Abstract
The concentration of low molecular weight compounds in tissues can yield valuable information about the metabolic state of an organism. Studies of changes in the metabolic state or metabonomics can reflect disease pathways, drug action, or toxicity. This research aims to develop a new approach, tissue targeted metabonomics. Microdialysis sampling and microcoil NMR analysis are employed to compare basal and ischemic metabolic states of various tissues (blood, brain, and heart) of Sprague–Dawley rats. Microdialysis sampling is localized, making the metabolic profile tissue specific. Coupling to NMR analysis is highly advantageous, because a complete metabolic profile is obtained in a single spectrum. However, small sample volumes and low analyte concentrations make analysis of microdialysis samples challenging. Microcoil NMR uses low sample volumes and has improved mass sensitivity, relative to standard 5mm probes. The coupling of these techniques is a potentially powerful tool for metabonomics analysis.
Keywords: Microcoil NMR, Metabonomics, Ischemia, NMR, Microdialysis
1. Introduction
Metabonomics has emerged as a powerful approach to systems biology, providing unique insights into the physiological effects of disease, drugs, toxicants, or genetic alterations [1–5]. Metabonomics-based investigations typically involve monitoring perturbations in the concentrations of endogenous metabolites in biological fluids, such as urine, plasma, cerebrospinal fluid, bile, or seminal fluid. Common methods of sampling of endogenous metabolites for metabonomics studies vary from biofluid collection to tissue excision. Biofluids, such as urine or plasma, are popular in metabonomics studies because they are fairly simple to collect, contain an abundance of metabolic information, and are minimally invasive to the animal during collection. However, these biofluids represent the average metabolic status of the organism and cannot give tissue-specific metabolic profiles. To address this, some studies have examined whole tissues or tissue extracts. Tissue excision can offer localized metabolic information, but it is conducted post-mortem or through biopsy and cannot give long-term, real-time information on the same animal.
Alternatively, microdialysis sampling allows for the acquisition of localized chemical information in nearly any tissue [6,7]. The technique is minimally invasive, allowing subjects to be awake and mobile during experimentation. Analyte sampling is accomplished by the implantation of a semipermeable membrane in the site of interest. Perfusion of a fluid through the dialysis probe facilitates diffusion of small hydrophilic molecules across the membrane, where they can be collected for analysis. Most dialysate samples are free of the macromolecules that can overshadow small molecules in NMR analysis. Problems arising from enzymatic degradation of the sample are also eliminated.
In this study, microdialysis sampling is applied to a metabonomics study of myocardial ischemia. Ischemia is the cessation of blood flow to a tissue or portion of tissue. Common clinical ischemic conditions are stroke and heart attack. Reperfusion is the period following ischemia when blood flow is restored. Both events drastically alter the metabolic state of the tissue in a specific location, making tissue targeted metabonomics studies important to truly understand the condition in vivo. This approach was previously applied to a metabonomics study of rat brain neurochemistry after the administration of tetradotoxin, a neurotoxin, by Khandelwal et al. [8,9].
Metabonomics analysis [5] is most commonly performed with 1H NMR [10,11]. For metabonomics studies of microdialysis samples, the sample volume is insufficient for traditional NMR analysis. Microcoil NMR probes can overcome these sample limitations. The microcoil probe used in these experiments (CapNMR probe produced by Protasis/MRM) is composed of a length of capillary with a bubble cell. The observed volume of this bubble cell is 1.5 µL and total probe volume is approximately 10 µL. Thus, sample volume requirements are reduced significantly when compared to traditional NMR analysis. A further advantage of microcoil NMR analysis is increased mass sensitivity, meaning that given the same mass of analyte detected, the microcoil probe will give a larger response than a traditional 5mm NMR probe [12]. Therefore, microcoil NMR is advantageous for analysis of mass and volume limited samples.
2. Experimental
2.1. Reagents
All deuterated solvents were obtained from Cambridge Isotope Laboratories, Andover, MA. Ringer’s solution was prepared in-house, with the salts obtained from Sigma Chemical Co., St. Louis, MO.
2.2. Animal protocols
For all animal studies, Sprague–Dawley rats (Sasco, Wilmington, MA) were pre-anesthetized with isoflurane, followed by full anesthesia by i.m. injection of a ketamine (100 mg/kg)/xylazine (10 mg/kg) mixture. The animals remained anesthetized and were closely monitored throughout the experimental procedures. Booster doses of one-fourth the original dose of ketamine were used as needed to maintain adequate anesthesia. Heated mats maintained the rats’ body temperature. All surgical techniques described below were performed separately on several rats, rather than simultaneously on a single rat. For the data presented in this communication, surgeries were not repeated.
2.3. Brain probe implantation
To insert microdialysis probes into the cerebral cortex of Sprague–Dawley rats, the hair on top of the rat skull was shaved and the region disinfected. The animal was then securely positioned in a stereotaxic apparatus and a midline 1-in. incision was made through the skin at the top of the skull parallel to the saggital suture. Adventitious tissue covering the skull was removed with cotton swabs. To provide a firm location to which dental adhesive could attach, two 1-mm diameter holes were hand-drilled approximately 2mm anterior to the insertion site of the guide cannula. Two stain-less steel anchor screws (1mm diameter, 2mm length) (BAS, West Lafayette, IN) were secured in these holes. Next, a 1 mm diameter hole was drilled through the skull at the insertion site and an intracerebral guide cannula was lowered into the brain using a micromanipulator attached to the stereotaxic apparatus. The guide cannula (BAS) was positioned 2 mm above the cerebral cortex, and then affixed to the skull with dental cement. The dummy probe in the guide cannula was replaced with a BAS microdialysis probe (BAS) after the guide cannula was glued securely in place. For all experiments, the molecular weight cut-off of the dialysis membrane was 5000 Da. To collect brain dialysate, Ringer’s solution was passed through the probe at 1 µL/min.
2.4. Heart probe implantation
To begin the heart probe implantation, the animal’s heart rate was monitored using a Digi-Med sinus rhythm analyzer (Louisville, KY). After the animal’s normal heart rate was established, a tracheotomy was performed by first exposing, and then isolating the trachea on a spatula. Cotton swabs removed any moisture from the exposed trachea. A scalpel was used to create an opening between the rings of the trachea, and a tube was inserted into the trachea and secured with sutures. The tube was then connected to a respirator. Artificial ventilation was performed with a constant-volume respirator using room air (Model 683 Rodent Respirator, Harvard Apparatus, Holliston, MA). To prepare for the microdialysis probe insertion into the heart, the thoracic cavity was shaved and cleansed. To expose the heart, with the animal in the lateral position, a left thoracotomy was performed by making an incision between the fifth and sixth ribs approximately 1.5–2 cm in diameter on the left side, and the pericardium was opened. A microdialysis probe (prepared in-house) was implanted into the myocardium through a small incision in the pericardium. Specifically, a needle was used to implant a microdialysis probe into the beating heart muscle adjacent to the left descending coronary artery along the longitudinal axis of the heart. Linear heart probes were made in-house from polyacrylonitrile membrane, and were approximately 2 mm in membrane length. Finally, after the lungs were fully expanded, Ringer’s solution was passed through the probe at 1 µL/min to collect heart dialysate.
2.5. Jugular probe implantation
A small midline skin incision was made on the neck and the jugular vein was located. The jugular vein was cleared from fine connective tissue by blunt dissection and cotton swabs. The vein was externalized by placing a spatula under it, perpendicular to the axis of the vein. Using a pair of fine spring scissors, a nick was made and a vascular probe (made in-house) with an introducer was placed into the vein towards the heart. Vascular probes were made in-house from polyacrylonitrile membrane and were approximately 10 mm in membrane length. Silk ligatures were used to secure the probe into place and the introducer was removed. To collect blood dialysate, Ringer’s solution was passed through the probe at 1 µL/min.
2.6. Blood sampling
Blood samples were collected from a cannula implanted into the femoral vein. To cannulate this vein, the anesthetized rat was positioned so that a small midline skin incision could be made on the inside of the leg and the femoral blood vessels located. The femoral vein was cleared from fine connective tissue, the femoral artery and nerve, by blunt dissection and cotton swabs, and then externalized by placing a spatula under it, perpendicular to the axis of the vein. A 1-cm section of femoral vein was temporarily ligated with silk ligatures. Using a pair of fine spring scissors, a nick was made between the ligatures, and PE-10 cannula was gently inserted into the femoral vein toward the vena cava lumen 2 in. towards the heart. The silk ligatures were secured around the cannula and blood was periodically drawn in 100 µL samples.
2.7. Ischemia
To create oxidative stress by ischemia in the heart, ischemia was induced by ligation of the left anterior descending coronary artery. The ischemia was maintained for 30 min.
2.8. Sample preparation
Dialysate samples were taken at 30 min intervals, corresponding to a sample volume of 30 µL. Samples were prepared by 50% dilution with 10 mM sodium trimethylsilyl propionate (TSP) or TSP-d4 in D2O. Samples were centrifuged at 12,400 rpm for 10 min immediately prior to analysis to pellet any particulates. The supernatant was removed for analysis. Whole blood samples were centrifuged for 15 min at 12,400 rpm. The supernatant was removed and diluted by 20% with D2O (because TSP is known to bind to plasma proteins, none was added to this sample).
2.9. NMR spectroscopy
All samples were analyzed by 1H NMR using a 600 MHz Varian INOVA spectrometer (Varian Instruments Inc., Palo Alto, CA) at 298 K. Dialysate samples were referenced to TSP at 0.00 ppm. Line broadening of 1Hz was applied to all spectra except the basal plasma dialysate, which had a line broadening of 5 Hz. Spectra were also zero-filled to 65,536 points.
Dialysate samples were analyzed using a capillary microcoil probe (Protasis/MRM Corp., Savoy, IL) equipped with proton and carbon-observe channels, deuterium lock, and z-gradients. Flow cell volume was 5 µL with an observe volume of 1.5 µL. The probe was prepared by rinsing with 50 µL acetone-d6 and 50 µL D2O. A 5 mL air plug was injected between rinses. Approximately 15 µL of sample was injected and both ends were capped to prevent siphoning. All dialysate samples were analyzed using WET solvent suppression [13] by coaddition of 3000 transients measured with a tip angle of 90° and an acquisition time of 1.5 s. The number of points was 25,188.
Plasma samples were analyzed with a 5 mm Varian triple-axis gradient probe. The solvent resonance was suppressed by pre-saturation [14], using a saturation delay of 2.0 s. The spectra were measured using a tip angle of 90°. A total of 26,868 data points were collected during the acquisition time of 1.6 s. To increase the signal-to-noise ratio of the spectra, 1024 transients were coadded. Resonances were assigned by comparison with standard spectra and literature chemical shift values.
3. Results/discussion
3.1. Whole plasma and plasma dialysate
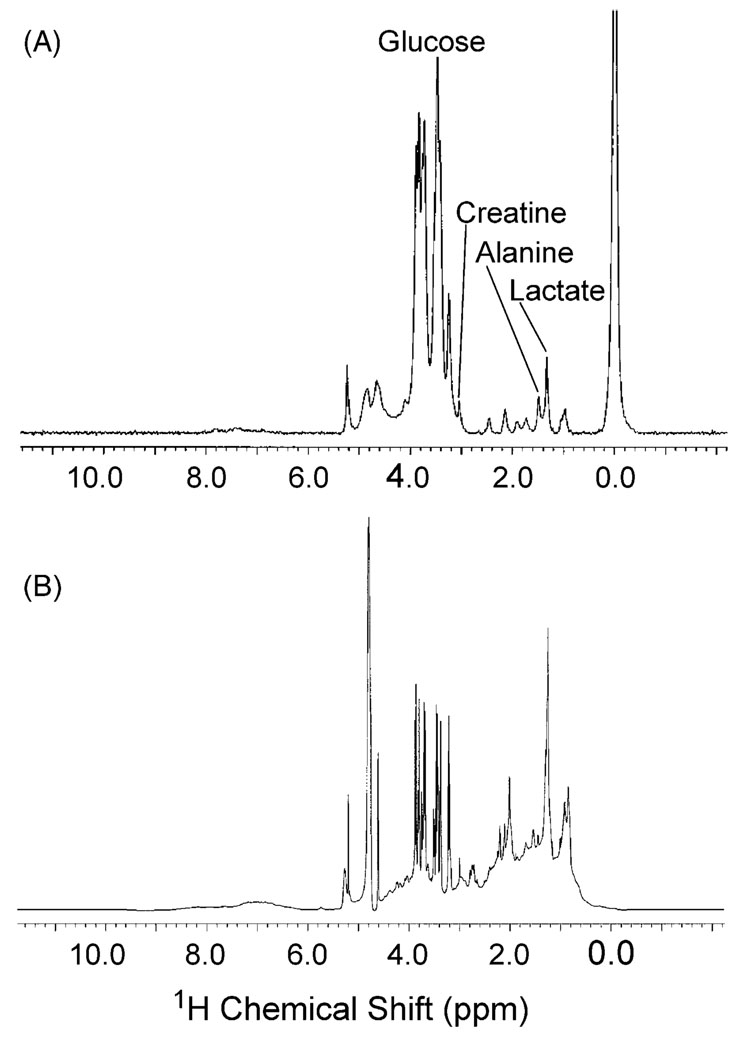
Differences in the resolution of small molecule signals are shown in Figs. 1A and B. Fig. 1A depicts the plasma dialysate spectrum and Fig. 1B depicts the whole plasma spectrum, both sampled from the same rat. The dialysate samples do not contain macromolecules that can obscure the detection of small molecule signals because they are beyond the molecular weight cut-off of the membrane. This is particularly apparent in the aliphatic region of the spectrum (1–3 ppm), where the lipid and protein resonances mask important small molecule resonances like lactate or alanine.
Fig. 1.
Spectral comparison of whole plasma and plasma dialysate from the same rat analyzed by NMR. (A) Plasma dialysate (50% D2O with TSP) analyzed by microcoil NMR. (B) Whole plasma (20% D2O) analyzed with a triple-axis gradient probe.
It should be noted that it is possible in NMR experiments to attenuate macromolecule resonances without removal of the macromolecule itself from the sample. Pulse sequences, such as CPMG, that incorporate relaxation editing can selectively detect small molecules by capitalizing on their slower T2 relaxation times [15,16]. However, this does nothing to prevent enzymatic degradation of the sample and can introduce quantitation problems due to differential relaxation of analyte resonances.
3.2. Myocardial ischemia
The effects of myocardial ischemia were considered from three perspectives—heart, plasma, and brain dialysate. In each tissue, a unique metabolic profile was observed, both in the basal and ischemic conditions, demonstrating the ability of this method to distinguish between different metabolic states and different tissue sites. In this paper, only results from the basal and ischemic periods are shown. Future studies are planned to include the reperfusion period.
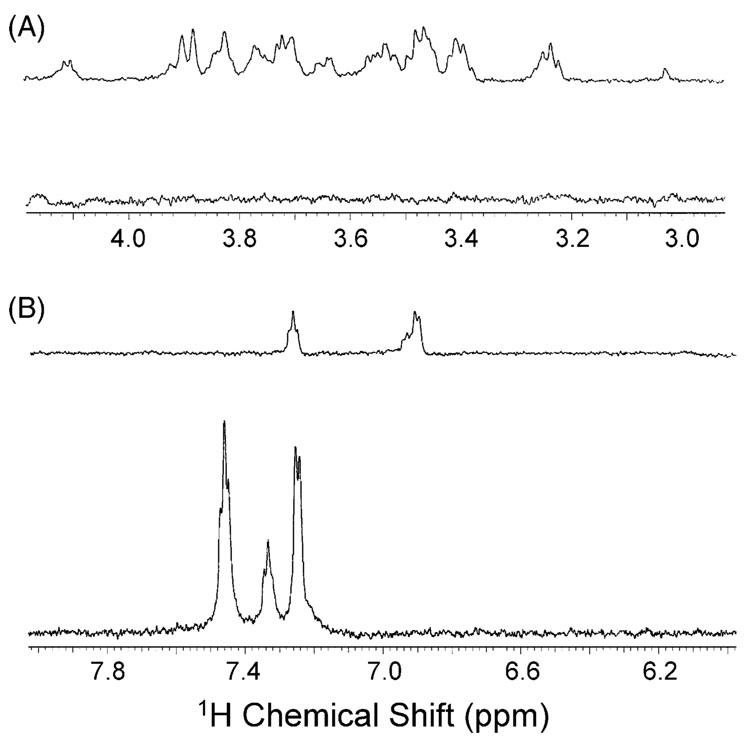
Basal heart dialysate was characterized by prominent glucose, lactate, and unassigned aromatic resonances. Lesser amounts of creatine and alanine were also observed. Ischemia causes dramatic changes in the metabolic profile of the heart tissue, as depicted in Fig. 2. For example, the glucose resonances (Fig. 2A) primarily in the region of 3.2–3.9 ppm, with anomeric resonances at 4.64 and 5.23 ppm, are no longer observed after ischemia was induced. This change was expected, as the cessation of blood flow to a tissue causes the depletion of localized energy stores, such as glucose and fatty acids. Additionally, the intensity of the lactate doublet at 1.33 ppm decreased, which was unexpected because during ischemia lactate is produced as a byproduct of anaerobic metabolism of glucose or amino acids [17]. Finally, it is interesting to note that the aromatic resonances (Fig. 2B) undergo a large change in chemical shift and multiplicity after ischemia. These resonances are as of yet unassigned, but the dramatic shift indicates an alteration in tissue metabolism, such as a dramatic pH change. Experiments are underway to determine the identity of these resonances.
Fig. 2.
Expansion of the heart dialysate spectra before and during myocardial ischemia to show the aliphatic (A) and aromatic (B) spectral regions. For each set of spectra, the upper spectrum was measured before ischemia and the bottom spectrum was measured using dialysis samples collected during ischemia. The depletion of glucose occurs during ischemia as the heart rapidly utilizes its tissue stores to meet the heart’s large energy demand. Shifts were observed in the aromatic region during ischemia, suggesting that pH fluctuations may have occurred.
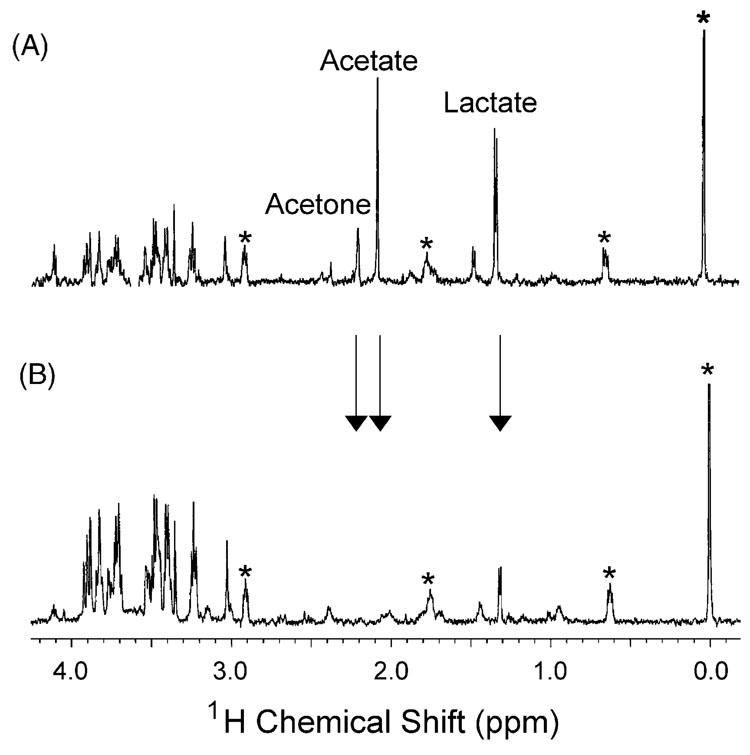
Changes were also observed in the spectra of plasma dialysate (Fig. 3). It should be noted that the plasma dialysate was sampled in close proximity to the heart. Different metabolic profiles would be expected by sampling plasma in a remote location, as plasma dialysate taken close to the heart will contain small molecules exported by cardiac tissue. When compared to basal heart dialysate, basal plasma dialysate has unique characteristics. Although both contain glucose and lactate resonances, plasma dialysate also has strong acetate (2.05 ppm) and acetone (2.15 ppm) signals. Upon inducing ischemia, a decrease in lactate is observed, consistent with observations in heart tissue but inconsistent with trends described in the literature. Additionally, the acetate and acetone resonances disappear during ischemia. Again, dramatic and tissue specific changes in the metabolic state of the organism are observed due to the ischemic condition.
Fig. 3.
Plasma dialysate before (A) and during (B) myocardial ischemia. Changes in the levels of lactate (1.40 ppm), acetate (2.05 ppm), and acetone (2.15 ppm) are observed during ischemia. The asterisk (*) indicates internal standard resonances. Arrows mark key metabolic changes observed during ischemia.
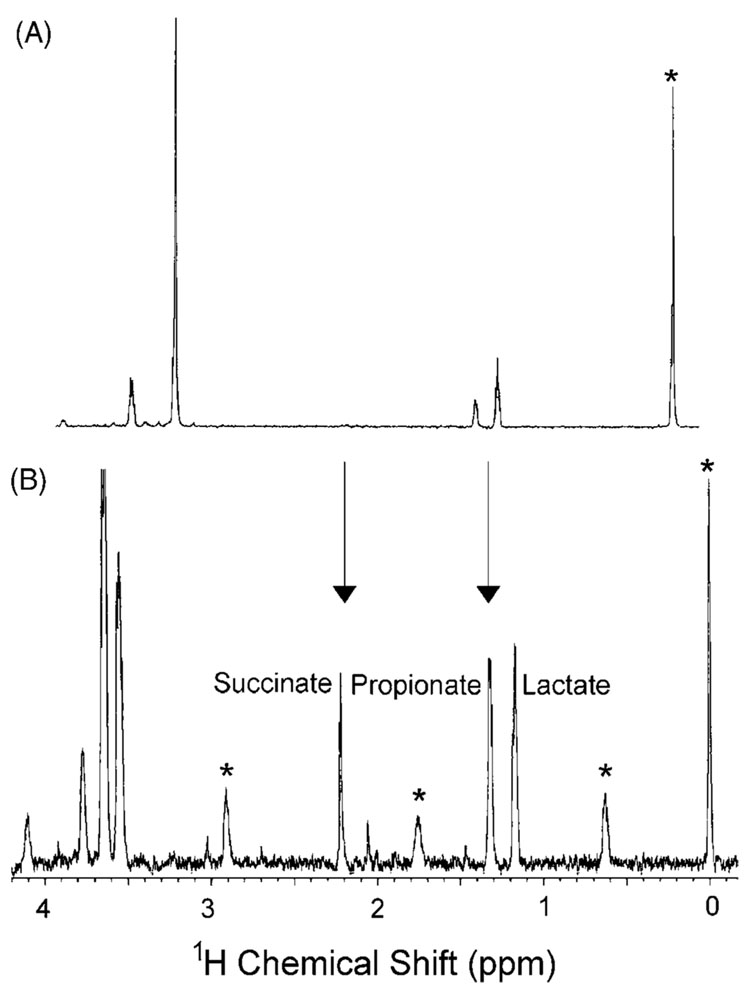
The final tissue studied was the cerebral cortex of the brain (Fig. 4). Brain dialysate, particularly from an anesthetized animal, does not contain the diversity of endogenous metabolites demonstrated in heart and plasma. Nevertheless, changes can be detected with this tissue targeted technique. Basal brain dialysate contains most noticeably lactate and proprionate. After a myocardial ischemic event, the metabolic profile changes dramatically. Succinate (2.2 ppm), one of the known byproducts of anaerobic respiration, appears in the dialysate during ischemia [17]. A peak tentatively assigned as glycerol is detected in the ischemia spectra, suggesting that fatty acid oxidation may be occurring as a means of energy production. This is unusual for brain tissue, which uses glucose and ketone bodies almost exclusively as an energy source.
Fig. 4.
Brain dialysate before (A) and during (B) myocardial ischemia. Note the appearance of glycerol (3.4–3.8 ppm) and succinate (2.2 ppm) during ischemia and changes in propionate (1.2 ppm) and lactate (1.3 ppm) levels, demonstrating that brain tissue metabolism is affected by myocardial ischemia. Arrows mark key metabolic changes observed during ischemia.
In summary, our approach to metabonomics studies, tissue targeted metabonomics, can provide minimally invasive, tissue specific metabolic information not available using current methodologies. This is accomplished by microdialysis sampling of tissues, coupled with analysis using microcoil NMR. Microdialysis sampling provides a means to continuously monitor multiple biochemical components at specific tissue sites in an experimental animal. Microdialysis experiments can, therefore, provide both temporal and spatial information about the status of the affected tissue. Metabolic profiling of these small volume dialysate samples using microcoil NMR can provide a unique perspective for metabonomics studies.
4. Conclusions
The preliminary data presented above indicate that microdialysis sampling with microcoil NMR detection is a promising approach for tissue targeted metabonomics studies. The ability of NMR to distinguish between metabolic profiles of different tissues is critical for successful experimentation. In the spectra presented, it is possible to differentiate between blood, heart, and brain dialysate. In addition, tissue targeted metabonomics can discriminate between metabolic states of individual tissues, that is, the basal and ischemic state for each tissue clearly show that changes have occurred in the small molecule levels within a localized area.
Further studies are planned to study myocardial ischemia in greater depth. The effects of myocardial ischemia-reperfusion on the heart directly and on other peripheral tissues and biofluids, such as urine and plasma, will be studied to identify biomarker patterns of this condition and to explain alterations of metabolic pathways that occur during this event. The time course of ischemia-reperfusion is important and should be studied in greater detail because the length of the ischemic period can cause different perturbations in metabolic pathways. In addition, comparisons between ischemia of different tissues could be conducted by similar studies of cerebral ischemia (stroke).
In conclusion, tissue targeted metabonomics will offer a unique perspective of localized metabolic information to metabonomics research. By expanding the current possibilities for in vivo study, a greater understanding of disease pathways and drug action will be gained.
Acknowledgements
Funding by NIH grant HL069014 (C.E.L.) and NSF grant CHE-0213407 (C.K.L.) is gratefully acknowledged. K.E.P. gratefully recognizes the Madison and Lila Self Graduate Fellowship for financial support.
References
- 1.Lindon JC, Holmes E, Nicholson JK. Anal. Chem. 2003;75:384A–391A. doi: 10.1021/ac031386+. [DOI] [PubMed] [Google Scholar]
- 2.Nicholson JK, Lindon JC, Holmes E. Xenobiotica. 1999;29:1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 3.Reily MD, Robertson DG, Delnomdedieu M, Baker JD. Am. Pharm. Rev. 2003;6:105–109. [Google Scholar]
- 4.Robertson DG, Bulera SJ. Curr. Opin. Drug Discov. Dev. 2000;3:42–47. [PubMed] [Google Scholar]
- 5.Lindon JC, Holmes E, Bollard ME, Stanley EG, Nicholson JK. Biomarkers. 2004;9:1–31. doi: 10.1080/13547500410001668379. [DOI] [PubMed] [Google Scholar]
- 6.Lunte CE, Scott DO, Kissinger PT. Anal. Chem. 1991;63:773A–774A. 776A–778A, 780A. [Google Scholar]
- 7.Weiss DJ, Lunte SM, Lunte CE. Trends Anal. Chem. 2000;19:606–616. [Google Scholar]
- 8.Khandelwal P, Beyer CE, Lin Q, McGonigle P, Schechter LE, Bach AC., II J. Neurosci. Methods. 2004;133:181–189. doi: 10.1016/j.jneumeth.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 9.Khandelwal P, Beyer CE, Lin Q, Schechter LE, Bach AC., II Anal. Chem. 2004;76:4123–4127. doi: 10.1021/ac049812u. [DOI] [PubMed] [Google Scholar]
- 10.Lindon JC, Nicholson JK, Holmes E, Everett JR. Concepts Magn. Reson. 2000;12:289–320. [Google Scholar]
- 11.Griffin JL. Curr. Opin. Chem. Biol. 2003;7:648–654. doi: 10.1016/j.cbpa.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 12.Olson DL, Norcross JA, O’Neil-Johnson M, Molitor PF, Detlefsen DJ, Wilson AG, Peck TL. Anal. Chem. 2004;76:2966–2974. doi: 10.1021/ac035426l. [DOI] [PubMed] [Google Scholar]
- 13.Smallcombe SH, Patt SL, Keifer PA. J. Magn. Reson. 1995;A 117:295–303. [Google Scholar]
- 14.Hoult DI. J. Magn. Reson. 1976;21:337–347. [Google Scholar]
- 15.Rabenstein DL, Millis KK, Strauss EJ. Anal. Chem. 1988;60:1380A–1391A. doi: 10.1021/ac00175a001. [DOI] [PubMed] [Google Scholar]
- 16.Van QN, Chmurny GN, Veenstra TD. Biochem. Biophys. Res. Commun. 2003;301:952–959. doi: 10.1016/s0006-291x(03)00079-2. [DOI] [PubMed] [Google Scholar]
- 17.Taegtmeyer H, King LM, Jones BE. Am. J. Cardiol. 1998;82:54K–60K. doi: 10.1016/s0002-9149(98)00538-4. [DOI] [PubMed] [Google Scholar]