Abstract
Identification of pre-B-cell colony-enhancing factor (PBEF) interacting partners may reveal new molecular mechanisms of PBEF in the pathogenesis of acute lung injury (ALI). The interactions between PBEF and NADH dehydrogenase subunit 1(ND1), ferritin light chain and interferon induced transmembrane 3 (IFITM3) in human pulmonary vascular endothelial cells were identified and validated. ND1, ferritin and IFITM3 are involved in oxidative stress and inflammation. Overexpression of PBEF increased its interactions and intracellular oxidative stress, which can be attenuated by rotenone. The interaction modeling between PBEF and ND1 is consistent with the corresponding experimental finding. These interactions may underlie a novel role of PBEF in the pathogenesis of ALI.
Structured summary
MINT-6538697:
PBEF (uniprotkb:P43490) physically interacts (MI:0218) with NADH1 (uniprotkb:P03886) by two hybrid (MI:0018)
MINT-6538811, MINT-6538868:
PBEF (uniprotkb:P43490) physically interacts (MI:0218) with interferon-induced transmembra (uniprotkb:Q01628) by anti bait coimmunoprecipitation (MI:0006)
MINT-6538787, MINT-6538841:
PBEF (uniprotkb:P43490) physically interacts (MI:0218) with NADH1 (uniprotkb:P03886) by anti bait coimmunoprecipitation (MI:0006)
MINT-6538755:
PBEF (uniprotkb:P43490) physically interacts (MI:0218) with gamma-glutamil-transferase (uniprotkb:P19440) by two hybrid (MI:0018)
MINT-6538799, MINT-6538862:
PBEF (uniprotkb:P43490) physically interacts (MI:0218) with Ferritin light chain (uniprotkb:P02792) by anti bait coimmunoprecipitation (MI:0006)
MINT-6538769:
PBEF (uniprotkb:P43490) physically interacts (MI:0218) with E2L6 (uniprotkb:O14933) by two hybrid (MI:0018)
MINT-6538741:
PBEF (uniprotkb:P43490) physically interacts (MI:0218) with Adenosine A2aR (uniprotkb:P29274) by two hybrid (MI:0018)
MINT-6538727:
PBEF (uniprotkb:P43490) physically interacts (MI:0218) with interferon-induced transmembra (uniprotkb:Q01628) by two hybrid (MI:0018)
MINT-6538712:
PBEF (uniprotkb:P43490) physically interacts (MI:0218) with Ferritin light chain (uniprotkb:P02792) by two hybrid (MI:0018)
Keywords: PBEF, Interaction, vascular permeability, acute lung injury, inflammation, oxidative stress
Introduction
Acute lung injury (ALI) and its more severe form, acute respiratory distress syndrome ( ARDS), are characterized by refractory hypoxemia associated with lung inflammation and increased pulmonary vascular permeability (1). Although ARDS was first described by Ashbaugh and colleagues in the Lancet in 1967 (2) and considerable progress has been made, mortality of patients with ALI and ARDS remains 30 to 50%. This is because molecular mechanisms underlying the susceptibility and the severity of ARDS are still incompletely understood. Therefore, more studies are clearly needed to identify novel biochemical and genetic markers and elucidate their molecular involvements in ARDS.
In our previous study on animal models of ALI, we identified preB-cell colony enhancing factor (PBEF) as a significantly upregulated gene in ALI (3). We also found a susceptible GC haplotype in the human PBEF gene promoter, which conferred a 7.7-fold higher risk of ALI (3). Our result was confirmed in a separate population by Bajwa et al (4). We further found that an inhibition of PBEF expression by PBEF siRNA significantly attenuated pulmonary vascular endothelial cell barrier dysfunction (5). Taken together, these results strongly indicate PBEF as a potential novel candidate gene and biomarker in ALI.
This study aims to address the molecular mechanisms by which PBEF contributes to pathogenesis of ALI. Since the interactions between proteins are important for many biological functions and pathological processes, we applied BacterioMatch Two-Hybrid System (Stratagene) (6) to identify potential human PBEF interacting proteins in the lung. With this system, several PBEF interacting partner proteins in the lung were identified and validated by coimmunoprecipitation experiments in pulmonary vascular endothelial cells. Several Interacting proteins to PBEF as well as effect of PBEF on intracellular oxidative stress were examined in the absence or presence of IL-1β-stimulation. The computer modeling was also employed to predict the interacting mode between PBEF and NADH dehydrogenase subunit 1 (ND1). These results may reveal a novel role of PBEF in the pathogenesis of ALI.
Materials and methods
Cell Culture
Human primary pulmonary artery endothelial cells (HPAEC) and human primary lung microvascular endothelial cells (HLMVEC) were obtained from Cambrex Bio Science Inc. (Walkersville, MD). Routine cell culture and maintenance were performed as described before in our lab (3, 5).
BacterioMatch Two-Hybrid System Screenning
The application of BacterioMatch Two-Hybrid System (Stratagene, La Jolla, CA) for screening PBEF interacting partners in lung tissues was carried out according to our established protocol (6). The PBEF bait was prepared by subcloning the human PBEF cDNA into the pBait vector, pBT. A human PBEF coding cDNA prepared before in our lab (7), was PCR amplified and inserted in frame into EcoRI and BamHI sites of the pBT bait plasmid. Primers for the PCR amplification are: 5’-CTAGAATTCATGAATCCTGCGGCAGAAGCCG-3’and 5’-TATGGATCCATGTGCTGCTTCCAGTTCAATAT-3’, respectively. Underlined sequences are EcoRI and BamHI adapters, respectively. The target lung protein cDNA library in the target vector, pTRG, was obtained from the Stratagene ((La Jolla, CA).
Immunoprecipitation and Complex Analysis
Immunoprecipitation was performed as described before (8). HPAEC and HLMVEC were grown to confluence and incubated in serum-free media with or without IL-1β (10 ng/ml) for 4 hours. The cell lysate samples were immunoprecipitated with either rabbit anti-PBEF antibody (Bethyl Laboratory, Inc.), mouse anti-NADH dehydrogenase subunit 1 antibody (Mitoscience), rabbit anti-Ferritin light chain antibody (ADI), mouse anti-IFITM3 antibody (Abnova), rabbit anti-Adenosine A2a receptor antibody (Chemicon), mouse anti-γ-glutamyl-transferase antibody (Lab Vision) or mouse anti-Ubiquitin conjugating enzyme E2L 6 antibody (Abnova) followed by incubation with either goat anti-rabbit or goat anti-mouse conjugated sepharose beads (Sigma). Immunoprecipitated material and cellular lysates were run on SDS-PAGE on 4–15% polyacrylamide gels, transfer onto Immobilo™ membranes, and developed with specific primary and secondary antibodies. Visualization of immunoreactive bands was achieved using enhanced chemiluminescence (Amersham Biosciences).
Reactive Oxygen Species Assay
HPAEC cells were transfected with the vehicle control (c), PBEF stealth siRNA (Si), scrambled RNA (Sc), PBEF overexpression (E) and vector control (V) for 48 h before subjected to the treatment without or with IL-1β (10 ng/ml) for 4 hours. A549 cells, a lung Type II epithelial cell line, were similarly transfected for 48 h before treated with rotenone (Sigma, MO, USA) (10 µM) for 3 hours. Reactive oxygen species assay in HPAEC cells was carried out according to the protocol of World Precision Instruments Superoxide/Reactive Oxygen Species Determination kit (Superluminal, Sarasota, FL, USA).
The binding mode modeling between PBEF and ND1
We further probed the interaction between PBEF and ND1 by modeling their binding mode. The structure of human PBEF was downloaded from the Protein Data Bank (9) [pdb code: 2GVG (10)]. Because no atomic structure is available for NADH dehydrogenase subunit 1 (ND1), ND1 was modeled as follows. First, the sequence of human ND1 was extracted from the GenBank [accession No: NP536843, (11)]. Based on the ND1 sequence, similarity search was then performed against the sequences in the Protein Data Bank by using the BLAST program (12). It was found that the C-terminal domain of the catalase-peroxidase KatG (pdb code: 1U2L) has a good sequence similarity with ND1 near the potential interacting residues (81–85). Next, the overall sequence alignment between 1U2L and ND1 was obtained through the FASTA program (13) provided on the PDB web site (see Figure 2, A). Based on the sequence alignment, the protein structure of ND1 was created from the template structure 1U2L by using the homology modeling program MODELLER (14). The constructed ND1 structure was then docked to the PBEF structure (2GVG) by using the protein-protein docking program ZDOCK 2.1 (15). The best-scored orientation was predicted as the binding mode (Figure 3).
Figure 2. Effect of PBEF expression on reactive oxygen species (ROS) production in HPAEC in the presence or absence of IL-1β treatment.
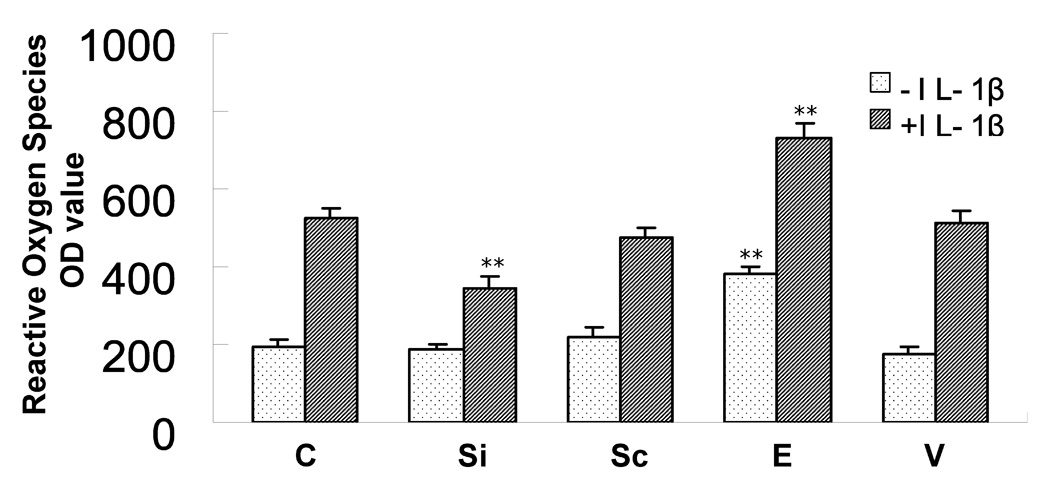
HPAEC culture, transfection, IL-1β treatment and ROS production assay were carried out as described in the method. 5 experimental groups are: vehicle control (C), PBEF stealth siRNA (Si), scrambled RNA (Sc), PBEF overexpression (E) and vector control (V). Each group has its baseline level compared to its IL-1β treatment. Results from each group are presented as mean ± SD of 4 samples from two separate experiments. Statistical comparative analyses of ROS production levels between the control and IL-1β treated group were performed using the unpaired t test. **, p<0.05.
Figure 3. Effect of PBEF expression on ROS production in A549 cells in the presence or absence of rotenone.
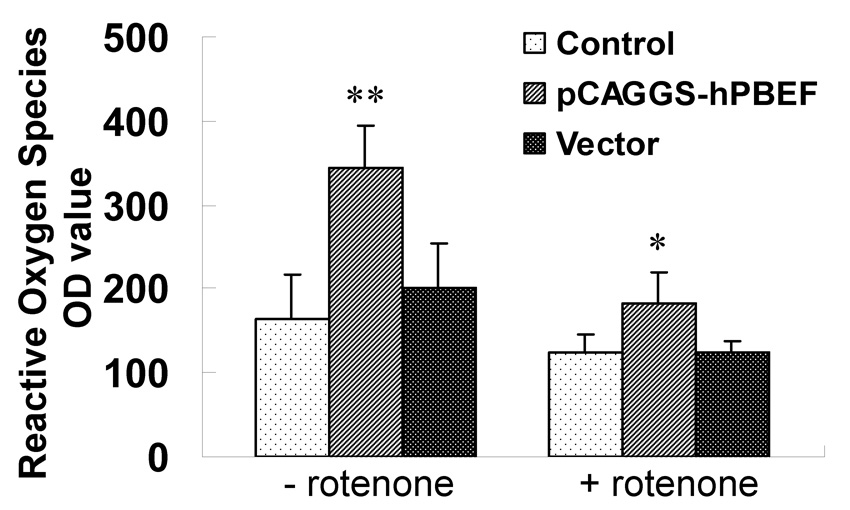
A549 cells were transfected with control, pCAGGS-hPBEF and pCAGGS vector only for 48 h, and then treated with Rotenone for 3 hours. The value of reactive oxygen species were measured TriStar Multimode Reader (LB 941, Berthold Technologies GnbH & Co.KG, Bad Wildbad, Germany) following instruction manual of the Superluminol kit (World Precision Instruments, Inc., Sarasota, FL, USA). Results from each group are presented as mean ± SD of 3 samples from two separate experiments. Statistical comparative analyses of ROS production levels between the control and rotenone treated group were performed using the unpaired t test. *, p<0.05 and **, p<0.01.
Results
Several potential PBEF interacting partners in expressed lung cDNA library identified by the BacterioMatch Two-Hybrid System
Because most proteins work as complexes to regulate biological and/or pathological processes in cells and tissues, we set out to determine whether PBEF has any interacting partner in lung tissues using the BacterioMatch Two-Hybrid System. Six potential PBEF-interacting proteins in the lung including their protein accession numbers, involved key biological processes, interacting fragments and references are presented in Table 1. They are NADH dehydrogenase subunit 1 (ND1), ferritin light chain, interferon induced transmembrance protein 3 (IFITM3), Adenosine A2a receptor (A2aR), γ-glutamyl transferase and ubiquitin conjugating enzyme E2L6 (UCE2L6)
Table 1.
Putative PBEF interacting proteins in the lung
| No. | Name | Protein Accessión# | Involved biological process | Interact. aa | Protein Length (aa) | Ref. |
|---|---|---|---|---|---|---|
| 1 | NADH dehydrogenase subunit 1 | NP_536843 | Involved in various metabolisms | 81–85 | 318 | Hattori Y et al. 2003 (16) |
| 2 | Ferritin Light chain | NP_000137 | Serum ferritin as a predictor of the acute respiratory distress syndrome | 1–175 | 175 | Connelly KG et al. 1997 (17) |
| 3 | Interferon induced transmembrane protein 3 | NP_066362 | Expression induced by interferon | 1–84 | 133 | Lewin AR et al. 1991 (18) |
| 4 | Adenosine A2a R | NP_000666 | Regulation of cytokine release, involved in inflammation | 113–187 | 412 | Lukashev DE et al. 2003 (19) |
| 5 | γ-glutamyl-transferase | NP_005256 | Its expression regulates reactive oxygen species and alters NF-kappaB activity | 557–569 | 569 | Carlisle IV et al. 2003 (20) |
| 6 | Ubiquitin conjugating Enzyme E2L 6 | NP_004214 | Essential element of the Ubiquitin pathway, mediating transfer of activated ubiquitin to target proteins with the aid of SCF ligases | 1–153 | 153 | Ardley HC et al. 2000 (21) |
Coimmunoprecipitation confirmation of PBEF interacting partners identified by the BacterioMatch Two-Hybrid System
We next employed a coimmunoprecipitation strategy to confirm whether those potential PBEF interacting partners identified above were true PBEF interacting partners. As presented in Figure 1, all six genes (ND1, ferritin light chain, IFITM3, A2aR, γ-glutamyl transferase and UCE2L6) are expressed in both HPAEC and HLMVEC (Figure 1, A). However, the coimmunoprecipitation experiments revealed only ND1, ferritin light chain and IFITM3 out of the 6 PBEF interacting partners identified by the BacterioMatch Two-Hybrid System were confirmed in the precipitation complexed with PBEF (Figure 1, B). These interactions were highly enhanced in the IL1-β-treated cells. The reverse coimmunoprecipitation experiments corroborated that only ND1, ferritin light chain and IFITM3 interacted with PBEF (Figure 1, C).
Figure 1. Coimmunoprecipitation confirmation of PBEF interacting partners identified by the BacterioMatch Two-Hybrid System.
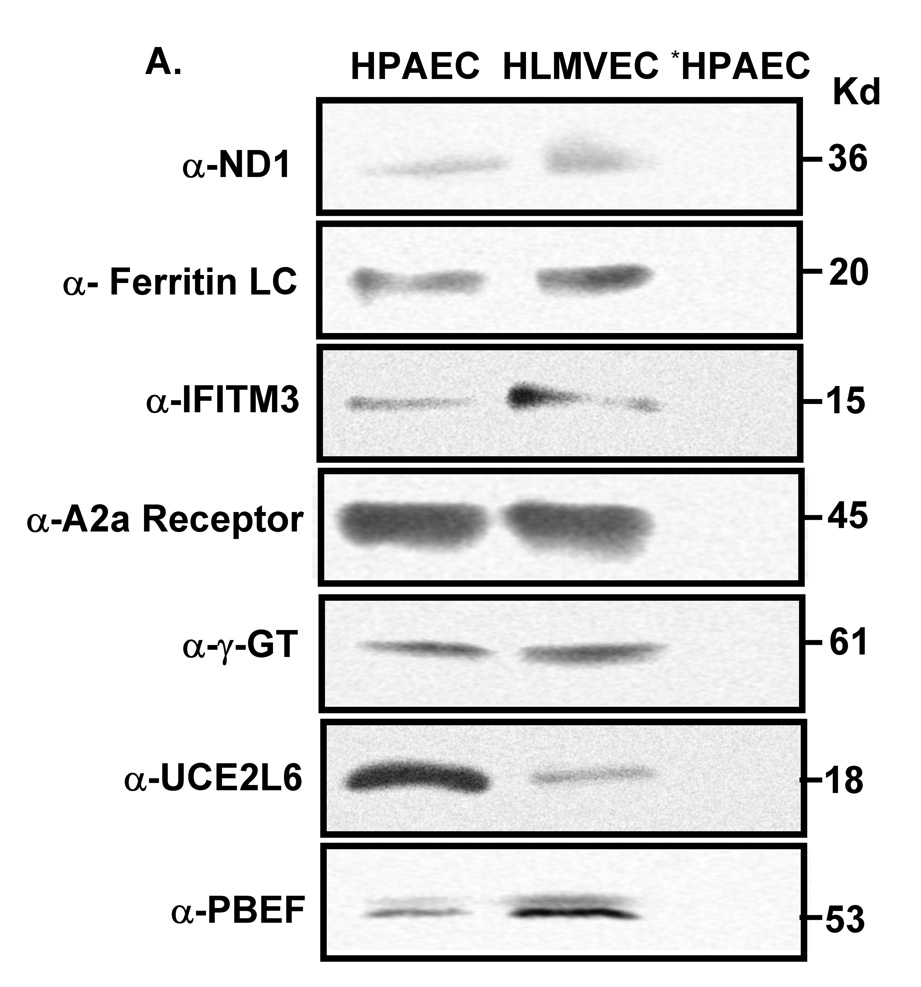
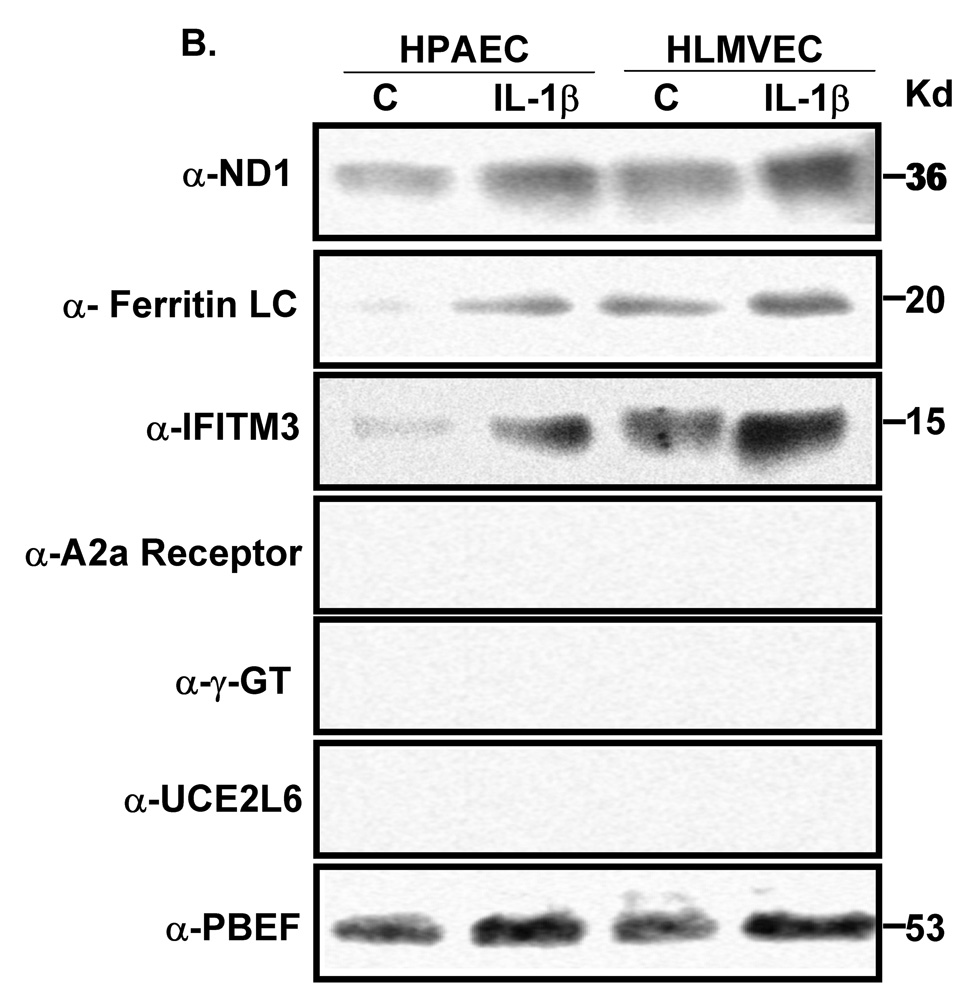
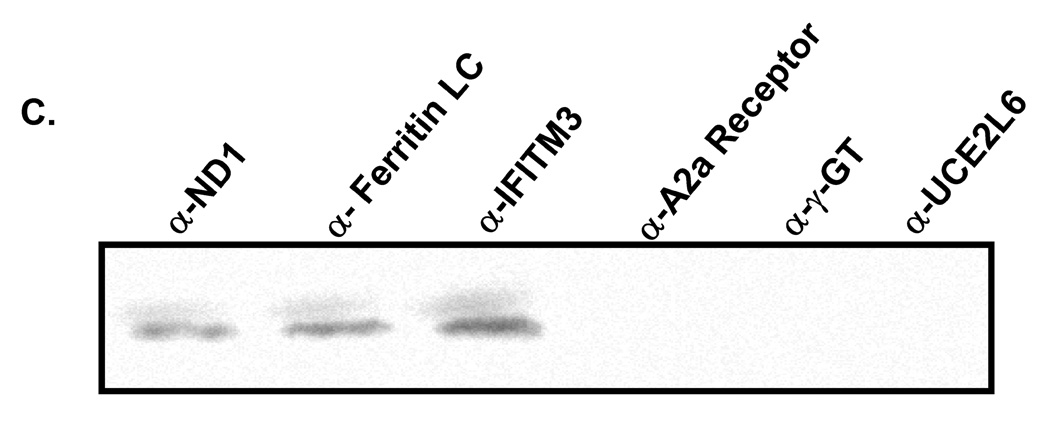
Both confluent HPAEC and HLMVEC cells were incubated for 4 hours in the presence and absence of IL1-β (10 ng/ml) and their cell lysates were subjected to coimmunoprecipitation analyses. A, Detection of protein expression. Cell lysate proteins were immunodetected with antibodies as indicated. *, preimmune IgG serves as a negative control. B, Coimmunoprecipitation. Cell proteins with or without the IL1-β treatment are immunoprecipitated using 6 different antibodies as indicated and then immunodetected using the antibody to PBEF. C, Reverse coimmunoprecipitation. Cell proteins with the IL1-β treatment are immunoprecipitated using the antibody to PBEF and then immunodetected using 6 different antibodies as indicated.
PBEF expression affected intracellular ROS production in HPAEC and A549 cells
Since above validated three interacting proteins to PBEF are involved in oxidative stress and inflammation, we examined whether PBEF expression affects intracellular ROS production in HPAEC in the presence or absence of IL1-β. As presented in Figure 2, Inhibition of PBEF expression by PBEF siRNA significantly blunted IL-1β induced ROS production compared to its control [343.43 ± 33.12 (Si + I) vs. 523.33 ± 25.17 (C + I), n = 4, p <0.05). Scramble RNA has no significant effect [475.67 ± 23.02 (Sc + I) vs. 523.33±25.17 (C + I), n=4, NS). Overexpression of PBEF significantly increased ROS production over the control [ 383.68 ± 16.80 (E) vs. 191 ± 18.52 (C), n=4, p<0.05]. Vector only control has no effect [178 ± 18.36 (V) vs. 191 ± 18.52 (C), n=4, NS]. Overexpression of PBEF plus IL1-β treatment also significantly augmented ROS production over vector control plus IL1-β treatment [730.33 ± 35.64 (E+I) vs. 513 ± 32.51 (V + I), n=4, p<0.05]. To examine whether the same phenomenon can be observed in A549 cells, a lung Type II epithelial cell line, we determined the effect of PBEF overexpression in A549 cells in the absence or presence of rotenone. As presented in Figure 3, overexpression of PBEF significantly augmented ROS production over vector control in A549 cells [345 ± 49.34 vs. 200 ± 53.54, n=3, p<0.05] (Figure 3). However, this effect was significantly attenuated by the treatment of rotenone, a NADH oxidase complex I inhibitor (22) [182.5 ± 35.94 (E + rotenone) vs 345 ± 49.34 (E-rotenone), n=3, p<0.01]
The binding mode modeling between PBEF and ND1
To explore the physical interaction between PBEF and ND1, we generated a hypothetical three dimensional structure of ND1 based on its sequence homology to the catalase-peroxidase KatG (Figure 4, A) before docking it to a known three dimensional structure of PBEF (Figure 4, B). Figure 3 A shows that the residues 81–85 of ND1 are indeed located in the interface between ND1 and PBEF. Our docking results predicted that the following residues of PBEF are within 4.5 A of the residues 81–85 of ND1: F193, D219, S241, V242, P243, A244, A245, E246, T249, I265, H264, Q268, and F269. These residues may directly interact with the residues 81–85 of ND1. Most of the above interacting residues within PBEF are hydrophobic.
Figure 4. The interacting modeling between PBEF and ND1.
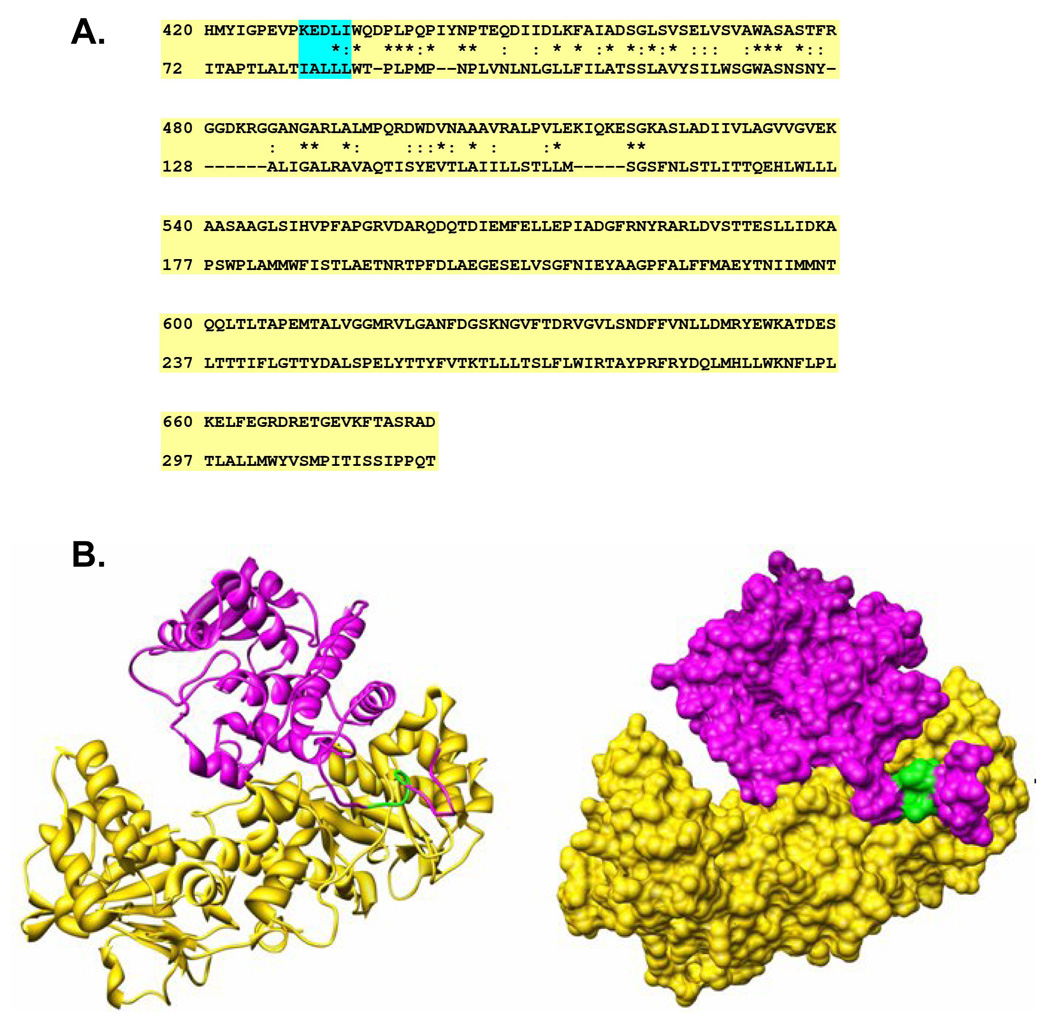
A. The sequence alignment between human NADH dehydrogenase subunit 1 (ND1) and the catalase-peroxidase KatG (pdb code: 1U2L). The sequence alignment was performed using the FASTA program (13). For each alignment row, the upper line is the sequence of 1U2L and the lower line is that of human ND1. The “*” symbols represent identical amino acid residues. The “:” symbols show residue pairs with positive matrix scores, representing similar sequence alignment according to the FASTA algorithm. B. The interacting mode between the PBEF and ND1. The left panel is a ribbon representation. The right panel shows the molecular surface of the same proteins. PBEF is shown in gold, and ND1 is in magenta. The green part represents residues 81–85 of ND1. This figure was prepared using the UCSF Chimera (34, 35)
Discussion
In this study, we have identified three interacting partners of PBEF in human pulmonary artery and microvascular endothelial cells: ND1, Ferritin light chain and IFITM3, using the BaterioMatch Two Hybrid System screening and a confirmative coimmunoprecipitation approach. These PBEF interactive proteins may provide insight into the PBEF-mediated signal transduction leading to the increased pulmonary vascular dysfunction and the pathogenesis in ALI.
Protein–protein interactions are fundamental to all biological processes. The determination of protein–protein interactions provides a framework for understanding pathophysiological functions of a unknown protein based on its known interacting partners (23). This is the so called ‘guilt-by-association’ principle (24). The Yeast two hybrid system, as originally described by Fields and Song (25), is the most commonly utilized system for identifying binary protein–protein interactions. We employed one of its derivatives, BaterioMatch Two Hybrid System from Stratagene, which is a faster, simpler and more efficient method for detecting protein-protein interactions in vivo (6). Using PBEF- pBT bait vector to bait potential interacting partners in a lung cDNA library in pTRG target vector, we identified several interesting proteins interacting with PBEF as listed in Table 1. Using an alternative immunoprecipitation approach, three out of 6 ‘positive’ interacting proteins, ND1, IFITM3 and Ferritin light chain were validated (Figure 1, B). This result was further corroborated by a reverse coimmunoprecipitation experiment (Figure 1, C). We initially focused on the validation of potential PBEF interacting partners in human pulmonary vascular endothelial cells. This is because the disruption of pulmonary endothelial cell barrier occurs during inflammatory disease states such as ALI/ARDS and results in the movement of fluid and macromolecules into the interstitium and pulmonary air spaces, which is the cardinal feature in the pathogenesis of ALI (26). Importantly, our previous findings provided evidence that PBEF is critically involved in thrombin-induced lung endothelial cell barrier dysregulation (5). Further exploration of underlying molecular mechanisms is warranted. These three identified PBEF interacting proteins might help shed light on the PBEF-mediated signal transduction leading to the increased pulmonary vascular dysfunction and the pathogenesis in ALI.
ND1 is among the three PBEF interacting partners validated in pulmonary vascular endothelial cells. ND1 is encoded by the mitochondrial DNA between nucleotide pairs 3307 and 4262 (27). It is one of essential components in the mammalian mitochondrial NADH dehydrogenase (complex I), which is the major entry point for the electron transport chain (ETC) of mitochondrial oxidative phosphorylation. ND1 is involved in electron transfer to ubiquinone. The ETC of the mitochondrial oxidative phosphorylation, the main mechanism for ATP production in mammalian cells, is also the major endogenous source of the ROS (O2 −, H2O2, and OH·), which are toxic by-products of respiration. This is because when the ETC is inhibited, the electrons accumulate in the early stages of the ETC (complex I and CoQ), where they can be donated directly to molecular oxygen to give superoxide anion (O2−) (28). Thus, inhibition of the ETC is likely to increase ROS production and oxidative stress. This study found that human PBEF physically interacts with ND1, a process enhanced by the IL1-β induction (Table 1 and Figure 1). It is possible that the increased interaction between PBEF and other proteins during the IL1-β treatment could simply be a consequence of increased amount of substrate proteins. Increased PBEF-ND1 interaction could consume ND1, reduce its availability from its normal function in the ETC of mitochondria oxidation and thus contribute to increase ROS production and oxidative stress. On the other hand, the possibility exists that increased ROS production is due to the enhancement of NADH oxidase complex I by the increased interaction between PBEF and ND1, which overwhelms the rest of ETC, leading to the overproduction of ROS. This hypothesis is supported by the fact that rotenone, an inhibitor of NADH oxidase complex I (22), significantly attenuated the production of ROS in PBEF overexpressing A549 cells (Figure 3). It warrants further exploration to clarify the precise mechanism. Oxidative stress is known to increase the permeability of the endothelial monolayer to fluid, macromolecules and inflammatory cells (29). Increase permeability of the endothelial monolayer leading to the pulmonary edema is the hallmark of the pathogenesis of ALI. Oxidative stress can lead to cell injury by various mechanisms (30), including modifications of proteins, lipids and DNA. Another PBEF interacting protein, ferritin, can contribute to maintain iron homeostasis in the lung (31). Constant exposure to the atmosphere results in significant exposure of the lungs to catalytically active iron. The lungs have a mechanism for detoxification to prevent its associated generation of oxidative stress. This includes transporting iron intracellularly and sequestering it in an inactive form within ferritin. Thus, ferritin serves as a protective gene by virtue of its antioxidant action. The homeostatic adjustments of ferritin have been shown effective in the protection of the endothelium against the damaging effects of exogenous heme and oxidants (32). By the same token, increased PBEF-Ferritin interaction could consume ferritin, reduce its availability from its normal function in the sequestration of an excess iron and thus contribute to increase iron-associated ROS production and oxidative stress. Indeed, PBEF expression affected intracellular ROS production (Figure 2 and 3). Inhibition of PBEF expression by PBEF siRNA significantly blunted the IL-1β induced ROS production. Overexpression of PBEF significantly increased ROS production over the control and overexpression of PBEF plus IL1-β treatment also significantly augmented ROS production over vector control plus IL1-β treatment. This result suggests that PBEF expression is involved in both baseline and IL-1β induced ROS production. PBEF interaction with ND1 and ferritin may underlie the PBEF involved ROS production. The third PBEF interacting partner, IFITM3, belongs to a human interferon-inducible gene family (33). These genes respond to type I and II interferons and encode for interferon-induced transmembrane protein thought to be involved with the homotypic cell adhesion functions of interferon. The significance of the PBEF-IFITM3 interaction remains to be explored.
The binding mode modeling between PBEF and ND1 (Figure 4) shows that the residues 81–85 of ND1 are indeed located as the interface between ND1 and PBEF, which is consistent with our experimental findings by the BaterioMatch Two Hybrid System screening (Table 1). Our docking results predicted that the following residues of PBEF are within 4.5 A of the residues 81–85 of ND1: F193, D219, S241, V242, P243, A244, A245, E246, T249, I265, H264, Q268, and F269. These residues may directly interact with the residues 81–85 of ND1. Because most of the above interacting residues are hydrophobic, it was predicted that the hydrophobic forces dominate the interactions between the residues 81–85 of ND1 and PBEF. Despite the consistency between our predicted interactions and experimental findings, there exists a limitation in the present model. That is, the crystal structure of ND1 is not available and the ND1 structure was constructed by homology modeling though currently 1U2L is the best template for modeling according to the BLAST program.
In summary, several PBEF interacting partner proteins in human lung were identified using the BacterioMatch Two-Hybrid System. Three of them: NADH dehydrogenase subunit 1 (ND1), ferritin light chain and interferon induced transmembrane 3 (IFITM3) in human pulmonary vascular endothelial cells were validate by coimmunoprecipitation experiments. Their interactions were enhanced in IL-1β-stimulated pulmonary endothelium. PBEF expression affected intracellular ROS production. PBEF interaction with ND1 and ferritin may underlie the PBEF involved ROS production, whose effect could be significantly attenuated by the treatment of rotenone, a NADH oxidase complex I inhibitor. The binding mode modeling between PBEF and ND1 indicates that the residues 81–85 of ND1 are located as the interface between ND1 and PBEF, which is consistent with our experimental findings. These studies suggest that interaction between PBEF and ND1, or ferritin or IFITM3) may in part explain the novel role of PBEF in the pulmonary inflammatory process and pathogenesis of ALI.
Acknowledgements
This work was in part supported by National Heart, Lung, and Blood Institute Grants HL 080042 (for Shui Qing Ye) and the University of Missouri-Columbia Start-up fund.
Abbreviation list
- ALI
acute lung injury
- ARDS
acute respiratory distress syndrome
- A2aR
Adenosine A2a receptor
- ETC
electron transport chain
- EC
endothelial cells
- HPAEC
Human primary pulmonary artery endothelial cells
- HLMVEC
human primary lung microvascular endothelial cells
- IFITM3
interferon induced transmembrane 3
- ND1
NADH dehydrogenase subunit 1
- PBEF
Pre-B-cell colony-enhancing factor
- UCE2L6
γ-glutamyl transferase and ubiquitin conjugating enzyme E2L6
- ROS
reactive oxygen species
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rubenfeld GD, Herridge MS. Epidemiology and outcomes of acute lung injury. Chest. 2007;131:554–562. doi: 10.1378/chest.06-1976. [DOI] [PubMed] [Google Scholar]
- 2.Ashbaugh DG, Bigelow DB, Petty TL, Levine BE. Acute respiratory distress in adults. Lancet. 1967;2:319–323. [Google Scholar]
- 3.Ye SQ, Simon B, Maloney JP, Zambelli-Weiner A, Gao L, Grant A, Easley RB, McVerry B, Tuder RM, Standiford T, Brower R, Barnes K, Garcia JGN. Pre-B-cell colony-enhancing factor as a potential novel biomarker in acute lung injury. Am. J. Respir. Crit. Care Med. 2005;171:361–370. doi: 10.1164/rccm.200404-563OC. [DOI] [PubMed] [Google Scholar]
- 4.Bajwa EK, Yu C, Gong MN, Thompson BT, Christiani DC. PBEF gene polymorphisms influence the risk of developing ARDS. Am J Respir Crit Care Med. 2006;3:A272. [Google Scholar]
- 5.Ye SQ, Zhang LQ, Adyshev D, Usatyuk PV, Garcia AN, Lavoie TL, Verin AD, Natarajan V, Garcia JG. Pre-B-cell-colony-enhancing factor is critically involved in thrombin-induced lung endothelial cell barrier dysregulation. Microvasc Res. 2005;70(3):142–151. doi: 10.1016/j.mvr.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Adyshev DM, Kolosova IA, Verin AD. Potential protein partners for the human TIMAP revealed by bacterial two-hybrid screening. Mol Biol Rep. 2006;33(2):83–89. doi: 10.1007/s11033-005-2311-y. [DOI] [PubMed] [Google Scholar]
- 7.McGlothlin JR, Gao L, Lavoie T, Simon BA, Easley RB, Ma SF, Rumala BB, Garcia JG, Ye SQ. Molecular cloning and characterization of canine pre-B-cell colony-enhancing factor. Biochem Genet. 2005;43(3–4):127–141. doi: 10.1007/s10528-005-1505-2. [DOI] [PubMed] [Google Scholar]
- 8.Singleton PA, Dudek SM, Chiang ET, Garcia JG. Regulation of sphingosine 1-phosphate-induced endothelial cytoskeletal rearrangement and barrier enhancement by S1P1 receptor, PI3 kinase, Tiam1/Rac1, and alpha-actinin. FASEB J. 2005;19(12):1646–1656. doi: 10.1096/fj.05-3928com. [DOI] [PubMed] [Google Scholar]
- 9.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khan JA, Tao X, Tong L. Molecular basis for the inhibition of human NMPRTase, a novel target for anticancer agents. Nat Struct Mol Biol. 2006;13:582–588. doi: 10.1038/nsmb1105. [DOI] [PubMed] [Google Scholar]
- 11.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2006;34:D16–D20. doi: 10.1093/nar/gkj157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lipman DJ, Pearson WR. Rapid and sensitive protein similarity searches. Science. 1985;227:1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- 14.Marti-Renom MA, Stuart A, Fiser A, Sanchez R, Melo F, Sali A. Comparative protein structure modelingof genes and genomes. Annu Rev Biophys Biomol Struct. 2000:29291–29325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 15.Chen R, Weng ZP. A novel shape complementarity scoring function for protein-protein docking. Proteins. 2003;51:397–408. doi: 10.1002/prot.10334. [DOI] [PubMed] [Google Scholar]
- 16.Hattori Y, Nakajima K, Eizawa T, Ehara T, Koyama M, Hirai T, Fukuda Y, Kinoshita M. Heteroplasmic mitochondrial DNA 3310 mutation in NADH dehydrogenase subunit 1 associated with type 2 diabetes, hypertrophic cardiomyopathy, and mental retardation in a single patient. Diabetes Care. 2003;26(3):952–953. doi: 10.2337/diacare.26.3.952. [DOI] [PubMed] [Google Scholar]
- 17.Connelly KG, Moss M, Parsons PE, Moore EE, Moore FA, Giclas PC, Seligman PA, Repine JE. Serum ferritin as a predictor of the acute respiratory distress syndrome. Am J Respir Crit Care Med. 1997;155(1):21–25. doi: 10.1164/ajrccm.155.1.9001283. [DOI] [PubMed] [Google Scholar]
- 18.Lewin AR, Reid LE, McMahon M, Stark GR, Kerr IM. Molecular analysis of a human interferon-inducible gene family. Eur J Biochem. 1991;199(2):417–423. doi: 10.1111/j.1432-1033.1991.tb16139.x. [DOI] [PubMed] [Google Scholar]
- 19.Lukashev DE, Smith PT, Caldwell CC, Ohta A, Apasov SG, Sitkovsky MV. Analysis of A2a receptor-deficient mice reveals no significant compensatory increases in the expression of A2b, A1, and A3 adenosine receptors in lymphoid organs. Biochem Pharmacol. 2003;65(12):2081–2090. doi: 10.1016/s0006-2952(03)00158-8. [DOI] [PubMed] [Google Scholar]
- 20.Carlisle ML, King MR, Karp DR. Gamma-glutamyl transpeptidase activity alters the T cell response to oxidative stress and Fas-induced apoptosis. Int Immunol. 2003;15(1):17–27. doi: 10.1093/intimm/dxg010. [DOI] [PubMed] [Google Scholar]
- 21.Ardley HC, Rose SA, Tan N, Leek JP, Markham AF, Robinson PA. Genomic organization of the human ubiquitin-conjugating enzyme gene, UBE2L6 on chromosome 11q12. Cytogenet Cell Genet. 2000;89(1–2):137–140. doi: 10.1159/000015593. [DOI] [PubMed] [Google Scholar]
- 23.Cusick ME, Klitgord N, Vidal M, Hill DE. Interactome: gateway into systems biology. Hum Mol Genet. 2005;14(Spec No 2):R171–R181. doi: 10.1093/hmg/ddi335. [DOI] [PubMed] [Google Scholar]
- 24.Oliver S. Guilt-by-association goes global. Nature. 2000;403:601–603. doi: 10.1038/35001165. [DOI] [PubMed] [Google Scholar]
- 25.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 26.Dudek SM, Garcia JG. Cytoskeletal regulation of pulmonary vascular permeability. J Appl Physiol. 2001;91(4):1487–1500. doi: 10.1152/jappl.2001.91.4.1487. [DOI] [PubMed] [Google Scholar]
- 27.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature. 1981;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 28.Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 29.Houle F, Huot J. Dysregulation of the endothelial cellular response to oxidative stress in cancer. Mol Carcinog. 2006;45(6):362–367. doi: 10.1002/mc.20218. [DOI] [PubMed] [Google Scholar]
- 30.Crimi E, Sica V, Slutsky AS, Zhang H, Williams-Ignarro S, Ignarro LJ, Napoli C. Role of oxidative stress in experimental sepsis and multisystem organ dysfunction. Free Radic Res. 2006;40(7):665–672. doi: 10.1080/10715760600669612. [DOI] [PubMed] [Google Scholar]
- 31.Ghio AJ, Turi JL, Yang F, Garrick LM, Garrick MD. Iron homeostasis in the lung. Biol Res. 2006;39(1):67–77. doi: 10.4067/s0716-97602006000100008. [DOI] [PubMed] [Google Scholar]
- 32.Balla J, Vercellotti GM, Jeney V, Yachie A, Varga Z, Eaton JW, Balla G. Heme, heme oxygenase and ferritin in vascular endothelial cell injury. Mol Nutr Food Res. 2005;49(11):1030–1043. doi: 10.1002/mnfr.200500076. [DOI] [PubMed] [Google Scholar]
- 33.Lewin AR, Reid LE, McMahon M, Stark GR, Kerr IM. Molecular analysis of a human interferon-inducible gene family. Eur J Biochem. 1991;199:417–423. doi: 10.1111/j.1432-1033.1991.tb16139.x. [DOI] [PubMed] [Google Scholar]
- 34.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera - A Visualization System for Exploratory Research and Analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 35.Sanner MF, Olson AJ, Spehner JC. Reduced surface: an efficient way to compute molecular surfaces. Biopolymers. 1996;38:305–320. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]


