Abstract
Inbred mouse strains are routinely used as genetically defined animal models for studying a wide assortment of biological and pathological processes, including immune system function. However, no studies have presented large-scale data on the immune cell populations among the inbred strains in physiological conditions. Here we present a systematic, quantitative analysis of peripheral blood cell phenotypes of 30 mouse strains assessed by flow cytometry. This cohort of mice represents a wide range of genetic origins and includes most of the strains used in genetic, physiological, and immunological studies. We evaluated the relative percentages of peripheral blood leukocyte subtypes (lymphocytes, granulocytes, and monocytes) and lymphocyte subpopulations (CD4+ T, CD8+ T, B220+ B, and natural killer cells) of mature (6-mo-old) mice. Our comprehensive study demonstrated: 1) marked differences in the relative proportions of blood cell populations among the strains at this age, 2) considerable variation of each immune trait with more than twofold difference between strains with the highest and the lowest trait values, and 3) haplotype analysis revealed a strong correlation between eosinophil percentage and a single region on chromosome 14 containing two candidate genes. The strain differences described here provide important information for researchers applying immunophenotyping of peripheral blood in immunological and genetic studies. The data from this study are available as part of the Mouse Phenome Database at http://www.jax.org/phenome.
Keywords: leukocytes, lymphocytes, flow cytometry, inbred mouse strains, immunophenotyping
leukocyte subpopulations and numbers are tightly regulated and remain relatively constant in the peripheral blood of animals of several mammalian species. This phenomenon, known as immune cell homeostasis, represents a balance between cell proliferation and programmed cell death of discrete leukocyte subsets (1, 14, 16). It is finely tuned and its disturbance could lead to immunodeficiency, transformation, or autoimmune disorders (4, 7). Peripheral blood provides an overall assessment of leukocyte homeostasis in circulation. Previous studies have shown that the relative percentage of peripheral blood leukocyte populations varies among mouse strains of a young age and also varies with age (2, 8). However, the power of those studies was limited in the number of strains analyzed. In this report, we use a flow cytometric approach to compare immune cells in males and females of 30 inbred strains. Our panel of mouse strains includes the most commonly used laboratory strains, as well as three representatives of wild-derived strains, MOLF/EiJ, PWD/PhJ, and WSB/EiJ, chosen for genetic diversity (for details see Table 1). The 30 strains were chosen so that all seven mouse groups comprising the mouse family tree were represented (13). Our cohorts were analyzed at 6 mo of age to profile mature mice.
Table 1.
List of selected inbred mouse strains (n = 30) included in the study
| Mouse Group | Strains |
|---|---|
| Group 1, Bagg albino derivatives | A/J, AKR/J, BALB/cByJ, C3H/HeJ, CBA/J, PL/J, MRL/MpJ |
| Group 2, Swiss mice | BUB/BnJ, FVB/NJ, RIIIS/J, SJL/J, SWR/J |
| Group 3, Japanese, New Zealand strains | KK/HlJ, NON/LtJ, NZO/HlLtJ, NZW/LacJ |
| Group 4, C57/58 strains | C57BL/10J, C57BL/6J, C57BLKS/J, C57BR/cdJ, C57L/J |
| Group 5, Castle's mice | 129S1/SvImJ, BTBR T+ tf/J, LP/J |
| Group 6, C.C. Little's DBA and related strains | DBA/2J, P/J, SM/J |
| Group 7, Wild-derived strains | MOLF/EiJ, PWD/PhJ, WSB/EiJ |
Mouse group numbers and names were adopted from Petkov et al. (13). Group 1 includes 7 strains (n = 7); group 2, n = 5, group 3, n = 4; group 4, n = 5; group 5, n = 3; group 6, n = 3; group 7, n = 3.
MATERIALS AND METHODS
Mice.
All strains were obtained from The Jackson Laboratory (TJL, Bar Harbor, ME): 129S1/SvImJ, A/J, AKR/J, BALB/cByJ, BTBR T+ tf/J, BUB/BnJ, C3H/HeJ, C57BL/10J, C57BL/6J, C57BLKS/J, C57BR/cdJ, C57L/J, CBA/J, DBA/2J, FVB/NJ, KK/HlJ, LP/J, MOLF/EiJ, MRL/MpJ, NON/LtJ, NZO/HlLtJ, NZW/LacJ, P/J, PL/J, PWD/PhJ, RIIIS/J, SJL/J, SM/J, SWR/J, and WSB/EiJ (strain names are listed in Table 1). In this study, eight females and eight males were analyzed for each strain at 6 mo of age.
Mice were housed in pressurized individually ventilated (PIV) cages (Thoren Caging Systems, Hazleton, PA) with HEPA filtered supply and exhaust in duplex polycarbonate cages measuring 31 × 31 × 214 cm that were divided into two pens with five mice/pen. Autoclaved white pine shavings (Crobb Box, Ellsworth, ME) were used as bedding. Cages and bedding were changed every 2 wk. Mice had ad libitum access to acidified water (pH 2.8–3.1) and autoclaved pelleted diet containing 6% fat (Lab Diet 5001; PMI Nutritional International, Brentwood, MO). The animal room was maintained at 23°C on a 12 h light-12 h dark cycle.
These colonies were regularly monitored for and are free of 15 viruses (mouse hepatitis virus, two mouse parvoviruses, reovirus, Theiler's mouse encephalomyelitis virus, ectromelia virus, mouse rotavirus, thymic virus, mouse pneumonia virus, Sendai virus, murine cytomegalovirus, lactic dehydrogenase-elevating virus, K virus, mouse adenovirus, and polyoma virus), 17 bacterial species (including Helicobacter spp. and P. pneumotropica), two Mycoplasma spp., external and internal parasites, and Encephalitozoon cuniculi. The mouse room was monitored for all these organisms regularly. The mice were housed in a barrier facility, the Genetic Resources Building, and room entry procedures required personnel to don caps, facemasks, disposable gowns, shoe covers, and gloves.
All manipulations with the mice were conducted according to the American Association of Laboratory Animal Care program and approved by the Animal Care and Use Committee at The Jackson Laboratory (TJL, Bar Harbor, ME).
Blood cell preparation.
Peripheral blood (∼250 μl) was collected from the retroorbital plexus of each mouse using glass microcapillary tubes in a tube containing K2EDTA as anticoagulant (BD Biosciences, San Jose, CA). Blood samples were centrifuged at 14,000 rpm for 10 min (4°C), and plasma was recovered and stored at −80°C for additional analyses. After centrifugation the cell pellet was resuspended in 0.5 ml FACS buffer (1× PBS containing 2% FBS, 2 mM EDTA, and 0.02% NaN3), and subjected twice to erythrocyte lysis using 3 ml of Gey's hemolytic solution. Cells were next washed with 4 ml of PBS containing 2 mM K2EDTA, resuspended with 100 μl PBS, divided into three groups, and subjected to cell staining using premixed antibodies (see antibodies) and flow cytometry. Peripheral blood obtained from 3-mo-old male C57BL6/J mice (n = 3) were included in each flow cytometry analysis over the course of study as assay control group.
Antibodies.
In this study we used monoclonal antibodies recognizing cell surface antigens expressed on different cell types. Several fluorochrome conjugated antibodies were combined in three separate mixtures to obtain comprehensive phenotyping of a variety of immune cell populations (Table 2). Mixture 1 was used to stain T cell subsets and included the following antibodies: anti-CD3e (clone 145-2C11) APC-Cy7, anti-CD4 (clone GK1.5) Cy3, and anti-CD8 (clone 53-6.7) PE. Mixture 2 was used to stain B cells, granulocytes, and monocytes and included: anti-CD45R/B220 (clone RA3-6B2) APC, anti-CD11b/Mac-1 (clone M1/70) PE, and anti-Ly6G/Gr-1 (clone RB6-8C5) APC-Cy7. Mixture 3 was generated to specifically stain natural killer (NK) cells and included: anti-CD3e (clone 145-2C11) APC-Cy7, anti-NK1.1 (clone PK136) PE, anti-Lgl-1 (clone 4D11) APC, and anti-NKG2A/C/E (clone 20d5) FITC. Antibodies used in the study were purchased from eBiosciences (San Diego, CA) and BD Biosciences (San Jose, CA).
Table 2.
Different populations of PB leukocytes
| Cell Population | Phenotype | Measure |
|---|---|---|
| 1. LY | low SSC vs. FSC profile | LY, % of leukocytes |
| 1.1 T cells | ||
| CD4+ T cells | CD3+CD4+ | CD4+ T, % of lymphocytes |
| CD8+ T cells | CD3+CD8+ | CD8+ T, % of lymphocytes |
| 1.2 B cells | ||
| B220+ B cells | B220+ | B220+ B, % of lymphocytes |
| 1.3 NK cells | CD3−NK1.1+ Lgl-1+ NKG2A/C/D+ | NK, % of lymphocytes |
| 2. Gr | SSChighCD11b+Gr-1high | Gr, % of leukocytes |
| 3. Eos | SSChighCD11b+Gr-1mid-to-low | Eos, % of leukocytes |
| 4. Mo | SSClowCD11b+ | Mo, % of leukocytes |
Blood lymphocytes are gated using dot-plots of granularity (SSC vs. FSC). PB, peripheral blood; SSC, side scatter; FSC, forward scatter; Gr, granulocytes; Eos, eosinophils; Mo, monocytes; LY, lymphocytes.
Flow cytometry.
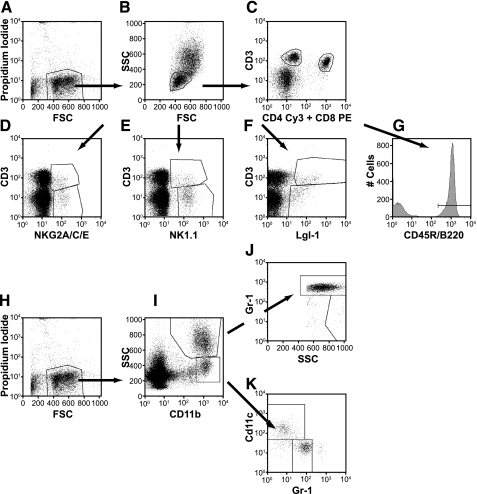
The cells were kept on ice until use. Propidium iodide (PI) was added prior to analysis to exclude dead cells and samples were run on the FACScan upgraded by Cytek Development (Cytek, Freemont, CA). At least 25,000 and up to 50,000 events were acquired per sample. Multicolor flow cytometry analyses were used to evaluate the proportions of different immune cell population. Acquired cell count files are analyzed with FlowJo software (Tree Star, Ashland, OR). Figure 1 shows raw data from one representative mouse with gating conditions. Viable cells are gated by forward scatter (FSC) and exclusion of PI (Fig. 1A). Red blood cells were excluded as they have lower FSC than lymphocytes. Lymphocytes (Fig. 1B) were gated from the viable cells by FSC vs. SSC (side scatter). They were subsequently gated for T cells, B cells, and NK cells. T cells are gated from the lymphocyte gate on the basis of CD3 and CD4 expression for CD4+ T cells (“T helper”), and CD3 and CD8 expression for CD8+ T cells (“cytotoxic”) (Fig. 1C). NK cells (9) were reported as CD3− cells divided into three subgroups: Lgl-1+, NK1.1+, and NKG2A/C/E+ (Fig. 1, D, E, F). Total NK cells are cells that fall in any of those three gates. B cells are gated from the lymphocytes as positive for the CD45R/B220 antigen (B220+ B cells) (Fig. 1G). Granulocytes are gated from viable cells (Fig. 1H) as SSChigh CD11b+ and Gr-1high (Fig. 1, I and J). Eosinophils (17) are gated as SSChigh CD11b+ and Gr-1mid-to-low (Fig. 1J). Monocytes (6) were CD11b+ and SSClow compared with granulocytes (Fig. 1I). Inflammatory monocytes are gated as CD11b+CD11c−Gr-1+ and resident monocytes are gated as CD11b+ CD11c+ Gr-1− cells (Fig. 1K).
Fig. 1.
An example of flow cytometry analysis with gating conditions. A: viable cells were gated by forward scatter (FSC) and exclusion of propidium iodide. B: lymphocytes were gated from the viable cells by FSC vs. SSC (side scatter). C: T cells were gated from the lymphocyte gate on the basis of CD3 and CD4 expression for CD4+ T cells, and CD3 and CD8 expression for CD8+ T cells. D–F: total natural killer (NK) cells were reported as CD3− cells expressing the following cell surface determinants: Lgl-1, NK1.1, and NKG2A/C/E. G: B cells were gated from the lymphocytes as positive for the CD45R/B220 antigen (B220+ B cells). H: viable cells. I: granulocytes were gated from viable cells as CD11b+SSChigh. J: granulocytes/neutrophils and eosinophils were gated as SSChigh Gr-1high and SSChigh Gr-1mid-to-low cells. K: monocytes were separated from granulocytes as CD11b+ and SSClow (as compared with granulocytes) and separated in 2 populations. Inflammatory monocytes were gated as CD11b+CD11c−Gr-1+ and resident monocytes were gated as CD11b+ CD11c+Gr-1− cells.
Haplotype association mapping.
The haplotype association mapping (HAM) analysis is employed in a two-step procedure. In the first step, genotype matrix of 63,222 loci across 29 inbred strains was imputed using Hidden Markov model (HMM) described in Szatkiewicz et al. (19). The same method was used to infer haplotype states under the assumption of moderate level of linkage disequilibrium in this dataset. The HMM inferred haplotype blocks ranged from a single nucleotide polymorphism to 10 or more. Within each block, strains were grouped by the inferred local haplotype states. The number of distinct haplotype groups ranged between 2 and 5 across the genome. In step 2, we calculate one-way analysis of variance (ANOVA) F statistics at each block to determine the haplotype and phenotype association. The nominal P value of the test statistic was computed and then transformed into logarithm-of-the-odds score using −log10. The final P value was determined after multiple testing adjustments using family-wide error rate control. The analysis of association was done in the MATLAB computing environment (http://www.mathworks.com).
Statistical analysis.
The 6-mo-old cohort consisted of 475 mice (240 females and 235 males). Raw data obtained from each blood sample were recorded using CellQuest (BD Biosciences, San Jose, CA) software, and the relative percentage of each cell population was calculated using FlowJo software (as described above). The statistical analysis was done using JMP software (SAS Institute, Cary, NC). Results were first calculated for each mouse, averaged per strain group (male and female groups were kept separately), and trimmed (if necessary) by removing values >3 SDs from the mean to moderate the effect of outliers. Results from the analysis were presented as mean percentage of each immune cell population ± SE. The data set can be found in the Mouse Phenome Database (http://www.jax.org/phenome).
Strain variations for each immune parameter between female and male groups were assessed using two-way ANOVA. A conventional value of P = 0.05 was considered statistically significant in all tests. Bonferroni correction of P value was used to reduce the number of falsely significant results. The Bonferroni adjustment (P value corrected) was done by multiplying the P value by number of tests carried out (30 × 8 = 240 tests). The significant threshold was set at 0.0002 (0.05/240 = 0.0002), and the corrected values less than the threshold are considered significant.
To assess the correlation between each pair of immune subsets among strains we used bivariate scatterplot fitting command of JMP and Pearson correlation coefficient (r) was calculated based on strain means. If there was an exact linear relationship between two variables, the correlation coefficient was ≤ 1 or ≥ −1, depending on whether the variables were positively or negatively correlated. R = 0 indicated absence of correlation.
RESULTS AND DISCUSSION
In this study we determined the relative percentages of leukocyte subpopulations in peripheral blood obtained from mature (6-mo-old) mice. The analysis included major leukocyte subpopulations of lymphocytes, granulocytes, eosinophils, monocytes, and lymphocyte subsets including CD4+, CD8+, B220+, and NK cells. Peripheral blood leukocytes from each mouse were analyzed independently according to the gating conditions presented in Fig. 1, averaged per strain group, and presented separately for males and females to assess the effect of sex and strain on the immune phenotypes (Figs. 2 and 3).
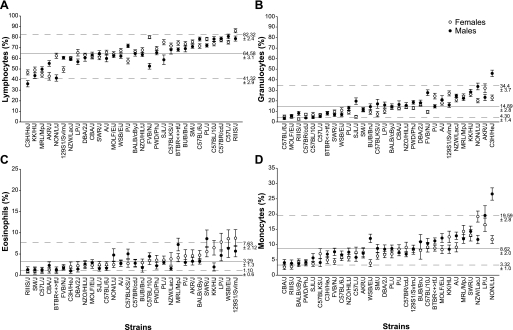
Fig. 2.
Strain- and sex-specific variations in peripheral blood leukocyte populations. Mean percentage of each immune cell population per strain per group is presented as a percentage of leukocytes ± SE. Female groups are shown as open symbols, and male groups as closed symbols. Bars indicate SE. The x-axis gives the strain names; y-axis represents the following immune subpopulations: lymphocytes (A), granulocytes (B), eosinophils (C), and monocytes (D). Mean percentages of each leukocyte group for all strains (female and male) are shown as solid lines, and minimum and maximum values are shown with dotted lines.
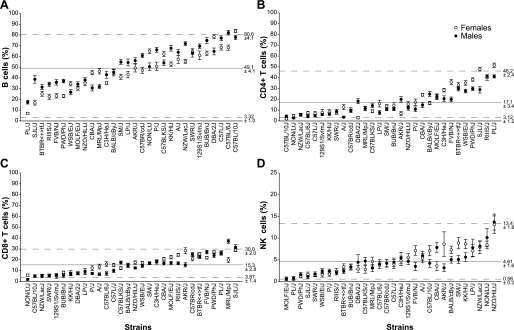
Fig. 3.
Strain- and sex-specific variations in peripheral blood lymphocyte subpopulations. Results are plotted as mean values (% of lymphocytes) per strain per group. Bars indicate SE. Female (open symbols) and male (closed symbols) groups for each strain were separated. The x-axis gives the strain names, and the y-axis gives the immune phenotype as follows: B220+ B cells (A), CD4+ T cells (B), CD8+ T cells (C), NK (D) cells. Lymphocyte subsets are presented as % of lymphocytes ± SE. The solid line in each panel represents sex and strain mean % ± SE, and the dotted lines represented minimum and maximum values.
The flow cytometry analysis of peripheral blood showed that the percentages of total lymphocytes (% LY) varied between 41.3 and 82.3% (Fig. 2A, Table 3). Both female and male groups of C3H/HeJ mice had the lowest percentage of lymphocytes at 6 mo of age (female 46.6 ± 2.3% and male 36.0 ± 2.9%). In contrast, female RIIIS/J mice and male C57L/J mice had the highest percentages of peripheral blood lymphocytes (respectively 86.0 ± 1.8% and 80.6 ± 2.2%). Results showed that the extremes (minimal %lymphocytes: female 44.3 and male 36.0%, maximum %lymphocytes: female 86.0% and male 80.6%) were twofold different from the overall strain means (female 65.8% and male 63.1%), suggesting moderate variations of lymphocytes among the inbred mouse strains. Our results were in agreement with the results described previously (8), showing that C3H/HeJ mice had the lowest proportion of PB lymphocytes among 15 inbred strains. However, the percentages of PB lymphocytes in C3H/HeJ mice in that study (female 75.7 ± 4.3%, male 72.3 ± 14.0%) were higher compared with our results (female 46.6 ± 2.3%, male 36.0 ± 2.9%). This discrepancy could be due to several reasons: 1) different method (complete blood cell count vs. flow cytometry) used in these two studies, 2) different age of the mice (3–4-mo-old vs. 6-mo-old), or 3) more pathogen exposure in the facility with higher lymphocytes compared with our specific pathogen-free barrier facility.
Table 3.
Strain variability of PB leukocyte populations
| Strain Name | Sex (n) | Lymphocytes | Granulocytes | Eosinophils | Monocytes | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Group 1 | ||||||||||
| A/J | F (8) | 65.5±2.0 | 17.9±2.0 | 2.73±0.6 | 9.10±1.5 | |||||
| M (8) | 59.8±3.1 | 21.0±3.8 | 2.80±0.7 | 12.8±1.2 | ||||||
| P value | P > 1.0 | P > 1.0 | P = 1.0 | P = 0.6 | ||||||
| AKR/J | F (7) | 43.3±3.6 | 31.9±4.0 | 4.54±1.4 | 7.46±1.5 | |||||
| M (8) | 55.1±2.6 | 23.1±2.9 | 3.27±0.8 | 7.13±1.5 | ||||||
| P value | P > 1.0 | P > 1.0 | P > 1.0 | P > 1.0 | ||||||
| BALB/cByJ | F (8) | 66.3±1.7 | 13.4±1.7 | 4.71±1.0 | 4.24±1.0 | |||||
| M (8) | 64.4±2.3 | 14.4±2.6 | 3.25±0.6 | 3.77±1.0 | ||||||
| P value | P > 1.0 | P >1.0 | P > 1.0 | P > 1.0 | ||||||
| C3H/HeJ | F (8) | 46.6±2.3 | 23.8±2.5 | 2.0±1.0 | 4.94±1.0 | |||||
| M (8) | 36.0±2.9 | 46.0±3.1 | 1.0±0.5 | 6.86±1.0 | ||||||
| P value | P > 1.0 | P = 0.01 | P > 1.0 | P > 1.0 | ||||||
| CBA/J | F (8) | 61.8±2.4 | 12.7±1.3 | 1.16±0.6 | 2.59±0.8 | |||||
| M (8) | 63.0±2.8 | 16.5±3.4 | 2.29±0.8 | 4.05±1.0 | ||||||
| P value | P > 1.0 | P > 1.0 | P = 0.3 | P > 1.0 | ||||||
| MRL/MpJ | F (8) | 44.3±2.0 | 24.1±2.3 | 4.11±0.9 | 14.2±2.0 | |||||
| M (8) | 49.6±2.3 | 22.4±2.3 | 7.22±1.4 | 11.8±1.4 | ||||||
| P value | P > 1.0 | P > 1.0 | P = 0.6 | P >1.0 | ||||||
| PL/J | F (7) | 78.1±1.6 | 8.95±1.5 | 3.54±0.9 | 6.90±1.2 | |||||
| M (7) | 72.1±1.5 | 12.9±1.9 | 3.02±0.4 | 8.74±1.3 | ||||||
| P value | P = 0.2 | P > 1.0 | P > 1.0 | P > 1.0 | ||||||
| Group 2 | ||||||||||
| BUB/BnJ | F (8) | 73.6±2.1 | 9.19±1.9 | 2.92±0.8 | 8.11±2.8 | |||||
| M (8) | 69.1±3.0 | 14.3±2.7 | 1.79±1.0 | 10.8±2.4 | ||||||
| P value | P > 1.0 | P > 1.0 | P > 1.0 | P > 1.0 | ||||||
| FVB/NJ | F (8) | 79.9±1.5 | 9.44±1.4 | 1.93±0.5 | 4.47±0.9 | |||||
| M (8) | 52.5±2.7 | 27.6±2.6 | 1.66±0.4 | 7.62±1.1 | ||||||
| P value | P = 0.00002 | P = 0.001 | P > 1.0 | P = 0.03 | ||||||
| RIIIS/J | F (8) | 86.0±1.8 | 2.73±1.1 | 0.93±0.4 | 2.80±0.8 | |||||
| M (8) | 78.6±2.3 | 8.12±1.9 | 1.27±0.6 | 3.85±1.0 | ||||||
| P value | P > 1.0 | P = 0.4 | P > 1.0 | P > 1.0 | ||||||
| SJL/J | F (8) | 75.1±2.4 | 4.87±1.4 | 2.45±0.7 | 3.69±0.8 | |||||
| M (6) | 58.6±4.4 | 19.6±3.1 | 1.55±0.6 | 5.65±1.3 | ||||||
| P value | P > 1.0 | P > 1.0 | P = 0.8 | P > 1.0 | ||||||
| SWR/J | F (8) | 61.0±2.3 | 9.75±1.5 | 5.55±1.2 | 13.0±1.8 | |||||
| M (8) | 64.5±2.6 | 6.74±1.5 | 8.58±2.0 | 14.2±1.7 | ||||||
| P value | P > 1.0 | P > 1.0 | P > 1.0 | P > 1.0 | ||||||
| Group 3 | ||||||||||
| KK/HlJ | F (8) | 48.9±2.7 | 26.2±2.2 | 6.44±1.7 | 12.5±1.8 | |||||
| M (8) | 43.7±2.6 | 28.3±2.5 | 1.92±0.9 | 8.49±1.3 | ||||||
| P value | P > 1.0 | P > 1.0 | P = 0.2 | P > 1.0 | ||||||
| NON/LtJ | F (8) | 62.4±2.3 | 20.2±2.1 | 2.47±0.7 | 11.6±1.2 | |||||
| M (8) | 41.4±3.0 | 33.5±3.1 | 4.75±1.4 | 26.5±2.0 | ||||||
| P value | P = 0.01 | P > 1.0 | P > 1.0 | P = 0.0001 | ||||||
| NZO/HlLtJ | F (8) | 67.8±2.1 | 14.1±1.4 | 2.33±0.8 | 5.68±1.0 | |||||
| M (8) | 63.5±2.0 | 15.6±2.6 | 2.63±0.7 | 7.48±1.3 | ||||||
| P value | P > 1.0 | P > 1.0 | P > 1.0 | P > 1.0 | ||||||
| NZW/LacJ | F (8) | 60.1±1.4 | 21.0±2.1 | 3.82±1.4 | 19.1±1.8 | |||||
| M (8) | 59.3±1.5 | 24.6±1.6 | 1.55±0.4 | 11.6±1.3 | ||||||
| P value | P > 1.0 | P > 1.0 | P > 1.0 | P = 0.01 | ||||||
| Group 4 | ||||||||||
| C57BL/10J | F (8) | 73.7±1.9 | 6.86±2.1 | 3.43±0.9 | 9.15±1.4 | |||||
| M (8) | 78.2±2.1 | 4.57±1.5 | 4.42±0.8 | 10.3±1.6 | ||||||
| P value | P > 1.0 | P > 1.0 | P > 1.0 | P > 1.0 | ||||||
| C57BL/6J | F (8) | 71.5±2.0 | 5.10±1.4 | 2.46±0.6 | 5.79±1.0 | |||||
| M (8) | 78.2±1.8 | 3.51±1.4 | 1.65±0.5 | 6.60±1.2 | ||||||
| P value | P = 0.92 | P > 1.0 | P = 0.23 | P > 1.0 | ||||||
| C57BLKS/J | F (8) | 71.7±2.6 | 7.36±2.4 | 2.75±0.8 | 6.99±1.6 | |||||
| M (8) | 68.1±4.1 | 17.1±4.3 | 5.02±1.1 | 4.11±1.3 | ||||||
| P value | P > 1.0 | P > 1.0 | P = 0.15 | P > 1.0 | ||||||
| C57BR/cdJ | F (8) | 77.1±1.5 | 7.08±1.1 | 2.81±0.9 | 9.06±1.7 | |||||
| M (8) | 78.4±1.7 | 3.78±0.7 | 2.82±0.7 | 6.85±0.8 | ||||||
| P value | P > 1.0 | P = 0.003 | P > 1.0 | P > 1.0 | ||||||
| C57L/J | F (8) | 75.1±1.7 | 6.64±1.3 | 1.13±0.7 | 7.48±1.6 | |||||
| M (8) | 80.6±2.2 | 5.59±1.5 | 1.29±0.5 | 5.80±1.2 | ||||||
| P value | P > 1.0 | P > 1.0 | P > 1.0 | P > 1.0 | ||||||
| Group 5 | ||||||||||
| 129S1/SvImJ | F (8) | 49.4±2.6 | 27.0±2.3 | 8.73±2.0 | 8.30±1.4 | |||||
| M (8) | 60.5±1.4 | 16.0±2.2 | 5.65±1.3 | 8.61±1.1 | ||||||
| P value | P = 0.16 | P = 0.30 | P > 1.0 | P > 1.0 | ||||||
| BTBRT+tf/J | F (8) | 74.4±2.0 | 8.18±1.7 | 1.17±0.7 | 8.83±0.9 | |||||
| M (8) | 68.2±1.6 | 8.07±1.5 | 1.31±0.6 | 11.1±0.9 | ||||||
| P value | P = 1.06 | P > 1.0 | P > 1.0 | P = 0.06 | ||||||
| LP/J | F (8) | 65.2±3.1 | 10.0±2.3 | 7.79±2.0 | 16.4±2.1 | |||||
| M (8) | 56.7±2.5 | 16.0±2.8 | 4.65±1.3 | 19.5±3.1 | ||||||
| P value | P > 1.0 | P > 1.0 | P > 1.0 | P > 1.0 | ||||||
| Group 6 | ||||||||||
| DBA/2J | F (8) | 63.0±1.9 | 16.5±1.6 | 2.24±0.9 | 6.70±1.0 | |||||
| M (8) | 60.5±3.2 | 18.0±2.6 | 1.43±0.6 | 8.49±1.4 | ||||||
| P value | P > 1.0 | P > 1.0 | P > 1.0 | P > 1.0 | ||||||
| P/J | F (8) | 57.4±2.7 | 24.4±2.6 | 4.53±1.4 | 8.53±1.4 | |||||
| M (8) | 71.7±1.7 | 14.7±1.6 | 2.98±0.6 | 7.38±1.0 | ||||||
| P value | P = 0.05 | P = 0.8 | P > 1.0 | P > 1.0 | ||||||
| SM/J | F (8) | 75.6±1.7 | 11.3±1.8 | 0.98±0.7 | 6.25±1.0 | |||||
| M (8) | 71.1±1.6 | 11.8±1.6 | 1.26±0.6 | 80.74±1.3 | ||||||
| P value | P > 1.0 | P > 1.0 | P > 1.0 | P = 0.86 | ||||||
| Group 7 | ||||||||||
| MOLF/EiJ | F (7) | 64.1±2.0 | 4.84±1.5 | 2.35±0.7 | 8.37±1.3 | |||||
| M (8) | 62.5±1.9 | 5.69±1.3 | 2.91±0.7 | 12.1±1.4 | ||||||
| P value | P > 1.0 | P > 1.0 | P > 1.0 | P = 0.5 | ||||||
| PWD/PhJ | F (8) | 69.1±2.0 | 13.1±1.4 | 3.52±1.1 | 3.89±0.8 | |||||
| M (8) | 65.3±2.0 | 18.5±1.7 | 2.20±0.7 | 4.18±0.8 | ||||||
| P value | P > 1.0 | P = 0.2 | P > 1.0 | P > 1.0 | ||||||
| WSB/EiJ | F (8) | 67.3±2.0 | 10.9±2.4 | 8.60±2.0 | 3.90±1.1 | |||||
| M (6) | 62.0±2.1 | 6.84±1.6 | 6.34±2.1 | 12.0±1.3 | ||||||
| P value | P > 1.0 | P > 1.0 | P > 1.0 | P = 0.0001 | ||||||
PB leukocyte populations are presented as % of viable cells. Data are mean % ± SE for each group of mice. The Bonferroni adjustment (P value corrected) is calculated to estimate the sex-specific differences between males and females. The significant threshold was set at 0.0002, and the corrected values less than the threshold are considered significant.
Interstrain comparison revealed a high degree of similarity in the percentage of lymphocytes among group 4 (C57/C58-related strains) used in our study including: C57BLKS/J, C57BL/6J, C57BL/10J, C57BR/cdJ, and C57BL/J (females 71.7, 71.5, 73.7, 77.1, 75.1% and males 68.1, 78.2, 78.2, 78.4, 80.6%) (Table 3). The same similarity of %lymphocytes existed among the wild-derived strains: MOLF/EiJ, PWD/PhJ, and WSB/EiJ. Female group had values of 64.1, 69.1, 67.3% and male group has values of 62.5, 65.3, 62.0%, respectively. These results suggested a genetic influence on lymphocyte proportions, most pronounced for group 4, which could be explained for example with the fact that C57/58 strains are derived from a common ancestor (13). In contrast, other strains belonging to the same group showed variability. For example, several strains including C3H/HeJ, MRL/MpJ, AKR /J belonging to mouse group 1 (Bagg albino derivatives) exhibited low percentage of lymphocytes (females 46.6, 44.3, 43.0, males 36, 49.6, 55.1), others (CBA/J, A/J, BALB/cByJ) had intermediate (females 61.8, 65.5, 66.3% and males 63.0, 59.8, 64.4%), and one (PL/J) had high proportions of lymphocytes (female 78.1%, male 72.1%). Strains belonging to groups 2, 3, 5, and 6 also showed substantial variability. This variability of lymphocyte proportions could be explained with the fact that some strains are derived from crosses of strains belonging to different mouse groups, which creates variability among the strains belonging to the same group.
To evaluate the effect of sex, the means of lymphocytes were compared for each mouse strain, and the P value was calculated. To validate our results and eliminate false-positive significant results, P values were subjected to Bonferroni correction. Significant difference (P value corrected <0.0002) between the two sexes was observed in 1 of 30 strains (Table 3).
Granulocytes and monocyte/macrophage lineage cells comprise the innate immune system, which is responsible for recognizing and destroying foreign pathogens. Because of the importance of the members of the innate immunity, we included granulocytes, eosinophils, and monocytes (Fig. 2, B, C, D and Table 3). In mice, blood granulocytes (eosinophils, neutrophils, and basophils) consist mainly of neutrophils, and their number in physiological conditions is tightly regulated (18). We separated the granulated cells based on the expression of Gr-1 and SSC in two groups: granulocytes/neutrophils (SSChighGr-1high) and eosinophils (SSChighGr-1mid-to-low). Our results demonstrated that the 30 inbred strains displayed significant variation in granulocytes (Fig. 2B). Extreme values were represented by RIIIS/J (females 2.7 ± 1.1%), C57BL/6J (males 3.5 ± 1.4%), AKR/J (females 31.9 ± 4.0%), and C3H/HeJ (males 46.0 ± 3.1%). C57BL/6J, MOLF/EiJ, RIIIS/J, C57BR/cdJ, C57BL/10J, and C57L/J strains had the lowest proportions of granulocytes (Fig. 2B). With the exception of MOLF/EiJ and RIIIS/J, these strains with low percentages of granulocytes belong to group 4 (C57/58 strains) of the mouse family tree. In contrast, AKR/J and C3H/HeJ, both members of group 1, had the highest proportions of granulocytes. Results also demonstrated that several strains including SJL/J, FVB/NJ, and C3H/HeJ demonstrated substantial but not significantly different (P value >0.0002) discrepancy between the two sexes (Table 3) with higher percentage of granulocyte in males compared with females. This result showed that SJL/ L male mice were enriched in granulocytes compared with females, which could be explained with their aggressiveness resulting in frequent scuffing, jumping and fighting.
Our comprehensive flow cytometry analysis allowed us to separate eosinophils from the granulocyte/neutrophil group. Eosinophil counts are presented in Fig. 2C and Table 3. The overall mean of this cell population is 3.29 ± 1.3% with lowest and highest percentage of 1.1 ± 0.6 and 7.6 ± 2.1%, respectively. There were no significant difference between males, and strains belonging to the same mouse group showed variations in eosinophil percentages. Note the marked variations between the strains with the lowest (female and male RIIIS/J) and the highest (female 129S1/SvImJ and male MRL/MpJ) counts (9.6-fold for 129S1/SvImJ vs. RIIIS/J and 6-fold difference for MRL/MpJ vs. RIIIS/J). These differences can be exploited for physiological studies and quantitative trait locus (QTL) analysis.
Mouse monocytes (5) were identified in this study as cells with low intracellular granularity and expressing CD11b/Mac-1 cell surface antigen (SSClowCD11b+). Figure 2D and Table 3 showed that 2 (WSB/EiJ and NON/LtJ) out of 30 of the inbred strains exhibited sexual dimorphism between females and males regarding monocyte percentages, however, the difference was not significant (P = 0.0001). Males exhibited slightly higher percentage of monocytes compared with female members of the same strain. The lowest monocyte percentage was measured in CBA/J (female 2.5 ± 0.8%) and RIIIS/J (male 3.8 ± 1.0%) strains and the highest in female LP/J (16.4 ± 4.7%) and male NON/LtJ (26.5 ± 2.0%) mice. Overall, the inbred strains (n = 30) also demonstrated substantial variations between the strains with the lowest and highest percentages of monocytes (females 4.6-fold and 6.9-fold for males). Moderate individual differences among the members of the same groups also existed.
Studies conducted at The Jackson Laboratory by Peters's group (12) have previously reported a set of hematolologic parameters, including %lymphocytes, neutrophils, and monocytes across 43 inbred strains. The authors showed marked variations in total white blood cell counts among the inbred strains (fourfold difference). Despite the different method used in their study, blood cell sedimentation assay, and the younger age of the mice (10 wk), a correlation between their data and ours has been found. For example, the average percentages of lymphocyte in their study are 79.2 ± 5.29% for females and 75.5 ± 6.94% for males. We report here an average percent of 63.1 ± 11.5% for females and 65.6 ± 10.9% for males. Lymphocyte values in our study are lower but not significantly different from Peters's data. Moreover, C57L/J, C57BR/cdJ, C57BL/10J, and RIIIS/J strains had the lowest proportions of granulocytes/neutrophils in contrast to KK/HlJ and NON/LiJ, which had the highest values in both studies. It should be noted that the mice used in these two studies are of different age, 10 wk vs. 6 mo, and aging has impact on immune cells proportions in peripheral blood. Importantly, the correlation with Peters's data indicates that the immunophenotyping approach used in the study is reliable and highly reproducible.
Lymphocytes specifically recognize and respond to foreign antigens and consist mainly of B and T lymphocytes. Our results showed that the overall mean percentage (calculated as percent of total lymphocytes) of B220+ B cells was 49.1 ± 4.1% (Fig. 3A). The relative percentages of B220+ B cells demonstrated substantial variation among the strains, the mean varying from 6.9 to 83.6% among females and 17.4 to 77.6% among males. PL/J strain had the lowest percentage of mature B cells (female 6.9 ± 0.8%, male 17.4 ± 1.8%) compared with female C57BL/10J and male C57BL/6J, which had the highest percentages (83.6 ± 1.5% and 82.0 ± 2.1%). Results also demonstrated that 2 out of 30 strains exhibited significantly different (P < 0.0002) disparity between the sexes (see Table 4). Among 30 strains, 22 strains had higher percentage of B220+ B cells in males compared with females, and only few strains demonstrated the prevalence of B220+ B cells in females. This result suggested that male mice were enriched in B cells compared with females. This observation was in concordance with previously published studies conducted using different mouse stocks but leading to the same conclusion (21).
Table 4.
Strain variability of PB lymphocyte subpopulations
| Strain Name | Sex (n) | B220+ B | CD4+ T | CD8+ T | NK | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Group 1 | ||||||||||
| A/J | F (8) | 57.1±3.7 | 12.7±2.3 | 13.7±2.2 | 5.30±1.0 | |||||
| M (8) | 63.7±2.7 | 3.52±0.7 | 6.72±1.2 | 5.90±1.0 | ||||||
| P value | P > 1.0 | P = 0.18 | P = 0.76 | P > 1.0 | ||||||
| AKR/J | F (7) | 49.1±3.6 | 6.83±1.8 | 28.3±2.6 | 8.61±2.8 | |||||
| M (8) | 56.1±2.5 | 19.8±2.4 | 15.3±1.9 | 3.16±0.8 | ||||||
| P value | P > 1.0 | P = 0.02 | P = 0.76 | P > 1.0 | ||||||
| BALB/cByJ | F (8) | 45.3±2.3 | 16.5±1.6 | 14.6±1.6 | 7.27±0.8 | |||||
| M (8) | 44.9±3.1 | 24.8±2.2 | 15.1±1.8 | 5.03±0.7 | ||||||
| P value | P > 1.0 | P = 0.23 | P = 0.11 | P = 0.003 | ||||||
| C3H/HeJ | F (8) | 35.1±1.2 | 28.3±1.6 | 18.3±1.0 | 4.76±1.0 | |||||
| M (8) | 42.9±3.2 | 20.4±2.6 | 18.0±1.9 | 5.42±1.2 | ||||||
| P value | P > 1.0 | P > 1.0 | P > 1.0 | P > 1.0 | ||||||
| CBA/J | F (8) | 44.8±1.9 | 24.1±1.2 | 20.9±1.4 | 3.57±1.0 | |||||
| M (8) | 30.6±2.7 | 14.5±2.2 | 16.8±1.5 | 7.83±1.0 | ||||||
| P value | P = 0.07 | P > 1.0 | P = 0.68 | P = 0.0006 | ||||||
| MRL/MpJ | F (8) | 46.1±2.4 | 11.4±1.9 | 19.8±2.4 | 4.58±1.0 | |||||
| M (8) | 31.8±1.9 | 11.3±1.7 | 37.1±1.8 | 3.10±0.8 | ||||||
| P value | P = 0.01 | P > 1.0 | P = 0.001 | P > 1.0 | ||||||
| PL/J | F (7) | 6.99±0.8 | 51.4±2.0 | 26.6±1.6 | 0.68±0.4 | |||||
| M (7) | 17.4±1.8 | 41.1±0.7 | 25.8±1.6 | 0.81±0.3 | ||||||
| P value | P = 0.0006 | P > 1.0 | P = 0.0001 | P > 1.0 | ||||||
| Group 2 | ||||||||||
| BUB/BnJ | F (8) | 62.9±2.1 | 14.4±2.4 | 8.29±1.3 | 3.37±0.6 | |||||
| M (8) | 74.7±2.8 | 10.8±1.4 | 5.47±1.7 | 3.29±0.6 | ||||||
| P value | P = 0.58 | P > 1.0 | P > 1.0 | P > 1.0 | ||||||
| FVB/NJ | F (8) | 23.3±2.0 | 35.9±2.2 | 21.9±1.2 | 7.15±1.5 | |||||
| M (8) | 35.8±2.5 | 19.9±2.1 | 27.0±1.6 | 3.49±0.9 | ||||||
| P value | P = 0.1 | P = 0.002 | P = 0.06 | P = 0.002 | ||||||
| RIIIS/J | F (8) | 22.3±2.4 | 40.0±2.4 | 24.5±1.4 | 2.28±07 | |||||
| M (8) | 34.1±1.8 | 41.1±1.5 | 16.8±1.2 | 2.03±0.5 | ||||||
| P value | P = 0.06 | P > 1.0 | P < 0.0002 | P > 1.0 | ||||||
| SJL/J | F (8) | 16.7±2.0 | 47.7±1.7 | 28.8±1.4 | 1.62±0.5 | |||||
| M (6) | 38.7±3.6 | 28.8±3.0 | 31.5±2.5 | 1.17±0.4 | ||||||
| P value | P = 0.8 | P > 1.0 | P > 1.0 | P > 1.0 | ||||||
| SWR/J | F (8) | 64.3±2.6 | 7.61±1.5 | 5.69±1.2 | 1.48±0.3 | |||||
| M (8) | 63.1±3.7 | 8.80±1.9 | 5.15±1.2 | 1.71±0.4 | ||||||
| P value | P > 1.0 | P > 1.0 | P > 1.0 | P = 0.02 | ||||||
| Group 3 | ||||||||||
| KK/HlJ | F (8) | 52.8±2.0 | 8.42±2.0 | 7.68±2.4 | 8.68±1.0 | |||||
| M (8) | 67.6±2.6 | 6.35±1.3 | 7.01±1.4 | 5.63±1.0 | ||||||
| P value | P = 0.04 | P > 1.0 | P > 1.0 | P = 0.01 | ||||||
| NON/LtJ | F (8) | 64.7±2.1 | 4.65±1.3 | 5.86±1.0 | 8.42±0.8 | |||||
| M (8) | 50.4±3.4 | 2.69±0.7 | 1.87±0.7 | 10.13±2.0 | ||||||
| P value | P > 1.0 | P > 1.0 | P < 0.0002 | P > 1.0 | ||||||
| NZO/HlLtJ | F (8) | 32.6±2.6 | 15.5±2.2 | 17.5±1.4 | 13.11±1.9 | |||||
| M (8) | 38.1±2.1 | 11.8±1.9 | 12.4±1.0 | 13.70±1.8 | ||||||
| P value | P > 1.0 | P > 1.0 | P = 0.01 | P > 1.0 | ||||||
| NZW/LacJ | F (8) | 55.3±1.7 | 5.65±1.4 | 4.88±1.3 | 7.91±1.4 | |||||
| M (8) | 71.9±1.6 | 5.62±0.9 | 5.55±1.0 | 8.16±1.0 | ||||||
| P value | P < 0.0002 | P > 1.0 | P > 1.0 | P > 1.0 | ||||||
| Group 4 | ||||||||||
| C57BL/10J | F (8) | 83.6±1.5 | 1.89±0.6 | 5.03±1.1 | 7.11±1.1 | |||||
| M (8) | 77.6±2.0 | 4.35±1.2 | 4.94±1.2 | 4.20±0.9 | ||||||
| P value | P = 0.6 | P = 0.10 | P > 1.0 | P = 0.04 | ||||||
| C57BL/6J | F (8) | 68.1±2.3 | 7.48±1.3 | 16.0±2.1 | 3.74±0.7 | |||||
| M (8) | 82.0±2.1 | 4.18±1.2 | 7.42±1.6 | 4.02±1.0 | ||||||
| P value | P = 0.02 | P = 0.46 | P = 0.08 | P > 1.0 | ||||||
| C57BLKS/J | F (8) | 62.1±2.2 | 8.91±1.4 | 12.3±1.4 | 2.77±0.8 | |||||
| M (8) | 54.1±2.5 | 14.1±1.7 | 17.5±1.3 | 4.71±0.8 | ||||||
| P value | P > 1.0 | P > 0.30 | P = 0.03 | P = 0.03 | ||||||
| C57BR/cdJ | F (8) | 47.0±2.7 | 9.97±2.1 | 21.0±2.0 | 4.18±0.8 | |||||
| M (8) | 60.8±2.0 | 9.30±1.4 | 25.0±1.5 | 4.21±08 | ||||||
| P value | P = 0.13 | P > 1.0 | P > 1.0 | P > 1.0 | ||||||
| C57L/J | F (8) | 68.3±2.4 | 7.12±1.4 | 15.1±1.5 | 4.02±1.0 | |||||
| M (8) | 76.9±2.2 | 4.70±1.1 | 9.88±1.6 | 4.45±0.9 | ||||||
| P value | P > 1.0 | P > 1.0 | P = 0.16 | P > 1.0 | ||||||
| Group 5 | ||||||||||
| 129S1/SvImJ | F (8) | 60.9±4.0 | 7.78±2.7 | 5.15±2.2 | 5.55±0.9 | |||||
| M (8) | 69.5±2.1 | 5.90±0.8 | 5.82±0.5 | 4.66±0.7 | ||||||
| P value | P > 1.0 | P > 1.0 | P > 1.0 | P > 1.0 | ||||||
| BTBRT+tf/J | F (8) | 25.0±2.6 | 33.1±2.2 | 25.3±1.3 | 3.37±0.6 | |||||
| M (8) | 31.3±2.3 | 29.5±1.8 | 22.6±1.3 | 2.36±0.5 | ||||||
| P value | P > 1.0 | P > 1.0 | P > 1.0 | P = 0.04 | ||||||
| LP/J | F (8) | 43.0±3.0 | 15.9±3.0 | 9.37±2.3 | 7.08±1.4 | |||||
| M (8) | 54.3±3.1 | 7.68±1.8 | 9.06±2.7 | 7.78±1.9 | ||||||
| P value | P > 1.0 | P > 1.0 | P > 1.0 | P > 1.0 | ||||||
| Group 6 | ||||||||||
| DBA/2J | F (8) | 78.3±1.9 | 2.84±1.0 | 7.96±1.2 | 2.90±0.7 | |||||
| M (8) | 64.6±3.2 | 18.2±2.2 | 7.25±1.1 | 4.45±1.7 | ||||||
| P value | P = 0.83 | P = 0.0002 | P > 1.0 | P > 1.0 | ||||||
| P/J | F (8) | 50.77±3.8 | 17.2±3.0 | 12.3±2.1 | 1.70±0.4 | |||||
| M (8) | 66.0±2.5 | 10.8±2.1 | 7.38±1.0 | 2.37±0.4 | ||||||
| P value | P > 1.0 | P > 1.0 | P > 1.0 | P = 0.02 | ||||||
| SM/J | F (8) | 40.9±2.4 | 13.5±1.5 | 16.8±0.9 | 8.86±1.3 | |||||
| M (8) | 54.7±2.2 | 10.7±1.3 | 16.8±1.3 | 5.08±0.9 | ||||||
| P value | P = 0.03 | P > 1.0 | P > 1.0 | P = 0.01 | ||||||
| Group 7 | ||||||||||
| MOLF/EiL | F (7) | 30.3±1.9 | 22.0±2.3 | 22.4±1.6 | 0.54±0.2 | |||||
| M (8) | 34.1±2.1 | 22.6±2.2 | 17.2±1.3 | 0.63±0.1 | ||||||
| P value | P > 1.0 | P > 1.0 | P = 0.1 | P > 1.0 | ||||||
| PWD/PhJ | F (8) | 23.1±1.5 | 38.2±1.5 | 27.3±1.0 | 1.17±0.3 | |||||
| M (8) | 37.0±1.7 | 29.7±1.6 | 24.0±1.8 | 1.58±0.5 | ||||||
| P value | P = 0.00001 | P = 0.003 | P > 1.0 | P = 0.2 | ||||||
| WSB/EiJ | F (8) | 34.5±2.0 | 35.0±2.1 | 17.3±1.3 | 1.55±0.5 | |||||
| M (6) | 26.7±2.4 | 27.5±1.6 | 14.3±1.3 | 2.45±0.5 | ||||||
| P value | P > 1.0 | P = 0.83 | P > 1.0 | P = 0.02 | ||||||
Results from the analysis were presented as mean percentages of each lymphocyte subpopulation. B, CD4 T, CD8 T, and natural killer (NK) cells are presented as % of lymphocytes ± SE. P value corrected is set at 0.0002, and the corrected values less than the threshold are considered significant.
The second large group of PB lymphocytes consists of T cells, which generate cell-mediated immune responses to antigens. In this study, we focused on CD4+ and CD8+ T cell subsets based on their important functional properties: CD4+ T cells are associated with helper function and CD8+ T cells are associated with cytotoxic function. The overall mean percentages of CD4+ T (Fig. 3B) and CD8+ T cell subsets (Fig. 3C) were similar for both females and males (18.4 vs. 16.0% for CD4+ and 15.7 vs. 14.5% for CD8+). Strain comparison revealed a large degree of variation in percentages of these two T cell subsets among the 30 inbred strains. CD4+ T cells demonstrated 28.5-fold (female) and 15.8-fold (male) difference and CD8+ T cells had 6-fold (female) and 20.6-fold (male) difference. C57BL/10J and NON/LtJ mice had the lowest percentages of CD4+ T and CD8+ T cells, whereas PL/J and SJL/J mice showed the highest values for both parameters (for exact values see Table 4). As expected, we observed a general inverse correlation between B220+ B cell and T cell (CD4+ and CD8+) percentages, best exemplified by PL/J (high percentages of CD4+ and CD8+ and low percentage of B220+ cells) and C57BL/10J (low percentages of CD4+ and CD8+ and high percentage of B220+ cells). Our data also demonstrated that mouse strains such as PWD/PhJ, SJL/J, and PL/J, which had high percentage of CD4+ T cells, tend to have more CD8+ T cells as well, demonstrating positive correlation between these two subsets.
The average overall mean value of NK cells was within the reference range of 5% (female 4.7 ± 1.7%; male 4.4 ± 1.6%), but there was substantial difference between the lowest and the highest values among the 30 strains (Fig. 3D). In both female and male groups, the wild-derived strain MOLF/EiJ and the laboratory strain PL/J and had the lowest percentage of NK cells (MOLF/EiJ female 0.5 ± 0.2%, and male 0.6 ± 0.4%; PL/J female 0.6 ± 0.4%, and male 0.8 ± 0.3%). In contrast, NZO/HlLtJ had the highest (female 13.1 ± 1.9%; male 13.7 ± 1.8%). MOLF/EiJ and PL/J exhibited low expression of all three NK specific cell surface markers such as NK1.1, Lgl-1, and NKG2A/C/E (data not included). Our results showing NK cell deficiency in peripheral blood obtained from PL/J mice correlated with previously reported decreased NK activity in the same strain (10). Interestingly, NZO/HlLtJ, NON/LtJ, and NZW/Lac/J, all members of group 3, had high percentages of NK cells (Table 4). No significant (P < 0.0002) sex-specific difference in NK percentages was detected between the female and male groups.
To determine the correlations between the immune cell populations and get an insight about genetic factors regulating immune cell homeostasis, we compared the strain means for each immune trait pair. Correlation studies shown in Table 5 revealed statistically significant (P < 0.0001) positive correlation between CD4+ and CD8+ T cells (r = 0.41), and eosinophils and monocytes (r = 0.07). Other combinations of two variables demonstrated negative correlation, such as: lymphocytes and granulocytes (r = −0.72), lymphocytes, and monocytes (r = −0.13), B220+ B cells and CD4+ T cells (r = −0.68), B220+ B cells and CD8+ T cells (r = −0.53), CD4+ T cells and NK cells (r = −0.19), and CD8+ T cells, and NK cells (r = −0.10). This data suggest genetic overlap in the regulation of CD4 and CD8 T cells, myeloid cells including monocytes, and eosinophils, and provide information for further investigation.
Table 5.
Correlation among PB cell populations
Values represent the Pearson correlation coefficient (r) for each pair of variables. Positive r indicates positive correlation, negative r indicates negative correlation.
Significant correlation (P ≤ 0.0001).
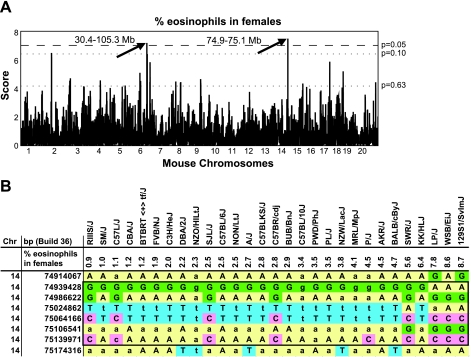
Our data showed compelling evidence for significant (>2-fold) interstrain variations in all immune parameters, suggesting genetic control of immune phenotypes. To associate this natural diversity of peripheral blood cell counts with discrete genomic regions we applied HAM (15, 19). The comprehensive phenotype data, large number of inbred strains (n = 30), and the availability of sequence data for these strains allowed us to expect sufficient statistical power of the HAM. The data obtained for all eight immune phenotype traits were used in the analysis. Results demonstrated that, three of the immune phenotypes including eosinophils, CD4, and CD8 had HAM peaks above genome-wide suggestive threshold level of P = 0.63. Female and male data were separated and run independently. Figure 4A shows the HAM scans of female eosinophils. Using our data (%eosinophils) we identified 16 HAM peeks above the genome-wide accepted threshold (P = 0.63), three peaks above the suggestive P value of 0.10, and two peaks above the significant value of P = 0.05. The same assay demonstrated a peak on chromosome 6 in males, however, the P value didn't reach the threshold of 0.63 (data not included). The most significant P values HAM peaks obtained for eosinophils (female group) were located on chromosomes 6 and 14 (Fig. 4A). The HAM peak on chromosome 6 is wide and includes several regions: 30.4–30.5, 35.5–35, 81.6–81.8, 82.0–82.1, 82.2–82.4, 84.2–84.4, 103.7, and 105.1–105.3 Mb. The region on chromosome 14 is located between 74.9 and 75.1 Mb (Fig. 4B) and contained two genes: ribosomal protein S2 and Tsc22d1. Tsc22d1 is a transcription regulator, member 1 of TSC22 (transforming growth factor-beta stimulated clone 22) domain family. Tsc22d1 is expressed in different tissues including hematopoietic system [Mouse Phenome Database (MPD), http://www.informatics.jax.org]. Recent studies have shown that TSC22 induces erythroid differentiation (3); however, the role of its member 1 in myeloid differentiation has not been thoroughly investigated. Future experiments should be conducted to determine whether Tsc22d1 indeed regulates the proportions of eosinophils in peripheral blood.
Fig. 4.
Haplotype association mapping (HAM) identifies highly significant association of eosinophil trait with 2 regions on chromosomes 6 and 14. A: HAM scan of eosinophils using female data were obtained from 29 inbred strains (MOLF/EiJ strain was not included). Single nucleotide polymorphism (SNP) data were used from Build 36 version 5.5. Significant P values (P > 0.05) were obtained for 2 regions located on chromosomes 6 and 14 (indicated by arrows). The peak on chromosome 6 located between 30.4 and 105.3 Mb. The genomic position of the second peak located on chromosome 14 was narrow, located between 74.9 and 75.1 Mb. P = 0.05 was considered significant, P = 0.10 suggestive, P = 0.63 threshold level. B: visualization of the 74.9–75.1 Mb region on chromosome 14. SNP alleles were shown in different colors. The strains were listed in specific order, from low to high percentage of eosinophils. The region containing informative SNPs has been framed.
In summary, we conducted an immune survey of 30 inbred mouse strains of different origins. Our data demonstrated: 1) high degree of strain-related variations in immune cell populations, 2) considerable variation of each immune trait with more than twofold difference between strains with the highest and the lowest trait values, 3) continuous distribution of the means of the immune cell populations for the majority of the strains, suggesting that multiple genes and or/alleles control the numbers of immune cell populations in peripheral blood, 4) a strong correlation, revealed by haplotype analysis, between eosinophil percentage and a single region on chromosome 14 containing two candidate genes.
Our data set provide the most complete assessment of relative leukocyte measurements of murine peripheral blood of inbred mouse strains using a standardized flow cytometry protocol. Several applications emerge from the current data. First, it could serve as reference data to study the genetic control of immune cell homeostasis using QTL analysis of a cross between inbred strains showing extreme phenotype values. Indeed, recent findings by Valdar at al. (20) have mapped QTLs for several complex immunological phenotypes (CD4+, CD8+, and CD4/CD8 ratio) using heterogeneous mouse stock. Second, it could be used in longitudinal cohort studies to monitor age-related changes in immune cell populations to define age-related biomarkers. Third, this immune survey will add substantial information to the comprehensive phenotype characterization of inbred mouse strains and could be used to correlate with other phenotype data using information stored at MPD (11).
GRANTS
This work was supported by The Ellison Medical Foundation Grants AG-1A-0049-04 (to B. Paigen), AG-1A-0204-05 (to R. Woychik), and Nathan Shock Center Grant AG-25707 from the National Institute of Aging.
Acknowledgments
We thank Theodore Duffy for the flow cytometry consultation; Neeta Kumari, Dana Godfrey, Millie So, and David Shultz for technical assistance; and David Serreze, Petko Petkov, and Luanne Peters for critical reading and helpful suggestions.
Address for reprint requests and other correspondence: S. Petkova, The Jackson Laboratory, 600 Main St., Bar Harbor, ME 04609 (e-mail: stefka.petkova@jax.org).
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Busslinger M, Nutt SL, Rolink AG. Lineage commitment in lymphopoiesis. Curr Opin Immunol 12: 151–158, 2000. [DOI] [PubMed] [Google Scholar]
- 2.Chen J, Harrison DE. Quantitative trait loci regulating relative lymphocyte proportions in mouse peripheral blood. Blood 99: 561–566, 2002. [DOI] [PubMed] [Google Scholar]
- 3.Choi SJ, Moon JH, Ahn YW, Ahn JH, Kim DU, Han TH. Tsc-22 enhances TGF-beta signaling by associating with Smad4 and induces erythroid cell differentiation. Mol Cell Biochem 271: 23–28, 2005. [DOI] [PubMed] [Google Scholar]
- 4.Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature 411: 342–348, 2001. [DOI] [PubMed] [Google Scholar]
- 5.Geissmann F, Jung S, Littman DR. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity 19: 71–82, 2003. [DOI] [PubMed] [Google Scholar]
- 6.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev 5: 953–964, 2005. [DOI] [PubMed] [Google Scholar]
- 7.Gregersen PK, Behrens TW. Genetics of autoimmune diseases–disorders of immune homeostasis. Nat Rev Genet 7: 917–928, 2006. [DOI] [PubMed] [Google Scholar]
- 8.Kile BT, Mason-Garrison CL, Justice MJ. Sex and strain-related differences in the peripheral blood cell values of inbred mouse strains. Mamm Genome 14: 81–85, 2003. [DOI] [PubMed] [Google Scholar]
- 9.Lanier LL Natural killer cells: from no receptors to too many. Immunity 6: 371–378, 1997. [DOI] [PubMed] [Google Scholar]
- 10.Lattime EC, Pecoraro G, Stutman O. Natural cytotoxic cells against solid tumors in mice. IV. Natural cytotoxic (NC) cells are not activated natural killer (NK) cells. Int J Cancer 30: 471–477, 1982. [DOI] [PubMed] [Google Scholar]
- 11.Paigen K, Eppig JT. A mouse phenome project. Mamm Genome 11: 715–717, 2000. [DOI] [PubMed] [Google Scholar]
- 12.Peters LL, Cheever EM, Ellis HR, Magnani PA, Svenson KL, Von Smith R, Bogue MA. Large-scale, high-throughput screening for coagulation and hematologic phenotypes in mice. Physiol Genomics 11: 185–193, 2002. [DOI] [PubMed] [Google Scholar]
- 13.Petkov PM, Ding Y, Cassell MA, Zhang W, Wagner G, Sargent EE, Asquith S, Crew V, Johnson KA, Robinson P, Scott VE, Wiles MV. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res 14: 1806–1811, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plas DR, Rathmell JC, Thompson CB. Homeostatic control of lymphocyte survival: potential origins and implications. Nat Immunol 3: 515–521, 2002. [DOI] [PubMed] [Google Scholar]
- 15.Pletcher MT, McClurg P, Batalov S, Su AI, Barnes SW, Lagler E, Korstanje R, Wang X, Nusskern D, Bogue MA, Mural RJ, Paigen B, Wiltshire T. Use of a dense single nucleotide polymorphism map for in silico mapping in the mouse. PLoS Biol 2: e393, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rathmell JC, Thompson CB. Pathways of apoptosis in lymphocyte development, homeostasis, and disease. Cell 109, Suppl: S97–S107, 2002. [DOI] [PubMed] [Google Scholar]
- 17.Rothenberg ME, Hogan SP. The eosinophil. Annu Rev Immunol 24: 147–174, 2006. [DOI] [PubMed] [Google Scholar]
- 18.Simon HU Neutrophil apoptosis pathways and their modifications in inflammation. Immunol Rev 193: 101–110, 2003. [DOI] [PubMed] [Google Scholar]
- 19.Szatkiewicz JP, Beane GL, Ding Y, Hutchins L, Pardo-Manuel de Villena F, Churchill GA. An imputed genotype resource for the laboratory mouse. Mamm Genome 19: 199–208, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valdar W, Solberg LC, Gauguier D, Burnett S, Klenerman P, Cookson WO, Taylor MS, Rawlins JN, Mott R, Flint J. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat Genet 38: 879–887, 2006. [DOI] [PubMed] [Google Scholar]
- 21.Valdar W, Solberg LC, Gauguier D, Cookson WO, Rawlins JN, Mott R, Flint J. Genetic and environmental effects on complex traits in mice. Genetics 174: 959–984, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]