Abstract
The mouse is a premiere experimental organism that has contributed significantly to our understanding of vertebrate biology. Manipulation of the mouse genome via embryonic stem (ES) cell technology makes it possible to engineer an almost limitless repertoire of mutations to model human disease and assess gene function. In this review we outline recent advances in mouse experimental genetics and provide a “how-to” guide for those people wishing to access this technology. We also discuss new technologies, such as transposon-mediated mutagenesis, and resources of targeting vectors and ES cells, which are likely to dramatically accelerate the pace with which we can assess gene function in vivo, and the progress of forward and reverse genetic screens in mice.
Keywords: gene targeting, transposons, mutagenesis, embryonic stem cells
technologies for modifying the mouse genome can be broadly split into two classes. These include technology for gene-driven analyzes such as those used for knocking out genes or precisely modifying them and technology used to generate random mutations across the genome, or in selected regions of the genome. In this review we will initially discuss gene-driven approaches of modifying the mouse genome and then discuss approaches that may be exploited for random mutagenesis.
The most commonly used gene-driven approach in mouse is gene targeting. The derivation of embryonic stem (ES) cells (21, 45) and the illustration that homology-directed recombination could be accomplished in these cells (19, 75) heralded a new era in biology. We will discuss how gene targeting can be used to generate gain- and loss-of-function alleles in mice (both in specific tissues or the whole animal) and also how to engineer large-scale rearrangements of entire mouse chromosomes. While loss-of-function alleles are usually the first step toward understanding gene function, only a subset of human disease causing alleles are loss-of-function alleles. In many cases disease causing mutations generate hypomorphic or neomorphic alleles. Recently oligo-based gene targeting approaches have emerged that potentially represent a significant advance in our ability to rapidly generate precise base pair changes or allelic series in a target gene. We will discuss these approaches and their application to modeling human disease. Finally we will discuss progress in the use of small hairpin RNA (shRNA) as a tool for the rapid assessment of “knockdown” phenotypes in the mouse and advances in the use of lentiviral and bacterial artificial chromosome (BAC) for transgenesis.
While many investigators have the ability to generate knockout mice by themselves or within core facilities, access to mouse mutants can still be a limiting and expensive. For this reason several large gene-trap and gene targeting programs have been established. These include existing gene-trap resources provided by several groups under the apices of the International Gene Trap Consortium (ITGC) (51) as well as the Knockout Mouse Project (KOMP) funded by the National Institutes of Health (NIH), the European Conditional Mouse Mutagenesis Program (EUCOMM) funded by the European Union, the North American Conditional Mouse Mutagenesis Project (NorCOMM) fund by Genome Canada, and the Sanger Institute's Mouse Genetics Programme (MGP), which is a Wellcome Trust Initiative. We will outline what reagents these resources have to offer and how to access them.
The abovementioned approaches represent reverse genetic (gene-driven) strategies for assessing gene function. However, all mouse geneticists are familiar with the disappointment of spending months or years painstakingly engineering a mouse mutant only to discover that the mouse lacks the desired phenotype or any obvious discernable phenotype at all. For this reason many researchers have taken to using forward genetic (phenotype-driven) strategies where the genome is mutated at random and mutant offspring are screened for phenotypes of interest, usually through a battery of tests. Classically these approaches have involved the use of chemical mutagens such as N-ethyl-N-nitrosourea (ENU), ethylmethanesulfonate (EMS), chlorambucil, or ionizing radiation. The limitation of these mutagenic strategies, however, is the ease with which mutations can be found and then proven to be causal. Powerful gene-trapping and transposon-based approaches have also been developed (17, 25, 80). The advantage of such approaches is that the transposon or gene trap represents a DNA “tag” so the phenotype of the mutant can be rapidly correlated with the presence of an insertion event within a mutated gene. Each of these mutagens has distinct advantages and disadvantages, which we will discuss in this review. We will also direct the reader to collections of mutants and provide links to facilitate access to repositories of phenotyping data.
REVERSE GENETIC (GENE-DRIVEN) APPROACHES OF MODIFYING THE MOUSE GENOME
Overview of ES Cell Technology and Gene Targeting
Genetic background of ES cells.
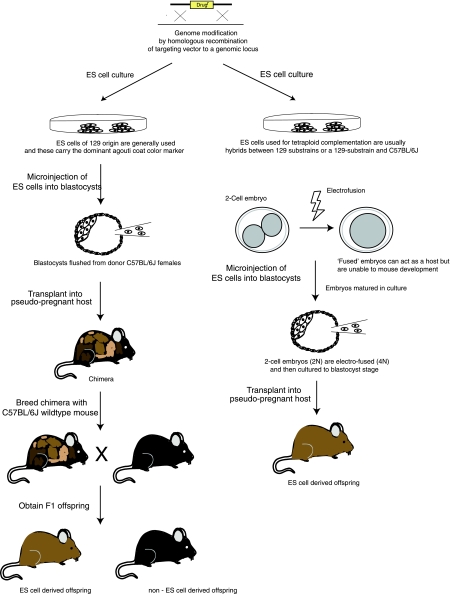
The development of ES cell technology whereby pluripotent ES cells can be removed from a preimplantation embryo, modified in culture, and then reintroduced into a host preimplantation embryo to colonize the germline has been a profound advance in mouse genetics. An overview of the procedure is provided in Fig. 1.
Fig. 1.
Mutant mice can be generated either by microinjection or by tetraploid complementation. Embryonic stem (ES) cells are derived from the inner cell mass of blastocysts and grown in culture. These cells can be modified by gene-targeting and then either microinjected into a host blastocyst to generate chimeras that are then bred for germline transmission (left) or alternatively used to make entirely ES cell-derived mice using tetraploid complementation (right). Usually the ES cells used are derived from a 129 mouse strain (or hybrids with a 129 strain) and carry the dominantly active agouti locus allowing coat color (brown) to be used for tracking both the contribution of ES cells to the formation of a chimera and for the identification of germline ES cell-derived offspring.
Historically the ES cell lines used for gene targeting experiments have been derived from substrains of 129 because ES cell lines from these genetic backgrounds have been shown to efficiently colonize the mouse germline (69). Commonly used 129-derived ES cell lines include E14 (129OlaHsd) (34), AB2.2 (129S7) (57), R1 (129X1/SvJ × 129S1) (50), and J1 (129SvJae) (43). However, when the mouse genome was sequenced C57BL/6J was selected as the reference strain because of its central role in mouse genetics, and whereas 129 substrains and C57BL/6J are both classed as “common laboratory strains” and are derived from a common pool of founders (Mus musculus, Mus domesticus, and Mus castaneous), there are distinct differences in their genomes both at the nucleotide level and when copy number variants are compared between them (30, 81). These genomic differences manifest as phenotypic differences for a range of parameters (73).
As shown in Fig. 1, during the process of making knockout mice it is usual to inject genetically modified 129-derived ES cells into host C57BL/6J blastocysts and to transmit the modified allele onto a C57BL/6J background. Although this approach is advantageous, because coat color markers can be used to score for germline transmission, the resultant offspring are 129-C57BL/6J hybrids. For this reason, and because the reference genome sequence is from C57BL/6J, many investigators have attempted to derive C57BL/6J ES cell lines. One commonly used C57BL/6J-derived line is Bruce4, although strictly speaking they are a congenic ES cell line because they were made from the mouse strain C57BL/6J-Thy1.1 (39). More recently, ES cells derived from a substrain of C57, termed C57BL/6N, were found to make reliable ES cells (63). The C57BL/6N strain was derived from C57BL/6J stock originating from the NIH and has been maintained in isolated breeding colonies for at least 160 generations. Jackson Labs, Charles River, and Taconic have maintained separate colonies of this strain for several decades. It is likely that each of these colonies have diverged significantly from C57BL/6J and each other, which should be taken into consideration given that important phenotypic differences between C57BL/6J and C57BL/6N have been reported (18, 71). When C57BL/6N ES cells are used for gene targeting they may be injected into albino C57BL/6J blastocysts and the male chimeras crossed with albino C57BL/6J females so the resultant F1 offspring, which will be black if they are ES cell derived, should be considered hybrids. It is also possible to breed male C57BL/6N chimeras with C57BL/6N females and transmit onto a “pure” C57BL/6N background, although if this strategy is employed all of the offspring will need to be genotyped because it will no longer be possible to score germline transmission by coat color. Several C57BL/6N ES cells lines have been made including B6/Blu-1 (unpublished; generated by Tim Ley from Washington University, St. Louis, MO). B6/Blu-1 ES cells have been elegantly designed to carry a LacZ marker, which allows chimerism to be scored by LacZ staining of peripheral blood smears. Modified B6/Blu-1 ES cells may be injected into C57BL/6N or C57BL/6J blastocysts and male chimeras mated with C57BL/6N females resulting in pure C57BL/6N offspring. Additional C57BL/6N ES cell lines have been made by investigators participating in the EUCOMM and KOMP programs.
The vast majority of the >5,000 knockouts that have been made over the last 20 yr have been generated by microinjection of modified ES cells into host blastocysts and then breeding the chimeric offspring for germline transmission. Over the last 10 yr an alternative approach has emerged, termed tetraploid complementation (50). As shown in Fig. 1, this approach involves passing an electric current through two-cell preimplantation mouse embryos to fuse them together. This renders these embryos 4N and, as such, able to contribute to trophoblast and primitive endoderm lineages but unable to participate in forming the developing embryo. After electrofusion, the embryos are cultured through to blastocysts, microinjected with ES cells, and then transplanted into pseudopregnant females. Alternatively a technique called “aggregation” can be used whereby ES cells are cocultured with electrofused two-cell embryos that have had their zona pellucida removed and they become part of the blastocyst as it forms. The ES cell lines most widely used for tetraploid complementation are either hybrids between 129 substrains such as R1 (47) or hybrids between a 129 substrain and C57BL/6J, such as G4 (26, 68). Analysis of mice generated by using tetraploid complementation is usually performed on a hybrid background. It is possible to “double target” ES cells to disrupt both alleles of a gene, or to target a transgene, and then generate totally ES cell-derived embryos (64, 68). With this approach it is possible to phenotype mice without the need for germline transmission. The drawback of this approach is that the rate of generation of tetraploid offspring can be low, and therefore it may be problematic to generate large cohorts of mice for phenotyping.
Types of targeting vectors.
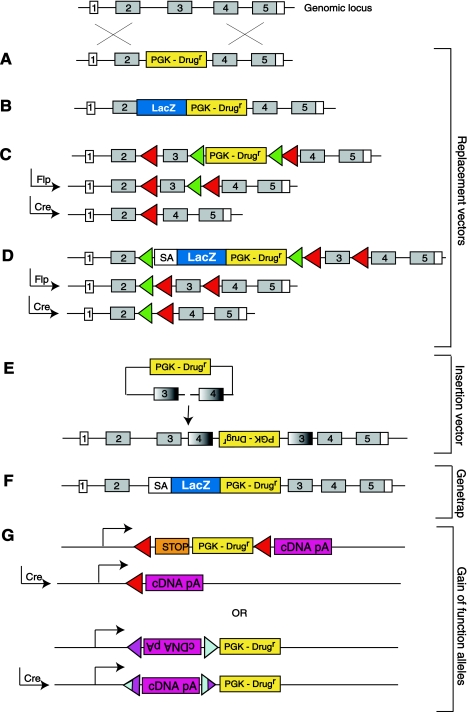
There are four classic types of targeting vectors, replacement, knock-in, conditional, and insertional, as shown in Fig. 2. To generate most alleles replacement vectors are used, and insertion vectors are usually employed only as part of chromosome engineering-based approaches. Whereas replacement vectors undergo double reciprocal recombination events when they are targeted into the genome, insertional targeting vectors undergo a single reciprocal recombination event and, as such, target the genome at a high frequency (32). However, the caveat of insertional targeting vectors is that they 1) generate complex rearrangements of the genome by duplicating their homology and 2) rely on gene rearrangement by interfering with splicing to disrupt a gene, as opposed to gene ablation which can be achieved with replacement vectors (Fig. 2, A and E). There are also elegant targeting vector designs that allow multiple aspects of a gene's function to be assessed with one targeting event, and these strategies are made possible by using the site-specific recombinase systems, Cre and Flp (58, 60) (Fig. 2, C and D).
Fig. 2.
There are various designs of targeting vector that can be used to generate loss- or gain-of-function alleles in ES cells. The structure of a generic gene is shown with numbered boxes representing the exons (either coding, shown in gray, or noncoding, shown in white). Homologous recombination (shown by crosses) results in different modifications of the targeted locus depending on the nature of the targeting vector used. A: when using a standard replacement vector, typically a drug resistance marker (PGK-Drugr) is inserted into the gene to disrupt a critical exon. B: knock-in vectors can be used to target expression markers such as LacZ into a locus. These vectors typically result in fusion of LacZ with an exon of the gene. With this approach the gene is disrupted and its expression tagged. C: elegant strategies can be used to generate both null and conditional alleles of a gene using the Cre/LoxP and Flp/FRT recombinase systems. LoxP sites are shown as red triangles, and FRT sites are shown as green triangles. Vectors of this design can be used to inactivate a gene somatically or in a particular cell or tissue type. D: “knockout first” alleles combine the features of both knock-in and conditional alleles to generate targeting events that result in null alleles that are tagged with LacZ to monitor gene expression. Endogenous transcripts of a gene are captured by the splice acceptor (SA) and terminated. These alleles can be treated with Flp to generate a conditional allele and then with Cre to generate a loss-of-function allele in a cell- or tissue-specific manner. E: insertion vectors, unlike replacement vectors, have a single length of homology and are linearized by cutting within the homologous sequence. When they target the genome they insert vector sequences such as the drug resistance marker at the insertion site. F: gene-trap vectors have a very similar design to knock-in vectors except that they are randomly introduced into the genome as opposed to being targeted to a specific locus. They usually contain SA sequences and fusions between drug resistance and expression markers such as neomycin and LacZ. G: it is also possible to generate gain-of-function alleles in mice. One common approach is to target a cDNA of interest behind a loxP-flanked STOP element, which prevents expression of the cDNA. Expression of the cDNA occurs after the STOP element is removed by Cre-mediated excision (41). Another strategy involves placing the cDNA of interest between inverted mutant loxP sites, shown as blue and purple triangles (62). After expression of Cre, the cDNA “flips” into the “on” position and is expressed. Importantly, recombination of these mutant loxP sites generates a variant loxP site that is unable to recombine again, locking the cDNA in the on position.
The first consideration when designing a targeting vector is the selection marker to be used. Most commonly the cDNA of neomycin (neo) is driven by the strong phosphoglycerate kinase (PGK) promoter. ES cells transfected with vectors carrying a neomycin cassette can be selected in the drug G418. Other commonly used selection markers include puromycin (puro), blastocidin (blasto/Bsd), and hygromycin (hygro), and cells transfected with vectors carrying these markers can be selected in the drugs puromycin, blastocidin, and hygromycin, respectively. Each selection agent has different properties, and ES cells behave differently under different drug selection conditions. For example G418 selection is slow, and it takes up to a week for resistant colonies to emerge. In contrast puromycin and blastocidin selection kills cells very quickly, and significant cell death is evident within 72 h of these drugs being added to the culture medium. In our hands a construct carrying a neomycin marker driven by PGK will give more colonies after selection than the same construct carrying a puromycin or blastocidin marker driven by PGK. Fusions between herpes simplex thymidine kinase (tk) and both neomycin and puromycin have been reported (8, 9, 78), and these allow the removal of selection cassettes by negative selection in either 1-(-2-deoxy-2-fluoro-1-b-d-arabino-furanosyl)-5-iodouracil (FIAU) or gancyclovir. Truncations of tk have been developed (termed Δtk) and fused to puromycin to circumvent the male sterility associated with expression of full-length tk (9).
One aspect worth considering in the design of a targeting vector is that selectable markers, particularly neomycin, have been reported to act as silencers of genes nearby the targeted locus (55). Thus it is critical that selection markers are removed from the genome before analyzing the phenotype of a mutant. While no ill effects have been reported from alleles carrying puromycin, blastocidin, or hygromycin there has been speculation that in some instances the PGK promoter used to drive most selection markers may function bidirectionally, and in at least one case this has been shown to result in the formation of an antisense transcript (59). Thus it is prudent to remove selection markers from a targeted locus before analysis of the mutant mice.
Source of DNA for the gene-targeting vector.
One of the critical determinants of gene targeting efficiency is the length of homology between the genome and the targeting vector. Generally speaking, the longer the homology the higher the targeting frequency (31), although larger targeting vectors can become unwieldy and may potentially become rearranged in Escherichia coli. In recent years significant resources have become available for generating targeting vectors. For example genome-wide end-sequenced bacterial artificial clone libraries now exist for C57BL/6J (53) and 129S7 strains (4), and both of these resources are available via the Ensembl genome browser (24) (www.ensembl.org). Other large insert libraries are also available for the 129/OlaHsd strain (52) from which the commonly used E14 ES cell line is derived, although 129S7 DNA has been successfully used to target E14 ES cells, at least at some loci. Similarly, we routinely use DNA from C57BL/6J BAC libraries to target genes in C57BL/6N ES cells. Other factors that are believed to influence targeting frequency include the level of expression of the target gene (with more highly expressed genes thought to be easier to target) and the chromatin environment surrounding the target locus, although neither of these theories has been formally tested.
Construction of targeting vectors.
In the past, targeting vectors were usually constructed by restriction digest of DNA fragments, followed by standard subcloning into a plasmid-based targeting vector. Some investigators prefer to use PCR-based approaches to generate homology arms for gene targeting, although there is always the concern that PCR may introduce errors into the amplified fragment. If PCR is used to generate a targeting vector it is wise to use a proofreading “high-fidelity” polymerase and to at least sequence the exons and splice junctions on the amplified fragment.
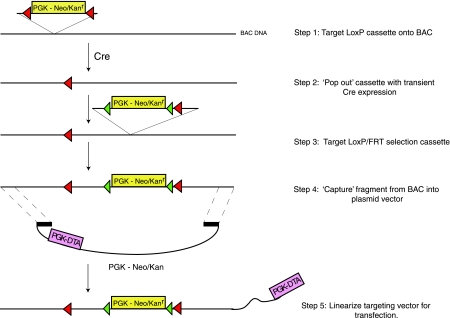
Recently, targeting vector construction has been revolutionized by the development of “recombineering” (11). This approach employs λ phage-derived recombinases to direct homologous recombination of selection markers onto a DNA fragment and gap repair for capturing a genomic segment that represents the targeting vector homology (Fig. 3). In our hands the λRed system developed by Neal Copeland and Don Court is extremely efficient and flexible and can be used to develop targeting vectors within 2 wk (42). Similarly the system developed by Francis Stewart and colleagues (79) works very efficiently and has the advantage that any bacterial cell can be converted to a recombineering proficient strain simply by introducing a plasmid carrying the Red operon (which includes the three components: Red∝, a 5′-to-3′ exonuclease; Redβ, an annealing protein; and Redγ, an inhibitor of the major E. coli exonuclease and recombination complex, RecBCD).
Fig. 3.
Recombineering is a powerful strategy for rapidly generating targeting vectors. Typically, bacterial artificial chromosomes (BACs) are modified using λ Red-mediated recombination in bacteria. Firstly a LoxP-flanked bacterial selection marker that carries kanamycin resistance (Kanr) is targeted onto the BAC. This marker is then “popped out” by expressing Cre recombinase in the bacteria, leaving behind a single loxP site. The BAC is then retargeted with a second cassette carrying a neomycin-kanamycin (Neo/Kanr) selection maker flanked by FRT sites and a single LoxP site. This vector carries a mammalian promoter such as phosphoglycerate kinase (PGK) and a bacterial promoter such as EM7, allowing drug resistance in ES cells and Escherichia coli, respectively. Finally, the modified fragment of DNA is “captured” from the BAC by a process called “gap-repair.” The backbone of the capture vector may contain a negative selectable marker such as diphtheria toxin (DTA), which suppresses random integration of the targeting vector in the genome. The resulting vector is a conditional allele.
If you are unable to generate a targeting vector by yourself it is also possible to have them made on a contract basis by several companies including GeneBridges and VectorBioLabs, which use recombineering and PCR-based approaches respectively.
Tissue-specific gene targeting.
In many cases it may be desirable to assess gene function in a particular organ or cell type. Furthermore, if a gene is essential for embryonic development, then germline loss-of-function in the whole animal can preclude such analysis. To circumvent this problem site-specific recombinase systems including Flp/FRT (47), Cre/LoxP (5) and phiC31 integrase (76) have been developed. These elements can be used to flank DNA segments for excision from the genome, and in the case of the Flp/FRT and Cre/LoxP systems inverted FRT and LoxP sites, respectively, can be used to “flip” (invert) segments of DNA, which can be useful in conditional gene targeting approaches (62). An outline of the various strategies that can be used to conditionally inactivate and activate genes is shown in Figs. 2, C and D, and 4. There are numerous tissue- and cell-specific Cre lines that are now available in “Cre-Zoos,” and Andras Nagy curates a database of these lines: http://www.mshri.on.ca/nagy/ (49).
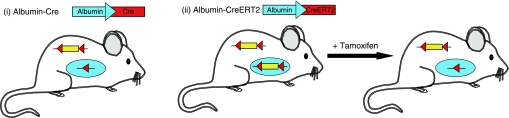
Fig. 4.
Cre recombinase can be used to activate or delete a gene in a tissue-specific manner. Tissue-specific gene recombination can be performed either by 1) breeding the mouse carrying the conditional allele with a mouse expressing a tissue-specific Cre (i.e., whereby Cre is only expressed in a specific tissue type, such as liver represented as a blue circle) or 2) somatically by breeding the mouse carrying the conditional allele with a mouse expressing an inducible tissue-specific Cre (i.e., whereby tissue-specific expression of the Cre only occurs after induction of the promoter via treatment with a chemical, such as tamoxifen). In both cases, expression of Cre or a fusion between Cre and a truncated estrogen receptor sequence is driven by a tissue-specific promoter (e.g., Albumin-Cre, which switches on Cre expression only in the liver).
In some cases it may be desirable to study a gene in the adult animal or to somatically alter gene function in vivo. This is possible by using inducible expression systems. There are several inducible expression systems including the tetracycline-inducible system (29), the LacZ-inducible system (13), and the tamoxifen-inducible system (22). The tamoxifen-inducible system has been extensively used and shown to be both tightly regulated and sensitive. This system employs a fusion between a recombinase and the estrogen receptor and has been developed for both the Flp/FRT and Cre/LoxP systems (6, 22). Expression of the chimeric fusion protein can be directed ubiquitously or tissue-specifically, depending on which promoter the fusion is expressed from. When mice are administered tamoxifen, or its derivative 4-OHT, the chimeric fusion protein translocates to the nucleus allowing the recombinase to act on FRT or LoxP sites integrated into the genome. Thus by using this approach it is possible to alter gene function in a temporal- and tissue-specific manner.
Resources of targeting vectors, mutant ES cells, and mice.
Even investigators expert in gene targeting would rather not have to develop a targeting vector and target ES cells if there were an alternative source for their mutant allele. For this reason, and to facilitate the dissemination of mutant lines to the research community, several large gene-trap and gene targeting resources have been developed. One such collection is the IGTC resource (http://www.genetrap.org/), which as of February 2008 contained 120,043 ES cell lines in 10,982 genes (17.2% coverage of the genome). These cell lines are largely 129-derived ES cell lines. The accumulated resource is composed of alleles generated by vector-based gene traps and also gene traps generated using viruses. Another large resource has been developed by the Texas Institute for Genomic Medicine and is composed of just over 270,000 ES cell lines. This resource contains both 129 and C57BL/6N ES cell lines. For many genes there are multiple gene-trap insertions in different regions of the gene in these resources meaning that it is possible to use these cell lines to generate allelic series. As shown in Fig. 2F the generic structure of gene traps includes a splice acceptor and βgeo selection marker, which hijacks a genes transcription machinery to express βgeo, a LacZ/neomycin fusion. In some alleles within the resource, FRT and LoxP sites have been included, which makes it possible to revert the allele or to use it as a substrate for recombinase-mediated cassette exchange (RMCE) (23). All of the cell lines generated by the IGTC are made freely available on a cost recovery basis. The Sanger Institute Gene Trap Resource represents ∼12,000 of the cell lines in the IGTC database. These gene-trap ES cell lines are E14Tg2a-derived (34). We have obtained 80% germline transmission of gene traps from this resource, including a gene trap in the gene Geminin (28).
Although gene traps have been shown to be very effective mutagens there have been concerns that some intragenic insertions may not “trap” all of a gene's transcripts and that there could be splicing over the gene-trap cassette. In the case of gene traps in the Tfeb gene this has been shown to be true (46), although the frequency with which splicing over the gene-trap cassette is a problem will vary significantly between gene-trap vector designs and each genomic locus and insertion. While this may represent a potentially undesirable aspect of gene traps, in some case, splicing around the gene trap may result in the formation of hypomorphic alleles that may allow the analysis of genes in mice that would be lethal if a null allele was generated.
Clearly the most desirable resource of mutant ES cell lines is one containing targeted conditional mutations. For this reason the KOMP, the EUCOMM, the NorCOMM, and the Sanger Institute's MGP were established. Each of these programs aims to produce conditional alleles containing a LacZ expression marker known as “knockout first” alleles (Fig. 2D) (74). Although still in their infancy, these programs have already generated an impressive list of targeting vectors and mutant cell lines that represent many lifetimes of work from a single laboratory. These programs are made possible by the use of an elegant recombineering/gateway based approach, which facilitates the rapid generation of targeting vectors. A summary of resources of targeting vectors, ES cell lines, BAC libraries, and gene traps is provided in Table 1.
Table 1.
Collections of genetrap ES cell lines, targeted ES cell lines, targeting vectors, and bacterial artificial chromosome libraries for targeting vector construction
| Reagent | URL | Comment |
|---|---|---|
| Gene traps | http://www.genetrap.org | International Gene Trap Consortium |
| IGTC cell lines in database: 132910 cell lines | ||
| IGTC cell line coverage: 8878 genes (51). | ||
| Texas Institute for Genomic Medicine Gene Traps | http://www.tigm.org | Extensive gene-trap library composed of 129 and C57BL/6N-derived ES cell lines. There are over 270,000 ES cell lines in this collection representing mutations in just over 9,000 genes. |
| Targeted ES cell lines & Targeting vectors | http://www.knockoutmouse.org | Collectively these projects will generate “knockout first” alleles (see Fig. 2D) for all genes in the mouse genome. This is a rapidly expanding resource. |
| http://www.eucomm.org | ||
| http://www.komp.org | ||
| http://norcomm.phenogenomics.ca/ | ||
| http://www.sanger.ac.uk/modelorgs/mouse.shtml | ||
| Insertion targeting vectors (mutagenic insertion and chromosome engineering resource [MICER] clones) | http://www.sanger.ac.uk/micer | These vectors are spaced on average every 100 kb across the genome. They are mainly useful for chromosome engineering but can be used for gene disruption (3). |
| http://www.geneservice.co.uk | ||
| http://www.ensembl.org | ||
| Indexed bacterial artificial libraries for targeting vector construction | http://www.geneservice.co.uk/home/ | Indexed libraries covering the entire mouse genome facilitate the construction of targeting vectorhttp://bacpac.chori.org/ |
| http://www.ensembl.org | ||
| http://bacpac.chori.org/ | ||
| Indexed libraries are available from the strains 129S7 and C57BL/6J (4, 53). |
ES, embryonic stem.
RMCE/“Plug and Socket” Targeting
RMCE (7, 67) and plug-and-socket targeting (84) are both methods that allow cassettes, which may contain cDNAs or expression markers, to be introduced into the same genomic locus at a “docking site.” This docking site contains either LoxP or FRT sites such that transient expression of either Cre or Flp recombinases will direct integration of a LoxP or FRT flanked cassette into the docking site. RMCE docking sites usually contain two LoxP or FRT sites, while plug and socket target employs just one LoxP or FRT site. Given that the site of integration of a transgene affects both the level and the spatial and temporal expression of genes expressed from the transgene, RMCE and plug-and-socket targeting allows the reproducible functional characterization of genetic elements in the same genetic and chromatin environment.
Lentiviral Transgenesis
Lentiviral vectors represent a potentially elegant tool for studying gene function in mice (54). The advantage of lentiviral vectors is that unlike standard “DNA-based” transgenesis lentiviral insertions are usually single copy, transgenesis is efficient with the majority of offspring carrying a lentiviral insertion, and viral insertions are generally introduced into active transcriptional units. Lentiviral vectors have been used for delivering cDNAs and also for delivering shRNAs. The issues that still need to be resolved with these vectors include transgene silencing, genetic mosaicism, and vector-related toxicity. Recent improvements in vector safety and the ability to produce ecotropic lentiviruses (61) now make lentiviral vectors a safe tool for use in most laboratory settings.
BAC Transgenesis
Large genomic fragments such as BACs may be used for transgenesis in mice. Given that the average size of a gene in the mouse genome is 34 kb, most genes and their regulatory elements can fit on a BAC. BAC transgenesis allows genes to be expressed from a BAC under the control of their endogenous promoter elements as the cis-splicing machinery and elements with the 5′- and 3′-untranslated regions of genes are present, ensuring that splicing occurs, that all transcripts of a gene are expressed, and that gene expression is appropriately posttranscriptional regulated. For these reasons BAC transgenesis is a particularly useful tool for genetic rescue experiments (12) and also for dissecting the functional elements of genes without having to go through the laborious process of targeting the endogenous locus (48). By combining BAC-transgenesis with RMCE into the Hprt locus, which is subject to random X-inactivation, it is also possible to generate genetic mosaics (56).
Generating Targeted Point Mutations
A significant proportion of genetic variation in humans is at the nucleotide level (1), and site-specific modification of the mouse genome is a powerful approach for functionally characterizing mutations and distinguishing between silent polymorphisms and pathogenic mutations. Although point mutations can be introduced by replacement vector targeting (or knock-in targeting) this is an extremely laborious approach and precludes the generation of large numbers of mutations. Oligonucleotide-mediated gene targeting is emerging as a powerful tool for the introduction of subtle gene modifications (such as point mutations) in mouse ES cells with numerous reports showing that oligonucleotides differing from the target locus by one or a few nucleotides can be used to introduce specific mutations into both episomally and chromosomally located genes (2, 14, 15). In most cases, chemically modified RNA-DNA chimeric oligonucleotides or single-stranded DNA oligonucleotides were used in which the chemical modifications served to protect the oligonucleotides from nucleolytic degradation. The mechanism of transfer of genetic information from the oligonucleotide to the target remains largely elusive, with numerous different cellular processes being involved [including transcription, DNA replication, homologous recombination, and DNA mismatch repair (MMR)].
Until recently, oligonucleotide-mediated gene targeting frequencies in mouse ES cells appeared to be relatively low. However, Hein te Riele and colleagues (2, 14, 15) developed a technique whereby suppression of the DNA MMR machinery by knockdown of Msh2 or Msh3 made it possible to introduce several specific point mutations into the Rb gene and Fancf, respectively. Transient suppression of Msh2 is likely to result in mismatch base changes elsewhere in the genome, and therefore knocking down Msh2 is not ideal. Msh3 null cells on the other hand do not show an overt mutator phenotype (14), suggesting that it will be possible to combine suppression of Msh3 and oligo-based gene targeting for the rapid generation of point mutants. To make oligo-based gene targeting approaches feasible for high-throughput applications it will necessary to increase the oligo targeting frequency or to scale up the identification of point mutant ES cell lines within the pool of transfected ES cell clones. The latter may be possible with new sequencing technologies.
shRNA Knockdown in the Mouse
RNA interference through the expression of shRNA molecules (where one strand is complementary to the coding region of the target gene) potentially holds great promise as a reverse genetics tool as it may allow inexpensive and rapid functional analysis in vivo. If combined with techniques such as tetraploid complementation (Fig. 1), it is possible to generate entirely ES cell-derived mice where a particular gene of interest is knocked down. Single copy shRNA transgenes under the control of either the U6 or the H1 promoter have been shown to mediate efficient and ubiquitous gene knockdown in mice when integrated at the Rosa26 locus (66), and shRNA knockdown has been shown to phenocopy a targeted mutation in the Fgf gene (40). However, shRNA knockdown approaches have gained only limited acceptance due largely to concerns over off-target effects. It has also emerged that it can be problematic to identify shRNAs that effectively knockdown a gene and that in some cases many hairpins may need to be screened to find one that is both specific and effective. For these reasons direct manipulation of the genome by gene targeting has remained the predominant technology. One very appealing aspect of shRNA knockdown in vivo is the ability to conditionally and reversibly silence a gene by combining shRNA expression with tet-regulatable promoters (65). This approach has made it possible to study the effect of Plk1 inhibition on tumor growth in vivo (36) and will be useful in applications where conditional inactivation and then reactivation of a gene are desirable. It has recently been determined that shRNA sequences within miRNA hosts are both more efficient at knocking down gene expression and also more specific (16) than hairpin designs used previously. With further evolution of this technology gene knockdown may become an increasingly feasible approach.
Chromosome Engineering for Generating Large-scale Chromosomal Rearrangements
Chromosome engineering refers to an approach whereby gene targeting in ES cells and the Cre/loxP site-specific recombination strategy is used to introduce defined chromosomal rearrangements into the genome such as a deletion, duplication, inversion, or translocation (83). In this technique, gene targeting is used to sequentially insert loxP sites into two different loci, and the doubly targeted ES cells are then exposed to Cre to induce recombination between the loxP sites and generate the rearranged chromosome (dependent upon the orientation of the targeted loxP sites and the relative position of selection cassettes used to recover the ES cells that have undergone recombination) (3, 57, 70, 85).
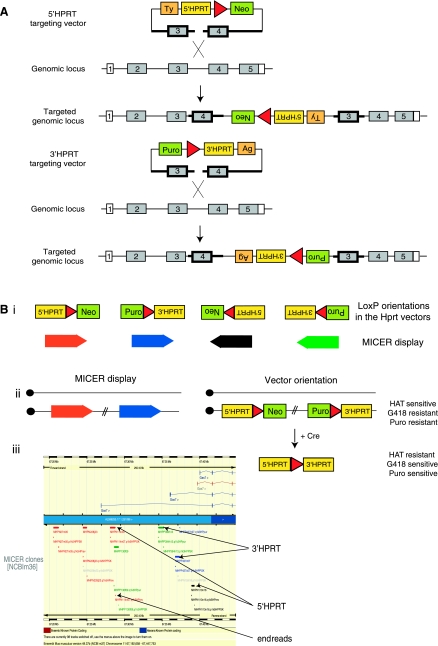
The most widely used approach for chromosome engineering employs two vectors called the 5′HPRT and 3′HPRT vectors: each of these vectors contain a loxP site, a portion of the HPRT mini-gene (5′HPRT or 3′HPRT), selection marker (neomycin or puromycin), and a coat-color tag (Agouti or Tyrosinase), shown in Fig. 5A (85). A single length of homologous genomic sequence in these vectors allows them to be used as insertion vectors (Fig. 2). Recently libraries of 5′HPRT and 3′HPRT targeting vectors were developed that contain randomly “shotgun cloned” genomic inserts (85). These libraries have been end-sequenced to generate the Mutagenic Insertion and Chromosome Engineering Resource, which allows a researcher to browse the mouse genome using Ensembl for 5′HPRT and 3′HPRT vectors that they can use to generate the rearrangement they desire (Fig. 5B). 5′HPRT and 3′HPRT vectors are displayed orientated on the genome and are color-coded to facilitate the easy selection of targeting vectors suitable for the generation of the desired chromosomal rearrangement, as shown in Fig. 5B (3).
Fig. 5.
Gene targeting in ES cells to engineer chromosomes. A: insertional targeting vectors, as shown, can be used to insert loxP sites (red triangles), positive selectable markers (such as the neomycin resistance cassette, Neo, and the puromycin resistance cassette, Puro) and HPRT gene fragments [5′ (exons 1–2) and 3′ (exons 3–9)] into the mouse genome for chromosome engineering. These vectors are called the 5′HPRT and 3′HPRT vectors, respectively. After the expression of Cre recombinase recombination between these vectors results in the formation of a functional HPRT mini-gene and allows ES cells to be grown in HAT (hypoxanthine, aminopterin, and thymidine)-containing medium. HAT resistance allows in vitro selection of ES cells carrying the desired chromosomal rearrangement (3, 85). B: the Mutagenic Insertion and Chromosome Engineering Resource (MICER) library is composed of 5′HPRT and 3′HPRT vectors. i: The orientation of the genomic insert in these vectors will dictate the orientation of the vector when it is targeted to the genome. These orientations are color coded to facilitate the selection of MICER vectors required to generate the desired rearrangement. ii: Shown is an example of the MICER vector selection required to generate a deletion by targeting the vectors in cis-. iii: Over 100,000 of these 5′HPRT and 3′HPRT vectors have been end-sequenced and are displayed on the Ensembl genome browser. Further details on MICER clone configurations to generate different types of rearrangements has been published previously (83).
Very large rearrangements can be generated by chromosome engineering (86); there is probably no limit to the size of inversions that can be generated, but the size of deletions that can be generated is limited by haploinsufficiency of genes within the deleted interval killing the ES cells. If a very large deletion is to be generated it is possible to circumvent potential haploinsuffiency by targeting 5′HPRT and 3′HPRT vectors in trans-, and after the expression of Cre a balanced deletion/duplication may be generated on homologous chromosomes (83). In most cases rearrangements of chromosomal regions are generated in vitro and then transmitted. It is, however, possible to generate rearrangements somatically by conditionally expressing Cre recombinase in a cell- or tissue-specific manner (86). This approach may be useful for studying tumor suppressor regions of the genome.
FORWARD GENETICS APPROACHES OF MODIFYING THE MOUSE GENOME
Large-scale ENU Mutagenesis
ENU is an alkylating agent that is a powerful mutagen in mouse spermatogonial stem cells and can be used to produce single locus mutations at a frequency equivalent in the range of one out of every 175–655 gametes screened. Analysis of sequenced germline mutations reveals that ENU predominantly modifies A.T base pairs, and when translated into a protein product, these changes predominantly result in missense mutations. Point mutations induced by ENU provide a unique mutant resource because they: 1) reflect the consequences of single base change independent of position effects, 2) dissect protein function at amino acid resolution, 3) generate many different types of alleles (loss-of-function mutations, viable hypomorphs of lethal complementation groups, antimorphs, and gain-of-function mutations), and 4) discover gene functions in an unbiased manner. Phenotype-driven ENU screens in the mouse are being performed to identify genes involved in a range of human disease areas including cardiology, physiology, neurology, immunity, hematopoiesis, and mammalian development (38). ENU-based approaches are extremely powerful in understanding complex human diseases and traits: the base-pair changes can accurately model base changes found in human diseases, and subtle mutant alleles in a standard genetic background provide the ability to analyze the consequences of compound genotypes. Ongoing mouse ENU mutagenesis experiments are generating a rich resource of new mutations allowing an in-depth study of a single gene, a chromosomal region, or a biological system. A summary of ENU mutagenesis and other mouse resources is available in Table 2.
Table 2.
Resources of live mice and phenotyping data
| Source | URL |
|---|---|
| Mouse Strains | |
| Federation of International Mouse Strain Resources | http://www.fimre.org/ |
| International Mouse Strain Resource | http://www.informatics.jax.org/imsr/index.jsp |
| The European Mutant Mouse Archive | http://www.emmanet.org |
| The Jackson Laboratory | http://www.jax.org |
| Mutant Mouse Regional Resource Centers | http://www.mmrrc.org |
| Mouse Models of Human Cancers Consortium | http://web.ncifcrf.gov/researchresources/mmhcc |
| Riken Bioresource Center | http://www.brc.riken.jp/lab/animal/en/ |
| Center for Animal Resources and Development Database | http://cardb.cc.kumamoto-u.ac.jp/transgenic/index.jsp |
| ENU Mouse Lines | |
| Baylor College of Medicine | http://www.mouse-genome.bcm.tmc.edu/enu/ENUHome.asp |
| Helmholtz Zentrum Mn̈chen | http://www.helmholtz-muenchen.de/en/ieg/group-genome-project/enu-screen/ |
| Harwell Mutagenesis Programme | http://www.har.mrc.ac.uk/ |
| The Jackson Laboratory | http://www.jax.org |
| The Oak Ridge National Laboratory | http://mouse.ornl.gov/mmdb |
| The McLaughlin Research Institute | http://www.montana.edu/wwwmri/ |
| Australian Phenomics Facility | http://www.apf.edu.au/ |
| Centre for Modeling Human Disease | http://www.cmhd.ca/ |
| Phenotyping Data | |
| Mouse Genome Informatics | http://www.informatics.jax.org/ |
| Deltagen and Lexicon Knockout Mice | http://www.informatics.jax.org/external/ko/ |
| PBMice | http://www.scbit.org/PBmice/ |
| European Mouse Disease Clinic | http://www.eumodic.org/ |
| Sanger Mouse Genetics Programme | http://www.sanger.ac.uk |
ENU, N-ethyl-N-nitrosourea.
One of the major limitations of approaches that use random mutagenesis (such as ENU) is that finding mutations and proving they are causal can be extremely laborious. Recently, new technology that allows the specific capture of DNA by array hybridization coupled with high-throughput parallel sequencing has been developed (33). These technologies have the potential to facilitate a resurgence in ENU screens because they will dramatically accelerate the recovery of mutations. Although it is also possible to generate random genome-wide mutations using radiation and other mutagens such as EMS or chlorambucil, these mutagens, each of which produces its own signature of mutations, are not frequently utilized.
Emerging Transposon-based Approaches
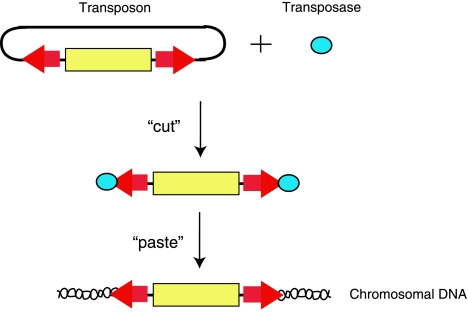
As mentioned above chemical mutagen-based approaches using agents such as ENU are very powerful because they can mutate a gene in many different ways, potentially generating a polychromatic view of a gene's function, albeit a view that may be difficult to interpret. Transposons represent an alternative random mutagenic approach. Transposons are DNA-based elements that can function as part of a bipartite system with a transposase to mutate genes both in vitro (in ES cells) and in vivo (in the germline or somatically) (10, 17, 20, 27, 35), as shown in Fig. 6.
Fig. 6.
The bipartite transposon system is composed of a transposon and transposase. Transposons may function by a “cut-and-paste” mechanism to insert sequence (yellow box) between the transposon repeats (red arrows) into the genome. This “jumping” is catalyzed by the transposase enzyme (blue circles). Transposons, like all insertional mutagens, have preferences for the sequence into which they like to insert.
The most widely studied transposon/transposase system for mammalian mutagenesis in vitro and in vivo is the Sleeping Beauty transposon of the Tc1-like mariner family (35). The Sleeping Beauty transposase was originally discovered in salmon, where it had been silenced through evolution, and was point mutated to “resurrect” its ability to transpose the genome (35). Subsequent additional mutations have been added to the Sleeping Beauty transposase to improve its ability to stimulate “jumping” of the Sleeping Beauty transposon (82). Although the Sleeping Beauty transposon system has been shown to function as an efficient germline mutagen analysis of mutant mice generated using this system has been complicated by gene mutations and genomic rearrangements due to transposon mobilization from chromosomal concatemers (27). This is due largely to the fact that the Sleeping Beauty transposon generates a double strand break when it is excised from the genome as part of a cut-and-paste procedure. Despite this the system has been used to generate several interesting mutants, and because the transposon acts as a tag the mutated genes were easily identified. The Sleeping Beauty transposon system has also been used to perform regional saturating screens of the mouse genome (37). These screens have been facilitated by the fact that the Sleeping Beauty transposon preferentially integrates close to the site in the genome from where it was initially mobilized (37).
More recently an alternative transposon called Piggybac, derived from the cabbage looper moth (Trichoplusia ni), has been employed for germline mutagenesis (17). This transposon is thought to more randomly generate mutations across the genome and has the advantage that it can carry large payloads between its transposon repeats, unlike Sleeping Beauty whose transposition efficiently drops dramatically when the size of the payload between its IR/DRs increases above 2 kb (44). A database of Piggybac mutant mouse lines and some phenotyping data of these lines exist on the web (http://www.scbit.org/PBmice/) (72). At present this database is limited but is set to expand over the next few years. Piggybac has also been used together with Cre/LoxP technology to generate large-scale rearrangements of the mouse genome including duplications, deletions, and translocations (80). Over the next few years it is likely that there will be a considerable amount of technology development pushing forward the use of Piggybac as a genome-wide insertional mutagen. There are many other transposons that are yet to be tested in mammalian cells, and it's likely that some of these will have features that make them a useful part of the toolkit for modifying the mouse genome.
CONCLUSIONS
The mouse has always been the experimental organism of choice for modeling most human diseases and for the discovery of gene function in vivo. The generation of large resources of gene traps, targeting vectors, targeted ES cells, and transposon mice, together with new technology such as the ability to rapidly engineer point mutations into the genome, will further accelerate the pace of discovery. In a previous review in Physiological Genomics (77), we wrote “In the postgenomic era the mouse will be central to the challenge of ascribing a function to the 40,000 or so genes that constitute our genome.” This statement is now almost certainly the case.
GRANTS
D. J. Adams is funded by Cancer Research-UK and the Wellcome Trust. L. van der Weyden is funded by a Fellowship from the Kay Kendell Leukaemia Foundation.
Address for reprint requests and other correspondence: D. J. Adams & L. van der Weyden, Experimental Cancer Genetics, Wellcome Trust Sanger Inst., Wellcome Trust Genome Campus, Hinxton, Cambs, CB10 1SA, UK (e-mail: da1@sanger.ac.uk & lvdw@sanger.ac.uk).
REFERENCES
- 1.International HapMap Consortium. A haplotype map of the human genome. Nature 437: 1299–1320, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aarts M, Dekker M, de Vries S, van der Wal A, te Riele H. Generation of a mouse mutant by oligonucleotide-mediated gene modification in ES cells. Nucleic Acids Res 34: e147, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams DJ, Biggs PJ, Cox T, Davies R, van der Weyden L, Jonkers J, Smith J, Plumb B, Taylor R, Nishijima I, Yu Y, Rogers J, Bradley A. Mutagenic insertion and chromosome engineering resource (MICER). Nat Genet 36: 867–871, 2004. [DOI] [PubMed] [Google Scholar]
- 4.Adams DJ, Quail MA, Cox T, van der Weyden L, Gorick BD, Su Q, Chan WI, Davies R, Bonfield JK, Law F, Humphray S, Plumb B, Liu P, Rogers J, Bradley A. A genome-wide, end-sequenced 129Sv BAC library resource for targeting vector construction. Genomics 86: 753–758, 2005. [DOI] [PubMed] [Google Scholar]
- 5.Austin S, Ziese M, Sternberg N. A novel role for site-specific recombination in maintenance of bacterial replicons. Cell 25: 729–736, 1981. [DOI] [PubMed] [Google Scholar]
- 6.Awatramani R, Soriano P, Rodriguez C, Mai JJ, Dymecki SM. Cryptic boundaries in roof plate and choroid plexus identified by intersectional gene activation. Nat Genet 35: 70–75, 2003. [DOI] [PubMed] [Google Scholar]
- 7.Bouhassira EE, Westerman K, Leboulch P. Transcriptional behavior of LCR enhancer elements integrated at the same chromosomal locus by recombinase-mediated cassette exchange. Blood 90: 3332–3344, 1997. [PubMed] [Google Scholar]
- 8.Candotti F, Agbaria R, Mullen CA, Touraine R, Balzarini J, Johns DG, Blaese RM. Use of a herpes thymidine kinase/neomycin phosphotransferase chimeric gene for metabolic suicide gene transfer. Cancer Gene Ther 7: 574–580, 2000. [DOI] [PubMed] [Google Scholar]
- 9.Chen YT, Bradley A. A new positive/negative selectable marker, puDeltatk, for use in embryonic stem cells. Genesis 28: 31–35, 2000. [DOI] [PubMed] [Google Scholar]
- 10.Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature 436: 272–276, 2005. [DOI] [PubMed] [Google Scholar]
- 11.Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nat Rev Genet 2: 769–779, 2001. [DOI] [PubMed] [Google Scholar]
- 12.Coutinho JM, Singaraja RR, Kang M, Arenillas DJ, Bertram LN, Bissada N, Staels B, Fruchart JC, Fievet C, Joseph-George AM, Wasserman WW, Hayden MR. Complete functional rescue of the ABCA1-/- mouse by human BAC transgenesis. J Lipid Res 46: 1113–1123, 2005. [DOI] [PubMed] [Google Scholar]
- 13.Cronin CA, Gluba W, Scrable H. The lac operator-repressor system is functional in the mouse. Genes Dev 15: 1506–1517, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dekker M, Brouwers C, Aarts M, van der Torre J, de Vries S, van de Vrugt H, te Riele H. Effective oligonucleotide-mediated gene disruption in ES cells lacking the mismatch repair protein MSH3. Gene Ther 13: 686–694, 2006. [DOI] [PubMed] [Google Scholar]
- 15.Dekker M, Brouwers C, te Riele H. Targeted gene modification in mismatch-repair-deficient embryonic stem cells by single-stranded DNA oligonucleotides. Nucleic Acids Res 31: e27, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, Lowe SW. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet 37: 1289–1295, 2005. [DOI] [PubMed] [Google Scholar]
- 17.Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 122: 473–483, 2005. [DOI] [PubMed] [Google Scholar]
- 18.Diwan BA, Blackman KE. Differential susceptibility of 3 sublines of C57BL/6 mice to the induction of colorectal tumors by 1,2-dimethylhydrazine. Cancer Lett 9: 111–115, 1980. [DOI] [PubMed] [Google Scholar]
- 19.Doetschman T, Gregg RG, Maeda N, Hooper ML, Melton DW, Thompson S, Smithies O. Targetted correction of a mutant HPRT gene in mouse embryonic stem cells. Nature 330: 576–578, 1987. [DOI] [PubMed] [Google Scholar]
- 20.Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature 436: 221–226, 2005. [DOI] [PubMed] [Google Scholar]
- 21.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature 292: 154–156, 1981. [DOI] [PubMed] [Google Scholar]
- 22.Feil R, Brocard J, Mascrez B, LeMeur M, Metzger D, Chambon P. Ligand-activated site-specific recombination in mice. Proc Natl Acad Sci USA 93: 10887–10890, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng YQ, Seibler J, Alami R, Eisen A, Westerman KA, Leboulch P, Fiering S, Bouhassira EE. Site-specific chromosomal integration in mammalian cells: highly efficient CRE recombinase-mediated cassette exchange. J Mol Biol 292: 779–785, 1999. [DOI] [PubMed] [Google Scholar]
- 24.Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, Down T, Dyer SC, Eyre T, Fitzgerald S, Fernandez-Banet J, Graf S, Haider S, Hammond M, Holland R, Howe KL, Howe K, Johnson N, Jenkinson A, Kahari A, Keefe D, Kokocinski F, Kulesha E, Lawson D, Longden I, Megy K, Meidl P, Overduin B, Parker A, Pritchard B, Prlic A, Rice S, Rios D, Schuster M, Sealy I, Slater G, Smedley D, Spudich G, Trevanion S, Vilella AJ, Vogel J, White S, Wood M, Birney E, Cox T, Curwen V, Durbin R, Fernandez-Suarez XM, Herrero J, Hubbard TJ, Kasprzyk A, Proctor G, Smith J, Ureta-Vidal A, Searle S. Ensembl 2008. Nucleic Acids Res 36: D707–D714, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Floss T, Schnutgen F. Conditional gene trapping using the FLEx system. Methods Mol Biol 435: 127–138, 2008. [DOI] [PubMed] [Google Scholar]
- 26.George SH, Gertsenstein M, Vintersten K, Korets-Smith E, Murphy J, Stevens ME, Haigh JJ, Nagy A. Developmental and adult phenotyping directly from mutant embryonic stem cells. Proc Natl Acad Sci USA 104: 4455–4460, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geurts AM, Collier LS, Geurts JL, Oseth LL, Bell ML, Mu D, Lucito R, Godbout SA, Green LE, Lowe SW, Hirsch BA, Leinwand LA, Largaespada DA. Gene mutations and genomic rearrangements in the mouse as a result of transposon mobilization from chromosomal concatemers. PLoS Genet 2: e156, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonzalez MA, Tachibana KE, Adams DJ, van der Weyden L, Hemberger M, Coleman N, Bradley A, Laskey RA. Geminin is essential to prevent endoreduplication and to form pluripotent cells during mammalian development. Genes Dev 20: 1880–1884, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science 268: 1766–1769, 1995. [DOI] [PubMed] [Google Scholar]
- 30.Graubert TA, Cahan P, Edwin D, Selzer RR, Richmond TA, Eis PS, Shannon WD, Li X, McLeod HL, Cheverud JM, Ley TJ. A high-resolution map of segmental DNA copy number variation in the mouse genome. PLoS Genet 3: e3, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hasty P, Rivera-Perez J, Bradley A. The length of homology required for gene targeting in embryonic stem cells. Mol Cell Biol 11: 5586–5591, 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hasty P, Rivera-Perez J, Chang C, Bradley A. Target frequency and integration pattern for insertion and replacement vectors in embryonic stem cells. Mol Cell Biol 11: 4509–4517, 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hodges E, Xuan Z, Balija V, Kramer M, Molla MN, Smith SW, Middle CM, Rodesch MJ, Albert TJ, Hannon GJ, McCombie WR. Genome-wide in situ exon capture for selective resequencing. Nat Genet 39: 1522–1527, 2007. [DOI] [PubMed] [Google Scholar]
- 34.Hooper M, Hardy K, Handyside A, Hunter S, Monk M. HPRT-deficient (Lesch-Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature 326: 292–295, 1987. [DOI] [PubMed] [Google Scholar]
- 35.Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell 91: 501–510, 1997. [DOI] [PubMed] [Google Scholar]
- 36.Kappel S, Matthess Y, Zimmer B, Kaufmann M, Strebhardt K. Tumor inhibition by genomically integrated inducible RNAi-cassettes. Nucleic Acids Res 34: 4527–4536, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keng VW, Yae K, Hayakawa T, Mizuno S, Uno Y, Yusa K, Kokubu C, Kinoshita T, Akagi K, Jenkins NA, Copeland NG, Horie K, Takeda J. Region-specific saturation germline mutagenesis in mice using the Sleeping Beauty transposon system. Nat Methods 2: 763–769, 2005. [DOI] [PubMed] [Google Scholar]
- 38.Kile BT, Hentges KE, Clark AT, Nakamura H, Salinger AP, Liu B, Box N, Stockton DW, Johnson RL, Behringer RR, Bradley A, Justice MJ. Functional genetic analysis of mouse chromosome 11. Nature 425: 81–86, 2003. [DOI] [PubMed] [Google Scholar]
- 39.Kontgen F, Suss G, Stewart C, Steinmetz M, Bluethmann H. Targeted disruption of the MHC class II Aa gene in C57BL/6 mice. Int Immunol 5: 957–964, 1993. [DOI] [PubMed] [Google Scholar]
- 40.Kunath T, Gish G, Lickert H, Jones N, Pawson T, Rossant J. Transgenic RNA interference in ES cell-derived embryos recapitulates a genetic null phenotype. Nat Biotechnol 21: 559–561, 2003. [DOI] [PubMed] [Google Scholar]
- 41.Lakso M, Sauer B, Mosinger B Jr, Lee EJ, Manning RW, Yu SH, Mulder KL, Westphal H. Targeted oncogene activation by site-specific recombination in transgenic mice. Proc Natl Acad Sci USA 89: 6232–6236, 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73: 56–65, 2001. [DOI] [PubMed] [Google Scholar]
- 43.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69: 915–926, 1992. [DOI] [PubMed] [Google Scholar]
- 44.Luo G, Ivics Z, Izsvak Z, Bradley A. Chromosomal transposition of a Tc1/mariner-like element in mouse embryonic stem cells. Proc Natl Acad Sci USA 95: 10769–10773, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin GR Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci USA 78: 7634–7638, 1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McClive P, Pall G, Newton K, Lee M, Mullins J, Forrester L. Gene trap integrations expressed in the developing heart: insertion site affects splicing of the PT1-ATG vector. Dev Dyn 212: 267–276, 1998. [DOI] [PubMed] [Google Scholar]
- 47.McLeod M, Craft S, Broach JR. Identification of the crossover site during FLP-mediated recombination in the Saccharomyces cerevisiae plasmid 2 microns circle. Mol Cell Biol 6: 3357–3367, 1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murisier F, Guichard S, Beermann F. A conserved transcriptional enhancer that specifies Tyrp1 expression to melanocytes. Dev Biol 298: 644–655, 2006. [DOI] [PubMed] [Google Scholar]
- 49.Nagy A, Mar L. Creation and use of a Cre recombinase transgenic database. Methods Mol Biol 158: 95–106, 2001. [DOI] [PubMed] [Google Scholar]
- 50.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci USA 90: 8424–8428, 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nord AS, Chang PJ, Conklin BR, Cox AV, Harper CA, Hicks GG, Huang CC, Johns SJ, Kawamoto M, Liu S, Meng EC, Morris JH, Rossant J, Ruiz P, Skarnes WC, Soriano P, Stanford WL, Stryke D, von Melchner H, Wurst W, Yamamura K, Young SG, Babbitt PC, Ferrin TE. The International Gene Trap Consortium Website: a portal to all publicly available gene trap cell lines in mouse. Nucleic Acids Res 34: D642–648, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ohtsuka M, Ishii K, Kikuti YY, Warita T, Suzuki D, Sato M, Kimura M, Inoko H. Construction of mouse 129/Ola BAC library for targeting experiments using E14 embryonic stem cells. Genes Genet Syst 81: 143–146, 2006. [DOI] [PubMed] [Google Scholar]
- 53.Osoegawa K, Tateno M, Woon PY, Frengen E, Mammoser AG, Catanese JJ, Hayashizaki Y, de Jong PJ. Bacterial artificial chromosome libraries for mouse sequencing and functional analysis. Genome Res 10: 116–128, 2000. [PMC free article] [PubMed] [Google Scholar]
- 54.Park F Lentiviral vectors: are they the future of animal transgenesis? Physiol Genomics 31: 159–173, 2007. [DOI] [PubMed] [Google Scholar]
- 55.Pham CT, MacIvor DM, Hug BA, Heusel JW, Ley TJ. Long-range disruption of gene expression by a selectable marker cassette. Proc Natl Acad Sci USA 93: 13090–13095, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prosser HM, Rzadzinska AK, Steel KP, Bradley A. Mosaic complementation demonstrates a regulatory role for myosin VIIa in actin dynamics of stereocilia. Mol Cell Biol 28: 1702–1712, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ramirez-Solis R, Liu P, Bradley A. Chromosome engineering in mice. Nature 378: 720–724, 1995. [DOI] [PubMed] [Google Scholar]
- 58.Sauer B, Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc Natl Acad Sci USA 85: 5166–5170, 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scacheri PC, Crabtree JS, Novotny EA, Garrett-Beal L, Chen A, Edgemon KA, Marx SJ, Spiegel AM, Chandrasekharappa SC, Collins FS. Bidirectional transcriptional activity of PGK-neomycin and unexpected embryonic lethality in heterozygote chimeric knockout mice. Genesis 30: 259–263, 2001. [DOI] [PubMed] [Google Scholar]
- 60.Schaft J, Ashery-Padan R, van der Hoeven F, Gruss P, Stewart AF. Efficient FLP recombination in mouse ES cells and oocytes. Genesis 31: 6–10, 2001. [DOI] [PubMed] [Google Scholar]
- 61.Schambach A, Galla M, Modlich U, Will E, Chandra S, Reeves L, Colbert M, Williams DA, von Kalle C, Baum C. Lentiviral vectors pseudotyped with murine ecotropic envelope: increased biosafety and convenience in preclinical research. Exp Hematol 34: 588–592, 2006. [DOI] [PubMed] [Google Scholar]
- 62.Schnutgen F, Doerflinger N, Calleja C, Wendling O, Chambon P, Ghyselinck NB. A directional strategy for monitoring Cre-mediated recombination at the cellular level in the mouse. Nat Biotechnol 21: 562–565, 2003. [DOI] [PubMed] [Google Scholar]
- 63.Schoonjans L, Kreemers V, Danloy S, Moreadith RW, Laroche Y, Collen D. Improved generation of germline-competent embryonic stem cell lines from inbred mouse strains. Stem Cells 21: 90–97, 2003. [DOI] [PubMed] [Google Scholar]
- 64.Schwenk F, Zevnik B, Bruning J, Rohl M, Willuweit A, Rode A, Hennek T, Kauselmann G, Jaenisch R, Kuhn R. Hybrid embryonic stem cell-derived tetraploid mice show apparently normal morphological, physiological, and neurological characteristics. Mol Cell Biol 23: 3982–3989, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seibler J, Kleinridders A, Kuter-Luks B, Niehaves S, Bruning JC, Schwenk F. Reversible gene knockdown in mice using a tight, inducible shRNA expression system. Nucleic Acids Res 35: e54, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seibler J, Kuter-Luks B, Kern H, Streu S, Plum L, Mauer J, Kuhn R, Bruning JC, Schwenk F. Single copy shRNA configuration for ubiquitous gene knockdown in mice. Nucleic Acids Res 33: e67, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seibler J, Schubeler D, Fiering S, Groudine M, Bode J. DNA cassette exchange in ES cells mediated by Flp recombinase: an efficient strategy for repeated modification of tagged loci by marker-free constructs. Biochemistry 37: 6229–6234, 1998. [DOI] [PubMed] [Google Scholar]
- 68.Seibler J, Zevnik B, Kuter-Luks B, Andreas S, Kern H, Hennek T, Rode A, Heimann C, Faust N, Kauselmann G, Schoor M, Jaenisch R, Rajewsky K, Kuhn R, Schwenk F. Rapid generation of inducible mouse mutants. Nucleic Acids Res 31: e12, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Seong E, Saunders TL, Stewart CL, Burmeister M. To knockout in 129 or in C57BL/6: that is the question. Trends Genet 20: 59–62, 2004. [DOI] [PubMed] [Google Scholar]
- 70.Smith AJ, De Sousa MA, Kwabi-Addo B, Heppell-Parton A, Impey H, Rabbitts P. A site-directed chromosomal translocation induced in embryonic stem cells by Cre-loxP recombination. Nat Genet 9: 376–385, 1995. [DOI] [PubMed] [Google Scholar]
- 71.Stiedl O, Radulovic J, Lohmann R, Birkenfeld K, Palve M, Kammermeier J, Sananbenesi F, Spiess J. Strain and substrain differences in context- and tone-dependent fear conditioning of inbred mice. Behav Brain Res 104: 1–12, 1999. [DOI] [PubMed] [Google Scholar]
- 72.Sun LV, Jin K, Liu Y, Yang W, Xie X, Ye L, Wang L, Zhu L, Ding S, Su Y, Zhou J, Han M, Zhuang Y, Xu T, Wu X, Gu N, Zhong Y. PBmice: an integrated database system of piggyBac (PB) insertional mutations and their characterizations in mice. Nucleic Acids Res 36: D729–734, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Svenson KL, Von Smith R, Magnani PA, Suetin HR, Paigen B, Naggert JK, Li R, Churchill GA, Peters LL. Multiple trait measurements in 43 inbred mouse strains capture the phenotypic diversity characteristic of human populations. J Appl Physiol 102: 2369–2378, 2007. [DOI] [PubMed] [Google Scholar]
- 74.Testa G, Schaft J, van der Hoeven F, Glaser S, Anastassiadis K, Zhang Y, Hermann T, Stremmel W, Stewart AF. A reliable lacZ expression reporter cassette for multipurpose, knockout-first alleles. Genesis 38: 151–158, 2004. [DOI] [PubMed] [Google Scholar]
- 75.Thomas KR, Capecchi MR. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell 51: 503–512, 1987. [DOI] [PubMed] [Google Scholar]
- 76.Thorpe HM, Smith MC. In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family. Proc Natl Acad Sci USA 95: 5505–5510, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Van der Weyden L, Adams DJ, Bradley A. Tools for targeted manipulation of the mouse genome. Physiol Genomics 11: 133–164, 2002. [DOI] [PubMed] [Google Scholar]
- 78.Van der Weyden L, Adams DJ, Harris LW, Tannahill D, Arends MJ, Bradley A. Null and conditional semaphorin 3B alleles using a flexible puroDeltatk loxP/FRT vector. Genesis 41: 171–178, 2005. [DOI] [PubMed] [Google Scholar]
- 79.Wang J, Sarov M, Rientjes J, Fu J, Hollak H, Kranz H, Xie W, Stewart AF, Zhang Y. An improved recombineering approach by adding RecA to lambda Red recombination. Mol Biotechnol 32: 43–53, 2006. [DOI] [PubMed] [Google Scholar]
- 80.Wu S, Ying G, Wu Q, Capecchi MR. Toward simpler and faster genome-wide mutagenesis in mice. Nat Genet 39: 922–930, 2007. [DOI] [PubMed] [Google Scholar]
- 81.Yang H, Bell TA, Churchill GA, Pardo-Manuel de Villena F. On the subspecific origin of the laboratory mouse. Nat Genet 39: 1100–1107, 2007. [DOI] [PubMed] [Google Scholar]
- 82.Yant SR, Park J, Huang Y, Mikkelsen JG, Kay MA. Mutational analysis of the N-terminal DNA-binding domain of sleeping beauty transposase: critical residues for DNA binding and hyperactivity in mammalian cells. Mol Cell Biol 24: 9239–9247, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yu Y, Bradley A. Engineering chromosomal rearrangements in mice. Nat Rev Genet 2: 780–790, 2001. [DOI] [PubMed] [Google Scholar]
- 84.Zhao R, Fahs SA, Weiler H, Duncan SA. An efficient method to successively introduce transgenes into a given genomic locus in the mouse. BMC Dev Biol 1: 10, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zheng B, Mills AA, Bradley A. A system for rapid generation of coat color-tagged knockouts and defined chromosomal rearrangements in mice. Nucleic Acids Res 27: 2354–2360, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zheng B, Sage M, Sheppeard EA, Jurecic V, Bradley A. Engineering mouse chromosomes with Cre-loxP: range, efficiency, and somatic applications. Mol Cell Biol 20: 648–655, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]