% confluence were transfected with short interfering KDM5b (siKDM5b) (ACG CAC TAA GTC ATA GCT ACG CTC T ), or siEGFP designed by IDTdna. Cells were transfected using Lipofectamine LTX for 24 h. Samples were then prepared for either ChIP or RT-PCR analysis.
Generation of Flag-EGFP-KDM5b and puromycin-stable mESC line.
To generate an mESC line expressing KDM5b (mESCKDM5b) or a puromycin-selected mESC line (mESCPURO), E14Tg mESCs were cotransfected with the mammalian expression plasmid carrying Flag-KDM5b and a selectable marker for puromycin resistance or with just a puromycin resistance vector (for puromycin control). mESCs were grown in the presence of 5 μg/ml puromycin, and individual colonies were isolated and analyzed for the stable expression of Flag-KDM5b by using both anti-Flag (Sigma) and anti-KDM5b antibodies (28).
EB assay.
The embryoid body (EB) differentiation assay was carried out by using a hanging drop method with a slight modification of the protocol described by Doble et al. (10). In brief, mESCs were rinsed with phosphate-buffered saline (PBS), trypsinized (Invitrogen), and disaggregated by manual pipetting to obtain a single-cell suspension. The EB medium used contains two changes from the mESC medium listed above: the FBS was decreased from 10% to 5%, and ESGRO was omitted. mESCs were resuspended in EB medium, counted with a hemacytometer, and diluted to a concentration of 45,000 cells/ml. The diluted cells were suspended in 20-μl drops on the lid of a 100-mm dish, which was inverted onto a dish containing 5 ml of EB medium, and incubated for 72 h. EBs were harvested and plated at a density of approximately six EBs per well of a 24-well ultra-low binding plate (Corning) containing EB medium. The EB medium was changed every 48 h. At day 9, EBs were harvested, and 12 to 15 EBs were combined for a single data point for RT-PCR and ChIP analysis. All experiments were done at least in triplicate.
Neural differentiation assay.
mESCs were grown in mESC medium to 75 to 85% confluence. mESC medium was then replaced with neural differentiation medium consisting of Neurobasal (Invitrogen), 5% serum, and 1× B-27 supplement and 1 mM l-glutamine. After 48 h, the cells were gently released from the tissue culture dish and replated in a nonadherent Petri dish with a 1:5 dilution of the same mESC medium. After 24 h, the neurospheres were transferred to a poly-d-lysine-coated tissue culture dish where they could be maintained for 3 to 10 additional days by changing the medium every 48 h.
Chromatin preparation.
Samples were lysed using lysis buffer (50 mM Tris-HCl [pH 7.9], 0.2 mM EDTA, 10% glycerol, 0.2% Triton X-100, 5 mM dithiothreitol [DTT], 0.2 mM phenylmethylsulfonyl fluoride [PMSF]) and centrifuged at 13,000 rpm for 20 min at 4°C. Supernatant was saved as the soluble fraction. The pellet was then resuspended in chromatin buffer (50 mM Tris [pH 7.9], 25% glycerol, 0.5 mM EDTA, 5 mM MgCl2, 5 mM DTT, 0.2 mM PMSF) and sonicated for 15 min with a Diagenode Biorupter (60 s on, 30 s off). Sodium dodecyl sulfate (SDS) loading buffer was then added, and samples were run on standard 4 to 12% gradient gels (Invitrogen) and transferred to polyvinylidene difluoride membranes, using standard protocols.
ChIP assay.
The mESCs, neurospheres, or EBs were washed twice with PBS, and cross-linking was performed using 1% paraformaldehyde for 5 to 10 min at 37°C. The adherent mESCs were scraped, washed twice with cold PBS, resuspended in 200 μl of SDS-lysis buffer containing protease inhibitor (1% SDS, 10 mM EDTA, 50 mM Tris [pH 8.1]), and incubated on ice for 10 min. The total volume was brought to 2 ml with ChIP dilution buffer (0.01% SDS, 1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl [pH 8.1], 167 mM NaCl), and samples were sonicated using a Diagenode Biorupter for 12 min (pulses of 30 s on and 30 s off). Samples were precleared using 25 μl of salmon sperm DNA-protein A-agarose (50%) beads (Calbiochem) for 1 h at 4°C on a Nutator unit. The salmon sperm DNA-protein A beads were removed by centrifugation at 2,000 rpm for 2 min at 4°C, and the supernatant was collected.
For the input control, 100 μl of precleared ChIP sample was removed and prepared similar to the method used for eluted ChIP DNA. For PCR standards, 100 μl total of precleared ChIP sample was combined from all samples and prepared by a method similar to that used for ChIP DNA. Samples were divided equally among anti-KDM5b (10 μl) antibody, anti-3meH3K4 (5 μl) antibody, or no antibody (immunoglobulin G [IgG] control) and incubated overnight at 4°C. Antibody-chromatin complexes were precipitated with 25 μl of salmon sperm DNA/protein A beads, agitated for 2 h at 4°C, and centrifuged at 2,000 rpm for 2 min at 4°C. The beads were washed five times in 1× low-salt immune complex wash buffer (0.1% SDS, 1%Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], 150 mM NaCl), 1× high-salt immune complex wash buffer (0.1% SDS, 1%Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], 500 mM NaCl), 1× LiCl immune complex wash buffer (0.25 M LiCl, 1% IGEPAL-CA630 [Sigma], 1% deoxycholic acid [sodium salt], 1 mM EDTA, 10 mM Tris [pH 8.1]), and 2× Tris-EDTA buffer. Chromatin was eluted from the antibody by using 100 μl of 1% SDS, 0.1 M NaHCO3 for 2 h at room temperature. The DNA-chromatin complexes were de-cross-linked by incubation at 65°C for 12 to 18 h and purified using a Qiagen PCR purification kit. The purified samples were analyzed by quantitative (q)PCR, performed with a Chroma4 (Bio-Rad) system with an iQSybr green qPCR kit (Bio-Rad). Samples were quantitated using Opticon software. Values shown represent percentages of DNA specifically normalized by ChIP analyses to the amount of DNA found in the input. These values were then compared between those of the control and those of the experimental samples. Samples where PCR products were seen in the IgG-negative control had this value subtracted from the input, anti-KDM5b and anti-3meH3K4, prior to analysis. The ChIP primers used are shown in Table S2 in the supplemental material.
svChIP.
The small-volume ChIP (svChIP) protocol was modified from the standard ChIP protocol as follows. Neurospheres were collected in 10 μl of medium. One microliter of 10% paraformaldehyde was added and incubated for 10 min at 37°C, and 20 μl of ChIP SDS lysis buffer containing protease inhibitors was added. Then, 100 μl of ChIP dilution buffer was added, and the sample was sonicated, similar to the standard ChIP protocol. The total volume was brought to 200 μl with ChIP dilution buffer and precleared with 20 μl of protein A/single-stranded DNA beads for 1 h. Twenty microliters of each sample was kept for input, and PCR standards and the remaining sample were divided equally among the anti-KDM5b (1 μl), the anti-3meH3K4 (1 μl), and the no-antibody solutions and incubated overnight at 4°C. Elution, de-cross-linking, and analysis were performed as described above (ChIP assay).
Quantitative RT-PCR.
RNA was prepared using a Qiagen RNeasy kit (Qiagen) and reverse transcribed using Invitrogen's Superscript III First Strand Synthesis kit (Invitrogen). qPCR was performed using Chroma4 (Bio-Rad) with an iQSybr green qPCR kit (Bio-Rad). Samples were quantitated using Opticon software. Values were expressed as the percentages of the control per unit of GAPDH. All experiments were done in triplicate (three separate experiments).
Flow cytometry.
Samples were analyzed using a BD FACScalibur for 3meH3K4 (Upstate Biotech), phospho-histone H3 (P-H3; Upstate Biotech), or Oct4 (BD biosciences). Cells were stained using a Caltag kit as per the protocol provided: 1 μg of 3meH3K4 or P-H3 antibody with Alexa 647 goat anti-rabbit secondary antibody (Invitrogen) was used. mESCs were gated on forward and side scatter, and the distribution of the 3meH3K4 or P-H3 population was assessed with FlowJo analysis software, based on the location of the peaks within the mESCPURO or the puromycin-selected neurospheres (NSPURO) control. This was applied to the mESCKDM5b or the neurospheres expressing KDM5b (NSKDM5b), and values were expressed as the (fold) change of the control. Double staining of Oct4 and P-H3 in EBs was performed using a method similar to that described above, with the addition of 1 μl of Oct4 (BD Biosciences) and Alexa 488 goat anti-mouse secondary antibody (Invitrogen). For 3meH3K4 and P-H3 assessment, samples from three separate dishes were pooled prior to analysis, stained together, and then run three temporally separate times.
Immunofluorescence.
For neurospheres and neural differentiation from EBs, chamber slides were coated with poly-d-lysine. Cells were blocked in 5% FBS, 0.1% Triton X-100 in PBS for 1 h at room temperature. All primary antibodies were incubated overnight at 4°C in the following the dilutions: anti-Oct4 (Santa Cruz), 1:200; anti-Sox1, 1:100; anti-TUJ-1 (Sigma-Aldrich), 1:500; antinestin (Chemicon), 1:200; and anti-Nanog (Abcam), 1:200. All secondary antibodies (anti-goat Alexa 488, anti-rabbit Alexa 647, and anti-mouse Alexa 488) were diluted 1:2,000. Slides were mounted with Prolong mounting medium with 4′,6′-diamidino-2-phenylindole (DAPI; Invitrogen). Images were taken with an Olympus IX81 system, with an X model camera.
RESULTS
KDM5b is a potent H3K4 demethylase and transcriptional repressor in ESCs.
To determine whether KDM5b is active in mESCs, we transiently transfected cells with either Flag-EGFP or Flag-KDM5b and extracted histones by a high-salt/high-detergent lysis. As seen in Fig. 1A, transfection with Flag-EGFP did not affect the overall levels of 3meH3K4, whereas Flag-KDM5b caused a large decrease in 3meH3K4 in mESCs. This is in contrast to data reported by Yamane et al., indicating mild decreases were seen in global demethylation in MCF7 cells (48), suggesting that in more primitive cell types, the necessary cofactors required for demethylation may be more abundant.
To further understand the role that KDM5b plays in mESCs, we performed RT-PCR after cells were transiently transfected. KDM5b's reported ability to repress cell cycle control genes is an important aspect of its function. As KDM5b is developmentally expressed early in the epiblast, when cells are choosing between self-renewal and differentiation, we chose to analyze genes that have previously been shown to have dual roles in cell cycle control and cell lineage choice, e.g., Egr1, an immediate early gene that is upregulated during differentiation (24); BMI1, an early cell lineage marker for both neural (31) and hematopoietic lineages (37) and a key control of self-renewal in progenitor populations (36); and p27KIP1, which controls developmental cell cycle exit (7). Additional genes including TRAIL, E2F1, microRNA-21, HoxA9, and the brachyury gene showed changes in expression but no KDM5b binding and were excluded as direct targets (data not shown). A third set of genes, GATA4, microRNA-17, NeuroD2, Nanog, and Oct4, showed KDM5b binding but no change in either 3meH3K4 or mRNA transcription and were also excluded (data not shown).
Genes in the subset that was directly effected (Egr1, BMI1, and p27) were significantly decreased when KDM5b was overexpressed (Fig. 1B) (by a mean n-fold decrease ± standard error of the mean [SEM] of 0.5% ± 20% for Egr1, 0.4% ± 10% for BMI1, and 0.4% ± 24% for p27 [P < 0.01 for all]). However, only Egr1 (Fig. 1B) (decrease of 1.5% ± 0% [P = 0.05]) appeared to be sensitive to the loss of KDM5b. This suggests that BMI1 and p27 are tightly regulated, presumably due to their strong oncogenic effects (1, 35), whereas Egr1 is an immediate early gene (46) and has a high degree of variability in expression in mESCs. To confirm the direct effect of KDM5b, we performed ChIP analysis of both 3meH3K4 and KDM5b. We saw significant decreases of at least 50% in 3meH3K4 at the promoters of all three genes. BMI1 showed the largest change in 3meH3K4, with a 90% decrease (Fig. 1C) (0.1% ± 15% [P < 0.01]), Egr1 showed a 70% decrease (Fig. 1D) (0.3% ± 7% [P < 0.05]), and p27 had 50% of the control 3meH3K4 levels (Fig. 1E) (P = 0.05). The binding of KDM5b showed a general correlation with the decrease seen for 3meH3K4, with Egr1 having the greatest increase (Fig. 1C) (11-fold ± 25% [P < 0.01]), BMI1 having a 10-fold increase (Fig. 1D) (SEM, 30% [P = 0.05]), and p27 having the least increase (Fig. 1E) (3% ± 6% [P = 0.05]). To understand whether the promoters of these genes are sensitive to KDM5b loss as well as increase, we performed ChIP analysis after an siRNA-mediated knockdown. After the knockdown of KDM5b, the promoters of both Egr1 and p27 showed significant increases in 3meH3K4 levels (Fig. 1F) (4.5% ± 0% and 3% ± 0%, respectively [P < 0.01 for both]), but no significant difference was seen for BMI1 (data not shown). The sensitivity of the p27 promoter to the loss of KDM5b without the correlated increase in transcript confirms that there are additional layers of regulation present in this system.
Constitutive expression of KDM5b modifies ESC proliferation and 3meH3K4 levels.
The transient transfection data suggest that KDM5b has the ability to repress genes involved in differentiation in addition to its recognized role in cell cycle control. This led us to test what effect constitutive KDM5b expression would have on ESCs by making a stable cell line using the Flag-KDM5b construct (mESCsKDM5b). We tested two clones, each showing approximately threefold overexpression of KDM5b (Fig. 2A, clone 2), by Western blotting. Data shown represent the combined data for both clones; all comparisons to the control were the combined data of parental mESCs and puromycin resistance-stable cell line (mESCsPURO). No significant differences were observed between the expression of KDM5b or any of the KDM5b target genes of the mESCsPURO and those of the parental mESCs. To complement these studies, we attempted to make a constitutive knockdown cell line. However, all of the drug-selected clones that could be passaged showed normal levels of KDM5b, and the clones that showed the knockdown of KDM5b were incapable of more than one passage and appeared to be senescent (data not shown).
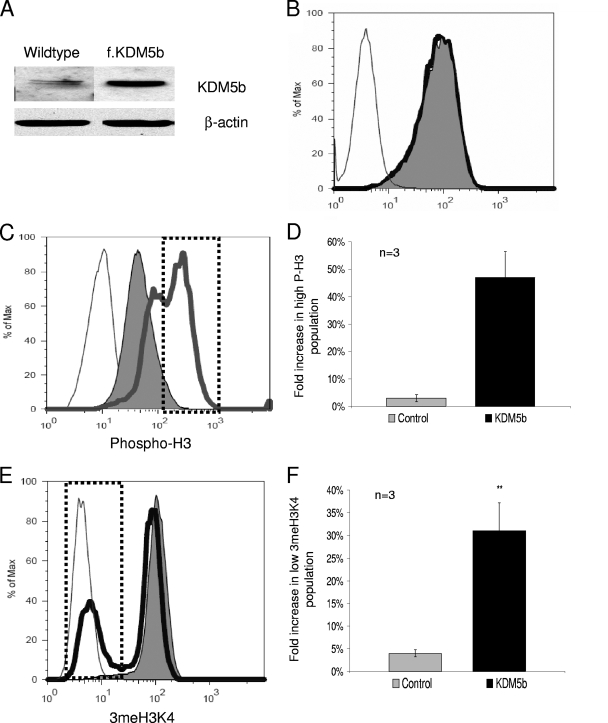
FIG. 2.
Constitutive KDM5b increases the mitotic rate and decreases global 3meH3K4. (A) Expression of KDM5b in wild-type and KDM5b-stable mESCs. (B) Representative measurement of the Oct4 protein in mESCsKDM5b and mESCsPURO as measured by flow cytometry with anti-Oct4 (BD Biosciences) and Alexa 488 secondary antibody (Invitrogen). (C) Representative measurement of the mitotic rates in mESCsKDM5b and mESCsPURO as measured by flow cytometry with anti-P-H3 (Ser10) (Upstate Biotechnology) and Alexa 647 secondary antibody (Invitrogen). (D) Flow cytometry data given in panel C from all three replicates. (E) Representative measurements of 3meH3K4 in mESCsKDM5b and mESCsPURO as measured by flow cytometry with anti-3meH3K4 (Upstate Biotechnology) and Alexa 647 secondary (Invitrogen). (F) Graph of flow cytometry data given in panel E from all three replicates. For all flow cytometry data, the thin line represents values of the secondary antibody control alone, the solid area is mESCPURO values, and the heavy line represents mESCKDM5b. *, P = 0.05; **, P < 0.01. All data are in triplicate except where noted.
To understand what effect the overexpression of KDM5b would have on stem cell properties, we used flow cytometry to measure the percentage of cells that maintained the stem cell marker Oct4. Using the expression of Oct4 as a marker allowed us to assess the population dynamics of the stable cultures. ESC cultures of either the wild-type cells, the mESCsPURO, or the mESCsKDM5b showed no differences in the percentages of Oct4-positive cells or in the fluorescence intensity levels of Oct4 expression, suggesting that constitutive KDM5b did not affect stem cell dynamics (Fig. 2B, compare the solid area to the black line data). The fact that Oct4 levels were not strongly affected suggests that KDM5b stable cells were staying as stem cells, as Oct4 must be maintained within tight parameters to assure self-renewal (34).
To further characterize the properties of the mESCsKDM5b, we used P-HP to measure the proliferation rate of the cells by assessing the percentage of cells in mitosis. Compared to the population of control cells, this population appeared to have shifted toward higher levels of P-H3 (Fig. 2C, compare the solid area to the heavy gray line data). Measurement of the number of cells at the extreme high range (Fig. 2C, dotted box) of P-H3 (as a measure of mitotic cells) showed that there was 16-fold increase in this fraction (Fig. 2D) (3% ± 60% versus 47% ± 5%, respectively [P < 0.001; n = 3]), demonstrating that the mESCsKDM5b had a higher rate of mitosis. To ensure that the global effects seen were directly attributable to KDM5b activity, we measured the 3meH3K4 in the mESCs by flow cytometry. In Fig. 2E, the mESCsKDM5b (Fig. 2E, black line) showed a bifurcated population in contrast to that of the control, which showed a single population (Fig. 2E, solid area). This second population represented 31% (versus 4% in the control [P < 0.001; n = 3]) (Fig. 2F) of the total cells, an eightfold increase in cells with low 3meH3K4. The division of the mESCsKDM5b into two populations of both P-H3 and 3meH3K4 suggests a link between stem cell proliferation and low H3K4 methylation; unfortunately, costaining of these two markers was not possible by flow cytometry analysis. The linkage between low H3K4 methylation and proliferation in stem cells was similar to that seen with a form of leukemia, where a variety of MLL1, an H3K4 methyltransferase, loss of activity translocations (23) caused an increase in proliferation rates.
mESCsKDM5b have aberrant neural differentiation and increased expression of self-renewal markers.
All three of the target genes selected have previously been shown to have an effect on neural differentiation. Therefore, we tested the ability of the mESCsKDM5b to differentiate into neurons. First, to test the efficiency of the early steps of differentiation, we tested day 3 neurospheres for downregulation of Oct4 and decreases in proliferation as indications that mESCKDM5b could differentiate in neurospheres (NSKDM5b). As shown in Table 1, while most mESCs did follow the normal differentiation pattern (Table 1, Oct− Mito−), the NSKDM5b showed twofold increased proliferation in both the Oct4− (2.2-fold [P = 0.0002]) and the Oct4+ (2.3-fold [P = 0.005]) fractions. This suggests that while mESCs are differentiating, the neural stem cell fraction (Mito+ Oct4−) had a greater proliferation and that the mESCs (Oct4+ Mito+) were not committing at the same rate as that of the control. To test this, we plated neurospheres from mESCPURO (NSPURO) and NSKDM5b onto poly-d-lysine to induce commitment to neural cell fate. To confirm the identity of the cells, we stained day 5 and day 7 neurospheres from both NSPURO (Fig. 3A and C) and NSKDM5b (Fig. 3B and D) with the neuronal marker TUJ-1 and the stem cell/neural crest marker Oct4. Many of the cells migrating out of or onto the surface of the neurospheres appeared to be TUJ-1 positive (Fig. 3A and C), confirming that they were differentiating toward the neural lineage. In general, we did not see Oct4-positive cells in the control neurospheres or in the cells that migrated outward. However, the majority of NSKDM5b did not plate down and continued to grow in suspension. When neurospheres did plate down, they had few TUJ-1-positive cells (Fig. 3B and D). There also appeared to be pockets of Oct4-positive cells within the NSKDM5b (Fig. 3B); since Oct4 expression is seen in the early neural crest (15), in addition to its stem cell expression, it is possible that these cells are neural crest cells, a multipotent progenitor cell type. A change of cell fate toward the neural crest would explain the TUJ-1/Oct4 double-positive cells that are seen (Fig. 3D). The presence of the neural crest would not explain the propensity of the neurospheres to proliferate and stay in suspension. It is likely that NSKDM5b is a mixture of neural crest and more primitive multipotent cell types.
TABLE 1.
KDM5b increases stem cell markers and proliferationa
| Marker(s) | mESC differentiation pattern
|
P value | ||
|---|---|---|---|---|
| % KDM5b | % Control | Fold change (KDM5b/control) | ||
| Oct4− Mito− | 89.74 | 95.28 | 0.9 | 7.5E-05 |
| Mito+ Oct4− | 9.26 | 4.25 | 2.2 | 0.0002 |
| Oct4+ | 0.08 | 0.08 | 1.1 | 0.9 |
| Oct4+ Mito+ | 0.93 | 0.40 | 2.3 | 0.005 |
Results of flow cytometry analysis of day 3 neurospheres showing the percentage of all live cells that stained with anti-Oct4 (Oct4−) primary/Alexa 488 secondary and/or (Mito+) anti-P-H3/Alexa 647 secondary.
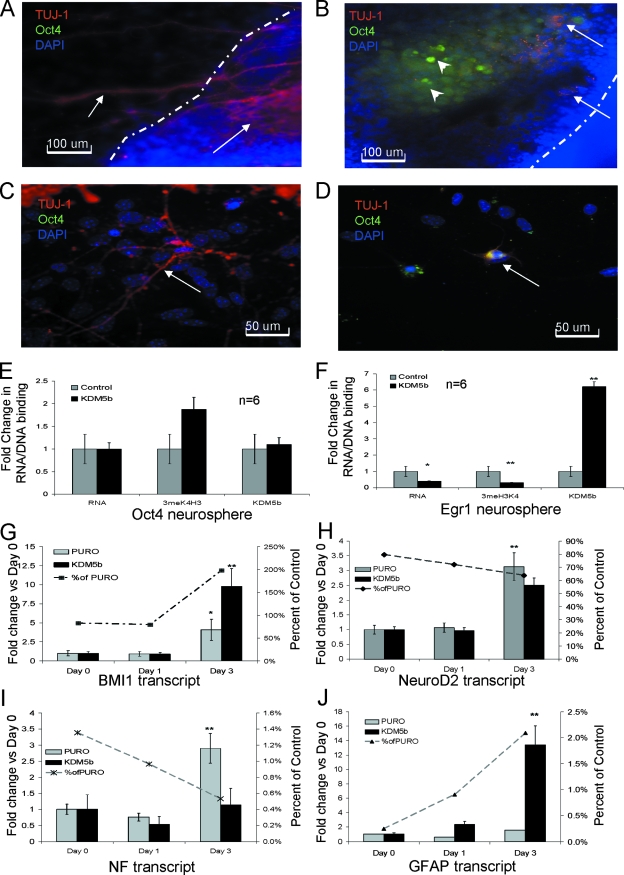
FIG. 3.
Terminal neural differentiation is blocked by constitutive KDM5b expression. (A) Day 5 neurospheres from control mESCs adhered to the dish. The dashed line shows the edge of the neurosphere. Arrows denote TUJ-1-positive cells. (B) Neurospheres from mESCsKDM5b. Dashed line shows the edge of the neurosphere. Arrows denote TUJc-positive cells; arrowheads denote Oct4-positive cells. (C) Day 7 neurospheres from control mESCs. Arrow shows TUJ-1-positive cell. (D) Day 7 neurospheres from mESCs KDM5b. Arrow shows TUJ-1/Oct4 double-positive cell. For panels A, B, C, and D, blue is DAPI, red is TUJ-1, and green is Oct4. (E and F) qRT-PCR and svChIP for Oct4 and Egr1 for day 3 neurospheres from control (mESCPURO and mESCs) and mESCKDM5b showing transcript, 3meH3K4 binding or KDM5b binding (primers are listed in Table S1 and S2 in the supplemental material). (G, H, I and J) qRT-PCR of BMI1, NeuroD2, NF, and GFAP transcript during neurosphere formation. Bar graph and left y axis show the change value for each day with respect to day 0 value. Line graph and right y axis denote the percentage value for NSKDM5b compared to that for NSPURO on the same day. *, P = 0.05; **, P < 0.01. All data are in triplicate except where noted.
Since only a subset of cells from NSKDM5b appeared to have differentiated into neurons, we decided to look at individual neurospheres by RT-PCR and ChIP analyses. To perform the ChIP analysis, we modified the ChIP assay. The modified version (svChIP) showed very little loss of signal compared to that of the standard ChIP protocol (see Fig. S1A in the supplemental material). This method gave consistent and similar results with mESC cultures, down to 5,000 cells (see Fig. S1B in the supplemental material), demonstrating that ChIP can be performed with sample sizes analogous to those for a neurosphere. We first tested the stem cell gene Oct4 to determine whether the neurospheres contained undifferentiated progenitors. RT-PCR results suggested that neurospheres from both the control and the KDM5b are repressing Oct4 at the transcript level and are differentiating (Fig. 3E). The svChIP analysis showed no change in KDM5b recruitment, although there appeared to be an increased amount of 3meH3K4 at the Oct4 promoter, possibly anticipating upregulation of the Oct4 transcript (Fig. 3E). In contrast, Egr1, a gene that is upregulated during early neural differentiation was significantly decreased in NSKDM5b (0.4% ± 7% [P = 0.05]); this decreased mRNA was coupled to both an 80% decrease in 3meH3K4 and a twofold increase in KDM5b localization (Fig. 3F).
To characterize differences in neural differentiation, we followed four neural markers during early differentiation, as follows: BMI1, a neural stem cell proliferation gene (31); NeuroD2, a definitive neural marker (21); neural filament (NF), a marker of committed neurons; and glial fibrillary acidic protein (GFAP), a glial marker. While BMI1 is a KDM5b target in mESCs, it did not appear that KDM5b regulates BMI1 expression during forced neural differentiation. Since the mESCKDM5b retained the ability to differentiate, the induction of BMI1 suggested that they are progressing from a pluripotent cell to a neural stem cell. BMI1 expression was similar to that of the control for the first 24 h, consistent with its required role during neural differentiation. Although both cells have significantly increased BMI1 expression at 72 h compared to that at day 0 (Fig. 3G) (4% ± 4% in control and 10% ± 4% in KDM5b), BMI1 was upregulated to a twofold higher level (Fig. 3G) in the NSKDM5b than in the NSPURO. An increase in BMI1 at 72 h suggests that the NSKDM5b had an increased progenitor proliferation rate, consistent with KDM5b's regulation of the cell cycle and the increased P-H3, as seen in Table 1. The expression of NeuroD2, NF, and GFAP showed induction patterns in the NSKDM5b similar to those in the NSPURO compared to that of the day 0 sample (Fig. 3H, I, and J). When values of the control were compared with those of KDM5b, the rate at which expression increased for these more definitive markers was slower in the NSKDM5b (Fig. 3H, I, and J). This was most pronounced at day 3 for both NF and GFAP levels, when the expression of these genes was only 0.5% and 2%, respectively, of that of the NSPURO (Fig. 3H, I, and J). This lack of committed neural markers suggests that while constitutive KDM5b expression did not block the initial cell fate decisions, it might have blocked or delayed terminal differentiation, allowing progenitor populations to proliferate.
EB assay reveals a role for KDM5b in the maintenance of stem and progenitor cells during differentiation.
To further understand the role of KDM5b in differentiation, we turned to the EB assay to assess the general role that KDM5b plays in early development. In addition to analyzing the mESCPURO and mESCKDM5b, we analyzed a KDM5b heterozygous cell line from BayGenomics (mESC5b+/−). The mESC5b+/− appeared morphologically normal, with a mitotic rate and an Oct4 expression profile similar to those of the mESCPURO, suggesting that a single allele was sufficient for maintaining the pluripotent state (data not shown). All three lines were capable of generating EBs; however, at day 9, the KDM5b EBs (EBKDM5b) appeared to be slightly larger than those of the control (Fig. 4, compare B versus A), and EBs generated from EB5b+/− appeared smaller, with poorly defined structures (Fig. 4, compare C versus A). The EB5b+/− expressed approximately 25% of the KDM5b seen in the controls (Fig. 4D), which correlated to extremely low levels of the stem cell markers Oct4 and Nanog (Fig. 4E). The EBKDM5b maintained an approximately twofold higher level of KDM5b during differentiation, and while Oct4 levels were only mildly increased at the mRNA level, Nanog transcription was increased by sixfold, suggesting that a percentage of cells within the EB retained stem cell markers (Fig. 4E).
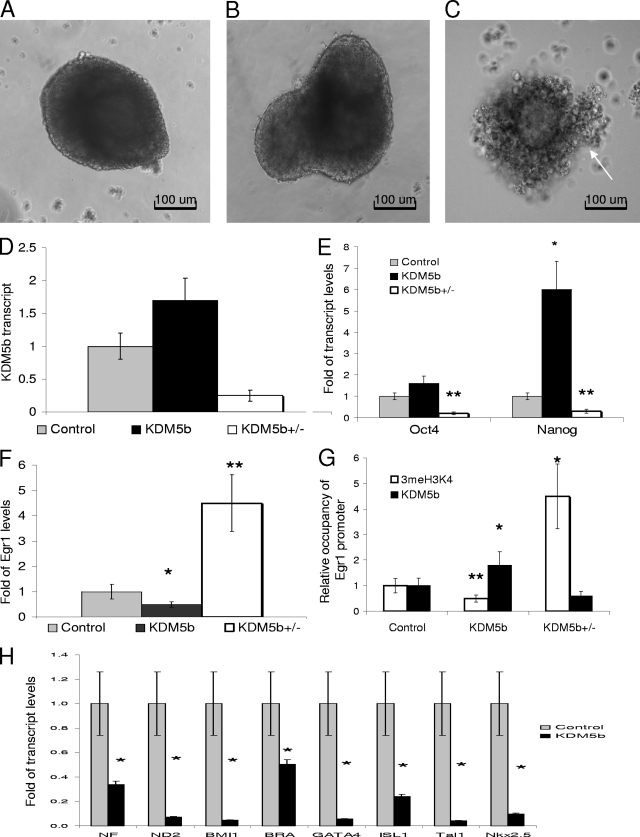
FIG. 4.
EB assay reveals a general blockage in cell specification. (A, B, and C) Day 9 EBs from mESCs, mESCKDM5b, and mESC5b+/−; arrow shows cells poorly attached to EBs. (D) qRT-PCR of KDM5b transcript from day 9 EBs. (E) qRT-PCR for Oct4 and Nanog for day 9 EBs from mESC, mESCKDM5b, and mESCs5b+/− (primers are listed in Table S1 in the supplemental material). (F) qRT-PCR for Egr1 for day 9 EBs from mESC, mESCKDM5b, and mESCs5b+/−. (G) qsvChIP analysis for 3meH3K4 binding or KDM5b binding at the Egr1 promoter for day 9 EBs from mESC, mESCKDM5b, and mESCs5b+/−. (H) qRT-PCR of early differentiation markers from day 9 EBs. *, P = 0.05; **, P < 0.01. All data are in triplicate except where noted.
In P19 EC cells, increased Egr1 levels cause spontaneous differentiation, whereas dominant-negative Egr1 strongly blocks differentiation (24). This suggests a conserved role for Egr1 in differentiation. This can also be seen in our model, where EBKDM5b had significantly less Egr1 transcript (Fig. 4F) and the EB5b+/− expressed significantly more Egr1 (Fig. 4F). The expression of Egr1 negatively correlated with the expression of Oct4 and Nanog in both the heterozygous and overexpression KDM5b mESCs. Since Egr1 is a direct target of KDM5b, we performed svChIP with groups of EBs from control (a combination of mESC and mESCPURO), KDM5b, and KDM5b5b+/−. svChIP analysis showed that the Egr1 promoter had lower 3meH3K4 levels and higher KDM5b levels in the EBKDM5b (Fig. 4G) (0.34% ± 3% [P = 0.01] and 2.1% ± 8% [P = 0.05], respectively) and higher 3meH3K4 levels and lower KDM5b levels in the EB5b+/− (Fig. 4G) (4.5% ± 9% [P = 0.05] and 0.6% ± 6%, respectively). In addition, the EB5b+/− showed low expression of cell lineage markers and tended to disintegrate shortly after day 9. The general lack of organization and extremely low levels of all cell type markers in EB5b+/− suggest a high level of apoptosis, and therefore these EBs may not be representative of actual KDM5b expression profiles. This led us to exclude the heterozygous EBs from further analysis. To determine whether the EBKDM5b were being directed toward a specific lineage, we analyzed eight different markers that collectively measure early developmental tissue types, as follows: ectodermal (NF, ND2, BMI1, and ISL1), endodermal (GATA4 and ISL1), and mesodermal (bra, GATA4, BMI1, Tal1, and Nkx2.5) (Fig. 4H). All eight of these genes were significantly downregulated at the transcript level, suggesting that at day 9, the EBKDM5b were not committing to any lineage at the same rate as the controls.
To further determine the ability of KDM5b to affect differentiation, we measured the proliferation rate of the day 9 EBs. We first calculated the percentage of cells in the EBs of the control or mESCsKDM5b that were proliferating and/or expressing Oct4. As shown in Fig. 5A, the percent of cells within the EB that did not express Oct (differentiated) was slightly less than that in the EBKDM5b (54% versus 60%, respectively; a 10% difference). Proliferative cells that downregulated the Oct4 protein, presumably uncommitted progenitors, were significantly increased in the EBKDM5b (Fig. 5A) (3.5% ± 18% versus 1.4% ± 4% [P = 0.01]), as were the Oct4-positive proliferative cells, which represent stem cells, and a minor population of Oct4-expressing progenitors (mesoderm and primitive ectoderm) (34) (Fig. 5A) (30% ± 6% versus 23% ± 2% [P < 0.05]). Additionally, nonproliferating Oct4-positive cells were decreased in the EBKDM5b (Fig. 5A) (11% ± 8% versus 17% ± 7% [P < 0.05]), although the total fraction of Oct4-positive cells was the same as those of the control and KDM5b. This suggests that similar to results for the neurosphere assay, KDM5b was not blocking differentiation but was delaying terminal differentiation through increased proliferation. This is corroborated by the lack of TRAIL transcript in day 9 EBKDM5b (Fig. 5B) (0.01% ± 1% versus 1% ± 0% [P < 0.01]).
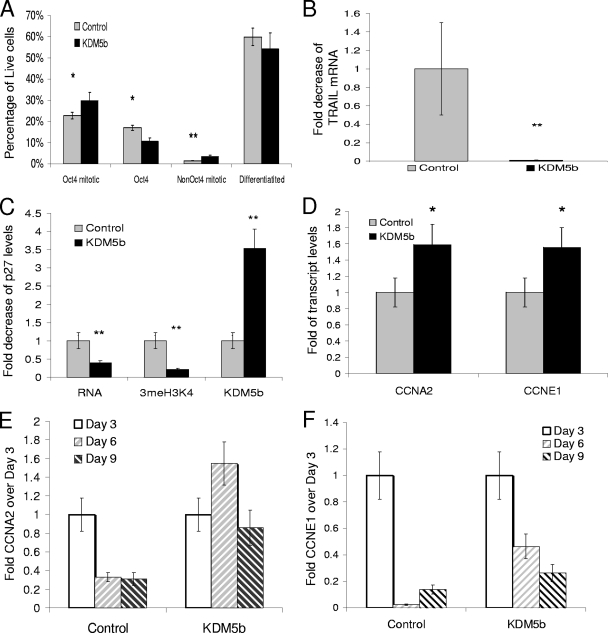
FIG. 5.
EBKDM5b display increased proliferation through increased expression of cyclin A2 and E1. (A) Graph of flow cytometry results showing the percentage of all live cells that stained with either anti-Oct4 primary/Alexa 488 secondary and/or anti-P-H3/Alexa647 secondary; “Differentiated” represents all live cells that were not stained by either antibody in either control (EB and EBPURO) or EBKDM5b at day 9. (B) Expression of TRAIL mRNA in day 9 EBs, either control (normal EBs) or EBKDM5b at day 9. (C) qRT-PCR and svChIP for p27 for day 9 EBs from control (mESCs) and mESCKDM5b of transcript, 3meH3K4 binding, or KDM5b binding. (D) qRT-PCR of cyclin A2 and cyclin E1 transcript from day 9 EBs (primers are listed in Table S1 in the supplemental material). (E and F) Comparison of qRT-PCR for cyclin A2 and cyclin E1 resulting from the EB assay. The baseline value is the amount of transcript in day 3 EBs. *, P = 0.05; **, P < 0.01. All data in triplicate except where noted.
Cell cycle control during development involves, in part, regulation of the G1 transition (27). The ability of p27KIP1 to inhibit CDK2-cyclin E complexes (13), as well as its additional functions (5), may be critical to the control of many progenitor cell types. Since p27 was a direct target of KDM5b, we determined whether p27 was downregulated in day 9 EBs and might allow EBKDM5b to continue proliferating. p27 mRNA was decreased by 60%, with an 80% reduction in 3meH3K4 and an approximately threefold increase in KDM5b binding (Fig. 5C) (0.4% ± 1% [P < 0.05], 0.22% ± 4% [P < 0.01], and 3.5% ± 4% [P = 0.01], respectively). The EBKDM5b showed an increased mitotic rate and decreased expression levels of a G1 control gene. We determined how this affected the expression of cyclin genes by testing the expression of an M-phase cyclin, CCNA2, and a G1-phase cyclin, CCNE1. Both of these cyclins showed mild but significant increases in their mRNA levels (Fig. 5D) (CCNA2, 1.6% ± 8% [P = 0.01]; and CCNE1, 1.6% ± 4% [P = 0.01]). Additionally, during differentiation, cyclin A2 did not appear to be downregulated, and cyclin E1 expression was not as strongly downregulated in the EBKDM5b (Fig. 5E and F, respectively). Maintenance of these cyclins might have allowed a larger fraction of cells to transition through G1, thereby bypassing G0 and terminal differentiation. This, coupled with the decreased expression of p27, might have caused progenitor cells to be refractory to differentiation signals, resulting in a net gain of undifferentiated cells.
mESCKDM5b retains proliferative capacity during differentiation.
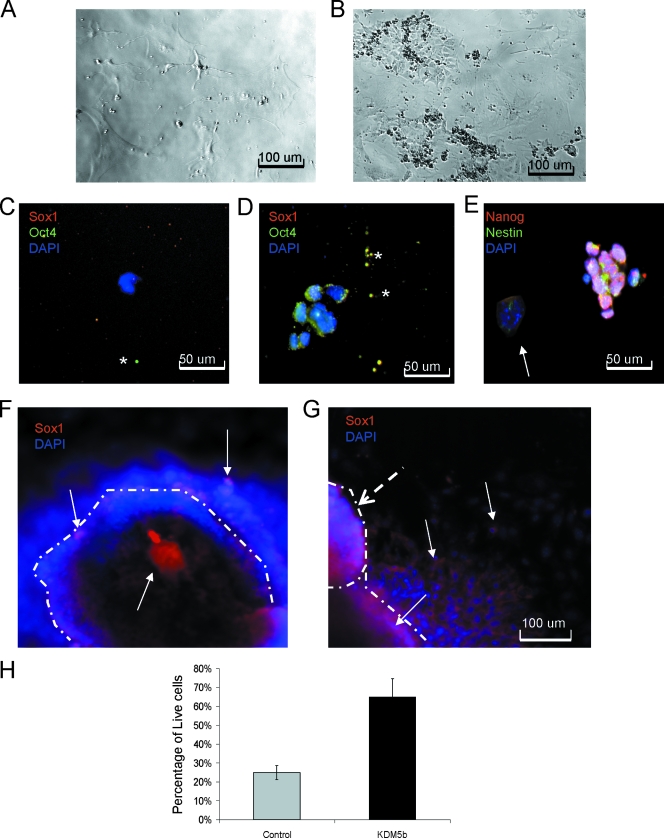
To determine whether there were more uncommitted cells in day 9 EBs, we dissociated the EBs and replated single cells at a medium density in mESC medium (approximately 24 EBs pooled from three replicates into 48 wells). Only a few of the replated cells from control EBs continued to proliferate and could be passaged (3/48), and most of these cells had the appearance of differentiated cell types (Fig. 6A). These cells failed to sustain the cultures by passage 5. In contrast, approximately half of the cells from EBKDM5b (23/48) sustained proliferation and could be serially passaged for a minimum of 20 passages (Fig. 6B). Using early passage cultures, we attempted to identify the cells that were proliferating in the culture. Since it appeared from the neural differentiation experiments that mESCKDM5b might maintain neural progenitors, we used Oct4 and Sox1 to test for the presence of stem cells and neural stem cells in the replated cells. In controls, neither Oct4 nor Sox1 was seen in the remaining cells (Fig. 6C). In comparison, the replated cells did not express Sox1 but expressed Oct4, although staining was not exclusively nuclear (Fig. 6D). To further understand the identity of these cells, we tested for the expression of the stem cell gene Nanog and the neural/neural crest progenitor gene Nestin. As shown in Fig. 6E, the majority of cells expressed Nanog, and some appeared to have Nestin expression as well. There were colonies of cells that did not express either of these genes and showed a very flat appearance, suggesting a fibroblast identity. Since Nanog is seen primarily in embryogenesis (17), we believe that the continued expression of Nanog was blocking differentiation of these cells and maintaining their continued renewal, similar to that seen by Chambers et al. (8), where Nanog was a key element in the ESCs' choice to reenter the cell cycle. The presence of both Oct4 and Nanog in conjunction with the data showing increased proliferation of cells in day 9 EBs strongly suggests that these cells were mESCs that were not differentiated. In addition to replating cells, we took day 9 EBs and differentiated them into neurons. Both the control and the KDM5b EBs were capable of early neural differentiation as seen by the presence of Sox1 (Fig. 6F and G). The cells in the control appeared to have downregulated Sox1 as they migrated out of the sphere, whereas the KDM5b cells appeared to maintain the expression of Sox1 after they left the sphere. This is consistent with the blockage of terminal differentiation that was suggested by the neural differentiation and the replating experiments. In addition, KDM5b cultures consistently showed the appearance of secondary neurospheres (Fig. 6G). The KDM5b cultures also had a greater amount of floating cells—65% of all floating debris was live cells—than the control cells, where only 25% were live cells as detected by 7-amino-actinomycin D staining (Fig. 6H). The ability of the day 9 EBs to give rise to multipassage cultures, the continued presence of cells in suspension, and the maintenance of stem cell markers suggest that during differentiation, undifferentiated progenitors were being maintained in a KDM5b-dependent manner.
FIG. 6.
EBKDM5b retain highly proliferative cells that are refractory to differentiation. (A) Cells from dissociated normal EBs after four passages. (B) Cells from dissociated EBKDM5b after two passages. (C) Cells from dissociated normal EBs after two passages stained with DAPI (blue), Oct4 (green), and Sox1 (red). *, shows background spots. (D) Cells from dissociated EBKDM5b after four passages stained with DAPI (blue), Oct4 (green), and Sox1 (red). *, shows background spots (C and D). Cells from dissociated EBKDM5b after four passages stained with DAPI (blue), Nestin (green), and Nanog (red). (E) Arrow shows Nanog, Nestin double-negative colony of cells. (F) Neural differentiation of day 9 EBs from control; the edge of the colony is marked by a dashed line. Arrows show Sox1-positive (red) cells and DAPI-stained (blue) cells. (G) Neural differentiation of day 9 EBsKDM5b; the edge of the colony is marked by dashed lines. Dashed arrow shows secondary colony forming. Arrows show Sox1-positive (red) cells and DAPI-stained (blue) cells. (H) Graph of 7-amino-actinomycin D staining of cells found in media during neural differentiation of day 9 EBs from control (normal mESCs) and mESCKDM5b. The experiment was done in triplicate.
The Nanog repressor mTcf3/hTcf7L1 is a direct target of KDM5b and is misexpressed during differentiation.
While the presence of proliferating cells might be attributed in part to the repression of Egr1 and p27KIP1, the apparently blocked differentiation suggested by the continued Nanog and Oct4 expression is not. To understand the significant increase in Nanog expression seen in EBKDM5b, we searched for transcription factors that have been shown to repress the Nanog transcription. One such gene was the Tcf3 (hTcf7L1) mouse gene, required for mESC differentiation through direct repression of Nanog (38). To determine whether mTcf3 was a direct target, we first measured the effect of a transient transfection of KDM5b on mESCs. The transient transfection of KDM5b in mESCs reduced expression of mTcf3 to 37% of the control transfection; however, similar to p27 and BMI1, mTcf3 was not affected by siRNA directed against KDM5b (Fig. 7A).
The promoter region for mTcf3 had not been previously characterized; therefore, we first mapped the promoter using ChIP. Due to the high GC content of the promoter, only three promoter segments produced PCR products suitable for quantitation by both ChIP and svChIP, as shown in Fig. 7B. To confirm that mTcf3 was a direct target of KDM5b, we determined the binding of KDM5b to all three regions and found that while binding was seen at all three regions, KDM5b preferentially bound to the region from −50 to −150 (Fig. 7C and D). This region was then used to analyze the effect of KDM5b on the mTcf3 promoter. During transient transfection, the mTcf3 promoter was significantly demethylated (Fig. 7E) (0.03% ± 5% [P = 0.01]), with only minor increases in KDM5b binding (1.4% ± 7%) (Fig. 7E). Due to the small increase in KDM5b binding, we tested the stable cell lines and EBs to ensure that KDM5b was a direct regulator of mTcf3.
In the mESCKDM5b cell lines, mTcf3 was constitutively repressed (Fig. 7F) (1% ± 6% versus 0.43% ± 4% [P < 0.05]), confirming that increased KDM5b expression can lead to decreased mTcf3 transcription. Since we hypothesized that KDM5b-mediated repression of mTcf3 was causal to the increase in the Nanog transcript, as seen in day 9 EBs, we then tested the transcript and promoter status of mTcf3 in the EBs. EBKDM5b expressed 13% of the mTcf3 gene of the control EBs (Fig. 7G) (1% ± 5% versus 0.13% ± 100% [P = 0.01]), and this was coupled to a 60% decrease in 3meH3K4 and a 2.4-fold increase in KDM5b binding at the mTcf3 promoter (0.4% ± 8% and 2.4% ± 10% [P = 0.01] for both) (Fig. 7G). The repression of mTcf3 and the upregulation of Nanog were consistent with results from previous reports, indicating that the mTcf3 null mESCs did not differentiate as a result of their inability to downregulate Nanog (38).
DISCUSSION
In this study, we identified a role for the histone demethylase KDM5b in cell fate decisions through its ability to directly regulate genes that control cell cycle, cell differentiation, and cell lineage. In mESCs, these genes include mTcf3, BMI1, Egr1, and p27. Although not all of these target genes display ESC null phenotypes, they appear to be functionally important in many tissues. Both the overexpression and the knockout of BMI1 confer developmental abnormalities, stemming from its role in lineage selection and progenitor survival (45). This role in cell lineage selection and survival suggests that tight control of BMI1 levels may be an essential element of pluripotency. The loss of Egr1 has effects in both hematopoietic and neural lineages and appears to be a potent inducer of differentiation in several multipotent cell types. In P19 cells, tight control of Egr1 is essential for maintaining the pluripotent state (22, 24).
A key element in maintaining both stem cell and progenitor populations is the control over reentry into the cell cycle. While p27's contribution to cell cycle control is unclear, it has been established that the loss of p27 increases cell number and blocks terminal differentiation (33). While these genes are not the only KDM5b targets, which contribute to the overall phenotype, they are proper and consistent model genes with which to understand KDM5b function during differentiation.
Previous studies of KDM5b have clearly implicated it in cell cycle control (48). While the mechanism of control has previously appeared to be indirect through BRCA1, here we report the direct control of cell cycle through its regulation of p27KIP1 and Egr1. Recent reports suggest that the loss of p27 may play a role in breast cancer progression (26), strongly suggesting that regulation of p27 may be a conserved function of KDM5b. Egr1's ability to effect apoptosis and growth arrest in several cell types through a regulatory network consisting of p53 and p73 demonstrates an important role for regulation of Egr1 in proliferating cells (49). In stem cells, control of these two genes could be key to their choice to exit the cell cycle. The constitutive expression of KDM5b in stem cells ensures that these genes, in addition to other gene targets, are repressed, and therefore the percentage of cells in mitosis at any given time is increased. This is also consistent with the published roles of p27KIP1 and Egr1 in differentiation.
While mESCsKDM5b show increased mitosis, there is no change in the expression profile of the stem cell markers under normal conditions. Although the majority of mESCsKDM5b can differentiate or at least lose pluripotency, a significant portion of cells remains undifferentiated, suggesting that some cells are refractory to environmental cues or are interpreting the signals in a different manner. Further studies need to be done to understand the signaling cascades that control KDM5b localization. As our results suggest, some targets may escape repression during development, based on which cell lineages are chosen, e.g., BMI1 is expressed during forced neural differentiation but remains repressed during EB assays. We further putatively identified the transcription factors Oct4, Nanog, NeuroD2, and GATA4 as gene targets, but additional studies need to be done to confirm them as targets and to understand what signaling pathways regulate KDM5b activity. It is possible that these factors represent cell type-specific targets that must be addressed in a specific progenitor population, e.g., NeuroD2 repression by KDM5b may be seen best in epithelial progenitors after cells have committed to ectodermal lineages.
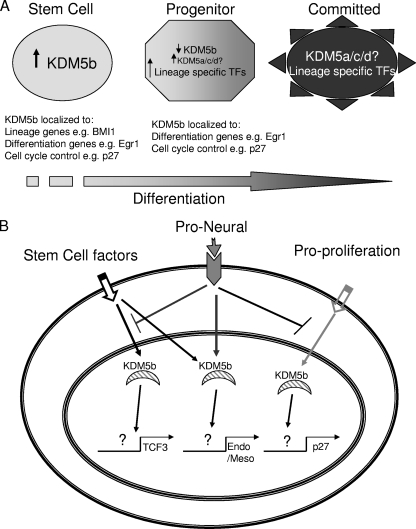
Our results are not dissimilar to those seen during morphogenesis, for which the best characterized pathway is Drosophila DPP (decapentaplegic) and its regulation of the transcription factor gene dBrinker (Bri). DPP negatively regulates the expression of bri in a graded fashion, i.e., the more DPP a cell has bound to its surface, the greater the repression of bri transcription. This graded loss of bri expression changes the target genes which are repressed, i.e., more DPP results in an increased number of different Brinker target genes but not necessarily an increased transcription of each target gene (32). This allows DPP to create a gradient for wing imaginal disc development (2). For KDM5b, this would be translated into a “commitment” gradient, where KDM5b is downregulated as cells lose pluripotency and proliferative capacity. This may occur by a hierarchal model, where the presence of KDM5b at the promoter is based on protein expression (Fig. 8A). When a cell commits to a cell lineage, it downregulates KDM5b expression, possibly by KDM5a, which has been shown to oppose KDM5b function in cell cycle regulation (16). In a developmental context, this would be intertwined with control over the recruitment of KDM5b in response to signal cascades (Fig. 8B). This would allow for fine tuning based on cell fate decisions. A combination of proneural with proliferation signals, for example, would allow for the maintenance of neural stem cells. Further support for this model is provided by the direct regulation of mTcf3, a repressor of Nanog that allows differentiation to proceed by downregulation of stem cell markers. The key finding of this study is that of a direct role for KDM5b in cell fate decisions through the regulation of cell cycle reentry and coregulation of prodifferentiation molecules. This dual role may provide a mechanism by which uncommitted cells coordinate cell cycle and lineage choice.
FIG. 8.
Model for KDM5b function during early development. (A) As differentiation progresses, KDM5b is downregulated, and other KDM5 family members are upregulated, thereby allowing different subsets of its target genes to be expressed in terminally differentiated cells. KDM5b is replaced by other KDM5 family members. (B) In stem cells, KDM5b may be directed to different types of target genes based on the extracellular signaling that the cell receives.
Supplementary Material
Footnotes
Published ahead of print on 30 June 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abdulkader, I., L. Sanchez, J. Cameselle-Teijeiro, F. Gude, J. E. Chavez, R. Lopez-Lopez, J. Forteza, and M. Fraga. 2005. Cell-cycle-associated markers and clinical outcome in human epithelial cancers: a tissue microarray study. Oncol. Rep. 141527-1531. [DOI] [PubMed] [Google Scholar]
- 2.Affolter, M., and K. Basler. 2007. The decapentaplegic morphogen gradient: from pattern formation to growth regulation. Nat. Rev. Genet. 8663-674. [DOI] [PubMed] [Google Scholar]
- 3.Benevolenskaya, E. V., H. L. Murray, P. Branton, R. A. Young, and W. G. Kaelin, Jr. 2005. Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol. Cell 18623-635. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein, B. E., T. S. Mikkelsen, X. Xie, M. Kamal, D. J. Huebert, J. Cuff, B. Fry, A. Meissner, M. Wernig, K. Plath, R. Jaenisch, A. Wagschal, R. Feil, S. L. Schreiber, and E. S. Lander. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125315-326. [DOI] [PubMed] [Google Scholar]
- 5.Besson, A., H. C. Hwang, S. Cicero, S. L. Donovan, M. Gurian-West, D. Johnson, B. E. Clurman, M. A. Dyer, and J. M. Roberts. 2007. Discovery of an oncogenic activity in p27Kip1 that causes stem cell expansion and a multiple tumor phenotype. Genes Dev. 211731-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer, L. A., K. Plath, J. Zeitlinger, T. Brambrink, L. A. Medeiros, T. I. Lee, S. S. Levine, M. Wernig, A. Tajonar, M. K. Ray, G. W. Bell, A. P. Otte, M. Vidal, D. K. Gifford, R. A. Young, and R. Jaenisch. 2006. Polycomb complexes repress developmental regulator in murine embryonic stem cells. Nature 441349-353. [DOI] [PubMed] [Google Scholar]
- 7.Carruthers, S., J. Mason, and N. Papalopulu. 2003. Depletion of the cell-cycle inhibitor p27(Xic1) impairs neuronal differentiation and increases the number of ElrC(+) progenitor cells in Xenopus tropicalis. Mech. Dev. 120607-616. [DOI] [PubMed] [Google Scholar]
- 8.Chambers, I., J. Silva, D. Colby, J. Nichols, B. Nijmeijer, M. Robertson, J. Vrana, K. Jones, L. Grotewold, and A. Smith. 2007. Nanog safeguards pluripotency and mediates germline development. Nature 4501230-1234. [DOI] [PubMed] [Google Scholar]
- 9.Christensen, J., K. Agger, P. A. Cloos, D. Pasini, S. Rose, L. Sennels, J. Rappsilber, K. H. Hansen, A. E. Salcini, and K. Helin. 2007. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell 1281063-1076. [DOI] [PubMed] [Google Scholar]
- 10.Doble, B. W., S. Patel, G. A. Wood, L. K. Kockeritz, and J. R. Woodgett. 2007. Functional redundancy of GSK-3alpha and GSK-3beta in Wnt/beta-catenin signaling shown by using an allelic series of embryonic stem cell lines. Dev. Cell 12957-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ernst, P., J. K. Fisher, W. Avery, S. Wade, D. Foy, and S. J. Korsmeyer. 2004. Definitive hematopoiesis requires the mixed-lineage leukemia gene. Dev. Cell 6437-443. [DOI] [PubMed] [Google Scholar]
- 12.Frankenberg, S., L. Smith, A. Greenfield, and M. Zernicka-Goetz. 2007. Novel gene expression patterns along the proximo-distal axis of the mouse embryo before gastrulation. BMC Dev. Biol. 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grimmler, M., Y. Wang, T. Mund, Z. Cilensek, E. M. Keidel, M. B. Waddell, H. Jakel, M. Kullmann, R. W. Kriwacki, and L. Hengst. 2007. Cdk-inhibitory activity and stability of p27Kip1 are directly regulated by oncogenic tyrosine kinases. Cell 128269-280. [DOI] [PubMed] [Google Scholar]
- 14.Guenther, M. G., S. S. Levine, L. A. Boyer, R. Jaenisch, and R. A. Young. 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 13077-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo, Y., R. Costa, H. Ramsey, T. Starnes, G. Vance, K. Robertson, M. Kelley, R. Reinbold, H. Scholer, and R. Hromas. 2002. The embryonic stem cell transcription factors Oct-4 and FoxD3 interact to regulate endodermal-specific promoter expression. Proc. Natl. Acad. Sci. USA 993663-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutierrez, G. M., E. Kong, and P. W. Hinds. 2005. Master or slave: the complex relationship of RBP2 and pRb. Cancer Cell 7501-502. [DOI] [PubMed] [Google Scholar]
- 17.Hart, A. H., L. Hartley, M. Ibrahim, and L. Robb. 2004. Identification, cloning and expression analysis of the pluripotency promoting Nanog genes in mouse and human. Dev. Dyn. 230187-198. [DOI] [PubMed] [Google Scholar]
- 18.Iwase, S., F. Lan, P. Bayliss, L. de la Torre-Ubieta, M. Huarte, H. H. Qi, J. R. Whetstine, A. Bonni, T. M. Roberts, and Y. Shi. 2007. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 1281077-1088. [DOI] [PubMed] [Google Scholar]
- 19.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 2931074-1080. [DOI] [PubMed] [Google Scholar]
- 20.Jorgensen, H. F., S. Giadrossi, M. Casanova, M. Endoh, H. Koseki, N. Brockdorff, and A. G. Fisher. 2006. Stem cells primed for action: polycomb repressive complexes restrain the expression of lineage-specific regulators in embryonic stem cells. Cell Cycle 51411-1414. [DOI] [PubMed] [Google Scholar]
- 21.Kanda, S., Y. Tamada, A. Yoshidome, I. Hayashi, and T. Nishiyama. 2004. Over-expression of bHLH genes facilitate neural formation of mouse embryonic stem (ES) cells in vitro. Int. J. Dev. Neurosci. 22149-156. [DOI] [PubMed] [Google Scholar]
- 22.Krishnaraju, K., B. Hoffman, and D. A. Liebermann. 2001. Early growth response gene 1 stimulates development of hematopoietic progenitor cells along the macrophage lineage at the expense of the granulocyte and erythroid lineages. Blood 971298-1305. [DOI] [PubMed] [Google Scholar]
- 23.Krivtsov, A. V., and S. A. Armstrong. 2007. MLL translocations, histone modifications and leukaemia stem-cell development. Nat. Rev. Cancer 7823-833. [DOI] [PubMed] [Google Scholar]
- 24.Lanoix, J., A. Mullick, Y. He, R. Bravo, and D. Skup. 1998. Wild-type egr1/Krox24 promotes and dominant-negative mutants inhibit, pluripotent differentiation of p19 embryonal carcinoma cells. Oncogene 172495-2504. [DOI] [PubMed] [Google Scholar]
- 25.Lee, M. G., J. Norman, A. Shilatifard, and R. Shiekhattar. 2007. Physical and functional association of a trimethyl H3K4 demethylase and Ring6a/MBLR, a polycomb-like protein. Cell 128877-887. [DOI] [PubMed] [Google Scholar]
- 26.Leivonen, M., S. Nordling, J. Lundin, K. von Boguslawski, and C. Haglund. 2001. p27 expression correlates with short-term, but not with long-term prognosis in breast cancer. Breast Cancer Res. Treat. 6715-22. [DOI] [PubMed] [Google Scholar]
- 27.Loyer, P., S. Cariou, D. Glaise, M. Bilodeau, G. Baffet, and C. Guguen-Guillouzo. 1996. Growth factor dependence of progression through G1 and S phases of adult rat hepatocytes in vitro. Evidence of a mitogen restriction point in mid-late G1. J. Biol. Chem. 27111484-11492. [DOI] [PubMed] [Google Scholar]
- 28.Lu, P. J., K. Sundquist, D. Baeckstrom, R. Poulsom, A. Hanby, S. Meier-Ewert, T. Jones, M. Mitchell, P. Pitha-Rowe, P. Freemont, and J. Taylor-Papadimitriou. 1999. A novel gene (PLU-1) containing highly conserved putative DNA/chromatin binding motifs is specifically up-regulated in breast cancer. J. Biol. Chem. 27415633-15645. [DOI] [PubMed] [Google Scholar]
- 29.Madsen, B., B. Spencer-Dene, R. Poulsom, D. Hall, P. J. Lu, K. Scott, A. T. Shaw, J. M. Burchell, P. Freemont, and J. Taylor-Papadimitriou. 2002. Characterisation and developmental expression of mouse Plu-1, a homologue of a human nuclear protein (PLU-1) which is specifically up-regulated in breast cancer. Gene Expr. Patterns 2275-282. [DOI] [PubMed] [Google Scholar]
- 30.Mikkelsen, T. S., M. Ku, D. B. Jaffe, B. Issac, E. Lieberman, G. Giannoukos, P. Alvarez, W. Brockman, T. K. Kim, R. P. Koche, W. Lee, E. Mendenhall, A. O'Donovan, A. Presser, C. Russ, X. Xie, A. Meissner, M. Wernig, R. Jaenisch, C. Nusbaum, E. S. Lander, and B. E. Bernstein. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448553-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molofsky, A. V., R. Pardal, T. Iwashita, I. K. Park, M. F. Clarke, and S. J. Morrison. 2003. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature 425962-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moser, M., and G. Campbell. 2005. Generating and interpreting the Brinker gradient in the Drosophila wing. Dev. Biol. 286647-658. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen, L., A. Besson, J. I. Heng, C. Schuurmans, L. Teboul, C. Parras, A. Philpott, J. M. Roberts, and F. Guillemot. 2006. p27kip1 independently promotes neuronal differentiation and migration in the cerebral cortex. Genes Dev. 201511-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niwa, H., J. Miyazaki, and A. G. Smith. 2000. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24372-376. [DOI] [PubMed] [Google Scholar]
- 35.Nowak, K., K. Kerl, D. Fehr, C. Kramps, C. Gessner, K. Killmer, B. Samans, B. Berwanger, H. Christiansen, and W. Lutz. 2006. BMI1 is a target gene of E2F-1 and is strongly expressed in primary neuroblastomas. Nucleic Acids Res. 341745-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park, I. K., S. J. Morrison, and M. F. Clarke. 2004. Bmi1, stem cells, and senescence regulation. J. Clin. Investig. 113175-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park, I. K., D. Qian, M. Kiel, M. W. Becker, M. Pihalja, I. L. Weissman, S. J. Morrison, and M. F. Clarke. 2003. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 423302-305. [DOI] [PubMed] [Google Scholar]
- 38.Pereira, L., F. Yi, and B. J. Merrill. 2006. Repression of Nanog gene transcription by Tcf3 limits embryonic stem cell self-renewal. Mol. Cell. Biol. 267479-7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi, Y., F. Lan, C. Matson, P. Mulligan, J. R. Whetstine, P. A. Cole, R. A. Casero, and Y. Shi. 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119941-953. [DOI] [PubMed] [Google Scholar]
- 40.Shilatifard, A. 2006. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 75243-269. [DOI] [PubMed] [Google Scholar]
- 41.Sims, R. J., III, K. Nishioka, and D. Reinberg. 2003. Histone lysine methylation: a signature for chromatin function. Trends Genet. 19629-639. [DOI] [PubMed] [Google Scholar]
- 42.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 40341-45. [DOI] [PubMed] [Google Scholar]
- 43.Szutorisz, H., C. Canzonetta, A. Georgiou, C. M. Chow, L. Tora, and N. Dillon. 2005. Formation of an active tissue-specific chromatin domain initiated by epigenetic marking at the embryonic stem cell stage. Mol. Cell. Biol. 251804-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan, K., A. L. Shaw, B. Madsen, K. Jensen, J. Taylor-Papadimitriou, and P. S. Freemont. 2003. Human PLU-1 Has transcriptional repression properties and interacts with the developmental transcription factors BF-1 and PAX9. J. Biol. Chem. 27820507-20513. [DOI] [PubMed] [Google Scholar]
- 45.van der Lugt, N. M., M. Alkema, A. Berns, and J. Deschamps. 1996. The polycomb-group homolog Bmi-1 is a regulator of murine Hox gene expression. Mech. Dev. 58153-164. [DOI] [PubMed] [Google Scholar]
- 46.Waters, C. M., D. C. Hancock, and G. I. Evan. 1990. Identification and characterisation of the egr-1 gene product as an inducible, short-lived, nuclear phosphoprotein. Oncogene 5669-674. [PubMed] [Google Scholar]
- 47.Xiang, Y., Z. Zhu, G. Han, X. Ye, B. Xu, Z. Peng, Y. Ma, Y. Yu, H. Lin, A. P. Chen, and C. D. Chen. 2007. JARID1B is a histone H3 lysine 4 demethylase up-regulated in prostate cancer. Proc. Natl. Acad. Sci. USA 10419226-19231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamane, K., K. Tateishi, R. J. Klose, J. Fang, L. A. Fabrizio, H. Erdjument-Bromage, J. Taylor-Papadimitriou, P. Tempst, and Y. Zhang. 2007. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol. Cell 25801-812. [DOI] [PubMed] [Google Scholar]
- 49.Yu, J., V. Baron, D. Mercola, T. Mustelin, and E. D. Adamson. 2007. A network of p73, p53 and Egr1 is required for efficient apoptosis in tumor cells. Cell Death Differ. 14436-446. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.