Abstract
Target-of-rapamycin proteins (TORs) are Ser/Thr kinases serving a central role in cell growth control. TORs function in two conserved multiprotein complexes, TOR complex 1 (TORC1) and TORC2; the mechanisms underlying their actions and regulation are not fully elucidated. Saccharomyces TORC2, containing Tor2p, Avo1p, Avo2p, Avo3p/Tsc11p, Bit61p, and Lst8p, regulates cell integrity and actin organization. Two classes of avo3 temperature-sensitive (avo3ts) mutants that we previously identified display cell integrity and actin defects, yet one is suppressed by AVO1 while the other is suppressed by AVO2 or SLM1, defining two TORC2 downstream signaling mechanisms, one mediated by Avo1p and the other by Avo2p/Slm1p. Employing these mutants, we explored Avo3p functions in TORC2 structure and signaling. By observing binary protein interactions using coimmunoprecipitation, we discovered that the composition of TORC2 and its recruitment of the downstream effectors Slm1p and Slm2p were differentially affected in different avo3ts mutants. These molecular defects can be corrected only by expressing AVO3, not by expressing suppressors, highlighting the role of Avo3p as a structural and signaling scaffold for TORC2. Phenotypic modifications of avo3ts mutants by deletion of individual Rho1p-GTPase-activating proteins indicate that two TORC2 downstream signaling branches converge on Rho1p activation. Our results also suggest that Avo2p/Slm1p-mediated signaling, but not Avo1p-mediated signaling, links to Rho1p activation specifically through the Rho1p-guanine nucleotide exchange factor Tus1p.
TOR (target of rapamycin) proteins are conserved Ser/Thr protein kinases found in diverse eukaryotes ranging from yeasts to mammals (11, 13, 28, 39). A large body of evidence points to TOR as a central regulator of a complex signaling network that controls cell growth, proliferation, and survival. TOR integrates signals from nutrients, energy status, growth factors, and various cellular stressors to regulate a myriad of processes. Diverse readouts of TOR functions include transcription, translation, tRNA and ribosome biogenesis, protein turnover, quiescence, autophagy, and actin cytoskeleton organization (13, 26, 57, 67, 69). Unlike most eukaryotic species, which possess a single TOR, the budding yeast Saccharomyces cerevisiae has two TOR proteins, Tor1p and Tor2p (24). Tor1p and Tor2p share a rapamycin-sensitive function, to control cell growth by regulating transcription, translation, and ribosome biogenesis (3, 5, 24, 48, 53). Tor2p has an additional, rapamycin-insensitive function that Tor1p is unable to perform, i.e., to control actin organization throughout cell cycle progression (22, 24, 59).
Consistent with the existence of multiple protein interaction modules in its structure (8), TOR appears to exert its functions through multiprotein complexes. There are two distinct TOR complexes, TORC1 and TORC2, identified first in budding yeast (37, 66). Either Tor1p or Tor2p can associate with Lst8p, Kog1p, and Tco89p to form TORC1, while TORC2 is composed of Tor2p, Lst8p, Avo1p, Avo2p, Avo3p (also designated Tsc11p), and Bit61p. The two TOR complexes are distinct not only in structure but also in function. Evidence supports the notion that TORC1 mediates the shared TOR function of regulating rapamycin-sensitive, cell growth-related processes, whereas TORC2 mediates the Tor2p-unique function of regulating actin organization (37). Remarkably, both TOR complexes are structurally and functionally conserved, and the presence of their mammalian counterparts, mTORC1 and mTORC2, has been demonstrated (16, 21, 29, 32, 33, 55, 70). Like yeast TORC1, the rapamycin-sensitive mTORC1, containing mTOR, GβL (mLST8), and Raptor (mKOG1), is involved in regulating ribosome biogenesis and protein synthesis. On the other hand, mTORC2, containing mSIN1 (mAVO1, the mammalian homolog of Avo1p) and Rictor (mAVO3, the mammalian homolog of Avo3p) in addition to mTOR and GβL, participates in the control of actin cytoskeletal organization.
The discovery of two distinct TOR-containing complexes adds to the challenge in understanding TOR signaling mechanisms. In Saccharomyces cerevisiae, TORC1 responds to nutrient conditions by acting on a phosphatase regulatory system that includes the yeast type 2A phosphatases (PP2A) Pph21p and Pph22p, the PP2A-related phosphatase Sit4p together with its associated proteins, and two regulatory proteins, Tap42p and Tip41p (27). TORC1 function also links to kinases, including the Ras/cyclic AMP (cAMP) signaling-related kinase Yak1p, the cAMP-regulated kinase protein kinase A (PKA), and Sch9p (another member of the AGC [PKA, PKG, and PKC] family of protein kinases) (9, 30, 56, 63, 72). It remains to be elucidated how TORC1 acts to coordinate these phosphatases and kinases in mediating the regulation of cell growth-related processes. To date, the upstream signals that serve to activate TORC2 remain elusive, yet studies have implicated several effector pathways downstream of TORC2. In budding yeast, Rho1p serves as a pivotal regulator of cell wall integrity and actin organization (7, 36). Once activated, Rho1p binds and activates Pkc1p, which in turn activates a downstream mitogen-activated protein kinase module cascade. Tor2p, presumably functioning from within TORC2, regulates the actin cytoskeleton by activating the Rho1p pathway via the guanine nucleotide exchange factor (GEF) Rom2p (58). Overexpression of RHO1, PKC1, and genes encoding components of the mitogen-activated protein kinase cascade or deletion of the GTPase-activating protein (GAP) gene SAC7 can rescue the phenotype of tor2 temperature-sensitive mutant (tor2ts) cells (6, 23, 58). Moreover, the GEF activity of Rom2p toward Rho1p is dramatically reduced in tor2ts cells (58). Despite evidence linking Tor2p and Rho1p functions, the mechanisms underlying the regulation of the Rho1p pathway by TORC2 are not well understood. In addition to Rho1p, two functionally redundant pleckstrin homology domain-containing phosphatidylinositol-4,5-bisphosphate-binding proteins, Slm1p and Slm2p, also act downstream of TORC2 to control actin cytoskeleton organization (2, 15). Slm1p and Slm2p physically interact with the TORC2 components Avo2p and Bit61p and are phosphorylated by Tor2p in vivo and in vitro. slm1ts slm2Δ cells demonstrate defects in cell wall integrity and actin organization, which can be rescued by overexpression of PKC1. It is still unclear whether the two Slm proteins regulate the actin cytoskeleton through Rho1p or other pathways. Another TORC2 downstream pathway involves the yeast protein kinases Ypk1p and Ypk2p, which are also AGC kinases (31). These Ypk proteins can be activated by the yeast PDK1 homologs Pkh1p and Pkh2p, and they serve to regulate actin organization and the cell wall (51, 52). Active Ypk2p (and also Ypk1p) can suppress the growth defect of cells depleted of TORC2. Furthermore, Ypk2p has been demonstrated to be phosphorylated by Tor2p in vitro, and the kinase activity of Ypk2p is decreased in tor2ts cells (31). The molecular mechanism by which TORC2 regulates Ypk proteins has not been elucidated yet.
The molecular organization of TORC2 has been characterized in more detail (68). TORC2 appears to exist in cells in oligomeric forms, most likely as dimers. Avo1p and Avo3p may bind cooperatively to the N-terminal HEAT (Huntingtin, elongation factor 3, A subunit of PP2A, and TOR1) repeat region of Tor2p. Depletion of either Avo1p or Avo3p disrupts the binary interactions between TORC2 components except for the Tor2p-Lst8p interaction, suggesting that Avo1p and Avo3p may act as scaffold proteins in TORC2. On the other hand, Lst8p binds to the C-terminal kinase domain region in Tor2p and may participate in modulating Tor2p kinase activity. Avo2p and Bit61p are nonessential proteins in TORC2, and they may function as adaptors for binding downstream effectors such as Slm1p and Slm2p (2, 15, 68). Despite the plethora of evidence for physical interactions in the TOR complex, no specific molecular actions of individual TORC2 components have been demonstrated except for the Ser/Thr kinase activity of Tor2p.
Our previous analyses using temperature-sensitive mutants of AVO3 suggest the existence of complex and probably multiple signaling mechanisms downstream of TORC2 (25). Although different avo3ts mutants similarly exhibit actin and cell wall integrity defects, they can be grouped into two distinct classes; AVO1 serves as a multicopy suppressor of one class, while the other class can be rescued by overexpression of AVO2 or SLM1. The allelic specificity in dosage suppression leads to the speculation that Avo3p may exert its functions through two distinct mechanisms: one mediated by Avo1p and the other mediated by Avo2p and Slm1p. Since the avo3ts suppressors AVO1 and AVO2 encode components of TORC2 and the SLM1 product is one of the TORC2 downstream effectors, it is conceivable that Avo3p may modulate protein interactions within TORC2 or the association with different downstream effectors to specify TORC2 signaling. Therefore, in this study we explore the molecular defects in the two classes of avo3ts mutants by examining their TORC2 composition and downstream signaling functions. We demonstrate that the integrity of TORC2 and its interaction with the downstream effectors Slm1p and Slm2p were perturbed differently in the two classes of avo3ts mutants, suggesting a possible role of Avo3p as a scaffold for the molecular architecture and downstream coupling of TORC2. By deleting various RhoGAPs (negative regulators of Rho family GTPases) in different avo3ts mutants, we provided genetic evidence supporting the notion that Avo3p functionally interacts with the Rho1p signaling pathway. Furthermore, we showed that Avo3p may coordinate Avo2p and Slm1p signaling to activate Rho1p through the Rho1p-GEF Tus1p.
MATERIALS AND METHODS
Yeast growth conditions.
Yeast-peptone-dextrose (YPD) medium and a synthetic complete minimal medium (SC) supplement with the appropriate nutrients for plasmid maintenance were prepared as described elsewhere (20). Yeast strains were cultured at 27°C unless indicated otherwise. For G418 selection, YPD medium was supplemented with 200 μg/ml of G418 (Sigma). For galactose induction, an overnight yeast culture in a medium supplemented with 2% raffinose was diluted 1:100 into 100 ml of 2% raffinose-containing medium and grown for another 12 h. Galactose was then added to a final concentration of 3%, and the culture was incubated for another 2 to 4 h to allow the induction of GAL1 promoter-driven gene expression.
Yeast strains.
Standard yeast genetic methods were performed as described elsewhere (20). A lithium acetate method was employed for yeast transformations (18). Yeast strains and oligonucleotide primers used in this work are listed in Tables S1 and S2 in the supplemental material, respectively.
To engineer a chromosomal allele expressing N-terminal triple-hemagglutinin (HA) epitope-tagged Tor2p, we followed procedures described previously (66). Briefly, a recombination cassette was amplified from the pHS8 plasmid by using primers TOR2-tagF4 and TOR2-tagR3 and was transformed into the desired yeast strains. Transformants were selected on G418-containing YPD plates. G418-resistant clones were restreaked twice to avoid false-positive clones, and candidate clones were checked for 3HA-Tor2p expression by Western blot analysis using anti-HA antibodies (Covance). To generate different yeast strains expressing C-terminal 13Myc-tagged TORC1 or TORC2 components, we performed PCR-based gene tagging as described elsewhere (38). Gene-specific primers F2 and R1 were used to amplify a cassette containing multiple copies of the Myc epitope and the HIS3 selection marker, and the PCR products were concentrated and transformed into yeast cells. After transformation, yeast cells were grown on SC-His plates to select for integrants. Candidate clones were checked for tagged protein expression by Western blot analysis using anti-Myc antibodies (Upstate).
Strains with individual gene deletions were generated based on a PCR-mediated gene disruption strategy (4). To disrupt each gene encoding RhoGAP and TUS1, a KanMX4 expression cassette-containing genomic knockout fragment was amplified from corresponding commercially available BY4741-based deletion strains (Open Biosystems) (17) using gene-specific primers S1 and A1. PCR products were introduced into yeast cells, and the transformants were selected on G418-containing YPD plates. Correct replacement of the target gene with the KanMX4 expression cassette was verified by PCR amplifications using KanB1 plus gene-specific primer S2 and KanC3 plus gene-specific primer A2. To delete AVO2, ROM1, and ROM2, gene-specific primers KO-S1 and KO-A1 were used to amplify the HIS3 or URA3 marker from pRS403 or pRS306. PCR products were transformed into yeast cells, and the chromosomal deletion in transformants was confirmed by PCR amplification using gene-specific primers CK-S1 and CK-A1.
Plasmids.
DNA manipulations and bacterial transformation were carried out by following standard protocols. The plasmids used in this study are listed in Table S3 in the supplemental material. pHS8 was constructed as described elsewhere (66).
To construct pHS9, a genomic fragment containing AVO1 with a C-terminal 13Myc epitope fusion, along with 491 nucleotides upstream and 760 nucleotides downstream of the gene, was amplified from the genomic DNA of YMY117 (see Table S1 in the supplemental material) using primers AVO1-S6 and FA6a-HIS3-A1. The PCR product was digested by BglII and subcloned into pYYL6, which contains the full-length AVO1 open reading frame along with its endogenous promoter (Y.-Y. Liao, unpublished data).
Plasmids expressing glutathione S-transferase (GST) fusion proteins were constructed as follows. For a GST-SLM1 fusion, a genomic fragment containing the SLM1 gene from the start codon to 208 bp downstream of the stop codon was amplified from YMY97 using primers SLM1-XbaI-S1 and SLM1-A1; the PCR product was digested by XbaI and subcloned into pGAL1-GST-URA3 (provided by J.-J. Lin at National Yang-Ming University), resulting in pHS10, which is a YEp-based plasmid containing the URA3 selection marker and a GAL1 promoter-driven GST-SLM1 expression cassette. For a GST-SLM2 fusion, a SLM2-containing genomic fragment was amplified from the yeast genome using primers Slm2-S2 and Slm2-A2 and was subjected to TA cloning into pCR2.1-TOPO (Invitrogen) to produce pCR2.1-TOPO-SLM2; the 2.7-kb KpnI/XhoI fragment from pCR2.1-TOPO-SLM2 was subsequently subcloned into pRS424 to produce pHS12. The SLM2 gene from the start codon to 215 bp downstream of the stop codon was amplified from pHS12 using primers SLM2-XbaI-S1 and SLM2-XbaI-A1; the PCR product was digested with XbaI and subcloned into pGAL1-GST-URA3 to generate pHS13. For a GST-RHO1 fusion, the full-length RHO1 gene was amplified from the yeast genome with primers RHO1-XbaI-S1 and RHO1-XbaI-A1, digested with XbaI, and subcloned into pGAL1-GST-URA3 to generate pHYS1.
Plasmids expressing Sac7p, Sac7p(R173A), and their HA-tagged versions were constructed as follows. To construct pHYS2 and pHYS3, the entire SAC7 open reading frame was amplified from the wild-type genomic DNA using primers SAC7-S2 and SAC7-A1 and was subcloned into pRS314 to generate pHYS2; pHYS3 was derived from pHYS2 by performing PCR site-directed mutagenesis using primers SAC7-R173A-Nru and SAC7-R173A-Anti. pHYS4 and pHYS5, which express N-terminal HA-tagged versions of Sac7p and Sac7p(R173A), respectively, were constructed by replacing the BclI/XhoI fragment of pHYS2 or pHYS3 with a BclI/XhoI PCR fragment generated by using SAC7-cHA-S1 and SAC7-cHa-XhoI-A1 as primers and pHYS2 as the template. To generate pHYS6 and pHYS7, ∼2-kb EcoRI/XhoI fragments from pHYS4 and pHYS5, respectively, were inserted into pRS416GAL (44).
The Tus1p-HA-expressing plasmid pHS11 was derived from BG1805-TUS1 (Open Biosystems), which contains URA3 and expresses Tus1p-HA under the control of the GAL1 promoter. To change the selection marker from URA3 to LEU2, a LEU2 fragment was amplified from pRS315 using primers StuI-LEU2-S1 and StuI-LEU2-A1, and the PCR product was digested with StuI and subcloned into BG1805-TUS1 to produce pHS11.
Coimmunoprecipitation.
Strains expressing two different epitope-tagged TORC2 component proteins were grown to an optical density at 600 nm (OD600) of 0.7 to 0.8 and divided into two aliquots, one of which was kept at 27°C while the other was shifted to 37°C for another 2 h. Harvested cells were broken by vortexing with glass beads in a lysis buffer containing 1× phosphate-buffered saline (PBS), 10% (wt/vol) glycerol, 0.5% (vol/vol) Tween 20, and various inhibitors including 10 mM NaF, 10 mM NaN3, 10 mM p-nitrophenylphosphate, 10 mM sodium pyrophosphate, 10 mM β-glycerophosphate, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1× protease inhibitor cocktail (Roche). Crude extracts were centrifuged at 500 × g for 15 min at 4°C to remove cell debris. Samples of lysates containing 3 mg of proteins were adjusted to 500 μl with lysis buffer. For immunoprecipitation, 4 μl of a monoclonal anti-HA (clone 16B12; Covance) or anti-Myc (clone 9E10; Upstate) antibody was added, and the reaction tubes were rotated at 4°C for 2 h. A protein G-Sepharose slurry (Sigma) was added, and tubes were rotated at 4°C for another 1 h. Beads were collected by centrifugation, washed three times with 1 ml lysis buffer, and resuspended in sample buffer. After heating at 100°C for 5 min, samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis using anti-HA or anti-Myc antibodies.
Spot assay for yeast growth.
In the semiquantitative plate assay for yeast growth, the same numbers of cells collected from 27°C overnight cultures of different strains were used to make 10-fold serial dilutions over a 10,000-fold range. Five microliters of each dilution was spotted onto appropriate plates and incubated at different temperatures until colonies formed.
GST pull-down assay.
To examine the interaction of TORC2 with Slm proteins, yeast strains carrying a tagged TORC2 component(s) were transformed with pGAL1-GST-URA3 (as a control) or a plasmid expressing GST-Slm1p (pHS10) or GST-Slm2p (pHS13) under the regulation of the GAL1 promoter. Cultures of transformants were subjected to galactose induction for 2 h, divided into two aliquots, one of which was kept at 27°C while the other was shifted to 37°C, and grown further for 2 h. Cells were harvested, and total lysates were prepared as for coimmunoprecipitation. Lysate samples containing 10 mg of proteins were each adjusted to 1 ml with lysis buffer, and 50 μl of glutathione-Sepharose 4B beads was added (Amersham Pharmacia). After incubation at 4°C for 2 h with rotation, proteins were pulled down with beads, washed five times with lysis buffer, resuspended in sample buffer, and subjected to Western blot analysis using anti-HA or anti-Myc antibodies.
To check the interaction between Rho1p and wild-type or mutant Sac7p, the GST pull-down assay was performed as described elsewhere, with modifications (60). Appropriate yeast strains were induced by galactose to express GST-Rho1p from the GAL1 promoter. Harvested cells were resuspended in extraction buffer I (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1 mM EDTA, 0.5% NP-40, 1 mM PMSF) and lysed by vortexing with glass beads. Crude extracts were centrifuged at 500 × g for 10 min to remove cell debris. GST-Rho1p was isolated by incubating the cell extracts with glutathione Sepharose 4B beads (Amersham Pharmacia) for 2 h. Beads were collected and washed five times with extraction buffer I containing 500 mM NaCl and 0.5% Triton X-100 and once with extraction buffer I. Pulled down GST or GST-Rho1p samples were resuspended in 50 μl extraction buffer I and preloaded with 1 mM GTPγS at 25°C for 15 min. Subsequently, extracts containing 3 mg proteins from cells expressing HA-Sac7p or HA-Sac7p(R173A) were added, and the reaction mixtures were incubated at 4°C for 1 h with rotation. Beads were collected, washed five times with wash buffer (50 mM Tris-HCl [pH 7.5], 500 mM NaCl, 0.1% NP-40, 0.5% Triton X-100, and 1 mM EDTA), resuspended in 50 μl sample buffer, and boiled at 100°C for 5 min. Pulled down proteins were subjected to SDS-PAGE and Western blot analysis with anti-HA antibodies to detect HA-Sac7p or HA-Sac7p(R173A).
To investigate the interaction between Tus1p and Slm1p, yeast cells were cotransformed with plasmids expressing Tus1p-HA (pHS11) and GST-Slm1p (pHS10) or GST (pGAL1-GST-URA3). Transformants were subjected to galactose induction, and cells were collected and lysed in a lysis buffer (100 mM NaCl, 1 mM sodium pyrophosphate, 5 mM NaF, 1 mM EDTA, 1% Triton X-100, 0.5% sodium deoxycholate, 50 mM Tris-HCl [pH 7.5], 1 mM PMSF, and 1 mM sodium orthovanadate) containing 1× protease inhibitor cocktail (Roche). After centrifugation at 12,000 rpm for 15 min to remove cell debris, the supernatant was subjected to GST pull-down procedures as described above. Pulled down samples were washed five times with lysis buffer, resuspended in 1× sample buffer, and subjected to SDS-PAGE and Western blot analysis using anti-HA antibodies to detect Tus1p-HA.
Trypan blue assay for cell wall integrity.
Yeast cell wall integrity was assessed by a trypan blue exclusion assay essentially as described elsewhere (25). Overnight cultures were diluted to an OD600 of 0.15 in 5 ml of appropriate liquid medium and were grown at 27°C for 2 h. Each culture was subsequently divided into two aliquots, one of which was kept at 27°C while the other was shifted to the nonpermissive temperature, and further cultured for 5 h. Harvested cells were washed with distilled water and stained with 0.02% trypan blue in water for 1 h. At least 200 cells from each sample were examined under the microscope to determine the proportion of stained cells.
Actin staining.
Overnight yeast cultures were diluted to an OD600 of 0.35, grown at 27°C for 2 h, and shifted to restrictive temperatures for 3 h. Cells harvested from 10 ml of culture were fixed in 4% formaldehyde for 1 h at room temperature, washed twice, and resuspended in 500 μl of 1× PBS. Ten microliters of 6.6 μM tetramethyl rhodamine isothiocyanate (TRITC)-phalloidin (Sigma) was added to each 100-μl cell suspension sample, and samples were then incubated at 27°C for 1.5 h. Cells were collected, washed once with 1× PBS, resuspended in 50 μl of mounting solution, and examined by fluorescence and differential interference contrast microscopy for actin distribution and cell morphology, respectively. Because small-budded cells normally show a polarized distribution of actin in the daughter cells, we considered small-budded cells with four or more actin patches in mother cells to be abnormal.
GAP activity assay.
The GAP activities of HA-Sac7p and HA-Sac7p(R173A) were examined as described elsewhere (60) with modifications. Yeast lysates containing HA-Sac7p or HA-Sac7p(R173A) expressed under the control of the GAL1 promoter were prepared as described for the GST pull-down experiments. Samples of lysates containing 2 mg of proteins were subjected to immunoprecipitation using anti-HA antibodies and protein G-Sepharose beads. Precipitated HA-Sac7p or HA-Sac7p(R173A) bound on the beads was washed four times with extraction buffer I. GST and GST-Rho1p were purified as described above. The pulled down GST and GST-Rho1p were preloaded with 1 μCi of [α-32P]GTP for 15 min at 25°C. The reaction mixtures were first mixed with the immune complex containing HA-Sac7p, HA-Sac7p(R173A), or beads only in extraction buffer I containing 2 mM dithiothreitol (DTT) and 2 mM MgCl2 and were then incubated at 25°C for 5 min. Beads were collected, washed three times with extraction buffer I, and then incubated with 15 μl of nucleotide elution buffer (1% SDS-20 mM EDTA) at 65°C for 5 min to elute the bound nucleotides. Eluates were loaded onto a polyethyleneimine cellulose F plate (Merck) and subjected to thin-layer chromatography along with nucleotide standards to identify the nucleotides.
Purification of GST-RBD and GST-Pkc1-RBD.
Escherichia coli plasmids expressing a GST-fused rhotekin binding domain (GST-RBD) or a GST-fused Rho1p-binding domain of Saccharomyces Pkc1p (GST-Pkc1-RBD) were provided by J.-Y. Chen at the Institute of Biomedical Sciences, Academia Sinica, and David Pellman at Dana-Farber Cancer Institute, respectively. E. coli cells expressing GST, GST-RBD, or GST-Pkc1-RBD were collected, resuspended in cold buffer (1 M Tris-HCl [pH 7.5], 5 M NaCl, 0.5 M EDTA) with 1% Triton X-100, and lysed by two passages through a French press at a pressure of 1,000 lb/in2. Lysates were centrifuged at 12,000 rpm for 30 min at 4°C, and glutathione-Sepharose 4B beads (Amersham Biosciences) were added to the supernatants. After incubation at 4°C for 1 h, the protein-bound beads were collected, washed five times, resuspended in cold buffer, and stored at −70°C for further use.
Pull-down assays for active Rho1p levels.
Assays for Rho1p activation were performed as described elsewhere (35, 64) with modifications. Transformed wild-type and avo3ts yeast cells were induced to express Rho1p-HA by galactose and were incubated at 37°C for 2 h before the preparation of lysates. For GST-RBD binding, harvested cells were lysed in GPLB buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM MgCl2, 0.5% NP-40, 5 mM glycerophosphate, 5 mM NaF, 1 mM DTT, 1 mM PMSF) with protease inhibitor cocktail, and the extracts were incubated with either bead-bound GST-RBD or GST alone for 1 h at 4°C. Beads were pulled down, washed three times with GPLB buffer, and resuspended in sample buffer. For GST-Pkc1-RBD binding, harvested cells were lysed in a lysis buffer {150 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 12 mM MgCl2, 1 mM DTT, 1 mM PMSF, 0.6% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS]} with protease inhibitor cocktail (Roche), and lysates containing 5 mg of proteins were incubated with bead-bound GST-Pkc1-RBD or GST at 4°C for 1 h. Beads were collected by centrifugation, washed five times with lysis buffer, and resuspended in sample buffer. Pulled down samples were analyzed by immunoblotting with anti-HA antibodies to detect the active form of Rho1p-HA.
RESULTS
The composition of TORC2 is differentially affected in two different avo3ts mutants.
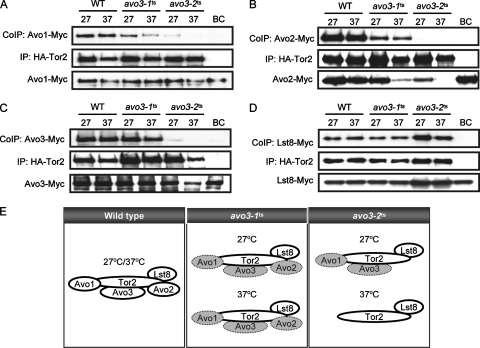
Our previous studies have identified two distinct temperature-sensitive avo3 mutants, avo3-1ts and avo3-2ts (originally referred to as tsB1 and tsD1 mutants, respectively) (25), and found that AVO1 was an avo3-1ts mutant-specific multicopy suppressor while AVO2 and SLM1 were avo3-2ts mutant-specific multicopy suppressors. Since AVO1 and AVO2 encode TORC2 components, the observed allele-specific suppression raised the possibility that the two avo3ts alleles might have affected TORC2 integrity differently. Hence, we set out to investigate TORC2 composition in these avo3ts mutants. To this end, we engineered chromosomal gene fusions that express epitope-tagged forms of TORC2 component proteins. By using strains with double tags (see Table S1 in the supplemental material), binary interactions of Tor2p with other TORC2 components were individually examined in wild-type, avo3-1ts, and avo3-2ts backgrounds by coimmunoprecipitation. Our results showed that in avo3-1ts cells, the Tor2p-Avo1p interaction was slightly affected at the permissive temperature (27°C) and was obviously decreased at the nonpermissive temperature (37°C) from that in wild-type cells; in avo3-2ts cells, the Tor2p-Avo1p interaction was barely detected at 27°C and undetectable at 37°C (Fig. 1A). The Tor2p-Avo2p interaction was reduced in avo3-1ts cells and completely undetectable in avo3-2ts cells at both 27°C and 37°C (Fig. 1B). We also noted a significant decrease in Avo2p expression in both mutants at 37°C and in avo3-2ts cells even at 27°C, suggesting that avo3ts mutations may have affected the stability of Avo2p. The Tor2p-Avo3p interaction was slightly decreased in avo3-1ts cells at both temperatures, while it was dramatically decreased at 27°C and undetectable at 37°C in avo3-2ts cells, compared to that in wild-type cells (Fig. 1C). On the other hand, the Tor2p-Lst8p interaction did not appear to be affected in either avo3ts mutant (Fig. 1D). Taken together, the results demonstrated that defects in Avo3p could indeed influence the molecular organization of TORC2. The deduced TORC2 compositions of different mutants are summarized in Fig. 1E. In avo3-1ts cells, TORC2 integrity was significantly perturbed, with weakened binary interactions between Tor2p and other TORC2 components. In avo3-2ts cells, TORC2 composition was greatly altered, lacking Avo2p even at permissive temperatures and losing all three Avo proteins at nonpermissive temperatures. The different effects of avo3 mutations on TORC2 structure in avo3-1ts and avo3-2ts cells are compatible with the possibility that Avo3p may serve as a scaffold protein in TORC2.
FIG. 1.
Interactions between Tor2p and other TORC2 components are differentially affected in different avo3ts mutants. Mid-log-phase cultures of strains simultaneously expressing HA-tagged Tor2p and another, Myc-tagged TORC2 component were divided into two aliquots and further incubated at 27°C or 37°C for 2 h. Cell lysates were prepared and subjected to immunoprecipitation (IP) using anti-HA antibodies. Immunoprecipitates were separated by SDS-PAGE and examined for coimmunoprecipitated (CoIP) partner proteins by Western blot analysis using anti-Myc antibodies. Expression of the Myc-tagged TORC2 component in the lysates is shown at the bottom of each panel. “Bead control” (BC) samples were prepared using lysates from double-epitope-tagged wild-type (WT) strains without adding anti-HA antibodies during immunoprecipitation. (A) Tor2p-Avo1p interaction detected in YMY120 (WT), YMY218 (avo3-1ts), and YMY318 (avo3-2ts). (B) Tor2p-Avo2p interaction detected in YMY121 (WT), YMY219 (avo3-1ts), and YMY319 (avo3-2ts). (C) Tor2p-Avo3p interaction detected in YMY123 (WT), YMY221 (avo3-1ts), and YMY321 (avo3-2ts). (D) Tor2p-Lst8p interaction detected in YMY122 (WT), YMY220 (avo3-1ts), and YMY320 (avo3-2ts). (E) Schematic summary of TORC2 composition in different strains at the indicated temperatures. Shaded ovals indicate proteins with weakened interaction with Tor2p.
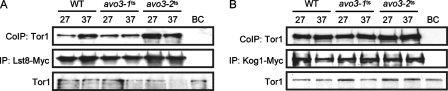
We also examined the composition of TORC1 in avo3-1ts and avo3-2ts cells. Consistent with the fact that Avo3p is a TORC2-specific component, neither the Tor1p-Lst8p (Fig. 2A) nor the Tor1p-Kog1p (Fig. 2B) interaction was affected in the two avo3ts mutants.
FIG. 2.
Interactions between TORC1 components are not affected in avo3ts mutants. Strains expressing a Myc-tagged TORC1 component were subjected to the same procedures as those for Fig. 1. Immunoprecipitates were examined by Western blot analysis using anti-Tor1p antibodies. Expression of Tor1p in the lysates is shown at the bottom of each panel. WT, wild type; BC, bead control; IP, immunoprecipitation; CoIP, coimmunoprecipitation. (A) Tor1p-Lst8p interaction detected in YMY118 (WT), YMY216 (avo3-1ts), and YMY316 (avo3-2ts). (B) Tor1p-Kog1p interaction detected in YMY119 (WT), YMY217 (avo3-1ts), and YMY317 (avo3-2ts).
Allele-specific multicopy suppressors do not restore TORC2 integrity in avo3ts mutants.
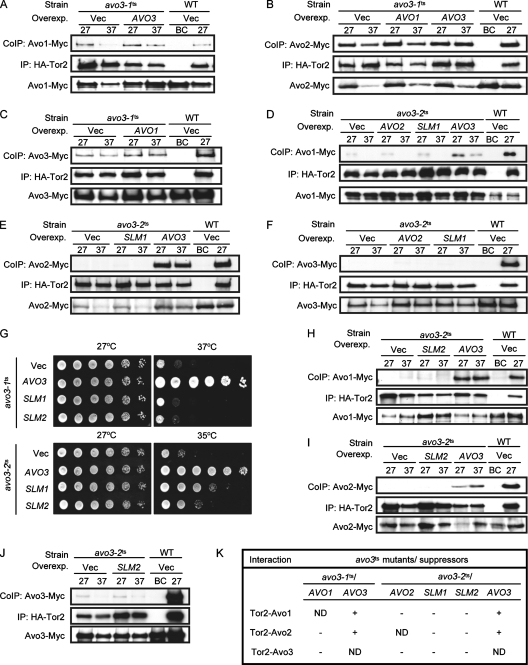
We next investigated if expression of avo3-1ts- or avo3-2ts-specific multicopy suppressors could restore TORC2 integrity in the corresponding avo3ts mutants. Vectors overexpressing untagged Avo1p, Avo2p, Slm1p, or Avo3p were respectively transformed into double-epitope-tagged strains to analyze interactions between Tor2p and other TORC2 components by coimmunoprecipitation. We did not examine the interaction between Tor2p and any specific tagged TORC2 component when its untagged version was overexpressed, since the untagged component could compete with the tagged form in interacting with Tor2p, and therefore the amounts of the coimmunoprecipitated tagged form might not be a good measure of the interaction. Our results showed that AVO3 overexpression could restore Tor2p-Avo1p and Tor2p-Avo2p interactions in both avo3ts mutants (Fig. 3A, B, D, and E). In avo3-1ts cells, overexpression of the suppressor AVO1 could not rescue Tor2p-Avo2p or Tor2p-Avo3p interactions (Fig. 3B and C), suggesting that the suppression effect of AVO1 does not work by restoring the integrity of TORC2. Likewise, in avo3-2ts cells, overexpression of AVO2 or SLM1 could not restore Tor2p-Avo1p and Tor2p-Avo3p interactions (Fig. 3D and F), and SLM1 also failed to rescue Tor2p-Avo2p interaction (Fig. 3E), indicating that neither AVO2 nor SLM1 suppresses avo3-2ts phenotypes by restoring TORC2 composition. Because Slm1p and Slm2p are two functionally redundant phosphatidylinositol-4,5-bisphosphate-binding proteins acting downstream of TORC2 to control actin organization (2, 15), we tested if SLM2 could also be a multicopy suppressor for avo3ts alleles. The results showed that SLM2 overexpression did not rescue the growth of avo3-1ts cells at the nonpermissive temperature but partially suppressed the temperature sensitivity of avo3-2ts cells (Fig. 3G), indicating that SLM2 is also an avo3-2ts-specific suppressor, albeit a slightly weaker one than SLM1. Despite its suppression of temperature sensitivity, SLM2 overexpression failed to restore the interaction of Tor2p with Avo1p, Avo2p, or Avo3p (Fig. 3H to J). Altogether, while AVO3 rescued the affected interactions of TORC2 components in avo3ts mutants, none of the suppressors tested could do so (Fig. 3K), suggesting that Avo3p plays a pivotal role in maintaining TORC2 integrity and that the multicopy suppressors of avo3-1ts and avo3-2ts alleles may exert their suppression effects by acting on downstream signaling.
FIG. 3.
Overexpression of AVO3 but not allele-specific multicopy suppressors restores interactions between Tor2p and other TORC2 components in avo3ts mutants. Plasmids expressing AVO3 (pTSS1), AVO1 (pHS2), AVO2 (pHS5), SLM1 (pHS6), or SLM2 (pHS12) and the control vector (pRS424) (Vec) were individually transformed into doubly tagged strains expressing HA-Tor2p and another, Myc-tagged TORC2 component. Lysates were prepared from transformants, and coimmunoprecipitation (CoIP) was performed as in the experiments for which results are shown in Fig. 1, by using anti-HA antibodies to immunoprecipitate (IP) HA-Tor2p and anti-Myc antibodies to detect the coprecipitated partner protein. Expression of the Myc-tagged TORC2 component in lysates is shown at the bottom of each panel. Overexp., overexpression; WT, wild type. (A) Tor2p-Avo1p interaction detected in transformants of YMY120 (WT) and YMY218 (avo3-1ts). (B) Tor2p-Avo2p interaction detected in transformants of YMY121 (WT) and YMY219 (avo3-1ts). (C) Tor2p-Avo3p interaction detected in transformants of YMY123 (WT) and YMY221 (avo3-1ts). (D and H) Tor2p-Avo1p interaction detected in transformants of YMY120 (WT) and YMY318 (avo3-2ts). (E and I) Tor2p-Avo2p interaction detected in transformants of YMY121 (WT) and YMY319 (avo3-2ts). (F and J) Tor2p-Avo3p interaction detected in transformants of YMY123 (WT) and YMY321 (avo3-2ts). (G) SLM2 serves as an avo3-2ts-specific multicopy suppressor like SLM1. Transformants of YMY99 (avo3-1ts) and YMY100 (avo3-2ts) were subjected to a spot assay for temperature sensitivity. (K) Summary of the effects of individual avo3ts mutant-specific suppressors on interactions between Tor2p and other TORC2 components. −, unable to rescue; +, able to rescue; ND, not determined.
Interactions between Tor2p and the downstream effectors Slm1p and Slm2p are differentially affected in avo3ts mutants.
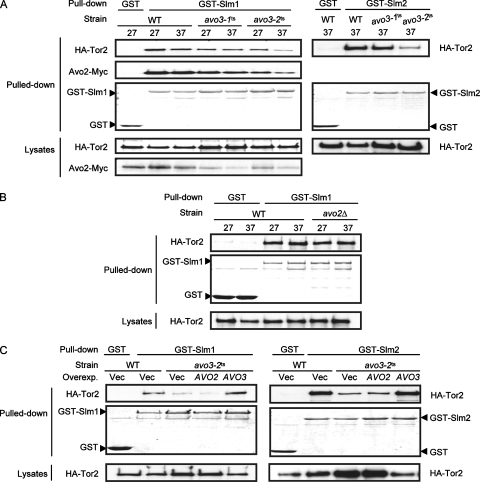
Slm1p and Slm2p have been found to physically interact with Avo2p and to serve as downstream effectors of TORC2 (2, 15). Given that SLM1 or SLM2 suppresses the avo3-2ts but not the avo3-1ts allele (25) (Fig. 3G), we investigated whether these avo3ts alleles differently affected the physical interaction of Slm proteins with TORC2. The interaction between Slm1p and TORC2 appeared rather weak; we were not able to observe their interaction at endogenous protein expression levels by coimmunoprecipitation (data not shown). We therefore examined the interaction by a GST pull-down assay in strains expressing GST-Slm1p and a tagged TORC2 component(s). In the wild-type background, Tor2p and Avo2p could be pulled down together with GST-Slm1p at 27°C or 37°C, demonstrating the interactions between Slm1p and these TORC2 components (Fig. 4A). Compared to those in wild-type cells, the amounts of Tor2p and Avo2p pulled down with GST-Slm1p were very slightly decreased, if at all, in avo3-1ts cells but markedly reduced in avo3-2ts cells at the nonpermissive temperature. When similar experiments were done using GST-Slm2p in place of GST-Slm1p, we also observed noticeably less pulled down Tor2p in avo3-2ts cells than in wild-type or avo3-1ts cells. These data suggest a weakened interaction of TORC2 with the two downstream effector proteins in avo3-2ts but not avo3-1ts cells, consistent with our finding that SLM1 and SLM2 serve as specific suppressors for the avo3-2ts but not the avo3-1ts allele.
FIG. 4.
Different avo3ts mutations affect TORC2 interaction with Slm proteins differently. (A) Yeast strains were respectively transformed with plasmids to express GST (pGAL1-GST-URA3), GST-Slm1p (pHS10), or GST-Slm2p (pHS13) under the control of the GAL1 promoter. On the left, doubly tagged strains expressing 3HA-Tor2p and Avo2p-13Myc, including YMY121 (wild type [WT]), YMY219 (avo3-1ts), and YMY319 (avo3-2ts), were used. On the right, strains expressing 3HA-Tor2p, including YMY116 (WT), YMY215 (avo3-1ts), and YMY315 (avo3-2ts), were used. Transformants were induced to express SLM1 or SLM2 for 2 h and were further grown at 27°C or shifted to 37°C for another 2 h. Total lysates were prepared and subjected to GST pull-down procedures. The pulled down proteins were separated by SDS-PAGE; stained with Coomassie blue to visualize the pulled down GST, GST-Slm1p, or GST-Slm2p; and blotted with anti-HA or anti-Myc antibodies to detect the copurified 3HA-Tor2p or Avo2p-13Myc. Expression levels of 3HA-Tor2p and Avo2p-13Myc in lysates were also examined by Western blot analysis. (B) WT (YMY116) and avo2Δ (YMY124) strains expressing 3HA-Tor2p were respectively transformed with pGAL1-GST-URA3 or pHS10 and subjected to the same procedures as those for panel A. (C) YMY315 (avo3-2ts) cells were respectively transformed with the control vector pRS424 (Vec), pHS5 (AVO2), or pTSS1 (AVO3), together with pHS10 or pHS13. The interaction of Tor2p with Slm1p or Slm2p was examined as described above and compared to that in YMY116 (WT) transformants.
No Tor2p-Avo2p interaction was detectable in avo3-2ts cells (Fig. 1B), yet some amount of Tor2p was still pulled down with GST-Slm1p (Fig. 4A), suggesting that Avo2p may not be required for the interaction between Tor2p and Slm1p. To address this question more directly, we examined Tor2p-Slm1p interaction when AVO2 was deleted. We found that similar amounts of Tor2p were pulled down together with GST-Slm1p in wild-type and avo2Δ cells (Fig. 4B), showing that Avo2p is not essential in order for Slm1p to interact physically with Tor2p. Besides Avo2p, Slm1p interacts with two other TORC2-interacting proteins, Bit61p and the YBR270C product (15). However, Bit61p and the YBR270C product also appeared dispensable for Tor2p-Slm1p interaction, since similar amounts of Tor2p were pulled down by GST-Slm1p in wild-type, bit61Δ, and ybr270cΔ cells (data not shown). Taken together, our data suggest that the decreased amount of Tor2p pulled down with GST-Slm1p in avo3-2ts cells reflects the perturbing effect of specific avo3ts mutations on TORC2-Slm1p interaction.
We tested the effect of overexpressing the allele-specific suppressor AVO2 on the recruitment of the two Slm proteins to TORC2 in avo3-2ts cells. While overexpression of AVO3 in avo3-2ts cells increased the amounts of Tor2p pulled down with GST-Slm1p or GST-Slm2p to a wild-type-like level, overexpression of AVO2 did not (Fig. 4C); these observations highlight the essential role of Avo3p in modulating Tor2p-Slm1p (or Tor2p-Slm2p) interaction. Altogether, our data demonstrated the differential effects of mutations in avo3-1ts and avo3-2ts cells on the physical interaction of Tor2p and its downstream effector proteins, raising the possibility that Avo3p may modulate the coupling of TORC2 to specific downstream signaling pathways.
AVO3 displays genetic interactions with RhoGAPs.
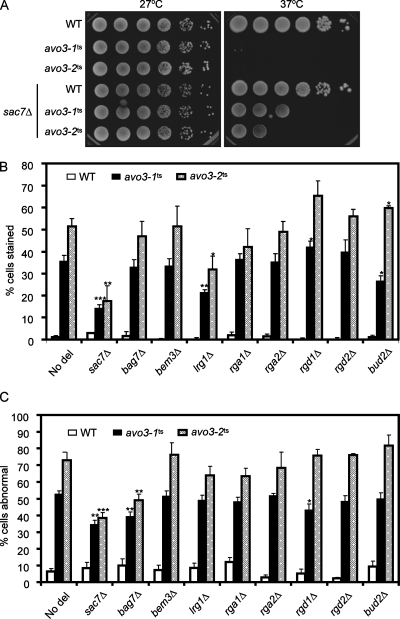
We investigated next whether different avo3 mutations exert differential effects on TORC2 downstream signaling. Although the exact molecular linkage remains unclear, the Rho1p pathway represents one of the TORC2 downstream signaling pathways, since tor2 mutants can be rescued by overexpressing components of the Rho1p signaling pathway (23, 58). Considering that the major defects of avo3ts mutants are in cell wall integrity and actin organization, and that most members of the Rho GTPase family regulate actin organization or cell wall modeling (41, 49), we decided to explore the possible involvement of different Rho family proteins in Avo3p signaling. Our strategy was to delete individual negative regulators of Rho family GTPases, i.e., RhoGAPs, in avo3ts mutants, thus activating the downstream pathway, and to look for modification of avo3ts phenotypes. Searching through the Saccharomyces cerevisiae genome database (http://www.yeastgenome.org/), we identified nine genes encoding RhoGAPs as targets for deletion. We also chose to delete two RasGAP genes, IRA2 and BUD2, as controls to check if the genetic interaction is specific to RhoGAPs. Individual deletions of these GAP genes were generated in wild-type, avo3-1ts, and avo3-2ts backgrounds. Assays for four phenotypes of avo3ts mutants, temperature sensitivity, caffeine sensitivity, actin organization, and cell wall defects, were performed to check for genetic interaction between each deletion and the avo3ts alleles.
We checked temperature sensitivity by comparing the growth of strains with or without a specific GAP gene deletion in different mutant backgrounds at 37°C. Among the 11 GAP deletions we examined, only SAC7 deletion was able to partially rescue the temperature sensitivity of avo3-1ts and avo3-2ts mutants (Fig. 5A).
FIG. 5.
The phenotypes of avo3ts mutants can be partially rescued by deletion of specific RhoGAP-encoding genes. (A) Deletion of SAC7 partially rescues the temperature sensitivity of avo3ts mutants. Tenfold serial dilutions of different yeast cultures were spotted onto YPD plates and incubated at different temperatures until colonies appeared. (B) The cell wall integrity of yeast strains carrying different GAP gene deletions was assessed by the trypan blue assay as described in Materials and Methods. The percentage of trypan blue-stained cells represents the extent of the cell wall defect of each strain. Shown are means ± standard deviations of results from three independent experiments. Statistical data were derived from Student's t test; each deletion strain was compared to its corresponding strain without any GAP gene deletion (No del). ***, P < 0.001; **, P < 0.005; *, P < 0.05. (C) Actin distribution in yeast strains carrying different GAP gene deletions was examined by TRITC-phalloidin staining and fluorescence microscopy. Small budded cells with four or more actin patches in the mother cell were counted as cells with abnormal actin distribution. Shown are means ± standard deviations of results from three independent experiments. Statistical data were obtained as for panel B.
We next assessed the effect of each RhoGAP gene deletion on the cell wall integrity of avo3ts mutants. Although the molecular targets for caffeine in signaling pathways serving to maintain cell integrity are still elusive, increased caffeine sensitivity correlates with defects in the cell integrity pathway (36, 43). We compared the cell growth of strains with or without deletions in medium containing 6 mM caffeine. In the wild-type background, none of the GAP gene deletions affected growth in the absence of caffeine (data not shown), and only BEM2 deletion rendered cells slightly more caffeine sensitive (Table 1). However, analyses done in the mutant backgrounds revealed two different groups of deletions interacting with avo3ts alleles, one ameliorating and the other aggravating caffeine sensitivity; in both avo3ts backgrounds, disruption of SAC7, LRG1, or BEM3 improved the growth of avo3ts cells in caffeine, while deletion of BEM2 or RGD1 worsened such growth. Unexpectedly, deletion of the RasGAP gene IRA2 also increased caffeine sensitivity in avo3ts mutants, suggesting an interaction between Avo3p signaling and the Ras pathway. The cross talk may be mediated by the Rom2p-Rho1p-Pkc1p pathway, since a previous report describes evidence for the negative regulation of the Ras-cAMP pathway by Rom2p (46). It is conceivable that the downregulation of the Rho1p pathway activity caused by avo3ts mutations and the hyperactivation of the Ras-cAMP pathway due to IRA2 deletion together lead to increased sensitivity to caffeine stress. A second test, the trypan blue staining assay, was used to check if any RhoGAP deletions could rescue the cell wall defect of avo3ts mutants. Because BEM2 deletion caused swollen cell bodies with large vacuoles and very little cytoplasm in all the yeast backgrounds we used (data not shown), making it difficult to differentiate normal and abnormal cell walls or actin organization, bem2Δ strains were excluded from this assay (and the actin assay). In agreement with the results of the caffeine plate assay, deletion of SAC7 or LRG1 in both avo3ts mutants significantly lowered the percentages of trypan blue staining, indicating improved cell wall integrity, while RGD1 deletion slightly increased the numbers of trypan blue-stained avo3ts cells, demonstrating the worsening of the cell integrity defect (Fig. 5B). Perhaps due to different sensitivities of the tests, trypan blue staining did not reveal an effect of BEM3 deletion on the cell integrity defect of avo3ts cells as the caffeine sensitivity assay did. Intriguingly, disruption of the RasGAP gene BUD2 had opposite effects on the trypan blue staining of avo3-1ts and avo3-2ts mutants, rescuing the avo3-1ts cells while aggravating the cell integrity defect in avo3-2ts cells. The molecular basis for this observation awaits further investigation.
TABLE 1.
Effects of RhoGAP deletions on the caffeine sensitivities of avo3ts mutants
| GAP deletion | Cellular target(s) of deleted GAP | Growtha of yeast cells
|
||
|---|---|---|---|---|
| Wild type | avo3-1ts | avo3-2ts | ||
| None | ++++++ | ++++ | +++ | |
| sac7Δ | Rho1, Rho2 | ++++++ | +++++ | +++++ |
| bag7Δ | Rho1 | ++++++ | ++++ | +++ |
| bem2Δ | Rho1, Cdc42 | +++++ | − | − |
| lrg1Δ | Rho1 | ++++++ | ++++++ | ++++++ |
| rgd1Δ | Rho3, Rho4 | ++++++ | ++ | + |
| bem3Δ | Cdc42, Rho1 | ++++++ | +++++ | +++++ |
| rga1Δ | Cdc42 | ++++++ | ++++ | +++ |
| rga2Δ | Cdc42 | ++++++ | ++++ | +++ |
| rgd2Δ | Cdc42, Rho5 | ++++++ | ++++ | +++ |
| ira2Δ | Ras1, Ras2 | ++++++ | ++ | − |
| bud2Δ | Rsr1 | ++++++ | ++++ | +++ |
At 27°C in 6 mM caffeine. The more plus signs, the greater the extent of growth. −, no growth.
To evaluate the effects of RhoGAP deletions on the actin phenotype of avo3ts mutants, yeast cells were stained by fluorophore-conjugated phalloidin to visualize the distribution of polymerized actin. Deletion of either SAC7 or BAG7 caused decreases in the percentages of cells with abnormal actin distribution in both avo3ts mutants (Fig. 5C), indicating a partial rescue of the actin phenotype. Deletion of RGD1, however, rescued the actin phenotype only in avo3-1ts cells, not in the avo3-2ts background (Fig. 5C).
Altogether, our genetic tests identified six RhoGAPs, including SAC7, BAG7, BEM2, LRG1, RGD1, and BEM3, whose deletion could modify the phenotypes of avo3ts mutants. Among these AVO3-interacting RhoGAPs, Sac7p, Bag7p, Bem2p, and Lrg1p regulate Rho1p, Rgd1p specifically inhibits Rho3p/Rho4p, and Bem3p acts on Cdc42p and Rho1p (Table 1) (14, 54, 60, 61). With the exception of BEM2, deletion of genes encoding GAPs for Rho1p significantly rescued the phenotypes of avo3ts mutants, implying that Rho1p acts as a main effector in Avo3p downstream signaling.
Deletion of SAC7 rescues avo3ts mutants by removing GAP activity.
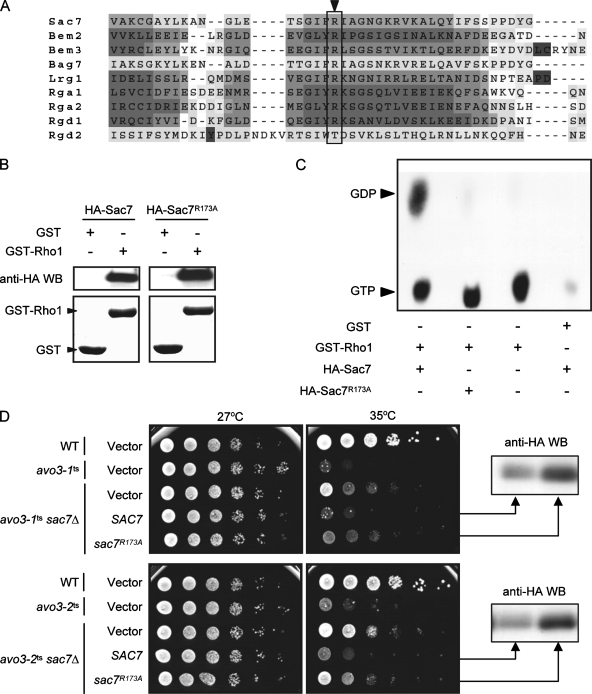
The Rho1p-GAP Bem2p has been reported to serve GAP-independent functions (42). It is not clear whether other Rho1p-GAPs, including Sac7p, Bag7p, and Lrg1p, also have such GAP-independent functions; thus, it remains possible that the rescue of avo3ts mutants was caused by the removal of GAP-independent functions of the deleted RhoGAP. To understand the mechanism underlying avo3ts suppression by RhoGAP deletions, we further analyzed Sac7p, since its deletion rescued most avo3ts phenotypes.
In order to investigate the effect of Sac7p GAP activity on avo3ts cells, we sought to generate a GAP-dead mutant allele, sac7-GD, expressing a mutant form of Sac7p that lacks GAP activity but retains other functions, by mutating the GAP domain. We aligned the GAP domains of different RhoGAPs in yeast by using T-Coffee analysis (http://www.ebi.ac.uk/t-coffee/) (45) to find conserved residues, which may more likely be essential for the GAP activity. An arginine at position 173 (R173) of Sac7p was one of the most conserved residues we found among GAP domains from different RhoGAPs (Fig. 6A); mutation at the corresponding Arg residue of Bem2p abolishes its GAP activity toward Cdc42p without disturbing the overall folding of the GAP domain (42). We therefore changed R173 of Sac7p into an alanine by site-directed PCR mutagenesis and tested if sac7(R173A) acts as a sac7-GD allele. In a GST pull-down experiment, the R173A mutant form of Sac7p was able to interact with Rho1p as well as wild-type Sac7p did (Fig. 6B), suggesting that the substitution did not affect the global protein structure or the Rho1p-interacting activity of Sac7p. We then tested Sac7p(R173A) for its GAP activity toward Rho1p in an assay where [α-32P]GTP-loaded GST-Rho1p was incubated with immunoprecipitated wild-type or mutant HA-tagged Sac7p. While wild-type HA-Sac7p was able to convert the Rho1p-bound GTP into GDP, no Rho1p-bound GDP could be detected when the reaction contained HA-Sac7p(R173A) (Fig. 6C), suggesting that the R173A substitution eliminated the GAP activity of Sac7p and indeed produced a GAP-dead mutant protein.
FIG. 6.
Removal of the Sac7p GAP activity contributes to the rescue of avo3ts mutants. (A) Comparison of GAP domains from different yeast RhoGAPs. The multiple sequence alignment was generated by T-Coffee analysis. The arrowhead indicates a conserved Arg residue at position 173 of Sac7p. (B) Mutation of Arg173 in Sac7p does not affect its Rho1p-binding activity. The interaction between Rho1p and Sac7p was examined using a GST pull-down assay. Glutathione Sepharose bead-bound GST or GST-Rho1p was incubated with yeast lysates containing HA-Sac7p or HA-Sac7p(R173A). Pulled down proteins were examined by Western blot analysis (WB) using anti-HA antibodies (upper panels) or Coomassie blue staining (lower panels). (C) Mutation of Arg173 in Sac7p affects its GAP activity. HA-Sac7p or HA-Sac7p(R173A) was incubated with [α-32P]GTP-loaded GST-Rho1p to assay for GAP activity in vitro. Bound guanine nucleotides were extracted and analyzed by thin-layer chromatography. An autoradiograph of the thin-layer chromatographic plate is shown. (D) Spot assay for temperature sensitivity. Tenfold serial dilutions of the indicated yeast cell suspensions were spotted onto plates and incubated at different temperatures until colonies formed. Western blot analysis using anti-HA antibodies confirmed the expression of HA-Sac7p or HA-Sac7p(R173A) in strains.
We next examined the effects of expressing wild-type or GAP-dead versions of Sac7p in yeast cells. Plasmids expressing SAC7 or sac7(R173A) from its endogenous promoter were transformed into wild-type and avo3ts strains with or without SAC7 deletion. Expressing SAC7 or sac7(R173A) in the wild-type background did not influence cell growth at permissive or nonpermissive temperatures (data not shown). Compared to avo3ts cells without SAC7 deletion, sac7Δ avo3ts cells displayed better viability at nonpermissive temperatures; reexpression of SAC7 in sac7Δ avo3ts cells eliminated the suppression effect of SAC7 deletion on temperature sensitivity (Fig. 6D). In contrast, although Sac7p(R173A) protein was expressed in sac7Δ avo3ts cells, it had little effect on growth at nonpermissive temperatures. These results indicate that expression of Sac7p(R173A) could not cover the function(s) served by the wild-type version of Sac7p in reversing the suppression effect of SAC7 deletion on avo3ts phenotypes. Together, these data suggest that removal of the GAP activity of Sac7p was the major reason why SAC7 deletion could rescue avo3ts mutants.
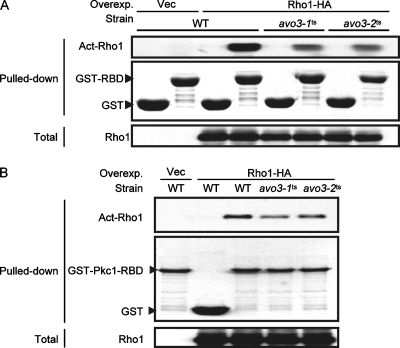
Both avo3-1ts and avo3-2ts mutants are defective in Rho1p activation.
Since our results described above point out Rho1p as a major Avo3p downstream effector and show that removal of Sac7p GAP activity toward Rho1p was sufficient to rescue avo3ts mutants, Rho1p activation may very likely be defective in avo3ts mutants. To address this question, we employed two recombinant proteins, GST-RBD and GST-Pkc1p-RBD, both of which bind specifically to the GTP-bound active form of Rho1p (35, 64, 71). Lysates prepared from wild-type or avo3ts cells expressing Rho1p-HA were mixed with glutathione-Sepharose bead-bound GST-RBD or GST-Pkc1p-RBD and subjected to pull-down procedures. We detected lower levels of active Rho1p in lysates from both avo3ts mutants cultured at nonpermissive temperatures than in lysates from wild-type cells by using either GST-RBD (Fig. 7A) or GST-Pkc1p-RBD (Fig. 7B), suggesting that Avo3p functions in modulating Rho1p activation.
FIG. 7.
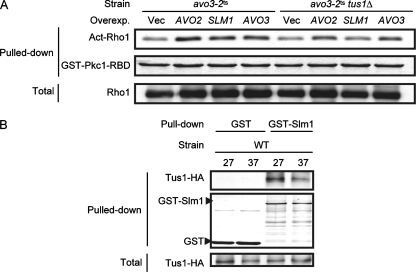
Mutations of AVO3 cause decreases in active Rho1p (Act-Rho1) levels in cells. Lysates were prepared from transformants of YMY97 (wild type [WT]), YMY99 (avo3-1ts), and YMY100 (avo3-2ts) induced to express Rho1p-HA at 37°C for 2 h. The GTP-bound form of Rho1p-HA in lysates was pulled down using either GST-RBD (A) or GST-Pkc1-RBD (B). Pulled down samples were separated by SDS-PAGE; GST fusion proteins were detected by Coomassie blue staining, and active Rho1p-HA was detected by Western blot analysis using anti-HA antibodies. Total Rho1p-HA levels in lysates were also examined by Western blot analysis. Overexp., overexpression; Vec, vector.
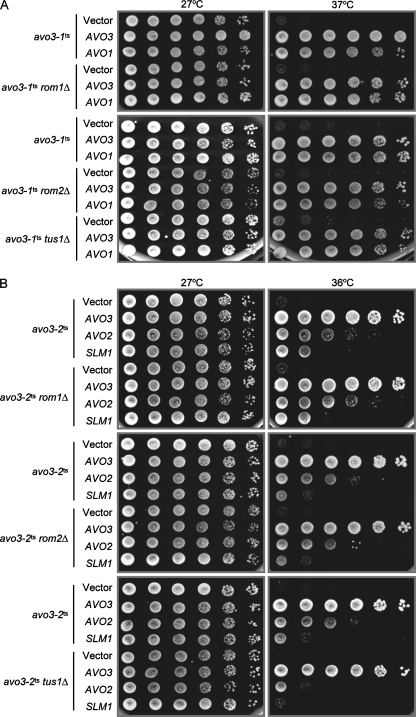
Avo2p and Slm1p signal to the Rho1p pathway through one of the Rho1p-GEFs, Tus1p.
To investigate whether avo3-1ts- and avo3-2ts-specific suppressors rescued avo3ts mutants by boosting Rho1p activation, we next tested if positive regulators, i.e., Rho1p-GEFs, are required for the dosage suppression. We deleted each of the known Rho1p-GEFs, ROM1, ROM2 and TUS1, in avo3ts backgrounds and examined the effect of each GEF deletion on avo3ts suppression by allele-specific suppressors. Our results revealed that in the avo3-1ts background, the allele-specific suppressor AVO1 still suppressed temperature sensitivity, even when ROM1, ROM2, or TUS1 was deleted (Fig. 8A). In the avo3-2ts background, deletion of either ROM1 or ROM2 had no effect, while TUS1 deletion considerably reduced the AVO2- and SLM1-rescued growth of mutant cells at nonpermissive temperatures (Fig. 8B). These results suggest that avo3-2ts-specific suppression by AVO2 or SLM1 is linked to Tus1p-mediated Rho1p activation. We then examined the effect of AVO2 or SLM1 overexpression in avo3-2ts cells on their levels of active Rho1p and found that overexpression of AVO2 or SLM1 indeed increased the amount of active Rho1p to a level similar to that in AVO3-complemented mutant cells (Fig. 9A) and that this increase was at least partially dependent on Tus1p, since TUS1 deletion reduced the effect. Since Avo2p exists in TORC2 and Slm1p is considered a TORC2 downstream effector, we tested if Slm1p interacts with Tus1p to link to the Rho1p pathway. Using lysates from wild-type yeast cells coexpressing GST-Slm1p and Tus1p-HA to perform a GST pull-down assay, we discovered that Tus1p-HA could copurify with GST-Slm1p (Fig. 9B), demonstrating a physical interaction between Tus1p and Slm1p. These observations together suggest that there exists an Avo2p-specific TORC2 downstream signaling pathway acting through Slm1p and Tus1p to activate Rho1p.
FIG. 8.
Tus1p participates in Avo2p- and Slm1p-mediated, but not Avo1p-mediated, suppression of avo3ts phenotypes. Three known Rho1p GEF genes, ROM1, ROM2, and TUS1, were individually deleted in avo3ts mutants. The resulting strains were transformed by allele-specific multicopy suppressors. Transformants were subjected to spot assays for growth to test the effects of GEF deletion on suppression. (A) Rho1p-GEF deletions have no effect on AVO1-mediated suppression of the avo3-1ts phenotype. Plasmid pRS424 (Vector), pTSS1 (AVO3), or pHS2 (AVO1) was transformed into YMY99 (avo3-1ts), YMY212 (avo3-1ts rom1Δ), YMY213 (avo3-1ts rom2Δ), or YMY214 (avo3-1ts tus1Δ). The resulting transformants were tested for growth at different temperatures. (B) Deletion of TUS1, but not ROM1 or ROM2, reduces AVO2- and SLM1-mediated suppression of the avo3-2ts phenotype. Plasmid pRS424 (Vector), pTSS1 (AVO3), pHS5 (AVO2), or pHS6 (SLM1) was transformed into YMY100 (avo3-2ts), YMY312 (avo3-2ts rom1Δ), YMY313 (avo3-2ts rom2Δ), or YMY314 (avo3-2ts tus1Δ). Transformants were tested for growth at different temperatures.
FIG. 9.
Tus1p links Avo2p and Slm1p signaling to Rho1p activation. (A) Overexpression (Overexp.) of AVO2 or SLM1 restores levels of active Rho1p in avo3-2ts cells. Plasmids expressing AVO3 (pTSS1), AVO2 (pHS5), or SLM1 (pHS6) from their endogenous promoters and the control vector (pRS424) were individually transformed into YMY100 (avo3-2ts) or YMY314 (avo3-2ts tus1Δ). After induction of Rho1p-HA expression at 37°C for 2 h, lysates were prepared and subjected to the pull-down assay for active Rho1p-HA (Act-Rho1) by using GST-Pkc1-RBD. Coomassie blue staining was used to detect GST-Pkc1p-RBD, while Western blot analysis with anti-HA antibodies was used to detect active and total Rho1p-HA. (B) Slm1p physically interacts with Tus1p. YMY97 (wild type [WT]) cells were cotransformed with plasmids expressing Tus1p-HA (pHS11) and GST (pGAL1-GST-URA3) or GST-Slm1p (pHS10) from the GAL1 promoter. Lysates were prepared from transformants induced by galactose and grown at 27°C or shifted to 37°C for 2 h; then the lysates were subjected to GST pull-down procedures. Pulled down proteins were separated by SDS-PAGE, stained by Coomassie blue to visualize the GST or GST-Slm1p, and examined by Western blot analysis with anti-HA antibodies for copurified Tus1p-HA. Expression levels of Tus1p-HA in lysates were also examined by Western blot analysis.
DISCUSSION
Considering the diverse cellular events influenced by TOR and the existence of TOR-containing multiprotein complexes within cells, it is plausible that TOR-binding partners participate in specifying target effectors and regulating TOR downstream signaling. Using the two classes of avo3ts mutants defective in different downstream signaling functions that were isolated previously (25), we explored the role of Avo3p as a scaffold protein in TORC2 structure and signaling in this study.
Supporting the notion that Avo3p provides a structural scaffold in TORC2, our results underscore the importance of Avo3p function in maintaining TORC2 integrity. First, different avo3ts mutations could affect TORC2 integrity differently. Second, expressing wild-type AVO3, but not any suppressor genes, in avo3ts cells could restore TORC2 integrity. A previous study in which GAL1-driven expression of individual TORC2 components was shut down (68) suggested that Avo1p and Avo3p bind cooperatively to Tor2p and are required for TORC2 integrity. Our data are compatible with a cooperative association of Avo1p and Avo3p with TORC2; in avo3ts mutants, we found Avo1p and Avo3p either both present or both absent in TORC2. However, in view of the fact that AVO1 overexpression in avo3ts cells could not correct defective protein interactions in TORC2 (Fig. 3; H.-L. Ho and M.-Y. Chen, unpublished data), our data seem to suggest that Avo3p plays a more important role than Avo1p in organizing TORC2 structure. Intriguingly, the mammalian counterpart of Avo3p, Rictor, also appears to play a role in modulating mTORC2 integrity. The mammalian ortholog of Avo1p, mSIN1, cannot interact with mTOR when Rictor is knocked down (16). In agreement with a previous finding that Lst8p binds apart from Avo1p or Avo3p to the C-terminal portion of Tor2p (68), we found the Tor2p-Lst8p association unchanged in avo3ts mutants, even under conditions where Avo1p and Avo3p no longer remained in TORC2.
In agreement with the hypothesis that Avo3p may serve to channel TORC2 signaling to various downstream effector pathways, our observations support the idea that different avo3ts mutations differentially affect TORC2 downstream signaling mechanisms. First, since avo3ts suppressors failed to restore TORC2 integrity in mutants and their suppression was allele specific, they could very likely exert their suppression effect by acting on differentially affected downstream signaling in different avo3ts mutants. Second, physical interactions between TORC2 and the two downstream effector proteins Slm1p and Slm2p were hardly affected in avo3-1ts cells but significantly inhibited in avo3-2ts cells; the fact that this inhibition could be eliminated by expressing wild-type AVO3 but not the suppressor gene AVO2 argues for a primary role of Avo3p in modulating the coupling of TORC2 to these downstream effectors. Third, TUS1 deletion specifically weakened the effect of avo3-2ts-specific but not avo3-1ts-specific suppressors in rescuing mutants, suggesting that Tus1p may participate only in specific TORC2 downstream functions, not in all those activated by suppressor expression.
The results of this study support and extend our previous understanding based on genetic data that TORC2 downstream signaling branches into an Avo1p-mediated pathway and an Avo2p/Slm1p-mediated pathway (25). In avo3-1ts cells, we found weakened but not missing binary interactions within TORC2, which we suspect was not the major cause for temperature sensitivity and actin defects. It is more likely that the mutated Avo3p in avo3-1ts cells influences the coupling of TORC2 to certain downstream signaling functions. Based on specific suppression by AVO1 overexpression and the unaffected recruitment of Slm proteins to TORC2, the defective function in avo3-1ts cells is mostly likely an Avo1p-mediated, Slm1p-independent function. The molecular components mediating this TORC2 downstream function await discovery. On the other hand, in avo3-2ts cells, Avo2p was missing in TORC2, a finding that correlates well with allele-specific suppression by AVO2, considering the still-detectable Tor2p-Avo2p interaction in avo3-1ts cells. Taking together the allele-specific suppression by SLM1 or SLM2 overexpression and the decreased physical interactions between TORC2 and the two Slm proteins, we suggest that the avo3-2ts mutant is defective in Avo2p- and Slm1p/Slm2p-dependent TORC2 downstream signaling. Whether Avo2p and Slm1p define a single downstream pathway or couple to separate pathways is still unclear; one line of evidence supporting a single Avo2p-Slm1p linked pathway is that Slm1p physically interacts with Avo2p (15). In addition, in view of the facts that expression of AVO2 or the two SLM genes could not restore TORC2 composition and that AVO2 expression failed to facilitate the recruitment of Slm proteins to the complex in avo3-2ts cells, these suppressors probably rescue the mutant by acting at a level downstream of Avo2p and Slm proteins. We discovered that overexpression of AVO2 or SLM1 could bring the reduced levels of active Rho1p in avo3-2ts cells back to wild-type-like levels, suggesting that Avo2p and Slm proteins act upstream of Rho1p activation.
Besides the differentially affected functions discussed above, our analyses also revealed a common defect, i.e., reduced activation of the Rho1p pathway, in both classes of avo3ts mutants. The reasons for this conclusion include the following: (i) lower levels of active Rho1p were detected in mutants; (ii) deletion of GAPs, including Sac7p, Bag7p, and Lrg1p, which are known to negatively regulate Rho1p (54, 60), resulted in the rescue of avo3ts phenotypes; and (iii) removal of the GAP activity of Sac7p, which should result in Rho1p activation, was sufficient to confer the rescuing effect on avo3ts phenotypes. Interestingly, other avo3ts allele-interacting Rho-GAPs that we found, including Bem2p and Rgd1p, deletion of which aggravated avo3ts phenotypes, have been reported to have negative effects on the cell integrity pathway when they are mutated. Bem2p is a GAP for Rho1p (47); a specific bem2 allele causes weakness of the cell wall and renders mutant cells more sensitive to the cell wall-degrading enzyme Zymolyase (10). Rgd1p is a Rho3p- and Rho4p-specific GAP (14); it has been shown that deletion of RGD1 causes attenuation of PKC pathway activity (12). Taken together, since avo3ts mutants are defective in Rho1p activation, it can be expected that any molecular defects diminishing the signaling functions linking to Rho1p may aggravate the phenotypes of avo3ts mutants.
Rho1p regulates a variety of cellular functions, including cell wall biogenesis, actin organization, bud growth, and polarized secretion (36). Consistent with reduced Rho1p activation in avo3ts mutants, they exhibit cell wall and actin defects, and suppressors capable of restoring wild-type-like levels of active Rho1p in avo3ts cells (Fig. 9) can also rescue their cell wall and actin phenotypes (25). Distinct functions of Rho1p are thought to be regulated via modulation of different Rho1p downstream pathways by different GAPs (36, 40, 54, 60, 62). Genetic data suggest that BEM2 and SAC7 mediate the activation of the Pkc1p-Mpk1p cell wall integrity pathway and that SAC7 and BAG7 regulate an uncharacterized effector pathway to control actin organization (60). LRG1 encodes a specialized RhoGAP regulating 1,3-β-glucan synthesis (65). Our results are in good agreement with these previous findings. Deletion of SAC7 or LRG1 partially rescued the cell wall integrity defects of avo3ts cells, while deletion of SAC7 or BAG7 partially corrected actin organization defects in mutants. Since Sac7p participates in more Rho1p functions, including actin and cell wall regulation, than the other the avo3ts allele-interacting RhoGAPs, only deletion of SAC7 could rescue all the avo3ts phenotypes examined.
Considering the putatively distinct signaling branches downstream of TORC2, one plausible explanation for the shared Rho1p activation defect in avo3ts mutants is that the Avo1p-mediated and Avo2p/Slm1p/Slm2p-mediated pathways converge on Rho1p. Following this line of reasoning, Rho1p regulators, including Rho1p-GAPs and Rho1p-GEFs, make good candidate targets for Avo3p action in modulating TORC2 coupling to different downstream signaling mechanisms. Our analyses did not reveal allele-specific modification of avo3ts phenotypes by deletion of any individual Rho1p-GAPs, suggesting that the branching of TORC2 downstream signaling is not mediated by different GAPs. On the other hand, overexpression of genes encoding Rho1p-GEFs such as Rom2p or Tus1p failed to rescue the temperature sensitivity of avo3ts mutants (Ho and Chen, unpublished). Since at least five effectors, including Fks1/2p, Pkc1p, Bni1p, Sec3p, and Skn7p, are known to act downstream of Rho1p (1, 19, 23, 34, 50), one possible explanation for our data is that the temperature sensitivity of avo3ts mutants may be a result of multiple defects in Rho1p downstream signaling, and thus overexpression of a single Rho1p-GEF may not be sufficient to restore all defective signaling functions. Alternatively, perhaps the molecular mechanism involved in activating Rom2p or Tus1p is defective in avo3ts cells, and therefore overexpression of these GEFs still could not lead to their activation. Finally, our investigation into the roles of Rho1p-GEFs in the suppression effect of avo3ts multicopy suppressors uncovered another molecular mechanism differentially affected in the two classes of avo3ts mutants. In avo3-1ts cells, individual deletion of ROM1, ROM2, or TUS1 did not influence AVO1 suppression; either the three GEFs function redundantly in Avo1p-mediated TORC2 downstream signaling or some unidentified components other than the three GEFs may be involved. Interestingly, deletion of the Rho1p-GEF gene TUS1, but not that of ROM2 or ROM1, diminished the effects of avo3-2ts-specific suppressors. Together with a demonstrated physical interaction between Tus1p and Slm1p, our results define a novel Avo2p- and Slm1p/Slm2p-dependent signaling mechanism acting through Tus1p to activate the Rho1p pathway.
Supplementary Material
Acknowledgments
We are grateful to F.-J. S. Lee at National Taiwan University for supplying strains carrying specific KanMX4 gene replacements as genomic DNA sources for PCR amplification of knockout modules in our construction of yeast deletion strains. We thank J.-J. Lin at National Yang-Ming University, J.-Y. Chen at the Institute of Biomedical Sciences, Academia Sinica, and David Pellman at the Dana-Farber Cancer Institute for providing plasmids.
This work was supported by grants NSC94-2311-B-010-010 and NSC95-2311-B-010-013 from the National Science Council, Taiwan, and a grant (“Aim for the Top University Plan”) from the Ministry of Education, Taiwan.
Footnotes
Published ahead of print on 13 June 2008.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Alberts, A. S., N. Bouquin, L. H. Johnston, and R. Treisman. 1998. Analysis of RhoA-binding proteins reveals an interaction domain conserved in heterotrimeric G protein beta subunits and the yeast response regulator protein Skn7. J. Biol. Chem. 2738616-8622. [DOI] [PubMed] [Google Scholar]
- 2.Audhya, A., R. Loewith, A. B. Parsons, L. Gao, M. Tabuchi, H. Zhou, C. Boone, M. N. Hall, and S. D. Emr. 2004. Genome-wide lethality screen identifies new PI4,5P2 effectors that regulate the actin cytoskeleton. EMBO J. 233747-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbet, N. C., U. Schneider, S. B. Helliwell, I. Stansfield, M. F. Tuite, and M. N. Hall. 1996. TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell 725-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baudin, A., O. Ozier-Kalogeropoulos, A. Denouel, F. Lacroute, and C. Cullin. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 213329-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berset, C., H. Trachsel, and M. Altmann. 1998. The TOR (target of rapamycin) signal transduction pathway regulates the stability of translation initiation factor eIF4G in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 954264-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bickle, M., P. A. Delley, A. Schmidt, and M. N. Hall. 1998. Cell wall integrity modulates RHO1 activity via the exchange factor ROM2. EMBO J. 172235-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cabib, E., J. Drgonova, and T. Drgon. 1998. Role of small G proteins in yeast cell polarization and wall biosynthesis. Annu. Rev. Biochem. 67307-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cafferkey, R., M. M. McLaughlin, P. R. Young, R. K. Johnson, and G. P. Livi. 1994. Yeast TOR (DRR) proteins: amino-acid sequence alignment and identification of structural motifs. Gene 141133-136. [DOI] [PubMed] [Google Scholar]
- 9.Chen, J. C., and T. Powers. 2006. Coordinate regulation of multiple and distinct biosynthetic pathways by TOR and PKA kinases in S. cerevisiae. Curr. Genet. 49281-293. [DOI] [PubMed] [Google Scholar]
- 10.Cid, V. J., R. Cenamor, M. Sanchez, and C. Nombela. 1998. A mutation in the Rho1-GAP-encoding gene BEM2 of Saccharomyces cerevisiae affects morphogenesis and cell wall functionality. Microbiology 14425-36. [DOI] [PubMed] [Google Scholar]
- 11.Crespo, J. L., and M. N. Hall. 2002. Elucidating TOR signaling and rapamycin action: lessons from Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 66579-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Bettignies, G., D. Thoraval, C. Morel, M. F. Peypouquet, and M. Crouzet. 2001. Overactivation of the protein kinase C-signaling pathway suppresses the defects of cells lacking the Rho3/Rho4-GAP Rgd1p in Saccharomyces cerevisiae. Genetics 1591435-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Virgilio, C., and R. Loewith. 2006. The TOR signalling network from yeast to man. Int. J. Biochem. Cell Biol. 381476-1481. [DOI] [PubMed] [Google Scholar]
- 14.Doignon, F., C. Weinachter, O. Roumanie, and M. Crouzet. 1999. The yeast Rgd1p is a GTPase activating protein of the Rho3 and Rho4 proteins. FEBS Lett. 459458-462. [DOI] [PubMed] [Google Scholar]
- 15.Fadri, M., A. Daquinag, S. Wang, T. Xue, and J. Kunz. 2005. The pleckstrin homology domain proteins Slm1 and Slm2 are required for actin cytoskeleton organization in yeast and bind phosphatidylinositol-4,5-bisphosphate and TORC2. Mol. Biol. Cell 161883-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frias, M. A., C. C. Thoreen, J. D. Jaffe, W. Schroder, T. Sculley, S. A. Carr, and D. M. Sabatini. 2006. mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr. Biol. 161865-1870. [DOI] [PubMed] [Google Scholar]
- 17.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418387-391. [DOI] [PubMed] [Google Scholar]
- 18.Gietz, D., A. St Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 201425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo, W., F. Tamanoi, and P. Novick. 2001. Spatial regulation of the exocyst complex by Rho1 GTPase. Nat. Cell Biol. 3353-360. [DOI] [PubMed] [Google Scholar]
- 20.Guthrie, C., and G. R. Fink. 1991. Guide to yeast genetics and molecular biology, p. 3-187. Academic Press, New York, NY.
- 21.Hara, K., Y. Maruki, X. Long, K. Yoshino, N. Oshiro, S. Hidayat, C. Tokunaga, J. Avruch, and K. Yonezawa. 2002. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110177-189. [DOI] [PubMed] [Google Scholar]
- 22.Helliwell, S. B., I. Howald, N. Barbet, and M. N. Hall. 1998. TOR2 is part of two related signaling pathways coordinating cell growth in Saccharomyces cerevisiae. Genetics 14899-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helliwell, S. B., A. Schmidt, Y. Ohya, and M. N. Hall. 1998. The Rho1 effector Pkc1, but not Bni1, mediates signalling from Tor2 to the actin cytoskeleton. Curr. Biol. 81211-1214. [DOI] [PubMed] [Google Scholar]
- 24.Helliwell, S. B., P. Wagner, J. Kunz, M. Deuter-Reinhard, R. Henriquez, and M. N. Hall. 1994. TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol. Biol. Cell 5105-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ho, H. L., Y. S. Shiau, and M. Y. Chen. 2005. Saccharomyces cerevisiae TSC11/AVO3 participates in regulating cell integrity and functionally interacts with components of the Tor2 complex. Curr. Genet. 47273-288. [DOI] [PubMed] [Google Scholar]
- 26.Inoki, K., and K. L. Guan. 2006. Complexity of the TOR signaling network. Trends Cell Biol. 16206-212. [DOI] [PubMed] [Google Scholar]
- 27.Inoki, K., H. Ouyang, Y. Li, and K. L. Guan. 2005. Signaling by target of rapamycin proteins in cell growth control. Microbiol. Mol. Biol. Rev. 6979-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacinto, E., and M. N. Hall. 2003. Tor signalling in bugs, brain and brawn. Nat. Rev. Mol. Cell Biol. 4117-126. [DOI] [PubMed] [Google Scholar]
- 29.Jacinto, E., R. Loewith, A. Schmidt, S. Lin, M. A. Ruegg, A. Hall, and M. N. Hall. 2004. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 61122-1128. [DOI] [PubMed] [Google Scholar]
- 30.Kaeberlein, M., R. W. Powers III, K. K. Steffen, E. A. Westman, D. Hu, N. Dang, E. O. Kerr, K. T. Kirkland, S. Fields, and B. K. Kennedy. 2005. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 3101193-1196. [DOI] [PubMed] [Google Scholar]
- 31.Kamada, Y., Y. Fujioka, N. N. Suzuki, F. Inagaki, S. Wullschleger, R. Loewith, M. N. Hall, and Y. Ohsumi. 2005. Tor2 directly phosphorylates the AGC kinase Ypk2 to regulate actin polarization. Mol. Cell. Biol. 257239-7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim, D. H., D. D. Sarbassov, S. M. Ali, J. E. King, R. R. Latek, H. Erdjument-Bromage, P. Tempst, and D. M. Sabatini. 2002. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110163-175. [DOI] [PubMed] [Google Scholar]
- 33.Kim, D. H., D. D. Sarbassov, S. M. Ali, R. R. Latek, K. V. Guntur, H. Erdjument-Bromage, P. Tempst, and D. M. Sabatini. 2003. GβL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol. Cell 11895-904. [DOI] [PubMed] [Google Scholar]
- 34.Kohno, H., K. Tanaka, A. Mino, M. Umikawa, H. Imamura, T. Fujiwara, Y. Fujita, K. Hotta, H. Qadota, T. Watanabe, Y. Ohya, and Y. Takai. 1996. Bni1p implicated in cytoskeletal control is a putative target of Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 156060-6068. [PMC free article] [PubMed] [Google Scholar]
- 35.Kono, K., S. Nogami, M. Abe, M. Nishizawa, S. Morishita, D. Pellman, and Y. Ohya. 2008. G1/S cyclin-dependent kinase regulates small GTPase Rho1p through phosphorylation of RhoGEF Tus1p in Saccharomyces cerevisiae. Mol. Biol. Cell 191763-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levin, D. E. 2005. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69262-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loewith, R., E. Jacinto, S. Wullschleger, A. Lorberg, J. L. Crespo, D. Bonenfant, W. Oppliger, P. Jenoe, and M. N. Hall. 2002. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10457-468. [DOI] [PubMed] [Google Scholar]
- 38.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14953-961. [DOI] [PubMed] [Google Scholar]
- 39.Lorberg, A., and M. N. Hall. 2004. TOR: the first 10 years. Curr. Top. Microbiol. Immunol. 2791-18. [DOI] [PubMed] [Google Scholar]
- 40.Lorberg, A., H. P. Schmitz, J. J. Jacoby, and J. J. Heinisch. 2001. Lrg1p functions as a putative GTPase-activating protein in the Pkc1p-mediated cell integrity pathway in Saccharomyces cerevisiae. Mol. Genet. Genomics 266514-526. [DOI] [PubMed] [Google Scholar]
- 41.Madden, K., and M. Snyder. 1998. Cell polarity and morphogenesis in budding yeast. Annu. Rev. Microbiol. 52687-744. [DOI] [PubMed] [Google Scholar]
- 42.Marquitz, A. R., J. C. Harrison, I. Bose, T. R. Zyla, J. N. McMillan, and D. J. Lew. 2002. The Rho-GAP Bem2p plays a GAP-independent role in the morphogenesis checkpoint. EMBO J. 214012-4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martín, H., J. M. Rodriguez-Pachon, C. Ruiz, C. Nombela, and M. Molina. 2000. Regulatory mechanisms for modulation of signaling through the cell integrity Slt2-mediated pathway in Saccharomyces cerevisiae. J. Biol. Chem. 2751511-1519. [DOI] [PubMed] [Google Scholar]
- 44.Mumberg, D., R. Muller, and M. Funk. 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 225767-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302205-217. [DOI] [PubMed] [Google Scholar]
- 46.Park, J. I., E. J. Collinson, C. M. Grant, and I. W. Dawes. 2005. Rom2p, the Rho1 GTP/GDP exchange factor of Saccharomyces cerevisiae, can mediate stress responses via the Ras-cAMP pathway. J. Biol. Chem. 2802529-2535. [DOI] [PubMed] [Google Scholar]
- 47.Peterson, J., Y. Zheng, L. Bender, A. Myers, R. Cerione, and A. Bender. 1994. Interactions between the bud emergence proteins Bem1p and Bem2p and Rho-type GTPases in yeast. J. Cell Biol. 1271395-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Powers, T., and P. Walter. 1999. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 10987-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pruyne, D., and A. Bretscher. 2000. Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J. Cell Sci. 113365-375. [DOI] [PubMed] [Google Scholar]
- 50.Qadota, H., C. P. Python, S. B. Inoue, M. Arisawa, Y. Anraku, Y. Zheng, T. Watanabe, D. E. Levin, and Y. Ohya. 1996. Identification of yeast Rho1p GTPase as a regulatory subunit of 1,3-β-glucan synthase. Science 272279-281. [DOI] [PubMed] [Google Scholar]
- 51.Roelants, F. M., P. D. Torrance, N. Bezman, and J. Thorner. 2002. Pkh1 and Pkh2 differentially phosphorylate and activate Ypk1 and Ykr2 and define protein kinase modules required for maintenance of cell wall integrity. Mol. Biol. Cell 133005-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roelants, F. M., P. D. Torrance, and J. Thorner. 2004. Differential roles of PDK1- and PDK2-phosphorylation sites in the yeast AGC kinases Ypk1, Pkc1 and Sch9. Microbiology 1503289-3304. [DOI] [PubMed] [Google Scholar]
- 53.Rohde, J. R., S. Campbell, S. A. Zurita-Martinez, N. S. Cutler, M. Ashe, and M. E. Cardenas. 2004. TOR controls transcriptional and translational programs via Sap-Sit4 protein phosphatase signaling effectors. Mol. Cell. Biol. 248332-8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roumanie, O., C. Weinachter, I. Larrieu, M. Crouzet, and F. Doignon. 2001. Functional characterization of the Bag7, Lrg1 and Rgd2 RhoGAP proteins from Saccharomyces cerevisiae. FEBS Lett. 506149-156. [DOI] [PubMed] [Google Scholar]
- 55.Sarbassov, D. D., S. M. Ali, D. H. Kim, D. A. Guertin, R. R. Latek, H. Erdjument-Bromage, P. Tempst, and D. M. Sabatini. 2004. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 141296-1302. [DOI] [PubMed] [Google Scholar]
- 56.Schmelzle, T., T. Beck, D. E. Martin, and M. N. Hall. 2004. Activation of the RAS/cyclic AMP pathway suppresses a TOR deficiency in yeast. Mol. Cell. Biol. 24338-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmelzle, T., and M. N. Hall. 2000. TOR, a central controller of cell growth. Cell 103253-262. [DOI] [PubMed] [Google Scholar]
- 58.Schmidt, A., M. Bickle, T. Beck, and M. N. Hall. 1997. The yeast phosphatidylinositol kinase homolog TOR2 activates RHO1 and RHO2 via the exchange factor ROM2. Cell 88531-542. [DOI] [PubMed] [Google Scholar]
- 59.Schmidt, A., J. Kunz, and M. N. Hall. 1996. TOR2 is required for organization of the actin cytoskeleton in yeast. Proc. Natl. Acad. Sci. USA 9313780-13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmidt, A., T. Schmelzle, and M. N. Hall. 2002. The RHO1-GAPs SAC7, BEM2 and BAG7 control distinct RHO1 functions in Saccharomyces cerevisiae. Mol. Microbiol. 451433-1441. [DOI] [PubMed] [Google Scholar]
- 61.Smith, G. R., S. A. Givan, P. Cullen, and G. F. Sprague, Jr. 2002. GTPase-activating proteins for Cdc42. Eukaryot. Cell 1469-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tcherkezian, J., and N. Lamarche-Vane. 2007. Current knowledge of the large RhoGAP family of proteins. Biol. Cell 9967-86. [DOI] [PubMed] [Google Scholar]
- 63.Urban, J., A. Soulard, A. Huber, S. Lippman, D. Mukhopadhyay, O. Deloche, V. Wanke, D. Anrather, G. Ammerer, H. Riezman, J. R. Broach, C. De Virgilio, M. N. Hall, and R. Loewith. 2007. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell 26663-674. [DOI] [PubMed] [Google Scholar]
- 64.Varelas, X., D. Stuart, M. J. Ellison, and C. Ptak. 2006. The Cdc34/SCF ubiquitination complex mediates Saccharomyces cerevisiae cell wall integrity. Genetics 1741825-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Watanabe, D., M. Abe, and Y. Ohya. 2001. Yeast Lrg1p acts as a specialized RhoGAP regulating 1,3-β-glucan synthesis. Yeast 18943-951. [DOI] [PubMed] [Google Scholar]
- 66.Wedaman, K. P., A. Reinke, S. Anderson, J. Yates III, J. M. McCaffery, and T. Powers. 2003. Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae. Mol. Biol. Cell 141204-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wullschleger, S., R. Loewith, and M. N. Hall. 2006. TOR signaling in growth and metabolism. Cell 124471-484. [DOI] [PubMed] [Google Scholar]
- 68.Wullschleger, S., R. Loewith, W. Oppliger, and M. N. Hall. 2005. Molecular organization of target of rapamycin complex 2. J. Biol. Chem. 28030697-30704. [DOI] [PubMed] [Google Scholar]
- 69.Yang, Q., and K. L. Guan. 2007. Expanding mTOR signaling. Cell Res. 17666-681. [DOI] [PubMed] [Google Scholar]
- 70.Yang, Q., K. Inoki, T. Ikenoue, and K. L. Guan. 2006. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 202820-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yoshida, S., K. Kono, D. M. Lowery, S. Bartolini, M. B. Yaffe, Y. Ohya, and D. Pellman. 2006. Polo-like kinase Cdc5 controls the local activation of Rho1 to promote cytokinesis. Science 313108-111. [DOI] [PubMed] [Google Scholar]
- 72.Zurita-Martinez, S. A., and M. E. Cardenas. 2005. Tor and cyclic AMP-protein kinase A: two parallel pathways regulating expression of genes required for cell growth. Eukaryot. Cell 463-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.