Abstract
A systematic examination of the impact of the yatakemycin left and right subunits and their substituents is detailed along with a study of its unique three subunit arrangement (sandwiched vs extended and reversed analogues). The examination of the ca. 50 analogues prepared illustrate that within the yatakemycin three subunit structure, the subunit substituents are relatively unimportant and that it is the unique sandwiched arrangement that substantially increases the rate and optimizes the efficiency of its DNA alkylation reaction. This potentiates the cytotoxic activity of yatakemycin and its analogues overcoming limitations typically observed with more traditional compounds in the series (CC-1065, duocarmycins). Moreover, a study of the placement of the alkylation subunit within the three subunit arrangement (sandwiched vs extended and reversed analogues) indicates that it has a profound impact not only on the rate and efficiency of DNA alkylation, but that it controls and establishes the DNA alkylation selectivity as well, where both enantiomers of such sandwiched agents alkylate the same adenine sites exhibiting the same DNA alkylation selectivity independent of their absolute configuration.
Introduction
Yatakemycin (1)1 is the newest and most potent (IC50, L1210 = 3–5 pM) member of a family of antitumor antibiotics that includes CC-1065 (2, IC50 = 20 pM),2 duocarmycin A (3, IC50 = 200 pM),3 and duocarmycin SA (4, IC50 = 10 pM, Figure 1).4 Each derives its properties through a characteristic sequence-selective alkylation of duplex DNA.5–9 The yatakemycin structure is a remarkable hybrid of the preceding natural products, incorporating a central DNA alkylation subunit identical to that of duocarmycin SA, a left subunit identical to the DNA-binding subunits found in CC-1065, and a right subunit similar to that found in the duocarmycins. Distinct from the preceding agents, yatakemycin is the first naturally occurring member possessing a DNA binding subunit flanking each side of the alkylation subunit in a characteristic “sandwiched” arrangement. Recently, we disclosed first and second generation total syntheses10–13 of yatakemycin that served to reassign its structure as 110 and reported preliminary evaluations of (+)- and ent-(−)-yatakemycin and key partial structures.11
Figure 1.

Natural products.
Prior to the discovery of yatakemycin, we had utilized the C-terminus methyl ester of duocarmycin SA as a site for attachment of a second DNA binding subunit providing the first, albeit limited, series of such sandwiched analogues that were found to exhibit potent cytotoxic activity, an enhanced DNA alkylation rate, and a uniquely altered alkylation selectivity.14 The subsequent isolation of a naturally occurring member of this class has focused a more intense interest in such sandwiched compounds and now provides the incentive to more thoroughly evaluate this unusual structural arrangement. Herein, we report efforts to systematically define the relationships between structure, reactivity, and biological activity of such sandwiched agents.
Extensive efforts have defined the structural features of the duocarmycins that contribute to their remarkable rate, selectivity, and efficiency of DNA alkylation.9 In these studies, the right-hand DNA binding subunit was identified as one that not only provides noncovalent DNA binding affinity and selectivity, but is also responsible for catalysis of the DNA alkylation reaction (Figure 2).14,15 The alkylation subunit cyclopropane is stabilized by the cross-conjugated delocalization of the N2 lone pair via a vinylogous amide. Upon binding in the minor groove, the compound is forced to adopt a twisted conformation to accommodate the helical pitch of DNA, which disrupts the vinylogous amide conjugation and activates the cyclopropane for nucleophilic attack. Through use of this DNA binding-induced activation (shape-dependent catalysis), the compound avoids reaction with competitive nucleophiles until being selectively activated by and at its biological target.16
Figure 2.

DNA alkylation catalysis and substituent effects on DNA alkylation efficiency, rate, and biological potency
Results and Discussion
Synthesis of Analogues with Modifications in the Right Subunit
Studies conducted on duocarmycin SA have indicated that only the C5-methoxy substituent of the DNA binding subunit has a pronounced effect on the rate and efficiency of DNA alkylation, and the biological potency of the compounds.9,14 These effects are independent of the substituent electronic properties and could be attributed simply to its presence and the resulting increase in rigid length of the agent.17 This rigid extension increases the extent of the DNA binding-induced conformational change, increasing the degree of vinylogous amide disruption and enhancing the rate of DNA alkylation. Consequently, a series of key yatakemycin analogues were synthesized to establish the impact of its right subunit substituents in such sandwiched structures.
In route to these key analogues and for comparison purposes, the partial structures 9a–9f were first prepared and lack the left subunit of yatakemycin (Scheme 1). As such, they represent not only key partial structures of yatakemycin, but also additional key analogues of duocarmycin SA itself. Boc deprotection of each enantiomer of 611 (only natural enantiomer shown, 4 N HCl–EtOAc, 70 °C, 1 h), followed by coupling with the corresponding indole-2-carboxylic acid (5a–f, 4 equiv of EDCI, 3 equiv of NaHCO3, DMF, 25 °C, 14 h, 62–78%) gave 7. Benzyl ether deprotection of 7 (1 atm H2, 10% Pd/C, 9:1 THF–MeOH, 2 h, 80–95%) provided 8, and subsequent spirocyclization (sat. aq. NaHCO3 in 2:1 DMF–H2O, 25 °C, 1 h, 64–89%) provided (+)- and ent-(−)-9a–f.
Scheme 1.

The synthesis of sandwiched analogues 14b–f was accomplished as shown in Scheme 2. Methyl ester hydrolysis of 7 (only natural enantiomer shown, 4 equiv of LiOH, 3:2:1 THF–MeOH–H2O, 25 °C, 14 h, 78–94%) followed by benzyl ether deprotection of 11 (1 atm H2, 10% Pd/C, 9:1 THF–MeOH, 2 h, 84–100%) gave 12. Coupling of 12 with the hydrochloride salt of 1011 (4 equiv of EDCI, DMF, 25 °C, 16 h, 42–77%) furnished 13, and subsequent spirocyclization (sat. aq. NaHCO3 in 2:1 DMF–H2O, 25 °C, 1 h, 63–98%) provided (+)- and ent-(−)-14b–f.
Scheme 2.
Synthesis of Analogues with Modifications in the Left Subunit
Yatakemycin is structurally unique among members of its class, possessing the additional left-hand DNA binding subunit. A series of left subunit analogues were prepared to establish its contribution to yatakemycin’s properties. In previous studies, the CDPI18 and indole subunits (Scheme 3) were found to constitute unsubstituted and simplified replacements for the PDE subunits found in CC-1065 that are readily accessible by chemical synthesis, maintain the full cytotoxic potency of the natural products and impart more efficacious in vivo activity.19 Thus, yatakemycin analogues incorporating these and related subunits were prepared for evaluation and constitute analogues in which the left-hand subunit substituents or key structural features are removed.
Scheme 3.

The synthesis of 16a–b incorporating a pyrrolidine in place of the left subunit was accomplished as shown in Scheme 3. Coupling of carboxylic acids 12a and 12c with pyrrolidine hydrochloride (1.0 equiv, 4 equiv of EDCI, DMF, 25 °C, 16 h, 49–65%) gave 15a–b, and subsequent spirocyclization (sat. NaHCO3 in 2:1 DMF–H2O, 25 °C, 1 h, 58–87%) provided (+)- and ent-(−)-16a–b.
The synthesis of yatakemycin analogues incorporating additional modifications in the left subunit was accomplished as shown in Schemes 4 and 5. Coupling of carboxylic acid 12a with the CDPI-based amine hydrochlorides derived from 1718 or 18 (1.0 equiv, 4 equiv of EDCI, DMF, 25 °C, 16 h, 37–41%) gave 19a–b, and subsequent spirocyclization (sat. NaHCO3 in 2:1 DMF–H2O, 25 °C, 0.7 h, 66–91%) provided (+)- and ent-(−)-20a–b. Similarly, coupling of carboxylic acid 12f with 21 (1.0 equiv, 4 equiv of EDCI, DMF, 25 °C, 24 h, 53%) gave 22, and subsequent spirocyclization (sat. NaHCO3 in 2:1 DMF–H2O, 25 °C, 1 h, 93%) provided (+)- and ent-(−)-23.
Scheme 4.
Scheme 5.

Design and Synthesis of Extended, Sandwiched, and Reversed Analogues
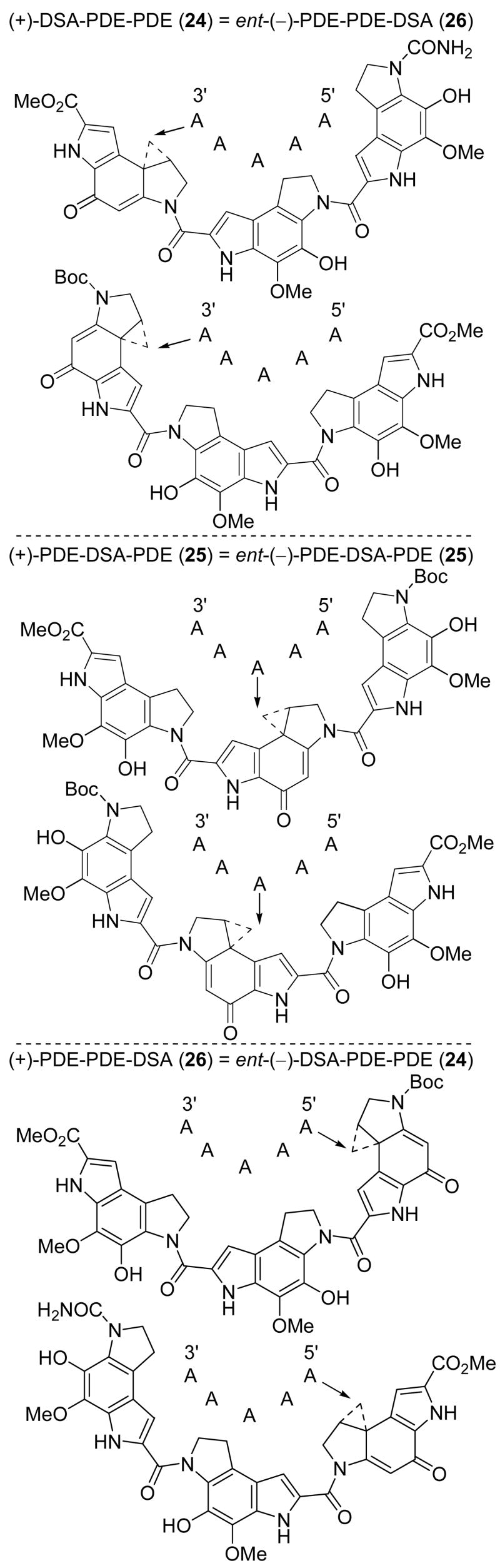
Extended analogue 24, sandwiched analogue 25, and reversed analogue 26 were prepared to establish the importance of the agent structure and the location of the DNA binding subunits on the DNA alkylation selectivity (Figure 3). Each analogue in this series is composed of the alkylation subunit of duocarmycin SA and two PDE subunits attached in each of three possible arrangements. Extended analogue 24 contains the same DNA binding subunits organized in the same arrangement as found in CC-1065, but incorporates the more stable (4–5-fold) and more potent (4–5-fold) duocarmycin SA alkylation subunit.20 As such, it would be expected to alkylate DNA with the same selectivity as CC-1065, but to be a more efficient DNA alkylating agent and a more potent cytotoxic compound.21 Moreover, the two enantiomers of 24, like CC-1065, would be expected to exhibit characteristic and distinguishable alkylation selectivities constituting an adenine N3 alkylation at the terminus of a five base-pair AT-rich sequence albeit with opposite bound orientations and with that of the unnatural enantiomer offset by one base pair (i.e., nat. 5′-AAAAA vs unnat. 5′-AAAAA).6d Sandwiched analogue 25 constitutes yatakemycin modified with incorporation of a PDE right-hand subunit. As such, the properties of 25 would be expected to be comparable to those of yatakemycin, both enantiomers of which exhibit identical alkylation selectivities (i.e., 5′-AAAAA)11 and indistinguishable cytotoxic potencies. Reversed analogue 26 has two PDE subunits bound to the left of the alkylation subunit, and is capped on the right as its Boc derivative. Unlike the extended and sandwiched agents, the reversed agent lacks an extended heteroaromatic N2 acyl substituent on the alkylation subunit to participate in a binding-induced activation of the alkylation reaction. Consequently, both its rate and efficiency of DNA alkylation would be expected to be diminished relative to 24 and 25, resulting in a diminished cytotoxic potency. Just as significantly and because the DNA binding subunits extend in opposite directions relative to those found in the extended analogue 24, they should exhibit a reversed enantiomeric alkylation selectivity if it is controlled by the noncovalent binding selectivity of the compounds. As such, the natural enantiomer of 26 would be expected to alkylate the same sites as the unnatural enantiomer of 24, whereas its unnatural enantiomer should alkylate DNA with a selectivity indistinguishable from the natural enantiomer of 24.14 Significantly, all three agents have comparable DNA-binding surfaces and the same inherent reactivity, thereby allowing a direct comparison of the impact of the subunit arrangement on their DNA alkylation profile and biological properties.
Figure 3.
Extended, reversed, and sandwiched analogues.
The synthesis of extended analogue 24 was accomplished as shown in Scheme 6. Acid-catalyzed Boc deprotection of 2711 (4 N HCl–EtOAc, 25 °C, 3 h) followed by coupling with 2822 (1.0 equiv, 4 equiv of EDCI, DMF, 25 °C, 22 h, 36%), and subsequent spirocyclization (sat. aq. NaHCO3 in 2:1 DMF–H2O, 25 °C, 0.5 h, 78%) provided (+)- and ent-(−)-24. Sandwiched analogue 25 was prepared by the strategy outlined in Scheme 7. Boc deprotection of 611 (4 N HCl–EtOAc, 25 °C, 3 h) followed by coupling with 3011,22 (1.0 equiv, 4 equiv of EDCI, DMF, 25°C, 14 h, 81%) provided 32. Methyl ester hydrolysis (4 equiv of LiOH, 3:2:1 THF–MeOH–H2O, 25 °C, 5 h, 96%) followed by benzyl ether deprotection of (1 atm H2, 10% Pd/C, 9:1 THF–MeOH, 1 h, 93%) gave 34. Coupling of 34 with the hydrochloride salt of 3111 (4 equiv of EDCI, DMF, 25 °C, 16 h, 69%) furnished 35, and subsequent spirocyclization (sat. aq. NaHCO3 in 2:1 DMF–H2O, 25 °C, 1 h, 89%) provided (+)- and ent-(−)-25. The reversed analogue 26 (Scheme 8) was prepared by Boc deprotection of 3622 (4 N HCl–EtOAc, 25 °C, 0.5 h) followed by coupling with 3714b (1.0 equiv, 4 equiv of EDCI, DMF, 25 °C, 16 h, 67%), and subsequent spirocyclization (sat. aq. NaHCO3 in 2:1 DMF–H2O, 25 °C, 40 m, 68%) to provide (+)- and ent-(−)-26. Partial structure 40 having the same two subunit arrangement as duocarmycin SA incorporating a single PDE subunit was also synthesized by benzyl ether deprotection of 32 (1 atm H2, 10% Pd/C, 9:1 THF–MeOH, 2 h, 93%) followed by spirocyclization (sat. aq. NaHCO3 in 2:1 DMF–H2O, 25 °C, 1 h, 96%) to provide (+)- and ent-(−)-40 (Scheme 9).
Scheme 6.
Scheme 7.
Scheme 8.
Scheme 9.
Cytotoxic Activity
Summarized in Figure 4 is the cytotoxic activity of the series of right subunit analogues against a cell line (L1210 mouse leukemia) for which we have extensive comparisons for this class. Compound 14b differs from yatakemycin only by switching the right subunit substituent locations, and exhibited cytotoxic potency that was indistinguishable from the natural product or its unnatural enantiomer. Analogue 14c is a hybrid structure consisting of yatakemycin’s three subunit sandwiched arrangement incorporating the 5,6,7-trimethoxyindole subunit found in the duocarmycins. Compound 14c (IC50 = 5 pM) and ent-(−)-14c (IC50 = 5 pM) were found to be potent cytotoxic agents, indistinguishable in the cellular assay and equipotent with (+)- and ent-(−)-yatakemycin.11 Both compounds, like yatakemycin, are 2-fold more potent than duocarmycin SA (4, IC50 = 10 pM) and 20-fold more potent than ent-(−)-duocarmycin SA. The substantial increase in potency for the unnatural enantiomer relative to duocarmycin SA is a characteristic feature observed with the sandwiched agents. Presumably, the added left subunit contributes sufficient additional binding affinity23 to compensate for the less effective DNA alkylation characteristic of the unnatural enantiomer of duocarmycins.
Figure 4.
L1210 Cytotoxic activity.
The contribution of each right subunit substituent was determined by evaluation of a series of key simplified analogues. Yatakemycin partial structure 9a (IC50 = 10 pM) served as a valuable baseline comparison for identifying the contribution of the left subunit for each analogue. Both enantiomers of 14d lacking only the C6-methoxy group were equipotent (IC50 = 5 pM) and indistinguishable from yatakemycin. In contrast, the corresponding partial structure 9d (IC50 = 30 pM) was 6-fold less potent than yatakemycin and 3-fold less potent than 9a, and its unnatural enantiomer (IC50 = 600 pM) was considerably less active. Similarly, 14e lacking the C5-hydroxy group (IC50 = 4 pM; ent-(−)-14e, IC50 = 10 pM) was essentially indistinguishable from yatakemycin, whereas the corresponding partial structure 9e (IC50 = 25 pM) was 5-fold less potent than yatakemycin, 2–3 times less active than 9a, and its unnatural enantiomer (IC50 = 1300 pm) was 260-fold less potent than the sandwiched structures.
A particularly interesting analogue was 14f, incorporating a right subunit indole lacking additional substituents. The natural enantiomer was essentially equipotent with yatakemycin (IC50 = 6 pM) despite lacking a C5 substituent. Distinct from the preceding sandwiched analogues, the unnatural enantiomer of 14f (IC50 = 35 pM) was 6-fold less potent than the natural enantiomer, and 7-fold less potent than the ent-(−)-yatakemycin. The corresponding partial structure 9f14b was found to be 10-fold less potent (IC50 = 60 pM) than 14f and 6-fold less potent than 9a, whereas the unnatural enantiomer ent-(−)-9f was nearly 50-fold less potent (IC50 = 1700 pM) than ent-(−)-14f and 4–5 fold less potent than ent-(−)-9a. Clear from these comparisons is the observation that the incorporation of a left subunit, which increases the DNA binding affinity of the sandwiched analogues, compensates for less effective right subunits, and substantially increases the cytotoxic potency of the unnatural enantiomers relative to the corresponding partial structures incorporating only the central and right-hand subunits. Only with the combination of a less effective right-hand subunit and the unnatural enantiomeric configuration in ent-(−)-14f was a detectable difference in cytotoxic activity observed between enantiomers. Even here, the difference in potency between the two enantiomers remains remarkably small (5-fold) relative to the typical partial structures incorporating only the central and right-hand subunits (10–40-fold). An immediate conclusion to these studies is that the right-hand subunit substituents significantly and predictably impact the cytotoxic activity of duocarmycin-like compounds (series 9) especially improving the potency of the unnatural enantiomers,14 but they have a minor impact on the activity of yatakemycin.
Compound 40 is the most potent compound in the partial structure series (IC50 = 5 pM), being approximately 2-fold more potent than duocarmycin SA and equipotent with yatakemycin and 25 (IC50 = 6 pM). However, the unnatural enantiomer of 40 (IC50 = 360 pM) is still 60-fold less potent than the unnatural enantiomer of the corresponding sandwiched agent (ent-(−)-25, IC50 = 6 pM).
As shown in Figure 5, a separate series of compounds were evaluated to establish the impact of the left subunit. Its removal altogether providing 9a afforded an analogue that was not distinguishable from duocarmycin SA and that was 2-fold (natural enantiomer) or 70-fold (unnatural enantiomer) less potent than yatakemycin and its unnatural enantiomer. The CDPI subunit lacking the methoxy and hydroxyl substituents was incorporated into analogue 20a (IC50 = 5 pM, ent-(−)-20a IC50 = 6 pM) which was found to be equipotent with yatakemycin. The thiomethyl ester derivative of CDPI was similarly incorporated into analogue 20b (IC50 = 7 pM, ent-(−)-20b IC50 = 6 pM) with no change in potency. Thus, like observations made with yatakemycin itself,11 no unique effect was detected for the presence of the methyl thioester. To probe the impact of the pyrrolidine ring alone, 16a was prepared which significantly reduces the size and length of the left subunit. The cytotoxic potency of the natural enantiomer (IC50 = 7 pM) was comparable to 1 and perhaps a bit more potent than duocarmycin SA and 9a. The unnatural enantiomer was 10-fold less potent (IC50 = 70 pM), but notably 5-fold more potent than the corresponding methyl ester ent-(−)-9a (IC50 = 400 pM). Pyrrolidine analogue 16b (IC50 = 7 pM; ent-(−)-16b IC50 = 80 pM) possessing a 5,6,7-trimethoxyindole right subunit was equipotent with 16a. The small increase in agent length and rigidity provided by the pyrrolidine amide results in an increase in the potency of the unnatural enantiomer although a significant difference between each enantiomer’s potency remains. The sandwiched analogue with two indole subunits 23 (IC50 = 15 pM) exhibited 3-fold reduced cytotoxic activity relative to yatakemycin or its indole analogue 14f, but it was still nearly as potent as duocarmycin SA (IC50 = 10 pM). Nonetheless, the indole left subunit contributes sufficiently to exceed the potency of 9f (IC50 = 60 pM). The unnatural enantiomer of 23 was approximately 20-fold less potent (IC50 = 275 pM) than the natural enantiomer, 50-fold less potent than yatakemycin and its enantiomer, and 8-fold less potent than the unnatural enantiomer of 14f. Clearly, an optimal right subunit can compensate for substantial simplifications in the left subunit and this is especially true in the natural enantiomer series.
Figure 5.

L1210 Cytotoxic activity.
The cytotoxic activity of the extended, sandwiched, and reversed agents is summarized in Figure 6. The unnatural enantiomers of CC-106522 and yatakemycin11 exhibit cytotoxic potency that is indistinguishable from the natural enantiomers, which is distinct from the behavior of duocarmycin SA where the unnatural enantiomer is approximately 10-fold less active than the natural enantiomer.20 The presence of two DNA binding subunits appears to compensate for the reduced potency that is characteristic of the unnatural enantiomer of duocarmycin SA. The extended agent 24 (IC50 = 4 pM) was essentially equivalent in potency with its unnatural enantiomer (IC50 = 6 pM), indistinguishable in potency with yatakemycin, but 4–5-fold more potent than CC-1065. The greater potency of 24 relative to CC-1065 can be attributed to the incorporation of the more stable and more potent duocarmycin SA alkylation subunit.21 The sandwiched analogue (+)-25 (IC50 = 6 pM) was equipotent with its unnatural enantiomer ent-(−)- 25 (IC50 = 6 pM) and both enantiomers of yatakemycin (IC50 = 5 pM). In contrast, the reversed analogues were approximately 10-fold less potent than the preceding analogues ((+)-26, IC50 = 40 pM; ent-(−)-26, IC50 = 60 pM) but displayed similar potencies. The reversed agents lack an extended N2-acyl substituent that has been shown to be largely responsible for catalysis of the DNA alkylation reaction15 such that its removal results in less effective DNA alkylating agents as shown below.
Figure 6.

Cytotoxic activity (L1210) and relative rates (krel) and efficiencies of DNA alkylation (w836).
Relative Rate and Efficiency of DNA Alkylation
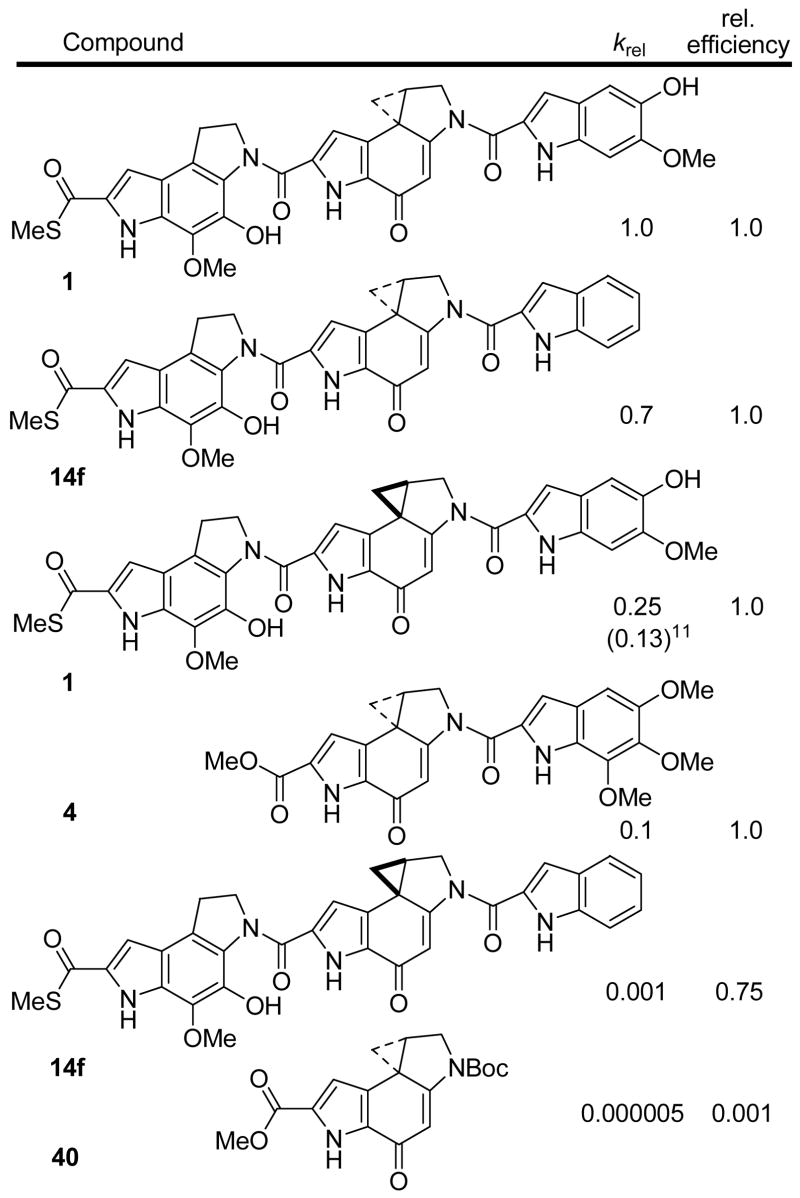
To date, the biological properties of members of this class of natural products have typically mirrored their relative efficiencies of DNA alkylation. For the sandwiched analogues explored herein which contain three subunits, little or no differences in the final efficiencies of DNA alkylation were observed even though they often incorporated suboptimal DNA binding subunits or even possess the unnatural absolute configuration. These observations mirror the near equivalent cytotoxic activities observed within the 14 and 20 series and even between enantiomeric pairs of such analogues. The exceptions to these generalizations include only the unnatural enantiomers of the compounds bearing the simplest, unsubstituted indole right-hand subunit which proved to alkylate DNA with a marginally reduced efficiency, Figure 7. However, these differences are magnified if one examines the relative rates versus final efficiencies of DNA alkylation. Here, readily detectable differences in the rates of DNA alkylation are observed with each enantiomeric pair (natural > unnatural enantiomer) as well as between pairs of analogues. Illustrative of these differences and in a comparison that highlights the importance and role of the right-hand subunit, the relative rates of DNA alkylation in w836 DNA conducted at 5 °C for each enantiomer of yatakemycin were compared with those of 14f bearing the unsubstituted indole as its right-hand subunit, Figure 7.
Figure 7.
Relative rates (krel) and efficiencies of DNA alkylation (w836 DNA).
As observed with yatakemycin, the natural enantiomer of 14f alkylates DNA much faster than its unnatural enantiomer (700-fold) and for each enantiomer, yatakemycin alkylates DNA faster than 14f (1.5-fold for natural enantiomer and ca. 200-fold for unnatural enantiomer). Consistent with its role in catalysis, the right-hand subunit substituents (C5 substituent)14,17 increase the rate of DNA alkylation and this effect is most pronounced with the unnatural enantiomers.17
Notably, the natural enantiomer of 14f incorporating a suboptimal right-hand subunit and even the unnatural enantiomer of yatakemycin alkylate DNA faster than the natural enantiomer of duocarmycin SA. In addition to highlighting the special characteristics of such sandwiched compounds, these studies illustrate that the incorporation of the yatakemycin left-hand subunit can compensate for suboptimal right-hand subunits (for catalysis) or structural features (unnatural enantiomer configuration) that would limit the behavior of typical two subunit members of this class including duocarmycin SA. So much so, that even the loss of 1,000-fold in the rate of DNA alkylation with the unnatural enantiomer of 14f relative to yatakemycin itself provides a very effective DNA alkylating agent whose DNA alkylation efficiency and resulting cytotoxic potency approach that of the natural product.
A second set of comparisons is summarized in Figure 6 entailing the PDE-based sandwiched, extended, and reversed analogues of yatakemycin which further illustrate the importance and role of the right-hand subunit to the catalysis of the DNA alkylation reaction. Both enantiomers of the sandwiched analogue 25 alkylated DNA at extraordinary rates comparable to yatakemycin itself, and the natural enantiomer of the extended analogue 24 also alkylated DNA at a rate that was intermediate of these two enantiomers. By contrast, the unnatural enantiomer of the extended analogue 24 was considerably slower (100-fold), but ultimately achieved the same efficiency of DNA alkylation. Although the reversed natural enantiomer was faster than its unnatural enantiomer, neither comes close to the rates exhibited by the sandwiched or extended analogues 24 and 25 and more closely approximate of the rates of DNA alkylation observed with N-Boc-DSA which lacks the DNA binding subunits altogether.8 Several important generalizations are illustrated with these comparisons including: 1) the observation of natural enantiomer > unnatural enantiomer rates within each series and this was especially pronounced for the extended analogue (100-fold) versus the sandwiched (2–4 fold) and reversed (3-fold) analogues, and 2) the relative DNA alkylation rates follow the order sandwiched > extended ≫ reversed analogues for each enantiomeric series. By far, the most striking observation is the extraordinarily slow rates (ca. 1,000-fold) and the reduced efficiency (10-fold) of DNA alkylation for the reversed analogues that may be attributed to their lack of an alkylation subunit N2-substituent that can participate in the binding-induced catalysis derived from disruption of the alkylation subunit vinylogous amide.14
DNA Alkylation Selectivity
The DNA alkylation selectivity of the yatakemycin analogues was examined within five 150 base-pair segments of DNA described previously.24 The alkylation site identification and the assessment of the relative selectivity among the available sites were obtained by thermally-induced strand cleavage of the singly 5′-end-labeled duplex DNA after exposure to the compounds as detailed.5–8 Since the DNA alkylation properties of members of each class of these agents have been established in preceding studies including a description of the unaltered alkylation selectivity of (+)- and ent-(−)-yatakemycin within nucleosome packaged DNA5b as well as a detailed comparison of the DNA alkylation properties of (+)- and ent-(−)-yatakemycin,11 we focused our analysis on a select set of the new yatakemycin analogues. Most representative of this set are the PDE-based sandwiched, extended, and reversed analogues of yatakemycin (24–26). Here, the placement of the alkylation subunit within the trimer structure has a profound and controlling impact on the DNA alkylation selectivity, as well as exerts an influence on the efficiency of DNA alkylation, and exhibits a unique impact on the rate of DNA alkylation that was detailed in the preceding section.
Illustrated in Figure 8 is a representative comparison of the DNA alkylation selectivity of the PDE-based sandwiched, extended, and reversed analogues of yatakemycin within a six base-pair A-rich site found in w836 DNA which beautifully highlights the distinctions in the compounds. As might be anticipated, the PDE-based sandwiched analogue 25 exhibited a DNA alkylation selectivity essentially identical to the natural product yatakemycin itself. Most significant in this comparison is the observation that both enantiomers of PDE-DSA-PDE (25), like both enantiomers of yatakemycin, alkylate the same site(s) (adenines central to the six base-pair A-rich site, 5′-AAAAAA-3′) with essentially the same relative efficiency.11
Figure 8.

Thermally-induced strand cleavage of w836 DNA (146 bp, nucleotide no. 5189-91) after DNA–agent incubation with DSA-PDE-PDE (24), PDE-DSA-PDE (25), and PDE-PDE-DSA (26, 23 h, 25 °C), removal of unbound agent by EtOH precipitation and 30 min thermolysis (100 °C), followed by denaturing 8% PAGE and autoradiography. Lanes 1 and 2, (+)-yatakemycin and ent-(−)-yatakemycin (1 × 10−5 M); lanes 3 and 4, (+)-PDE-DSA-PDE (25) and ent-(−)-PDE-DSA-PDE (25, 1 × 10−5 M); lane 5, control DNA; lanes 6–9, Sanger G, C, A, and T sequencing standards; lanes 10 and 11, (+)-DSA-PDE-PDE (24) and ent-(−)-DSA-PDE-PDE (24, 1 × 10−5 M); lanes 12 and 13, (+)-PDE-PDE-DSA (26) and ent-(−)-PDE-PDE-DSA (26, 1 × 10−4 M).
In contrast, the two enantiomers of the extended analogue 24, a close analogue of CC-1065 incorporating the yatakemycin alkylation subunit, exhibited very distinct alkylation selectivities which were both markedly different from yatakemycin and its enantiomer. The natural enantiomer (+)-DSA-PDE-PDE (24) alkylated the two adenines at the 3′ end of six base-pair A-rich site (5′-AAAAAA-3′) consistent with alkylation only at the 3′-end of a five base-pair AT-rich site.6 In contrast, the unnatural enantiomer alkylated the three adenines at the opposite 5′-end of the site (5′-AAAAAA-3′). These non overlapping alkylated adenines for the two enantiomers of 24 represent opposite bound orientations (3′→5′ for nat. enantiomer and 5′→3′ for unnat. enantiomer) of the compounds across a five base-pair AT-rich site starting with (nat. enantiomer)6 or preceding (unnat. enantiomer)6c the alkylated adenine.
Most remarkable in this set of comparisons is the behavior of the two enantiomers of the reversed analogues PDE-PDE-DSA (26). Although much less efficient than preceding analogues even with extended reaction times (≥10-fold, requiring 10−4 vs 10−5 M concentrations), the two enantiomers of the reversed analogues exhibited distinct DNA alkylation selectivities that were not only different from that of the sandwiched compounds and their enantiomers, but were also opposite that of extended analogues 24. That is, the natural enantiomer of the reversed analogue exhibited the identical selectivity as the unnatural enantiomer of the extended analogue 24 alkylating the adenines at the 5′-end of the site, whereas its unnatural enantiomer alkylated the same 3′ adenines as the extended natural enantiomer 24, Figure 8 and 9. This complete reversal of the enantiomeric selectivity14,25 between the extended versus reversed analogues indicates that it is not the alkylation subunit and its absolute configuration that dominates the alkylation selectivity, rather that it is the compound’s AT-rich noncovalent binding selectivity23 that delivers the modestly reactive electrophile to accessible nucleophiles within a noncovalent binding site.14
Figure 9.
Alkylation selectivities.
This distinct reversed enantiomer behavior of the extended versus reversed analogues is also and even more clearly illustrated in Figure 10. Within w794 DNA, the natural and unnatural enantiomers of the extended analogues 24 each alkylate a single distinct site within the 150 base-pair segment of DNA consistent with observations first made while examining CC-1065 and its unnatural enantiomer.25 Examination of the unnatural enantiomer of the reversed analogue 26 alongside both enantiomers of the extended analogue 24 illustrate that it alkylates the natural enantiomer site (ent-(−)-PDE-PDE-DSA = (+)-DSA-PDE-PDE) beautifully highlighting this reversal of the enantiomer selectivity between the extended and reversed analogues.
Figure 10.

Thermally-induced strand cleavage of w794 DNA (144 bp, nucleotide no. 5238-138) after DNA–agent incubation with DSA-PDE-PDE (24) (25 °C, 5 days) and PDE-PDE-DSA (26) (37 °C, 5 days), removal of unbound agent by EtOH precipitation and 30 min. thermolysis (100 °C), followed by denaturing 8% PAGE and autoradiography. Lane 1, control DNA; lanes 2-5, Sanger G, C, A, and T sequencing standards; lane 6, (+)-DSA-PDE-PDE (24, 25 °C, 1×10−5 M); lane 7, ent-(−)-DSA-PDE-PDE (24, 25 °C, 1×10−5 M); lane 8, ent-(−)-PDE-PDE-DSA (26, 37 °C, 1×10−4 M).
Models of the Alkylation Products
Presented in Figure 11 are models of the sandwiched, extended, and reversed PDE-based analogues alkylation of their predominant w836 sites (see Figure 8) that visually illustrate the selectivity features discussed earlier. Most notable in the comparisons is the fact that all six compounds (two enantiomers of three compounds) bind across the identical five base-pair A-rich site with each enantiomer pair binding with opposite orientations and undergoing alkylation at the resulting adenine proximal to the reactive cyclopropane. The two enantiomers of the sandwiched analogues 25 alkylate the same adenine central to the site, the natural enantiomer of the extended analogue 24 and the unnatural enantiomer of the reversed analogue 26 alkylate the same adenine at the 3′ end of the site, whereas the unnatural enantiomer of the extended analogue 24 and the natural enantiomer of the reversed analogue 26 alkylate an adenine at the opposite 5′ end of the site (Figure 12).
Figure 11.

a) Models of (+)-DSA-PDE2 (left) and ent-(−)-DSA-PDE2 (right). b) Models of (+)-PDE-DSA-PDE (left) and ent-(−)-PDE-DSA-PDE (right). c) Models of (+)-PDE2-DSA (left) and ent-(−)-PDE2-DSA (right). See Supporting Information for larger images of the models.
Figure 12.
Graphic representation of bound orientations and alkylation sites for the extended, sandwiched, and reversed analogues.
Conclusions
A systematic examination of the yatakemycin left and right-hand subunits and their substituents is detailed establishing their impact on the cytotoxic potency and DNA alkylation properties of such sandwiched compounds. Within the context of this three subunit structural arrangement, the studies illustrate that the subunit substituents are now relatively unimportant and that the yatakemycin left-hand subunit, as well as those incorporating significant simplifications, can compensate for suboptimal right-hand subunits and substituents or structural features (i.e. unnatural enantiomer configuration) that limit the more traditional compounds in the series. Significantly, this sandwiched subunit arrangement substantially increases the rate of DNA alkylation, maximizes the efficiency of DNA alkylation, and potentiates the cytotoxic potency of the compounds. Moreover, the placement of the alkylation subunit within the three subunit structure has a profound impact on not only the rate (PDE-DSA-PDE > DSA-PDE2 ≫ PDE2-DSA) and efficiency (PDE-DSA-PDE≥DSA-PDE2 > PDE2-DSA) of DNA alkylation, but controls the DNA alkylation selectivity as well. Most notable in this regard is the remarkable observation that both enantiomers of such sandwiched compounds alkylate the same sites exhibiting the same DNA alkylation selectivity independent of the enantiomeric configuration ((+)-PDE-DSA-PDE = ent-(−)-PDE-DSA-PDE). Contrasting this behavior is the distinct DNA alkylation selectivities of the enantiomeric pairs of extended or reversed analogues which are not only different from that of the sandwiched compounds, but which exhibit reversed enantiomeric selectivities (e.g., (+)-DSA-PDE2 = ent-(−)-PDE2-DSA). These remarkable observations, which are easily rationalized and visualized based on prevailing models of the DNA alkylation reaction,26 are consistent with preferential noncovalent binding of the compounds across a five base-pair AT-rich site delivering the natural enantiomer of a modestly reactive electrophile to adenine positioned central (sandwiched) or at the 3′ terminus (extended) and 5′ terminus (reversed) of such sites, whereas the unnatural enantiomers bind within the same sites but with reversed orientations delivering the electrophile to adenine central (sandwiched) or at the opposite 5′ terminus (extended) and 3′ terminus (reversed) of the sites. Superimposed on this binding-derived alkylation selectivity is the impact of catalysis attributable to the alkylation subunit N2 substituent. The subunit arrangements incorporating an extended heteroaromatic N2 acyl substituent on the alkylation subunit (sandwiched, extended analogues) alkylate DNA with remarkable rates and efficiencies for such a modest electrophile, whereas those that lack this structural feature (reversed analogues) do so only at markedly reduced rates (≥1,000-fold). We have attributed this pronounced impact on the rate to a unique source of catalysis for the DNA alkylation reaction that is derived from a DNA binding-induced conformation change in the molecule that activates them for nucleophilic attack. Thus, the adoption of a DNA bound helical conformation disrupts the N2 vinylogous amide conjugation stabilizing the alkylation subunit (sandwiched and extended, but not reversed) activating the cross-conjugated cyclopropane for nucleophilic attack.
Tucked into these studies, we have also prepared and evaluated a key analogue of CC-1065. The extended analogue 24, (+)- and ent-(−)-DSA-PDE2, constitutes the substitution of the duocarmycin SA/yatakemycin alkylation subunit into the CC-1065 structure replacing its CPI alkylation subunit. This substitution provided a hybrid analogue of CC-1065 and its unnatural enantiomer (L1210 IC50 = 20 pM, both enantiomers) that exhibited an unaltered DNA alkylation selectivity relative to CC-1065, but which proved to be 5-fold more potent (IC50 = 4 pM, Figure 6) than the natural product precisely in line with expectations21 based on its 5-fold enhanced stability.27
Supplementary Material
Full experimental details are provided. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
We gratefully acknowledge the financial support of the National Institutes of Health (CA41986 and CA42056), the Skaggs Institute for Chemical Biology, and predoctoral fellowship support from the American Chemical Society (2005–2006 M.S.T., sponsored by Roche Pharmaceuticals) and American Society for Engineering Education (2003–2005 J.D.T., NDSEG). J.D.T., K.S.M. and M.S.T. are Skaggs Fellows.
References
- 1.Igarashi Y, Futamata K, Fujita T, Sekine A, Senda H, Naoki H, Furumai T. J Antibiot. 2003;56:107–113. doi: 10.7164/antibiotics.56.107. [DOI] [PubMed] [Google Scholar]
- 2.Martin DG, Biles C, Gerpheide SA, Hanka LJ, Krueger WC, McGovren JP, Mizsak SA, Neil GL, Stewart JC, Visser J. J Antibiot. 1981;34:1119–1125. doi: 10.7164/antibiotics.34.1119. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi I, Takahashi K, Ichimura M, Morimoto M, Asano K, Kawamoto I, Tomita F, Nakano H. J Antibiot. 1988;41:1915–1917. doi: 10.7164/antibiotics.41.1915. [DOI] [PubMed] [Google Scholar]
- 4.Ichimura M, Ogawa T, Takahashi K, Kobayashi E, Kawamoto I, Yasuzawa T, Takahashi I, Nakano H. J Antibiot. 1990;43:1037–1038. doi: 10.7164/antibiotics.43.1037. [DOI] [PubMed] [Google Scholar]
- 5.Yatakemycin: Parrish JP, Kastrinsky DB, Wolkenberg SE, Igarashi Y, Boger DL. J Am Chem Soc. 2003;125:10971–10976. doi: 10.1021/ja035984h.Trzupek JD, Gottesfeld JM, Boger DL. Nature Chem Biol. 2006;2:79–82. doi: 10.1038/nchembio761.
- 6.(a) CC-1065: Hurley LH, Lee C-S, McGovren JP, Warpehoski MA, Mitchell MA, Kelly RC, Aristoff PA. Biochemistry. 1988;27:3886–3892. doi: 10.1021/bi00410a054. [DOI] [PubMed] [Google Scholar]; (b) Hurley LH, Warpehoski MA, Lee CS, McGovren JP, Scahill TA, Kelly RC, Mitchell MA, Wicnienski NA, Gebhard I, Johnson PD, Bradford VS. J Am Chem Soc. 1990;112:4633–4649. [Google Scholar]; (c) Boger DL, Johnson DS, Yun W, Tarby CM. Bioorg Med Chem. 1994;2:115–135. doi: 10.1016/s0968-0896(00)82007-6. [DOI] [PubMed] [Google Scholar]; (d) Boger DL, Coleman RS, Invergo BJ, Sakya SM, Ishizaki T, Munk SA, Zarrinmayeh H, Kitos PA, Thompson SC. J Am Chem Soc. 1990;112:4623–4632. [Google Scholar]
- 7.Duocarmycin A: Boger DL, Ishizaki T, Zarrinmayeh H, Munk SA, Kitos PA, Suntornwat O. J Am Chem Soc. 1990;112:8961–8971.Boger DL, Ishizaki T, Zarrimayeh H. J Am Chem Soc. 1991;113:6645–6649.Boger DL, Yun W, Terashima S, Fukuda Y, Nakatani K, Kitos PA, Jin Q. Bioorg Med Chem Lett. 1992;2:759–765.
- 8.Duocarmycin SA: Boger DL, Johnson DS, Yun W. J Am Chem Soc. 1994;116:1635–1656.
- 9.Reviews: Boger DL, Johnson DS. Angew Chem Int Ed Engl. 1996;35:1438–1474.Boger DL. Acc Chem Res. 1995;28:20–29.Boger DL, Johnson DS. Proc Natl Acad Sci USA. 1995;92:3642–3649. doi: 10.1073/pnas.92.9.3642.Boger DL, Garbaccio RM. Acc Chem Res. 1999;32:1043–1052.
- 10.Tichenor MS, Kastrinsky DB, Boger DL. J Am Chem Soc. 2004;126:8396–8398. doi: 10.1021/ja0472735. [DOI] [PubMed] [Google Scholar]
- 11.Tichenor MS, Trzupek JD, Kastrinsky DB, Futoshi S, Hwang I, Boger DL. J Am Chem Soc. 2006;128:15683–15696. doi: 10.1021/ja064228j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.An independent route to the natural product has been reported: Okano K, Tokuyama H, Fukuyama T. J Am Chem Soc. 2006;128:7136–7137. doi: 10.1021/ja0619455.
- 13.Review of related efforts: Boger DL, Boyce CW, Garbaccio RM, Goldberg JA. Chem Rev. 1997;97:787–828. doi: 10.1021/cr960095g.
- 14.(a) Boger DL, Bollinger B, Hertzog DL, Johnson DS, Cai H, Mesini P, Garbaccio RM, Jin Q, Kitos PA. J Am Chem Soc. 1997;119:4987–4998. [Google Scholar]; (b) Boger DL, Hertzog DL, Bollinger B, Johnson DS, Cai H, Goldberg J, Turnbull P. J Am Chem Soc. 1997;119:4977–4986. [Google Scholar]
- 15.Boger DL, Garbaccio RM. Bioorg Med Chem. 1997;5:263–276. doi: 10.1016/s0968-0896(96)00238-6. [DOI] [PubMed] [Google Scholar]
- 16.Reviews: Wolkenberg SE, Boger DL. Chem Rev. 2002;102:2477–2496. doi: 10.1021/cr010046q.Tse WC, Boger DL. Chem Biol. 2004;11:1607–1617. doi: 10.1016/j.chembiol.2003.08.012.
- 17.Parrish JP, Kastrinsky DB, Stauffer F, Hedrick MP, Hwang I, Boger DL. Bioorg Med Chem. 2003;11:3815–3838. doi: 10.1016/s0968-0896(03)00194-9. [DOI] [PubMed] [Google Scholar]
- 18.(a) Boger DL, Coleman RS. J Org Chem. 1984;49:2240–2245. [Google Scholar]; (b) Boger DL, Coleman RS, Invergo BJ. J Org Chem. 1987;52:1521–1530. [Google Scholar]; (c) Boger DL, Coleman RS. J Org Chem. 1988;53:695–698. [Google Scholar]
- 19.(a) Warpehoski MA, Gebhard I, Kelly RC, Krueger WC, Li LH, McGovern JP, Praire MD, Wienienski N, Wierenga W. J Med Chem. 1988;31:590–603. doi: 10.1021/jm00398a017. [DOI] [PubMed] [Google Scholar]; (b) Boger DL, Ishizaki T, Sakya SM, Munk SA, Kitos PA, Jin Q, Besterman JM. Bioorg Med Chem Lett. 1991;1:115–120. [Google Scholar]; (c) Boger DL, Yun W, Han N. Bioorg Med Chem. 1995;3:1429–1453. doi: 10.1016/0968-0896(95)00130-9. [DOI] [PubMed] [Google Scholar]
- 20.(a) Boger DL, Machiya K. J Am Chem Soc. 1992;114:10056–10058. [Google Scholar]; (b) Boger DL, Machiya K, Hertzog DL, Kitos PA, Holmes D. J Am Chem Soc. 1993;115:9025–9036. [Google Scholar]
- 21.(a) Parrish JP, Hughes TV, Hwang I, Boger DL. J Am Chem Soc. 2004;126:80–81. doi: 10.1021/ja038162t. [DOI] [PubMed] [Google Scholar]; (b) Boger DL, Munk SA, Ishizaki T. J Am Chem Soc. 1991;113:2779–2780. [Google Scholar]
- 22.(a) Boger DL, Coleman RS. J Am Chem Soc. 1987;109:2717–2727. [Google Scholar]; (b) Coleman DLRS. J Am Chem Soc. 1988;110:1321–1323. [Google Scholar]; 1988;110:4796–4807. [Google Scholar]
- 23.(a) Boger DL, Coleman RS, Invergo BJ, Zarrinmayeh H, Kitos PA, Thompson SC, Leong T, McLaughlin LW. Chem-Biol Interact. 1990;73:29–52. doi: 10.1016/0009-2797(90)90107-x. [DOI] [PubMed] [Google Scholar]; (b) Boger DL, Zarrinmayeh H, Munk SA, Kitos PA, Suntornwat O. Proc Natl Acad Sci USA. 1991;88:1431–1435. doi: 10.1073/pnas.88.4.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boger DL, Munk SA, Zarrinmayeh H, Ishizaki T, Haught J, Bina M. Tetrahedron. 1991;47:2661–2682. [PubMed] [Google Scholar]
- 25.Boger DL, Johnson DS. J Am Chem Soc. 1995;117:1443–1444. [Google Scholar]
- 26.Boger DL, Johnson DS, Yun W. J Am Chem Soc. 1994;116:1635–1656. [Google Scholar]
- 27.This stability was measured by assessing solvolysis reactivity of the respective N-Boc derivatives at pH 3: N-Boc-CPI (t1/2 = 37 h, k = 5.26 × 10−6s−1) versus N-Boc-DSA (t1/2 = 177 h, k = 1.08 × 10−6s−1). See: Boger DL, Yun W. J Am Chem Soc. 1994;116:5523–5524.Boger DL, Yun W. J Am Chem Soc. 1994;116:7996–8006.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Full experimental details are provided. This material is available free of charge via the Internet at http://pubs.acs.org.