Abstract
Resident membrane proteins of the trans-Golgi network (TGN) of Saccharomyces cerevisiae are selectively retrieved from a prevacuolar/late endosomal compartment. Proper cycling of the carboxypeptidase Y receptor Vps10p between the TGN and prevacuolar compartment depends on Vps35p, a hydrophilic peripheral membrane protein. In this study we use a temperature-sensitive vps35 allele to show that loss of Vps35p function rapidly leads to mislocalization of A-ALP, a model TGN membrane protein, to the vacuole. Vps35p is required for the prevacuolar compartment-to-TGN transport of both A-ALP and Vps10p. This was demonstrated by phenotypic analysis of vps35 mutant strains expressing A-ALP mutants lacking either the retrieval or static retention signals and by an assay for prevacuolar compartment-to-TGN transport. A novel vps35 allele was identified that was defective for retrieval of A-ALP but functional for retrieval of Vps10p. Moreover, several other vps35 alleles were identified with the opposite characteristics: they were defective for Vps10p retrieval but near normal for A-ALP localization. These data suggest a model in which distinct structural features within Vps35p are required for associating with the cytosolic domains of each cargo protein during the retrieval process.
INTRODUCTION
The secretory pathway consists of a series of membrane-enclosed compartments that are distinct from each other in terms of morphology, molecular composition, and function. The establishment and maintenance of the unique identity of each organelle depend on the correct localization and retention of its resident proteins (Rothman and Wieland, 1996). One mechanism used by secretory pathway organelles to retain their resident proteins is to prevent entry of the resident proteins into transport vesicles that form from an organelle. A second mechanism that can also be used is the selective retrieval of resident proteins after they have left the organelle with other proteins. For example, the endoplasmic reticulum (ER) retains its resident proteins by a sorting mechanism that ensures that ER-derived vesicles are enriched for certain secretory proteins and depleted of resident proteins (Rexach et al., 1994; Bednarek et al., 1995). Any ER resident proteins that aberrantly enter these vesicles and are delivered to the cis-Golgi are transported back to the ER by selective retrieval machinery (Dean and Pelham, 1990; Gaynor et al., 1994; Townsley et al., 1994).
The mechanism of protein retention in the Golgi apparatus is less clear, although recent studies are beginning to illuminate this process. Several studies of the retention of membrane proteins in the earlier regions of the Golgi support a model in which the Golgi enzymes form complexes too large to enter transport vesicles, thus leading to static retention (Weisz et al., 1993; Nilsson et al., 1994). Since the transmembrane domains of Golgi enzymes have often been found to contain features necessary for proper retention, it has also been proposed that Golgi enzymes achieve residence by partitioning into distinct lipid domains within Golgi stacks (Pelham and Munro, 1993).
In contrast to the earlier regions of the Golgi, the localization of membrane proteins to the trans-Golgi network (TGN) appears to be a highly dynamic process. In animal cells, membrane proteins such as TGN38 (Stanely and Howell, 1993), furin (Chapman and Munro, 1994b; Molloy et al., 1994), and the mannose 6-phosphate receptor (Duncan and Kornfeld, 1988) are known to continuously cycle between the TGN, plasma membrane, and endosomes. The sorting signals responsible for trafficking between these compartments reside on the cytosolic domains of these proteins and include aromatic amino acid, dileucine, and acidic cluster-based signals (Kornfeld, 1992; Stanely and Howell, 1993; Schäfer et al., 1995; Takahashi et al., 1995; Voorhees et al., 1995). Endocytosis of these proteins from the plasma membrane and exit from the TGN appear to be mediated by interactions of their cytosolic domain signals with clathrin/adaptor coat complexes (for a review see Kirchhausen et al., 1997). Retrograde transport from the late endosome to the TGN appears to be vesicle mediated and requires the small GTP-binding protein Rab9 at a postvesicle formation step (Riederer et al., 1994). Although little is known about components of the vesicle coat that act in retrieval of TGN proteins from late endosomes, two recently identified peripheral membrane proteins, PACS-1 (Wan et al., 1998) and TIP47 (Diaz and Pfeffer, 1998), are good candidates for such components. PACS-1 and TIP47 may play roles in cargo sorting at the late endosome because they localize to endosomal structures and bind in a sorting-signal–specific manner to the cytosolic domains of furin and the mannose 6-phosphate receptor, respectively.
Resident membrane proteins of the yeast TGN (sometimes referred to as the late Golgi) are also localized in a dynamic manner. The yeast TGN contains membrane proteins Kex2p, Kex1p, and dipeptidyl aminopeptidase (DPAP) A, which are involved in proteolytic processing of the secreted mating pheromone, α-factor (Fuller et al., 1988). These proteins are known to cycle between the TGN and a prevacuolar endosomal compartment but, in contrast to animal cell TGN proteins, do not appear to visit the cell surface (Cooper and Bussey, 1992; Roberts et al., 1992; Wilcox et al., 1992; Bryant and Stevens, 1997). Localization of DPAP A and Kex2p is achieved, in part, by retrieval from the prevacuolar compartment, a process mediated by aromatic amino acid signals in their cytosolic domains (Wilcox et al., 1992; Nothwehr et al., 1993; Brickner and Fuller, 1997; Bryant and Stevens, 1997). In DPAP A, this signal consists of a FXFXD motif in which both F residues are absolutely required. Recently, a second signal in the DPAP A cytosolic domain required for reducing its rate of transport from the TGN to the prevacuolar compartment was identified (Bryant and Stevens, 1997).
The yeast TGN is also the site of sorting of vacuolar hydrolases, a process mediated by Vps10p. Vps10p functions as a membrane-bound sorting receptor in the TGN for the vacuolar hydrolase, carboxypeptidase Y (CPY) (Marcusson et al., 1994; Cooper and Stevens, 1996). Analogous to the function of the mannose 6-phosphate receptor of animal cells, Vps10p/CPY complexes at the TGN are thought to enter vesicles that are transported to the prevacuolar compartment. CPY then dissociates from its receptor and is transported via a default pathway to the vacuole, the yeast equivalent of the lysosome. The receptor is then transported back to the TGN via a transport mechanism that is probably also vesicle mediated (Piper et al., 1995; Seaman et al., 1997; Seaman et al., 1998). Vps10p also contains an aromatic-amino-acid–based signal in its cytosolic domain necessary for retrieval (Cereghino et al., 1995; Cooper and Stevens, 1996). Genetic analysis of the molecular machinery for transport between the TGN and prevacuolar compartment indicates that, for the most part, DPAP A, Kex2p, and Vps10p require the same machinery, although some notable exceptions have been observed (Rothman and Stevens, 1986; Robinson et al., 1988; Nothwehr et al., 1996; Voos and Stevens, 1998).
Recent experimental advances in yeast have shed light on the mechanism of retrograde traffic between the prevacuolar endosomal compartment and the TGN. In strains containing mutations in the VPS29, VPS30, and VPS35 genes, Vps10p is mislocalized to the vacuole by a pathway dependent upon the prevacuolar compartment t-SNARE Pep12p but independent of late-secretory-pathway functions (Seaman et al., 1997). This observation suggests that these three genes play a role in retrieval of Vps10p. More recently, the products of these three genes, along with the products of three other genes known to be involved in trafficking between the TGN and prevacuolar compartment, Vps5p, Vps17p, and Pep8p/Vps26p, have been shown to form a complex (Horazdovsky et al., 1997; Nothwehr and Hindes, 1997; Seaman et al., 1998). This complex, termed the “retromer complex,” localizes to the cytosolic face of the prevacuolar compartment and vesicles. This group of proteins may therefore act as a coat for retrograde vesicles that originate from the prevacuolar compartment and have a role in cargo-protein sorting. Homologues of the retromer proteins have been identified in higher eukaryotes, including humans, suggesting that they may perform an analogous role in cargo retrieval from an endosomal compartment in these systems. Implicit in the working model of yeast retromer function is that one or more of the retromer proteins binds to the retrieval signals on the cytosolic domains of Vps10p, DPAP A, and Kex2p and mediates their selective entry into forming vesicles. However, little is known about the nature of such interactions.
In this study we have investigated the role of Vps35p in localization of a model TGN-membrane protein, A-ALP, and Vps10p. Using a variety of experimental approaches, we show that Vps35p is required for retrieval of both proteins from the prevacuolar compartment. Surprisingly, mutant alleles of VPS35 were found that exhibited specific defects in either DPAP A (A-ALP) or Vps10p retrieval. Thus, distinct structural features within Vps35p are necessary for retrieval of each protein, an observation suggesting that Vps35p has a direct role in cargo sorting.
MATERIALS AND METHODS
Materials
Restriction enzymes and other enzymes used in subcloning procedures were obtained from New England Biolabs (Beverly, MA), United States Biochemical (Cleveland, OH), or Promega (Madison, WI). [35S]express label and [α-35S]dATP were purchased from New England Nuclear (Boston, MA). Oxalyticase was obtained from Enzogenetics (Corvallis, OR). All secondary antibodies used for immunofluorescence experiments were from Jackson ImmunoResearch Labs. (West Grove, PA). Other reagents were obtained from Sigma Chemical (St. Louis, MO) or as indicated.
Genetic and Nucleic Acid Manipulations
Most of the plasmids and all yeast strains used in this study are indicated in Tables 1 and 2, respectively. A centromeric (CEN), TRP1-based plasmid containing the A-ALP construct was generated by subcloning the 2.37-kilobase pair (kbp) EagI–EcoRI fragment from pSN55 into the EagI/EcoRI sites of pRS314, resulting in pAH16. Deletion of codons 2–11 of the STE13 gene in plasmid pCJR71 (Roberts et al., 1992) was performed (Kunkel et al., 1987) to generate pAH48. To incorporate the deletion into the context of the STE13–PHO8 fusion construct that expresses the A-ALP fusion protein, an EagI–BglII fragment from pAH48 was swapped for the corresponding EagI–BglII fragment present in pSN55, resulting in plasmid pAH49.
Table 1.
Plasmids used in this study
| Plasmid | Description | Reference |
|---|---|---|
| pAH16 | pRS314 containing a DNA fragment encoding A-ALP | This study |
| pSN54 | pRS313 containing a DNA fragment encoding A-ALP | Nothwehr et al. (1993) |
| pSN55 | pRS316 containing a DNA fragment encoding A-ALP | Nothwehr et al. (1993) |
| pAH49 | pSN55 with codons 2-11 of the A-ALP construct deleted | This study |
| pSN100 | pSN55 with the Phe85 and Phe87 codons of A-ALP mutated to Ala | Nothwehr et al. (1993) |
| pLS13 | VPS35 gene in pRS316 | This study |
| pLS12 | vps35-101 allele in pRS316 | This study |
| pPB7-9.7 | vps35-103 allele in pRS316 | This study |
| pPB7-10.1 | vps35-104 allele in pRS316 | This study |
| pPB7-42.8 | vps35-105 allele in pRS316 | This study |
| pPB7-108.5 | vps35-107 allele in pRS316 | This study |
| pPB7-010.4 | vps35-108 allele in pRS316 | This study |
| pHY5 | pRS316 containing the VPS27 gene under GAL1 promoter control | T. H. Stevens |
| pPB6 | vps35Δ∷URA3 allele in Bluescript KS+ | This study |
| pGPY55 | vps35Δ∷HIS3 allele in a CEN plasmid | Paravicini et al. (1992) |
Table 2.
Saccharomyces cerevisiae strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| SNY36-9A | MATa ura3-52 leu2-3,112 his3-Δ200 trp1-901 suc2-Δ9 pho8Δ∷ADE2 | Nothwehr et al. (1995) |
| SNY17 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ∷LEU2 | Nothwehr et al. (1995) |
| SNY17-190 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ∷LEU2 vps35-101 | This study |
| SNY38 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ∷ADE2 vps1Δ∷LEU2 | Nothwehr et al. (1995) |
| SNY79 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ∷LEU2 vps35Δ∷HIS3 | This study |
| SNY94 | MATa ura3-52 leu2-3,112 his3-Δ200 trp1-901 suc2-Δ9 pho8Δ∷ADE2 end3-ts | This study |
| SNY96-9D | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ∷ADE2 vps1Δ∷LEU2 end3-ts | This study |
| LSY6-2A | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 pho8∷ste13-pho8 vps35Δ∷HIS3 | This study |
| LSY9-5B | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ∷ADE2 end3-ts vps35Δ∷HIS3 | This study |
| PBY1 | MATα ra3-52 leu2-3,112 his3-Δ200 trp1-901 suc2-Δ9 lys2-801 pho8Δ∷LEU2 pep4Δ∷TRP1 vps35Δ∷HIS3 | This study |
| PBY3 | MATa ura3-52 leu2-3,112 his3-Δ200 trp1-901 suc2-Δ9 pho8Δ∷ADE2 vps35Δ∷URA3 | This study |
| PBY4 | MATa ura3-52 leu2-3,112 his3-Δ200 trp1-901 suc2-Δ9 pho8Δ∷ADE2 vps35-109 (ts) | This study |
| PBY15 | MATa ura3-52 leu2-3,112 his3-Δ200 trp1-901 suc2-Δ9 pho8Δ∷ADE2 pep4-ΔH3 vps35Δ∷HIS3 | This study |
| W303-1A | MATa ura3-1 leu2-3,112 his3-11 trp1-1 ade2-1 can1-100 | R. Rothstein |
| AHY65 | MATa ura3-1 leu2-3,112 his3-11 trp1-1 ade2-1 can1-100 pho8Δ∷ADE2 pep4-ΔH3 vps27Δ∷LEU2 | This study |
| AHY68 | MATa ura3-1 leu2-3,112 his3-11 trp1-1 ade2-1 can1-100 pho8Δ∷ADE2 pep4-ΔH3 vps27Δ∷LEU2 vps35Δ∷HIS3 | This study |
| AHY69 | MATa ura3-1 leu2-3,112 his3-11 trp1-1 ade2-1 can1-100 pho8Δ∷ADE2 pep4-ΔH3 vps27Δ∷LEU2 vps35-101 | This study |
All strains are derived from SNY17 or SNY36-9A with the exception of AHY65, AHY68, and AHY69, which are derived from W303-1A.
A construct designed to integrate the STE13–PHO8 fusion at the PHO8 locus was made by subcloning the 2.37-kbp EagI–EcoRI fragment from pSN55 into the EagI/EcoRI sites of pRS306 (Sikorski and Hieter, 1989). The resulting plasmid, pSN282, was digested with SacII, blunt-ended with Klenow enzyme and dNTPs, digested with EagI, and then ligated to a 1.4-kbp SmaI–EagI fragment from the 5′-untranslated region of the STE13 gene, resulting in pAH17. pAH17 was digested with SacI, blunt-ended with Klenow enzyme and dNTPs, and was ligated to a blunt-ended 0.83-kbp EcoRI–HindIII fragment from pSN92 (containing the 5′-region of the PHO8 gene), resulting in plasmid pSN288. The STE13–PHO8 construct was integrated into the PHO8 locus by transforming yeast strains with pSN288 digested with SacI and selecting for Ura+ prototrophs. Ura+ clones were then grown on media containing 5-fluoroorotic acid (Boeke et al., 1984) to select for clones that looped out wild-type PHO8.
A vps35Δ::URA3 gene replacement construct was made by subcloning the 1.1-kbp SmaI fragment from pJJ242 (Jones and Prakash, 1990) containing URA3 into the SnaBI sites of pLS9, a plasmid consisting of the 3.9-kbp MscI fragment containing VPS35 inserted into the SmaI site of Bluescript KS+. The resulting plasmid, pPB6, was digested with BseRI/EcoNI, transformed into yeast strain SNY36–9A, and Ura+ prototrophs were screened for the Vps− phenotype, resulting in strain PBY3. Likewise, to generate yeast strains carrying the vps35Δ::HIS3 allele, plasmid pGPY55 (Paravicini et al., 1992) was digested with NciI/XbaI, the 4.5-kbp fragment was purified and transformed into yeast, and His+ Vps− transformants were identified.
To construct a CEN-VPS35 plasmid, a 3.9-kbp MscI fragment containing the VPS35 gene was obtained from p35–1, a YCp50-based plasmid isolated from a yeast-genomic library (Rose et al., 1987). This fragment was subcloned into the SacI/KpnI sites of pRS316 that had been made blunt by Klenow DNA polymerase, resulting in pLS13. The vps35–101 allele was cloned by gap-repairing plasmid p35–1 digested with SnaBI/AflII in yeast strain SNY17–190 (Table 2), resulting in plasmid pLS11. The 3.9-kbp MscI fragment from pLS11 was subcloned into pRS316 (using the same approach used for pLS13), resulting in plasmid pLS12.
Random Mutagenesis of the VPS35 Gene
The VPS35 gene was subjected to random PCR mutagenesis using an in vivo gap-repair method. Using a VPS35-containing plasmid as a template, a 3.272-kbp PCR fragment was amplified under mutagenic PCR conditions (Cadwell and Joyce, 1992). This fragment corresponded to positions −177 to +3095 of the VPS35 gene, where the first nucleotide of the open reading frame (ORF) is defined as the +1 nucleotide. The PCR fragment was cotransformed into yeast strain LSY6–2A, along with linearized pLS13 plasmid DNA that had been digested with BseRI/EcoNI, so that most of the VPS35 ORF was removed. Yeast transformants containing circular plasmids generated via homologous recombination were selected on minimal media lacking uracil. A total of 14,000 transformants were screened for defects in CPY sorting using a colony-blotting assay (Roberts et al., 1991) and for A-ALP retention defects using a plate-activity assay (Nothwehr et al., 1996). Plasmids were isolated from mutants that exhibited either the A-ALP retention phenotype or the CPY secretion phenotype (but not both), and the linkage of the phenotype to the plasmid was verified. Mutants were screened separately for temperature-sensitive vps35 alleles by assessing CPY secretion at 22 and 35°C. Plasmid p11G-2 containing the vps35–109 allele was isolated from one of the temperature-sensitive mutants.
To replace the VPS35 allele with the temperature-sensitive vps35–109 allele in yeast, the insert from p11G-2 was released by digestion with BseRI/EcoNI, gel purified, and cotransformed with pRS315 (Sikorski and Hieter, 1989) into strain PBY3. Leu+ transformants in which the mutant vps35 allele from the plasmid insert replaced the vps35Δ::URA3 allele were identified by growth on 5-fluoroorotic acid. The presence of the vps35–109 allele was confirmed by analyzing CPY secretion at the nonpermissive and permissive temperatures, resulting in yeast strain PBY4. The introduction of the vps35–101 allele into yeast, resulting in strain AHY69, was achieved using a similar approach.
Mutations causing the CPY secretion or A-ALP retention phenotypes were initially mapped to intervals within the VPS35 ORF contained within the pLS13-derived mutant plasmids. This was achieved by swapping of restriction fragments with nonmutagenized pLS13 and analyzing the phenotypes of the resulting plasmids when transformed into a vps35Δ yeast strain. The region within the interval containing the critical mutation was then DNA sequenced and compared with the wild-type VPS35 sequence. If an interval contained more than one mutation, site-directed mutagenesis was used to introduce each single mutation into pLS13, and the phenotypic effects of each mutation were assessed, thus leading to identification of a single-residue change responsible for the phenotype.
Radiolabeling, Immunoprecipitation, and Subcellular Fractionation
The procedure for immunoprecipitation of CPY was performed using a rabbit antibody against CPY as described previously (Vater et al., 1992). Likewise, immunoprecipitation of A-ALP was performed using previously generated rabbit anti-ALP serum (Nothwehr et al., 1996) and a procedure previously described (Nothwehr et al., 1993). Radioactively labeled proteins were quantified from gels using a Phosphorimager system (Fuji Photo Film). Half-times of processing were determined by linear-regression analysis followed by plotting the percentage of total and processed protein as a function of time.
Subcellular fractionation was carried out by incubating 5 OD600 U of cells grown in minimal media lacking methionine in 1 ml of 0.1 M Tris, pH 9.4, 10 mM DTT for 10 min at room temperature. The cells were pelleted and spheroplasted in a 1 ml solution containing 20 mM Tris, pH 7.5, minimal media lacking methionine, 1 M sorbitol, and 25 μg/ml oxalyticase at 30°C. The spheroplasts were pelleted, resuspended in 1 ml minimal media lacking methionine and containing 1 M sorbitol, and pulsed at 30°C by adding 200 μCi of [35S]express label. After a 30-min pulse, a 45-min chase was initiated by adding 250 μl of a solution containing 25 mM methionine and 5 mM cysteine. The labeled spheroplasts were harvested and lysed by incubating in 1 ml of 25 mM sodium phosphate, pH 7.4, 200 mM mannitol, 1 mM EDTA on ice for 20 min. Unlysed cells were removed by centrifugation at 450 × g for 5 min. The 450 × g supernatant was then centrifuged at 13,000 × g for 15 min to generate pellet (P13) and supernatant (S13) fractions. The S13 was then centrifuged at 150,000 × g for 60 min to generate P150 and S150 fractions. The S150 fractions were trichloroacetic acid precipitated and pellets were washed with acetone. The S150 trichoroacetic acid pellets, as well as the P13 and P150 membrane pellets, were resuspended in 100 μl of 8 M urea, 5% SDS, and were heated at 65°C for 10 min. Immunoprecipitation of Vps35p from 20 μl of each fraction was carried out as previously described (Nothwehr et al., 1993) using 1.2 μl of anti-Vps35p rabbit serum (a generous gift from Scott Emr).
Fluorescence Microscopy
The procedures for preparation of fixed spheroplasted yeast cells, attachment to microscope slides, and costaining of the A-ALP fusion protein and Vma2p using an anti-ALP polyclonal antibody and anti-Vma2p monoclonal antibody 13D11-B2 (Molecular Probes, Eugene, OR) were previously described (Roberts et al., 1991; Nothwehr et al., 1995).
For induction of VPS27 in yeast strains via the GAL1 promoter before analysis by fluorescence microscopy, strains were propagated overnight in minimal media containing 2% (wt/vol) raffinose. Log-phase cultures were then adjusted to 2% galactose and, after 0 and 90 min, aliquots of 10 ml were removed and fixed by addition to 1.2 ml of 37% formaldehyde.
To simultaneously detect Vps10p and A-ALP, fixed cells on slides were incubated with the following solutions followed by extensive washing using 5 mg/ml BSA in PBS after every step: 1) a 1:30 dilution of rabbit anti-Vps10 (a generous gift of T.H. Stevens) and a 1:3 dilution of mouse anti-ALP 1D3 monoclonal supernatant (Nothwehr et al., 1996); 2) 1:500 dilution of biotin-conjugated donkey anti-rabbit IgG (H+L); and 3) a 1:500 dilution of FITC–streptavidin and a 1:2000 dilution of Texas Red-conjugated goat anti-mouse IgG (H+L). To simultaneously detect Vps10p and Vma2p, the following incubations were used: 1) a 1:30 dilution of rabbit anti-Vps10 and a 1:10 dilution of mouse anti-Vma2p; 2) a 1:500 dilution of biotin-conjugated goat anti-rabbit IgG (H+L); and 3) 1:500 dilutions of FITC–streptavidin and of Texas Red-conjugated goat anti-mouse IgG (H+L).
Yeast cells were photographed using an Olympus BX-60 fluorescence microscope (Olympus, Lake Success, NY). Film negatives were digitized using a Umax Powerlook III scanner and adjusted using Adobe Photoshop 3.0 (Adobe Systems, Mountain View, CA).
RESULTS
Vps35p Has a Direct Role in Retention of a TGN-Membrane Protein
A-ALP is a model TGN-membrane protein consisting of a fusion between the N-terminal cytosolic domain of DPAP A and the transmembrane and lumenal domains of alkaline phosphatase (ALP). A failure to retain A-ALP in the TGN results in its delivery to the vacuole where its C-terminal propeptide is removed by a vacuolar protease (Klionsky and Emr, 1989; Nothwehr et al., 1993). A-ALP processing can be detected either by a mobility change on SDS-PAGE gels or as an increase in enzymatic activity. This is because Golgi-localized pro-A-ALP is inactive, and vacuole-localized mature A-ALP is active. Thus, the processing/activity of A-ALP serves as a convenient assay for TGN retention.
We previously reported a genetic screen for grd mutants that exhibit vacuolar processing of A-ALP (Nothwehr et al., 1996). Mutants from the grd9/vps35 complementation group exhibited a very strong A-ALP processing phenotype and also secreted unprocessed α-factor, suggesting a defect in retention of Kex2p as well. In addition, vps35 null mutants aberrantly secrete most of their newly synthesized CPY (Paravicini et al., 1992).
The retention defect in vps35 mutants could reflect a direct role for Vps35p in some aspect of A-ALP retention or could be an indirect consequence of the prolonged absence of Vps35p in vps35 null mutants. To distinguish between these possibilities, we analyzed Golgi-retention phenotypes after rapid inactivation of Vps35p expressed from a temperature-sensitive allele.
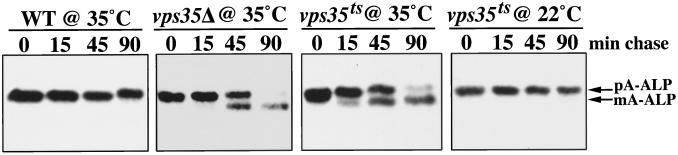
A library of vps35Δ strains containing centromeric (CEN) plasmids with randomly mutagenized VPS35 inserts was screened for mutants that exhibited temperature-sensitive defects. One such allele, vps35–109 (hereafter referred to as vps35-ts), exhibited normal TGN retention and CPY sorting at 22°C but severe defects in these processes at 35°C. The rate of appearance of the A-ALP–processing defect was assessed after shifting vps35-ts cells from 22 to 35°C. Figure 1 shows the results of an experiment in which wild-type, vps35Δ, and vps35-ts cells were propagated overnight at 22°C, preincubated at 35°C for 10 min, pulse labeled for 10 min, chased for the indicated times, and subjected to immunoprecipitation of A-ALP. Whereas little or no processing is observed in the wild-type strain even after a 90-min chase, the vps35Δ strain exhibits a processing half-time of ∼50–60 min. A severe processing phenotype was also observed in the vps35-ts strain at 35°C that was virtually indistinguishable from that of the null strain. However, at the permissive temperature of 22°C, the vps35-ts strain exhibits no detectable processing during the time course. The observation that A-ALP retention is rapidly lost after inactivating Vps35p suggests that Vps35p performs a function intimately involved in the mechanism of TGN localization.
Figure 1.
A-ALP is processed rapidly in a vps35-ts strain after shifting to the nonpermissive temperature. Wild-type (SNY36–9A), vps35Δ (SNY79), and vps35-ts (PBY4) yeast strains carrying a CEN plasmid directing expression of A-ALP (pSN55) were analyzed. The strains were grown for several doublings at 22°C, preincubated at 35°C for 10 min, and then pulsed with [35S]methionine/cysteine for 10 min before unlabeled amino acids were added to initiate the chase. At the indicated times A-ALP was immunoprecipitated from the cultures and analyzed by SDS-PAGE and fluorography. The positions of unprocessed pro-A-ALP (pA-ALP) and mature A-ALP (mA-ALP) are indicated.
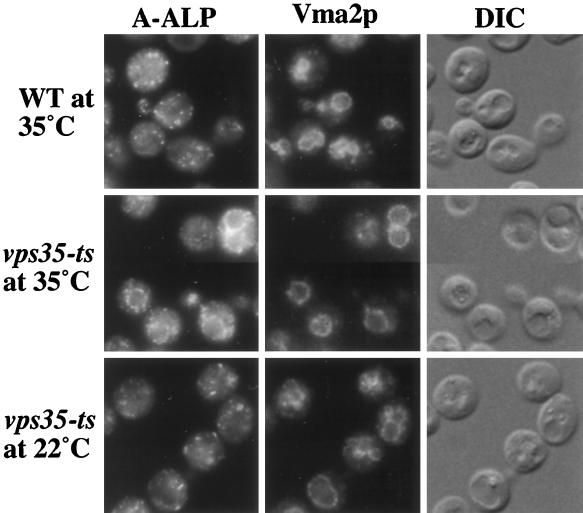
Immunofluorescence microscopy was used to address whether A-ALP is indeed mislocalized to the vacuole in vps35 mutants. The localization of A-ALP and Vma2p, a vacuolar membrane marker, in wild-type and vps35-ts cells was analyzed at the permissive and nonpermissive temperatures (Figure 2). In wild-type cells, A-ALP exhibits a nonvacuolar, punctate staining pattern characteristic of the yeast Golgi (Redding et al., 1991; Nothwehr et al., 1993). However, in vps35-ts cells incubated at the nonpermissive temperature for 40 min, most of the A-ALP colocalizes with the Vma2p marker, indicating that A-ALP has been mislocalized to the vacuole. The remaining A-ALP that is localized to nonvacuolar structures in these cells could still be retained in the TGN or could be in the prevacuolar compartment awaiting retrieval. Consistent with the pulse–chase experiment in Figure 1, at 22°C A-ALP expressed in the vps35-ts strain appears to be localized normally to the Golgi apparatus.
Figure 2.
A-ALP is mislocalized to the vacuole in vps35-ts mutant cells. Wild-type (SNY17) and vps35-ts (PBY4) strains carrying a CEN plasmid directing expression of A-ALP (pSN55) were propagated for several doublings at 22°C before being shifted to 35°C for 40 min or incubated at 22°C for 40 min (as indicated). The cells were then fixed, spheroplasted, and costained with antibodies against ALP and Vma2p. After subsequent treatment with fluorochrome-conjugated secondary antibodies, the cells were viewed by DIC optics and by epifluorescence through filters specific for FITC and Texas Red.
A-ALP Reaches the Vacuole in vps35Δ Cells Independent of the Endocytic Pathway
The observation that a loss of Vps35p function caused A-ALP to be mislocalized to the vacuole suggested that Vps35p could act at the level of the TGN or it could act in retrieval from the prevacuolar compartment. In the latter case, A-ALP in vps35 mutants would be expected to be directly transported from the TGN to the prevacuolar compartment and then on to the vacuole. However, if Vps35p acts at the TGN, it is possible that, in the absence of Vps35p function, A-ALP may be initially mislocalized to the plasma membrane and then travel to the vacuole via the endocytic pathway, as has been observed in vps1 mutants (Nothwehr et al., 1995).
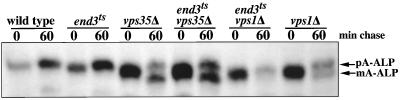
To distinguish between these possibilities, we asked whether the transport of A-ALP to the vacuole in vps35 mutant cells was dependent on the END3 gene that encodes a protein necessary for the internalization step of endocytosis (Bénédetti et al., 1994). Immunoprecipitation of A-ALP from a vps35Δ end3-ts strain incubated at the nonpermissive temperature for the end3-ts allele (37°C) shows that, after a 60-min chase, A-ALP is processed with kinetics similar to that of the vps35Δ single mutant (Figure 3). As expected, under these conditions no processing is observed in the wild-type and end3-ts single-mutant strains. A-ALP in a vps1Δ mutant is ∼50% processed after 60 min, but this processing is blocked if the end3-ts allele is introduced, consistent with prior observations (Nothwehr et al., 1995). Because A-ALP is known to depend on the early-endocytic pathway for delivery to the vacuole in vps1 cells, this control experiment shows that under these conditions the endocytic pathway is blocked in the strains carrying the end3-ts allele. These data are consistent with the view that, in the absence of Vps35p function, A-ALP is transported from the TGN to the prevacuolar compartment and then to the vacuole.
Figure 3.
The delivery of A-ALP to the vacuole in vps35Δ cells does not require the early endocytic pathway. Wild-type (SNY36–9A), end3-ts (SNY94), vps35Δ (SNY79), end3-ts vps35Δ (LSY9–5B), vps1Δ (SNY38), and end3-ts vps1Δ (SNY96–9D) cells carrying a plasmid expressing A-ALP (pSN55) were analyzed. Each strain was propagated at 22°C for several doublings before being preincubated at 37°C for 10 min, and then pulsed with [35S]methionine/cysteine for 10 min and chased by the addition of unlabeled amino acids. At the indicated times A-ALP was immunoprecipitated from the cultures and analyzed by SDS-PAGE and fluorography. The positions of unprocessed pro-A-ALP (pA-ALP) and mature A-ALP (mA-ALP) are indicated.
VPS35 Is Involved in Retrieval of A-ALP from the Prevacuolar Compartment but Not in Slowing Exit from the TGN
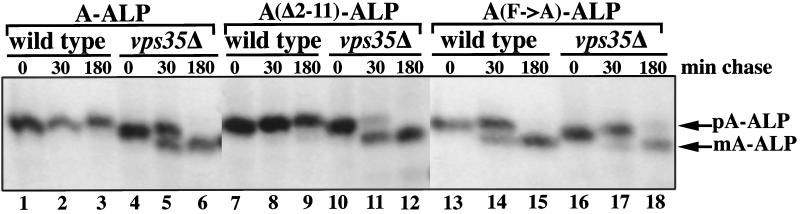
The observation that A-ALP is not transported to the vacuole via the plasma membrane in vps35 mutant cells is consistent with a role for Vps35p in retrieval of A-ALP from the prevacuolar compartment. If this is the case, a strain containing both a mutation in A-ALP that specifically prohibits retrieval and a vps35Δ mutation should have no stronger a retention defect than strains containing either mutation alone. To test this prediction, we analyzed the processing kinetics of A-ALP and a retrieval-defective A-ALP mutant, A(F85A; F87A)-ALP (Nothwehr et al., 1993; Bryant and Stevens, 1997), wild-type and vps35Δ strains (Figure 4). Consistent with a role for Vps35p in retrieval, the processing rate of A(F85A; F87A)-ALP in vps35Δ cells was no more severe than processing of A(F85A; F87A)-ALP in a wild-type strain or A-ALP in a vps35Δ strain and was, in fact, slightly less severe (compare lanes 16–18 with lanes 13–15 and 4–6).
Figure 4.
Phenotypes resulting from a loss of Vps35p function combined with a retrieval-defective, or static retention-defective, A-ALP mutant indicate a role for Vps35p in retrieval. Wild-type (SNY17) and vps35Δ (SNY79) strains carrying plasmids expressing A-ALP (pSN55; lanes 1–6), A(Δ2–11)-ALP (pAH49; lanes 7–12), or A(F85A; F87A)-ALP (pSN100; lanes 13–18) were propagated at 30°C, radioactively pulsed for 10 min and chased, and at the indicated times wild-type or mutant A-ALP was immunoprecipitated. Samples were analyzed using SDS-PAGE and fluorography.
Similarly, if Vps35p is involved in retrieval, then a strain containing both a mutation in A-ALP that specifically prohibits static retention and a vps35Δ mutation should exhibit a much more dramatic A-ALP retention defect than strains containing either mutation alone. We indeed observed that the processing rate of static retention-defective A(Δ2–11)-ALP (Bryant and Stevens, 1997) in a vps35Δ strain was much more rapid (half-time of 15–20 min) than in a wild-type strain that expressed A(Δ2–11)-ALP (half-time of >180 min) or a vps35Δ strain that expressed A-ALP (half-time of ∼40 min; compare lanes 10–12 with lanes 4–6 and 7–9). Taken together, these double-mutant analyses indicate that Vps35p is involved in retrieval of A-ALP from the prevacuolar compartment but is not required for static retention.
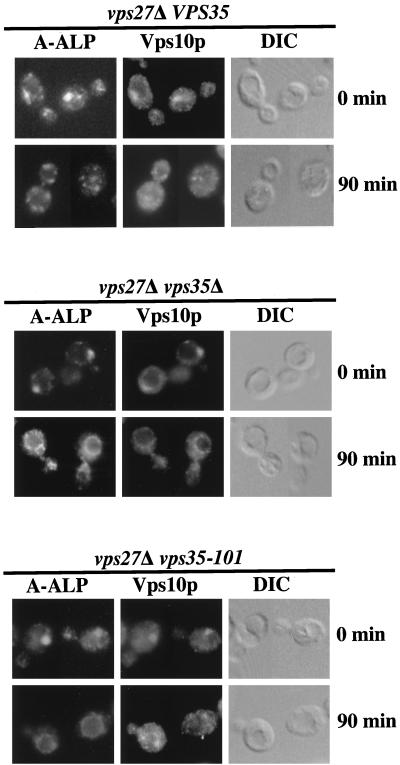
To extend these results we used a recently developed assay (Bryant and Stevens, 1997) to simultaneously analyze retrieval of both A-ALP and Vps10p from the prevacuolar compartment by indirect-immunofluorescence microscopy. Mutations in the VPS27 gene are known to cause both a block in the prevacuolar compartment to TGN-retrieval pathway and in the anterograde pathway between the prevacuolar compartment and the vacuole (Piper et al., 1995). Thus, in vps27 mutant cells, proteins such as A-ALP and Vps10p that normally cycle between the TGN and the prevacuolar compartment are trapped in an exaggerated prevacuolar compartment. Rapid induction of synthesis of Vps27p in a vps27Δ strain, using a plasmid containing the VPS27 gene under control of the galactose-inducible GAL1 promoter, allows retrieval of Vps10p and A-ALP from the prevacuolar compartment back to the TGN after 90 min of induction (Voos and Stevens, 1998). We therefore tested whether loss of Vps35p function would prevent the retrieval observed in a vps27Δ strain after induction of Vps27p synthesis.
Before induction of Vps27p synthesis (0 min time point), A-ALP and Vps10p are localized to one to two prominent, nonvacuolar structures per cell for both the vps27Δ VPS35 and vps27Δ vps35Δ strains (Figure 5), typical of the staining pattern for these antigens in vps27 mutants (Raymond et al., 1992; Piper et al., 1995). After 90 min of induction, both A-ALP and Vps10p in the vps27Δ VPS35 strain exhibit a punctate staining pattern, indicating that they have redistributed back to the TGN. We observed a less-than-complete overlap of the staining patterns of A-ALP and Vps10p, perhaps because of the fact that Vps10p cycles between the prevacuolar compartment and TGN much more frequently than A-ALP (Bryant and Stevens, 1997). This would suggest that a much greater fraction of Vps10p might be localized to the prevacuolar compartment in the steady state as compared with A-ALP. In contrast, in the strain lacking Vps35p function (vps27Δ vps35Δ), both A-ALP and Vps10p exhibited vacuolar membrane staining after 90 min of Vps27p induction (Figure 5; compare staining patterns with the crater-like appearance of vacuoles observed under differential–interference contrast [DIC] optics). These data indicate that, upon induction of Vps27p synthesis, retrieval of A-ALP and Vps10p was blocked because of the absence of Vps35p function, and instead they were transported from the prevacuolar compartment to the vacuole by default. In summary, these experiments indicate that Vps35p performs a function essential for retrieval of both A-ALP and Vps10p from the prevacuolar compartment.
Figure 5.
Vps35p is required for retrieval of A-ALP and Vps10p from the prevacuolar compartment to the Golgi. vps27Δ (AHY65), vps27Δ vps35Δ (AHY68), and vps27Δ vps35–101 (AHY69) strains containing CEN plasmids directing expression of A-ALP (pAH16) and containing the VPS27 gene under GAL1 promoter control (pHY5) were propagated in media containing raffinose as a carbon source. At the 0-min time point the cultures were adjusted to 2% galactose to induce expression of VPS27. After 0 and 90 min after galactose addition, cells were fixed, spheroplasted, and costained with antibodies against ALP and Vps10p. After subsequent treatment with fluorochrome-conjugated secondary antibodies, the cells were viewed by DIC optics and by epifluorescence through filters specific for FITC and Texas Red.
Distinct Domains in Vps35p Are Involved in Regulating Retrieval of A-ALP and Vps10p
In addition to the grd mutants reported to comprise 18 complementation groups (Nothwehr et al., 1996), we have recently characterized additional grd mutants. One of these mutant strains (SNY17–190) exhibited a substantial A-ALP-processing phenotype but showed no CPY-missorting defect. Genetic analysis revealed that the mutation responsible for the phenotype was contained within the GRD9/VPS35 gene. This was surprising because vps35 null mutants are severely defective for both CPY sorting (Paravicini et al., 1992) and retention of A-ALP (Figure 1). The discovery of a vps35 allele defective for A-ALP retention but fully functional for CPY sorting suggests that Vps35p may contain a structural feature necessary for retention of A-ALP distinct from a structural feature necessary for proper trafficking of the CPY receptor, Vps10p.
The identification of additional vps35 alleles with cargo-specific defects would further support the idea that Vps35p contains distinct structural features specific for retrieval of different cargo proteins. Mapping the corresponding mutations would provide insight about structure–function relationships. With these goals in mind, we screened a library of vps35Δ strains carrying CEN plasmids with randomly mutagenized VPS35 inserts. Mutants were tested for A-ALP retention defects using an ALP plate activity assay (Chapman and Munro, 1994a; Nothwehr et al., 1996) and for CPY secretion. No additional alleles defective for A-ALP retention and normal for CPY sorting were identified, but five new alleles were identified that exhibited CPY-sorting defects and little or no A-ALP–retention defect.
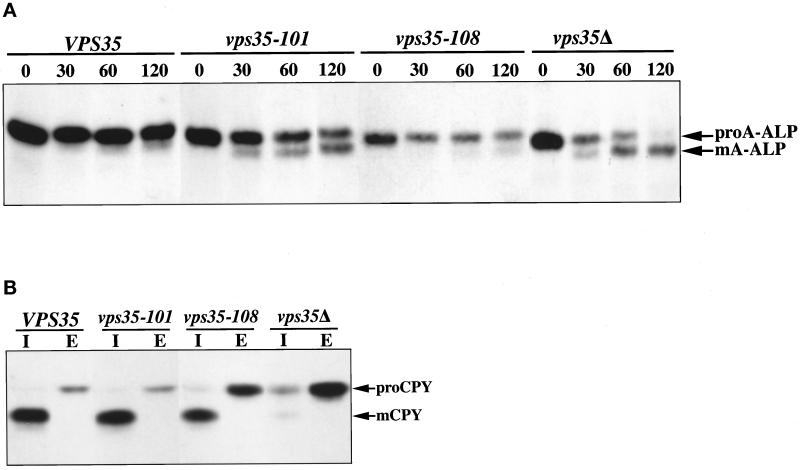
Yeast strains carrying the six new VPS35 alleles were pulsed with [35S]methionine/cysteine and chased for various time points, and A-ALP was immunoprecipitated to allow determination of the half-time of A-ALP processing. In addition, CPY was separately immunoprecipitated from the media and from the cells after a 45-min chase to determine the severity of CPY-sorting defects. Representative data for the vps35–101 and vps35–108 alleles are shown in Figure 6. The vps35–101 allele exhibits a pronounced A-ALP retention defect (Figure 6A) with a processing half-time of 115 min. CPY is correctly sorted to the vacuole in the vps35–101 strain, as indicated by the vast majority of CPY being found in the intracellular fraction in the mature form, similar to the results seen with the wild-type VPS35 strain (Figure 6B). In contrast, the vps35–108 allele exhibits near-normal retention of A-ALP with only a very minor amount of processing detected after a 120-min chase and an overall half-time exceeding 180 min (Figure 6A and Table 3). A substantial portion of the CPY in the vps35–108 strain was aberrantly secreted into the extracellular fraction as Golgi-modified pro-CPY (Figure 6B). Thus, these two alleles exhibit a high level of specificity toward the trafficking of the two cargo proteins, A-ALP and the CPY receptor Vps10p.
Figure 6.
The vps35–101 and vps35–108 alleles exhibit specific defects in A-ALP and Vps10p trafficking, respectively. The VPS35, vps35–101, vps35–108, and vps35Δ yeast strains correspond to the following strain/plasmid combinations, respectively: SNY79/pSN54/pLS13, SNY79/pSN54/pLS12, SNY79/pSN54/pPB7–010.4, and PBY3/pSN54 (please refer to Tables 1 and 2 for strain and plasmid information). (A) The strains were pulsed for 10 min, chased for the indicated times, and subjected to A-ALP immunoprecipitation as detailed in the legend of Figure 4. (B) The strains were pulsed with [35S]methionine/cysteine for 10 min, and then chased for 45 min with unlabeled amino acids. CPY was immunoprecipitated from intracellular (I) and extracellular (E) fractions and analyzed by SDS-PAGE and fluorography. The positions of precursor (proCPY) and mature (mCPY) forms of CPY are indicated.
Table 3.
Phenotypes of VPS35 alleles and nature of the responsible mutations
| Allele | A-ALP processing half-time (min) | % CPY secretion | Responsible mutation |
|---|---|---|---|
| VPS35 | >180 | 18 | Not applicable |
| vps35Δ | 65 | 74 | Deletion of ORF |
| vps35-101 | 115 | 16 | D123N |
| vps35-103 | >180 | 50 | L568P |
| vps35-104 | >180 | 50 | 247-498 |
| vps35-105 | >180 | 32 | D528G |
| vps35-107 | >180 | 40 | 733-935 (stop codon) |
| vps35-108 | >180 | 41 | L563M |
The half-time of A-ALP processing and the percent CPY secretion were determined from vps35Δ strains carrying the indicated VPS35 alleles on CEN plasmids (Table 1) as described in the legend to Figure 6 and in MATERIALS AND METHODS. The data shown for the percent CPY secretion is an average of data from two independent experiments. The values from each experiment for each strain differed by <5% secretion. Mapping of the mutations responsible for the phenotype of each allele was performed as described in MATERIALS AND METHODS. For the vps35-104 and vps35-107 alleles the mutation was narrowed down to the indicated region given in terms of codon number.
Experiments such as those shown in Figure 6 were also performed on the remaining four vps35 alleles, and the quantified results for all the alleles are shown in Table 3. The alleles fall into two groups: those having a specific defect in A-ALP retention (vps35–101) and those specifically defective for CPY sorting (vps35–103, 104, 105, 107, and 108). Although the vps35–101 allele has a somewhat less severe A-ALP retention defect than the vps35Δ allele, it exhibits the cleanest separation of the two phenotypes because it is indistinguishable from the VPS35 strain for CPY sorting.
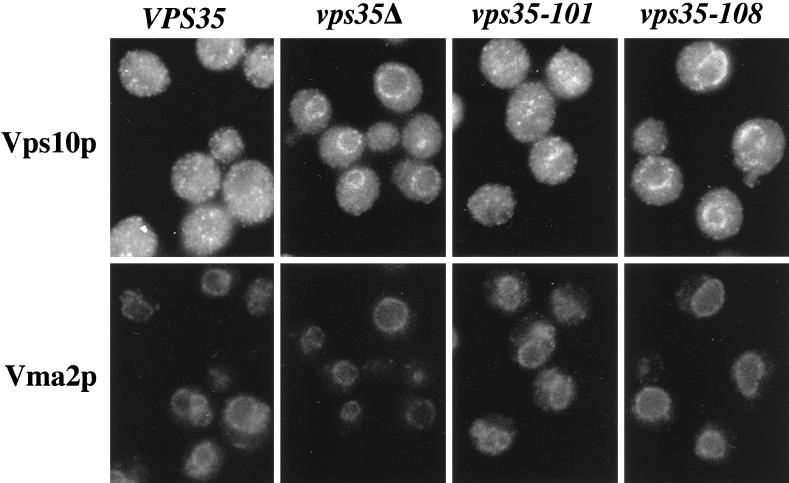
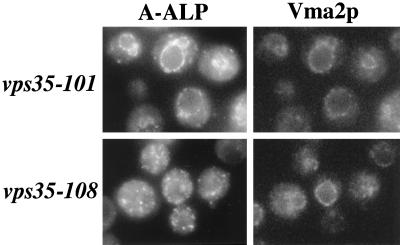
Additional experiments were performed to determine whether the phenotypic assays reflect an inability to retrieve either A-ALP or Vps10p from the prevacuolar compartment. Consistent with the CPY-sorting data in Figure 6B, the vps35–101 strain exhibited a normal Golgi-like punctate staining pattern for Vps10p, whereas the vps35Δ and vsp35–108 strains exhibited a predominantly vacuolar membrane-staining pattern for Vps10p (Figure 7; compare the staining patterns of the vacuolar membrane marker Vma2p with Vps10p). Since the data in Figure 5 demonstrate that Vps35p is required for retrieval of Vps10p, the data in Figure 7 strongly suggest that the vps35–108 allele is defective for retrieval of Vps10p. Moreover, A-ALP predominantly exhibited a punctate, Golgi-like staining pattern in vps35–108 cells, whereas A-ALP was clearly localized to the vacuole in vps35–101 cells (Figure 8). These results further emphasize the A-ALP retention defect in vps35–101 cells and near-normal A-ALP trafficking in vps35–108 cells.
Figure 7.
Vps10p is properly localized in a vps35–101 strain but is mislocalized to the vacuole in a vps35–108 strain. VPS35 (LSY6–2A/pLS13), vps35Δ (LSY6–2A), vps35–101 (LSY6–2A/pLS12), and vps35–108 (LSY6–2A/pPB7–010.4) strains were fixed, spheroplasted, and costained with antibodies against Vps10p and Vma2p. After subsequent treatment with fluorochrome-conjugated secondary antibodies, the cells were viewed by DIC optics and by epifluorescence through filters specific for FITC and Texas Red.
Figure 8.
A-ALP is mislocalized to the vacuole in a vps35–101 strain but exhibits Golgi staining in a vps35–108 strain. vps35–101 (SNY79/pLS12/pAH16) and vps35–108 (SNY79/pPB7–010.4/pAH16) strains were fixed, spheroplasted, and costained with antibodies against A-ALP and Vma2p. After treatment with fluorochrome-conjugated secondary antibodies, the cells were viewed by DIC optics and by epifluorescence through filters specific for FITC and Texas Red.
To test whether a vps35–101 strain is functional for retrieval of Vps10p and defective for A-ALP retrieval in a more direct manner, we analyzed a vps35–101 strain for its ability to retrieve these proteins in the VPS27 induction assay. After induction of VPS27 for 90 min, A-ALP has clearly been transported from the prevacuolar compartment to the vacuole, whereas Vps10p has instead been retrieved to punctate structures consistent with Golgi staining (Figure 5). Thus, the vps35–101 allele is defective for retrieval of A-ALP but not Vps10p.
Membrane Association of the Mutant Vps35 Proteins
If Vps35p associates with membranes as a member of the protein complex including cargo proteins such as Vps10p and A-ALP, it is possible that Vps35p mutants that fail to retrieve either Vps10p or A-ALP may exhibit altered membrane association relative to wild-type Vps35p. We therefore carried out subcellular fractionation by pulse labeling spheroplasts with [35S]methionine/cysteine for 30 min and chasing with unlabeled amino acids for 45 min. Lysates were centrifuged at 13,000 × g to generate a pellet (P13) and supernatant (S13). The S13 fraction was then centrifuged at 150,000 × g to generate a P150 fraction and a supernatant (S150) fraction containing soluble proteins. Immunoprecipitation of Vps35p and other marker proteins was then carried out from protein extracts made from the P13, P150, and S150 fractions. Given the fractionation of various markers in similar types of subcellular fractionation schemes performed previously (Marcusson et al., 1994; Nothwehr and Hindes, 1997), the P13 would be expected to contain vacuoles, ER, and plasma membrane, whereas the P150 would contain Golgi, endosomes, and vesicles. Accordingly, we observed under these conditions that ∼80% of the vacuolar membrane marker Vph1p fractionated in the P13, whereas ∼90% of the Golgi marker DPAP A fractionated in the P150 (our unpublished data).
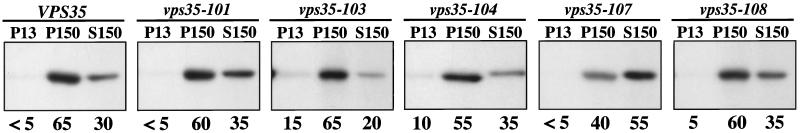
In the vps35Δ strain PBY1 carrying a CEN plasmid containing wild-type VPS35, most of the Vps35p (65%) was found in the P150 with 30% in the soluble S150 fraction (Figure 9). These results are similar to previously published fractionation data for Vps35p (Paravicini et al., 1992). Overall, the proteins expressed from the vps35 mutant alleles exhibited membrane association that was quite similar to wild-type Vps35p, with a few subtle differences. For example, a small-but-reproducible increase in the amount of Vps35p found in the P13 fraction was observed in the vps35–103 and vps35–104 strains compared with wild type. Vps35p was previously shown to fractionate in the P13 fraction containing vacuolar membranes under conditions in which Vps10p was mislocalized to the vacuole (Seaman et al., 1997). Interestingly, the vps35–103 and vps35–104 alleles show the most severe CPY-sorting defect (Table 3) and would be expected to have the most severe mislocalization of Vps10p to the vacuole. In addition, one of the alleles, vps35–107, encodes Vps35p with somewhat reduced membrane association. Nevertheless, the general trend that mutant Vps35 proteins exhibit near-normal membrane association is consistent with the view that Vps35p membrane association does not depend solely on interaction with the cytosolic domains of cargo proteins.
Figure 9.
Membrane association of wild-type and mutant Vps35 proteins. Spheroplasts from a vps35Δ strain (PBY1) carrying plasmids encoding either the wild-type (pLS13), vps35–101 (pLS12), vps35–103 (pPB7–9.7), vps35–104 (pPB7–10.1), vps35–107 (pPB7–108.5), or vps35–108 (pPB7–010.4) alleles were pulsed for 45 min with [35S]methionine/cysteine and chased for 15 min with unlabeled amino acids. The spheroplasts were then lysed, and lysates were centrifuged at 13,000 × g to generate a pellet fraction (P13). The supernatant was then centrifuged at 150,000 × g to generate pellet (P150) and supernatant (S150) fractions. Wild-type and mutant Vps35p were immunoprecipitated from each fraction and analyzed by SDS-PAGE and fluorography. The relative percentage of Vps35p present in each fraction as determined by Phosphorimager analysis is indicated below each panel.
Mapping of Mutations Responsible for Cargo-specific Phenotypes
Homology searches using the S. cerevisiae Vps35p sequence have identified protein sequences in Caenorhabditis elegans, Mus musculus, and Homo sapiens that exhibit a high degree of similarity to the yeast sequence. Comparison of the sequences allowed division of the 937-residue yeast Vps35p sequence into three domains based on extent of identity (Seaman et al., 1997). Domain I lies between residues 1 and 352 and exhibits the most conservation (43–46% identity), whereas domains II and III lie between 353 and 660 and 661 and 937 and exhibit 20–22% and 23–35% homology, respectively.
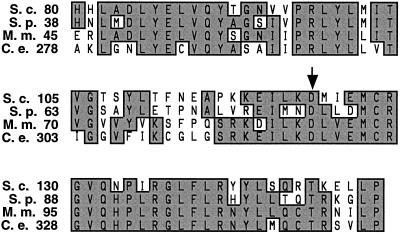
Mapping of the mutations responsible for the phenotype in the mutant alleles was carried out to provide information regarding the position of cargo-specific motifs within the Vps35p polypeptide sequence (Table 3). The A-ALP–retrieval defect of the vps35–101 allele was caused by a point mutation that changed the Asp codon at position 123 to Asn. Interestingly, Asp123 lies within the most highly conserved region of domain I and is itself conserved between S. cerevisiae Vps35p and S. pombe, M. musculus, and C. elegans homologues (Figure 10), consistent with this region being necessary for a function common to all four of the proteins. Three of the five alleles specific for Vps10p-retrieval defects have been mapped to single residue changes, whereas the others have been mapped to intervals of ≤251 amino acids. In contrast to the vps35–101 allele, all five of the Vps10p-specific alleles roughly map within the C-terminal two-thirds of the ORF. The mutations responsible for the phenotype in the vps35–103, vps35–105, and vps35–108 alleles map to residues in regions that exhibit little conservation with the homologues from other organisms (Table 3 and our unpublished data), suggesting that these regions may mediate a function specific to yeast Vps35p. Likewise, the other two alleles map to intervals that generally exhibit weak similarity among the Vps35p homologues. Taken together, these data indicate that 1) a conserved structural motif within domain I of the protein is necessary for A-ALP retrieval but is expendable for Vps10p retrieval and 2) one or more motifs in domains II/III are specifically necessary for efficient Vps10p retrieval.
Figure 10.
The mutation responsible for the vps35–101 phenotype eliminates a conserved Asp residue. A comparison of the most conserved region of the S. cerevisiae Vps35p (S.c.) sequence with its S. pombe (S.p.), M. musculus (M.m.), and C. elegans (C.e.) homologues is shown. The numbers refer to the amino acid residue number of each sequence. The shaded boxes indicate the most highly conserved residues. The arrowhead indicates the conserved D123 residue that was mutated to N in the vps35–101 allele.
DISCUSSION
Several membrane proteins are transported from the TGN to the prevacuolar/endosomal compartment of yeast. A subset of these, such as Vps10p and DPAP A, are then retrieved back from the prevacuolar compartment to the TGN, whereas others, such as carboxypeptidase S (Cowles et al., 1997), are transported by default from the prevacuolar compartment to the vacuole. These results imply the existence of a cargo-sorting machinery at the prevacuolar compartment. In this article we characterize the role of Vps35p in retrieval of two cargo proteins, A-ALP and Vps10p. The data strongly suggest that Vps35p plays a role at the prevacuolar compartment in selection of cargo to be transported to the TGN via a retrograde pathway.
A Role for Vps35p in Retrieval of Both A-ALP and Vps10p from the Prevacuolar Compartment
Previous work has supported the proposal that Vps35p is involved in retrieval of Vps10p from the prevacuolar compartment. A loss of Vps35p function caused Vps10p to be mislocalized to the vacuolar membrane in a manner independent of late-secretory-pathway functions (Seaman et al., 1997). However, the delivery of Vps10p to the vacuole in vps35 mutants was reduced by mutations in PEP12, which encodes a late endosomal, prevacuolar compartment t-SNARE (Becherer et al., 1996).
Our present data provide additional evidence that Vps35p is required for retrieval of Vps10p, a protein that frequently cycles between the TGN and prevacuolar compartment as part of its role in CPY sorting. In addition, we present the first direct evidence that Vps35p is required for retrieval of a resident TGN enzyme (A-ALP). Inactivation of Vps35p using a rapid-onset, temperature-sensitive mutant showed that the A-ALP-retention defect was exhibited very rapidly after shifting to the nonpermissive temperature. This result argues against an indirect role for Vps35p in retention of A-ALP. A-ALP was transported to the vacuole in vps35Δ and vps35-ts mutant cells with a half-time of ∼60 min, which is about the same rate as transport of an A-ALP mutant lacking its retrieval signal (Nothwehr et al., 1993). In contrast to its transport in vps1 mutant cells, A-ALP is transported to the vacuole in vps35 cells in a manner independent of the early endocytic pathway, consistent with a TGN-to-prevacuolar compartment-to-vacuole route.
A-ALP employs two independent mechanisms to maintain localization to the TGN: a mechanism for reducing its rate of exit from the TGN and a retrieval mechanism from the prevacuolar compartment (Bryant and Stevens, 1997). We took advantage of A-ALP mutants specifically defective in either static retention or retrieval to determine which mechanism depends on Vps35p function. By assessing the phenotypes of combining the A-ALP mutations with the vps35Δ mutation, only the retrieval mechanism that utilizes the FXFXD signal on A-ALP was shown to depend on Vps35p function. Thus, Vps35p and the FXFXD signal must act at the same step to mediate retrieval from the prevacuolar compartment.
An immunofluorescence microscopy assay capable of assessing transport of membrane proteins out of an exaggerated prevacuolar compartment was used to further assess the role of Vps35p in trafficking of Vps10p and A-ALP. Both proteins were retrieved in cells expressing wild-type Vps35p, but A-ALP and Vps10p were transported to the vacuole by default in cells lacking Vps35p. The inability of Vps10p and A-ALP to be retrieved in this assay provides additional evidence that Vps35p is involved in the retrieval pathway.
Although Vps10p and resident TGN enzymes, such as DPAP A (A-ALP) and Kex2p, are generally thought to be retrieved from the same late endosomal, prevacuolar compartment, it has recently been suggested that this may not be the case. Holthuis et al. (1998) recently characterized two new t-SNARE family members, Tlg1p and Tlg2p, that appear to be localized to a putative early endosome and the TGN, respectively. In tlg1Δ and tlg2Δ mutants, Kex2p and DPAP A are destabilized in a manner dependent on vacuolar proteases. In addition, both Tlg1p and Tlg2p interact with Vti1p, a v-SNARE thought to target TGN-derived vesicles to the recycling compartment as well as to mediate other vesicle-targeting events in the endomembrane system (Fischer von Mollard et al., 1997). Little or no CPY-sorting defects were observed in tlg1Δ and tlg2Δ mutants (Holthuis et al., 1998). This observation, taken together with the above mentioned data for Tlg1p, led to the proposal that Kex2p and DPAP A cycle between the TGN and a Tlg1p-containing early endosome, whereas Vps10p cycles to and from the prevacuolar endosome (Holthuis et al., 1998). However, an assignment of Tlg1p localization to the early endosome must be taken as tenuous since in yeast this compartment is not well characterized, and no membrane protein markers are available.
Other studies are more consistent with the idea that DPAP A (A-ALP), Kex2p, and Vps10p are all recycled from the same prevacuolar compartment. For example, Vps10p, Kex2p, and A-ALP have been shown to be trapped in an exaggerated prevacuolar endosome in class E vps mutants (Raymond et al., 1992; Cereghino et al., 1995; Piper et al., 1995), strongly suggesting that they are transported to and from this compartment in wild-type cells. In addition, a more recent analysis of CPY sorting using an immunoprecipitation strategy indicated that tlg2Δ cells missort 20% of their CPY (Abeliovich et al., 1998) contrary to previous results (Holthuis et al., 1998). This raises the question of whether Tlg1p may also be involved in CPY sorting and, by extension, trafficking of Vps10p. Clearly, additional characterization of Tlg1p and Tlg2p should help resolve these issues.
vps35 Mutant Alleles Exhibit Cargo-specific Defects in Retrieval
Because a complete loss of Vps35p function results in strong defects in retrieval of both Vps10p and A-ALP, we were surprised to find an allele, vps35–101, that has a cargo-specific defect. This allele exhibited entirely-normal CPY sorting and Vps10p retrieval but had a substantial defect in retrieval of A-ALP. Random mutagenesis of VPS35 led to identification of several other alleles that exhibited the opposite type of phenotype: a defect in Vps10p retrieval and near-normal function for A-ALP retrieval.
If Vps35p were to function in the retrieval pathway downstream of the cargo-sorting step, then all mutations that alter its activity would be expected to have the same effect on different cargo proteins. The discovery of cargo-specific alleles is more consistent with Vps35p having a direct role in sorting of the cargo proteins. We propose that Vps35p may directly interact with the cytosolic domains of DPAP A and Vps10p. The discovery of cargo-specific alleles would reflect the existence of distinct structural features in Vps35p necessary for interaction with certain cargo proteins but expendable for others. The mapping data suggest that an N-terminal domain in Vps35p is specifically involved with DPAP A interaction, whereas Vps10p-specific structural features reside in the C-terminal two-thirds of the protein.
Previous data also support the idea that Vps35p associates with the cytosolic domains of cargo proteins retrieved from the prevacuolar compartment. Seaman et al. (1997) localized both Vps10p and Vps35p by subcellular fractionation in wild-type and vps29 mutant cells. In wild-type cells, Vps10p and Vps35p fractionated with endosomal and Golgi membranes, but in vps29 cells both proteins were present in a fraction containing vacuolar membranes. These data led to the suggestion that in the absence of Vps29p function, retrieval of Vps10p was blocked, and a complex containing Vps10p and Vps35p was mislocalized to the vacuolar membrane (Seaman et al., 1997).
An alternative, but more complex, model to explain the existence of the cargo-specific alleles would propose that distinct protein complexes associate with the cytosolic domains of each cargo protein. The complexes would be involved in sorting of the cargo proteins into newly forming vesicles at the prevacuolar compartment. Each protein complex would have subunits in common such as Vps35p and Vps29p and other subunits not shared between the complexes. The existence of cargo-specific Vps35p alleles could then be explained by the use of distinct structural features that mediate association with each complex, the nature of which would vary somewhat between cargo proteins. In a related model, Vps35p could act as a scaffold that interacts with different cargo-recognizing proteins such as Grd19p (see below). Such an interaction with Vps35p would mediate binding of a given cargo-recognizing protein with the cytosolic domain of the appropriate cargo protein. In this model, distinct domains in Vps35p could be used for interaction with different cargo-recognizing proteins. The recent characterization of Grd19p (Voos and Stevens, 1998), a PX domain-containing protein, is consistent with the idea that the protein machinery that associates with the cytosolic domains of Vps10p and DPAP A is not identical. Grd19p has been shown to exhibit a very mild defect in Vps10p retrieval while exhibiting a strong defect in A-ALP retrieval. Grd19p has been observed to interact with the cytosolic domain of DPAP A under certain in vitro conditions, although it is not known whether the two proteins interact in vivo. Under the same in vitro conditions, no interaction of Grd19p with Kex2p or Vps10p was observed. Thus Grd19p may represent a component of the sorting machinery specific to DPAP A.
New insight into the vesicle formation step of prevacuolar compartment-to-TGN retrieval has been gained by characterization of a protein complex termed the “retromer” (Seaman et al., 1998). A subcomplex containing Pep8p, Vps29p, and Vps35p was found to associate with another subcomplex containing Vps5p and Vps17p at the prevacuolar compartment membrane. Based primarily on cell fractionation and immunoelectron microscopy data, the complete retromer complex has been proposed to function as a vesicle coat and have a role in cargo selection. Interestingly, pep8, vps29, and vps35 mutants exhibit little or no vacuolar morphology defect, whereas vps5 and vps17 exhibit highly fragmented vacuoles (Raymond et al., 1992; Köhrer and Emr, 1993; Horazdovsky et al., 1997; Nothwehr and Hindes, 1997). Based primarily on this observation and on the severity of vacuolar hydrolase missorting, it has been proposed that Vps5p and Vps17p are part of general machinery for retrieval. Our results with Vps35p are thus consistent with the proposal that the Pep8p/Vps29p/Vps35p subcomplex is involved in sorting of certain cargo, whereas Vps5p/Vps17p serves a structural role in vesicle formation (Seaman et al., 1998).
In accordance with the role of vesicle coats in other transport steps (Rothman and Wieland, 1996; Schmid, 1997), the coat present on retrograde vesicles originating from the yeast prevacuolar compartment would probably be shed shortly after vesicle budding and before fusion with the TGN. Thus, the association between Vps35p and the cytosolic domains of cargo proteins would be transient in nature and relatively unstable. Indeed, our efforts at detecting interactions between Vps35p and the cytosolic domains of cargo proteins using biochemical approaches have been unsuccessful to date. However, novel alleles, such as vps35–101, that have subtle structural alterations resulting in cargo-specific defects may provide tools for detecting such interactions by using a genetic approach.
ACKNOWLEDGMENTS
We acknowledge the excellent technical assistance of Anna Hindes throughout this work and Mona Aujla for her role in generating the vps35 temperature-sensitive mutant. Antibodies against Vps35p and plasmid pGPY55 were gifts of Scott Emr, who we also thank for fruitful discussions regarding this work. Finally, we thank Liz Conibear and Steve Alexander for comments on the manuscript. This work was supported by a grant from the Howard Hughes Medical Institute awarded to the University of Missouri, Columbia, for undergraduate research; a grant awarded to L.A.S. from the Tri β Foundation, a grant awarded to L.A.S. from the University of Missouri Arts and Sciences undergraduate research program; and a grant awarded to S.F.N. from the National Institutes of Health (GM-53449).
Abbreviations used:
- CEN
yeast centromere
- CPY
carboxypeptidase Y
- DPAP
dipeptidyl aminopeptidase
- ER
endoplasmic reticulum
- ORF
open reading frame
- TGN
trans-Golgi network
REFERENCES
- Abeliovich H, Grote E, Novick P, Ferronovick S. Tlg2p, a yeast syntaxin homolog that resides on the Golgi and endocytic structures. J Biol Chem. 1998;273:11719–11727. doi: 10.1074/jbc.273.19.11719. [DOI] [PubMed] [Google Scholar]
- Becherer KA, Rieder SE, Emr SD, Jones EW. Novel syntaxin homologue, Pep12p, required for the sorting of lumenal hydrolases to the lysosome-like vacuole in yeast. Mol Biol Cell. 1996;7:579–594. doi: 10.1091/mbc.7.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarek SY, Ravazzola M, Hosobuchi M, Amherdt M, Perrelet A, Schekman R, Orci L. COPI- and COPII-coated vesicles bud directly from the endoplasmic reticulum in yeast. Cell. 1995;83:1183–1196. doi: 10.1016/0092-8674(95)90144-2. [DOI] [PubMed] [Google Scholar]
- Bénédetti H, Raths S, Crausaz F, Riezman H. The END3 gene encodes a protein that is required for the internalization step of endocytosis and for actin cytoskeleton organization in yeast. Mol Biol Cell. 1994;5:1023–1037. doi: 10.1091/mbc.5.9.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoroorotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Brickner JH, Fuller RS. SOI1 encodes a novel, conserved protein that promotes TGN-endosomal cycling of Kex2p and other membrane proteins by modulating the function of two TGN localization signals. J Cell Biol. 1997;139:23–26. doi: 10.1083/jcb.139.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant NJ, Stevens TH. Two separate signals act independently to localize a yeast late Golgi membrane protein through a combination of retrieval and retention. J Cell Biol. 1997;136:287–297. doi: 10.1083/jcb.136.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell RC, Joyce GG. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- Cereghino JL, Marcusson EG, Emr SD. The cytoplasmic tail domain of the vacuolar protein sorting receptor Vps10p and a subset of VPS gene products regulate receptor stability, function, and localization. Mol Biol Cell. 1995;6:1089–1102. doi: 10.1091/mbc.6.9.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman RE, Munro S. The functioning of the yeast Golgi apparatus requires an ER protein encoded by ANP1, a member of a new family of genes affecting the secretory pathway. EMBO J. 1994a;13:4896–4907. doi: 10.1002/j.1460-2075.1994.tb06817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman RE, Munro S. Retrieval of TGN proteins from the cell surface requires endosomal acidification. EMBO J. 1994b;13:2305–2312. doi: 10.1002/j.1460-2075.1994.tb06514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A, Bussey H. Yeast Kex1p is a Golgi-associated membrane protein: deletions in a cytoplasmic targeting domain result in mislocalization to the vacuolar membrane. J Cell Biol. 1992;119:1459–1468. doi: 10.1083/jcb.119.6.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper AA, Stevens TH. Vps10p cycles between the late-Golgi and prevacuolar compartments in its function as the sorting receptor for multiple yeast vacuolar hydrolases. J Cell Biol. 1996;133:529–541. doi: 10.1083/jcb.133.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles CR, Snyder WB, Burd CG, Emr SD. Novel Golgi to vacuole delivery pathway in yeast — identification of a sorting determinant and required transport component. EMBO J. 1997;16:2769–2782. doi: 10.1093/emboj/16.10.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean N, Pelham HR. Recycling of proteins from the Golgi compartment to the ER in yeast. J Cell Biol. 1990;111:369–377. doi: 10.1083/jcb.111.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz E, Pfeffer SR. TIP47 — a cargo selection device for mannose 6-phosphate receptor trafficking. Cell. 1998;93:433–443. doi: 10.1016/s0092-8674(00)81171-x. [DOI] [PubMed] [Google Scholar]
- Duncan JR, Kornfeld S. Intracellular movement of two mannose 6-phosphate receptors: return to the Golgi apparatus. J Cell Biol. 1988;106:617–628. doi: 10.1083/jcb.106.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer von Mollard G, Nothwehr SF, Stevens TH. The yeast v-SNARE Vti1p mediates two vesicle transport pathways through interactions with the t-SNAREs Sed5p and Pep12p. J Cell Biol. 1997;137:1511–1524. doi: 10.1083/jcb.137.7.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller RS, Sterne RE, Thorner J. Enzymes required for yeast prohormone processing. Annu Rev Physiol. 1988;50:345–362. doi: 10.1146/annurev.ph.50.030188.002021. [DOI] [PubMed] [Google Scholar]
- Gaynor EC, te Heesen S, Graham TR, Aebi M, Emr SD. Signal-mediated retrieval of a membrane protein from the Golgi to the ER in yeast. J Cell Biol. 1994;127:653–665. doi: 10.1083/jcb.127.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holthuis JCM, Nichols BJ, Dhruvakumar S, Pelham HRB. Two syntaxin homologues in the TGN/endosomal system of yeast. EMBO J. 1998;17:113–126. doi: 10.1093/emboj/17.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horazdovsky BF, Davies BA, Seaman MNJ, McLaughlin SA, Yoon S, Emr SD. A sorting nexin-1 homologue, Vps5p, forms a complex with Vps17p and is required for recycling the vacuolar protein-sorting receptor. Mol Biol Cell. 1997;8:1529–1541. doi: 10.1091/mbc.8.8.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JS, Prakash L. Yeast Saccharomyces cerevisiae selectable markers in pUC18 polylinkers. Yeast. 1990;6:363–366. doi: 10.1002/yea.320060502. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T, Bonifacino JS, Riezman H. Linking cargo to vesicle formation — receptor tail interactions with coat proteins. Curr Opin Cell Biol. 1997;9:488–495. doi: 10.1016/s0955-0674(97)80024-5. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. Membrane protein sorting: biosynthesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO J. 1989;8:2241–2250. doi: 10.1002/j.1460-2075.1989.tb08348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhrer K, Emr SD. The yeast VPS17 gene encodes a membrane-associated protein required for the sorting of soluble vacuolar hydrolases. J Biol Chem. 1993;268:559–569. [PubMed] [Google Scholar]
- Kornfeld S. Structure and function of the mannose 6-phosphate/insulin-like growth factor II receptors. Annu Rev Biochem. 1992;61:307–330. doi: 10.1146/annurev.bi.61.070192.001515. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Marcusson EG, Horazdovsky BF, Cereghino JL, Gharakhanian E, Emr SD. The sorting receptor for yeast vacuolar carboxypeptidase Y is encoded by the VPS10 gene. Cell. 1994;77:579–586. doi: 10.1016/0092-8674(94)90219-4. [DOI] [PubMed] [Google Scholar]
- Molloy SS, Thomas L, Vanslyke JK, Stenberg PE, Thomas G. Intracellular trafficking and activation of the furin proprotein convertase — localization to the TGN and recycling from the cell surface. EMBO J. 1994;13:18–33. doi: 10.1002/j.1460-2075.1994.tb06231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Hoe MH, Slusarewicz P, Rabouille C, Watson R, Hunte F, Watzele G, Berger EG, Warren G. Kin recognition between medial Golgi enzymes in HeLa cells. EMBO J. 1994;13:562–574. doi: 10.1002/j.1460-2075.1994.tb06294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Bryant NJ, Stevens TH. The newly identified yeast GRD genes are required for retention of late-Golgi membrane proteins. Mol Cell Biol. 1996;16:2700–2707. doi: 10.1128/mcb.16.6.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Conibear E, Stevens TH. Golgi and vacuolar membrane proteins reach the vacuole in vps1 mutant yeast cells via the plasma membrane. J Cell Biol. 1995;129:35–46. doi: 10.1083/jcb.129.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Hindes AE. The yeast VPS5/GRD2 gene encodes a sorting nexin-1-like protein required for localizing membrane proteins to the late Golgi. J Cell Sci. 1997;110:1063–1072. doi: 10.1242/jcs.110.9.1063. [DOI] [PubMed] [Google Scholar]
- Nothwehr SF, Roberts CJ, Stevens TH. Membrane protein retention in the yeast Golgi apparatus: dipeptidyl aminopeptidase A is retained by a cytoplasmic signal containing aromatic residues. J Cell Biol. 1993;121:1197–1209. doi: 10.1083/jcb.121.6.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paravicini G, Horazdovsky BF, Emr SD. Alternative pathways for the sorting of soluble vacuolar proteins in yeast: a vps35 null mutant missorts and secretes only a subset of vacuolar hydrolases. Mol Biol Cell. 1992;3:415–427. doi: 10.1091/mbc.3.4.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham HR, Munro S. Sorting of membrane proteins in the secretory pathway. Cell. 1993;75:603–605. doi: 10.1016/0092-8674(93)90479-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper RC, Cooper AA, Yang H, Stevens TH. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J Cell Biol. 1995;3:603–617. doi: 10.1083/jcb.131.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol Biol Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding K, Holcomb C, Fuller RS. Immunolocalization of Kex2 protease identifies a putative late Golgi compartment in the yeast Saccharomyces cerevisiae. J Cell Biol. 1991;113:527–538. doi: 10.1083/jcb.113.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexach MF, Latterich M, Schekman RW. Characteristics of endoplasmic reticulum-derived transport vesicles. J Cell Biol. 1994;126:1133–1148. doi: 10.1083/jcb.126.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riederer MA, Soldati T, Shapiro AD, Lin J, Pfeffer SR. Lysosome biogenesis requires Rab9 function and receptor recycling from endosomes to the trans-Golgi network. J Cell Biol. 1994;125:573–582. doi: 10.1083/jcb.125.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CJ, Nothwehr SF, Stevens TH. Membrane protein sorting in the yeast secretory pathway: evidence that the vacuole may be the default compartment. J Cell Biol. 1992;119:69–83. doi: 10.1083/jcb.119.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CJ, Raymond CK, Yamashiro CT, Stevens TH. Methods for studying the yeast vacuole. Methods Enzymol. 1991;194:644–661. doi: 10.1016/0076-6879(91)94047-g. [DOI] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Novick P, Thomas JH, Botstein D, Fink GR. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Rothman JE, Wieland FT. Protein sorting by transport vesicles. Science. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- Rothman JH, Stevens TH. Protein sorting in yeast: mutants defective in vacuole biogenesis mislocalize vacuolar proteins into the late secretory pathway. Cell. 1986;47:1041–1051. doi: 10.1016/0092-8674(86)90819-6. [DOI] [PubMed] [Google Scholar]
- Schäfer W, Stroh A, Berghofer S, Seiler J, Vey M, Kruse ML, Kern HF, Klenk HD, Garten W. Two independent targeting signals in the cytoplasmic domain determine trans-Golgi network localization and endosomal trafficking of the the proprotein convertase furin. EMBO J. 1995;14:2424–2435. doi: 10.1002/j.1460-2075.1995.tb07240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid SL. Clathrin-coated vesicle formation and protein sorting — an integrated process. Annu Rev Biochem. 1997;66:511–548. doi: 10.1146/annurev.biochem.66.1.511. [DOI] [PubMed] [Google Scholar]
- Seaman MNJ, Marcusson EG, Cereghino JL, Emr SD. Endosome to Golgi retrieval of the vacuolar protein sorting receptor, Vps10p, requires the function of the Vps29, Vps30, and Vps35 gene products. J Cell Biol. 1997;137:79–92. doi: 10.1083/jcb.137.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MNJ, McCaffery JM, Emr SD. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J Cell Biol. 1998;142:665–681. doi: 10.1083/jcb.142.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanely KK, Howell KE. TGN38/41: a molecule on the move. Trends Cell Biol. 1993;3:252–255. doi: 10.1016/0962-8924(93)90046-4. [DOI] [PubMed] [Google Scholar]
- Takahashi S, Nakagawa T, Banno T, Watanabe T, Murakami K, Nakayama K. Localization of furin to the trans-Golgi network and recycling from the cell surface involves Ser and Tyr residues in the cytoplasmic domain. J Biol Chem. 1995;270:28397–28401. doi: 10.1074/jbc.270.47.28397. [DOI] [PubMed] [Google Scholar]
- Townsley FM, Frigerio G, Pelham HR. Retrieval of HDEL proteins is required for growth of yeast cells. J Cell Biol. 1994;127:21–28. doi: 10.1083/jcb.127.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vater CA, Raymond CK, Ekena K, Howald SI, Stevens TH. The VPS1 protein, a homolog of dynamin required for vacuolar protein sorting in Saccharomyces cerevisiae, is a GTPase with two functionally separable domains. J Cell Biol. 1992;119:773–786. doi: 10.1083/jcb.119.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees P, Deignan E, van Donselaar E, Humphrey J, Marks MS, Peters PJ, Bonifacino JS. An acidic sequence within the cytoplasmic domain of furin functions as a determinant of trans-Golgi network localization and internalization from the cell surface. EMBO J. 1995;14:4961–4975. doi: 10.1002/j.1460-2075.1995.tb00179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voos W, Stevens TH. Retrieval of resident late-Golgi membrane proteins from the prevacuolar compartment of Saccharomyces cerevisiae is dependent on the function of Grd19p. J Cell Biol. 1998;140:577–590. doi: 10.1083/jcb.140.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Molloy SS, Thomas L, Liu GP, Xiang Y, Rybak SL, Thomas G. PACS-1 defines a novel gene family of cytosolic sorting proteins required for trans-Golgi network localization. Cell. 1998;94:205–216. doi: 10.1016/s0092-8674(00)81420-8. [DOI] [PubMed] [Google Scholar]
- Weisz OA, Swift AM, Machamer CE. Oligomerization of a membrane protein correlates with its retention in the Golgi complex. J Cell Biol. 1993;122:1185–1196. doi: 10.1083/jcb.122.6.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox CA, Redding K, Wright R, Fuller RS. Mutation of a tyrosine localization signal in the cytosolic tail of yeast Kex2 protease disrupts Golgi retention and results in default transport to the vacuole. Mol Biol Cell. 1992;3:1353–1371. doi: 10.1091/mbc.3.12.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]