Abstract
Glc7, the type1 serine/threonine phosphatase in the yeast Saccharomyces cerevisiae, is targeted by auxiliary subunits to numerous locations in the cell, where it regulates a range of physiological pathways. We show here that the accumulation of Glc7 at mating projections requires Afr1, a protein required for the formation of normal projections. AFR1-null mutants fail to target Glc7 to projections, and an Afr1 variant specifically defective in binding to Glc7 [Afr1(V546A F548A)] forms aberrant projections. The septin filaments in mating projections of AFR1 mutants initiate normally but then rearrange asymmetrically as the projection develops, suggesting that the Afr1-Glc7 holoenzyme may regulate the maintenance of septin complexes during mating. These results demonstrate a previously unknown role for Afr1 in targeting Glc7 to mating projections and in regulating the septin architecture during mating.
How cells make appropriate morphological changes in response to external and internal cues is a central question in biology. The budding yeast Saccharomyces cerevisiae has been used extensively as a model to investigate the regulation of morphological changes. In dividing cells, growth is largely restricted to the bud, the site of which is determined by a complex set of internal cues that depend on cell type and the position of the previous bud (9). Once the site of polarization is established, a complex of septin proteins forms a ring at the polarization site through which new growth and cell wall expansion are directed. In mating competent cells, mating pheromones induce growth into mating projections rather than into buds. The site of polarization and the overall shape of the projection are determined both by internal cues and by a concentration gradient of pheromone (9). This program of directional growth allows cells of opposite mating types to fuse efficiently in order to form a zygote, which initiates the diploid vegetative cycle.
Septins, a collection of related structural proteins, play a central role in morphological regulation during both vegetative growth and the mating response. They are evolutionarily conserved and have physiological roles in cytokinesis, vesicle trafficking, and the secretory pathway in many eukaryotes (15, 30, 57). There are seven septin proteins in Saccharomyces cerevisiae, of which five, Cdc3, Cdc10, Cdc11, Cdc12, and Shs1, are expressed in vegetatively growing cells (42). Four of these, Cdc3, Cdc10, Cdc11, and Cdc12, form stoichiometric complexes in vivo and in vitro (22, 59, 60). Septins form a ring at the site of polarization prior to bud formation. The ring transforms into an hourglass-shaped collar at bud initiation and separates into two rings prior to cytokinesis (reviewed in reference 42). This collar acts both as a diffusion barrier to separate membrane-associated proteins in the mother cell from those in the bud (3, 16, 56) and as a scaffold for proteins that direct chitin synthesis, actin nucleation, cell cycle control, and other signaling pathways (reviewed in reference 27). The precise structure of septin complexes at the bud neck has been controversial. Filaments running either parallel or perpendicular to the mother-daughter axis have been proposed for structures at the neck (reviewed in reference 61), and recent electron microscopic studies suggest a more elaborate architecture made up of filaments that form a gauze- or net-like complex (46). Recent experiments with septin-green fluorescent protein (GFP) fusion proteins using polarized fluorescence microscopy indicate that septin filaments run parallel to the mother-daughter axis throughout most of the cell cycle but flip to become circumferential when the collar splits at cytokinesis (62).
Septins acquire a different morphology during mating. Rather than a well-formed ring or collar, septins form a diffuse collar at the base of the projection that often appears as bars or filaments that run parallel to the axis of the projection (20, 34). Loss of septin function due to incubation of a temperature-sensitive septin mutant at the restrictive temperature results in a serious defect in projection formation (25). Although a large number of septin-binding proteins have been identified at the bud neck in vegetatively growing cells (27), much less is known about their occurrence and function in mating projections. One septin binding protein present in mating projections is Afr1, first identified from a screen for genes that induce resistance to mating pheromone when expressed at high dosages (37). Afr1 is induced during the mating response and accumulates at mating projections in a septin-dependent manner (37, 38). A two-hybrid interaction between Afr1 and the septin Cdc12 suggests that Afr1 binds directly to septins (38). Mating projections in AFR1-null mutants are malformed, but the role of Afr1 in this process is not known.
This study was motivated by the observation that protein phosphatase type 1 (PP1) (Glc7 in yeast) is localized to mating projections in a septin-dependent manner (5). Glc7 is a highly conserved phosphatase with physiological roles ranging from the activation of glycogen biosynthesis to chromosome segregation (reviewed in reference 53). The activity of Glc7 is regulated by targeting/regulatory subunits that direct the phosphatase to a specific location and otherwise regulate its activity. We show here that Afr1 is responsible for targeting Glc7 to mating projections. Furthermore, AFR1-null mutants show aberrant septin structures at the base of mating projections. These results and other data suggest that Afr1, along with Glc7, is required for the maintenance of septin architecture during mating.
MATERIALS AND METHODS
Yeast strains, media, and reagents.
The yeast strains used in this work are listed in Table 1 and are congenic to JC482 (8) except for strain CTY10-5d (19) (a gift from R. Sternglanz, State University of New York, Stony Brook). Yeast strains were grown at 30°C on YPD medium (2% Bacto peptone, 1% yeast extract, and 2% glucose) or synthetic complete (SC) medium unless stated otherwise. Diploid strains were sporulated at 24°C on medium containing 2% Bacto peptone, 1% yeast extract, and 2% potassium acetate. SC medium and media lacking specific amino acids were made as described by Sherman et al. (51). Yeast transformations, manipulation of Escherichia coli, and the preparation of bacterial growth media were performed as described previously (24, 33, 48). Mating projections were induced by incubating cells in SC medium containing 10−7 M α-factor (Sigma Chemical Co.) for 2 h at 30°C unless stated otherwise.
TABLE 1.
Yeast strains
| Strain | Genotype | Source or reference |
|---|---|---|
| CTY10-5d | MATaade2 trp1-901 leu2-3,112 his3-200 gal4 URA3::lexA-lacZ | R. Sternglanz |
| KT1112 | MATaleu2 ura3-52 his3 | 54 |
| KT1113 | MATα leu2 ura3-52 his3 | 23 |
| KT1193 | MATaleu2 ura3-52 his3 bar1-1 | This study |
| KT1357 | MATaleu2 ura3-52 his3 trp1-1 | 5 |
| KT1358 | MATα leu2 ura3-52 his3 trp1-1 | 5 |
| KT2792 | MATaleu2 ura3-52 his3 bar1-1 afr1VA/FA-EmCitrine::SpHis5::URA3::afr1::KanMX4 GLC7-mCherry::SpHis5 | This study |
| KT2793 | MATaleu2 ura3-52 his3 bar1-1 AFR1-EmCitrine::SpHis5::URA3::afr1::KanMX4 GLC7-mCherry::SpHis5 | This study |
| KT2794 | MATaleu2 ura3-52 his3 bar1-1 AFR1-13Myc::SpHis5 GLC7-mCherry::SpHis5 | This study |
| KT2795 | MATaleu2 ura3-52 his3 bar1-1 afr1-Δ545-13Myc::SpHis5 GLC7-mCherry::SpHis5 | This study |
| JB66-2D | MATaleu2 ura3-52 his3 bar1-1 afr1::KanMX4 | This study |
| JB75-11C | MATaleu2 ura3-52 his3 bar1-1 ura3::GFP-GLC7::URA3 | This study |
| JB75-12D | MATaleu2 ura3-52 his3 bar1-1 afr1::KanMX4 ura3::GFP-GLC7::URA3 | This study |
| JB142-9C | MATaleu2 ura3-52 his3 bar1-1 afr1::KanMX4:AFR1:URA3 ura3::GFP-GLC7::URA3 | This study |
| JB144-9C | MATaleu2 ura3-52 his3 bar1-1 afr1::KanMX4:afr1VA/FA::URA3 ura3::GFP-GLC7::URA3 | This study |
| JB173-12B | MATaleu2 ura3-52 his3 bar1-1 bni4Δ::TRP1 ura3::GFP-GLC7::URA3 | This study |
| JB229-2B | MATaleu2 ura3-52 his3 bar1-1 GLC7-EmCFP::SpHIS5 | This study |
| JB241-5B | MATaleu2 ura3-52 his3 bar1-1 GLC7-EmCFP::SpHIS5 AFR1-EmCitrine::SpHIS5 | This study |
| JB241-19A | MATaleu2 ura3-52 his3 bar1-1 AFR1-EmCitrine::SpHIS5 | This study |
| JB250-12B | MATaleu2 ura3-52 his3 bar1-1 CDC10-EmCFP::SpHIS5 | This study |
| JB251-10B | MATaleu2 ura3-52 his3 bar1--1 CDC10-EmCFP::SpHIS5 AFR1-EmCitrine::SpHIS5 | This study |
| JB254-17C | MATaleu2 ura3-52 his3 bar1-1 CDC10-EmCFP::SpHIS5 afr1::KanMX4 | This study |
| JB254-19C | MATaleu2 ura3-52 his3 bar1-1 CDC10-EmCFP::SpHIS5 | This study |
| JB255-8A | MATaleu2 ura3-52 his3 bar1-1 CDC10-EmCFP::HIS5 afr1::KanMX4::afr1VA/FA::URA3 | This study |
To generate gene deletion, 13Myc-tagged, and GFP fusion strains by PCR, each amplified cassette was transformed into a KT1112 × KT1113 or KT1357 × KT1358 diploid strain, unless otherwise noted. Transformants were sporulated, and haploid meiotic segregants were isolated by tetrad analysis. Primers are listed in Table 2. Deletion cassettes were amplified using afr1::KanMX4 and primers JB25-F and JB26-R, yer158c::KanMX6 and primers JB50-F and JB51-R, and DNA from the strains from the Research Genetics deletion panel (64). bni4::KanMX6 was made as previously described (40). Nourseothricin (Nat)-resistant strains were created by digesting pAG25 (28) with EcoRI, transforming the fragment into the G418-resistant haploids, and selecting for growth on YPD medium containing 0.1 mg/ml nourseothricin (Werner BioAgents, Germany).
TABLE 2.
Primers used in this study
| Name | Sequence (5′-3′) |
|---|---|
| SP63-F | GGAAGTACACGATGATTCGCGATGTTACACACATTTTTATGGTGACGGTGCTGGTTTA |
| SP66-R | TGTATGATTTGATTCATTTCCATTTCTCCCAGTTTTCTGCTTCGATGAATTCGAGCTCG |
| JB25-F | CATTCGTAATTAGAGCACCAGC |
| JB26-R | GTACGTCTTATTTTTGATTATATG |
| JB45-F | TGTCCATGAGAACAGCAAGTGCTTTACACATTATTTAATTCGGATCCCCGGGTTAATTAA |
| JB46-R | CTATTGCAAAAAATAAACTGGATTATGTTCGTTAATAACGAATTCGAGCTCGTTTAAAC |
| JB47-F | TGATAAAGTCGAAACACCTCCAAAACCAATAAAAAAAGATCGGATCCCCGGGTTAATTAA |
| JB50-F | GTTAAGCAAGCATCCATCAATAA |
| JB51-R | CACCGCGTTGCGTTCGAGACACC |
| JB52-F | CCAATAAAAAAAGATGCCCGGGCTGCAAAAGAAGTATG |
| JB53-R | CATACTTCTTTTGCAGCCCGGGCATCTTTTTTTATTGG |
| JB63-F | CGTACGCGTCGACATGGAGGGCTCATATCTATC |
| JB64-R | CGCGCGGATCCTCAAATTAAATAATGTGTAAAGC |
| JB67-F | TGTCCATGAGAACAGCAAGTGCTTTACACATTATTTAATTGGTCGACGGATCCCCGGG |
| JB68-R | CTATTGCAAAAAATAAACTGGATTATGTTACGTTAATAACATCGATGAATTCGAGCTCG |
| JB77-F | TGTCCATGAGAACAGCAAGTGCTTTACACATTATTTAATTGGTGACGGTGCTGGTTTA |
All integrated 13Myc, GFP, cyan fluorescent protein (CFP), and yellow fluorescent protein (YFP) fusions were constructed according to the method described by the Yeast Resource Center (University of Washington, Seattle, WA; http://depts.washington.edu/yeastrc/), which is based on the method described by Wach et al. (62a), using pFA6a-13Myc-HIS3MX6 (43), pLK3 (39), pKT211, or pKT210 (50) as a template for 13Myc, GFP, mCitrine, and mCFP fusions, respectively. Construction of CDC10-yEmCFP and GLC7-yEmCitrine has been described previously (4, 40). The 13Myc integration cassettes were amplified using primers JB45-F and JB46-R for AFR1-13Myc and primers JB47-F and JB46-R for afr1-Δ546-13Myc. The fluorescent protein integration cassettes were amplified using primer JB67-F or JB77-F with JB68-R for AFR1-GFP and AFR1-yEmCitrine, respectively, and primers SP63-F and SP66-R for BNI4-yEmCitrine. To integrate wild-type AFR1 and afr1(V546A F548A) (referred to below as afr1VA/FA) at the AFR1 locus, strain JB66-2D was transformed with MscI-digested pJB13 or pJB14 (described below). These strains were then transformed with the AFR1-yEmCitrine PCR products to generate AFR1-yEmCitrine and afr1VA/FA-yEmCitrine, respectively. All deletions, truncations, and integrations were confirmed by genomic PCR or immunoblotting.
Site-directed mutagenesis and plasmid construction.
Plasmids are listed in Table 3. Standard techniques were used for DNA manipulation (48). Restriction and modification enzymes were used as recommended by the manufacturers (Promega, Madison, WI; New England Biolabs, Beverly, MA; Roche, Germany). The sequences of mutant alleles were confirmed at Retrogen Inc., San Diego, CA. The AFR1 gene was subcloned as a SalI-BamHI fragment from pLG146 (25) into pUC19 (66) to create pJB10. Site-directed mutagenesis of the VXF motif of AFR1 in pJB10 to V546A F548A (VA/FA) was carried out using the QuikChange kit (Stratagene, La Jolla, CA) and primers JB52-F and JB53-R, generating pJB11. For the yeast two-hybrid assay, the sequence encoding afr1VA/FA was amplified by PCR using primers JB63-F and JB64-R with pJB11 as the template and was cloned into pGEM-T (pGEM-T Easy vector systems kit; Promega) to generate pJB19. A SalI-NdeI fragment of afr1VA/FA from pJB19 was subcloned into plexA-AFR1 (38), replacing wild-type AFR1, to generate pJB20. SalI-BamHI fragments from pJB10 and pJB11 were cloned into pRS306 (52) to create pJB13 and pJB14, respectively.
TABLE 3.
Plasmids used in this study
| Name | Description | Source |
|---|---|---|
| pACTII | LEU2, 2μm two-hybrid vector with GAL4 activation domain | 18 |
| PBTM116-AFR1 | plexA-DBD-AFR1 | 38 |
| PBTM116-afr1-Δ474 | plexA-DBD-afr1-Δ474 | 25 |
| pJB20 | pBTM116-afr1VA/FA | This study |
| pHH148 | GLC7 in pACTII | This study |
| pCDV471 | GLC7 in pEG202 (ADH1-LexA-DBD) | 41 |
| pAR16 | pGADC1-BNI4 | 39 |
| pJK52 | YEplac181-AFR1 | 25 |
| YEp181-afr1-C7 | YEplac181-afr1-Δ474-620 | 25 |
| pRS425 | LEU2, 2μm vector | 10 |
Yeast two-hybrid assay.
The system used is based on the method of Fields and Song (19), modified to use a lexA DNA-binding domain fusion (7) to AFR1 (38) and GLC7 fused to the GAL4 activation domain in pACTII (65). The yeast strain CTY10-5d was cotransformed with two hybrid plasmids, and transformants were assayed for lacZ reporter gene activity by measuring β-galactosidase activity qualitatively on nitrocellulose filters or quantitatively using crude extracts as described elsewhere (32).
Biochemical procedures.
To assess Afr1-mCitrine and 13Myc-tagged protein levels, total protein was prepared from cultures treated with α-factor by lysis in trichloroacetic acid (12, 54). Unless otherwise stated, cells were treated with 10−7M α-factor (Sigma Chemical Co.) for 2 h. Proteins were electrophoresed on 8% polyacrylamide-sodium dodecyl sulfate gels. Immunoblot analysis was performed as described elsewhere (54) using the anti-Myc ascites antibody 9E10 or the BD Living Colors A.v. monoclonal anti-GFP antibody JL-8, with subsequent detection using the enhanced chemiluminescence system (Amersham ECL Plus; GE Healthcare, United Kingdom). Phosphoglycerate kinase (Pgk1) expression was used as a loading control.
Microscopy.
Cells were imaged for GFP, CFP, and YFP as described previously (39). Fluorescence resonance energy transfer (FRET) was calculated as FRETR, the relative increase of signal in the FRET channel over the background from spillover, and is described elsewhere (45) using a tutorial available at http://depts.washington.edu/yeastrc/pages/FRET_1.html. Indirect immunofluorescence was performed as described previously (39). Cells were stained with monoclonal anti-Myc antibody 9E10, followed by a tetramethyl rhodamine isocyante-conjugated goat anti-mouse secondary antibody (Sigma Chemical Co.), or with polyclonal anti-Cdc12 (kindly provided by M. Versele and J. Thorner, University of California, Berkeley) and an Alexa Fluor 488-conjugated anti-rabbit secondary antibody. Fluorescence levels along the sides of mating projections were quantitated as previously described (39) using the average of 4 adjacent pixels and subtracting the cytoplasmic signal from that at the edges of mating projections. The signal for the wild-type strain was set arbitrarily to 1.0. At least 100 cells of each genotype were assayed. For time lapse microscopy of mating projection formation, cells were grown to early-log phase in SC medium and placed on a pad of 2% agarose in SC medium containing 10−7 M α-factor.
RESULTS
Afr1 interacts with Glc7.
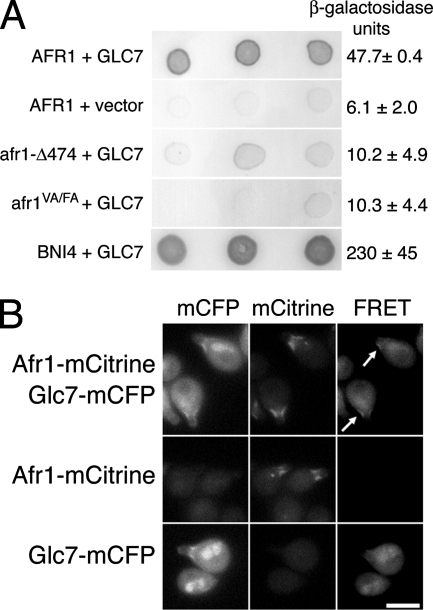
Glc7 localizes to the necks of mating projections (5), but its role in mating is unknown. A potential clue comes from the observation that Bni4, a Glc7-targeting subunit that is responsible for tethering Glc7 to the bud neck, is related to Afr1 and another protein, Yer158c (Fig. 1). Yer158c is ∼49% similar to Afr1 but does not appear to play a role in the mating response (14). The region of similarity between the yeast proteins also extends to the COOH-terminal region of a family of mammalian PP1 and β-actin binding proteins termed phactrs, for phosphatase and actin regulators (1, 47). The conserved domain in Bni4 and phactrs is necessary for binding Glc7 and PP1, respectively (1, 39). We confirmed an interaction between Afr1 and Glc7 using a lexA-based two-hybrid assay (Fig. 2A). Afr1 and Glc7 consistently show an interaction above the level for the negative controls.
FIG. 1.
ClustalW alignment of the amino acid sequences of the COOH termini of Afr1, Bni4, Yer158c, Candida albicans Bni4 (AAT73074), human phactr-3/scapinin (NM080672), and proteins of unknown function from Drosophila melanogaster (LD44321) and Caenorhabditis elegans (NP_492478). Identical residues are highlighted in black and similar residues in gray. The PP1 binding motif is amino acids 831 to 833 in Bni4.
FIG. 2.
Afr1 is a Glc7-targeting subunit. (A) Two-hybrid analysis of Afr1 and Glc7. Three representative transformants are shown for each interaction tested. Dark color reflects activation of the lacZ reporter gene. The plasmids used are as follows: for AFR1, pBTM116-AFR1; for afr1-Δ474, pBTM116-afr1-Δ474; for afr1VA/FA, pJB20; for GLC7, pHH148 or pCDV471 (used for BNI4-GLC7 two-hybrid analysis); vector, pACTII; for BNI4, pAR16. Units of β-galactosidase activity are reported on the right. (B) FRET analysis of Afr1 and Glc7. MATa strains JB241-5B (Afr1-mCitrine, Glc7-mCFP), JB241-19A (Afr1-mCitrine), and JB229-2B (Glc7-mCFP) were treated with 10−7 M α-factor for 2 h and imaged with CFP, YFP, and FRET filter sets. Bar, 5 μm. Arrows mark regions where FRET signal was observed.
FRET was used to confirm the Glc7-Afr1 interaction. Glc7 and Afr1 were tagged with codon-optimized monomeric variants of CFP (mCFP) and YFP (mCitrine), respectively (50). The fusions were integrated at the chromosomal loci for GLC7 and AFR1, and each is functional, as determined by the localization of each fusion to the necks of mating projections and by the ability of each fusion strain to grow at wild-type rates and with normal morphologies (data not shown). As expected, Afr1-mCitrine and Glc7-mCFP appeared to colocalize at the necks of mating projections (Fig. 2B). However, in some cells, Afr1-mCitrine also resided in a small spot that appeared to be in or near the nucleus, as judged by the location of the nuclear Glc7-mCFP (Fig. 2B). We do not know the significance of this spot or what, if any, cellular structure it is associated with, but Glc7 does not appear to colocalize with Afr1 in these spots. FRET between Afr1-mCitrine and Glc7-mCFP was measured as the increase in YFP emission after excitation of CFP using a metric defined by Muller et al. (45). To assay FRETR, strains containing one or both of the fluorescent protein fusions were treated with α-factor for 2 h prior to imaging. A FRETR signal at mating projections was observed only when Afr1-mCitrine and Glc7-mCFP were coexpressed (Fig. 2B). Quantitation of the FRETR signal between Afr1-mCitrine and Glc7-mCFP gave a value of 2.2 ± 0.5 (n = 22 cells) compared with 2.5 ± 0.7 (n = 15 cells) for Bni4-mCitrine and Glc7-mCFP, another pair of known interacting proteins (39). The relatively high FRETR value for Afr1-mCitrine and Glc7-mCFP indicates that these proteins are in close proximity, i.e., no more than 75 Å apart (49).
Afr1 contains a putative PP1/Glc7-binding consensus RVXF motif near its COOH terminus (Fig. 1). To determine if this motif is necessary for its interaction with Glc7, we constructed an AFR1 mutant, afr1(V546A F548A) (referred to below as afr1VA/FA), in which the conserved valine (V546) and phenylalanine (F548) residues in the consensus-binding site were changed to alanine. Afr1VA/FA and a previously constructed COOH-terminal deletion, Afr1-C7 (Afr1-Δ474) (25), fail to interact with Glc7 in the two-hybrid assay (Fig. 2A). These results indicate that Afr1, Bni4, mammalian phactrs, and related proteins in other eukaryotes contain a highly conserved COOH-terminal PP1 binding domain.
Afr1 is the limiting factor for Glc7 localization to the necks/bases of mating projections.
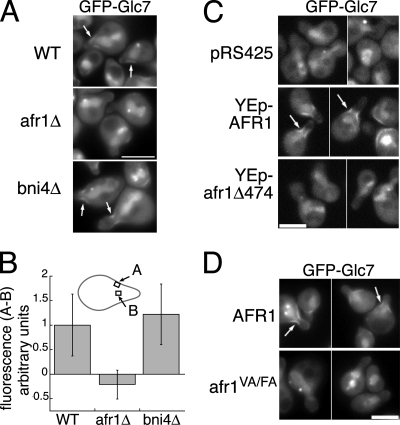
To determine whether Afr1 is necessary for the targeting of Glc7 to the necks of mating projections, GFP-Glc7 was imaged in strains treated with α-factor. GFP-Glc7 localizes to the sides of mating projections in wild-type cells but not to mating projections in an afr1Δ strain (Fig. 3A). As reported elsewhere (39), GFP-Glc7 localizes to mating projections in bni4-null mutants (Fig. 3A), confirming that Bni4 does not directly target Glc7 to mating projections, even though it contains a Glc7 binding domain related to Afr1 and binds to septins. To quantitate GFP-Glc7 levels along the sides of mating projections, we subtracted GFP fluorescence in the cytoplasm of projections from GFP fluorescence along the edge, as described in Materials and Methods. This value was set to 1.0 for wild-type cells. The fluorescence levels in the bni4Δ strain were similar to those found in wild-type cells but much greater than those in afr1Δ cells (Fig. 3B). When an afr1Δ strain was transformed with a high-copy-number plasmid encoding AFR1, we observed a 2.1-fold elevation in GFP-Glc7 levels at the sides of mating projections of such cells relative to those for wild-type cells (Fig. 3C) (n = 40 cells; P < 0.0001 by an unpaired Student t test). Consistent with the results of the two-hybrid assay between Glc7 and Afr1VA/FA (Fig. 2A), GFP-Glc7 does not associate with mating projections in strains expressing afr1VA/FA (Fig. 3D) or afr1-Δ474 (Fig. 3C). These results indicate that Afr1 is limiting for the recruitment of Glc7 to the necks of mating projections, and they confirm that the RVXF motif of Afr1 is required to target Glc7.
FIG. 3.
Afr1 targets Glc7 to mating projections. (A) GFP-Glc7 fails to localize to the necks of mating projections in afr1Δ cells. Wild-type (WT) (JB75-11C), bni4Δ (JB173-12B), and afr1Δ (JB75-12D) strains containing GFP-GLC7 were treated with 10−7 M α-factor for 2 h and imaged with a GFP filter set. (B) Quantitation of GFP-Glc7 fluorescence at the necks of mating projections in the strains listed in the legend to panel A. The diagram depicts where GFP signal was measured. (C) afr1Δ GFP-GLC7 (JB75-12D) cells carrying pRS425, pJK52, or afr1-C7 were treated as described above and imaged with a GFP filter set. (D) The RVXF motif is essential for Glc7 binding. afr1VA/FA GFP-GLC7 (JB144-9C) and control AFR1 GFP-GLC7 (JB142-9C) strains were treated as described above. Bars, 5 μm. Arrows mark GFP-Glc7 at mating projections.
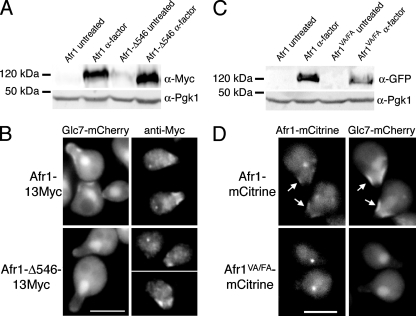
Bni4(V831A F833A), a variant of Bni4 that fails to interact with Glc7 due to mutation of the RVXF motif, accumulates at low levels at the neck in vegetative cells, indicating that Bni4 and Glc7 are at least partially interdependent for their localization to the neck (39). We tested for a similar interdependence of Afr1 and Glc7 in mating projections by assaying the localization of Afr1 proteins that are unable to interact with Glc7. We first assayed the location of a COOH-terminal truncation of AFR1 lacking residues 546 to 620 and tagged with 13Myc epitopes, referred to below as afr1-Δ546. As a control, the full-length Afr1 protein was also tagged with 13Myc. Immunoblot analysis and indirect immunofluorescence with the anti-Myc antibody 9E10 indicate that Afr1-Δ546-13Myc is expressed normally in the presence of α-factor (Fig. 4A) and accumulates at mating projections but fails to recruit Glc7 to the projections (Fig. 4B). We also visualized an Afr1VA/FA-mCitrine fusion in mating projections. AFR1-EmCitrine and afr1VA/FA-EmCitrine fusions were expressed from the AFR1 promoter at the AFR1 locus. As shown in Fig. 4C, Afr1-mCitrine and Afr1VA/FA-mCitrine are expressed at similar levels after α-factor induction. However, Afr1VA/FA-mCitrine accumulates to only low levels on mating projections (Fig. 4D). As noted for Afr1-mCitrine, a single fluorescent spot is observed in many Afr1VA/FA-mCitrine cells. This spot is brighter than that found in the Afr1-mCitrine-expressing cells. We do not know why Afr1-Δ546-13Myc localizes to mating projections but Afr1VA/FA-mCitrine does not. It is possible that the fluorescent protein tag on the C terminus of the Afr1VA/FA protein could influence localization. Nevertheless, these data indicate that Glc7 binding may be required for normal localization of Afr1 to mating projections.
FIG. 4.
Afr1 localization to the necks of projections may be partially dependent on Glc7. (A) Expression of 13Myc-tagged AFR1 confirmed by immunoblot analysis of extracts from AFR1-13Myc (KT2794) and afr1-Δ546-13Myc (KT2795) strains. Protein extracts were prepared from cells growing in logarithmic phase in YPD medium (untreated) or after α-factor treatment. (B) Fluorescence microscopy of Glc7-mCherry and indirect immunofluorescence of Afr1-13Myc in AFR1-13Myc (KT2794) and afr1-Δ546-13Myc (KT2795) strains. (C) Immunoblot analysis of extracts prepared from AFR1-EmCitrine (KT2793) and afr1VA/FA-EmCitrine (KT2792) strains. Protein extracts were prepared from cells growing in logarithmic phase in YPD medium (untreated) or after α-factor treatment. (D) Fluorescence microscopy of Afr1-mCitrine and Glc7-mCherry in AFR1-EmCitrine (KT2793) and afr1VA/FA-EmCitrine (KT2792) strains. Cultures were induced with α-factor. Bars, 5 μm. Arrows mark mating projections where Afr1 and Glc7 colocalize.
Glc7 binding may be required for Afr1 function in regulating polarized morphogenesis during mating.
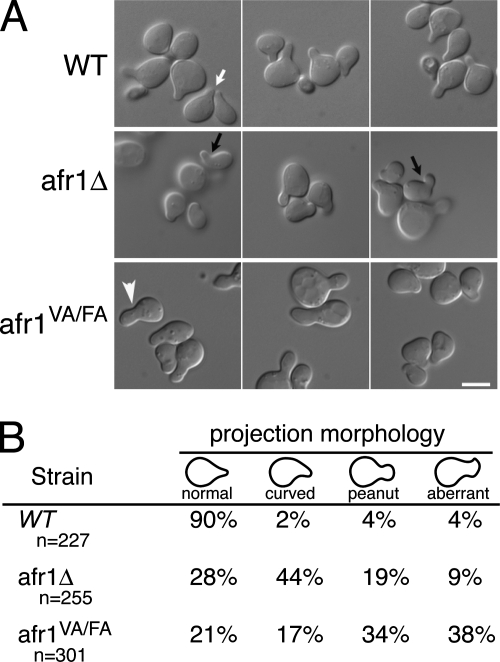
AFR1-null cells display normal sensitivity to mating pheromones but form aberrant mating projections (37, 38). At the concentrations of α-factor used in our experiments (10−7M), AFR1-null mutants form projections that are most often curved (Fig. 5A). In contrast, wild-type cells form mostly acute projections (Fig. 5A). Mating projections of the afr1VA/FA mutant are abnormal (Fig. 5A), but they usually have either a peanut-like shape (Fig. 5A) or a more complex aberrant shape rather than the curved projections of the afr1Δ strain. To quantify differences in morphology, we arrested cells with α-factor, imaged the cells in multiple focal planes, and then placed cells into four categories based on the shapes of mating projections (Fig. 5B). Ninety percent of projections from wild-type cells were classified as normal, acute projections. Only 28% of projections from the AFR1-null mutant were classified as normal. A majority of the projections were classified as curved, and the remainder were either a peanut shape or an odd shape that we simply defined as “aberrant.” In contrast, 72% of afr1VA/FA cells form projections that are either peanut shaped or aberrant. These results suggest that Glc7 plays a role in regulating the morphology of mating projections, but we cannot rule out the possibility that reduced levels of Afr1VA/FA-mCitrine in mating projections are responsible for the distinct morphological phenotype of afr1VA/FA projections.
FIG. 5.
Effect of Afr1-Glc7 on pheromone-induced morphogenesis. (A) Differential interference contrast images of wild-type (WT) (JB75-11C), afr1Δ (JB75-12D), and afr1VA/FA (JB144-9C) cells treated with α-factor. Representative wild-type (white arrow), curved (black arrow), and peanut-shaped (arrowhead) projections are shown. Bar, 5 μm. (B) Quantitation of the morphology of mating projections.
Afr1 regulates septin architecture in polarized morphogenesis during mating.
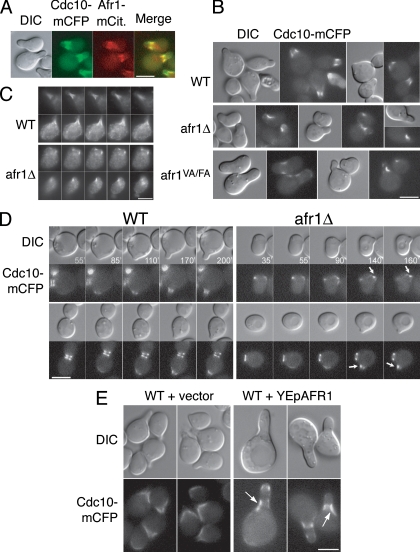
Cells carrying the temperature-sensitive cdc12-6 mutation are defective in forming acute mating projections and fail to properly localize Afr1 to projections, even at the permissive temperature (25). GFP-Glc7 fails to localize to projections in cdc3-1 and cdc10-1 mutants at the nonpermissive temperature (5). These results, together with two-hybrid interaction between Afr1 and Cdc12 (25, 38), suggest that septins are necessary for Afr1 localization. This relationship is similar to that of septins and Bni4 in vegetatively growing cells, where septin complexes are necessary to target Bni4 to the bud neck (13). To investigate the relationship between Afr1 and septins in more detail, we first imaged Cdc10-mCFP and Afr1-mCitrine in strains that contained chromosomal fluorescent protein fusions. As shown in Fig. 6A, Cdc10-mCFP and Afr1-mCitrine colocalize at the bases of mating projections, consistent with the two-hybrid interaction data (25).
FIG. 6.
Afr1 regulates septin organization during mating. (A) Colocalization of Cdc10-mCFP and Afr1-mCitrine in strain JB251-10B. Differential interference contrast (DIC), fluorescence, and merged images are shown. Cdc10-mCFP and Afr1-mCitrine are pseudocolored green and red, respectively. (B) Cdc10-mCFP is asymmetric in AFR1 mutants. DIC and fluorescence images of wild-type (WT) (JB254-19C), afr1Δ (JB254-17C), and afr1VA/FA (JB255-8A) strains induced with α-factor are shown. (C) Cdc12 distribution is asymmetric in afr1Δ cells. Indirect immunofluorescence of Cdc12 with anti-Cdc12 antibody in WT (KT1193) and afr1Δ (JB66-2D) strains was performed. Images from five consecutive focal planes, 0.5 μm apart, are shown. (D) Time lapse microscopy of Cdc10-mCFP in WT (JB254-19C) and afr1Δ (JB254-17C) cells placed on agarose pads containing α-factor. (E) Cdc10-mCFP distribution in AFR1-overexpressing cells. Strain JB250-12B was transformed with pRS425 or pJK52. Transformants were treated with α-factor and visualized as described in Materials and Methods. DIC and fluorescence images are shown. Bars, 5 μm. Arrows indicate aberrant septin localization.
In vegetatively growing cells, septins are necessary for the targeting of Bni4 and Glc7 to the bud neck (5, 13). BNI4-null mutants have a normal septin architecture, as assayed by indirect immunofluorescence (13) or with septin-fluorescent protein fusions (L. Kozubowski and J. Larson, unpublished data), but a bni4-null mutation was found, by use of a sensitive genetic assay for septin function, to influence septins (26). To determine the functional relationship between Afr1 and septin complexes in mating projections, we imaged septin complexes by indirect immunofluorescence and with septin-mCFP fusions and then quantitated the fluorescence distribution at mating projections (Fig. 6B and C; Table 4). In wild-type cells, septins accumulate at the bases of mating projections as a diffuse collar (Fig. 6A) (20, 34) or as broad bars that are distributed uniformly around the base of the complex (44). In afr1Δ cells with curved projections, Cdc10-mCFP is located in the crook or concave surface of the projection (Fig. 6B). Even in cells without curved projections, Cdc10-mCFP is asymmetrically distributed over the surface of the projection (Table 4). The distribution of Cdc10-mCFP in afr1VA/FA cells is also perturbed (Fig. 6B), suggesting that Glc7 activity may be necessary for normal septin localization.
TABLE 4.
Cdc10-mCFP location at the necks of mating projections
| Strain | No. of cells | Time in α-factor (min) | % of cells with:
|
|||
|---|---|---|---|---|---|---|
| Signal on one side of neck | Signal on both sides of neck
|
No signal | ||||
| Symmetric | Asymmetric | |||||
| Wild type | 194 | 60 | <1 | 91 | 8 | 1 |
| 314 | 120 | 0 | 88 | 12 | 0 | |
| afr1Δ strain | 403 | 60 | 54 | 18 | 16 | 12 |
| 63 | 120 | 50 | 19 | 27 | 4 | |
| afr1VA/FA strain | 96 | 60 | 45 | 20 | 6 | 29 |
| 336 | 120 | 36 | 29 | 24 | 4 | |
We also detected Cdc12 in projections by indirect immunofluorescence. In wild-type cells, Cdc12 was observed as bars or filaments that were symmetrically distributed over the projections in 95% of the cells (n = 203). This pattern is illustrated in Fig. 6C. In contrast, in afr1Δ cells, Cdc12 was observed in patches that are asymmetrically localized on the surfaces of projections in ∼65% of cells (n = 356) (Fig. 6C).
We next followed the development of the underlying septin architecture during projection formation by time lapse microscopy. CDC10-EmCFP cells were placed on a pad of medium containing α-factor, and images were collected periodically as the cells formed projections. In wild-type cells, Cdc10-mCFP fluorescence appears at the site of the incipient projection concomitant with projection formation (Fig. 6D, left panel, 85-min time point). As the projection enlarges, the Cdc10-mCFP remains at the base as either a broad collar or a ring. In the afr1Δ cells (Fig. 6D, right panels), the timing of projection appearance and the pattern of Cdc10-mCFP fluorescence are similar to those observed for wild-type cells (note the 35- and 55-min time points for the top row and the 55- and 90-min time points for the bottom row in Fig. 6D). However, at later times, Cdc10-mCFP appears to reorganize around the base of the projection, such that it becomes more concentrated on one side of the projection than on the other (Fig. 6D). Time lapse imaging of 37 afr1Δ cells revealed that 73% (27 cells out of 37) underwent the reorganization of Cdc10-mCFP that is shown in Fig. 6D. In 19% of cells (7/37), the Cdc10-mCFP signal remained symmetric at the end of the experiment, but these cells had small projections. The remaining three cells (8%) had no detectable Cdc10-mCFP signal at the necks of the projections. In contrast, the Cdc10-mCFP signal in 100% of wild-type cells (n = 46) remained symmetric around the base of the mating projection throughout the time course.
The redistribution of septin complexes in the AFR1-null mutant is consistent with the possibility that Afr1 has a role in regulating septin architecture. If this is the case, it might be expected that increased dosage of Afr1 would also alter septin structures. To test this, we imaged a CDC10-EmCFP strain transformed with a high-copy-number AFR1 plasmid. Two effects on the Cdc10-mCFP fluorescent pattern were observed. First, Cdc10-mCFP levels appear higher in the AFR1-overexpressing cells, although quantitation of the fluorescent signal revealed an increase of only ∼1.2-fold. This difference was limited to mating projections, since Cdc10-mCFP levels are the same at the bud necks of vegetatively growing YEpAFR1 and vector control cells (data not shown). Second, Cdc10-mCFP is asymmetrically distributed over the projection (Fig. 6E). Together, these results suggest that Afr1 directly or indirectly regulates the architecture of septin filaments in mating cells.
DISCUSSION
Glc7 is an essential phosphoserine/threonine phosphatase that participates in a wide range of cellular processes and pathways, but substrates of Glc7 for many of these have not been identified. One successful approach to identifying these pathways and substrates has been to identify specific Glc7-binding proteins that target Glc7 to a specific location or biochemical complex. Examples of this approach have been studies on Reg1, Gac1, Bud14, and Pta1, which have illuminated roles of Glc7 in glucose repression (58), glycogen synthesis (21), microtubule dynamics (36), and mRNA 3′ processing (31, 63), respectively. Our identification of Afr1 as a targeting subunit of Glc7 identifies new roles for Glc7 during both vegetative growth and mating.
Afr1 was identified as a promoter of α-factor resistance when overexpressed (37). The mechanism of this resistance is not well understood, although it is thought to be independent of receptor phosphorylation and/or endocytosis (11). Giot and Konopka (25) noted that an Afr1 variant, Afr1-C7, localizes normally to the necks of mating projections but fails to induce α-factor resistance when overexpressed. This variant lacks the COOH-terminal 146 residues of Afr1, which include the Glc7-binding domain. It is therefore possible that the inability to induce resistance to α-factor could be due in part to the failure to recruit Glc7 to the projection neck, and this suggests a role for Glc7 in the negative regulation of pheromone signaling.
The C-terminal Glc7-binding domain of Afr1 is related to PP1/Glc7 binding domains found in yeast Bni4, the mammalian phactr/scapinin proteins, and related proteins in other metazoans. The physiological relevance of this C-terminal domain is illustrated by the observation that a mouse humdy mutant, which has serious developmental defects of the brain and nerve cord, is due to a missense mutation in the PP1-binding domain of the Phactr4 gene (35). This mutation blocks the ability of the phactr4 protein to bind PP1. It has also been shown recently that the Glc7-binding domain of Bni4, when fused directly to the septin protein Cdc10, is sufficient to target chitin synthase III to the bud neck (40). Although the substrates for these proteins are likely to be diverse, the common and essential Glc7/PP1 binding domain reinforces the importance of phosphatase activity for their biological functions.
Afr1 interacts with the septin Cdc12 in the two-hybrid assay (25, 38) and requires normal septin function for its localization to mating projections (25). Interestingly, Afr1 is the third Glc7 binding protein that is targeted to the septin cytoskeleton. Bni4 and Glc7 bind to the septin ring in budding cells, where they regulate chitin deposition (13, 39). bni4-null mutants have morphologically normal septin structures ((13; Larson and Kozubowski, unpublished), but loss of BNI4 has been reported to influence septin assembly in a sensitive screen designed to identify septin regulators (26). Gip1, together with Glc7, is required for septin organization and spore wall formation during meiotic spore maturation (55). However, Afr1, Bni4, and Gip1 are largely unrelated in sequence, other than a Glc7-binding VXF motif and a more extensive region of similarity in the COOH-terminal domains of Afr1 and Bni4. As in the case of Gip1, and possibly Bni4, Afr1 regulates the organization of septins. Septin filaments are mislocalized in the mating projections of afr1Δ cells, where they tend to accumulate asymmetrically on one side of the cell. Furthermore, increased expression of AFR1 leads to increased deposition of septins in mating projections. The enforced expression of AFR1 in vegetative cells results in elongated buds (38), also suggesting an influence on septins.
One mechanism to explain the influence of Afr1 on septin assembly is that Afr1 acts to keep septins diffusely distributed around the bases of mating projections. Loss of Afr1 could result in a collapse of the septin network into a more tightly packed complex, which cannot extend uniformly around the projection base. Given the possible requirement of Glc7 binding for Afr1 function, we speculate that at least part of the regulation of septin architecture could occur by altering the phosphorylation state of septins or other septin regulators. Septins are phosphorylated during vegetative growth, and a key regulatory mechanism required for septin ring formation in unbudded cells involves septin phosphorylation by the protein kinase Cla4 (60). Later in the cell cycle, septin dynamics are regulated by Cla4 and Gin4 protein kinases, together with PP2A (17). It is not known if these mechanisms are important for the regulation of the septin architecture in mating projections, but it is noteworthy that a screen for phosphorylated peptides that change in abundance following pheromone treatment revealed that the levels of phosphopeptides from three septins, Shs1, Cdc11, and Cdc3, decrease upon pheromone treatment (29). In the same study, Afr1 was found to be highly phosphorylated after pheromone treatment, but the significance of this observation is not known. Two phosphopeptides from Gin4 are also less abundant following pheromone treatment. Gin4 is a protein kinase implicated in regulating septin architecture (6, 44) that can be phosphorylated and activated in vitro by the Elm1 protein kinase (2). It should be noted that mutants lacking the protein kinase Gin4 often have diffuse septin structures that at least superficially resemble the bars of septins sometimes observed in mating projections (44). Is it possible that Afr1-Glc7 maintains septins in a hypophosphorylated state necessary to maintain the diffuse septin structures at the necks/bases of mating projections? Further work on septins in mating cells will be necessary to test this hypothesis.
Acknowledgments
We thank Lucy Robinson for reading the manuscript and Matthias Versele for the anti-Cdc12 antibody.
This work was supported by National Institutes of Health grant GM-47789.
Footnotes
Published ahead of print on 13 June 2008.
REFERENCES
- 1.Allen, P. B., A. T. Greenfield, P. Svenningsson, D. C. Haspeslagh, and P. Greengard. 2004. Phactrs 1-4: a family of protein phosphatase 1 and actin regulatory proteins. Proc. Natl. Acad. Sci. USA 1017187-7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asano, S., J. E. Park, L. R. Yu, M. Zhou, K. Sakchaisri, C. J. Park, Y. H. Kang, J. Thorner, T. D. Veenstra, and K. S. Lee. 2006. Direct phosphorylation and activation of a Nim1-related kinase Gin4 by Elm1 in budding yeast. J. Biol. Chem. 28127090-27098. [DOI] [PubMed] [Google Scholar]
- 3.Barral, Y., V. Mermall, M. S. Mooseker, and M. Snyder. 2000. Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol. Cell 5841-851. [DOI] [PubMed] [Google Scholar]
- 4.Bharucha, J. P., J. R. Larson, L. Gao, L. K. Daves, and K. Tatchell. 2008. Ypi1, a positive regulator of nuclear protein phosphatase type 1 activity in Saccharomyces cerevisiae. Mol. Biol. Cell 191032-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloecher, A., and K. Tatchell. 2000. Dynamic localization of protein phosphatase type 1 in the mitotic cell cycle of Saccharomyces cerevisiae. J. Cell Biol. 149125-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouquin, N., Y. Barral, R. Courbeyrette, M. Blondel, M. Snyder, and C. Mann. 2000. Regulation of cytokinesis by the Elm1 protein kinase in Saccharomyces cerevisiae. J. Cell Sci. 1131435-1445. [DOI] [PubMed] [Google Scholar]
- 7.Brent, R., and M. Ptashne. 1985. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell 43729-736. [DOI] [PubMed] [Google Scholar]
- 8.Cannon, J. F., and K. Tatchell. 1987. Characterization of Saccharomyces cerevisiae genes encoding subunits of cyclic AMP-dependent protein kinase. Mol. Cell. Biol. 72653-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chant, J. 1999. Cell polarity in yeast. Annu. Rev. Cell Dev. Biol. 15365-391. [DOI] [PubMed] [Google Scholar]
- 10.Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110119-122. [DOI] [PubMed] [Google Scholar]
- 11.Davis, C., P. Dube, and J. B. Konopka. 1998. Afr1p regulates the Saccharomyces cerevisiae α-factor receptor by a mechanism that is distinct from receptor phosphorylation and endocytosis. Genetics 148625-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis, N. G., J. L. Horecka, and G. F. Sprague, Jr. 1993. cis- and trans-acting functions required for endocytosis of the yeast pheromone receptors. J. Cell Biol. 12253-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeMarini, D. J., A. E. M. Adams, H. Fares, C. De Virgilio, G. Valle, J. S. Chuang, and J. R. Pringle. 1997. A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J. Cell Biol. 13975-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeMattei, C. R., C. P. Davis, and J. B. Konopka. 2000. Point mutations identify a conserved region of the Saccharomyces cerevisiae AFR1 gene that is essential for both the pheromone signaling and morphogenesis functions. Genetics 15543-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dent., J., K. Kato, X. R. Peng, C. Martinez, M. Cattaneo, C. Poujol, P. Nurden, A. Nurden, W. S. Trimble, and J. Ware. 2002. A prototypic platelet septin and its participation in secretion. Proc. Natl. Acad. Sci. USA 993064-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dobbelaere, J., and Y. Barral. 2004. Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science 305393-396. [DOI] [PubMed] [Google Scholar]
- 17.Dobbelaere, J., M. S. Gentry, R. L. Hallberg, and Y. Barral. 2003. Phosphorylation-dependent regulation of septin dynamics during the cell cycle. Dev. Cell 4345-357. [DOI] [PubMed] [Google Scholar]
- 18.Durfee, T., K. Becherer, P. L. Chen, S. H. Yeh, Y. Yang, A. E. Kilburn, W. H. Lee, and S. J. Elledge. 1993. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 7555-569. [DOI] [PubMed] [Google Scholar]
- 19.Fields, S., and O.-K. Song. 1989. A novel genetic system to detect protein-protein interactions. Nature 340245-246. [DOI] [PubMed] [Google Scholar]
- 20.Ford, S. K., and J. R. Pringle. 1991. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC11 gene product and the timing of events at the budding site. Dev. Genet. 12281-292. [DOI] [PubMed] [Google Scholar]
- 21.François, J. M., S. Thompson-Jaeger, J. Skroch, U. Zellenka, W. Spevak, and K. Tatchell. 1992. GAC1 may encode a regulatory subunit for protein phosphatase type 1 in Saccharomyces cerevisiae. EMBO J. 1187-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frazier, J. A., M. L. Wong, M. S. Longtine, J. R. Pringle, M. Mann, T. J. Mitchison, and C. Field. 1998. Polymerization of purified yeast septins: evidence that organized filament arrays may not be required for septin function. J. Cell Biol. 143737-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frederick, D. L., and K. Tatchell. 1996. The REG2 gene of Saccharomyces cerevisiae encodes a type1 protein phosphatase-binding protein that functions with Reg1p and the Snf1p protein kinase to regulate growth. Mol. Cell. Biol. 162922-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gietz, D., A. St. Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 201425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giot, L., and J. B. Konopka. 1997. Functional analysis of the interaction between Afr1p and the Cdc12p septin, two proteins involved in pheromone-induced morphogenesis. Mol. Biol. Cell 8987-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gladfelter, A. S., L. Kozubowski, T. R. Zyla, and D. J. Lew. 2005. Interplay between septin organization, cell cycle and cell shape in yeast. J. Cell Sci. 1181617-1628. [DOI] [PubMed] [Google Scholar]
- 27.Gladfelter, A. S., J. R. Pringle, and D. J. Lew. 2001. The septin cortex at the yeast mother-bud neck. Curr. Opin. Microbiol. 4681-689. [DOI] [PubMed] [Google Scholar]
- 28.Goldstein, A. L., and J. H. McCusker. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 151541-1553. [DOI] [PubMed] [Google Scholar]
- 29.Gruhler, A., J. V. Olsen, S. Mohammed, P. Mortensen, N. J. Faergeman, M. Mann, and O. N. Jensen. 2005. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol. Cell. Proteomics 4310-327. [DOI] [PubMed] [Google Scholar]
- 30.Hall, P. A., and S. E. Russell. 2004. The pathobiology of the septin gene family. J. Pathol. 204489-505. [DOI] [PubMed] [Google Scholar]
- 31.He, X., and C. Moore. 2005. Regulation of yeast mRNA 3′ end processing by phosphorylation. Mol. Cell 19619-629. [DOI] [PubMed] [Google Scholar]
- 32.Kaiser, C., S. Michaelis, and A. Mitchell. 1994. Methods in yeast genetics: a Cold Spring Harbor Laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 33.Kaiser, P., R. A. Sia, E. G. Bardes, D. J. Lew, and S. I. Reed. 1998. Cdc34 and the F-box protein Met30 are required for degradation of the Cdk-inhibitory kinase Swe1. Genes Dev. 122587-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim, H. B., B. K. Haarer, and J. R. Pringle. 1991. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC3 gene product and the timing of events at the budding site. J. Cell Biol. 112535-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim, T. H., J. Goodman, K. V. Anderson, and L. Niswander. 2007. Phactr4 regulates neural tube and optic fissure closure by controlling PP1-, Rb-, and E2F1-regulated cell-cycle progression. Dev. Cell 1387-102. [DOI] [PubMed] [Google Scholar]
- 36.Knaus, M., E. Cameroni, I. Pedruzzi, K. Tatchell, C. De Virgilio, and M. Peter. 2005. The Bud14p-Glc7p complex functions as a cortical regulator of dynein in budding yeast. EMBO J. 243000-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Konopka, J. B. 1993. AFR1 acts in conjunction with the α-factor receptor to promote morphogenesis and adaptation. Mol. Cell. Biol. 136876-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Konopka, J. B., C. DeMattei, and C. Davis. 1995. AFR1 promotes polarized apical morphogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 15723-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kozubowski, L., H. Panek, A. Rosenthal, A. Bloecher, D. J. DeMarini, and K. Tatchell. 2003. A Bni4-Glc7 phosphatase complex that recruits chitin synthase to the site of bud emergence. Mol. Biol. Cell 1426-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larson, J. R., J. P. Bharucha, S. Ceaser, J. Salamon, C. J. Richardson, S. M. Rivera, and K. Tatchell. 2008. Protein phosphatase type 1 directs chitin synthesis at the bud neck in Saccharomyces cerevisiae. Mol. Biol. Cell 293040-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lenssen, E., N. James, I. Pedruzzi, F. Dubouloz, E. Cameroni, R. Bisig, L. Maillet, M. Werner, J. Roosen, K. Petrovic, J. Winderickx, M. A. Collart, and C. De Virgilio. 2005. The Ccr4-Not complex independently controls both Msn2-dependent transcriptional activation—via a newly identified Glc7/Bud14 type I protein phosphatase module—and TFIID promoter distribution. Mol. Cell. Biol. 25488-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Longtine, M. S., and E. Bi. 2003. Regulation of septin organization and function in yeast. Trends Cell Biol. 13403-409. [DOI] [PubMed] [Google Scholar]
- 43.Longtine, M. S., A. McKenzie III, D. J. DeMarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14953-961. [DOI] [PubMed] [Google Scholar]
- 44.Longtine, M. S., H. Fares, and J. R. Pringle. 1998. Role of the yeast Gin4p protein kinase in septin assembly and the relationship between septin assembly and septin function. J. Cell Biol. 143719-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muller, E. G., B. E. Snydsman, I. Novik, D. W. Hailey, D. R. Gestaut, C. A. Niemann, E. T. O'Toole, T. H. Giddings, Jr., B. A. Sundin, and T. N. Davis. 2005. The organization of the core proteins of the yeast spindle pole body. Mol. Biol. Cell 163341-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodal, A. A., L. Kozubowski, B. L. Goode, D. G. Drubin, and J. H. Hartwig. 2005. Actin and septin ultrastructures at the budding yeast cell cortex. Mol. Biol. Cell 16372-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sagara, J., T. Higuchi, Y. Hattori, M. Moriya, H. Sarvotham, H. Shima, H. Shirato, K. Kikuchi, and S. Taniguchi. 2003. Scapinin, a putative protein phosphatase-1 regulatory subunit associated with the nuclear nonchromatin structure. J. Biol. Chem. 27845611-45619. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 49.Selvin, P. R. 1995. Fluorescence resonance energy transfer. Methods Enzymol. 246300-334. [DOI] [PubMed] [Google Scholar]
- 50.Sheff, M. A., and K. S. Thorn. 2004. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21661-670. [DOI] [PubMed] [Google Scholar]
- 51.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 52.Sikorksi, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 12219-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stark, M. J. R. 1996. Yeast protein serine/threonine phosphatases: multiple roles and diverse regulation. Yeast 121647-1675. [DOI] [PubMed] [Google Scholar]
- 54.Stuart, J. S., D. L. Frederick, C. M. Varner, and K. Tatchell. 1994. The mutant type 1 protein phosphatase encoded by glc7-1 from Saccharomyces cerevisiae fails to interact productively with the GAC1-encoded regulatory subunit. Mol. Cell. Biol. 14896-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tachikawa, H., A. Bloecher, K. Tatchell, and A. M. Neiman. 2001. A Gip1p-Glc7p phosphatase complex regulates septin organization and spore wall formation. J. Cell Biol. 155797-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takizawa, P. A., J. L. DeRisi, J. E. Wilhelm, and R. D. Vale. 2000. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science 290341-344. [DOI] [PubMed] [Google Scholar]
- 57.Trimble, W. S. 1999. Septins: a highly conserved family of membrane-associated GTPases with functions in cell division and beyond. J. Membr. Biol. 16975-81. [DOI] [PubMed] [Google Scholar]
- 58.Tu, J., and M. Carlson. 1995. REG1 binds to protein phosphatase type 1 and regulates glucose repression in Saccharomyces cerevisiae. EMBO J. 145939-5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Versele, M., B. Gullbrand, M. J. Shulewitz, V. J. Cid, S. Bahmanyar, R. E. Chen, P. Barth, T. Alber, and J. Thorner. 2004. Protein-protein interactions governing septin heteropentamer assembly and septin filament organization in Saccharomyces cerevisiae. Mol. Biol. Cell 154568-4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Versele, M., and J. Thorner. 2004. Septin collar formation in budding yeast requires GTP binding and direct phosphorylation by the PAK, Cla4. J. Cell Biol. 164701-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Versele, M., and J. Thorner. 2005. Some assembly required: yeast septins provide the instruction manual. Trends Cell Biol. 15414-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vrabioiu, A. M., and T. J. Mitchison. 2006. Structural insights into yeast septin organization from polarized fluorescence microscopy. Nature 443466-469. [DOI] [PubMed] [Google Scholar]
- 62a.Wach, A., A. Brachat, C. Alberti-Segui, C. Rebischung, and P. Philippsen. 1997. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast 131065-1075. [DOI] [PubMed] [Google Scholar]
- 63.Walsh, E. P., D. J. Lamont, K. A. Beattie, and M. J. Stark. 2002. Novel interactions of Saccharomyces cerevisiae type 1 protein phosphatase identified by single-step affinity purification and mass spectrometry. Biochemistry 412409-2420. [DOI] [PubMed] [Google Scholar]
- 64.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, R. W. Davis, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285901-906. [DOI] [PubMed] [Google Scholar]
- 65.Wu, X., and K. Tatchell. 2001. Mutations in yeast protein phosphatase type 1 that affect targeting subunit binding. Biochemistry 407410-7420. [DOI] [PubMed] [Google Scholar]
- 66.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33103-119. [DOI] [PubMed] [Google Scholar]