Abstract
Using cryo-electron tomography, we are developing a refined description of native cellular structures in the pathogenic spirochete Treponema denticola. Tightly organized bundles of periplasmic flagella were readily observed in intact plunge-frozen cells. The periplasmic space was measured in both wild-type and aflagellate strains, and found to widen by less than the diameter of flagella when the latter are present. This suggests that a structural change occurs in the peptidoglycan layer to accommodate the presence of the flagella. In dividing cells, the flagellar filaments were found to bridge the cytoplasmic cylinder constriction site. Cytoplasmic filaments, adjacent to the inner membrane, run parallel to the tightly organized flagellar filaments. The cytoplasmic filaments may be anchored by a narrow plate-like structure. The tapering of the cell ends was conserved between cells, with a patella-shaped structure observed in the periplasm at the tip of each cytoplasmic cylinder. Several incompletely characterized structures have been observed in the periplasm between dividing cells, including a cable-like structure linking two cytoplasmic cylinders and complex foil-shaped structures.
Keywords: Intermediate-like filament, flagella, motility, cell division, spirochetes, electron microscopy
1. Introduction
Treponema sp. are causative agents of diseases such as syphilis, yaws, and pinta. Treponemes are also involved in polymicrobial diseases in human and animals, including periodontal diseases, endodontic infection, and digital dermatitis (Simonson et al., 1988; Choi et al., 1997; Siqueira and Rocas, 2004). The Treponema denticola load has been correlated with the severity of both periodontal tissue destruction (Simonson et al., 1988), and endodontic infection (Siqueira et al., 2000). Pathogenicity among spirochetes and treponemes varies genetically.
While phylogenetically distant from Gram-negative bacteria (Paster et al., 1991), Treponema sp., Borrelia sp., Brachyspira sp. and Leptospira sp. share a common membrane organization (Hovind-Hougen, 1976). Spirochetes have outer and inner membranes, defining a peptidoglycan-containing periplasmic space that contains a species-characteristic number of flagellar filaments (Hovind-Hougen, 1976). All spirochetes have the ability to penetrate into dense media and some species can penetrate into tissues (Lux et al., 2001), a capability associated with the periplasmic location of the flagellar filaments (Limberger, 2004). The organization of the periplasm, particularly in relationship to the arrangement of the flagellar filaments in the presence of the peptidoglycan layer, is poorly understood. Spirochetes have the smallest ratio of cell diameter to length among pathogenic bacteria, and consequently are extremely sensitive to the dehydration and fixation techniques conventionally used in prior ultrastructural studies.
Cryo-electron tomography of vitreously frozen cell whole-mounts allows 3-D study of cellular ultrastructure in the native state, and the in situ visualization of macromolecular complexes (Koster et al., 1997; Frank, 2006). The main advantage of cryo-electron microscopy resides in the ability to observe the sample in a fully hydrated physiological state in the absence of stain or chemical treatment (Dubochet et al., 1988). Improvements in electron tomography techniques now allow the visualization of intact prokaryotic cells and associated structures. From the first cryo-tomograms of Archaea (Grimm et al., 1998) to the present, the resolution has greatly improved. It has become possible to observe the organization of cytoskeleton filaments (Izard et al., 2004; Kurner et al., 2005), cellular organization (Matias et al., 2003), membrane invaginations (Zhang et al., 2004), molecular motors (Murphy et al., 2006), and extra-cellular structures (Nickell et al., 2003). In this report, cells of T. denticola wild-type and aflagellate mutant strains were observed fully hydrated and “caught in action” (flash-frozen) in their growth medium.
Comparison of the flagellate (wild-type) and aflagellate (mutant) strains of the pathogen T. denticola has allowed us to analyze the organization of the periplasm in the presence and absence of flagellar filaments, and to clarify the relationship between flagellar and cytoplasmic filaments. Study of wild-type dividing cells has revealed organizational details of overlapping flagellar filaments and led to the discovery of periplasmic structural elements that will need to be characterized at the molecular level.
2. Materials and Methods
2.1. Strains and culture
T. denticola ATCC 33520 wild-type and the flgE-interrupted aflagellate mutant (Limberger et al., 1999) were grown in New Oral Spirochete medium (NOS) with 10% heat-inactivated rabbit serum and 10 μg of cocarboxylase per ml at 36 °C in an anaerobic chamber (Coy Laboratory Products Inc., Grass Lake, MI) under an atmosphere of 85% nitrogen, 10% carbon dioxide, and 5% hydrogen (Limberger et al., 1999).
2.2. Sample preparation
Quantifoil EM specimen grids (type R2/1 or R3.5/1; Quantifoil Microtools, Jena, Germany) were pre-treated with a 10-nm colloidal gold suspension, which was dried down to provide fiducial markers for alignment of the tomographic tilt series (Penczek et al., 1995). T. denticola cells in anaerobic liquid culture (reduced medium), maintained at 36°C, were applied to the grid without dilution or washing. The excess medium was blotted with filter paper. The grid was immediately plunge-frozen into liquid ethane at liquid-nitrogen temperature (Dubochet et al., 1988). This procedure avoided the step of mixing the gold solution with the cells, and allowed plunge-freezing after only several seconds of exposure to aerobic conditions. The endogenous O2 utilization by T. denticola cells (Caldwell and Marquis, 1999; Lai and Chu, 2007) may also provide moderate protection against damage by reactive oxygen species. Grids were stored under liquid nitrogen for future examination.
2.3. Electron microscopy and tomographic reconstruction
Images were recorded at −178°C, using a JEOL JEM4000FX equipped with a Gatan GIF2002 energy filter. The microscope was operated at 400 kV acceleration voltage, in zero-loss energy-filtered mode. Single-axis tilt series were collected with a 1° increment and 120° angular range. The thickness of the ice layer was 250–400 nm, as measured by electron energy-loss spectroscopy (Egerton, 1996). This was sufficient to avoid flattening of the cells, as evidenced by the cross-sections from the tomograms. The total electron dose for a tilt series was 70–90 e−/Å2, with the higher dose used for thicker specimens. The calculated limit to resolution (Crowther et al., 1970) in the x-y plane (parallel to the grid) was 6–10 nm, with the better resolution corresponding to thinner specimens. The calculated z (depth) resolution was 1.6 times the x-y resolution due to the limited tilt range in these single-tilt axis reconstructions (Radermacher and Hoppe, 1980). The underfocus value, 15 μm, was chosen to maximize the transfer of information in images at the expected resolution limit (McEwen et al., 2002). All image processing was done using SPIDER (Frank et al., 1996), and the reconstructions were computed by weighted back-projection (Radermacher, 1992). Twelve wild-type cells, including three dividing cells, were present in six independent tomograms. Fourteen aflagellate mutant cells were present in four independent tomograms.
2.4. Measurements and visualization
Reconstructed 3-D volumes were viewed slice-by-slice in ImageJ (http://rsb.info.nih.gov/ij/). The line tool was used to measure the distance between two points, or the width of a feature. The original pixel size of the tilt-series images, as well as the voxel size of the reconstructions, was 1.8 nm. All measurements were made on x-y plane slices, to avoid effects of elongation due to limited tilt range. Averages were calculated using 0.1 nm precision, and rounded to the nearest integer for inclusion in text and tables. Values are based on nominal instrument magnification with an expected accuracy of ±5%. The standard deviation of measurements made on consecutive slices was less than the pixel size.
Diameters of cells (delimited by the outer membrane) and of cytoplasmic cylinders (delimited by the inner membrane) were measured in consecutive slices of cell cross-sections, then averages were calculated. The measurements were made in the mid-section of the cell in the horizontal plane, avoiding regions containing flagella. The diameter of each flagellar filament was taken using the greatest width within a single slice, thus avoiding the effects of curvature. The measurements were repeated in slices above and below.
Surface-rendered models were created using segmentation routines in Sterecon (Marko and Leith, 1996) and SPIDER (Frank et al., 1996). Final rendering was done with NAG Iris Explorer (NAG, Oxford, UK), and Amira (Mercury Computer Systems, San Diego, CA.). Animations were created using Amira.
3. Results and Discussion
3.1. Organization of flagella in the periplasm
The periplasmic location of the flagellar filaments is a unique feature common to all spirochetes. Cryo-electron tomography of whole-cell mounts was of great benefit in understanding the structure of this region of the cell. The periplasmic space was widened above the flagellar filaments, creating a dome- or tent-like effect when seen in cross-section (Figs. 1A, 3D–E and 5A).
Figure 1.

Widening of the periplasmic space in the presence of flagellar filaments, and position of the peptidoglycan layer. A. Cross-section of wild-type cell (WT-4) with widened periplasmic space, accommodating two flagellar filaments on the upper right (arrows). B. Cross section of aflagellate mutant cell (Afla-1) showing uniform periplasmic space and a peptidoglycan layer (arrowhead). The membranes at the top and bottom are poorly defined due to the tomographic missing wedge. C. Peptidoglycan layer (arrowhead, inset) in longitudinal slice of another wild-type cell. D. Peptidoglycan layer (arrowhead, inset) in aflagellate mutant; longitudinal slice represented by cross-section in B. Tomographic slices 1.8 nm (A, B) and 9 nm (C, D) thick; scale bars = 100 nm.
Figure 3.
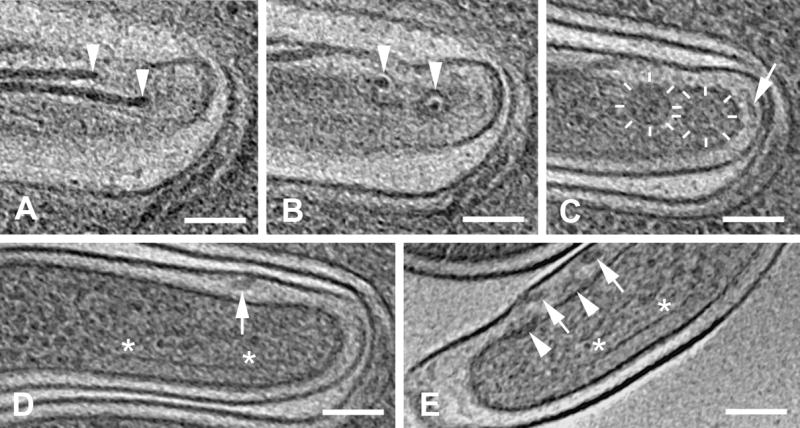
Flagellar filament basal bodies and profiles of a plate-like structure. (A) Slice at the level of the hooks (arrowheads), attaching the flagellar filaments to the basal bodies. (B) Slice 18 nm below A, showing upper rings of the flagellar basal bodies (arrowheads). (C) Slice 32 nm below B showing the larger lower rings of the flagellar basal bodies (indicated by radial line segments). Also seen in C is a cell-end patella-shaped structure (also shown in Fig. 4). (D) Slice 34 nm below C, showing a flagellar filament (arrow) in a widened periplasmic space, and a profile of a plate-like structure (shown in Fig. 4), the two ends of which are marked (*). A–D are from cell WT-10. (E) Side views of basal bodies and hooks from cell WT-8. The periplasmic rings (arrowheads) are below the hooks (barely visible), giving rise to the flagellar filaments (arrows). This cell also has a profile (indicated by * at both ends) of a plate-like structure. Additional membranes from an adjacent cell are seen above and below the cell end of interest in A–D (see Figure S2 for a larger field of view). Tomographic slices, 1.8 nm thick; scale bars = 100 nm.
Figure 5.
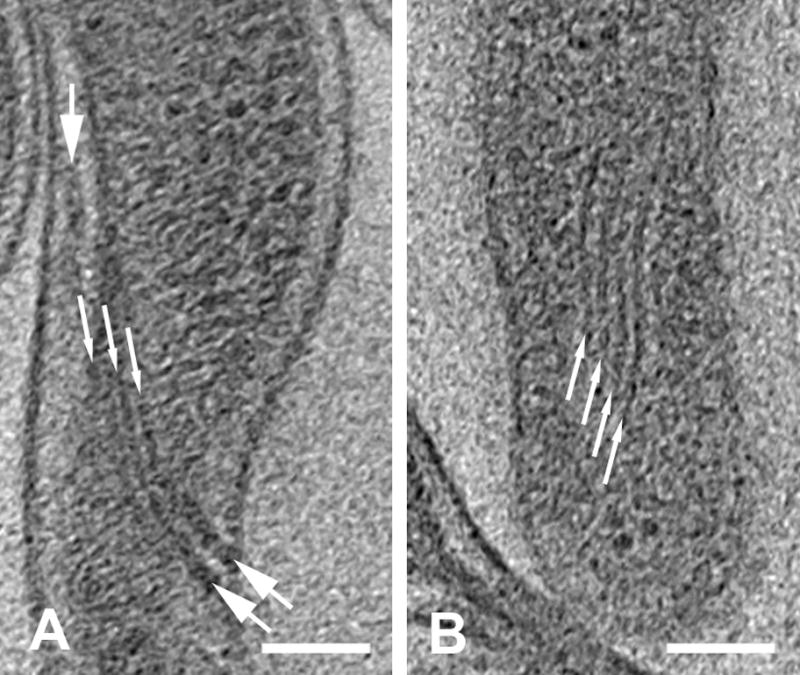
Cytoplasmic filaments in wild-type and aflagellate strains. (A) Cytoplasmic filaments (thin arrows) and flagellar filaments (arrows) in wild-type cell WT-4. The flagella are parallel and above the cytoplasmic filaments, so only the downward-curving portion is seen in the periplasmic space. (B) Cytoplasmic filaments (thin arrows) in the aflagellate strain (cell Afla-4). Tomographic z-slices, 1.8 nm thick; scale bars = 100 nm.
Four characteristics of the flagellar filament organization could be discerned. First, after the first turn of the cell helix, the flagellar filaments are organized in parallel into a tightly packed bundle. This organization persists for the length of the flagella, even when crossing the constriction area of a dividing cell (see below). Second, the measured diameter of a flagellar filament (18–20 nm; Table S1) was somewhat smaller than the 23 nm previously reported by conventional plastic-section electron microscopy (Ruby et al., 1997). The difference is likely due to the preparation, but could also be due to the use of a different strain of T. denticola. Third, there is a symmetrical arrangement of the flagella in newly formed cells. Two flagellar basal bodies are visible on either side of the cytoplasmic cylinder constriction site (Fig. 2). Fourth, there is a uniform electron-lucent space, about 9 nm wide, between the flagellar filaments along the length of the bundle. This inter-filament spacing may be maintained by glycosylation of flagellar proteins (Wyss, 1998); the sugars would not be visible because they scatter electrons in a similar way as the surrounding water. While glycosylation of flagellar proteins is not required for spirochetal motility (Wyss, 1998), it is characteristic of pathogenic treponemes. Glycosylation may play a role in mechanical dynamics during the rotations of the flagellar filaments within a bundle and between overlapping bundles, and it may also be important in protein stability (Mer et al., 1996).
Figure 2.
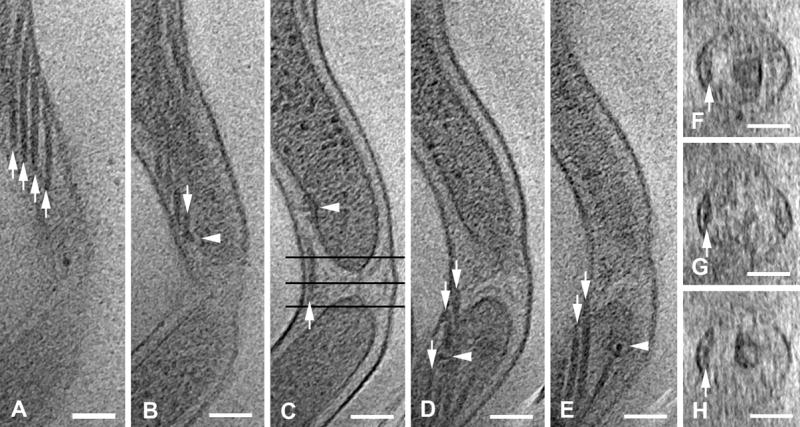
Overlap of periplasmic flagellar filaments at the cell constriction site (cell WT-11). (A) Four flagellar filaments (arrows). (B) Basal body ring (arrowhead), giving rise to flagellar filament (arrow); depth at 27 nm below A. (C) Flagellar filament (arrow) passing through cell constriction site; side view of the second basal body ring (arrowhead); depth 63 nm below B. Black lines indicate position of cross-sections shown in F, G, and H. (D) Crossing of the two flagellar filaments (upper arrows); basal body ring (arrowhead) giving rise to flagellar filament (lower arrow); depth 59 nm below C. (E) Continuation of the two flagellar filaments that crossed the constriction site (arrows); second basal body ring (arrowhead); depth 13 nm below (D). (F) Cross section at tip of upper cytoplasmic cylinder (black line in C), showing two closely-apposed flagellar filaments (arrow); separation between filaments, and continuity of the outer membrane are poorly defined due to the tomographic missing wedge. (G) Cross section midway between the cytoplasmic cylinders, with continuation of flagellar filaments (arrow). (H) Cross section at the tip of the lower cytoplasmic cylinder, with continuation of flagellar filaments (arrow). Tomographic slices, 1.8 nm thick; scale bars = 100 nm.
3.2. Flagellar basal body organization
In T. denticola, the flagellar filaments are attached at one end of the cell, the other being free in the periplasm; the flagellar filaments originating at opposite ends of the same cell overlap at mid-cell (Chan et al., 1993). The formation and rotation of the flagellar filament is associated with its attachment to the flagellar basal body, consisting of the motor, regulator, and export machinery, which is embedded in the cytoplasmic membrane, and protrudes from the cell end (Limberger, 2004).
The basal body is composed of a cytoplasmic ring (Fig. 3C) located on the cytoplasmic face of the inner membrane, the rotor-stator element embedded in the inner membrane, and a periplasmic ring located on the periplasmic face of the inner membrane (Fig. 3E) (Limberger, 2004). The average diameter, from six periplasmic rings, was 29 nm, while the rotor-stator elements and the cytoplasmic ring were 65 nm and 55 nm in diameter, respectively. These results are in agreement with the described substructures of the flagellar basal bodies of Treponema primitia (Murphy et al., 2006). In contrast with T. primitia, T. denticola has, on average, two flagella at each cell end.
At the cell end, basal bodies were often paired, with a spacing of at least 20 nm and a center-to-center distance of 90–100 nm. Organized placement of the basal bodies was also observed in Treponema phagedenis cells (J. Izard, L. L. Slivienski-Gebhardt, W. A. Samsonoff and R. L. Limberger, unpublished data), which have on average five flagella per cell end (Izard et al., 1999). This organization within the cell volume may be associated with a structural framework at the ends of the cytoplasmic cylinder (see below).
3.3. Overlapping of flagellar filaments during cell division
When a cell divides, the cytoplasmic cylinder generates two cytoplasmic cylinders, through constriction by the FtsZ ring. In T. denticola, the two generated cytoplasmic cylinders remain under a unique outer membrane for an undetermined period of time (Figs. 2 and S1). In Figure 2, two flagellar filaments arise from basal bodies at the upper end of the top cell cylinder (outside the field of view). These extend downwards past basal bodies at the bottom of the top cell (Figs. 2B and 2C), then through the constriction area, and continue past the basal bodies at the top end of the lower cell cylinder (Figs. 2D and E). The flagella extending upwards from the basal bodies at the lower end of the upper cytoplasmic cylinder form the four-flagella bundle seen in Fig. 2A, and the flagella arising at the upper end of the bottom cell extend downward form a four-flagella bundle just out of the field of view in Fig. 2E.
In order to produce cell-end gyration, the flagellar filaments rotate in a small fraction of the periplasmic space, in either clockwise or counter-clockwise directions (Charon and Goldstein, 2002), while maintaining a tightly organized bundle (Fig. 2). Intracellular hydrodynamic events, as well as mechanical constraints, may be responsible for the limited lateral movement of the flagellar filament bundle within the periplasmic space.
Control of the length of flagella relative to that of the cell body during cell division would seem to be critically important to avoid interference of the flagellar filaments with outer membrane constriction or even release of flagella outside the periplasm. Yet, T. denticola flagellar filament length is not predetermined (ranging from 1 to 6.1 μm; mean 3.5 μm; Slivienski-Gebhardt et al., 2004), and varies within a single cell (data not shown). Bacterial flagellar filament extension is directed by a cap-like terminal structure of the flagellar filament (Yonekura et al., 2000). Released flagellar filaments from the periplasm have been observed in some cells (Charon et al., 1992), but most of the cells within a population keep all their flagella within the periplasm. It is possible that flagellar filaments are severed prior to or during outer membrane constriction but any such severing mechanism would have to be compatible with the flagellar filaments’ rotation.
In wild-type and mutant strains, no features were observed in the periplasm that might restrict or limit flagellar motion. Such limiting structure, if it existed, would have to be located between the polar attachment sites of the flagella (basal bodies) and follow the helical pattern of the flagellar filaments. A structure that could resist the flagellar rotation and tension within the periplasm would have to be well organized along the length of the cell, and would be expected to be visible in the tomograms. Such a structure was also not seen in negative-stain preparations of T. denticola (L. L. Slivienski-Gebhardt and W. A. Samsonoff, personal communication).
3.4. Periplasmic space organization
The periplasmic space contains two critical components of cell architecture: the flagellar filaments and the peptidoglycan. As previously mentioned, the presence of the flagellar filaments is accommodated by a dome-like bulging of the periplasmic space. To evaluate the effect of such displacement, the width of the periplasmic space was estimated by measuring the distance from the outer edge of the inner membrane to the outer edge of the outer membrane in cell cross-sections (Table 1). The width of the periplasmic space was estimated by subtracting the average width of the outer membrane (see below) from the measurement of the distance between the outer edges of the inner and outer membranes. We found this technique to be a more practical convention than trying to measure directly the gap between the two membranes. The combination of a weakly contrasted inner edge of the outer membrane, and the close apposition of the peptidoglycan in some cross-sections, would adversely affect the accuracy of the latter measurements.
Table 1.
Measurements* of cell dimensions based on tomographic slices of wild-type and aflagellate cells.
| Cell (a) | Cell diameter (nm) | Cytoplasmic cylinder diameter (nm) | Ratio (b) | Measured P and OM (Left side) (nm) (c) | Measured P and OM (Right side) (nm) (c) |
|---|---|---|---|---|---|
| WT-1 | 293 | 244 | 1.20 | 25 | 24 |
| WT-2 | 270 | 211 | 1.28 | 30 | 28 |
| WT-3 | 269 | 211 | 1.28 | 31 | 28 |
| Afla-1 | 241 | 192 | 1.26 | 24 | 25 |
| Afla-2 | 233 | 180 | 1.29 | 28 | 25 |
| Afla-3 | 230 | 178 | 1.29 | 26 | 26 |
Pixel size was 1.8 nm. Standard deviation of the measurements made on consecutive slices was smaller than the pixel size.
WT = wild-type, Afla = Aflagellate mutant.
Cell diameter/cytoplasmic cylinder diameter.
Combined width of periplasm (P) and outer membrane (OM), measured between the outer edges of the inner and outer membranes. Left and right indicate independent measurements on the cell cross-section.
The width of the outer membrane (edge to edge) was measured to be 9–11 nm. Assuming an average outer membrane width of 10 nm, the width of the periplasmic space in wild-type cells in the absence of flagella is 15–22 nm (including the peptidoglycan layer) (Tables 1 and 2). This space widens by 10–16 nm in the presence of flagella (Table 2). The enlargement of the periplasmic space was in all cases less than the flagellar filament diameter (18–20 nm) (Table S1). This apparent discrepancy may be accounted for by physical change in the peptidoglycan layer adjacent to the flagellar filaments, such as localized stretching that has been postulated in current models of peptidoglycan organization (Dmitriev et al., 2003; Vollmer and Holtje, 2004; Meroueh et al., 2006).
Table 2.
Cell characteristics associated with enlargement of the periplasmic space by the flagellar filaments in the wild-type strain as measured on a single tomographic slice. Values are relative to the nominal pixel size.
| Cell | Cell diameter (nm) (a)(b) | Cytoplasmic cylinder diameter (nm) (a) | Width of P and OM with flagella present (nm) (a)(c) | Width of P and OM no flagella (nm) (a)(c) | Increase of width associated with flagella (nm) |
|---|---|---|---|---|---|
| WT-3 | 283 | 203 | 45 | 31 | 14 |
| WT-4 | 301 | 228 | 44 | 31 | 14 |
| WT-5 | 280 | 209 | 42 | 32 | 10 |
| WT-12 | 286 | 214 | 45 | 29 | 16 |
The measurements were made on 1.8 nm thick tomographic z-slices, at the level of the flagellar filaments.
The irregularity of the outer membrane in the presence of flagella influences the cell diameter.
Combined width of periplasm (P) and outer membrane (OM), measured between the outer edges of the inner and outer membranes.
In absence of the flagellar filaments, the peptidoglycan can be observed in the periplasm mid-section closer to the outer membrane in both wild-type and aflagellate strains (Figs. 1 B–D). The width of the peptidoglycan layer is at or below the tomographic resolution limit (6 nm in the best direction). The peptidoglycan layer was not observed in large sections of the tomograms in periplasm sections containing or adjacent to flagella. However, the spacing between the outer membrane, the peptidoglycan layer (when visible) and the flagella, observed in cell cross-sections, suggests a position of the peptidoglycan close to the outer membrane. Genomic analysis of T. denticola coding sequences (Seshadri et al., 2004) reveals no ortholog to FlgJ, a muramidase which allows the flagellar rod of Gram-negative bacteria to cross the peptidoglycan layer (Nambu et al., 1999). The absence of this enzyme is consistent with a location of flagella between the inner membrane and the peptidoglycan layer (Chevance et al., 2007).
The periplasmic dimensions of an aflagellate mutant strain T. denticola flgE (Li et al., 1996; Limberger et al., 1999) were compared to the wild-type. The width of the periplasmic space, 14 to 18 nm, was in the same range as that of the wild-type strain in regions lacking flagella (15 to 22 nm) (Fig. 1B; Table 1).
3.5. Novel periplasmic structures at the cell end
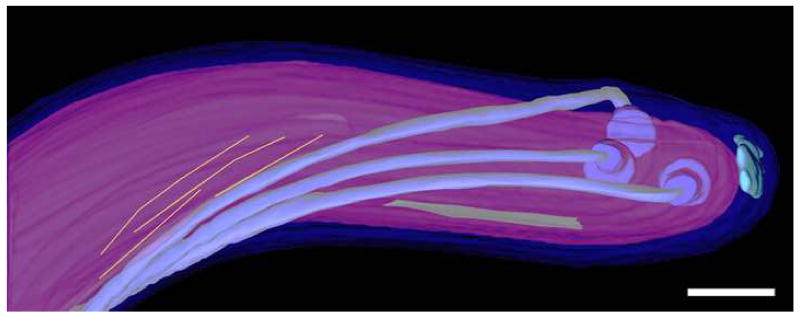
Patella-like structures were observed facing the tip of each of the seven fully-septated cells studied (Figs. 3C, 4 and S1C; Movie S1). A comparable but more cone-shaped structure was recently found at the cell tip of T. primitia (Murphy et al., 2008). The latter structure was described as porous, while in T. denticola the patella-like structure appears to have a more uniform density.
Figure 4.
Surface-rendered model of WT-10 cell end showing three flagellar filaments arising from basal bodies (blue), a periplasmic patella-shaped structure (light blue), a plate-like structure (green), and cytoplasmic filaments (yellow). The outer membrane is dark blue, and the cytoplasmic cylinder is purple. An animation of this surface-rendered model is presented in supplemental material on the publisher’s website (Movie S1). Scale bar = 100 nm.
3.6. Shaping of the cell ends
In both wild-type cells and cells of the aflagellate mutant strain, the major portion of the cytoplasmic cylinder has a constant diameter (Table 1). Tapering begins about 200 nm from the end of the cylinder, and ends with a cap facing the patella-shaped structure at the tip of the cell (Fig. 3). The cytoplasmic cylinder diameter was plotted against the distance from the tip, and the curves (not shown) were found to have a nearly constant slope, which varied little among cells. This is in contrast with the shape of the outer membrane at the cell ends, which differs from cell to cell, in part due to the bulges caused by the flagella. The tapered shape of the cytoplasmic cylinder end may be maintained by a framework that anchors the flagellar basal bodies and maintains distance between them, as well as anchors the cytoplasmic filaments through an associated attachment-plate (see below).
3.7. Cytoplasmic filaments and attachment plate
Cytoplasmic filaments, formed by CfpA monomers (Izard et al., 2001), can be observed in both flagellate and aflagellate strains (Fig. 5). In wild-type cells, the orientation of the filament bundle follows the path of the flagellar filament bundle (Fig. 5A). While this alignment was inferred using a freeze-etch technique (Zemper and Black, 1978), tomographic analysis of native cells clearly shows the location of the cytoplasmic filaments directly below the flagellar filaments. The alignment of the cytoplasmic filaments and the flagellar filaments may be imposed by the overall geometry of the cells and mechanical constraints. Mutagenesis experiments indicated the absence of a relationship between those two structures (Izard et al., 2001). In a mutant strain lacking the cytoplasmic filament formed by CfpA proteins, the motility of single cells was not altered (Izard et al., 2001). Possible physical contact between the two structures may exist through the anchor proteins of the cytoplasmic filaments located in the inner membrane of the cell (Izard et al., 2004). Those inner-membrane anchor proteins were observed to localize along the cytoplasmic filaments, and are still to be identified at the genetic level (Izard et al., 2004).
In the aflagellate mutant strain, the width of the cytoplasmic filament bundle was 57 nm for a four-filament bundle, and 21 nm for a two-filament bundle. In the wild-type strain, the width of the cytoplasmic filament bundle was 36–38 nm for the three-filament bundles measured. These data are consistent with previously obtained measurements of the filament width (5 nm) and interfilament spacing (10 nm) based on tomography of negatively stained cytoplasmic cylinders (Izard et al., 2004). In wild-type cells, the number of cytoplasmic filaments varies from cell to cell and is uncorrelated with the number of flagella which is often lower (data not shown; Izard et al., 1999).
A cytoplasmic plate-like structure was observed in the wild-type cells within the tapered regions at the cell ends (Figs. 3 D–E and 4; Movie S1). Its orientation along the cytoplasmic membrane, and its proximity to the cell tip, is consistent with it representing the attachment point for the cytoplasmic filaments, previously observed only in cells treated to disrupt the membranes (Izard et al., 1999). Cytoplasmic filaments in tomograms of intact frozen-hydrated cells displayed reduced contrast at the cell ends (due in part to their direction relative to that of the missing wedge in these tomograms), and so could not reliably traced in this region, as in models like that of Figure 4. Yet, in some tomograms, short segments of the cytoplasmic filaments could be seen to approach and terminate within 20 nm of the densest part of the plate-like structures (data not shown), suggesting possible interaction with the plate itself or associated material. A similar plate-like structure was observed in T. primitia (Murphy et al., 2008), although no cytoplasmic filaments were described. Whether the absence of cytoplasmic filaments in T. primitia is due to the absence of the gene encoding for the protein constituting the filaments (cfpA) or to poor detectability due to their small size, or low number of filaments is unknown at this time.
3.8. Cell to cell contact
When two cells were apposed, a space of 7–10 nm was observed between the outer membranes. This minimal distance was relatively constant in both wild-type flagellated cells and aflagellate mutant cells (data not shown). The space is likely filled by the sugar moieties of lipoteichoic acids (Schultz et al., 1998), which are not visible in tomograms because they scatter electrons in a similar way as the surrounding water. In the same manner, the sugar moieties of Escherichia coli lipopolysaccharides have poor contrast compared to membrane proteins (Matias et al., 2003).
3.9. Novel structures in dividing cells
A cable-like structure, extending between the two cytoplasmic cylinders, was observed in two independent cells undergoing cytoplasmic cylinder septation. The cable-like structure had a cross-section of approximately 10x15 nm (width x depth) and a length of approximately 60 nm (Figs. 6 and S2 A–B; Movies S2 and S3). The cable-like structure may be a characteristic of late-stage cell division in which the cytoplasmic cylinders are not yet separated, although the flagellar apparatus are fully formed on both newly shaped cell ends (Izard et al., 1999; Izard and Limberger, 2006). The presence of connecting structures during the separation of the cytoplasmic cylinders has been described in C. crescentus (Judd et al., 2005) and in a FtsZ temperature-sensitive mutant in E. coli (Addinall and Lutkenhaus, 1996).
Figure 6.
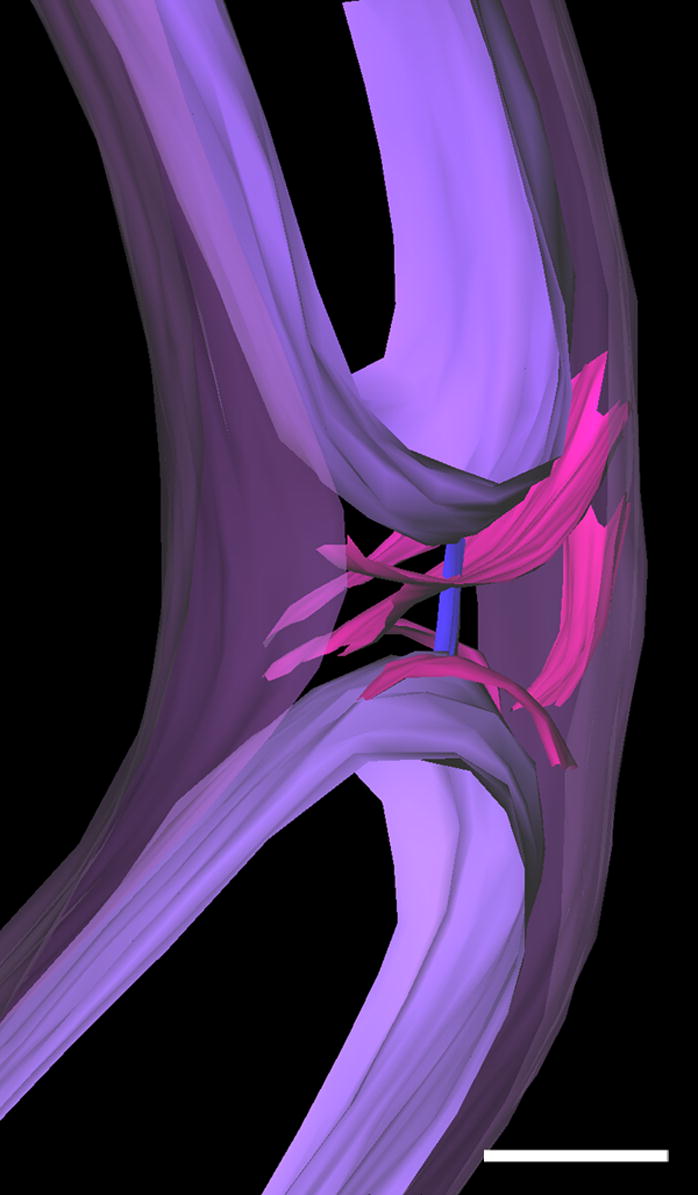
Surface-rendered model of the cell constriction area of cell WT-11, as presented in Figures S1 A and B. The transient bridging cable linking the two cytoplasmic cylinders is in blue. Foil-shaped structures (pink) are seen in the periplasmic space located at the tip of each new cytoplasmic cylinder. The outer membrane is purple, and the cytoplasmic cylinder is blue. An animation of this surface-rendered model (Movie S2), and the associated raw data (Movie S3) are presented in supplemental material on the publisher’s website. Scale bar = 100 nm.
Foil-shaped structures within the periplasm were also observed in tomograms of cells in the late stage of cell division where the two newly formed cytoplasmic cylinders are almost independent under a common outer membrane (Figs. 6 and S2; Movies S2 and S3). These structures may be related to the periseptal annulus observed prior to the separation of the cytoplasmic cylinders in E. coli and Salmonella typhimurium (Macalister et al., 1983; Cook et al., 1987). More recently, slower diffusion of periplasmic proteins was observed at the constriction site at a late stage of C. crescentus cell division (Judd et al., 2005); consistent with a the transient occurrence of periplasmic periseptal structures. Note that the foil-shaped structures in T. denticola were not observed in the cell that had fully septated cytoplasmic cylinders under a single outer membrane (Fig. S2 C).
Cell division is a complex, multi-step process. It will be a challenge to record the large number of tomogram needed to assign the different observed structures to a precise timeline in non-synchronized bacterial cultures undergoing active cell division. At this time, the presence and number of structural components (constriction ring, separation of cytoplasmic filament bundle, flagellar component present at mid-cell, and presence of independent cytoplasmic cylinders) are used as guide to provide milestones to the cell division processes (Izard et al., 1999; Izard and Limberger, 2006).
Resolution of finer structures will require cryo-electron tomography of thinner specimens, which can be obtained by cryo-ultramicrotomy (Hsieh et al., 2006) or focused-ion-beam cryo-milling (Marko et al., 2007). Proteomic analysis of cellular membrane fractions, and the development of methods for the construction of conditional mutants in this difficult-to-grow anaerobe, as well as of a reliable in vivo protein labeling method for treponemes, will be required for the molecular identification of the novel structural components observed in this study.
Supplementary Material
Novel structures associated with late stages of cytoplasmic cylinder constriction. (A) Cable joining the cytoplasmic cylinders, and profiles of foil-like structures (small arrows). The cable is shown in an orthogonal slice in (B), taken at the position of the vertical black lines. Also shown in A is a side view of a periplasmic basal body ring (arrowhead) and a flagellar filament (large arrow) that is continuous across the constriction space (see Fig. 2). (C) Two patella-shaped structures (arrowheads) at the tip of independent cytoplasmic cylinders after cell division (cell WT-5). Tomographic z-slices, 1.8 nm thick; scale bars = 100 nm.
Overview images of wild-type cells. Details for the boxed areas in A and B are shown in Figures 2 and 3, respectively. Tomographic slices 1.8 nm thick; scale bars = 100 nm.
Animation showing the z-slices in the tomogram of cell WT-10, followed by superposition and rotation of the surface-rendered model shown in Figure 4.
Rotation animation of the surface-rendered model in Figure 6 (cell WT-11), combined with the 1.8 nm thick tomographic z-slice.
Animation showing the z-slices in the tomogram of the cell WT-11 showing the mid-cell featuring the cable-like structure, the foil-shaped structures, and the overlap of flagellar filaments.
Acknowledgments
We would like to thank William A. Samsonoff and Linda L. Slivienski-Gebhardt (Wadsworth Center) for sharing electron micrographs of negatively stained T. denticola, and the Wadsworth Center’s Core Facility for Electron Microscopy. This work was supported by the John W. Hein Fellowship of The Forsyth Institute, the National Institutes of Health (NIH) Public Health Service grants AI34354 and DE017106, and the NIH-NCRR grant P41 RR01219 that supports the Wadsworth Center’s Resource for Biological Complexity as a National Biotechnological Resource.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Addinall SG, Lutkenhaus J. FtsZ-spirals and -arcs determine the shape of the invaginating septa in some mutants of Escherichia coli. Mol Microbiol. 1996;22(2):231–237. doi: 10.1046/j.1365-2958.1996.00100.x. [DOI] [PubMed] [Google Scholar]
- Caldwell CE, Marquis RE. Oxygen metabolism by Treponema denticola. Oral Microbiol Immunol. 1999;14(1):66–72. doi: 10.1034/j.1399-302x.1999.140109.x. [DOI] [PubMed] [Google Scholar]
- Chan EC, Siboo R, Keng T, Psarra N, Hurley R, Cheng SL, Iugovaz I. Treponema denticola (ex Brumpt 1925) sp. nov., nom. rev., and identification of new spirochete isolates from periodontal pockets. Int J Syst Bacteriol. 1993;43(2):196–203. doi: 10.1099/00207713-43-2-196. [DOI] [PubMed] [Google Scholar]
- Charon NW, Goldstein SF. Genetics of motility and chemotaxis of a fascinating group of bacteria: the spirochetes. Annu Rev Genet. 2002;36:47–73. doi: 10.1146/annurev.genet.36.041602.134359. [DOI] [PubMed] [Google Scholar]
- Charon NW, Goldstein SF, Block SM, Kurci K, Ruby JD, Kreiling JA, Limberger RJ. Morphology and dynamics of protruding spirochete periplasmic flagella. J Bacteriol. 1992;174:832–840. doi: 10.1128/jb.174.3.832-840.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevance FF, Takahashi N, Karlinsey JE, Gnerer J, Hirano T, Samudrala R, Aizawa S, Hughes KT. The mechanism of outer membrane penetration by the eubacterial flagellum and implications for spirochete evolution. Genes Dev. 2007;21(18):2326–2335. doi: 10.1101/gad.1571607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi BK, Nattermann H, Grund S, Haider W, Göbel UB. Spirochetes from digital dermatitis lesions in cattle are closely related to treponemes associated with human periodontitis. Int J Syst Bacteriol. 1997;47(1):175–181. doi: 10.1099/00207713-47-1-175. [DOI] [PubMed] [Google Scholar]
- Cook WR, Kepes F, Joseleau-Petit D, MacAlister TJ, Rothfield LI. Proposed mechanism for generation and localization of new cell division sites during the division cycle of Escherichia coli. Proc Natl Acad Sci U S A. 1987;84(20):7144–7148. doi: 10.1073/pnas.84.20.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowther RA, De Rosier DJ, Klug A. The reconstruction of a three-dimesional structure from the projections and its application to electron microscopy. Proc R Soc London. 1970;A317:319–340. [Google Scholar]
- Dmitriev BA, Toukach FV, Schaper KJ, Holst O, Rietschel ET, Ehlers S. Tertiary structure of bacterial murein: the scaffold model. J Bacteriol. 2003;185(11):3458–3468. doi: 10.1128/JB.185.11.3458-3468.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubochet J, Adrian M, Chang JJ, Homo JC, Lepault J, McDowall AW, Schultz P. Cryo-electron microscopy of vitrified specimens. Q Rev Biophys. 1988;21(2):129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- Egerton RF. Electron energy-loss spectroscopy in the electron microscope. New York, NY: Plenum; 1996. [Google Scholar]
- Frank J. Electron tomography of cells and tissue. New York, NY: Springer; 2006. [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116(1):190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Grimm R, Singh H, Rachel R, Typke D, Zillig W, Baumeister W. Electron tomography of ice-embedded prokaryotic cells. Biophys J. 1998;74(2 Pt 1):1031–1042. doi: 10.1016/S0006-3495(98)74028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovind-Hougen K. Determination by means of electron microscopy of morphological criteria of value for classification of some spirochetes, in particular treponemes. Acta Pathol Scand Sect B. 1976;255:1–41. [PubMed] [Google Scholar]
- Hsieh CE, Leith A, Mannella CA, Frank J, Marko M. Towards high-resolution three-dimensional imaging of native mammalian tissue: electron tomography of frozen-hydrated rat liver sections. J Struct Biol. 2006;153(1):1–13. doi: 10.1016/j.jsb.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Izard J, Limberger RJ. Structural and genomic features of treponemal architecture. In: Radolf JD, Lukehart SA, editors. Treponema molecular and cellular biology. Norfolk, UK: Horizon Bioscience; 2006. [Google Scholar]
- Izard J, McEwen BF, Barnard RM, Portuese T, Samsonoff WA, Limberger RJ. Tomographic reconstruction of treponemal cytoplasmic filaments reveals novel bridging and anchoring components. Mol Microbiol. 2004;51(3):609–618. doi: 10.1046/j.1365-2958.2003.03864.x. [DOI] [PubMed] [Google Scholar]
- Izard J, Samsonoff WA, Kinoshita MB, Limberger RJ. Genetic and structural analysis of the cytoplasmic filaments of wild-type and flagellar filament mutant of Treponema phagedenis. J Bacteriol. 1999;181(21):6739–6746. doi: 10.1128/jb.181.21.6739-6746.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izard J, Samsonoff WA, Limberger RJ. Cytoplasmic filament-deficient mutant of Treponema denticola has pleiotropic defects. J Bacteriol. 2001;183(3):1078–1084. doi: 10.1128/JB.183.3.1078-1084.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judd EM, Comolli LR, Chen JC, Downing KH, Moerner WE, McAdams HH. Distinct constrictive processes, separated in time and space, divide Caulobacter inner and outer membranes. J Bacteriol. 2005;187(20):6874–6882. doi: 10.1128/JB.187.20.6874-6882.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster AJ, Grimm R, Typke D, Hegerl R, Stoschek A, Walz J, Baumeister W. Perspectives of molecular and cellular electron tomography. J Struct Biol. 1997;120(3):276–308. doi: 10.1006/jsbi.1997.3933. [DOI] [PubMed] [Google Scholar]
- Kurner J, Frangakis AS, Baumeister W. Cryo-electron tomography reveals the cytoskeletal structure of Spiroplasma melliferum. Science. 2005;307(5708):436–438. doi: 10.1126/science.1104031. [DOI] [PubMed] [Google Scholar]
- Lai Y, Chu L. A novel mechanism for conditional aerobic growth of anaerobic Treponema denticola. Appl Environ Microbiol. 2008;74(1):73–79. doi: 10.1128/AEM.01972-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruby J, Charon N, Kuramitsu H. Gene inactivation in the oral spirochete Treponema denticola: construction of an flgE mutant. J Bacteriol. 1996;178(12):3664–3667. doi: 10.1128/jb.178.12.3664-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limberger RJ. The periplasmic flagellum of spirochetes. J Mol Microbiol Biotechnol. 2004;7:30–40. doi: 10.1159/000077867. [DOI] [PubMed] [Google Scholar]
- Limberger RJ, Slivienski LL, Izard J, Samsonoff WA. Insertional inactivation of Treponema denticola tap1 results in a nonmotile mutant with elongated flagellar hooks. J Bacteriol. 1999;181(12):3743–3750. doi: 10.1128/jb.181.12.3743-3750.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux R, Miller JN, Park NH, Shi W. Motility and chemotaxis in tissue penetration of oral epithelial cell layers by Treponema denticola. Infect Immun. 2001;69(10):6276–6283. doi: 10.1128/IAI.69.10.6276-6283.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macalister TJ, Macdonald B, Rothfield LI. The periseptal annulus: An organelle associated with cell division in Gram-negative bacteria. Proc Natl Acad Sci U S A. 1983;80(5):1372–1376. doi: 10.1073/pnas.80.5.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marko M, Hsieh C, Schalek R, Frank J, Mannella C. Focused-ion-beam thinning of frozen-hydrated biological specimens for cryo-electron microscopy. Nat Methods. 2007;4(3):215–217. doi: 10.1038/nmeth1014. [DOI] [PubMed] [Google Scholar]
- Marko M, Leith A. Sterecon--three-dimensional reconstructions from stereoscopic contouring. J Struct Biol. 1996;116(1):93–98. doi: 10.1006/jsbi.1996.0016. [DOI] [PubMed] [Google Scholar]
- Matias VR, Al-Amoudi A, Dubochet J, Beveridge TJ. Cryo-transmission electron microscopy of frozen-hydrated sections of Escherichia coli and Pseudomonas aeruginosa. J Bacteriol. 2003;185(20):6112–6118. doi: 10.1128/JB.185.20.6112-6118.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BF, Marko M, Hsieh CE, Manella CA. Use of frozen-hydrated axonemes to assess imaging parameters and resolution limits in cryo-electron tomography. J Struct Biol. 2002;138(1–2):47–57. doi: 10.1016/s1047-8477(02)00020-5. [DOI] [PubMed] [Google Scholar]
- Mer G, Hietter H, Lefevre JF. Stabilization of proteins by glycosylation examined by NMR analysis of a fucosylated proteinase inhibitor. Nat Struct Biol. 1996;3(1):45–53. doi: 10.1038/nsb0196-45. [DOI] [PubMed] [Google Scholar]
- Meroueh SO, Bencze KZ, Hesek D, Lee M, Fisher JF, Stemmler TL, Mobashery S. Three-dimensional structure of the bacterial cell wall peptidoglycan. Proc Natl Acad Sci U S A. 2006;103(12):4404–4409. doi: 10.1073/pnas.0510182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy GE, Leadbetter JR, Jensen GJ. In situ structure of the complete Treponema primitia flagellar motor. Nature. 2006;442(7106):1062–1064. doi: 10.1038/nature05015. [DOI] [PubMed] [Google Scholar]
- Murphy GE, Matson EG, Leadbetter JR, Berg HC, Jensen GJ. Novel ultrastructures of Treponema primitia and their implications for motility. Mol Microbiol. 2008;67(6):1184–1195. doi: 10.1111/j.1365-2958.2008.06120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambu T, Minamino T, Macnab RM, Kutsukake K. Peptidoglycan-hydrolyzing activity of the FlgJ protein, essential for flagellar rod formation in Salmonella typhimurium. J Bacteriol. 1999;181(5):1555–1561. doi: 10.1128/jb.181.5.1555-1561.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickell S, Hegerl R, Baumeister W, Rachel R. Pyrodictium cannulae enter the periplasmic space but do not enter the cytoplasm, as revealed by cryo-electron tomography. J Struct Biol. 2003;141(1):34–42. doi: 10.1016/s1047-8477(02)00581-6. [DOI] [PubMed] [Google Scholar]
- Paster BJ, Dewhirst FE, Weisburg WG, Tordoff LA, Fraser GJ, Hespell RB, Stanton TB, Zablen L, Mandelco L, Woese CR. Phylogenetic analysis of the spirochetes. J Bacteriol. 1991;173:6101–6109. doi: 10.1128/jb.173.19.6101-6109.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penczek P, Marko M, Buttle K, Frank J. Double-tilt electron tomography. Ultramicroscopy. 1995;60(3):393–410. doi: 10.1016/0304-3991(95)00078-x. [DOI] [PubMed] [Google Scholar]
- Radermacher M. Weighted back-projection methods. In: Frank J, editor. Electron Tomography. New York, NY: Plenum Press; 1992. pp. 91–115. [Google Scholar]
- Radermacher M, Hoppe W. Properties of 3-D reconstructions from projections from conical tilting compared to single-axis tilting. Proc 7th Int Cong Electron Microsc. 1980;1:132–133. [Google Scholar]
- Ruby JD, Li H, Kuramitsu H, Norris SJ, Goldstein SF, Buttle KF, Charon NW. Relationship of Treponema denticola periplasmic flagella to irregular cell morphology. J Bacteriol. 1997;179(5):1628–1635. doi: 10.1128/jb.179.5.1628-1635.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz CP, Wolf V, Lange R, Mertens E, Wecke J, Naumann D, Zahringer U. Evidence for a new type of outer membrane lipid in oral spirochete Treponema denticola. Functioning permeation barrier without lipopolysaccharides. J Biol Chem. 1998;273(25):15661–15666. doi: 10.1074/jbc.273.25.15661. [DOI] [PubMed] [Google Scholar]
- Seshadri R, Myers GS, Tettelin H, Eisen JA, Heidelberg JF, Dodson RJ, Davidsen TM, DeBoy RT, Fouts DE, Haft DH, Selengut J, Ren Q, Brinkac LM, Madupu R, Kolonay J, Durkin SA, Daugherty SC, Shetty J, Shvartsbeyn A, Gebregeorgis E, Geer K, Tsegaye G, Malek J, Ayodeji B, Shatsman S, McLeod MP, Smajs D, Howell JK, Pal S, Amin A, Vashisth P, McNeill TZ, Xiang Q, Sodergren E, Baca E, Weinstock GM, Norris SJ, Fraser CM, Paulsen IT. Comparison of the genome of the oral pathogen Treponema denticola with other spirochete genomes. Proc Natl Acad Sci U S A. 2004;101(15):5646–5651. doi: 10.1073/pnas.0307639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonson LG, Goodman CH, Bial JJ, Morton HE. Quantitative relationship of Treponema denticola to severity of periodontal disease. Infect Immun. 1988;56(4):726–728. doi: 10.1128/iai.56.4.726-728.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siqueira JF, Jr, Rocas IN. Treponema species associated with abscesses of endodontic origin. Oral Microbiol Immunol. 2004;19(5):336–339. doi: 10.1111/j.1399-302x.2004.00156.x. [DOI] [PubMed] [Google Scholar]
- Siqueira JF, Jr, Rocas IN, Favieri A, Santos KR. Detection of Treponema denticola in endodontic infections by 16S rRNA gene-directed polymerase chain reaction. Oral Microbiol Immunol. 2000;15(5):335–337. doi: 10.1034/j.1399-302x.2000.150512.x. [DOI] [PubMed] [Google Scholar]
- Slivienski-Gebhardt LL, Izard J, Samsonoff WA, Limberger RJ. Development of a novel chloramphenicol resistance expression plasmid used for genetic complementation of a fliG deletion mutant in Treponema denticola. Infect Immun. 2004;72(9):5493–5497. doi: 10.1128/IAI.72.9.5493-5497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W, Holtje JV. The architecture of the murein (peptidoglycan) in gram-negative bacteria: vertical scaffold or horizontal layer(s)? J Bacteriol. 2004;186(18):5978–5987. doi: 10.1128/JB.186.18.5978-5987.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss C. Flagellins, but not endoflagellar sheath proteins, of Treponema pallidum and of pathogen-related oral spirochetes are glycosylated. Infect Immun. 1998;66(12):5751–5754. doi: 10.1128/iai.66.12.5751-5754.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura K, Maki S, Morgan DG, DeRosier DJ, Vonderviszt F, Imada K, Namba K. The bacterial flagellar cap as the rotary promoter of flagellin self- assembly. Science. 2000;290(5499):2148–2152. doi: 10.1126/science.290.5499.2148. [DOI] [PubMed] [Google Scholar]
- Zemper ED, Black SH. Morphology of freeze-etched Treponema refringens (Nichols) Arch Microbiol. 1978;117(3):227–238. doi: 10.1007/BF00738540. [DOI] [PubMed] [Google Scholar]
- Zhang P, Bos E, Heymann J, Gnaegi H, Kessel M, Peters PJ, Subramaniam S. Direct visualization of receptor arrays in frozen-hydrated sections and plunge-frozen specimens of E. coli engineered to overproduce the chemotaxis receptor Tsr. J Microsc. 2004;216(Pt 1):76–83. doi: 10.1111/j.0022-2720.2004.01395.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Novel structures associated with late stages of cytoplasmic cylinder constriction. (A) Cable joining the cytoplasmic cylinders, and profiles of foil-like structures (small arrows). The cable is shown in an orthogonal slice in (B), taken at the position of the vertical black lines. Also shown in A is a side view of a periplasmic basal body ring (arrowhead) and a flagellar filament (large arrow) that is continuous across the constriction space (see Fig. 2). (C) Two patella-shaped structures (arrowheads) at the tip of independent cytoplasmic cylinders after cell division (cell WT-5). Tomographic z-slices, 1.8 nm thick; scale bars = 100 nm.
Overview images of wild-type cells. Details for the boxed areas in A and B are shown in Figures 2 and 3, respectively. Tomographic slices 1.8 nm thick; scale bars = 100 nm.
Animation showing the z-slices in the tomogram of cell WT-10, followed by superposition and rotation of the surface-rendered model shown in Figure 4.
Rotation animation of the surface-rendered model in Figure 6 (cell WT-11), combined with the 1.8 nm thick tomographic z-slice.
Animation showing the z-slices in the tomogram of the cell WT-11 showing the mid-cell featuring the cable-like structure, the foil-shaped structures, and the overlap of flagellar filaments.