Abstract
Trichomonas vaginalis, the protist that causes vaginal itching, has a huge genome with numerous gene duplications. Recently we found that Trichomonas has numerous genes encoding putative dolichyl-phosphate-glucose (Dol-P-Glc) synthases (encoded by ALG5 genes) despite the fact that Trichomonas lacks the glycosyltransferases (encoded by ALG6, ALG8, and ALG10 genes) that use Dol-P-Glc to glucosylate dolichyl-PP-linked glycans. In addition, Trichomonas does not have a canonical DPM1 gene, encoding a dolichyl-P-mannose (Dol-P-Man) synthase. Here we show Trichomonas membranes have roughly 300 times the Dol-P-Glc synthase activity of Saccharomyces cerevisiae membranes and about one-fifth the Dol-P-Man synthase activity of Saccharomyces membranes. Endogenous Dol-P-hexoses of Trichomonas are relatively abundant and contain 16 isoprene units. Five paralogous Trichomonas ALG5 gene products have Dol-P-Glc synthase activity when expressed as recombinant proteins, and these Trichomonas Alg5s correct a carboxypeptidase N glycosylation defect in a Saccharomyces alg5 mutant in vivo. A recombinant Trichomonas Dpm1, which is deeply divergent in its sequence, has Dol-P-Man synthase activity. When radiolabeled Dol-P-Glc is incubated with Trichomonas membranes, Glc is incorporated into reducing and nonreducing sugars of O-glycans of endogenous glycoproteins. To our knowledge, this is the first demonstration of Dol-P-Glc as a sugar donor for O-glycans on glycoproteins.
UDP-glucose:dolichyl-phosphate β-d-glucosyltransferase (Dol-P-Glc synthase; EC 2.4.1.117), which is encoded by the ALG5 gene in Saccharomyces cerevisiae, converts cytoplasmic UDP-Glc and endoplasmic reticulum (ER) membrane-bound Dol-P to Dol-P-Glc (9, 11). On the luminal face of the ER, Dol-P-Glc serves as the sugar donor for transfer of three glucose residues to the Man9GlcNAc2-PP-dolichol precursor to Asn-linked glycans (N-glycans) (10). The glucosyltransferases, which are called Alg6, Alg8, and Alg10 in Saccharomyces, make the 14-sugar N-glycan precursor (Glc3Man9GlcNAc2-PP-dolichol), which is transferred by the oligosaccharyltransferase to Asn residues on nascent glycoproteins in the ER lumen (15).
GDP-mannose:dolichyl-phosphate β-d-mannosyltransferase (Dol-P-Man synthase; EC 2.4.1.83), which is encoded by DPM1 in Saccharomyces (a paralog of ALG5), converts cytoplasmic GDP-mannose and Dol-P to Dol-P-Man (22, 23). On the luminal face of the ER, Dol-P-Man serves as a mannose donor for three separate groups of enzymes. (i) Mannosyltransferases, encoded by ALG3, ALG9, and ALG12 genes in Saccharomyces, use Dol-P-Man to convert the N-glycan precursor Man5GlcNAc2-PP-dolichol to Man9GlcNAc2-PP-dolichol (which is subsequently glucosylated to Glc3Man9GlcNAc2-PP-dolichol) (10). (ii) Mannosyltransferases, encoded by GPI genes in Saccharomyces and PIG genes in mammals, use Dol-P-Man to make glycosylphosphatidylinositol (GPI) anchors for GPI-linked membrane proteins (24). (iii) Mannosyltransferases, encoded by PMT genes in Saccharomyces and POMT genes in mammals, use Dol-P-Man to add single O-linked mannose residues to serine/threonine residues of glycoproteins in the ER lumen (20, 35).
Trichomonas vaginalis is the protist that causes vaginitis and increases the risk of transmission of human immunodeficiency virus (28, 34). The 150-Mb genome of Trichomonas, which predicts 60,000 proteins, is by far the largest of any protist (4). We are interested in protein glycosylation in Trichomonas for the following reasons. First, Trichomonas is missing glycosyltransferases, which use Dol-P-Man and Dol-P-Glc to add mannose and glucose residues to N-glycan precursors in the lumen of the ER (25). Therefore, Trichomonas makes a seven-sugar N-glycan precursor (Man5GlcNAc2-PP-dolichol), which is transferred by the oligosaccharyltransferase to Asn on nascent peptides.
Second, Trichomonas has multiple putative ALG5 genes, which likely synthesize Dol-P-Glc, even though there is no obvious use for Dol-P-Glc by the protist (25). Third, Trichomonas is the only eukaryote we have examined that does not have a canonical Dol-P-Man synthase gene (DPM1) (25). Fourth, Trichomonas is missing the enzymatic machinery for making GPI anchors, an important posttranslational modification in all other eukaryotes examined to date (4, 24). Fifth, Trichomonas makes a unique lipophosphoglycan (LPG), which is antigenic and is involved in pathogenesis (3, 8, 29).
Several specific questions were asked here. Does Trichomonas synthesize Dol-P-Glc and Dol-P-Man? Which candidate ALG5 genes of Trichomonas synthesize Dol-P-Glc and/or Dol-P-Man when expressed in a heterologous system? What is the in vitro product when Dol-P-Glc is incubated with Trichomonas membranes?
MATERIALS AND METHODS
Materials.
UDP-glucose, GDP-mannose, and [1-14C]isopentenyl pyrophosphate (60 mCi/mmol) were purchased from Sigma-Aldrich. UDP-[6-3H]glucose (60 Ci/mmol), UDP-[U-14C]glucose (300 mCi/mmol), GDP-[2-3H)]mannose (20 Ci/mmol), GDP-[U-14C]mannose (298.1 mCi/mmol), and sodium [3H]borotritide were from American Radiolabeled Chemicals. Antibodies used in Western blots included mouse anti-S tag (Novagen), mouse anti-carboxydase Y (Molecular Probes), and horseradish peroxidase-conjugated secondary antibodies (Promega). Peptide-N-glycosidase F (PNGase F) was from New England Biolabs. All other chemicals were of the highest quality available.
Dol-P-Glc and Dol-P-Man synthase activities of Trichomonas membranes.
The UR1 isolate of Trichomonas vaginalis (8) was grown in liquid TYM medium supplemented with 10% heat-inactivated bovine serum (Invitrogen) (5). Cultured Trichomonas cells were chilled, washed three times with phosphate-buffered saline, resuspended in a buffer composed of 50 mM Tris-HCl (pH 7.4), 5 mM 2-mercaptoethanol, and 0.15 mM MgCl2, and sonicated on ice for 30 seconds. Large debris was removed by low-speed centrifugation, and Trichomonas membranes, which were obtained by centrifugation for 1 h at 100,000 × g, were used fresh or were stored at −80°C in the same buffer plus 3.5 mM MgCl2. As a control, fresh and frozen membranes were prepared from the BY4741 strain of Saccharomyces cerevisiae or its derivative (Euroscarf), which was grown in synthetic complete medium and disrupted with glass beads. In our experience, freezing decreases the enzyme activities of Trichomonas and Saccharomyces membranes by 10 to 15%.
The Dol-P-Glc synthase activity of 100 μg of Trichomonas or Saccharomyces membranes was determined in 50-μl reaction mixtures containing 10 μg of exogenous Dol-P as a sugar acceptor, 0.1% NP-40, 10 mM MgCl2, 50 mM Tris-HCl (pH 7.4), and radiolabeled UDP-Glc. Conditions for these assays were determined by varying the concentration of Trichomonas membranes and by varying the pH with recombinant Trichomonas enzymes (see Fig. S1 in the supplemental material). For Dol-P-Man synthase activities, membranes were incubated with radiolabeled GDP-Man (17). After 5 to 15 min at 30°C, reactions were stopped by addition of 4 ml of chloroform-methanol (3/2, vol/vol). Organic phases containing radiolabeled Dol-P-Glc or Dol-P-Man were washed once with 1 ml of 4 mM MgCl2 and twice with 0.8 ml of chloroform-methanol-4 mM MgCl2 (3/48/47). The radioactivity incorporated into the lipid fraction was measured by liquid scintillation counting.
The identity of the synthesized lipid was confirmed by comigration with a Saccharomyces Dol-P-Glc standard by thin-layer chromatography (TLC) on silica gel plates developed in chloroform-methanol-water (65/25/4). The sensitivity of the product to mild acid treatment (10 min of hydrolysis in 40 mM trifluoroacetic acid at 90°C) was tested, and the identity of the released sugar as Glc or Man was confirmed by comigration with an internal standard by high-pH anion-exchange chromatography as described previously (33).
Purification and characterization by mass spectrometry of endogenous Trichomonas Dol-P-hexose.
Membranes from Trichomonas grown in a 2-liter culture were extracted with chloroform-methanol (3/2). The organic phase was washed once with 1/5 volume of 4 mM MgCl2 and twice with chloroform-methanol-4 mM MgCl2 (3/48/47). Dried lipids were dissolved in 5 ml of chloroform-methanol-water (2/3/1) and separated on a 5-ml DEAE-cellulose column equilibrated in the acetate form, as described previously (14).
Liquid chromatography (LC)/mass spectrometry of lipids was performed using a Shimadzu LC system (comprising a solvent degasser, two LC-10A pumps, and an SCL-10A system controller) coupled to a QSTAR XL quadrupole time of flight tandem mass spectrometer (Applied Biosystems, Foster City, CA) equipped with an electrospray source. LC was operated at a flow rate of 200 μl per min with a linear gradient as follows: 100% of mobile phase A was held isocratically for 2 min and then linearly increased to 100% mobile phase B over 14 min and held at 100% B for 4 min. Mobile phase A consisted of methanol-acetonitrile-aqueous 1 mM ammonium acetate (60/20/20). Mobile phase B consisted of 100% ethanol containing 1 mM ammonium acetate. A Zorbax SB-C8 reversed-phase column (5 μm, 2.1 by 50 mm) was obtained from Agilent (Palo Alto, CA). The postcolumn splitter diverted ∼10% of the LC flow to the electrospray ionization source of the mass spectrometer. Negative-ion mass spectra were acquired with an electrospray voltage of −4,500 V.
The LC/mass spectrometry results were compared with those of an in vitro cis-prenyltransferase assay (32), in which Trichomonas membranes were incubated with radiolabeled isopentenyl pyrophosphate and exogenous farnesyl pyrophosphate in the presence of 0.1% Triton X-100. The length of the dehydrodolichol product was determined by reverse-phase TLC using dehydrodolichol produced in vitro by Saccharomyces as a standard.
In silico identification of Dol-P-Glc and Dol-P-Man synthases from Trichomonas.
Saccharomyces cerevisiae Alg5 (Dol-P-Glc synthase) and Dpm1 (Dol-P-Man synthase) were used to search predicted proteins of Trichomonas vaginalis using BLASTP or TBLASTN in the databases managed by The J. Craig Venter Institute (http://www.tigr.org/tdb/e2k1/tvg/) (1, 4, 21). This resulted in five putative Trichomonas ALG5 genes (TvALG5A to TvALG5E), as well as weak hit (TvALG5G). TvALG5G was in turn used to identify the TvDPM1 gene, which was present in two overlapping protein predictions. The corrected TvDPM1 gene sequence was determined by sequencing a TvDPM1 PCR product made with primers to the 5′ and 3′ ends of the coding sequence. Comparison of PCR products obtained with genomic DNA and cDNA as a template revealed that none of the Trichomonas Alg5 or Dpm1 genes contain an intron.
Transmembrane helices in the Trichomonas Dol-P-Glc and Dol-P-Man synthases were predicted by using the Phobius combined transmembrane topology and signal peptide predictor (13). Alignments of protein sequences were made using Clustal W (http://www.ebi.ac.uk/clustalw), and manual adjustments and trimming of the alignments were performed with jalview (7, 18). Phylogenetic trees were constructed from the positional variation with maximum likelihood by using quartet puzzling (12, 27).
Cloning and recombinant expression of Trichomonas Dol-P-Glc and Dol-P-Man synthase genes.
The coding sequences of putative Trichomonas glycosyltransferase genes and Saccharomyces ALG5 and DPM1 genes were amplified using genomic DNA as a template and a pair of primers listed in Table S1 in the supplemental material. PCR products were cloned into the pGEM-T Easy vector and sequenced (Promega).
To express Trichomonas genes in the Saccharomyces cells, NotI-surrendered inserts were subcloned into the pNEV-N plasmid under the control of the PMA1 promoter and terminator (see Table S1 in the supplemental material) (26). pNEV constructs containing Trichomonas ALG5 genes, as well as the Saccharomyces ALG5 gene, were transformed into the BY4741 derivative alg5Δ yeast strain (Euroscarf). As a second positive control, wild-type BY4741 cells transformed with empty vector pNEV-N were also tested. Membrane fractions were isolated from transformed yeast and tested for Dol-P-Glc synthase activity as described above.
To examine the functionality of TvAlg5 proteins in vivo, we tested the N glycosylation status of carboxypeptidase Y in Scalg5Δ strains transformed with plasmids carrying Trichomonas ALG5 genes in comparison with wild-type and Scalg5Δ mutant Saccharomyces strains (9). Yeast proteins were obtained by alkaline lysis and resolved on sodium dodecyl sulfate (SDS)-NuPAGE-Novex Bis-Tris 4 to 12% gel under reducing conditions. Proteins were transferred to polyvinylidene difluoride membranes, which were blocked in 5% milk and probed with antibodies against Saccharomyces carboxypeptidase Y.
To express Trichomonas genes in Escherichia coli, BamI/NotI fragments were cloned into the pET30a vector (Novagen) in such a way that the protein was tagged at the N terminus with a polyhistidine-S-tag fusion. pET30a vectors containing the ScDPM1, TvALG5E, or TvDPM1 gene were each transformed into Escherichia coli Rosetta 2 cells (Novagen). E. coli cells in the logarithmic growth phase were induced to express heterologous protein by incubation with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 h at 30°C. The harvested cells were lysed by sonication, and His-tagged proteins were purified on a nickel column according to the Invitrogen protocol. Protein purity was judged by SDS-polyacrylamide gel electrophoresis and Western blotting using monoclonal mouse antibody against the S tag. The amount of the purified protein was estimated using the S•Tag rapid assay kit (Novagen), and 0.1 pmol of purified enzyme was added to the Dol-P-Glc or Dol-P-Man synthase assay mixtures.
Real-time PCR.
Total RNA was isolated from mid-log-phase Trichomonas cells using the RNeasy kit (Qiagen), and RNA was treated with DNA-free reagent (Ambion) in accordance with the manufacturer's instructions. Reverse transcription of RNA was carried out with the RETROscript kit (Ambion) using oligo(dT)18. Levels of specific mRNA species were measured by real-time PCR using the Sybr green method in a 96-well plate format with a Stratagene MX4000 cycler. Reaction mixtures contained 12.5 μl 2× Brilliant Sybr green quantitative PCR master mix (Stratagene), primers (100 nM), and a template in a total volume of 25 μl. Master mix was made with reverse-transcribed RNA to minimize the sampling error. The thermal profile for amplification was 95°C for 10 min, followed by 40 cycles of 95°C for 30 s, 52°C for 30 s, and 72°C for 30 s. Primers for real-time PCR were designed (see Table S1 in the supplemental material) using OligoPerfect software (Invitrogen). Gel electrophoresis was carried out on representative samples to confirm product size. Amplification plots were analyzed using MX4000 software, version 4.20 (Stratagene), to determine the relative quantity of mRNA species using a Trichomonas actin gene as the calibrator.
In vitro glucosylation of Trichomonas proteins using radiolabeled Dol-P-Glc.
Radiolabeled dolichyl-[14C]glucose was synthesized by incubating commercially available UDP-[14C]Glc and Dol-P with recombinant TvAlg5E, which was purified from E. coli as described above. Radiolabeled Dol-P-Glc was extracted into chloroform, and its identity was confirmed by its mobility in TLC and by its sensitivity to mild acid hydrolysis. The reaction mixture, which was incubated at 30°C for 60 min in a volume of 250 μl, contained 0.5% octyl-d-thioglucoside, 10 mM MnCl2, 50 mM Tris-HCl, pH 7.4, 20,000 cpm DolP-[14C]Glc, and 500 μg of Trichomonas membranes. Control reaction mixtures were incubated for 0 min, incubated at 0°C, incubated at pH 3 or pH 10, or incubated in the presence of EDTA (see Fig. S2 in the supplemental material). An additional control used membranes heated to 100°C for 2 min to denature the Dol-P-Glc-dependent glucosyltransferase.
Reactions were stopped with 4 ml of chloroform-methanol (3/2), and the combined pellets from 20 reactions were delipidated four times with 10 ml of chloroform-methanol (3/2), four times with 10 ml of chloroform-methanol-water (10/10/3), and four times with 10 ml of solvent E, which contains water-ethanol-diethylether-pyridine-NH4OH (15/15/5/10/0.017) (29). The protein pellet was subjected to mild acid hydrolysis (40 mM trifluoroacetic acid treatment for 10 min at 90°C), PNGase F treatment, overnight digestion with α-amylase from Bacillus subtilis (Fluka), and several washes with 50% methanol. Finally, O-glycans were released by alkali-induced β-elimination in the presence of sodium [3H]borotritide.
The labeled reduced O-glycans were separated by Bio-Gel P-2 chromatography (110- by 1-cm column) in 0.1 M acetic acid-1% butanol. Fractions of 400 μl were collected, and aliquots were taken for scintillation counting. Peak fractions were recovered and dried. To determine whether Dol-P-Glc is a donor for the reduced sugar in the O-glycan chain, samples were taken from each peak and hydrolyzed in 2 M HCl for 2 h at 90°C. After the samples were cooled on ice, acid was removed by evaporation. The radiolabeled sugars were chromatographed in a Dionex instrument, using a CarboPac MA1 column and hexose and alditol standards, as described previously (33).
Nucleotide sequence accession numbers.
Nucleotide sequences for TvDPM1 and derived amino acid sequences have been submitted to GenBank with accession number EU477838. All other Trichomonas ALG5 genes have been previously annotated (see Table S1 in the supplemental material).
RESULTS AND DISCUSSION
Trichomonas membranes have ∼300 times the Dol-P-Glc synthase activity of Saccharomyces cerevisiae membranes.
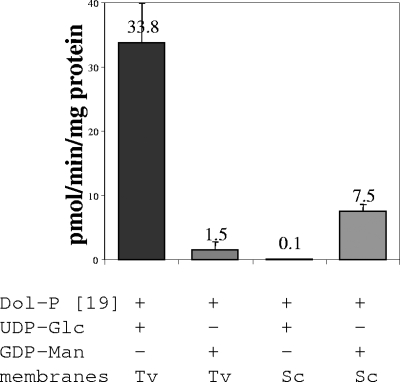
Trichomonas membranes have ∼300 times the Dol-P-Glc synthase activity of membranes prepared from Saccharomyces (Fig. 1). The Dol-P-Glc synthase activity of Trichomonas membranes is ∼20 times the Dol-P-Man synthase activity of Trichomonas membranes, which was quite variable from experiment to experiment (Fig. 1). In contrast, the Dol-P-Man synthase activity of Trichomonas membranes is one-fifth that of Saccharomyces membranes, which in turn is ∼75 times the Dol-P-Glc synthase activity of Saccharomyces membranes. In summary, Trichomonas membranes have a very high Dol-P-Glc synthase activity and a somewhat low Dol-P-Man synthase activity relative to Saccharomyces membranes.
FIG. 1.
Dol-P-Glc and Dol-P-Man synthase activities of Trichomonas and Saccharomyces membranes. Error bars represent standard deviations of three experiments. Membranes isolated from Trichomonas (Tv) and Saccharomyces (Sc) were incubated with exogenous Dol-P containing 19 isoprene units and either radiolabeled UDP-Glc or GDP-Man. Radiolabeled glycolipids, which partitioned into the organic fraction, were counted and confirmed by TLC in Fig. 2A. Conditions for these assays were determined by varying the pH and the concentration of Trichomonas membranes (see Fig. S1 in the supplemental material).
Endogenous Dol-P-hexoses of Trichomonas contain 16 isoprene units.
Dolichol chain length, which is determined by properties of the cis-prenyltransferase that catalyzes the formation of the dehydrodolichol pyrophosphate precursor, varies among the eukaryotes examined (16). Dolichols of mammals contain 15 to 23 isoprene units, those of Saccharomyces cerevisiae in logarithmic growth phase contain 14 to 18 isoprene units, and those of protists (Plasmodium, Trypanosoma, and Leishmania spp.) contain 11 or 12 isoprene units (2, 6, 19, 31).
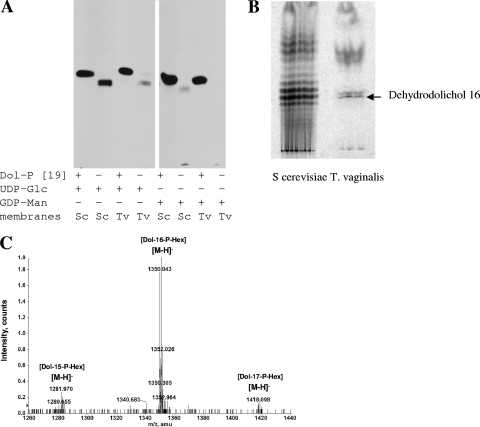
The length of endogenous dolichol of Trichomonas vaginalis was estimated in three ways. First, Dol-P-Glc made from endogenous Dol-P of Trichomonas has the same mobility in TLC as Saccharomyces Dol-P-Glc, which is made from an endogenous Dol-P that predominantly contains 16 isoprenoid units (Fig. 2A). In contrast, on the basis of Rfs, Dol-P-Glc made from endogenous Trichomonas dolichol runs more slowly than that made with addition of exogenous Dol-P containing 19 isoprenoid units.
FIG. 2.
Characterization of the dolichol portion of Trichomonas Dol-P-hexoses. (A) Dol-P-Glc and Dol-P-Man synthase assays were performed as described for Fig. 1, except that (i) exogenous Dol-P was omitted from some of the reactions and (ii) Saccharomyces strains used for Dol-P-Glc synthase assays overexpressed ScALG5. Radiolabeled Dol-P-Glc and Dol-P-Man from Trichomonas (Tv) and Saccharomyces (Sc) membranes were extracted and analyzed by TLC, where the faster-migrating species contain longer dolichol chains. (B) Reverse-phase TLC of dehydrodolichols made by membranes of Saccharomyces and Trichomonas, which were incubated with radiolabeled isopentenyl pyrophosphate and exogenous farnesyl pyrophosphate (in vitro cis-prenyltransferase assay). (C) Mass spectrum of Dol-P-hexose isolated from cultured Trichomonas, showing a major ion peak ([M-H]−) with 16 isoprene units and minor peaks with 15 and 17 isoprene units.
Second, dehydrodolichols produced in an in vitro assay of cis-prenyltransferase, in which Trichomonas membranes were incubated with radiolabeled isopentenyl pyrophosphate and exogenous farnesyl pyrophosphate, contain 15 or 16 isoprenoid units, like the predominant species of Saccharomyces (Fig. 2B).
Third, mass spectrometry showed that the predominant Trichomonas Dol-P-hexose, extracted from cultured parasites and fractionated on the acetate form of a DEAE-cellulose column, contains 16 isoprene units. As shown in Fig. 2C, the observed [M-H]− ion at m/z 1,350.043 (monoisotopic peak) is consistent with the expected value of 1,350.040 for the [M-H]− ion of the Dol-P-hexose with 16 isoprene units.
Five paralogous Trichomonas Alg5s have strong Dol-P-Glc synthase activities in transformed Saccharomyces.
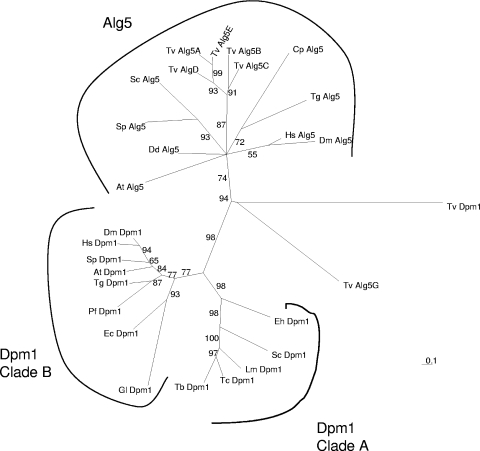
Phylogenetic analyses of Alg5 and Dpm1 demonstrate three clades, which include Alg5s that have an N-terminal signal anchor, clade A Dpm1s that have a C-terminal transmembrane helix, and clade B Dpm1s that lack a C-terminal transmembrane helix and are part of a multisubunit Dol-P-Man synthase complex (22). Trichomonas has five putative ALG5 genes (TvALG5A to TvALG5E), which encode proteins with N-terminal signal anchors that are part of the large Alg5 clade (Fig. 3; see Table S1 and Fig. S3 in the supplemental material). The five predicted Trichomonas Alg5 proteins are 325 to 337 amino acids long and show 52 to 80% identity to each other. Because these Trichomonas Alg5 proteins are much more similar to each other than to Alg5 proteins of other eukaryotes, they appear to be the product of numerous gene duplications after Trichomonas branched from the main eukaryotic tree.
FIG. 3.
Phylogenetic reconstruction using the maximum likelihood method of representative Dol-P-Glc synthases (Alg5s) and Dol-P-Man synthases (Dpm1s). Branch lengths are proportionate to differences between sequences, and numbers at nodes indicate bootstrap values for 100 replicates. TvAlg5A to TvAlg5E, which result from gene duplications in Trichomonas, belong to the Alg5 clade of other protists, fungi, and metazoa. TvAlg5G and TvDPM1 belong to no clade, while eukaryotic Dpm1s are split between two groups with (clade A) and without (clade B) a C-terminal transmembrane helix. In addition to Trichomonas vaginalis (Tv), protists include Cryptosporidium parvum (Cp), Plasmodium falciparum (Pf), Toxoplasma gondii (Tg), Leishmania major (Lm), Trypanosoma cruzi (Tc), Trypanosoma brucei (Tb), Dictyostelium discoideum (Dd), and Giardia lamblia (Gl). Fungi include Saccharomyces cerevisiae (Sc), Schizosaccharomyces pombe (Sp), and Encephalitozoon cuniculi (Ec), while metazoa include Homo sapiens (Hs) and Drosophila melanogaster (Dm). Arabidopsis thaliana (At) is the single plant.
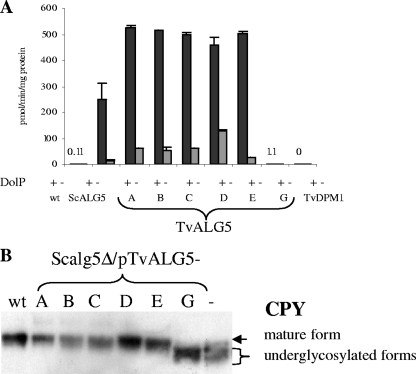
All five of these Trichomonas ALG5 genes complement a Saccharomyces alg5Δ mutant (Fig. 4A). In each case, the Dol-P-Glc synthase activity of the membranes from the Saccharomyces alg5Δ mutant transformed with TvALG5A to TvALG5E genes is greater than that of the wild-type Saccharomyces or that of the alg5Δ mutant expressing ScALG5 on the same promoter (Fig. 4A). The recombinant Trichomonas Dol-P-Glc synthases, which are encoded by TvALG5A to TvALG5E, also correct a carboxypeptidase Y N glycosylation defect in the Saccharomyces alg5Δ mutant in vivo (Fig. 4B).
FIG. 4.
Complementation of Saccharomyces alg5Δ mutant with ALG5 genes of Trichomonas. (A) Dol-P-Glc synthase activities with (black bars) or without (gray bars) exogenous Dol-P19 of wild-type (wt) Saccharomyces, as well as a Saccharomyces alg5Δ mutant transformed with the ScALG5 gene and the TvALG5A to TvALG5E, TvALG5G, and TvDPM1 genes. The Dol-P-Glc activity of wild-type Saccharomyces is difficult to detect, as is the Dol-P-Glc synthase activity of the Saccharomyces alg5Δ mutant transformed with TvALG5G. (B) N glycosylation of carboxypeptidase Y (CPY) by the same set of transformants. In Saccharomyces, Dol-P-Glc is used to add three Glc residues to the N-glycan precursor, which is transferred to the nascent peptide. In the absence of Dol-P-Glc synthase activity (TvALG5G and empty vector), the N-glycan precursor lacking Glc is less efficiently transferred to glycoproteins, so CPY, which is detected with an anti-CPY monoclonal antibody, is underglycosylated and runs more quickly on SDS-polyacrylamide gel electrophoresis.
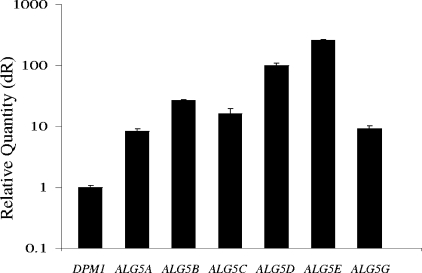
While mRNAs for all five Trichomonas Dol-P-Glc synthase genes (TvALG5A to TvALG5E) were detected by reverse transcription-PCR with total RNA from Trichomonas trophozoites, those of TvALG5D and TvALG5E were by far the most abundant (Fig. 5). These results suggest that the abundant Dol-P-Glc synthase activity of Trichomonas membranes is caused by expression of multiple TvALG5A to TvALG5E gene products.
FIG. 5.
Use of real time PCR to estimate the relative abundances of ALG5 and DPM1 mRNAs in cultured Trichomonas (see Table S1 in the supplemental material for the list of primers). A log scale shows that the TvALG5D and TvALG5E mRNAs are much more abundant than those of TvALG5A to -C and TvALG5G, which in turn are much more abundant than that of TvDPM1. Calibrations were relative to a Trichomonas actin gene.
A deeply divergent Trichomonas DPM1 gene product has Dol-P-Man synthase activity in vitro.
Trichomonas has two other genes (TvDPM1 and TvALG5G) that encode deeply divergent proteins, which are not members of the Alg5 clade or Dpm1 clades A or B (Fig. 3; see Fig. S3 in the supplemental material). TvALG5G encodes a 334-amino-acid protein that shows 19 to 24% identify to those encoded by TvALG5A to TvALG5E. TvAlg5G has very weak Dol-P-Glc activity in the transformed Saccharomyces alg5Δ mutant (Fig. 4A), and TvAlg5G does not correct the carboxypeptidase N glycosylation defect (Fig. 4B). These results make it unclear what the function of TvAlg5G is in vivo.
The TvDPM1 gene, which was named for the activity of its recombinant protein in E. coli, was identified based upon a weak identity with TvALG5G (see Fig. S3 in the supplemental material). The TvDPM1 gene, which was reconstructed from two overlapping protein predictions in the NCBI database, encodes a 345-amino-acid protein with a C-terminal transmembrane helix. With the exception of TvAlg5G, TvDpm1 aligns poorly with all the other eukaryotic Dpm1s and Alg5s (see Fig. S3 in the supplemental material).
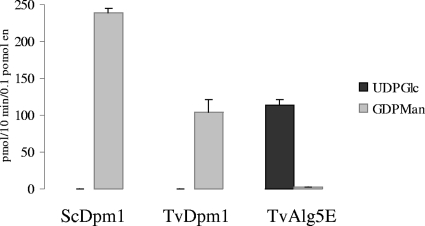
TvDpm1 shows Dol-P-Man synthase activity when expressed as a recombinant protein in E. coli (Fig. 6). The TvDPM1 gene product is slightly less active than Saccharomyces Dol-P-Man synthase (ScDPM1 gene product) expressed in E. coli, and its activity is similar to the Dol-P-Glc synthase activity of the TvALG5E gene product expressed in E. coli (Fig. 6). The Trichomonas DPM1 gene product then is the most deeply divergent Dol-P-Man synthase identified to date. The TvDPM1 mRNAs are much less abundant than those of the Trichomonas ALG5s (Fig. 5), which is consistent with the variable Dol-P-Man synthase activity of Trichomonas membranes in Fig. 1.
FIG. 6.
Dol-P-Man synthase of TvDpm1 expressed as a recombinant protein in E. coli. E. coli cells were transformed with ScDPM1, TvDPM1, and TvALG5E; recombinant proteins were purified; and their Dol-P-Glc synthase and Dol-P-Man synthase activities were determined by addition of radiolabeled UDP-Glc or GDP-Man, respectively.
Dol-P-Glc is incorporated into reducing and nonreducing sugars of Trichomonas protein O-glycans.
Because Trichomonas makes an N-glycan precursor that is not glucosylated (Man5GlcNAc2-PP-dolichol) (10, 25), it is not clear what Dol-P-Glc is used for in Trichomonas trophozoites. Trichomonas membranes incubated with radiolabeled Dol-P-Glc incorporate radiolabeled Glc into glycolipids and glycoproteins. Control reactions, which demonstrate the dependence of this reaction on pH, on time, and on temperature, are shown in Fig. S2 in the supplemental material. Radiolabeled Glc, which is present in trace amounts in glycolipids extracted in chloroform-methanol-water (10/10/3), is also present in solution E extracts that contain Trichomonas LPG (3, 8, 29). However, the counts were so small that we were unable to further characterize radiolabeled Glc from Dol-P-Glc in the Trichomonas LPG fraction.
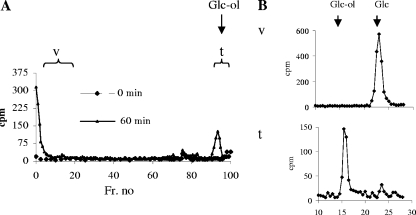
Radiolabeled Glc, which is associated with the glycoprotein pellet isolated from Trichomonas membranes incubated with Dol-P-Glc, is not released by PNGase F or mild acid but is released by β-elimination. This result suggests that Dol-P-Glc is used for synthesis of glycoproteins with O-glycans but not those with N-glycans or O-phosphodiester-linked glycans. On a Bio-Gel P-2 sizing column, glycans containing radiolabeled Glc from the Dol-P-Glc incubation with Trichomonas membranes run in the void (large sugars) or near the total volume (single sugar) (Fig. 7A). Large sugars are predominantly composed of Glc, while the single sugar is glucitol (the reducing sugar) (Fig. 7B). As far as we know, this is the first time that Dol-P-Glc has been shown to be involved in the synthesis of O-glycans on glycoproteins.
FIG. 7.
Characterization of O-glycans attached to endogenous glycoproteins when radiolabeled Dol-P-Glc is added to Trichomonas membranes. Control reactions, which demonstrate the dependence of this reaction on pH, on time, and on temperature, are shown in Fig. S2 in the supplemental material. (A) Radiolabeled glycans released by β-elimination from Trichomonas glycoproteins after incubation of membranes with radiolabeled Dol-P-Glc run with the void (v) volume and total (t) volume of a Bio-Gel P-2 column. (B) Monosaccharide analysis of the radiolabeled O-glycans shows that the large glycan running at the void volume is predominantly Glc, while the single sugar running with the total volume is glucitol (the reducing sugar).
Major conclusions and unresolved questions.
Despite the fact that Trichomonas has no apparent need for either Dol-P-Glc or Dol-P-Man, Trichomonas membranes have synthase activities for both activated sugars. Trichomonas has many ALG5 genes, which are active in transformed Saccharomyces and likely account for the unusually large amount of Dol-P-Glc synthase activity of Trichomonas membranes. Conversely, Trichomonas has a deeply divergent DPM1 gene, which encodes a Dol-P-Man synthase that likely is responsible for the Dol-P-Man synthase activity of Trichomonas membranes.
While we have not yet identified the uses for Dol-P-Man by Trichomonas, Dol-P-Glc may be used to make O-linked glycans in Trichomonas glycoproteins. This is similar to the use of Dol-P-Man by protein mannosyltransferases (PMTs) of yeast and protein O-mannosyltransferases (POMTs) of metazoa to make glycoproteins containing O-linked mannose (20, 35). Remarkably, Trichomonas has >30 genes which encode PMT- or POMT-like proteins that might use Dol-P-Glc or Dol-P-Man to make protein O-linked glycans (4).
While the use Dol-P-Glc to glucosylate N-glycan precursors in the lumen of the ER has been known for a long time (9-11), this is the first report of the use of Dol-P-Glc in the synthesis of O-linked glycans of glycoproteins. Dol-P-Glc may also be used to make ceramide (30). We speculate that Trichomonas will not be the only organism that uses Dol-P-Glc to make O-glycans on glycoproteins, and we do not rule out the possibility that Trichomonas uses Dol-P-Glc or Dol-P-Man to make glycolipids or LPG.
Supplementary Material
Acknowledgments
This work was supported in part by NIH grants AI48082 (J.S.) and GM31318 (P.W.R.). Z.G. and the mass spectrometry facility in the Department of Biochemistry, Duke University Medical Center, were supported by a LIPID MAPS glue grant (GM-069338) from the National Institutes of Health.
We are grateful to Ewa Swiezewska (Institute of Biochemistry and Biophysics, PAS, Warsaw, Poland) for providing dolichyl phosphate and Paula Magnelli (Boston University) for help with high-pH anion-exchange chromatography.
Footnotes
Published ahead of print on 13 June 2008.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arruda, D. C., F. L. D'Alexandri, A. M. Katzin, and S. L. Uliana. 2005. Antileishmanial activity of the terpene nerolidol. Antimicrob. Agents Chemother. 491679-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bastida-Corcuera, F. D., C. Y. Okumura, A. Colocoussi, and P. J. Jonson. 2005. Trichomonas vaginalis lipophosphoglycan mutants have reduced adherence and cytotoxicity to human ectocervical cells. Eukaryot. Cell 41951-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlton, J. M., R. P. Hirt, J. C. Silva, A. L. Delcher, M. Schatz, Q. Zhao, et al. 2007. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 315207-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark, C. G., and L. S. Diamond. 2002. Methods for cultivation of luminal parasitic protists of clinical importance. Clin. Microbiol. Rev. 15329-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Couto, A. C., E. A. Kimura, V. J. Peres, M. L. Uhrig, and A. M. Katzin. 1999. Active isoprenoid pathway in the intra-erythrocytic stages of Plasmodium falciparum: presence of dolichols of 11 and 12 isoprene units. Biochem. J. 341629-637. [PMC free article] [PubMed] [Google Scholar]
- 7.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 321792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fichorova, R. N., R. T. Trifonova, R. O. Gilbert, C. E. Costello, G. R. Hayes, J. J. Lucas, and B. N. Singh. 2006. Trichomonas vaginalis lipophosphoglycan triggers a selective upregulation of cytokines by human female reproductive tract epithelial cells. Infect. Immun. 745773-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heesen, S., L. Lehle, A. Weissmann, and M. Aebi. 1994. Isolation of the ALG5 locus encoding the UDP-glucose:dolichyl-phosphate glucosyltransferase from Saccharomyces cerevisiae. Eur. J. Biochem. 22471-79. [DOI] [PubMed] [Google Scholar]
- 10.Helenius, A., and M. Aebi. 2004. Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 731019-1049. [DOI] [PubMed] [Google Scholar]
- 11.Huffaker, T., and P. W. Robbins. 1983. Yeast mutants deficient in protein glycosylation. Proc. Natl. Acad. Sci. USA 807466-7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8275-282. [DOI] [PubMed] [Google Scholar]
- 13.Käll, L., A. Krogh, and E. L. L. Sonnhammer. 2004. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 3381027-1036. [DOI] [PubMed] [Google Scholar]
- 14.Kanjilal-Kolar, S., and C. R. Raetz. 2006. Dodecaprenyl phosphate-galacturonic acid as a donor substrate for lipopolysaccharide core glycosylation in Rhizobium leguminosarum. J. Biol. Chem. 28112879-12887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelleher, D. J., and R. Gilmore. 2006, An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 1647R-62R. [DOI] [PubMed] [Google Scholar]
- 16.Kharel, Y., S. Takahashi, S. Yamashita, and T. Koyama. 2006. Manipulation of prenyl chain length determination mechanism of cis-prenyltransferases. FEBS J. 273647-657. [DOI] [PubMed] [Google Scholar]
- 17.Kruszewska, J. S., A. H. Butterweck, W. Kurzatkowski, A. Migdalski, C. P. Kubicek, and G. Palamarczyk. 1999. Overexpression of the Saccharomyces cerevisiae mannosylphosphodolichol synthase-encoding gene in Trichoderma reesei results in an increased level of protein secretion and abnormal cell ultrastructure. Appl. Environ. Microbiol. 652382-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 232947-2948. [DOI] [PubMed] [Google Scholar]
- 19.Low, P., G. Dallner, S. Mayor, S. Cohen, B. T. Chait, and A. K. Menon. 1991. The mevalonate pathway in the bloodstream form of Trypanosoma brucei. Identification of dolichols containing 11 and 12 isoprene residues. J. Biol. Chem. 26619250-19257. [PubMed] [Google Scholar]
- 20.Manya, H., A. Chiba, A. Yoshida, X. Wang, Y. Chiba, Y. Jigami, R. Margolis, and T. Endo. 2004. Demonstration of mammalian protein O-mannosyltransferase activity: coexpression of POMT1 and POMT2 required for enzymatic activity. Proc. Natl. Acad. Sci. USA 101500-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mewes, H. W., K. Albermann, M. Bahr, D. Frishman, A. Gleissner, J. Hani, K. Heumann, K. Kleine, A. Maierl, S. G. Oliver, F. Pfeiffer, and A. Zollner. 1997. Overview of the yeast genome. Nature 3877-65. [DOI] [PubMed] [Google Scholar]
- 22.Oriol, R., I. Martinez-Duncker, I. Chantret, R. Mollicone, and P. Codogno. 2002. Common origin and evolution of glycosyltransferases using Dol-P-monosaccharides as donor substrate. Mol. Biol. Evol. 191451-1463. [DOI] [PubMed] [Google Scholar]
- 23.Orlean, P., C. Albright, and P. W. Robbins. 1988. Cloning and sequencing of the yeast gene for dolichol phosphate mannose synthase, an essential protein. J. Biol. Chem. 26317499-17507. [PubMed] [Google Scholar]
- 24.Orlean, P., and A. K. Menon. 2007. Thematic review series: lipid posttranslational modifications. GPI anchoring of protein in yeast and mammalian cells, or: how we learned to stop worrying and love glycophospholipids. J. Lipid Res. 48993-1011. [DOI] [PubMed] [Google Scholar]
- 25.Samuelson, J., S. Banerjee, P. Magnelli, J. Cui, D. J. Kelleher, R. Gilmore, and P. W. Robbins. 2005. The diversity of protist and fungal dolichol-linked precursors to Asn-linked glycans likely results from secondary loss of sets of glycosyltransferases. Proc. Natl. Acad. Sci. USA 1021548-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sauer, N., and J. Stolz. 1994. SUC1 and SUC2: two sucrose transporters from Arabidopsis thaliana; expression and characterization in baker's yeast and identification of the histidine-tagged protein. Plant J. 667-77. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt, H. A., K. Strimmer, M. Vingron, and A. von Haeseler. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18502-504. [DOI] [PubMed] [Google Scholar]
- 28.Schwebke, J. R., and D. Burgess. 2004. Trichomoniasis. Clin. Microbiol. Rev. 17794-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh, B. N. 1993. Lipophosphoglycan-like glycoconjugate of Trichomonas foetus and Trichomonas vaginalis. Mol. Biochem. Parasitol. 57281-294. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki, Y., C. P. Ecker, and H. A. Blough. 1984. Enzymatic glucosylation of dolichol monophosphate and transfer of glucose from isolated dolichyl-D-glucosyl phosphate to ceramides by BHK-21 cell microsomes. Eur. J. Biochem. 143447-453. [DOI] [PubMed] [Google Scholar]
- 31.Swiezewska, E., and W. Danikiewicz. 2005. Polyisoprenoids: structure, biosynthesis and function. Prog. Lipid Res. 44235-258. [DOI] [PubMed] [Google Scholar]
- 32.Szkopinska, A., K. Grabinska, D. Delourme, F. Karst, J. Rytka, and G. Palamarczyk. 1997. Polyprenol formation in the yeast Saccharomyces cerevisiae: effect of farnesyl diphosphate synthase overexpression. J. Lipid Res. 38962-968. [PubMed] [Google Scholar]
- 33.Van Dellen, K. L., A. Chatterjee, D. M. Ratner, P. E. Magnelli, J. F. Cipollo, M. Steffen, P. W. Robbins, and J. Samuelson. 2006. Unique posttranslational modifications of chitin-binding lectins of Entamoeba invadens cyst walls. Eukaryot. Cell 5836-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Der Pol, B., C. Kwok, B. Pierre-Louis, A. Rinaldi, R. A. Salata, P. L. Chen, J. van de Wijgert, F. Mmiro, R. Mugerwa, T. Chipato, and C. S. Morrison. 2008. Trichomonas vaginalis infection and human immunodeficiency virus acquisition in African women. J. Infect. Dis. 197548-554. [DOI] [PubMed] [Google Scholar]
- 35.Willer, T., M. Brandle, M. Sipiczki, and S. Strahl. 2005. Protein O-mannosylation is crucial for cell wall integrity, septation and viability in fission yeast. Mol. Microbiol. 57156-170. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.