Abstract
We have investigated the regulation of translation during the period of rapid liver growth that occurs at the end of gestation in the rat. This work was based on our prior observation that fetal hepatocyte proliferation is resistant to the inhibitory effects of rapamycin, an inhibitor of the mammalian target of rapamycin (mTOR), a nutrient-sensing kinase that controls ribosome biogenesis and protein translation. We hypothesized that translation control in late-gestation fetal liver differs from that in adult liver. We first examined the ability of rapamycin to inhibit the translation of mRNAs encoding ribosomal proteins. Consistent with the effect of rapamycin on proliferation, the activation of adult liver 5′-terminal oligopyrimidine tracts (5′-TOP) translation that occurred during refeeding after food deprivation was sensitive to rapamycin. Fetal liver 5′-TOP translation was insensitive. We went on to examine the eukaryotic initiation factor (eIF) 4F cap-binding complex that controls global protein synthesis. The molecular weights of the multiple eIF4G1 isoforms present in fetal and adult liver eIF4F complexes differed. In addition, fetal liver expressed the eIF4A1 form of the eIF4A helicase, whereas adult liver contained eIF4A1 and eIF4A2. Rapamycin administration before refeeding in adult rats inhibited formation of the preinitiation complex to a much greater degree than rapamycin administration to fetal rats in situ. We conclude that there are major structural and functional differences in translation control between late-gestation fetal and adult liver. These differences may confer differential sensitivity to the growth inhibitory effects of rapamycin.
Keywords: liver, hepatocyte, mammalian target of rapamycin, signal transduction, ribosome
the fetal rat grows at an extraordinarily rapid rate during the latter stage of gestation, tripling its somatic mass during the 3 days before term (22). This increase in size is accompanied by a proportionate increase in liver mass, which is a function of a high rate of hepatocyte proliferation (20). Our laboratory has focused on late-gestation liver development in the rat as a model of hepatocyte growth regulation, one that contrasts with the synchronized hepatocyte proliferation that occurs after partial hepatectomy in the adult rat. We have observed a number of differences between the two models, including the activation state of mitogenic signaling pathways (7, 8), the expression of cell cycle-regulating genes (4, 9), the mechanisms controlling apoptosis (15), and, most relevant to the present studies, the sensitivity of hepatocyte proliferation in vivo to the growth inhibitory effects of rapamycin (6).
Rapamycin is an inhibitor of the nutrient-sensing protein kinase referred to as the mammalian target of rapamycin (mTOR). On the basis of its ability to control global protein translation and ribosome biogenesis, mTOR has been assigned a central role in the regulation of cell growth and proliferation (17, 25, 37). Given that cell growth is a requirement for proliferation, it is not surprising that rapamycin is a potent inhibitor of cell cycle progression. Its effects are exerted largely during G1. We interpreted the observation that fetal hepatocyte proliferation in the rat is not inhibited by rapamycin (6) as relating to the known resistance of many cancer cell types to its growth inhibitory properties (28).
The mechanisms by which mTOR controls global protein translation and ribosome biogenesis have been the subject of intense investigation in recent years. Its ability to induce the phosphorylation of the eukaryotic initiation factor 4E (eIF4E) binding-protein 1 (4E-BP1) is an established mechanism, whereby mTOR can upregulate global protein translation. This regulatory activity is in large part a function of the formation of the mRNA cap-binding complex eIF4F (31, 38). When 4E-BP1 is in the hyperphosphorylated state, it is, as a consequence, unbound to eIF4E, thereby allowing eIF4E to interact with eIF4G. The ability of mTOR to activate a second pathway leading to the phosphorylation of ribosomal protein S6 was long considered to be central to the translational activation of other ribosomal proteins that are encoded by mRNAs that have 5′-terminal oligopyrimidine tracts (5′-TOP mRNAs) (27, 29, 40). However, recent studies indicate that there is not a direct functional relationship between the phosphorylation of ribosomal protein S6 and 5′-TOP mRNA translation. Among these studies is our own that demonstrated the rapamycin resistance of fetal hepatocyte proliferation (6). These studies showed a dissociation between S6 phosphorylation and hepatocyte proliferation. Among other published studies, perhaps the most compelling was the creation of a knockin mouse model in which the phosphorylation sites in S6 were deleted. Hepatocytes from these mice showed normal 5′-TOP mRNA translation (41). However, the possibility remains that S6 phosphorylation is involved in regulation of cell size, ribosome biogenesis, and/or translation initiation in some cell types (40).
The present studies were intended as an extension of our previous observation that the in situ administration of rapamycin to late-gestation fetal rats did not affect hepatocyte proliferation (6). By demonstrating a potent inhibitory effect of rapamycin on S6 phosphorylation in this in vivo model, we were able to confirm mTOR inhibition resulting from rapamycin administration. We interpreted our results as indicating that the mechanisms regulating ribosome biogenesis and global protein translation in the developing liver might differ from those in adult liver. We have therefore undertaken an analysis of the effects of rapamycin on the translation of mRNA encoding ribosomal proteins in the developing fetal liver as well as an examination of the cap-binding complex in fetal vs. adult liver.
EXPERIMENTAL PROCEDURES
Materials.
Antibodies to S6, phosphorylated S6 (Ser235/236 and Ser240/244), phospho-eIF4G (Ser1108) and eIF4E were purchased from Cell Signaling Technology (Beverly, MA). Antibodies to eIF4G1, L28, L11, eIF4A1, and eIF4A2 were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). 7-Methyl-GTP (7mGTP)-Sepharose 4B was purchased from GE Healthcare Life Sciences (Piscataway, NJ).
Animals.
Male and timed-pregnant female Sprague-Dawley rats were obtained from Charles River Laboratories (Wilmington, MA). Pregnant rats of known gestational age (term being 21 days) were used for all fetal studies. Dams were fed standard laboratory chow ad libitum. For fetal studies, a laparotomy was performed under pentobarbital sodium anesthesia (35 mg/kg, ip injection) on embryonic day 19 (E19). Fetuses were exteriorized under sterile conditions. Rapamycin (2.5 μg; LC Laboratories, Woburn, MA) or an equivalent amount of dimethyl sulfoxide (DMSO) vehicle was administered by intraperitoneal injection to each fetus in situ. The fetuses were replaced, laparotomy incisions were closed, and gestation was allowed to continue for either 6 or 24 h as specified for individual experiments. At the end of this period time, a cesarean section was performed, and fetal livers were harvested.
Male rats, 140–180 g, were used for fasting/refeeding studies, which were carried out as described previously (3). Following withdrawal of food for 48 h and just before refeeding, DMSO vehicle or 2.5 μg/g rapamycin were administered to the animals by intraperitoneal injection. After injection (15 min), the refeeding period was initiated by replacing the laboratory chow in their cage covers. Ad libitum feeding was allowed to proceed for times ranging from 5 min to 6 h at which time animals were killed under isoflurane anesthesia. Partial hepatectomy was performed as previously described (6). DMSO or rapamycin was administered 1 h before the surgical procedure as for the fasting/refeeding experiments. Animals were killed 24 h after surgery by exsanguination under pentobarbital sodium anesthesia (50 mg/kg by ip injection).
All animals studies were approved by the Rhode Island Hospital Institutional Animal Care and Use Committee.
Biochemical analyses.
For all studies, liver tissue was flash-frozen in liquid nitrogen then stored at −70°C until use. Liver homogenates for Western immunoblotting were prepared as described previously (2). Sample preparation and conditions for affinity purifying the 7mGTP cap-binding components have also been described previously (3). Western immunoblotting was done using standard electrophoresis and transfer methods with chemiluminescent detection (1). Protein determinations were made using the bicinchoninic acid method (Pierce Chemical, Rockford, IL) using BSA as the standard.
Polysome preparation and fractionation was carried out using a modification of the methods we have described previously (3). A 10–50% sucrose gradient was developed in each tube by ultracentrifugation after which tube piercing was accomplished using a Brandel apparatus. Fraction collection started with the lightest portion of the gradient at the top of the tube. Monitoring of absorbance at 254 nm was used to identify samples containing free RNA, monosomes, and polysomes. RNA was extracted and analyzed by RT-PCR for abundance of specific RNAs in the various pools (3). RNA (1.5 μg) was used to generate the first-strand cDNA using the SuperScript First-Strand Synthesis System (Invitrogen, Carlsbad, CA).
Rat-derived sequence data were used for primer design except when not available (eIF4G1), in which case the human sequence was used. PCR primers were designed using the MIT Primers3 design program (www.genome.wi.mit.edu//cgi-bin/primer3.egi). Ribosomal protein S8 primer sequences were provided by Dr. Scot Kimball (Pennsylvania State College of Medicine, Hershey, PA). RT-PCR was performed in a semiquantitative manner, as described previously (3, 4, 23). Primer sequences, PCR conditions, and expected product sizes are shown in Table 1. The specificity of the primers for the three forms of eIF4G was confirmed by sequencing the PCR products.
Table 1.
Primers and conditions used for RT-PCR
| Forward Primer | Reverse Primer | TA, °C | No. of Cycles | Product Size | |
|---|---|---|---|---|---|
| eIF4G1 | 5′-GCCAGCACTTCTACCCTAGC-3′ | 5′-GTAGGGCTTGCACCTGGATA-3′ | 59 | 26 | 581 |
| eIF4G2 | 5′-GCTTCCCAAAAGGCATGTTA-3′ | 5′-AATAGACTGCCTCCGCTTGA-3′ | 59 | 25 | 476 |
| eIF4G3 | 5′-CCACTGTTCACTGCTGAGGA-3′ | 5′-GTCGGCGTACATTCCACTTT-3′ | 57 | 30 | 323 |
| rpS6 | 5′-ACTGGCTGTCAGAAACTCAT-3′ | 5′-CCACATAACCCTTCCACTCT-3′ | 55 | 28 | 120 |
| rpS8 | 5′-CGTGCTCTGAGATTGGATGT-3′ | 5′-CGGACAAGCTCGTTATTGG-3′ | 55 | 28 | 109 |
| rpL28 | 5′-TACGTGAGGACCACCATCAA-3′ | 5′-CAGATCAGGGCGGTACTTGT-3′ | 55 | 28 | 90 |
| β-Actin | 5′-TTGTAACCAACTGGGACGATATGG-3′ | 5′-GATCTTGATCTTCATGGTGCTAG-3′ | 59 | 25 | 764 |
TA, annealing temperature; eIF, eukaryotic initiation factor; rp, ribosomal protein.
RESULTS
To test the hypothesis that late-gestation fetal hepatic 5′-TOP mRNA translation is insensitive to rapamycin, fetuses were injected in situ with DMSO vehicle or rapamycin and then allowed to recover for varying periods of time before repeat cesarean section. At the end of the experiment, fetal livers were harvested and processed to prepare ribosomes. For a comparison group, we used fasted adult rats that were injected with DMSO vehicle or rapamycin before refeeding.
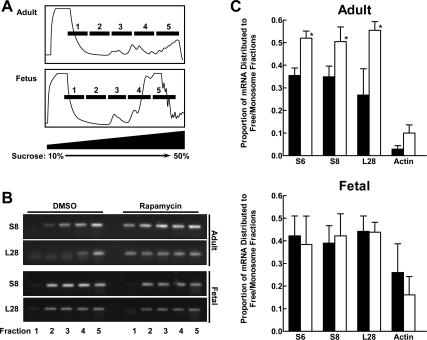
In all fetal and adult animals that were administered rapamycin, we observed nearly complete inhibition of S6 phosphorylation, as demonstrated by phosphospecific immunoblotting (data not shown). Sucrose gradient fractionation of ribosomes (Fig. 1) for adult and fetal liver showed that polysomes were present in higher abundance relative to total RNA in the fetal samples. This was a consistent finding. Analysis of mRNA distribution showed a modest but consistent rapamycin-induced shift from the polysome to nonpolysome fractions for three 5′-TOP mRNA species encoding ribosomal proteins S6, S8, and L28 in adult rats killed 6 h after rapamycin administration. A non-5′-TOP mRNA, β-actin, was unaffected. In contrast, administration of rapamycin to E19 fetuses 6 h before death had no effect on the distribution of the S6, S8, and L28 5′-TOP mRNAs, as illustrated by the absence of a shift to the left in the polysome profiles.
Fig. 1.
Effect of rapamycin on hepatic 5′-terminal oligopyrimidine tracts (5′-TOP) mRNA translation in refed adult rats and late-gestation fetal rats. Adult rats underwent starvation for 48 h. Before refeeding (15 min), dimethyl sulfoxide (DMSO) vehicle or rapamycin was administered by ip injection. Refeeding was allowed to proceed for 6 h. DMSO or rapamycin were also administered to embryonic day 19 (E19) fetal rats in situ. Fetal livers were recovered by cesarean section 6 h later. A: total RNA fractions were resolved by sucrose density centrifugation. These representative profiles showing absorbance at 254 nm indicate how fractions were pooled to obtain free mRNA (fractions 1 and 2), 40S monosomes (fraction 3), 60S, 80S, and short polysomes (fraction 4), and long polysomes (fraction 5). B: adult and fetal liver samples from control and rapamycin-injected animals were analyzed by RT-PCR for the distribution of mRNA species encoding ribosomal proteins S6, S8, and L28 and β-actin. Representative analyses of S8 and L28 distribution are shown. C: RT-PCR results for triplicate, paired adult/fetal analyses are shown. Filled bars, results for control animals; open bars, results for the rapamycin group. Data are shown as means and SE. *P < 0.05 by paired t-test.
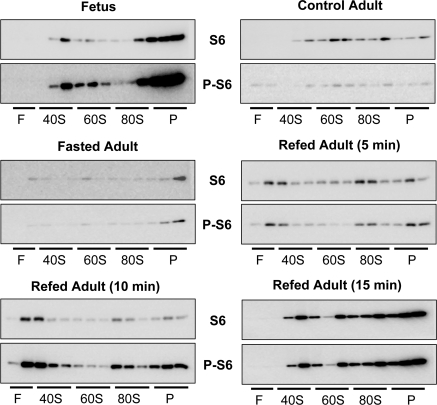
Although its role in translation control is uncertain, S6 phosphorylation still retains a purported physiological role in ribosome assembly. We reasoned that involvement of S6 phosphorylation in ribosome assembly would be reflected in the distribution of phospho-S6 in nascent ribosomes at various stages and that loss of such a function could contribute to rapamycin resistance. To examine this possibility, we studied the distribution of phospho-S6 in ribosomes fractionated in sucrose density gradients (Fig. 2).
Fig. 2.
Distribution of ribosomal protein S6 and phosphorylated S6 (P-S6) in fetal and adult liver ribosomes fractionated by sucrose density centrifugation. Liver ribosomes were prepared from fetal and adult rats. The adult rats were fed ad libitum (control), fasted for 48 h, or fasted for 48 h and then refed. Refed animals were killed at 5, 10, or 15 min after they began eating the food reintroduced into their cages. Sucrose gradients were fractionated into 14 samples, all of which were analyzed by immunoblotting for S6 and phosphorylated-S6. F, free; P, polysomes.
Phospho-S6 levels (Ser235/236) were considerably higher in fetal rats than in adult rats relative to total S6. In both fetal and adult liver ribosomes, the distribution of phospho-S6 in the sucrose density profiles coincided with the distribution of S6 itself. Fasting of adult rats reduced total S6 content and phospho-S6 levels, although the distribution of phospho-S6 again coincided with S6 abundance. To detect a role for S6 phosphorylation in ribosome assembly, animals were fasted for 48 h and then refed for periods (5, 10, or 15 min) that were sufficiently short to observe the accumulation of preinitiation complexes and polysomes, as indicated by the distribution of S6. In all cases, phosphorylation of S6 in individual fractions was in proportion to S6 content. Results using an antibody directed toward Ser240/244 gave similar results (data not shown). This experiment was replicated in several additional groups of animals. The results were interpreted as indicating that S6 phosphorylation is not involved in ribosome assembly in liver.
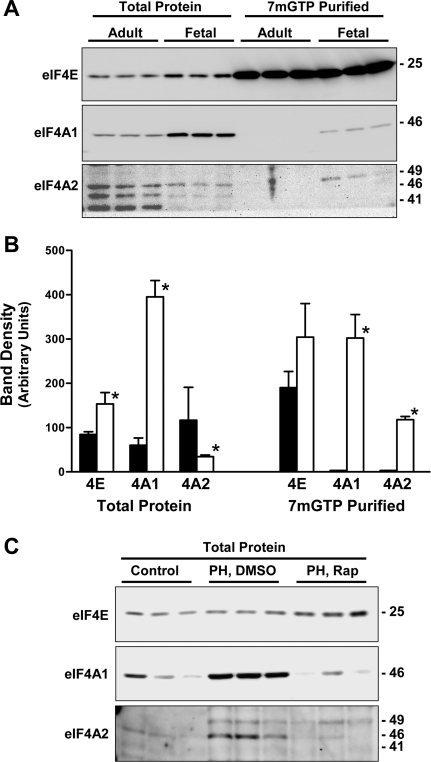
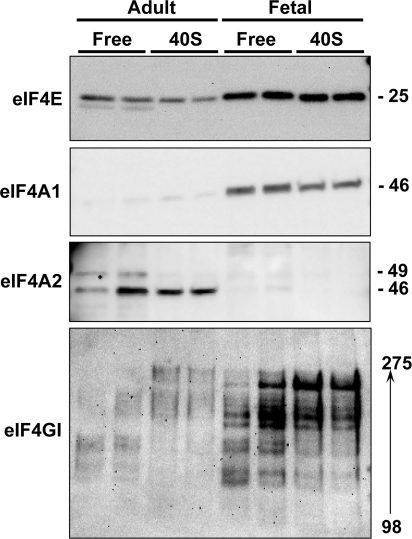
We proceeded to compare the composition of fetal and adult liver eIF4F complexes. Triplicate fetal and adult liver homogenates were analyzed by direct immunoblotting and by 7mGTP affinity purification followed by immunoblotting (Fig. 3, A and B). For these experiments, 7mGTP affinity purification of eIF4F complexes was carried out by incubating homogenates with 7mGTP beads for 18 h.
Fig. 3.
Cap-binding components in fetal vs. adult liver. A: triplicate adult and fetal liver samples were processed for direct immunoblotting and for 7-methyl-GTP (7mGTP) affinity purification of cap-binding complexes. Numbers on right in immunoblots represent apparent molecular mass in kDa. Note that an overexposed version of the panel on top is shown to make the total protein analysis visible. B: results from A quantified as the mean and 1 SD for each set of triplicate analyses. The eukaryotic initiation factor (eIF) 4A2 results are for the 46-kDa band that was found in subsequent experiments to be the translationally active form. The use of chemiluminescent imaging allowed us to optimize the quantification for each moiety. Filled bars, adult; open bars, fetal. *P < 0.05 for fetal results vs. corresponding adult results. C: direct immunoblotting for eIF4E, eIF4A1, and eIF4A2 was carried out on triplicate liver homogenates prepared from adult animals (control: basal, fed conditions), animals that underwent partial hepatectomy (PH: DMSO vehicle administered 1 h before partial hepatectomy), and regenerating liver from animals that were administered rapamycin 1 h before partial hepatectomy. Liver tissue was obtained from animals that underwent partial hepatectomy 24 h after surgery.
The level of total eIF4E was slightly higher in fetal liver homogenates than in adult liver homogenates. However, the amounts of eIF4E recovered in 7mGTP-purified fractions were similar. eIF4A1 levels in unfractionated homogenates were again somewhat higher in fetal liver than in adult liver. In contrast, levels in the 7mGTP-purified fraction were far higher in fetal liver than in adult liver. Examination of eIF4A2 showed that multiple immunoreactive bands were present in unfractionated homogenates. The levels of all were higher in adult than in fetal liver. The largest of the immunoreactive proteins (49 kDa), the main species that was recovered in the 7mGTP-purified fractions, was present in considerably higher levels in fetal samples than in adult samples. Incubation of samples with control Sepharose beads lacking the 7mGTP did not yield any of the eIF4F components, as indicated by immunoblotting (data not shown).
Given that fetal hepatocytes are actively proliferating while adult hepatocytes are quiescent under basal conditions, we examined the effect of partial hepatectomy on the content of eIF4F components, including the two isoforms of eIF4A (Fig. 3C). Regenerating liver obtained 24 h after partial hepatectomy was associated with an increase in both eIF4A1 and eIF4A2, whereas eIF4E levels were unchanged. Inhibition of liver regeneration by preadministration of rapamycin prevented the increases in both eIF4A isoforms. This was not the case for eIF4E, the content of which was slightly increased by rapamycin pretreatment. This result raised the possibility that eIF4A1 and eIF4A2 were behaving like 5′-TOP mRNAs. However, examination of the distribution of eIF4A mRNAs in fractionated polysomes did not demonstrate an effect of rapamycin (data not shown).
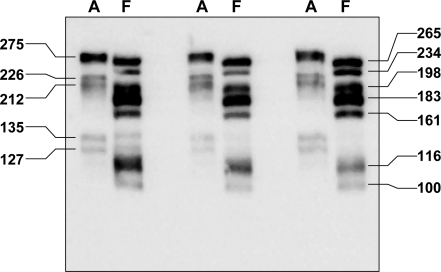
The results of the 7mGTP affinity purification of cap-binding proteins indicated a far higher level of eIF4F formation and cap-dependent transcription initiation in fetal liver relative to adult liver. As the scaffold for eIF4F formation, eIF4G is required for formation of the cap-binding complex. Levels of eIF4G1 in unfractionated homogenates of adult and fetal liver were similar but were low and difficult to detect. We therefore employed 7mGTP affinity purification to compare eIF4G1 content and isoform expression in adult and fetal liver (Fig. 4). Results showed that the multiple molecular weight forms of eIF4G1 in adult and fetal liver differ.
Fig. 4.
eIF4G1 in 7mGTP affinity-purified preparations from adult and fetal liver. Liver homogenates, prepared from adult (A) and fetal (F) liver samples, were analyzed by 7mGTP affinity purification followed by immunoblotting for eIF4G1. Numbers on left and right of the immunoblot represent apparent molecular mass in kDa.
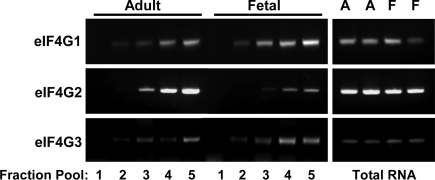
We did not observe markedly higher levels of 7mGTP-bound eIF4G1 in fetal samples relative to adult samples as would have been predicted by the eIF4A results. We therefore examined the possibility that other forms of eIF4G could account for this observation. The functional roles of eIF4G2 and eIF4G3 have not been fully characterized. We hypothesized that either or both might account for high levels of eIF4F recovered from fetal liver. Available antibody reagents proved to not be useful for examining eIF4G2 and eIF4G3 content in liver samples. As an alternative, we examined the translation of eIF4G1, eIF4G2, and eIF4G3 by analyzing for mRNA distribution in adult and fetal liver polysome profiles. Three sets of analyses were completed (representative results shown in Fig. 5). We found that eIF4G1 and eIF4G3 translation was similar in fetal and adult liver. eIF4G2 translation was higher in adult liver relative to fetal liver. We did not observe differences in total RNA content for any of the three forms. Based on these results, we could not attribute higher levels of eIF4F formation in fetal liver to either eIF4G2 or eIF4G3.
Fig. 5.
Expression and translation of hepatic eIF4G1, eIF4G2, and eIF4G3. Total RNA and ribosomes were prepared from adult (A) and fetal (F) livers. The ribosome preparations were fractionated by sucrose density centrifugation as for Fig. 1. Fraction pools and total RNA were analyzed by RT-PCR for abundance of mRNA for eIF4G1, eIF4G2, and eIF4G3. Cycle number was 26, 25, and 30 for the three forms, respectively. Results are representative for single adult and fetal ribosome profiles and for duplicate adult and fetal total RNA preparations. The results shown here were confirmed in a second, independent experiment.
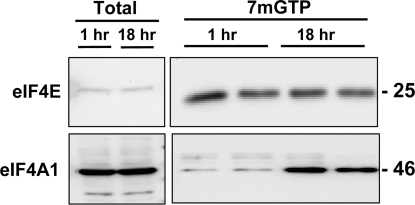
Given the disparity between eIF4G and eIF4A levels in 7mGTP-purified fractions, we investigated a possible caveat in the interpretation of these results. An overnight period of incubation with 7mGTP beads had been used to optimize recovery of eIF4F complexes from homogenates. We considered the possibility that the complex was forming posthomogenization during the affinity purification process. A time course of recovery of eIF4F components by 7mGTP beads was performed (Fig. 6). Results showed that recovery of eIF4E was rapid, being as high after a 1-h incubation period as it was after an overnight incubation. However, the recovery of eIF4A1 was far higher after the longer incubation period, indicating posthomogenization formation of eIF4F. On the basis of this observation, we reinterpreted the 7mGTP purification results as indicating that the content of eIF4F components available for eIF4F formation was higher in fetal than in adult liver. We also concluded that the eIF4F complexes recovered from fetal liver did not necessarily represent translationally active complexes.
Fig. 6.
Formation of the cap-binding complex in fetal liver homogenates over time during 7mGTP affinity purification. A fetal liver homogenate was held at 4°C for 1 or 18 h and then analyzed by immunoblotting for total eIF4E and eIF4A1 to serve as a control for the period of incubation with 7mGTP beads (left). Duplicate homogenates were incubated with 7mGTP beads for 1 or 18 h before analysis for eIF4E and eIF4A1 content (right) to ascertain the effect of time on affinity purification of cap-binding complexes. Numbers on right in immunoblots represent apparent molecular mass in kDa.
To address this issue, we employed protein-RNA cross-linking to examine translationally active eIF4F complexes in liver homogenates. Preliminary experiments (not shown) to determine the concentration of formaldehyde and incubation time needed to cross-link eIF4F components to ribosomes confirmed that ribosome preparation and fractionation resulted in the complete dissociation of eIF4F components from the preinitiation (40S) complex without cross-linking. Recovery of eIF4E in the 40S fraction was maximal when ribosomes were incubated with 3% formaldehyde for 1 h.
A comparison of cross-linked adult and fetal ribosome profiles (Fig. 7) showed higher levels of eIF4E, eIF4A1, and eIF4G1 in the fetal 40S fractions. These results, which are consistent with a primary role for eIF4G1 in promoting cap-dependent translation in fetal liver, were replicated in a second, independent experiment. Most striking was the absence of eIF4A2 from the fetal 40S fractions. In adult 40S fractions, the form of eIF4A2 that was present had a molecular mass of 46 kDa. The 49- and 41-kDa forms seen in total homogenates and the 49-kDa form present in 7mGTP-purified preparations (Fig. 3) were not identified in cross-linking experiments, indicating that the immunoreactive 46-kDa moiety may be the only form that is translationally active. Thus the use of cross-linking and sucrose density fractionation disclosed a marked difference in the form of eIF4A that was translationally active in adult vs. fetal liver.
Fig. 7.
Analysis of translationally active cap-binding complexes in adult and fetal liver. Liver ribosomes were prepared from adult and fetal liver. Before fractionation by sucrose density centrifugation, cross-linking was accomplished by incubating samples with 3% formaldehyde for 1 h. Fractions corresponding to free (nonribosomal bound) proteins and the 40S preinitiation complex were analyzed by immunoblotting for cap-binding components. Numbers on right in immunoblots represent apparent molecular mass in kDa. Numbers to right of the eIF4G1 immunoblot represent the molecular mass range of the isoforms that were detected.
We showed previously that rapamycin administration before refeeding of adult rats did not affect eIF4F formation even though rapamycin induced hypophosphorylation of 4E-BP1 (3). However, those analyses used 7mGTP affinity purification. Given the evidence that eIF4F complexes can form posthomogenization, we repeated this experiment using formaldehyde cross-linking followed by sucrose-density fractionation. Adult rats were fasted for 48 h. DMSO vehicle or rapamycin was administered 15 min before a refeeding period that was allowed to continue for 1 h. Ribosomes were prepared, cross-linked, and fractionated on sucrose gradients. The fractions were analyzed by Western immunoblotting.
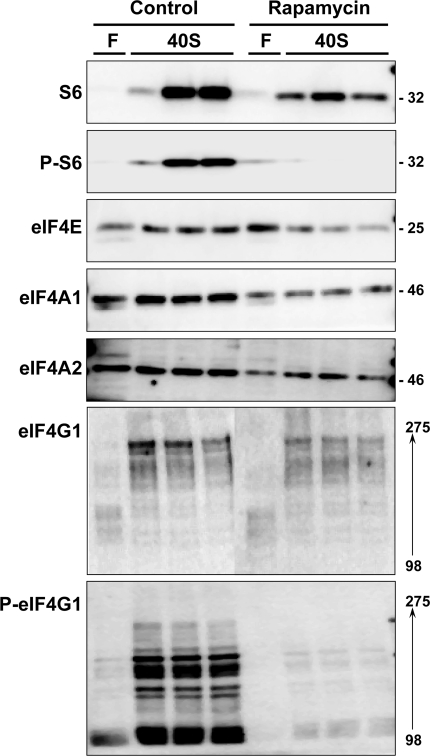
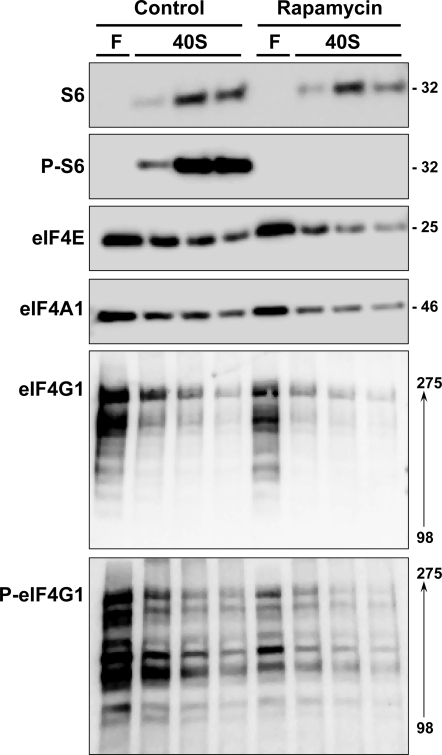
A representative analysis is shown in Fig. 8. Immunoblotting for P-S6 was markedly reduced in the animal that was administered rapamycin, thus confirming the efficacy of the drug in blocking mTOR signaling via S6 kinase. There was a modest reduction in the content of S6 in the 40S fraction. The contents of eIF4E, eIF4A1, eIF4A2, and eIF4G1 in the 40S fraction were also reduced.
Fig. 8.
Effect of rapamycin on translationally active cap-binding complexes in adult liver. Liver ribosomes were prepared from adult rats that were fasted for 48 h. DMSO and rapamycin were administered 15 min before refeeding, and refeeding was allowed to continue for 1 h before death. After cross-linking, ribosomes were fractionated by sucrose density centrifugation. For this analysis and for the analysis in Fig. 9, one fraction representing unbound proteins was analyzed while three fractions were collected across the 40S peak. Fractions were analyzed by immunoblotting for S6, phospho-S6 (Ser235/236), and for cap-binding components, including phospho-eIF4G1 (Ser1108). Numbers to right of the immunoblots represent apparent molecular mass in kDa. Numbers to right of the eIF4G1 immunoblot represent the molecular mass range of the isoforms that were detected. Results were replicated in a second experiment.
The animal experiment was replicated three times. The aggregate results of the three experiments showed that rapamycin induced a 40% decrease in S6 content in the 40S peak. This is consistent with our previously reported results on total S6 in unfractionated liver homogenates (3). The content of eIF4E in the 40S fraction relative to the content in the free fractions was reduced by ∼75% [ratio of 40S to free, 3.82 ± (SD) 1.31 in control animals and 1.21 ± 0.50 in the rapamycin group, P = 0.032]. The triplicate analyses showed a reduction in eIF4A1 and eIF4A2 content in the 40S fraction that followed the reduction in S6 content.
Samples were also analyzed for phosphorylated eIF4G1 (Ser1108). Results (Fig. 8) showed a disproportionate reduction in eIF4G1 phosphorylation in response to rapamycin. Of note, lower-molecular-weight forms of eIF4G1 showed the highest stoichiometry of phosphorylation.
An analogous experiment was carried out to examine the effect of rapamycin on cap-binding complex formation in fetal liver (representative results shown in Fig. 9). Rapamycin administration to fetuses in situ 6 h before repeat cesarean section and recovery of fetal livers was associated with a small decrease in S6 abundance in the 40S fractions. As expected, rapamycin induced a marked decrease in S6 phosphorylation. Rapamycin also induced a loss of the hyperphosphorylated form of 4E-BP1 (data not shown). Abundance of eIF4E, eIF4A1, and eIF4G1 in the 40S fraction was minimally affected by rapamycin.
Fig. 9.
Effect of rapamycin on translationally active cap-binding complexes in fetal liver. Liver ribosomes were prepared from fetal rats that were given DMSO vehicle or rapamycin by in situ injection. Fetuses and mother were allowed to recover for 6 h before death. After cross-linking, ribosomes were fractionated by sucrose density centrifugation. Fractions corresponding to unbound proteins and the 40S preinitiation complex were analyzed by immunoblotting for S6, phospho-S6 (Ser235/236), and for cap-binding components, including phospho-eIF4G1 (Ser1108). Fractions were also analyzed for immunoreactive eIF4A2, but none was detected. Numbers to right of the immunoblots represent apparent molecular mass in kDa. Numbers to right of the eIF4G1 immunoblot represent the molecular mass range of the isoforms that were detected.
The fetal analyses were also carried out in triplicate. The aggregate results confirmed a modest reduction in 40S ribosomal S6 content [28 ± 6% (SD) in 3 paired analyses]. eIF4E content in the 40S fractions relative to free eIF4E was reduced in the rapamycin samples (ratio of 40S to free: 0.65 ± 0.25 in control animals and 0.27 ± 0.08 in rapamycin group), but the results did not reach significance (P = 0.064). Of note, eIF4A1 was not reduced (40S-free: 0.89 ± 0.31 in control animals and 1.04 ± 0.69 in rapamycin group).
As was the case in adult animals, rapamycin inhibited eIF4G1 phosphorylation (Fig. 9). Again, the highest relative levels of phosphorylation were present in lower-molecular-weight forms of eIF4G1, all of which were detected in 40S fractions.
DISCUSSION
The model of liver growth in the late-gestation fetal rat is relevant to several areas of liver biology. The ability of fetal rat hepatocytes to repopulate the adult liver (13) and to proliferate independent of their differentiation status (21) indicates their potential for repair of liver injury. Their mitogen-independent signaling phenotype (7, 8) and resistance to apoptosis (15) suggest a relationship to liver carcinogenesis. A potential relationship to mechanisms involved in the development of hepatocellular carcinoma is perhaps best exemplified by the resistance of fetal hepatocytes in vivo to the growth inhibitory effects of rapamycin (6, 39). That being said, we examined a panel of cell lines that we have used previously to examine the relationship between hepatic cell transformation and regulation of protein phosphatase type 2A (44). There was no correlation between the tumorigenicity of the cells and their expression of eIFA1 or eIFA2, nor was there a relationship with the pattern of eIF4G1 isoform expression. We are left to conclude that fetal hepatocytes in vivo may share some characteristics with hepatocellular carcinoma cells but that the specifics of fetal liver translation control are not recapitulated in these cell lines.
The process of cell replication involves a balance between cell growth and cell cycle control. During the G1 phase of the cell cycle, a cell doubles in size. This process requires the synthesis of ribosomes and an increase in global protein translation. Progression beyond the late G1 restriction point involves the ability of a cell to detect its size (14, 19, 42). Recent evidence points to a capacity for cells to detect an adequate complement of ribosomes, the primary determinant of which is rRNA content (34). A number of mechanisms that result in the dysregulation of ribosome biogenesis and translation initiation have been described in cancer cells (39). These mechanisms involve control by mTOR, thus the relationship between rapamycin resistance and cancer. Through its ability to inhibit mTOR signaling, rapamycin is thought to block the processes of rRNA transcription and global protein translation, thus preventing progression from late G1 to S. Studies by Fingar and Blenis (16) indicate that the ability of rapamycin to inhibit cell cycle progression is mediated by its inhibitory effect on cell growth.
One of the first roles assigned to mTOR was the control of ribosome biogenesis. mTOR mediates the activation of ribosomal protein S6 kinases, which in turn catalyze the phosphorylation of ribosomal protein S6, a constituent of the 40S ribosomal subunit. In the present studies, we found that fetal hepatocytes in vivo are resistant to the inhibitory effect of rapamycin on 5′-TOP mRNA translation even though rapamycin administration potently inhibited S6 phosphorylation. Although the signal transduction mechanisms that control the translation of ribosomal proteins have not been elucidated, candidate proteins that bind to 5′-TOP sequences have been identified (24). Our data are consistent with the hypothesis that the expression and/or function of such 5′-TOP-binding proteins differ in fetal and adult hepatocytes.
Studies dissociating S6 phosphorylation from 5′-TOP translation do not preclude a role for S6 in controlling cell size (33, 40, 41). We hypothesized that such a mechanism could involve a role for S6 phosphorylation in ribosome assembly and that such a functional role would result in the preferential distribution of phospho-S6 to nascent ribosomes. Our studies showed that the distribution of phospho-S6 was identical to that of S6 itself, consistent with the absence of a role for S6 phosphorylation in the formation of mature ribosomes. We are left to conclude that the ability of fetal hepatocytes to proliferate in vivo under conditions associated with S6 hypophosphorylation is not related to a role for S6 phosphorylation in ribosome biogenesis.
In addition to resistance to the inhibitory effects of rapamycin on 5′-TOP translation, we hypothesized that rapamycin resistance might also involve changes in the global control of protein synthesis by formation of the eIF4F complex that binds to the 5′-cap of most mRNA species. We (3) and others (30) had reported that hepatic translation control via 4E-BP1 in the rat is insensitive to rapamycin. To address the mechanism of control in fetal liver, we performed direct immunoblotting for 4E-BP1 on liver homogenates from fetal rats that were first injected with either DMSO vehicle or rapamycin. Fetal liver 4E-BP1 phosphorylation was markedly downregulated by administration of rapamycin. Thus any rapamycin insensitivity of cap-binding complex formation in fetal liver could not be attributed to the absence of 4E-BP1 or the absence of an effect of rapamycin on its phosphorylation.
To compare the effect of rapamycin on cap-binding complex formation in fetal and adult liver, we first undertook an analysis of the constituents of the eIF4F complex using 7mGTP affinity purification. This approach yielded no apparent effect of rapamycin on the cap-binding complex in adult rats that underwent refeeding after starvation. Although consistent with our previous observations (3), the comparison of fetal and adult liver eIF4F raised questions about the results obtained using this approach. An experiment to determine if the eIF4F complex can form posthomogenization indicated that this is indeed the case.
To examine the translationally active components of the eIF4F complex, we used formaldehyde cross-linking and ribosome fractionation by sucrose gradient centrifugation. Our initial experiments disclosed differences between fetal and adult liver eIF4F complexes. We confirmed a markedly higher level of eIF4F components in the 40S fraction derived from fetal liver compared with that recovered from adult liver. Whereas the affinity purification approach gave similar content of eIF4G1 isoforms in the two, the cross-linking approach demonstrated a much higher level in fetal liver. The level was sufficiently high to warrant an assignment of the scaffolding function in the cap-binding complex to eIF4G1. The differences in eIF4E and eIF4A1 were more pronounced with cross-linking. Most impressive was the result obtained for eIF4A2, which showed the presence of this helicase only in the adult liver 40S fraction.
The cross-linking analysis also demonstrated a rapamycin-induced reduction in the eIF4F components in the 40S peak in adult liver. The decreases in the 40S content of S6, eIF4E, eIF4A1, eIF4A2, and eIF4G1 were roughly equivalent. Unlike the results obtained using 7mGTP affinity purification, these results supported the conclusion that rapamycin reduces the content of the preinitiation complex in refeeding. The results for the fetal animals also showed a modest reduction in S6 and eIF4E content in the 40S peak. However, eIF4A1 content was unchanged. This dissociation between eIF4A1 and eIF4E distribution might be accounted for by the cycling of the helicase through the eIF4F complex during translation such that there need not be a stoichiometric relationship between eIF4E and eIF4A (36). Indeed, the absence of an effect of rapamycin on eIF4A1 content in the fetal liver preinitiation complex may reflect an alternative means of regulating translation initiation and the relative lack of a rapamycin effect on fetal liver translation initiation.
Although 7mGTP purification proved unsuitable for detecting effects of rapamycin on translation initiation, it did allow us to detect differences in the isoforms of eIF4G1 that were present in adult and fetal liver. Because we did not see a difference in eIF4G1 levels in 7mGTP-purified complexes, we investigated a possible role for eIF4G2 and eIF4G3 in eIF4F formation. eIF4G1, also referred to as eIF4GI, is the prototype for the eIF4G family. eIF4G2, also referred to as p97, DAP5, or NAT1, was initially found to be converted to an active component of the translation initiation complex upon caspase cleavage in apoptotic cells. More recent studies (35) indicate that it can function as a translation activator in unstressed cells. eIF4G3, which is also referred to as eIF4GII, shares 66% sequence homology with eIF4G1 at the level of primary structure. It was originally thought to functionally complement eIF4G1, although some studies have shown that it has a physiological role distinct from that of eIF4G1 (10, 11). We found that neither eIF4G2 nor eIF4G3 mRNA was expressed or translated at higher levels in fetal than in adult liver. We were left to conclude that eIF4G1 is the primary form of eIF4G in both fetal and adult complexes, a conclusion that was supported by the results of the cross-linking studies.
eIF4G1 isoforms, which are largely derived from alternative start sites, may function differently in translation initiation (12). The full-length form of the protein seems to have the highest translation initiation activity. Although the possibility has been raised by several authors that the different forms of eIF4G1 regulate the translation of different mRNAs, this hypothesis has not yet been tested. Hinton et al. (26) have mapped the domains in eIF4G1 responsible for individual binding activities, but the investigators found that these domains did not predict specific functional roles for the different forms of eIF4G1. In fact, mutation of critical binding sites did not completely abrogate eIF4G1 function, thus leading the authors to speculate that the protein-protein interactions in eIF4F may be more regulatory than they are essential.
eIF4GI undergoes reversible phosphorylation on multiple residues that can contribute to the regulation of global translation (5, 32). The best-characterized site is Ser1108, which is said to augment the translation activity of eIF4G1. Goggin et al. (18) recently showed that liver regeneration was associated with eIF4G1 phosphorylation at Ser1108 and that this signaling event was rapamycin sensitive. We found that the lower-molecular-weight forms of eIF4G1 are phosphorylated to a greater degree than the longer forms of the protein. The phosphorylation of all eIF4G1 isoforms was inhibited in both fetal and adult liver by rapamycin. Because fetal hepatocytes in vivo are resistant to rapamycin-induced cell cycle inhibition, the phosphorylation of eIF4G1 may not be essential to maintain the translation initiation function of this factor.
As noted above, we observed a significant difference in the forms of eIF4A that were expressed and translationally active in adult and fetal liver. In contrast to fetal liver, adult liver contained significant levels of eIF4A2. Few data are available on the functional roles of eIF4A1 vs. eIF4A2. Some years ago, it was shown that the expression of the two isoforms was dependent on cell growth status in the adult mouse, with eIF4A1 being expressed in all tissues while eIF4A2 was expressed only in tissues with a low rate of cell proliferation (43). Our observations seem to indicate that a developmental change in the in vivo proliferation rate of hepatocytes is associated with a switch in eIF4A isoform expression that is consistent with the previous observation. The use of partial hepatectomy to activate the proliferation of adult hepatocytes did not result in a shift in the relative expression of eIF4A1 and eIF4A2. This indicates that the expression of the two isoforms is not a simple function of hepatocyte proliferation. We detected three immunoreactive species with eIF4A2. Only the 46-kDa form was recovered in the polysomal fraction following cross-linking. We therefore presume that this is a genuine, translationally active form of eIF4A2. We cannot assign a functional role to the other two immunoreactive species in the eIF4A2 immunoblots.
The functional importance of the developmental expression of eIF4A1 and eIF4A2 is unknown. There is one mention in the literature of an unpublished observation that the two are functionally equivalent (43), but data to this effect have not been published. More recently, the interactions between eIF4A2 and a regulator of cell transformation, Pdcd4, were characterized (45). The findings were consistent with an antiproliferative effect of eIF4A2. Interestingly, the authors did not discuss the significance of their having studied the eIF4A2 isoform as opposed to eIF4A1.
Perspectives and Significance
Our studies illustrate the complexity of translational control during liver development. We hypothesize that the relative lack of rapamycin sensitivity of 5′-TOP translation and eIF4F formation in fetal liver contributes to the resistance of fetal hepatocytes to the growth inhibitory effects of rapamycin. The developmental differences in at least two components of the eIF4F complex, eIF4G1 and eIF4A, may contribute to this difference in rapamycin sensitivity. In contrast, mTOR signaling to S6 phosphorylation and 4E-BP1 does not appear to contribute to the developmental changes in hepatocyte response to rapamycin in vivo. Our findings indicate the presence of multiple and subtle regulatory mechanisms that must be characterized and their role elucidated if we are to understand the determinants of rapamycin sensitivity at the translation control level.
GRANTS
This work was supported by National Institute of Child Health and Human Development Grants HD-035831 and HD-024455.
Acknowledgments
We thank Theresa C. Bienieki for assistance with the animal studies.
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Anand P, Boylan JM, Ou Y, Gruppuso PA. Insulin signaling during perinatal liver development in the rat. Am J Physiol Endocrinol Metab 283: E844–E852, 2002. [DOI] [PubMed] [Google Scholar]
- 2.Anand P, Gruppuso PA. The regulation of hepatic protein synthesis during fasting in the rat. J Biol Chem 280: 16427–16436, 2005. [DOI] [PubMed] [Google Scholar]
- 3.Anand P, Gruppuso PA. Rapamycin inhibits liver growth during refeeding in rats via control of ribosomal protein translation but not cap-dependent translation initiation. J Nutr 136: 27–33, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Awad MM, Sanders JA, Gruppuso PA. A potential role for p15(Ink4b) and p57(Kip2) in liver development. FEBS Lett 483: 160–164, 2000. [DOI] [PubMed] [Google Scholar]
- 5.Bolster DR, Vary TC, Kimball SR, Jefferson LS. Leucine regulates translation initiation in rat skeletal muscle via enhanced eIF4G phosphorylation. J Nutr 134: 1704–1710, 2004. [DOI] [PubMed] [Google Scholar]
- 6.Boylan JM, Anand P, Gruppuso PA. Ribosomal protein S6 phosphorylation and function during late gestation liver development in the rat. J Biol Chem 276: 44457–44463, 2001. [DOI] [PubMed] [Google Scholar]
- 7.Boylan JM, Gruppuso PA. A comparative study of the hepatic mitogen-activated protein kinase and Jun-NH2-terminal kinase pathways in the late-gestation fetal rat. Cell Growth Diff 7: 1261–1269, 1996. [PubMed] [Google Scholar]
- 8.Boylan JM, Gruppuso PA. Uncoupling of hepatic, epidermal growth factor-mediated mitogen-activated protein kinase activation in the fetal rat. J Biol Chem 273: 3784–3790, 1998. [DOI] [PubMed] [Google Scholar]
- 9.Boylan JM, Gruppuso PA. D-type cyclins and G1 progression during liver development in the rat. Biochem Biophys Res Commun 330: 722–730, 2005. [DOI] [PubMed] [Google Scholar]
- 10.Caron S, Charon M, Cramer E, Sonenberg N, Dusanter-Fourt I. Selective modification of eukaryotic initiation factor 4F (eIF4F) at the onset of cell differentiation: recruitment of eIF4GII and long-lasting phosphorylation of eIF4E. Mol Cell Biol 24: 4920–4928, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castello A, Alvarez E, Carrasco L. Differential cleavage of eIF4GI and eIF4GII in mammalian cells. Effects on translation. J Biol Chem 281: 33206–33216, 2006. [DOI] [PubMed] [Google Scholar]
- 12.Coldwell MJ, Morley SJ. Specific isoforms of translation initiation factor 4GI show differences in translational activity. Mol Cell Biol 26: 8448–8460, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dabeva MD, Petkov PM, Sandhu J, Oren R, Laconi E, Hurston E, Shafritz DA. Proliferation and differentiation of fetal liver epithelial progenitor cells after transplantation into adult rat liver. Am J Pathol 156: 2017–2031, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dolznig H, Grebien F, Sauer T, Beug H, Mullner EW. Evidence for a size-sensing mechanism in animal cells. Nat Cell Biol 6: 899–905, 2004. [DOI] [PubMed] [Google Scholar]
- 15.Embree-Ku M, Gruppuso PA. The role of nuclear factor kappaB in late-gestation liver development in the rat. Hepatology 42: 326–334, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fingar DC, Blenis J. Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene 23: 3151–3171, 2004. [DOI] [PubMed] [Google Scholar]
- 17.Gingras AC, Raught B, Sonenberg N. mTOR signaling to translation. Curr Top Microbiol Immunol 279: 169–197, 2004. [DOI] [PubMed] [Google Scholar]
- 18.Goggin MM, Nelsen CJ, Kimball SR, Jefferson LS, Morley SJ, Albrecht JH. Rapamycin-sensitive induction of eukaryotic initiation factor 4F in regenerating mouse liver. Hepatology 40: 537–544, 2004. [DOI] [PubMed] [Google Scholar]
- 19.Grebien F, Dolznig H, Beug H, Mullner EW. Cell size control: new evidence for a general mechanism. Cell Cycle 4: 418–421, 2005. [DOI] [PubMed] [Google Scholar]
- 20.Gruppuso PA, Awad M, Bienieki TC, Boylan JM, Fernando S, Faris RA. Modulation of mitogen-independent hepatocyte proliferation during the perinatal period in the rat. In Vitro Cell Dev Biol Anim 33: 562–568, 1997. [DOI] [PubMed] [Google Scholar]
- 21.Gruppuso PA, Bienieki TC, Faris RA. The relationship between differentiation and proliferation in late gestation fetal rat hepatocytes. Pediatr Res 46: 14–19, 1999. [DOI] [PubMed] [Google Scholar]
- 22.Gruppuso PA, Brautigan DL. Induction of hepatic glycogenesis in the fetal rat. Am J Physiol Endocrinol Metab 256: E49–E54, 1989. [DOI] [PubMed] [Google Scholar]
- 23.Gruppuso PA, Vaslet CA, Boylan JM. Identification of candidate growth-regulating genes that are over-expressed in late gestation fetal liver in the rat. Biochim Biophys Acta 1494: 242–247, 2000. [DOI] [PubMed] [Google Scholar]
- 24.Hamilton TL, Stoneley M, Spriggs KA, Bushell M. TOPs and their regulation. Biochem Soc Trans 34: 12–16, 2006. [DOI] [PubMed] [Google Scholar]
- 25.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev 18: 1926–1945, 2004. [DOI] [PubMed] [Google Scholar]
- 26.Hinton TM, Coldwell MJ, Carpenter GA, Morley SJ, Pain VM. Functional analysis of individual binding activities of the scaffold protein eIF4G. J Biol Chem 282: 1695–1708, 2007. [DOI] [PubMed] [Google Scholar]
- 27.Hornstein E, Tang H, Meyuhas O. Mitogenic and nutritional signals are transduced into translational efficiency of TOP mRNAs. Cold Spring Harbor Symp Quantit Biol LXVI: 477–484, 2001. [DOI] [PubMed] [Google Scholar]
- 28.Hosoi H, Dilling MB, Liu LN, Danks JK, Shikata T, Sekulic A, Abraham RT, Lawrence JC, Houghton PJ. Studies on the mechanism of resistance to rapamycin in human cancer cells. Mol Pharmacol 54: 815–824, 1998. [DOI] [PubMed] [Google Scholar]
- 29.Jefferies HBJ, Fumagalli S, Dennis PB, Reinhard C, Pearson RB, Thomas G. Rapamycin suppresses 5′TOP mRNA translation through inhibition of p70s6k. EMBO J 16: 3693–3704, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang YP, Ballou LM, Lin RZ. Rapamycin-insensitive regulation of 4E-BP1 in regenerating rat liver. J Biol Chem 276: 10943–10951, 2001. [DOI] [PubMed] [Google Scholar]
- 31.Lawrence JC, Abraham RT. PHAS/4E-BPs as regulators of mRNA translation and cell proliferation. Trends Biochem Sci 22: 345–349, 1997. [DOI] [PubMed] [Google Scholar]
- 32.Ling J, Morley SJ, Traugh JA. Inhibition of cap-dependent translation via phosphorylation of eIF4G by protein kinase Pak2. EMBO J 24: 4094–4105, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Montagne J, Stewart MJ, Stocker H, Hafen E, Kozma SC, Thomas G. Drosophila S6 kinase: a regulator of cell size. Science 285: 2126–2129, 1999. [DOI] [PubMed] [Google Scholar]
- 34.Moss T At the crossroads of growth control; making ribosomal RNA. Curr Opin Genet Dev 14: 210–217, 2004. [DOI] [PubMed] [Google Scholar]
- 35.Nousch M, Reed V, Bryson-Richardson RJ, Currie PD, Preiss T. The eIF4G-homolog p97 can activate translation independent of caspase cleavage. RNA 13: 374–384, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pause A, Methot N, Svitkin Y, Merrick WC, Sonenberg N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J 13: 1205–1215, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Proud CG Signalling to translation: how signal transduction pathways control the protein synthetic machinery. Biochem J 403: 217–234, 2007. [DOI] [PubMed] [Google Scholar]
- 38.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433: 477–480, 2005. [DOI] [PubMed] [Google Scholar]
- 39.Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat Rev Cancer 3: 179–192, 2003. [DOI] [PubMed] [Google Scholar]
- 40.Ruvinsky I, Meyuhas O. Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem Sci 31: 342–348, 2006. [DOI] [PubMed] [Google Scholar]
- 41.Ruvinsky I, Sharon N, Lerer T, Cohen H, Stolovich-Rain M, Nir T, Dor Y, Zisman P, Meyuhas O. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev 19: 2199–2211, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Umen JG The elusive sizer. Curr Opin Cell Biol 17: 435–441, 2005. [DOI] [PubMed] [Google Scholar]
- 43.Williams-Hill DM, Duncan RF, Nielsen PJ, Tahara SM. Differential expression of the murine eukaryotic translation initiation factor isogenes eIF4A(I) and eIF4A(II) is dependent upon cellular growth status. Arch Biochem Biophys 338: 111–120, 1997. [DOI] [PubMed] [Google Scholar]
- 44.Yoo SJ, Jimenez RH, Sanders JA, Boylan JM, Brautigan DL, Gruppuso PA. The α4-containing form of protein phosphatase 2A in liver and hepatic cells. J Cell Biochem In press. [DOI] [PMC free article] [PubMed]
- 45.Zakowicz H, Yang HS, Stark C, Wlodawer A, Laronde-Leblanc N, Colburn NH. Mutational analysis of the DEAD-box RNA helicase eIF4AII characterizes its interaction with transformation suppressor Pdcd4 and eIF4GI. RNA 11: 261–274, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]