Abstract
A gene (NhKIN1) encoding a kinesin was cloned from Nectria haematococca genomic DNA by polymerase chain reaction amplification, using primers corresponding to conserved regions of known kinesin-encoding genes. Sequence analysis showed that NhKIN1 belongs to the subfamily of conventional kinesins and is distinct from any of the currently designated kinesin-related protein subfamilies. Deletion of NhKIN1 by transformation-mediated homologous recombination caused several dramatic phenotypes: a 50% reduction in colony growth rate, helical or wavy hyphae with reduced diameter, and subcellular abnormalities including withdrawal of mitochondria from the growing hyphal apex and reduction in the size of the Spitzenkörper, an apical aggregate of secretory vesicles. The effects on mitochondria and Spitzenkörper were not due to altered microtubule distribution, as microtubules were abundant throughout the length of hyphal tip cells of the mutant. The rate of spindle elongation during anaphase B of mitosis was reduced 11%, but the rate was not significantly different from that of wild type. This lack of a substantial mitotic phenotype is consistent with the primary role of the conventional kinesins in organelle motility rather than mitosis. Our results provide further evidence that the microtubule-based motility mechanism has a direct role in apical transport of secretory vesicles and the first evidence for its role in apical transport of mitochondria in a filamentous fungus. They also include a unique demonstration that a microtubule-based motor protein is essential for normal positioning of the Spitzenkörper, thus providing a new insight into the cellular basis for the aberrant hyphal morphology.
INTRODUCTION
Motility events within living cells involve components of the cytoskeleton acting as tracks along which motor proteins transport various cargos (Allan, 1995; Cole and Lippincott-Schwartz, 1995; Vallee and Sheetz, 1996). Microtubule (MT)-associated motor proteins are primarily of two types, dynein and kinesin. The kinesin superfamily is divided into various subfamilies comprising conventional kinesin and several kinesin-related proteins (KRPs) (Bloom and Endow, 1995; Moore and Endow, 1996). Conventional kinesins are mainly involved in the intracellular transport of membranous organelles, whereas most KRPs function in nuclear division (Brady, 1995; Barton and Goldstein, 1996; Vallee and Sheetz, 1996).
Filamentous fungi, including the ascomycete Nectria haematococca, grow as tubular filaments called “hyphae,” which are compartmentalized into cytoplasmically connected cells by perforated cross-walls. Hyphae grow only at the apex, or apical dome, of the leading cell, the hyphal tip cell. This highly localized and polarized growth process, here referred to as “elongation,” is generally acknowledged to involve the subapical production of secretory vesicles that are transported to the apex where they fuse with the plasma membrane, thus contributing to the locally and rapidly expanding cell wall and plasma membrane (Heath, 1994). Transport of the secretory vesicles from their site of production by Golgi bodies, or Golgi equivalents, to the growing apex is believed to involve MTs, F-actin, or both (Heath, 1994). Evidence for a role of the motor proteins – dynein, kinesin, and myosin – in this vesicle transport process is reviewed briefly in the DISCUSSION. Once the secretory vesicles arrive in the vicinity of the apex, they are apparently shuttled into a (usually) visible, primary cluster of vesicles referred to as the “Spitzenkörper” (Bartnicki-Garcia et al., 1989) or into smaller satellite Spitzenkörper, which then fuse with the primary Spitzenkörper (López-Franco et al., 1995). According to the hyphoid model of hyphal tip growth, the Spitzenkörper is envisaged to act as a vesicle supply center from which the secretory vesicles are released and move radially in all directions to the cell surface, where they fuse with the plasma membrane (Bartnicki-Garcia et al., 1995a). Furthermore, it has been demonstrated that the position and movement of the Spitzenkörper determines the shape and form of the hypha (Bartnicki-Garcia et al., 1995b). What controls the location of the Spitzenkörper has, therefore, become a question of great importance in the study of fungal morphogenesis.
In addition to their role in the transport of membranous organelles, motor proteins are also involved in generating mitotic forces (Vallee and Sheetz, 1996). During anaphase B, for example, KRPs may generate force within the spindle that helps to elongate the spindle and separate the spindle poles (Saunders, 1993).
We are examining the roles of various MT-associated motor proteins during growth, morphogenesis, and mitosis in N. haematococca. This initial report focuses on the cloning of a gene (NhKIN1) encoding a member of the conventional kinesin family and the phenotypic characterization of a mutant deleted for this motor protein. Similar phenotypes of an unstable mutant of the same gene were reported elsewhere (Aist et al., 1996).
MATERIALS AND METHODS
Strains and Media
Wild-type (WT) isolate T213 of N. haematococca Berk. and Br. mating population VI was the progenitor of mutants generated for this study. Transformant TSN25 was deleted for NhKIN1 by transformation with linearized vector pTMS5 (Table 1 and Figure 2) which resulted in a double cross-over, homologous recombination event that replaced NhKIN1 with the selectable marker gene hygB (which confers resistance to hygromycin β). Transformant TSN20 carried pTMS5 at an ectopic site, leaving NhKIN1 intact. All strains were stored in 25% glycerol at −80°C and freshly recovered for each experiment. Culture media included complete with xylose (CMX, Leach et al., 1982; Tzeng et al., 1992), yeast extract and 1% glucose (YEG, Aist and Bayles, 1988), 1% yeast extract plus 0.5% glucose and 1.0% Gelrite (LH-YEGGR), yeast extact-peptone plus 2% dextrose (YEPD, Sherman et al., 1986), V-8 and glucose-asparagine (GA) (VanEtten and Stein, 1978).
Table 1.
Plasmids
| Name | Size (kb) | Characteristics | Reference |
|---|---|---|---|
| λZAPII | Vector for construction of subgenomic library | Stratagene | |
| pBS (SK−) | 2.96 | Excised phagemid vector from λZAPII | Stratagene |
| pUC18 | 2.7 | Cloning vector | Messing, 1983 |
| pCRII | 3.9 | Cloning vector | Invitrogen |
| pBS (KS+) | 2.96 | Cloning vector | Stratagene |
| pklp73 | pBluescript SK− phagemid with genomic DNA including NhKIN1 | This study | |
| pTA16 | 4.5 | 601 bp NhKIN1 fragment inserted into pCRII | This study |
| pC1-42 | 3.86 | 0.9 kb NhKIN1 cDNA fragment | This study |
| Accession no. U86521 2400-3310 bp cloned into pBS (KS+) | |||
| pC4-1 | 5.96 | 3.0 kb NhKIN1 cDNA fragment | This study |
| Accession no. U86521 2710-5810 bp cloned into pBS (KS+) | |||
| pBSklp6 | 11.96 | 9.0 kb EcoRI genomic DNA fragment from pklp73 containing NhKIN1 inserted into pBS (KS+) | This study |
| pCWhyg1 | 5.0 | PUC19 containing hygB cassette for fungal transformation | Wasmann, University of AZ |
| pMR1003 | 6.4 | 3.4 kb Pst1-BamHI fragment carrying KAR3 cloned into pBS M13 KS− (Stratagene) | Meluh and Rose, 1990 |
| pTMS4 | 8.1 | 5.4 kb SphI-ApaLI (blunt ended) genomic NhKIN1 cloned into SphI-SmaI site of Puc18 | This study |
| pTMS5 | 7.4 | 2.3 kb hygB SalI (blunt ended)-XbaI fragment of pCWhyg1 cloned into the SacII (blunt ended)-NheI site of pTMS4 | This study |
Figure 2.
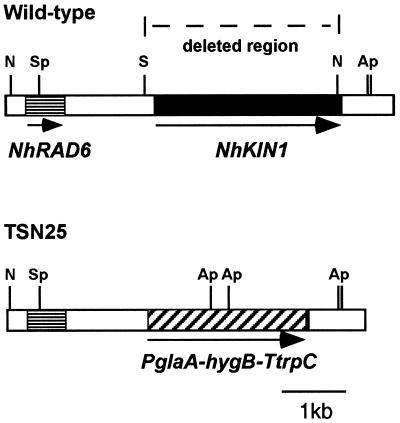
Deletion of NhKIN1 from the wild-type genome. The wild-type chromosome shows NhKIN1, the deleted region, and RAD6, an apparent homologue of genes encoding ubiquitin-conjugating proteins. The SphI-KpnI fragment of pTMS5 was used to delete most of NhKIN1. A double cross-over event resulted in the recombinant chromosome, which is deleted for a region starting 182 bp 5′ of the translational start site and stopping 26 bp 5′ of the translation stop site of NhKIN1. The deleted sequence was replaced with hygB. Certain restriction enzyme sites mentioned in the text are noted: N, NheI; Ap, ApaLI; K, KpnI; S, SacII; and Sp, SphI.
Fungal Nucleic Acid Isolation
For DNA extraction, plugs bearing mycelium grown on LH-YEGGR were inoculated into a 125-ml flask containing 50 ml YEG plus 0.1% MgCl2. Flasks were shaken (125 rpm) at 25°C; mycelium was harvested after 3 d, lyophilized, and ground in liquid nitrogen in a mortar, and DNA was isolated as described previously (Yoder, 1988).
An alternative DNA isolation procedure employed the Wizard Genomic DNA Purification Kit (Promega, Madison, WI), with the following modifications. Lyophilized mycelium (300 μl) was added to a 2.0-ml microcentrifuge tube containing 150 μl glass beads (0.45–0.5 mm diameter). Mycelium was crushed to a powder using a glass rod. Cells were lysed by adding 1.5 ml Tris-EDTA buffer (50 mM Tris/HCl, pH 7.5, 50 mM EDTA pH 8.0, 3% SDS, 1% mercaptoethanol) and inverting the tube several times, followed by a 30-min incubation at 65°C and centifugation at 12,000 × g in a microcentrifuge (Microfuge 12, Beckman, Fullerton, CA) for 10 min. The supernatant was transferred to a new tube, 500 μl of Wizard protein precipitation solution were added, and the tube was vortexed for 20 s. After 5 min on ice, the mixture was centrifuged at 12,000 × g for 10 min. The supernatant was divided equally between two 1.5-ml microcentrifuge tubes, and 0.7 volume absolute alcohol was added to each tube and mixed gently by inverting the tube until the DNA formed a visible mass. The DNA was pelleted by centrifugation for 10 min at 12,000 × g, washed with 1 ml 70% ethanol, recentrifuged for 2 min at 12,000 × g, and dissolved in 100 μl Tris-EDTA after air drying for 10–15 min.
To isolate RNA, 290 mg of lyophilized mycelium were ground in a mortar in 9 ml guanidine hydrochloride-containing buffer and extracted with phenol and chloroform. After a 45-min centrifugation at 20,000 rpm in a SM34 rotor, total RNA was precipitated from the supernatant with ethanol and acetic acid (Logemann et al., 1987). Poly(A)+ RNA was purified from total RNA using oligo(dT)+ cellulose (Sambrook et al., 1989).
Blotting and Hybridization Procedures
DNA blots were prepared and hybridizations were done as described by Turgeon et al. (1993).
PCR
Degenerate primers, 5′ tacgaattc (T/C)T(C/I)GC(C/I)TA(T/C)GG(I/C)CA(A/G)AC (C/I)GG 3′ (primer 1) encoding amino acids (F/L)AYGQTG and 5′ gctgaattc (A/T)(C/I)T(C/T)(C/I) C(G/T)(A/G)(A/T)A(C/I)GG(A/G)AT(A/G)TG 3′ (primer 3) encoding amino acids HIP(Y/F)R(N/E)S were synthesized by the Cornell University Oligonucleotide Synthesis Facility (Ithaca, NY). Lower case letters indicate an EcoRI site added for subsequent cloning and three additional nucleotides to promote efficient cleavage. For amplification from genomic DNA, reaction mixtures (100 μl) contained approximately 1 μg of genomic DNA as template, 100 pmol of each primer, 5 U of Taq polymerase (Promega), 10 mM Tris-HC1, pH 8.3, 50 mM KC1, 12.5 mM MgCl2, and 200 mM each dNTP. The thermal program included one cycle at 94°C (2 min), 45 cycles of [95°C (1 min), 40°C (2 min)] and 72°C (1 min)] and one cycle at 72°C (10 min).
PCR products were resolved by agarose (3% NuSieve, FMC Bio Products, Rockland, ME) gel electrophoresis. A gel slice containing a band of the expected size (∼600 base pair [bp]) that hybridized to KAR3 (Table 1) (encoding a kinesin-related protein from Saccharomyces cerevisiae [Meluh and Rose, 1990]) was excised, and DNA was eluted with GENECLEAN (Bio101, Vista, CA), and ligated into the pCRII cloning vector (Invitrogen, San Diego, CA). One clone (pTA16, Table 1) containing the 601-bp insert was sequenced from both ends with primers SP6 and M13–40 using a Sequenase 2.0 kit (United States Biochemical, Cleveland, OH).
For reverse transcriptase (RT)-PCR, 1.2 μg poly(A)+ RNA were used as template to synthesize first-strand cDNA in a 20-μl reaction containing 1× Taq DNA polymerase buffer (Promega), 1 mM each dNTP, 1 U RNAsin/μl, 100 pmol primer 3 (Figure 1), and 200 U of Moloney MuLV reverse transcriptase (Life Technologies, Grand Island, NY). The reaction was carried out for 10 min at room temperature, followed by 60 min at 42°C, 5 min at 95°C, and then quick-chilled on ice. Eighty microliters of [1× Taq DNA polymerase buffer (Promega), 100 pmoles primer 1, 100 pmoles primer 3 and 5 U of Taq DNA polymerase (Promega)] were added to this mixture, and thermal cycling was done as described above.
Figure 1.
Predicted amino acid sequence of NhKIN1. Filled arrowheads denote positions of introns. Splice junctions were verified by sequencing of NhKIN1 cDNA. Conserved amino acids from which degenerate primers were made are underlined. Asterisks indicate the first amino acid in each of three potential nucleotide-binding motifs, identified by the program PROSITE (Bairock, 1992). Boxed amino acids designate a highly conserved region shared among conventional kinesins, as noted for Nkin (Steinberg and Schliwa, 1995). Open arrowheads depict positions of leucines in the leucine zipper motif.
Construction and Screening of a Subgenomic Library
To isolate the genomic copy of NhKIN1, genomic DNA (40 μg) was digested with EcoRI overnight and separated in 0.7% agarose. One lane containing the digest was blotted and probed with the 601-bp insert of pTA16 (Table 1); an ∼9-kilobase (kb) fragment hybridized. A band of this size was excised from the unblotted portion of the gel, eluted with GENECLEAN, and ligated with EcoRI-digested vector (l μl ZAPII; Stratagene, La Jolla, CA) and a 5-μl aliquot was packaged into phage according to the Gigapack II packaging extract instruction manual (Stratagene). The phage library was probed at 65°C with the 601-bp insert of pTA16.
Construction and Screening of a cDNA Library
PolyA+ RNA (6 μg) from mycelium cultured for 3 d was used in cDNA library construction as described in the ZAP-cDNA Gigapack II Gold Cloning Kit (Stratagene).
Sequence Analysis
pBSklp6 (Table 1) DNA was prepared by alkaline lysis and further purified by CsCl-ethidium bromide gradient (Sambrook et al., 1989). Nested deletions of the insert were made with a KpnI/ClaI enzyme combination (Promega), and sequencing was performed using Sequenase 2.0 kit (United States Biochemical). In addition, oligonucleotides (18–22 mers) were synthesized every 350 bp based on sequence obtained and used as primers for further sequencing at the Cornell DNA Sequencing Facility using TaqCycle automated sequencing with DyeDeoxy terminators (Applied Biosystems, Norwalk, CT) Both strands of the genomic clone of NhKIN1 in pBSklp6 were completely sequenced. cDNA clone pC1–42 (Table 1) was sequenced using the −20 universal primer and the reverse universal primer. pC4–1 (Table 1) was sequenced using the −20 universal and two NhKIN1-specific primers. Amino acid sequence comparison was performed using MegAlign DNASTAR program version 3.14a (DNASTAR, Madison, WI).
Construction of a Plasmid for Deletion of NhKIN1
To subclone NhKIN1, pBSklp6 (Table 1) was digested with ApaLI, blunt ended, and then digested with SphI. A 5.4-kb fragment, containing the entire NhKIN1 coding region and flanking DNA, was ligated into the SphI-SmaI sites of pUC18, generating pTMS4 (Table 1). A deletion construct, pTMS5 (Table 1 and Figure 2), was made by removing a SacII-NheI fragment from pTMS4 and replacing it with the hygB marker, as follows: pCWhyg1 (Table 1) (carrying hygB controlled by the Aspergillus nidulans glucoamylase promoter [PglaA] and tryptophan synthase terminator [TtrpC], provided by C. Wasmann and H. VanEtten, University of Arizona, Tucson, AZ) was digested with SalI and XmnI, blunt ended, and digested with XbaI. A 2.3-kb fragment containing hygB was isolated, ligated to SacII-digested, blunt ended, NheI-digested pTMS4 to yield pTMS5. Plasmid DNA was purified from Escherichia coli using a Qiagen midi column according to the manufacturer’s protocol (Qiagen, Chatsworth, CA).
Fungal Transformation
pTMS5 (Table 1) was linearized with SphI and KpnI, precipitated, resuspended in sorbitol-Tris-calcium buffer (1.2 M sorbitol, 10 mM Tris-HCl, 50 mM CaCl2), pH 7.5 (STC), and transformed into wild-type isolate T213 of N. haematococca as described by Wasmann and VanEtten (1996), with modifications adapted from Turgeon et al. (1987). Two enzymes (both from InterSpex, Foster City, CA), Novozyme 234 (10 mg/ml) and Driselase (10 mg/ml), were dissolved in osmoticum (1.2 M MgSO4, 10 mM sodium phosphate, pH 5.8) and stored overnight at 4°C; 10 μl β-glucuronidase/ml (Sigma, St. Louis, MO) was added the day of use and the solution was filtered. Aurintricarboxylic acid and spermidine were not used.
Colonies derived from single spores were grown on YEPD medium containing 50 μg hygromycin B/ml, and then transferred to V8 medium containing 50 μg hygromycin B/ml and incubated for 7 d under long-wave UV light at 20°C. Conidia were placed in 35 ml GA medium in 50-ml Falcon tubes and grown for 4 d with shaking at room temperature before harvesting of mycelium for DNA isolation.
Growth Rate Analysis
Strains T213, TSN20, and TSN25 were first grown on CMX plates at 27°C for 4 d. A piece of agar (4 mm2) bearing actively growing mycelium was cut 1 cm from the edge of the colony, transferred to the center of a new CMX plate, and incubated in the dark at 30°C. Colony diameters were measured at various intervals, and data were plotted using Cricket Graph III 1.53f.
Microscopic Analyses
Effects of NhKIN1 deletion on hyphal morphology and the organization and behavior of organelles in hyphal tip cells were determined by growing the isolates on microscope slides coated with LH-YEGGR. Microscopic images were recorded on videotape using the previously described videomicroscopy system (Aist and Bayles, 1991) upgraded with a Hamamatsu Argus-20 Image Processor (Hamamatsu Photonic Systems, Bridgewater, NJ), employing dark-field, epifluorescence, and phase-contrast optics. Images from the videotapes were captured and processed using Image-Pro Plus (Media Cybernetics, Silver Spring, MD). Prints of composite figures were made with a Codonics NP-1600 Photographic Network Printer (Codonics, Middleburg Heights, OH).
Visualization of MTs by immunofluorescence videomicroscopy was done according to the procedures described by Aist et al. (1991).
A new system for acquiring and plotting mitotic data from the videotapes was used. It was operated by a customized software program designed and compiled by William Schubarg (Empire Imaging Systems, Cicero, NY). The software uses Image-Pro Plus to capture images, and then makes and calculates selected linear measurements that are down-loaded to Excel for Windows spreadsheets. Problems of synchronization between the frame-grabber board and the VCR were solved by connecting a Hotronic model AP41 TBC/Frame Synchronizer (Hotronic, Campbell, CA) between the two. From the Excel files, plots of spindle elongation and movements of the spindle pole bodies and mitotic apparatus (MA) of each analyzed mitosis were made using Sigma Plot for Windows and printed on a Hewlett-Packard Laserjet 4 laser printer (Hewlett-Packard, Boise, ID). The rate of spindle elongation was determined from a 40- to 60-s period of linear elongation during the first half of anaphase B in each mitosis.
To quantify and compare the amount of migratory activity of the MAs in the three isolates, we used the plots generated as described above. A migration was defined as a one-way movement of the MA of ≥ 2 μm occurring in ≤ 20 s.
Measurements of elongation rate and diameter of hyphae, median cross-sectional area (size) of Spitzenkörper, and distance of mitochondria from the apical cell wall (Table 1) were made using the morphometric capabilities of Image-Pro Plus. Sizes of Spitzenkörper were measured using images “zoomed” to 200%, to enhance the precision of tracing the outlines of the Spitzenkörper. Data were down-loaded to Excel, and statistical analyses were performed using Minitab for Windows.
RESULTS
A portion of NhKIN1 (601 bp) was initially isolated by PCR amplification using degenerate primers corresponding to conserved regions of kinesin motor domains. Identity of the product was verified by sequencing, after which the cloned PCR product in pTA16 (Table 1) was used to probe a subgenomic library to obtain the entire coding region plus flanking DNA. The initial screening yielded four positive plaques among 50,000. Two remained positive after a second and third screening. Phage DNA was isolated from one positive plaque and digested with EcoRI to release the 9-kb insert, which was subcloned into the EcoRI site of pBS(KS+) (Table 1) to create two identical clones, pBSklp6 and pBSklp10 (Table 1). pBSklp6 was used for nested deletion reactions; 6050 bp were sequenced (GenBank accession number U86521).
NhKIN1 is predicted to encode a 929-amino acid polypeptide, with a molecular mass of 103 kDa and 77% identity to Neurospora crassa conventional kinesin encoded by NKIN (Steinberg and Schliwa, 1995). The NhKIN1 coding region is interrupted by two introns (Figure 1) whose relative positions are identical to those of NKIN. cDNA clones were obtained by probing a cDNA library (∼3 × 105 plaques) with the 601-bp insert of pTA16. Three positive plaques were identified, two of which (pC1–42 and pC4–1, Table 1) were analyzed further. The sequence of the 0.9-kb insert of pC1–42, which corresponds to nucleotide positions 2400–3310 (GenBank U86521), verified that the first intron includes nucleotides 2613–2680. Partial sequencing of the 3-kb insert of pC4–1, which corresponds to nucleotides 2710–5810, verified that the second intron includes nucleotides 5104–5161 (Figure 1).
Conventional kinesins have three distinct domains, an N-terminal motor domain containing a conserved P-loop motif required for ATP hydrolysis and an MT-binding site, an α-helical coiled-coil stalk domain, important for dimerization, and a globular tail region that is thought to interact with organelles or other cargo (Bloom and Endow, 1995). Within the putative motor domain of fungal conventional kinesins, NhKIN1 (amino acids 3–337) is 93.4% identical to N. crassa Nkin (amino acids 4–338) but only 71% identical to Ustilago maydis Kin2 (amino acids 2–340) (Lehmler et al., 1997). The C-terminal region (stalk and tail) of NhKIN1 (amino acids 338–929) is less conserved with only 67.9% identity to Nkin (amino acids 339–926) and 40.8% identity to Kin2 (amino acids 341–968). Like Nkin and Kin2, NhKIN1 contains a second P-loop motif in the motor domain, G/A(4X)GKT/S (Saraste et al., 1990), beginning at amino acid position 237 (see Figure 1). However, unlike Nkin, NhKIN1 contains a third P-loop motif toward the end of the presumptive stalk domain starting at position 723. Although the second and third P-loop motifs follow the general consensus, they do not contain a G in the fourth position of the motif (G is often in the fourth position G/AXXGXGKT/S), and the third motif lacks bulky hydrophobic residues within the five amino acids prior to the P-loop motif, common to most proteins that bind to ATP/GTP via the P-loop motif (Saraste et al., 1990; E. V. Koonin, National Center for Biotechnology Information, personal communication). Particularly interesting is the fact that NhKIN1 contains a leucine zipper motif (see Figure 1) that overlaps with a region in the C-terminal domain, which shows high similarity to an analogous area of Nkin and all other conventional kinesins (Hurst, 1995; Steinberg and Schliwa, 1995). Furthermore, the periodicity of the leucines in this region is conserved in kinesin heavy chains from human, Caenorhabditis elegans, Drosophila melanogaster, N. crassa, and U. maydis (Yang et al., 1989; Navone et al., 1992, Patel et al., 1993; Steinberg and Schliwa, 1995, Lehmler et al., 1997).
Upstream of NhKIN1 is another open reading frame, NhRAD6 (deposited with NhKIN1 under accession number U86521), encoding a protein with homology to the ubiquitin-conjugating proteins from S. cerevisiae RAD6 and N. crassa MUS8 (Reyonds et al., 1985; Soshi et al., 1996). The peptides derived from NhRAD6 and NcMus8 are 98% identical: both are 151 amino acids long and both genes have three introns at exactly the same positions. NhRAD6 is 66% identical to its homologue in wheat, suggesting a high level of conservation (Sullivan and Vierstra, 1991).
Deletion of NhKIN1
Loss of NhKIN1 function was achieved by replacing the majority of the coding region of NhKIN1 with the selectable marker hygB (Figure 2). Transformation of N. haematococca wild-type isolate T213 with linearized pTMS5 yielded 12 hygromycin-resistant transformants. Two different types of growth pattern were apparent on regeneration plates — normal wild-type growth rate and hyphal morphology, and a slower growth rate with a distinctly helical hyphal morphology. Gel blot analysis was performed on NheI/ApaLI-digested genomic DNA from three normal-growth and seven slow-growth transformants probed with the wild-type NhKIN1 insert of pTMS4. All seven slow-growth transformants sustained homologous integration of pTMS5 at the NhKin1 locus via a double cross-over recombination event, thus deleting most of NhKIN1 and resulting in a shift in size of the native 5.3- and 0.6-kb NhKIN1 bands to 3.1 and 1.7 kb, respectively (our unpublished results). Two normal-growth transformants showed banding patterns consistent with a homologous single-cross-over integration of two copies of pTMS5, generating fragments of 5.1, 3.1, 2.9, 2.4, and 2.4 kb and leaving an intact copy of NhKIN1; the third normal growth transformant had a banding pattern similar to the other two, plus an additional band of approximately 2.0 kb (possibly the result of an integration of pTMS5 at an ectopic site). The observation that several independent Nhkin1 deletion mutants displayed the same slow-growth phenotype suggested that slow growth results from loss of NhKIN1 function. One Nhkin1 deletion mutant (TSN25) and one transformant with an intact NhKIN1 gene (TSN20) were chosen for comparison with wild-type isolate T213 in microscopic and growth rate analyses.
We considered the possibility that gene redundancy could explain the apparent nonessential nature of NhKIN1; several observations suggest that NhKIN1 is present as a single copy. First, if more than one copy of NhKIN1 were to exist in the genome, then probing with a fragment encoding NhKIN1 should yield at least two distinct bands, unless the restriction digest were to yield the same banding pattern. However, in all seven of the slow-growth transformants tested, a probe encoding the entire NhKIN1 protein, plus additional flanking DNA, hybridized strongly only to fragments of the predicted size for disruption at the locus corresponding to the cloned NhKIN1 gene (described in detail above). Second, a probe containing sequence encoding a highly conserved region of the NhKIN1 motor domain (pTA16 insert, Table 1) hybridized to a single band in SalI-digested N. haematococca DNA, and the size of the band shifted in SalI-digested genomic DNA from an Nhkin1− disrupted transformant (our unpublished results; Aist et al., 1996), eliminating the possibility of an additional hybridizing band of the same size. Third, if there is a second, but highly diverged, conventional kinesin-encloding gene, it could not be detected by PCR with primers corresponding to highly conserved regions of the KHC motor domain. Using such primers (see MATERIALS AND METHODS) we generated and sequenced 27 PCR products; all were identical to NhKIN1. And fourth, using genomic DNA from the Nhkin1− deletion strain as template for PCR, we successfully amplified genes for two additional kinesin-related proteins belonging to the BIMC and KAR3 subfamilies (our unpublished results). No other conventional kinesin genes were identified. These results, taken together, argue persuasively that there is a single genomic copy of the conventional kinesin gene in N. haematococca and, therefore, that NhKIN1 is nonessential. However, a second conventional kinesin cannot be formally eliminated.
Growth Phenotypes Caused by Deletion of NhKIN1
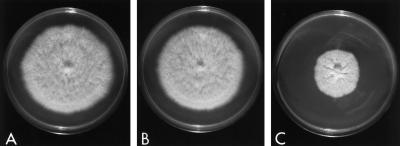
As shown in Figure 3, colonies of NhKIN1+ transformant TSN20 grew at about the same rate as did those of the wild type. Deletion of NhKIN1 reduced the rate of colony growth to about 50% that of the controls (Figure 3). Plots of the radial growth revealed that WT and TSN20 colonies grew at an average rate of about 5.2 mm/d, whereas colonies of the Nhkin1− mutant TSN25 grew at an average rate of 2.5 mm/d. Similarly, the average growth rate of individual hyphal tip cells of the mutant was only 41–45% that of the controls (Table 2).
Figure 3.
Growth of wild-type isolate T213 (A), NhKIN1+ transformant TSN20 (B), and Nhkin1− mutant TSN25, which lacks KIN1 (C), on CMX at 30°C for 6 d.
Table 2.
Effects of NhKIN1 deletion on hyphal tip cells, organelles, and anaphase B in Nectria haematococca
| Fungal isolate | Elongation rate of hyphae (μm/min) n = 8 | Diameter of hyphae (μm) n = 8 | Area of Spitzenkörper (μm2) n = 8 | Distance to mitochondria (μm) n = 8 | Elongation rate of spindle (μm/min) n = 15 |
|---|---|---|---|---|---|
| WT | 3.06(±0.68)a | 4.19(±0.37)a | 1.99(±0.37)a | 2.37(±0.65)a | 5.43(±1.49)a |
| TSN25 Nhkin1− | 1.27(±0.33)b | 3.19(±0.71)b | 1.00(±0.39)b | 23.77(±7.27)b | 4.85(±1.26)a |
| TSN20 NhKIN1+ | 2.84(±0.36)a | 4.01(±0.22)a | 1.67(±0.28)a | 2.27(±0.75)a | ---c |
Data are means (± 1 SD), and n = sample size. Values in each column not followed by the same letter were significantly different (P < 0.02) by the two-sample t test. For the spindle elongation rate, P = 0.26.
Not determined.
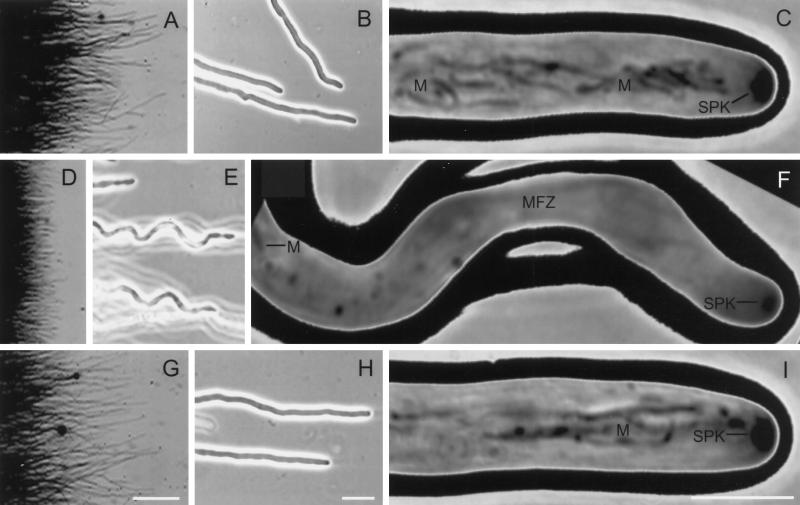
In addition, the Nhkin1− mutant exhibited a wavy or helical hyphal morphology (Figure 4, E and F), compared with the straight or slightly wavy hyphal morphology of either the NhKIN1+ strain TSN20 (Figure 4, H and I) or the WT (Figure 4, B and C). Nhkin1− colonies had discrete colony margins (Figures 3B and 4D) and densely packed (Figure 4D), wavy or helical hyphal tip cells with reduced diameter (Table 2). Careful focusing at high magnification revealed that the majority of the hyphal tip cells were actually in the shape of a right-handed helix (Figure 4E). Nhkin1− colonies growing on solid media were further distinguished from the controls by radial creases in the mycelial mat (Figure 3C) and indentation of the medium surface at colony margins.
Figure 4.
Videomicrographs depicting the development of distinctive mycelial, hyphal, and cellular characteristics of Nhkin1− deletion mutant TSN25 relative to those of NhKIN1+ transformant TSN20 and WT. (A), (D), and (G), Reverse dark-field images of colony margins of WT (A), TSN25 (D), and TSN20 (G), illustrating the relatively larger number of hyphal tip cells arranged more uniformly in the mutant (D) than in the other two isolates. (B), (E), and (H) Phase-contrast images. At low magnification, the mutant tip cells (E) exhibit a regular undulation that describes a spiral in three dimensions, whereas the WT (B) and TSN20 (H) exhibit relatively straight growth. (C), (F), and (I) Phase-contrast images. High magnification of apical portions of living hyphal tip cells, to illustrate some of the effects of NhKIN1 deletion on organelles. In the mutant cell (F), mitochondria (M) do not occupy the apical region (MFZ, mitochondria-free zone) as they do in the WT (C) and TSN20 (I) cells. Also, the Spitzenkörper (SPK) in the mutant is smaller than in either the WT or TSN20 cells. Bars: (A), (D), and (G), 0.5 mm; (B), (E), and (H), 20 μm; (C), (F), and (I), 5 μm.
Effects of NhKIN1 Deletion on Organelle Distribution and Vesicle Transport
When growing hyphal tip cells were examined at high magnification (Figure 4, C, F, and I), striking effects of NhKIN1 deletion on organelle distribution were observed. Mitochondria in growing hyphal tips of filamentous, ascomycetous fungi are well known to be highly elongated, phase-dark, undulating organelles that are readily identified in living cells (Girbardt, 1969; Grove and Bracker, 1970). The mitochondria in the mutant, although retaining this characteristic morphology and appearance, did not occupy their normal subapical position behind the Spitzenkörper (Figure 4, C and I), but instead there was a long (16–30 μm) gap of mitochondrion-free cytoplasm (Table 2 and Figure 4F). Within this gap was a normal array of cytoplasmic MTs, extending almost to the apex of the hyphal tip, as in control cells (Figure 5).
Figure 5.
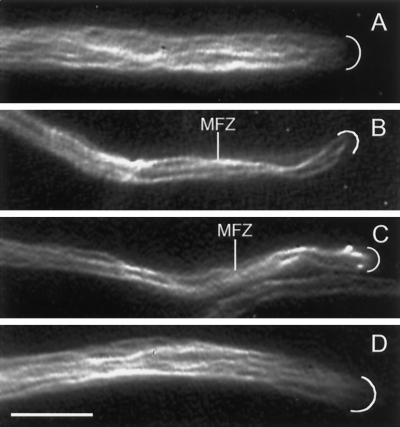
Fluorescence videomicrographs showing immunocytochemical localization of MTs in hyphal tip cells of the wild type (A), the NhKIN1− deletion mutant TSN25 (B and C), and the NhKIN1+ transformant TSN20 (D). The MT distribution in the mutant cells appears normal, with MTs present throughout the mitochondrion-free zone (MFZ). White arcs mark the positions of the hyphal apices. Bar, 5 μm.
Spitzenkörper in the mutant hyphal tip cells were smaller (median cross-sectional area) compared with those in control cells (Table 2 and Figure 4, C, F, and I). As growth of the hyphal tips of the mutant was characteristically wavy or helical, we looked for indications that aberrant Spitzenkörper behavior might be involved in producing this morphology. We observed that changes in the position of the Spitzenkörper, from central to eccentric, within the apical dome of mutant cells were associated with subsequent and corresponding changes in the direction of hyphal tip growth, whereas the Spitzenkörper of control cells typically remained centrally located (Figure 6).
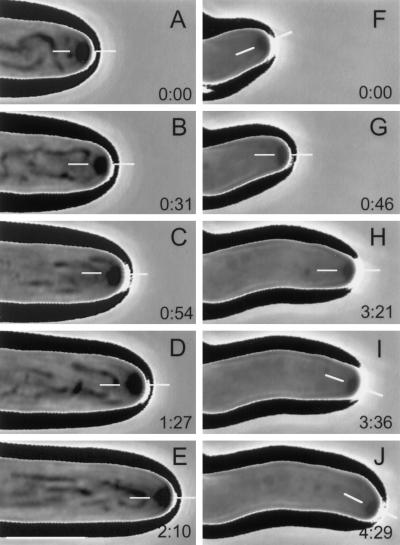
Figure 6.
Phase-contrast, time-lapse videomicrographs of growing hyphal tip cells illustrating the effects of kinesin deletion on positioning of the Spitzenkörper and, consequently, on hyphal morphogenesis. Elapsed time (min:sec) is shown in the lower right corner of each panel, and thin, white bars in each panel indicate the position of the Spitzenkörper relative to the hyphal apex. (A–E) A representative tip cell of the ectopic transformant-control. The central position of the Spitzenkörper was maintained throughout, resulting in a straight hypha. (F–J) A representative tip cell of the kinesin mutant. Central positioning of the Spitzenkörper produced a short, straight segment of hyphae (F). Then the position of the Spitzenkörper was shifted (G) and maintained long enough to produce another straight segment of hypha (H) with a different growth orientation. Finally, another shift of the Spitzenkörper (I) produced another short segment of hypha (J) with yet another growth orientation. Bar (in the lower left corner of panel E), 5 μm.
Normal Mitosis in the Nhkin1 Deletion Mutant
All stages of mitosis in the Nhkin1− mutant appeared indistinguishable (or nearly so) from those in both the NhKIN1+ transformant and WT, even when observed at 10,000×. The duration of metaphase was not determined. Although the time spent in anaphase A was shorter for Nhkin1− (37 s) than for WT (44 s), this difference was not statistically significant (p = 0.10, with the two-sample t test). Moreover, time in anaphase A was identical for Nhkin1− and the NhKIN1+ transformant. There was no difference between Nhkin1− and the WT in the number of cells exhibiting migrations of the MA. The sample size was 15 mitoses for each isolate. Precise measurement of spindle elongation, using image processing and computer-assisted data acquisition and analysis (Aist and Bayles, 1991), allowed us to detect a small (11%), but statistically insignificant (p = 0.26), reduction in the rate at which the anaphase B spindle elongated in Nhkin1− (Table 2).
DISCUSSION
Phylogenetic analyses of NhKIN1 and other kinesins suggest that Nkin and NhKIN1 root out at the base of the authentic kinesins and are distinct from kinesin-like proteins (Steinberg and Schliwa, 1995; data not shown). In view of the high level of amino acid sequence identity between NhKIN1 and the N. crassa organelle motor, Nkin (Steinberg and Schliwa, 1995), as well as the homology between NhKIN1 and conventional kinesin organelle motors, it is not surprising that the most striking phenotypes in the Nhkin1− mutant are apparently related to transport of membranous organelles. Retraction of the mitochondria from an apical to a subapical position within the hyphal tip cell, diminution in size of the Spitzenkörper, and a drastically reduced rate of hyphal growth are all indicative, or at least suggestive, of impaired organelle transport in the mutant. Our immunocytochemical localizations of cytoplasmic MTs in hyphal tip cells demonstrated that, in the Nhkin1− mutant, the MT distribution was normal. Thus, the effects on transport and/or position of membranous organelles was due to a primary effect of the absence of a functional organelle motor rather than to a secondary effect on MT distribution within the cells. That MTs and kinesin are involved in the transport and positioning of membranous organelles in animal cells is well established (Allan, 1995; Cole and Lippincott-Schwartz, 1995; Hirokawa, 1996; Moore and Endow, 1996). In fungi, also, there is evidence that MTs and KRPs play similar roles (Howard and Aist, 1980; Steinberg and Schliwa, 1993; Heath, 1994; Hoyt, 1994; Allan, 1995; Lehmler et al., 1997; Seiler et al., 1997). However, this report provides experimental evidence by specific gene mutation indicating that a member of the kinesin superfamily is responsible for the apical transport of mitochondria in a filamentous fungus. This result is in stark contrast to the lack of such a phenotype in similar kinesin mutants of other filamentous fungi (Lehmler et al., 1997; Seiler et al., 1997) and is further discussed below.
A slow-growth or colonial phenotype has been reported for dynein mutants of A. nidulans (Xiang et al., 1995) and N. crassa (Plamann et al., 1994), for dynein (Li et al., 1993) and KRP (Meluh and Rose, 1990) mutants of yeast, and for kinesin mutants of N. crassa (Seiler et al., 1997) and U. maydis (Lehmler et al., 1997). As Plamann et al. (1994) pointed out, this phenotype most likely results from the absence of a motor protein that is essential for efficient transport of secretory vesicles to the growing apex. In fact, one would infer, according to the mathematical equation that defines the hyphoid model of hyphal tip growth (Bartnicki-Garcia et al., 1989), that the Spitzenkörper of a narrower hypha would be releasing secretory vesicles at a slower rate, especially if the hyphal elongation rate were also reduced. Because the Spitzenkörper in our mutant cells were much smaller than those in control cells, we infer that vesicle transport to the growing apex was reduced in the mutant cells, an inference that was also drawn from similar results with kinesin mutants of other filamentous fungi (Lehmler et al., 1997; Seiler et al., 1997). Thus, our results — slower growth rate, narrower hyphae, and smaller Spitzenkörper — are consistent with predictions of the hyphoid equation, which describes hyphal tip growth mathematically (Bartnicki-Garcia et al., 1989). By contrast, Seiler et al. (1997) reported a two- to threefold increase in hyphal diameter of their kinesin mutant of N. crassa, a result that would not be anticipated solely on the basis of limited apical transport of secretory vesicles. As shown by Seiler et al. (1997), the N. crassa mutant has small clusters of apical vesicles at various sites in the apical cell, rather than a well-defined Spitzenkörper, and this “… defect in the formation of the vesicle supply center” apparently accounts for the wider hyphae.
That polarized growth continues at all in such slow-growth mutants may indicate that apical vesicle transport in fungi involves either functionally redundant, MT-associated motor proteins (Goldstein, 1993) or redundant transport mechanisms involving different cytoskeletal components (Lillie and Brown, 1992; Barton and Goldstein, 1996), because our results suggest that there is no gene redundancy involved. Indeed, there are several lines of evidence indicating that F-actin–based transport of membranous organelles occurs in fungi (Betina et al., 1972; Allen et al., 1980; Grove and Sweigard, 1980; Novick and Botstein, 1985; Heath, 1994; McGoldrick et al., 1995). Because an actin-myosin mechanism is capable of supporting hyphal tip growth, it seems reasonable to postulate that, in the absence of either MTs or an MT-associated motor protein that is important in vesicle transport, the fungal cell will continue to grow, albeit abnormally, by utilizing the actin-based motility mechanism.
The morphogenetic effects of mutating the NhKIN1 gene included the development of wavy or helical hyphae. A similar phenotype was reported for the colonial (cot 1) (Steele and Trinci, 1977) and dynein (Plamann et al., 1994) mutants of N. crassa, and gnarled or otherwise distorted hyphal morphology was reported for kinesin mutants of N. crassa (Seiler et al., 1997) and U. maydis (Lehmler et al., 1997). A Spitzenkörper that typically oscillates back and forth or moves in circles around the advancing apical dome, rather than staying in a more-or-less central position, would be expected to produce wavy or helical growth, respectively (Bartnicki-Garcia et al., 1995a; Sherwood-Higham et al., 1995), and we demonstrated this relationship in our kinesin mutant. What was surprising in the present study, however, was that the absence of an MT-associated organelle motor affected the positioning of the Spitzenkörper. This unique result compels us to consider the possibility that NhKIN1 may function, along with MTs and F-actin (Howard, 1981; Bartnicki-Garcia et al., 1989), to maintain the central location of the Spitzenkörper in control cells.
In addition to their similarity to phenotypes of other mutants, the morphogenetic effects we report here for N. haematococca are reminiscent of those obtained by treatment of filamentous fungi with anti-MT drugs (Howard and Aist, 1977, 1980; Hoch et al., 1987; Rossier et al., 1989; Raudaskoski et al., 1994) and with cytochalasins (as referenced above). What all of these studies have in common is that in every case a cytoskeletal motor protein transport system was inhibited, by disrupting either the transporter (motor protein) or the tracks (MTs or F-actin) along which the secretory vesicles are presumably moved. Gooday (1995) and Sherwood-Higham et al. (1995) pointed out that hyphae of many fungal species exhibit helical growth on surfaces in response to various environmental factors including rigidity of the substrate, nutrient concentrations, and temperature. Thus, “… a helical element appears to be inherent in apical growth of hyphae” (Gooday, 1995). Inhibition or elimination of a component of a cytoskeleton–motor protein system (e.g., kinesin) may remove a controlling element, allowing this inherent hyphal helicity to be more fully expressed.
The role of the MT-kinesin transport system in maintaining the apical position of mitochondria is not restricted to fungal cells. Rodionov et al. (1993) reported that antibodies to kinesin heavy chain caused mitochondria in human skin fibroblasts to withdraw from the cell periphery to the central, perinuclear region. Morris and Hollenbeck (1995) found that depolymerization of MTs in vertebrate neurons resulted in net movement of the mitochondria toward the cell body, with the extremities becoming devoid of mitochondria. These responses would seem to be analogous to the retraction of mitochondria from the hyphal apex that we observed in the Nhkin1− mutant of N. haematococca and to the similar effects of MT-depolymerizing agents, MBC in Fusarium acuminatum (Howard and Aist, 1977) and griseofulvin in Uromyces appendiculatus (Hoch et al., 1987). The normal distribution of MTs in hyphal tip cells of the Nhkin1− mutant shows that mitochondria were absent from the apex because a functional motor, NhKIN1, was missing, rather than because the MTs were missing from the apical region. Thus, experimental results with both fungal and animal cells are in accord with other evidence indicating that mitochondria move along MTs in living cells (Steinberg and Schliwa, 1993; Nangaku et al., 1994; Baumann and Murphy, 1995; Yaffe et al., 1996) and that kinesin is attached to the outer membrane of mitochondria (Leopold et al., 1992; Jellali et al., 1994; Nangaku et al., 1994). Morris and Hollenbeck (1995) further showed that the retrograde mitochondrial movement in neurons, which resulted in mitochondrial retreat to the cell body, was mediated by F-actin. In view of the presence of an actin–myosin system for membranous organelle transport in fungi (discussed above) and the recent report of actin-dependent mitochondrial motility in yeast (Simon et al., 1995), it is reasonable to speculate that the mitochondrial retraction induced in N. haematococca by deletion of NhKIN1 may also be mediated by F-actin.
Our analysis of mitosis in isolates of N. haematococca indicated that deletion of NhKIN1 resulted in a small, but statistically insignificant, reduction in the rate of spindle elongation during anaphase B and that other aspects of the mitotic process were apparently also unaffected. This result is consistent with the primary role of conventional kinesins, to which NhKIN1 is most closely related, in organelle motility rather than mitosis (Brady, 1995; Barton and Goldstein, 1996; Hirokawa, 1996).
That NhKIN1 is an important transporter of membranous organelles in N. haematococca is implied by morphological and other cytological phenotypes of the mutant. The smaller Spitzenkörper and the reduced rate of hyphal tip cell elongation can be accounted for by partial inhibition of apical transport of secretory vesicles. Moreover, the sequence homology of NhKIN1 to animal kinesins involved in organelle transport and to the Neurospora organelle motor, Nkin, would predict a major role for NhKIN1 in vesicle motility. Although this suggested role of NhKIN1 requires confirmation by its immunocytochemical localization to the vesicles and by electron microscopic demonstration of altered vesicle distribution in the mutant (cf. Howard and Aist, 1980), our present results lend support to the concept that such transport in filamentous fungi involves an MT-mediated mechanism, while in no way excluding the concomitant or potential participation of other transport mechanisms. They also show that an MT-associated motor protein may play an important role in maintaining the normal positioning of the fungal Spitzenkörper. Thus, cytoskeletal motor proteins may play a key role in the process by which fungal morphogenesis is controlled.
ACKNOWLEDGMENTS
Helpful suggestions for improving the manuscript were provided by Harvey Hoch. We thank Pamela Meluh and Cathy Wasmann for providing plasmids and Satoshi Inoue for processing specimens for immunocytochemical localization of microtubules. This research was funded by National Science Foundation Research grants DCB-8916338 (to J.R.A.) and MCB-9305703 and MCB-9408249 (to J.R.A., O.C.Y., and B.G.T.).
REFERENCES
- Aist JR, Bayles CJ. Video motion analysis of mitotic events in living cells of the fungus Fusarium solani. Cell Motil Cytoskel. 1988;9:325–336. [Google Scholar]
- Aist JR, Bayles CJ. Detection of spindle pushing forces in vivo during anaphase B in the fungus Nectria haematococca. Cell Motil Cytoskel. 1991;19:18–24. [Google Scholar]
- Aist JR, Bayles CJ, Tao W, Berns MW. Direct experimental evidence for the existence, structural basis and function of astral forces during anaphase B in vivo. J Cell Sci. 1991;100:279–288. doi: 10.1242/jcs.100.2.279. [DOI] [PubMed] [Google Scholar]
- Aist JR, Wu Q, Wirsel SG, Turgeon BG, Yoder OC. Disruption of a fungal kinesin gene affects both morphogenesis and mitosis. Mol Biol Cell. 1996;7:408a. doi: 10.1091/mbc.9.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan V. Membrane traffic motors. FEBS Lett. 1995;369:101–106. doi: 10.1016/0014-5793(95)00615-g. [DOI] [PubMed] [Google Scholar]
- Allen ED, Aiuto R, Sussman AS. Effects of cytochalasins on Neurospora crassa Growth, I., and ultrastructure. Protoplasma. 1980;102:63–75. doi: 10.1007/BF01276948. [DOI] [PubMed] [Google Scholar]
- Bairoch, A. (1992). PROSITE: a dictionary of sites and patterns in proteins. Nucleic Acids Res. 20(suppl), 2013–2018. [DOI] [PMC free article] [PubMed]
- Bartnicki-Garcia S, Bartnicki DD, Gierz G. Determinants of fungal cell wall morphology: the vesicle supply center. Can J Bot. 1995a;73:S372–S378. [Google Scholar]
- Bartnicki-Garcia S, Gierz G, Bartnicki DD, Lopez-Franco R, Bracker CE. Evidence that Spitzenkörper behavior determines the shape of a fungal hypha: a test of the hyphoid model. Exp Mycol. 1995b;19:153–159. doi: 10.1006/emyc.1995.1017. [DOI] [PubMed] [Google Scholar]
- Bartnicki-Garcia S, Hergert F, Gierz G. Computer simulation of fungal morphogenesis and the mathematical basis for hyphal (tip) growth. Protoplasma. 1989;153:46–57. [Google Scholar]
- Barton NR, Goldstien LSB. Going mobile: Microtubule motors and chromosome segregation. Proc Natl Acad Sci USA. 1996;93:1735–1742. doi: 10.1073/pnas.93.5.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann O, Murphy DB. Microbubule-associated movement of mitochondria and small particles in Acanthamoeba castellanii. Cell Motil Cytoskel. 1995;32:305–317. doi: 10.1002/cm.970320407. [DOI] [PubMed] [Google Scholar]
- Betina V, Mic̆eková D, Nemec P. Antimicrobial properties of cytochalasins and their alteration of fungal morphology. J Gen Microbiol. 1972;71:343–349. [Google Scholar]
- Bloom GS, Endow SA. Motor Proteins 1: kinesins. Protein Profile. 1995;2:1109–1171. [PubMed] [Google Scholar]
- Brady ST. A kinesin medley: biochemical and functional heterogeneity. Trends Cell Biol. 1995;5:159–164. doi: 10.1016/s0962-8924(00)88980-1. [DOI] [PubMed] [Google Scholar]
- Cole NB, Lippincott-Schwartz J. Organization of organelles and membrane traffic by microtubules. Curr Opin Cell Biol. 1995;7:55–64. doi: 10.1016/0955-0674(95)80045-x. [DOI] [PubMed] [Google Scholar]
- Girbardt M. Die Ultrastruktur der Apikalregion von Pilzhyphen. Protoplasma. 1969;67:413–441. [Google Scholar]
- Goldstein LSB. Functional redundancy in mitotic force generation. J Cell Biol. 1993;120:1–3. doi: 10.1083/jcb.120.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooday GW. The dynamics of hyphal growth. Mycol Res. 1995;99:385–394. [Google Scholar]
- Grove SN, Bracker CE. Protoplasmic organization of hyphal tips among fungi: vesicles and Spitzenkörper. J Bacteriol. 1970;104:989–1009. doi: 10.1128/jb.104.2.989-1009.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove SN, Sweigard JA. Cytochalasin A inhibits spore germination and hyphal tip growth in Gilbertella persicaria. Exp Mycol. 1980;4:239–250. [Google Scholar]
- Heath IB. The cytoskeleton in hyphal growth, organelle movements, and mitosis. In: Wessels JGH, Meinhardt F, editors. The Mycota I Growth—Differentiation and Sexuality. Berlin: Springer-Verlag; 1994. pp. 43–65. [Google Scholar]
- Hirokawa N. Organelle transport along microtubules: the role of KIFs. Trends Cell Biol. 1996;6:135–141. doi: 10.1016/0962-8924(96)10003-9. [DOI] [PubMed] [Google Scholar]
- Hoch HC, Tucker BE, Staples RC. An intact microtubule cytoskeleton is necessary for mediation of the signal for cell differentiation in Uromyces. Eur J Cell Biol. 1987;45:209–218. [Google Scholar]
- Howard RJ. Ultrastructural analysis of hyphal tip cell growth in fungi: Spitzenkörper, cytoskeleton and endomembranes after freeze-substitution. J Cell Sci. 1981;48:89–103. doi: 10.1242/jcs.48.1.89. [DOI] [PubMed] [Google Scholar]
- Howard RJ, Aist JR. Effects of MBC on hyphal tip organization, growth, and mitosis of Fusarium acuminatum, and their antagonism by D2O. Protoplasma. 1977;92:195–210. doi: 10.1007/BF01279458. [DOI] [PubMed] [Google Scholar]
- Howard RJ, Aist JR. Cytoplasmic microtubules and fungal morphogenesis: ultrastructural effects of methyl benzimidazole-2-ylcarbamate determined by freeze-substitution of hyphal tip cells. J Cell Biol. 1980;87:55–64. doi: 10.1083/jcb.87.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA. Cellular roles of kinesin and related proteins. Curr Opin Cell Biol. 1994;6:63–68. doi: 10.1016/0955-0674(94)90117-1. [DOI] [PubMed] [Google Scholar]
- Hurst HC. Transcription factors 1: bZIP proteins. Protein Profile. 1995;2:101–168. [PubMed] [Google Scholar]
- Jellali A, Metz-Boutique M-H, Surgucheva I, Janczik V, Schwartz C, Filliol D, Gelfand VI, Rendon A. Structural and biochemical properties of kinesin heavy chain associated with rat brain mitochondria. Cell Motil Cytoskel. 1994;28:79–93. doi: 10.1002/cm.970280108. [DOI] [PubMed] [Google Scholar]
- Leach J, Lang BR, Yoder OC. Methods for selection of mutants and in vitro culture of Cochliobolus heterostrophus. J Gen Microbiol. 1982;128:1719–1729. [Google Scholar]
- Lehmler C, Steinberg G, Snetselaar KM, Schliwa M, Kahmann R, Bölker M. Identification of a motor protein required for filamentous growth in Ustilago maydis. EMBO J. 1997;16:3464–3473. doi: 10.1093/emboj/16.12.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leopold PL, McDowell AW, Pfister KK, Bloom GS, Brady ST. Association of kinesin with characterized membrane-bounded organelles. Cell Motil Cytoskel. 1992;23:19–33. doi: 10.1002/cm.970230104. [DOI] [PubMed] [Google Scholar]
- Li Y-Y, Yeh E, Harp T, Bloom K. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc Natl Acad Sci USA. 1993;90:10096–10100. doi: 10.1073/pnas.90.21.10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillie SH, Brown SS. Suppression of a myosin defect by a kinesin-related gene. Nature. 1992;356:358–361. doi: 10.1038/356358a0. [DOI] [PubMed] [Google Scholar]
- Logemann J, Schell J, Willmitzer L. Improved method for the isolation of RNA from plant tissues. Anal Biochem. 1987;163:16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- Lopez-Franco R, Howard RJ, Bracker CE. Satellite Spitzenkörper in growing hyphal tips. Protoplasma. 1995;188:85–103. [Google Scholar]
- McGoldrick CA, Gruver C, May GS. myoA of Aspergillus nidulans encodes an essential myosin I required for secretion and polarized growth. J Cell Biol. 1995;128:577–587. doi: 10.1083/jcb.128.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Rose MD. KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell. 1990;60:1029–1041. doi: 10.1016/0092-8674(90)90351-e. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Moore JD, Endow SA. Kinesin proteins: a phylum of motors for microtubule-based motility. Bioessays, 1996;18:207–219. doi: 10.1002/bies.950180308. [DOI] [PubMed] [Google Scholar]
- Morris RL, Hollenbeck PJ. Axonal transport of mitochondria along microtubules and F-actin in living vertebrate neurons. J Cell Biol. 1995;131:1315–1326. doi: 10.1083/jcb.131.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nangaku M, Sato-Yoshitake R, Okada Y, Noda Y, Takemura R, Yamazaki H, Hirokawa N. KIFlB, a novel microtubule plus end-directed monomeric motor protein for transport of mitochondria. Cell. 1994;79:1209–1220. doi: 10.1016/0092-8674(94)90012-4. [DOI] [PubMed] [Google Scholar]
- Navone F, Niclas J, Hom BN, Sparks L, Bernstein HD, Maccaffre G, Vale RD. Cloning and expression of a human kinesin heavy chain gene: Interactions of the cooh-terminal domain with cytoplasmic microtubules in transfected CV-1 cells. J Cell Biol. 1992;117:1263–1275. doi: 10.1083/jcb.117.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Botstein D. Phenotypic analysis of temperature-sensitive yeast actin mutants. Cell. 1985;40:405–416. doi: 10.1016/0092-8674(85)90154-0. [DOI] [PubMed] [Google Scholar]
- Patel N, Thierry-Mieg D, Mancillas JR. Cloning by insertional mutagenesis of a cDNA encoding Caenorhabditis elegans kinesin heavy chain. Proc Natl Acad Sci USA. 1993;90:9181–9185. doi: 10.1073/pnas.90.19.9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plamann M, Minke PF, Tinsley JH, Bruno KS. Cytoplasmic dynein and actin-related protein Arp1 are required for normal nuclear distribution in filamentous fungi. J Cell Biol. 1994;127:139–149. doi: 10.1083/jcb.127.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudaskoski M, Mao W-Z, Yli-Mattila T. Microtubule cytoskeleton in hyphal growth. Response to nocodazole in a sensitive and a tolerant strain of the homobasidiomycete Schizophyllum commune. Eur J Cell Biol. 1994;64:131–141. [PubMed] [Google Scholar]
- Reyonds P, Weber S, Prakash L. RAD6 gene of Saccharomyces cerevisiae encodes a protein containing a tract of 13 consecutive aspartates. Proc Natl Acad Sci USA. 1985;82:168–172. doi: 10.1073/pnas.82.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov VI, Gyoeva FK, Tanaka E, Bershadsky AD, Vasiliev JM, Gelfand VI. Microtubule-dependent control of cell shape and pseudopodial activity is inhibited by the antibody to kinesin motor domain. J Cell Biol. 1993;123:1811–1820. doi: 10.1083/jcb.123.6.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossier C, Hoang-Van K, Turian G. Secretion of an Mr 60000 protein by benomyl-treated cells of Neurospora crassa. Eur J Cell Biol. 1989;50:333–339. [PubMed] [Google Scholar]
- Sambrook J, Fitsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Saraste M, Sibbald PR, Wittinghofer A. The P-loop – a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- Saunders WS. Mitotic spindle pole separation. Trends Cell Biol. 1993;3:432–437. doi: 10.1016/0962-8924(93)90032-v. [DOI] [PubMed] [Google Scholar]
- Seiler S, Nargang FE, Steinberg G, Schliwa M. Kinesin is essential for cell morphogenesis and polarized secretion in Neurospora crassa. EMBO J. 1997;16:3025–3034. doi: 10.1093/emboj/16.11.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks J. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- Sherwood-Higham J, Zhu W-Y, Devine CA, Gooday GW, Gow NAR, Gregory DW. Helical growth of hyphae of Candida albicans. J Med Vet Mycol. 1995;32:437–445. doi: 10.1080/02681219480000591. [DOI] [PubMed] [Google Scholar]
- Simon VR, Swayne TC, Pon LA. Actin-dependent mitochondrial motility in mitotic yeast and cell-free systems: identification of a motor activity on the mitochondrial surface. J Cell Biol. 1995;130:345–354. doi: 10.1083/jcb.130.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soshi T, Sakuraba Y, Kafer E, Inoue H. The MUS-8 gene of Neurospora crassa encodes a structural and functional homolog of the Rad6 protein of Saccharomyces cerevisiae. Curr Genet. 1966;30:224–231. doi: 10.1007/s002940050125. [DOI] [PubMed] [Google Scholar]
- Steele GC, Trinci APJ. Effect of temperature and temperature shifts on growth and branching of a wild type and a temperature sensitive colonial mutant (Cot1) of Neurospora crassa. Arch Microbiol. 1997;113:43–48. doi: 10.1007/BF00428578. [DOI] [PubMed] [Google Scholar]
- Steinberg G, Schliwa M. Organelle movements in the wild type and wall-less fz;sg;os-1 mutants of Neurospora crassa are mediated by cytoplasmic microtubules. J Cell Sci. 1993;106:555–564. doi: 10.1242/jcs.106.2.555. [DOI] [PubMed] [Google Scholar]
- Steinberg G, Schliwa M. The Neurospora organelle motor: a distant relative of conventional kinesin with unconventional properties. Mol Biol Cell. 1995;6:1605–1618. doi: 10.1091/mbc.6.11.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan ML, Vierstra RD. Cloning of a 16-KDa ubiquitin carrier protein from wheat and Arabidopsis thaliana. Identification of functional domains by in vitro mutagenesis. J Biol Chem. 1991;266:23878–23885. [PubMed] [Google Scholar]
- Turgeon BG, Bohlmannn H, Ciuffetti LM, Christiansen SK, Yang G, Shafer W, Yoder OC. Cloning and analysis of the mating type genes from Cochliobolus heterostrophus. Mol Gen Genet. 1993;238:270–284. doi: 10.1007/BF00279556. [DOI] [PubMed] [Google Scholar]
- Turgeon BG, Garber RC, Yoder OC. Development of a fungal transformation system based on selection of a sequence with promoting activity. Mol Cell Biol. 1987;7:3297–3305. doi: 10.1128/mcb.7.9.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng TH, Lyngholm LK, Ford CF, Bronson CR. A restriction fragment length polymorphism map and electrophoretic karyotype of the fungal maize pathogen Cochliobolus heterostrophus. Genetics. 1992;130:81–96. doi: 10.1093/genetics/130.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee RB, Sheetz MP. Targeting of motor proteins. Science. 1996;271:1539–1544. doi: 10.1126/science.271.5255.1539. [DOI] [PubMed] [Google Scholar]
- VanEtten HD, Stein JI. Differential response of Fusarium solani isolates to pisatin and phaseollin. Phytopathology. 1978;68:1276–1283. [Google Scholar]
- Wasmann CC, VanEtten HD. Transformation-mediated chromosome loss and disruption of a gene for pisatin demethylase decreases the virulence of Nectria haematococca on pea. Mol Plant-Microbe Interact. 1996;9:793–803. [Google Scholar]
- Xiang X, Roghi C, Morris NR. Characterization and localization of the cytoplasmic dynein heavy chain in Aspergillus nidulans. Proc Natl Acad Sci USA. 1995;92:9890–9894. doi: 10.1073/pnas.92.21.9890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MP, Harata D, Verdes F, Eddison M, Toda T, Nurse P. Microtubules mediate mitochondrial distribution in fission yeast. Proc Natl Acad Sci USA. 1996;93:11664–11668. doi: 10.1073/pnas.93.21.11664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JT, Laymon RA, Goldstein LSB. A three-domain structure of kinesin heavy chain revealed by DNA sequence and microtuble binding assays. Cell. 1989;56:879–889. doi: 10.1016/0092-8674(89)90692-2. [DOI] [PubMed] [Google Scholar]
- Yoder OC. Cochliobolus heterostrophus, cause of Southern Corn Leaf Blight. In: Sidhu GS, editor. Genetics of Plant Pathogenic Fungi. San Diego, CA: Academic Press; 1988. pp. 93–112. [Google Scholar]