Abstract
The differentiation of neurons and the outgrowth of neurites depends on microtubule-associated proteins such as tau protein. To study this process, we have used the model of Sf9 cells, which allows efficient transfection with microtubule-associated proteins (via baculovirus vectors) and observation of the resulting neurite-like extensions. We compared the phosphorylation of tau23 (the embryonic form of human tau) with mutants in which critical phosphorylation sites were deleted by mutating Ser or Thr residues into Ala. One can broadly distinguish two types of sites, the KXGS motifs in the repeats (which regulate the affinity of tau to microtubules) and the SP or TP motifs in the domains flanking the repeats (which contain epitopes for antibodies diagnostic of Alzheimer’s disease). Here we report that both types of sites can be phosphorylated by endogenous kinases of Sf9 cells, and that the phosphorylation pattern of the transfected tau is very similar to that of neurons, showing that Sf9 cells can be regarded as an approximate model for the neuronal balance between kinases and phosphatases. We show that mutations in the repeat domain and in the flanking domains have opposite effects. Mutations of KXGS motifs in the repeats (Ser262, 324, and 356) strongly inhibit the outgrowth of cell extensions induced by tau, even though this type of phosphorylation accounts for only a minor fraction of the total phosphate. This argues that the temporary detachment of tau from microtubules (by phosphorylation at KXGS motifs) is a necessary condition for establishing cell polarity at a critical point in space or time. Conversely, the phosphorylation at SP or TP motifs represents the majority of phosphate (>80%); mutations in these motifs cause an increase in cell extensions, indicating that this type of phosphorylation retards the differentiation of the cells.
INTRODUCTION
The formation and growth of neurites from a neuronal cell and the differentiation into an axon and several dendrites depend on the polymerization of microtubules and microtubule-dependent transport. The growth and stability of microtubules is regulated by microtubule-associated proteins (MAPs).1 A number of MAPs have been characterized (for review, see Hirokawa, 1994). Several of them are prominent in neurons and tend to be compartmentalized; the best known examples are MAP2 (largely dendritic) and tau protein (largely axonal; Cleveland et al., 1977; Binder et al., 1985; Drubin et al., 1985; Drubin and Kirschner, 1986). The formation of neuronal processes and the selective targeting of cytoskeletal components to their destinations is currently an area of active research. Much of what we know today has been derived from neuronal cells in culture in which individual components can be tracked (e.g., by fluorescence methods) and manipulated, for example, increased by transfection or microinjection or suppressed by antibodies or antisense oligonucleotides (for review, see Mandell and Banker, 1995; Ludin and Matus, 1998). However, neuronal cells can be transfected only with low efficiency and yield protein quantities that are often below the limit for biochemical analysis. This has led to the development of cell models in which certain aspects of neuronal behavior can be studied more directly and in molecular detail (Kanai et al., 1989; Caceres et al., 1990, 1991, 1992; Chen et al., 1992; Barlow et al., 1994). One such system is that of the insect ovary cell line Sf9, which can be transfected by baculovirus vectors. When transfected with tau or other MAPs, these cells develop extensions analogous to those of differentiating neurons, stabilized by bundles of parallel microtubules and their associated proteins (Baas et al., 1991; Knops et al., 1991; Frappier et al., 1994; Kosik and McConlogue, 1994). This system is therefore suitable for studying biochemical and morphological issues, such as the regulation of the microtubule cytoskeleton via the phosphorylation of tau.
The phosphorylation of tau protein has received particular attention because the protein precipitates in a highly phosphorylated form in Alzheimer’s disease, an age-related dementia. The physiological function of tau is not well understood, but gradients of phosphorylation of tau or other MAPs suggest a role in the development and maintenance of neuronal processes (Burack and Halpain, 1996; Mandell and Banker, 1996). In Alzheimer’s disease it is generally thought that excess phosphorylation reflects an imbalance of cellular signal transduction pathways and is a prelude to neuronal degeneration (for review, see Trojanowski and Lee, 1995; Mandelkow and Mandelkow, 1998). This emphasizes the need for understanding the origin and role of tau’s phosphorylation and the kinases and phosphatases controlling it. Tau can be phosphorylated at many different sites and by a number of kinases. We broadly distinguish two types of phosphorylation. The first comprises the phosphorylation in the repeats, specifically at the KXGS motifs; their phosphorylation (notably of Ser262) strongly inhibits tau-microtubule interactions. These sites can be phosphorylated by MAP/microtubule affinity–regulating kinase (MARK) and (with lower efficiency) by cAMP-dependent protein kinase (PKA) (Biernat et al., 1993; Brandt et al., 1994; Drewes et al., 1995). The second type of phosphorylation occurs mostly in the domains flanking the repeats, which contain a number of SP or TP motifs and can be phosphorylated by several proline-directed kinases; these have only a weak influence on tau–microtubule interactions (Drechsel et al., 1992; Biernat et al., 1993). Both types of phosphorylation sites are elevated in Alzheimer tau (Morishima-Kawashima et al., 1995), and the SP or TP motifs are recognized by several phosphorylation-dependent antibodies, which are diagnostic for the “Alzheimer-like” state of tau protein (for review, see Goedert et al., 1994). To analyze the roles of these sites, we have therefore made mutants either in the repeat domain or in the flanking domains, eliminating phosphorylation sites by replacing Ser or Thr residues with Ala.
We show here that the different types of phosphorylation have opposite effects on process formation in Sf9 cells. The expression of wild-type tau induces processes in ∼60% of the cells. This critically requires the phosphorylation in the repeats: if two or more KXGS motifs are turned into KXGA (especially Ser262 and Ser356), process formation is largely suppressed. This suggests that a detachment of tau from microtubules and their destabilization is necessary for inducing cell processes. At the same time the extent of phosphorylation at these sites is remarkably low (∼5% of all sites), indicating that the detachment of tau may be needed only at a special place or time. On the other hand, phosphorylation in the flanking domains (at SP or TP motifs) inhibits process formation but only to a moderate extent (∼30%), despite the fact that these sites account for ∼80% of the total phosphorylation. Thus, if all SP or TP mutated into AP, process frequency increases by ∼30%. This argues that phosphorylation at SP or TP sites plays a generalized but weak role. Overall, the phosphorylation pattern of tau in transfected Sf9 cells is surprisingly similar to that of neuronal cells or other cells transfected with tau (Illenberger et al., 1998). This shows that the balance between kinases and phosphatases is comparable in different cell types and suggests that the Sf9 cell is a good model for studying the role of tau phosphorylation in neurite outgrowth.
MATERIALS AND METHODS
Cells and Viruses
Sf9 cells were obtained from Invitrogen (San Diego, CA) and were grown at 27°C in monolayer culture Grace’s medium (Life Technologies, Gaithersburg, MD) supplemented with 10% fetal bovine serum, 50 μg/ml Gentamycin, and 2.5 μg/ml Amphotericin. BaculoGold was obtained from PharMingen (San Diego, CA); pVL1392 was from Invitrogen.
Tau Transfection and Mutations
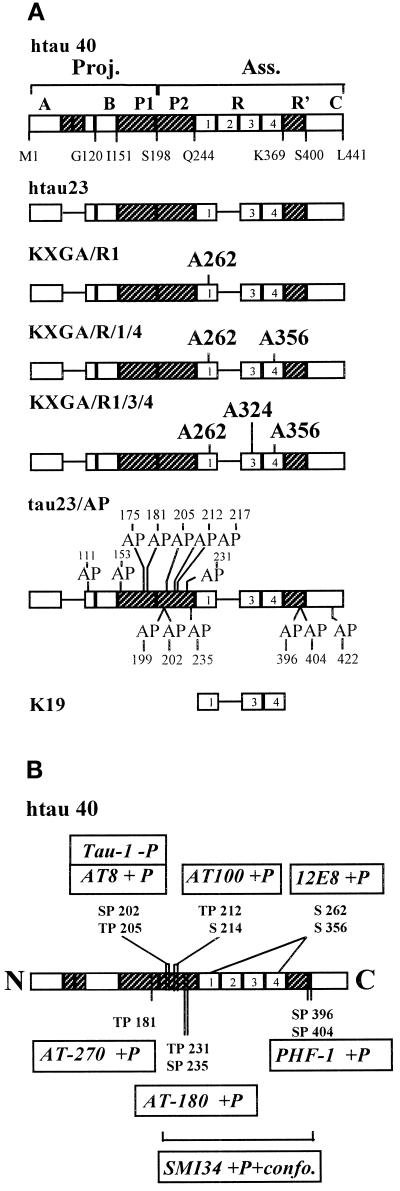
All mutated tau genes used in this study were derived from htau23, the shortest human tau isoform described by Goedert et al. (1989). Point mutants were made by PCR as described (Biernat et al., 1993). The mutant tau23/AP is a derivative of htau23 with all 14 Ser-Pro and Thr-Pro sites mutated into Ala-Pro. KXGA/R1 (htau23/A262) is a derivative of htau23 with a Ser262-Ala mutation, KXGA/R1/3/4, (htau23/A262/A324/A356) is htau23 containing Ser262-Ala, Ser324-Ala, and Ser356-Ala mutations. The recombinant tau genes were excised from the bacterial expression vector pNG2 (Biernat et al. 1993) with XbaI and BamHI, and inserted into the baculovirus transfer vector pVL1392 cut with the same restriction endonucleases. For the construction of tau-containing baculovirus vectors we used the BaculoGold system. The BaculoGold DNA is a modified type of baculovirus containing a lethal deletion and does not code for viable virus itself. Cotransfection of the BaculoGold DNA with a complementing baculovirus transfer vector rescued the lethal deletion of this virus DNA and reconstituted viable virus particles carrying the htau23 coding sequence. Plasmid DNA used for transfections was purified using Qiagen (Hilden, Germany) cartridges. Sf9 cells grown in monolayers (2 × 106 cells in a 60-mm cell culture dish) were cotransfected with baculovirus DNA (0.5 μg BaculoGold DNA) and with vector derivatives of pVL1392 (2 μg) using a calcium phosphate coprecipitation method. The following baculovirus strains were obtained: BaculoGold-htau23, -tau23/AP, -KXGA/R1, -KXGA/R1/4, -KXGA/R1/3/4, and -tau23/AP/R1/4. The presence of recombinant proteins was examined in the infected Sf9 cells 5 d after infection by SDS-PAGE and Western blotting.
Western Blotting
Sf9 cells were infected with either wild-type virus or recombinant virus at a multiplicity of infection (MOI) of 1–5. Cell lysates were prepared either directly with hot (100°C) SDS sample buffer (Laemmli, 1970) or in hypotonic lysis buffer (HLB; 50 mM Tris-HCl, pH 7.4, 120 mM NaCl, 10% glycerol, 1% Nonidet P-40, 5 mM DTT, 1 mM EGTA, 20 mM NaF, 1 mM orthovanadate, 5 μM microcystin, 100 μg/ml protease inhibitors leupeptin, aprotinin, and pepstatin). The cells lysed in HLB were centrifuged at 16,000 × g for 15 min, and the supernatant and pellet were separated. Proteins were then electrophoresed by SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and blotted with the following mAbs: AT-8 (1:2000), AT-180 (1:1000), AT-270 (1:4000), AT-100 (1:500) (AT antibodies from Innogenetics, Ghent, Belgium), PHF-1 (1:600; a gift from P. Davies, Albert Einstein College, Bronx, NY), 5E2 (1:500; a gift from K. Kosik, Harvard Medical School, Boston, MA), Tau-1 (1:500; a gift from L. Binder, Northwestern University, Chicago, IL), 12E8 (1:5000; a gift from P. Seubert, Athena Neurosciences, South San Francisco, CA) and SMI-34 (1:300; Sternberger Monoclonals, Baltimore, MD). The immunostaining was visualized using the ECL chemiluminescence system (Amersham, Braunschweig, Germany).
Immunofluorescence
Tau constructs were visualized with the rabbit polyclonal anti-tau antibody K9JA (Dako, Hamburg, Germany), and microtubules were visualized with monoclonal anti-α-tubulin antibody DM1A (Sigma, Deisenhofen, Germany). The cells were fixed for 5 min in −20°C MeOH, washed with PBS, and extracted for 5 min with 0.1% Triton X-100. Fixed cells were blocked by incubation in 10% FCS in PBS, incubated in the primary antibody, washed three times with PBS, incubated with anti-rabbit TRITC– or anti-mouse FITC–conjugated antibody (Dianova, Hamburg, Germany), washed again, mounted in Permafluor (Immunotech, Marseilles, France), and visualized by fluorescence microscopy.
Quantitation of Process Morphology Induced by tau23 and Its Derivatives
The frequency of process formation was determined in the monolayer culture. Cells (3 × 106) were grown on a 60-mm Petri dish, infected with recombinant baculoviruses at an MOI of 1–5, and incubated at 27°C. The morphological analysis was performed on unfixed cells because the cells have the tendency to detach, and processes are lost during the fixation procedure. Process morphology was quantitated in three independent experiments, scoring 200 cells each. The number of cells bearing the processes was plotted against time and analyzed by linear regression. At the end of the experiment the cells were harvested, resuspended in equal volumes of HLB, and incubated 30 min on ice. After that the concentration of NaCl in HLB was adjusted to 500 mM, and tau protein was isolated, making use of its heat stability. After boiling for 15 min and centrifugation at 16,000 × g for 30 min at 4°C, the supernatants were analyzed by gel electrophoresis and Western blotting. The protein concentrations were determined by the bicinchoninic acid method (Sigma). For precise quantitation of tau in the boiled supernatants, equal volumes were electrophoresed on 10% SDS-polyacrylamide gels and quantitated by scanning and densitometry, and the small fraction of other heat-stable proteins in the supernatant was subtracted. Evaluation was done using the program TINA 2.0 (Raytest, Straubenhardt, Germany). For some tau constructs the cell morphology was also determined with Sf9 cells in suspension. In this case the cell density and total cell number can be determined using a hemocytometer. Cell processes grow in suspension but are more prone to breakage so that cells with stable processes become overrepresented. Although the quantitation of cell morphology from suspensions is less reliable than from the monolayer cultures, it is useful for assessing the relative mechanical strengths provided by the different tau constructs.
In Vivo Labeling of tau Derivatives in Sf9 Cells and Phosphopeptide Mapping
Metabolic labeling of Sf9 cells with 32P was performed using 0.5 mCi 32Pi/ml TNM FH cell medium lacking phosphate and supplemented with dialyzed fetal calf serum. Sf9 cells were infected with recombinant baculovirus at an MOI of 10. At 36 h after infection cells were supplemented with 0.5 mCi of 32Pi/ml medium and incubated for 3 h. After labeling cells were washed, resuspended in lysis-boiling buffer, and immediately boiled. After centrifugation the supernatant was subjected to SDS-PAGE, and radioactive tau bands were eluted and precipitated by trichloroacetic acid. The radioactive tau derivatives were further used for peptide mapping. Two-dimensional (2D) phosphopeptide mapping (on thin-layer cellulose plates) was performed according to the methods of Boyle et al. (1991) and Illenberger et al. (1998). For quantitation of radioactivity the image plates were scanned with a BAS2000 phosphoimager (Raytest) and processed using the TINA 2.0 software (Raytest). The phosphorylation of tau construct K19 by MARK2 and PKA was carried out at 37°C in 40 mM HEPES, pH 7.2, 3 mM MgCl2, 5 mM EGTA, 1 mM PMSF, 2 mM [γ-32P]ATP (100–200 Ci/mol; Amersham) and a mixture of protease inhibitors (leupeptin, aprotinin, and pepstatin A at 10 μg/μl each and 1 mM PMSF) in the presence of heparin for 16 h. Protein kinases were removed by boiling the samples in 0.5 M NaCl and 5 mM DTT and centrifugation. Tau construct K19 remains in the supernatant and was precipitated by 15% trichloroacetic acid on ice. The precipitated K19 samples were digested with trypsin and processed by 2D phosphopeptide mapping. PKA was obtained from Promega (Madison, WI). Recombinant MARK2 (Drewes et al.1997) was prepared by G. Schmitt-Ulms (Max-Planck-Unit for Structural Molecular Biology). Note that the absolute degree of tau phosphorylation in Sf9 cells cannot be obtained from the radioactive labeling experiments, because the efficiency of conversion of 32P to [γ-32P]ATP in the cells is not known; however, it can be estimated from the Mr shifts and the known specificities of the phosphorylation-sensitive antibodies.
RESULTS
Process Formation Induced by tau Protein in Sf9 cells Is Enhanced or Suppressed by Different Phosphorylation Sites
Our aim in this study was to use the baculovirus-transfected Sf9 cell system to probe the role of tau phosphorylation in establishing cell polarity. Tau is a neuronal MAP that is involved in supporting the outgrowth of axons and in stabilizing them (Drubin and Kirschner, 1986; Lee et al., 1988; Barlow et al., 1994; Esmaeli-Azad et al., 1994). Cell processes are also induced when tau is transfected into nonneuronal cells, e.g., COS cells (Kanai et al., 1989) or Sf9 insect cells (Baas et al., 1991, 1994; Knops et al., 1991). The efficiency of the baculovirus transfection system makes it an attractive model for studying cellular reactions. The use of this system for studying MAPs and their variants is well established (for review, see Kosik and McConlogue, 1994). However, to extend these studies into the area of phosphorylation, we had to ascertain first that Sf9 cells contain endogenous kinases capable of phosphorylating tau after transfection.
We chose several constructs of tau as probes that had been characterized extensively in vitro, using several criteria such as affinity for microtubules and capacity to nucleate or stabilize microtubules, to alter their dynamic instability, or to induce microtubule bundling (Butner and Kirschner, 1991; Gustke et al., 1994; Panda et al., 1995; Trinczek et al., 1995; Goode et al., 1997). Tau has many phosphorylation sites and can be phosphorylated by various kinases, but there are two classes of phosphorylation sites that are particularly interesting. One class comprises the SP and TP motifs, which are the targets of several proline-directed kinases. This type of phosphorylation is developmentally regulated; i.e., it is enhanced in fetal tissue (Bramblett et al., 1993), and it is prominent in the pathological conditions of Alzheimer’s disease (for review, see Johnson and Jenkins, 1996; Mandelkow and Mandelkow, 1998). We therefore made constructs in which some or all SP or TP sites were mutated into AP and thus were no longer phosphorylatable. The smallest (fetal) human tau isoform contains 14 such sites, all of which were turned into AP in construct tau23/AP (Figure 1A). Some of these SP and TP motifs can be monitored conveniently, because there are a number of mAbs that recognize them in a phosphorylation-dependent manner (several of these antibodies were originally generated against Alzheimer tau, e.g., AT-8 [Mercken et al., 1992] and PHF-1 [Greenberg et al., 1992; for review, see Friedhoff and Mandelkow, 1999]). Another class of phosphorylation sites comprises the KXGS motifs in the repeat domain (X = I or C). The phosphorylation of the first of these (at Ser262) has a pronounced effect on the binding of tau to microtubules (Biernat et al., 1993) and is elevated in Alzheimer tau (Morishima-Kawashima et al., 1995; Seubert et al., 1995). The KXGS motifs are targets of the microtubule affinity–regulating kinase MARK as well as PKA (Drewes et al., 1995, 1997; Zheng-Fischhöfer et al., 1998). We therefore made constructs in which the KXGS motif in repeat 1 of tau23 was turned into KXGA (construct KXGA/R1), in repeats 1 and 4 (KXGA/R1/4), or in all three repeats, R1, R3, and R4, of tau23 (KXGA/R1/3/4; Figure 1A; note that the nomenclature of repeats and the sequence numbering is derived from the longest isoform, tau40).
Figure 1.
Diagrams of tau isoforms or constructs and antibody epitopes. (A) Constructs: 1) htau40, the largest isoform of tau in the human CNS, containing four 31-residue repeats in the C-terminal half (numbered boxes) and two inserts near the N-terminus (411 residues). The regions flanking the repeats are labeled P1-P2 and R′ (hatched). 2) htau23, the smallest of the six isoforms generated by alternative splicing (352 residues). It lacks the N-terminal inserts and the second repeat. 3) Construct KXGA/R1, in which Ser262 in the KXGS motif of the first repeat is replaced by Ala. 4) Construct KXGA/R1/4, in which the serines in the KXGS motifs of repeats 1 and 4 are replaced by Ala (S262A and S356A). 5) Construct KXGA/R1/3/4, in which the serines in the KXGS motifs of repeats 1, 3, and 4 are replaced by Ala (S262A, S324A, and S356A). 6) Construct tau23/AP, in which all SP and TP motifs are replaced by AP. Construct tau23/AP/R1/4 is similar, but in addition Ser262 and Ser356 are changed into Ala. 7) Construct K19 represents the repeats only. (B) Antibody epitopes. Most phosphorylation-dependent antibodies react with SP and TP motifs in the flanking domains before or after the repeats.
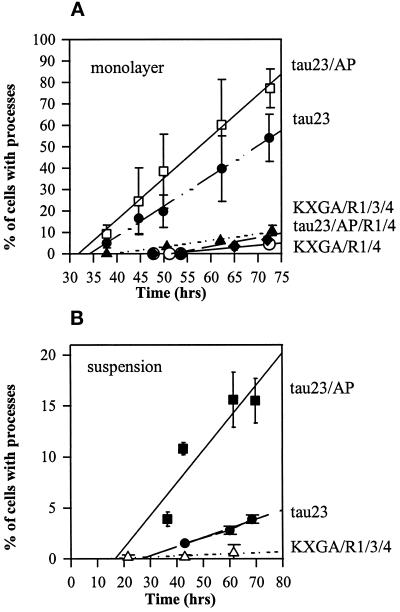
Cells were infected with tau-expressing baculovirus and observed in a monolayer under the microscope. The cell bodies have a round shape with a diameter of ∼16–22 μm (determined in a hemocytometer; corresponding volume, ∼2–4 pl), which remains roughly constant throughout their life time. Processes begin to appear after an incubation time of >30 h. Typically there is a single process per cell, of uniform diameter (1–2 μm) and up to 100 μm long (Figure 2). The frequency of processes increases linearly with time (Figure 3A), parallel to the increase in tau protein concentration. However, the efficiency of process induction varies considerably. The case of tau23 serves as a standard; process formation starts at ∼34 h and increases at a pace of 1.4%/h, reaching a level of ∼60% after 75 h. With construct tau23/AP, in which the SP and TP motifs cannot be phosphorylated, the efficiency is much higher (t0 = 32 h; slope, 1.9%/h; final level, ∼80%). Conversely, when two or three KXGS motifs in the repeats of tau23 are nonphosphorylatable, as in KXGA/R1/4 or KXGA/R1/3/4, the efficiency drops steeply (late onset, t0 = 38 h; and fivefold slower rise, 0.3%/h). These different behaviors are not related to protein expression, which reaches similar levels in all cases (∼45 μg/106 cells; Table 1). Thus the differences are due to the nature of the transfected proteins.
Figure 2.
Immunofluorescence of Sf9 cells transfected with different tau constructs. Cells were transfected with htau23 (A), KXGA/R1 (B), or KXGA/R1/3/4 (C) and immunostained 60 h after infection with antibodies DM1A (for tubulin, left) and K9JA (for tau, right). Transfection with htau23 (A) leads to many processes; when Ser262 is mutated to Ala (B), one still finds processes similar to tau23, but when all three KXGS motifs are changed into KXGA (C), the formation of processes is almost completely inhibited. Note that in the case of tau staining the cell bodies appear overexposed to image the more weakly stained processes. Bar, 50 μm.
Figure 3.
Time course of process formation. (A) Quantitation from monolayer culture. Processes begin to appear at ∼30–40 h after infection; their frequency rises linearly thereafter. For tau23 (filled circles), the extrapolated onset is at t0 = 34 h; the increase of process-bearing cells is 1.4% cells/h. Construct tau23/AP (open squares) is more efficient; processes appear earlier and increase faster (t0 = 32 h; slope, 1.9%/h), showing that when the SP and TP sites remain unphosphorylated, the formation of processes is enhanced. Construct KXGA/R1/3/4 (filled triangles) is much less efficient (t0 = 38 h; slope, 0.3%/h), showing that the phosphorylation of KXGS motifs enhances process formation. Construct KXGA/R1 behaves similar to tau23, whereas KXGA/R1/4 (open circles) behaves similar to KXGA/R1/3/4, indicating that two or more KXGS motifs must cooperate to change the response of the cells. Similarly, process formation is also inhibited with construct tau23/AP/R1/4 (diamonds), showing that the dominant effect of phosphorylation is in the repeats, not in the flanking regions. (B) Quantitation of process formation from suspension culture. This overemphasizes cells with stable processes, because labile ones are lost more easily by shear forces. Thus processes are less numerous, but qualitatively the trends are the same as in A; i.e., construct tau23/AP is most efficient, whereas KXGA/R1/3/4 induces almost no processes. Note the fourfold difference between tau23 and tau23/AP, indicating the greater stability of the processes, because the SP and TP motifs cannot be phosphorylated. Bars, standard deviations.
Table 1.
Tau protein concentrations in Sf9 cells as a function of time after infection
| Protein | Concentration (μg/106 cells)
|
||
|---|---|---|---|
| 48 h | 66 h | 73.5 h | |
| tau23 | 14.3 | 41.0 | 45.6 |
| KXGA/R1 | 15.7 | 41.3 | 46.4 |
| KXGA/R1/3/4 | 14.0 | 40.3 | 47.5 |
| tau23/AP | 10.5 | 36.0 | 45.6 |
Note that the concentrations are comparable for all constructs so that the differences in cell extensions are accounted for by mutations rather than the concentration.
The strong suppression of processes described above was initially observed when the three KXGS motifs were mutated to KXGA simultaneously. Because a single site (Ser262 in the first repeat) had a predominant effect on tau’s affinity for microtubules (Biernat et al., 1993), we asked whether this is also the case in the assays used here. We find, however, that the single site mutation to Ala262 (construct KXGA/R1) has no effect, whereas two such mutations in repeats 1 and 4 (Ala262 and Ala356, construct KXGA/R1/4) have the same inhibitory effect as mutations in all three motifs (Ala262, Ala324, and Ala356, construct KXGA/R1/3/4; Figure 3A). We conclude that the KXGA mutations in repeats 1 and 4 are necessary and sufficient for the full inhibitory effect on process formation. These sites coincide with the strongest phosphorylation sites for the kinase MARK (Ser262 and Ser356; Drewes et al., 1995); they can be phosphorylated by PKA as well and occur in Sf9 cells (see Figure 6, A and D).
Figure 6.
Two-dimensional phosphopeptide maps of tau constructs expressed in Sf9 cells and phosphorylated by endogenous kinases. (A) htau23 (higher magnification); (B) htau23/AP; (C) KXGA/R1/3/4; (D) construct K19 phosphorylated with PKA; (E) mixture of tau23 and K19 phosphorylated with PKA. In A the majority of spots are generated by phosphopeptides containing one or more of the 14 SP and TP motifs. The four phosphorylation sites in the repeats are highlighted and underlined (three KXGS motifs with S262, S324, and S356, plus S320). In B all SP or TP motifs are changed into AP so that the remaining major phosphorylation sites are Ser214 and those in the repeats, plus some unknown spots. In C the KXGS motifs in the repeats are changed into KXGA and are therefore absent from the map; besides the phosphorylated SP and SP motifs one observes S214 and S320. In D the repeat construct K19 was phosphorylated with PKA, showing only the sites S262, S324, S356, and S320. This sample was run together with tau23 in E to identify the sites. For details on analysis of phosphopeptides, see Illenberger et al. (1998).
Because the mutations at SP or TP sites have the opposite effect to the KXGA mutants, we also asked which of these mutations dominate when they are mixed. In one mutant, all SP or TP sites were mutated into AP, plus the KXGA sites in repeats 1 and 4 (construct tau23/AP/R1/4). Process formation was as strongly suppressed as in construct KXGA/R1/4 (Figure 3A). This shows that the KXGA mutations in the repeats dominate over the AP mutations in the flanking regions.
When the cell processes are determined from suspensions, one obtains a qualitatively similar picture, except that the differences between the tau constructs become much more pronounced (Figure 3B). For example, at 70 h after infection, ∼3% of the cells transfected with tau23 develop processes. For cells with the mutant tau23/AP the frequency is fivefold higher (∼15%), whereas cells with the mutant KXGA/R1/3/4 have almost none. Because the processes in suspension are subject to shear forces, which tend to break them, these data argue that constructs that generate processes more efficiently (in terms of early onset and rapid increase) also make them mechanically more stable.
Phosphorylation of tau in Sf9 Cells
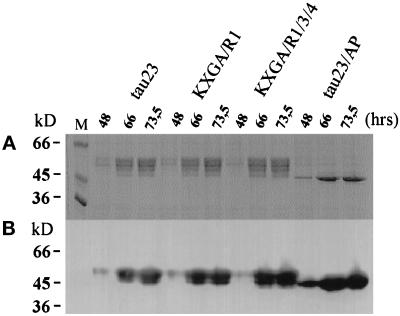
The interpretation that process formation is influenced by tau phosphorylation depends on which sites are actually targeted by the endogenous kinases of Sf9 cells. This issue was determined by several methods: gel shift, antibody reactions, and phosphopeptide analysis (Figures 4–6). An approximate survey can be obtained from the upward shift of tau in the SDS gel, which can reach an apparent increase of ≥5 kDa, depending on the phosphorylation site (Biernat et al., 1993). This shift is also characteristic of Alzheimer tau (A68 protein, Lee et al., 1991). Figure 4 shows that tau from transfected Sf9 cells also displays a pronounced shift (distributed over several bands), indicating that tau is phosphorylated by endogenous kinases in Sf9 cells. The shift is visible with wild-type tau23, with the “repeat” mutants KXGA/R1 and KXGA/R1/3/4 but not with the “flank” mutant tau23/AP (which runs as a homogeneous band, essentially as unphosphorylated, bacterially expressed tau). This confirms our previous observations that SP and TP sites are mainly involved in the shift and shows that endogenous, proline-directed kinases are active in Sf9 cells.
Figure 4.
Phosphorylation of tau and tau constructs transfected in Sf9 cells. (A) 10% SDS-PAGE. Tau protein or mutants (tau23, KXGA/R1, KXGA/R1/3/4, and tau23/AP) were isolated from equal quantities of Sf9 cells transfected with the appropriate tau constructs and prepared in equal volumes of sample buffer. Equal volumes of protein solution representing the different harvest time points were loaded onto the SDS gel (10%). The amounts of loaded proteins correspond approximately to 0.4 μg (48 h), 2.5 μg (66 h), and 3 μg (73.5 h). As seen from the multiple and strongly shifted bands, tau23 is highly phosphorylated in a heterogeneous manner. The same applies to contructs KXGA/R1 and KXGA/R1/3/4 (which lack only one or three phosphorylation sites at KXGS motifs but retain all SP or TP sites). By contrast, construct tau23/AP shows very little phosphorylation and no shift, because it lacks most phosphorylation sites (note that the KXGS sites do not induce a shift; Drewes et al., 1995). By implication, the strong shift sites S409 and S416 (targets of PKA or CaMKII) are also not phosphorylated in Sf9 cells. (B) The same samples were immunoblotted with antibody 5E2, which reacts independently of phosphorylation and reflects the total amount of tau protein. Lane M, marker proteins.
A number of phosphorylation-sensitive antibodies against tau are available, which can be used as diagnostic tools (Figure 1B). As a reference, the cell extracts of transfected Sf9 cells were immunoblotted with the phosphorylation-independent antibody 5E2 (Kosik et al., 1988), which indicates the total amount of tau. As seen in the Western blots (Figure 5), Tau-1 recognizes weakly all tau23 derivatives expressed in Sf9 cells, indicating that a small fraction is not phosphorylated around residue 200. The complementary antibody AT-8 reveals a strong signal by htau23, KXGA/R1, and KXGA/R1/3/4. These repeat mutants are also clearly recognized by other antibodies sensitive to proline-directed phosphorylation (AT-270, AT-180, SMI-34, and PHF-1), indicating phosphorylation at Thr181, Thr231, Ser235, Ser396, and Ser404 (Figure 1B). A particularly interesting example is that of the antibody AT-100, which is uniquely specific for Alzheimer tau (Matsuo et al., 1994). The epitope is formed by sequential phosphorylation first of Thr212 by GSK-3 and then of Ser214 by PKA and requires a PHF-like conformation induced by polyanions (Zheng-Fischhöfer et al., 1998). This epitope is present on tau in the transfected Sf9 cells (Figure 5). Similarly, Ser262/Ser356 is clearly phosphorylated, as seen from the reaction with antibody 12E8. The phosphorylation at the epitopes of AT-100 and 12E8 is particularly sensitive to phosphatases because they disappear rapidly during the initial steps of preparation.
Figure 5.
Phosphorylation sites of baculovirus-expressed tau and tau constructs. Sf9 cell lysates (72 h after infection) were blotted and incubated with different phosphorylation-dependent and -independent mAbs. The tau constructs are, from left to right: 1) tau23/AP, 2) KXGA/R1, 3) htau23, 4) KXGA/R1/3/4, 5) untransfected Sf9 cell extract (control without tau), 6) Sf9 cells transfected with wild-type baculovirus (control without tau), 7) htau23 expressed in E. coli (unphosphorylated control), and 8) htau23 expressed in E. coli and phosphorylated with brain extract kinase activity (affecting largely the SP and TP sites and inducing a strong Mr shift; see Biernat et al., 1993). Antibodies, from top to bottom: 5E2 (a pan-tau antibody) recognizes all preparations that contain tau. Tau-1 shows only a moderate reaction with tau constructs expressed in Sf9 cells, because a single P site in the vicinity of residue 200 suffices to reduce the binding, but there is a strong reaction with unphosphorylated tau expressed in E. coli or tau23/AP. The antibodies AT-8 to SMI-34 are specific for different phosphorylated SP and TP motifs and therefore show no reaction with the tau23/AP mutant or with E. coli–expressed htau23. Antibody 12E8 recognizes only those constructs that contain Ser262 and/or Ser356 in the repeats. AT-100 recognizes tau phosphorylated at Thr212 and Ser214; this reaction is highly specific for Alzheimer tau but occurs in Sf9 cells as well, provided both sites are phosphorylatable (e.g., not in the tau23/AP mutant).
Metabolic Labeling of tau and Analysis of Phosphopeptides
The detection of phosphorylation sites by antibodies suffers from two drawbacks: 1) there may be sites for which there are no antibodies, and 2) the antibody staining is not a reliable indicator of the extent of the phosphorylation (because the antibody affinities are variable and often unknown). For further characterization of phosphorylation sites of htau23 and its derivatives, we performed metabolic labeling of Sf9 cells using [32P]orthophosphoric acid. The labeled tau protein was isolated, digested with trypsin, and then processed for 2D peptide analysis (Boyle et al., 1991). To identify the peptides, tau expressed in Escherichia coli was phosphorylated radioactively using different kinases in vitro and digested with trypsin, and the peptides were purified by HPLC and identified by matrix-assisted laser desorption and ionization, phosphopeptide sequencing, and phosphopeptide mapping (for details, see Drewes et al., 1995; Illenberger et al., 1998; Zheng-Fischhöfer et al., 1998). Figure 6A shows the phosphopeptide map found with tau23 phosphorylated in Sf9 cells, in which the main spots are identified by their phosphorylation site (for details on the identification, see Illenberger et al., 1998). The experiments with the mutants enabled us to define the phosphorylation sites by exclusion of the corresponding spots from the 2D map (Figure 6, B and C). The majority of spots represent SP or TP sites, containing ∼80% of the total radioactivity. These spots disappear in the case of the tau23/AP mutant, in which only the non-SP or TP sites remain, among them S214, S262, S320, S356, and two unidentified spots (Figure 6B). It is remarkable that the distribution and intensity of phosphorylation sites of tau23 is quite similar to that of other cultured cells during interphase, including neuronal ones (Illenberger et al., 1998). This shows that the balance of kinases and phosphatases is similar in these cells, and it provides an additional rationale for using baculovirus-transfected Sf9 cells as a model system.
Figure 6C shows the phosphopeptide map for the mutant KXGA/R/1/3/4, where the three sites S262, S324, and S356 had been replaced by Ala and therefore no longer appear on the map (these spots are normally weak compared with the SP and TP sites and visible only at longer exposures; cf. Figure 6A). We confirmed the phosphorylation at the sites in the repeats by mixing the phosphopeptides from metabolically labeled tau23 in Sf9 cells with phosphopeptides from construct K19 (three repeats only) phosphorylated in vitro with MARK or PKA, which resulted in the overlapping of spots representing the repeat phosphorylation sites (S262, S324, and S356 plus S320; Figure 6, D and E).
DISCUSSION
Sf9 Cells Represent a model for Process Outgrowth Induced by tau and Phosphorylate tau at Similar Sites as Neurons
Several previous studies have established the model of baculovirus-transfected insect cells as a suitable system to study the development of cell processes (for review, see Kosik and McConlogue, 1994). The interplay between cytoskeletal fibers (microtubules or microfilaments) and the dependence on different MAPs during process formation are remarkably similar to those of neurite outgrowth from neurons (Mandell and Banker, 1995), even though the neuronal signaling machinery is absent. The major additional advantage of Sf9 insect cells is the high efficiency of baculovirus-mediated transfection, which makes it possible to analyze them biochemically. Thus we have been able to take the earlier studies one step further and ask how the process formation induced by tau depends on the phosphorylation by endogenous kinases. We have transfected Sf9 cells with different tau isoforms or mutants, quantitated their development of processes, and correlated this with the phosphorylation of tau as determined by gel shift, antibody reactions, and phosphopeptide analysis after metabolic labeling.
The choice of mutations was made on the basis of earlier results on the role of phosphorylation sites. We broadly distinguish two types of phosphorylation sites, in the repeat domain (Figure 1, numbered boxes) and in the domains flanking the repeats (Figure 1, hatched boxes). The phosphorylation of the repeats, especially at KXGS motifs, strongly reduces tau’s affinity for microtubules so that tau detaches and microtubules become unstable in vitro (Biernat et al., 1993); the same holds for other related MAPs (MAP2 and MAP4; Illenberger et al., 1996). The repeats are phosphorylated most efficiently by the kinase MARK (Drewes et al., 1995, 1997) but can be phosphorylated by other kinases as well, especially when activated with heparin (notably PKA; Scott et al., 1993; Drewes et al., 1995; Zheng-Fischhöfer et al., 1998). Particularly, Ser262 in the first KXGS motif shows higher phosphorylation in Alzheimer’s disease tau (Morishima-Kawashima et al., 1995), suggesting that the detachment of tau from microtubules after phosphorylation is an important step in the formation of Alzheimer PHFs. The phosphorylation at SP and TP motifs in the flanking domains is a target of several proline-directed kinases (e.g., MAP kinase, GSK-3, and cdk5). Phosphorylation at these sites also tends to diminish the interaction with microtubules, but the effect is much less pronounced than for the repeats (Drechsel et al., 1992; Trinczek et al., 1995). This phosphorylation is strongly enhanced in Alzheimer tau, and a number of diagnostic antibodies react with SP and TP motifs in a phosphorylation-dependent manner (for review, see Friedhoff and Mandelkow, 1999). In the “jaws” model of tau–microtubule interactions, the flanking domains act as “targeting domains” for positioning tau on the microtubule surface, whereas the repeats act as a “catalytic domain” for enhancing microtubule assembly (Gustke et al., 1994). One would therefore expect that the domains would respond differently to phosphorylation in a cellular context, and this is indeed borne out by the experiments presented here.
The first question to be answered was whether the insect cells contained a similar pool of kinases and phosphatases as neurons or other cells. As shown previously, the phosphorylation pattern of tau in neuronal or other cell types is remarkably similar, as judged from metabolic labeling and phosphopeptide analysis (Illenberger et al., 1998). The same is the case for the Sf9 cells (Figure 6), showing that we can indeed take these cells as a model for the “neuron-like” phosphorylation of tau. The majority of the phosphorylation sites (∼80%) are distributed over the SP and TP motifs, mostly in the flanking regions, whereas a minor fraction was in the repeats, mostly in KXGS motifs and Ser320. The occurrence of multiple phosphorylation sites, especially at SP and TP motifs, can be visualized by antibody labeling, using the same phosphorylation-dependent antibodies that have been developed for diagnosing Alzheimer PHFs (Figure 5). As a consequence of this phosphorylation, the tau bands are shifted upward in the SDS gel, in a manner reminiscent of Alzheimer tau. This shift depends mostly on proline-directed phosphorylation and is abolished when SP and TP motifs are mutated into AP (Figures 4 and 5). Thus, all Alzheimer-diagnostic antibodies against SP or TP motifs, as well against Ser262 (e.g., 12E8), recognize tau in Sf9 cells. The overall degree of phosphorylation is estimated at ∼3–4 Pi per tau, judging from the five or more shift stages and their antibody reactivities, each of which corresponds to one or two phosphates incorporated (see Figure 5; Lichtenberg-Kraag et al., 1992).
Phosphorylation in the Repeats and in the Flanking Domains Has Opposite Effects on Process Outgrowth
It is generally assumed that one of the major physiological functions of tau phosphorylation is to regulate its affinity with microtubules and, hence, microtubule stability (Butler and Shelanski, 1986; Drubin and Kirschner, 1986). Indeed, the phosphorylation states of tau studied so far all show a reduced affinity for microtubules, although the magnitude of the effect varies (strong inhibition for phosphorylation in the repeats, weak for the flanking domains). We would therefore have expected that in Sf9 cells the effects of repeat and flank phosphorylation would go in the same direction, possibly differing in magnitude. Surprisingly this was not the case: suppression of repeat phosphorylation (at KXGS motifs) nearly abrogated tau’s ability to induce processes; suppression of flank phosphorylation (at SP and TP sites) enhanced process formation (Figure 3). This evidence clearly shows that the phosphorylation in these two domains has very different physiological roles, although both reduce the tau–microtubule interaction in vitro. It appears that the phosphorylation at the KXGS motifs in the repeats is necessary for some crucial step in process formation (stimulatory), whereas proline-directed phosphorylation of the flanking domains is moderately inhibitory (∼30%).
The opposing effects become even more puzzling when one correlates them with the extent of tau phosphorylation. Despite the strong dependence of process formation on phosphorylation at KXGS motifs in the repeats, these motifs carry only a minor fraction of the total sites (∼5%). In contrast, the inhibition of process formation by phosphorylation at SP or TP sites is only moderate despite the fact that it accounts for ∼80% of the phosphate on tau. These data argue that the phosphorylation at KXGS motifs has some important catalytic role, whereas the regulating effects of phosphorylation at SP and TP sites are more subtle. We note, however, that overall extent of phosphorylation is only ∼3–4 Pi per tau molecule, most of which is on SP and TP motifs. Thus most tau molecules carry three or four sites whose combination is not clear at present, but in general they are distributed over >11 sites. To understand the inhibition of process outgrowth by SP or TP phosphorylation, we would have to assume that the different observed sites have roughly similar effects. Viewed in this light, the strong dependence on a minute fraction of KXGS phosphorylation sites in the repeat becomes even more remarkable; it can be rationalized only if we assume that the effect takes place in a small compartment or in a short period. An obvious candidate would be the immediate vicinity of the point where a process emerges.
Crosstalk between Phosphorylation Sites of tau Influences Process Outgrowth
We can carry this analysis one step further by considering certain phosphorylation sites in detail. To observe the inhibitory effect of mutated KXGS sites, we need two of them acting in concert, the motifs in repeats 1 and 4 (Ser262 and Ser356). One of them alone will not suffice, so that a mutation at Ser262 behaves like wild-type tau23. We had shown previously that phosphorylation at Ser262 alone can detach tau from microtubules in vitro (Biernat et al., 1993). Therefore, it appears that cellular effects depend on a more strictly regulated interaction between different sites. We note in this context that Ser262 and Ser356 are also the preferred targets of the kinase MARK (Drewes et al., 1995). Because kinases related to MARK are involved in the establishment of cellular polarity (e.g., par-1 in Caenorhabditis elegans; Guo and Kemphues 1995, Drewes et al., 1997), one can speculate that a similar MARK-like kinase operates in insect ovary cells during process formation.
The opposite effects of the two types of phosphorylation sites (repeats vs. flanking domains) are not additive. In general, mutations at KXGS motifs override those at SP and TP motifs. This means that when all sites are phosphorylatable in wild-type tau, the phosphorylation at SP and TP motifs (which is general and moderately inhibitory) does not matter vis-a-vis the specific phosphorylation at KXGS motifs, which strongly promotes processes.
One of the most interesting phosphorylation sites is Ser214. This is a non-SP site in the flanking domain that is embedded in a neighborhood of SP and TP motifs, which are epitopes of antibodies diagnostic of Alzheimer PHFs. In vitro, Ser214 can be phosphorylated by PKA. Next to the Ser262 site in the repeats, Ser214 is the second-most efficient site that can detach tau from microtubules (Brandt et al., 1994; Illenberger et al., 1998). This site shows strong phosphorylation in the phosphopeptide maps (Table 2). Phosphorylation at Ser214 is selectively enhanced in Chinese hamster ovary cells transfected with tau when they enter mitosis, resulting in a detachment of tau from microtubules (Illenberger et al., 1998). Most significantly, tau in Sf9 cells shows a reaction with antibody AT-100, one of the most specific antibodies against Alzheimer PHFs known to date. This is in contrast to Chinese hamster ovary or N2a cell models, in which reaction with other Alzheimer-diagnostic antibodies can be induced (Preuss et al., 1995). AT-100 requires the sequential phosphorylation of Thr212 by GSK-3 and of Ser214 by PKA (Zheng-Fischhöfer et al., 1998). However, testing the function of Ser214 with the single-site mutation tau23/S214A did not show a significant change in process formation. It is possible that phosphorylation at this site cooperates with the KXGS to influence process formation.
Table 2.
Relative intensities of phosphopeptides
| Phosphorylation site | Fraction (%) |
|---|---|
| Flanking regions: SP and TP sites | |
| Thr181 | 3.4 |
| Ser235 (spot 1) | 27.5 |
| Ser235 (spot 2) | 5.0 |
| Thr231/S235 | 3.8 |
| Ser199 | 1.4 |
| Ser202 | 5.5 |
| Ser202/Thr205 | 4.9 |
| Thr212 | 3.8 |
| Thr212/Thr217 | 4.9 |
| Ser404 | 9.5 |
| Ser396/Ser404 | 6.4 |
| Flanking regions: non-SP or TP site | |
| Ser214 | 7.9 |
| Repeats: KXGS motifs | |
| Ser262 | 3.8 |
| Ser356 | 0.15 |
| Ser324 | 0.26 |
| Repeats: non-KXGS motifs | |
| Ser320 | 2.4 |
| Unidentified spots | 9.4 |
The quantitation was done using a phosphoimage plate reader. The phosphorylation in the repeats accounts for ∼6% of the intensity; the SP and TP motifs in the flanking regions account for ∼80%. Note that several phosphopeptides are doubly phosphorylated. S235 is split into two spots because of an unknown chemical modification.
A Model Relating tau Phosphorylation to Process Formation
For a cell process to become visible it must be born and maintained. In both cases, microtubules and tau are involved. Once a process is born the cell tends to block others, so that most cells contain a single process. This is reminiscent of axonogenesis (Mandell and Banker, 1995), and it is characteristic of tau, in contrast to other MAPs such as MAP2, which tends to support several processes of somewhat different morphology (Leclerc et al., 1993, 1996; Leger et al., 1994). The birth of a process is preceded by subcellular changes at the appropriate location and at the right time (e.g., accumulation of material and softening of actin cortex; Knowles et al., 1994; Bradke and Dotti, 1997). The distinction among birth, life, and eventually death of a process is conceptually convenient, because it would explain why some phosphorylation sites that have apparently substoichiometric occupancies (when averaged over the whole cell population) could still play decisive roles at particular time points. Moreover, previous studies have shown that in neurons the generation or degeneration of neurites is accompanied by changes in tau phosphorylation (Sadot et al., 1995; Mandell and Banker, 1996).
In this framework the phosphorylation at KXGS sites in the repeat domain of tau would play a role during the prenucleation or budding events of a process. This would be consistent with their strong effect on promoting process outgrowth and on their substoichiometric average extent of phosphorylation. Thus the concerted action of phosphorylation events at two or more KXGS motifs (e.g., Ser262 and Ser356) would occur only in a small fraction of tau molecules (presumably at the budding extension) and during a short period. This type of phosphorylation weakens the tau–microtubule interaction and makes microtubules more labile; thus one could conclude that dynamic microtubules (rather than stable ones) matter for the onset of process extension. This would be roughly comparable to the temporary increase in the dynamic instability of microtubules at the onset of mitosis (Belmont et al., 1990; Hyman and Karsenti, 1996). Because the KXGS sites are phosphorylated by MARK, and because MARK-like kinases are involved in establishing and maintaining cell polarity (Drewes et al., 1998), it is likely that the polarity-defining events at the onset of process budding require a local destabilization of microtubules, as diagrammed in Figure 7A. Conversely, when the KXGS motifs are not phosphorylatable, tau superstabilizes microtubules so that cell processes are not observed either (Figure 7C).
Figure 7.
Diagram illustrating the effects of tau phosphorylation on process formation. The left side shows the interaction between a microtubule and tau in different states of phosphorylation. The right side shows a cell body and a cell process whose length represents the efficiency of process formation. (A) Tau with unphosphorylated KXGS motifs in the repeats binds to microtubules (left), but if it becomes phosphorylated at KXGS motifs (filled circles), it detaches from microtubules, and these become dynamic. This is necessary, at least temporarily, for the nucleation of a cell process (independently of whether the SP or TP sites are phosphorylated; filled or empty diamonds in the flanking domains). (B) Tau phosphorylatable at the KXGS motifs in the repeats but not at the SP and TP motifs (Ala mutations in the flanking domains) allows the formation of long and stable processes once they are initiated. (C) If tau cannot be phosphorylated at KXGS motifs (Ala mutations in repeats) it remains tightly attached to microtubules. This blocks the initiation of cell processes and therefore inhibits their growth, independently of the phosphorylation at SP or TP motifs.
A different scenario is envisaged for the role of the SP and TP motifs in the flanking domains. Here the extent of phosphorylation is higher so that we can assume a more general occurrence, not just at restricted points. Fluctuations in this type of phosphorylation are suggested by observations that dividing cells, fetal tissue, cells stimulated by external signaling, or degenerating cells often show enhanced phosphorylation at SP and TP sites, detectable by diagnostic antibodies (e.g., Bramblett et al., 1993; Braak et al., 1994; Preuss et al., 1995; Burack and Halpain, 1996; Sadot et al., 1996). Although this type of phosphorylation has been considered to regulate tau’s interaction with microtubules, the limited extent of the effect in vitro (Biernat et al., 1993) and observed here (Figure 3) begs the question of other possible roles. Besides microtubule dynamics, MAPs are known to regulate the spacing between microtubules or their stiffness (Chen et al., 1992; Dye et al., 1993; Frappier et al., 1994; Matus, 1994; Felgner et al., 1997) and interactions with enzymes or cytoskeletal elements (Obar et al., 1989; Sontag et al., 1995). All of these are important in process formation and could conceivably be affected by phosphorylation at SP and TP sites, resulting in the observed inhibition of process outgrowth. Conversely, when the SP and TP motifs are nonphosphorylatable, the inhibition is relieved, resulting in longer and more numerous cell processes (Figure 7B).
Finally, an emerging role of tau and other MAPs, distinct from microtubule stabilization, is the regulation of vesicle and organelle transport. There is increasing evidence that MAPs retard the movement of vesicles (Hamm-Alvarez et al., 1993; Bulinski et al., 1997), and tau inhibits particularly the kinesin-dependent and plus end–directed transport, which would be required for moving material down a growing cell process (Ebneth et al., 1998). Vesicle transport is in turn facilitated by enhanced phosphorylation (Lopez and Sheetz, 1995; Sato-Harada et al., 1996). This would be consistent with a view that MAPs obstruct the path of vesicles but can be moved out of the way by phosphorylation. In the case of tau, this could be achieved by the local phosphorylation at KXGS motifs by kinases such as MARK.
ACKNOWLEDGMENTS
We thank Heike Niebuhr and Katja Alm for excellent technical assistance, P. Friedhoff for help with statistical evaluation, R. Godemann for help with some phosphopeptide maps, and E. Mandelkow for stimulating discussions. Antibodies were generously provided by P. Davies (PHF-1), E. van Mechelen and A. van de Voorde (AT series), K. Kosik (5E2), P. Seubert (12E8), and L. Binder (Tau-1). This research was supported by the Deutsche Forschungsgemeinschaft.
Abbreviations used:
- GSK-3
glycogen-synthase-kinase-3
- HLB
hypotonic lysis buffer
- MAP
microtubule-associated protein
- MARK
MAP/microtubule affinity–regulating kinase
- MOI
multiplicity of infection
- PHF
paired helical filament
- PKA
cAMP-dependent protein kinase
- 2D
two-dimensional
REFERENCES
- Baas PW, Pienkowski TP, Kosik KS. Processes induced by tau expression in Sf9-cells have an axon-like microtubule organization. J Cell Biol. 1991;115:1333–1344. doi: 10.1083/jcb.115.5.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas P, Pienkowski T, Cimbalnik K, Toyama K, Bakalis S, Ahmad F, Kosik K. Tau confers drug stability but not cold stability to microtubules in living cells. J Cell Sci. 1994;107:135–143. doi: 10.1242/jcs.107.1.135. [DOI] [PubMed] [Google Scholar]
- Barlow S, Gonzalez-Garay ML, West R, Olmsted JB, Cabral F. Stable expression of heterologous microtubule-associated proteins (MAPs) in Chinese hamster ovary cells: evidence for differing roles of MAPs in microtubule organization. J Cell Biol. 1994;126:1017–1029. doi: 10.1083/jcb.126.4.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont LD, Hyman AA, Sawin KE, Mitchison TJ. Real-time visualization of cell-cycle dependent changes in microtubule dynamics in cytoplasmic extracts. Cell. 1990;62:579–589. doi: 10.1016/0092-8674(90)90022-7. [DOI] [PubMed] [Google Scholar]
- Biernat J, Gustke N, Drewes G, Mandelkow E-M, Mandelkow E. Phosphorylation of serine 262 strongly reduces the binding of tau protein to microtubules: distinction between PHF-like immunoreactivity and microtubule binding. Neuron. 1993;11:153–163. doi: 10.1016/0896-6273(93)90279-z. [DOI] [PubMed] [Google Scholar]
- Binder LI, Frankfurter A, Rebhun L. The distribution of tau in the mammalian CNS. J Cell Biol. 1985;101:1371–1378. doi: 10.1083/jcb.101.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle WJ, van der Geer P, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin layer cellulose plates. Methods Enzymol. 1991;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- Braak E, Braak H, Mandelkow E-M. A sequence of cytoskeleton changes related to the formation of neurofibrillary tangles and neuropil threads. Acta Neuropathol. 1994;87:554–567. doi: 10.1007/BF00293315. [DOI] [PubMed] [Google Scholar]
- Bradke F, Dotti CG. Neuronal polarity: vectorial cytoplasmic flow precedes axon formation. Neuron. 1997;19:1175–1186. doi: 10.1016/s0896-6273(00)80410-9. [DOI] [PubMed] [Google Scholar]
- Bramblett GT, Goedert M, Jakes R, Merrick SE, Trojanowski JQ, Lee VMY. Abnormal tau phosphorylation at Ser(396) in Alzheimer’s disease recapitulates development and contributes to reduced microtubule binding. Neuron. 1993;10:1089–1099. doi: 10.1016/0896-6273(93)90057-x. [DOI] [PubMed] [Google Scholar]
- Brandt R, Lee G, Teplow D, Shalloway D, Abdelghany M. Differential effect of phosphorylation and substrate modulation on tau’s ability to promote microtubule growth and nucleation. J Biol Chem. 1994;269:11776–11782. [PubMed] [Google Scholar]
- Bulinski JC, McGraw TE, Gruber D, Nguyen H-L, Sheetz MP. Overexpression of MAP4 inhibits organelle motility and trafficking in vivo. J Cell Sci. 1997;110:3055–3064. doi: 10.1242/jcs.110.24.3055. [DOI] [PubMed] [Google Scholar]
- Burack MA, Halpain S. Site-specific regulation of Alzheimer-like tau-phosphorylation in living neurons. Neuroscience. 1996;72:167–184. doi: 10.1016/0306-4522(95)00546-3. [DOI] [PubMed] [Google Scholar]
- Butler M, Shelanski ML. Microheterogeneity of microtubule-associated tau-proteins is due to differences in phosphorylation. J Neurochem. 1986;47:1517–1522. doi: 10.1111/j.1471-4159.1986.tb00788.x. [DOI] [PubMed] [Google Scholar]
- Butner KA, Kirschner MW. Tau-protein binds to microtubules through a flexible array of distributed weak sites. J Cell Biol. 1991;115:717–730. doi: 10.1083/jcb.115.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres A, Kosik KS. Inhibition of neurite polarity by tau antisense oligonucleotide in primary cerebellar neurons. Nature. 1990;343:461–463. doi: 10.1038/343461a0. [DOI] [PubMed] [Google Scholar]
- Caceres A, Mautino J, Kosik KS. Suppression of MAP2 in cultured cerebellar macroneurons inhibits minor neurite formation. Neuron. 1992;9:607–618. doi: 10.1016/0896-6273(92)90025-9. [DOI] [PubMed] [Google Scholar]
- Caceres A, Potrebic S, Kosik KS. The effect of tau-antisense oligonucleotides on neurite formation of cultured cerebellar macroneurons. J Neurosci. 1991;11:1515–1523. doi: 10.1523/JNEUROSCI.11-06-01515.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Kanai Y, Cowan N, Hirokawa N. Projection domains of MAP2 and tau determine spacings between microtubules in dendrites and axons. Nature. 1992;360:674–677. doi: 10.1038/360674a0. [DOI] [PubMed] [Google Scholar]
- Cleveland DW, Hwo S-Y, Kirschner MW. Physical and chemical properties of purified tau factor and the role of tau in microtubule assembly. J Mol Biol. 1977;116:227–247. doi: 10.1016/0022-2836(77)90214-5. [DOI] [PubMed] [Google Scholar]
- Drechsel DN, Hyman AA, Cobb MH, Kirschner MW. Modulation of the dynamic instability of tubulin assembly by the microtubule-associated protein tau. Mol Biol Cell. 1992;3:1141–1154. doi: 10.1091/mbc.3.10.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewes G, Ebneth A, Mandelkow E-M. MAPs, MARKs, and microtubule dynamics. Trends Biochem Sci. 1998;23:307–311. doi: 10.1016/s0968-0004(98)01245-6. [DOI] [PubMed] [Google Scholar]
- Drewes G, Ebneth A, Preuss U, Mandelkow E-M, Mandelkow E. MARK—a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell. 1997;89:297–308. doi: 10.1016/s0092-8674(00)80208-1. [DOI] [PubMed] [Google Scholar]
- Drewes G, Trinczek B, Illenberger S, Biernat J, Schmitt-Ulms G, Meyer HE, Mandelkow E-M, Mandelkow E. MAP/microtubule affinity regulating kinase (p110-MARK): a novel protein kinase that regulates tau-microtubule interactions and dynamic instability by phosphorylation at the Alzheimer-specific site Serine 262. J Biol Chem. 1995;270:7679–7688. doi: 10.1074/jbc.270.13.7679. [DOI] [PubMed] [Google Scholar]
- Drubin DG, Feinstein SC, Shooter EM, Kirschner MW. Nerve growth factor-induced neurite outgrowth in PC12 cells involves the coordinate induction of microtubule assembly and assembly-promoting factors. J Cell Biol. 1985;101:1799–1807. doi: 10.1083/jcb.101.5.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin D, Kirschner M. Tau protein function in living cells. J Cell Biol. 1986;103:2739–2746. doi: 10.1083/jcb.103.6.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye RB, Fink SP, Williams RC. Taxol-induced flexibility of microtubules and its reversal by MAP2 and tau. J Biol Chem. 1993;268:6847–6850. [PubMed] [Google Scholar]
- Ebneth A, Godemann R, Stamer K, Illenberger S, Trinczek B, Mandelkow E-M, Mandelkow E. Overexpression of tau protein alters vesicle trafficking, distribution of mitochondria and organization of the endoplasmic reticulum in living cells: implications for Alzheimer’s disease. J Cell Biol. 1998;143:777–794. doi: 10.1083/jcb.143.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmaeli-Azad B, Mccarty JH, Feinstein SC. Sense and antisense transfection analysis of tau-function: tau influences net microtubule assembly, neurite outgrowth and neuritic stability. J Cell Sci. 1994;107:869–879. doi: 10.1242/jcs.107.4.869. [DOI] [PubMed] [Google Scholar]
- Felgner H, Frank R, Biernat J, Mandelkow EM, Mandelkow E, Ludin B, Matus A, Schliwa M. Domains of neuronal microtubule-associated proteins and flexural rigidity of microtubules. J Cell Biol. 1997;138:1067–1075. doi: 10.1083/jcb.138.5.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frappier T, Georgieff I, Brown K, Shelanski ML. Tau regulation of microtubule-microtubule spacing and bundling. J Neurochem. 1994;63:2288–2294. doi: 10.1046/j.1471-4159.1994.63062288.x. [DOI] [PubMed] [Google Scholar]
- Friedhoff P, Mandelkow E. Tau protein. In: Kreis T, Vale R, editors. Guidebook to the Cytoskeletal and Motor Proteins. Oxford, United Kingdom: Oxford University Press; 1999. (in press). [Google Scholar]
- Goedert M, Jakes R, Crowther RA, Cohen P, Vanmechelen E, Vandermeeren M, Cras P. Epitope mapping of monoclonal antibodies to the paired helical filaments of Alzheimers disease: identification of phosphorylation sites in tau protein. Biochem J. 1994;301:871–877. doi: 10.1042/bj3010871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M, Spillantini M, Jakes R, Rutherford D, Crowther RA. Multiple isoforms of human microtubule-associated protein-tau: sequences and localization in neurofibrillary tangles of Alzheimers disease. Neuron. 1989;3:519–526. doi: 10.1016/0896-6273(89)90210-9. [DOI] [PubMed] [Google Scholar]
- Goode B, Denis P, Panda D, Radeke M, Miller H, Wilson L, Feinstein S. Functional interactions between the proline-rich and repeat regions of tau enhance microtubule-binding and assembly. Mol Biol Cell. 1997;8:353–365. doi: 10.1091/mbc.8.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg SG, Davies P, Schein JD, Binder LI. Hydrofluoric acid-treated tau-PHF proteins display the same biochemical properties as normal tau. J Biol Chem. 1992;267:564–569. [PubMed] [Google Scholar]
- Guo S, Kemphues KJ. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- Gustke N, Trinczek B, Biernat J, Mandelkow E-M, Mandelkow E. Domains of tau protein and interactions with microtubules. Biochemistry. 1994;33:9511–9522. doi: 10.1021/bi00198a017. [DOI] [PubMed] [Google Scholar]
- Hamm-Alvarez SF, Kim PY, Sheetz MP. Regulation of vesicle transport in CV-1 cells and extracts. J Cell Sci. 1993;106:955–966. doi: 10.1242/jcs.106.3.955. [DOI] [PubMed] [Google Scholar]
- Hirokawa N. Microtubule organization and dynamics dependent on microtubule-associated proteins. Curr Opin Cell Biol. 1994;6:74–81. doi: 10.1016/0955-0674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- Hyman A, Karsenti E. Morphogenetic properties of microtubules and mitotic spindle assembly. Cell. 1996;84:401–410. doi: 10.1016/s0092-8674(00)81285-4. [DOI] [PubMed] [Google Scholar]
- Illenberger S, Drewes G, Trinczek B, Biernat J, Meyer HE, Olmsted JB, Mandelkow E-M, Mandelkow E. Phosphorylation of microtubule-associated proteins MAP2 and MAP4 by the protein kinase p110-MARK: phosphorylation sites and regulation of microtubule dynamics. J Biol Chem. 1996;271:10834–10843. doi: 10.1074/jbc.271.18.10834. [DOI] [PubMed] [Google Scholar]
- Illenberger S, Zheng-Fischhöfer Q, Preuss U, Stamer K, Godemann R, Baumann K, Mandelkow E-M, Mandelkow E. The endogenous and cell cycle-dependent phosphorylation of the microtubule-associated protein tau in neuroblastoma and CHO cells: implications for protein kinases cdc2 and PKA. Mol Biol Cell. 1998;9:1495–1512. doi: 10.1091/mbc.9.6.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GV, Jenkins SM. Tau protein and Alzheimer’s disease brain. Alzheimer Dis Rev. 1996;1:38–54. doi: 10.3233/jad-1999-14-511. [DOI] [PubMed] [Google Scholar]
- Kanai Y, Takemura R, Oshima T, Mori H, Ihara Y, Yanagisawa M, Masaki T, Hirokawa N. Expression of multiple tau isoforms and microtubule bundle formation in fibroblasts transfected with a single tau cDNA. J Cell Biol. 1989;109:1173–1184. doi: 10.1083/jcb.109.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knops J, Kosik K, Lee G, Pardee J, Cohen-Gould L, McConlogue L. Overexpression of tau in a nonneuronal cell induces long cellular processes. J Cell Biol. 1991;114:725–733. doi: 10.1083/jcb.114.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles R, Leclerc N, Kosik KS. Organization of actin and microtubules during process formation in tau-expressing Sf9 cells. Cell Motil Cytoskeleton. 1994;28:256–264. doi: 10.1002/cm.970280308. [DOI] [PubMed] [Google Scholar]
- Kosik K, Orecchio L, Binder L, Trojanowski J, Lee V, Lee G. Epitopes that span the tau molecule are shared with paired helical filaments. Neuron. 1988;1:817–825. doi: 10.1016/0896-6273(88)90129-8. [DOI] [PubMed] [Google Scholar]
- Kosik KS, McConlogue L. Microtubule-associated protein function: lessons from expression in Spodoptera frugiperda cells. Cell Motil Cytoskeleton. 1994;28:195–198. doi: 10.1002/cm.970280302. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leclerc N, Baas PW, Garner CC, Kosik KS. Juvenile and mature MAP2 isoforms induce distinct patterns of process outgrowth. Mol Biol Cell. 1996;7:443–455. doi: 10.1091/mbc.7.3.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerc N, Kosik KS, Cowan N, Pienkowski TP, Baas P. Process formation in Sf9 cells induced by the expression of a microtubule-associated protein 2c-like construct. Proc Natl Acad Sci USA. 1993;90:6223–6227. doi: 10.1073/pnas.90.13.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Cowan N, Kirschner M. The primary structure and heterogeneity of tau protein from mouse brain. Science. 1988;239:285–288. doi: 10.1126/science.3122323. [DOI] [PubMed] [Google Scholar]
- Lee VMY, Balin BJ, Otvos L, Trojanowski JQ. A68—a major subunit of paired helical filaments and derivatized forms of normal tau. Science. 1991;251:675–678. doi: 10.1126/science.1899488. [DOI] [PubMed] [Google Scholar]
- Leger JG, Brandt R, Lee G. Identification of tau protein regions required for process formation in PC12 cells. J Cell Sci. 1994;107:3403–3412. doi: 10.1242/jcs.107.12.3403. [DOI] [PubMed] [Google Scholar]
- Lichtenberg-Kraag B, Mandelkow E-M, Biernat J, Steiner B, Schroeter C, Gustke N, Meyer HE, Mandelkow E. Phosphorylation dependent interaction of neurofilament antibodies with tau protein: epitopes, phosphorylation sites, and relationship with Alzheimer tau. Proc Natl Acad Sci USA. 1992;89:5384–5388. doi: 10.1073/pnas.89.12.5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez LA, Sheetz MP. A microtubule-associated protein (MAP2) kinase restores microtubule motility in embryonic brain. J Biol Chem. 1995;270:12511–12517. doi: 10.1074/jbc.270.21.12511. [DOI] [PubMed] [Google Scholar]
- Ludin B, Matus A. GFP illuminates the cytoskeleton. Trends Cell Biol. 1998;8:72–77. [PubMed] [Google Scholar]
- Mandelkow E, Mandelkow E-M. Tau in Alzheimer’s disease. Trends Cell Biol. 1998;8:425–427. doi: 10.1016/s0962-8924(98)01368-3. [DOI] [PubMed] [Google Scholar]
- Mandell JW, Banker GA. The microtubule cytoskeleton and the development of neuronal polarity. Neurobiol Aging. 1995;16:229–237. doi: 10.1016/0197-4580(94)00164-v. [DOI] [PubMed] [Google Scholar]
- Mandell JW, Banker GA. A spatial gradient of tau-protein phosphorylation in nascent axons. J Neurosci. 1996;16:5727–5740. doi: 10.1523/JNEUROSCI.16-18-05727.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo ES, Shin RW, Billingsley ML, Vandevoorde A, Oconnor M, Trojanowski JQ, Lee VMY. Biopsy-derived adult human brain tau is phosphorylated at many of the same sites as Alzheimer’s disease paired helical filament tau. Neuron. 1994;13:989–1002. doi: 10.1016/0896-6273(94)90264-x. [DOI] [PubMed] [Google Scholar]
- Matus A. Stiff microtubules and neuronal morphology. Trends Neurosci. 1994;17:19–22. doi: 10.1016/0166-2236(94)90030-2. [DOI] [PubMed] [Google Scholar]
- Mercken M, Vandermeeren M, Lübke U, Six J, Boons J, Van de Voorde A, Martin J-J, Gheuens J. Monoclonal antibodies with selective specificity for Alzheimer tau are directed against phosphatase-sensitive epitopes. Acta Neuropathol. 1992;84:265–272. doi: 10.1007/BF00227819. [DOI] [PubMed] [Google Scholar]
- Morishima-Kawashima M, Hasegawa M, Takio K, Suzuki M, Yoshida H, Titani K, Ihara Y. Proline-directed and non-proline-directed phosphorylation of PHF tau. J Biol Chem. 1995;270:823–829. doi: 10.1074/jbc.270.2.823. [DOI] [PubMed] [Google Scholar]
- Obar RA, Dingus J, Bayley H, Vallee RB. The RII subunit of cAMP-dependent protein-kinase binds to a common amino-terminal domain in microtubule-associated proteins 2a, 2b, and 2c. Neuron. 1989;3:639–645. doi: 10.1016/0896-6273(89)90274-2. [DOI] [PubMed] [Google Scholar]
- Panda D, Goode BL, Feinstein SC, Wilson L. Kinetic stabilization of microtubule dynamics at steady state by tau and microtubule-binding domains of tau. Biochemistry. 1995;34:11117–11127. doi: 10.1021/bi00035a017. [DOI] [PubMed] [Google Scholar]
- Preuss U, Döring F, Illenberger S, Mandelkow E-M. Cell cycle dependent phosphorylation and microtubule binding of tau protein stably transfected into Chinese hamster ovary cells. Mol Biol Cell. 1995;6:1397–1410. doi: 10.1091/mbc.6.10.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadot E, Barg J, Rasouly D, Lazarovici P, Ginzburg I. Short-term and long-term mechanisms of tau regulation in PC12 cells. J Cell Sci. 1995;108:2857–2864. doi: 10.1242/jcs.108.8.2857. [DOI] [PubMed] [Google Scholar]
- Sadot E, Gurwitz D, Barg J, Behar L, Ginzburg I, Fisher A. Activation of m(1) muscarinic acetylcholine-receptor regulates tau phosphorylation in transfected PC12 cells. J Neurochem. 1996;66:877–880. doi: 10.1046/j.1471-4159.1996.66020877.x. [DOI] [PubMed] [Google Scholar]
- Sato-Harada R, Okabe S, Umeyama T, Kanai Y, Hirokawa N. Microtubule-associated proteins regulate microtubule function as the track for intracellular membrane organelle transports. Cell Struct Funct. 1996;21:283–295. doi: 10.1247/csf.21.283. [DOI] [PubMed] [Google Scholar]
- Scott C, Spreen R, Herman J, Chow F, Davison M, Young J, Caputo C. Phosphorylation of recombinant tau by cAMP-dependent protein kinase: identification of phosphorylation sites and effect on microtubule assembly. J Biol Chem. 1993;268:1166–1173. [PubMed] [Google Scholar]
- Seubert P, et al. Detection of phosphorylated Ser(262) in fetal tau, adult tau, and paired helical filament tau. J Biol Chem. 1995;270:18917–18922. doi: 10.1074/jbc.270.32.18917. [DOI] [PubMed] [Google Scholar]
- Sontag E, Nunbhakdicraig V, Bloom GS, Mumby MC. A novel pool of protein phosphatase 2a is associated with microtubules and is regulated during the cell cycle. J Cell Biol. 1995;128:1131–1144. doi: 10.1083/jcb.128.6.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinczek B, Biernat J, Baumann K, Mandelkow E-M, Mandelkow E. Domains of tau protein, differential phosphorylation, and dynamic instability of microtubules. Mol Biol Cell. 1995;6:1887–1902. doi: 10.1091/mbc.6.12.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojanowski J, Lee VMY. Phosphorylation of paired helical filament tau in Alzheimer’s disease neurofibrillary lesions. FASEB J. 1995;9:1570–1576. doi: 10.1096/fasebj.9.15.8529836. [DOI] [PubMed] [Google Scholar]
- Zheng-Fischhöfer Q, Biernat J, Mandelkow E-M, Illenberger S, Godemann R, Mandelkow E. Sequential phosphorylation of tau-protein by GSK-3b and protein kinase A at Thr212 and Ser214 generates the Alzheimer-specific epitope of antibody AT-100 and requires a paired helical filament-like conformation. Eur J Biochem. 1998;252:542–552. doi: 10.1046/j.1432-1327.1998.2520542.x. [DOI] [PubMed] [Google Scholar]