Malaria is caused by protozoan parasites of the genus Plasmodium. Five species of Plasmodium cause infection in humans, with the majority of lethal cases caused by Plasmodium falciparum. This species is responsible for more than 500 million clinical cases of malaria each year (59). In the past 2 decades, efforts to combat this disease have been met with the emergence of widespread resistance to most of the commonly used antimalarial drugs (68). The poor efficacy of current drugs and the lack of a promising vaccine have resulted in alarming increases in the rates of malaria morbidity and mortality worldwide, with the major toll felt in the developing world.
P. falciparum is transmitted to humans via the bite of an infected female anopheline mosquito. In humans, the parasite undergoes one cycle of asexual multiplication in hepatocytes, followed by several cycles of infection and multiplication in red blood cells. Whereas the hepatocytic stage is asymptomatic, the erythrocytic stage is accompanied by the destruction of the host erythrocytes, resulting in anemia and, in the absence of treatment, death. Extensive efforts are presently under way to develop a vaccine, and advances in our understanding of the biology of the parasite and its metabolic and nutritional needs offer new routes for chemotherapy. In this regard, purine metabolism holds significant promise as a target for drug development.
It has long been recognized that protozoan parasites, including Plasmodium spp., are unable to synthesize purine rings de novo (4). Consistent with this observation, the sequencing of protozoan genomes has failed to uncover any genes encoding enzymes involved in the biosynthesis of purine nucleosides or nucleobases (12). Protozoan parasites rely instead on salvage of purines from the host. The strategies used in acquiring purines vary significantly among parasite genera. This review will focus primarily on the purine salvage pathways present in the Plasmodium parasite. Recent reviews have examined the metabolic pathways for purine salvage in other protozoa (11, 17, 29).
Initial studies on purine and pyrimidine synthesis in Plasmodium parasites were performed on erythrocytic stages of the rodent malaria species P. berghei (7, 8, 65), the macaque monkey malaria species P. knowlesi (46), and the avian malaria parasite P. lophurae (61, 66). Work by Sherman and colleagues indicated a functional purine salvage pathway in P. lophurae (61, 66). Bungener and Neilsen demonstrated that P. berghei can incorporate the purines [3H]adenosine and [3H]hypoxanthine, but not the pyrimidines [3H]uridine and [3H]thymidine, into nucleic acids (7, 8). Van Dyke and colleagues showed that uptake and incorporation (into nucleic acid) occur for the purine adenosine but not for the pyrimidines uridine and cytidine and that adenosine is incorporated far more efficiently than adenine, guanosine, and deoxyadenosine (65). Polet and Barr demonstrated that monkey erythrocytes infected with P. knowlesi were unable to incorporate [14C]thymine, [14C]thymidine, [14C]uracil, or [14C]uridine but could incorporate [14C]- and [3H]orotate (a pyrimidine precursor) and [14C]adenine into nucleic acids (46). Following on from these findings, Gutteridge and Trigg found that, in P. knowlesi, all radiolabeled purines were significantly incorporated into nucleic acids while none of the pyrimidines tested were (28). These early biochemical studies demonstrated that Plasmodium parasites lack the ability to metabolize exogenous pyrimidines and instead are entirely dependent on de novo synthesis. Conversely, Plasmodium parasites are entirely reliant upon the salvage of extracellular purines (4) and are capable of metabolizing a wide variety of exogenous purine nucleobases and nucleosides.
The continuous culture of P. falciparum in serum-free media is dependent upon the supply of exogenous purines (1, 43), suggesting that the erythrocyte adenylate nucleotide pool is not a sufficient purine source and that the parasite is dependent upon extraerythrocytic purine salvage for survival. The first step in purine salvage is, therefore, the transport of purines into the infected cell and, from there, into the intracellular parasite. We provide a short description of this process below; more-thorough reviews of purine transport in Plasmodium spp. have been provided elsewhere (2, 29).
PURINE TRANSPORT
Within the erythrocyte, the Plasmodium parasite resides within a parasitophorous vacuole created upon parasite invasion by the invagination of the host cell membrane. The parasite cytosol is therefore separated from the nutrient-rich extracellular medium by a series of three membranes: the host cell membrane, the parasitophorous vacuole membrane, and the parasite plasma membrane.
Nucleosides and nucleobases cross the host cell membrane via a combination of a high-affinity transport process and, in mature trophozoite-stage parasites, nonsaturable, broad-specificity new permeability pathways induced by the parasite in the erythrocyte membrane (25, 26, 36, 37, 64). The high-affinity transport process is sensitive to inhibition by nitrobenzylthioinosine and probably represents transporters native to the erythrocyte (25), such as human ENT1. The infected erythrocyte also displays the ability to transport l-enantiomers of nucleosides, and this uptake is inhibited by classical new permeability pathway blockers, such as furosemide and 5-nitro-2-(3-phenylpropylamino)benzoic acid (25, 27, 64). Once inside the infected erythrocyte, the nucleosides are presumed to cross the parasitophorous vacuole membrane via the large-diameter, nonselective pores present on this membrane (18).
From early work by Gero and colleagues, it was clear that the uptake of at least one nucleoside, the purine adenosine, across the parasite plasma membrane is extremely rapid (27, 64). In the first study of the functional expression of P. falciparum-encoded transporters in Xenopus laevis oocytes, cells injected with total mRNA from blood-stage parasites showed elevated transport of adenosine and hypoxanthine, consistent with the expression of one or more parasite-encoded nucleoside transporters (45); however, the proteins responsible were not identified. Subsequently, two independent studies reported the cloning of a P. falciparum nucleoside transporter, designated either P. falciparum NT1 (PfNT1) (10) or PfENT1 (44). In both cases, the protein was identified by searching for homologues of known nucleoside transporters in the P. falciparum genome sequence. The gene was cloned and expressed in Xenopus oocytes and shown to transport a range of nucleosides and nucleobases (10, 44). There were significant differences between the results of the two studies with regard to the reported characteristics of the transporter. Carter et al. (10) described the transporter (referred to hereafter as PfNT1) as having a high affinity for adenosine (Km = 13 μM) and being sensitive to inhibition by 10 μM dipyridamole, whereas Parker et al. (44) described it as having a much lower affinity for adenosine (Km = 320 μM) and being insensitive to 10 μM dipyridamole. These differences notwithstanding, it is clear that PfNT1 is a nucleoside transporter. In a subsequent study, Rager et al. used immunoelectron microscopy to obtain strong evidence that the protein is localized predominantly, if not exclusively, to the parasite plasma membrane (and not to the parasitophorous vacuole or host erythrocyte membrane) (51). Transport experiments on parasites isolated from their host cells by saponin treatment (which renders the host cell membrane freely permeable to macromolecules while leaving the parasite plasma membrane intact) revealed that nucleosides (20) and nucleobases (19) are transported across the parasite plasma membrane by a fast, low-affinity process, similar to that seen for the transport of these substrates by PfNT1 in Xenopus oocytes (19, 20). The absolute dependence of the parasite on PfNT1 expression was demonstrated in experiments by El Bissati and colleagues, in which it was shown that parasites harboring a genomic deletion of the PfNT1 gene (pfnt1Δ) were viable only when the purines adenosine, inosine or hypoxanthine were present at super-physiological concentrations (21). While initial transport studies in pfnt1Δ parasites (21) suggested that pfnt1Δ parasites exhibited residual (PfNT1-independent) adenosine and inosine transport, further studies (taking into account the metabolic properties of the parasite) have shown that pfnt1Δ parasites were unable to transport adenosine, inosine and hypoxanthine (21a). Further studies with this parasite strain have indicated that pfnt1Δ parasites were unable to grow when guanine, guanosine, xanthine, or adenine were present at physiological concentrations (21a) and that pfnt1Δ parasites were unable to transport guanine, guanosine, or xanthine but were able to transport adenine, albeit at a lower rate (21a).
Recently, Quashie and colleagues have proposed the presence at the parasite plasma membrane of a high-affinity adenosine transport mechanism, which they have equated with PfNT1 (48). Their experimental design and subsequent interpretation of the data, however, do not take into consideration the rapid metabolism of purines, which is a high-affinity process. Taking into account the difficulties of distinguishing between transport and metabolism, a comprehensive analysis of all the available PfNT1 transport data (10, 19-21a, 44, 48) would suggest that PfNT1 is a low-affinity transporter that mediates the rapid uptake of adenosine, inosine, hypoxanthine, adenine, guanine, guanosine, and xanthine. There is some evidence that for adenine there is, in addition to PfNT1, a PfNT1-independent mechanism (21a, 48), yet to be characterized.
While available studies demonstrate an essential function for PfNT1, the completion of the P. falciparum genome sequencing project revealed three additional nucleoside transporters (PfNT2, PfNT3, and PfNT4) (24, 40). Like PfNT1, these proteins are expressed during the intraerythrocytic stage of the parasite life cycle as well as in gametocytes (5, 40, 70). Their cellular localization, as well as their role in parasite physiology, is yet to be elucidated.
PURINE METABOLISM
Studies of P. falciparum parasites isolated from host erythrocyte membranes by mechanical rupture showed the presence of the purine salvage enzymes adenosine deaminase (ADA), purine nucleoside phosphorylase (PNP), and phosphoribosyl transferase (PRT) (52). Only minimal adenosine kinase activity was observed in this study (52). Subsequent studies have indicated that the parasite lacks adenosine kinase activity (55), and no adenosine kinase gene has been found in the genome (24). This is consistent with the idea that the majority of purine salvage occurs via the sequential conversion of adenosine and inosine to hypoxanthine, which is then phosphoribosylated to form IMP (52).
P. FALCIPARUM ADENOSINE DEAMINASE
The enzyme ADA catalyzes the irreversible hydrolytic cleavage of adenosine to produce inosine and ammonia. As native human ADA (hADA) activity is high in uninfected erythrocytes (15, 69), parasite-encoded ADA (PfADA) was initially characterized by infecting erythrocytes taken from a human donor genetically deficient in ADA with P. falciparum (15). Parasite growth levels were comparable in normal and deficient host cells, and infected deficient host cells exhibited a level of ADA activity twice that seen in normal, uninfected erythrocytes. The ADA activity in parasite protein preparations exhibited essentially the same Km for adenosine as the ADA activity in protein preparations from uninfected, normal erythrocytes (15). Both erythrocyte and parasite ADA enzymes are inhibited by nanomolar concentrations of deoxycoformycin (15). PfADA, however, has a much lower sensitivity than hADA to the inhibitor erythro-9-(2-hydroxy-3-nonyl)adenine (15). Recombinant PfADA (expressed in Escherichia coli) has a Km of 11 ± 2 μM for adenosine (22), a figure similar to that reported for free P. falciparum parasite lysates (7 μM) (6) and to that seen for lysates from the duck parasite P. lophurae (23 μM) (69). The P. falciparum enzyme is also reportedly sensitive to two l-nucleoside structural analogues of deoxy-d-coformycin that have no effect on hADA (6) as well as to 5′-methylthio coformycins (as detailed below) (63).
P. FALCIPARUM PURINE NUCLEOSIDE PHOSPHORYLASE
PNP enzymes are responsible for the phosphorolysis of nucleosides into the corresponding nucleobases and ribose groups. This reaction is reversible and is carried out in the presence of inorganic phosphate (9). There are two major families of nucleoside phosphorylases: the low-molecular-mass homotrimers, which include all mammalian PNPs and some bacterial PNPs, and the high-molecular-mass homohexamers, which include most bacterial PNPs, along with mammalian and bacterial uridine phosphorylases (9). Most PNPs contain consensus sequences specific to the relevant family (33). Initial studies of P. falciparum PNP (PfPNP; utilizing red blood cells taken from a child deficient in PNP) (16) showed that PfPNP is capable of utilizing inosine, guanosine, and deoxyguanosine but not adenosine or xanthosine. Expression of the gene encoding PfPNP in E. coli revealed that PfPNP is not a typical member of either of the two major classes of PNPs, having low sequence homology and a distinct substrate specificity to PNPs from other species (33). The purified enzyme has a Km for inosine of 5.0 ± 0.9 μM and can be inhibited by immucillins with dissociation constants as low as 0.6 nM (33). Immucillins are PNP transition state analogues, some of which have Kd values as low as 7 pM (54). Immucillin-H is currently in clinical trials for treating human T-cell malignancies (23, 54) and has been shown to inhibit PfPNP, as well as inhibiting P. falciparum growth, in vitro (33, 34). This in vitro inhibition of parasite growth can be reversed by the addition of hypoxanthine, but not inosine, to the culture medium, indicating that PNP is the target for this drug (34).
PfADA AND PfPNP UTILIZE UNIQUE SUBSTRATES
The crystal structure of PfPNP in complex with immucillin-H revealed a homo-hexamer, organized as a trimer of dimers, with an immucillin-H molecule bound at each of the catalytic sites (58). The structure also revealed that the catalytic site has a capacity for binding 5′-substituted nucleosides, and kinetic studies have confirmed that 5′-methylthioinosine (MTI) is a good substrate for PfPNP, being converted to hypoxanthine, indicating that this protein has dual catalytic functions in the parasite (58). Methylthioadenosine (MTA) is a common by-product of polyamine metabolism, and like PfPNP, PfADA is able to utilize this 5′-substituted compound, converting it to MTI (58, 60). The abilities of PfADA and PfPNP to utilize methylthiopurines allow the salvage of purines from both the traditional purine nucleoside pathways and the polyamine synthesis pathway (Fig. 1).
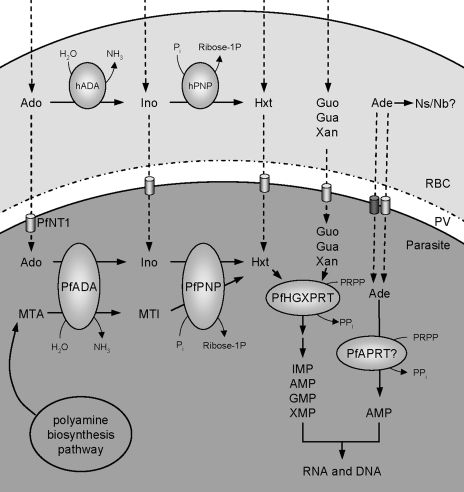
FIG. 1.
Purine metabolism within the infected erythrocyte. Inosine is produced from adenosine by ADA. Inosine is then converted to hypoxanthine by PNP). hADA and hPNP are present in the red blood cell cytoplasm, while the parasite ADA (PfADA) and PNP (PfPNP) are active in the parasite cytosol. Both PfADA and PfPNP are capable of acting on 5′-substituted compounds, and so the parasite is able to salvage purines from the polyamine biosynthesis pathway (58, 60). Ado, Ino, Hxt, Guo, Gua, Xan, and Ade are transported across the parasite plasma membrane by PfNT1. Adenine also enters the cell via a second, PfNT1-independent mechanism. Adenine may be metabolized to other nucleosides/nucleobases (Ns/Nb) in the erythrocyte cytoplasm. PRT activity converts hypoxanthine to IMP, guanine to GMP, and xanthine to XMP. IMP, GMP, and XMP are then converted to guanylate and adenylate nucleotides by the action of several more enzymes, outlined in the text and in Fig. 2. There is some evidence to suggest that the parasite can also convert adenine to AMP by using a parasite-encoded APRT (PfAPRT). Abbreviations: Ado, adenosine; Ino, inosine; Hxt, hypoxanthine; Guo, guanosine; Gua, guanine; Xan, xanthine; Ade, adenine; PRPP, phosphoribosylpyrophosphate; RBC, red blood cell; PV, parasitophorous vacuole; MTA, methylthioadenosine; MTI, methylthioinosine.
When supplied in vitro, [14C]MTI is metabolized by P. falciparum and incorporated into nucleic acids (60). A reaction mixture of purified PfADA and PfPNP efficiently converted MTA to hypoxanthine, but replacement of either enzyme with its mammalian counterpart stopped the reaction (60). Likewise, purified E. coli PNP had no detectable activity against MTI or MTA, despite phosphorolysing both adenosine and inosine (60).
PfADA AND PfPNP AS DRUG TARGETS
The abilities of PfADA and PfPNP to recognize methylthio-substituted purines led to the development of 5′-methylthio coformycins (63) and methylthio-immucillin-H (MT-immucillin-H) (60). The 5′-methylthio coformycins inhibit PfADA at very low concentrations but do not inhibit hADA (63). The abilities of these compounds to inhibit parasite growth in the presence of physiologically relevant concentrations of inosine and hypoxanthine remain to be established. The validity of ADA as an antiplasmodial target is questionable, however. While early studies showed that 2′-deoxycoformycin can clear an in vivo P. knowlesi infection (67), the in vitro growth of P. falciparum was not inhibited by 10 μM 2′-deoxycoformycin, despite complete inhibition of (both host and parasite) ADA enzyme activity at this concentration (15, 53). Whether this discrepancy is due to species-specific differences or due to in vitro culture conditions is unknown.
The addition of a 5′-methylthio group to immucillin-H allows the preferential binding of MT-immucillin-H to PfPNP over human PNP (hPNP) by a factor of 112 (58). MT-immucillin-H kills P. falciparum in vitro with a 50% inhibitory concentration of 50 nM, compared to a 50% inhibitory concentration of 63 nM for immucillin-H (60). It should be noted, however, that these values were derived from inhibition curves carried out with culture media containing no hypoxanthine, under which conditions parasite growth would be completely reliant upon PNP activity (60).
The potential of PfADA and PfPNP as therapeutic targets is yet to be validated by genetic means. While the extent to which purine concentrations fluctuate in mammalian blood is unknown and the amounts of adenosine, inosine, and hypoxanthine available in human plasma are a source of contention, several groups report that hypoxanthine is the major purine available in human serum in vivo (42, 62). Therefore, it seems likely that a block in the purine salvage pathway at the ADA or PNP step could be circumvented by the downstream salvage of hypoxanthine, guanine, or xanthine from the host, all of which are transported into the parasite by PfNT1 and phosphoribosylated by P. falciparum hypoxanthine-guanine-xanthine PRT (PfHGXPRT). There is a clear need for in vitro studies in which the serum concentrations of all purines are replicated or, ideally, in vivo animal studies on the effects of these compounds on Plasmodium growth. Despite the fact that recombinant Toxoplasma gondii PNP is sensitive to immucillin-H, the in vitro growth of the parasite is unaffected due to the presence of a redundant purine salvage pathway (the direct phosphorylation of adenosine by adenosine kinase). While T. gondii parasites deficient in adenosine kinase are sensitive to immucillin-H, this sensitivity can be rescued by addition of hypoxanthine to the medium (13).
P. FALCIPARUM PHOSPHORIBOSYLTRANSFERASE ENZYMES
Nucleobases can be converted directly to nucleoside monophosphates by the action of PRT enzymes. PRT enzymes catalyze the reversible transfer of a phosphoribosyl group from phosphoribosylpyrophosphate to a nucleobase, resulting in the production of a nucleotide and pyrophosphate (14). In P. falciparum, PRT activity is catalyzed by a single enzyme, HGXPRT (49), while a second enzyme was shown to account for adenine PRT (APRT) activity (Fig. 1) (50), as is the case for mammalian cells. The ability of the enzyme to utilize xanthine as a substrate distinguishes PfHGXPRT from its counterpart enzyme in the human red blood cell.
Although P. falciparum has only low levels of APRT activity (high-performance liquid chromatography data indicated that the amount of HGXPRT activity is about 1,500 times greater than that present for APRT) (50), this activity does exhibit properties that differ from those of the APRT present in human erythrocytes. These include a lower apparent molecular weight and a differing inhibition profile (50). Studies on duck erythrocytes infected with P. lophurae indicated that the avian parasite also has APRT activity with kinetic properties different from those of the host cell APRT enzyme (Plasmodium Km = 3.4 μM, and host Km = 69 μM) (69). Pollack and colleagues (47) reported the cloning of P. falciparum APRT, although the gene sequence was not determined. Mouse cell lines transfected with this gene exhibited APRT activity with a Km value similar to that seen for lysates from isolated P. falciparum (47). However, the presence of APRT activity in Plasmodium parasites remains an area of contention. A recent study suggested (based on genetic, biochemical, and genomic data) that APRT enzymes are not present in Plasmodium, Eimeria, or Cryptosporidium spp. (12). Although wild-type parasites are able to grow on adenine as a sole purine source, pfnt1Δ parasites are unable to complete the intraerythrocytic life cycle on physiological concentrations of adenine, despite measurable uptake of adenine by saponin-isolated pfnt1Δ parasites (21a). This suggests either that all adenine is converted in the red blood cell such that the parasite cytosol never sees adenine or that adenine cannot be metabolized by the parasite, a hypothesis consistent with the suggestion that the parasite lacks an APRT enzyme.
The PfHGXPRT enzyme displays high activity for all three purine substrates, with some variation in affinity for them (Km of hypoxanthine = 0.46 μM, Km of guanine = 0.30 μM, and Km of xanthine = 29 μM) (49). King and Melton reported the first cloning and sequencing of PfHGXPRT (35). When expressed in E. coli (31), the recombinant enzyme was shown to differ from the human enzyme in several respects, including an increased ability to utilize allopurinol as a substrate. There are many good inhibitors of PRT enzymes, including phosphorylated versions of the previously described immucillins (e.g., immucillin-HP and immucillin-GP). The cell permeability of phosphorylated inhibitors is, however, an obvious concern. The available crystal structures of PfHGXPRT (56) and human HGPRT (57), each in complex with these compounds, should aid in the design of species-specific inhibitors (38). Recently, several purine nucleobase analogues were identified that, while being good substrates for PfHGXPRT, were comparatively weak substrates for the human enzyme (32). The discovery of compounds that can interact preferentially with the parasite enzyme, together with the reported lack of nucleobase phosphoribosylation redundancy in P. falciparum, makes PfHGXPRT a promising target for the development of antimalarial therapies.
NUCLEOTIDE PRODUCTION
As detailed above, the great majority of purines salvaged by P. falciparum are funneled through hypoxanthine to IMP. To generate adenylate nucleotides within the parasite, IMP is first converted to adenylosuccinate (by adenylosuccinate synthetase), which is then converted by adenylosuccinate lyase to AMP (Fig. 2) (30, 39). In order to obtain guanylate nucleotides, IMP dehydrogenase converts IMP to XMP, which is then converted to GMP by GMP synthetase (41). A recent biochemical characterization of this enzyme suggests that it differs from its mammalian counterpart on several levels, including differences in ligand specificity (3). HGXPRT can also convert xanthine base directly to XMP and guanine base to GMP. AMP and GMP are then converted to the final nucleotides used in nucleic acid synthesis (Fig. 2).
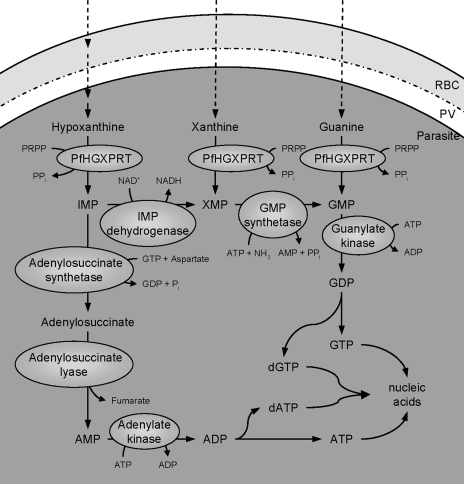
FIG. 2.
Nucleotide production in P. falciparum. Hypoxanthine, which is both transported into the cytoplasm from plasma and produced from adenosine and inosine metabolism (indicated by multiple arrows; see Fig. 1 for more detail), is converted to IMP by PfHGXPRT. IMP can then be converted to adenylate or guanylate nucleotides. To make adenylate nucleotides, IMP is converted to adenylosuccinate by adenylosuccinate synthetase. Adenylosuccinate lyase then converts adenylosuccinate to AMP. AMP is phosphorylated to ADP (by adenylate kinase). Alternatively, IMP can be converted to XMP by IMP dehydrogenase. XMP (which can also be produced directly from xanthine by PfHGXPRT) is converted to GMP by GMP synthetase. GMP can also be produced from guanine by PfHGXPRT. Guanylate kinase phosphorylates GMP to form GDP. Both GDP and ADP are then converted to the NTP or deoxynucleoside triphosphates used in nucleic acid synthesis. PRPP, phosphoribosylpyrophosphate; RBC, red blood cell; PV, parasitophorous vacuole.
SUMMARY
Purines enter the intraerythrocytic malaria parasite via a fast, low-affinity, broad-capacity process that can be attributed primarily to the parasite plasma membrane transporter PfNT1. The reliance of the parasite on PfNT1 makes this protein a worthy target for further investigation, while the determination of the roles played by PfNT2, PfNT3, and PfNT4 in parasite physiology should also prove interesting. Within the parasite, the metabolism of purines appears to be funneled through hypoxanthine by the sequential actions of ADA and PNP, prior to phosphoribosylation of hypoxanthine by PRT. The fact that hypoxanthine is the main purine source found in human serum suggests that PfADA and PfPNP are unlikely to play an essential function under physiological conditions or be considered good drug targets. However, the apparent dependence of the parasite on PfHGXPRT for nucleotide synthesis makes this protein a promising drug target. Efforts to develop compounds that can distinguish between this enzyme and the human counterpart are well justified and may well form the basis for future antimalarial drug strategies.
Acknowledgments
This research was supported by National Institute of Health and Department of Defense grants AI51507 and PR033005 as well as by Burroughs Wellcome Award 1006267 to C.B.M. and by grants from the Australian Research Council and the Australian National Health and Medical Research Council to K.K. C.B.M. is a recipient of the Burroughs Wellcome Award, Investigators of Pathogenesis of Infectious Diseases.
Footnotes
Published ahead of print on 20 June 2008.
REFERENCES
- 1.Asahi, H., T. Kanazawa, Y. Kajihara, K. Takahashi, and T. Takahashi. 1996. Hypoxanthine: a low molecular weight factor essential for growth of erythrocytic Plasmodium falciparum in a serum-free medium. Parasitology 11319-23. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin, S. A., G. A. McConkey, C. E. Cass, and J. D. Young. 2007. Nucleoside transport as a potential target for chemotherapy in malaria. Curr. Pharm. Des. 13569-580. [DOI] [PubMed] [Google Scholar]
- 3.Bhat, J. Y., B. G. Shastri, and H. Balaram. 2008. Kinetic and biochemical characterization of Plasmodium falciparum GMP synthetase. Biochem. J. 409263-273. [DOI] [PubMed] [Google Scholar]
- 4.Booden, T., and R. W. Hull. 1973. Nucleic acid precursor synthesis by Plasmodium lophurae parasitizing chicken erythrocytes. Exp. Parasitol. 34220-228. [DOI] [PubMed] [Google Scholar]
- 5.Bozdech, Z., M. Llinas, B. L. Pulliam, E. D. Wong, J. Zhu, and J. L. DeRisi. 2003. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 1E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, D. M., A. G. Netting, B. K. Chun, Y. Choi, C. K. Chu, and A. M. Gero. 1999. L-nucleoside analogues as potential antimalarials that selectively target Plasmodium falciparum adenosine deaminase. Nucleosides Nucleotides 182521-2532. [DOI] [PubMed] [Google Scholar]
- 7.Bungener, W., and G. Nielsen. 1967. Nucleic acid metabolism in experimental malaria. 1. Studies on the incorporation of thymidine, uridine, and adenosine in the malaria parasite (Plasmodium berghei and Plasmodium vinckei). Z. Tropenmed. Parasitol. 18456-462. (In German.) [PubMed] [Google Scholar]
- 8.Bungener, W., and G. Nielsen. 1968. Nucleic acid metabolism in experimental malaria. 2. Incorporation of adenosine and hypoxanthine into the nucleic acids of malaria parasites (Plasmodium berghei and Plasmodium vinckei). Z Tropenmed Parasitol. 19185-197. (In German.) [PubMed] [Google Scholar]
- 9.Bzowska, A., E. Kulikowska, and D. Shugar. 2000. Purine nucleoside phosphorylases: properties, functions, and clinical aspects. Pharmacol. Ther. 88349-425. [DOI] [PubMed] [Google Scholar]
- 10.Carter, N. S., C. Ben Mamoun, W. Liu, E. O. Silva, S. M. Landfear, D. E. Goldberg, and B. Ullman. 2000. Isolation and functional characterization of the PfNT1 nucleoside transporter gene from Plasmodium falciparum. J. Biol. Chem. 27510683-10691. [DOI] [PubMed] [Google Scholar]
- 11.Carter, N. S., P. Yates, C. S. Arendt, J. M. Boitz, and B. Ullman. 2008. Purine and pyrimidine metabolism in Leishmania. Adv. Exp. Med. Biol. 625141-154. [DOI] [PubMed] [Google Scholar]
- 12.Chaudhary, K., J. A. Darling, L. M. Fohl, W. J. Sullivan, Jr., R. G. Donald, E. R. Pfefferkorn, B. Ullman, and D. S. Roos. 2004. Purine salvage pathways in the apicomplexan parasite Toxoplasma gondii. J. Biol. Chem. 27931221-31227. [DOI] [PubMed] [Google Scholar]
- 13.Chaudhary, K., L. M. Ting, K. Kim, and D. S. Roos. 2006. Toxoplasma gondii purine nucleoside phosphorylase biochemical characterization, inhibitor profiles, and comparison with the Plasmodium falciparum ortholog. J. Biol. Chem. 28125652-25658. [DOI] [PubMed] [Google Scholar]
- 14.Craig, S. P., III, and A. E. Eakin. 2000. Purine phosphoribosyltransferases. J. Biol. Chem. 27520231-20234. [DOI] [PubMed] [Google Scholar]
- 15.Daddona, P. E., W. P. Wiesmann, C. Lambros, W. N. Kelley, and H. K. Webster. 1984. Human malaria parasite adenosine deaminase. Characterization in host enzyme-deficient erythrocyte culture. J. Biol. Chem. 2591472-1475. [PubMed] [Google Scholar]
- 16.Daddona, P. E., W. P. Wiesmann, W. Milhouse, J. W. Chern, L. B. Townsend, M. S. Hershfield, and H. K. Webster. 1986. Expression of human malaria parasite purine nucleoside phosphorylase in host enzyme-deficient erythrocyte culture. Enzyme characterization and identification of novel inhibitors. J. Biol. Chem. 26111667-11673. [PubMed] [Google Scholar]
- 17.de Koning, H. P., D. J. Bridges, and R. J. Burchmore. 2005. Purine and pyrimidine transport in pathogenic protozoa: from biology to therapy. FEMS Microbiol. Rev. 29987-1020. [DOI] [PubMed] [Google Scholar]
- 18.Desai, S. A., D. J. Krogstad, and E. W. McCleskey. 1993. A nutrient-permeable channel on the intraerythrocytic malaria parasite. Nature 362643-646. [DOI] [PubMed] [Google Scholar]
- 19.Downie, M. J., K. J. Saliba, S. Broer, S. M. Howitt, and K. Kirk. 2008. Purine nucleobase transport in the intraerythrocytic malaria parasite. Int. J. Parasitol. 38203-209. [DOI] [PubMed] [Google Scholar]
- 20.Downie, M. J., K. J. Saliba, S. M. Howitt, S. Bröer, and K. Kirk. 2006. Transport of nucleosides across the Plasmodium falciparum parasite plasma membrane has characteristics of PfENT1. Mol. Microbiol. 60738-748. [DOI] [PubMed] [Google Scholar]
- 21.El Bissati, K., R. Zufferey, W. H. Witola, N. S. Carter, B. Ullman, and C. Ben Mamoun. 2006. The plasma membrane permease PfNT1 is essential for purine salvage in the human malaria parasite Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 1039286-9291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21a.El Bissati, K., M. J. Downie, S.-K. Kim, M. Horowitz, N. Carter, B. Ullman, and C. Ben Mamoun. 3 July 2008. Genetic evidence for the essential role of PfNT1 in the transport and utilization of xanthine, guanine, guanosine and adenine by Plasmodium falciparum. Mol. Biochem. Parasitol. doi: 10.1016/j.molbiopara.2008.06.012. [DOI] [PMC free article] [PubMed]
- 22.Engelhardt, B. E., M. I. Jordan, K. E. Muratore, and S. E. Brenner. 2005. Protein molecular function prediction by bayesian phylogenomics. PLoS Comput. Biol. 1e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gandhi, V., J. M. Kilpatrick, W. Plunkett, M. Ayres, L. Harman, M. Du, S. Bantia, J. Davisson, W. G. Wierda, S. Faderl, H. Kantarjian, and D. Thomas. 2005. A proof-of-principle pharmacokinetic, pharmacodynamic, and clinical study with purine nucleoside phosphorylase inhibitor immucillin-H (BCX-1777, forodesine). Blood 1064253-4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gardner, M. J., N. Hall, E. Fung, O. White, M. Berriman, R. W. Hyman, J. M. Carlton, A. Pain, K. E. Nelson, S. Bowman, I. T. Paulsen, K. James, J. A. Eisen, K. Rutherford, S. L. Salzberg, A. Craig, S. Kyes, M. S. Chan, V. Nene, S. J. Shallom, B. Suh, J. Peterson, S. Angiuoli, M. Pertea, J. Allen, J. Selengut, D. Haft, M. W. Mather, A. B. Vaidya, D. M. Martin, A. H. Fairlamb, M. J. Fraunholz, D. S. Roos, S. A. Ralph, G. I. McFadden, L. M. Cummings, G. M. Subramanian, C. Mungall, J. C. Venter, D. J. Carucci, S. L. Hoffman, C. Newbold, R. W. Davis, C. M. Fraser, and B. Barrell. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419498-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gati, W. P., A. N. Lin, T. I. Wang, J. D. Young, and A. R. Paterson. 1990. Parasite-induced processes for adenosine permeation in mouse erythrocytes infected with the malarial parasite Plasmodium yoelii. Biochem. J. 272277-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gero, A. M., E. M. Bugledich, A. R. Paterson, and G. P. Jamieson. 1988. Stage-specific alteration of nucleoside membrane permeability and nitrobenzylthioinosine insensitivity in Plasmodium falciparum infected erythrocytes. Mol. Biochem. Parasitol. 27159-170. [DOI] [PubMed] [Google Scholar]
- 27.Gero, A. M., and S. T. Hall. 1997. Plasmodium falciparum: transport of enantiomers of nucleosides into Sendai-treated trophozoites. Exp. Parasitol. 86228-231. [DOI] [PubMed] [Google Scholar]
- 28.Gutteridge, W. E., and P. I. Trigg. 1970. Incorporation of radioactive precursors into DNA and RNA of Plasmodium knowlesi in vitro. J. Protozool. 1789-96. [DOI] [PubMed] [Google Scholar]
- 29.Hyde, J. E. 2007. Targeting purine and pyrimidine metabolism in human apicomplexan parasites. Curr. Drug Targets 831-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jayalakshmi, R., K. Sumathy, and H. Balaram. 2002. Purification and characterization of recombinant Plasmodium falciparum adenylosuccinate synthetase expressed in Escherichia coli. Protein Expr. Purif. 2565-72. [DOI] [PubMed] [Google Scholar]
- 31.Keough, D. T., A. L. Ng, D. J. Winzor, B. T. Emmerson, and J. de Jersey. 1999. Purification and characterization of Plasmodium falciparum hypoxanthine-guanine-xanthine phosphoribosyltransferase and comparison with the human enzyme. Mol. Biochem. Parasitol. 9829-41. [DOI] [PubMed] [Google Scholar]
- 32.Keough, D. T., T. Skinner-Adams, M. K. Jones, A. L. Ng, I. M. Brereton, L. W. Guddat, and J. de Jersey. 2006. Lead compounds for antimalarial chemotherapy: purine base analogs discriminate between human and P. falciparum 6-oxopurine phosphoribosyltransferases. J. Med. Chem. 497479-7486. [DOI] [PubMed] [Google Scholar]
- 33.Kicska, G. A., P. C. Tyler, G. B. Evans, R. H. Furneaux, K. Kim, and V. L. Schramm. 2002. Transition state analogue inhibitors of purine nucleoside phosphorylase from Plasmodium falciparum. J. Biol. Chem. 2773219-3225. [DOI] [PubMed] [Google Scholar]
- 34.Kicska, G. A., P. C. Tyler, G. B. Evans, R. H. Furneaux, V. L. Schramm, and K. Kim. 2002. Purine-less death in Plasmodium falciparum induced by immucillin-H, a transition state analogue of purine nucleoside phosphorylase. J. Biol. Chem. 2773226-3231. [DOI] [PubMed] [Google Scholar]
- 35.King, A., and D. W. Melton. 1987. Characterisation of cDNA clones for hypoxanthine-guanine phosphoribosyltransferase from the human malarial parasite, Plasmodium falciparum: comparisons to the mammalian gene and protein. Nucleic Acids Res. 1510469-10481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirk, K., H. A. Horner, B. C. Elford, J. C. Ellory, and C. I. Newbold. 1994. Transport of diverse substrates into malaria-infected erythrocytes via a pathway showing functional characteristics of a chloride channel. J. Biol. Chem. 2693339-3347. [PubMed] [Google Scholar]
- 37.Lauer, S. A., P. K. Rathod, N. Ghori, and K. Haldar. 1997. A membrane network for nutrient import in red cells infected with the malaria parasite. Science 2761122-1125. [DOI] [PubMed] [Google Scholar]
- 38.Li, C. M., P. C. Tyler, R. H. Furneaux, G. Kicska, Y. Xu, C. Grubmeyer, M. E. Girvin, and V. L. Schramm. 1999. Transition-state analogs as inhibitors of human and malarial hypoxanthine-guanine phosphoribosyltransferases. Nat. Struct. Biol. 6582-587. [DOI] [PubMed] [Google Scholar]
- 39.Marshall, V. M., and R. L. Coppel. 1997. Characterisation of the gene encoding adenylosuccinate lyase of Plasmodium falciparum. Mol. Biochem. Parasitol. 88237-241. [DOI] [PubMed] [Google Scholar]
- 40.Martin, R. E., R. I. Henry, J. L. Abbey, J. D. Clements, and K. Kirk. 2005. The ‘permeome’ of the malaria parasite: an overview of the membrane transport proteins of Plasmodium falciparum. Genome Biol. 6R26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McConkey, G. A. 2000. Plasmodium falciparum: isolation and characterisation of a gene encoding protozoan GMP synthase. Exp. Parasitol. 9423-32. [DOI] [PubMed] [Google Scholar]
- 42.Murray, A. W. 1971. The biological significance of purine salvage. Annu. Rev. Biochem. 40811-826. [DOI] [PubMed] [Google Scholar]
- 43.Ofulla, A. V., V. C. Okoye, B. Khan, J. I. Githure, C. R. Roberts, A. J. Johnson, and S. K. Martin. 1993. Cultivation of Plasmodium falciparum parasites in a serum-free medium. Am. J. Trop. Med. Hyg. 49335-340. [DOI] [PubMed] [Google Scholar]
- 44.Parker, M. D., R. J. Hyde, S. Y. Yao, L. McRobert, C. E. Cass, J. D. Young, G. A. McConkey, and S. A. Baldwin. 2000. Identification of a nucleoside/nucleobase transporter from Plasmodium falciparum, a novel target for anti-malarial chemotherapy. Biochem. J. 34967-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Penny, J. I., S. T. Hall, C. J. Woodrow, G. M. Cowan, A. M. Gero, and S. Krishna. 1998. Expression of substrate-specific transporters encoded by Plasmodium falciparum in Xenopus laevis oocytes. Mol. Biochem. Parasitol. 9381-89. [DOI] [PubMed] [Google Scholar]
- 46.Polet, H., and C. F. Barr. 1968. DNA, RNA, and protein synthesis in erythrocytic forms of Plasmodium knowlesi. Am. J. Trop. Med. Hyg. 17672-679. [DOI] [PubMed] [Google Scholar]
- 47.Pollack, Y., R. Shemer, S. Metzger, D. T. Spira, and J. Golenser. 1985. Plasmodium falciparum: expression of the adenine phosphoribosyltransferase gene in mouse L cells. Exp. Parasitol. 60270-275. [DOI] [PubMed] [Google Scholar]
- 48.Quashie, N. B., D. Dorin-Semblat, P. G. Bray, G. A. Biagini, C. Doerig, L. C. Ranford-Cartwright, and H. P. De Koning. 2008. A comprehensive model of purine uptake by the malaria parasite Plasmodium falciparum: identification of four purine transport activities in intraerythrocytic parasites. Biochem. J. 411287-295. [DOI] [PubMed] [Google Scholar]
- 49.Queen, S. A., D. Vander Jagt, and P. Reyes. 1988. Properties and substrate specificity of a purine phosphoribosyltransferase from the human malaria parasite, Plasmodium falciparum. Mol. Biochem. Parasitol. 30123-133. [DOI] [PubMed] [Google Scholar]
- 50.Queen, S. A., D. L. Vander Jagt, and P. Reyes. 1989. Characterization of adenine phosphoribosyltransferase from the human malaria parasite, Plasmodium falciparum. Biochim. Biophys. Acta 996160-165. [DOI] [PubMed] [Google Scholar]
- 51.Rager, N., C. B. Mamoun, N. S. Carter, D. E. Goldberg, and B. Ullman. 2001. Localization of the Plasmodium falciparum PfNT1 nucleoside transporter to the parasite plasma membrane. J. Biol. Chem. 27641095-41099. [DOI] [PubMed] [Google Scholar]
- 52.Reyes, P., P. K. Rathod, D. J. Sanchez, J. E. Mrema, K. H. Rieckmann, and H. G. Heidrich. 1982. Enzymes of purine and pyrimidine metabolism from the human malaria parasite, Plasmodium falciparum. Mol. Biochem. Parasitol. 5275-290. [DOI] [PubMed] [Google Scholar]
- 53.Roth, E., Jr., N. Ogasawara, and S. Schulman. 1989. The deamination of adenosine and adenosine monophosphate in Plasmodium falciparum-infected human erythrocytes: in vitro use of 2′deoxycoformycin and AMP deaminase-deficient red cells. Blood 741121-1125. [PubMed] [Google Scholar]
- 54.Schramm, V. L. 2004. Immucillins as antibiotics for T-cell proliferation and malaria. Nucleosides Nucleotides Nucleic Acids 231305-1311. [DOI] [PubMed] [Google Scholar]
- 55.Sherman, I. W. 1998. Purine and pyrimidine metabolism of asexual stages, p. 177-184. In I. W. Sherman (ed.), Malaria: parasite biology, pathogenesis and protection. ASM Press, Washington, DC.
- 56.Shi, W., C. M. Li, P. C. Tyler, R. H. Furneaux, S. M. Cahill, M. E. Girvin, C. Grubmeyer, V. L. Schramm, and S. C. Almo. 1999. The 2.0 A structure of malarial purine phosphoribosyltransferase in complex with a transition-state analogue inhibitor. Biochemistry 389872-9880. [DOI] [PubMed] [Google Scholar]
- 57.Shi, W., C. M. Li, P. C. Tyler, R. H. Furneaux, C. Grubmeyer, V. L. Schramm, and S. C. Almo. 1999. The 2.0 A structure of human hypoxanthine-guanine phosphoribosyltransferase in complex with a transition-state analog inhibitor. Nat. Struct. Biol. 6588-593. [DOI] [PubMed] [Google Scholar]
- 58.Shi, W., L. M. Ting, G. A. Kicska, A. Lewandowicz, P. C. Tyler, G. B. Evans, R. H. Furneaux, K. Kim, S. C. Almo, and V. L. Schramm. 2004. Plasmodium falciparum purine nucleoside phosphorylase: crystal structures, immucillin inhibitors, and dual catalytic function. J. Biol. Chem. 27918103-18106. [DOI] [PubMed] [Google Scholar]
- 59.Snow, R. W., C. A. Guerra, A. M. Noor, H. Y. Myint, and S. I. Hay. 2005. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature 434214-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ting, L. M., W. Shi, A. Lewandowicz, V. Singh, A. Mwakingwe, M. R. Birck, E. A. Ringia, G. Bench, D. C. Madrid, P. C. Tyler, G. B. Evans, R. H. Furneaux, V. L. Schramm, and K. Kim. 2005. Targeting a novel Plasmodium falciparum purine recycling pathway with specific immucillins. J. Biol. Chem. 2809547-9554. [DOI] [PubMed] [Google Scholar]
- 61.Tracy, S. M., and I. W. Sherman. 1972. Purine uptake and utilization by the avian malaria parasite Plasmodium lophurae. J. Protozool. 19541-549. [DOI] [PubMed] [Google Scholar]
- 62.Traut, T. W. 1994. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 1401-22. [DOI] [PubMed] [Google Scholar]
- 63.Tyler, P. C., E. A. Taylor, R. F. Frohlich, and V. L. Schramm. 2007. Synthesis of 5′-methylthio coformycins: specific inhibitors for malarial adenosine deaminase. J. Am. Chem. Soc. 1296872-6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Upston, J. M., and A. M. Gero. 1995. Parasite-induced permeation of nucleosides in Plasmodium falciparum malaria. Biochim. Biophys. Acta 1236249-258. [DOI] [PubMed] [Google Scholar]
- 65.Van Dyke, K., G. C. Tremblay, C. H. Lantz, and C. Szustkiewicz. 1970. The source of purines and pyrimidines in Plasmodium berghei. Am. J. Trop. Med. Hyg. 19202-208. [DOI] [PubMed] [Google Scholar]
- 66.Walsh, J. C., and I. W. Sherman. 1968. Purine and pyrimidine synthesis by the avian malaria parasite Plasmodium lophurae. J. Protozool. 15763-770. [DOI] [PubMed] [Google Scholar]
- 67.Webster, H. K., W. P. Wiesman, and C. S. Pavia. 1984. Adenosine deaminase in malaria infection: effect of 2′-deoxycoformycin in vivo. Adv. Exp. Med. Biol. 165225-229. [DOI] [PubMed] [Google Scholar]
- 68.World Health Organization. 2007. A safer future: global public health security in the 21st century. World Health Report 2007. World Health Organization, Geneva, Switzerland.
- 69.Yamada, K. A., and I. W. Sherman. 1981. Purine metabolizing enzymes of Plasmodium lophurae and its host cell, the duckling (Anas domesticus) erythrocyte. Mol. Biochem. Parasitol. 2349-358. [DOI] [PubMed] [Google Scholar]
- 70.Young, J. A., Q. L. Fivelman, P. L. Blair, P. de la Vega, K. G. Le Roch, Y. Zhou, D. J. Carucci, D. A. Baker, and E. A. Winzeler. 2005. The Plasmodium falciparum sexual development transcriptome: a microarray analysis using ontology-based pattern identification. Mol. Biochem. Parasitol. 14367-79. [DOI] [PubMed] [Google Scholar]