Abstract
Mob1p is an essential Saccharomyces cerevisiae protein, identified from a two-hybrid screen, that binds Mps1p, a protein kinase essential for spindle pole body duplication and mitotic checkpoint regulation. Mob1p contains no known structural motifs; however MOB1 is a member of a conserved gene family and shares sequence similarity with a nonessential yeast gene, MOB2. Mob1p is a phosphoprotein in vivo and a substrate for the Mps1p kinase in vitro. Conditional alleles of MOB1 cause a late nuclear division arrest at restrictive temperature. MOB1 exhibits genetic interaction with three other yeast genes required for the completion of mitosis, LTE1, CDC5, and CDC15 (the latter two encode essential protein kinases). Most haploid mutant mob1 strains also display a complete increase in ploidy at permissive temperature. The mechanism for the increase in ploidy may occur through MPS1 function. One mob1 strain, which maintains stable haploidy at both permissive and restrictive temperature, diploidizes at permissive temperature when combined with the mps1–1 mutation. Strains containing mob2Δ also display a complete increase in ploidy when combined with the mps1-1 mutation. Perhaps in addition to, or as part of, its essential function in late mitosis, MOB1 is required for a cell cycle reset function necessary for the initiation of the spindle pole body duplication.
INTRODUCTION
Proper cell division requires precise coordination and execution of several events in the cell cycle, including centrosome duplication, DNA replication, mitotic spindle assembly, chromosome segregation, and cytokinesis. A failure in the execution or proper timing of any of these events could lead to chromosome segregation defects resulting in aneuploidy or polyploidy. Such genomic instability is the hallmark of transformed cells but has also been observed in various mutant strains of yeast (Tlsty et al., 1995; Romanowski and Madine, 1996; Stillman, 1996). Analysis of mutant yeast strains that give rise to aneuploid or polyploid cells has led to an understanding of some of the mechanisms that cause genomic instability.
Endomitosis, DNA replication in the absence of karyokinesis and cytokinesis, is one mechanism that causes an increase in ploidy to yield polyploid cells. In some cells, such as developing endosperm of plant seeds (Grafi and Larkins, 1995) or mammalian megakaryocytes (Jackson, 1992), endomitosis is normal and appears to require the inhibition of M phase-promoting kinases and the activation of S phase-promoting kinases (Grafi and Larkins, 1995; Datta et al., 1996). In rapidly dividing cells such as yeast, overreplication can be induced by a number of conditions that decrease G2 cyclin/p34cdc2/Cdc28 activity or keep the levels of certain components of the DNA replication complex high (Chevalier and Blow, 1996; Romanowski and Madine, 1996). This suggests that the mechanism preventing rereplication of DNA is dependent on G2 cyclin/p34cdc2/Cdc28 activity. It is thought that in normal cells, the decrease in G2 cyclin/p34cdc2/Cdc28 activity at the end of mitosis stimulates the reassembly of a prereplicative complexes on DNA origins, thereby rendering the DNA competent for replication (Romanowski and Madine, 1996). In Saccharomyces cerevisiae, the assembly of prereplicative complexes and origin recognition complexes requires the activity of CDC14 and CDC5, both of which also have roles in the completion of mitosis (Hardy, 1996; Hardy and Pautz, 1996; Kroll et al., 1996).
A second class of defects that can give rise to an increase in ploidy are those that affect processes required for chromosome segregation such as centrosome duplication, kinetochore attachment and mitotic motor function, among others. Mutations causing defects in each of these processes can be found in yeast. Failure in proper spindle pole body (SPB, the yeast centrosome equivalent) duplication can lead to the generation of monopolar or multipolar spindles, both of which can give rise to aneuploid or polyploid cells (Schild et al., 1981; Thomas and Botstein, 1986; Rose and Fink, 1987; Baum et al., 1988; Levine et al., 1991; Winey et al., 1991). Improper kinetochore attachment, if left uncorrected, leads to aneuploidy (Doheny et al., 1993; Goh and Kilmartin, 1993). Defects in mitotic motors responsible for chromosome segregation or nuclear migration may also result in aneuploidy, polyploidy, or, in some cases, multinucleate cells (Hoyt et al., 1993; Saunders et al., 1995; Yeh et al., 1995). This is also true for defects in proteins that affect microtubule function or integrity (Hoyt and Geiser, 1997). One yeast gene required for the maintenance of ploidy is IPL1, which encodes an essential protein kinase that is required, in some way, for mitotic spindle function (Chan and Botstein, 1993; Francisco et al., 1994). Strains defective in this kinase become aneuploid.
Checkpoints are an important line of defense against genomic instability. They are signaling pathways that elicit temporary delays in cell cycle progression in response to a defect in DNA integrity, DNA synthesis, spindle function, or other cellular functions (for review see Elledge, 1996; Paulovich et al., 1997). The transient arrest allows time for the completion of some delayed process or for activation of repair mechanisms to attempt to correct the defects. If the defect is irreparable or there is a failure in the checkpoint or repair pathways, cells can suffer a loss in genomic stability. This is manifested by the segregation of defective chromosomes (i.e., unrepaired DNA) or the generation of aneuploid and polyploid cells (i.e., misattached kinetochore or defective spindle). Checkpoints that respond to DNA damage, under-replication, and defects in the mitotic spindle have been identified in budding yeast (Elledge, 1996; Rudner and Murray, 1996; Stewart and Enoch, 1996). Several of the genes involved in these checkpoint pathways have homologs in other eukaryotes, including mammals (Rudner and Murray, 1996; Stewart and Enoch, 1996; Paulovich et al., 1997).
MPS1 is an essential yeast gene that has two important functions with regard to genomic stability. It is required for SPB duplication and for mitotic checkpoint regulation (Winey et al., 1991; Weiss and Winey, 1996). Consequently, mps1 mutant strains, at the restrictive temperature, not only fail to duplicate their SPBs, but also fail to arrest the cell cycle in response to the defective spindle, leading to aneuploidy and a rapid loss in viability. However, a transient loss in MPS1 activity can lead to cells with twice the DNA content, attributable to a failure in one round of SPB duplication (Winey et al., 1991). The resultant monopolar mitotic spindle is incapable of chromosome segregation, thereby leaving all the chromatin in one cell.
MPS1 encodes an essential dual specificity protein kinase (Lauzéet al., 1995). Mps1p kinase activity is required for both SPB duplication and for the mitotic checkpoint. Proper activation of the Mps1p kinase requires Cdc37p, which is thought to act as a molecular chaperone (Schutz et al., 1997). To date, no substrate for Mps1p relevant to SPB duplication has been identified. However, Mps1p is required for the increased phosphorylation of the checkpoint gene product, Mad1p, which correlates with the activation of the mitotic checkpoint (Hardwick and Murray, 1995; Hardwick et al., 1996). Mad1p is also phosphorylated by purified Mps1p in vitro, but it is not known whether Mad1p is a direct substrate for Mps1p in vivo (Hardwick et al., 1996).
To identify potential substrates or effectors that interact with Mps1p kinase, we have employed the two-hybrid system. Here we describe MOB1 (Mps One Binder), which encodes an essential 314-amino acid protein containing no known structural motifs. MOB1 is a member of a large, conserved gene family with members present in a wide variety of eukaryotes. Its gene product, Mob1p, is a phosphoprotein in vivo and is an in vitro substrate of Mps1p. Unlike MPS1, MOB1 has no apparent role in the spindle assembly checkpoint; however, it is required for the completion of mitosis and maintenance of ploidy, the latter perhaps through MPS1 function. Strains harboring conditional mob1 mutations arrest in late nuclear division at the restrictive temperature. Several alleles of mob1 also cause a complete increase in ploidy at permissive temperature, which may suggest a role in SPB duplication (see DISCUSSION). In these cells, haploid strains appear to become diploid, a phenotype observed in cells defective in SPB duplication (Schild et al., 1981; Thomas and Botstein, 1986; Rose and Fink, 1987; Winey et al., 1991).
MATERIALS AND METHODS
Strains, Cell Culture, and Genetic Techniques
The yeast strains used in this study are listed in Table 1. Yeast media, growth conditions, and genetic and molecular techniques were as described (Guthrie and Fink, 1991; Lunblad, 1997). Selected yeast cultures were arrested in α-factor, hydroxyurea, or nocodazole (Sigma Chemical, St. Louis, MO) as previously described (Weiss and Winey, 1996). Counterselection for yeast harboring URA3-containing plasmids was achieved by growth in media containing 1 mg/ml 5-fluoroorotic acid (5-FOA) (United States Biological, Swampscott, MA) as described (Sikorski and Boeke, 1991). The DH5α strain of Escherichia coli (Bethesda Research Laboratories, Betheseda, MD) was transformed with plasmid DNA by electroporation using a Bio-Rad Gene Pulser electroporator (Bio-Rad, Hercules, CA) as described by the manufacturer.
Table 1.
Yeast strains
| Name | Genotype | Source |
|---|---|---|
| D8BX5CA | a/α, ura3-52/ura3-52, trp1Δ1/trp1Δ1, his3Δ200/his3Δ200, leu2-3,112/leu2-3,112 | Winey |
| WX257-5c | a ura3-52 trp1Δ1 his3Δ200 leu2-3,112 | Winey |
| WX257-8b | α ura3-52 trp1Δ1 his3Δ200 leu2-3,112 | Winey |
| FLY10 | a/α mob1Δ::HIS3/MOB1 ura3-52/ura3-52 trp11/trp1Δ1 his3Δ200/his3Δ200 leu2-3,112/leu2-3,112 | This study |
| FLY12A | a mob1Δ::HIS3 ura3-52 trp1Δ1 his3Δ200 leu2-3,112 [pGST-MOB1] | This study |
| FLY15A | a mob2Δ::HIS3 ura3-52 trp1Δ1 his3Δ200 leu2-3,112 | This study |
| FLY15B | α mob2Δ::HIS3 ura3-52 trp1Δ1 his3Δ200 leu2-3,112 | This study |
| FLY26 | a/a mob1-34/mob1-34 ura3-52/ura3-52 trp1Δ1/trp1Δ1 his3Δ200/his3Δ200 leu2-3,112/leu2-3,112 | This study |
| FLY29 | a/a mob1-46/mob1-46 ura3-52/ura3-52 trp1Δ1/trp1Δ1 his3Δ200/his3Δ200 leu2-3,112/leu2-3,112 | This study |
| FLY30 | a mob1-77 ura3-52 trp1Δ1 his3Δ200 leu2-3,112 | This study |
| FLY31 | a/a mob1-83/mob1-83 ura3-52/ura3-52 trp1Δ1/trp1Δ1 his3Δ200/his3Δ200 leu2-3,112/leu2-3,112 | This study |
| FLY32 | a mob1-95 ura3-52 trp1Δ1 his3Δ200 leu2-3,112 | This study |
| FLY33 | a mob1-67 ura3-52 trp1Δ1 his3Δ200 leu2-3,112 | This study |
| FLY40 | a/a mob1-55/mob1-55 ura3-52/ura3-52 trp1Δ1/trp1Δ1 his3Δ200/his3Δ200 leu2-3,112/leu2-3,112 | This study |
| FLY59 | a mob1Δ::HIS3 ura3-52 trp1Δ1 his3Δ200 leu2-3,112 [pRS316-MOB1] | This study |
| FLT60 | a mob1-46::LEU2::mob1Δ::HIS3 ura3-52 trp1Δ1 his3Δ200 leu2-3,112 [pRS316-MOB1] | This study |
| FLY61 | a mob1-55::LEU2::mob1Δ::HIS3 ura3-52 trp1Δ1 his3Δ200 leu2-3,112 [pRS316-MOB1] | This study |
| FLY62 | a mob1-83::LEU2::mob1Δ::HIS3 ura3-52 trp1Δ1 his3Δ200 leu2-3,112 [pRS316-MOB1] | This study |
| FLY64 | α mob1-77 ura3-52 trp1Δ1 his3Δ200 leu2-3,112 | This study |
| FLY66 | α mob1-95 ura3-52 trp1Δ1 his3Δ200 leu2-3,112 | This study |
| FLY67 | a mob1-77 mps1-1 ura3-52 trp1Δ1 his3Δ200 leu2-3,112 | This study |
| FLY69 | α mob1-95 mps1-1 ura3-52 trp1Δ1 his3Δ200 | This study |
| FLY70 | a mob1-95 mps1-1 ura3-52 trp1Δ1 his3Δ200 leu2-3,112 | This study |
| FLY71 | a mob2Δ::HIS3 mps1-1 ura3-52 trp1Δ1 his3Δ200 | This study |
| FLY72 | a mob2Δ::HIS3 mps1-1 ura3-52 trp1Δ1 his3Δ200 leu2-3,112 | This study |
| FLY111 | a lte1Δ::HIS3 ura3-52 trp1Δ1 his3Δ200 leu2-3,112 | This study |
| FLY112 | α lte1Δ::HIS3 ura3-52 trp1Δ1 his3Δ200 leu2-3,112 | This study |
| FLY113 | a mob1-95 lte1Δ::HIS3 ura3-52 trp1Δ1 his3Δ200 leu2-3,112 [pRS316-MOB1] | This study |
| FLY114 | a mob1-77 cdc15-2 ura3-52 trp1Δ1 leu2-3,112 [pRS426-MOB1] | This study |
| FLY115 | a mob1-95 cdc15-2 ura3-52 trp1Δ1 leu2-3,112 [pRS426-MOB1] | This study |
| H5C1A2 | a cdc5-1 ura1 his7 | Raymond |
| H5C1B1 | α cdc5-1 ura1 his7 | Raymond |
| WR142 | α cdc14-1 his7 leu2 gal1 can1 | Raymond |
| 4078-15-3 | a cdc15-2 ura3 his7 leu2 tyr1 gal1 can1 | Raymond |
| 4078-15-2 | α cdc15-2 trp1 his7 leu2 gal1 can1 | Raymond |
| WX241-7a | α mps1-1 ura3-52 trp1Δ1 his3Δ200 | Winey |
| AS132-10a | α mps1-737 ura3-52 his3Δ200 leu2-3,112 lys | Schutz |
| AS131-5b | α mps1-3796 ura3-52 trp1Δ1 his | Schutz |
| AS181-12b | α mps1-6 ura3-52 trp1 leu2-3,112 | Schutz |
| EGY40 | α ura3-52 his3 leu2 | Brent |
| BJ2168 | a ura3-52 trp1 leu2 pep4-3 prb1-1122 prc1-407 | Jones |
Two-Hybrid Screen
Plasmids and yeast strains for the two-hybrid system were provided by Roger Brent and used as described (Golemis et al., 1997). This version of the two-hybrid system employs the use of LexA-Mps1p fusion protein as “bait” encoded by pLexA-Δ63 MPS1 (Lauzéet al., 1995) to isolate and identify MOBs (Mps One Binders). The LexA protein is the DNA-binding domain that specifically binds the LexA operon upstream of the LacZ reporter gene, encoded by pSH18–34 (Golemis et al., 1997). pLexA-Δ63 MPS1 does not activate transcription of the reporter by itself. The “prey” genomic DNA library was constructed and provided by P. Watt (Watt et al., 1995). It consists of S. cerevisiae genomic DNA inserted into a plasmid downstream of a transcriptional activation domain. The yeast strain, EGY40, harboring pSH18–34 and pLexA-Δ63 MPS1, was transformed with the yeast prey library. Activation of the reporter gene was monitored either by plating transformants onto media containing X-gal or by colony-filter-lift assays (Golemis et al., 1997). The prey plasmids were rescued and introduced into E. coli as described (Hoffman and Winston, 1987). Rescued prey plasmids were screened for specificity of interaction by introduction into yeast harboring the reporter plasmid and pRHJM (encoding LexA-Bicoid [Golemis et al., 1997]), or pLexA-Δ63 MPS1. Only prey plasmids that were specifically dependent on pLexA-Δ63 MPS1 for activation of the reporter gene were kept for further analysis.
MOB1 Sequence Analysis
MOB (Mps One Binder) DNA was sequenced from pAD-MOB using oligonucleotide AD-1 (Table 2), and the Sequenase II DNA sequencing kit (United States Biochemical, Cleveland, OH) performed according to the manufacturer’s instructions. Five different MOB genes were obtained from this screen. Only MOB1 was novel. MOB1 DNA was excised from the prey plasmid pAD-MOB1 with EcoRI, purified and labeled with α-32P-dCTP (New England Nuclear, Boston, MA) as described by Brown (1997). Labeled MOB1 DNA was hybridized on S. cerevisiae genomic DNA-mapping filters (Riles et al., 1993). The hybridization pattern revealed that MOB1 resides on chromosome IX. The entire MOB1 open reading frame (ORF) and flanking sequences was provided by Bart Barrell as part of the Sanger Centre yeast genome project. MOB1 is the same as YIL106w listed in the Saccharomyces Genome Database (SGD) and has the GenBank accession number Z47147x72. It shares sequence similarity with YFL035c on chromosome VI (GenBank accession number D50617x34), which we have named MOB2. MOB2 was not isolated in this two-hybrid screen. Various databases, including dbEST and GenBank, were scanned for genes that share sequence similarity to MOB1. Sequence comparisons and manipulations were performed using GCG sequence analysis program (Genetics Computer Group, Madison, WI).
Table 2.
PCR primers
| Name | DNA sequence |
|---|---|
| 1-A | ACA AAC AAT CCA GTA AAC GGA TAT AAT GAA AGC GAC CAT GGA |
| AGG TCG TTC AGA ATG ACA CG | |
| 1-B | AAG AAT ACA ACC TAC AAG CAG ACT TAT ATA AAT ATA CAA TAC TAC |
| CTC TTG GCC TCC TCT AG | |
| 1-C | CGG AAT TCC AAG ATC AAG TTC CC |
| 1-D | CGG GAT CCA GCA TGT CTC CCG TCC TCA CTA CGC |
| 1-E | CCC GGA TCC AAA CCC TTC TTC TAC GCC |
| 1-G | GCG TTT ATC GTT TGC AT |
| 1-I | ACT TCA ATT TCC ATG TC |
| 1-K | CAT TTT GTA ATT GTT CAG CG |
| VI-B | GAA AAA TAA AAA CCA GCC CCT TAA TGT TGC TCA GCC GCC AGC ACT |
| CTT GGC CTC CTC TAG | |
| VI-C | ACC AAG CAT TTA TCA GC |
| VI-D | GCG TTT ATC GTT TGC AT |
| VI-E | TAA TTT TGC CCT GTT TTT CAA AGC TTT CAA TTA ACG GTA GCA GTC |
| GTT CAG AAT GAC ACG | |
| VI-F | CAA AGG ATC CTT CTA TCA AGT CAC CCC |
| VI-G | CCG GGA AGG TAC CGG TG |
| VI-H | GTT TGA GAA TGC CTC TTA GG |
| LTE1-A | GCT TTA CAT AGC CTC TAG CCA TTC AAA GAT TCG CTA CCC TGA ACC |
| CTC TTG GCC TCC TCT AG | |
| LTE1-B | GGA AAG TGG CAC AAT ACC TCA TGT GGA TAG TTC ATT AAT CTC TTC |
| TCG TTC AGA ATG ACA CG | |
| LTE1-C | CTC CTT ACT CAT CTT TGC |
| HIS3A | GAT TAG CGA CCA GCC GG |
| AD-1 | AAT TCG GCA CGA GGC G |
| T3 | ATT AAC CCT CAC TAA AGG GA |
Polymerase Chain Reaction (PCR) Conditions
Oligonucleotides were obtained from United States Biochemical (Cleveland, OH), Life Technologies/BRL (Grand Island, NY), or Operon (Alameda, CA) and are listed in Table 2. Deoxynucleoside triphosphates were from Life Technologies/BRL. Genomic DNA was prepared from yeast strains as described by Hoffman and Winston (1987). High-fidelity PCR was performed using Vent DNA polymerase (New England Biolabs, Beverly, MA) to amplify yeast genomic DNA. Mutagenic PCR was performed using Taq DNA polymerase (Promega, Madison, WI, or Life Technologies/BRL) as described (Fromant et al., 1995).
To determine whether MOB1 has a functional intron, MOB1 cDNA was amplified from a cDNA library (provided by S. Elledge) using 1-I and 1-G oligonucleotide primers and sequenced using the 1-K oligonucleotide primer and the Sequenase PCR sequencing kit (United States Biochemical, Cleveland, OH). MOB2 cDNA was amplified and sequenced from a cDNA library (Liu et al., 1992) in a similar manner using T3 and VI-G oligonucleotide primers and sequenced using the VI-H primer.
MOB1, MOB2, and LTE1 Deletions
MOB1, MOB2, or LTE1 (Wickner et al., 1987; Keng et al., 1994) were deleted from the wild-type diploid strain D8BX5CA (Table 1) using a one-step gene replacement technique (Baudin et al., 1993). HIS3 flanked by sequences of the targeted gene was amplified by PCR from plasmid DNA, such as pEG202 (Golemis et al., 1997), using oligonucleotides 1-A and 1-B for MOB1, VI-B and VI-E for MOB2, and LTE1-A and LTE1-B for LTE1 (Table 2). This method yields HIS3 DNA flanked by the 5′ and 3′ sequences of MOB1, MOB2, or LTE1. Coding regions upstream of the MOB1 and MOB2 introns were not deleted using these PCR products, leaving coding regions intact for the first 78 and 28 codons of MOB1 and MOB2, respectively. The PCR products were individually transformed into D8BX5CA to yield integrative recombinants that were selected for growth in media lacking histidine. Correct integration was confirmed by PCR amplification of genomic DNA with an internal HIS3 primer, HIS3A, and a primer external to the MOB1, MOB2, or LTE1 ORF: oligonucleotide 1-C, VI-C, and LTE1-C, respectively (Table 2). PCR product was only generated by these oligonucleotides when HIS3 was integrated at the targeted locus.
MOB1, MOB2, and MPS1 Plasmids
Restriction nucleases and T4 DNA ligase were obtained from New England Biolabs. Most of the coding region for MOB1 (amino acids 79–314) was amplified by PCR using oligonucleotides 1-C and 1-D. The PCR product was purified and cut with BamHI and EcoRI and ligated to the yeast expression vector pEG(kt) (provided by R. Deschenes [Mitchell et al., 1993]) to yield pGST-MOB1. The same PCR product was ligated to the E. coli expression vector, pRSETA (Invitrogen, San Diego, CA) to create pH6-MOB1. MOB1 and MOB2 were subcloned into low-copy and high-copy yeast shuttle vectors as follows. pRS314-MOB1, pRS316-MOB1, pRS424-MOB1, and pRS426-MOB1 were constructed by insertion of the entire ORF and flanking sequences of MOB1, amplified by oligonucleotides 1-C and 1-E and cut with BamHI and EcoRI, into the same sites of pRS314, pRS316, pRS424, and pRS426 (Sikorski and Heiter, 1989; Christianson et al., 1992). The entire ORF and flanking sequences of MOB2 were amplified with oligonucleotides VI-E and VI-F, cut with BamHI and SalI, and inserted into the same series of vectors to yield pRS314-MOB2, pRS424-MOB2, pRS316-MOB2, and pRS426-MOB2. MPS1 was subcloned into the high-copy yeast shuttle vector pRS424 as follows. MPS1 was excised from pEcoΔMPS1 (Lauzéet al., 1995) with EcoRI and SalI and ligated to the same sites of pRS424 to yield pRS424-MPS1.
Generation of Conditional Alleles of MOB1
To create a library of mutagenized MOB1 DNA, MOB1 was amplified from genomic DNA using oligonucleotides 1-C and 1-E under conditions of poor fidelity (above). The PCR product was purified, cut with BamHI and EcoRI, and ligated to the same sites of pRS314. After electroporation into the E. coli strain, DH5α, transformants were pooled and grown for 3 h in Luria-Bertani broth (Lech and Brent, 1997) supplemented with 100 μg/ml ampicillin. The pRS314-mob1 DNA library was introduced into FLY12A (Table 1). Yeast transformants were selected for growth in media lacking tryptophan. The URA3-containing plasmid also harboring MOB1 was counterselected by replica plating the yeast colonies onto media containing 1 mg/ml 5-FOA (United States Biochemical, Swampscott, MA) to give rise to mob1Δ cells kept alive by the pRS314-mob1 plasmids. These cells were then screened for conditional growth at 15° and 37°C. pRS314-mob1 plasmids causing conditional lethality were rescued as above and retransformed into FLY12A to confirm the mutant phenotype. Mutant mob1- containing plasmids were sequenced using the Sequenase PCR sequencing kit (United States Biochemical, Cleveland, OH) as described by manufacturer.
Targeted integration of mutant alleles of MOB1 was carried out as described (Rothstein, 1991). Conditional alles of MOB1 were subcloned from pRS314-mob1 plasmids and ligated to the URA3-marked pRS306 integrating vector (Sikorski and Heiter, 1989; Christianson et al., 1992). The pRS306-mob1 plasmids were linearized with PmlI and transformed into FLY12A. Transformants were grown on 5-FOA plates and were selected for colonies that were uracil auxotrophs and exhibited the mob1 temperature-sensitive phenotype.
SDS-PAGE and Immunoblots
SDS-PAGE was performed as described (Anderson et al., 1973). Immunoblots were carried out in a Hoefer Transphor transfer apparatus (Hoefer Scientific Instruments, San Francisco, CA) based on the method of Towbin (Towbin and Gordon, 1984). Immunoblots were blocked 30 min or greater within blocking buffer (3% bovine serum albumin, 2% nonfat dry milk, 0.1% Tween-20, 50 mM Tris, 150 mM NaCl, pH 7.4) at room temperature. They were probed with primary antibody diluted in blocking buffer for 1 h followed by four 15-min washes in TBST (0.1% Tween-20, 50 mM Tris, 150 mM NaCl, pH 7.4). Secondary antibody was also diluted in blocking buffer and reacted with the immunoblots for 1 h. After four more washes in TBST, the immunoblots, depending on the secondary antibody, were visualized by Enhanced Chemiluminescence System (ECL, Amersham, Arlington Heights, IL) according to manufacturer’s instruction or reacted with nitro blue tetrazolium/bromochloroindolyl phosphate (Promega) as described (Harlow and Lane, 1988). Immunoblots of glutathione-S-transferase (GST) and GST-Mob1p were probed with goat anti-GST antibody (Pharmacia, Piscataway, NJ) followed by alkaline phosphatase-conjugated rabbit anti-goat IgG (Pierce Chemical, Rockford, IL). Myc-Mps1p was probed with mouse monoclonal anti-myc antibody, 9E10 (provided by M. Klymkowsky) followed by peroxidase-conjugated anti-mouse IgG (Amersham). Autoradiaograms and ECL-treated immunoblots were visualized by exposure to Hyperfilm-MP film (Amersham).
Expression and Purification of Recombinant Proteins
Six histidine-tagged Mob1p (H6-Mob1p) encoded by pH6-MOB1 was expressed in the BL21(DE3) strain of E. coli (Studier et al., 1990). Cells were grown to an OD600 of 1 and induced for 4 h at 37°C with 1 mM IPTG (Sigma Chemical). Cells were harvested by centrifugation and lysed by sonication in urea-containing buffer as described (Luca et al., 1991). The cell lysate was centrifuged at 15,000 × g in a SS-34 rotor (Dupont, Wilmington, DE) to rid it of cell debris and unlysed cells. Recombinant H6-Mob1p was precipitated from the clarified cell extract by incubation with 1 ml of a 1:1 slurry of Ni-agarose (Qiagen, Chatsworth, CA). H6-Mob1p–bound resin was washed several times in lysis buffer followed by several washes in TBS (50 mM Tris, 150 mM NaCl, pH 7.4). Bound protein was eluted in TBS containing 1 mM EDTA and dialysed in TBS. Estimates of protein concentration were obtained by comparison to known amounts of bovine serum albumin on an SDS-PAGE.
GST, GST-Mob1p, and myc-Mps1p were inducibly expressed under the control of the GAL10 promoter in yeast and purified as previously described for GST-MPS1 with a few modifications (Lauzéet al., 1995). A 1-l culture of BJ2168 harboring pEG(kt), pGST-MOB1, or pELW325, encoding myc-Mps1p (Hardwick et al., 1996), was grown in selective media containing 2% raffinose to OD600 of 0.5. The cells were induced with 4% galactose (Sigma Chemical) for 6–8 h at 30°C, harvested by centrifugation, pooled, and washed one time in sorbitol break buffer (0.3 M sorbitol, 0.1 M NaCl, 5 mM MgCl2, 10 mM Tris, pH 7.4) at 4°C (Mitchell et al., 1993). They were then resuspended in 10 ml cold sorbitol buffer and lysed in a French press under 900 pounds per square inch. Cell debris was pelleted by centrifugation at 15,000 × g at 4°C to yield clarified extract. GST, GST-Mob1p, or GST-Mps1p was precipitated from the extract by incubation with glutathione-Sepharose (Pharmacia) (1 ml packed resin per 10 ml cell extract) for 1 h at 4°C. Bound protein was washed six times in phosphate-buffered saline (PBS) (0.15 mM NaCl, 50 mM sodium phosphate, pH 7.4.) + 0.1% Tween 20 followed by six washes in PBS or kinase buffer (50 mM HEPES, pH 7.5, 10 mM MgCl2, 2 mM MnCl2, 0.5 mM dithiothreitol) (Lauzéet al., 1995).
Myc-Mps1p was immunoprecipitated from yeast extracts with the 9E10 mouse monoclonal antibody using the following protocol: 0.5 ml of 1:10 diluted extract was first “precleared” by incubation for 1 h at 4°C with 0.1 ml gamma-bind G Sepharose (Pharmacia). The Sepharose beads were pelleted and the supernatant was incubated with 5 μg/ml 9E10 mouse monoclonal antibody for 1 h at 4°C. The antibody–antigen complex was precipitated with 50 μl of gamma-bind G Sepharose for 1 h at 4°C. Protein-bound resin was washed as above and analyzed by in vitro kinase assays or by immunoblots probed with 9E10 antibody.
Mob1p/Mps1p Coprecipitation and In Vitro Kinase Assays
Yeast extract (0.15 ml) containing overexpressed myc-MPS1 was diluted 10-fold and precleared with 0.15 ml GST-bound glutathione-Sepharose. The precleared extract was then split into three equal aliquots and incubated for 1 h at 4°C with either 5 μg 9E10 antibody and gamma-bind G Sepharose as outlined above, 50 μl GST immobilized onto glutathione-Sepharose, or 50 μl GST-MOB1 immobilized onto glutathione-Sepharose. All were washed six times in PBS + 0.1% Tween-20 and three times in PBS, followed by a final wash in kinase buffer; 3.5 μl of each were then loaded onto SDS-PAGE for Coomassie brilliant blue staining and immunoblot analysis. A portion of resin was used for in vitro kinase assay performed as described (Lauzéet al., 1995). Resin (12.5 μl) was added to 12.5 μl of kinase reaction buffer ± 2 μg myelin basic protein (MBP) (Sigma Chemical) and incubated for 30 min at 30°C. The kinase reactions were terminated by boiling in an equal volume of protein sample buffer. Ten microliters of kinase reaction were loaded onto 15% SDS-PAGE (Anderson et al., 1973) and processed for autoradiography.
GST-Mps1p in vitro kinase assays were performed as described (Lauzéet al., 1995). GTS-Mps1p (2.5 μl) bound to glutathione Sepharose was incubated in a total of 25 μl kinase reaction buffer ± 2.5 μg MBP or 5 μg H6-Mob1p. Reactions were terminated and analyzed as above.
In Vivo Labeling
32P labeling of yeast was done as described (Lauzéet al., 1995). Cultures (50 ml) of BJ2168 cells harboring pEG(kt) or pGST-MOB1 were grown to OD600 of 0.5. The cells were then transferred to phosphate-depleted media (Lauzéet al., 1995) and grown for 1.5 h. The cells were labeled for 3 h at 30°C with 5 mCi of [32P]orthophosphate (Amersham) while induced for expression of GST or GST-Mob1p by the addition of 4% galactose. 32P-labeled cells were harvested by centrifugation and lysed by vortexing in 0.4 ml sorbitol break buffer and 0.2 ml of acid-washed glass beads. Extracts were centrifuged and GST-Mob1p precipitated on glutathione-Sepharose as above. Mob1p was cleaved from GST by incubating 10 μl of immobilized GST-Mob1p with 4 U thrombin (Boehringer Mannheim, Indianapolis, IN) for 1 h at 37°C in TBS, pH 8.8. Free Mob1p was separated from the immobilized GST-Mob1p by centrifugation. 32P-labeled proteins were analyzed on an SDS-PAGE processed for autoradiography.
Cytological Techniques
Yeast cells were prepared for flow cytometry as described by Weiss and Winey (1996) using the DNA stain propidium iodide (Sigma Chemical). Stained cells were analyzed on a FACScan flow cytometer using CELLFIT and LYSYS software packages (Becton Dickinson, San Jose, CA) to obtain and analyze data. Budding indices were performed as described (Weiss and Winey, 1996). Viability of yeast cells was done by counting colony forming units from serial dilutions of a briefly sonicated cell culture. Immunofluorescence was performed as described by Winey et al. (1991) using rat monoclonal antitubulin, YOL 1/34, as primary antibody and fluorescein isothiocyanate-conjugated goat anti-rat IgG as secondary antibody (both from Accurate Biochemical, Westbury, NY). DNA was stained with 1 μg/ml 4,6-diamidino-2-phenylindole. Cells affixed to polylysine-coated (1 mg/ml) microscope slides and visualized with a CCD camera (Empix Imaging, Missisauga, Ontario, Canada) on a Zeiss photomicroscope (Carl Zeiss, Thornwood, NY). Images were captured and manipulated using Metamorph software (Universal Imaging, West Chester, PA).
RESULTS
Screen for Mps One Binding Proteins
Although S. cerevisiae Mps1p protein kinase is known to be essential for SPB duplication and for mitotic checkpoint regulation, its molecular interactions remain largely unknown. In an effort to identify Mps1p-interacting proteins, potential substrates, or effectors, we performed a two-hybrid screen (see MATERIALS AND METHODS). We screened a yeast genomic DNA library (provided by R. Brent and P. Watt) producing hybrid-activating proteins for interactions with a LexA-Mps1p fusion protein. We had previously shown that LexA-Mps1p is functional by its ability to complement mps1 mutations and by its activity in in vitro kinase assays of immunoprecipitated protein (Lauzéet al., 1995). We identified five genes encoding LexA-Mps1p–interacting proteins that failed to interact with an unrelated fusion protein, LexA-Bicoid (our unpublished results).
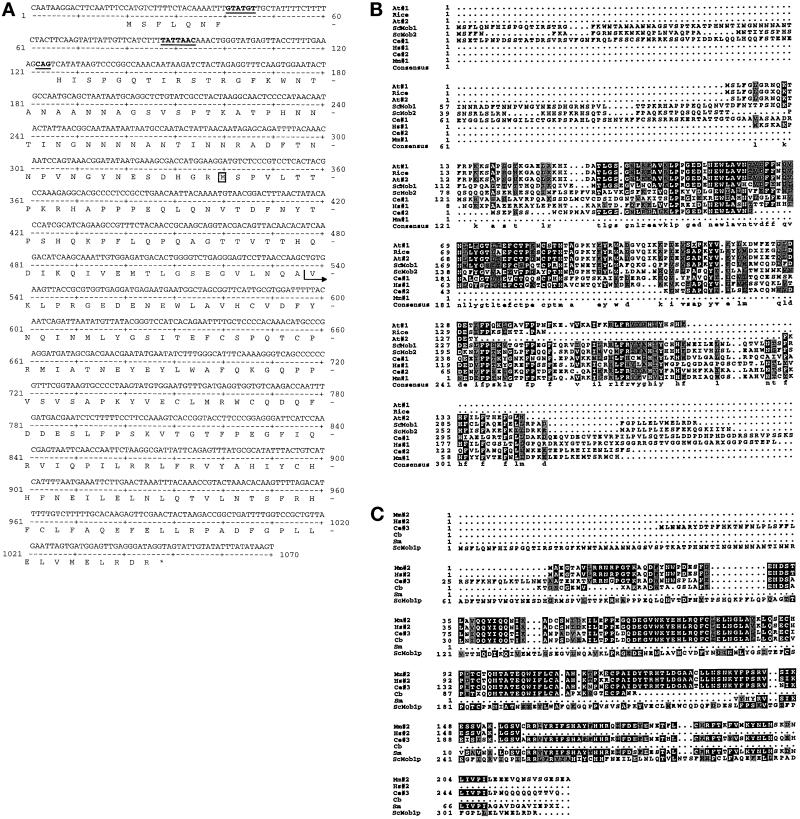
One of the genes encoding a protein that interacts with LexA-Mps1p is MOB1. We mapped MOB1 to chromosome IX using an S. cerevisiae physical mapping filter (provided by L. Riles and M. Olson). We then sequenced a portion of the MOB1 DNA and obtained the complete ORF and flanking sequences from Bart Barrel of the Sanger Centre Sequencing Group, Cambridge, United Kingdom (SGD ORF = YIL106w). The MOB1 prey plasmid from the two-hybrid screen encodes an N-terminal truncation (starting at valine 145, VKLP… ) (Figure 1A). MOB1 was independently identified by K. Chun and M. Goebl in a screen designed to identify essential yeast genes (Chun and Goebl, 1996). Initial analysis of MOB1 genomic DNA revealed a putative 85-nucleotide intron with canonical donor and acceptor sites but a noncanonical branch site, TATTAAC instead of the canonical TACTAAC sequence (Ruby and Abelson, 1991) (Figure 1A). If the intron were functional, spliced MOB1 mRNA would encode a 314-amino acid protein (35,859 Da) (Figure 1A). An unspliced MOB1 mRNA would encode a 236-amino acid protein (27,405 Da), assuming that translation starts at codon 79. We sequenced MOB1 cDNA amplified by PCR and found that the intron is spliced. However, using this strategy, we cannot distinguish whether MOB1 mRNA is differentially spliced.
Figure 1.
MOB1 sequence analysis. (A) MOB1 DNA sequence and ORF. Underlined sequences represent the donor, branch, and acceptor sites for mRNA splicing. Note that the branch site is noncanonical. The postion of the initiator methionine of an unspliced MOB1 is boxed. The arrow indicates the fusion junction in the two-hybrid prey plasmid. (B) Aligned sequences of a subset of class 1 Mob1p-related ORFs from various genome databases. Comparison of Mob1p (ScMob1) and Mob2p (ScMob2) to predicted amino acid sequences from A. thaliania (At#1 represents a contig of GenBank accession numbers H77164 and T04732; At#2, GenBank accession no. Z46539); Rice (GenBank accession no. C19722); human (Hs#1 represents a contig of GenBank accession numbers F11288, F11866, R19220, R13096, and R59435); mouse (Mm#1, GenBank accession no. AA118366); and C. elegans (Ce#1, GenBank accession no. Z69788; Ce#2, GenBank accession no. Z77660). Sequences shaded in black are identical. Sequences shaded in gray are conserved. X’s represent ambiguous sequence. Note that many of the sequences derive from expressed sequence tags (ESTs) and therefore may be incomplete. (C) Aligned sequences of a subset of Class 2 Mob1p-related ORFs from various genome databases. Mob1p (ScMob1) sequence is compared with predicted amino acid sequences of the class 2 Mob1p-related sequences isolated from mouse (Mm#2 represents a contig of GenBank accession numbers U01138, AA125007, and AA208960); human (Hs#2 represents a contig of GenBank accession numbers W76032, D59087, N55945, and T79518); C. elegans (Ce #3, GenBank accession no. L10990); C. briggsae (Cb, GenBank accession no. R03084); and S. masoni (Sm, GenBank accession no. AA1468).
Sequence analysis of Mob1p reveals no known structural motifs. MOB1 shares sequence similarity with another predicted S. cerevisiae ORF located on chromosome VI (SGD ORF = YFL035c), which we have named MOB2. Like MOB1, MOB2 also contains a putative intron; however its donor, acceptor, and branch sequences are canonical. We have confirmed that MOB2’s intron is functional by sequencing MOB2 cDNA. Mob1p shares 43% similarity and 33% identity with Mob2p at the amino acid level, as determined by BESTFIT analysis using GCG sequence analysis program. Mob1p also shares sequence similarity with a variety of predicted ORFs of unknown functions, from a variety of organisms, as diverse as Arabidopsis and humans (Figure 1, B and C). The predicted Mob1p-like proteins fall into two similar, but distinct, classes by sequence comparisons (Figure 1, B and C). Proteins of the class I of Mob1p-like sequences share sequence similarity with Mob1p ranging from 37–68% similarity and 27–56% identity at the amino acid level. Mob1p shares about 45% similarity and 29% identity to proteins of the class II Mob1p-like proteins. Strikingly, proteins of this class share about 75% identity to each other.
MOB1 Is an Essential Gene, whereas MOB2 Is not Essential
To determine whether MOB1 is an essential gene, we replaced most of one of the MOB1 genes in a wild-type diploid cell with the HIS3 gene (FLY10, Table 1; see MATERIALS AND METHODS). Upon sporulation and dissection of the tetrads, we found that only the two spores containing the wild-type MOB1 gene were viable (n = 10 tetrads). No HIS3-containing spores were viable. Spores containing mob1Δ::HIS3 could be recovered only if they contained a plasmid, such as pRS316-MOB1, encoding wild-type MOB1. Furthermore, no viable mob1Δ::HIS3-containing cells can be recovered after counter selection of the URA3 gene containing pRS316-MOB1 plasmid using 5-FOA (see MATERIALS AND METHODS). These results indicate that MOB1 is essential, which is in agreement with previous data (Chun and Goebl, 1996). We used the same strategy to delete the MOB2 gene and determined that MOB2 is not essential. Spores containing deletions of MOB2 were viable and displayed no temperature sensitivity for growth (n = 20 tetrads).
Mob1p Is a Phosphoprotein and Precipitates Active Mps1p Protein Kinase
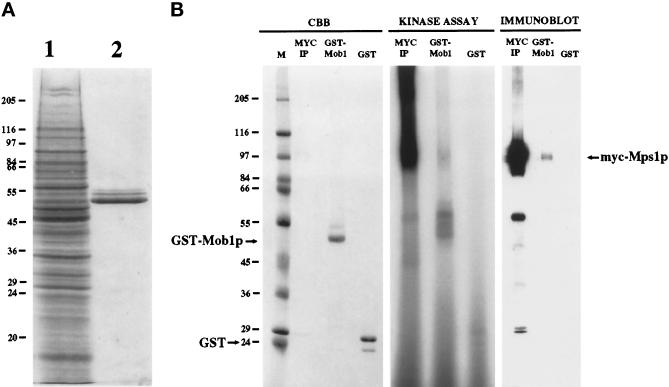
We expressed the majority of Mob1p (amino acids 79–314) in yeast as a GST-Mob1p fusion protein under control of the inducible GAL10 promoter in yeast. GST-Mob1p can be easily purified by precipitation with glutathione-bound Sepharose (Figure 2A). We determined the fusion protein to be functional in vivo because the encoding plasmid complements the mob1Δ strain both in glucose (repressing)- and in galactose (inducing)-containing media. This indicates that the 78 N-terminal amino acids of Mob1p encoded by spliced MOB1 mRNA are nonessential. We were unable to detect any growth or cell cycle defect as a result of the overexpression of this GST-Mob1p.
Figure 2.
GST-Mob1p precipitates Mps1p protein kinase. (A) Expression and purification of GST-Mob1p from yeast (see MATERIALS AND METHODS). Lane 1, total yeast protein containing expressed GST-Mob1p stained with Coomassie brilliant blue on SDS-PAGE. Lane 2, GST-Mob1p purified from yeast extract on glutathione-Sepharose. Positions of molecular weight markers are shown. (B) Protein from yeast extract containing myc-epitope- tagged Mps1p was precipitated with anti-myc antibody (MYC IP), GST-Mob1p-bound Sepharose (GST-Mob1p), or GST-bound Sepharose (GST). CBB, Coomassie brilliant blue-stained gel of precipitants. Kinase assay, autoradiogram showing 32P-labeled proteins in corresponding precipitants after performance of an Mps1p protein kinase assay. Immunoblot, precipitated protein were immunoblotted and probed with 9E10. The prominent ∼ 60 kDa band below myc-Mps1p on the immunoblot of MYC-IP is IgG heavy chain of the primary antibody recognized by the secondary antibody. Note that active Mps1p protein kinase is present in the anti-myc and GST-Mob1p precipitants but not in the GST precipitants. Also note that in addition to myc-Mps1p autophosphorylation, GST-Mob1p is phosphorylated in vitro. Molecular weight markers (M) and the positions of GST, GST-Mob1p, and myc-Mps1p are shown.
To confirm the two-hybrid interaction between Mps1p and Mob1p, we used GST-Mob1p immobilized on glutathione-Sepharose as an affinity reagent to precipitate active Mps1p protein kinase from cell extracts. Extracts made from yeast overexpressing Mps1p that bore the myc epitope were first precleared with GST bound to glutathione-Sepharose and then incubated with GST or GST-Mob1p–bound glutathione-Sepharose. After extensive washing, we assayed the Sepharose beads for the presence of bound Mps1p by immunoblot and by in vitro kinase assays. GST-bound Sepharose did not precipitate any detectable Mps1p; however, GST-Mob1p–bound Sepharose precipitated active Mps1p protein kinase (Figure 2B). Immunoblot analysis revealed that myc-Mps1p was present on the GST-Mob1p–bound Sepharose. The corresponding in vitro kinase assay indicated that the precipitated myc-Mps1p protein kinase was active as noted by Mps1p autophosphorylation (Figure 2B). Note that only a portion of myc-Mps1p binds the GST-Mob1p-Sepharose (compare levels of myc-Mps1p in myc-Ip to GST-Mob1p on the immunoblot probed with anti-myc antibody, Figure 2B). Perhaps Mob1p binds additional proteins that preclude the binding of Mps1p or maybe only a portion of Mob1p is posttranslationally modified in such a way to interact with Mps1p. Conversely, it is possible that only a subset of Mps1p is modified in such a way to render it capable of binding Mob1p. In fact, myc-Mps1p containing cell extracts was made from cells predominantly in late G2 early M phase, as overproduction of active Mps1p kinase causes a G2/M arrest (Hardwick et al., 1996). Perhaps the Mob1p/Mps1p interaction is not so prominent at this stage of the cell cycle. Note also that GST-Mob1p was phosphorylated during the in vitro kinase assay, (50–60 kDa region, center lane of kinase assay, Figure 2B); however, we cannot rule out the possibility of a coprecipitating kinase in addition to Mps1p.
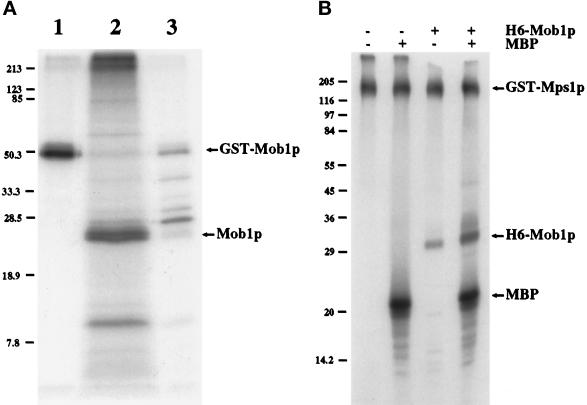
GST-Mob1p, purified on glutathione-Sepharose, appears as multiple electrophoretic forms on an SDS-PAGE, a feature suggestive of phosphorylation (Figure 2A). In contrast, GST alone migrates as a single electrophoretic band (our unpublished results). Some of the electrophoretic forms of GST-Mob1p can be eliminated by treatment with protein phosphatase 2A or calf intestinal alkaline phosphatase, suggesting that in vivo phosphorylation alters Mob1p’s mobility on SDS-PAGE (our unpublished results). To directly test whether Mob1p is phosphorylated in vivo, we labeled yeast expressing GST-Mob1p with [32P]orthophosphate. We then precipitated GST-Mob1p on glutathione-Sepharose (Figure 3A, lane 1), cleaved Mob1p from GST (which remains immobilized on glutathione-Sepharose) with thrombin (Figure 3A, lane 2), and analyzed the soluble Mob1p by SDS-PAGE and processed for autoradiography. The resultant autoradiograph revealed that thrombin-cleaved Mob1p was 32P-labeled revealing that Mob1p is phosphorylated in vivo.
Figure 3.
Mob1p is a phosphoprotein. (A) Autoradiogram of precipitated 32P-labeled GST-Mob1p from extracts of cells grown in the presence of [32P]orthophosphate (see MATERIALS AND METHODS). Lane 1, 32P-labeled GST-Mob1p precipitated by glutathione-Sepharose; lane 2, 32P-labeled Mob1p separated from GST by thrombin cleavage; lane 3, remainder of 32P-labeled protein on glutathione-Sepharose post-thrombin cleavage. (B) H6-Mob1p is phosphorylated by GST-Mps1p in vitro (see MATERIALS AND METHODS). Mps1p kinase assays of GST-Mps1p ± H6-Mob1p and ± MBP were separated by SDS-PAGE and processed for autoradiography. Positions of molecular weight markers are shown.
To address whether Mob1p is phosphorylated by Mps1p protein kinase in vitro, we expressed 6H-Mob1p, (six histidine N-terminally tagged to amino acids 79–314 of Mob1p), in E. coli and purified it to use as substrate in an in vitro Mps1p kinase assay (performed as described by Lauzéet al., 1995). We chose 6H-Mob1p for these experiments instead of yeast-made GST-Mob1p because Mob1p synthesized in E. coli migrates as a single electrophoretic band on an SDS-PAGE and is not coexpressed with any yeast kinases (our unpublished results). Upon incubation with GST-Mps1p and [γ32P]ATP, 6H-Mob1p is phosphorylated in vitro (Figure 3B). 6H-Mob1p had no effect on the ability of GST-Mps1p to autophosphorylate or to phosphorylate an equivalent amount of MBP, an exogenous substrate of Mps1p. We feel it is unlikely that H6-Mob1p is phosphorylated by a kinase that coprecipitates with GST-Mps1p. Previous work suggests that an inactive form of GST-Mps1p, when expressed and purified using the same procedure used for active GST-Mps1p, harbors no detectable coprecipitating kinase activity (Lauzéet al., 1995). These results suggest that Mob1p is phosphorylated by Mps1p in vitro; however, further analysis is required to determine whether Mob1p is a substrate of Mps1p in vivo.
MOB1 Is Required for Completion of Mitosis and Maintenance of Ploidy
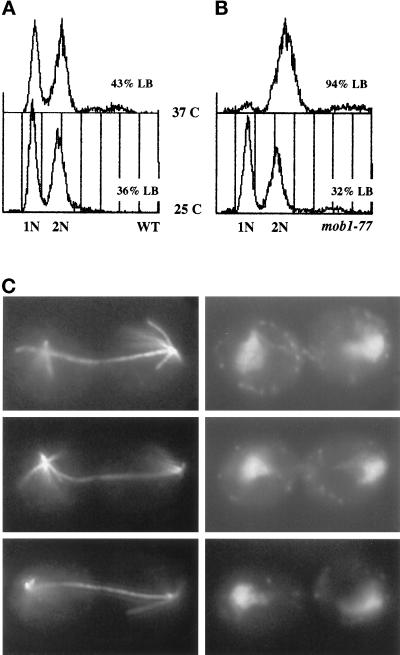
To explore the essential function(s) of MOB1 in vivo, we generated conditional alleles of MOB1. We used random PCR-mediated mutagenesis to create a library of mutagenized mob1 genes in a yeast shuttle vector (see MATERIALS AND METHODS). We introduced the mob1 library into a mob1Δ strain (FLY12A, Table 1) using a plasmid shuffle strategy and identified several temperature-sensitive alleles of MOB1, all of which have been sequenced. Most of the alleles are point mutations that cause amino acid changes in the conserved domains of Mob1p (Figure 4). Upon shift to the restrictive temperature, 37°C, all of the mob1 mutants arrest as large budded cells with 2 N DNA content and separated chromatin as shown for mob1–77 (Figure 5). Analysis of microtubule structures via antitubulin immunofluorescence reveals that the mutants arrest with long bipolar mitotic spindles (Figure 5C). This cell cycle arrest is reversible with high viability (>95%) if returned to the permissive temperature within 3–4 h. These data suggest that MOB1 is required for the completion of mitosis.
Figure 4.
Positions of mob1 lesions on Mob1p. Amino acid substitutions in mob1 alleles are noted under wild-type Mob1p sequence. Amino acids represented in bold print are conserved among the class 1 Mob1p-like proteins (see Figure 1). Underlined sequences represent the amino acids changed in mob1 mutants.
Figure 5.
Cells harboring mob1 mutants arrest in late nuclear division. (A) Flow cytometric analysis of DNA content of a wild-type yeast strain grown at 25°C or 37°C for 3 h. The biphasic peaks represent cells with G1 (1N) and G2/M (2N) DNA content. (B) Flow cytometric analysis of DNA content of yeast harboring the mob1–77 allele grown at 25°C or 37°C for 3 h. At 37°C cells harboring mob1–77 mutation arrest as large budded cells with G2/M DNA content. Percent large-budded (LB) cells are noted. (C) Arrested mob1–77 cells (3 h at 37°C) were stained with anti-tubulin antibody (left panels) and 4,6-diamidino-2-phenylindole (right panels). The cells arrest with long mitotic spindles and separated chromatin masses. Three representative cells are shown.
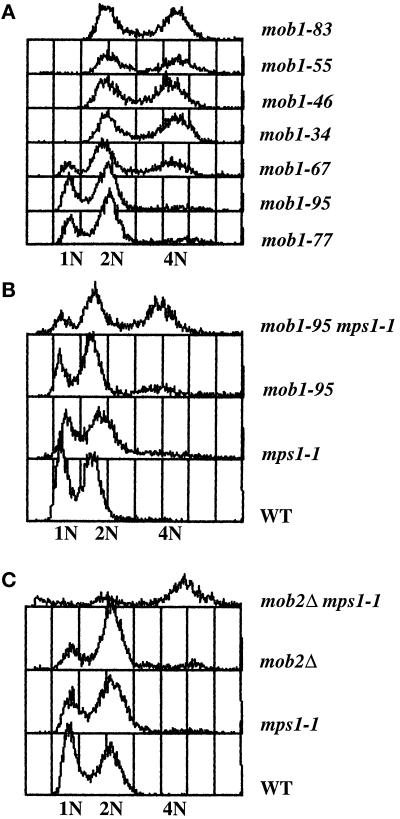
When the mob1 mutations are present on low-copy centromeric plasmids in a mob1Δ yeast strain, all of the cells are maintained as stable haploids at permissive temperature. However, when present as a single copy integrated into the genome, five mob1 alleles (mob1–34, mob1–46, mob1–55, mob1–67, mob1–83) cause an increase in ploidy at the permissive temperature (Figure 6A). The cells appear to have twice the amount of DNA, suggesting that they have become diploids. In agreement, when the diploidized mob1 strains are mated to haploid cells of the opposite mating type, the mating products appear triploid by flow cytometric analysis and produce very few viable spores (our unpublished results). The observed increase-in-ploidy phenotype at permissive temperature is unusual; however, it has been seen in several mutants defective in SPB duplication (Schild et al., 1981; Thomas and Botstein, 1986; Rose and Fink, 1987; Winey et al., 1991) and in mutants defective in other cellular processes (Chan and Botstein, 1993). All mob1 integrants, whether haploid or diploid, continue to arrest in late mitosis at the restrictive temperature but display a lower minimal restrictive temperature than when present on low-copy plasmids (34°C instead of 37°C).
Figure 6.
MOB1 and MOB2 are required for the maintenance of ploidy. (A) Flow cytometric analysis of DNA content of yeast harboring various alleles of MOB1 grown at 25°C. Cells harboring mob1–77 or mob1–95 mutations contain haploid biphasic peaks representing G1 (1N) and G2/M (2N) DNA content. Cells harboring mob1–34, mob1–46, mob1–55, or mob1–83 mutations contain diploid biphasic peaks (2N and 4N) indicating that they have diploidized. Populations of yeast harboring mob1–67 mutation contain both haploid and diploid cells. (B) Flow cytometric analysis of DNA content of tetratype tetrad resulting from crossing mob1–95 and mps1–1 strains. Populations of wild-type cells and cells harboring mob1–95 or mps1–1 mutations are haploids containing both G1 (1N) and G2/M (2N) DNA content. Cells harboring both mob1–95 and mps1–1 have diploid DNA content (2N and 4N). (C) Flow cytometric analysis of DNA content of tetratype tetrad resulting from crossing mob2Δ and mps1–1 strains. Populations of wild-type cells and cells harboring mob2Δ or mps1–1 mutations are haploids containing both G1 (1N) and G2/M (2N) DNA content. Cells harboring both mob2Δ and mps1–1 have diploid DNA content (2N and 4N).
MOB1 and MOB2 Share Genetic Interactions with MPS1
The late nuclear division arrest of mob1 mutants suggests that the execution point for MOB1 occurs in late mitosis, a time for which there is no evidence of MPS1 function. In an effort to identify the role for the Mob1p/Mps1p interaction, we looked for genetic interactions between MOB1 and MPS1. We crossed the two integrated alleles of mob1 that behave as stable haploids at permissive temperature (mob1–77 and mob1–95), to mps1 mutants in order to examine the phenotypes of the double mutants. We detected no synthetic lethality between either mob1–77 or mob1–95 and several mps1 alleles (summarized in Table 3). All of the mob1 mps1 double mutants grew without any detectable growth defect. However, mob1–95 mps1–1 double mutants diploidized at permissive temperature (Figure 6B). The degree of diploidization within a population of cells harboring both mutations is variable. This interaction appears to be allele specific for both mps1–1 and mob1–95 (Table 3). The increase-in-ploidy phenotype observed in the double mutants may suggest that the mechanism for the increase in ploidy displayed in some mob1 single mutants occurs through MPS1 function. Similarly, populations of mob2Δ mps1–1 double mutants display an increase in ploidy, suggesting a role for MOB2 in the maintenance of ploidy (Figure 6C; Table 3).
Table 3.
Synthetic growth phenotypes of double mutants
| mps1-1 | mps1-737 | mps1-3796 | mps1-6 | cdc5-1 | cdc14-1 | cdc15-2 | lte1Δ | |
|---|---|---|---|---|---|---|---|---|
| mob1-77 | + H | + H | + H | + H | SL | + | SL | SL |
| mob1-95 | + D | + H | + H | + H | SL | + | SL | SL |
| mob2Δ | + D | + H | + H | + H | + | + | + | nd |
+, no detectable synthetic growth defect; SL, synthetic lethality; H, haploid; D, diploid; nd, not determined.
The observation that both mob1–95 mps1–1 and mob2Δ mps1–1 double mutants display an increase in ploidy may suggest that MOB1 and MOB2 perform similar or overlapping roles. In an attempt to establish additional genetic interactions shared between MOB1 and MOB2, we looked for synthetic phenotypes displayed by mob1 mob2Δ double mutants and we found none. mob1–77 mob2Δ and mob1–95 mob2Δ double mutants are maintained as stable haploids and grow at wild-type rates at permissive temperature. They arrest in late mitosis, as do the mob1 single mutants, at restrictive temperature (our unpublished results). Furthermore, high-copy plasmids harboring wild-type MOB2 are unable to suppress either the increase-in-ploidy phenotype or the late nuclear division arrest of mob1 mutants at restrictive temperature (our unpublished results).
The Cell Cycle Arrest of mob1 Cells Does Not Require MPS1
Although MPS1 has no known function in late mitosis, it was possible that MOB1 requires MPS1 for its role in late mitosis. To address this possibility, we tested whether cells harboring mob1 mutations require MPS1 to arrest in late nuclear division at restrictive temperature. We first synchronized mob1–77 mps1–1 and mob1–95 mps1–1 double mutants in G1 with the mating pheromone, α factor, and released them into media containing hydroxyurea. This allows cells to execute MPS1’s SPB duplication function and arrest in early S phase. The cells were then released into fresh media without hydroxyurea at the restrictive temperature to inactivate MPS1’s checkpoint function (Weiss and Winey, 1996). If mob1 mutant cells require MPS1 to arrest in late mitosis, then mob1 mps1 double mutants should fail to arrest at the restrictive temperature. This is not the case. Both mob1–77 mps1–1 and mob1–95 mps1–1 double mutants arrest as large budded cells with G2 DNA content, similar to the mob1 single mutants (our unpublished results). Furthermore, high-copy plasmids harboring wild-type MPS1 were unable to suppress mob1 mutations (our unpublished results). Together, these data suggest that the late nuclear division arrest, caused by mob1 mutations, is independent of MPS1 function, as has been shown for other late nuclear division cdc mutants (Weiss and Winey, 1996; E. Weiss, personal communication).
MOB1 and MOB2 Are Not Required for the Spindle Assembly Checkpoint
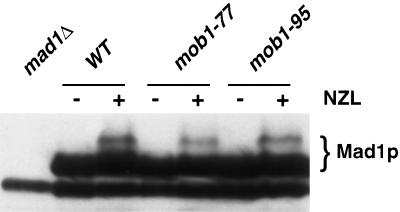
MPS1 is required for SPB duplication and for mitotic checkpoint control (Winey et al., 1991; Hardwick et al., 1996; Weiss and Winey, 1996). To test whether MOB1 and MOB2 are required for MPS1 function in the spindle assembly checkpoint, we assayed mob1 mutants and mob2Δ for their ability to arrest in nocodazole, a microtubule-disrupting agent that triggers the checkpoint pathway causing a cell cycle arrest. All of the mob1 and mob2Δ strains arrest in nocodazole at 25°C, indicating that they are not defective in the spindle assembly checkpoint at permissive temperature (our unpublished results). These results do not completely rule out the involvement of MOB1 in mitotic checkpoint regulation as it is possible that mob1 is defective at mitotic checkpoint control at the restrictive temperature. However, it cannot be distinguished cytologically whether nocodazole-treated mob1 cells arrest at restrictive temperature in response to nocodazole treatment or as a consequence of the defect in MOB1’s essential function, executed later in mitosis. To assay mitotic checkpoint activation in mob1 mutants at restrictive temperature, we monitored Mad1p hyperphosphorylation as a molecular marker for checkpoint activation. When the mitotic checkpoint is triggered, Mad1p becomes hyperphosphorylated, an event dependent on active Mps1p protein kinase and is easily detectable by a shift in mobility by SDS-PAGE (Hardwick and Murray, 1995; Hardwick et al., 1996). We first synchronized mob1–77 and mob1–95 cells at permissive temperature in early G1 with a mating pheromone, α factor. The cells were then released into media containing nocodazole at the restrictive temperature. Samples of mob1 mutants were taken after nocodazole treatment at restrictive temperature and were analyzed for Mad1p hyperphosphorylation by immunoblot as described by Hardwick and Murray (1995). By this analysis, we observed that Mad1p is hyperphosphorylated in nocodazole-treated mob1–77 and mob1–95 cells at restrictive temperature (Figure 7). The arrested cells also contained elevated levels of Clb2p (our unpublished results). Mad1p from cells treated similarly but in the absence of nocodazole was not hyperphosphorylated (our unpublished results). These results suggest that MOB1 is not required for the spindle assembly checkpoint. However, it remains possible that we do not possess the appropriate mob1 mutant alleles to demonstrate a checkpoint role. It is also possible, although unlikely, given its association with Mps1p, that Mob1p is required for a step in the spindle assembly checkpoint that is downstream of Mad1p hyperphosphorylation.
Figure 7.
Mad1p is hyperphosphorylated in mob1 mutants. Cells were synchronized in α-factor at 25°C and released in media containing 15 μg nocodozole (NZL) and 30 μg benomyl at 36°C. Samples were taken before (−) and after (+) treatment with nocododole and processed for 10% SDS PAGE and immunoblotted The immunoblot was probed with affinity- purified antiMad1p antibody. The lane loaded with extract from a strain deleted for MAD1 (mad1Δ) reveals an immunoreactive background protein. Lanes loaded with extract from wild type (WT), mob1–77, and mob1–95 cells are indicated. Mad1p hyperphosphorylation is detectable only in nocodazole-treated cells by the presence of slower migrating forms.
MOB1 Has Genetic Interactions with CDC5, CDC15, and LTE1
The late nuclear division arrest phenotype of mob1 mutants is similar to that described for cells harboring mutations in a number of genes, including CDC5, CDC14, CDC15, and LTE1 (Pringle and Hartwell, 1981; Wickner et al., 1987). CDC5 and CDC15 encode essential protein kinases, CDC14 encodes an essential protein tyrosine phosphatase, and LTE1 encodes a GDP/GTP exchange protein (Schweitzer and Philippsen, 1991; Wan et al., 1992; Kitada et al., 1993; Keng et al., 1994). Deletion of LTE1 yields cells that are cold sensitive for growth. To investigate whether MOB1 function is linked to the function of these four genes, we looked for genetic interactions between MOB1 and those four genes. We assayed for synthetic interactions between double mutants and observed no synthetic lethality or growth defects in cells harboring mob1 (using mob1–77 and mob1–95) and cdc14–1 mutations (Table 3). However, we were unable to obtain double mutants between either allele of mob1 (mob1–77, mob1–95) and cdc5–1, cdc15–2, or lte1Δ strains, suggesting that there are synthetic lethal interactions between mob1 and the three genes. To confirm these results, we crossed mob1 strains harboring pRS316-MOB1 or pRS426-MOB1 with cdc15–2 or lte1Δ strains. The resultant double mutants were plasmid dependent for viability. We conclude that both the mob1–77 and mob1-95 alleles are synthetically lethal with cdc5–1, cdc15–2 and lte1Δ strains, suggesting that MOB1 functions in the same pathway as CDC5, CDC15, and LTE1. Cells harboring mob2Δ did not display any synthetic lethality or growth defects when combined with cdc5–1, cdc14–1, or cdc15–2 (summarized in Table 3).
DISCUSSION
MOB1 is an essential yeast gene required for the completion of mitosis and for the maintenance of ploidy. It is a member of a conserved, and previously unrecognized, gene family, likely to be present in all eukaryotes. In addition to MOB1, budding yeast contain MOB2, a nonessential member of the MOB1 gene family. We have been unable to detect any direct genetic interactions (high-copy suppression and synthetic lethality) between MOB1 and MOB2. Although conserved, the predicted amino acid sequences of the MOB1-like gene products offer no insight to their molecular function. We have demonstrated by two independent methods, two-hybrid and coprecipitation, that Mob1p binds Mps1p, an essential protein kinase required for SPB duplication and mitotic checkpoint regulation. In addition, Mob1p is a phosphoprotein in vivo and a substrate of Mps1p protein kinase in vitro, raising the possibility that Mob1p is a substrate of Mps1p in vivo.
Cells harboring defective MOB1 genes arrest in late mitosis at restrictive temperature. This role of MOB1 in the completion of mitosis is probably mediated through its association with genes other than Mps1p. There are a number of genes, encoding regulatory proteins, that, when mutated, cause a cell cycle arrest similar to mob1 mutants. There is an extensive array of genetic interactions linking them together, suggesting that they belong to a common pathway (Pringle and Hartwell, 1981; Wickner et al., 1987; Johnston et al., 1990; Parkes and Johnston, 1992; Kitada et al., 1993; Molero et al., 1993; Spevak et al., 1993; Keng et al., 1994; Shirayama et al., 1994a, 1994b, 1996; Toyn and Johnston, 1994). We have begun to establish that MOB1 shares genetic interactions with several members of this class of genes. Mutations in MOB1 exhibit synthetic lethality with mutants in genes encoding two protein kinases (CDC15, CDC5) and a GDP/GTP exchange factor (LTE1). In addition, high-copy plasmids encoding MOB1 suppress cells lacking DBF2, a gene encoding another protein kinase required for the completion of mitosis. Furthermore, Mob1p physically interacts with Dbf2p protein kinase (C. Denis, personal communication). These results suggest MOB1 participates in a common regulatory event required to complete mitosis. Perhaps Mob1p is a substrate or effector for one or more of the kinases required for completion of mitosis (i.e., Cdc5p, Cdc15p, Dbf2p).
The specific roles for MOB1 and the other genes required for the completion of mitosis are unknown. However, the late nuclear division arrest has been characterized, to some extent, as the failure to complete a number of events. These events include the inactivation of the M phase-promoting protein kinase complex, Clb2p/Cdc28p, the disassembly of the mitotic spindle, the formation of prereplicative complexes on DNA origins, the lowering of cAMP levels, and the initiation of events responsible for cytokinesis (Smith et al., 1990; Russell et al., 1993; Spevak et al., 1993; Romanowski and Madine, 1996; Juang et al., 1997). These events may be regulated by the same or separate pathways that are triggered by a master switch at the end of anaphase.
Perhaps MOB1 and the other genes required for completion of mitosis mediate the transition from M phase to G1 by regulating the anaphase promoting complex (APC)/cyclosome (King et al., 1996). The APC/cyclosome targets several substrates for ubiquitin-mediated proteolysis, carried out by the 26S proteasome, which is crucial for the completion of mitosis. It is responsible for targeting degradation of the B-type cyclin, Clb2p, which is the regulatory subunit of the M phase-inducing kinase, Cdc28p. This event is a prerequisite for the completion of mitosis, as expression of an indestructible form of Clb2p in yeast causes a late nuclear division arrest (Surana et al., 1993). Strains harboring mutations in DBF2, CDC5, CDC15, CDC14, TEM1 or harboring triple deletions in the RAS1Δ, RAS2Δ, RSR1Δ genes all arrest in late nuclear division with high Clb2p/Cdc28p kinase activity, suggesting they may be required for cyclin degradation and/or Cdc28p inactivation (Surana et al., 1993; Shirayama et al., 1994; Toyn and Johnston, 1994; Morishita et al., 1995). The APC/cyclosome is also necessary for targeting the spindle- associated protein, Ase1p, for degradation at the end of mitosis (Juang et al., 1997). Ase1p degradation is a prerequisite for the disassociation of the mitotic spindle at the end of mitosis, and it is probable that Ase1p degradation is inhibited in the late mitotic mutants. Recently, a component of the 26S proteasome has been implicated in an early step in SPB duplication, suggesting that regulated proteolysis is also required for SPB duplication (McDonald and Byers, 1997). Perhaps all of the above mentioned regulated proteolytic events are coordinated to trigger the exit from mitosis and prepare cells to enter the next cell cycle (see below).
The role in the completion of mitosis for MOB1 may be independent of MPS1. There is no evidence for any genetic interactions between MPS1 and any of the genes, other than MOB1, required for the completion of mitosis (E. Weiss, personal communication). Moreover, cells harboring mutations in both MPS1 and MOB1 arrest with phenotypes similar to mob1 single mutants when shifted to the restrictive temperature. This suggests that the mitotic checkpoint function of MPS1 is not required for cells to arrest in late mitosis. Likewise, we have no evidence that MOB1 is required for the spindle assembly checkpoint, as determined by monitoring Mad1p hyperphosphorylation as molecular marker for checkpoint activation.
In addition to its role in late nuclear division, MOB1 is required for the maintenance of ploidy, which may reflect its interaction with MPS1. Most mob1 mutants display a complete increase in ploidy at permissive temperature, i.e., haploid cells become diploid. The increase in ploidy phenotype is unique to MOB1 among the collection of genes required for the completion of mitosis and may suggest an additional, perhaps separable, role for MOB1. The increase in ploidy phenotype in the mob1 mutants is sensitive to gene dosage. When present as a single integrated copy, many of the conditional alleles of MOB1 cause an increase in ploidy at a temperature permissive for growth. However, when the mutant mob1 alleles are present on low-copy number plasmids, which presumably express more Mob1p than the integrated gene, cells remain haploid. To date, we have been unable to study the increase in ploidy in isolation using the available alleles in synchronized cells or using other methods designed to inactivate Mob1p.
Although this type of increase-in-ploidy phenotype, complete diploidization, is unusual, there are several possible mechanisms that may bring it about. One such mechanism is a failure to undergo budding and cytokinesis. Such a defect would yield binucleate cells, which we have not observed for mob1 mutants. Another possible mechanism for a complete increase in ploidy is a failure to properly segregate the duplicated DNA at mitosis. This could occur as a result of a defect in nuclear migration, chromosome segregation, or mitotic spindle formation. Perhaps proper mitotic spindle formation is affected in select mob1 mutants (see below). Cells that overreplicate their DNA, meaning that DNA replication occurs twice in one cell cycle, may also lead to a complete increase in ploidy. Such a defect might occur in mob1 mutants if MOB1 were required to limit DNA replication to once per cell cycle.
There is a precedent for genes that are required for completion of mitosis that also have a role in initiation of DNA synthesis. Several genes required for the completion of late mitosis are also required for the formation of prereplicative complexes. Prereplicative complexes are essential for “licensing” DNA for replication in the following S phase and form in late mitosis (Chevalier and Blow, 1996; Romanowski and Madine, 1996; Stillman, 1996). The formation of prereplicative complexes requires the activities of late-division gene CDC14 and probably the late-division gene CDC5, which encode a protein phosphatase and a protein kinase, respectively (Hardy, 1996; Hardy and Pautz, 1996; Kroll et al., 1996). Furthermore, several genetic interactions have been established between CDC5 and CDC14 and components of the origin recognition complex (ORC) (Hardy, 1996; Hardy and Pautz, 1996; Kroll et al., 1996). Recently it has been shown that Cdc5p physically interacts with Dbf4p, the regulatory subunit of the S phase-activating protein kinase, Cdc7p, supporting previous evidence of a genetic interaction (Kitada et al., 1993; Hardy and Pautz, 1996). Whether this signifies an S phase execution point for Cdc5p or a late mitotic role for Dbf4p remains to be seen. Nonetheless, at least some of the late nuclear division genes are required to reset the DNA synthesis cycle in late mitosis. Although it is possible that, in addition to a role in exiting mitosis, MOB1 is required for maintaining proper ploidy through a role in DNA replication, it is the only gene reported of this class that, when mutated, causes a complete increase in ploidy.
Perhaps the mechanism of the increase in ploidy of mob1 mutants derives from the association between MOB1 and MPS1. Strains harboring both mob1–95 and mps1–1 become diploid while strains harboring either mutation alone remain haploid. The complete increase in ploidy could be the result of synergistic damage to parallel pathways, one being the control of the initiation DNA synthesis and the other being the regulation of SPB duplication; or it could be caused by additive damage to a common pathway. This common pathway could be the regulation of SPB duplication. Although MPS1 has no role in DNA synthesis, it has been well established that Mps1p is required for SPB duplication (Winey et al., 1991). A complete increase in ploidy can occur in mutants that affect SPB duplication (Schild et al., 1981; Thomas and Botstein, 1986; Rose and Fink, 1987; Winey et al., 1991). In these mutants, cells progress through the cell cycle in the absence of SPB duplication. The cells complete S phase and undergo mitosis with a monopolar spindle, which is incapable of properly segregating the replicated DNA. After completion of mitosis, all of the replicated genome localizes to one cell, while the sister cell becomes aploid. This mechanism for an increase in ploidy also occurs in cells that have transiently lost Mps1p activity (Winey et al., 1991). Given the association of MOB1 and MPS1, reported here, a plausible mechanism for the increase in ploidy for mob1 single mutants and mob1–95 mps1–1 double mutants is the failure of SPB duplication. Further, Mob1p may be required to physically associate with Mps1p for proper SPB duplication. Support for this comes from the observation that, in a two-hybrid assay, Mps1p binds poorly to a mutant Mob1p (from mob1–55) that causes strains to become diploid. However, Mps1p binds strongly to two other mutant Mob1p proteins that do not cause an increase in ploidy (C. Denis, personal communication).
Genetic interactions between MOB1 and MPS1, other than the diploidization of mob1–95 mps1–1 mutants, have not been detected. Nonetheless, the increase-in-ploidy phenotype is also observed in mob2Δ mps1–1 double mutants, but not in the single-mutant strains. This suggests a role for the MOB1 family in the maintenance of ploidy and may suggest an overlapping role for MOB2. However, more work will be required to understand the elusive relationship between MOB1 and MOB2.
All of the genes required for the completion of mitosis, discussed above, may be collectively viewed as performing cell cycle resetting functions. These cell cycle resetting functions may be necessary not only to allow the cell to exit mitosis, but may also be required for progression into and through the next cell cycle. Support for this concept comes from data indicating that prereplicative complexes, required to render chromosomes competent for replication, are assembled in late mitosis (Chevalier and Blow, 1996; Romanowski and Madine, 1996; Stillman, 1996). In this vein, there may be an event that occurs at the M/G1 transition responsible for rendering SPBs competent for duplication. That event may involve targeted proteolysis, since a defect in the cap subunit of the 26S proteasome causes a failure in SPB duplication (McDonald and Byers, 1997). Moreover, the hypothetical SPB “licensing” may require MOB1 function through its association with MPS1.
Whether the mechanism of the complete increase in ploidy of mob1 mutants occurs through a failure in SPB duplication, over-replication of DNA, or some other mechanism, MOB1 remains unique in having an additional requirement for the completion of mitosis. MOB1 appears to reveal a link between the completion of the cell cycle and events in the subsequent cell cycle. Further work on MOB1 will be necessary to understand how its highly conserved gene product and associated proteins coordinate the timing and execution of these different cell cycle events.
ACKNOWLEDGMENTS
We are grateful to R. Brent and P. Watt for providing the two-hybrid reagents and S cerevisiae library. We thank B. Barrell for providing us with S. cerevisiae chromosome IX sequence before it was available at the SGD and K. Hardwick and A. Murray for providing antibodies to Mad1p. Thanks to A. Schutz, W. Raymond and B. Jones for yeast strains. We also thank E. Weiss, A. McBride, H. Chial and S. Jones for technical advice and valuable discussion and especially to J. Paal for sequencing the mob1 alleles. This work was supported by grants to M.W. from National Sscience Foundation (MCB-9357033), National Institutes of Health (GM51312), and the Pew Scholars Program in the Biomedical Sciences (P0020SC). F.C.L. was supported by a postdoctoral fellowship from the Leukemia Society of America.
REFERENCES
- Anderson CW, Baum PR, Gesteland RF. Processing of adenovirus-2 induced proteins. J Virol. 1973;12:241–252. doi: 10.1128/jvi.12.2.241-252.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Sacchoromyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum P, Yip C, Goetsch L, Byers B. A yeast gene essential for regulation of spindle pole body duplication. Mol Cell Biol. 1988;8:5386–5397. doi: 10.1128/mcb.8.12.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown T. Hybridization analysis of DNA blots. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smoth JA, editors. Current Protocols in Molecular Biology. K. Struhl, New York: John Wiley & Sons, Inc.; 1997. pp. 2.10.1–2.10.16. [Google Scholar]
- Chan C, Botstein D. Isolation and characterization of chromosome-gain and increase-in-ploidy mutants in yeast. Genetics. 1993;135:677–691. doi: 10.1093/genetics/135.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier S, Blow JJ. Cell cycle control of replication initiation in eukaryotes. Curr Opin Cell Biol. 1996;8:815–821. doi: 10.1016/s0955-0674(96)80082-2. [DOI] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Heiter P. Multifunctional yeast high-copy shuttle vectors. Genetics. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Chun K, Goebl M. The identification of transposon-tagged mutations in essential genes that affect cell morphology in Saccharomyces cerevisiae. Genetics. 1996;142:39–50. doi: 10.1093/genetics/142.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta N, Williams L, Caldwell J, Curry A, Ashcraft E, Long M. Novel alterations in CDK1/cyclin B1 kinase complex formation occur during the acquisition of a polyloid DNA content. Mol Biol Cell. 1996;7:209–223. doi: 10.1091/mbc.7.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doheny K, Sorger P, Hyman A, Tugendreich S, Spencer F, Hieter P. Identification of essential components of the S. cerevisiae kinetochore. Cell. 1993;73:761–774. doi: 10.1016/0092-8674(93)90255-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge S. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1671. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- Francisco L, Wang W, Chan CSM. Type 1 protein phosphatase acts in opposition to IPL1 protein kinase in regulating yeast chromosome segregation. Mol Cell Biol. 1994;14:4731–4740. doi: 10.1128/mcb.14.7.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromant M, Blanquet S, Plateau P. Direct random mutagenesis of gene-sized DNA fragments using polymerase chain reaction. Anal Biochem. 1995;224:347–353. doi: 10.1006/abio.1995.1050. [DOI] [PubMed] [Google Scholar]
- Goh P, Kilmartin J. NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J Cell Biol. 1993;121:503–512. doi: 10.1083/jcb.121.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golemis E, Gyuris J, Brent R. Interaction trap/two hybrid system to identify interacting proteins. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smoth JA, editors. Current Protocols in Molecular Biology. K. Struhl, New York: John Wiley & Sons; 1997. pp. 13.14.1–13.14.17. [Google Scholar]
- Grafi G, Larkins B. Endoreduplication in maize endosperm: involvement of M phase-promoting factor inhibition and induction of S phase-related kinases. Science. 1995;269:1262–1664. doi: 10.1126/science.269.5228.1262. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink G. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:1–933. [PubMed] [Google Scholar]
- Hardwick K, Murray A. Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J Cell Biol. 1995;131:709–720. doi: 10.1083/jcb.131.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick K, Weiss E, Luca FC, Winey M, Murray A. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 1996;273:953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- Hardy CF. Characterization of an essential Orc2p-associated factor that plays a role in DNA replication. Mol Cell Biol. 1996;16:1832–1841. doi: 10.1128/mcb.16.4.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy CF, Pautz A. A novel role for Cdc5p in DNA replication. Mol Cell Biol. 1996;16:6775–6782. doi: 10.1128/mcb.16.12.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Hoffman CS, Winston F. A ten minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Genetics. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Hoyt A, He L, Totis L, Saunders W. Loss of function of Saccharomyces cerevisiae kinesin-related CIN8 and KIP1 is suppressed by KAR3 motor domain mutations. Genetics. 1993;135:35–44. doi: 10.1093/genetics/135.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, Geiser JR. Genetic analysis of the mitotic spindle. Annu Rev Genet. 1997;30:7–33. doi: 10.1146/annurev.genet.30.1.7. [DOI] [PubMed] [Google Scholar]
- Jackson CW. Megakaryocyte endomitosis: a review. Int J Cell Cloning. 1992;8:224–226. doi: 10.1002/stem.5530080405. [DOI] [PubMed] [Google Scholar]
- Johnston LH, Eberly SL, Chapman JW, Araki H, Sugino A. The product of the Saccharomyces cerevisiae cell cycle gene DBF2 has homology with protein kinases and is periodically expressed in the cell cycle. Mol Cell Biol. 1990;10:1358–1366. doi: 10.1128/mcb.10.4.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juang Y-L, Huang J, Peters J-M, McLaughlin ME, Tai C-Y, Pellman D. APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science. 1997;275:1311–1314. doi: 10.1126/science.275.5304.1311. [DOI] [PubMed] [Google Scholar]
- Keng T, Clark MW, Storms RK, Fortin N, Zhong W, Ouellette BF, Barton AB, Kaback DB, Bussey H. LTE1 of Saccharomyces cerevisiae is a 1435 codon open reading frame that has sequence similarities to guanine nucleotide releasing factors. Yeast. 1994;10:953–958. doi: 10.1002/yea.320100710. [DOI] [PubMed] [Google Scholar]
- King R, Deshaies R, Peters J, Kirschner M. How proteolysis drives the cell cycle. Science. 1996;274:1652–1658. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- Kitada K, Johnson AL, Johnston LH, Sugino A. A multicopy suppressor gene of the Saccharomyces cerevisiae G1 cell cycle mutant gene dbf4 encodes a protein kinase and is identified as CDC5. Mol Cell Biol. 1993;13:4445–4457. doi: 10.1128/mcb.13.7.4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll ES, Hyland KM, Hieter P, Li JJ. Establishing genetic interactions by a synthetic dosage lethality phenotype. Genetics. 1996;143:95–102. doi: 10.1093/genetics/143.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauzé E, Stoelcker B, Luca FC, Weiss E, Schutz A, Winey M. Yeast spindle pole duplication gene MPS1 encodes an essential dual specificity protein kinase. EMBO J. 1995;14:1655–1663. doi: 10.1002/j.1460-2075.1995.tb07154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lech K, Brent R. Escherichia coli: media preparation and biological tools. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smoth JA, editors. Current Protocols in Molecular Biology. K. Struhl, New York: John Wiley & Sons; 1997. pp. 1.1.1–1.1.6. [Google Scholar]
- Levine D, Sanchez C, Rabinovitch P, Reid B. Formation of the tetraploid intermediate is associated with the development of cells with more than four centrioles in the elastas-simian virus 40 tumor antigen transgenic mouse model of pancreatic cancer. Proc Natl Acad Sci USA. 1991;88:6427–6431. doi: 10.1073/pnas.88.15.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Krizek J, Bretscher A. Construction of a GAL1-regulated yeast cDNA expression library and its application to the identification of genes whose overexpression causes lethality in yeast. Genetics. 1992;132:665–673. doi: 10.1093/genetics/132.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca FC, Shibuya EK, Dohrmann CE, Ruderman JV. Both cyclin AΔ60 and BΔ97 are stable and arrest cells in M-phase, but only BΔ97 turns on cyclin degradation. EMBO J. 1991;109:4311–4320. doi: 10.1002/j.1460-2075.1991.tb05009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunblad V. Saccharomyces cerevisiae. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smoth JA, editors. Current Protocols in Molecular Biology, Chap. 13. K. Struhl, New York: John Wiley & Sons; 1997. [Google Scholar]
- McDonald HB, Byers B. A proteasome cap subunit required for spindle pole body duplication in yeast. J Cell Biol. 1997;137:539–553. doi: 10.1083/jcb.137.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DA, Marshall TK, Deschenes RJ. Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast. 1993;9:715–723. doi: 10.1002/yea.320090705. [DOI] [PubMed] [Google Scholar]
- Molero G, Yuste-Rojas M, Montesi A, Vazquez A, Nombela C, Sanchez M. A cdc-like autolytic Saccharomyces cerevisiae mutant altered in budding site selection is complemented by SPO12, a sporulation gene. J Bacteriol. 1993;175:6562–6570. doi: 10.1128/jb.175.20.6562-6570.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita T, Mitsuzawa H, Nakafuku M, Nakamura S, Hattori S, Anraku Y. Requirement of Saccharomyces cerevisiae ras for completion of mitosis. Science. 1995;270:1213–1215. doi: 10.1126/science.270.5239.1213. [DOI] [PubMed] [Google Scholar]
- Parkes V, Johnston LH. SPO12 and SIT4 suppress mutations in DBF2, which encodes a cell cycle protein kinase that is periodically expressed. Nucleic Acids Res. 1992;20:5617–5623. doi: 10.1093/nar/20.21.5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulovich AG, Toczyski DP, Hartwell LH. When checkpoints fail. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Hartwell LH. The Saccharomyces cerevisiae cell cycle. In: Atrathern JN, Jones EW, Braoch JR, editors. The Molecular Biology of the Yeast Saccharomyces cerevisiae. Life Cycle and Inheritance. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1981. pp. 97–142. [Google Scholar]
- Riles L, Dutchik J, Baktha A, McCauley B, Thayer E, Leckie M, Braden V, Depke J, Olson M. Physical maps of the six smallest choromosomes of Saccharomyces cerevisiae at a resolution of 2.6 kilobase pairs. Genetics. 1993;134:81–150. doi: 10.1093/genetics/134.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski P, Madine M. Mechanisms restricting DNA replication to once per cell cycle: MCMs, pre-replicative complexes and kinases. Trends Biochem Sci. 1996;6:184–188. doi: 10.1016/0962-8924(96)10015-5. [DOI] [PubMed] [Google Scholar]
- Rose M, Fink G. KAR1, a gene required for function of both intranuclear and extranuclear microtubules in yeast. Cell. 1987;48:1047–1060. doi: 10.1016/0092-8674(87)90712-4. [DOI] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Ruby SW, Abelson J. Pre-mRNA splicing in yeast. Trends Genet. 1991;7:79–85. doi: 10.1016/0168-9525(91)90276-V. [DOI] [PubMed] [Google Scholar]
- Rudner AD, Murray AW. The spindle assembly checkpoint. Curr Opin Cell Biol. 1996;8:773–780. doi: 10.1016/s0955-0674(96)80077-9. [DOI] [PubMed] [Google Scholar]
- Russell M, Bradshaw-Rouse J, Markwardt D, Heideman W. Changes in gene expression in the Ras/Adenylate cyclase system of Saccharomyces cerevisiae: correlation with cAMP levels and growth arrest. Mol Biol Cell. 1993;4:757–765. doi: 10.1091/mbc.4.7.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W, Koshland D, Eshel D, Gibbons I, Hoyt A. Saccharomyces cerevisiae kinesin- and dynein-related protein required for anaphase chromosome segregation. J Cell Biol. 1995;128:617–624. doi: 10.1083/jcb.128.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild D, Ananthaswamy H, Mortimer R. An endomitotic effect of a cell cycle mutation of Saccharomyces cerevisiae. Genetics. 1981;97:551–562. doi: 10.1093/genetics/97.3-4.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutz AR, Giddings TH, Steiner E, Winey M. The yeast CDC37 gene interacts with MPS1 and is required for proper execution of spindle pole body duplication. J Cell Biol. 1997;136:969–982. doi: 10.1083/jcb.136.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer B, Philippsen P. CDC15, an essential cell cycle gene in Saccharomyces cerevisiae, encodes a protein kinase domain. Yeast. 1991;7:265–273. doi: 10.1002/yea.320070308. [DOI] [PubMed] [Google Scholar]
- Shirayama M, Matsui Y, Tanaka K, Toh-e A. Isolation of a CDC25 family gene, MSI2/LTE1, as a multicopy suppressor of ira1. Yeast. 1994a;10:451–461. doi: 10.1002/yea.320100404. [DOI] [PubMed] [Google Scholar]
- Shirayama M, Matsui Y, Toh EA. The yeast TEM1 gene, which encodes a GTP-binding protein, is involved in termination of M phase. Mol Cell Biol. 1994b;14:7476–7482. doi: 10.1128/mcb.14.11.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Matsui Y, Toh-e A. Dominant mutant alleles of yeast protein kinase gene CDC15 suppress the lte1 defect in termination of M phase and genetically interact with CDC14. Mol Gen Genet. 1996;251:176–185. doi: 10.1007/BF02172916. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Boeke JD. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Heiter P. A system of shuttle vectors and yeast host stains for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ME, Dickinson JR, Wheals AE. Intracellular and extracellular levels of cyclic AMP during the cell cycle of Saccharoyces cervisiae. Yeast. 1990;6:53–60. doi: 10.1002/yea.320060106. [DOI] [PubMed] [Google Scholar]
- Spevak W, Keiper BD, Stratowa C, Castanon MJ. Saccharomyces cerevisiae cdc15 mutants arrested at a late stage in anaphase are rescued by Xenopus cDNAs encoding N-ras or a protein with beta-transducin repeats. Mol Cell Biol. 1993;13:4953–4966. doi: 10.1128/mcb.13.8.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart E, Enoch T. S-phase and DNA-damage checkpoints: a tale of two yeasts. Curr Biol. 1996;8:781–787. doi: 10.1016/s0955-0674(96)80078-0. [DOI] [PubMed] [Google Scholar]
- Stillman B. Cell cycle control of DNA replication. Science. 1996;274:1659–1663. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- Studier WF, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Annu Rev Biochem. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Surana U, Amon A, Dowzer C, McGrew J, Byers B, Nasmyth K. Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO J. 1993;12:1969–1978. doi: 10.1002/j.1460-2075.1993.tb05846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J, Botstein D. A gene required for the separation of chromosomes on the spindle apparatus in yeast. Cell. 1986;44:65–76. doi: 10.1016/0092-8674(86)90485-x. [DOI] [PubMed] [Google Scholar]
- Tlsty TD, Briot A, Gualberto A, Hall I, Hess S, Hixon M, D, Ki, Romanov S, Sage M, White A. Genomic stability and cancer. Mutat Res. 1995;337:1–7. doi: 10.1016/0921-8777(95)00016-d. [DOI] [PubMed] [Google Scholar]
- Towbin H, Gordon J. Immunoblotting and dot immunobinding- current status and outlook. J Immunol Methods. 1984;72:313–340. doi: 10.1016/0022-1759(84)90001-2. [DOI] [PubMed] [Google Scholar]
- Toyn JH, Johnston LH. The Dbf2 and Dbf20 protein kinases of budding yeast are activated after the metaphase to anaphase cell cycle transition. EMBO J. 1994;13:1103–1113. doi: 10.1002/j.1460-2075.1994.tb06359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Xu H, Grunstein M. CDC14 of Saccharomyces cerevisiae. cloning, sequence analysis, and transcription during the cell cycle. J Biol Chem. 1992;267:11274–11280. [PubMed] [Google Scholar]
- Watt PM, Loouis EJ, Borts RH, Hickson ID. SGS1: a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell. 1995;81:253–260. doi: 10.1016/0092-8674(95)90335-6. [DOI] [PubMed] [Google Scholar]
- Weiss E, Winey M. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J Cell Biol. 1996;132:111–123. doi: 10.1083/jcb.132.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB, Koh TJ, Crowley JC, O’Neil J, Kaback DB. Molecular cloning of chromosome I DNA from Saccharomyces cerevisiae: Isolation of the MAK16 gene and analysis of an adjacent gene essential for growth at low temperatures. Yeast. 1987;3:51–57. doi: 10.1002/yea.320030108. [DOI] [PubMed] [Google Scholar]
- Winey M, Goetsch L, Baum P, Byers B. MPS1 and MPS2: novel yeast genes defining distinct steps of spindle pole body duplication. J Cell Biol. 1991;114:745–754. doi: 10.1083/jcb.114.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Skibbens R, Cheng J, Salmon E, Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]