Abstract
In many organisms, there are multiple isoforms of cytoplasmic dynein heavy chains, and division of labor among the isoforms would provide a mechanism to regulate dynein function. The targeted disruption of somatic genes in Tetrahymena thermophila presents the opportunity to determine the contributions of individual dynein isoforms in a single cell that expresses multiple dynein heavy chain genes. Substantial portions of two Tetrahymena cytoplasmic dynein heavy chain genes were cloned, and their motor domains were sequenced. Tetrahymena DYH1 encodes the ubiquitous cytoplasmic dynein Dyh1, and DYH2 encodes a second cytoplasmic dynein isoform, Dyh2. The disruption of DYH1, but not DYH2, resulted in cells with two detectable defects: 1) phagocytic activity was inhibited, and 2) the cells failed to distribute their chromosomes correctly during micronuclear mitosis. In contrast, the disruption of DYH2 resulted in a loss of regulation of cell size and cell shape and in the apparent inability of the cells to repair their cortical cytoskeletons. We conclude that the two dyneins perform separate tasks in Tetrahymena.
INTRODUCTION
Dynein is a molecular motor that transduces chemical energy into mechanical motion along microtubules. There are two functional classes of dynein: axonemal dynein that produces the propagated bending of cilia and of eukaryotic flagella and nonaxonemal or “cytoplasmic” dynein that performs functions other than ciliary beating. Cytoplasmic dynein is implicated in a variety of intracellular movements, including fast retrograde transport (Schroer et al., 1989; Muresan et al., 1996), slow axonal transport (Cleveland and Hoffman, 1991; Dillman et al., 1996), mediation of the trafficking of membrane-bounded organelles (Corthesy-Theulaz et al., 1992; Aniento et al., 1993; Fath et al., 1994; Oda et al., 1995; Holleran et al., 1996), the organization and separation of the bipolar mitotic spindle (Verde et al., 1991; Vaisberg et al., 1993; Saunders et al., 1995; Echeverri et al., 1996), and postmitotic nuclear migrations in yeast and filamentous fungi (Eshel et al., 1993; Li et al., 1993; Plamann et al., 1994; Xiang et al., 1994; Inoue et al., 1998).
The realization that dynein performs so many different tasks and the discovery that many organisms express multiple dynein heavy chain genes lead to the hypothesis that different dynein isoforms perform different cellular tasks (reviewed in Asai, 1996). Cilia and eukaryotic flagella provide clear examples of dynein functional specialization (Piperno, 1990). The close coordination of ∼12 distinct axonemal dynein heavy chains produces axonemal bending (e.g., Piperno and Ramanis, 1991; Mastronarde et al., 1992; reviewed in Brokaw, 1994).
The functional specialization of the cytoplasmic dyneins is not well understood. All eukaryotes examined express homologues of the ubiquitous Dyh1 (also called MAP1C, cDHC1, DHC1a), and some organisms, including Saccharomyces, Aspergillus, Neurospora, and Dictyostelium, apparently have only this dynein. In the analysis of the family of dynein genes expressed in sea urchin, the catalytic domain of a second putative cytoplasmic dynein heavy chain, originally called DYH1b and here referred to as Dyh2, was identified and partly sequenced (Gibbons et al., 1994). Homologues of Dyh2 have subsequently been identified in mammals, in which it has been called DLP4, DHC1b, and cDHC2 (Tanaka et al., 1995; Criswell et al., 1996; Criswell and Asai, 1998; Vaisberg et al., 1996). This second cytoplasmic dynein isoform also has been identified in Chlamydomonas (Pazour et al., 1999; Porter et al., 1999) and Caenorhabditis elegans (Wilson et al., 1994). In this manuscript, we describe two Tetrahymena dynein heavy chain genes: DYH1 encodes Dyh1 protein that is the homologue of sea urchin 1a, and DYH2 encodes Dyh2 that is the homologue of sea urchin 1b. To simplify the following discussion, we use “Dyh1” and “Dyh2” to describe these isoforms that, in the original manuscripts, were referred to by other names.
Previous studies provide compelling evidence that Dyh2 is a bona fide cytoplasmic dynein. The gene encoding Dyh2 is expressed in unciliated tissues (Tanaka et al., 1995; Criswell et al., 1996; Vaisberg et al., 1996). The Dyh2 protein was immunolocalized to the Golgi apparatus in unciliated fibroblasts (Vaisberg et al., 1996), and injection of antibodies to Dyh2 caused the dispersal of the Golgi apparatus (Vaisberg et al., 1996). The expression of the gene encoding Dyh2 but not Dyh1 was shown to be upregulated during ciliogenesis in primary cultures of rat tracheal epithelial cells, and isoform-specific antibodies revealed that Dyh2 protein accumulated at the apical ends of these cells but appeared to be excluded from the cilia (Cris-well et al., 1996).
As do larger eukaryotes, the ciliated protozoan Tetrahymena thermophila expresses ∼15 separate dynein heavy chain genes, including ones encoding Dyh1 and Dyh2 (Lee et al., 1997). The Tetrahymena microtubule cytoskeleton includes a cortical cage that helps to determine cell size and shape and provides the framework for the rows of ciliary basal bodies and other cytoplasmic microtubules that mediate intracellular movements including micronuclear mitosis and meiosis (reviewed in Frankel, 1999). Each cell has two functionally distinct nuclei: the diploid germline micronucleus is transcriptionally silent and therefore not required for vegetative growth, and the somatic macronucleus contains ∼45 copies of each gene and determines the phenotype of the cell. During vegetative growth, the cell divides every ∼2.5 h during which an intranuclear mitotic spindle mediates the accurate separation of the five micronuclear chromosomes. However, unlike the micronucleus, the macronucleus divides amitotically, being pinched apart during cytokinesis. The amitotic division of the macronucleus separates the somatic genome imperfectly and can lead to phenotypic assortment of a macronuclear allele (Sonneborn, 1974). Because the micronucleus is not transcribed, the accurate segregation of micronuclear chromosomes is not required for vegetative growth. Indeed, many species of Tetrahymena are amicronucleate; they are propagated vegetatively but cannot undergo sexual reproduction (Nanney and Simon, 1999).
Recent advances provide efficient methods to achieve macronuclear gene disruption in which a selectable marker is inserted into the targeted chromosome exclusively by homologous recombination (Gaertig and Gorovsky, 1992; Cassidy-Hanley et al., 1997). Phenotypic assortment leads to the rapid and complete replacement of the wild-type gene if the targeted gene is not required for growth or to the incomplete elimination of an essential gene. In both cases, the transformants remain viable, and even in the case of an incomplete replacement, a phenotype may be observed. Thus Tetrahymena presents the unique opportunity to focus on the cellular contributions of an individual dynein in a cell with many dyneins. In the present study, we have disrupted the macronuclear DYH1 and DYH2 genes individually. These disruptions reveal that the two cytoplasmic dyneins are functionally specialized.
MATERIALS AND METHODS
Nomenclature Used in This Manuscript
The nomenclature originally introduced in the sea urchin study (Gibbons et al., 1994) is useful because it organizes the dynein heavy chains in a hierarchy based on the sequences of the catalytic domains (see Gibbons, 1995). However, similarity between sequences does not necessarily mean that the two dyneins are also functionally related. Thus, we have adopted a nomenclature that is neutral in terms of dynein function and is in keeping with the recently adopted rules for naming Tetrahymena genes (Allen et al., 1998).
Cloning and Characterization of DYH1 and DYH2
Degenerate oligonucleotide primers corresponding to conserved sequences near the dynein P1 loop were used in RNA-directed PCR (reverse transcription [RT]-PCR) (Asai and Criswell, 1995). Sequencing of the 180-bp RT-PCR products identified several different dynein heavy chain fragments, including ones whose deduced amino acid sequences corresponded to those of Dyh1 and Dyh2 described in other organisms. Genomic clones of Tetrahymena DYH1 and DYH2 were obtained by screening a phage library constructed from wild-type (B2086) macronuclear DNA partially digested with BglII and ligated into the XhoI-half site of the lambda arms (LambdaGem-11, Promega, Madison, WI). Double-stranded DNA was purified (plasmid columns, Qiagen, Chatsworth, CA) and sequenced using Thermo Sequenase (Amersham, Cleveland, OH) or by the Purdue Center for DNA sequencing using cycle-sequencing methods. Sequences were analyzed with the GCG software package (version 9.1 [1997], University of Wisconsin, Madison, WI). Southern blots of genomic DNA and Northern blots of total RNA (Chomczynski and Sacchi, 1987) were processed by standard methods (Sambrook et al., 1989). The Northern blot of RNA from reciliating cells (see Figure 2) was probed, stripped by treating the blot for 5 min with hot 0.1% SDS, and then reprobed. The intensities of the signals were estimated by densitometry using ImageQuant (Molecular Dynamics, Sunnyvale, CA).
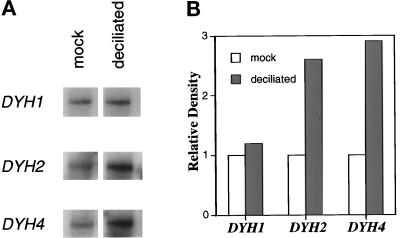
Figure 2.
Stimulation of DYH2 in response to deciliation. Total RNA was isolated from mock- and twice-deciliated wild-type cells. (a) The Northern blots were repeatedly probed, exposed to x-ray film, stripped, and then reprobed to obtain the data shown. The autoradiography signals were measured by densitometry. (b) The relative densities are plotted. The steady-state concentration of DYH2 RNA, but not that of DYH1, increased during reciliation. The positive control in this experiment was ciliary dynein β heavy chain DYH4 (accession number AF072878) that also increased in expression in response to deciliation.
The intron–exon organizations of the genes were determined by RNA-directed PCR. RT-PCR using primers whose sequences were derived from the genomic sequence was used to produce overlapping cDNAs spanning the catalytic domains of DYH1 and DYH2. Approximately 20 μg of total RNA from wild-type (B2086) cells was primed with 100 pmol of random hexamers and reverse-transcribed (Superscript II, Life Technologies, Gaithersburg, MD). Approximately 1 μg of the resulting cDNA was then used as the template for each PCR reaction. The amplified cDNAs were purified and sequenced.
Targeted Disruption of Macronuclear DYH1 and DYH2
The neomycin-resistance gene was inserted into the coding region of each dynein gene using the p4T2–1 disruption plasmid (the gift of Dr. Marty Gorovsky, University of Rochester, Rochester, NY) that contains the histone 4 promoter region, the neomcyin-resistance coding region, and the 3′-untranslated region of β tubulin. The DYH1 disruption construct was made by inserting the neo gene at the KpnI site, and the DYH2 disruption construct was made by deleting the 1.8-kb EcoRV-EcoRV fragment and replacing it with the neo gene (see Figure 3, a and b). Transformation was achieved by biolistic bombardment of mating cells (Cassidy-Hanley et al., 1997). Wild-type B2086 and CU428.1CSHL cells (from Dr. Peter Bruns, Cornell University, Ithaca, NY) were starved in 10 mM Tris-Cl, pH 7.5, before their mating. Ten to twelve hours after initiation of conjugation, the paired cells were bombarded with the disruption plasmids coated onto 1-μm gold particles (∼3 mg of DNA per 1 g of particles) using the PDS-1000/He Biolistic Particle Delivery System particle gun (Bio-Rad, Richmond, CA). After bombardment, the cells were recovered for 3 h in Neff medium (0.25% proteose peptone, 0.25% yeast extract, 0.5% dextrose, 0.03 mM FeCl3) and then plated into Neff medium supplemented with paromomycin at 0.1 mg/ml. Transformants were identified after 3 d and then replated in progressively higher concentrations of paromomycin. Chemicals were purchased from Sigma (St. Louis, MO).
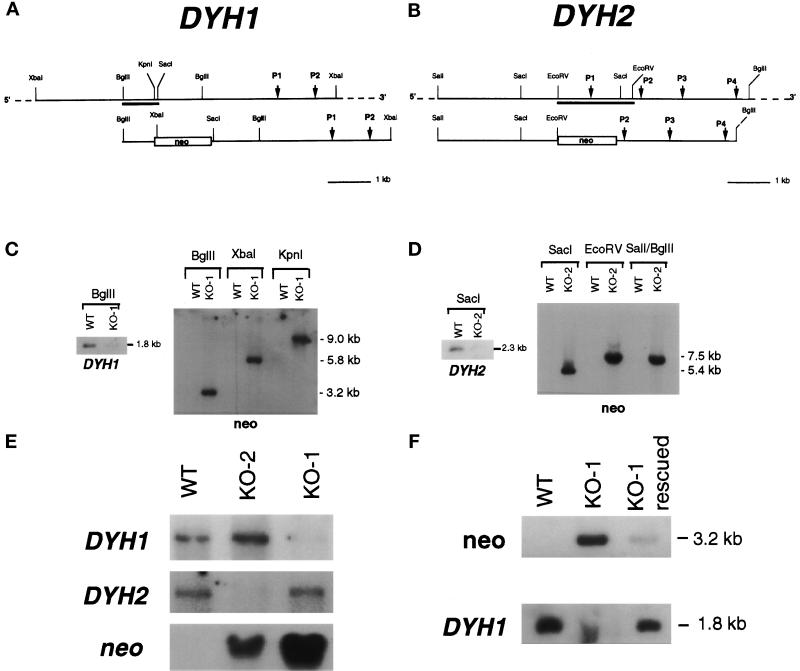
Figure 3.
Targeted disruptions of DYH1 and DYH2. (a and b) Diagrams of portions of the macronuclear wild-type (WT) genes (top) and the disruption constructs (bottom) for DYH1 (a) and DYH2 (b). The DYH1 disruption construct was made by inserting the neomycin-resistance gene at the chromosomal KpnI site. The DYH2 disruption construct was made by deleting the EcoRV-EcoRV fragment and replacing it with the neomycin-resistance gene. In each diagram, the positions of the genes encoding the P-loops (P1–P4) are indicated. The locations of the gene-specific probes used in the Southern and Northern blots are underlined. (c and d) Southern blots demonstrating the targeted disruptions of the DYH1 (c) and DYH2 (d) genes. In each panel, the blot on the left was probed with the gene-specific probe and showed the loss of the appropriately sized hybridizing fragment in the KO cell lines. The blots on the right of each panel were probed with the coding region of the neomycin-resistance gene and showed that the neo gene was inserted in the appropriate locations. In each case, the neo probe hybridized with a single band. (e) Northern blots of total RNAs obtained from wild-type (B2086), KO-2, and KO-1 cells. The ∼14.5-kb dynein heavy chain bands and the 1.4-kb neo bands were identified with gene-specific probes. The disruption of each dynein gene affected the expression of only the targeted gene. (f) Southern blots of DNAs from wild-type (B2086) and KO-1 cells probed with neo and DYH1. Genomic DNA digested with BglII was probed in this experiment. The paromomycin-selected KO-1 cells (lane labeled KO-1) possessed a large amount of the neo gene and very little of the wild-type version of DYH1. After growth for 3 d in the absence of paromomycin, the cells (lane labeled KO-1 rescued) possessed a high copy number of wild-type DYH1 and only a small amount of the neomycin-resistance gene. This experiment demonstrates that the KO-1 cells were incomplete knockouts of the DYH1 gene and that the copy number of the DYH1 gene could be manipulated by changing the selection pressure.
Evaluation of the Phenotypes by Microscopy
Phagocytosis.
Living cells were fed 2.16-μm fluorescent carboxylated polystyrene beads (Sigma, St. Louis, MO) by the use of the method described by Batz and Wunderlich (1976). After 1 h of incubation with the beads, the cells were washed, fixed in formaldehyde, and visualized by confocal fluorescence microscopy.
4′,6-Diamidino-2-phenylindole (DAPI) Staining.
Cells were fixed in 3.7% formaldehyde and stained with DAPI. The DAPI-stained cells were examined and photographed with an epi-illumination fluorescence microscope using a 50-W Hg lamp.
Indirect Immunofluorescence Microscopy.
Cells were permeabilized and fixed in microtubule-stabilizing buffer (Cole and Stuart, 1999). To visualize the mitotic apparatuses, we double-stained the cells with mouse monoclonal anti-tubulin antibodies (Asai et al., 1982) and with rabbit anti-phosphorylated histone antibody (Upstate Biotechnology, Lake Placid, NY), which stains only condensed chromatin (Lu et al., 1994). To visualize the surface cilia, we stained the cells with monoclonal anti-tubulin antibodies. The antibody-stained cells were viewed by confocal laser fluorescence microscopy using a Bio-Rad MRC1024 instrument equipped with a Kr/Ar laser.
Surface Areas.
Cells were fixed in 2.5% glutaraldehyde in 100 mM NaCacodylate, pH 7.3. The cells were photographed using differential interference contrast optics, and the images were captured using a Dage (Michigan City, IN) Nuvicon camera and Scion (Frederick, MD) frame-grabber. Measurements were made using the particle analysis program in National Institutes of Health Image 1.61 (http://rsb.info.nih.gov/nih-image/).
Scanning Electron Microscopy.
Cells were fixed in 2.5% glutaraldehyde and 1% OsO4 in 100 mM NaCacodylate, pH 7.3, for 10 min, washed in water, stained with 1% uranyl acetate for 1 h, and critical point dried.
Reciliation Experiments
Cells were deciliated by a modification of standard methods (Rosenbaum and Carlson, 1969). Exponentially growing cultures of cells were harvested by centrifugation (1000 × g, 2 min, room temperature) in oil tubes. The cell pellet was resuspended in two volumes of ice-cold buffer A (10 mM EDTA, 50 mM sodium acetate, pH 6.0) and transferred to a sterile 50-ml tube containing 10 ml of ice-cold sterile water and 0.5 ml of 0.2 M CaCl2. The cell suspension was gently agitated by end-over-end mixing for 45 s. The deciliated cells were then transferred to 600 ml of fresh Neff medium and allowed to reciliate. In a Northern blot experiment (see Figure 2), the cells were allowed to reciliate for 2 h and then deciliated again. Total RNA was isolated from the cells after 45 min of recovery from the second deciliation. Total RNA was also isolated from mock-deciliated cells that were subjected to the same treatments except that the calcium was omitted from the deciliation steps. The deciliation and reciliation of the cells were confirmed by observing the cells by bright-field microscopy.
RESULTS
Characterization of Tetrahymena DYH1 and DYH2
Short fragments of several Tetrahymena dynein heavy chain genes were obtained by the RT-PCR method using degenerate oligonucleotide primers that correspond to conserved sequences. Comparison of these sequences with those of dynein isoforms from other organisms identified the Tetrahymena genes encoding Dyh1 and Dyh2. The RT-PCR fragments were used to screen a Tetrahymena macronuclear genomic library that yielded single lambda clones containing large portions of DYH1 and DYH2. The DYH1 clone is ∼17 kb in length, begins near the 5′ end of the coding region, and extends past the 3′ end of the gene. The DYH2 clone is ∼15 kb in length, includes the 5′ end of the coding region, but lacks 3–4 kb of the 3′ end of the gene. The lambda inserts were mapped with restriction enzymes, and the central motor domains, which include the four P-loops, were sequenced. These sequences have been deposited in the database (GenBank accession numbers AF025312 and AF025313). To identify unambiguously the open-reading frames, we sequenced overlapping cDNAs obtained by RNA-directed PCR. The comparisons between the genomic sequences and the cDNA sequences identified several introns in the central regions encoding the catalytic domains of both dynein genes (see GenBank deposits). DYH1 has 10 introns of 50–64 nucleotides (average length, 56); DYH2 has 12 introns of 50–108 nucleotides (average length, 75). The intron–exon organizations of the two genes are distinct.
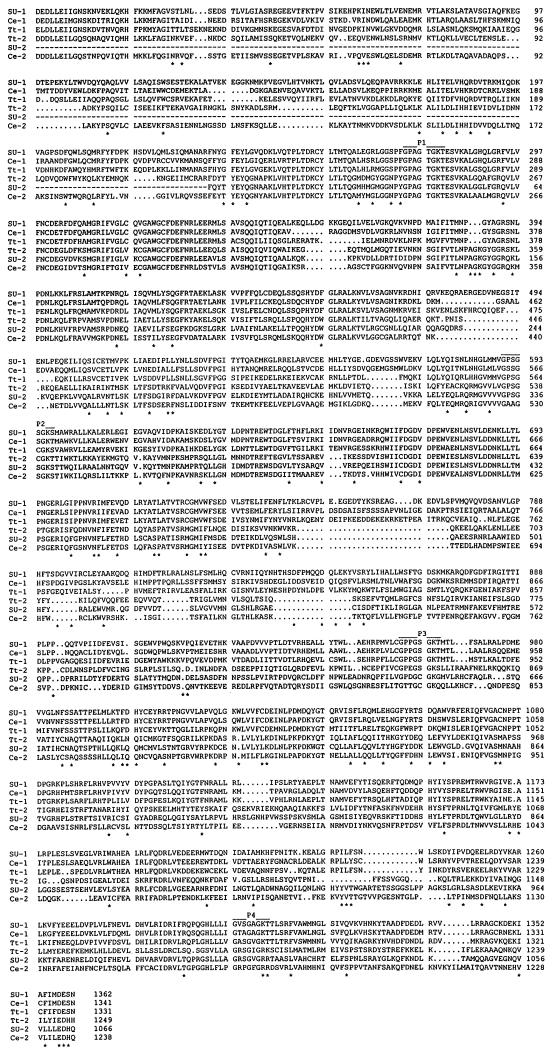
There are three organisms from which substantial sequence information for both Dyh1 and Dyh2 has been determined: sea urchin (Gibbons et al., 1992, 1994), nematode (Wilson et al., 1994; Lye et al., 1995), and Tetrahymena. The aligned catalytic domain sequences of the six dyneins are shown in Figure 1. An asterisk marks a position at which there is a conserved amino acid in the three Dyh1 sequences and a different conserved amino acid in the three Dyh2 sequences. The three Dyh1 sequences are more similar to one another than to any Dyh2 sequence, and the three Dyh2 sequences are more similar to one another than to any Dyh1 sequence.
Figure 1.
Alignment of catalytic domain sequences of Dyh1 and Dyh2. The deduced amino acid sequences of Dyh1 and Dyh2 from Tripneustes gratilla (SU-1 and SU-2; accession numbers Z21941 and U03969), C. elegans (Ce-1 and Ce-2; accession numbers L33260 and Z75536), and Tetrahymena thermophila (Tt-1 and Tt-2; accession numbers AF025312 and AF025313) were aligned by the PILEUP program (GCG, University of Wisconsin). The four P-loops are identified. Each position (*) at which there is a conserved residue in the three Dyh1 sequences that is different from a conserved residue in the three Dyh2 sequences is identified.
Similar to what was found in sea urchin embryos (Gibbons et al., 1994) and rat tracheal epithelial cells (Criswell et al., 1996), DYH2 expression increased during ciliogenesis in Tetrahymena. Total RNA was obtained from mock- and twice-deciliated wild-type (B2086) cells. Northern blots revealed that the steady-state concentration of DYH2 RNA, but not of DYH1 RNA, was increased in response to deciliation. The results of one such experiment are shown in Figure 2. To circumvent potential variations caused by uneven loading of the gel, we repeatedly probed and stripped the same blot. As a positive control, the blot was also probed with a cDNA fragment of ciliary dynein β heavy chain (the DYH4 gene). Deciliation interrupts the growth of the cells, and a gene whose expression is reliably unaltered is unavailable. Thus, this experiment provides a comparison of the changes after deciliation in DYH1 and DYH2 expression but does not measure absolute levels of expression.
KO-1 and KO-2 Are Targeted Knockouts of DYH1 and DYH2
The two cytoplasmic dynein genes were separately disrupted by inserting the neomycin-resistance gene into the coding region, which resulted in two gene knockout cell lines referred to as KO-1 and KO-2 (see Figure 3, a and b). Southern blots using dynein-specific probes showed the loss of the appropriately sized fragment in the KO cell lines, and the neo probe, which hybridized with only a single band in each case, demonstrated that the neomycin-resistance gene was inserted only in the targeted genes (Figure 3, c and d). Northern blots showed the decreased expression of the targeted gene, but not of the untargeted gene, in each transformed cell line (Figure 3e). On the basis of the Southern and Northern analyses, the wild-type version of DYH2 appeared to be completely eliminated in the KO-2 transformants. However, the wild-type version of DYH1 could not be completely eliminated in the KO-1 transformants even after prolonged selection in paromomycin; thus, KO-1 is an incomplete knockout. Culturing the KO-1 cells without paromomycin for 3 d resulted in an increase in the copy number of the wild-type version of DYH1 (Figure 3f). Therefore, it was possible to modulate the level of DYH1 by adjusting the selection pressure, and this modulation was exploited in the following experiments.
Dyh1, but Not Dyh2, Is Required for Phagocytic Activity in Tetrahymena
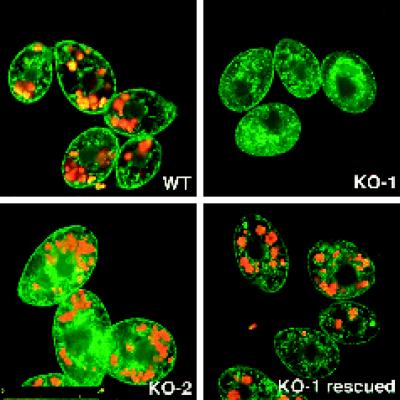
Normal feeding behavior in Tetrahymena begins at the oral apparatus where food particles are phagocytosed. The engulfed material is then transported along the length of the cell, and the unused material is eventually expelled at the cytoproct located at the posterior end. This process requires ∼2 h and can be easily visualized by feeding the cells ∼2-μm fluorescent latex beads (Batz and Wunderlich, 1976). Using fluorescent beads, we performed a simple phagocytosis assay, and the results are shown in Figure 4. Cells were allowed to “feed” on the beads for 1 h and then washed, fixed, and counterstained with the nonspecific membrane dye 1,1′-dioctadecyl-3,3,3′,3′-tetramethyloxycarbocyanine perchlorate (DiOC6, Sigma). The cells were then examined by confocal fluorescence microscopy. The KO-1 cells, but not the KO-2 cells, were mostly unable to take up the beads. The defect in phagocytosis exhibited by the KO-1 cells could be reversed by first growing the KO-1 cells for 3 d in no paromomycin, a treatment that restores the wild-type copy number of the DYH1 gene (see Figure 3f, above). The inability of the drug-selected KO-1 cells to undergo phagocytosis was not simply a side effect of the paromomycin because the KO-2 cells, which continued to phagocytose the beads, were grown in the same drug concentration as were the KO-1 cells (paromomycin at 10 mg/ml). Thus, Dyh1 is required for normal phagocytosis activity.
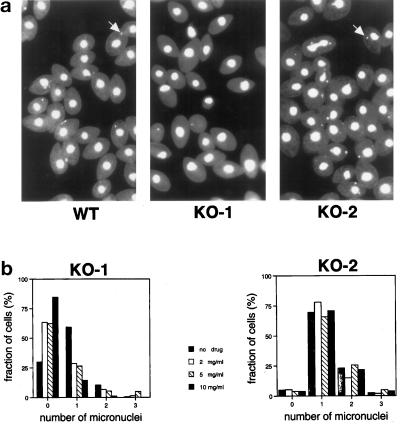
Figure 4.
Dyh1 is required for phagocytosis. Cells were incubated for 1 h with fluorescent latex beads, washed, fixed with formaldehyde, and counterstained with DiOC6. In these confocal fluorescence micrographs, the beads are red, and the membranes stained with DiOC6 are green. The KO-1 cells were unable to phagocytose the beads. However, KO-1 cells grown for 3 d in the absence of paromomycin (KO-1 rescued) possessed phagocytic activity. All photographs are at the same magnification (bar, in micrometers, in lower left corner of KO–2). Note the larger size of the KO-2 cells.
Dyh1, but Not Dyh2, Is Required for Chromosome Segregation during Micronuclear Mitosis
Cultures of vegetatively growing cells were fixed and stained with DAPI to assess their nuclear phenotypes. When the KO-1 cells were grown in the presence of a high concentration of drug (paromomycin at 10 mg/ml), many of the cells lacked a distinct micronucleus (Figure 5a). It is possible that these cells retained a fragment of the micronucleus, which was obscured by the brightly stained macronucleus. In contrast, the KO-2 cells grown under the same selection conditions possessed a normal-appearing micronucleus, thus demonstrating that the micronuclear effect in the KO-1 cells was not a side effect of the paromomycin treatment. The relationship between DYH1 and the micronuclear phenotype was further explored by culturing the cells in different concentrations of paromomycin, staining the cells with DAPI, and scoring the cells for the presence of a distinct micronucleus (Figure 5b). In this experiment, samples were coded and then scored (n > 200 for each treatment) by a student who did not know the code. Increasing the paromomycin concentration resulted in a reduction in the proportion of KO-1 cells with a distinct micronucleus. In contrast, the same treatments had no effect on the presence or appearance of the micronuclei in KO-2 cells. Wild-type cells are sensitive to paromomycin and so would not be a particularly meaningful control in this experiment. However, wild-type cells grown with no drug, fixed, and stained with DAPI revealed numbers of micronuclei per cell similar to what was found with KO-2 cells. Because the copy number of the wild-type version of DYH1 could be manipulated by adjusting the drug concentration (see Figure 3f), this result demonstrates that Dyh1 is required for the maintenance of a normal micronucleus during vegetative growth.
Figure 5.
Many KO-1 cells lack a distinct micronucleus. (a) Fields of cells stained with DAPI and viewed by fluorescence microscopy. Most of the wild-type (B2086) and KO-2 cells contained a micronucleus (examples indicated with arrows), but most of the KO-1 cells did not. The KO-1 and KO-2 cells were grown for several weeks in paromomycin at 10 mg/ml before this experiment. (b) Effects of paromomycin concentration on the number of visible micronuclei per cell in KO-1 and KO-2. Cells were grown in paromomycin at 10 mg/ml and then transferred to medium containing different concentrations of the drug (0–10 mg/ml). After 3 d of vegetative growth, the cells were fixed and stained with DAPI. Approximately 200 cells at each drug concentration were viewed by fluorescence microscopy in a blind protocol. The number of distinct micronuclei in the KO-1 cells was affected by the paromomycin. At high concentrations of the drug, most of the KO-1 cells lacked a visible micronucleus, whereas in the absence of drug, most of the KO-1 cells had one micronucleus. The different paromomycin concentrations did not affect the micronuclear phenotype of the KO-2 cells, demonstrating that the effect in the KO-1 cells was not simply attributable to the drug. The KO-2 cells with two micronuclei included dividing cells in these nonsynchronized cultures.
The apparent loss of the micronucleus in KO-1 cells during vegetative growth suggested that these cells were defective in some aspect of micronuclear mitosis. Normal cell division in Tetrahymena occurs in three discrete steps and is illustrated in Figure 6, top: 1) The condensed micronuclear chromosomes undergo an early anaphase A in which the two sets of daughter chromosomes are drawn to opposite ends of the micronucleus, 2) this is followed by an extensive anaphase B in which the two sets of chromosomes are separated ∼30 μm, and 3) after anaphase B, the cell undergoes cytokinesis that results in one daughter micronucleus in each cell and the pinching apart of the (amitotic) macronucleus.
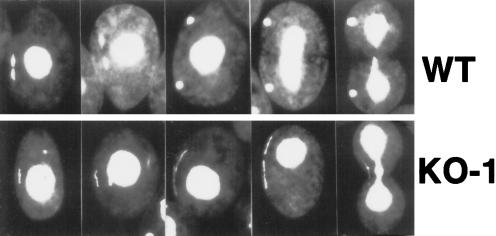
Figure 6.
Dyh1 is required for normal micronuclear chromosome distribution during mitosis. Wild-type and KO-1 cells were fixed with formaldehyde, stained with DAPI, and viewed by fluorescence microscopy. To capture KO-1 cells with micronuclei, we grew the cells for 3 d in the absence of paromomycin, returned the cells to drug at 10 mg/ml for 1 d, and then fixed and stained the cells. A gallery of cells is shown in this figure; in each photograph, the dividing micronucleus is at the viewer’s left. Top, wild-type micronuclear mitosis. Bottom, KO-1 micronuclear mitosis. In the wild-type cells, the two sets of chromosomes separated very early in mitosis. In the KO-1 cells, the chromosomes never segregated to opposite poles. Instead, chromosomal DNA was spread through the micronucleus as mitosis proceeded. This resulted in one or more thread-like DAPI-stained structures.
To determine how the micronuclei were lost in KO-1 cells, we grew the cells for 3 d in the absence of selection drug to restore to the culture a significant number of cells with micronuclei and then returned the cells to paromomycin at 10 mg/ml and grew them for another day in the drug to deplete them of wild-type DYH1 gene. The cells were then fixed and stained with DAPI. Most of the KO-1 cells examined lacked distinctly stained micronuclei. However, the few KO-1 cells with a micronucleus and fixed during cell division displayed an aberrant micronuclear mitosis. In contrast to what was observed in wild-type cells, the KO-1 cells failed to segregate their chromosomes to opposite poles (Figure 6, bottom). Instead, the condensed chromosomes appeared to fill the length of the micronucleus, resulting in thread-like DAPI-stained material as the micronuclei elongated. Occasionally more than one thread-like micronucleus was observed in the same cell, suggesting that the cell had more than one micronucleus (or more than one micronuclear fragment). Wild-type cells also were occasionally multimicronucleated.
To confirm the continued presence of spindle microtubules in the dividing KO-1 cells, we fixed and double-stained the cells with monoclonal mouse anti-tubulin antibodies and rabbit anti-phosphorylated histone antibody that reacts only with condensed chromatin (Lu et al., 1994). The micronuclei of the dividing KO-1 cells possessed what appeared to be normal spindles except that the chromosomes were intertwined with the microtubules throughout the micronucleus instead of being segregated at the poles (Figure 7).
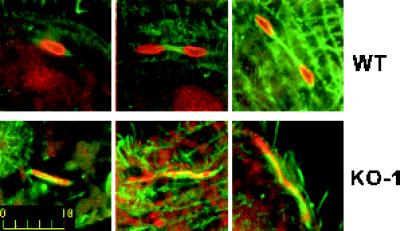
Figure 7.
Double immunofluorescence microscopy of micronuclear mitotic apparatuses. Cells were permeabilized, fixed, and stained with anti-tubulin and anti-phosphorylated histone antibodies. The cells were viewed by confocal scanning laser fluorescence microscopy. The microtubules are green, the chromatin is red, and yellow is the superposition of the two stains. Top, wild-type micronuclear mitosis. Bottom, KO-1 micronuclear mitosis. Bar, 10 μm.
Dyh2 Contributes to the Maintenance of the Cortical Cytoskeleton
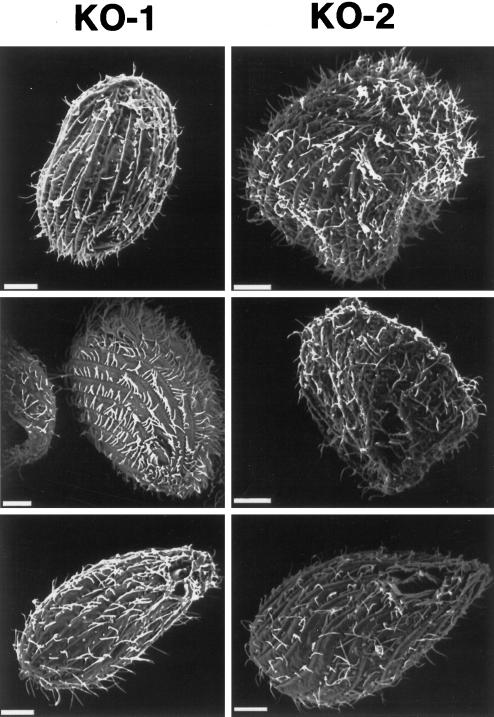
In contrast to the KO-1 cells, the KO-2 transformants underwent normal phagocytosis and micronuclear mitosis. However, the KO-2 cells were often aberrant in size and shape. Unlike wild-type and KO-1 cells, the KO-2 cells varied widely in size, and many of the KO-2 cells were significantly larger than the wild-type and KO-1 cells (Figure 8). Scanning electron microscopy revealed that many of the KO-2 cells were misshapen (Figure 9).
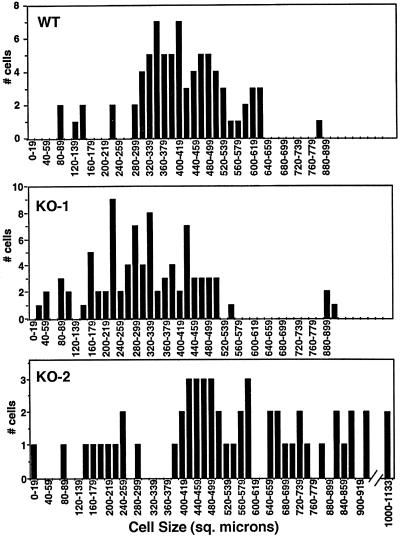
Figure 8.
Dyh2 contributes to cell size. Surface areas of wild-type (B2086), KO-1, and KO-2 cells were measured from cells photographed using differential interference contrast microscopy. The distributions by surface area for each cell type are shown in the graphs. The KO-2 cells were found to have a wide range of sizes, with many cells being significantly larger than wild-type and KO-1 cells. The slightly smaller size of the KO-1 cells relative to wild-type cells may be related to the involvement of Dyh1 in phagocytosis.
Figure 9.
Dyh2 contributes to cell shape. A gallery of KO-1 and KO-2 cells viewed by scanning electron microscopy are shown. Many KO-2 cells were misshapen. Bars, 6 μm.
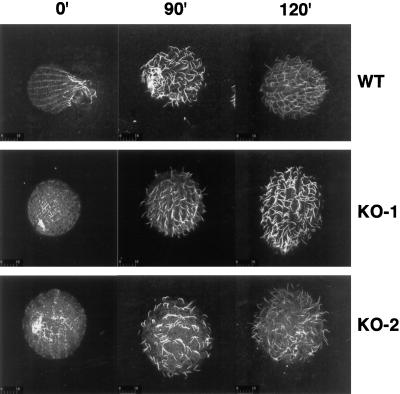
The abnormal shapes of many of the KO-2 cells suggested possible defects in the integrity of the cortical cytoskeleton. Because the cortical cytoskeleton is responsible for the positioning of the ciliary basal bodies, we reasoned that the pattern of surface cilia would reflect the organization of the underlying cortical microtubules. Wild-type, KO-1, and KO-2 cells were deciliated with a calcium shock and then returned to fresh media and allowed to reciliate. All three cell lines regrew cilia at approximately the same rate. However, unlike the normal pattern of cilia on the wild-type and KO-1 cells, the cilia on the KO-2 cells often emerged randomly and not in the normal rows (Figure 10). This result suggests that the KO-2 cortical microtubules were disrupted during the calcium deciliation step and did not reform correctly during the time course of this experiment.
Figure 10.
Disruption of surface ciliary pattern in reciliated KO-2 cells. Wild-type (B2086), KO-1, and KO-2 cells were deciliated with calcium and then returned to Neff medium. At the indicated times after deciliation, the cells were fixed and stained with anti-tubulin antibodies and then viewed by confocal laser scanning fluorescence microscopy. The wild-type and KO-1 cells regrew their cilia in rows corresponding to the longitudinal cortical microtubules. In contrast, the KO-2 cells regrew their cilia in a disorganized manner.
DISCUSSION
In this study, we have taken advantage of the nuclear dimorphism and targeted replacement of somatic genes in Tetrahymena thermophila to dissect the roles of two cytoplasmic dyneins in the same cell. The disruption of DYH1 resulted in a loss of phagocytic activity and a mitotic defect in which the chromosomes failed to segregate during anaphase A. In contrast, the disruption of DYH2 affected the ability of the cell to maintain the cortical microtubule cytoskeleton. We conclude that the two dyneins perform distinct functions in Tetrahymena.
Heretofore, most of the genetics of cytoplasmic dynein has focused on the ubiquitous cytoplasmic Dyh1. In organisms with only one dynein gene, including budding yeast and filamentous fungi, disruption of Dyh1 produces defects in mitotic spindle positioning and postmitotic nuclear movements, but the cells remain viable (Eshel et al., 1993; Li et al., 1993; Plamann et al., 1994; Xiang et al., 1994; Inoue et al., 1998). In contrast, in organisms with multiple dyneins–Drosophila (Gepner et al., 1996), mouse (Harada et al., 1998), and now Tetrahymena–Dyh1 appears to be required for normal growth.
Dyh1 and Phagocytosis
Our finding that KO-1 cells were defective in the phagocytosis of fluorescent beads is consistent with previous results obtained in other systems in which cytoplasmic dynein has been implicated in the retrograde trafficking of membrane-bounded organelles and vesicles (e.g., Schroer et al., 1989; Oda et al., 1995; Muresan et al., 1996). If the KO-1 cells were able to complete phagocytosis but were then blocked in the subsequent transport of the vesicles to the cytoproct, we would expect to find an accumulation of fluorescent beads at the anterior end of the cells, but this was not observed. Thus, our results demonstrate a requirement for Dyh1 in the engulfment of the beads but do not reveal whether dynein is also involved in vesicle transport. Because Tetrahymena can take up nutrients by alternate mechanisms (Rasmussen and Orias, 1975), it is not clear whether the effect on phagocytosis is the reason DYH1 is apparently an essential gene. The lack of phagocytic activity was not simply caused by a loss of the oral apparatus, which can occur when the micronucleus is completely eliminated (Haremaki et al., 1996), because anti-tubulin immunofluorescence microscopy revealed the presence of an oral apparatus in the KO-1 and KO-2 cells.
Dyh1 and Chromosome Distribution
During micronuclear anaphase A in wild-type cells, the two sets of chromosomes are drawn to opposite ends of the micronucleus by kinetochore microtubules (LaFountain and Davidson, 1979, 1980). After chromosome segregation, the micronucleus travels from the center of the cell to near the cortex where it undergoes a dramatic elongation, growing from a length of 1–2 μm to >30 μm. Micronuclear elongation is driven by the extension and rearrangement of the intranuclear spindle microtubules that are located between the two sets of chromosomes and by the stretching of the micronucleus surface along longitudinally oriented cortical microtubules (Jaeckel-Williams, 1978). In our experiments, the disruption of DYH1 blocked chromosome distribution but did not affect micronuclear elongation, and the disruption of DYH2 had no discernible effect on any aspect of mitosis. Thus, in Tetrahymena, dynein appears not to be involved in the migration of the dividing micronucleus from the center of the cell to near the surface, in the extension of the mitotic spindle, or in the stretching of the micronucleus along cortical microtubules.
In the dividing KO-1 cells, the micronuclear chromosomes failed to segregate, implying that Dyh1 is required for the effective attachment of the chromosomes to the kinetochore microtubules or for normal chromosome movement to the opposite poles after kinetochore attachment. Experiments in other organisms have shown that dynein activity is associated with the kinetochore (Pfarr et al., 1990; Rieder and Alexander, 1990; Steuer et al., 1990; Echeverri et al., 1996). We are currently developing reagents with which to visualize the Dyh1 protein in Tetrahymena cells; the location of dynein relative to the mitotic apparatus will be important in understanding the mechanism by which Dyh1 participates in anaphase A.
Because the micronuclear genome is not transcribed, the inaccurate segregation of chromosomes during micronuclear mitosis would not be expected to affect the phenotype of the daughter cells, and micronuclear aneuploidy would be detected only after mating when a new macronucleus is formed. Such a cell might be analogous to nullisomic or “star” strains that lack substantial portions of the micronuclear genome (Bruns et al., 1983). However, in experiments in which the entire micronucleus is eliminated at one time, the progeny do not survive (Haremaki et al., 1996). Thus, a culture of KO-1 cells is likely to be unstable because the micronuclei are randomly fragmented at every cell division; if too much of the micronucleus is lost, then the cell may not survive. The KO-1 cells illustrate the potential of Tetrahymena as a model system with which to dissect mitotic mechanisms: 1) anaphase A and B are temporally well resolved, 2) mutations can be generated by targeted gene disruptions, and 3) mutations affecting mitosis do not immediately affect the viability of the cell.
Dyh2 and the Maintenance of the Cytoskeleton
The disruption of DYH2 produced an intriguing phenotype, quite distinct from the effects of disrupting DYH1. The KO-2 cells were defective in the maintenance of their cortical cytoskeletons. This apparent loss of regulation led to wide variations in cell size and shape. One interpretation of these results is that Dyh2 participates in the repair and remodeling of the Tetrahymena cytoskeleton, which must be reformed at each cell division (every 2.5 h at 30°C). The putative repair function of Dyh2 was most evident in the reciliation experiment. The cells were deciliated with a brief exposure to a high concentration of calcium ion, a treatment that disrupted the cortical microtubules that arrange the ciliary basal bodies or kinetids. The pattern of reciliation revealed that the underlying cytoskeleton remained disorganized in the KO-2 cells. An important future problem will be to determine what cargoes Dyh2 carries to perform this task. The result in Tetrahymena is reminiscent of findings in Dictyostelium discoideum and Saccharomyces cerevisiae in which overexpression of the dynein globular head domain affected the microtubule cytoskeleton and cell polarity (Koonce and Samso, 1996; Shaw et al., 1997). Slime mold and yeast have only one dynein, Dyh1, whereas Tetrahymena uses a separate dynein, Dyh2, to organize the cytoskeleton, thus underscoring the division of labor achieved with multiple dynein isoforms.
In nonciliated fibroblasts, Dyh2 was shown to participate in the organization of the Golgi apparatus (Vaisberg et al., 1996). In Tetrahymena thermophila, the Golgi apparatus is located at and organized by the cortical cytoskeleton (Kurz and Tiedtke, 1993). If Dyh2 is important in maintaining the cortical cytoskeleton, then the absence of Dyh2 might have an indirect effect on Golgi function. However, we have observed no noticeable defect in regulated exocytosis (Chilcoat et al., 1996) in the KO-2 cells.
In sea urchin embryos, rat tracheal epithelial cells, and Tetrahymena, the steady-state concentration of transcripts encoding Dyh2 increased during cilia formation. This implies that Dyh2 is either a component of the cilia or is involved in ciliogenesis, perhaps carrying ciliary precursors to the site of their incorporation. We therefore expected that the elimination of DYH2 expression would have an effect on cilia formation in Tetrahymena. However, the KO-2 cells were able to regenerate their cilia with approximately normal kinetics, and no gross abnormality in the KO-2 ciliary axonemes was detected by electron microscopic analysis. This result is particularly intriguing because of the recent reports that disruption of the gene encoding Dyh2 in Chlamydomonas results in abnormal flagella (Pazour et al., 1999; Porter et al., 1999). If Dyh2 is used for cytoskeletal remodeling as we suggest, its impact may be observed in different places depending on the organism. In Tetrahymena, most of the cilia are terminal organelles that do not shorten or elongate, and remodeling occurs mostly in the cortical cytoskeleton. However, in Chlamydomonas, remodeling occurs in the flagella, which must be able to change in length constantly in response to environmental cues and for cell division (see Tuxhorn et al., 1998).
Complexity of Dynein Functions: Genes and Environment
The techniques of modern molecular biology have provided the means to identify rapidly the dynein heavy chain genes in several model organisms. Although we do not yet have a complete understanding of the structure–function relationships of dynein heavy chains in organisms with multiple dyneins, it is reasonable to expect that there is functional specialization among the dynein isoforms, as has been documented for the kinesin family of microtubule motors (reviewed in Cole and Scholey, 1995; Vale and Fletterick, 1997). The genetic dissection of the several axonemal dyneins demonstrates their individual specializations (reviewed in Piperno, 1990; Asai and Brokaw, 1993; Brokaw, 1994), and isoform-specific antibodies suggest separate tasks for three different cytoplasmic dyneins in vertebrate cells (Vaisberg et al., 1996). The present study extends this idea by providing a clear example of the division of labor between two cytoplasmic dyneins within the same cell.
Although there is specialization among dynein isoforms within a single cell, it is appropriate to consider cellular context as a second important element in the determination of dynein function. A particular dynein isoform may do different things in different contexts. For example, Dyh2 appears to perform distinct tasks in unciliated fibroblasts versus ciliated epithelial cells and in Tetrahymena versus Chlamydomonas. Another striking example is the apparent involvement of dynein β heavy chain in the establishment of visceral handedness at a time when the embryonic cells may not be ciliated (Supp et al., 1997). The contextual regulation of dynein function suggests that it is not simply the heavy chain isoform sequence that determines function. Rather, what a dynein does in a particular cell may also be controlled by other factors, including interaction with specific dynein subunits and dynein accessory proteins, and by posttranslational modifications of dynein subunits.
ACKNOWLEDGMENTS
We are grateful for the helpful advice of many persons, including Kerry Bloom, Peter Bruns, Jim Forney, Joe Frankel, Marty Gorovsky, Mike Koonce, Mary Porter, Conly Rieder, Aaron Turkewitz, and Norm Williams. David Wilkes and Jifan Chen began the Tetrahymena RNA-directed PCR analysis, Dana Ahn helped with the experiment summarized in Figure 5, and Sunkyung Lee contributed to the results shown in Figure 10. This work was supported by research grants from the National Institutes of Health (to W.L.D.) and the American Cancer Society and National Science Foundation (to D.J.A.).
REFERENCES
- Allen SL, Altschuler MA, Bruns PJ, Cohen J, Doerder FP, Gaertig J, Gorovsky M, Orias E, Turkewitz A. Proposed genetic nomenclature rules for Tetrahymena thermophila, Paramecium primaurelia and Paramecium tetraurelia. Genetics. 1998;149:459–462. doi: 10.1093/genetics/149.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aniento F, Emans N, Griffiths G, Gruenberg J. Cytoplasmic dynein-dependent vesicular transport from early to late endosomes. J Cell Biol. 1993;123:1373–1387. doi: 10.1083/jcb.123.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai DJ. Functional and molecular diversity of dynein heavy chains. Semin Cell Dev Biol. 1996;7:311–320. [Google Scholar]
- Asai DJ, Brokaw CJ. Dynein heavy chain isoforms and axonemal motility. Trends Cell Biol. 1993;3:398–402. doi: 10.1016/0962-8924(93)90090-n. [DOI] [PubMed] [Google Scholar]
- Asai DJ, Brokaw CJ, Harmon RC, Wilson L. Monoclonal antibodies to tubulin and their effects on the movement of reactivated sea urchin spermatozoa. Cell Motil. 1982;1:175–180. doi: 10.1002/cm.970020733. [DOI] [PubMed] [Google Scholar]
- Asai DJ, Criswell PS. Identification of new dynein heavy chain genes by RNA-directed PCR. Methods Cell Biol. 1995;47:579–585. doi: 10.1016/s0091-679x(08)60863-8. [DOI] [PubMed] [Google Scholar]
- Batz W, Wunderlich F. Structural transformation of the phagosomal membrane in Tetrahymena cells endocytosing latex beads. Arch Microbiol. 1976;109:215–220. doi: 10.1007/BF00446631. [DOI] [PubMed] [Google Scholar]
- Brokaw CJ. Control of flagellar bending: a new agenda based on dynein diversity. Cell Motil Cytoskeleton. 1994;28:199–204. doi: 10.1002/cm.970280303. [DOI] [PubMed] [Google Scholar]
- Bruns PJ, Brussard TB, Merriam EV. Nullisomic Tetrahymena. II. A set of nullisomics define the germinal chromosomes. Genetics. 1983;104:257–270. doi: 10.1093/genetics/104.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy-Hanley D, Bowen J, Lee JH, Cole E, VerPlank LA, Gaertig J, Gorovsky MA, Bruns PJ. Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics. 1997;146:135–147. doi: 10.1093/genetics/146.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilcoat ND, Melia SM, Haddad A, Turkewitz AP. Granule lattice protein 1 (Grl1p), an acidic, calcium-binding protein in Tetrahymena thermophila dense-core secretory granules, influences granule size, shape, content organization, and release but not protein sorting or condensation. J Cell Biol. 1996;135:1775–1787. doi: 10.1083/jcb.135.6.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Cleveland DW, Hoffman PN. Slow axonal transport models come full circle: evidence that microtubule sliding mediates axon elongation and tubulin transport. Cell. 1991;67:453–456. doi: 10.1016/0092-8674(91)90518-4. [DOI] [PubMed] [Google Scholar]
- Cole DG, Scholey JM. Structural variations among the kinesins. Trends Cell Biol. 1995;5:259–262. doi: 10.1016/s0962-8924(00)89028-5. [DOI] [PubMed] [Google Scholar]
- Cole ES, Stuart KR. Nuclear and cytoskeletal fluorescence microscopy techniques. In: Asai DJ, Forney JD, editors. Methods in Cell Biology: Tetrahymena thermophila. San Diego: Academic Press; 1999. (in press). [DOI] [PubMed] [Google Scholar]
- Corthesy-Theulaz I, Pauloin A, Pfeffer SR. Cytoplasmic dynein participates in the centrosomal localization of the Golgi complex. J Cell Biol. 1992;118:1333–1345. doi: 10.1083/jcb.118.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criswell PS, Asai DJ. Evidence for four cytoplasmic dynein heavy chain isoforms in rat testis. Mol Biol Cell. 1998;9:237–247. doi: 10.1091/mbc.9.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criswell PS, Ostrowski LE, Asai DJ. A novel cytoplasmic dynein heavy chain: expression of DHC1b in mammalian ciliated epithelial cells. J Cell Sci. 1996;109:1891–1898. doi: 10.1242/jcs.109.7.1891. [DOI] [PubMed] [Google Scholar]
- Dillman JF, III, Dabney LP, Pfister KK. Cytoplasmic dynein is associated with slow axonal transport. Proc Natl Acad Sci USA. 1996;93:141–144. doi: 10.1073/pnas.93.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri CJ, Paschal BM, Vaughan KT, Vallee RB. Molecular characterization of the 50-kDa subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J Cell Biol. 1996;132:617–633. doi: 10.1083/jcb.132.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshel D, Urrestarazu LA, Vissers S, Jauniaux J-C, van Vliet-Reedijk JC, Planta RJ, Gibbons IR. Cytoplasmic dynein is required for normal nuclear segregation in yeast. Proc Natl Acad Sci USA. 1993;90:11172–11176. doi: 10.1073/pnas.90.23.11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fath KR, Trimbur GM, Burgess DR. Molecular motors are differentially distributed on Golgi membranes from polarized epithelial cells. J Cell Biol. 1994;126:661–675. doi: 10.1083/jcb.126.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel J. Cell biology of Tetrahymena thermophila. In: Asai DJ, Forney JD, editors. Methods in Cell Biology: Tetrahymena thermophila. San Diego: Academic Press; 1999. (in press). [Google Scholar]
- Gaertig J, Gorovsky M. Efficient mass transformation of Tetrahymena thermophila by electroporation of conjugants. Proc Natl Acad Sci USA. 1992;89:9196–9200. doi: 10.1073/pnas.89.19.9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gepner J, Li M-g, Ludmann S, Kortas C, Boylan K, Iyadurai SJP, McGrail M, Hays TS. Cytoplasmic dynein function is essential in Drosophila melanogaster. Genetics. 1996;142:865–878. doi: 10.1093/genetics/142.3.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons BH, Asai DJ, Tang W-JY, Hays TS, Gibbons IR. Phylogeny and expression of axonemal and cytoplasmic dynein genes in sea urchins. Mol Biol Cell. 1994;5:57–70. doi: 10.1091/mbc.5.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons IR. Dynein family of motor proteins: present status and future questions. Cell Motil Cytoskeleton. 1995;32:136–144. doi: 10.1002/cm.970320214. [DOI] [PubMed] [Google Scholar]
- Gibbons IR, Asai DJ, Tang W-JY, Gibbons BH. A cytoplasmic dynein heavy chain in sea urchin embryos. Biol Cell. 1992;76:303–309. doi: 10.1016/0248-4900(92)90432-z. [DOI] [PubMed] [Google Scholar]
- Harada A, Takei Y, Kanai Y, Tanaka Y, Nonaka S, Hirokawa N. Golgi vesiculation and lysosome dispersion in cells lacking cytoplasmic dynein. J Cell Biol. 1998;141:51–59. doi: 10.1083/jcb.141.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haremaki T, Sugai T, Takahashi M. Involvement of active cellular mechanisms on the disorganization of oral apparatus in amicronucleate cells in Tetrahymena thermophila. Cell Struct Funct. 1996;21:73–80. doi: 10.1247/csf.21.73. [DOI] [PubMed] [Google Scholar]
- Holleran EA, Tokito MK, Karki S, Holzbaur ELF. Centractin (ARP1) associates with spectrin revealing a potential mechanism to link dynactin to intracellular organelles. J Cell Biol. 1996;135:1815–1829. doi: 10.1083/jcb.135.6.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S, Turgeon BG, Yoder OC, Aist JR. Role of fungal dynein in hyphal growth, microtubule organization, spindle pole body motility and nuclear migration. J Cell Sci. 1998;111:1555–1566. doi: 10.1242/jcs.111.11.1555. [DOI] [PubMed] [Google Scholar]
- Jaeckel-Williams R. Nuclear divisions with reduced numbers of microtubules in Tetrahymena. J Cell Sci. 1978;34:303–319. doi: 10.1242/jcs.34.1.303. [DOI] [PubMed] [Google Scholar]
- Koonce MP, Samso M. Overexpression of dynein’s globular head causes a collapse of the interphase microtubule network in Dictyostelium. Mol Biol Cell. 1996;7:935–948. doi: 10.1091/mbc.7.6.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz S, Tiedtke A. The Golgi apparatus of Tetrahymena thermophila. J Euk Microbiol. 1993;40:10–13. doi: 10.1111/j.1550-7408.1993.tb04874.x. [DOI] [PubMed] [Google Scholar]
- LaFountain JR, Jr, Davidson LA. An analysis of spindle ultrastructure during prometaphase and metaphase of micronuclear division in Tetrahymena. Chromosoma. 1979;75:293–308. doi: 10.1007/BF00293473. [DOI] [PubMed] [Google Scholar]
- LaFountain JR, Jr, Davidson LA. An analysis of spindle ultrastructure during anaphase of micronuclear division in Tetrahymena. Cell Motil. 1980;1:41–61. doi: 10.1002/cm.970010105. [DOI] [PubMed] [Google Scholar]
- Lee S-w, Wisniewski JC, Forney JD, Asai DJ. Gene knockouts demonstrate that cytoplasmic dyneins 1A and 1B perform separate functions in Tetrahymena. Mol Biol Cell. 1997;8:160a. doi: 10.1091/mbc.10.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y-Y, Yeh E, Hays T, Bloom K. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc Natl Acad Sci USA. 1993;90:10096–10100. doi: 10.1073/pnas.90.21.10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu MJ, Dadd CA, Mizzen CA, Perry CA, McLachlan DR, Annunziato AT, Allis CD. Generation and characterization of novel antibodies highly selective for phosphorylated linker histone H1 in Tetrahymena and HeLa cells. Chromosoma. 1994;103:111–121. doi: 10.1007/BF00352320. [DOI] [PubMed] [Google Scholar]
- Lye RJ, Wilson RK, Waterston RH. Genomic structure of a cytoplasmic dynein heavy chain gene from the nematode Caenorhabditis elegans. Cell Motil Cytoskeleton. 1995;32:26–36. doi: 10.1002/cm.970320104. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN, O’Toole ET, McDonald KL, McIntosh JR, Porter ME. Arrangement of inner dynein arms in wild-type and mutant flagella of Chlamydomonas. J Cell Biol. 1992;118:1145–1152. doi: 10.1083/jcb.118.5.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muresan V, Godek CP, Reese TS, Schnapp BJ. Plus-end motors override minus-end motors during transport of squid axon vesicles on microtubules. J Cell Biol. 1996;135:383–397. doi: 10.1083/jcb.135.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanney DL, Simon EM. Laboratory and evolutionary history of Tetrahymena thermophila. In: Asai DJ, Forney JD, editors. Methods in Cell Biology: Tetrahymena thermophila. San Diego: Academic Press; 1999. (in press). [DOI] [PubMed] [Google Scholar]
- Oda H, Stockert RJ, Collins C, Wang H, Novikoff PM, Satir P, Wolkoff AW. Interaction of the microtubule cytoskeleton with endocytic vesicles and cytoplasmic dynein in cultured rat hepatocytes. J Biol Chem. 1995;270:15242–15249. doi: 10.1074/jbc.270.25.15242. [DOI] [PubMed] [Google Scholar]
- Pazour, G.J., Dickert, B.L., and Witman, G.B. (1999). The DHC1B (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J. Cell Biol. (in press). [DOI] [PMC free article] [PubMed]
- Pfarr CM, Coue M, Grissom PM, Hays TS, Porter ME, McIntosh JR. Cytoplasmic dynein is localized to kinetochores during mitosis. Nature. 1990;345:263–265. doi: 10.1038/345263a0. [DOI] [PubMed] [Google Scholar]
- Piperno G. Functional diversity of dyneins. Cell Motil Cytoskeleton. 1990;17:147–149. doi: 10.1002/cm.970170302. [DOI] [PubMed] [Google Scholar]
- Piperno G, Ramanis Z. The proximal portion of Chlamydomonas flagella contains a distinct set of inner dynein arms. J Cell Biol. 1991;112:701–709. doi: 10.1083/jcb.112.4.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plamann M, Minke PF, Tinsley JH, Bruno KS. Cytoplasmic dynein and actin-related protein arp1 are required for normal nuclear distribution in filamentous fungi. J Cell Biol. 1994;127:139–149. doi: 10.1083/jcb.127.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ME, Bower R, Knott JA, Byrd P, Dentler WL. Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol Biol Cell. 1999;10:693–712. doi: 10.1091/mbc.10.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen L, Orias E. Tetrahymena: growth without phagocytosis. Science. 1975;190:464–465. doi: 10.1126/science.1166313. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Alexander SP. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J Cell Biol. 1990;111:81–95. doi: 10.1083/jcb.110.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JL, Carlson K. Cilia regeneration in Tetrahymena and its inhibitions by colchicine. J Cell Biol. 1969;40:415–425. doi: 10.1083/jcb.40.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Saunders WS, Koshland D, Eshel D, Gibbons IR, Hoyt MA. Saccharomyces cerevisiae kinesin- and dynein-related proteins required for anaphase chromosome segregation. J Cell Biol. 1995;128:617–624. doi: 10.1083/jcb.128.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer TA, Steuer ER, Sheetz MP. Cytoplasmic dynein is a minus end-directed motor for membranous organelles. Cell. 1989;56:937–946. doi: 10.1016/0092-8674(89)90627-2. [DOI] [PubMed] [Google Scholar]
- Shaw SL, Yeh E, Maddox P, Salmon ED, Bloom K. Astral microtubule dynamics in yeast: a microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J Cell Biol. 1997;139:985–994. doi: 10.1083/jcb.139.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneborn TM. Handbook of Genetics: Plants, Plant Viruses and Protists. R.C. King, New York: Plenum Press; 1974. Tetrahymena pyriformis; pp. 433–467. [Google Scholar]
- Steuer ER, Wordeman L, Schroer TA, Sheetz MP. Localization of cytoplasmic dynein to mitotic spindles and kinetochores. Nature. 1990;345:266–268. doi: 10.1038/345266a0. [DOI] [PubMed] [Google Scholar]
- Supp DM, Witte DP, Potter SS, Brueckner M. Mutation of an axonemal dynein affects left-right asymmetry in inversus viscerum mice. Nature. 1997;389:963–966. doi: 10.1038/40140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Zhang Z, Hirokawa N. Identification and molecular evolution of new dynein-like protein sequences in rat brain. J Cell Sci. 1995;108:1883–1893. doi: 10.1242/jcs.108.5.1883. [DOI] [PubMed] [Google Scholar]
- Tuxhorn J, Daise T, Dentler WL. Regulation of flagellar length in Chlamydomonas. Cell Motil Cytoskeleton. 1998;40:133–146. doi: 10.1002/(SICI)1097-0169(1998)40:2<133::AID-CM3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Vaisberg EA, Grissom PM, McIntosh JR. Mammalian cells express three distinct dynein heavy chains that are localized to different cytoplasmic organelles. J Cell Biol. 1996;133:831–842. doi: 10.1083/jcb.133.4.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisberg EA, Koonce MP, McIntosh JR. Cytoplasmic dynein plays a role in mammalian mitotic spindle formation. J Cell Biol. 1993;123:849–858. doi: 10.1083/jcb.123.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD, Fletterick RJ. The design plan of kinesin motors. Annu Rev Cell Dev Biol. 1997;13:745–777. doi: 10.1146/annurev.cellbio.13.1.745. [DOI] [PubMed] [Google Scholar]
- Verde F, Berrez J-M, Antony C, Karsenti E. Taxol-induced microtubule asters in mitotic extracts of Xenopus eggs: requirement for phosphorylated factors and cytoplasmic dynein. J Cell Biol. 1991;112:1177–1187. doi: 10.1083/jcb.112.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R, et al. 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature. 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- Xiang X, Beckwith SM, Morris NR. Cytoplasmic dynein is involved in nuclear migration in Aspergillus nidulans. Proc Natl Acad Sci USA. 1994;91:2100–2104. doi: 10.1073/pnas.91.6.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]