Abstract
Controlled, transient cytokine production by monocytes depends heavily upon rapid mRNA degradation, conferred by 3′ untranslated region-localized AU-rich elements (AREs) that associate with RNA-binding proteins. The ARE-binding protein AUF1 forms a complex with cap-dependent translation initiation factors and heat shock proteins to attract the mRNA degradation machinery. We refer to this protein assembly as the AUF1- and signal transduction-regulated complex, ASTRC. Rapid degradation of ARE-bearing mRNAs (ARE-mRNAs) requires ubiquitination of AUF1 and its destruction by proteasomes. Activation of monocytes by adhesion to capillary endothelium at sites of tissue damage and subsequent proinflammatory cytokine induction are prominent features of inflammation, and ARE-mRNA stabilization plays a critical role in the induction process. Here, we demonstrate activation-induced subunit rearrangements within ASTRC and identify chaperone Hsp27 as a novel subunit that is itself an ARE-binding protein essential for rapid ARE-mRNA degradation. As Hsp27 has well-characterized roles in protein ubiquitination as well as in adhesion-induced cytoskeletal remodeling and cell motility, its association with ASTRC may provide a sensing mechanism to couple proinflammatory cytokine induction with monocyte adhesion and motility.
Many mRNAs encoding proteins transiently required for inflammatory responses, cell proliferation, and intracellular signaling are labile due to AU-rich elements (AREs) in their 3′ untranslated regions (UTRs) (14, 21, 57). ARE association by ELAV-like (embryonic lethal, abnormal vision) proteins, such as HuR, blocks ARE-mediated mRNA decay (AMD) (31). By contrast, association of proteins such as AUF1, tristetraprolin (TTP), BRF1 (butyrate-responsive factor-1), K-homology splicing regulatory protein (KSRP), ring finger K-homology domain 1 (RKHD1), polymyositis-scleroderma 75-kDa antigen (PM-Scl75), or microRNA miR16 or miR289 with an ARE promotes AMD (6, 8, 12, 18, 24, 34, 43). The phosphorylation state of TTP, BRF1, and AUF1 affects AMD efficiency (3, 37, 51, 56), indicating that signal transduction networks regulate this pathway.
AUF1 has four protein isoforms—p37, p40, p42, and p45—generated by alternative pre-mRNA splicing (50). Based upon extensive biochemical studies of AUF1, we proposed an integrated, three-step model for induction of AMD by AUF1 via assembly of a trans-acting complex that targets the mRNA for degradation (52). The first step is dynamic AUF1 dimer binding to an ARE and formation of an oligomeric AUF1 complex (7, 52). Stabilizing ARE-binding proteins (AUBPs) may compete with AUF1 for binding to the ARE during this step, thus preventing AUF1 oligomerization and subsequent factor recruitment (25). Binding of AUF1 to an ARE then permits the second step involving recruitment of additional trans-acting factors including eukaryotic translation initiation factor eIF4G, poly(A)-binding protein, dual-functional heat shock/AUBPs Hsp/Hsc70 (27), and additional unknown proteins, forming a multisubunit AUF1- and signal transduction-regulated complex (ASTRC) on ARE-bearing mRNAs (ARE-mRNAs). The third step, mRNA degradation, involves two linked catabolic steps—ubiquitin-dependent degradation of AUF1 by proteasomes and mRNA destruction by mRNA degradation enzymes (27, 28). Most observations indicate that 3′-5′ exoribonucleolytic cleavage of the poly(A) tract is the initial catabolic step during AMD (4). Decapping and 5′-3′ and additional 3′-5′ degradation follow (11, 34, 44).
In circulating monocytes, ARE-bearing mRNAs (ARE-mRNAs) encoding proinflammatory cytokines and chemokines are maintained at very low, basal levels. This is due in large part to their rapid degradation. Upon monocyte adhesion to extracellular matrix components at sites of tissue damage, these mRNAs undergo rapid stabilization, which increases their levels 50- to 100-fold within 1 to 2 h (40). In vitro ARE-binding experiments with extracts of nonadherent monocytes showed that they support assembly of RNP complexes containing AUF1. Parallel experiments with extracts of adherent monocytes demonstrated both qualitative and quantitative differences in the assembly of AUF1-RNP complexes, indicative of protein-ARE remodeling. We hypothesized that these RNP remodeling events contribute to stabilization of cytokine ARE-mRNAs in adherent monocytes. In addition, inhibition of a number of signal transduction pathways blocked both adhesion-induced mRNA stabilization and protein-ARE remodeling (40). In subsequent work, we utilized the human promonocyte cell line THP-1. Activation of THP-1 cells by acute treatment with phorbol ester 12-O-tetradecanoylphorbol-13-acetate (TPA) mimics cytokine ARE-mRNA stabilization of adherent monocytes (41), activates protein kinase C, and promotes adhesion to extracellular matrix components (38). In nonactivated THP-1 cells, p40AUF1 is phosphorylated on Ser83 and Ser87, and ARE-mRNAs encoding interleukin-1β and tumor necrosis factor alpha (TNF-α) are unstable. Activation with TPA leads to robust transcript stabilization coincident with dephosphorylation of p40AUF1 on both serines (56). Fluorescence resonance energy transfer (FRET) experiments revealed that binding of nonphosphorylated AUF1 induces transition of an ARE-RNA from a flexible, open conformation to a spatially condensed structure that exhibits restricted backbone flexibility (55). By contrast, p40AUF1 phosphorylated on Ser83 and Ser87 does not induce this structural transition. Thus, the AUF1 phosphorylation state influences local ARE-RNA structure (51). Within the context of the three-step model of AUF1 function noted above, these studies led us to hypothesize that activation of THP-1 cells may induce subunit rearrangements within ASTRC concomitant with cytokine mRNA stabilization.
To better understand the role of ASTRC in control of proinflammatory cytokine AMD in monocytes, we examined AUF1-containing complexes in nonactivated and activated THP-1 cells. We first found that cell activation results in ASTRC subunit reorganization and ARE-mRNA stabilization. Secondly, we identified chaperone Hsp27 as a subunit of ASTRC and found it to possess high-affinity ARE-binding activity. Knockdown of Hsp27 expression led to dramatic stabilization of a cytokine ARE-mRNA. Taken together, these studies indicate that Hsp27 functions as a novel trans-acting modulator of AMD.
MATERIALS AND METHODS
Materials.
THP-1, a human promonocytic leukemia cell line, was provided by Charles McCall (Wake Forest University School of Medicine). K562, a human chronic myelogenous leukemia cell line, was from the American Type Culture Collection. Anti-eIF4G-I was a kind gift from Nahum Sonenberg. Hsp27 and Hsp70 antibodies were from Stressgen (SPA-803 and SPA-812). Anti-Hsc70 was from Santa Cruz (sc-7298). Horseradish peroxidase-conjugated secondary antibodies were from Promega Corporation (Madison, WI). All primer oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA).
Cell culture.
THP-1 and K562 cell lines were maintained in RPMI 1640 medium (Cellgro Mediatech, Herndon, VA) supplemented with 10% defined, endotoxin-free, fetal bovine serum (HyClone, Logan, UT), and 1× penicillin-streptomycin-glutamine (Gibco) at 37°C in 5% CO2. In some experiments noted below, cells were cultured in the absence of antibiotics.
THP-1 cell fractionation.
The pellet fraction from centrifugation of cytoplasm at 130,000 × g (P130) was prepared from THP-1 control (treated with dimethyl sulfoxide [DMSO] vehicle) or TPA-treated (10 nM for 1 h) cells by lysis in buffer A (10 mM Tris [pH 7.4], 1 mM potassium acetate, 1.5 mM magnesium acetate, 2 mM dithiothreitol and protease inhibitors [10 μg/ml leupeptin, 10 μg/ml pepstatin A, 1 mM phenylmethylsulfonyl fluoride]) as described previously (2).
Immunopurifications.
Affinity-purified AUF1 antibody was isolated from crude serum with immobilized His6-p37AUF1. Eighty million cell equivalents of P130 fraction was treated with RNase A (Qiagen, Hilden, Germany) at a concentration of 4 mg/ml at 30°C for 15 min, and 4 μg of affinity-purified antibody was used for immunoprecipitation with a Catch and Release Immunoprecipitation System (Upstate, Charlottesville, VA). Two purifications were combined, fractionated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and stained with Sypro Ruby (Molecular Probes, Eugene, OR). Alternatively, proteins were transferred to nitrocellulose membranes and detected by Western blotting with chemiluminescence reagent (Pierce, Rockford, IL). Protein band intensities from films were quantified with the Kodak EDAS 120 gel documentation system and software (Eastman Kodak Co.).
Protein identification by mass spectrometry.
A ∼26-kDa band was excised from a Sypro Ruby-stained gel containing immunoprecipitated proteins. The gel slice was subject to in-gel tryptic digestion, and peptide fragments in the mass range of 810 to 2,000 Da were analyzed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) with a PerkinElmer Biosystems DE-PRO mass spectrometer (PerSeptive Biosystems, Framingham, MA) linked to a Voyager-DE PRO work station as described previously (56). Peptide mass/charge (m/z) ratios were used for their identification utilizing MS-Fit and MS-Digest programs (P. R. Baker and K. R. Clauser, Mass Spectrometry Facility, University of California, San Francisco, CA [http://prospector.ucsf.edu]).
Plasmid constructs, transfections, and spectrum deconvolution for live-cell FRET analyses.
The cDNAs encoding enhanced cyan fluorescent protein (ECFP) and enhanced yellow fluorescent protein (EYFP), each carrying an A206K mutation, were obtained from plasmids pcDNA3-FL-IFN-γR2/[A206K]EYFP and pcDNA3-FL-IFN-γR2/[A206K]EYFP (23) (where IFN-γR2 is gamma interferon receptor chain 2). The A206K modification prevents autodimerization of the fluorescent proteins (58). For the experiments described in this report, the ECFP and EYFP cDNAs were fused to the N terminus of Hsp27 and to the C terminus of p37AUF1. To fuse ECFP and EYFP to Hsp27, the coding regions of ECFP and EYFP were amplified by PCR with the following primers: 5′-GCACGGTACCGCCACCATGGTGAGCAAGGGCGAGG-3′ (C/YFP forward) and 5′-GCACGGATCCGGAAACTTGTACAGCTCGTCCATGCC-3′ (reverse). The resulting PCR products contain a KpnI restriction site (underlined) and a BamHI site (underlined). PCR fragments were digested with KpnI and BamHI and ligated into expression vector pEF3 (22, 23), also digested with KpnI and BamHI. A SacI-truncated human EF1A promoter drives transgene expression in pEF3. The coding region of Hsp27 was amplified by PCR with the following primers: 5′-GCACGGATCCCACCGAGCGCCGCGTCCC-3′ (forward) and 5′-GCACGAATTCTTACTTGGCGGCAGTCTCATC-3′ (reverse). The forward primer contains a BamHI site (underlined), and the reverse primer contains an EcoRI site (underlined). Amplified Hsp27 cDNA was digested with BamHI and EcoRI and ligated into plasmids pEF3-ECFP and pEF3-EYFP digested with the same enzymes.
To fuse ECFP and EYFP to p37AUF1, the coding regions of ECFP and EYFP were amplified by PCR with the following primers: 5′-GCACGGATCCGTGAGCAAGGGCGAGGAG-3′ (forward) and 5′-GCACGATATCTTACTTGTACAGCTCGTCCATG-3′ (C/YFP reverse). The resulting PCR products contain a BamHI site (underlined) and an EcoRV site (underlined). PCR fragments were digested with BamHI and EcoRV and ligated into expression vector pEF3, also digested with BamHI and EcoRV. The coding region of p37AUF1 was amplified with the following primers: 5′-GCACGGTACCGCCACCATGTCGGAGGAGCAGTTCGG-3′ (forward) and 5′-GCACGGATCCGTATGGTTTGTAGCTATTTTGATG-3′ (reverse). The forward primer contains a KpnI restriction site (underlined) at the 5′ end, and the reverse primer contains a BamHI site at the 3′ end. The amplified p37AUF1 was digested with KpnI and BamHI and ligated into the similarly digested pEF3-ECFP/EYFP vector.
To synthesize pEF3-ECFP (i.e., ECFP alone, not as a fusion protein), the coding region of ECFP was amplified with the C/YFP forward and C/YFP reverse primers listed above. Amplified ECFP cDNA was digested with KpnI and EcoRV and ligated into the similarly digested pEF3 vector.
Hsp90 cDNA was amplified by reverse transcription-PCR (RT-PCR) from HeLa cell total RNA and modified for placement of EYFP on its C terminus. The following primers were used to amplify Hsp90: 5′-GCACGTTTAAACATGCCTGAGGAAGTGCACC-3′ (forward) and 5′-GCACACTAGTATCGACTTCTTCCATGCGAG-3′ (reverse). The forward primer contains a PmeI site (underlined) and the reverse primer contains a SpeI site (underlined). Plasmid pEF3/Hsp27-EYFP was digested with Acc65I, blunted, and then digested with SpeI to release the Hsp27 cDNA. Amplified Hsp90 cDNA was digested with PmeI and SpeI and then ligated with the digested vector, yielding plasmid pEF3/Hsp90-EYFP.
One million THP-1 cells were transiently cotransfected with the ECFP and EYFP plasmid pairs with Effectene reagent (Qiagen) according to the manufacturer's protocol. Twenty-four hours after transfection, cells were harvested and prepared as described below for live-cell FRET assays. Twenty million K562 cells were electroporated with 10 μg each of the ECFP and EYFP plasmid pairs in RPMI 1640 medium supplemented with 10% fetal calf serum in the absence of antibiotics. After 48 to 72 h, whole-cell lysates of K562 cells were prepared in 1× SDS loading buffer for Western blot analyses of fusion protein expression.
Transiently transfected THP-1 cells were washed with phosphate-buffered saline and water mounted onto coverslips at 24 h posttransfection. Protein-protein interactions between p37AUF1-p37AUF1 and p37AUF1-Hsp27 were determined by confocal fluorescence spectroscopy (22, 23). To demonstrate EYFP/ECFP fluorescence and FRET, a custom-built confocal microscope adapted to include a monochrometer interfaced with a cooled charge-coupled-device camera was used so that both confocal fluorescence images and fluorescence emission spectra could be obtained from optical sections. EYFP was directly excited with a 488-nm argon laser rather than a 514-nm laser to permit separation of the laser spectrum from the EYFP emission spectrum (i.e., 527 nm) and to minimize photobleaching of EYFP. ECFP was excited with a 442-nm helium-cadmium laser. Images of ECFP emission were obtained with a 480- ± 10-nm band-pass filter and Olympus FluoView software. EYFP emission images were obtained with a 520- ± 10-nm band-pass filter whether excited by 442 or 488 nm light so that either direct EYFP emission or EYFP emission indicative of FRET, respectively, could be measured.
Accurate calculation of FRET efficiency depends on reliable estimation of the amount of donor and acceptor fluorescence present in a cell. Total fluorescence emission is the combined contributions of ECFP, EYFP, and endogenous cellular fluorescent components (e.g., nicotinamide and riboflavin derivates). Thus, a recently developed algorithm for spectral deconvolution was employed to separate an emission spectrum into contributions by these three components (22) for subtraction of endogenous fluorescence. This procedure permits a more accurate estimate of donor fluorescence in the presence of acceptor (FDA) and acceptor fluorescence in the presence of donor (FAD). The efficiency of energy transfer (EFRET) was then calculated from a deconvoluted spectrum by the following equation (22):
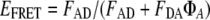 |
(1) |
where ΦA is the quantum yield of acceptor fluorescence emission; ΦA is 0.61 for EYFP. The distance between the two proteins (R) was estimated by the following equation (22):
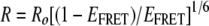 |
(2) |
where Ro is the Förster distance, defined as the radius between freely rotating donor and acceptor molecules that yields an EFRET value of 0.5; Ro is 49.2 Å for the ECFP-EYFP pair. Spectra were obtained with Andor charge-coupled device control software by analyzing a specific area within the cytoplasm chosen with emission images.
FRET efficiencies vary at different points within a cell and from cell to cell. Some of the cell-to-cell variation can be attributed to different relative expression levels of ECFP- and EYFP-tagged fusion proteins in each cell within a transfected population. Thus, each cell has a different acceptor/donor fluorophore ratio. Consequently, FRET variability can be attributed to underrepresentation of acceptor fluorescence protein in some cells, resulting in lower EFRET. As a result of this observation, deconvoluted EYFP fluorescence intensities resulting from 488-nm excitation of at least 40 cells were plotted versus their respective FRET efficiencies obtained upon 442-nm excitation. Data were fit to a hyperbola, and only those cells that exhibited EYFP fluorescence higher than 10,000 fluorescence intensity units (at which EFRET was asymptotic and no longer increased with increasing EYFP fluorescence) were analyzed statistically for comparison among plasmid pairs by a Student's t test. P values of <0.05 were considered significantly different.
To verify that FRET signatures corresponded to bona fide protein-protein interactions, control photobleaching experiments were performed. Specific photobleaching of the acceptor (EYFP) was performed by continuously scanning a region of a fluorescent cell with 514-nm light set at the highest intensity to photobleach a minimum of ∼85% of the EYFP fluorescence. Spectra were obtained before and after photobleaching of the acceptor EYFP. The resulting spectra were deconvoluted, and fluorescence intensities of the donor were obtained from the deconvoluted spectra before and after photobleaching of the acceptor. For both p37AUF1-p37AUF1 and p37AUF1-Hsp27 protein pairs, three independent cells were analyzed by this method.
Preparation of recombinant Hsp27.
PCR was utilized with plasmid pCMVtag2B-FLAG-Hsp27 (29) to prepare Hsp27 devoid of the FLAG tag. This fragment was subcloned into pBAD/HisB (Invitrogen, Carlsbad, CA) to generate plasmid pBAD/HisB-Hsp27. Recombinant His6-Hsp27 was purified from Escherichia coli TOP10 cells transformed with pBAD/HisB-Hsp27 as described previously (54). When necessary, cleavage of the His6 tag from Hsp27 was achieved with an Enterokinase Cleavage Capture Kit (Novagen-EMD Biosciences).
RNA oligoribonucleotides.
RNAs containing the 38-nucleotide (nt) core ARE from TNF-α mRNA or a fragment of similar size from the rabbit β-globin (Rβ) coding region were synthesized by Dharmacon (Lafayette, CO) as described previously (53). Fl-TNF-α ARE and Fl-Rβ substrates contain 5′-fluorescein (Fl) and were tested and quantified spectrophotometrically as described previously (54). For electrophoretic mobility shift assays (EMSAs), TNF-α ARE or Rβ RNAs were labeled at their 5′ termini with [γ-32P]ATP and T4 polynucleotide kinase (52).
EMSAs.
EMSAs with His6-Hsp27 and 5′-32P-TNF-α or -Rβ RNA substrates were performed as described previously (52). Reaction products were visualized and analyzed with a Typhoon 9410 PhosphorImager (Molecular Dynamics, Amersham Biosciences).
Analysis of RNA-protein binding by fluorescence polarization.
Equilibrium His6-Hsp27-RNA binding activity was assessed by monitoring interactions between a constant amount of fluorescein-conjugated RNA (0.15 nM) and a titration of His6-Hsp27 protein by fluorescence polarization with a Beacon 2000 Variable Temperature Fluorescence Polarization System (Panvera, Madison, WI) as described previously (52, 54). Where indicated on the figures, reaction mixtures included 5 mM MgCl2; all mixtures without Mg2+ contained 0.5 mM EDTA. Fluorescence polarimetry data were resolved by a variant of the Hill equation (42):
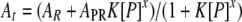 |
(3) |
where [P] is the Hsp27 protein concentration. Total measured anisotropy (At) and intrinsic anisotropy of free RNA (AR) were determined experimentally. The Hill coefficient (x), intrinsic anisotropy of the protein-associated RNA (APR), and equilibrium constant (K) were solved by nonlinear least squares regression with the PRISM program, version 3.03 (GraphPad, San Diego, CA). Data are presented as means ± standard deviations. Student's t test was performed, and a P value of <0.05 was considered statistically significant (GraphPad InStat, version 3.05, San Diego, CA).
Plasmids for shRNA expression and cell transfections.
pSilencer 2.1-U6-hygro vector (Ambion, Austin TX) was modified by replacing the U6 promoter with a U6 promoter/tetracycline (Tet)-operator combination (32). Oligonucleotides encoding short hairpin RNAs (shRNAs) were annealed, phosphorylated with T4 polynucleotide kinase, and ligated into the BamHI-HindIII sites, creating pSilencer/U6/tetO/shHsp27 (5′-GATCCCGCTAGCCACGCAGTCCAACTTCAAGAGAGTTGGACTGCGTGGCTAGCTTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAAGCTAGCCACGCAGTCCAACTCTCTTGAAGTTGGACTGCGTGGCTAGCGG-3′), pSilencer/U6/tetO/shAUF1 (5′-GATCCCGTTGTAGACTGCACTCTGATTCAAGAGATCAGAGTGCAGTCTACAACTTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAAGTTGTAGACTGCACTCTGATCTCTTGAATCAGAGTGCAGTCTACAACGG-3′), or a negative control not found in the human genome which was supplied with the Ambion kit. Vectors expressing shRNAs were linearized with XmnI and transfected into THP-1 cells with Effectene reagent (Qiagen, Hilden, Germany). Stably transfected cells were selected with 250 units/ml hygromycin B (Calbiochem). To assess knockdown, proteins were visualized and quantified as described above, with α-tubulin as a loading control.
Determination of TNF-α mRNA half-life.
THP-1 cells were treated with vehicle (DMSO; J. T. Baker) or 10 nM TPA (Sigma) for 1 h, and then actinomycin D (Calbiochem, La Jolla, CA) at a final concentration of 5 μg/ml was added to inhibit transcription. Time courses were limited to 3 h to avoid affecting cellular mRNA decay pathways by actinomycin D-enhanced apoptosis (46). Cells were harvested at each time point, lysed with QiaShredders (Qiagen, MD), and purified with an RNeasy kit (Qiagen, MD). Molecular beacons for β-actin [5′-6-FAM-d(CGCGATCATGGAGTCCTGTGGCATCCACGAAGATCGCG)-DABCYL-3′, where FAM is carboxyfluorescein and DABCYL is 4-(4′-dimethylaminophenylazo)benzoic acid] and TNF-α [5′-QUASAR 670-d(CGCGATCACTCCCAGGTCCTCTTCAAGGGCGATCGCG)-BHQ-2-3′, where BHQ is Black Hole quencher] and primers (for β-actin, 5′-TTGGCAATGAGCGGTTCC-3′ and 5′-AGCACTGTGTTGGCGTAC-3′; for TNF-α, 5′-ATGGCGTGGAGCTGAGAG-3′ and 5′-GATGCGGCTGATGGTGTG-3′) were designed with Premier Biosoft Beacon Designer Software (Stratagene, Cedar Creek, TX) purchased from Biosearch Technologies (Novato, CA). Melting temperatures were determined as described previously (47). Total RNA (1.25 μg) was reverse transcribed with an Access RT-PCR kit (Promega, Madison WI) and primers specific to β-actin and TNF-α. Real-time quantitative PCR (qPCR) was performed with a Stratagene MX3005P qPCR System with a Stratagene Brilliant qPCR MasterMix kit. Reaction mixtures were assembled in triplicate with 0.5 mM primer, 100 ng of molecular beacon, and 0.25 ng of the RNA equivalent of cDNA for β-actin or 25 ng of the RNA equivalent of cDNA for TNF-α. Relative mRNA levels were calculated from a standard curve. TNF-α levels were normalized with β-actin and plotted as a percentage of the time zero value. Data were analyzed by nonlinear regression, and half-life was calculated from the first-order decay constant (k) obtained with PRISM software, version 3.03 (GraphPad, San Diego, CA). Standard error about the regression solution was calculated by the software using n − 2 degrees of freedom and is linear about k (and therefore hyperbolic about the mRNA half-life, ln2/k). Suitability of each regression solution was evaluated with the runs test for random distribution of residuals (P < 0.05 cutoff). Assignment of explicit half-lives to very stable mRNAs is rarely informative, since the hyperbolic nature of half-life relative to the first-order mRNA decay constant inflates errors in the half-life value when k approaches zero. Accordingly, half-life values are given as >5 h for mRNAs where k is at least two standard errors below 0.139 h−1 (equal to ln2/5 h). Comparisons between mRNA decay constants were performed with an unpaired two-tailed t test, with differences yielding a P value of <0.05 considered significant.
Reporter mRNA half-life determinations.
Plasmid pTRE (Clontech) expressing the 1.7-kb Rβ gene, pTRE/Rβ-wt (where wt is wild type), was used as a stable control mRNA for reporter assays. The 38-bp core ARE from the human TNF-α gene (51) was inserted into the 3′ UTR of the Rβ gene at the unique BglII site to create plasmid pTRE/Rβ-ARE. pTRE/Rβ-wt or pTRE/Rβ-ARE reporter plasmids, pTet-Off (encoding Tet-responsive transcriptional activator tTA), and pEGFP-C2 (encoding internal control mRNA) were cotransfected into THP-1 cell lines with Effectene reagent (Qiagen). Two days posttransfection, doxycycline (Sigma) was added to culture medium to a final concentration of 2 μg/ml. Cells were harvested at each time point and lysed with Qiagen QiaShredder cartridges, and RNA was purified with a Qiagen RNeasy kit. Multiplex reactions were assembled with the SuperScriptIII Platinum One-Step qRT-PCR kit (Invitrogen, Carlsbad, CA) with 15 picomoles of each primer (for Rβ, 5′-GTGAACTGCACTGTGACAAGC-3′ and 5′-ATGATGAGACAGCACAATAACCAG-3′; for EGFP, 5′-GCGACACCCTGGTGAACC-3′ and 5′-GATGTTGTGGCGGATCTTGAAG-3′), 5 picomoles of each probe [for Rβ, 5′-(56-FAM)-CGTTGCCCAGGAGCCTGAAGTTCTCA(3BHQ_1)-3′; for EGFP, 5′-(5Cal610)-CACCTTGATGCCGTTCTTCTGCTTGTCG-(3BHQ_2)-3′], and 1 μg of total RNA. Reactions were run with the Stratagene MX3005P thermocycler. Relative mRNA levels were calculated based upon standard curves. Reporter mRNA levels were normalized with EGFP mRNA and plotted as a percentage of the levels at time zero. Data were analyzed as described above.
RESULTS
Hsp27 is an ASTRC-associated protein.
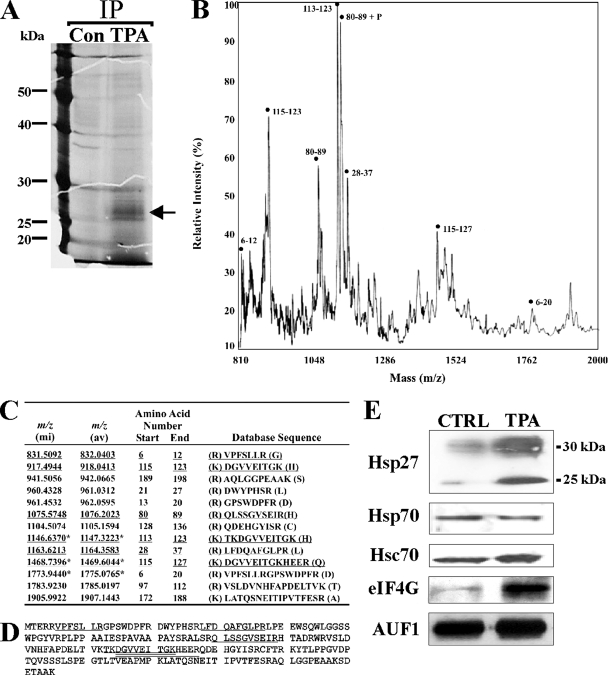
The balance of stabilizing and destabilizing trans-acting factors within ASTRC, their posttranslational modifications, and overall subunit composition and stoichiometry might collectively dictate the rate of AMD in response to extracellular signals (56). A P130 fraction of cytoplasmic extracts contains ribosome/polyribosome-associated mRNAs, mRNA degradation enzymes, and AUF1 (35, 59). The P130 fraction from activated versus nonactivated control THP-1 cells (Fig. 1A) was thus used for immunopurification of AUF1-containing complexes with affinity-purified anti-AUF1 immunoglobulin G. Fractionation by SDS-PAGE and staining revealed an additional 26-kDa polypeptide in complexes from activated cells compared to control cells (Fig. 1A). MALDI-TOF analysis of this polypeptide (Fig. 1B) indicated the presence of several fragments with masses predicted from an in silico trypsin digest of Hsp27 within the mass range examined (Fig. 1C, underlined). Six of the predicted 13 trypsin fragments in the 810 to 2,000 m/z range were resolved, representing 25% of the Hsp27 polypeptide (Fig. 1D). Western blot analysis confirmed the 26-kDa polypeptide to be Hsp27 (Fig. 1E, top) (see below).
FIG. 1.
Hsp27 is an ASTRC-associated protein. (A) Cytoplasmic P130 fractions from nonactivated (Con) and activated (TPA) THP-1 cells were immunoprecipitated with anti-AUF1. Proteins were resolved by SDS-PAGE and detected with Sypro Ruby. The arrow denotes the band from the TPA sample excised and analyzed by MALDI-TOF mass spectrometry. (B) MALDI-TOF analysis of the 26-kDa polypeptide. (C) Predicted tryptic peptides for Hsp27. One-letter codes for amino acids are shown, and letters in parentheses are amino acids at the trypsin cleavage site. An asterisk denotes a fragment containing a tryptic site not cleaved by trypsin. Underlined peptides were detected experimentally in panel B, where peptides with mass/charge ratios (m/z) corresponding to Hsp27 are indicated by amino acid numbers within the Hsp27 sequence. mi, monoisotopic mass; av, average mass. Monoisotopic mass is calculated with the lowest common isotope for each element (e.g., 12C, 1H, 14N, 16O, 32S, and 31P). Average mass is calculated with isotopes for each element with abundances reflecting their normal proportion in the biosphere (http://prospector.ucsf.edu). (D) Amino acid sequence of Hsp27. Underlined residues represent peptides from panels B and C that were resolved by MALDI-TOF mass spectrometry. Some regions were contained in up to three fragments. (E) Western blot analyses of ASTRC subunits. AUF1-associated proteins were immunopurified from equal cell equivalents of cytoplasmic P130 fraction from untreated (CTRL) and activated (TPA) (10 nM TPA for 1 h) THP-1 cells. The indicated proteins were detected.
Analyses of ASTRC subunits.
Western blot analyses of immunopurified ASTRC from control and activated THP-1 cells revealed Hsp27 and, as expected, Hsp70, Hsc70, eIF4G, and AUF1 (Fig. 1E). Additionally, Hsp27 antibody was immunoreactive with several polypeptides ranging from 26 to 33 kDa, and their association with ASTRC increased upon activation (Fig. 1E, top). With the AUF1 signal for normalization, the 26-kDa polypeptide increased ∼6.5-fold upon activation, and polypeptides in the 29- to 33-kDa range increased ∼2.5-fold. (MALDI-TOF mass spectrometry was performed with the 26-kDa polypeptide) (Fig. 1A). We speculate that the higher-molecular-weight polypeptides may be posttranslationally modified Hsp27. Hsp27 can be phosphorylated on Ser15, Ser78, and/or Ser82 (10, 26). Indeed, MALDI-TOF analysis identified a fragment with an m/z consistent with phospho-Ser82 (Fig. 1B). However, detailed analysis of Hsp27 phosphorylation is beyond the scope of this work. Activation also increased association of eIF4G with ASTRC by approximately fivefold, but the levels of Hsp70, Hsc70, and AUF1 varied less than 50% following activation from values for control cells (Fig. 1E). These results resemble the increases in eIF4G association with ASTRC that occurs upon heat shock-induced ARE-mRNA stabilization (27). In contrast to heat shock, however, activation of THP-1 cells did not significantly alter Hsp70 association with ASTRC. We conclude that Hsp27 resides in one or more complexes with AUF1, Hsc/Hsp70, and eIF4G in the cytoplasm of THP-1 cells. These results also suggest that ASTRC subunit levels are dynamic in response to specific cellular stimuli that alter mRNA degradation dynamics.
Analyses of AUF1-AUF1 and AUF1-Hsp27 interactions by live-cell FRET.
To complement the biochemical approach employed to identify association of Hsp27 with ASTRC, we utilized a combination of confocal fluorescence spectroscopy and FRET to examine protein-protein interactions in live cells expressing p37AUF1 and Hsp27 fused to fluorescent proteins. FRET is a noninvasive, spectroscopic technique that allows real-time analysis of protein-protein interactions in live cells. FRET is a nonquantum transfer of energy from a fluorescent donor protein to a fluorescent acceptor protein, the efficiency of which is directly related to the intermolecular distance between the two proteins. As p37AUF1-p37AUF1 interactions are known to occur in vitro (7) and in cells (5, 27), this interaction pair was examined in live cells prior to experiments with the p37AUF1-Hsp27 interaction pair.
Plasmids expressing full-length p37AUF1 fused at its N or C terminus with ECFP (the donor) or EYFP (the acceptor) and Hsp27 fused at its N or C terminus with ECFP or EYFP were used for experiments. Unlike endogenous p37AUF1, which is localized in the nucleus and cytoplasm (59), p37AUF1 with N-terminal ECFP or EYFP was restricted to the nucleus (data not shown). By contrast, p37AUF1 with C-terminal ECFP or EYFP (p37AUF1-ECFP and p37AUF1-EYFP, respectively) localized in the nucleus and cytoplasm (see below). Thus, FRET experiments were performed only with p37AUF1-ECFP and p37AUF1-EYFP. Western blot analyses of lysates from transfected cells verified that all fluorescently tagged proteins had the expected apparent molecular weights (see Fig. S1 in the supplemental material).
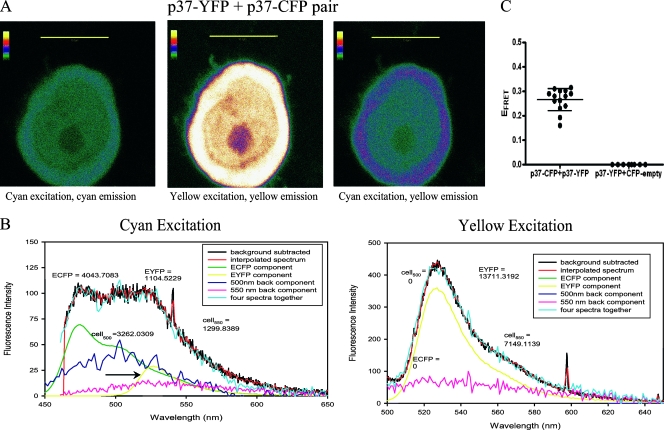
To examine interactions between p37AUF1-ECFP and p37AUF1-EYFP, plasmids encoding these proteins were transiently cotransfected into THP-1 cells. Both p37AUF1-ECFP (Fig. 2A, left panel) and p37AUF1-EYFP (Fig. 2A, middle panel) were located in the nucleus and cytoplasm, just as endogenous p37AUF1 is (5, 36, 59). Excitation of ECFP at 442 nm also led to emission of EYFP at 527 nm (Fig. 2A, right panel). EYFP emission, observed primarily in the cytoplasm, is indicative of a FRET signature and implied p37AUF1-ECFP-p37AUF1-EYFP interactions.
FIG. 2.
Analysis of p37AUF1-p37AUF1 interactions in live cells by FRET. (A) Confocal fluorescence imaging of THP-1 cells coexpressing p37AUF1-ECFP and p37 AUF1-EYFP. Emission of p37AUF1-ECFP upon excitation at 442 nm (left), emission of p37AUF1-EYFP upon excitation at 488 nm (middle), and emission of p37AUF1-EYFP upon excitation of p37AUF1-ECFP at 442 nm (right), indicative of a FRET signature. Images are shown as thermal gradients, where white indicates the strongest signal. Scale bar, 10 μm. (B) Deconvolutions of fluorescence emission spectra of the cell shown in panel A. The cell was irradiated with 442 nm light, and the spectrum was deconvoluted into its major components (left). After the raw spectrum (black line) was transformed so that it possessed even wavelength intervals (red line), it was separated into the following components: ECFP (i.e., p37AUF1-ECFP expression, green line), EYFP (FRET signature from p37AUF1-EYFP, yellow line and arrow), nicotinamide background at 500 nm (navy blue line), and riboflavin background at 550 nm (pink line). Addition of the four components (light blue line) closely approximated the observed spectrum, as it should. For this cell, EFRET = 0.31, with a calculated distance of 56 Å between the two fluorescent-tagged p37AUF1 proteins. At right is shown the deconvoluted spectrum of the same cell upon direct EYFP excitation at 488 nm. The yellow line represents p37AUF1-EYFP expression. For both panels and all deconvolutions shown in subsequent figures, fluorescence units are listed for ECFP, EYFP, and the cell at 550 nm (cell550); these were determined by integration of the curves at the respective emission wavelength maxima ± 10 nm (the band-pass width of the filters). (C) Statistical analysis of p37AUF1-EYFP/p37AUF1-ECFP interactions and the control ECFP/p37AUF1-EYFP interaction. Scatter plots of deconvoluted spectra of cells coexpressing p37AUF1-EYFP with p37AUF1-ECFP or ECFP with p37AUF1-EYFP were derived from FRET analyses of 42 and 8 cells, respectively. Cells that exhibited EYFP fluorescence of 10,000 units or higher (n = 14 and 8, respectively) (see Materials and Methods) were used in subsequent statistical analyses. Each point represents EFRET calculated from one deconvoluted spectrum. P < 0.0001 for this analysis.
To accurately quantify FRET efficiencies for comparisons between protein pairs, we employed a twofold strategy (described in detail in Materials and Methods). First, spectral scans of a selected region of cytoplasm were performed from 450 to 650 nm, and fluorescence data were deconvoluted to determine the contributions of ECFP and EYFP to total fluorescence; these corrected fluorescence values were used for calculation of FRET efficiency (EFRET) by equation 1 (see Materials and Methods). Second, EFRET was determined for multiple cells within a transfected population to permit statistical comparisons of differences between two protein pairs (interactions of p37AUF1-ECFP and p37AUF1-EYFP versus interactions of control ECFP and p37AUF1-EYFP). Figure 2B shows a spectral deconvolution analysis of a section of cytoplasm in the THP-1 cell shown in Fig. 2A. Excitation of ECFP at 442 nm produced a peak at 475 nm indicative of p37AUF1-ECFP expression (Fig. 2B, left panel); excitation of EYFP at 488 nm produced a peak at 527 nm indicative of p37AUF1-EYFP expression (Fig. 2B, right panel). These data indicate that p37AUF1-ECFP and p37AUF1-EYFP were indeed coexpressed in the cell. Excitation of ECFP at 442 nm in this cell also produced a peak of yellow emission at 527 nm, consistent with a FRET signature (Fig. 2B, left panel). EFRET was calculated from deconvoluted spectra to be 0.31 for this cell. Analyses of multiple cells provided an average EFRET of 0.27 ± 0.05 for the transfected cell population (Fig. 2C) with a mean distance of 58 Å between the proteins (calculated by equation 2) (see Materials and Methods).
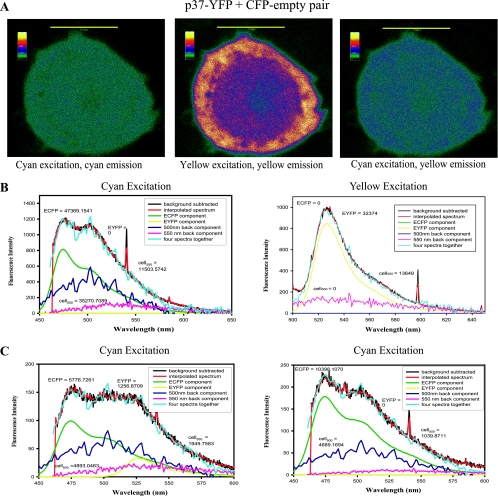
To confirm bona fide p37AUF1-p37AUF1 interactions, two control experiments were performed. First, EFRET was determined for cells cotransfected with plasmids expressing ECFP alone (i.e., not fused to p37AUF1) and p37AUF1-EYFP. Second, photobleaching of p37AUF1-EYFP was performed to examine the effect on fluorescence intensity of p37AUF1-ECFP. For a true protein-protein interaction, photobleaching p37AUF1-EYFP should increase p37AUF1-ECFP fluorescence as photobleached p37AUF1-EYFP is less capable of acting as an energy acceptor. THP-1 cells were cotransfected with plasmids expressing ECFP (not fused to p37AUF1) and p37AUF1-EYFP. Fluorescent images were obtained, and spectral scans were performed. Excitation at 442 nm revealed ECFP expression (Fig. 3B, left panel) and homogenous distribution of ECFP across the cell (Fig. 3A, left panel). Likewise, excitation at 488 nm revealed p37AUF1-EYFP expression (Fig. 3B, right panel) and both nuclear and cytoplasmic p37AUF1-EYFP localization (Fig. 3A, middle panel). However, excitation of ECFP at 442 nm did not produce significant yellow fluorescence at 527 nm (Fig. 3A, compare right panel to middle panel); a deconvoluted spectrum had no detectable fluorescence component originating from EYFP (Fig. 3B, left panel). Thus, EFRET was essentially zero. Analyses of deconvoluted data from cells coexpressing p37AUF1-ECFP and p37AUF1-EYFP (Fig. 2) and cells coexpressing ECFP and p37AUF1-EYFP (Fig. 3) revealed a highly significant difference in mean EFRET between the two populations (Fig. 2C) (P < 0.0001). For a second control, selective photobleaching of p37AUF1-EYFP was performed in cells coexpressing p37AUF1-ECFP. Fluorescence spectra from cytoplasmic regions that exhibited FRET were obtained prior to and after photobleaching. The photobleached region had higher ECFP fluorescence than it did before photobleaching (∼10,000 versus 5,776 fluorescence units, respectively) (Fig. 3C, compare right and left panels), demonstrating that EYFP emission from 442-nm (ECFP) excitation arose from FRET and not from direct excitation of EYFP. Three independent cells showed similar results. Taken together, the data in Fig. 2 and 3 indicate that p37AUF1-p37AUF1 interactions occur in live THP-1 cells. We should note, however, that since p37AUF1-p37AUF1 interactions occur both in solution and when bound to an ARE, the FRET experiments cannot distinguish between these two states. Indeed, it is highly likely that within the cell, p37AUF1-p37AUF1 dimers are present in RNA-bound and unbound populations.
FIG. 3.
Control FRET experiments for p37AUF1-p37AUF1 interactions. (A) THP-1 cells were cotransfected with plasmids expressing ECFP alone (not linked to p37AUF1) and p37AUF1-EYFP. Emission of ECFP upon excitation at 442 nm (left), emission of p37AUF1-EYFP upon excitation at 488 nm (middle), and emission of p37AUF1-EYFP upon excitation of ECFP at 442 nm (right), indicative of no FRET signature. Images are shown as thermal gradients. Scale bar, 10 μm. (B) Deconvolutions of fluorescence emission spectra from the cell shown in panel A. The cell was irradiated with 442 nm light, and the spectrum was deconvoluted into its major components (left) as described in the legend of Fig. 2B. For this cell, EFRET was not detectable. Figure 2C contains a scatter plot analysis of eight control cells analyzed. At right is shown the deconvoluted spectrum of the same cell upon direct EYFP excitation at 488 nm. The yellow line represents p37AUF1-EYFP expression. (C) Photobleaching control. Deconvolution analyses of a THP-1 cell coexpressing p37AUF1-ECFP and p37AUF1-EYFP showed a value of 5,779 fluorescence units for ECFP (left panel) and an increase to 10,398 fluorescence units after photobleaching of EYFP (right panel), indicative of a true FRET signature. Three cells were analyzed with comparable results. cell550, fluorescence of the cell at 550 nm.
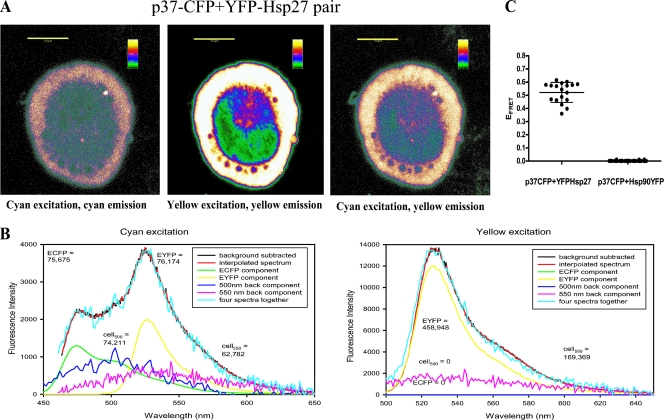
Similar experiments were performed to identify p37AUF1-Hsp27 interactions in live THP-1 cells. Cells were cotransfected with plasmids encoding p37AUF1-ECFP and EYFP-Hsp27. Coexpression of p37AUF1-ECFP (Fig. 4A, left panel) and EYFP-Hsp27 (Fig. 4A, middle panel) produced a FRET signature (Fig. 4A, right panel) indicative of interactions between these proteins. Spectral scans from 450 to 650 nm of a selected region of cytoplasm in multiple cells and data deconvolution permitted determinations of the contributions of ECFP and EYFP to total fluorescence and calculations of EFRET as described above for AUF1-AUF1 interactions. For example, excitation of ECFP at 442 nm produced a peak at 475 nm indicative of p37AUF1-ECFP expression (Fig. 4B, left panel); excitation of EYFP at 488 nm produced a peak at 527 nm, indicative of EYFP-Hsp27 expression (Fig. 4B, right panel). Excitation of ECFP at 442 nm in this cell also produced a peak of yellow emission at 527 nm, consistent with a FRET signature (Fig. 4B, left panel). EFRET was calculated from deconvoluted spectra to be 0.62 in this cell. Analyses of multiple cells provided a mean EFRET of 0.52 ± 0.06 for the transfected cell population (Fig. 4C) with an average distance of 49 Å between the proteins (calculated by equation 2; see Materials and Methods). Similar results were obtained with p37 AUF1-EYFP and ECFP-Hsp27 pairs (data not shown).
FIG. 4.
Analysis of p37AUF1-Hsp27 interactions in live cells by FRET. (A) Confocal fluorescence imaging of THP-1 cells coexpressing p37AUF1-ECFP and EYFP-Hsp27. Emission of p37AUF1-ECFP upon excitation at 442 nm (left), emission of EYFP-Hsp27 upon excitation at 488 nm (middle), and emission of EYFP-Hsp27 upon excitation of p37AUF1-ECFP at 442 nm (right), indicative of a FRET signature. Images are shown as thermal gradients. Scale bar, 5 μm. (B) Deconvolutions of fluorescence emission spectra from the cell shown in panel A. The cell was irradiated with 442 nm light, and the spectrum was deconvoluted into its major components (left) as described in the legend for Fig. 2B. For this cell, EFRET = 0.62, with a distance of 45 Å between the two fluorescently tagged proteins. At right is shown the deconvoluted spectrum of the same cell upon direct EYFP excitation at 488 nm. The yellow line represents p37AUF1-EYFP expression. (C) Statistical analysis of interactions of p37AUF1-ECFP with EYFP-Hsp27 and the interaction of the negative control p37AUF1-ECFP with Hsp90-EYFP. Scatter plots of deconvoluted spectra of cells coexpressing p37AUF1-ECFP and EYFP-Hsp27 or p37AUF1-ECFP and Hsp90-EYFP were derived from FRET analyses of >30 cells for each protein pair, as described in the legend to Fig. 2C. cell550, fluorescence of the cell at 550 nm.
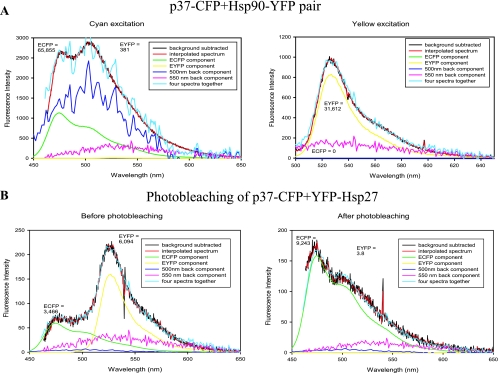
As a control, EFRET was determined for cells cotransfected with plasmids expressing p37AUF1-ECFP and another chaperone, Hsp90-EYFP. Excitation at 442 nm indicated p37AUF1-ECFP expression (Fig. 5A, left panel) and excitation at 488 nm indicated Hsp90-EYFP expression (Fig. 5A, right panel). However, the deconvoluted spectrum had no significant EYFP fluorescence upon excitation of ECFP at 442 nm (Fig. 5A, left panel), indicating near-zero EFRET. Analyses of deconvoluted data from cells coexpressing p37AUF1-ECFP and EYFP-Hsp27 (Fig. 4) and cells coexpressing p37AUF1-ECFP and Hsp90-EYFP (Fig. 5) demonstrated a highly significant difference in mean EFRET between the two populations (Fig. 4C) (P < 0.0001). This result demonstrates that AUF1 does not indiscriminately associate with heat shock/chaperone proteins, consistent with previous biochemical experiments (27).
FIG. 5.
Control FRET experiments for p37AUF1-Hsp27 interactions. (A) THP-1 cells were cotransfected with plasmids expressing p37AUF1-ECFP and Hsp90-EYFP (a negative control). Deconvolutions of fluorescence emission spectra from at least 30 cells were performed as described in the legend to Fig. 2B. The cell was irradiated with 442-nm light, and the spectrum was deconvoluted into its major components to permit calculation of EFRET, which was not detectable (left). Figure 4C contains a scatter plot analysis of 30 cells analyzed for each transfection pair. At right is shown the deconvoluted spectrum of the same cell upon direct EYFP excitation at 488 nm. The yellow line represents Hsp90-EYFP expression. (B) Photobleaching control. Deconvolution analyses of a THP-1 cell coexpressing p37AUF1-ECFP and EYFP-Hsp27 showed a value of 3,466 fluorescence units for ECFP before photobleaching and an increase to 9,423 fluorescence units after photobleaching of EYFP, indicative of a true FRET signature. Three cells were analyzed with comparable results.
For a second control, selective photobleaching of EYFP-Hsp27 was performed in cells coexpressing p37AUF1-ECFP. As expected, photobleached regions exhibited more ECFP fluorescence than they did prior to photobleaching (9,243 versus 3,466 fluorescence units, respectively) (Fig. 5B, compare right and left panels). This demonstrates that EYFP emission upon 442-nm (ECFP) excitation was due to FRET and not to excitation of EYFP. Three independent cells showed similar results. In conclusion, the data in Fig. 4 and 5 indicate that p37AUF1-Hsp27 interactions occur in live THP-1 cells. This has likely implications for AMD.
Binding of Hsp27 to the TNF-α ARE.
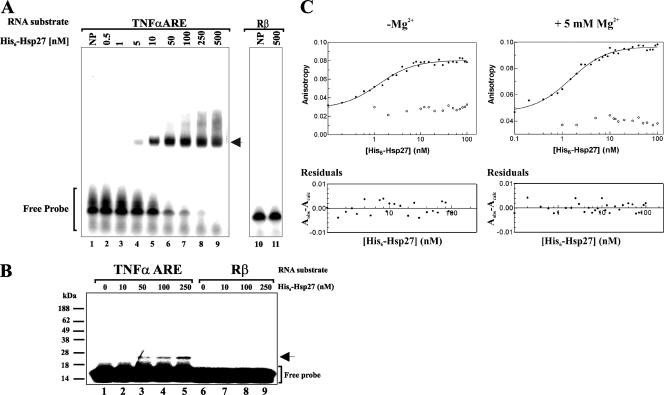
Since Hsp70, Hsc70, and AUF1 all display high-affinity ARE-binding activity (6, 16, 53), we hypothesized that Hsp27 might also bind AREs with high affinity. RNA EMSAs were performed with purified recombinant His6-Hsp27 and a 38-nt RNA containing the core ARE from TNF-α mRNA. A major RNA-protein complex was observed with increasing amounts of recombinant Hsp27 (Fig. 6A). Hsp27 did not bind a control 31-nt RNA derived from the Rβ coding region (Fig. 6A, lanes 10 to 11). Thus, Hsp27 displays high ARE-binding affinity. UV cross-linking confirmed direct ARE contact by Hsp27 (Fig. 6B, lanes 3 to 5). Lack of UV cross-linking to the Rβ coding region again confirmed specificity for ARE binding (Fig. 6B, lanes 6 to 9).
FIG. 6.
Hsp27 is an AUBP. (A) EMSA for Hsp27-ARE interaction. 32P-labeled TNF-α ARE (lanes 1 to 9) or a fragment of the Rβ coding region (lanes 10 and 11) was incubated with the indicated concentrations of His6-Hsp27 and fractionated by native gel electrophoresis. Free RNA is indicated with a bracket, and the protein-RNA complex is indicated by the arrow. NP, no protein added. (B) UV cross-linking analysis of Hsp27-ARE interaction. Binding reactions contained increasing concentrations of His6-Hsp27 and 0.2 nM 5′-32P-labeled TNF-α ARE (lanes 1 to 5) or Rβ (lanes 6 to 9). Reactions were irradiated with UV and fractionated by SDS-PAGE. The arrowhead indicates the protein-RNA complex. The migration positions of markers are indicated on the left of the gel. (C) Evaluation of Hsp27-RNA equilibrium binding by fluorescence polarization. Individual binding reactions containing fluorescein-labeled RNA substrates Fl-TNF-α ARE (filled circles) or Fl-Rβ (open circles) were assembled across a titration of His6-Hsp27 concentrations in the absence (left) or the presence of 5 mM Mg2+ (right). The anisotropy value for each binding reaction is plotted versus protein concentration. Data were described by nonlinear regression by equation 3. Residuals plots of these solutions depict the calculated anisotropy (Acalc) subtracted from the observed anisotropy value (Aobs) and demonstrate that solutions are not biased (lower panels). The same 38-nt core ARE-RNA was used for all three binding assays.
To quantitatively evaluate equilibrium association between Hsp27 and RNA, fluorescence polarization assays were performed with 5′ fluorescein-conjugated RNA substrates. A fluorescent RNA alone yields a low anisotropy value since its small molecular volume allows rapid tumbling and, hence, significant light depolarization. By contrast, protein-RNA association increases molecular volume, slowing RNA mobility, resulting in a higher anisotropy value. Consequently, increasing concentrations of His6-Hsp27 under conditions of limiting Fl-TNF-α RNA increased total measured anisotropy (At) (Fig. 6C, top panel). These data were well described by the Hill equation, equation 3 (see Materials and Methods) (Fig. 6C, top left panel), as data points were randomly distributed around this solution, indicated by the residuals plot (Fig. 6C, bottom left panel). The Hill coefficient (x) resolved to x = 1.15 ± 0.08 (n = 3), indicating that association of Hsp27 with the TNF-α ARE did not significantly deviate from a single-site binding model. The association binding constant (K) resolved to (6.8 ± 0.9) × 108 M−1, corresponding to a dissociation binding constant of 1.5 nM (Kd = 1/K), which is comparable in magnitude to the AUBPs AUF1 (Kd = 0.8 nM) and HuR (Kd = 0.5 nM) (9, 55). Similar results were obtained with Hsp27 in which the His6 tag was enzymatically removed prior to binding assays (data not shown), indicating no contribution of the His6 tag. Addition of increasing amounts of His6-Hsp27 to the Fl-Rβ RNA substrate had no effect upon fluorescence anisotropy, indicative of no binding (Fig. 6C, top left panel). We conclude that Hsp27 binds specifically to the TNF-α ARE-RNA with high affinity.
Mg2+ or other multivalent cations stabilize the TNF-α ARE-RNA in a folded, condensed structure that significantly inhibits association with AUF1 (54). To a lesser extent Mg2+ impedes ARE binding by Hsp70 but has no influence on HuR binding to the ARE (55). These observations imply that ARE presentation within mRNAs may favor binding of some AUBPs. To determine whether RNA folding affects ARE binding by Hsp27, fluorescence polarimetry experiments were repeated with 5 mM Mg2+. Consistent with earlier experiments, Mg2+ increased the intrinsic anisotropy of free RNA substrates (AR; equation 3) due to cation-induced or cation-stabilized RNA structures (54). His6-Hsp27 binding to the TNF-α ARE was again well described by a single-site binding model (Fig. 6C, top right panel), as indicated by the residuals plot (Fig. 6C, bottom right panel). The Hill coefficient (x = 1.17 ± 0.05) and association constant [K = (6.2 ± 0.1) × 108 M−1, corresponding to a Kd of 1.6 nM] resolved for these data did not differ significantly from values obtained in the absence of Mg2+. His6-Hsp27 did not bind to Rβ in the presence of 5 mM Mg2+ (Fig. 6C, top right panel). Based upon these results, we conclude that Hsp27 binds specifically to the TNF-α ARE with high affinity, and unlike AUF1, binding appears independent of RNA structural influences. Multiple AUBPs possessing different binding preferences may permit ASTRC to associate with ARE-mRNAs that present a broad array of secondary structures and perhaps dictate competitive binding equilibria between stabilizing and destabilizing trans-acting factors in response to extracellular stimuli.
Degradation of TNF-α mRNA requires both Hsp27 and AUF1.
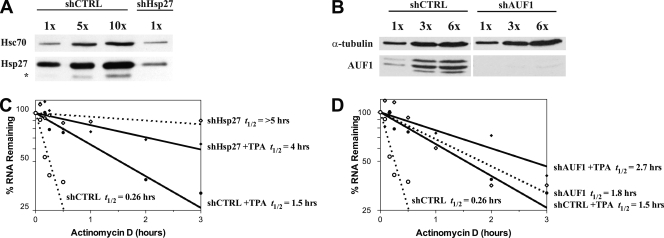
To examine the biological significance of Hsp27-ARE interactions, we established THP-1 cells stably expressing either a scrambled control shRNA (shCTRL) or shRNA directed against Hsp27 (shHsp27) and measured effects upon TNF-α mRNA degradation. The Hsp27 level was reduced 63% compared to cells expressing control shRNA (Fig. 7A, compare lane 4 to lane 1). Likewise, we established THP-1 cells expressing shRNA against all four AUF1 isoforms (shAUF1); knockdown was greater than 90% (Fig. 7B). The half-life of TNF-α mRNA was determined by the actinomycin D time course normalized to β-actin mRNA levels for each time point following an acute treatment with vehicle (DMSO) or TPA (see Materials and Methods for details). Consistent with earlier observations (56), activation with TPA of cells expressing shCTRL stabilized TNF-α mRNA approximately sixfold (Fig. 7C, shCTRL versus shCTRL+TPA) (0.26 h versus 1.5 h, respectively; P = 0.0007). Thus, shRNA expression has no effect upon stabilization of TNF-α mRNA following activation with TPA. Knockdown of Hsp27 stabilized TNF-α mRNA more than 10-fold to a half-life of >5 h, compared to cells expressing shCTRL (i.e., >5 h versus 0.26 h, respectively; P = 0.0003) (Fig. 7C). The TNF-α mRNA half-life was approximately 4 h upon activation of cells expressing shHsp27 with TPA (Fig. 7C), indicating little effect of activation on stabilization of TNF-α mRNA in cells with reduced Hsp27 expression. We conclude that (i) proper Hsp27 expression is essential for rapid degradation of TNF-α mRNA in nonactivated cells and (ii) TPA-mediated activation may reduce Hsp27 activity to stabilize TNF-α mRNA.
FIG. 7.
Knockdown of Hsp27 or AUF1 stabilizes TNF-α mRNA. THP-1 cells were stably transfected with plasmids expressing shCTRL, shHsp27, or shAUF1 to elicit RNA interference. Knockdown of Hsp27 (A) and AUF1 (B) was assessed by Western blotting with various amounts of cell extracts from cells expressing the indicated shRNAs and antibodies to Hsp27 and AUF1. Hsc70 or α-tubulin served as internal controls. Analyses of multiple exposures indicated 63% knockdown of Hsp27 and >90% knockdown of AUF1. *, nonspecific band in panel A. (C and D) Cells expressing the indicated shRNAs were treated with vehicle or 10 nM TPA for 1 h, and then 5 μg/ml actinomycin D was added to culture medium to block transcription. RNA was purified at the indicated time points, quantified by quantitative RT-PCR, and analyzed with nonlinear regression to determine mRNA half-life. (C) Analysis of shHsp27-expressing cells. (D) Analysis of shAUF1-expressing cells. t1/2, half-life.
Similar to AUF1−/− mice (30), knockdown of AUF1 in THP-1 cells stabilized TNF-α mRNA, in this case, approximately sevenfold (0.26 h versus 1.8 h, respectively; P = 0.0007) (Fig. 7D, shCTRL versus shAUF1). However, activation of cells expressing shAUF1 did not lead to further statistically significant mRNA stabilization compared to nonactivated cells (Fig. 7D, shAUF1 versus shAUF+TPA) (P = 0.08). Taken together, the knockdown experiments indicated that (i) proper expression of both Hsp27 and AUF1 is necessary for rapid degradation of TNF-α mRNA in nonactivated cells and (ii) activation reduces AUF1 and/or Hsp27 activities to effect mRNA stabilization.
Hsp27 and AUF1 promote AMD via the TNF-α ARE.
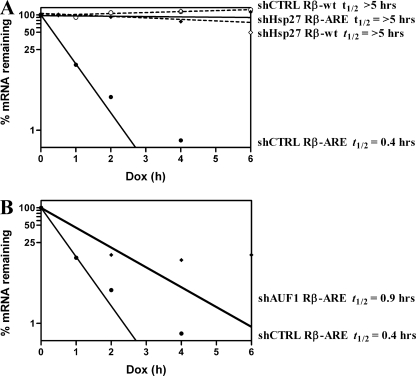
Next, we determined whether Hsp27 and AUF1 affect TNF-α mRNA, at least in part, through the ARE. Tet-responsive reporter plasmids containing either an unmodified rabbit β-globin gene (Rβ-wt) or β-globin linked to the core TNF-α ARE in the 3′ UTR (Rβ-ARE) were cotransfected into THP-1 cells expressing shCTRL, shHsp27, or shAUF1 together with a plasmid encoding a fusion protein of the Tet repressor DNA-binding domain and VP16 trans-activation domain (13). After 2 days, doxycycline was added to the culture medium to block reporter transcription, and RNA was analyzed at each time point. In shCTRL-expressing cells, Rβ mRNA was relatively stable with a half-life of >5 h; by contrast, the Rβ-ARE mRNA half-life was 0.4 h, indicating that the TNF-α ARE potently induces AMD (Fig. 8A) (P < 0.0001). However, the Rβ-ARE mRNA half-life was >5 h in shHsp27-expressing cells (Fig. 8A) (P = 0.0001 compared to shCTRL); Rβ mRNA was still relatively stable with a half-life of >5 h. Likewise, knockdown of AUF1 stabilized the Rβ-ARE reporter mRNA over twofold to 0.9 h (Fig. 8B) (P = 0.0024 compared to shCTRL). We note, however, that the time points appear to plateau between 2 to 6 h with <15% reporter mRNA remaining in cells with AUF1 knockdown. While this might suggest biphasic kinetics, single-population decay kinetics were employed to analyze these data, and as such, the 0.9-h half-life represents an average decay rate for the ARE-reporter mRNA upon knockdown of AUF1. This does not preclude that multiple subpopulations may exist with different decay kinetics. However, the data suggest that any such a stable population, if it exists, must be <15% of the total reporter mRNA. In any event, the twofold stabilization observed here is consistent with AUF1−/− mice and AUF1 knockdown experiments examining other AREs (25, 30, 45).
FIG. 8.
Knockdown of Hsp27 or AUF1 reduces AMD efficiency. Transcription of the indicated Rβ reporter constructs was blocked by adding 2 μg/ml doxycycline (Dox) to the media of shCTRL-, shHsp27-, and shAUF1-expressing THP-1 cells. Reporter mRNA half-lives were determined as described in the legend of Fig. 7. (A) shHsp27-expressing cells. (B) shAUF1-expressing cells. t1/2, half-life.
Since wild-type β-globin mRNA is relatively stable in cells regardless of the shRNA expressed, we examined an additional control for specificity of mRNA stabilization in shHsp27- and shAUF1-expressing cells; the constitutive decay element, an AMD-independent destabilizing sequence in TNF-α mRNA (45), was inserted into the 3′ UTR of Rβ. This reporter mRNA was unstable in shCTRL-, shHsp27-, and shAUF1-expressing cells (data not shown). Taken together, these results indicate that Hsp27 and AUF1 can promote AMD via the TNF-α ARE.
DISCUSSION
Proper regulation of innate immune responses is essential as prolonged production of proinflammatory cytokines is highly deleterious. AUBPs and microRNAs play indispensable roles in immune regulation. For example, the AUBPs HuR, TIA-1, and TTP act in concert to limit proinflammatory cytokine biosynthesis in macrophages at the levels of mRNA decay and translation (20). In addition, TTP collaborates with miR16 to promote degradation of a reporter mRNA containing the TNF-α ARE in both HeLa and Drosophila cells (18). Moreover, AUF1 knockout mice are highly susceptible to endotoxemia due to compromised degradation of ARE-mRNAs encoding TNF-α and interleukin-1β (30). Recent work has unveiled yet another layer of complexity in TNF-α regulation: cell cycle-dependent activation of TNF-α translation involving Ago, FXRI, and miR396-3, a microRNA that activates TNF-α translation in nonproliferating cells (48, 49). Clearly, to attain a comprehensive understanding of proinflammatory cytokine gene expression in innate immunity, it is important to identify all the effectors of AMD. Toward this goal, we focused on the ASTRC protein ensemble, as AUF1 knockout mice revealed its central requirement for AMD and proinflammatory gene regulation in vivo (30). In this work, our key finding was that chaperone Hsp27 is a novel ASTRC subunit critical for cytokine AMD.
Hsp27 has diverse cellular functions including, but not limited to, molecular chaperoning (17), actin polymerization (1), and protection from oxidative stress via modulation of glutathione levels (33). Our results demonstrate that Hsp27 is also a novel, high-affinity AUBP that associates with AUF1-containing protein complexes in vivo and is essential for AMD in monocytes. Three previous observations indicated a possible role for Hsp27 in mRNA stability. (i) Shchors and colleagues found that AUF1 and Hsp27 associate with cell death-inhibiting RNA, a U-rich transcript derived from the 3′ UTR of a gene with unknown function. Binding of the AUF1-Hsp27 complex was associated with reduced AMD and an antiapoptotic phenotype (39). (ii) Lasa and colleagues showed that overexpression of an Hsp27 phosphomimetic (glutamic acid substitutions at serines 15, 78, and 82), but not wild-type, protein resulted in stabilization of a β-globin/COX-2 ARE reporter transcript (29). (iii) Sommer and colleagues showed that overexpression of Hsp27 reduced levels of the ARE-mRNA encoding the c-Yes oncoprotein (42). However, in these studies, there was no clear mechanistic connection between Hsp27 and mRNA stability. Our data suggest that these previous results might be explained by the association of Hsp27 with ASTRC and the ability of Hsp27 to bind and modulate the stability of ARE-mRNAs.
What are potential roles for Hsp27 in AMD? Consideration of a previous observation may provide clues. During heat shock, increased association of Hsp70 and eIF4G with ASTRC occurs coincident with inactivation of AMD (27). Indeed, activation of THP-1 cells with TPA increased association of both eIF4G and Hsp27, but not Hsp70, with ASTRC (Fig. 1). These observations together suggest that activation-induced association of Hsp27 with ASTRC may permit coupling of monocyte activation and AMD. By contrast, increased association of Hsp70 with ASTRC following heat shock might be indicative of a heat shock-specific signal to the AMD machinery. Clearly, future work, including the identification of novel ASTRC components and additional AUBPs, will be required to adequately address the hypothesis that ASTRC is adaptive and that its individual subunits can serve as intermediaries between specific stimuli and AMD.
For reasons that are not yet clear, ASTRC contains at least four AUBPs: AUF1, Hsp70, Hsc70, and Hsp27. We offer two hypotheses. (i) HuR, Hsp70, and p37AUF1 display distinct binding affinities for stabilized ARE secondary structures (9). For example, secondary structure has a modest effect upon Hsp70 and no effect upon HuR binding but impairs ARE-binding affinity of p37AUF1 by >10-fold. By contrast, ARE binding by Hsp27 is unaffected by RNA structure (Fig. 6C). As such, it does not favor particular ARE-secondary structure presentations, distinguishing it from AUF1. Thus, multiple AUBPs possessing different binding preferences may permit ASTRC to associate with ARE-mRNAs that present broad arrays of secondary structures. (ii) Reporter assays utilizing either a control ARE or a folded ARE indicated that a strongly folded ARE slows AMD (9), likely due to the favored binding of stabilizing AUBPs. Therefore, competition between stabilizing and destabilizing trans-acting factors for ARE occupancy is likely regulated in part by ARE conformation. Thus, regulated mRNA stabilization may be mediated by factors that have the potential to modulate ARE presentation, such as flanking sequences, association of RNA-binding proteins at adjacent sequences, microRNAs, or local cation concentrations. Another observation worth noting is that despite the fact that many ASTRC subunits are RNA-binding proteins, their association with the complex may not be simply due to RNA bridging as proteins were coimmunoprecipitated following exhaustive RNase digestion (Fig. 1). Although this experiment does not rule out the possibility that complex assembly may be ARE dependent, this evidence, together with the ARE-binding activity of Hsp27, suggests that association of Hsp27 with ASTRC may occur via both protein-protein and protein-mRNA interactions. Nonetheless, a protein complex consisting of several AUBPs, such as ASTRC, that possesses both various preferences for ARE-structure and various contributions to ARE-mRNA stability would permit rapid alterations in AMD in response to extracellular stimuli.
In conclusion, our experiments have unveiled chaperone Hsp27 as both a new subunit of ASTRC and essential for cytokine AMD. We note that among its many functions, Hsp27 is also an actin-binding protein involved in cell motility (19). As monocyte activation by adhesion to endothelium and motility stimulate cytoskeletal reorganization and stabilize many cytokine ARE-mRNAs, it is tempting to speculate that Hsp27 association with ASTRC provides a new piece to an age-old puzzle as to how proinflammatory cytokine biosynthesis is coupled with cell adhesion/motility (15, 40, 41). As extracellular stimuli drive Hsp27 into numerous signaling complexes as well (60), future elucidation of their interactions with ASTRC will add additional details to our understanding of the multilayered control systems required to initiate, maintain, and limit the innate immune response.
Supplementary Material
Acknowledgments
We thank Andy Clark and Daiya Takai for plasmids, Nahum Sonenberg for eIF4G antibodies, Daniel Sinsimer for assistance with molecular beacon design and methods, and Gerald Wilson for reporter quantitative RT-PCR methods and assistance with statistical analyses. We thank Lori Covey for comments on the manuscript.
This work was supported by grants P01 AI057596 from the NIH to S.P. and G.B. and R01 AI059465 from the NIH to S.P. K.S. was supported by training grant T32 AI00743 from the NIH to S.P. F.M.G. and A.M.K. were supported by Integrative Graduate Education and Research Traineeship DGE0333196 from the NSF to Prabhas Moghe (Department of Biomedical Engineering, Rutgers University). F.M.G. was also supported by Initiative for Minority Students R25 GM058389 from the NIH to Michael Leibowitz (UMDNJ).
Footnotes
Published ahead of print on 23 June 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Benndorf, R., K. Hayess, S. Ryazantsev, M. Wieske, J. Behlke, and G. Lutsch. 1994. Phosphorylation and supramolecular organization of murine small heat shock protein HSP25 abolish its actin polymerization-inhibiting activity. J. Biol. Chem. 26920780-20784. [PubMed] [Google Scholar]
- 2.Brewer, G., and J. Ross. 1990. Messenger RNA turnover in cell-free extracts. Methods Enzymol. 181202-209. [DOI] [PubMed] [Google Scholar]
- 3.Carballo, E., H. Cao, W. S. Lai, E. A. Kennington, D. Campbell, and P. J. Blackshear. 2001. Decreased sensitivity of tristetraprolin-deficient cells to p38 inhibitors suggests the involvement of tristetraprolin in the p38 signaling pathway. J. Biol. Chem. 27642580-42587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, C. Y., R. Gherzi, S. E. Ong, E. L. Chan, R. Raijmakers, G. J. Pruijn, G. Stoecklin, C. Moroni, M. Mann, and M. Karin. 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107451-464. [DOI] [PubMed] [Google Scholar]
- 5.David, P. S., R. Tanveer, and J. D. Port. 2007. FRET-detectable interactions between the ARE binding proteins, HuR and p37AUF1. RNA 131453-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeMaria, C. T., and G. Brewer. 1996. AUF1 binding affinity to A+U-rich elements correlates with rapid mRNA degradation. J. Biol. Chem. 27112179-12184. [DOI] [PubMed] [Google Scholar]
- 7.DeMaria, C. T., Y. Sun, L. Long, B. J. Wagner, and G. Brewer. 1997. Structural determinants in AUF1 required for high affinity binding to A+U-rich elements. J. Biol. Chem. 27227635-27643. [DOI] [PubMed] [Google Scholar]
- 8.Donnini, M., A. Lapucci, L. Papucci, E. Witort, A. Jacquier, G. Brewer, A. Nicolin, S. Capaccioli, and N. Schiavone. 2004. Identification of TINO: a new evolutionarily conserved BCL-2 AU-rich element RNA-binding protein. J. Biol. Chem. 27920154-20166. [DOI] [PubMed] [Google Scholar]
- 9.Fialcowitz, E. J., B. Y. Brewer, B. P. Keenan, and G. M. Wilson. 2005. A hairpin-like structure within an AU-rich mRNA-destabilizing element regulates trans-factor binding selectivity and mRNA decay kinetics. J. Biol. Chem. 28022406-22417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaestel, M., W. Schroder, R. Benndorf, C. Lippmann, K. Buchner, F. Hucho, V. A. Erdmann, and H. Bielka. 1991. Identification of the phosphorylation sites of the murine small heat shock protein Hsp25. J. Biol. Chem. 26614721-14724. [PubMed] [Google Scholar]
- 11.Gao, M., C. J. Wilusz, S. W. Peltz, and J. Wilusz. 2001. A novel mRNA-decapping activity in HeLa cytoplasmic extracts is regulated by AU-rich elements. EMBO J. 201134-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gherzi, R., K. Y. Lee, P. Briata, D. Wegmuller, C. Moroni, M. Karin, and C. Y. Chen. 2004. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell 14571-583. [DOI] [PubMed] [Google Scholar]
- 13.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 895547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guhaniyogi, J., and G. Brewer. 2001. Regulation of mRNA stability in mammalian cells. Gene 26511-23. [DOI] [PubMed] [Google Scholar]
- 15.Haskill, S., C. Johnson, D. Eierman, S. Becker, and K. Warren. 1988. Adherence induces selective mRNA expression of monocyte mediators and proto-oncogenes. J. Immunol. 1401690-1694. [PubMed] [Google Scholar]
- 16.Henics, T., E. Nagy, H. J. Oh, P. Csermely, A. von Gabain, and J. R. Subjeck. 1999. Mammalian Hsp70 and Hsp110 proteins bind to RNA motifs involved in mRNA stability. J. Biol. Chem. 27417318-17324. [DOI] [PubMed] [Google Scholar]
- 17.Jakob, U., M. Gaestel, K. Engel, and J. Buchner. 1993. Small heat shock proteins are molecular chaperones. J. Biol. Chem. 2681517-1520. [PubMed] [Google Scholar]
- 18.Jing, Q., S. Huang, S. Guth, T. Zarubin, A. Motoyama, J. Chen, P. F. Di, S. C. Lin, H. Gram, and J. Han. 2005. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell 120623-634. [DOI] [PubMed] [Google Scholar]
- 19.Jog, N. R., V. R. Jala, R. A. Ward, M. J. Rane, B. Haribabu, and K. R. McLeish. 2007. Heat shock protein 27 regulates neutrophil chemotaxis and exocytosis through two independent mechanisms. J. Immunol. 1782421-2428. [DOI] [PubMed] [Google Scholar]
- 20.Katsanou, V., O. Papadaki, S. Milatos, P. J. Blackshear, P. Anderson, G. Kollias, and D. L. Kontoyiannis. 2005. HuR as a negative posttranscriptional modulator in inflammation. Mol. Cell 19777-789. [DOI] [PubMed] [Google Scholar]
- 21.Knapinska, A. M., P. Irizarry-Barreto, S. Adusumalli, I. Androulakis, and G. Brewer. 2005. Molecular mechanisms regulating mRNA stability: physiological and pathological significance. Curr. Genomics 6471-486. [Google Scholar]
- 22.Krause, C. D., E. Mei, O. Mirochnitchenko, N. Lavnikova, J. Xie, Y. Jia, R. M. Hochstrasser, and S. Pestka. 2006. Interactions among the components of the interleukin-10 receptor complex. Biochem. Biophys. Res. Commun. 340377-385. [DOI] [PubMed] [Google Scholar]
- 23.Krause, C. D., E. Mei, J. Xie, Y. Jia, M. A. Bopp, R. M. Hochstrasser, and S. Pestka. 2002. Seeing the light: preassembly and ligand-induced changes of the interferon γ receptor complex in cells. Mol. Cell. Proteomics 1805-815. [DOI] [PubMed] [Google Scholar]
- 24.Lai, W. S., E. Carballo, J. R. Strum, E. A. Kennington, R. S. Phillips, and P. J. Blackshear. 1999. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol. Cell. Biol. 194311-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lal, A., K. Mazan-Mamczarz, T. Kawai, X. Yang, J. L. Martindale, and M. Gorospe. 2004. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 233092-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landry, J., H. Lambert, M. Zhou, J. N. Lavoie, E. Hickey, L. A. Weber, and C. W. Anderson. 1992. Human HSP27 is phosphorylated at serines 78 and 82 by heat shock and mitogen-activated kinases that recognize the same amino acid motif as S6 kinase II. J. Biol. Chem. 267794-803. [PubMed] [Google Scholar]
- 27.Laroia, G., R. Cuesta, G. Brewer, and R. J. Schneider. 1999. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science 284499-502. [DOI] [PubMed] [Google Scholar]
- 28.Laroia, G., B. Sarkar, and R. J. Schneider. 2002. Ubiquitin-dependent mechanism regulates rapid turnover of AU-rich cytokine mRNAs. Proc. Natl. Acad. Sci. USA 991842-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lasa, M., K. R. Mahtani, A. Finch, G. Brewer, J. Saklatvala, and A. R. Clark. 2000. Regulation of cyclooxygenase 2 mRNA stability by the mitogen-activated protein kinase p38 signaling cascade. Mol. Cell. Biol. 204265-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu, J. Y., N. Sadri, and R. J. Schneider. 2006. Endotoxic shock in AUF1 knockout mice mediated by failure to degrade proinflammatory cytokine mRNAs. Genes Dev. 203174-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma, W. J., S. Cheng, C. Campbell, A. Wright, and H. Furneaux. 1996. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 2718144-8151. [DOI] [PubMed] [Google Scholar]
- 32.Matsukura, S., P. A. Jones, and D. Takai. 2003. Establishment of conditional vectors for hairpin siRNA knockdowns. Nucleic Acids Res. 31e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mehlen, P., C. Kretz-Remy, X. Preville, and A. P. Arrigo. 1996. Human hsp27, Drosophila hsp27 and human αB-crystallin expression-mediated increase in glutathione is essential for the protective activity of these proteins against TNFα-induced cell death. EMBO J. 152695-2706. [PMC free article] [PubMed] [Google Scholar]
- 34.Mukherjee, D., M. Gao, J. P. O'Connor, R. Raijmakers, G. Pruijn, C. S. Lutz, and J. Wilusz. 2002. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 21165-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ross, J., and G. Kobs. 1986. H4 histone messenger RNA decay in cell-free extracts initiates at or near the 3′ terminus and proceeds 3′ to 5′. J. Mol. Biol. 188579-593. [DOI] [PubMed] [Google Scholar]
- 36.Sarkar, B., J. Y. Lu, and R. J. Schneider. 2003. Nuclear import and export functions in the different isoforms of the AUF1/heterogeneous nuclear ribonucleoprotein protein family. J. Biol. Chem. 27820700-20707. [DOI] [PubMed] [Google Scholar]
- 37.Schmidlin, M., M. Lu, S. A. Leuenberger, G. Stoecklin, M. Mallaun, B. Gross, R. Gherzi, D. Hess, B. A. Hemmings, and C. Moroni. 2004. The ARE-dependent mRNA-destabilizing activity of BRF1 is regulated by protein kinase B. EMBO J. 234760-4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwende, H., E. Fitzke, P. Ambs, and P. Dieter. 1996. Differences in the state of differentiation of THP-1 cells induced by phorbol ester and 1,25-dihydroxyvitamin D3. J. Leukoc. Biol. 59555-561. [PubMed] [Google Scholar]
- 39.Shchors, K., F. Yehiely, R. K. Kular, K. U. Kotlo, G. Brewer, and L. P. Deiss. 2002. Cell death inhibiting RNA (CDIR) derived from a 3′-untranslated region binds AUF1 and heat shock protein 27. J. Biol. Chem. 27747061-47072. [DOI] [PubMed] [Google Scholar]
- 40.Sirenko, O. I., A. K. Lofquist, C. T. DeMaria, J. S. Morris, G. Brewer, and J. S. Haskill. 1997. Adhesion-dependent regulation of an A+U-rich element-binding activity associated with AUF1. Mol. Cell. Biol. 173898-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sirenko, O., U. Bocker, J. S. Morris, J. S. Haskill, and J. M. Watson. 2002. IL-1β transcript stability in monocytes is linked to cytoskeletal reorganization and the availability of mRNA degradation factors. Immunol. Cell Biol. 80328-339. [DOI] [PubMed] [Google Scholar]
- 42.Sommer, S., Y. Cui, G. Brewer, and S. A. Fuqua. 2005. The c-Yes 3′-UTR contains adenine/uridine-rich elements that bind AUF1 and HuR involved in mRNA decay in breast cancer cells. J. Steroid Biochem. Mol. Biol. 97219-229. [DOI] [PubMed] [Google Scholar]
- 43.Stoecklin, G., M. Colombi, I. Raineri, S. Leuenberger, M. Mallaun, M. Schmidlin, B. Gross, M. Lu, T. Kitamura, and C. Moroni. 2002. Functional cloning of BRF1, a regulator of ARE-dependent mRNA turnover. EMBO J. 214709-4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stoecklin, G., T. Mayo, and P. Anderson. 2006. ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 772-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stoecklin, G., M. Lu, B. Rattenbacher, and C. Moroni. 2003. A constitutive decay element promotes tumor necrosis factor alpha mRNA degradation via an AU-rich element-independent pathway. Mol. Cell. Biol. 233506-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki, A., Y. Tsutomi, K. Akahane, T. Araki, and M. Miura. 1998. Resistance to Fas-mediated apoptosis: activation of caspase 3 is regulated by cell cycle regulator p21WAF1 and IAP gene family ILP. Oncogene 17931-939. [DOI] [PubMed] [Google Scholar]
- 47.Tyagi, S., and F. R. Kramer. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14303-308. [DOI] [PubMed] [Google Scholar]
- 48.Vasudevan, S., and J. A. Steitz. 2007. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell 1281105-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vasudevan, S., Y. Tong, and J. A. Steitz. 2007. Switching from repression to activation: microRNAs can up-regulate translation. Science 3181931-1934. [DOI] [PubMed] [Google Scholar]
- 50.Wagner, B. J., C. T. DeMaria, Y. Sun, G. M. Wilson, and G. Brewer. 1998. Structure and genomic organization of the human AUF1 gene: alternative pre-mRNA splicing generates four protein isoforms. Genomics 48195-202. [DOI] [PubMed] [Google Scholar]
- 51.Wilson, G. M., J. Lu, K. Sutphen, Y. Suarez, S. Sinha, B. Brewer, E. Villanueva-Feliciano, R. M. Ysla, S. Charles, and G. Brewer. 2003. Phosphorylation of p40AUF1 regulates binding to A+U-rich mRNA-destabilizing elements and protein-induced changes in ribonucleoprotein structure. J. Biol. Chem. 27833039-33048. [DOI] [PubMed] [Google Scholar]
- 52.Wilson, G. M., Y. Sun, H. Lu, and G. Brewer. 1999. Assembly of AUF1 oligomers on U-rich RNA targets by sequential dimer association. J. Biol. Chem. 27433374-33381. [DOI] [PubMed] [Google Scholar]
- 53.Wilson, G. M., K. Sutphen, S. Bolikal, K. Y. Chuang, and G. Brewer. 2001. Thermodynamics and kinetics of Hsp70 association with A+U-rich mRNA-destabilizing sequences. J. Biol. Chem. 27644450-44456. [DOI] [PubMed] [Google Scholar]
- 54.Wilson, G. M., K. Sutphen, K. Chuang, and G. Brewer. 2001. Folding of A+U-rich RNA elements modulates AUF1 binding. Potential roles in regulation of mRNA turnover. J. Biol. Chem. 2768695-8704. [DOI] [PubMed] [Google Scholar]
- 55.Wilson, G. M., K. Sutphen, M. Moutafis, S. Sinha, and G. Brewer. 2001. Structural remodeling of an A+U-rich RNA element by cation or AUF1 binding. J. Biol. Chem. 27638400-38409. [DOI] [PubMed] [Google Scholar]
- 56.Wilson, G. M., J. Lu, K. Sutphen, Y. Sun, Y. Huynh, and G. Brewer. 2003. Regulation of A+U-rich Element-directed mRNA turnover involving reversible phosphorylation of AUF1. J. Biol. Chem. 27833029-33038. [DOI] [PubMed] [Google Scholar]
- 57.Wilusz, C. J., M. Wormington, and S. W. Peltz. 2001. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2237-246. [DOI] [PubMed] [Google Scholar]
- 58.Zacharias, D. A., J. D. Violin, A. C. Newton, and R. Y. Tsien. 2002. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296913-916. [DOI] [PubMed] [Google Scholar]
- 59.Zhang, W., B. J. Wagner, K. Ehrenman, A. W. Schaefer, C. T. DeMaria, D. Crater, K. DeHaven, L. Long, and G. Brewer. 1993. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol. Cell. Biol. 137652-7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng, C., Z. Lin, Z. J. Zhao, Y. Yang, H. Niu, and X. Shen. 2006. MAPK-activated protein kinase-2 (MK2)-mediated formation and phosphorylation-regulated dissociation of the signal complex consisting of p38, MK2, Akt, and Hsp27. J. Biol. Chem. 28137215-37226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.