Abstract
Fission yeast Cdc18, a homologue of Cdc6 in budding yeast and metazoans, is periodically expressed during the S phase and required for activation of replication origins. Cdc18 overexpression induces DNA rereplication without mitosis, as does elimination of Cdc2-Cdc13 kinase during G2 phase. These findings suggest that illegitimate activation of origins may be prevented through inhibition of Cdc18 by Cdc2. Consistent with this hypothesis, we report that Cdc18 interacts with Cdc2 in association with Cdc13 and Cig2 B-type cyclins in vivo. Cdc18 is phosphorylated by the associated Cdc2 in vitro. Mutation of a single phosphorylation site, T104A, activates Cdc18 in the rereplication assay. The cdc18-K9 mutation is suppressed by a cig2 mutation, providing genetic evidence that Cdc2-Cig2 kinase inhibits Cdc18. Moreover, constitutive expression of Cig2 prevents rereplication in cells lacking Cdc13. These findings identify Cdc18 as a key target of Cdc2-Cdc13 and Cdc2-Cig2 kinases in the mechanism that limits chromosomal DNA replication to once per cell cycle.
INTRODUCTION
DNA replication must be stringently controlled to guarantee that the genome is duplicated exactly once during each cell cycle—failure to maintain this control would create havoc with the genome. Thus, once S phase is initiated, control mechanisms ensure that all chromosomal DNA is replicated and a new round of replication does not occur before chromosomes are segregated into the two daughter cells at mitosis. The mechanisms that limit DNA replication to once per cell cycle have been the focus of major research efforts in the last few years (Muzi-Falconi et al., 1996a; Stillman, 1996; Wuarin and Nurse, 1996), but they remain poorly understood.
The fission yeast Schizosaccharomyces pombe has served as an outstanding model organism for studying cell cycle controls. Recent studies of fission yeast have suggested that the Cdc18 protein plays an important role in regulating chromosomal DNA replication. The cdc18+ gene is essential for initiation of DNA replication (Nasmyth and Nurse, 1981; Kelly et al., 1993). High constitutive expression of Cdc18 protein causes cells to undergo continuous DNA replication without intervening mitosis, whereas cells deleted for the cdc18+ gene bypass S phase and enter mitosis directly from G1 (Kelly et al., 1993; Nishitani and Nurse, 1995). The levels of cdc18+ mRNA and protein are highly periodic during the cell cycle, being absent in G1, peaking during S phase and returning to a very low level during G2 and M (Nishitani and Nurse, 1995; Muzi-Falconi et al., 1996b). These data suggest that Cdc18 activity is rate limiting for initiation of DNA replication and that accurate control of Cdc18 activity may be required to prevent rereplication.
Many studies have implicated cyclin dependent kinases (CDKs) in the cell cycle controls regulating DNA replication (Muzi-Falconi et al., 1996a; Stillman, 1996; Wuarin and Nurse, 1996). In S. pombe the cyclin-dependent kinase Cdc2 is required for both the G1-S and G2-M cell cycle transitions (Nurse and Bisset, 1981). Cig2 and Cdc13 are the major B-type cyclins that associate with and activate Cdc2. The G1-S activity is predominantly provided by Cdc2-Cig2, whereas Cdc2-Cdc13 is required for the initiation of mitosis. However, in cells lacking the cig2+ gene, Cdc2-Cdc13 is sufficient to promote the onset of S phase with only a modest delay relative to wild-type cells (Fisher and Nurse, 1996; Martin-Castellanos et al., 1996; Mondesert et al., 1996). Cdc2 is also implicated in the mechanism that prevents reinitiation of S phase from G2 phase. Thus, deletion of the cdc13+ gene or overexpression of Rum1, an inhibitor of Cdc2, induces successive rounds or continuous DNA replication without intervening mitosis (Hayles et al., 1994; Nishitani and Nurse, 1995). Furthermore, expression of high amounts of Cdc2 and Cdc13 in G1 represses the onset of S phase and induces entry into M directly from G1 (Hayles et al., 1994). In the budding yeast Saccharomyces cerevisiae, inactivation of Clb-Cdc28p kinases is required to generate a permissive period that allows association at the origins of proteins essential for initiation of replication and the activation of Clb-Cdc28p during late G1 inhibits further association (Dahmann et al., 1995; Piatti et al., 1996). Similarly, in Xenopus egg extracts, Cdk2 kinase is required to initiate replication of exogenously added chromatin while on the contrary high amounts of added Cdk2-cyclin A or E inhibits DNA replication (Fang and Newport, 1991; Jackson et al., 1995; Hua et al., 1997). These findings lead to a model in which low CDK activity is required to activate replication origins at the G1-S transition, whereas high CDK activity later in the cell cycle represses origin activation, thereby preventing reinitiation of replication.
The Cdc2 substrates involved in control of initiation of DNA replication are unknown, although a clue was recently provided through the discovery of Orp2 as a protein that interacts with Cdc2 in vivo (Leatherwood et al., 1996). Orp2 is required for DNA replication in S. pombe and is related to Orc2p, a component of origin recognition complex in budding yeast (Gavin et al., 1995). Orp2 also interacts with Cdc18 protein (Leatherwood et al., 1996). The physical association between these proteins suggests that Orp2 and/or Cdc18 may be important substrates of Cdc2. In this article, we present data which indicate that this is the case for Cdc18. We demonstrate that Cdc18 is associated with Cdc2-Cdc13 and Cdc2-Cig2 and that these kinases phosphorylate Cdc18 on multiple sites. We have found that elimination of one of the major phosphorylation sites moderately activates Cdc18 in the rereplication assay without changing Cdc18 abundance. Although previous work suggested that Cig2 B-type cyclin is exclusively involved in the induction of S phase, our new genetic studies have revealed that Cdc2-Cig2 kinase also contributes to the inhibition of Cdc18 activity. We propose that inhibitory phosphorylation of Cdc18 is an important part of the mechanism by which Cdc2 prevents the reactivation of replication origins following the onset of S phase.
MATERIALS AND METHODS
Plasmids, Strains, and General Methods
Schizosaccharomyces pombe strains expressing glutathione S-transferase (GST) or GST-Cdc18 were created by targeted integration of plasmids pAL24 and pAL27 at the ura4–294 locus. Plasmid pAL24 was constructed by inserting the 2.8-kb PstI–SacI fragment from pJL205 plasmid, containing the nmt1 promoter upstream of the GST open reading frame, into pJK210, an integrative vector (Keeney and Boeke, 1994). The cdc18 open reading frame was amplified by polymerase chain reaction from pRep4-Cdc18-HA using primers AL19 (5′-CGGGATCCAT GGGCCATGTGTC-3′) and oJL19 (5′-CTAGCAGTAC TGGCAAGGGA GAC-3′) (Leatherwood et al., 1996). The polymerase chain reacction product was digested with BamHI and the 1.8-kb fragment was ligated into BamHI-digested pAL24 to create pAL27. pAL24 and pAL27 were linearized with StuI and integrated at the ura4–294 locus in strain OM1603 (h+ leu1–32 ura4–294) to generate strains AL1853 and AL1854, respectively. The strain AL1856 [cdc18-k9 leu1–32 ura4–294:nmt1-GST-cdc18+(ura4+)] was used for the mutant complementation studies. The rum1+ coding region was amplified using primers RUM1 NDE (5′-CGCATATGGA ACCTTCAACA CC-3′) and RUM1 NOT (5′-CAGCGGCCGC TATTTCGTAA TAAATTGTGC-3′), digested with NdeI and NotI, and ligated into NdeI–NotI-digested pGEX-KG expression vector (Guan and Dixon, 1991), creating pGEX-KG-rum1+. GST-Rum1 was expressed in Escherichia coli strain BL21-DE3 and purified with reduced glutathione (GSH)-Sepharose. The strain expressing GST-Atf1 was constructed by transformation of pREP1-KZ-atf1 into PR109 (leu1–32 ura4-D18). Strains JL1290 (cdc18-K9 leu1–32 ura4-D18), PR1008 (cig2::ura4+ leu1–32 ura4-D18), PR617 (cig1::ura4+ leu1–32 ura4-D18), AL1994 and AL1995 (cdc18-K9 cig2::ura4+ leu1–32 ura4-D18), and AL1996 and AL1997 (cdc18-K9 cig1::ura4+ leu1–32 ura4-D18) were used for the genetic interaction studies.
Expression from the nmt1 promoter was induced by thiamine depletion (Maundrell, 1993). General genetic and biochemical procedures relevant to fission yeast, including analysis of DNA content by fluorescence-activated cell sorting analysis of cells stained with propidium iodide and 4′,6-diamino-2-phenylindole, have been described (Moreno et al., 1991; Alfa et al., 1993).
Protein Methods
Purification of GST fusion proteins expressed in fission yeast was performed as described previously (Leatherwood et al., 1996). Cells were disrupted with glass beads in buffer L (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Nonidet P-40, 5 mM EDTA, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, 5 μg/ml pepstatin, 5 μg/ml leupeptin, and 5 μg/ml aprotinin), and lysates were centrifuged at 16,000 × g for 5 min at 4°C. GSH-Sepharose (50 μl; Pharmacia, Pistcataway, NJ) was added to 1 ml of supernatant (5–10 mg/ml protein concentration), incubated at 4°C for 2 h, and then washed three times in buffer L. Associated proteins were separated by gradient SDS-PAGE. Immunoblot detection was performed using an enhanced Luminol reagent kit (Pierce, Rockfors, IL) and film or a Vistra ECF Western blotting kit (Amersham, Arlington Heights, IL) and Molecular Dynamics Storm 840. The phosphorylation reaction by the associated protein kinases was performed in KBC buffer (50 mM Tris-HCl, pH 7.2, 10 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride, 5 μg/ml pepstatin, 5 μg/ml leupeptin, and 5 μg/ml aprotinin) containing 40 μCi of [γ-32P]ATP and 100 μM unlabeled ATP for 15 min at 30°C. For the Cdc2 kinase precipitation, S. pombe extracts were prepared by glass bead lysis in buffer L and incubated with p13suc1-Sepharose or with E7 (anti-Cdc13) antibody for 1 h. Antibodies were recovered with 20 μl of protein A-Sepharose. The beads were washed three times with buffer L and incubated in 50 μl of KBC buffer supplemented with [γ-32P]ATP and the indicated substrates (histone H1 was added at 1 mg/ml protein concentration). For the in vitro phosphorylation of GST-Cdc18 by Cdc2, GST-Cdc18 precipitated with GSH-Sepharose was treated with 1 mM FSBA in KBC buffer four times at 30°C for 15 min and then washed in KBC buffer containing 1 mM dithiothreitol. Two-dimensional tryptic phosphopeptide mapping and phospho-amino acid analysis were performed as described (Boyle et al., 1991). For peptide mapping, pH 1.9 buffer was used for the first dimension electrophoresis and phosphochromatography buffer for the second dimension ascending chromatography. Radioactive signals were detected using film or a Molecular Dynamics Storm 840.
In Vitro Mutagenesis
Cdc18 threonine codons corresponding to residues 10, 46, 60, 104, 134, and 374 were changed to alanine and residue 104 to serine by Altered Sites II in vitro mutagenesis system (Promega, Madison, WI). Plasmid pALTER-cdc18 was constructed by inserting the 1.8-kb BamHI fragment from pAL27 into pATLER-1. This plasmid was used as template for the mutagenesis reaction. Primer sequences introducing the mutations were: AL39 (5′-GGTTGTCATG CACCTCGAAG-3′); AL40 (5′-ATTCCGACTG CACCC AGCAG-3′); A43 (5′-GCTCACATTT CCAAGCACCC ACAAAAAG-3′); AL33 (5′-ACTCCTAAAG CCCCCAAAAG-3′); AL35 (5′-TTGCAATCGG CACCTCACCG-3′); AL44 (5′-CAGAAAAAAA CAATCCTTTTG CTCCTATTAAA TCAATCTCTG-3′); and AL45 (5′-ACTCCTAAAT CCCCCAAAAGG-3′). DNA sequence analysis confirmed the mutations. The mutated genes were cloned into the integration plasmid pAL24, creating pAL27-T10A, -T46A, -T60A, -T104A, -T134A, T374A and, -T104S. These plasmids were integrated at the ura4–294 locus in OM1603 to create strains AL1988-AL1992 and AL2049. The strain AL2129 [cdc18-k9 leu1–32 ura4–294:nmt1-GST-cdc18T104A(ura4+)] was used for the mutant complementation studies.
nmt1+ Promoter Turn-off Experiment
The strains AL1854 and AL1991 were grown in minimal medium for 14 h before adding back 5 μl/ml thiamine to repress the expression of the nmt1+ promoter. Aliquots were taken at the indicated time points and processed for immunoblot analysis as described above.
RESULTS
High Expression of GST-Cdc18 Induces Continuous DNA Replication
Biochemical investigations of Cdc18 were facilitated by construction of strains that used the nmt1 promoter to regulate expression of GST-Cdc18 fusion protein. Two experiments were carried out to analyze the functional properties of the GST-Cdc18 fusion protein in vivo. The first experiment tested the ability of GST-Cdc18 to rescue the cell cycle defect of a cdc18-K9 mutant. This analysis revealed that GST-Cdc18 expressed at very low levels from the repressed nmt1 promoter was sufficient to rescue the temperature-sensitive cell cycle defect of a cdc18-K9 mutant, indicating that GST-Cdc18 was functional in vivo. The second experiment evaluated whether GST-Cdc18 was able to induce cell cycle arrest and continuous DNA replication when highly overexpressed from the induced nmt1 promoter. Cells expressing unfused GST or GST-Cdc18 were incubated for 2 d on plates. Cells expressing GST appeared to be normal, whereas cells expressing GST-Cdc18 became extremely elongated, exhibiting a cell division cycle arrest phenotype (Figure 1A). Next, we measured the DNA content of these cells during the time course of induction. Cells expressing GST had 2C DNA content throughout the experiment, whereas DNA content increased steadily in cells expressing GST-Cdc18 (Figure 1B). These experiments show that GST-Cdc18 is functional and when produced at high levels it induces continuous replication without intervening mitosis.
Figure 1.
GST-Cdc18 is functional in vivo and induces rereplication when overexpressed. (A) Cells expressing GST (strain AL1853) or GST-Cdc18 (strain AL1854) from the thiamine-repressible nmt1 promoter were photographed after 2 d of incubation at 30°C on agar medium lacking thiamine. Cells expressing GST grew well, whereas cells expressing GST-Cdc18 underwent several divisions and then became cdc arrested. (B) DNA content was measured by FACS in cells grown in liquid medium lacking thiamine. Note that the nmt1 promoter becomes active approximately 10 h following depletion of thiamine from the growth medium, reaching maximum activity at approximately 16 h (Maundrell, 1993). 2C and 4C positions were determined using wild-type haploid and diploid strains growing exponentially in YES medium at 30°C.
GST-Cdc18 Interacts with Orp2, Cdc2 and B-type Cyclins
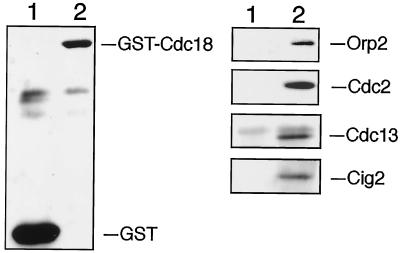
A series of experiments were carried out to investigate the relationship between Cdc18 and Cdc2-cyclin B kinases. As a first step in this analysis, we asked whether GST-Cdc18 associates with Cdc2 and the replication factor Orp2. GST-Cdc18 and GST were purified from cell lysates using GSH-Sepharose and the bound proteins were separated by SDS-PAGE. Immunoblotting detection revealed that both Cdc2-Cdc13 and Cdc2-Cig2 kinases coprecipitated with GST-Cdc18, whereas none of these proteins associated with GST (Figure 2). Orp2 also coprecipitated specifically with GST-Cdc18. We previously reported an interaction between Cdc18 and Orp2 when both proteins were overexpressed (Leatherwood et al., 1996). In the experiment shown in Figure 2, endogenous Orp2 associates with GST-Cdc18, suggesting that the association between Orp2 and Cdc18 might be requisite for Cdc18 function in vivo.
Figure 2.
GST-Cdc18 interacts with Orp2, Cdc2, and B-type cyclins. GST and GST-Cdc18 expression was induced for 18 h. Lysates from those cells, GST (lane 1) and GST-Cdc18 (lane 2), were incubated with GSH-Sepharose beads. The bound proteins were separated by SDS-PAGE and analyzed by immunoblotting with antibodies specific for GST, Orp2, Cdc2, Cdc13, or Cig2, as indicated. GST-Cdc18 copurified specifically with Orp2, Cdc2, Cdc13, and Cig2.
GST-Cdc18 Is Phosphorylated by a Rum1-sensitive Kinase
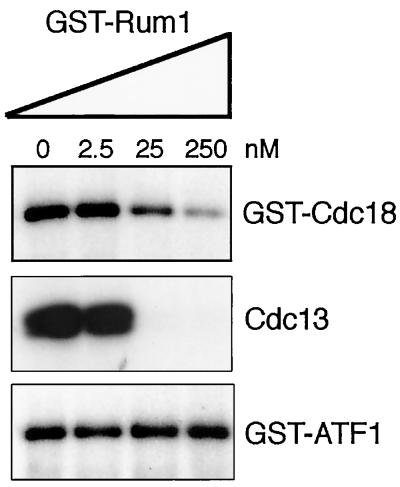
We next explored the possibility that Cdc18 is a substrate of the kinase Cdc2. GST-Cdc18 was purified from S. pombe and incubated in the presence of [γ-32P]ATP. In this assay, GST-Cdc18 became heavily phosphorylated, indicating that GST-Cdc18 copurifies with a protein kinase that phosphorylates GST-Cdc18 efficiently (Figure 3, upper panel, lane 0). The potential involvement of Cdc2 was investigated by adding GST-Rum1 to the kinase reactions. Rum1 is a potent inhibitor of Cdc2-Cdc13 and a weaker inhibitor of Cdc2-Cig2 (Moreno and Nurse, 1994; Correa-Bordes and Nurse, 1995; Jallepalli and Kelly, 1996; Martin-Castellanos et al., 1996). Phosphorylation of GST-Cdc18 was readily inhibited by GST-Rum1 (Figure 3, upper panel). The residual activity remaining in the reactions containing 250 nM GST-Rum1 was consistent with the association of GST-Cdc18 with Cdc2-Cig2, as this kinase is only partially inhibited by Rum1 (Correa-Bordes and Nurse, 1995). As a positive control it was found that phosphorylation of histone H1 by Cdc2-Cdc13 kinase, purified from S. pombe lysates by antiCdc13 immunoprecipitation, was inhibited by GST-Rum1 (Figure 3, middle panel). As a negative control we examined the phosphorylation of GST-Atf1 by Spc1, a stress-activated kinase that associates with Atf1 bZIP transcription factor in fission yeast (Shiozaki and Russell, 1996). GST-Rum1 had no effect on GST-Atf1 phosphorylation (Figure 3, lower panel).
Figure 3.
GST-Cdc18 purified from fission yeast is phosphorylated by an associated kinase that is inhibited by Rum1. GST, GST-Cdc18, and GST-Atf1 expression was induced for 18 h in medium lacking thiamine. GST, GST-Cdc18, and GST-Atf1 were precipitated using GSH-Sepharose and incubated with Mg2+ and [γ-32P]ATP in the presence of increasing concentrations of GST-Rum1 purified from bacteria. Autoradiography of samples subjected to gel electrophoresis revealed that GST-Cdc18 became phosphorylated by a kinase that was inhibited by GST-Rum1 (upper panel). As a positive control it was confirmed that phosphorylation of histone H1 by Cdc2-Cdc13 kinase, purified from S. pombe lysates by anti-Cdc13 immunoprecipitation, was inhibited by GST-Rum1 (middle panel). As a negative control it was found that phosphorylation of GST-Atf1 by associated Spc1 kinase was insensitive to GST-Rum1 (lower panel).
Cdc2 Phosphorylates Cdc18
The results described above supported the conclusion that Cdc2 is the kinase responsible for GST-Cdc18 phosphorylation. We further explored this question by determining whether GST-Cdc18 was phosphorylated by purified Cdc2 in vitro. Protein kinases that associated with GST-Cdc18 were inactivated by treatment with FSBA (ρ-fluorosulfonyl-benzoyl 5′-adenosine), an irreversible protein kinase inhibitor (Zoller and Taylor, 1979). FSBA treatment was followed by addition of Cdc2 isolated by affinity purification using p13Suc1-Sepharose. Suc1 is a 13-kDa protein that has a high affinity for active Cdc2 associated with various B-type cyclins (Dunphy et al., 1988). As shown in Figure 4A, GST-Cdc18 was readily phosphorylated by Suc1-associated Cdc2. This phosphorylation was largely inhibited by GST-Rum1 (left panel), as was phosphorylation of histone H1 by Cdc2 (right panel). Unfused GST was not phosphorylated by Cdc2. As additional proof, two-dimensional tryptic phosphopeptide maps were performed with GST-Cdc18 phosphorylated by its associated kinase and with GST-Cdc18 phosphorylated by purified Cdc2 (Figure 4B). The maps were quite complex and evidently very similar, suggesting a minimum of five major phosphorylation sites and many minor sites. In total these studies strongly suggest that GST-Cdc18 associates with active Cdc2-Cig2 and Cdc2-Cdc13 kinases and undergoes phosphorylation at multiple sites.
Figure 4.
GST-Cdc18 is an in vitro substrate of Cdc2 kinase. (A) Precipitated GST-Cdc18 was treated with 1 mM FSBA to irreversibly inactivate the copurified protein kinases (compare lane 1, not treated GST-Cdc18, with lane 2, FSBA-treated GST-Cdc18). GST-Cdc18 treated with FSBA was mixed with active p13suc1-Sepharose-purified Cdc2 kinase in [γ-32P]ATP kinase assay buffer in the presence (lane 4) or absence of 250 nM GST-Rum1 (lane 3). GST-Cdc18 phosphorylation was restored by the addition of Cdc2 kinases in the reaction. The ability of Cdc2 kinases to phosphorylate GST-Cdc18 or histone H1 (right panel, lanes 5 and 6) was largely inhibited by GST-Rum1. (B) Two-dimensional tryptic phosphopeptide maps of GST-Cdc18 phosphorylated by associated kinases (left panel) or by Cdc2 kinase (right panel) were very similar. (C) Phospho-amino acid analysis of GST-Cdc18 phosphorylated by associated kinases detected only phosphothreonine.
Cdc2 Phosphorylates Cdc18 on T104
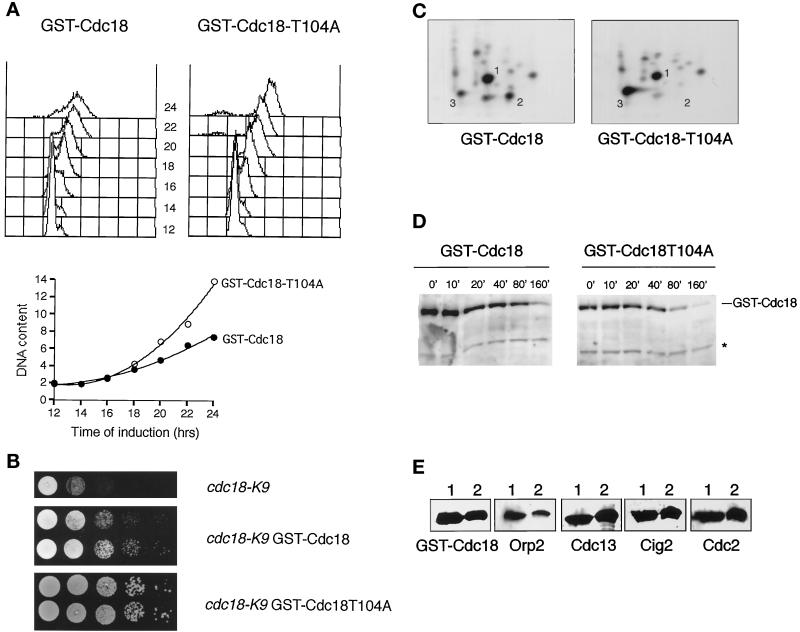
Cdc2 prefers to phosphorylate serine or threonine in the motif S/T-P-X-K/R, in which X is any amino acid (Holmes and Solomon, 1996). Phospho-amino acid analysis indicated that GST-Cdc18 was phosphorylated almost exclusively on threonine by its associated kinase (Figure 4C). Remarkably, the N-terminal domain of Cdc18 has five T-P-X-K/R motifs starting at positions 10, 46, 60, 104, and 134. A sixth T-P-X-K/R motif starts at 374. Accordingly, the threonine codons were individually mutated to alanine in six nmt1:GST:cdc18 constructs. The constructs were integrated into S. pombe and tested in the rereplication assay. Five of the mutant constructs induced rereplication in a manner that was indistinguishable from wild-type GST-Cdc18. However, rereplication was moderately enhanced in cells that expressed the fifth mutant, GST-Cdc18T104A (Figure 5A). In agreement with these data, at low levels of expression cdc18-K9 was rescued better by GST-Cdc18T104A than GST-Cdc18 (Figure 5B). Tryptic phosphopeptide mapping revealed that GST-Cdc18T104A lacked one of the two major phosphopeptides (labeled 2 in Figure 5C). Note that these phosphopeptide patterns are less complex than those shown in Figure 2 due to more complete trypsin digestion achieved in this experiment. Mutation of T104 to serine did not alter the appearance of peptide 2, indicating that T104 was an authentic phosphorylation site. Curiously, loss of peptide 2 coincided with increased phosphorylation of peptide 3. The tryptic peptide containing T104 is located immediately C-terminal to a peptide that contains two T-P dipeptides in contexts that do not exactly match the Cdc2 consensus sequence but are nevertheless potential Cdc2 phosphorylation sites. Accordingly, threonine codons 98 and 101 were mutated to alanine and the mutant nmt1:GST:cdc18 constructs were integrated and expressed. Both constructs induced rereplication in a manner indistinguishable from wild-type GST-Cdc18. Tryptic phosphopeptide mapping revealed that the T98A and T101A mutants lacked phosphopeptide 3, indicating that one and perhaps both sites were phosphorylated. These findings strongly suggest that the T104A phenotype is a specific consequence of the absence of phosphorylation at residue 104.
Figure 5.
GST-Cdc18 is activated by mutation of the threonine-104 phosphorylation site. (A) DNA content was examined in cells expressing GST-Cdc18 or GST-Cdc18-T104A constructs from the nmt1 promoter. FACS analysis revealed that the DNA content of cells expressing GST-Cdc18-T104A increased more rapidly following derepression of the nmt1 promoter (upper panels). The mean DNA content is plotted in the lower panel. (B) Serial dilutions of the strains cdc18-K9 (upper panel), cdc18-K9 having a single integrated copy of pAL27 (nmt1:GST:cdc18+, middle panel) or cdc18-K9 having a single integrated copy of pAL27-T104A (nmt1:GST:cdc18T104A, bottom panel). The cells were spotted on a growth medium (YES) that allows only low-level expression from the nmt1 promoter and then tested for their ability to form colonies at the restrictive temperature of 35.5°C. The cdc18-K9 mutation was rescued better by GST-Cdc18T104A than GST-Cdc18. (C) The T104A mutation eliminates phosphopeptide 2. GST-Cdc18 and GST-Cdc18-T104A were expressed in S. pombe, purified with GSH-Sepharose, and incubated with Mg2+ and [γ-32P]ATP as described above. Phosphorylated GST-Cdc18 and GST-Cdc18-T104A were analyzed by two-dimensional tryptic phosphopeptide mapping. The maps of both proteins were very similar except that one of the major phosphopeptides detected within GST-Cdc18, labeled 2, was absent in GST-Cdc18-T104A. Note that these phosphopeptide patterns are less complex than those shown in Figure 2 due to more complete trypsin digestion achieved in this experiment. (D) The stability of GST-Cdc18 and GST-Cdc18-T104A proteins was determined by an nmt1 turn-off experiment. GST-Cdc18 or GST-Cdc18-T104A was expressed for 14 h before the nmt1 promoter was repressed by addition of thiamine. The levels of GST-Cdc18 and GST-Cdc18-T104A were analyzed by immunoblot analysis at the indicated time points and quantified using a cross-reacting band (*) as a loading control. The half-life of both proteins was very similar (∼50 min). Note that the half-life of GST-Cdc18 was quite long compared with HA epitope-tagged Cdc18 (∼5 min) (Muzi-Falconi et al., 1996b), presumably due to the stabilizing property of GST. (E) GST-Cdc18-T104A interacts with Cdc2-cyclin B complexes and Orp2 in vivo. S. pombe lysates derived from cells expressing GST-Cdc18 (lane 1) or GST-Cdc18-T104A (lane 2) were precipitated with GSH-Sepharose and subjected to immunoblot analysis. Orp2, Cdc2, Cdc13, and Cig2 were found in association with GST-Cdc18 and GST-Cdc18-T104A.
The T104A mutant phenotype may be attributable to increased activity or abundance of Cdc18. We addressed the latter possibility by immunoblot analysis of GST-Cdc18 and GST-Cdc18T104A following induction in thiamine-free medium. The abundance of GST-Cdc18 and GST-Cdc18T104A underwent a parallel and equal increase following induction in thiamine-free medium. Indeed, an nmt1 promoter turn-off experiment showed that GST-Cdc18 and GST-Cdc18T104A did not differ in stability (Figure 5D). This result strongly suggests that the T104A mutation enhances the intrinsic activity of GST-Cdc18 as opposed to altering GST-Cdc18 stability. Immunoblotting revealed that GST-Cdc18T104A coprecipitated with Orp2, Cdc2, Cdc13, and Cig2, indicating that the T104A mutation did not alter the association between Cdc18 and these interacting proteins (Figure 5E).
Role of Cig2 in the Negative Regulation of Cdc18
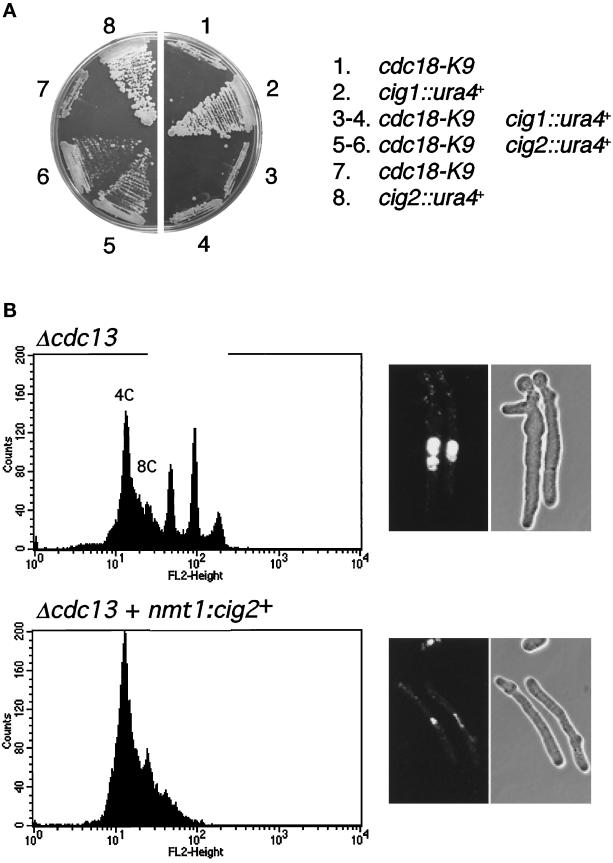
Our findings are fully consistent with studies showing that Cdc2-Cdc13 kinase is required to prevent reactivation of replication origins during G2 phase (Hayles et al., 1994). However, previous studies indicated that Cdc2-Cig2 kinase is exclusively involved in promoting the onset of S phase from G1 phase (Fisher and Nurse, 1996; Martin-Castellanos et al., 1996; Mondesert et al., 1996). Therefore, the association involving Cdc18 and Cig2 was inconsistent with the hypothesis that phosphorylation of Cdc18 catalyzed by Cdc2 has a purely inhibitory role. We therefore reopened studies of the role of Cig2 in regulating DNA replication by examining the interaction involving the Δcig2 mutation and the temperature-sensitive mutation cdc18-K9. If Cdc2-Cig2 kinase activates Cdc18, the model predicted that Δcig2 might exacerbate cdc18-K9. On the other hand, if Cdc2-Cig2 inhibits Cdc18, the model predicted that Δcig2 might suppress cdc18-K9. We found that Δcig2 suppressed the cdc18-K9 growth defect at 35.5°C, whereas Δcig1 mutation failed to rescue cdc18-K9, indicating that Cdc2-Cig2 inhibits Cdc18 (Figure 6A).
Figure 6.
Cig2 B-type cyclin contributes to the inhibition of Cdc18 and DNA replication. (A) The cdc18-K9 temperature-sensitive mutation is rescued by deletion of the cig2+ gene. Strains harboring Δcig1, Δcig2, and cdc18-K9 mutations alone or in combinations were tested for their ability to form colonies in YES medium at the restrictive temperature of 35.5°C. The cig1+ gene encodes a B-type cyclin of unknown function. The Δcig1 and Δcig2 mutants grow as well as wild-type at 35.5°C. The Δcig1 mutation failed to rescue cdc18-K9. (B) Overexpression of Cig2 inhibits rereplication in cells lacking Cdc13. Cells lacking Cdc13 were obtained by selective germination of spores from strain PR1334 (cdc13::his7+/cdc13+ ura4+/ura4-D18, top) or JLP162 (cdc13:: his7+/cdc13+ nmt1:cig2+(ura4+)/ura4-D18, bottom). Rereplication was strongly inhibited by expression of cig2+ from the nmt1 promoter. Germinated cells were analyzed by FACS (left) or photographed after staining with DAPI (right). Representative data for cells after 16 h at 32°C are shown.
We also tested whether the Cig2 cyclin was capable of inhibiting rereplication. For this, cig2+ was expressed from the nmt1 promoter to uncouple cig2+ expression from cell cycle effects and to provide high levels of Cig2 that might overcome negative regulation by inhibitors or proteolysis. Selective spore germination was used to obtain cells lacking the essential B-cyclin Cdc13. As previously reported, Δcdc13 spores having only the wild-type allele of cig2+ are unable control replication and rereplicate their DNA without entering mitosis. Overexpression of cig2+ prevented the rereplication but was not sufficient to drive mitosis (Figure 6B). Germination experiments of Δcdc13 nmt1:cig2+ spores in presence of B1 gave an intermediate result, thus the level of cig2+ expression is responsible for the activity to inhibit rereplication. The observed effects are unlikely to be caused by cig2+ toxicity because overexpression of cig2+ has little effect on wild-type cells.
DISCUSSION
Genetic studies of fission yeast have established that the cyclin-dependent kinase Cdc2 is required for both positive and negative regulation of DNA replication. Put simply, these studies have shown that elimination of Cdc2 activity in G1 phase prevents the onset of S phase, whereas elimination of Cdc2 kinase in G2 phase causes the reinitiation of S phase without an intervening M phase (Broek et al., 1991; Nurse and Bisset, 1981; Hayles et al., 1994; Moreno et al., 1994). The goal of the our investigations has been to discover substrates of Cdc2 that are involved in regulating the initiation of S phase. We have focused our attention of Cdc18 DNA replication factor for two key reasons. First, high expression of Cdc18 induces continuous DNA replication, suggesting that repression of Cdc18 may be important for preventing inappropriate activation of replication origins (Nishitani and Nurse, 1995). The second reason for concentrating on Cdc18 is that both it and Cdc2 interact with Orp2, a presumptive component of origin recognition complex in fission yeast (Leatherwood et al., 1996).
Herein we have described a series of physical and genetic interactions between Cdc2 and Cdc18. Foremost among these findings is the fact that functional GST-Cdc18 produced in fission yeast is associated with Cdc2 and two B-type cyclins, Cdc13 and Cig2. The GST-Cdc18 is isolated in association with an active protein kinases that phosphorylate Cdc18 and which are strongly inhibited by GST-Rum1. Rum1 is a highly specific inhibitor of Cdc2-Cdc13, as shown both by in vitro protein kinase assays and by the remarkably similar phenotypes which arise from high expression of rum1+ or deletion of cdc13+ (Hayles et al., 1994; Moreno et al., 1994; Nishitani and Nurse, 1995). Therefore, the potent inhibition of the Cdc18-associated kinase by Rum1 strongly suggests that Cdc2-Cdc13 is the major kinase that copurifies with GST-Cdc18. This conclusion receives further support from the close similarity of the two-dimensional tryptic phosphopeptide maps derived from GST-Cdc18 phosphorylated by its associated kinase as compared with Cdc2. Moreover, the context of one of the major phosphorylation sites, T104, exactly matches the consensus phosphorylation site sequence that is preferred by Cdc2. These findings constitute strong biochemical evidence that Cdc18 is a substrate of Cdc2 kinase, although it remains to be formally proven that Cdc2 phosphorylates Cdc18 in vivo. These observations are entirely consistent with another recent study (Brown et al., 1997).
Having discovered a close physical relationship involving Cdc18 and Cdc2, the questions remains as whether the interaction is important for regulating DNA replication and if so, is it concerned with the positive or negative regulation of S phase, or perhaps both. One approach to this problem is to begin with the hypothesis that Cdc2-Cdc18 association exists because Cdc18 is phosphorylated by Cdc13, hence mapping and mutating phosphorylation sites may reveal the role of Cdc2 in regulating Cdc18. As a first step in this investigative process we have mutated the six threonine residues in Cdc18 that are most likely to be phosphorylated by Cdc2. High expression of all six mutant forms of GST-Cdc18 induces continuous DNA replication at rate that is undiminished from that caused by high expression of wild-type GST-Cdc18. Therefore, in this type of assay, individual phosphorylation of the six sites is not required for Cdc18 activity. None of the mutant GST-Cdc18 proteins are deficient at inducing overreplication, but one of the mutants, T104A, consistently causes a moderate increase in the rate of DNA replication relative to wild-type. Importantly, T104 appears to be one of the major sites that is phosphorylated by Cdc2. Thus, at least in the Cdc18-induced DNA rereplication assay (Nishitani and Nurse, 1995), Cdc18 activity is enhanced by elimination of a site phosphorylated by Cdc2. Accordingly, at low levels of expression Cdc18-T104A exhibited an enhanced ability to rescue a cdc18-K9 mutant strain at restrictive temperature. Future experiments will reveal whether wild-type levels of expression of Cdc18-T104A induce replication abnormalities. Experiments are underway to perform gene replacement analysis of each of the mutations individually and in combination, as well as to determine which of the sites other than T104 are actually phosphorylated by Cdc2.
The biochemical data described above, which suggest that Cdc2 may catalyze the negative regulation of Cdc18, are consistent with the genetic data showing that Cdc2-Cdc13 represses the reinitiation of S phase in cells that are in the G2 phase. These data are also consistent with the observation that a moderate increase in Rum1 abundance will suppress, at least partially, the temperature-sensitive phenotype of a cdc18-K9 mutant (Jallepalli and Kelly, 1996). The Rum1 rescue of cdc18-K9 was proposed to occur through the inhibition of Cdc2-Cdc13 kinase. In this article, we have presented new biochemical and genetic evidence indicating that Cdc2-Cig2 kinase also contributes to the inhibition of Cdc18. The biochemical evidence is that Cig2 kinase coprecipitates with GST-Cdc18. We presume that Cdc2-Cig2 kinase also phosphorylates Cdc18, in fact our recent studies show that Cdc2-Cig2 kinase, purified by anti-Cig2 immunoprecipitation, phosphorylates GST-Cdc18 in vitro and produces a tryptic phosphopeptide map that closely matches the map generated from GST-Cdc18 that has been phosphorylated by its associated kinases.
The genetic data indicating that Cdc2-Cig2 contributes to the inhibition of Cdc18 is derived from two experiments. The first experiment showed that elimination of the cig2+ gene rescues cdc18-K9, whereas the second experiment revealed that constitutive overproduction of Cig2 suppressed inappropriate DNA replication in Δcdc13 cells. The rescue of cdc18-K9 by Δcig2 is reminiscent of the rescue of cdc18-K9 by Rum1 overproduction (Jallepalli and Kelly, 1996). Our new findings raise the question of whether the Rum1 effect is due to inhibition of Cdc2-Cdc13 or Cdc2-Cig2. We favor the latter possibility, largely because Cdc2-Cig2 is more active than Cdc2-Cdc13 during S phase, at least when both complexes are assayed as histone H1 kinases in vitro (Mondesert et al., 1996). We propose that there is a rather brief window during S phase in which reduction of Cdc2 kinase activity will rescue cdc18-K9, and it is during this window that Cdc2-Cig2 kinase is most active. Since the onset of S is normally triggered by Cdc2-Cig2 kinase, our new data suggest that in wild-type cells a single kinase, Cdc2-Cig2, may be primarily responsible for first activating primed replication origins and inhibiting reactivation of replication origins during S. The responsibility for inhibiting inappropriate initiation of DNA replication is then transferred to Cdc2-Cdc13 kinase as cells enter G2 phase. Of course, in Δcig2 cells, Cdc2-Cdc13 kinase is fully capable of performing both function of Cdc2-Cig2, therefore there appears to be no intrinsic difference between Cig2 and Cdc13 with respect to the positive and negative regulation of S phase. Further experiments will be required to test these hypotheses.
As mentioned above, a limitation of the phosphorylation site mutation studies presented in this report is that they have relied on high expression of GST-Cdc18 to assay changes in Cdc18 activity. Overexpression studies have been used in the past to illuminate the role of Cdc18 in regulating DNA replication (Nishitani and Nurse, 1995), and we believe they may be equally useful for dissecting the mechanisms of negative regulation of Cdc18. It is reasonable to propose that the negative regulation of Cdc18 that is catalyzed by Cdc2 kinase may be only one part of the mechanism by which Cdc2 kinase prevents reactivation of replication origins following the onset of S phase. In the normal cell cycle, cdc18+ mRNA expression is limited to S phase by a process that may also be controlled by Cdc2 (Kelly et al., 1993; Nishitani and Nurse, 1995; Muzi-Falconi et al., 1996b). This transcriptional regulation of cdc18+, coupled with the short half-life of Cdc18 protein that may also be regulated by Cdc2, may collaborate to provide a fail-safe mechanism that prevents inappropriate activation of replication origins following the onset of S phase.
In budding yeast Cdc6p associates with Cdc28p, a Cdc2 homologue, and Cdc28p is able to phosphorylate Cdc6p in vitro (Elsasser et al., 1996). Cdc6p is required for DNA replication but it can only promote initiation of replication when cyclin B-Cdc28 levels are very low, between the end of anaphase and the beginning of the next S phase (Piatti et al., 1996). Activation of Cdc28p-Cyclin B in late G1 defines a point at which Cdc6p synthesis can no longer promote DNA replication. The mechanism by which Clb-Cdc28p prevents Cdc6p function is unknown but is likely to involve phosphorylation.
The strong physical association between Cdc18 and Cdc2 is unusual for a kinase-substrate pair and is remarkably similar to the interaction of the Sic1p with Saccharomyces cerevisiae Cdc28p (Mendenhall, 1993; Schwob et al., 1994). Sic1p is an inhibitor of Cdc28. Sic1p readily associates with Cdc28 in vivo and is heavily phosphorylated by Cdc28 in vivo. In an analogous way, perhaps the tight association of Cdc18 with the Cdc2 kinase accounts for the cell cycle arrest when Cdc18 is overexpressed, either by Cdc2 or by preventing association of Cdc2 with other relevant targets. In this regard, it is interesting that S. cerevisiae CDC6 appeared as a high-copy suppressor of an S. pombe mutant strain that undergoes lethal mitosis due to premature activation of Cdc2 kinase (Bueno and Russell, 1992). High expression of CDC6 delays the onset of mitosis in both S. pombe and S. cerevisiae wild-type strains. These finding suggest that the interaction involving Cdc6p and Cdc18 with their cognate cyclin-dependent kinases may be conserved between the two yeasts. These observations also raise the question of whether high expression of Cdc18 results in direct inhibition of Cdc2 kinase activity and whether this plays an important role in producing the continuous DNA replication phenotype. Therefore, we should introduce a note of caution by stating that we cannot exclude the possibility that Cdc18-T104A possesses an enhanced ability to inhibit Cdc2.
ACKNOWLEDGMENTS
We thank G. Degols, C. McGowan, N. Rhind, and K. Shiozaki for technical advice; K. Gould and S. Reed for antibodies; the Scripps Cell Cycle Labs for general support; T. Kelly for communicating results before publication; Ministerio de Educación y Ciencia (Spain) (A.L.-G.), American Cancer Society (J.L.), and the National Institutes of Health (P.R.).
REFERENCES
- Alfa C, Fantes P, Hyams J, McLeod M, Warbrick E. Experiments with Fission Yeast. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- Boyle WJ, Van Der Geer P, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin layer cellulose plates. Methods Enzymol. 1991;210:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- Broek D, Bartlett R, Crawford K, Nurse P. Involvement of p34cdc2 in establishing the dependency of S phase on mitosis. Nature. 1991;349:388–393. doi: 10.1038/349388a0. [DOI] [PubMed] [Google Scholar]
- Brown G, Jallepalli P, Huneycutt B, Kelly T. Interaction of the S phase regulator Cdc18 with cyclin-dependent kinase in fission yeast. Proc Natl Acad Sci. 1997;94:6142–6147. doi: 10.1073/pnas.94.12.6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno A, Russell P. Dual functions of CDC6, a yeast protein required for DNA replication also inhibits nuclear division. EMBO J. 1992;11:2167–2176. doi: 10.1002/j.1460-2075.1992.tb05276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa-Bordes J, Nurse P. p25rum1 orders S phase and mitosis by acting as an inhibitor of the p34cdc2 mitotic kinase. Cell. 1995;83:1001–1009. doi: 10.1016/0092-8674(95)90215-5. [DOI] [PubMed] [Google Scholar]
- Dahmann C, Diffley J, Masmyth K. S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Curr Biol. 1995;5:1257–1269. doi: 10.1016/s0960-9822(95)00252-1. [DOI] [PubMed] [Google Scholar]
- Dunphy WG, Brizuela L, Beach D, Newport J. The Xenopus cdc2 protein is a component of MPF, a cytoplasmic regulator of mitosis. Cell. 1988;54:423–431. doi: 10.1016/0092-8674(88)90205-x. [DOI] [PubMed] [Google Scholar]
- Elsasser S, Lou F, Wang B, Campbell JL, Jong A. Interaction between yeast Cdc6 protein and B-type cyclin/Cdc28 kinases. Mol Biol Cell. 1996;7:1723–1735. doi: 10.1091/mbc.7.11.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F, Newport J. Evidence that the G1-S and G2-M transitions are controlled by different cdc2 proteins in higher eukaryotes. Cell. 1991;66:731–742. doi: 10.1016/0092-8674(91)90117-h. [DOI] [PubMed] [Google Scholar]
- Fisher DL, Nurse P. A single fission yeast mitotic cyclin B p34cdc2 promotes both S-phase and mitosis in the absence of G1-cyclins. EMBO J. 1996;15:850–860. [PMC free article] [PubMed] [Google Scholar]
- Gavin KA, Hidaka M, Stillman B. Conserved initiator proteins in eukaryotes. Science. 1995;270:1667–1671. doi: 10.1126/science.270.5242.1667. [DOI] [PubMed] [Google Scholar]
- Guan K, Dixon JE. Eukaryotic proteins expressed in Escherichia coli: An improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- Hayles J, Fisher D, Woollard A, Nurse P. Temporal order of S phase and mitosis in fission yeast is determined by the state of the p34cdc2-mitotic B cyclin complex. Cell. 1994;78:813–822. doi: 10.1016/s0092-8674(94)90542-8. [DOI] [PubMed] [Google Scholar]
- Holmes JK, Solomon MJ. A predictive scale for evaluating cyclin-dependent kinase substrates. A comparison of p34cdc2 and p33cdk2. J Biol Chem. 1996;271:25240–25246. doi: 10.1074/jbc.271.41.25240. [DOI] [PubMed] [Google Scholar]
- Hua X, Yan H, Newport J. A role for Cdk2 kinase in negatively regulating DNA replication during S phase of cell cycle. J Cell Biol. 1997;137:183–192. doi: 10.1083/jcb.137.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson P, Chevalier S, Philippe M, Kirschner M. Early events in DNA replication require Cyclin E and are blocked by p21Cip1. J Cell Biol. 1995;130:755–769. doi: 10.1083/jcb.130.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jallepalli PV, Kelly TJ. Rum1 and Cdc18 link inhibition of cyclin-dependent kinase to the initiation of DNA replication in Schizosaccharomyces pombe. Genes Dev. 1996;10:541–552. doi: 10.1101/gad.10.5.541. [DOI] [PubMed] [Google Scholar]
- Keeney JB, Boeke JD. Efficient targeted integration at leu1–32 and ura4–294 in Schizosaccharomyces pombe. Genetics. 1994;136:849–856. doi: 10.1093/genetics/136.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly TJ, Martin GS, Forsburg SL, Stephen RJ, Russo A, Nurse P. The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell. 1993;74:371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- Leatherwood J, Lopez-Girona A, Russell P. Interaction of Cdc2 and Cdc18 with a fission yeast ORC2-like protein. Nature. 1996;379:360–364. doi: 10.1038/379360a0. [DOI] [PubMed] [Google Scholar]
- Martin-Castellanos C, Labib K, Moreno S. B-type cyclins regulate G1 progression in fission yeast in opposition to the p25rum1 cdk inhibitor. EMBO J. 1996;15:839–849. [PMC free article] [PubMed] [Google Scholar]
- Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- Mendenhall MD. An inhibitor of p34CDC28 protein kinase activity from Saccharomyces cerevisiae. Science. 1993;259:216–219. doi: 10.1126/science.8421781. [DOI] [PubMed] [Google Scholar]
- Mondesert O, McGowan CH, Russell P. Cig2, a B-type cyclin, promotes the onset of S in Schizosaccharomyces pombe. Mol Cell Biol. 1996;16:1527–1533. doi: 10.1128/mcb.16.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Moreno S, Labib K, Correa J, Nurse P. Regulation of the cell cycle timing of start in fission yeast by the rum1+ gene. J Cell Sci Suppl. 1994;18:63–68. doi: 10.1242/jcs.1994.supplement_18.9. [DOI] [PubMed] [Google Scholar]
- Moreno S, Nurse P. Regulation of progression through the the G1 phase of the cell cycle by the rum1+ gene. Nature. 1994;367:236–242. doi: 10.1038/367236a0. [DOI] [PubMed] [Google Scholar]
- Muzi-Falconi M, Brown G, Kelly T. DNA replication: Controlling initiation during the cell cycle. Curr Biol. 1996a;6:229–233. doi: 10.1016/s0960-9822(02)00464-5. [DOI] [PubMed] [Google Scholar]
- Muzi-Falconi M, Brown GW, Kelly TJ. cdc18+ regulates initiation of DNA replication in Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 1996b;93:1566–1570. doi: 10.1073/pnas.93.4.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Nurse P. Cell division cycle mutants altered in DNA replication and mitosis in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1981;182:119–124. doi: 10.1007/BF00422777. [DOI] [PubMed] [Google Scholar]
- Nishitani H, Nurse P. p65cdc18 plays a major role controlling the initiation of DNA replication in fission yeast. Cell. 1995;83:397–405. doi: 10.1016/0092-8674(95)90117-5. [DOI] [PubMed] [Google Scholar]
- Nurse P, Bisset Y. Gene required in G1 for commitment to the cell cycle and G2 for control of mitosis in fission yeast. Nature. 1981;292:558–560. doi: 10.1038/292558a0. [DOI] [PubMed] [Google Scholar]
- Piatti S, Bohm T, Cocker JH, Diffley JF, Nasmyth K. Activation of S-phase-promoting CDKs in late G1 defines a “point of no return” after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes Dev. 1996;10:1516–1531. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- Schwob E, Bohm T, Mendenhall MD, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- Shiozaki K, Russell P. Conjugation, meiosis and the osmotic stress response are regulated by Spc1 kinase via Atf1 transcription factor in fission yeast. Genes Dev. 1996;10:2276–2288. doi: 10.1101/gad.10.18.2276. [DOI] [PubMed] [Google Scholar]
- Stillman B. Cell cycle control of DNA replication. Science. 1996;274:1659–1664. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- Wuarin J, Nurse P. Regulating S phase: CDKs, licensing and proteolysis. Cell. 1996;85:785–787. doi: 10.1016/s0092-8674(00)81261-1. [DOI] [PubMed] [Google Scholar]
- Zoller MJ, Taylor SS. Affinity labelling of the nucleotide binding site of cAMP-dependent protein kinase using ρ-fluorosulfonyl-[14C]benzoyl 5′-adenosine. Identification of a modified lysine residue. J Biol Chem. 1979;254:8363–8368. [PubMed] [Google Scholar]